Abstract
Introduction
The population structure of Mycobacterium tuberculosis complex (MTBC) in Ethiopia is diverse but dominated by Euro-American (Lineage 4) and East-African-Indian (Lineage 3) lineages. The objective of this study was to describe the genetic diversity of MTBC isolates in Central, Eastern and Southeastern Ethiopia.
Methods
A total of 223 MTBC culture isolates obtained from patients referred to Adama and Harar TB reference laboratories were spoligotyped. Demographic and clinical characteristics were collected.
Results
Six major lineages: Euro-American (Lineage 4), East-African-Indian (Lineage 3), East Asian (Lineage 2), Indo-Oceanic (Lineage 1), Mycobacterium africanum (Lineage 5 and Lineage 6) and Ethiopian (Lineage 7) were identified. The majority (94.6 %) of the isolates were Euro-American and East-African-Indian, with proportions of 75.3 % and 19.3 %, respectively. Overall, 77 different spoligotype patterns were identified of which 42 were registered in the SITVIT2 database. Of these, 27 spoligotypes were unique, while 15 were clustered with 2–49 isolates. SIT149/T3_ETH (n = 49), SIT53/T1 (n = 33), SIT21/CAS1_Kili (n = 24) and SIT41/Turkey (n = 11) were the dominant spoligotypes. A rare Beijing spoligotype pattern, SIT541, has also been identified in Eastern Ethiopia. The overall clustering rate of sub-lineages with known SIT was 71.3 %. Age group (25–34) was significantly associated with clustering.
Conclusion
We found a heterogeneous population structure of MTBC dominated by T and CAS families, and the Euro-American lineage. The identification of the Beijing strain, particularly the rare SIT541 spoligotype in Eastern Ethiopia, warrants a heightened surveillance plan, as little is known about this genotype. A large-scale investigation utilizing a tool with superior discriminatory power, such as whole genome sequencing, is necessary to gain a thorough understanding of the genetic diversity of MTBC in the nation, which would help direct the overall control efforts.
Keywords: Spoligotyping, Genetic diversity, Lineage, Sub-lineage, Mycobacterium tuberculosis complex, Ethiopia
1. Introduction
Tuberculosis (TB) and its consequences afflict millions of individuals worldwide. The World Health Organization (WHO) [1] estimated that TB killed 1.3 million people in 2020. Ethiopia is one of the 30 countries with the highest TB and TB/HIV infection rates, with an estimated 151,000 TB cases in 2020 [1].
Molecular typing of Mycobacterium tuberculosis complex (MTBC) has become a vital public health tool helping researchers and TB control programs better understand how specific strains emerge and spread, as well as measure the overall impact of genetic diversity on the outcome of TB infection and disease [2]. Some biological aspects of MTBC strains from distinct genetic lineages show variation, such as in vitro growth rate, pathogenicity in animal models, and the ability to acquire drug resistance [3].
Lineages known to cause TB in humans are divided into seven major TB lineages [4,5]: Indo-Oceanic (Lineage 1), East-Asian (Lineage 2), East-African-Indian (Lineage 3), Euro-American (Lineage 4), West-Africa 1 (Lineage 5), West-Africa 2 (Lineage 6) and Ethiopian (Lineage 7). Most recently, Lineage 8 [6] and Lineage 9 [7] were reported from the Central and Eastern Africa regions, respectively. Among these, the most common lineage on the planet is lineage 4 (L4). A collection of L4 was sequenced and further classified into ten sub-lineages as generalists (globally distributed) and specialists (geographically restricted) based on the ecological niche they occupy [8,9]. A report by Comas et al. (2015) [10] indicated that L4 predominates across Ethiopia, L3 is widespread but more common in the north, and L7 is mostly found in the northern Ethiopian highlands. Numerous studies have identified spoligotypes SIT 149 and SIT 53 as major clades circulating in Ethiopia [[11], [12], [13], [14]].
The number of studies on the genetic diversity of MTBC in Ethiopia has been increasing. However, most of the studies reviewed elsewhere in the literature [15,16] were from Addis Ababa, the Southwestern and the Northwestern parts of Ethiopia. Studies conducted in other regions of the country are scarce or nonexistent. The lack of such information prevents the capacity of the national TB control program to implement targeted interventions, identify transmission patterns, prevent further transmission of the disease, and impede the design of novel control tools. Consequently, the findings of this study not only add to the scientific literature in the field of TB epidemiology, but also hold significant implications for improving TB control efforts in Ethiopia. Therefore, the objective of this study was to describe the population structure of MTBC isolates collected from a large geographical area that mainly included the central, eastern and southeastern parts of Ethiopia.
2. Materials and methods
2.1. Study setting and source of the isolates
This is a health facility-based cross-sectional study conducted at two TB referral diagnostic laboratories found in Harar and Adama, Ethiopia. From August 2018 to January 2019, 232 MTB culture isolates were obtained from pulmonary TB patients referred to Adama and Harar TB regional laboratories located in the premises of Adama Public Health Research and Referral Laboratory Center and Harar Health Research and Regional Laboratory, respectively. Demographic (age, sex, and address) and clinical (history of treatment) characteristics were collected. The regional TB laboratories in Adama and Harar serve as referral centers for Mycobacteria culture and drug susceptibility testing. The Adama TB regional laboratory is located in Adama city, which is 99 km southeast of Addis Ababa, on the main trade corridor of the country. It serves nine zones in Oromia, as well as the surrounding Amhara and Afar regions. The Harar TB regional laboratory, located in Harar city 500 km from the capital, provides referral diagnostic services to Diredawa, east Oromia, Somali, and Harari regions. Once the isolates were confirmed to be Mycobacterium tuberculosis complex (MTBC) using Capilia TB-Neo (Tauns Laboratories, Japan) and AFB smear staining [17], they were subcultured onto Lowenstein Jensen media and harvested within 3–4 weeks. In duplicate, two loopful colonies of the culture were transferred to 2 ml sterile cryo-vials containing 1 ml 7H9 Middlebrook liquid medium. The isolates were subsequently transported to the University of Pretoria's Medical Microbiology Department, TB laboratory for further testing.
2.2. DNA extraction
Isolates were stored at −80 °C until processed. The stored isolates were then thawed and mixed vigorously. DNA was extracted by heating the isolates at 80 °C for 60 min in a water bath. After centrifugation, the supernatant was collected for further use.
2.3. Isolate characterization
The isolates were characterized using spoligotyping following the procedure described by Kamerbeek et al. [18]. Briefly, the set of spacers in the isolates was amplified by PCR using DRa and DRb primers. The amplicons were then hybridized on a reference set of 43 spacers obtained from Mycobacterium tuberculosis H37Rv and Mycobacterium bovis BCG impregnated on a membrane (Animal and Plant Health Agency, Great Britain). The presence or absence of spacers was visualized on a film as black and white squares, which were later converted to binary codes (1/0) for analysis. Positive and negative controls were included in each batch of the test run.
2.4. Spoligotype assignment and database comparison
The spoligotype patterns entered into the MS Excel spreadsheet were converted into binary and octal formats using the SITVITWEB website (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/tools.jsp#) which were later compared with online international databases such as SITVIT2 [19], SpolLineages [5] and RUN TB-lineage [20,21]. Sub-lineages and International Shared Types (SIT) were obtained using the updated version of SPOLDB4 and SITVITWEB (http://www.pasteur-guadeloupe.fr:8081/SITVIT2/batch.jsp). Conformal Bayesian Network (CBN) major lineages, and SNP-based lineages were determined using online tools RUN TB-lineage (https://tbinsight.cs.rpi.edu/run_tb_lineage.html) and SpolLineages (http://www.pasteur-guadeloupe.fr:8081/SpolLineages/spol.jsp), respectively. CBN does not distinguish between the West African (L6 and L5) and Ethiopian (L7) lineages, classifying them as Mycobacterium africanum. We used SpolLineages to avoid this, further verify the CBN classification of major lineages and explore the classification of lineages as generalists and specialists [8,9].
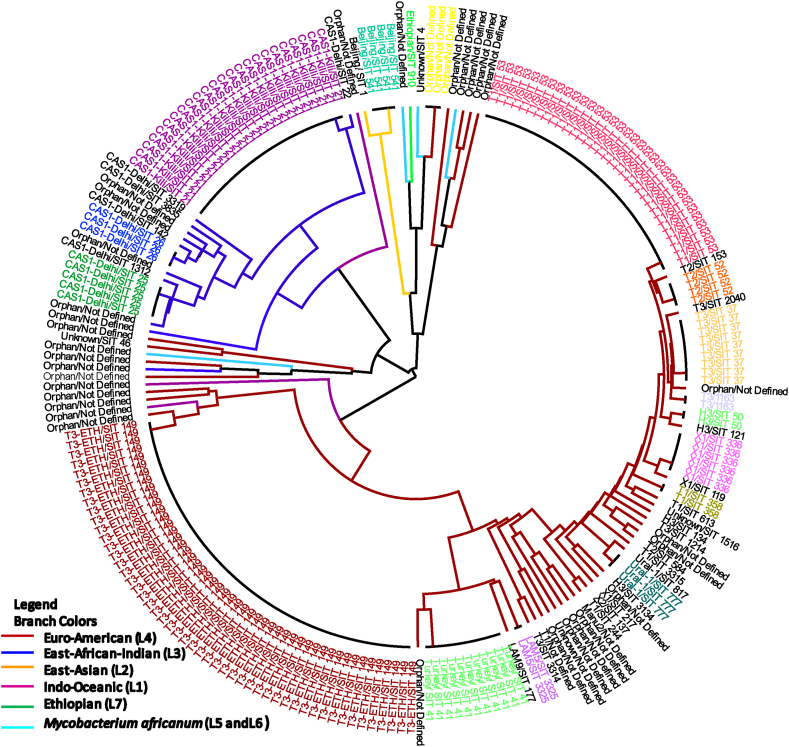
A dendrogram was constructed based on the UPGMA algorithm using the MIRU-VNTRplus identification database to determine the molecular clustering of the isolates. The UPGMA tree was then retouched using FigTree v1.4.4 (Fig. 1). A cluster was defined as two or more isolates with similar spoligotype patterns. A spoligotype pattern that was not previously reported in an international database was defined as orphan or new whereas a spoligotype with SIT reported once in the study was defined as unique.
