Abstract
Vibrio alginolyticus contained two adjacent genes, ktrA and ktrB, which encode a new type of bacterial K+-uptake system. KtrA and KtrB are peripheral and integral membrane proteins, respectively. Six of the nine sequenced bacterial genomes contain homologs to both ktrA and ktrB, suggesting that KtrAB is widespread.
In prokaryotes, K+ uptake is essential for the homeostatic processes of turgor pressure regulation and maintenance of cytoplasmic pH (5, 30). Escherichia coli K-12 contains two major types of K+-uptake systems (Trk and Kdp) and one minor K+-uptake system (Kup) (22, 30). The inducible Kdp system belongs to the family of P-type ATPases. It transports K+ with high affinity (26, 27). TrkH and TrkG are two constitutive, rapid K+-uptake systems with a relatively low affinity for K+ (6). They consist of several subunits: an integral membrane protein, TrkH or TrkG (25); a membrane surface protein, TrkA, that binds 32P-NAD(H) in vitro (23); and the sapDF gene products from the sapABCDF operon (9), which encodes an ABC transporter of unknown function (20). Little is known about K+ uptake in bacteria other than E. coli. Enterococcus hirae contains an inducible system, K+-transport system II (KtrII), which accepts Rb+ poorly (low affinity and low rate) (10). KtrII requires the ntpJ gene product for activity. NtpJ is an integral membrane protein. It exhibits weak sequence similarity to portions of both the TrkG and TrkH proteins and the K+-uptake proteins Trk1 and Trk2 from yeasts (31). It has been speculated that NtpJ is not the only component of KtrII (13). Several bacterial genomes contain an ntpJ homolog, suggesting that KtrII is widespread among bacteria (4).
Vibrio alginolyticus is a marine bacterium that grows at neutral to alkaline pH. K+ transport is particularly important for cytoplasmic pH homeostasis of this bacterium at an alkaline external pH (16). It accumulates K+ via at least two systems: a low-affinity, Trk-like, constitutive system and an inducible high-affinity system different from Kdp (15). We have previously cloned and sequenced the trkAH gene cluster from V. alginolyticus (14, 17). These genes are expressed in E. coli and form active hybrids with other components of the E. coli TrkH and TrkG systems (17). Here we report on the cloning of genes encoding a new type of high-affinity K+-uptake system, KtrAB from V. alginolyticus. It consists of two gene products, one of which (KtrB) is homologous to NtpJ from E. hirae.
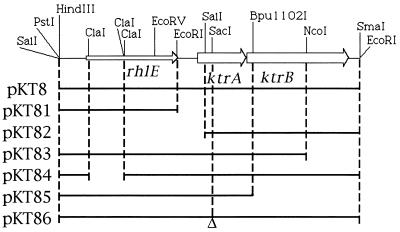
The strains and plasmids used in this study are given in Table 1 and Fig. 1, respectively. Plasmid pKT8 was selected from a V. alginolyticus 138-2 gene bank in plasmid pHG165 (28) by allowing the K+-uptake-negative E. coli strain TK2450/pKT8 to grow on plates at 3 mM K+ and on the basis of having a nucleotide sequence different from that of the V. alginolyticus trkAH-containing plasmid pKT6 (17). Plasmid pKT8 contained a chromosomal insert of 4,004 bp with three complete open reading frames of the same orientation (Fig. 1). (Coordinates in this publication are identical to those of the database record.) The first gene started at nucleotide 351 and terminated at nucleotide 1584. It encoded a protein of 411 amino acid residues with a molecular mass of 45,623 Da that was similar to the RNA helicase-like protein RhlE from E. coli (18). Putative −35 and −10 regions and a putative ribosome binding site were found 84 and 7 nucleotides upstream of rhlE, respectively. The second gene started at nucleotide 1837 and terminated at nucleotide 2497. It encoded a protein with a predicted molecular mass of 23,804 Da. Cell fractionation studies of minicells (24) expressing gene 2 showed that its product occurred both in the soluble protein fraction and in the membrane (results not shown), suggesting that this protein is a peripheral membrane protein. Putative −35 and −10 regions and a putative ribosome binding site were found 56 and 5 nucleotides upstream of this gene, respectively. The third gene overlapped with gene 2 by 1 nucleotide. It started at position 2496 and ended at nucleotide 3861. A putative ribosome binding site was found 10 nucleotides upstream of the third gene. It encoded a hydrophobic protein with a calculated molecular mass of 49,675 Da and with 36% identity to NtpJ from E. hirae (31). A putative ρ-independent termination signal was found 65 bases downstream of the ntpJ-like gene.
