Highlights
-
•
Drug repurposing screens identify Tubercidin as a potent antiviral agent against PEDV.
-
•
Tubercidin exhibited significant antiviral activity against both classical and variant PEDV on Vero and LLC-PK1 cells.
-
•
Tubercidin displayed a significant inhibitory effect on viral post-entry events but not other periods.
-
•
Tubercidin had better docking efficiency and formed hydrophobic interactions with the active pocket of RdRp of nidoviruses.
-
•
Tubercidin can also suppress SADS-CoV and PRRSV infections.
Keywords: Coronavirus, Porcine epidemic diarrhea virus, Tubercidin, Antiviral, RNA-dependent RNA polymerase
Abstract
The emergence of new coronaviruses poses a significant threat to animal husbandry and human health. Porcine epidemic diarrhea virus (PEDV) is considered a re-emerging porcine enteric coronavirus, which causes fatal watery diarrhea in piglets. Currently, there are no effective drugs to combat PEDV. Drug repurposing screens have emerged as an attractive strategy to accelerate antiviral drug discovery and development. Here, we screened 206 natural products for antiviral activity using live PEDV infection in Vero cells and identified ten candidate antiviral agents. Among them, Tubercidin, a nucleoside analog derived from Streptomyces tubercidicus, showed promising antiviral activity against PEDV infection. Furthermore, we demonstrated that Tubercidin exhibited significant antiviral activity against both classical and variant PEDV. Time of addition assay showed that Tubercidin displayed a significant inhibitory effect on viral post-entry events but not during other periods. Molecular docking analysis indicated that Tubercidin had better docking efficiency and formed hydrophobic interactions with the active pocket of RNA-dependent RNA polymerase (RdRp) of PEDV and other nidoviruses. Additionally, Tubercidin can effectively suppress other porcine nidoviruses, such as SADS-CoV and PRRSV, demonstrating its broad-spectrum antiviral properties. In summary, our findings provide valuable evidence for the antiviral activity of Tubercidin and offer insights into the development of new strategies for the prevention and treatment of coronavirus infections.
1. Introduction
Nidoviruses, including arteriviruses, coronaviruses, and roniviruses, are a phylogenetically compact but diverse group of positive-strand RNA viruses that include important human and animal pathogens (Gorbalenya et al., 2006). Coronaviruses are important viral pathogens that infect a broad range of vertebrates, including humans, mammalian and avian species (Fehr and Perlman, 2015). Porcine epidemic diarrhea virus (PEDV), an important member of the genus Alphacoronavirus, can cause vomiting, watery diarrhea, dehydration, and weight loss in piglets, with mortality rates of up to 90–100 %. In the 1970s, PEDV was first detected in Europe and subsequently spread to Asia (Wood, 1977). In late 2010, a highly variable and virulent strain of PEDV caused a large-scale outbreak in China, resulting in significant economic losses (Li et al., 2012a; Sun et al., 2012). Since then, the virus has continued to spread rapidly across countries and undergo frequent mutations, resulting in different evolutionary branches (Huang et al., 2013; Marthaler et al., 2013). Based on the spike (S) gene sequence, PEDV was classified into classical strains (GI subtype) that first emerged in the 1970s and new variant strains (GII subtype) that have been prevalent worldwide since 2010 (Chen et al., 2013; Li et al., 2012b). These two groups have been subdivided into five subgroups (GIa, GIb, GIIa, GIIb, and GIIc) that differ in molecular characteristics, antigenicity, and pathogenicity (Zhang et al., 2022). Recent studies revealed that the GIIa subgroup has been increasing rapidly and has become dominant in recent years (Li et al., 2022). Unfortunately, PEDV frequently undergoes mutations that allow it to escape the effects of vaccines, resulting in immunization failure (Gerdts and Zakhartchouk, 2017). Despite the severity of PEDV infections, there are currently no effective drugs available to combat the virus. Consequently, there is an urgent need to discover antiviral drugs that can treat PEDV infections.
The genetic material of coronaviruses is positive-sense single-stranded RNA, which can directly act as a viral messenger RNA (mRNA) and translate into viral proteins in the host cell (V'Kovski et al., 2021). Before producing the progeny viral genome based on the single-stranded RNA template, viral RNA-dependent RNA polymerase (RdRp) must first be synthesized, which implies that it is an indispensable protein in the process of viral replication (Venkataraman et al., 2018). The RdRp which is the core component of the multisubunit replication and transcription complexes of non-structural proteins, plays a significant role in both viral RNA replication and transcription (Subissi et al., 2014). RdRp is highly conserved in the life cycle of RNA viruses, including influenza virus, Hepatitis C virus (HCV), Zika virus (ZIKV), and Coronavirus (CoV) (Wang et al., 2021; Yin et al., 2020). The conserved protein structure allows RdRp to have a common function across different virus genera. Given the approval of Remdesivir, a therapeutic drug acting on RdRp of SARS-CoV-2, it has become a promising therapeutic target for developing broad-spectrum antiviral drugs, particularly against RdRp of coronaviruses (Picarazzi et al., 2020; Xu et al., 2022).
In recent years, drug repurposing screens have become an attractive strategy to discover antiviral agents (Wang et al., 2023). Natural products, which offer a wide range of structures and chemical diversity along with relatively low toxicity, have had a significant impact on drug development and pharmacotherapy. In this study, a natural compound library consisting of 206 compounds was screened for its antiviral efficacy against PEDV, resulting in the identification of ten effective antiviral agents. Among these agents, Tubercidin (7-deazaadenosine), a nucleoside analog derived from Streptomyces tubercidicus, exhibited the most potent anti-PEDV activity without causing obvious cytotoxicity. Besides, we also demonstrated that Tubercidin inhibits viral infection by affecting the period of virus replication. Furthermore, potential interactions between Tubercidin and RdRp proteins of various nidoviruses were identified through molecular docking and homolog model-based analysis. In addition, the specific antiviral activity of Tubercidin against SADS-CoV and PRRSV was examined. This research provides valuable insights into the potential of Tubercidin as a broad-spectrum antiviral agent against PEDV and other nidovirus infections.
2. Materials and methods
2.1. Cells, viruses and compounds
African green monkey kidney cell (Vero), Porcine kidney proximal tubular epithelial cell (LLC-PK1) and Marc145 cell were maintained in our laboratory. All cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, HyClone, Logan, UT, United States) supplemented with 10% fetal bovine serum, 100 U/mL penicillin–streptomycin and maintained at 37 °C in a humidified 5% CO2 incubator. The PEDV strain CV777 (GIa, GenBank accession number: AF353511.1), HNAY (S insertion, GenBank accession number: MT338518.1), HNXX (GIIa, GenBank accession number: MT338517.1), HB (GIIb, GenBank accession number: KY928065.1), recombinant DR13-GFP (GIa, GenBank accession number: JQ023162.1) and GDU-GFP (GIIb, GenBank accession number: KU985230.1), and PRRSV-GFP (GenBank accession number: JX087437.1) were stored in our laboratories. All viruses were propagated on cells with 6 μg/mL trypsin except for DR13-GFP and PRRSV-GFP. The natural compound library, which contained 206 compounds, was provided by Henan University of Chinese Medicine School of Medicine. Tubercidin with a purity of ≥ 98.0 % was purchased from MedChemExpress.
2.2. Antiviral drug screening
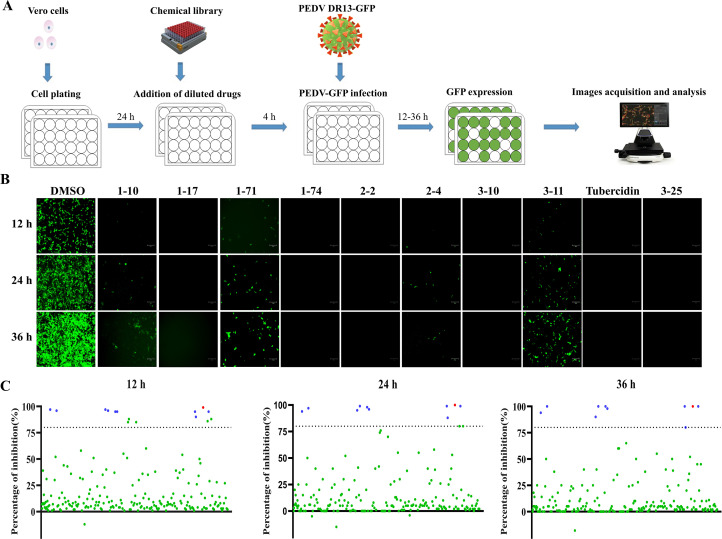
Vero cells were grown as a monolayer in 24-well plates, the cells were treated with 10 μM compound or 0.1% DMSO for 4 h. The cells were washed with PBS, then infected with DR13-GFP (0.1 MOI). At 12, 24 and 36 h post infection (hpi), CPE were observed and recorded suing an EVOSTM M5000 Imaging System (invitrogen-EVOS M5000, USA). Subsequently, the fluorescence intensity was measured using ImageJ software. The percentage of fluorescence intensity inhibition was calculated by comparing drug treatment to DMSO treatment. The formula used was: (DMSO sample - drug sample) / DMSO sample × 100%. In the primary screening round, compounds were eliminated if they exhibited visible cytotoxicity or showed less than a 50% reduction of CPE compared to the 0.1 % DMSO control group. In the second and third screening rounds, cell viability and PEDV inhibition needed to be 80 % or higher.
