Abstract
Chromosomal inversions are usually balanced structural chromosomal rearrangements that do not have an impact on the clinical phenotype of a carrier. The main clinical consequence of inversions is the risk for unbalanced gametes and offspring with severe phenotypes. Rarely though, inversions are associated with a phenotype, mainly due to submicroscopic Copy Number Variants (CNVs) or disruption at the breakpoints of a functionally important gene and/or genomic elements. In this study, a paracentric inversion of chromosome 16 [inv(16)(q22.3q24.1)] was identified in a three-generation family with discordant phenotypes with/without epilepsy and/or intellectual impairment, as well as with an unaffected carrier. This finding was confirmed by fluorescence in situ hybridization (FISH). Genetic investigation, initially with chromosomal microarray (CMA), did not reveal any copy number variants. Finally, Clinical Exome Sequencing (CES), detected the presence of a pathogenic nonsense variant (rs797044912) in the Chromodomain Helicase DNA-binding protein 2 (CHD2) gene [NM_001271.4:c.5035C>T p.(Arg1679Ter)]. CHD2 pathogenic variants have been associated with Developmental and Epileptic Encephalopathy-94 (DEE-94), a rare yet severe condition, characterized by developmental delay, seizures with an early onset, intellectual impairment, autism spectrum disorder, and sometimes behavioral issues. Family testing showed that the variant segregated with phenotypic heterogeneity in the affected individuals and appears to be causative. To the best of our knowledge, this is the first CHD2 pathogenic variant segregating in a three-generation family and the fourth familial case reported. These results further support our previous findings that familial, balanced rearrangements with discordant phenotypes in the same family are, in the vast majority, coincidental.
Keywords: Paracentric inversion 16, CHD2, Familial, Developmental epileptic encephalopathy, Inherited, Seizure, Clinical exome sequencing (CES)
1. Introduction
Inversions are balanced chromosomal rearrangements, which result in no loss or gain of chromosomal material and are usually not associated with a pathological phenotype. Nevertheless, certain mechanisms have been proposed to explain a pathological phenotype associated with a balanced rearrangement, such as the “position effect”, where the expression of genes located in the inverted DNA segment is altered [1]. Additionally, gene disruption on one allele may unmask a pathogenic variant on the second allele of a recessive gene. The presence of cryptic submicroscopic imbalances at/near the breakpoints can also affect the phenotype of the carrier.
Elucidating the phenotypic outcome lies heavily on the determination of the exact breakpoints, which is mostly done through cytogenetic techniques, such as G-banding and chromosomal microarray (CMA). However, these techniques are limited in terms of resolution and cannot identify all types of variants. Therefore, Next Generation Sequencing (NGS)-based technologies are often utilized. Depending on the application employed, NGS can not only be used to detect single nucleotide variants, small insertions and deletions, but also the exact positions of copy number variants and other structural rearrangements that may be missed by another method.
In this study we report a female pediatric patient with a familial paracentric inversion on chromosome 16 segregating in affected and unaffected family members with discordant phenotypes. The proband also carries a pathogenic nonsense variant, which segregates only in affected family members and seems to explain the phenotype. The presence of this variant highlights the need for additional investigation in cases of familial, balanced rearrangements, as previously stated in Ref. [1], where the inversion does not seem to explain the pathological phenotype.
