Abstract
Growing evidence suggests that exposure to fine particulate matter (PM2.5) may reduce life expectancy; however, the causal pathways of PM2.5 exposure affecting life expectancy remain unknown. Here, we assess the causal effects of genetically predicted PM2.5 concentration on common chronic diseases and longevity using a Mendelian randomization (MR) statistical framework based on large-scale genome-wide association studies (GWAS) (>400,000 participants). After adjusting for other types of air pollution and smoking, we find significant causal relationships between PM2.5 concentration and angina pectoris, hypercholesterolaemia and hypothyroidism, but no causal relationship with longevity. Mediation analysis shows that although the association between PM2.5 concentration and longevity is not significant, PM2.5 exposure indirectly affects longevity via diastolic blood pressure (DBP), hypertension, angina pectoris, hypercholesterolaemia and Alzheimer’s disease, with a mediated proportion of 31.5, 70.9, 2.5, 100, and 24.7%, respectively. Our findings indicate that public health policies to control air pollution may help improve life expectancy.
Subject terms: Risk factors, Epidemiology, Ageing
Introduction
Most of the world’s population is affected by air pollution1. Air pollutants, especially fine particulate matter pollution (PM2.5), pose a major threat to human health2. Previous studies have confirmed that long-term exposure to PM2.5 increases the risk of a wide range of chronic diseases and may cause premature death in parts of the population2–7. A noteworthy large cross-sectional study conducted in the United States revealed that a reduction of 1 μg/m3 in PM2.5 exposure corresponded to an increase in life expectancy of 0.12 years8. Another population-based cohort of 2.7 million adults in Canada supported the potential public health benefits of air quality interventions4. However, the plethora of confounding factors affecting life expectancy makes it difficult to make causal inferences9. For instance, socioeconomic factors, such as socioeconomic status (SES), educational attainment, income, and occupation, play a significant role as confounding variables in longevity studies10–12. SES can have a substantial impact on various aspects of individuals’ lives, including access to healthcare, living conditions, lifestyle choices, and exposure to environmental hazards, all of which can influence longevity outcomes13. Other environmental factors, such as noise pollution and access to green spaces may influence longevity outcomes independently of the exposure being studied13. In addition, the mediating pathway by which PM2.5 exposure affects longevity remains unknown.
Growing evidence suggests that air pollution affects individuals to varying degrees, and genetic polymorphisms play a significant role in this phenomenon14,15. Certain genetic variants may increase susceptibility to the detrimental effects of air pollutants, while others may offer some level of protection. It has been discovered that alleles in the human genome, which mitigate smoke damage, have been present for at least 550,000 years16. The impact of air pollution on longevity can be influenced by gene polymorphism in candidate genes associated with longevity, such as SIRT1 and FOXO315,17. Additionally, genetic variations can affect an individual’s inflammatory and oxidative stress responses, which are critical mechanisms in the body’s reaction to air pollution and, therefore, can influence the risk of adverse health outcomes and potentially reduce longevity18,19. Moreover, the heritability of longevity is considerably high, with genetic factors accounting for approximately one-third of the variability in human lifespan20. Consequently, genetic data can be employed to investigate the causal relationship between PM2.5 concentration and longevity outcomes.
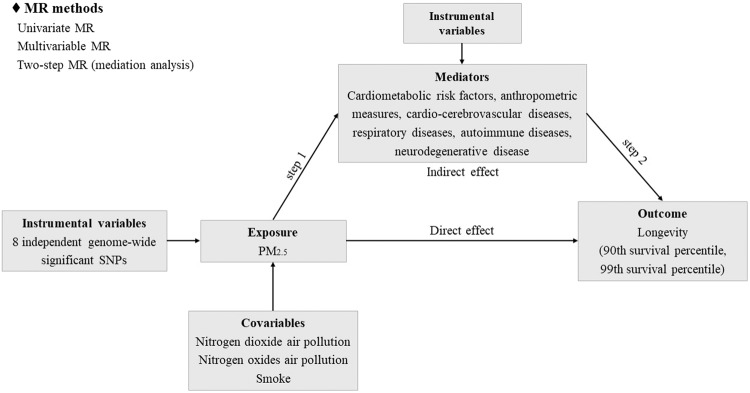
In the absence of randomized controlled trials (RCTs), we designed a Mendelian randomization (MR) statistical framework using large-scale genome-wide association studies (GWAS). MR utilized genetic variants as instrumental variables to assess the causal relationship between exposure and outcome21–26. The randomly allocated process of alleles of genetic variants was used to simulate RCTs27,28. The alleles of these instruments were determined at the time of meiosis and fertilization, thereby minimizing issues of confounding factors and reverse causality27. Here, we used univariate MR to assess the causal relationship between genetically predicted PM2.5 concentration and longevity. We utilized multivariate MR to adjust for the effects of other types of air pollution and smoking on causal estimates. Finally, we conducted a two-step MR analysis (mediation analysis) to explore whether PM2.5 concentration influenced longevity via mediating factors (cardiometabolic risk factors, cardio-cerebrovascular diseases, respiratory diseases, autoimmune diseases, and neurodegenerative disease). The conceptual framework was depicted in Fig. 1.
Fig. 1.
Study design.
Results
Univariable MR
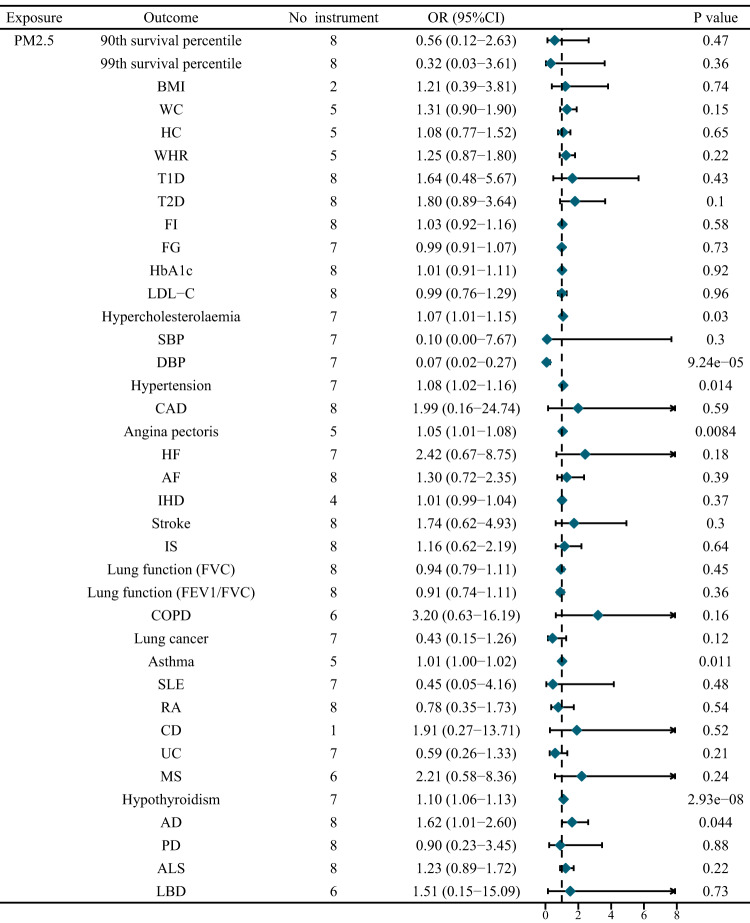
We performed a univariate MR analysis to investigate the total causal effect of genetically predicted PM2.5 concentration on longevity. A total of eight independent genome-wide significant genetic variants were used as instrumental variables, with no weak instruments (F statistic < 10) (Supplementary Table 1). Univariable MR analysis showed a non-significant causal relationship between genetically predicted PM2.5 concentration and longevity [90th percentile: Odds Ratio (OR) = 0.56, 95% CI = 0.12 to 2.63, P = 0.47; 99th percentile: OR = 0.32, 95% CI = 0.03 to 3.61, P = 0.36] (Fig. 2, Supplementary Table 2, Supplementary Figure 1). Genetically predicted PM2.5 concentration was also not associated with longevity in sensitivity analysis.