2.3. Cell viability and CC50 determination
Firstly, Vero cells were seeded into 96-well plates containing Tubercidin (0.1–30 μM). Afterwards, the cells were incubated with Tubercidin for 16 h. Following the manufacturer's instructions, cell viability was measured using the cell counting kit-8 (CCK-8, Abbkine Wuhan, China) assay. Subsequently, the culture medium in each well was replaced with DMEM containing 10 % CCK-8 solution and then incubated for an additional 1 h. Finally, the optical density (OD 450) of the culture was measured using a microplate reader (BMG Labtech POLAR star Omega). The maximum nontoxic concentration of a drug and the half maximal cytotoxic concentration (CC50) of cells can be calculated using the following formula: cell viability = (As−Ab) ÷ (Ac−Ab) × 100 % (As represents OD value of incubated Tubercidin, CCK-8, and cell solution; Ab represents OD value of well without cells; Ac represents OD value of well without Tubercidin but with incubated cells and CCK-8).
2.4. IC50 determination
Vero cells were pre-treated with Tubercidin or 0.1 % DMSO for 4 h in a 24-well plate. After the pre-treatment, DR13-GFP (0.1 MOI) was used to infect the cells in the presence or absence of Tubercidin. After 12 h, the infected cells were digested with 2.5 % trypsin and centrifuged at 200 × g for 5 min. The resulting cell pellet was washed three times with 300 μL of PBS. Finally, the cells were resuspended in PBS phosphate buffer. Flow cytometry was then utilized to determine the percentage of green fluorescent cells in the overall cell population.
2.5. Antiviral activity study of Tubercidin in different drug-addition approaches
To evaluate the antiviral activity of Tubercidin under different drug-addition approaches, Vero cells were infected with DR13-GFP (0.1 MOI). Different concentrations of Tubercidin were added to cells by four kinds of treatments: all-treatment (before and after infection), pre-treatment (4 h before infection), co-treatment (simultaneous incubation), and post-treatment (1 h after infection). Virus infection was determined using TCID50 and RT-qPCR assay. Each experimental group was independently replicated three times.
2.6. Virus inactivation assay
Tubercidin (1 μM) or 0.1 % DMSO was incubated with PEDV (0.1 MOI) at 37 °C for 2 h. A mixture of Tubercidin (1 μM) or 0.1 % DMSO and PEDV was added to Vero cells in 24-well plates. After incubation at 37 °C for an additional 1 h, the cells were washed with PBS. The culture supernatants were then replaced with fresh DMEM and incubated for an additional 12 h. The control group consisted of cells simultaneously exposed to the virus and Tubercidin.Finally, the RNA expression level of PEDV was determined by RT-qPCR.
2.7. Virus attachment assay
Vero cells were seeded in 24-well plates, and then treated with a mixture of Tubercidin (1 μM) or 0.1 % DMSO and PEDV (1 MOI) for 2 h at 4 °C to allow virus attachment and prevent virus entry. After that, the cells were washed three times with pre-cold PBS and harvested to determine the RNA expression level of PEDV using RT-qPCR.
2.8. Virus internalization assay
Vero cells were seeded in 24-well plates, then incubated with PEDV (1 MOI) for 2 h at 4 °C. After washing with PBS to remove noninternalized virus, the cells were treated with Tubercidin (1 μM) or 0.1 % DMSO for 2 h at 37 °C. The cells were washed three times with PBS and harvested to determine the RNA expression level of PEDV using RT-qPCR.
2.9. Virus replication assay
Vero cells were seeded in 24-well plates, then incubated with PEDV (0.1 MOI) for 1 h at 37 °C. The cells were washed with PBS to remove free virus. At 6 hpi, the cells were cultured in DMEM containing Tubercidin (1 μM) or 0.1 % DMSO. At 8, 10 and 12 hpi, the cells were washed three times with PBS and harvested to determine the RNA expression level of PEDV using RT-qPCR.
2.10. Virus release assay
Vero cells were seeded in 24-well plates, then incubated with PEDV (0.1 MOI) for 1 h at 37 °C. The cells were washed with PBS to remove free virus. At 12 hpi, the cells were cultured in DMEM containing Tubercidin (1 μM) or 0.1 % DMSO. At 16 hpi, The cells and the culture supernatants were harvested to determine the RNA expression level of PEDV using RT-qPCR.
2.11. Antiviral activity study of different drug-addition times
Vero cells were infected with two different genotypes of PEDV, DR13-GFP and HB. At −2, 2, 4, 6, 8, 10, or 12 hpi, Tubercidin (1 μM) was added to cells. After 16 h, the cell and culture supernatant mixture was lysed using TRIzol reagent (TIANGEN, Beijing, China). Finally, the RNA expression level of PEDV was determined by RT-qPCR.
2.12. TCID50 assay
Vero cells were inoculated into 96-well plates and grown to monolayer. Vero cells in 96-well plates was infected with 10-fold serial dilutions of PEDV virus solution, with a total of 6 repetitions. The changes of the virus-infected cells were recorded through continuous observation for 96 h. The TCID50 was calculated using the Reed and Munch method.
2.13. Total RNA extraction and RT-qPCR
Total RNA was extracted using a TRNzol Reagent (Tiangen, Beijing, China) following the manufacturer's instructions, and the concentration of extracted RNA was measured by ultramicron spectrophotometer. cDNA was synthesized using HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China) as per the manufacturer's instructions. The absolute RT-qPCR assay for quantification of PEDV or SADS-CoV genomic material was performed using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China). The Ct values of each sample represented the copy number of PEDV RdRp, SADS-CoV N and PRRSV M gene on the standard curves. The primer sequences for RT-qPCR of PEDV were as follows: PEDV-RdRp-F: 5′-CCGAACACGATTTCTTCACT-3′ and PEDV-RdRp-R: 5′-ACTCCTCACAAGCA CCTACCT-3′. The primer sequences for RT-qPCR of SADS-CoV were as follows: SADS-CoV-N-F: 5′-CTCAACAGCCGTCACAGTCT-3′ and SADS-CoV-N-R: 5′-GCTGAACGAGGTCACTGTCA-3′. The primer sequences for RT-qPCR of PRRSV were as follows: PRRSV-M-F: 5′-TTGCTAGGCCGCAAGTAC-3′ and PRRSV-M-R: 5′-ACGCCGGACGACAAATGC-3′.
2.14. Indirect immunofluorescence assay (IFA)
The cells were rinsed with PBS and then fixed in 4 % paraformaldehyde (PFA, Solarbio, Beijing, China) at room temperature for 15 min. They were washed with PBS three times. Next, the cells were permeabilized with Triton X-100 (Solarbio, Beijing, China) for 15 min and washed with PBS three times. The cells were blocked with PBS containing 1 % bovine serum albumin for 30 minutes at RT, and washed with PBS three times. Following that, the cells were incubated with a mouse anti-PEDV-S monoclonal antibody (prepared by our laboratory at a 1:1000 dilution) for 1 h at 37 °C. They were washed with PBS three times. Afterwards, the cells were incubated with CoraLite 488 coupled with Goat anti-mouse IgG (Proteintech, Rosemont, IL, USA) for 30 min at 37 °C. The antibody was prepared at 1:1000 dilution and the cells were washed again with PBS three times. Finally, the cells were stained with DAPI for 15 min at RT to label the nucleus while avoiding light. The immunofluorescence images were taken with EVOSTM M5000 Imaging System.
