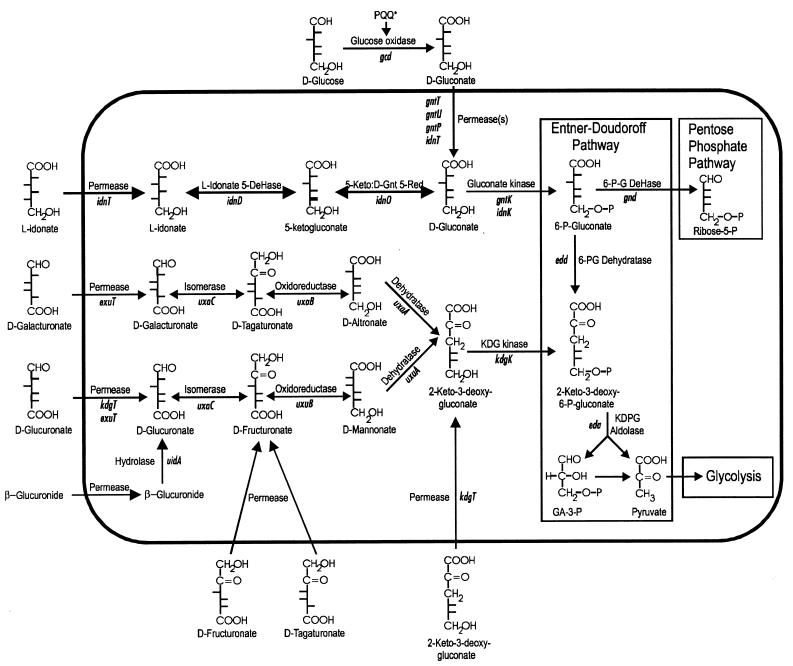
The Entner-Doudoroff (ED) pathway was first discovered in 1952 in Pseudomonas saccharophila (21) and several years later was shown to be present in Escherichia coli (23). Although generally considered to be restricted to gram-negative bacteria, the ED pathway is present in all three phylogenetic domains, including the most deeply rooted Archaea (18). The ubiquity of the ED pathway suggests that it is of far greater importance in nature than was previously recognized. In fact, a recent essay on the evolution of glycolytic pathways suggested that the ED pathway predates the Embden-Meyerhof-Parnas pathway (60). E. coli is generally recognized as a member of the normal gastrointestinal microbiota and is extremely versatile, with the ability to grow on many of the nutrients that can be found there (45). But what exactly are the nutrients preferred by E. coli to support its growth in the intestine? And what about growth of E. coli outside of the intestine? Certainly E. coli spends periods of time in aquatic, aerobic habitats when it is in between host intestinal tracts. Mounting evidence suggests that sugar acid metabolism via the ED pathway is important for growth of E. coli in both intestinal and aquatic habitats. The role of the ED pathway, especially with regard to colonization of the mammalian large intestine, will be discussed below. The biochemistry and physiology of sugar acid metabolism in E. coli are very well understood, and several newly discovered genes encoding sugar acid transporters, associated regulatory proteins, and a novel pathway for sugar acid catabolism have been identified as a result of the E. coli genome project (12). Homology searches reveal that the genes of the ED pathway are present on several other genomes (Table 1), and it is becoming clear that there will be additional, perhaps even novel, sugar acid catabolic pathways revealed by analysis of these genomes.
TABLE 1.
Comparison of E. coli gluconate enzyme, amino acid query sequences against the published genome databasesa
| Organism | GntT | GntK | Edd | Eda |
|---|---|---|---|---|
| Actinobacillus actinomycetemcomitans | xd | x | x | 100b; contig478c |
| Bacillus subtilis | 482; GntP | 107; GntK | x | 264; KdgA |
| Deinococcus radiodurans | x | 101; gdr52 | x | x |
| Haemophilus influenzae | 671; HI1015 | x | x | 419; HI0047 |
| Helicobacter pylori | x | x | 602; HP1100 | 208; HP1099 |
| Neisseria meningitidis | x | x | x | 120; GNMAA15F |
| Neisseria gonorrhoeae | 194; contig269 | 89; contig269 | 722; contig279 | 432; contig279 |
| Pseudomonas aeruginosa | 250; contig1173 | 87; contig1173 | 723, contig1775 | 157; contig1856 |
| Rhodobacter capsulatus | x | 86; RRC00167 | 1785; RRC01198 | 690; RRC01197 |
| Saccharomyces cerevisiae | 89; Ch XIV, SCR14 | 267; ChIV, CHR4 | x | x |
| Streptococcus pneumoniae | x | x | x | 73; stp4167 |
| Streptococcus pyogenes | x | x | x | 89; contig213 |
| Synechocystis sp.strain PCC6803 | x | x | x | 240; s110107 |
| Thermotoga maritima | x | x | x | 92; BTMAD55F |
| Treponema pallidum | x | x | x | 179; ORF00668 |
| Vibrio cholerae | 113; GVCDJ90F | x | x | 83; GVCDJ90R |
No hits for any of these queries were found for genomes of the following species: Archaeoglobus fulgidus, Borrelia burgdorfei, Enterococcus faecalis, Methanobacterium thermoautotrophicum, Methanococcus janaschii, Mycobacterium tuberculosis, Mycoplasm pneumoniae, Mycoplasma genitalium, Plasmodium falciparum, Pyrococcus hoikoshii, and Staphylococcus aureus.
BLASTP 2.0 score (3).
Locus on genome.
x, No significant hits were found.