TABLE 1.
Strains used in this study
| Straina | Genotype, property, or V. alginolyticus genes present | Source | Refer- ence |
|---|---|---|---|
| LB650 | TK1001 trkH::CamrtrkG::Kanr | Lab collection | 17 |
| LB670 | TK1001 sapABCDF::Kanr | Lab collection | 17 |
| LB680 | TK1001 trkH::CamrsapABCDF::Kanr | Lab collection | 17 |
| LB700 | LB2003 sapABCDF::Kanr | Lab collection | 17 |
| LB2003 | TK1001 ΔtrkA | Lab collection | 29 |
| TK1001 | F−thi lacZ gal rha ΔkdpFABC5 trkD1 | W. Epstein | 21 |
| TK2450 | F−thi rha lacZ gal ΔkdpFABC5 trkD1 trkH::CamrtrkG::Kanr | W. Epstein | 25 |
| TK2693 | ΔkdpFABC5 trkD1 trkE80 trkG90 nadA | W. Epstein | 6 |
All strains are derivatives of E. coli K-12.
FIG. 1.
Deletion plasmids derived from plasmid pKT8. Left- and right-end restriction sites are in multiple cloning sites of pHG165.
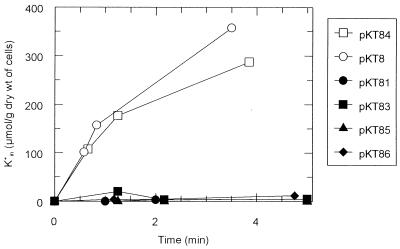
A series of deletion plasmids was constructed in order to examine the role of the three cloned genes in K+ transport (Fig. 1). Various E. coli strains lacking kdpFABC genes, a functional Kup system, and one or more trk genes were transformed with the pKT8 series of plasmids (Table 2). Only plasmid pKT8 and its ΔrhlE derivative, pKT84, allowed these strains to grow at low K+ concentrations (Table 2), suggesting that the cloned genes 2 and 3 are both required for K+ uptake. This notion was confirmed in a test in which net uptake by K+-depleted, energized E. coli cells was measured (1). Only plasmids pKT8 and pKT84 conferred K+-uptake activity to cells of the ΔkdpFABC5 kup-1 ΔtrkA strain, LB2003 (Fig. 2). K+ uptake by these cells was rapid (about 200 μmol min−1 g−1) and comparable to that of the E. coli TrkH and TrkG systems assayed under similar conditions (3). At a K+ concentration as low as 50 μM, K+ uptake occurred at the same rate as at 0.5 mM K+, suggesting that the system has a high affinity for K+. At K+ concentrations of less than 50 μM, K+ measurements of uptake become inaccurate with the assay method used. Neither the presence nor the absence of rhlE on the plasmid (Fig. 2) nor growth of the cells at 0.3 instead of 30 mM K+ affected the Vmax of the system (results not shown). The cells took up 86Rb+ with a Vmax similar to that for K+ (about 240 μmol min−1 g−1) and with a much higher Km (about 1 mM) than for K+ (less than 50 μM; see above).
TABLE 2.
Effect of plasmids with V. alginolyticus genes on growth of K+-uptake-defective E. coli strains at low K+ concentrations
| Plasmid | V. alginolyticus gene(s) present | Minimal K+ concn (mM) required for growth of straina:
|
|||||
|---|---|---|---|---|---|---|---|
| LB2003 ΔtrkA | LB670 ΔsapABCDF (“ΔtrkE”) | LB700 ΔtrkA ΔsapABCDF | LB650 ΔtrkH ΔtrkG | TK2693 ΔtrkG1 ΔtrkH92 | LB680 ΔtrkH ΔsapABCDF | ||
| pKT8 | rhlE ktrA ktrB | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| pKT81 | rhlE | 30 | 3 | 30 | 30 | 30 | 3 |
| pKT82 | ktrB | 30 | 3 | 30 | 30 | 30 | 3 |
| pKT83 | rhlE ktrA | 30 | 3 | 30 | 30 | 30 | 3 |
| pKT84 | ktrA ktrB | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| pKT85 | rhlE ktrA | 30 | 3 | 30 | 30 | 30 | 3 |
| pKT86 | rhlE ktrB | 10 | 1 | ND | 10 | ND | ND |
| pHG165 or no plasmid | 30 | 3 | 30 | 30 | 30 | 3 | |
Genotype with respect to trk genes. All strains are ΔkdpFABC5 kup-1. ND, not determined.