2. Patient data
2.1. Study participants
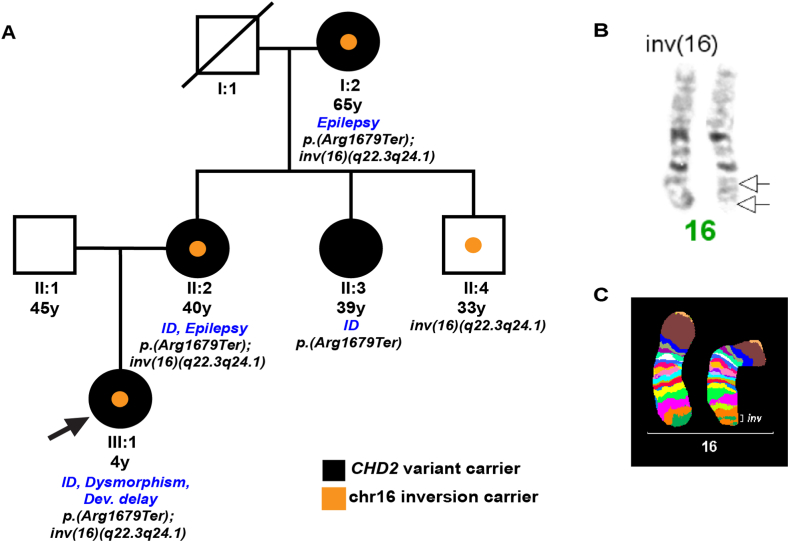
The family included in the study involved affected and non-affected members. Written informed consent was obtained from the uncle; guardian of the family. The family members and clinical description are as follows (see Fig. 1A).
Fig. 1.
(A) Family pedigree. Filled black symbols represent carriers of the CHD2 pathogenic variant, whereas orange symbols represent carriers of the chromosome 16 inversion. The arrow indicates the proband. Clinical information is also included. (B) Partial karyogram of proband's chromosomes 16. The black arrows indicate the inversion breakpoints on chromosome 16. (C) Multicolor banding probe set for chromosome 16. The result is depicted as pseudocolor banding and the inversion is highlighted (inv).
2.2. Clinical description
2.2.1. Proband (sample III:1)
The proband, today a 4-year-old female, was initially referred to the Genetics department at 20 days of age, due to the presence of dysmorphic features, hypotonia, and global developmental delay. She was admitted to the hospital again at the age of 21 months with reported, weekly recurrent episodes of fixed gaze, absences-like loss of contact, myoclonic-like movements of upper and lower arms with a duration of a few seconds to 15 minutes in the last 2 months and frequent falls. Pediatric and neurological evaluation revealed normal tonus and reflexes, mild psychomotor delay, and broad-based gait.
According to the Denver test, gross motoric development was adequate, but the fine motoric, the language, and the sociability skills were at the level of 16 months. A sleep-deprived electroencephalogram (EEG) was normal but there was no follow-up. Until now, the proband has not developed epilepsy. Brain magnetic resonance imaging (MRI) showed mild frontal colpocephaly. The proband's weight was 12,600 g (80th centile), length 89 cm (90th centile), and OFC 46.6 cm (25th centile, i.e. relative microcephaly). Status was normal except for hemangiomas on the left side of the chest and on the inside of the left upper arm. All routine hematological, biochemical, thyroid, otolaryngological, and cardiological tests were normal. Ophthalmological examination revealed mild hyperopia, intermittent strabismus and nystagmus of congenital origin.
The dysmorphic features of the proband included a hypotonic face, high frontal hairline and thin, long eyebrows with lateral extension and medial flare. She also presented with marked epicanthus inversus, upslanting palpebral fissures, round nostrils, full cheeks, protruding lower lip, apparently low-set and posteriorly rotated ears with up-lifted ear lobes. Other features included simple descending helix, protruding descending anthelix, increased intermamillary distance, mild pectus excavatum, atypical hemangiomas over the chest left and on the inside of left upper arm, 5th-finger clinodactyly, and a broad 1st toe.
2.2.2. Mother (sample II:2)
The patient is the only child of a mother (40 years old) with moderate intellectual disability and childhood-onset epilepsy. She is treated with valproic acid 500 mg twice a day and she is free of seizures. The mother also presented with bitemporal narrowing, sloping forehead, bushy arched eyebrows, divergent strabismus right, mildly upslanting palpebral fissures, facial hirsutism, anteverted ears, simple helix and anthelix, short deeply-grooved philtrum, full cheeks, central obesity, and mild hallux valgus.
2.2.3. Father (sample II:1)
The father is 45 years old and apparently healthy.