2.15. Molecular docking
To analyze the binding energy and interaction mode of Tubercidin and different viruses RdRp proteins, molecular docking was used for analysis. The molecular structure of Tubercidin was obtained from PubChem Compound (https://pubchem.ncbi.nlm.nih.gov/). The SWISS-MODEL homology modeling method was utilized to predict the three-dimensional structure models of the PEDV RdRp, SADS-CoV, TGEV RdRp, FCoV RdRp, CCoV RdRp, SARS-CoV RdRp, SARS-CoV-2 RdRp, MERS-CoV RdRp, IBV RdRp, PDCoV RdRp and PRRSV RdRp proteins. To conduct the docking analysis, all protein and molecular files were converted to PDBQT format. During this process, all water molecules were excluded, and polar hydrogen atoms were added. Lastly, Autodock Vina was employed for molecular docking studies and the compounds, as well as key active sites, were visualized using Pymol. MOE software was used for drawing the 2D depictions.
2.16. Statistical analysis
GraphPad Prism 8.0.2 was used to analyze the statistics in this study. Statistical analyses were performed using one-way ANOVA or student's t-test. All results were expressed as means and standard deviations (SD). The above experiments were performed with three independent replicates. Statistical significances were defined as P < 0.05 (*), and with higher significance was denoted by P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) and P < 0.0001 (****) .
3. Results
3.1. Drug screen for compounds inhibiting PEDV infection
To identify novel antiviral agents against PEDV infection, Vero cells were first treated with 206 natural compounds at the concentration of 10 μM for 4 h. After this treatment, the cells were infected with DR13-GFP at 0.1 MOI and incubated with the compounds for an additional 36 h. The GFP expression at 12, 24, and 36 hpi was recorded and statistically analyzed (Fig. 1A, 1B). Following three rounds of screening, ten natural compounds, including Oleanolic acid (1–10), Moxidectin (1–17), Octyl gallate (1–71), 1,2,3,4,6-Pentagalloylglucose (1–74), Theaflavin 3,3′- digallate (2–2), Curcumin (2–4), Baicalin (3–10), Xanthohumol (3–11), Tubercidin (3–19), Rubitecan (3–25). These compounds inhibited infection by more than 80 % in each screen (Fig. 1C). Due to its low price and effective inhibition, Tubercidin was selected for further exploration.
Fig. 1.
Schematic of PEDV inhibitors screening from a natural product library. Vero cells were pre-treated with natural compounds or 0.1 % DMSO for 4 h, and then infected with DR13-GFP in the presence of compounds. The CPE was observed and recorded at 12, 24 and 36 hpi, scale bars = 150 μm. (A) Diagram of the initial antiviral screening model. (B) Ten drugs, including Oleanolic acid (1–10), Moxidectin (1–17), Octyl gallate (1–71), 1,2,3,4,6-Pentagalloylglucose (1–74), Theaflavin 3,3′- digallate (2–2), Curcumin (2–4), Baicalin (3–10), Xanthohumol (3–11), Tubercidin (3–19), Rubitecan (3–25), displayed inhibitory effects on PEDV DR13-GFP infection. (C) PEDV replication inhibition rates after treatment. The blue dot represents the natural compounds with effective antiviral activity, while the red dot represents Tubercidin.
3.2. The antiviral activity of Tubercidin against PEDV on Vero and LLC-PK1 cells
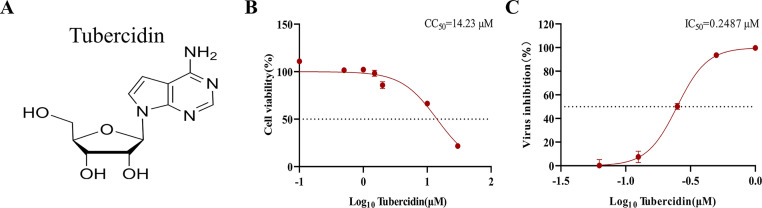
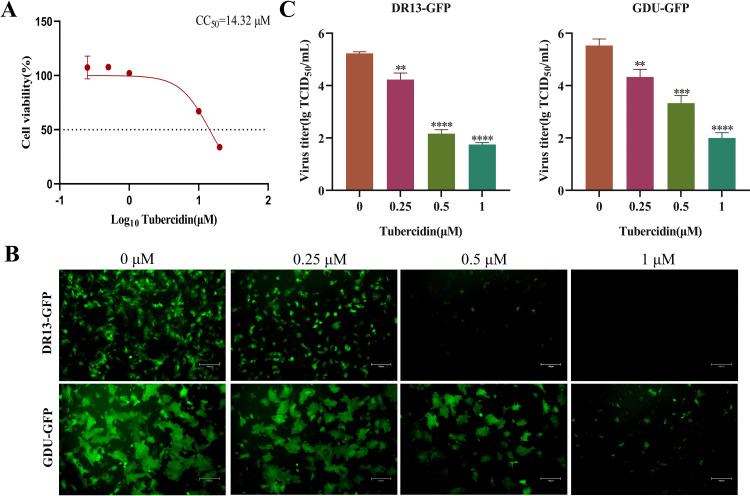
To determine the maximum nontoxic concentration of Tubercidin, Vero cells were treated with a concentration gradient range of 0.1–30 μM for 16 h and then cytotoxicity was determined using the CCK-8 assay. Results showed that the CC50 value of Tubercidin on Vero cells was determined to be 14.23 μM, and did not show obvious toxicity within 1 μM concentration (Fig. 2A, 2B). Furthermore, flow cytometry analysis on Vero cells revealed an IC50 value of 0.2487 μM (Fig. 2C). Similarly, LLC-PK1 cells were treated with Tubercidin to determine the maximum nontoxic concentration (1 μM) and the CC50 value (14.32 μM) (Fig. 3A). In order to further verify the antiviral activity of Tubercidin, LLC-PK1 cells were treated with Tubercidin and then infected by PEDV DR13-GFP and GDU-GFP. The GFP expression results showed a gradual decrease in GFP intensity as the drug concentration increased (Fig. 3B). TCID50 results also confirmed the dose-dependent antiviral effect of Tubercidin against DR13-GFP and GDU-GFP, with a 3.5 log10 inhibition effect observed for DR13-GFP (Fig. 3C). Therefore, it can be concluded that Tubercidin effectively inhibits PEDV infection on Vero and LLC-PK1 cells within a concentration of 1 μM.
Fig. 2.
Evaluation of cytotoxicity on Vero cells and antiviral efficacy of Tubercidin against PEDV. (A) Tubercidin structural formula. (B) Cytotoxicity of Tubercidin on Vero cells. Evaluation of cell viability using CCK-8 assay. Vero cells were treated or not treated with different concentrations of Tubercidin for 16 h. (C) IC50 determination of Tubercidin. Vero cells were pre-treated or not treated with Tubercidin, infected with DR13-GFP at 0.1 MOI in the presence or absence of Tubercidin. After 16 h, flow cytometry was used to assess the proportion of GFP-positive cells in the cell population. The IC50 and CC50 values were calculated by a best-fit Log(dose)-response curve-fitting using GraphPad Prism 8.0.2.
Fig. 3.
Evaluation of cytotoxicity on LLC-PK1 cells and antiviral efficacy of Tubercidin against PEDV. (A) Cytotoxicity of Tubercidin in LLC-PK1 cells. The cytotoxicity was determined by CCK-8 assay. The LLC-PK1 cells were treated or not treated with different concentrations of Tubercidin for 16 h. (B and C) Microscopic images of LLC-PK1 cells infected with DR13-GFP and GDU-GFP (0.1 MOI) treated with different concentrations of Tubercidin (0.25–1 μM). At the same time, viral fluids were collected and virus titer was determined by TCID50 assay. The experiment was performed three times independently, images were representative of results obtained from three independent experiments, scale bars = 150 μm. Differences were considered significant at P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) and P < 0.0001 (****).