OVERVIEW OF THE ED PATHWAY
The ED pathway can be properly considered as one of three pathways found in nature, in addition to the Embden-Meyerhof-Parnas and pentose phosphate pathways, that feed into the “bottom half” of glycolysis, which is central to all of intermediary metabolism (18, 28). The overall schemes of the ED and Embden-Meyerhof-Parnas pathways are quite similar: 6-carbon sugars are primed by phosphorylation and subsequently cleaved by aldolase enzymes into two 3-carbon intermediates (Fig. 1). The primary distinction between the two pathways lies in the nature of the 6-carbon metabolites that serve as substrates for aldol cleavage: for the ED pathway this intermediate is 2-keto-3-deoxy-6-phosphogluconate (KDPG). The first of the two key enzymes is unique to the ED pathway, 6-phosphogluconate dehydratase (Edd), which catalyzes a dehydration of 6-phosphogluconate to form KDPG. The second enzyme of the ED pathway, KDPG aldolase (Eda), catalyzes an aldol cleavage of KDPG to form pyruvate and glyceraldehyde-3-phosphate. The triose phosphate intermediate is further metabolized by the glycolytic pathway and provides energy via substrate-level phosphorylation.
FIG. 1.
Pathways for catabolism of several sugar acids by E. coli.
ECOLOGY OF THE MAMMALIAN LARGE INTESTINE
Since E. coli has almost certainly evolved to grow in the intestine, the ecology of the gut is considered. The intestinal habitat contains some 400 to 500 different bacterial species (13, 26, 31), but relatively little is known about which substrates support their growth in the intestine, which metabolic pathways are important, and which genes are required (64). In fact, we do not know for certain which nutrient(s) allows for intestinal colonization by any individual bacterium. The diversity of microorganisms in the gut ecosystem is believed to reflect their abilities to occupy different ecological niches. The nutrient/niche theory, developed by Freter (29), postulates that the numerous ecological niches within the intestine are defined by nutrient availability. According to this hypothesis, individual species have a preference for one or a very few of the plethora of substrates that arise in the intestine from ingested food, epithelial and bacterial cell debris, and the mucus lining of the epithelium. The result is a balanced ecosystem in which each of the numerous intestinal niches is occupied by an individual bacterium, with individual population sizes being determined by the available concentration of the preferred nutrient.
The composition of the intestinal microflora is normally quite stable (29). Nevertheless, there appears to be a continuous succession of E. coli strains in the mammalian intestine. Some strains are present for extended periods of time (months to years), while others are only transiently (a few days) detected, with an average of five E. coli biotypes found in the feces of individual humans (4). Thus, diversity exists even among normal commensal E. coli strains in the intestine, suggesting that different E. coli strains may possess different capacities for utilizing growth-limiting nutrients.
The large intestine is both rich and diverse in terms of nutrient availability. The majority of intestinal bacteria require a fermentable carbohydrate for growth, so it has generally been assumed that carbohydrate metabolism is necessary for colonization by most species (62). In the large intestine, carbohydrates arise from partially digested food and from host secretions (29, 40). Naturally occurring sugar acids such as galacturonate from pectin (40, 62) and gluconate and ketogluconate from muscle tissues (25) are present in the foods we eat. The mucus layer covering epithelial tissues is also recognized as an important source of carbohydrates in the intestine (29, 62). Mucus is a complex gel of glycoproteins and glycolipids; the sugar substituents of mucus include N-acetylglucosamine, N-acetylgalactosamine, galactose, fucose, sialic acids, and lesser amounts of glucuronate and galacturonate (2).
In situ hybridization using an E. coli-specific, fluorescent rRNA-directed probe revealed that E. coli cells grow while embedded within the mucus layer overlying intestinal epithelial cells (56). This result is compatible with experiments which indicate that E. coli can specifically bind to mouse colonic mucus glycoprotein (17). It should be noted that studies of E. coli fim mutants demonstrated that type 1 pili are not necessary for colonization of the mouse large intestine (44). Other studies indicate that E. coli can grow rapidly in mouse cecal mucus, but not in cecal luminal contents (68). This suggests that E. coli does not necessarily grow on nutrients ingested by the host but rather grows on nutrients secreted by the host in the form of mucus.
The measured generation time of E. coli in the large intestine of a gnotobiotic mouse is 30 min, significantly faster than the 3-h half-life of the intestinal contents in mice (29). Since in situ hybridization and growth studies indicate that E. coli probably grows in the mucus layer, it may be more appropriate to consider the turnover rate of the mucus itself. These calculations predict that E. coli needs to grow with a generation time of roughly 1 h in order to achieve a stable population size of 108 CFU per g of feces (55), which is normal in the mouse model (44). By using ribosome numbers as a measure of the growth rate of E. coli in the mouse large intestine, a generation time of 40 to 80 min was verified (55). Since E. coli can grow rapidly in mucus, but not in feces, Poulsen et al. (55) proposed that there are at least two populations of E. coli in the large intestine, one embedded in the mucus layer with an apparent generation time of approximately 1 h and another which is essentially static with respect to growth, sloughed into the luminal contents, and excreted in the feces.
SUBSTRATES OF THE ED PATHWAY MAY PROVIDE A NUTRITIONAL NICHE FOR E. COLI IN THE LARGE INTESTINE
The ED pathway was recently shown to be important for E. coli to colonize the mammalian large intestine (63). All 72 of the standard (ECOR) reference strains of E. coli isolated from natural populations (48) possess the ED pathway and grow on gluconate and glucuronate (46a), indicating that the ED pathway is highly conserved. The colonization properties of one typical strain, E. coli F-18, isolated from human feces, has been studied in detail (17, 44, 63, 64, 68). When E. coli F-18, a natural gntP mutant strain, was transduced with the wild-type gntP gene from E. coli K-12, it gained the ability to occupy a distinct niche in the mouse large intestine (64). That is, E. coli F-18 carrying the wild-type gntP gene is able, when ingested in small numbers, to grow and colonize in the presence of high numbers of its parent. It was confirmed that only a single gene, gntP, transduced from E. coli K-12 was required for E. coli F-18 to occupy the distinct niche. Since gntP apparently encodes a gluconate transporter (38), it was suggested that the distinct niche occupied by E. coli F-18 carrying the wild-type gntP gene is nutritionally defined by the presence of gluconate.