2.2.4. Maternal grandmother (sample I:2)
The maternal grandmother is 65 years old and is the second family member with late-onset epilepsy, for which she is administered with phenytoin at a dosage of 100 mg 3 times per day. Her medical record included controlled diabetes mellitus with a history of ketoacidosis, hypertension, hypercholesterolemia, hyperthyroidism, and microangiopathic ischemic changes. Brain MRI showed an empty sella turcica. She is also on antipsychotic medication. She has a highly problematic social communication.
2.2.5. Maternal aunt (sample II:3)
The maternal aunt (39 years old) presented with mild intellectual impairment, but no overt epilepsy. An EEG could not be performed.
2.2.6. Maternal uncle (sample II:4)
The maternal uncle is apparently healthy.
3. Methods
3.1. Cytogenetic analysis
Chromosomes were obtained from peripheral blood and prepared according to standard procedures. Karyotype analysis was performed by GTG high resolution banding according to standard techniques. In all cases, 20 metaphases were analyzed from two independent cultures.
3.2. FISH analysis
Following GTG-banding, FISH was performed using a multicolor banding probe set for chromosome 16 [2]. This was applied according to standard procedures [3]. Evaluation was done on a Axioplan microscope equipped with a suited software (ISIS, MetaSystems, Altlussheim, Germany).
3.3. Microarray analysis
Array-based Comparative Genomic Hybridization was performed using SurePrint ISCA array (Agilent-version 2.0) with 60,000 oligos using build GRCh37 (hg19).
3.4. Clinical exome sequencing
Library preparation for CES was performed for the family trio, using the TruSight One sequencing panel (Illumina, San Diego, CA, USA; part number FC-141-1007) according to the manufacturer's protocol. Paired-end sequencing of the pooled libraries was performed on a NextSeq 500 system (Illumina) by using the NextSeq 500/550 High Output Kit v2.5 (300 Cycles) and following the manufacturer's guidelines (NextSeq System Denature and Dilute Libraries Guide, Document #15048776, v02, and NextSeq System Guide, Illumina, Document #15046563, v02). Demultiplexing and adapter trimming was performed automatically using BaseSpace Sequencing Hub Apps (Illumina).
Bioinformatic processing, analysis, annotation and interpretation was performed by an in-house pipeline using the human reference genome build hg19. CES was used to detect only single nucleotide variants and small insertions and deletions within the target region.
In-house bioinformatics pipeline: Fastq reads produced by sequencing were aligned to the human reference genome (GRCh37/hg19) with the BWA-MEM algorithm (0.7.15) (<http://bio-bwa.sourceforge.net). Variant calling and filtering followed the GATK best practices with GATKv3.6-0 [4]. Variant effect prediction was performed by VEP (release101) [5], and annotation and filtering by GEMINI tool (v0.30.2) [6]. The pathogenicity potential of the identified variants was assessed considering the predicted consequence, the biochemical properties of codon change, the degree of evolutionary conservation as well as frequency based on a number of reference population databases, such as gnomAD genome and exome (v 2.1) and a local dataset of screened individuals referred to the lab. The variant classification followed the ACMG guidelines [7] and was assisted by VarSome classification [8].
4. Results
4.1. Cytogenetic analysis
Cytogenetic analysis of the peripheral blood of the proband (III:1) revealed a rearranged chromosome 16, which was confirmed by FISH to be a paracentric inversion inherited by the mother (II:2), 46,XX,inv(16)(q22.3q24.1)mat (Fig. 1B and C). The karyotype of the father (II:1) was normal 46,XY. The maternal grandmother (I:2) presented with the same structural rearrangement. The maternal aunt (II:3) had a normal karyotype of 46,XX, whereas the maternal unaffected uncle (II:4) shared the same inversion 46,XY,inv(16)(q22.3q24.1).