3.3. Antiviral activity of Tubercidin against different genotypes of PEDV
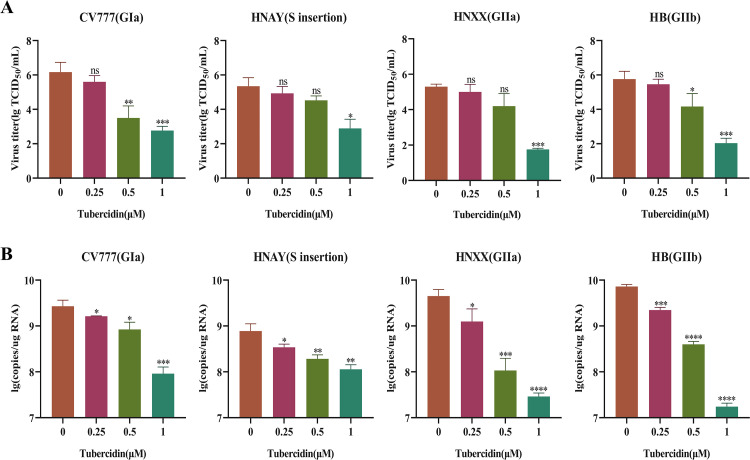
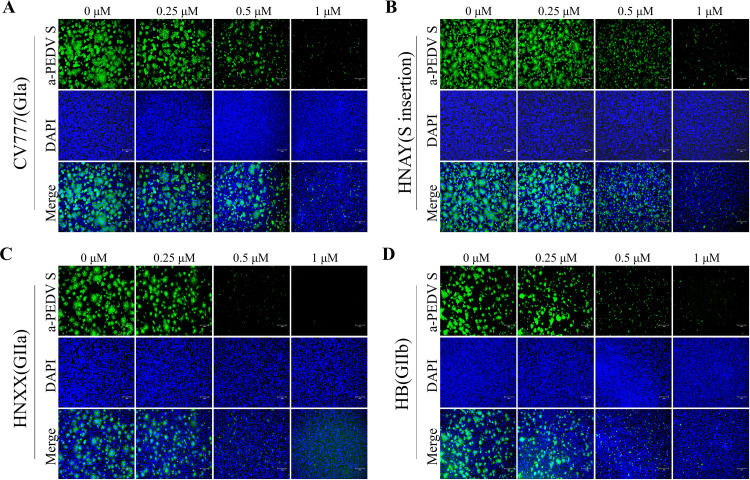
Currently, based on their molecular characteristics and pathogenicity, PEDV strains can be divided into classical (GI) and variant (GII) strains. Given the emergence of PEDV variants and the potential of antivirals to target diverse strains of PEDV, we conducted a study to determine if Tubercidin was effective against PEDV variants. In our experiment, Vero cells were treated with Tubercidin (0.25–1 μM) and then infected with different PEDV strains, including CV777 (GIa), HNAY(S insertion), HNXX (GIIa), and HB (GIIb). The TCID50 results demonstrated that Tubercidin exhibited a dose-dependent inhibitory effect on the proliferation of all four PEDV strains, with the highest inhibition observed on HB, up to approximately 3.5 log10 at 1 μM (Fig. 4A). Similarly, RT-qPCR results revealed that Tubercidin treatment led to a dose-dependent reduction in viral RNA levels (Fig. 4B). These findings demonstrated that Tubercidin effectively restricts viral genome replication and virion production in different genotypes of PEDV. To further assess the impact of Tubercidin on PEDV protein synthesis, we performed IFA to examine the expression of the S protein in infected cells. The results showed a gradual decrease in PEDV-S specific immunofluorescence in PEDV-infected cells with increasing Tubercidin concentration (Fig. 5A-D). These results suggest that Tubercidin can inhibit the synthesis of the PEDV S protein in PEDV-infected cells.
Fig. 4.
Effect of Tubercidin on different genotypes of PEDV strains. Vero cells were infected with CV777, HNAY, HNXX, or HB strains at 0.1 MOI with Tubercidin (0.25–1 μM) treatment. At 16 hpi, viral fluids were collected. (A) The PEDV titers were measured by TCID50 assay. (B) The genomic RNA level of PEDV was measured by RT-qPCR assay. All experiments were performed with three independent replicates. Differences were considered significant at P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) and P < 0.0001 (****).
Fig. 5.
Effect of Tubercidin on S protein Synthesis of PEDV strains. (A-D) Vero cells were infected with CV777, HNAY, HNXX or HB strains (0.1 MOI) with Tubercidin (0.25–1 μM) treatment. At 12 hpi, the S protein expression of four PEDV strains on Vero cells was determined by IFA. Images were representative of results obtained from three independent experiments, scale bars = 150 μm.
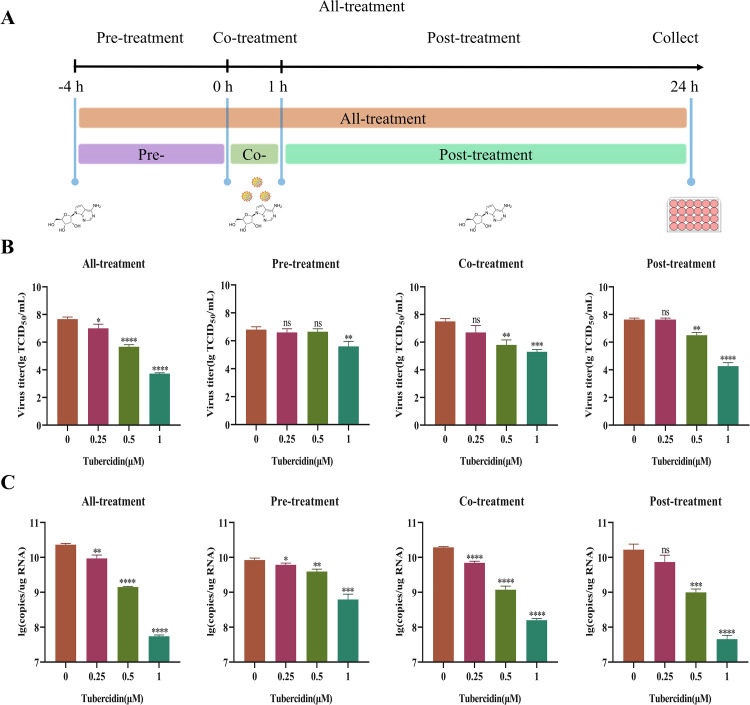
3.4. Antiviral activity of Tubercidin on DR13-GFP infections at different kinds of drug-addition approaches
To explore the possible antiviral mechanism of Tubercidin, four different treatments were applied on Vero cells, including all-treatment (before and after infection), pre-treatment (4 h before infection), co-treatment (simultaneous incubation), and post-treatment (1 h after infection) using DR13-GFP as a model (Fig. 6A). The antiviral activity of Tubercidin was assessed at 16 hpi using TCID50 (Fig. 6B) and RT-qPCR (Fig. 6C). In the pre-treatment assay, Tubercidin demonstrated a significant inhibitory effect on viral infection only at 1 μM, resulting in a reduction of viral infection by approximately 1 log10. Both the TCID50 and RT-qPCR results indicated a significant reduction in DR13-GFP infection under co-treatment and post-treatment conditions, showing a dose-dependent effect. However, the all-treatment condition exhibited the most significant inhibition, with a reduction of up to about 4 log10 at 1 μM. These findings demonstrate that Tubercidin effectively inhibits PEDV infection through different drug-addition approaches.
Fig. 6.
Antiviral activity of Tubercidin on DR13-GFP infections at different kinds of drug-addition approaches. (A) Schematics of Tubercidin addition experiments. Different concentrations of Tubercidin (0.25–1 μM) were added to Vero cells prior to infection with DR13-GFP (−4 h), as well as at 0 or 1 h post-infection. At 16 hpi, the samples were collected. (B) The PEDV titers were measured by TCID50 assay. (C) The genomic RNA level of PEDV was measured by RT-qPCR assay. All experiments were performed with three independent replicates. Differences were considered significant at P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) and P < 0.0001 (****).