Fig. 1.
Radial UPGMA tree based on spoligotyping data of 223 MTB isolates from central, eastern and southeastern Ethiopia. Annotations with similar colored fonts in the outer ring represent clustered sub-lineages/spoligotypes. The ones written in black fonts are unique/orphan isolates. The colors of branches in the inner circle indicate the major lineages. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.5. Spatial distribution of lineages and sub-lineages analyses
Geographic mapping of MTBC lineage and sub-lineages was performed using QGIS v3.22.6. The shape files of study sites from where the isolates collected were obtained from UNOCHA website (https://data.humdata.org/dataset/cod-ab-eth).
2.6. Statistical analysis
Data were entered into an Excel spreadsheet; cleared, and analyzed using the SPSS statistical software package, V20 (SPSS Inc., Chicago, IL, USA). Sociodemographic variables were depicted using descriptive statistics. A logistic regression model was used to assess variables associated with clustering in terms of the odds ratio and 95 % confidence interval (CI). The chi-squared test was applied to compare categorical data. A p-value less than or equal to 0.05 was considered significant.
2.7. Ethical consideration
The Ethical Review Board of Natural Sciences at Addis Ababa University granted ethical approval for this study. The Ethiopian Food, Medicine and Health Care Administration and Control Authority (now known as the Ethiopian Food and Drug Administration) and the Health Department of South Africa approved the transfer of mycobacterial isolates to South Africa for further molecular analysis. Permission to perform the research was also acquired from the Harari and Adama Public Health Research and Referral Laboratories. Only isolates routinely obtained from patients for diagnostic and therapeutic purposes were used in the study. Patients' personal information was not collected.
3. Result
3.1. Characteristics of the study population
Of the 232 isolates collected, 9 were either lost or did not have successful spoligotyping result. Hence, this study examined a total of 223 isolates from TB patients who were presumed to have DR TB, with a mean age of 30.4 years. Most of the study subjects (58.3 %) were males and in the age group-15-34 years (58.3 %). The majority (80.8 %) were from the Central (Arsi; East, North, Southeast and West Shewa) and Eastern (Diredawa, Harar, Jigjiga, East and West Hararge) parts of Ethiopia. The remaining were from the Southeastern part of Ethiopia, specifically from Bale, Borena, Guji, West Arsi and West Guji zones. There was an equal proportion of patients in relation to treatment history (Table 1).
Table 1.
Characteristics of study subjects.
| Variable | Frequency (Percent) | |
|---|---|---|
| Age, years | ||
| ≤15 | 19 (8.5) | |
| 16–24 | 56 (25.1) | |
| 25–34 | 74(33.2) | |
| 35–44 | 38(17.0) | |
| >45 | 34(15.2) | |
| Missing | 2(.9) | |
| Sex | ||
| Male | 130 (58.3) | |
| Female | 93 (41.7) | |
| History of anti-TB drug treatment | ||
| New | 112 (50.2) | |
| Previously treated | 111 (49.8) | |
| Region | ||
| Central | 94 (42.2) | |
| Southeastern | 43 (19.3) | |
| Eastern | 86 (38.6) | |
3.2. Genetic diversity of Mycobacterium tuberculosis lineages/sub-lineages
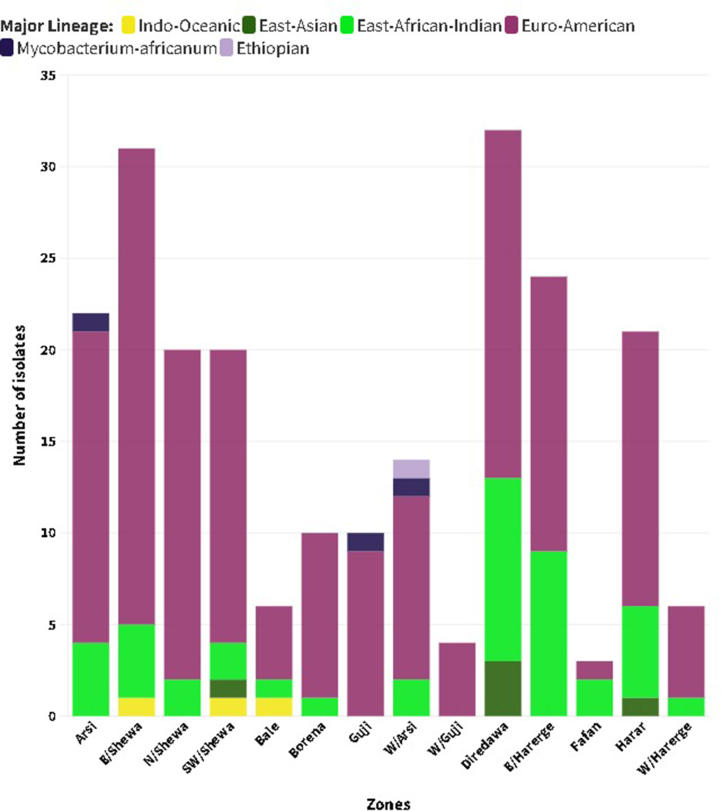
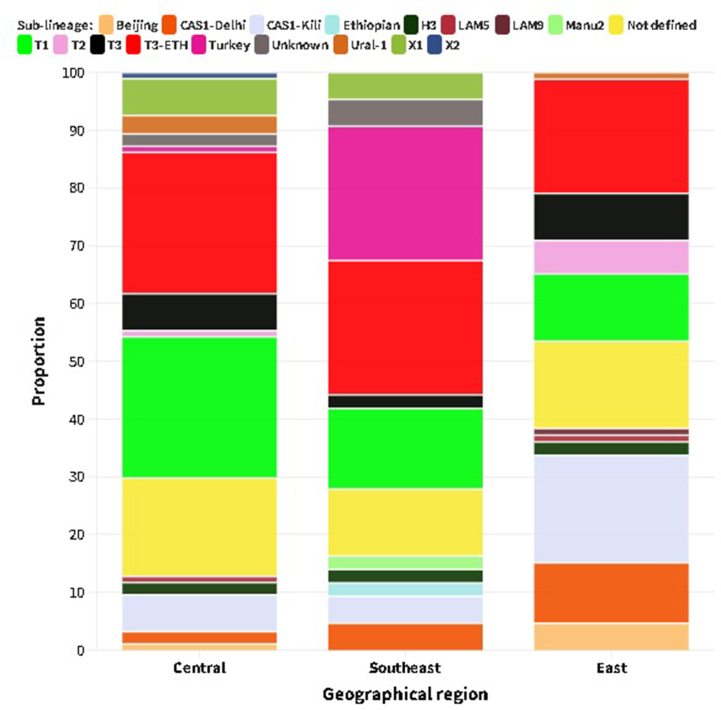
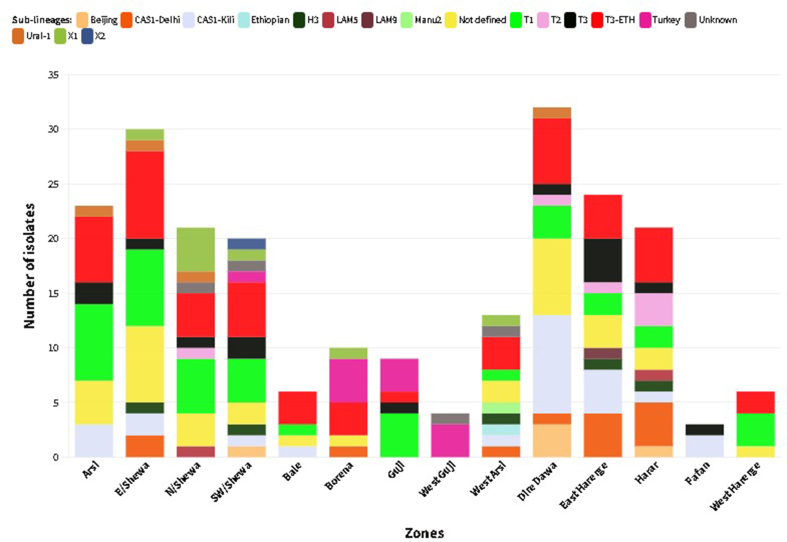
Spoligotyping was used to genotype 223 isolates, and 77 different spoligotype patterns were identified, of which 42 spoligotype patterns comprising 186 isolates were registered in the SITVIT2 database. Of these, 27 were unique while 15 were clustered with 2–49 isolates that accounted for 85.5 % (159/186) of all isolates with known SIT. The remaining 35 orphan patterns, representing 16.6 % (37/223) of the total isolates, were not found in the SITVIT2 database (Table 2, Table 3). The overall clustering rate of sub-lineages with known SIT was 71.3 %. The proportion of clustered isolates was higher in each geographic region: Central (72.3 %), Eastern (72.1 %) and Southeastern (67.4 %). There were five dominant spoligotypes with SIT, which accounted for over half of the genotyped isolates 117 (52.5 %): SIT149/T3_ETH (n = 49), SIT53/T1 (n = 33), SIT21/CAS1_Kili (n = 24) and SIT41/Turkey (n = 11). T and CAS were the dominant families, accounting for 48.9 % and 16.6 % of the isolates, respectively. Of the 49 isolates with SIT149/T3_ETH sub-lineage, 8 (16.3 %) were children of 15 years or younger. According to the CBN analysis, 94.6 % of the total 223 isolates belonged to two major lineages: EA/L4 (75.3 %) and EAI/L3 (19.3 %). The remaining 12/223 (5.4 %) isolates were represented by EAS/L2, IO/L1, ETH/L7 and MA (L5 and L6) which were represented by five, three, one and three isolates, respectively. Of those classified by CBN as EA and EAI, 8.3 % (EA) and 4.7 % (EAI) were not known by SNP-based lineage analysis of the SpolLineages online tool. Additionally, we found two generalist sub-lineages [8,9], L4.1.2/Haarlem (n = 5) and L4.3/LAM (n = 5), and most (9/10) were isolated from the central and eastern regions (Table 2).