Since ED metabolism was implicated as playing a role in colonization of the mouse large intestine, Sweeney et al. (63) decided to investigate the role of this pathway in a more systematic way. Mutants of E. coli F-18 and E. coli K-12 carrying an insertion in the eda gene are unable to grow on laboratory media containing gluconate, glucuronate, or galacturonate (Fig. 1). More importantly, the eda mutants were found to be incapable of colonizing the mouse large intestine (63). Genetic complementation of the E. coli F-18 eda mutant strain with the wild-type eda gene by transformation with pTC190 (20) rescued the ability to colonize the large intestine. Furthermore, while E. coli F-18 can grow on cecal mucus in vitro, an E. coli F-18 eda mutant cannot (63). Unfortunately, these results did not distinguish which of the three ED substrates, gluconate, glucuronate, or galacturonate, is most important for colonization.
Further studies suggested that glucuronate provides a niche for E. coli to colonize. A mutant derivative of E. coli F-18 with a deletion of the uxuA gene, which specifically prevents glucuronic acid metabolism (Fig. 1), was tested in the mouse model and found to colonize at a 100-fold-lower level than the wild-type parent strain (64). Interestingly, experiments designed to specifically investigate the role of gluconate catabolism showed that E. coli F-18 edd eda and E. coli K-12 edd eda double mutants are able to colonize when fed to mice alone, though not quite as well as the parent strains (63). However, when fed simultaneously with their parent strains, E. coli F-18 edd eda and E. coli K-12 edd eda double mutants colonized very poorly. This result indicates that the parent strains are able to occupy a niche which cannot be occupied by the edd eda double mutants. Since gluconate is the only carbon source known to require the edd gene product for efficient catabolism, Sweeney et al. (63) concluded that gluconate is a major carbon source for both E. coli F-18 and E. coli K-12 to colonize the mouse large intestine. While these colonization studies are provocative, they are inconclusive with respect to the importance of gluconate. Further experiments designed to specifically investigate the roles of the individual sugar acid catabolic pathways in colonization are clearly necessary.
Recently, we determined the concentration of sugar acids in mouse cecal mucus by high-performance liquid chromatography (HPLC). In addition to significant amounts of glucuronate and galacturonate (2), gluconate is present in mouse cecal mucus at a concentration of 0.69 mM (unpublished results). In vitro growth experiments showed that 0.69 mM gluconate in minimal medium allows formation of 1.5 × 108 E. coli cells, which is precisely the population of E. coli cells in colonized mice (17a).
GLUCONATE CATABOLISM
In order for E. coli to grow on gluconate, it must first be transported and phosphorylated to form 6-phosphogluconate. The apparent redundancy of the gluconate kinases, of which there are two (65), and of the gluconate transporters, of which there are at least four (50), has led to much confusion. Many of the open questions regarding the multiple systems for gluconate transport and phosphorylation will be answered below.
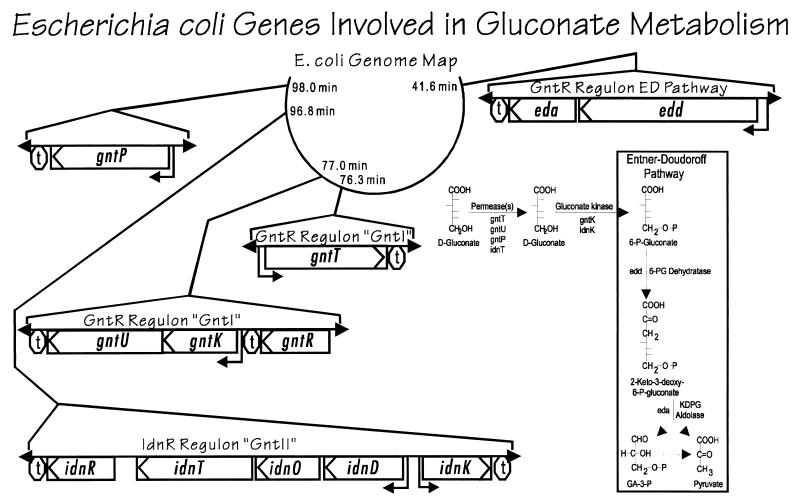
Catabolism of gluconate via the ED pathway in E. coli is controlled by the GntR regulon (Table 2; Fig. 2). The edd-eda operon is fully induced for growth on gluconate (20). High basal expression of eda occurs regardless of carbon source (28), which is consistent with the role of Eda as a key enzyme of the catabolic pathways for hexuronate metabolism (Fig. 1). GntI, the main system for gluconate transport and phosphorylation (Fig. 2), contains gntT and gntU, which code for high- and low-affinity gluconate transporters (apparent Kms of 6 and 212 μM, respectively), and gntK, a thermoresistant gluconokinase (52, 65). The edd, eda, gntT, gntU, and gntK genes are all under negative control by the gntR product (Table 2), and it was recently proven that the true inducer is gluconate (51). The regulatory regions of all of the known gluconate-inducible genes in E. coli contain one or two copies of a highly conserved operator sequence. GntR binding to these sites is inactivated by gluconate, and a single base change anywhere within the consensus operator sequence significantly affects GntR binding (51).
TABLE 2.