3.5. Explore the antiviral activity of Tubercidin at different stages of PEDV infection
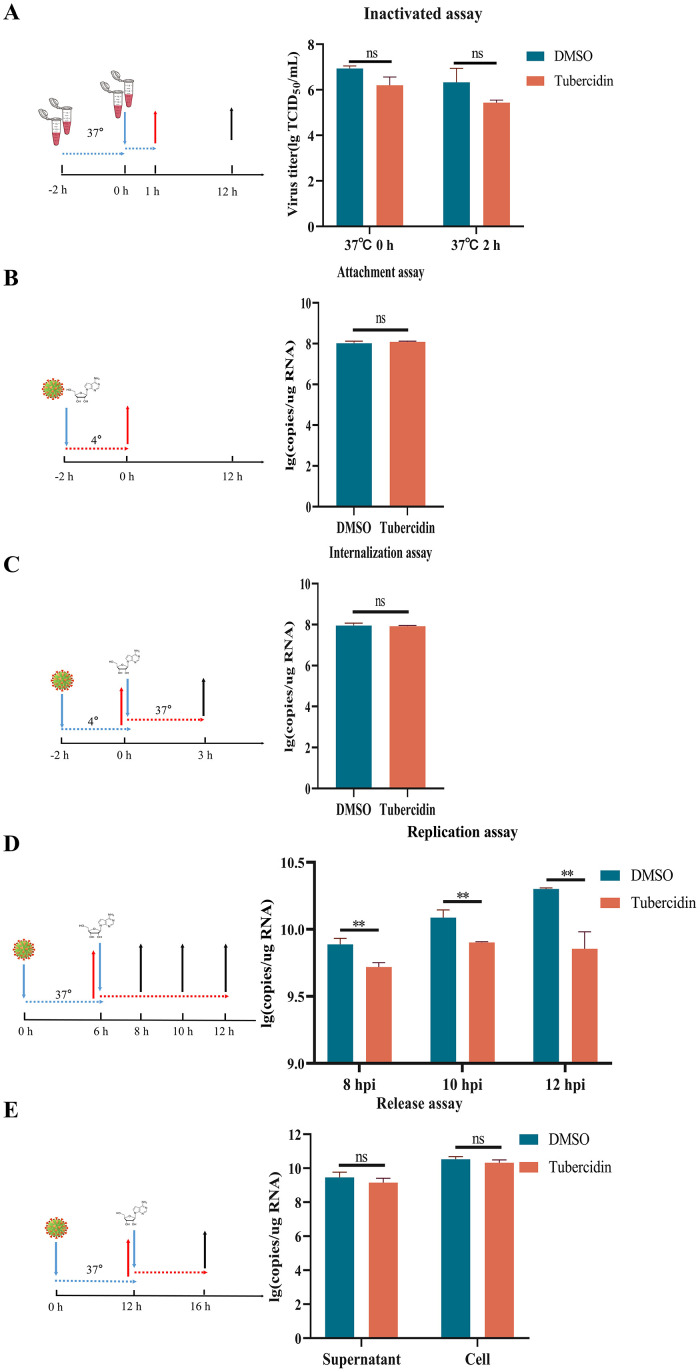
To further explore the effect of Tubercidin on the PEDV life cycle, we first tested whether Tubercidin could directly inactivate PEDV. The results show that Tubercidin failed to directly inactivate PEDV (Fig. 7A). Next, we examined the effect of Tubercidin on the attachment and internalization of PEDV to Vero cells. The relative amounts of PEDV RNA expression were detected by RT-qPCR. The results showed that Tubercidin treatment did not significantly block viral attachment and internalization compared to DMSO treatment (Fig. 7B, 7C). Then, we investigated the effect of Tubercidin on PEDV replication. Tubercidin was added during the replication phase and the levels of PEDV RNA were measured. As shown in Fig. 7D, Tubercidin treatment significantly reduced PEDV replication compared to DMSO treatment, indicating that Tubercidin inhibits PEDV replication. Additionally, we assessed the effect of Tubercidin on viral release. The viral release assay indicated that Tubercidin treatment did not lead to a significant decrease in PEDV RNA in the cells and supernatants (Fig. 7E). These findings suggest that Tubercidin primarily hinders PEDV infection by targeting the stage of viral replication.
Fig. 7.
Antiviral activity of Tubercidin at different stages of PEDV infection. Vero cells were infected with PEDV and treated with Tubercidin at indicated time points, which represented the stage of viral inactivation (A), attachment (B), internalization (C), replication (D) or release (E), respectively. All experiments were performed with three independent replicates. After 16 h, the samples were collected for determining the RNA expression level of PEDV by RT-qPCR.
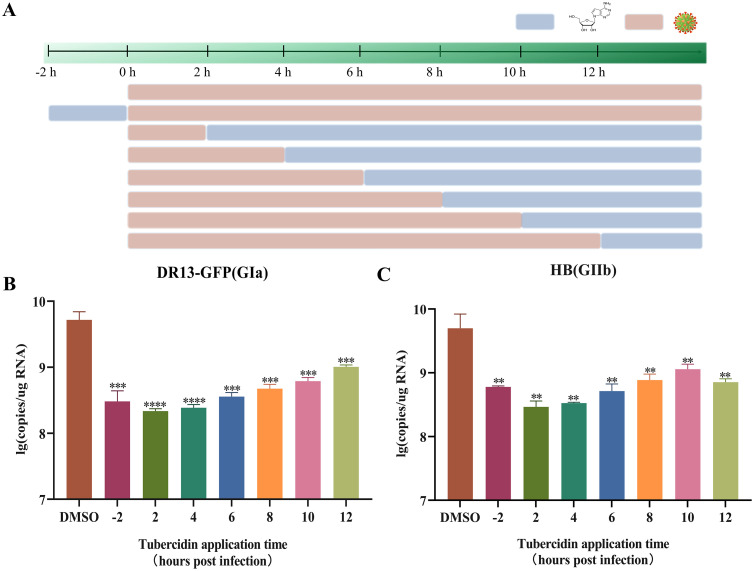
Furthermore, to evaluate its effect on virus replication at various intervals pre- and post-PEDV infection, Tubercidin at 1 μM was added to Vero cells at 2 h before infection, as well as at 2, 4, 6, 8, 10 and 12 hpi. After 16 h, samples were collected for RT-qPCR assay (Fig. 8A). Results showed that Tubercidin treatment significantly reduced the RNA levels of both classical and variant PEDV in all time periods, even at 12 hpi, compared with the control group (Fig. 8B, 8C). In conclusion, these findings imply that Tubercidin may have a significant prophylactic and therapeutic effects against PEDV infection.
Fig. 8.
Effect of Tubercidin on virus replication at various intervals pre- and post-PEDV infection. (A) Add Tuberculin to Vero cells pre- and post-infection with PEDV at the specific time points. Light brown bars represent PEDV infection, light blue bars represent Tubercidin treatment. (B and C) Vero cells were infected with DR13-GFP and HB variant and treated with Tubercidin (1 μM) or 0.1 % DMSO at 2 h before infection, as well as at 2, 4, 6, 8, 10 and 12 h post-infection. After 16 h, the samples were collected for determining viral RNA copies by RT-qPCR assay. All experiments were performed with three independent replicates. Differences were considered significant at P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) and P < 0.0001 (****).
3.6. Molecular docking of Tubercidin with RdRp of nidoviruses
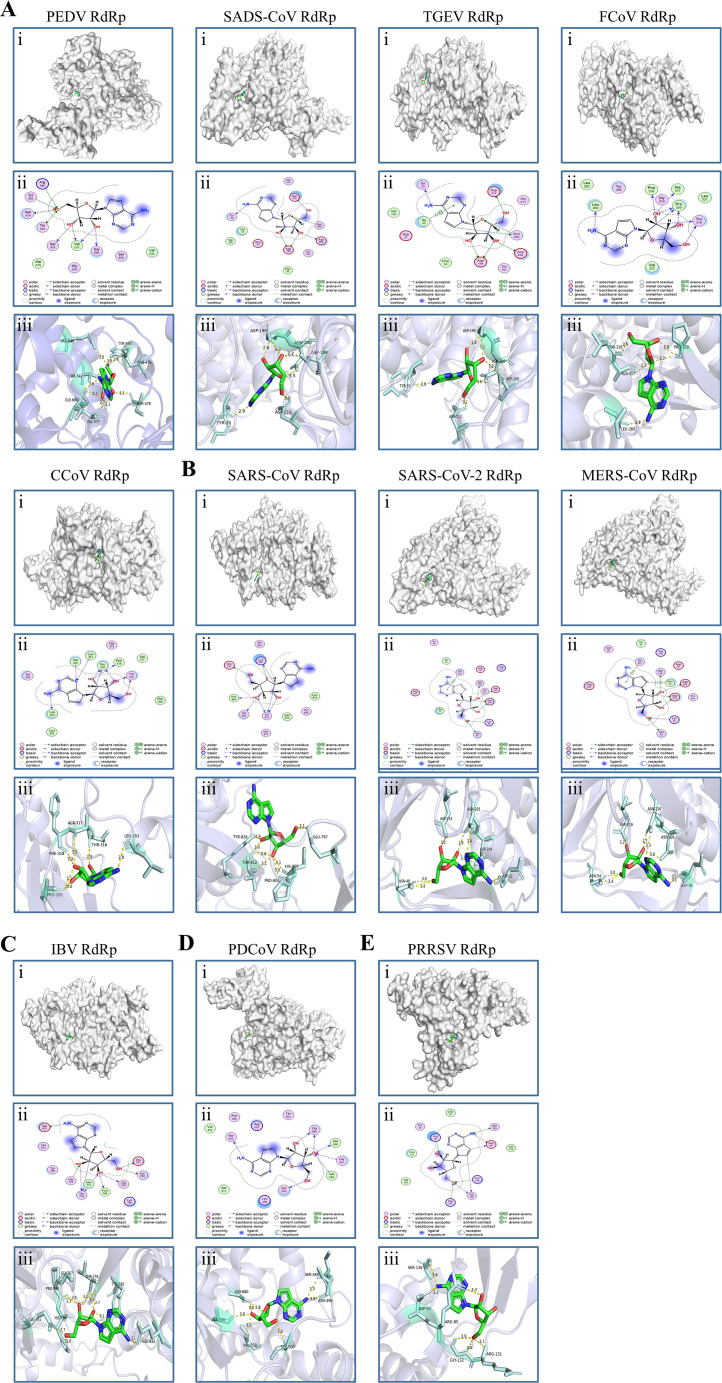
A molecular docking analysis was performed to assess the affinity of Tubercidin across RdRp of nidoviruses. Autodock Vina was used to obtain the binding poses of Tubercidin interactions with RdRp of eleven nidoviruses (Fig. 9A-9E). The resulting binding energy was then used to evaluate the molecular binding capacity (Table 1). Hydrogen bonding, strong electrostatic, and hydrophobic interactions, which have been widely recognized as crucial contributors to protein-ligand binding (Chen et al., 2016), were found to play a significant role in the binding of drugs with their protein targets. Our molecular docking results revealed that Tubercidin established prominent interactions primarily through hydrogen bonding, strong electrostatic, and hydrophobic interactions with the binding site residues of eleven nidoviruses. Specifically, significant interactions were observed between PEDV RdRp and Asn476, Ser478, Arg546, Tyr493, Thr542, Val537, and Gly660. Similarly, SADS-CoV RdRp and TGEV RdRp exhibited prominent interactions with Tyr33, Asn200, Asp199, Asp209, and Asp212, except for Lle32. FCoV RdRp displayed significant interactions involving Leu263, Thr316, Ala317, Phe318, and Pro320. On the other hand, CCoV RdRp shared similar binding sites with FCoV, except for Leu321 and Val322. Furthermore, SARS-CoV RdRp demonstrated prominent interactions at Glu797, Lys802, Pro804, His805, Thr812, and Tyr826. Similarly, SARS-CoV-2 RdRp displayed significant interactions at Asp32, Lys46, Asn48, Thr202, Asp204, Asn205, and Asp214. In the case of MERS-CoV RdRp, the prominent interactions occurred at Phe35, Asp36, Lys52, Asn54, Asn210, Thr207, Asp209, and Asp219. IBV RdRp showed Tubercidin in significant interactions at Glu761, Gly767, Pro768, His769, His775, Thr776, Met777, Tyr790, and Glu832. Additionally, PDCoV RdRp exhibited prominent interactions at Ser495, Asn496, Ser500, Ala556, Gly557, Val558, and Gly680 binding sites. Lastly, PRRSV RdRp showed significant interactions with Asp84, Arg85, Lys130, Arg131, Gly132, and Ser136. Notably, these docking results consistently yielded low binding energies of approximately −6 kcal/mol, indicating a remarkably high degree of binding stability. Consequently, the molecular docking results strongly suggest that Tubercidin inhibits the replication of PEDV and other nidoviruses by obstructing the binding pocket of RdRp, thereby demonstrating its broad-spectrum antiviral properties.