4.2. Investigation of inversion breakpoints
The breakpoints of the inversion were at 16q22.3 and 16q24.1 (Fig. 1B and C), but the OMIM disease genes found herein were not related to the phenotype of the proband. In more detail, region 16q22.3 contains only the ZFHX3 gene (OMIM #104155) which is associated with early onset prostate cancer. Region 16q24.1 contains CO4I1 (OMIM #123864), which is associated with mitochondrial complex IV deficiency nuclear type 16 (MC4DN16) and is an autosomal recessive disease; IRF8 (OMIM# 601565), variants in which are implicated in immunodeficiency in an autosomal dominant manner; FOXF1 gene, which is associated with alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV, #601089); and FOXC2 gene (OMIM #602402), which is associated with lymphedema-distichiasis syndrome (LPHDST) and is caused by heterozygous variations.
4.2.1. Microarray analysis
The microarray analysis of the individuals did not reveal any cryptic microdeletions or microduplications at or near the breakpoints at the resolution of 200kb.
4.3. Clinical Exome Sequencing analysis
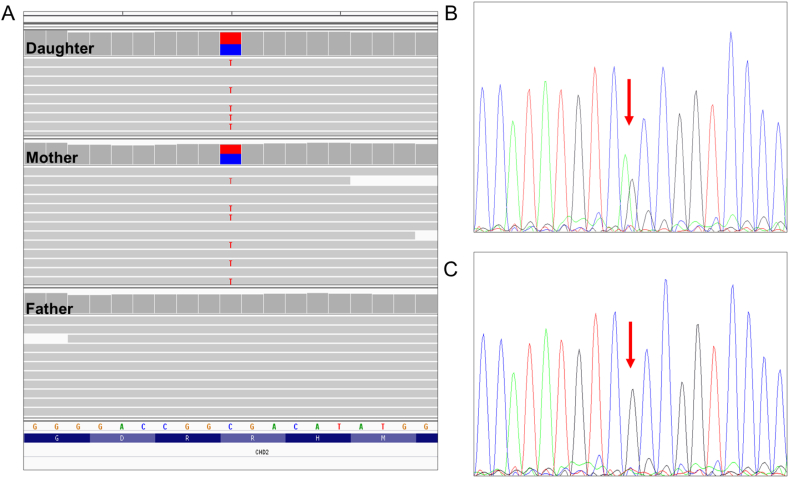
CES analysis revealed a known (rs797044912) pathogenic nonsense variant in the CHD2 gene NM_001271.4:c.5035C>T p.(Arg1679Ter) in heterozygosity, which was maternally inherited and absent from the father (Fig. 2A). The variant was found in the penultimate exon 38 of the transcript at the C terminal domain, it is predicted to undergo nonsense mediated decay, and loss-of function is a known mechanism of disease for this gene. The CHD2 gene consists of a chromodomain and an ATPase Helicase domain at the N terminal, however, the function of the C-terminal domain remains unknown. The variant is absent from the gnomAD frequency database and the local frequency dataset. Additionally, the variant was previously submitted in ClinVar (five submissions) consistent as pathogenic and associated with Developmental Epileptic Encephalopathy 94(OMIM #615369). It has been previously described as pathogenic [9,10].
Fig. 2.
Candidate variant investigation. (A) Integrative Genomics Viewer screenshot of the genomic region overlapping the nonsense CHD2 variant in the proband, mother and father. (B) Sanger sequencing confirmed that the affected proband, mother, maternal aunt and grandmother are heterozygous for the mutated allele while, (C) the non-affected maternal uncle is homozygous for the reference allele. The positions are indicated by the red arrows.
Sanger sequencing was carried out, confirming the presence of the variant in the proband and her mother. Segregation analysis further supported the pathogenicity of this variant, as affected maternal grandmother and aunt carry the variant (Fig. 2B), but it is absent from the apparently healthy maternal uncle (Fig. 2C).
5. Discussion
Cytogenetically detected inversions by definition are balanced rearrangements with no adverse phenotypic effects [11]. However, there is limited literature on the genetic causality of familial paracentric inversions where one member is clinically affected and the other(s) is/are phenotypically normal. We have previously shown that there are families with apparently balanced translocations, which are considered coincidental and unrelated to the phenotype [1,12]. Similarly, in this study we conclude that the current inversion on chromosome 16 is unrelated to the clinical phenotype of the proband and the affected individuals in the family under study. This is supported by the absence of cryptic imbalances at the breakpoints and, most importantly, the presence of an unaffected carrier in the family.