Sugar acid regulons on the E. coli genome
| Regulon | Operon | Location (kb) | Gene | Function | Reference |
|---|---|---|---|---|---|
| GntR | gntR | 3575.9 | gntR | Repressor: gluconate induction | 65 |
| edd-eda | 1931.7 | edd | 6-Phosphogluconate dehydratase | 20 | |
| 1930.6 | eda | KDPG aldolase | 20 | ||
| gntT | 3544.8 | gntT | High-affinity gluconate transport | 52 | |
| gntKU | 3574.9 | gntK | Gluconate kinase | 65 | |
| 3574.2 | gntU | Low-affinity gluconate transport | 65 | ||
| IdnR | idnDOTR | 4488.2 | idnR | Regulator: l-idonate induction | 8, 9 |
| idnK (gntV) | 4492.5 | idnK | Gluconate kinase | 8, 9 | |
| idnDOTR (yjgVUTS) | 4491.8 | idnD | l-Idonate 5-dehydrogenase | 8, 9 | |
| 4490.6 | idnO | Gluconate 5-dehydrogenase | 8, 9 | ||
| 4489.3 | idnT | l-Idonate transport | 8, 9 | ||
| KdgR | kdgR | 1908.2 | kdgR | Repressor: KDG induction | 57 |
| kdgK | 3676.4 | kdgK | KDG kinase | 57 | |
| kdgT | 4099.8 | kdgT | KDG transport | 39 | |
| eda | 1930.6 | eda | KDPG aldolase | 20 | |
| ExuR | exuR | 3244.7 | exuR | Repressor: hexuronate induction | 36 |
| exuT | 3243.5 | exuT | Glucuronate/galacturonate transport | 46 | |
| uxaCA | 3241.7 | uxaC | Glucuronate/galacturonate isomerase | 35 | |
| 3240.3 | uxaA | Altronate dehydratase (galacturonate) | 35 | ||
| uxaB | 1608.0 | uxaB | Altronate oxidoreductase (galacturonate) | 35 | |
| UxuR | uxuR | 4552.6 | uxuR | Repressor: glucuronate induction | 58 |
| uxuAB | 4549.5 | uxuA | Mannonate dehydratase (glucuronate) | 58 | |
| 4551.2 | uxuB | Mannonate oxidoreductase (glucuronate) | 58 | ||
| gntP | 4548.3 | gntP | Hexonate/hexuronate transport? | 38 | |
| UidR | uidR | 1694.8 | uidR | Repressor: β-glucuronide induction | 11 |
| uidAB | 1693.2 | uidA | β-Glucuronide hydrolase | 11 | |
| 1691.6 | uidB | β-Glucuronide transport | 11 |
FIG. 2.
Genetic map of E. coli K-12 showing locations of the genes and operons of gluconate metabolism.
There are four known gluconate transporters and three other E. coli orthologs of unknown function (DsdX, ORFo454, and YjhF); together these seven proteins constitute a novel transporter family (50). There is controversy as to why E. coli possesses so many gluconate transporters, and it is not yet known whether the two gluconate transporters of the GntI system, GntT and GntU, play different roles during growth on gluconate. Previous reports indicated that the high-affinity gluconate transporter (GntT) is induced for growth on gluconate, while the low-affinity transporter (GntU) is not (24, 53). On the other hand, it is clear that gntU and gntT are simultaneously induced by very low gluconate concentrations in vivo (2 to 10 μM) (52, 65), and kinetic analysis suggests the operation of at least two gluconate transporters during growth on gluconate (65). In contrast to gntU, however, gntT expression is highest as stationary-phase cells initiate log growth on gluconate (52), suggesting that GntT is more important during growth at low cell density. This pattern of expression is similar to that of the Fis protein, and a putative Fis binding site was found upstream of the gntT promoter, but expression of the gntT::lacZ fusion was unaffected in a fis background (51). Thus, the regulatory mechanism involved in the early peak of gntT expression remains obscure.
The gntT and gntKU operons, but not the edd-eda operon, are subject to cyclic AMP (cAMP)-dependent catabolite repression (20, 52, 65). Yet paradoxically, E. coli can cometabolize mixtures of gluconate (or glucuronate) and glucose (23, 35). Furthermore, gluconate itself is nearly as effective as glucose for catabolite repression (22). It is generally thought that changes in the cAMP concentration, as mediated by a component of the phosphoenolpyruvate:phosphotransferase system (PTS) (54), are responsible for catabolite repression. Thus, a problem is encountered when trying to explain how non-PTS substrates such as glucose-6-phosphate, lactose, and gluconate are able to exert catabolite repression. More recently it was realized that gluconate lowers both cAMP receptor protein (CRP) and cAMP to nearly the same extent as glucose (32). Furthermore, since non-PTS sugars still exert strong catabolite repression in crr mutant strains (lacking EIIAGlc), it appears that EIIAGlc of the PTS is not involved (32). Presumably, gluconate or a component of the gluconate-regulatory system interacts either directly or indirectly with adenylate cyclase and crp by mechanisms which have yet to be defined. Thus, gluconate acts both as an inducer, releasing the repressor GntR from the operator site, and as a repressor, lowering the CRP and cAMP pools. In fact, growth on gluconate in the presence of cAMP results in accumulation of methylglyoxal and subsequent growth inhibition (7). Also, intracellular accumulation of KDPG, the key intermediate of the ED pathway, is bacteriostatic (30). These results suggest an important physiological role for balanced induction and catabolite repression by gluconate, preventing the buildup of toxic metabolites, a principle which might perhaps be extended to other non-PTS sugars.
OTHER PROPOSED ROLES FOR THE ED ENZYMES
A growing number of studies have shown that eda expression is regulated in response to several conditions in addition to its role in sugar acid catabolism. The high basal expression of eda has been suggested to reflect its importance for detoxification of reactive compounds such as glyoxylate (49). Higher levels of the eda gene product are detected in cells starved for nitrogen and phosphate, in cells exposed to dinitrophenol, and in stationary-phase cells (47, 67). Synthesis of Eda is induced threefold during phosphate limitation, and a putative PhoB box was identified within the eda promoter region (67). Nystrom showed that induction of the RelA protein results in high expression of eda (47). Interestingly, Eda might also play a role in the SOS response (15). It was shown by two-dimensional gel electrophoresis that the Eda protein is induced 200-fold following treatment with DNA-damaging agents. This induction occurs only in the presence of RecA protein and was shown to be independent of eda transcription. In addition, an eda mutant was found to be UV sensitive and unable to recover from respiration inhibition, leading to the hypothesis that Eda is necessary for the recovery of respiration following the SOS response (15). Lastly, edd expression, or perhaps a portion of the Edd protein itself, has been postulated to play a role in ameliorating the toxicity that results from overexpression of the chaperone DnaK (59).