Table 2.
Description of 35 orphan strains (n = 37) and the corresponding spoligotyping defined lineages/sub‐lineages recorded among MTB strains isolated from Central, Eastern and Southeastern Ethiopia.
| Octal code | Binary code | SITVIT2 | CBN | SNP-based lineage |
|---|---|---|---|---|
| 525252525240521 | ■□■□■□■□■□■□■□■□■□■□■□■□■□■□■□■□□□□□■□■□■□■ | ND | EA | EA (L4.1.1) |
| 007777777760771 | □□□□□□■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | T1 | EA | EA (L4) |
| 500177740003171 | ■□■□□□□□□□□■■■■■■■■■■■□□□□□□□□□□□□■■□□■■■■■ | ND | EAI | EAI (L3) |
| 501252500002121 | ■□■□□□□□■□■□■□■□■□■□■□□□□□□□□□□□□□■□□□■□■□■ | ND | EAI | EAI (L3) |
| 777000377777771 | ■■■■■■■■■□□□□□□□□□□■■■■■■■■■■■■■■■■■■■■■■■■ | ND | IO | UNK |
| 700006037177661 | ■■■□□□□□□□□□□□□■■□□□□□■■■■■□□■■■■■■■■■□■■□■ | ND | MA | WA (L5 and L6) |
| 703757740003171 | ■■■□□□□■■■■■■□■■■■■■■■□□□□□□□□□□□□■■□□■■■■■ | ND | EAI | EAI (L3) |
| 713247664001661 | ■■■□□■□■■□■□■□□■■■■■□■■□■□□□□□□□□□□■■■□■■□■ | ND | IO | UNK |
| 777777777000171 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□□□□□□□□■■■■■ | UKN | EA | UNK |
| 771046024100261 | ■■■■■■□□■□□□■□□■■□□□□□■□■□□□□■□□□□□□□■□■■□■ | ND | EA | UNK |
| 773777776000771 | ■■■■■■□■■■■■■■■■■■■■■■■■■■□□□□□□□□□□■■■■■■■ | ND | EA | UNK |
| 774242525042000 | ■■■■■■■□□□■□■□□□■□■□■□■□■□■□□□■□□□■□□□□□□□□ | ND | MA | UNK |
| 703357740003071 | ■■■□□□□■■□■■■□■■■■■■■■□□□□□□□□□□□□■■□□□■■■■ | ND | EAI | EAI (L3) |
| 703777710003771 | ■■■□□□□■■■■■■■■■■■■■■□□■□□□□□□□□□□■■■■■■■■■ | ND | EAI | UNK |
| 777737777360771 | ■■■■■■■■■■■■□■■■■■■■■■■■■■■□■■■■□□□□■■■■■■■ | ND | EA | EA (L4) |
| 777775477760731 | ■■■■■■■■■■■■■■■■□■■□□■■■■■■■■■■■□□□□■■■□■■■ | ND | EA | EA (L4) |
| 777777404060771 | ■■■■■■■■■■■■■■■■■■■□□□□□■□□□□□■■□□□□■■■■■■■ | ND | EA | EA (L4.3) |
| 477777376413771 | ■□□■■■■■■■■■■■■■■■□■■■■■■■□■□□□□■□■■■■■■■■■ | ND | IO | IO (L1) |
| 777737377750771 | ■■■■■■■■■■■■□■■■■■□■■■■■■■■■■■■□■□□□■■■■■■■ | ND | EA | UNK |
| 510047236343661 | ■□■□□■□□□□□□■□□■■■□■□□■■■■□□■■■□□□■■■■□■■□■ | ND | MA | WA (L5 and L6) |
| 525252500000000 | ■□■□■□■□■□■□■□■□■□■□■□□□□□□□□□□□□□□□□□□□□□□ | ND | EA | UNK |
| 551246236340261 | ■□■■□■□□■□■□■□□■■□□■□□■■■■□□■■■□□□□□□■□■■□■ | ND | EA | EA (L4) |
| 511047236340261 | ■□■□□■□□■□□□■□□■■■□■□□■■■■□□■■■□□□□□□■□■■□■ | ND | EA | EA (L4) |
| 777347777763771 | ■■■■■■■■■□■■■□□■■■■■■■■■■■■■■■■■□□■■■■■■■■■ | Manu2 | EA | UNK |
| 511046637561671 | ■□■□□■□□■□□□■□□■■□■■□□■■■■■■□■■■□□□■■■□■■■■ | ND | EA | UNK |
| 603777700003771 | ■■□□□□□■■■■■■■■■■■■■■□□□□□□□□□□□□□■■■■■■■■■ | ND | EAI | UNK |
| 311000377760771 | □■■□□■□□■□□□□□□□□□□■■■■■■■■■■■■■□□□□■■■■■■■ | ND | EA | EA (L4) |
| 777000377730771 | ■■■■■■■■■□□□□□□□□□□■■■■■■■■■■■□■■□□□■■■■■■■ | ND | EA | UNK |
| 713357776363771 | ■■■□□■□■■□■■■□■■■■■■■■■■■■□□■■■■□□■■■■■■■■■ | ND | EA | UNK |
| 777347636361771 | ■■■■■■■■■□■■■□□■■■■■□□■■■■□□■■■■□□□■■■■■■■■ | ND | EA | UNK |
| 777775777760721 | ■■■■■■■■■■■■■■■■□■■■■■■■■■■■■■■■□□□□■■■□■□■ | ND | EA | EA (L4) |
| 774177770000021 | ■■■■■■■□□□□■■■■■■■■■■■■■□□□□□□□□□□□□□□□□■□■ | ND | EA | UNK |
| 520000377760771 | ■□■□■□□□□□□□□□□□□□□■■■■■■■■■■■■■□□□□■■■■■■■ | ND | EA | EA (L4) |
| 700000017760771* | ■■■□□□□□□□□□□□□□□□□□□□□■■■■■■■■■□□□□■■■■■■■ | ND | EA | EA (L4) |
| 777357677761771 | ■■■■■■■■■□■■■□■■■■■■□■■■■■■■■■■■□□□■■■■■■■■ | ND | EA | UNK |
EA = Euro-American, EA = East-Asian, EAI = East-African-Indian, IO=Indo-Oceanic, UKN=Unknown, WA=West Africa, MA = Mycobacterium Africanum, ND=Not defined,*n=3.
Table 3.