Fig. 2. Univariate MR results for the causal relationship of PM2.5 concentration on potential mediators and longevity.
AD Alzheimer’s disease, AF atrial fibrillation, ALS amyotrophic lateral sclerosis, BMI body mass index, CAD coronary artery disease, CD Crohn’s disease, COPD chronic obstructive pulmonary disease, DBP diastolic blood pressure, FEV1/FVC 1 s forced expiratory volume/FVC, FG fasting glucose, FI fasting insulin, FVC forced vital capacity, HbA1c glycated hemoglobin, HC hip circumference, HF heart failure, IHD ischemic heart disease, IS ischemic stroke, LBD lewy body dementia, MS multiple sclerosis, PD Parkinson’s disease, RA rheumatoid arthritis, SBP systolic blood pressure, SLE Systemic lupus erythematosus, T1D type 1 diabetes, T2D type 2 diabetes, UC Ulcerative colitis, WC waist circumference, WHR: waist-hip ratio.
Although PM2.5 concentration did not directly affect human life span, it significantly affected several potential mediators and might further indirectly affect human life span. As a result, genetically predicted PM2.5 concentration was causally associated with diastolic blood pressure (DBP), hypercholesterolaemia, hypertension, angina pectoris, asthma, hypothyroidism, and Alzheimer’s disease (AD) (Fig. 2, Supplementary Table 3). Genetically predicted PM2.5 concentration had no causal effect estimate consistent with body mass index (BMI), hip circumference (HC), waist circumference (WC), waist-hip ratio (WHR), type 1 diabetes (T1D), type 2 diabetes (T2D), fasting glucose (FG), fasting insulin (FI), glycated hemoglobin (HbA1c), low density lipoprotein cholesterolsystolic (LDL-C), blood pressure (SBP), coronary artery disease (CAD), heart failure (HF), atrial fibrillation (AF), ischemic heart disease (IHD), stroke, ischemic stroke (IS), forced vital capacity (FVC), 1 s forced expiratory volume (FEV1)/FVC, chronic obstructive pulmonary disease (COPD), lung cancer, systemic lupus erythematosus (SLE), ulcerative colitis (UC), Crohn’s disease (CD), rheumatoid arthritis (RA), multiple sclerosis (MS), amyotrophic lateral sclerosis (ALS), Parkinson’s disease (PD) and lewy body dementia (LBD). Specifically, for each unit (1.06 micro-g/m3) increase in PM2.5 exposure, the risk of hypercholesterolaemia increased by 7% (OR = 1.07, 95% CI = 1.01–1.15, P = 0.03), the risk of hypertension increased by 8% (OR = 1.08, 95% CI = 1.02–1.16, P = 0.014), the risk of angina pectoris increased by 5% (OR = 1.05, 95% CI = 1.01–1.08, P = 0.0084), the risk of asthma increased by 1% (OR = 1.01, 95% CI = 1.00–1.02, P = 0.011), the risk of hypothyroidism increased by 10% (OR = 1.10, 95% CI = 1.06–1.13, P = 2.93E − 08), and the risk of AD increased by 10% (OR = 1.62, 95% CI = 1.01 to 2.60, P = 0.044) (Fig. 2, Supplementary Table 3). Genetically predicted PM2.5 concentration was significantly associated with reduced DBP (OR = 0.07, 95% CI = 0.02–0.27, P = 9.24E − 05).
The sensitivity tests essentially replicated the results of inverse variance weighted (IVW) analysis (Supplementary Table 3). Notably, genetically predicted PM2.5 concentration was associated with an increased risk of T2D (OR = 1.86, 95%CI = 1.20–2.87, P = 0.039) and AF (OR = 1.06, 95%CI = 1.01–1.10, P = 0.046) after the removal of pleiotropic SNPs using MR-PRESSO method.
Multivariable MR
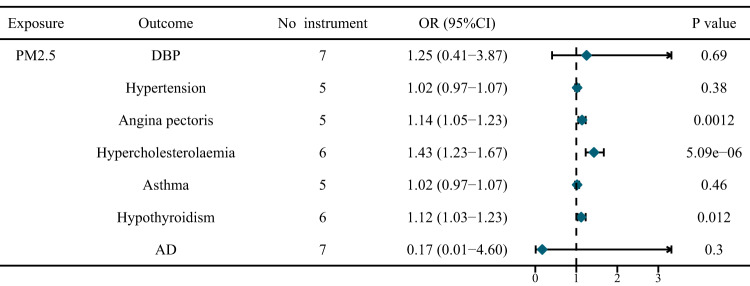
After adjusting for nitrogen dioxide air pollution, nitrogen oxides air pollution, and smoking, genetically predicted PM2.5 concentration was associated with an elevated risk of angina pectoris (OR = 1.14, 95%CI = 1.05–1.23, P = 0.0012), hypercholesterolaemia (OR = 1.43, 95%CI = 1.23–1.67, P = 5.09E-06), and hypothyroidism (OR = 1.12, 95%CI = 1.03–1.23, P = 0.012) (Fig. 3, Supplementary Table 4–10). The causal effect sizes of them in multivariable model were all larger than those of univariable model. Interestingly, nitrogen oxides air pollution reduced the risk of hyperemployeolaemia and hypothyroidism in multivariable MR analysis (Supplementary Table 6, 8). Additionally, the multivariate MR showed no direct causal relationship between air pollution and longevity (Supplementary Table 11–12).
Fig. 3.
Multivariable MR results adjusting for the effects of other types of air pollution and smoking.