FIG. 2.
Net K+ uptake by K+-depleted cells of strain LB2003 containing plasmid pKT8 or one of its derivatives. Cells were grown at 30 mM K+ in the minimal medium described in reference 7. K+-depleted, energized cells were prepared as described in reference 1. At t = 0, 0.5 mM KCl was added to the cell suspension. At the time points indicated on the abscissa, cells from a 1-ml sample were centrifuged through silicone oil (1). The K+ content of the pellet was analyzed by flame photometry (1). Symbols: ○, cells carrying plasmid pKT8; •, cells carrying plasmid pKT81; ▪, cells carrying plasmid pKT83; □, cells carrying plasmid pKT84; ▴, cells carrying plasmid pKT85; ⧫, cells carrying plasmid pKT86.
E. coli K-12 does not contain genes homologous to the cloned genes 2 and 3 from V. alginolyticus (2). Moreover, none of the known E. coli K+-uptake genes were required for K+ uptake via the products of the cloned genes (Table 2 and Fig. 2). Hence we conclude that the two overlapping genes 2 and 3 together encode a new type of K+-uptake system. Since NtpJ (standing for natrium [sodium] transport) was a misnomer (31) and since this protein is involved in K+-transport system II (KtrII) from E. hirae (13), we propose to call the cloned genes 2 and 3 ktrA and ktrB, respectively, and the new system KtrAB. NtpJ was suspected to function together with some other protein(s) (13). We therefore examined whether the sequenced genomes containing ktrB (ntpJ) also contained a ktrA homolog. This is indeed the situation. Genes homologous to both ktrA and ktrB (ntpJ) are found in Mycoplasma genitalium, Mycoplasma pneumoniae, Synechocystis sp. strain PCC 6803, Bacillus subtilis (two copies of ktrA and ktrB each), Borrellia burgdorferi, and Aquifex aeolicus. Moreover, partial ktrA and ktrB sequences are available for Thermoanaerobacter ethanolicus (see the legend to Fig. 3 for the accession numbers). In M. genitalium, M. pneumoniae, B. burgdorferi, T. ethanolicus, and A. aeolicus, ktrA and ktrB are adjacent genes. The same is true for one set of B. subtilis ktr genes (ktrA1 and ktrB1), except that in the latter case the gene organization is the same as that in V. alginolyticus. In the two mycoplasmas, ktrA and ktrB are transcribed in opposite directions, whereas in B. burgdorferi, T. ethanolicus, and A. aeolicus the gene order is ktrB ktrA instead of the order ktrA ktrB, which is the case in V. alginolyticus (Fig. 1). Prokaryote genomes that did not contain a ktrB gene homolog also did not contain a ktrA homolog (i.e., Haemophilus influenzae, E. coli [2], Helicobacter pylori, and the three archaea Methanococcus jannaschii, Archaeoglobus fulgidus, and Methanobacterium thermoautotrophicum). Our results are significant in showing that KtrB (NtpJ) homologs usually occur with KtrA and that this hitherto unknown combination of components produces K+-uptake systems of a novel type.
FIG. 3.