Description of 42 shared types (SITs; n = 186 isolates) registered in the SITVIT2 or SpolDB4 database and the corresponding spoligotyping defined lineages/sub‐lineages isolated from the Central, Eastern and Southeastern Ethiopia.
| Octal code | Binary code | SITVIT2 | CBN | SNP-based lineage | SIT | Number of isolates |
|---|---|---|---|---|---|---|
| 777000377760771 | ■■■■■■■■■□□□□□□□□□□■■■■■■■■■■■■■□□□□■■■■■■■ | T3-ETH | EA | EA (L4) | 149 | 49 |
| 777777777760771 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | T1 | EA | EA (L4) | 53 | 33 |
| 703377400001771 | ■■■□□□□■■□■■■■■■■■■□□□□□□□□□□□□□□□□■■■■■■■■ | CAS1-Kili | EAI | EAI (L3) | 21 | 24 |
| 777777404760771 | ■■■■■■■■■■■■■■■■■■■□□□□□■□□■■■■■□□□□■■■■■■■ | Turkey | EA | EA (L4.2.2.1) | 41 | 11 |
| 777737777760771 | ■■■■■■■■■■■■□■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | T3 | EA | EA (L4) | 37 | 9 |
| 777776777760731 | ■■■■■■■■■■■■■■■■■□■■■■■■■■■■■■■■□□□□■■■□■■■ | X1 | EA | EA (L4.1.1) | 336 | 6 |
| 703777740003171 | ■■■□□□□■■■■■■■■■■■■■■■□□□□□□□□□□□□■■□□■■■■■ | CAS1-Delhi | EAI | EAI (L3) | 25 | 5 |
| 777777777760731 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■□■■■ | T2 | EA | EA (L4) | 52 | 4 |
| 000000000003711 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■■□□■■ | Beijing | EAS | EAS (L2) | 541 | 4 |
| 703777740003771 | ■■■□□□□■■■■■■■■■■■■■■■□□□□□□□□□□□□■■■■■■■■■ | CAS1-Delhi | EAI | EAI (L3) | 26 | 3 |
| 777777777420771 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□■□□□□■■■■■■■ | Ural-1 | EA | EA (L4.2.1) | 777 | 3 |
| 677737777760771 | ■■□■■■■■■■■■□■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | T3 | EA | EA (L4) | 1163 | 2 |
| 776737407760771 | ■■■■■■■■□■■■□■■■■■■□□□□□■■■■■■■■□□□□■■■■■■■ | LAM5 | EA | EA (L4.3) | 3325 | 2 |
| 717777777760771 | ■■■□□■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | T1 | EA | EA (L4) | 358 | 2 |
| 777777777720771 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□■□□□□■■■■■■■ | H3 | EA | EA (L4.1.2) | 50 | 2 |
| 000000000003771 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■■■■■■ | Beijing | EAS | EAS (L2) | 1 | 1 |
| 777776777760771 | ■■■■■■■■■■■■■■■■■□■■■■■■■■■■■■■■□□□□■■■■■■■ | X1 | EA | EA (L4.1.1) | 119 | 1 |
| 777777775720771 | ■■■■■■■■■■■■■■■■■■■■■■■■■□■■■■□■□□□□■■■■■■■ | H3 | EA | EA (L4.1.2) | 121 | 1 |
| 777617777760771 | ■■■■■■■■■■■□□□■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | T3 | EA | EA (L4) | 1214 | 1 |
| 703777740003131 | ■■■□□□□■■■■■■■■■■■■■■■□□□□□□□□□□□□■■□□■□■■■ | CAS1-Delhi | EAI | EAI (L3) | 1312 | 1 |
| 777777777720631 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□■□□□□■■□□■■■ | H3 | EA | EA (L4.1.2) | 134 | 1 |
| 777776777760601 | ■■■■■■■■■■■■■■■■■□■■■■■■■■■■■■■■□□□□■■□□□□■ | X2 | EA | EA (L4.1.1) | 137 | 1 |
| 703777700003771 | ■■■□□□□■■■■■■■■■■■■■■□□□□□□□□□□□□□■■■■■■■■■ | CAS1-Delhi | EAI | EAI (L3) | 142 | 1 |
| 777377777761771 | ■■■■■■■■■□■■■■■■■■■■■■■■■■■■■■■■□□□■■■■■■■■ | UKN | EA | UNK | 1516 | 1 |
| 757777777760731 | ■■■■□■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■□■■■ | T2 | EA | EA (L4) | 153 | 1 |
| 377777607760771 | □■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■■□□□□■■■■■■■ | LAM9 | EA | EA (L4.3) | 177 | 1 |
| 777737777760371 | ■■■■■■■■■■■■□■■■■■■■■■■■■■■■■■■■□□□□□■■■■■■ | T3 | EA | EA (L4) | 2040 | 1 |
| 777736777760771 | ■■■■■■■■■■■■□■■■■□■■■■■■■■■■■■■■□□□□■■■■■■■ | X1 | EA | EA (L4.1.1) | 217 | 1 |
| 703777400001771 | ■■■□□□□■■■■■■■■■■■■□□□□□□□□□□□□□□□□■■■■■■■■ | CAS1_Dehi | EAI | EAI (L3) | 22 | 1 |
| 777777777760601 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■□□□□■ | T1 | EA | EA (L4) | 244 | 1 |
| 777737377720771 | ■■■■■■■■■■■■□■■■■■□■■■■■■■■■■■□■□□□□■■■■■■■ | H3 | EA | EA (L4.1.2) | 3134 | 1 |
| 177000377760771 | □□■■■■■■■□□□□□□□□□□■■■■■■■■■■■■■□□□□■■■■■■■ | T3-ETH | EA | EA (L4) | 3141 | 1 |
| 006737777760771 | □□□□□□■■□■■■□■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | T3 | EA | EA (L4) | 3314 | 1 |
| 376777737760771 | □■■■■■■■□■■■■■■■■■■■■□■■■■■■■■■■□□□□■■■■■■■ | T1 | EA | EA (L4) | 3315 | 1 |
| 703417740003771 | ■■■□□□□■■■□□□□■■■■■■■■□□□□□□□□□□□□■■■■■■■■■ | CAS1-Delhi | EAI | EAI (L3) | 3319 | 1 |
| 703177740003771 | ■■■□□□□■■□□■■■■■■■■■■■□□□□□□□□□□□□■■■■■■■■■ | CAS1-Delhi | EAI | EAI (L3) | 3835 | 1 |
| 000000007760771 | □□□□□□□□□□□□□□□□□□□□□□□□■■■■■■■■□□□□■■■■■■■ | UKN | EA | EA (L4.3) | 4 | 1 |
| 777777770000000 | ■■■■■■■■■■■■■■■■■■■■■■■■□□□□□□□□□□□□□□□□□□□ | UKN | EA | UKN | 46 | 1 |
| 777775777760731 | ■■■■■■■■■■■■■■■■□■■■■■■■■■■■■■■■□□□□■■■□■■■ | T2 | EA | EA (L4) | 584 | 1 |
| 777177777760771 | ■■■■■■■■■□□■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | T1 | EA | EA (L4) | 613 | 1 |
| 777777777420731 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□■□□□□■■■□■■■ | Ural-1 | EA | EA (L4.2.1) | 817 | 1 |
| 700000007177771 | ■■■□□□□□□□□□□□□□□□□□□□□□■■■□□■■■■■■■■■■■■■■ | ETH | MA | ETH (L7) | 910 | 1 |
EA = Euro-American, EA = East-Asian, EAI = East-African-Indian, IO=Indo-Oceanic, ETH = Ethiopian, UKN=Unknown, WA=West Africa, MA = Mycobacterium Africanum, ND=Not-defined.