Fig. 9.
Molecular docking of Tubercidin with RdRp of nidoviruses. Tubercidin was docked with RdRp proteins from viruses including (A) α-coronaviruses (PEDV, SADS-CoV, TGEV, FCoV and CCoV), (B) β-coronaviruses (SARS-CoV, SARS-CoV-2, and MERS-CoV), (C) γ-coronavirus (IBV), (D) δ-coronavirus (PDCoV) and (E) Arteritis virus (PRRSV) using Autodock. (i) Cartoon representation, overlay of the crystal structures of Tubercidin and viruses RdRp protein were illustrated. (ii) 2D interactions of Tubercidin and viruses RdRp protein. (iii) Three-dimensional structures of the binding pockets were showed by PyMOL software. The proteins and compound are represented as cartoons and sticks, respectively. The RdRp proteins' three-dimensional structures are colored light white. The Tubercidin's two-dimensional structure is colored green, red, and blue. The Pymol software displays the sticks structure (Pale cyan) of amino acids residues in the virus RdRp protein. These amino acids residues are connected through H-bonds (yellow dotted lines) to Tubercidin, which is stabilized in the active pocket. The binding energy of the Tubercidin–viruses RdRp protein, calculated using Autodock, is listed (Table 1).
Table 1.
Binding energy for RdRp protein of nidoviruses with Tubercidin.
| Virus | Binding Energy (kcal/mol) |
|---|---|
| PEDV RdRp | −6.367 |
| SADS-CoV RdRp | −6.424 |
| TGEV RdRp | −6.661 |
| FCoV RdRp | −5.878 |
| CCoV RdRp | −5.681 |
| SARS-CoV RdRp | −6.181 |
| SARS-CoV-2 RdRp | −6.268 |
| MERS-CoV RdRp | −6.373 |
| IBV RdRp | −6.681 |
| PDCoV RdRp | −6.008 |
| PRRSV RdRp | −5.998 |
3.7. Antiviral activity of Tubercidin against SADS-CoV and PRRSV
SADS-CoV, an emerging porcine enteric coronavirus since 2017, can cause vomiting, watery diarrhea, dehydration, and weight loss in piglets, with mortality rates of up to 90–100 % (Pan et al., 2017). To further explore Tubercidin's potential antiviral activity against SADS-CoV, we treated Vero cells with Tubercidin at concentrations ranging from 0.25 to 1 μM. Then, we infected the cells with SADS-CoV at 0.1 MOI. The results of the cytopathic effect analysis showed a significant decrease in SADS-CoV-induced syncytium with the increase in drug concentration (Fig. 10A). Furthermore, the TCID50 assay results demonstrated that Tubercidin effectively inhibits SADS-CoV, reducing it by 2.5 log10 at 1 μM (Fig. 10B). Additionally, in a dose-dependent manner, Tubercidin treatment significantly reduced the RNA levels of SADS-CoV (Fig. 10C). These findings support the potential antiviral activity of Tubercidin against SADS-CoV.
Fig. 10.
Antiviral effect of Tubercidin on SADS-CoV. (A) Vero cells were pre-treated with Tubercidin (0.25–1 μM) for 4 h, and then infected them with SADS-CoV in the presence or absence of Tubercidin. At 16 hpi, CPE results were observed and viral fluids were collected. (B) The SADS-CoV titers were measured by TCID50 assay. (C) The genomic RNA levels of SADS-CoV were measured by RT-qPCR assay. The experiment was performed three times independently, images were representative of results obtained from three independent experiments, scale bars = 150 μm. Differences were considered significant at P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) and P < 0.0001 (****).
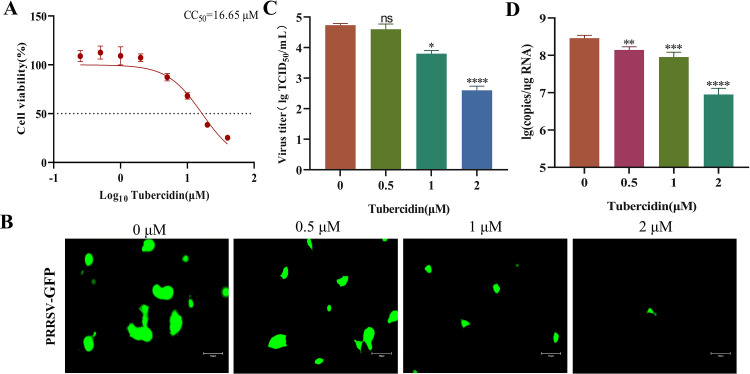
PRRSV, the most economically important infectious disease of pigs worldwide, is the cause of severe reproductive and respiratory disease in swine. In order to investigate Tubercidin's antiviral potential against PRRSV on Marc145 cells, we determined its maximum nontoxic concentration at 2 μM using the CCK-8 assay, with a CC50 value of 16.65 μM (Fig. 11A). Moreover, the PRRSV-expressed GFP gradually decreased as the drug concentration increased (Fig. 11B). The TCID50 results confirmed the antiviral effect of Tubercidin on PRRSV infection, with an inhibitory effect of approximately 2 log10 at 2 μM (Fig. 11C). Additionally, Tubercidin treatment significantly reduced the RNA levels of PRRSV, in a dose-dependent manner (Fig. 11D). In conclusion, Tubercidin effectively inhibits PRRSV infection on Marc145 cells.
Fig. 11.
Antiviral effect of Tubercidin on PRRSV. (A) Cytotoxicity of Tubercidin on Marc145 cells. Marc145 cells were treated or not treated with different concentrations of Tubercidin for 16 h. The cytotoxicity was determined by CCK-8 assay. (B) Microscopic images of Marc145 cells infected with PRRSV-GFP (0.1 MOI) treated with different concentrations of Tubercidin (0.5–2 μM). (B) The PRRSV titers were measured by TCID50 assay. (C) The genomic RNA levels of PRRSV were measured by RT-qPCR assay. Each experiment was performed three times independently, images were representative of results obtained from three independent experiments, scale bars = 150 μm. Differences were considered significant at P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) and P < 0.0001 (****).