Phylograms of the family of proteins or protein domains to which KtrA (A) and KtrB (B) belong. Alignment of protein sequences was first done with the program CLUSTAL W and was then improved by hand before a phylogram was calculated (32). Phylograms were drawn with the program TREEVIEW (19). For the different entries, the gene or protein and the GenBank accession number are given within parentheses. (A) KtrA Ssp, KtrA Val, KtrA2 Bsu, KtrA1 Bsu, KtrA Aae, KtrA Mge, KtrA Mpn, and KtrA Bbu, complete KtrA sequences from Synechocystis strain PCC6803 (sll049 in D64006), V. alginolyticus (D89592, this work), B. subtilis (YkqB in Z99111), B. subtilis (YuaA in Z99119), A. aeolicus (Aq1503 in AE000743), M. genitalium (MG323 in U39714), M. pneumoniae (H08-orf 231 in AE001046), and B. burgdorferi (BB075 in AE001172), respectively; KtrA Tet, an incomplete N-terminal KtrA sequence of 198 residues from T. ethanolicus (AF001974); TrkAN Eco, TrkAN Val, TrkA Mja, and TrkAC Eco and TrkAC Val, the N-terminal TrkA half from E. coli (X52114, residues 1 to 232) (23) and V. alginolyticus (D86411, residues 1 to 232), the complete TrkA sequence from Methanococcus jannaschii (MJ1105), and the C-terminal TrkA half from E. coli (X52114, residues 233 to 458) (23) and V. alginolyticus (D86411, residues 233 to 458), respectively; KchX Ssp and KefC Eco, C-terminal putative NAD+-binding domain from putative K+ channel from Synechocystis strain PC6803 (sllo993, residues 127 to 3365) and from KefC of K+-efflux channel from E. coli (P03819 in X56742, residues 399 to 620) (12), respectively. (B) KtrB Bbu, KtrB Ssp, KtrB Aae, KtrB Ehi, KtrB Val, KtrB1 Bsu, KtrB2 Bsu, KtrB Mge, and KtrB Mpn, KtrB from B. burgdorferi (BB0724 in AE001172), Synechocystis strain PCC6803 (sll1509 in D90911), A. aeolicus (Aq1504 in AE000743), E. hirae (NtpJ, D17462) (31), V. alginolyticus (D89592, this work), B. subtilis (YubG in Z99119), B. subtilis (YkrM in Z99111), M. genitalium (MG322 in U39714), and M. pneumoniae (H08-orf 565 in AE000036), respectively; KtrB Tet, a 350-residue C-terminal KtrB fragment from T. ethanolicus (AF001974); Trk1 Sce and Trk2 Sce, Trk membrane domains from Saccharomyces cerevisiae (P12685, residues 1 to 138 and 755 to 1235 [8], and P28584, residues 1 to 141 and 442 to 889 [11], respectively); TrkH Val and TrkH Mja, TrkH from V. alginolyticus (D86411) (17) and M. jannaschii (MJ1485), respectively.
Sequence alignments showed that KtrA and KtrB belong to a broader family of proteins and protein domains from microbial K+ transporters and K+ channels, respectively (Fig. 3). KtrA is distantly related to one half of TrkA, which is a fused dimer (23). Like TrkA, KtrA contains a putative NAD+-binding domain similar to that of NAD+-dependent dehydrogenases (from K2 to G128 of KtrA) (23). A C-terminal cytoplasmic domain of various types of putative K+ channels from both bacteria and archaea forms two additional subgroups of this family. Figure 3A gives a rooted tree (phylogram) for the different types of subunits/domains belonging to the family. The KtrA proteins of the different species identified on the basis of sequence similarity with KtrA from V. alginolyticus form one cluster, which is distinct from that of the three other subgroups of this protein (domain) family, of which only a few examples are shown (Fig. 3A). This finding supports the notion that the putative KtrA proteins of the different species have been identified correctly and that KtrA is indeed a novel type of bacterial K+-transport protein.
It has been recognized before that parts of NtpJ align both with parts of the TrkH subunits from the Trk system and with the membrane domain C-terminal to the large cytoplamic domain of the K+-uptake systems Trk1 and Trk2 from yeast (8, 25, 31). We observed that the N-terminal part of NtpJ (KtrB) aligns also with the two putative N-terminal transmembrane helices of Trk1 and Trk2, which precede the soluble domain of these proteins. Figure 3B gives a rooted tree based on the alignment of the complete KtrB sequences, the complete membrane domains of representative Trk1 and Trk2 proteins from yeast, and representative TrkH and TrkG proteins from prokaryotes. Once again, all KtrB proteins, identified on the basis of high sequence similarity to each other, form a cluster separate from those of the two other, distantly related, subgroups of this protein family, suggesting that KtrB also is a K+-transport protein of a new type (Fig. 3B [only a few examples of the yeast Trk1 and Trk2 and TrkH proteins are shown]).