OXIDATIVE GLUCOSE METABOLISM
The discovery of a pyrroloquinoline quinone (PQQ)-dependent glucose dehydrogenase, which catalyzes the oxidation of glucose to gluconate (Fig. 1) in the periplasm, suggested an alternate route for glucose catabolism in E. coli (33). Mutants defective in enzyme I of the PTS are able to grow on glucose in the presence of exogenous PQQ (16). Paradoxically, wild-type E. coli does not synthesize PQQ (43). Results showed that under aerobic, but not anaerobic, conditions functional glucose dehydrogenase allowed growth on glucose of double mutants blocked at phosphoglucose isomerase and glucose-6-phosphate dehydrogenase (1). Subsequent experiments provided direct evidence that the ED pathway is turned on by oxidation of glucose to gluconate in the periplasm (27). The oxidative glucose pathway might be important for survival of E. coli in aerobic, aquatic environments. Similarly, Klebsiella pneumoniae and Pseudomonas aeruginosa preferentially oxidize as much as 80% of the glucose they consume to gluconate, which is catabolized via the ED pathway (34, 41).
L-IDONATE CATABOLISM
E. coli is able to grow on l-idonate by first converting it to d-gluconate (8, 9). The natural occurrence of l-idonate is apparently limited to its involvement as an intermediate in catabolism of 2,5-diketogluconate by Erwinia (66) and Gluconobacter (61), as well as tartaric acid formation from ascorbic acid in grapes (42). We have looked for l-idonate in mouse mucus by using HPLC but have not found it (unpublished results). In addition to E. coli, the only other organism reported to grow on l-idonate is Erwinia sp. strain ATCC 39140 (66), and it will be interesting to find out whether other microorganisms can grow on l-idonate.
The IdnR regulon (Table 2; Fig. 2) encodes the enzymes necessary for catabolism of l-idonate (8, 9). The metabolic sequence (Fig. 1) is as follows: IdnT allows the uptake of l-idonate; IdnD catalyzes a reversible oxidation of l-idonate to form 5-ketogluconate; IdnO catalyzes a reversible reduction of 5-ketogluconate to form d-gluconate; IdnK is the subsidiary gluconate kinase, which forms 6-phosphogluconate; the 6-phosphogluconate is metabolized via the ED pathway.
For many years it was believed that the genes of the IdnR regulon encoded the so-called GntII (or subsidiary) system for gluconate transport and phosphorylation (Fig. 2). The GntII system was discovered in a GntI deletion mutant which, after a long lag phase, began to grow on gluconate (6). This pseudorevertant strain was found to induce a second gluconate kinase (encoded by idnK, formerly gntV) and an alternative gluconate transporter (encoded by idnT, formerly gntW) when grown on gluconate (6, 37, 65). It was never established in these previous studies how the GntII system is regulated. It is now clear that the natural inducer of the IdnR regulon is l-idonate (8, 9). This suggests that growth of the pseudorevertants on gluconate may be the result of autoinduction, that is, l-idonate formation from gluconate, leading to induction of the IdnR regulon. Since gluconate is an intermediate of the l-idonate pathway, there must be cross talk with the GntR regulon which leads to induction of the ED pathway. There is also the possibility that the GntR regulon cross-talks with the IdnR regulon by way of two highly conserved GntR binding sites located within the divergent promoter region of the l-idonate regulon (52).
CATABOLISM OF HEXURONATES AND HEXURONIDES
The UidR regulon (Table 2) allows for the growth of E. coli on the d-glucuronate and d-galacturonate moieties of the β-glucuronides and β-galacturonides present in mucus (10). After entering the cell, glucuronate and galacturonate are degraded via parallel pathways (Fig. 1) involving consecutive isomerization, reduction, and dehydration steps (5). The ExuR regulon (36) governs expression of the genes involved in galacturonate catabolism (Table 2), although ExuT (46) and UxaC (36) allow for the uptake of both galacturonate and glucuronate and their isomerization to fructuronate and tagaturonate, respectively. The latter intermediates can also be utilized as exogenous substrates by E. coli (46) and appear to be the inducers of the ExuR regulon (36). The uxaA and uxaB genes are specific for galacturonate catabolism (35). The UxuR regulon (Table 2) governs the catabolism of glucuronate (58). The uxuA and uxuB genes are specifically involved in glucuronate catabolism (58). The ExuR and UxuR repressors apparently act together to mediate regulation of the UxuR regulon by the true inducer, fructuronate (57). These controls are thought to allow induction of the shared gene, uxaC, by glucuronate or galacturonate, as well as repression of the glucuronate genes in the presence of galacturonate only. The two hexuronate pathways form the common intermediate 2-keto-3-deoxygluconate (KDG). Catabolism of KDG is governed by the KdgR regulon (57). KDG is phosphorylated by KDG kinase to form KDPG, the substrate of Eda (Fig. 1). Interestingly, eda is also a member of the KdgR regulon and is induced approximately fourfold by growth on glucuronate (20). Glucuronate can enter the cell via KdgT (in addition to ExuT), but kdgT is only weakly induced by KDG, and in fact, E. coli can grow on KDG only if a mutation renders expression of kdgT constitutive (39).
HOMOLOGS OF THE ED PATHWAY IN E. COLI
d-Galactonate is catabolized via a pathway which is analogous to the ED pathway (19). Galactonate is transported by a specific permease and then phosphorylated and dehydrated to the 2-keto-3-deoxy form and cleaved by an aldolase to form pyruvate and glyceraldehyde-3-phosphate. In contrast to gluconate, galactonate is first dehydrated and then phosphorylated by 2-keto-3-deoxygalactonate kinase. Both of the phosphorylated 2-keto-3-deoxy compounds are toxic when allowed to accumulate in the E. coli cell (19, 30). The dgo operon, encoding the enzymes of galactonate metabolism, is negatively regulated by the dgoR product and specifically induced by galactonate (19). Several related pathways may exist in E. coli. For example, edd is similar (28% identical) to yjhG and yagF, and in both cases these edd orthologs are immediately adjacent to putative aldolases and transporters (BLASTP search).