Further investigation with CES revealed the presence of a previously described pathogenic nonsense variant in the CHD2 gene, which may explain the phenotype of the patient and the affected family members and further support that the inversion is coincidental. The variant was classified as pathogenic based on the evidence and the phenotypes presented in the family.
CHD2 has been associated with a broad spectrum of neurodevelopmental disorders, such as autism spectrum disorders and intellectual disability [13]. Most of the reported cases occurred de novo [14]. Nevertheless, to date, this is the fourth report of a familial CHD2 pathogenic variant and the first one that the variant is present in three generations. The first familial case was of a 5-year-old female with developmental delay, microcephaly, and seizures who had a maternally inherited nonsense heterozygous pathogenic CHD2 variant (c.628G>T, p. (E210X)). Her mother had a history of infantile meningitis, bipolar disorder, attention deficit hyperactivity disorder, language delay, and generalised tonic-clonic epilepsy with seizure onset at the age of 5 [15]. Furthermore, another patient was reported with eyelid myoclonia with absence (EMA) that inherited from the clinically-unaffected mother (whose EEG was not performed) a missense CHD2 variant (c.653C>T p.(P218L)) [16]. Nevertheless, with current ACMG guidelines [7] and updated ClinGen recommendations, this variant is considered of unclear significance (VUS). Chen et al. (2020), also, presented three more patients with paternally inherited CHD2 variants [17]. Two of the patients were dizygotic twins that had the same seizure type (GTCS). These patients inherited the same variation (c.5232G>A, p.(M1744I)) from their affected father, who had a history of febrile seizures in infancy. However, based on ACMG guidelines [7] and updated ClinGen recommendations, this variant is classified as VUS and further evidence is needed to support its pathogenicity. The third patient inherited a pathogenic splicing CDH2 variant (c.5153+2T>C) from an unaffected father.
The c.5035C>T variant reported herein was previously described as de novo in monozygotic twins with identical manifestations, both presented with tonic-clonic seizures starting at age 3, intellectual impairment, and a pathological EEG (spike and sharp wave) [9]. Moreover, Helbig et al. (2016) detected the variant in a patient suffering from epileptic encephalopathy, with childhood-onset [10]. Although most CHD2 variants are associated with developmental delay, intellectual impairment and seizures, there is great phenotypic heterogeneity regarding the latter. Chen et al. (2020) found that 63 out of 70 patients with a CHD2 variant suffered from epileptic seizures [17]. Nevertheless, not every patient had the same epilepsy form; they presented with EMA, Dravet syndrome, juvenile myoclonic epilepsy, Jeavons syndrome, febrile seizures plus, LGS, West syndrome, or non-specific epileptic encephalopathy. Interestingly, some patients did not have seizures, similar to our proband's maternal aunt. There is limited knowledge regarding patients with a CHD2 pathogenic variant and the absence of seizures, but Lebrun et al. (2017) presented a case of two male siblings with autism spectrum disorder, both of whom had the same CHD2 variant but only one had seizures [18]. We speculate that this heterogeneity could also explain why the maternal aunt carrying the variant does not suffer from epilepsy. Moreover, the proband in this study has not had a documented epilepsy so far, but she has had some suspect episodes anamnestically, thus a follow-up is needed.
Pathogenic variants in CHD2 were reported to have a wide range of phenotypic variability but mainly known to cause brain-restricted phenotypes in humans. Patients with pathogenic CHD2 variants were previously described by Wang et al., with additional clinical features such as microcephaly, hypotonia, and slender fingers [19]. The authors also reviewed previously described patients with pathogenic CHD2 variants and more dysmorphic features, however a clear genotype-phenotype correlation has not been established yet. At this stage, we cannot exclude the possibility that dysmorphic features present in the proband may not be related with the CHD2 variant. Detailed evaluation and description of additional patients with CHD2 variants is needed and further investigation with whole genome sequencing may reveal additional causative variants.