Mediation analysis
PM2.5 exposure may affect longevity via potential mediators. Among the 36 potential mediators, we identified the causal relationships between genetically predicted PM2.5 concentration and 7 mediators in the first step, and identified the causal effects of DBP, hypertension, hypercholesterolaemia, angina pectoris, hypothyroidism, and AD on longevity in the second step (Table 1, Supplementary Table 13–14). We further evaluated the proportion of indirect effects to the overall effects. The mediation effect of DBP was 0.18 (95%CI = 0.083–0.28, P = 0.00029) with a mediated proportion of 31.5% (95% CI, 14.4% to 48.5%) in 90th survival percentile subgroup and 0.21 (95%CI = 0.088 to 0.32, P = 0.0006) with a mediated proportion of 28.4% (95% CI, 7.8–18.1%) in 99th survival percentile subgroup (Table 1). The mediation effect of hypertension was −0.41 (95%CI = −0.74 to −0.070, P = 0.018) with a mediated proportion of 70.9% (95% CI, 12.2–100%) in 90th percentile subgroup and −0.46 (95%CI = −0.86 to −0.065, P = 0.023) with a mediated proportion of 40.9% (95% CI, 5.7–76.0%) in 99th percentile subgroup (Table 1). For every unit increase (1.06 micro-g/m3) in PM2.5 exposure, the possibility of longevity (top 5%) decreased by 1% via angina pectoris risk (OR = 0.99, 95%CI = 0.97 to 1.00; mediated proportion = 2.5%, 95% CI = 0.4–4.5%; P = 0.02) and the possibility of longevity (top 1%) decreased by 2% via angina pectoris risk (OR = 0.98, 95%CI = 0.97–1.00; mediated proportion = 1.5%, 95% CI = 0.2 to 2.9%; P = 0.026). The mediation effects of hypercholesterolaemia and AD were significant only in 90th survival percentile subgroup.
Table 1.
The mediation effect of PM2.5 concentration on longevity via potential mediators.
| Mediator | Longevity | Total effect β (95% CI) | Direct effect A β (95% CI) | Direct effect B β (95% CI) | Mediation effect β (95% CI) | P value | Mediated proportion (%) |
|---|---|---|---|---|---|---|---|
| DBP | 90th | −0.57 (−2.12 to 0.97) | −2.65 (−3.97 to -1.32) | -0.068 (−0.082 to −0.054) | 0.18 (0.083 to 0.28) | 0.00029 | 31.48 (14.44 to 48.51) |
| DBP | 99th | −1.13 (−3.55 to 1.28) | −2.65 (−3.97 to −1.32) | −0.077 (−0.099 to −0.056) | 0.21 (0.088 to 0.32) | 0.00060 | 28.42 (7.76 to 18.09) |
| Hypertension | 90th | −0.57 (−2.12 to 0.97) | 0.081 (0.016 to 0.15) | −5.01 (−6.09 to −3.92) | −0.41(−0.74 to −0.070) | 0.018 | 70.86 (12.23 to 100) |
| Hypertension | 99th | −1.13 (−3.55 to 1.28) | 0.081 (0.016 to 0.15) | −5.70 (−7.52 to −3.87) | −0.46 (−0.86 to −0.065) | 0.023 | 40.86 (5.70 to 76.02) |
| Hypercholesterolaemia | 90th | −0.57 (−2.12 to 0.97) | 0.072 (0.0070 to 0.14) | −9.61 (−12.95 to −6.27) | −0.69 (−1.36 to −0.023) | 0.043 | 100 (3.92 to 100) |
| Hypercholesterolaemia | 99th | −1.13 (−3.55 to 1.28) | 0.072 (0.0070 to 0.14) | −13.13 (−18.87 to −7.38) | −0.94 (−1.89 to 0.0029) | 0.051 | 83.18 (0 to 100) |
| Angina pectoris | 90th | −0.57 (−2.12 to 0.97) | 0.045 (0.011 to 0.078) | −0.36 (−0.50 to −0.21) | −0.014 (−0.026 to −0.0022) | 0.020 | 2.45 (0. 39 to 4.50) |
| Angina pectoris | 99th | −1.13 (−3.55 to 1.28) | 0.045 (0.011 to 0.078) | −0.39 (−0.58 to −0.20) | −0.017 (−0.033 to −0.0020) | 0.026 | 1.53 (0. 18 to 2.88) |
| Hypothyroidism | 90th | −0.57 (−2.12 to 0.97) | 0.092 (0.060 to 0.13) | −3.69 (−7.27 to −0.11) | −0.34 (−0.69 to 0.011) | 0.058 | 59.37 (0 to 100) |
| Hypothyroidism | 99th | −1.13 (−3.55 to 1.28) | 0.092 (0.060 to 0.13) | −1.48 (−7.23 to 4.27) | −0.14 (−0.67 to 0.40) | 0.62 | 12.05 (0 to 59.16) |
| AD | 90th | −0.57 (−2.12 to 0.97) | 0.48 (0.014 to 0.96) | −0.29 (−0.36 to −0.22) | −0.14 (−0.28 to −0.00016) | 0.05 | 24.65 (0 to 49.27) |
| AD | 99th | −1.13 (−3.55 to 1.28) | 0.48 (0.014 to 0.96) | −0.39 (−0.50 to −0.28) | −0.19 (−0.38 to 0.0026) | 0.053 | 16.61 (0 to 33.46) |
Discussion
Long-term exposure to air pollutants has been shown to have a detrimental effect on human life expectancy, potentially leading to premature death29. However, little is known about the mediating pathway by which PM2.5 affects longevity. In the absence of large-scale RCTs, MR studies that are qualitatively consistent with the results of RCTs can be used for causal inference. To investigate the causal relationship between genetically predicted PM2.5 exposure and longevity, we used genetic instrumental variables as proxies. Our primary analyses indicated that, although the association between PM2.5 concentration and longevity was not significant, genetically predicted PM2.5 increased the risk of hypertension, hypercholesterolaemia, angina pectoris, hypothyroidism and AD, and thus decreased the likelihood of longevity. To account for potential confounders such as other types of air pollution and smoking, we conducted a multivariate MR model and further identified three significant mediators: angina pectoris, hypercholesterolaemia, and hypothyroidism.