3.3. Geographical distribution of the lineages/sub-lineages
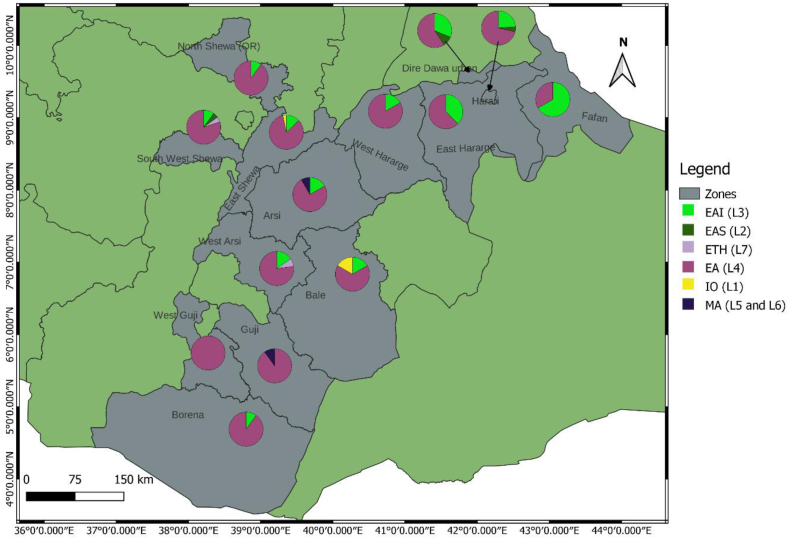
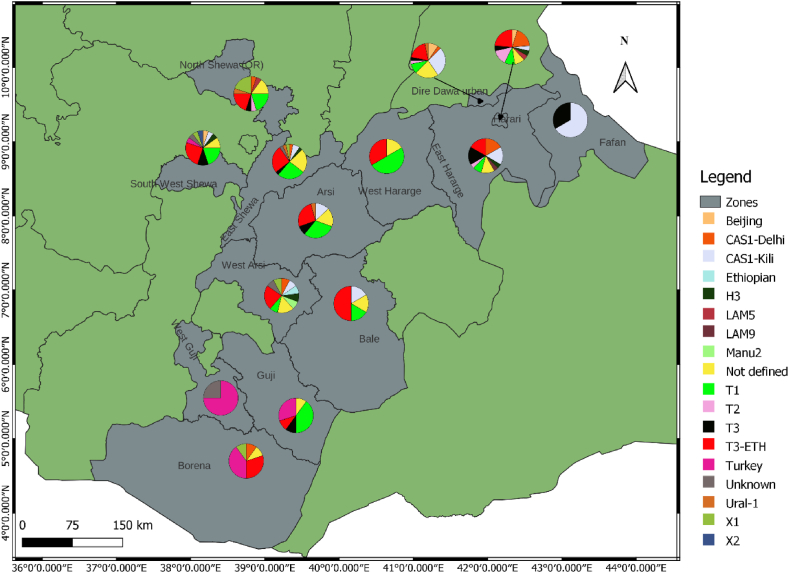
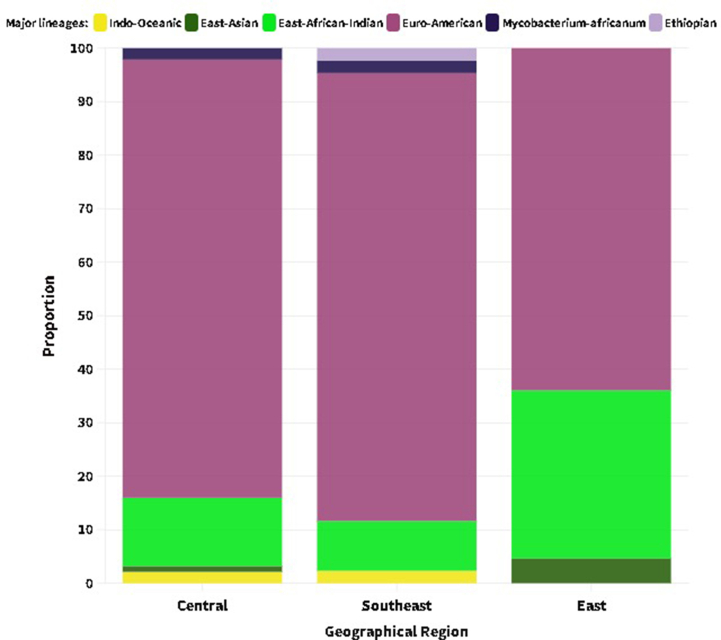
EA (L4) contributed a significantly high proportion to the lineage distribution across the study sites: 83.7 % (Southeastern), 81.9 % (Central) and 64.0 % (Eastern) of the isolates collected in the respective parts of Ethiopia (Supplementary material 1). EAI (L3) was reported mainly in the eastern regions, followed by the central regions, with proportions of 62.8 % and 27.9 % of the total isolates, respectively. With the exception of Guji/West Guji, where no EAI was recorded, EA and EAI were identified in all zones from where the isolates were acquired. MTBC isolates obtained from Arsi (n = 2) and West Guji (n = 1) zones were identified as Mycobacterium africanum (Fig. 2, Supplementary material 2). The SIT910 spoligotype (ETH/L7), which is confined to Ethiopia, was isolated from West Arsi. There were five isolates with Beijing sub-lineage (EAS/L2), four from the eastern (three from Diredawa and one from Harar) and one from the central part (Southwest Shewa) of Ethiopia. Of note, 10/11 (90.9 %) of EA (L4) lineage, SIT41 spoligotype/Turkey sub-lineage were reported from southeastern Ethiopia (Borena and Guji). On the contrary, the SIT149 spoligotype/T3-ETH sub-lineage and SIT53 spoligotype/T1 sub-lineages were reported from most of the sites where the isolates were collected (Fig. 3, Supplementary materials 3 and 4).
Fig. 2.
Distribution of major lineages in central, eastern and southeastern Ethiopia. Pie charts show the proportions of the six lineages among MTBC isolates in each zone from where isolates were obtained. The size of the circle does not correspond to the number of isolates analyzed. The actual numbers are shown in Supplementary material 2: color codes are as in Fig. 2. A total of 223 MTBC isolates were included. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
Distribution of MTBC sub-lineages in central, eastern and southeastern Ethiopia. Pie charts show the proportions of the sub-lineages among MTBC isolates in each zone from where isolates were obtained. The size of the circle does not correspond to the number of isolates analyzed. The actual numbers are shown in Supplementary material 4: color codes are as shown in Fig. 3. A total of 223 MTBC isolates were included. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Comparison of factors associated with clustering
In a bivariate logistic regression analysis, factors such as age, treatment history, and lineage were statistically associated with isolate clustering at p-values 0.2. However, there was no statistically significant association (p-value>0.05) of sex and region with clustering. Age group was the only variable statistically associated with clustering of MTBC isolates (p-value<0.05) in multivariable analysis (Table 4).
Table 4.
Comparison of patient characteristics with clustering of MTBC isolates from the central, eastern and southeastern Ethiopia.
| Variable | Category | Clustered (N/%) | Unique (N/%) | COR (95 % CI) | P-value | AOR (95 % CI) | P-value |
|---|---|---|---|---|---|---|---|
| Age | ≤15 | 17 (89.5) | 4 (10.5) | 2.975 (0.823–10.760) | 0.096 | 3.055 (0.831–11.230) | 0.093 |
| 16–24 | 36 (64.3) | 20 (35.7) | 1.260 (0.525–3.022) | 0.605 | 1.393 (0.572–3.390) | 0.465 | |
| 25–34 | 61 (82.4) | 13 (17.6) | 3.285 (1.324–8.146) | 0.01 | 3.728 (1.467–9.470) | 0.006 | |
| 35–44 | 25 (65.8) | 13 (34.2) | 1.346 (0.517–3.505) | 0.543 | 1.363 (0.517–3.593) | 0.531 | |
| ≥45 | 20 (58.8) | 14 (41.2) | 1 | 1 | |||
| Sex | Male | 95 (73.1) | 35 (26.9) | 1.230 (0.685–2.209) | 0.488 | ||
| Female | 64 (68.8) | 29 (31.2) | 1 | ||||
| Region | Central | 68 (72.3) | 26 (27.9) | 1.012 (0.527–1.945) | 0.97 | ||
| Southeast | 29 (67.4) | 14 (32.6) | 0.802 (0.363–1.772) | 0.585 | |||
| East | 62 (72.1) | 24 (27.9) | 1 | ||||
| Treatment | New | 74 (66.7) | 37 (33.3) | 0.635 (0.354–1.141) | 0.129 | 0.644 (0.350–1.185) | 0.158 |
| Previously treated | 85 (75.9) | 27 (24.1) | 1 | 1 | |||
| Lineage | EA (L4) | 123 (73.2) | 45 (26.8) | 1.443 (0.751–2.770) | 0.201 | 1.842 (0.919–3.689) | 0.061 |
| Others | 36 (65.5) | 19 (34.5) | 1 |
4. Discussion
A review on molecular epidemiology of MTBC in Ethiopia [16] highlighted the scarcity of data from certain regions such as Harari and pastoralist communities of Oromia which include the Borena and Guji zones. The low number of isolates identified from these regions and other less investigated areas would restrict the scientific community's ability to observe the full picture of the transmission dynamics and population structure of MTBC in Ethiopia. In line with this, the current study examined the genomic diversity of MTBC isolates obtained from patients in a large geographical area spanning from the central part of Ethiopia to the far Southeast (Moyale) bordering the northern part of Kenya and the East (Diredawa, Harar, and Jigjiga) doorway to Djibouti and Somalia.
Supporting previous studies summarized in two recent reviews [15,16], a heterogeneous MTBC population structure dominated by the T and CAS families with the corresponding EA (L4) and EAI (L3) lineages was shown in the current study. Three-quarters (75.3 %) of the isolates in this study were members of EA (L4). A similar or higher proportion of EA (L4) has been reported from different parts of Ethiopia, including the current study sites [14,[22], [23], [24], [25], [26], [27]]. However, a lower proportion (40.1 %) of EA (L4) has been reported in northwest Ethiopia [28]. We also identified the globally predominant sub-lineages L4.1.2/Haarlem (n = 5) and L.4.3/LAM (n = 5). L4.1.2/Haarlem was previously shown to be connected to a high rate of transmission clusters [29] in Ethiopia. It is also the third major sub-lineage in Ethiopia [15,16], albeit isolated in modest proportions in the current study.
Various studies conducted in different parts of Ethiopia reported an overall low prevalence of IO (L1) and ETH (L7) [15,16]. Accordingly, the current study identified three IO (L1) isolates from Bale, East, and Southwest Shewa and one ETH (L7) isolate from West Arsi. ETH (L7), first reported from Woldia [30], is predominant in the northern highlands of Ethiopia [10,31]. It has also been identified in different parts of Ethiopia [11,14,[32], [33], [34], [35]]. Notably, this lineage is known to progress toward disease at a slower rate than other lineages [36] and is mainly limited to Ethiopia and Ethiopian immigrants [37,38]. Consequently, extensive investigation is required to comprehend why it is unique to Ethiopia and Ethiopians.