4. Discussions
Emerging and re-emerging viral infectious diseases have been found to have a significant negative impact on global human and animal health. Since the emergency of novel variants in 2010, PEDV has spread to many countries, causing substantial economic losses in the global pork industry. Effective prophylactic and therapeutic agents against this virus are urgently needed as there are no fully effective specific drugs directed against PEDV. In recent antiviral research, much attention has been given to natural products, which have undergone extensive study. Our drug-repurposing goals were to identify natural compounds specific for individual viruses but to also focus on those which might have a broad spectrum of activity against a range of viral families. From a natural product library consisting of 206 compounds, we identified ten candidates as hit compounds, such as Oleanolic acid, Moxidectin, Octyl gallate, 1,2,3,4,6-Pentagalloylglucose, Theaflavin 3,3′- digallate, Curcumin, Baicalin, Xanthohumol, Tubercidin, Rubitecan that showed excellent antiviral activity. One of them, Tubercidin (7-Deazaadenosine), showed excellent antiviral activity against both classical and variant PEDV, with a selectivity index of 57.2 on Vero cells. Molecular docking analysis indicated that Tubercidin had better docking efficiency and formed hydrophobic interactions with the active pocket of RdRp of PEDV and other nidoviruses. Furthermore, we also demonstrated that Tubercidin can effectively inhibit SADS-CoV and PRRSV infection. Our results provide valuable insights into the potential of Tubercidin as a broad-spectrum antiviral agent against nidovirus infections.
The conservative structure and crucial role of RdRp make it an optimal target for developing potent antiviral agents against RNA viruses (Xu et al., 2022). RdRp inhibitors are generally classified into nucleoside analogs and non-nucleoside analogs based on their structure and binding location on RdRp (Tian et al., 2021). Currently, several nucleoside analogs, including remdesivir (Yin et al., 2020), molnupiravir (EIDD-2801)(Cox et al., 2021), favipiravir (Kisoet al., 2010), GS-441,524 (Amirian and Levy, 2020) and ATV006 (Cao et al., 2022), have been found to be effective against RNA viruses, making great contributions to fight the emerging and re-emerging viral infectious. As an FDA-approved drug, remdesivir has been widely reported to exhibit antiviral activities against various virus families, including Filoviridae, Paramyxoviridae, Pneumoviridae, Flaviviridae, and the Coronaviridae by inhibiting viral RdRp activity (Eastman et al., 2020). Tubercidin, a nucleoside analog obtained from the bacteria Streptomyces tubercidicus (Suzuki and Marumo, 1960), has been identified as having anti-parasite, anti-cancer, and antiviral effects (Chen et al., 2022; Drew et al., 2003; Schultz et al., 2022). Tubercidin and its derivatives have exhibited antiviral activity against various viruses, such as vesicular stomatitis virus (VSV), coxsackie virus, and ZIKV (De Clercq et al., 1986; Eyer et al., 2016; Milisavljevic et al., 2021). Recent studies have reported the antiviral activities of 5-substituted tubercidin derivatives against emerging RNA viruses (Milisavljevic et al., 2021). Moreover, Tubercidin and its derivatives have demonstrated potent effectiveness against SARS-CoV-2 infection in vitro without causing cytotoxicity (Uemura et al., 2021). Furthermore, these derivatives have shown inhibition of viral RNA replication, particularly during the late stages of the SARS-CoV-2 infection process (Zhao et al., 2022). In our study, we observed that Tubercidin did not significantly affect the inactivation, attachment, internalization, and release stages of PEDV infection, but it did have a significant inhibitory effect on viral replication. Molecular docking analysis revealed that Tubercidin showed better docking efficiency and formed hydrophobic interactions with RdRp of PEDV ( −6.367 kcal/mol), SADS-CoV (−6.424 kcal/mol), PRRSV(−5.998 kcal/mol), and other nidoviruses, including α-coronaviruses: TGEV, FCoV, and CCoV; β-coronaviruses: SARS-CoV, SARS-CoV-2, and MERS-CoV; γ-coronavirus: IBV; and δ-coronavirus such as PDCoV. These findings suggest that Tubercidin has the potential to be a lead compound for the development of a broad spectrum anti-coronavirus agents, which can be further explored for new drug discovery.
Recently, increasing studies have found that Tubercidin, in addition to targeting RdRp of RNA viruses, has other antiviral targets or side effects for host cells. Recently, Tsukamoto et al. demonstrated that Tubercidin can directly bind to host 2′-O-ribose methyltransferase 1 (MTr1), which is essential for influenza A and B virus infections, suppressing viral replication (Tsukamoto et al., 2023). Furthermore, Tubercidin derivatives have shown a strong immunomodulatory effect in curbing SARS-CoV-induced hyperinflammation (Bergant et al., 2022). However, there are also reports indicating that Tubercidin strongly inhibits cell proliferation by interfering with various cellular processes, leading to significant cytotoxicity (Chen et al., 2019). Nonetheless, in our study, we did not observe strong cytotoxicity of Tubercidin at certain concentrations in Vero, LLC-PK1, and Marc145 cells. However, the safety, effectiveness, and potential clinical applications of Tubercidin in different coronavirus hosts need to be further evaluated through in vivo antiviral activities and in vivo kinetics studies.
5. Conclusions
In summary, we discovered ten antiviral compounds against PEDV from a natural-product library. Among them, Tubercidin exhibited a promising prospect for the prevention and treatment of PEDV infection. It is also meaningful to investigate the broad-spectrum antiviral potential of Tubercidin against PEDV and other emerging coronavirus infections.
CRediT authorship contribution statement
Tianliang Wang: Conceptualization, Methodology, Software, Investigation, Data curation, Validation, Writing – original draft, Writing – review & editing. Guanmin Zheng: Investigation, Methodology, Validation, Formal analysis, Writing – review & editing. Zilu Chen: Methodology, Resources, Investigation, Data curation, Validation. Yue Wang: Methodology, Resources, Investigation, Data curation, Validation. Chenxu Zhao: Data curation, Software, Formal analysis. Yaqin Li: Data curation, Software, Formal analysis. Yixin Yuan: Data curation, Software, Formal analysis. Hong Duan: Data curation, Software, Formal analysis. Hongsen Zhu: . Xia Yang: Methodology, Visualization, Supervision. Wentao Li: Resources, Writing – review & editing, Supervision. Wenjuan Du: Resources, Writing – review & editing, Supervision. Yongtao Li: Conceptualization, Resources, Project administration, Writing – review & editing, Supervision, Funding acquisition. Dongliang Li: Conceptualization, Resources, Project administration, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was funded by the National Natural Science Foundation of China (Grant No.32172839, 31602033 and 32202826), the China Scholarship Council (201908410129), Innovation and Entrepreneurship Program for College Students in Henan Province (202210471045), Henan Province Science Foundation for Youths (Grant No. 222300420216) and Key Scientific Research Projects of Higher Education Institutions in Henan Province in 2024 (24A230011).
Contributor Information
Yongtao Li, Email: yongtaole@126.com.
Dongliang Li, Email: 497222038@sina.com.
Data availability
Data will be made available on request.