Previous observational evidence has demonstrated the gene-environment interaction between longevity genes and air pollution, with certain alleles being more vulnerable to air pollution. For instance, SIRT1_391 (rs3758391) allele carriers counteract the detrimental effect of PM2.5 exposure and reduce the risk of premature mortality by 26.1%15. Similarly, FOXO3 rs2802292 G allele carriers are protected from the dangers of PM2.5 exposure17. Since both air pollution and longevity are affected by genetic factors, the association between them can be inferred using genetic variants as proxy. Compared to observational studies, MR method is novel and effective, but needs to meet the three assumptions27. Choosing genetic instruments associated with PM2.5 concentration is easily achievable, but the other two assumptions need to be tested using statistical methods. Assumption 2 requires that genetic instruments be independent of confounding factors. These PM2.5-related SNP instruments may be associated with other types of air pollution and smoking, although the alleles of these SNP established during meiosis and fertilization have avoided the effects of acquired confounding factors. Therefore, we used multivariate MR to adjust for potential confounders (nitrogen dioxide air pollution, nitrogen oxides air pollution, and smoking). For assumption 3, we used MR-Egger to perform a pleiotropy test, and used MR-PRESSO to delete the potential pleiotropic SNP instruments. After adjustment, the results of IVW analysis were basically replicated. Among the non-significant causal estimates in IVW analysis, genetically predicted PM2.5 concentration significantly increased the risk of T2D (OR = 1.86, 95%CI = 1.20 to 2.87, P = 0.039) and AF (OR = 1.06, 95%CI = 1.01 to 1.10, P = 0.046), consistent with previous observational findings.
PM2.5 exposure has been shown to be associated with an increased risk of cardiometabolic risk factors and cardio-cerebrovascular diseases9. Cross-sectional evidence from the China Health and Retirement Longitudinal Study (CHARLS) among 19,529 participants has demonstrated that an increase in PM2.5 concentration is significantly associated with a higher prevalence of hypertension (OR = 1.07, 95% CI = 1.03 to 1.11) and diabetes (OR = 1.15, 95% CI = 1.10 to 1.20), with the impact being relatively stronger in nonsmokers than smokers30. Additionally, exposure to both household and outdoor air pollutants has been linked to an increased risk of angina pectoris and myocardial infarction (MI)31,32. Recent evidence has also suggested a significant association between PM2.5 concentration and dementia3,33. A nationwide population-based cohort study conducted by Shi et al. revealed that an interquartile range increase in PM2.5 exposure was associated with a 9% increase in AD risk3. Among the constituents of PM2.5, black carbon (BC) and sulfate (SO42−) showed the strongest associations. Interestingly, even improving ambient air quality in late life was associated with a significant reduction in dementia risk, indicating that PM2.5 exposure-induced damage to the aging brain may be reversible33.
Nevertheless, the exact mechanism by which PM2.5 affects chronic disease and longevity in humans remains unclear. It is likely that PM2.5 deposits in the lungs, promoting aging through oxidative stress and immune responses6,34. Additionally, smaller air pollution particles may enter the bloodstream through different transport routes and mechanisms, leading to toxicity beyond the lung, including potential neurotoxic consequences35. Thus, long-term exposure to PM2.5 may have a significant impact on the health of a variety of human tissues and organs.
We acknowledge that our study has several limitations. The GWAS for several potential mediators are derived from UK biobank, which have certain sample overlap with the cohort of air pollution. However, there are no other data sources for these phenotypes. Next, genetically predicted PM10 and PM2.5-10 do not have genome-wide significant genetic variants as instruments, making it difficult to perform MR analysis. Another limitation is that there may be quantitative differences between MR and observational studies or RCTs that should not be interpreted directly as the estimated impact of interventions36. Burgess et al. suggest that MR estimates are usually larger than those of observational studies36. Additionally, the IVW method usually provides the most effective causal estimates. Deletion of pleiotropic genetic instruments by MR-PRESSO method may result in reduced power or overly precise causal estimates. Therefore, the causal effects of genetically predicted PM2.5 on T2D and AF may only be considered suggestive. Finally, power represents a common challenge in the investigation of gene-environment interactions37,38. The genetically determined impact of PM2.5 on longevity might only constitute a fraction of the total influence. Consequently, it is anticipated that forthcoming research endeavors will address this limitation by augmenting the sample size and enhancing the application of statistical methodologies.
In conclusion, exposure to PM2.5 has been linked to an increased risk of hypertension, hypercholesterolaemia, angina pectoris, hypothyroidism and AD, thus having a detrimental effect on longevity. Interventions to reduce environmental PM2.5 concentrations are likely to have a significant impact on public health.
Methods
MR model
We applied a MR design to investigate the causal effect of PM2.5 concentration on longevity and whether potential mediators played a mediating role (Fig. 1). To ensure that the causal estimate is valid, three assumptions must be met: (1) the SNP instruments are significantly associated with exposure, (2) the SNP instruments are not associated with any potential confounder, and (3) the SNP instruments do not affect outcome independently of exposure.
Data sources
The GWAS summary statistics in this study was publicly available and ethical approval was obtained in all original studies.
Exposure
GWAS data for PM2.5 concentration was obtained from UK Biobank and was as a part of the European Study of Cohorts for Air Pollution Effects (ESCAPE)39. A land use regression (LUR) model was used to model for each address between 26 Jan 2010 to 18 January 2011, and air pollution estimates were representative for the year 201039,40. By 2010, the study included 423,796 samples. The mean value of PM2.5 concentration was 9.99 micro-g/m3 and the standard deviation (SD) was 1.06.
Outcome
Longevity outcome was derived from a GWAS meta-analysis for age of survival of participants from 20 cohorts of European, East Asian, and African American populations41. Cases were participants who lived to an age above the 90th (11,262 cases) or 99th percentile (3,484 cases) based on cohort life tables41. Controls were participants who died at or before the age at the 60th percentile. and the 99th survival percentile (25,483 controls)41. For instance, the 60th, 90th, and 99th percentile correspond to ages of 83, 94, and 102 years for women in the United States cohort41.
Potential confounders
Other types of air pollution and smoking were identified as potential confounding factors. Data for nitrogen dioxide air pollution, nitrogen oxides air pollution, PM10, and PM2.5-10 were obtained from UK biobank39. GWAS for PM10 and PM2.5-10 had no genome-wide significant genetic variants. GWAS for cigarettes smoked per day was derived from GWAS & Sequencing Consortium of Alcohol and Nicotine use (GSCAN) containing 337,334 participants42.