Three isolates of the West African-bound MA (L5 and L6) lineages were also identified from Arsi and Guji. When verified using the SNP-based lineage identification tool [5], one isolate was identified as unknown, whereas the other two were confirmed as MA (L5 and L6). There is an evidence that the MA (L5 and L6) are less virulent and are being replaced by EA (L4) in West Africa [10]. Conversely, in Ethiopia where EA (L4) is predominant, MA (L5 and L6) lineages have been isolated sporadically [22,23,32,39,40]. Therefore, it is crucial to carefully examine the impact of these lineages on the epidemiology of TB in Ethiopia, as this may have implications for TB control strategies and treatment approaches.
Consistent with other investigations carried out in Ethiopia [[11], [12], [13]], the T3-ETH sub-lineage/SIT149 spoligotype, reported to be more likely in a cluster [36,41,42], was the most frequent in our study, followed by the T1 sub-lineage/SIT53 spoligotype. A study conducted in Oromia region [43], which included the zones from which two-thirds of our isolates were obtained, found a high proportion of clustered ST149 spoligotype followed by orphan spoligotypes rather than the commonly reported T or CAS sub-lineages. This could be attributed to several factors including differences in sampling techniques, the broad geographic area and high population of the region and the relatively small sample size used in the current study.
The high proportion of clustered spoligotypes, particularly SIT149, identified in different parts of the country magnifies its key role in TB transmission dynamics and disease burden in Ethiopia. Furthermore, the finding that 16.3 % of the 49 isolates with spoligotype SIT149 in the current study were from children aged ≤15 years may indicate an ongoing TB transmission from adults to children. A similar proportion (16.6 %) of the total isolates were not registered in the online SITVIT2/SPOLDB4 database suggesting that there is a need further clarify the population structure of MTBC causing TB in Ethiopia.
A relatively large cluster (n = 11) of the SIT41 spoligotype, which is phylogeographically specific to Turkey [44], was reported in this study with the majority (10/11) being from patients in southeastern (Borena and Guji) Ethiopia. This sub-lineage, although in small numbers (n ≤ 5), was previously reported [23,28,30,32,43,[45], [46], [47], [48]] in different parts of Ethiopia. Similarly, it was reported to be less common in the African continent [49]. Clusters of larger size (n = 6) from Guji and Liben area pastoralists [50] and most recently from Arsi zone (n = 7) [29] were reported. The findings of our study strengthen the increasing importance of this clade in the pastoralist communities of the southeastern Ethiopia and call for further investigation to assess its role in TB disease burden of these regions and to implement appropriate interventions.
The widely studied Beijing sub-lineage of the (EAS) L2 lineage characterized by its worldwide geographical distribution and virulent properties [38] has previously been documented in some parts of Ethiopia [25,48,51,52]. In this study, in addition to the classical Beijing strain-SIT1, we identified a rare spoligotype cluster, SIT541, characterized by the absence of two more spacers (40 and 41) compared to the predominant SIT1. At the time of our analysis, only 16 isolates were reported worldwide, representing 0.1 % of the total registered Beijing lineage, as per the SITVIT2 database. Interestingly, in the present study, all SIT541 isolates were detected in eastern Ethiopia, specifically in Diredawa (three isolates) and Harar (one isolate). The exclusive detection of this spoligotype in eastern Ethiopia and its limited global distribution may suggest a localized emergence and transmission of this strain in the region. Furthermore, the findings of the current study, together with a previous study reporting a similar frequency of SIT1 from Diredawa [24], may indicate a unique epidemiological pattern and stress the evolving nature of the Beijing strain in the region, highlighting the need for enhanced surveillance and monitoring. A large scale longitudinal study is required to better understand the pressure that this lineage might put on the community in the region and on the overall TB control effort of the region.
A high rate (71.3 %) of clustered sub-lineages with known SIT was recorded in the current study. Besides, the proportion of clustered isolates was higher in each geographic region: Central (72.3 %), Eastern (72.1 %) and Southeastern (67.4 %). A comparable rate of clustering has been reported from different regions across Ethiopia [11,13,23,53,54]. The high rate of clustering rate, although not with epidemiological link information, could indicate an ongoing transmission of TB in the respective regions. Variables, such as sex, geographic region, treatment history, and lineage were not significantly associated with an isolate being in a cluster. However, patients aged 25–34 years (AOR = 3.728, 95 % CI (1.467–9.470)) were more likely to be part of a cluster compared to those aged >45 years. This may indicate that individuals in this age group had a higher risk of recent TB and therefore, could be an entry point for targeted interventions.
Convenient selection of isolates from specific diagnostic centers may pose selection bias and the relatively small number of isolates could affect the representativeness of the study result. Besides, the low discriminatory power of spoligotyping limits our conclusion regarding MTBC transmission dynamics. In contrast, whole-genome sequencing (WGS) provides a more comprehensive understanding of TB transmission patterns and strain diversity owing to its higher resolution, single nucleotide polymorphisms identification capabilities, better discrimination and provision of additional information compared to traditional typing methods such as spoligotyping [55]. Additionally, it prevents unnecessary false cluster investigations and allows more targeted interventions [56]. Additionally, it enables more focused actions to avoid pointless cluster investigations.
5. Conclusion
Though dominated by T (L4) and CAS (L3) families, our findings indicate that the genetic diversity of MTBC in the study area is diverse. The predominance of EA (L4) in the study area, comprising three-fourths of the isolates analyzed, shows its impact on the TB landscape of the country. Moreover, the identification of the Beijing strain, particularly the rare SIT541 spoligotype cluster in Eastern Ethiopia, calls for enhanced surveillance. Understanding the genetic diversity and transmission dynamics of EA (L4) could reveal the factors contributing to its predominance in Ethiopia. Hence, it is imperative to undertake a large scale study using advanced tools such as WGS to gather a comprehensive information and guide the national TB control program in implementing targeted interventions to combat TB in Ethiopia.
Data availability statement
Data related to the current study are all included in the results section.
CRediT authorship contribution statement
Mulualem Agonafir: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing - original draft, Writing - review & editing. Gurja Belay: Methodology, Supervision, Validation, Writing - review & editing. Nontuthuko E. Maningi: Investigation, Methodology, Validation, Writing - review & editing. Adey Feleke: Methodology, Validation, Writing - review & editing. Melese Abate Reta: Investigation, Validation, Writing - review & editing. Sharon L. Olifant: Formal analysis, Investigation, Validation, Writing - review & editing. Mohammed Suaudi Hassen: Investigation, Resources, Validation, Writing - review & editing. Tewodros Girma: Formal analysis, Investigation, Resources, Validation, Writing - review & editing. P. Bernard Fourie: Investigation, Methodology, Resources, Supervision, Validation, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank University of Pretoria, Medical Microbiology Department for supporting the laboratory work. We are grateful to Dr. Giorgio Roscigno for facilitating the process of collaborative work and Dr. Abel Gizaw for technical support during the manuscript preparation. We acknowledge the comprehensive assistance provided by the Department of Microbial, Cellular, and Molecular Biology, Addis Ababa University. We also express our gratitude to the regional TB reference laboratories at Adama and Harar.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e22898.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
References
- 1.World Health Organization . World Health Organization; Geneva: 2021. Global Tuberculosis Report 2021.https://apps.who.int/iris/handle/10665/346387 [Google Scholar]