References
- Amirian E.S., Levy J.K. Current knowledge about the antivirals remdesivir (GS-5734) and GS-441524 as therapeutic options for coronaviruses. One Health. 2020;9 doi: 10.1016/j.onehlt.2020.100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergant V., Yamada S., Grass V., Tsukamoto Y., Lavacca T., Krey K., Muhlhofer M.T., Wittmann S., Ensser A., Herrmann A., Vom Hemdt A., Tomita Y., Matsuyama S., Hirokawa T., Huang Y., Piras A., Jakwerth C.A., Oelsner M., Thieme S., Graf A., Krebs S., Blum H., Kummerer B.M., Stukalov A., Schmidt-Weber C.B., Igarashi M., Gramberg T., Pichlmair A., Kato H. Attenuation of SARS-CoV-2 replication and associated inflammation by concomitant targeting of viral and host cap 2′-O-ribose methyltransferases. EMBO J. 2022;41(17) doi: 10.15252/embj.2022111608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Li Y., Yang S., Li G., Zhou Q., Sun J., Xu T., Yang Y., Liao R., Shi Y., Yang Y., Zhu T., Huang S., Ji Y., Cong F., Luo Y., Zhu Y., Luan H., Zhang H., Chen J., Liu X., Luo R., Liu L., Wang P., Yu Y., Xing F., Ke B., Zheng H., Deng X., Zhang W., Lin C., Shi M., Li C.M., Zhang Y., Zhang L., Dai J., Lu H., Zhao J., Zhang X., Guo D. The adenosine analog prodrug ATV006 is orally bioavailable and has preclinical efficacy against parental SARS-CoV-2 and variants. Sci. Transl. Med. 2022;14(661):eabm7621. doi: 10.1126/scitranslmed.abm7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Breslin M.B., Guidry J.J., Lan M.S. 5′-Iodotubercidin represses insulinoma-associated-1 expression, decreases cAMP levels, and suppresses human neuroblastoma cell growth. J. Biol. Chem. 2019;294(14):5456–5465. doi: 10.1074/jbc.RA118.006761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Oezguen N., Urvil P., Ferguson C., Dann S.M., Savidge T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016;2(3) doi: 10.1126/sciadv.1501240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Barrett L., Lin Z., Kendrick S., Mu S., Dai L., Qin Z. Identification of natural compounds tubercidin and lycorine HCl against small-cell lung cancer and BCAT1 as a therapeutic target. J. Cell. Mol. Med. 2022;26(9):2557–2565. doi: 10.1111/jcmm.17246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu X., Shi D., Shi H., Zhang X., Li C., Chi Y., Feng L. Detection and molecular diversity of spike gene of porcine epidemic diarrhea virus in China. Viruses. 2013;5(10):2601–2613. doi: 10.3390/v5102601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 2021;6(1):11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E., Bernaerts R., Bergstrom D.E., Robins M.J., Montgomery J.A., Holy A. Antirhinovirus activity of purine nucleoside analogs. Antimicrob. Agents Chemother. 1986;29(3):482–487. doi: 10.1128/aac.29.3.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew M.E., Morris J.C., Wang Z., Wells L., Sanchez M., Landfear S.M., Englund P.T. The adenosine analog tubercidin inhibits glycolysis in Trypanosoma brucei as revealed by an RNA interference library. J. Biol. Chem. 2003;278(47):46596–46600. doi: 10.1074/jbc.M309320200. [DOI] [PubMed] [Google Scholar]
- Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent Sci. 2020;6(5):672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyer L., Smidkova M., Nencka R., Neca J., Kastl T., Palus M., De Clercq E., Ruzek D. Structure-activity relationships of nucleoside analogues for inhibition of tick-borne encephalitis virus. Antiviral Res. 2016;133:119–129. doi: 10.1016/j.antiviral.2016.07.018. [DOI] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts V., Zakhartchouk A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet. Microbiol. 2017;206:45–51. doi: 10.1016/j.vetmic.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4(5):e00737. doi: 10.1128/mBio.00737-13. -00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiso M., Takahashi K., Sakai-Tagawa Y., Shinya K., Sakabe S., Le Q.M., Ozawa M., Furuta Y., Kawaoka Y. T-705 (favipiravir) activity against lethal H5N1 influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 2010;107(2):882–887. doi: 10.1073/pnas.0909603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Lu H., Geng C., Yang K., Liu W., Liu Z., Yuan F., Gao T., Wang S., Wen P., Song H., Tian Y., Zhou D. Epidemic and evolutionary characteristics of swine enteric viruses in South-Central China from 2018 to 2021. Viruses. 2022;14(7) doi: 10.3390/v14071420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18(8):1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.L., Zhu L., Ma J.Y., Zhou Q.F., Song Y.H., Sun B.L., Chen R.A., Xie Q.M., Bee Y.Z. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field strains in south China. Virus Genes. 2012;45(1):181–185. doi: 10.1007/s11262-012-0735-8. [DOI] [PubMed] [Google Scholar]
- Marthaler D., Jiang Y., Otterson T., Goyal S., Rossow K., Collins J. Complete genome sequence of porcine epidemic diarrhea virus strain USA/Colorado/2013 from the United States. Genome Announc. 2013;1(4) doi: 10.1128/genomeA.00555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milisavljevic N., Konkolova E., Kozak J., Hodek J., Veselovska L., Sykorova V., Cizek K., Pohl R., Eyer L., Svoboda P., Ruzek D., Weber J., Nencka R., Boura E., Hocek M. Antiviral activity of 7-substituted 7-deazapurine ribonucleosides, monophosphate prodrugs, and triphoshates against emerging RNA viruses. ACS Infect. Dis. 2021;7(2):471–478. doi: 10.1021/acsinfecdis.0c00829. [DOI] [PubMed] [Google Scholar]
- Pan Y., Tian X., Qin P., Wang B., Zhao P., Yang Y.L., Wang L., Wang D., Song Y., Zhang X., Huang Y.W. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picarazzi F., Vicenti I., Saladini F., Zazzi M., Mori M. Targeting the RdRp of emerging RNA viruses: the structure-based drug design challenge. Molecules. 2020;25(23) doi: 10.3390/molecules25235695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D.C., Johnson R.M., Ayyanathan K., Miller J., Whig K., Kamalia B., Dittmar M., Weston S., Hammond H.L., Dillen C., Ardanuy J., Taylor L., Lee J.S., Li M., Lee E., Shoffler C., Petucci C., Constant S., Ferrer M., Thaiss C.A., Frieman M.B., Cherry S. Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2. Nature. 2022;604(7904):134–140. doi: 10.1038/s41586-022-04482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R.Q., Cai R.J., Chen Y.Q., Liang P.S., Chen D.K., Song C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18(1):161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Marumo S. Chemical Structure of Tubercidin:communications to the Editors. J. Antibiotics Ser. A. 1960;13(5):360. [Google Scholar]
- Tian L., Qiang T., Liang C., Ren X., Jia M., Zhang J., Li J., Wan M., YuWen X., Li H., Cao W., Liu H. RNA-dependent RNA polymerase (RdRp) inhibitors: the current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 2021;213 doi: 10.1016/j.ejmech.2021.113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y., Hiono T., Yamada S., Matsuno K., Faist A., Claff T., Hou J., Namasivayam V., Vom Hemdt A., Sugimoto S., Ng J.Y., Christensen M.H., Tesfamariam Y.M., Wolter S., Juranek S., Zillinger T., Bauer S., Hirokawa T., Schmidt F.I., Kochs G., Shimojima M., Huang Y.S., Pichlmair A., Kummerer B.M., Sakoda Y., Schlee M., Brunotte L., Muller C.E., Igarashi M., Kato H. Inhibition of cellular RNA methyltransferase abrogates influenza virus capping and replication. Science. 2023;379(6632):586–591. doi: 10.1126/science.add0875. [DOI] [PubMed] [Google Scholar]
- Uemura K., Nobori H., Sato A., Sanaki T., Toba S., Sasaki M., Murai A., Saito-Tarashima N., Minakawa N., Orba Y., Kariwa H., Hall W.W., Sawa H., Matsuda A., Maenaka K. 5-Hydroxymethyltubercidin exhibits potent antiviral activity against flaviviruses and coronaviruses, including SARS-CoV-2. iScience. 2021;24(10) doi: 10.1016/j.isci.2021.103120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman S., Prasad B., Selvarajan R. RNA Dependent RNA Polymerases: insights from Structure, Function and Evolution. Viruses. 2018;10(2) doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Anirudhan V., Du R., Cui Q., Rong L. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J. Med. Virol. 2021;93(1):300–310. doi: 10.1002/jmv.26264. [DOI] [PubMed] [Google Scholar]
- Wang Y., Huang H., Li D., Zhao C., Li S., Qin P., Li Y., Yang X., Du W., Li W., Li Y. Identification of niclosamide as a novel antiviral agent against porcine epidemic diarrhea virus infection by targeting viral internalization. Virol Sin. 2023;38(2):296–308. doi: 10.1016/j.virs.2023.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977;100(12):243–244. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- Xu X., Chen Y., Lu X., Zhang W., Fang W., Yuan L., Wang X. An update on inhibitors targeting RNA-dependent RNA polymerase for COVID-19 treatment: promises and challenges. Biochem. Pharmacol. 2022;205 doi: 10.1016/j.bcp.2022.115279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen Y., Zhou J., Wang X., Ma L., Li J., Yang L., Yuan H., Pang D., Ouyang H. Porcine epidemic diarrhea virus: an updated overview of virus epidemiology, virulence variation patterns and virus-host interactions. Viruses. 2022;14(11) doi: 10.3390/v14112434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Liu Q., Yi D., Li Q., Guo S., Ma L., Zhang Y., Dong D., Guo F., Liu Z., Wei T., Li X., Cen S. 5-Iodotubercidin inhibits SARS-CoV-2 RNA synthesis. Antiviral Res. 2022;198 doi: 10.1016/j.antiviral.2022.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.