Potential mediators
Previous observational studies have reported some potential outcomes of PM2.5 exposure, including cardiometabolic risk factors, cardio-cerebrovascular diseases, lung function, autoimmune diseases, and dementia (Table 2)2–6,27,40,43–46. These phenotypes were considered as potential mediators. GWAS data for BMI (N = 693,529), WHR (N = 693,529) after adjusting for BMI, HC (N = 142,762) after adjusting for BMI, and WC (N = 142,762) after adjusting for BMI were obtained from the genetic investigation of anthropometric traits (GIANT) consortium47,48. GWAS for T1D were obtained from the Common Metabolic Diseases Knowledge Portal (CMDKP) and T2D from the Diabetes Meta-Analysis of Trans-Ethnic association studies (DIAMANTE) Consortium49,50. GWAS for fasting glucose (FG), fasting insulin (FI) and glycated hemoglobin (HbA1c) were obtained from a meta-analysis, including 281,416 individuals without diabetes (~70% were of European ancestry)51. GWAS for LDL-C was obtained from a GWAS meta-analysis of UK Biobank (n = 431,167) and the Global Lipids Genetics Consortium (n = 188,577)52. GWAS for DBP and SBP were obtained from the International Consortium of Blood Pressure (ICBP) (N = 757,601). Data for CAD were obtained from a GWAS meta-analysis of nine European cohorts (86,847 cases and 417,789 controls)53. Data for HF was derived from a GWAS comprising 47,309 cases and 930,014 controls across 26 studies from the Heart Failure Molecular Epidemiology for Therapeutic Targets (HERMES) Consortium54. GWAS for AF (55,114 cases and 482,295 controls) contained more than 50 studies and most of the studies were part of the Atrial Fibrillation Genetics (AFGen) consortium and the Broad AF Study (Broad AF)55. Data for stroke was derived from a GWAS meta-analysis of 29 population-based cohorts or biobanks by Mishra et al.56. Data for lung function (FVC and FEV1/FVC ratio) were obtained from a meta-analysis of UK biobank and SpiroMeta Consortium (N = 321,047)57. Data for COPD in never smokers were obtained from UK biobank and were replicated in COPDGene and SpiroMeta Consortium by Kim et al.58. GWAS for lung cancer were obtained from a meta-analysis of four GWAS cohorts of lung cancer by Wang et al. from the International Lung Cancer Consortium (ILCCO) (11,348 cases and 15,861 controls)59. GWAS for Inflammatory bowel diseases (IBD), including UC and CD, were derived from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) (86,640 European individuals)60. GWAS data for SLE was obtained from a GWAS meta-analysis including 7,219 cases and 15,991 controls of European ancestry61. GWAS for MS was derived from International Multiple Sclerosis Genetics Consortium (IMSGC) including 47,429 cases and 68,374 controls62. GWAS for RA were obtained from a GWAS meta-analysis by Ha et al., including 14,361 cases and 43,923 controls63. GWAS for AD was derived from a GWAS meta-analysis of GWAS-by-proxy (GWAX) for family history of AD in UK Biobank (53,042 cases and 355,900 controls) with the latest GWAS for diagnosed AD (21,982 cases and 41,944 controls)64. GWAS data for ALS was derived from a GWAS analysis from Nicolas et al. containing 20,806 ALS cases and 59,804 controls65. GWAS for LBD came from a cohort of 2,981 patients diagnosed with LBD (1,789 autopsy-confirmed LBD cases and 802 clinical LBD cases) and 4,391 controls from 17 European and 27 North American sites/consortia66. GWAS data for PD was derived from International Parkinson’s Disease Genomics Consortium (IPDGC), including 33,674 cases and 449,056 controls excluding the 23andMe samples67. GWAS for hypercholesterolaemia, hypertension, angina pectoris, IHD, asthma, and hypothyroidism were derived from UK biobank39. All the participants of above studies were of European ancestry, and more details were shown in original studies and Supplementary materials. All the participants of above studies were of European ancestry.
Table 2.
GWAS summary statistics: source and description.
| Type | Phenotype | Data source | Sample | Ethnicity |
|---|---|---|---|---|
| Exposure | PM2.5 | UK Biobank/ ESCAPE | 423,796 | European |
| Outcome | Longevity | Longevity Genomics research group | 11,262/3,484 cases and 25,483 controls | European, East Asian, and African American |
| Potential confounders | nitrogen dioxide air pollution | UK Biobank/ ESCAPE | 456,380 | European |
| nitrogen oxides air pollution | UK Biobank/ ESCAPE | 456,380 | European | |
| PM10 | UK Biobank/ ESCAPE | 423,796 | European | |
| PM2.5-10 | UK Biobank/ ESCAPE | 423,796 | European | |
| cigarettes smoked per day | GSCAN | 337,334 | European | |
| Potential mediators | BMI | GIANT | 693,529 | European |
| WHR | GIANT | 693,529 | European | |
| HC | GIANT | 142,762 | European | |
| WC | GIANT | 142,762 | European | |
| T1D | CMDKP | 9,358 cases/15,705 controls | European | |
| T2D | DIAMANTE | 80,154 cases /853,816 controls | European | |
| FG, FI, HbA1c | GWAS Catalog | 281,416 | European | |
| LDL-C | UK Biobank, Global Lipids Genetics Consortium | 431,167 | European | |
| Hypercholesterolaemia | UK Biobank | 22,622 cases/440,388 controls | European | |
| DBP | ICBP | 757,601 | European | |
| SBP | ICBP | 757,601 | European | |
| Hypertension | UK Biobank | 2095 cases/460,838 controls | European | |
| CAD | CMDKP | 86,847 cases/417,789 controls | European | |
| Angina pectoris | UK Biobank | 4256 cases/458,754 controls | European | |
| HF | HERMES | 47,309 cases/930,014 controls | European | |
| AF | CMDKP | 55,114 cases/482,295 controls | European | |
| IHD | UK Biobank | 1195 cases/461,815 controls | European | |
| Stroke | GWAS Catalog | 73,652 cases/1,234,808 controls | European | |
| IS | GWAS Catalog | 62,100 cases/1,234,808 controls | European | |
| FVC | SpiroMeta Consortium, UK Biobank | 321,047 | European | |
| FEV1/FVC | SpiroMeta Consortium, UK Biobank | 321,047 | European | |
| COPD | UK Biobank, COPDGene Study, SpiroMeta Consortium | 21,077 cases/179,689 controls | European | |
| Lung cancer | ILCCO | 11,348 cases/15,861 controls | European | |
| Asthma | UK Biobank | 1877 cases/461,133 controls | European | |
| Hypothyroidism | UK Biobank | 22,687 cases/440,246 controls | European | |
| SLE | GWAS Catalog | 7219 cases/15,991 controls | European | |
| CD | IIBDGC | 17,897 cases/33,977 controls | European | |
| UC | IIBDGC | 13,768 cases/33,977 controls | European | |
| RA | GWAS Catalog | 14,361 cases/43,923 controls | European | |
| MS | IMSGC | 47,429 cases/68,374 controls | European | |
| ALS | GWAS Catalog | 20,806 cases/59,804 controls | European | |
| PD | IPDGC | 33,674 cases/449,056 controls | European | |
| AD | UK Biobank, IGAP | 75,024 cases/397,844 controls | European | |
| LBD | GWAS Catalog | 2981 cases/4,391 controls | European |
AD Alzheimer’s disease, AF atrial fibrillation, ALS amyotrophic lateral sclerosis, BMI body mass index, CAD coronary artery disease, CD Crohn’s disease, COPD chronic obstructive pulmonary disease, DBP diastolic blood pressure, FEV1/FVC 1 s forced expiratory volume/FVC, FG fasting glucose, FI fasting insulin, FVC forced vital capacity, HbA1c glycated hemoglobin, HC hip circumference, HF heart failure, IHD ischemic heart disease, IS ischemic stroke, LBD lewy body dementia, MS multiple sclerosis, PD Parkinson’s disease, RA rheumatoid arthritis, SBP systolic blood pressure, SLE Systemic lupus erythematosus, T1D type 1 diabetes, T2D type 2 diabetes, UC Ulcerative colitis, WC waist circumference, WHR waist-hip ratio.