- 2.Comas I., Gagneux S. The past and future of tuberculosis research. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mokrousov I., Pasechnik O., Vyazovaya A., Yarusova I., Gerasimova A., Blokh A., Zhuravlev V. Impact of pathobiological diversity of Mycobacterium tuberculosis on clinical features and lethal outcome of tuberculosis. BMC Microbiol. 2022;22:50. doi: 10.1186/s12866-022-02461-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brites D., Loiseau C., Menardo F., Borrell S., Boniotti M.B., Warren R., Dippenaar A., Parsons S.D.C., Beisel C., Behr M.A., Fyfe J.A., Coscolla M., Gagneux S. A new phylogenetic framework for the animal-adapted Mycobacterium tuberculosis complex. Front. Microbiol. 2018;9:2820. doi: 10.3389/fmicb.2018.02820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couvin D., Segretier W., Stattner E., Rastogi N. 2020. Novel Methods Included in SpolLineages Tool for Fast and Precise Prediction of Mycobacterium tuberculosis Complex Spoligotype Families, Database. 2020. baaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngabonziza J.C.S., Loiseau C., Marceau M., Jouet A., Menardo F., Tzfadia O., Antoine R., Niyigena E.B., Mulders W., Fissette K., Diels M., Gaudin C., Duthoy S., Ssengooba W., André E., Kaswa M.K., Habimana Y.M., Brites D., Affolabi D., Mazarati J.B., de Jong B.C., Rigouts L., Gagneux S., Meehan C.J., Supply P. A sister lineage of the Mycobacterium tuberculosis complex discovered in the African Great Lakes region. Nat. Commun. 2020;11:2917. doi: 10.1038/s41467-020-16626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coscolla M., Gagneux S., Menardo F., Loiseau C., Ruiz-Rodriguez P., Borrell S., Otchere I.D., Asante-Poku A., Asare P., Sánchez-Busó L., Gehre F., Sanoussi C.N., Antonio M., Affolabi D., Fyfe J., Beckert P., Niemann S., Alabi A.S., Grobusch M.P., Kobbe R., Parkhill J., Beisel C., Fenner L., Böttger E.C., Meehan C.J., Harris S.R., de Jong B.C., Yeboah-Manu D., Brites D. Phylogenomics of Mycobacterium africanum reveals a new lineage and a complex evolutionary history. Microb. Genom. 2021;7 doi: 10.1099/mgen.0.000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coll F., McNerney R., Guerra-Assunção J.A., Glynn J.R., Perdigão J., Viveiros M., Portugal I., Pain A., Martin N., Clark T.G. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat. Commun. 2014;5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stucki D., Brites D., Jeljeli L., Coscolla M., Liu Q., Trauner A., Fenner L., Rutaihwa L., Borrell S., Luo T., Gao Q., Kato-Maeda M., Ballif M., Egger M., Macedo R., Mardassi H., Moreno M., Tudo Vilanova G., Fyfe J., Globan M., Thomas J., Jamieson F., Guthrie J.L., Asante-Poku A., Yeboah-Manu D., Wampande E., Ssengooba W., Joloba M., Henry Boom W., Basu I., Bower J., Saraiva M., Vaconcellos S.E.G., Suffys P., Koch A., Wilkinson R., Gail-Bekker L., Malla B., Ley S.D., Beck H.-P., de Jong B.C., Toit K., Sanchez-Padilla E., Bonnet M., Gil-Brusola A., Frank M., Penlap Beng V.N., Eisenach K., Alani I., Wangui Ndung’u P., Revathi G., Gehre F., Akter S., Ntoumi F., Stewart-Isherwood L., Ntinginya N.E., Rachow A., Hoelscher M., Cirillo D.M., Skenders G., Hoffner S., Bakonyte D., Stakenas P., Diel R., Crudu V., Moldovan O., Al-Hajoj S., Otero L., Barletta F., Jane Carter E., Diero L., Supply P., Comas I., Niemann S., Gagneux S. Mycobacterium tuberculosis Lineage 4 comprises globally distributed and geographically restricted sublineages. Nat. Genet. 2016;48:1535–1543. doi: 10.1038/ng.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comas I., Hailu E., Kiros T., Bekele S., Mekonnen W., Gumi B., Tschopp R., Ameni G., Hewinson R.G., Robertson B.D., Goig G.A., Stucki D., Gagneux S., Aseffa A., Young D., Berg S. Population genomics of Mycobacterium tuberculosis in Ethiopia contradicts the virgin soil hypothesis for human tuberculosis in sub-saharan Africa. Curr. Biol. 2015;25:3260–3266. doi: 10.1016/j.cub.2015.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ameni G., Tadesse K., Hailu E., Deresse Y., Medhin G., Aseffa A., Hewinson G., Vordermeier M., Berg S. Transmission of Mycobacterium tuberculosis between farmers and cattle in Central Ethiopia. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garedew L., Mihret A., Ameni G. Molecular typing of mycobacteria isolated from extrapulmonary tuberculosis patients at Debre Birhan Referral Hospital, central Ethiopia. Scand. J. Infect. Dis. 2013;45:512–518. doi: 10.3109/00365548.2013.773068. [DOI] [PubMed] [Google Scholar]
- 13.Zewdie O., Mihret A., Ameni G., Worku A., Gemechu T., Abebe T. Molecular typing of mycobacteria isolated from tuberculous lymphadenitis cases in Addis Ababa, Ethiopia. Int. J. Tuberc. Lung Dis. Off. J. Int. Union Tuberc. Lung Dis. 2016;20:1529–1534. doi: 10.5588/ijtld.15.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merid Y., Hailu E., Habtamu G., Tilahun M., Abebe M., Hailu M., Hailu T., Datiko D.G., Woldeamanuel Y., Aseffa A. Molecular Epidemiology of Mycobacterium tuberculosis strains isolated from pulmonary tuberculosis patients in south Ethiopia. J. Infect. Dev. Ctries. 2021;15:1299–1307. doi: 10.3855/jidc.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mekonnen D., Derbie A., Chanie A., Shumet A., Biadglegne F., Kassahun Y., Bobosha K., Mihret A., Wassie L., Munshea A., Nibret E., Yimer S.A., Tønjum T., Aseffa A. Molecular epidemiology of M. tuberculosis in Ethiopia: a systematic review and meta-analysis. Tuberculosis. 2019;118 doi: 10.1016/j.tube.2019.101858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tulu B., Ameni G. Spoligotyping based genetic diversity of Mycobacterium tuberculosis in Ethiopia: a systematic review. BMC Infect. Dis. 2018;18:140. doi: 10.1186/s12879-018-3046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen G.-H., Chen C.-H., Hung C.-H., Wu K.-M., Lin C.-F., Sun Y.-W., Chen J.-H. Combining the Capilia TB assay with smear morphology for the identification of Mycobacterium tuberculosis complex. Int. J. Tubercul. Lung Dis. 2009;13:371–376. [PubMed] [Google Scholar]
- 18.Kamerbeek J., Schouls L., Kolk A., van Agterveld M., van Soolingen D., Kuijper S., Bunschoten A., Molhuizen H., Shaw R., Goyal M., van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couvin D., David A., Zozio T., Rastogi N. Macro-geographical specificities of the prevailing tuberculosis epidemic as seen through SITVIT2, an updated version of the Mycobacterium tuberculosis genotyping database. Infect. Genet. Evol. 2019;72:31–43. doi: 10.1016/j.meegid.2018.12.030. [DOI] [PubMed] [Google Scholar]
- 20.Aminian M., Shabbeer A., Bennett K.P. A conformal Bayesian network for classification of Mycobacterium tuberculosis complex lineages. BMC Bioinf. 2010;11:S4. doi: 10.1186/1471-2105-11-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aminian M., Couvin D., Shabbeer A., Hadley K., Vandenberg S., Rastogi N., Bennett K.P. Predicting Mycobacterium tuberculosis complex clades using knowledge-based bayesian networks. BioMed Res. Int. 2014;2014:1–11. doi: 10.1155/2014/398484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abebe G., Abdissa K., Abdella K., Tadesse M., Worku A., Ameni G. Spoligotype-based population structure of Mycobacterium tuberculosis in the Jimma Zone, southwest Ethiopia. MicrobiologyOpen. 2019;8 doi: 10.1002/mbo3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedewi Z., Worku A., Mekonnen Y., Yimer G., Medhin G., Mamo G., Pieper R., Ameni G. Molecular typing of Mycobacterium tuberculosis complex isolated from pulmonary tuberculosis patients in central Ethiopia. BMC Infect. Dis. 2017;17:184. doi: 10.1186/s12879-017-2267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mekonnen A., Merker M., Collins J.M., Addise D., Aseffa A., Petros B., Ameni G., Niemann S. Molecular epidemiology and drug resistance patterns of Mycobacterium tuberculosis complex isolates from university students and the local community in Eastern Ethiopia. PLoS One. 2018;13 doi: 10.1371/journal.pone.0198054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobie T.A., Woldeamanuel Y., Asrat D., Beyene D., Bjørås M., Aseffa A. Genetic diversity and drug resistance pattern of Mycobacterium tuberculosis strains isolated from pulmonary tuberculosis patients in the Benishangul Gumuz region and its surroundings, Northwest Ethiopia. PLoS One. 2020;15 doi: 10.1371/journal.pone.0231320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haile B., Tafess K., Zewude A., Yenew B., Siu G., Ameni G. Spoligotyping and drug sensitivity of Mycobacterium tuberculosis isolated from pulmonary tuberculosis patients in the Arsi Zone of southeastern Ethiopia. New Microbes New Infect. 2020;33 doi: 10.1016/j.nmni.2019.100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welekidan L.N., Yimer S.A., Skjerve E., Dejene T.A., Homberset H., Tønjum T., Brynildsrud O. Whole genome sequencing of drug resistant and drug susceptible Mycobacterium tuberculosis isolates from tigray region, Ethiopia. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.743198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ejo M., Torrea G., Uwizeye C., Kassa M., Girma Y., Bekele T., Ademe Y., Diro E., Gehre F., Rigouts L., de Jong B.C. Genetic diversity of the Mycobacterium tuberculosis complex strains from newly diagnosed tuberculosis patients in Northwest Ethiopia reveals a predominance of East-African-Indian and Euro-American lineages. Int. J. Infect. Dis. 2021;103:72–80. doi: 10.1016/j.ijid.2020.11.129. [DOI] [PubMed] [Google Scholar]