The present study adds towards the highly variable phenotypic spectrum of Developmental Epileptic Encephalopathy 94, since it is shown that members of the same family, carrying the same pathogenic variant, may have discordant phenotypes. The candidate variant identified provided evidence that the genetic cause of the patient's phenotype lays elsewhere in the genome unrelated to the inversion detected initially. Exome sequencing and segregation analysis is, therefore, recommended in cases where the inversion is familial and the known disrupted genes cannot explain the phenotype. This is in agreement with a recent study on cases of balanced translocations, which did not explain the phenotype, and the importance of exome sequencing in finding the cause of the pathologies of the patients [12]. We anticipate that additional investigation of more familial inversions and balanced rearrangements will shed more light towards understanding the underlying genetic mechanisms of the disease and provide better genetic counselling to the patients.
Disclosure conflict of interest
The authors declare no conflict of interest.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank all the family members participating in this study and the Bioinformatics group at CING for providing the NGS pipelines used herein.
References
- 1.Aristidou C., et al. Accurate breakpoint mapping in apparently balanced translocation families with discordant phenotypes using whole genome mate-pair sequencing. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0169935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liehr T., et al. Microdissection based high resolution multicolor banding for all 24 human chromosomes. Int. J. Mol. Med. 2002;9(4):335–339. [PubMed] [Google Scholar]
- 3.Liehr T. Fluorescence in Situ Hybridization (FISH) – Application Guide. second ed. Springer; 2017. Two- to three-color FISH. [Google Scholar]
- 4.DePristo M.A., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLaren W., et al. The ensembl variant effect predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paila U., et al. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput. Biol. 2013;9(7) doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richards S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopanos C., et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35(11):1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., et al. Genetic variants identified from epilepsy of unknown etiology in Chinese children by targeted exome sequencing. Sci. Rep. 2017;7 doi: 10.1038/srep40319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helbig K.L., et al. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet. Med. 2016;18(9):898–905. doi: 10.1038/gim.2015.186. [DOI] [PubMed] [Google Scholar]
- 11.Thomas N.S., et al. Investigation of the origins of human autosomal inversions. Hum. Genet. 2008;123(6):607–616. doi: 10.1007/s00439-008-0510-z. [DOI] [PubMed] [Google Scholar]
- 12.Aristidou C T.A., Alexandrou A., Papaevripidou I., Evangelidou P., Kosmaidou-Aravidou Z., Behjati F., Christophidou-Anastasiadou V., Tanteles G.A., Sismani C. Exploring the genetic causality of discordant phenotypes in familial apparently balanced translocation cases using whole exome sequencing. Genes. 2023;14(1) doi: 10.3390/genes14010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvill G.L., et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat. Genet. 2013;45(7):825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suls A., et al. De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am. J. Hum. Genet. 2013;93(5):967–975. doi: 10.1016/j.ajhg.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen A.K., et al. The first reported case of an inherited pathogenic CHD2 variant in a clinically affected mother and daughter. Am. J. Med. Genet. 2018;176(7):1667–1669. doi: 10.1002/ajmg.a.38835. [DOI] [PubMed] [Google Scholar]
- 16.Galizia E.C., et al. CHD2 variants are a risk factor for photosensitivity in epilepsy. Brain. 2015;138(Pt 5):1198–1207. doi: 10.1093/brain/awv052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J., et al. CHD2-related epilepsy: novel mutations and new phenotypes. Dev. Med. Child Neurol. 2020;62(5):647–653. doi: 10.1111/dmcn.14367. [DOI] [PubMed] [Google Scholar]
- 18.Lebrun N., et al. Autism spectrum disorder recurrence, resulting of germline mosaicism for a CHD2 gene missense variant. Clin. Genet. 2017;92(6):669–670. doi: 10.1111/cge.13073. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., et al. Novel loss-of-function variants in CHD2 cause childhood-onset epileptic encephalopathy in Chinese patients. Genes. 2022;13(5):908. doi: 10.3390/genes13050908. [DOI] [PMC free article] [PubMed] [Google Scholar]