Selection of instrumental variables
We selected independent genome-wide significant single-nucleotide polymorphisms (SNPs) associated with exposures as genetic instruments (P < 5 E-08). The instruments were clumped based on the European 1000 genomes reference panel using PLINK (r2 < 0.001). The instruments of palindromic and incompatible alleles were removed when harmonizing exposure and outcome. F statistic < 10 indicated a weak instrument bias in MR analysis.
Univariable MR
We used IVW as a primary approach to assess the causal effect of genetically predicted PM2.5 concentration on longevity, namely combining the Wald ratio estimates of each SNP instrument27,68–71. We supplemented IVW method with weighted median estimators, which allowed more powerful instruments to contribute more27.
Sensitivity analysis
If the SNP instruments show horizontal pleiotropy, the MR assumptions may be violated and the MR results may be severely biased36. We performed conservative analyses (including fewer variants) to remove the influence of pleiotropy using MR-Egger and MR-PRESSO27,72. MR-Egger allows all SNP instruments to have pleiotropic effects, but the pleiotropy effects should be independent of the SNP-exposure association36. MR-PRESSO method removes SNP instruments from the analysis whose causal estimates differ substantially from those of other instruments and then continues to perform IVW analysis36.
Multivariable MR
Other types of air pollution and smoking may be confounding factors for the effect of PM2.5 on longevity. Multivariable MR allows SNP instruments to be associated with more than one exposure, and estimates the direct causal effect of each exposure in a single MR model27,36. We performed multivariate MR analysis to assess the independent causal effect of genetically predicted PM2.5 concentration on potential mediators and longevity. The multivariate IVW was used as the primary analysis.
Mediation analysis
We applied a two-step MR model to calculate the mediation effect of potential mediators. In the first step, we used SNP instruments for PM2.5 to estimate the causal effect of PM2.5 concentration on potential mediators. In the second step, we used SNP instruments for potential mediating phenotypes to estimate the causal effect of potential mediators on longevity. We assessed the indirect effect of PM2.5 concentration on longevity via each mediating factor using product of coefficients method73–75. The standard error for the indirect effect was derived by using the delta method76.
R packages TwoSampleMR (version 0.5.6) and MRPRESSO (version 1.0) were used for MR analyses. The statistically significant association is defined to be P < 0.05/36 = 0.0014 after multiple testing.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work has been supported by the National Key Research and Development Program of China (No: 2021YFF1200100). We would like to acknowledge the UK Biobank, Longevity Genomics research group, GSCAN, GIANT, CMDKP, DIAMANTE, GWAS Catalog, Global Lipids Genetics Consortium, ICBP, HERMES, CMDKP, SpiroMeta Consortium, COPDGene Study, SpiroMeta Consortium, ILCCO, IIBDGC, IMSGC, IPDGC, IGAP, and other consortiums or authors for providing GWAS summary statistics.
Author contributions
Y.H. and G.Y.L. designed the study; S.Z.Q. analyzed the data. All authors contributed to the interpretation of the results and critical revision of the manuscript for important intellectual content and approved the final version of the manuscript.
Data availability
GWAS for longevity: https://www.longevitygenomics.org/downloads;
GWAS for PM2.5: https://gwas.mrcieu.ac.uk/datasets/ukb-b-10817/;
GWAS for PM10: https://gwas.mrcieu.ac.uk/datasets/ukb-b-18469/;
GWAS for PM2.5-10: https://gwas.mrcieu.ac.uk/datasets/ukb-b-12963/;
GWAS for BMI, WHR, HC, and WC: https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files;
GWAS for T1D and T2D: https://hugeamp.org/downloads.html;
GWAS for FG, FI, HbA1c, LDL-C, hypercholesterolaemia, DBP, SBP, hypertension, IHD, stroke, IS, FVC, FEV1/FVC, COPD, lung cancer, asthma, hypothyroidism, SLE, CD, UC, RA, MS, ALS, PD, AD, and LBD were obtained from IEU OpenGWAS project: https://gwas.mrcieu.ac.uk/;
GWAS for angina pectoris, HF, CAD, and AF: https://cd.hugeamp.org/downloads.html
GWAS Catalog: https://www.ebi.ac.uk/gwas/home.
Code availability
The codes for MR and MR-PRESSO are publicly available at https://github.com/MRCIEU/TwoSampleMR and https://github.com/rondolab/MR-PRESSO, respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yang Hu, Email: huyang@hit.edu.cn.
Guiyou Liu, Email: liuguiyou1981@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41514-023-00126-0.