The data in Fig. 3 show convincingly that both the putative KtrA proteins and the putative KtrB proteins from the different species form distinct clusters of proteins within a broader family of K+-transport proteins and putative K+ channels. This conclusion supports the notion that KtrAB is a new type of bacterial K+-uptake system distinct from Kdp, Trk, and Kup.
Nucleotide sequence accession number.
The sequence of the 4,004-bp chromosomal insert of pKT8 is listed in the EMBL, GenBank, and DDBJ databases under the accession no. D89592.
Acknowledgments
We thank Eva Limpinsel for expert technical assistance, Nancy Tholema for carrying out the experiment with 86Rb+ uptake by E. coli cells, and A. Lipski for his help with the programs CLUSTAL W and TREEVIEW.
This work was supported by the Fonds der Chemischen Industrie and by the Deutsche Forschungsgemeinschaft (SFB171).
REFERENCES
- 1.Bakker E P, Mangerich W E. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J Bacteriol. 1981;147:820–826. doi: 10.1128/jb.147.3.820-826.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Bossemeyer D, Borchard A, Dosch D C, Helmer G C, Epstein W, Booth I R, Bakker E P. K+ transport protein TrkA of Escherichia coli is a peripheral membrane protein that requires other trk gene products for attachment to the cytoplasmic membrane. J Biol Chem. 1989;264:16403–16410. [PubMed] [Google Scholar]
- 4.Clayton R A, White O, Ketchum K A, Ventor J C. The first genome from the third domain of life. Nature. 1997;387:459–462. doi: 10.1038/387459a0. [DOI] [PubMed] [Google Scholar]
- 5.Csonka L N, Epstein W. Osmoregulation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1210–1223. [Google Scholar]
- 6.Dosch D C, Helmer G L, Sutton S H, Salvacion F F, Epstein W. Genetic analysis of potassium transport loci in Escherichia coli: evidence for three constitutive systems mediating uptake of potassium. J Bacteriol. 1991;173:687–696. doi: 10.1128/jb.173.2.687-696.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein W, Kim B S. Potassium transport loci in Escherichia coli K-12. J Bacteriol. 1971;108:639–644. doi: 10.1128/jb.108.2.639-644.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaber R F, Styles C A, Fink G R. TRK1 encodes a plasma membrane protein required for high-affinity potassium transport in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2848–2859. doi: 10.1128/mcb.8.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harms, C., R. Tewes, E. Nölker, S. Stumpe, W. Epstein, and E. P. Bakker. Unpublished data.
- 10.Kakinuma Y. K+ transport in Enterococcus hirae. In: Bakker E P, editor. Alkali cation transport in prokaryotes. Boca Raton, Fla: CRC Press; 1993. pp. 277–290. [Google Scholar]
- 11.Ko C H, Gaber R F. TRK1 and TRK2 encode structurally related K+ transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4266–4273. doi: 10.1128/mcb.11.8.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munro A W, Ritchie G Y, Lamb A J, Douglas R M, Booth I R. The cloning and DNA sequence of the gene for the glutathione-regulated potassium-efflux system KefC of Escherichia coli. Mol Microbiol. 1991;5:607–616. doi: 10.1111/j.1365-2958.1991.tb00731.x. [DOI] [PubMed] [Google Scholar]
- 13.Murata T, Takase K, Yamato I, Igarashi K, Kakinuma Y. The ntpJ gene in the Enterococcus hirae ntp operon encodes a component of KtrII potassium transport system functionally independent of vacuolar Na+-ATPase. J Biol Chem. 1996;271:10042–10047. doi: 10.1074/jbc.271.17.10042. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura T, Matsuba Y, Yamamuro N, Booth I R, Unemoto T. Cloning and sequencing of a K+ transport gene (trkA) from the marine bacterium Vibrio alginolyticus. Biochim Biophys Acta. 1994;1219:701–705. doi: 10.1016/0167-4781(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura T, Suzuki F, Abe M, Matsuba Y, Unemoto T. K+ transport in Vibrio alginolyticus: isolation of a mutant defective in an inducible K+ transport system. Microbiology. 1994;140:1781–1785. [Google Scholar]