ED GENES ON OTHER GENOMES
A survey of published genome sequences, as well as a number of partially completed genomes, indicates that the ED pathway is present in several of these organisms (Table 1). This is not surprising given that a biochemical survey indicated that the ED enzymes are widely distributed amongst the Bacteria and are present in all three phylogenetic domains, including the most deeply rooted Archaea (18, 60). Paralogs of edd were found on only four of the genomes. The lack of gntT and gntK paralogs in Helicobacter pylori suggests that the pathway is not employed for gluconate but rather for catabolism of glucose via 6-phosphogluconate. The presence of the ED pathway in Neisseria gonorrhoeae has been reported previously and is, quite interestingly, known to be induced by serum in the growth medium (14). The presence of gntT and gntK paralogs in organisms lacking edd suggests that gluconate is metabolized via the pentose phosphate pathway in these species. Most widespread of the genes surveyed is eda, with paralogs on 14 genomes. The presence of eda in organisms lacking edd suggests that these species grow on sugar acids that lead to KDPG. It is interesting to speculate that the widespread distribution of eda also has something to do with the role of Eda in SOS recovery, starvation responses, or detoxification of glyoxylate, as described above.
CONCLUSIONS
The ED pathway serves as a “funnel,” receiving metabolites derived from the catabolism of several sugar acids, including gluconate, l-idonate, glucuronate, and galacturonate, as well as other hexonates and hexuronates (Fig. 1). Each of these compounds is found in nature and in several cases is known to be present in significant concentrations in the mucus layer that covers the epithelial cells of mammalian large intestines. The regulation of the sugar acid regulons allows E. coli to cometabolize the sugar acids, even in the presence of glucose. It is still not known which substrates E. coli grows on in the large intestine and what pathways provide it with the metabolic advantage necessary for it to compete with the hundreds of other bacteria with which it shares this habitat. New evidence suggests that sugar acid metabolism is an important aspect of the ecology of E. coli. As a result of the E. coli genome project, the function and regulation of new genes and operons involved in sugar acid metabolism are being described. There is a very good chance that additional sugar acid pathways remain to be found amongst the 38% of genes with unknown functions on the E. coli genome (12). We hypothesize that gluconate and other sugar acids are the daily bread of E. coli.
ACKNOWLEDGMENTS
We thank Paul Cohen for many helpful discussions.
Work on this project has been supported by grants from the DOE (DE-FG02-95ER20178) and NSF (MCB-9723593).
REFERENCES
- 1.Adamowicz M, Conway T, Nickerson K. Nutritional restoration of oxidative glucose metabolism in Escherichia coli by addition of the glucose dehydrogenase cofactor PQQ. Appl Environ Microbiol. 1991;57:2012–2015. doi: 10.1128/aem.57.7.2012-2015.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen A. The structure and function of gastrointestinal mucous. In: Boedeker I E C, editor. Attachment of organisms to the gut mucosa. II. Boca Raton, Fla: CRC Press, Inc.; 1984. pp. 3–11. [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apperloo-Renkema H Z, van der Waaij B D, van der Waaij D. Determination of colonization resistance of the digestive tract. Epidemiol Infect. 1990;105:355–361. doi: 10.1017/s0950268800047944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashwell G. Enzymes of glucuronic and galacturonic acid metabolism in bacteria. Methods Enzymol. 1962;5:190–208. [Google Scholar]
- 6.Bächi B, Kornberg H L. Genes involved in the uptake and catabolism of gluconate by Escherichia coli. J Gen Microbiol. 1975;90:321–335. doi: 10.1099/00221287-90-2-321. [DOI] [PubMed] [Google Scholar]
- 7.Bächi B, Kornberg H L. Utilization of gluconate by Escherichia coli. A role of adenosine 3′:5′-cyclic monophosphate in the induction of gluconate catabolism. Biochem J. 1975;150:23–128. doi: 10.1042/bj1500123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bausch C, Peekhaus N, Blais T, Conway T. Abstracts of the 97th General Meeting of the American Society for Microbiology, abstr. K-75. Washington, D.C: American Society for Microbiology; 1997. p. 354. [Google Scholar]
- 9.Bausch C, Peekhaus N, Utz C, Blais T, Murray E, Lowary T, Conway T. Sequence analysis of the GntII (subsidiary) system for gluconate metabolism reveals a novel pathway for l-idonic acid catabolism in Escherichia coli. J Bacteriol. 1998;180:3704–3710. doi: 10.1128/jb.180.14.3704-3710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco C, Ritzenthaler P, Mata-Gilsinger M. Cloning and endonuclease restriction analysis of uidA and uidR genes in Escherichia coli K-12: determination of transcription direction for the uidA gene. J Bacteriol. 1982;149:587–594. doi: 10.1128/jb.149.2.587-594.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanco C, Ritzenthaler P, Mata-Gilsinger M. Negative dominant mutations of the uidR gene in Escherichia coli: genetic proof for a cooperative regulation of uidA expression. Genetics. 1986;112:173–182. doi: 10.1093/genetics/112.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Grgor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1473. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 13.Borrelio S P. Microbial flora of the gastro-intestinal tract. In: Hill M J, editor. Microbial metabolism in the digestive tract. Boca Raton, Fla: CRC Press, Inc.; 1986. pp. 2–16. [Google Scholar]
- 14.Britigan B E, Chai Y, Cohen M S. Effects of human serum on growth and metabolism of Neisseria gonorrhoeae: an alternative view of serum. Infect Immun. 1985;50:738–744. doi: 10.1128/iai.50.3.738-744.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cayrol C, Petit C, Raynaud B, Capdevielle J, Guillemont J-C, Defais M. Recovery of respiration following the SOS response of Escherichia coli requires RecA-mediated induction of 2-keto-4-hydroxyglutarate aldolase. Proc Natl Acad Sci USA. 1995;92:11806–11809. doi: 10.1073/pnas.92.25.11806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleton-Jansen A, Goosen N, Fayet O, van de Putte P. Cloning, mapping, and sequencing of the gene encoding Escherichia coli quinoprotein glucose dehydrogenase. J Bacteriol. 1990;172:6308–6315. doi: 10.1128/jb.172.11.6308-6315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen P S, Rossoll R, Cabelli V J, Yang S-L, Laux D C. Relationship between the mouse colonizing ability of a human fecal Escherichia coli strain and its ability to bind a specific mouse colonic mucous gel protein. Infect Immun. 1983;40:62–69. doi: 10.1128/iai.40.1.62-69.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Cohen, P. S. Personal communication.