References
- 1.Li, Z. et al. Air pollution interacts with genetic risk to influence cortical networks implicated in depression. Proc. Natl. Acad. Sci. USA118, 10.1073/pnas.2109310118 (2021). [DOI] [PMC free article] [PubMed]
- 2.Collaborators, G. B. D. R. F. Global burden of 87 risk factors in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396:1223–1249. doi: 10.1016/S0140-6736(20)30752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi L, et al. Incident dementia and long-term exposure to constituents of fine particle air pollution: A national cohort study in the United States. Proc. Natl. Acad. Sci. USA. 2023;120:e2211282119. doi: 10.1073/pnas.2211282119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, et al. Impact of lowering fine particulate matter from major emission sources on mortality in Canada: A nationwide causal analysis. Proc. Natl. Acad. Sci. USA. 2022;119:e2209490119. doi: 10.1073/pnas.2209490119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yun, X. et al. Residential solid fuel emissions contribute significantly to air pollution and associated health impacts in China. Sci. Adv.6, 10.1126/sciadv.aba7621 (2020). [DOI] [PMC free article] [PubMed]
- 6.Brook RD, et al. Particulate matter air pollution and cardiovascular disease: An update to the scientific statement from the American Heart Association. Circulation. 2010;121:2331–2378. doi: 10.1161/CIR.0b013e3181dbece1. [DOI] [PubMed] [Google Scholar]
- 7.Wen M, Gu D. Air pollution shortens life expectancy and health expectancy for older adults: the case of China. J. Gerontol. A Biol. Sci. Med. Sci. 2012;67:1219–1229. doi: 10.1093/gerona/gls094. [DOI] [PubMed] [Google Scholar]
- 8.Allen RT, et al. Countervailing effects of income, air pollution, smoking, and obesity on aging and life expectancy: population-based study of U.S. Counties. Environ Health. 2016;15:86. doi: 10.1186/s12940-016-0168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen G, Gerber Y. Air Pollution and Successful Aging: Recent Evidence and New Perspectives. Curr. Environ. Health Rep. 2017;4:1–11. doi: 10.1007/s40572-017-0127-2. [DOI] [PubMed] [Google Scholar]
- 10.Payne CF, Xu KQ. Life Course Socioeconomic Status and Healthy Longevity in China. Demography. 2022;59:629–652. doi: 10.1215/00703370-9830687. [DOI] [PubMed] [Google Scholar]
- 11.Boing AF, deSouza P, Boing AC, Kim R, Subramanian SV. Air Pollution, Socioeconomic Status, and Age-Specific Mortality Risk in the United States. JAMA Netw. Open. 2022;5:e2213540. doi: 10.1001/jamanetworkopen.2022.13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu GG, Kwon O, Xue X, Fleisher BM. How Much Does Social Status Matter to Longevity?-Evidence from China’s Academician Election. Health Econ. 2017;26:292–304. doi: 10.1002/hec.3300. [DOI] [PubMed] [Google Scholar]
- 13.Yitshak-Sade, M. et al. Neighborhood Greenness Attenuates the Adverse Effect of PM(2.5) on Cardiovascular Mortality in Neighborhoods of Lower Socioeconomic Status. Int. J. Environ. Res. Public Health16, 10.3390/ijerph16050814 (2019). [DOI] [PMC free article] [PubMed]
- 14.Madaniyazi L, Li S, Li S, Guo Y. Candidate gene expression in response to low-level air pollution. Environ Int. 2020;140:105610. doi: 10.1016/j.envint.2020.105610. [DOI] [PubMed] [Google Scholar]
- 15.Yao Y, Liu L, Guo G, Zeng Y, Ji JS. Interaction of Sirtuin 1 (SIRT1) candidate longevity gene and particulate matter (PM2.5) on all-cause mortality: a longitudinal cohort study in China. Environ. Health. 2021;20:25. doi: 10.1186/s12940-021-00718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prufer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji JS, Liu L, Yan LL, Zeng Y. Comparing Effects of FOXO3 and Residing in Urban Areas on Longevity: A Gene-Environment Interaction Study. J. Gerontol. A Biol. Sci. Med. Sci. 2022;77:1549–1556. doi: 10.1093/gerona/glab362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haghani A, et al. Air Pollution Alters Caenorhabditis elegans Development and Lifespan: Responses to Traffic-Related Nanoparticulate Matter. J. Gerontol. A Biol. Sci. Med. Sci. 2019;74:1189–1197. doi: 10.1093/gerona/glz063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martens DS, et al. Prenatal Air Pollution and Newborns’ Predisposition to Accelerated Biological Aging. JAMA Pediatr. 2017;171:1160–1167. doi: 10.1001/jamapediatrics.2017.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks-Wilson AR. Genetics of healthy aging and longevity. Hum. Genet. 2013;132:1323–1338. doi: 10.1007/s00439-013-1342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23:R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu S, Wang D, Zhang Y, Hu Y. Mendelian randomization reveals potential causal candidates for COVID-19 in 123 blood metabolites. J. Infect. 2022;84:248–288. doi: 10.1016/j.jinf.2021.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu S, Zheng K, Hu Y, Liu G. Genetic correlation, causal relationship, and shared loci between vitamin D and COVID-19: A genome-wide cross-trait analysis. J. Med. Virol. 2023;95:e28780. doi: 10.1002/jmv.28780. [DOI] [PubMed] [Google Scholar]
- 24.Abaturov A, Nikulina A. Obesity in Children with Leptin Receptor Gene Polymorphisms. Acta. Medica. (Hradec Kralove) 2021;64:158–164. doi: 10.14712/18059694.2021.27. [DOI] [PubMed] [Google Scholar]
- 25.Hu Y, et al. Mendelian randomization highlights causal association between genetically increased C-reactive protein levels and reduced Alzheimer’s disease risk. Alzh. Dement. 2022;18:2003–2006. doi: 10.1002/alz.12687. [DOI] [PubMed] [Google Scholar]
- 26.Hu Y, et al. Cognitive performance protects against Alzheimer’s disease independently of educational attainment and intelligence. Mol. Psychiatry. 2022;27:4297–4306. doi: 10.1038/s41380-022-01695-4. [DOI] [PubMed] [Google Scholar]
- 27.Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife7, 10.7554/eLife.34408 (2018). [DOI] [PMC free article] [PubMed]
- 28.Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 29.Pope CA, 3rd, Ezzati M, Dockery DW. Fine-particulate air pollution and life expectancy in the United States. N. Engl. J. Med. 2009;360:376–386. doi: 10.1056/NEJMsa0805646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Z, Li X, Han Y, Wu Y, Fang Y. Association of long-term exposure to PM(2.5) with hypertension and diabetes among the middle-aged and elderly people in Chinese mainland: a spatial study. BMC Public Health. 2022;22:569. doi: 10.1186/s12889-022-12984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiwari, I., Herr, R. M., Loerbroks, A. & Yamamoto, S. S. Household Air Pollution and Angina Pectoris in Low- and Middle-Income Countries: Cross-Sectional Evidence from the World Health Survey 2002-2003. Int. J. Environ. Res. Public Health17, 10.3390/ijerph17165802 (2020). [DOI] [PMC free article] [PubMed]
- 32.Medina S, et al. Air pollution and doctors’ house calls: results from the ERPURS system for monitoring the effects of air pollution on public health in Greater Paris, France, 1991-1995. Evaluation des Risques de la Pollution Urbaine pour la Sante. Environ. Res. 1997;75:73–84. doi: 10.1006/enrs.1997.3773. [DOI] [PubMed] [Google Scholar]
- 33.Wang, X. et al. Association of improved air quality with lower dementia risk in older women. Proc. Natl. Acad. Sci. USA119, 10.1073/pnas.2107833119 (2022). [DOI] [PMC free article] [PubMed]
- 34.Wang X, et al. Exposure to Concentrated Ambient PM2.5 Shortens Lifespan and Induces Inflammation-Associated Signaling and Oxidative Stress in Drosophila. Toxicol. Sci. 2017;156:199–207. doi: 10.1093/toxsci/kfw240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terzano C, Di Stefano F, Conti V, Graziani E, Petroianni A. Air pollution ultrafine particles: toxicity beyond the lung. Eur. Rev. Med. Pharmacol. Sci. 2010;14:809–821. [PubMed] [Google Scholar]