- 29.Tafess K., Beyen T.K., Girma S., Girma A., Siu G. Spatial clustering and genetic diversity of Mycobacterium tuberculosis isolate among pulmonary tuberculosis suspected patients, Arsi Zone, Ethiopia. BMC Pulm. Med. 2021;21:206. doi: 10.1186/s12890-021-01567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Firdessa R., Berg S., Hailu E., Schelling E., Gumi B., Erenso G., Gadisa E., Kiros T., Habtamu M., Hussein J., Zinsstag J., Robertson B.D., Ameni G., Lohan A.J., Loftus B., Comas I., Gagneux S., Tschopp R., Yamuah L., Hewinson G., Gordon S.V., Young D.B., Aseffa A. Mycobacterial lineages causing pulmonary and extrapulmonary tuberculosis, Ethiopia. Emerg. Infect. Dis. 2013;19:460–463. doi: 10.3201/eid1903.120256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nebenzahl-Guimaraes H., Yimer S.A., Holm-Hansen C., de Beer J., Brosch R., van Soolingen D. Genomic characterization of Mycobacterium tuberculosis lineage 7 and a proposed name: ‘Aethiops vetus. Microb. Genom. 2016;2 doi: 10.1099/mgen.0.000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wondale B., Keehwan K., Medhin G., Teklu T., Mohammed T., Tolosa S., Zewude A., Amsalu F., Pieper R., Ameni G. Molecular epidemiology of clinical Mycobacterium tuberculosis complex isolates in South Omo, Southern Ethiopia. BMC Infect. Dis. 2020;20:750. doi: 10.1186/s12879-020-05394-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayalew S., Wegayehu T., Taye H., Wassie L., Girma S., Berg S., Mihret A. Drug resistance conferring mutation and genetic diversity of Mycobacterium tuberculosis isolates in tuberculosis lymphadenitis patients; Ethiopia. Infect. Drug Resist. 2021;14:575–584. doi: 10.2147/IDR.S298683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mengistu A., Enquselassie F., Hailu E., Aseffa A., Beyene D. Identification and characterization of Mycobacterium tuberculosis isolates from cattle owners in north western and north eastern parts of rural Ethiopia. Int. J. Innovat. Appl. Stud. 2015;10:85–94. [PMC free article] [PubMed] [Google Scholar]
- 35.Mihret A., Bekele Y., Loxton A.G., Jordan A.M., Yamuah L., Aseffa A., Howe R., Walzl G. Diversity of Mycobacterium tuberculosis isolates from new pulmonary tuberculosis cases in Addis Ababa, Ethiopia. Tuberc. Res. Treat. 2012;2012:1–7. doi: 10.1155/2012/892079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yimer S.A., Norheim G., Namouchi A., Zegeye E.D., Kinander W., Tønjum T., Bekele S., Mannsåker T., Bjune G., Aseffa A., Holm-Hansen C. Mycobacterium tuberculosis lineage 7 strains are associated with prolonged patient delay in seeking treatment for pulmonary tuberculosis in Amhara region, Ethiopia. J. Clin. Microbiol. 2015;53:1301–1309. doi: 10.1128/JCM.03566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blouin Y., Hauck Y., Soler C., Fabre M., Vong R., Dehan C., Cazajous G., Massoure P.-L., Kraemer P., Jenkins A., Garnotel E., Pourcel C., Vergnaud G. Significance of the identification in the horn of Africa of an exceptionally deep branching Mycobacterium tuberculosis clade. PLoS One. 2012;7 doi: 10.1371/journal.pone.0052841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coscolla M., Gagneux S. Consequences of genomic diversity in Mycobacterium tuberculosis. Semin. Immunol. 2014;26:431–444. doi: 10.1016/j.smim.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haile B., Tafess K., Zewude A., Yenew B., Siu G., Ameni G. Spoligotyping and drug sensitivity of Mycobacterium tuberculosis isolated from pulmonary tuberculosis patients in the Arsi Zone of southeastern Ethiopia. New Microbes New Infect. 2020;33 doi: 10.1016/j.nmni.2019.100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nuru A., Mamo G., Worku A., Admasu A., Medhin G., Pieper R., Ameni G. Genetic diversity of Mycobacterium tuberculosis complex isolated from tuberculosis patients in bahir dar city and its surroundings, northwest Ethiopia. BioMed Res. Int. 2015;2015:1–9. doi: 10.1155/2015/174732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tessema B., Beer J., Merker M., Emmrich F., Sack U., Rodloff A.C., Niemann S. Molecular epidemiology and transmission dynamics of Mycobacterium tuberculosis in Northwest Ethiopia: new phylogenetic lineages found in Northwest Ethiopia. BMC Infect. Dis. 2013;13:131. doi: 10.1186/1471-2334-13-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tadesse M., Abebe G., Bekele A., Bezabih M., de Rijk P., Meehan C.J., de Jong B.C., Rigouts L. The predominance of Ethiopian specific Mycobacterium tuberculosis families and minimal contribution of Mycobacterium bovis in tuberculous lymphadenitis patients in Southwest Ethiopia. Infect. Genet. Evol. 2017;55:251–259. doi: 10.1016/j.meegid.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 43.Hussien B., Zewude A., Wondale B., Hailu A., Ameni G. Spoligotyping of clinical isolates of Mycobacterium tuberculosis complex species in the Oromia region of Ethiopia. Front. Public Health. 2022;10 doi: 10.3389/fpubh.2022.808626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kisa O., Tarhan G., Gunal S., Albay A., Durmaz R., Saribas Z., Zozio T., Alp A., Ceyhan I., Tombak A., Rastogi N. Distribution of spoligotyping defined genotypic lineages among drug-resistant Mycobacterium tuberculosis complex clinical isolates in ankara, Turkey. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agonafir M., Lemma E., Wolde-Meskel D., Goshu S., Santhanam A., Girmachew F., Demissie D., Getahun M., Gebeyehu M., van Soolingen D. Phenotypic and genotypic analysis of multidrug-resistant tuberculosis in Ethiopia. Int. J. Tubercul. Lung Dis. 2010;14:1259–1265. [PubMed] [Google Scholar]
- 46.Diriba G., Kebede A., Tola H.H., Yenew B., Moga S., Addise D., Alemu A., Mohammed Z., Getahun M., Fantahun M., Tadesse M., Dagne B., Amare M., Assefa G., Abera D., Desta K. Molecular characterization and drug resistance patterns of Mycobacterium tuberculosis complex in extrapulmonary tuberculosis patients in Addis Ababa, Ethiopia. PLoS One. 2020;15 doi: 10.1371/journal.pone.0243493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garedew L., Mihret A., Mamo G., Abebe T., Firdessa R., Bekele Y., Ameni G. Strain diversity of mycobacteria isolated from pulmonary tuberculosis patients at Debre Birhan Hospital, Ethiopia. Int. J. Tubercul. Lung Dis. 2013;17:1076–1081. doi: 10.5588/ijtld.12.0854. [DOI] [PubMed] [Google Scholar]
- 48.Taye H., Alemu K., Mihret A., Ayalew S., Hailu E., Wood J.L.N., Shkedy Z., Berg S., Aseffa A. The ETHICOBOTS consortium, Epidemiology of Mycobacterium tuberculosis lineages and strain clustering within urban and peri-urban settings in Ethiopia. PLoS One. 2021;16 doi: 10.1371/journal.pone.0253480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chihota V.N., Niehaus A., Streicher E.M., Wang X., Sampson S.L., Mason P., Källenius G., Mfinanga S.G., Pillay M., Klopper M., Kasongo W., Behr M.A., Gey van Pittius N.C., van Helden P.D., Couvin D., Rastogi N., Warren R.M. Geospatial distribution of Mycobacterium tuberculosis genotypes in Africa. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gumi B., Schelling E., Berg S., Firdessa R., Erenso G., Mekonnen W., Hailu E., Melese E., Hussein J., Aseffa A., Zinsstag J. Zoonotic transmission of tuberculosis between pastoralists and their livestock in south-east Ethiopia. EcoHealth. 2012;9:139–149. doi: 10.1007/s10393-012-0754-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ali S., Beckert P., Haileamlak A., Wieser A., Pritsch M., Heinrich N., Löscher T., Hoelscher M., Niemann S., Rachow A. Drug resistance and population structure of M.tuberculosis isolates from prisons and communities in Ethiopia. BMC Infect. Dis. 2016;16:687. doi: 10.1186/s12879-016-2041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biadglegne F., Merker M., Sack U., Rodloff A.C., Niemann S. Tuberculous lymphadenitis in Ethiopia predominantly caused by strains belonging to the Delhi/CAS lineage and newly identified Ethiopian clades of the Mycobacterium tuberculosis complex. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belay M., Ameni G., Bjune G., Couvin D., Rastogi N., Abebe F. Strain diversity of Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients in Afar pastoral region of Ethiopia. BioMed Res. Int. 2014;2014:1–12. doi: 10.1155/2014/238532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maru M., Mariam S.H., Airgecho T., Gadissa E., Aseffa A. Prevalence of tuberculosis, drug susceptibility testing, and genotyping of mycobacterial isolates from pulmonary tuberculosis patients in dessie, Ethiopia. Tuberc. Res. Treat. 2015;2015:1–10. doi: 10.1155/2015/215015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Satta G., Lipman M., Smith G.P., Arnold C., Kon O.M., McHugh T.D. Mycobacterium tuberculosis and whole-genome sequencing: how close are we to unleashing its full potential? Clin. Microbiol. Infect. 2018;24:604–609. doi: 10.1016/j.cmi.2017.10.030. [DOI] [PubMed] [Google Scholar]
- 56.Nikolayevskyy V., Niemann S., Anthony R., van Soolingen D., Tagliani E., Ködmön C., van der Werf M.J., Cirillo D.M. Role and value of whole genome sequencing in studying tuberculosis transmission. Clin. Microbiol. Infect. 2019;25:1377–1382. doi: 10.1016/j.cmi.2019.03.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data related to the current study are all included in the results section.