- 18.Conway T. The Entner-Doudoroff pathway: history, physiology, and molecular biology. FEMS Microbiol Rev. 1992;103:1–27. doi: 10.1111/j.1574-6968.1992.tb05822.x. [DOI] [PubMed] [Google Scholar]
- 19.Cooper R A. The utilisation of d-galactonate and d-2-oxo-3-deoxygalactonate by Escherichia coli. K-12. Arch Microbiol. 1978;118:199–206. doi: 10.1007/BF00415730. [DOI] [PubMed] [Google Scholar]
- 20.Egan S, Fliege R, Tong S, Shibata A, Wolf R E, Jr, Conway T. Molecular characterization of the Entner-Doudoroff pathway in Escherichia coli: sequence analysis and localization of promoters for the edd-eda operon. J Bacteriol. 1992;174:4638–4646. doi: 10.1128/jb.174.14.4638-4646.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Entner N, Doudoroff M. Glucose and gluconic acid oxidation of Pseudomonas saccharophila. J Biol Chem. 1952;196:853–862. [PubMed] [Google Scholar]
- 22.Epstein W, Rothman-Denes L B, Hesse J. Adenosine 3′:5′-cyclic monophosphate as mediator of catabolite repression in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:2300–2304. doi: 10.1073/pnas.72.6.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esienberg R C, Dobrogosz W J. Gluconate metabolism in Escherichia coli. J Bacteriol. 1967;93:941–949. doi: 10.1128/jb.93.3.941-949.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faik P, Kornberg H L. Isolation and properties of E. coli mutants affected in gluconate uptake. FEBS Lett. 1973;32:260–264. doi: 10.1016/0014-5793(73)80847-6. [DOI] [PubMed] [Google Scholar]
- 25.Farber J M, Idziak E S. Detection of glucose oxidation products in chilled fresh beef undergoing spoilage. Appl Environ Microbiol. 1982;44:521–524. doi: 10.1016/s0315-5463(82)72428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finegold S M, Sutter V L, Mathisen G E. Normal indigenous intestinal microflora. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press, Inc.; 1983. pp. 3–31. [Google Scholar]
- 27.Fliege R, Tong S, Shibata A, Nickerson K W, Conway T. The Entner-Doudoroff pathway in Escherichia coli is induced for oxidative glucose metabolism via pyrroloquinoline quinone-dependent glucose dehydrogenase. Appl Environ Microbiol. 1992;58:3826–3829. doi: 10.1128/aem.58.12.3826-3829.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraenkel D G. Glycolysis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 189–198. [Google Scholar]
- 29.Freter R. Mechanisms that control the microflora in the large intestine. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press, Inc.; 1983. pp. 33–54. [Google Scholar]
- 30.Fuhrman L K, Wanken A, Nickerson K W, Conway T. Rapid accumulation of intracellular 2-keto-3-deoxy-6-phosphogluconate in an Entner-Doudoroff aldolase mutant results in bacteriostasis. FEMS Microbiol Lett. 1998;159:261–266. doi: 10.1111/j.1574-6968.1998.tb12870.x. [DOI] [PubMed] [Google Scholar]
- 31.Hill M J. The normal gut bacterial flora. In: Hill M J, editor. Role of gut bacteria in human toxicology and pharmacology. London, United Kingdom: Taylor and Francis; 1995. pp. 3–18. [Google Scholar]
- 32.Hogema B M, Arents J C, Inada T, Aiba H, van Dam K, Postma P W. Catabolite repression by glucose 6-phosphate, gluconate and lactose in Escherichia coli. Mol Microbiol. 1997;24:857–867. doi: 10.1046/j.1365-2958.1997.3991761.x. [DOI] [PubMed] [Google Scholar]
- 33.Hommes R W, Postma P W, Neijssel O M, Tempest D W, Kokter P, Duine J A. Evidence for quinoprotein glucose dehydrogenase apoenzyme in several strains of Escherichia coli. FEMS Microbiol Lett. 1984;24:329–333. [Google Scholar]
- 34.Hommes R W, Postma P W, Tempest D W, Neijssel O M. The influence of the culture pH value on the direct glucose oxidative pathway in Klebsiella pneumoniae. Arch Microbiol. 1989;151:261–267. doi: 10.1007/BF00413140. [DOI] [PubMed] [Google Scholar]
- 35.Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Isolation of fusions between the lac genes and several genes of the exu regulon: analysis of their regulation, determination of the transcription direction of the uxaC-uxaA operon, in Escherichia coli K-12. Mol Gen Genet. 1981;182:279–287. doi: 10.1007/BF00269671. [DOI] [PubMed] [Google Scholar]
- 36.Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Regulation and transcription direction of exuR, a self-regulated repressor in Escherichia coli K-12. J Mol Biol. 1982;156:221–228. doi: 10.1016/0022-2836(82)90468-5. [DOI] [PubMed] [Google Scholar]
- 37.Istúriz T, Palmero E, Vitelli-Flores J. Mutations affecting gluconate catabolism in Escherichia coli. Genetic mapping of the locus for the thermosensitive gluconokinase. J Gen Microbiol. 1986;132:3209–3212. doi: 10.1099/00221287-132-11-3209. [DOI] [PubMed] [Google Scholar]
- 38.Klemm P, Tong S, Nielsen H, Conway T. The gntP gene of Escherichia coli involved in gluconate uptake. J Bacteriol. 1996;178:61–67. doi: 10.1128/jb.178.1.61-67.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaGarde A E, Stoeber F R. Escherichia coli K-12 structural kdgT mutants exhibiting thermosensitive 2-keto-3-deoxy-d-gluconate uptake. J Bacteriol. 1977;129:606–615. doi: 10.1128/jb.129.2.606-615.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee A. Neglected niches. The microbial ecology of the gastrointestinal tract. Adv Microb Ecol. 1985;8:115–162. [Google Scholar]