- 36.Burgess S, et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 2019;4:186. doi: 10.12688/wellcomeopenres.15555.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melbourne CA, et al. Genome-wide gene-air pollution interaction analysis of lung function in 300,000 individuals. Environ. Int. 2022;159:107041. doi: 10.1016/j.envint.2021.107041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward-Caviness CK. A review of gene-by-air pollution interactions for cardiovascular disease, risk factors, and biomarkers. Hum. Genet. 2019;138:547–561. doi: 10.1007/s00439-019-02004-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doiron, D. et al. Air pollution, lung function and COPD: results from the population-based UK Biobank study. Eur. Respir. J.54, 10.1183/13993003.02140-2018 (2019). [DOI] [PubMed]
- 41.Deelen J, et al. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat. Commun. 2019;10:3669. doi: 10.1038/s41467-019-11558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu M, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verhoeven JI, Allach Y, Vaartjes ICH, Klijn CJM, de Leeuw FE. Ambient air pollution and the risk of ischaemic and haemorrhagic stroke. Lancet. Planet Health. 2021;5:e542–e552. doi: 10.1016/S2542-5196(21)00145-5. [DOI] [PubMed] [Google Scholar]
- 44.Glencross DA, Ho TR, Camina N, Hawrylowicz CM, Pfeffer PE. Air pollution and its effects on the immune system. Free Radic. Biol. Med. 2020;151:56–68. doi: 10.1016/j.freeradbiomed.2020.01.179. [DOI] [PubMed] [Google Scholar]
- 45.Sollis E, et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2023;51:D977–D985. doi: 10.1093/nar/gkac1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lyon MS, et al. The variant call format provides efficient and robust storage of GWAS summary statistics. Genome Biol. 2021;22:32. doi: 10.1186/s13059-020-02248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yengo L, et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum. Mol. Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahajan A, et al. Multi-ancestry genetic study of type 2 diabetes highlights the power of diverse populations for discovery and translation. Nat. Genet. 2022;54:560–572. doi: 10.1038/s41588-022-01058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forgetta V, et al. Rare Genetic Variants of Large Effect Influence Risk of Type 1 Diabetes. Diabetes. 2020;69:784–795. doi: 10.2337/db19-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J, et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 2021;53:840–860. doi: 10.1038/s41588-021-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klimentidis YC, et al. Phenotypic and Genetic Characterization of Lower LDL Cholesterol and Increased Type 2 Diabetes Risk in the UK Biobank. Diabetes. 2020;69:2194–2205. doi: 10.2337/db19-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aragam KG, et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat. Genet. 2022;54:1803–1815. doi: 10.1038/s41588-022-01233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shah S, et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat. Commun. 2020;11:163. doi: 10.1038/s41467-019-13690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roselli C, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 2018;50:1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mishra A, et al. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature. 2022;611:115–123. doi: 10.1038/s41586-022-05165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shrine N, et al. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat. Genet. 2019;51:481–493. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim W, et al. Genome-Wide Gene-by-Smoking Interaction Study of Chronic Obstructive Pulmonary Disease. Am. J. Epidemiol. 2021;190:875–885. doi: 10.1093/aje/kwaa227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat. Genet. 2014;46:736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.International Multiple Sclerosis Genetics, C. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science365, 10.1126/science.aav7188 (2019). [DOI] [PMC free article] [PubMed]
- 63.Ha E, Bae SC, Kim K. Large-scale meta-analysis across East Asian and European populations updated genetic architecture and variant-driven biology of rheumatoid arthritis, identifying 11 novel susceptibility loci. Ann. Rheum. Dis. 2021;80:558–565. doi: 10.1136/annrheumdis-2020-219065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartzentruber J, et al. Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer’s disease risk genes. Nat. Genet. 2021;53:392–402. doi: 10.1038/s41588-020-00776-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicolas A, et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron. 2018;97:1268–1283.e1266. doi: 10.1016/j.neuron.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chia R, et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat. Genet. 2021;53:294–303. doi: 10.1038/s41588-021-00785-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nalls MA, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qiu S, Cao P, Guo Y, Lu H, Hu Y. Exploring the Causality Between Hypothyroidism and Non-alcoholic Fatty Liver: A Mendelian Randomization Study. Front. Cell Dev. Biol. 2021;9:643582. doi: 10.3389/fcell.2021.643582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu S, Li M, Jin S, Lu H, Hu Y. Rheumatoid Arthritis and Cardio-Cerebrovascular Disease: A Mendelian Randomization Study. Front. Genet. 2021;12:745224. doi: 10.3389/fgene.2021.745224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bahrami S, et al. Dissecting the shared genetic basis of migraine and mental disorders using novel statistical tools. Brain. 2022;145:142–153. doi: 10.1093/brain/awab267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qiu S, Hu Y, Cheng L. A genome-wide cross-trait analysis highlights the shared genetic structure between COVID-19 and Alzheimer’s disease. J. Infect. 2022;84:e1–e2. doi: 10.1016/j.jinf.2021.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burgess S, Daniel RM, Butterworth AS, Thompson SG, Consortium, E. P.-I. Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int. J. Epidemiol. 2015;44:484–495. doi: 10.1093/ije/dyu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yao S, et al. Bidirectional two-sample Mendelian randomization analysis identifies causal associations between relative carbohydrate intake and depression. Nat. Hum. Behav. 2022;6:1569–1576. doi: 10.1038/s41562-022-01412-9. [DOI] [PubMed] [Google Scholar]
- 75.Carter AR, et al. Mendelian randomisation for mediation analysis: current methods and challenges for implementation. Eur. J. Epidemiol. 2021;36:465–478. doi: 10.1007/s10654-021-00757-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheung MW. Comparison of methods for constructing confidence intervals of standardized indirect effects. Behav. Res. Methods. 2009;41:425–438. doi: 10.3758/BRM.41.2.425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GWAS for longevity: https://www.longevitygenomics.org/downloads;
GWAS for PM2.5: https://gwas.mrcieu.ac.uk/datasets/ukb-b-10817/;
GWAS for PM10: https://gwas.mrcieu.ac.uk/datasets/ukb-b-18469/;
GWAS for PM2.5-10: https://gwas.mrcieu.ac.uk/datasets/ukb-b-12963/;
GWAS for BMI, WHR, HC, and WC: https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files;
GWAS for T1D and T2D: https://hugeamp.org/downloads.html;
GWAS for FG, FI, HbA1c, LDL-C, hypercholesterolaemia, DBP, SBP, hypertension, IHD, stroke, IS, FVC, FEV1/FVC, COPD, lung cancer, asthma, hypothyroidism, SLE, CD, UC, RA, MS, ALS, PD, AD, and LBD were obtained from IEU OpenGWAS project: https://gwas.mrcieu.ac.uk/;
GWAS for angina pectoris, HF, CAD, and AF: https://cd.hugeamp.org/downloads.html
GWAS Catalog: https://www.ebi.ac.uk/gwas/home.
The codes for MR and MR-PRESSO are publicly available at https://github.com/MRCIEU/TwoSampleMR and https://github.com/rondolab/MR-PRESSO, respectively.