- 16.Nakamura T, Tokuda H, Unemoto T. K+/H+ antiporter functions as a regulator of cytoplasmic pH in a marine bacterium, Vibrio alginolyticus. Biochim Biophys Acta. 1984;776:330–336. doi: 10.1016/0005-2736(84)90222-0. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura, T., N. Yamamuro, S. Stumpe, T. Unemoto, and E. P. Bakker. Cloning of trkAH gene cluster and characterization of the Trk K+ uptake system from Vibrio alginolyticus. Microbiology, in press. [DOI] [PubMed]
- 18.Ohmori H. Structural analysis of the rhlE gene of Escherichia coli. Jpn J Genet. 1994;69:1–12. doi: 10.1266/jjg.69.1. [DOI] [PubMed] [Google Scholar]
- 19.Page R M D. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 20.Parra-Lopez C, Baer M T, Groisman E A. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 1993;12:4053–4062. doi: 10.1002/j.1460-2075.1993.tb06089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhoads D B, Waters F B, Epstein W. Cation transport in Escherichia coli. VIII. Potassium transport mutants. J Gen Physiol. 1976;67:325–341. doi: 10.1085/jgp.67.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schleyer M, Bakker E P. Nucleotide sequence and 3′-end deletion studies indicate that the K+-uptake protein Kup from Escherichia coli is composed of a hydrophobic core linked to a large and partially essential hydrophilic C terminus. J Bacteriol. 1993;175:6925–6931. doi: 10.1128/jb.175.21.6925-6931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlösser A, Hamann A, Bossemeyer D, Schneider E, Bakker E P. NAD+ binding to the Escherichia coli K+-uptake protein TrkA and sequence similarity between TrkA and domains of a family of dehydrogenases suggest a role of NAD+ in bacterial transport. Mol Microbiol. 1993;9:533–543. doi: 10.1111/j.1365-2958.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 24.Schlösser A, Kluttig S, Hamann A, Bakker E P. Subcloning, nucleotide sequence, and expression of trkG, a gene that encodes an integral membrane protein involved in potassium uptake via the Trk system of Escherichia coli. J Bacteriol. 1991;173:3170–3176. doi: 10.1128/jb.173.10.3170-3176.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlösser A, Meldorf M, Stumpe S, Bakker E P, Epstein W. TrkH and its homologue, TrkG, determine the specificity and kinetics of cation transport by the Trk system of Escherichia coli. J Bacteriol. 1995;177:1908–1910. doi: 10.1128/jb.177.7.1908-1910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siebers A, Altendorf K. K+-translocating Kdp-ATPases and other bacterial P-type ATPases. In: Bakker E P, editor. Alkali cation transport systems in prokaryotes. Boca Raton, Fla: CRC Press; 1993. pp. 225–252. [Google Scholar]
- 27.Silver S. Transport of inorganic cations. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1091–1102. [Google Scholar]
- 28.Stewart G S A B, Lubinsky-Mink S, Jackson C G, Kuhn J. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid. 1986;15:172–181. doi: 10.1016/0147-619x(86)90035-1. [DOI] [PubMed] [Google Scholar]
- 29.Stumpe S, Bakker E P. Requirement of a large K+-uptake capacity and of extracytoplasmic protease activity for protamine resistance of Escherichia coli. Arch Microbiol. 1997;167:126–136. [PubMed] [Google Scholar]
- 30.Stumpe S, Schlösser A, Schleyer M, Bakker E P. K+ circulation across the prokaryotic cell membrane: K+-uptake systems. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Vol. 2. Amsterdam, The Netherlands: Elsevier Science B.V.; 1996. pp. 474–499. [Google Scholar]
- 31.Takase K, Kakinuma S, Yamato I, Konishi K, Igarashi K, Kakinuma Y. Sequencing and characterization of the ntp gene cluster for vacuolar-type Na+-translocating ATPase of Enterococcus hirae. J Biol Chem. 1994;269:11037–11044. [PubMed] [Google Scholar]
- 32.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positive-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]