- 41.Lessie T G, Phibbs P V. Alternative pathways of carbohydrate utilization in pseudomonads. Annu Rev Microbiol. 1984;38:359–387. doi: 10.1146/annurev.mi.38.100184.002043. [DOI] [PubMed] [Google Scholar]
- 42.Malipiero V, Ruffner H P, Rast D M. Ascorbic to tartaric acid conversion in grapevines. J Plant Physiol. 1987;129:33–40. [Google Scholar]
- 43.Matsushita K, Arents J C, Bader R, Yamada M, Adachi O, Postma P W. Escherichia coli is unable to produce pyrroloquinoline quinone (PQQ) Microbiology. 1997;143:3149–3156. doi: 10.1099/00221287-143-10-3149. [DOI] [PubMed] [Google Scholar]
- 44.McCormick B A, Franklin D P, Laux D C, Cohen P S. Type 1 pili are not necessary for colonization of the streptomycin treated mouse large intestine by type 1-piliated Escherichia coli F-18 and E. coli K-12. Infect Immun. 1989;57:3022–3029. doi: 10.1128/iai.57.10.3022-3029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neidhardt F C. The enteric bacterial cell and the age of bacteria. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1–3. [Google Scholar]
- 46.Nemoz G, Robert-Baudouy J, Stoeber F. Physiological and genetic regulation of the aldohexuronate transport system in Escherichia coli. J Bacteriol. 1976;127:706–718. doi: 10.1128/jb.127.2.706-718.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Nickerson, K. Personal communication.
- 47.Nystrom T. The glucose-starvation stimulon of Escherichia coli: induced and repressed synthesis of enzymes of central metabolic pathways and role of acetyl phosphate in gene expression and starvation survival. Mol Microbiol. 1994;12:833–843. doi: 10.1111/j.1365-2958.1994.tb01069.x. [DOI] [PubMed] [Google Scholar]
- 48.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patil R V, Dekker E E. Cloning, nucleotide sequence, overexpression, and inactivation of the Escherichia coli 2-keto-4-hydroxyglutarate aldolase gene. J Bacteriol. 1992;174:102–107. doi: 10.1128/jb.174.1.102-107.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peekhaus N, Tong S, Reizer J R, Saier M, Murray E, Conway T. Characterization of a novel transporter family that includes multiple Escherichia coli gluconate transporters and their homologues. FEMS Microbiol Lett. 1997;147:233–238. doi: 10.1111/j.1574-6968.1997.tb10247.x. [DOI] [PubMed] [Google Scholar]
- 51.Peekhaus N, Conway T. Positive and negative transcriptional regulation of the Escherichia coli gluconate regulon gene gntT by GntR and the cyclic AMP (cAMP)-cAMP receptor protein complex. J Bacteriol. 1998;180:1777–1785. doi: 10.1128/jb.180.7.1777-1785.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porco A, Peekhaus N, Bausch C, Tong S, Isturiz T, Conway T. Molecular genetic characterization of the Escherichia coli gntT gene of GntI, the main system for gluconate metabolism. J Bacteriol. 1997;179:1584–1590. doi: 10.1128/jb.179.5.1584-1590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porco A, Isturiz T. Selection of lacZ operon fusions in genes of gluconate metabolism in E. coli. Characterization of a gntT::lacZ fusion. Acta Cientifica Venezolana. 1991;42:270–275. [PubMed] [Google Scholar]
- 54.Postma P W, Lengeler J W, Jacobson G R. Phosphenolpyruvate: carbohydrate phosphotransferase systems. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1149–1174. [Google Scholar]
- 55.Poulsen L K, Licht T R, Rang C, Krogfelt K A, Molin S. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J Bacteriol. 1995;177:5840–5845. doi: 10.1128/jb.177.20.5840-5845.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poulsen L K F L, Kristensen C S, Hobolth P, Molin S, Krogfelt K A. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect Immun. 1994;62:5191–5194. doi: 10.1128/iai.62.11.5191-5194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pouyssegur J, Stoeber F. Genetic control of the 2-keto-3-deoxy-d-gluconate metabolism in Escherichia coli K-12: kdg regulon. J Bacteriol. 1974;117:641–651. doi: 10.1128/jb.117.2.641-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robert-Baudouy J, Portalier R, Stoeber F. Regulation of hexuronate system genes in Escherichia coli K-12: multiple regulation of the uxu operon by exuR and uxuR gene products. J Bacteriol. 1981;145:211–220. doi: 10.1128/jb.145.1.211-220.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rockabrand D, Blum P. Multicopy plasmid suppression of stationary phase chaperone toxicity in Escherichia coli by phosphogluconate dehydratase and the N-terminus of DnaK. Mol Gen Genet. 1995;249:498–506. doi: 10.1007/BF00290575. [DOI] [PubMed] [Google Scholar]
- 60.Romano A H, Conway T. Evolution of carbohydrate metabolic pathways. Res Microbiol. 1996;147:448–455. doi: 10.1016/0923-2508(96)83998-2. [DOI] [PubMed] [Google Scholar]
- 61.Saito Y, Ishii Y, Hayashi H, Imao Y, Akashi T, Yoshikawa K, Noguchi Y, Soeda S, Yoshida M, Niwa M, Hosoda J, Shimomura K. Cloning of genes coding for l-sorbose and l-sorbosone dehydrogenases from Gluconobacter oxydans and microbial production of 2-keto-l-gulonate, a precursor of l-ascorbic acid, in a recombinant G. oxydans strain. Appl Environ Microbiol. 1997;63:454–460. doi: 10.1128/aem.63.2.454-460.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salyers A A, Leedle J A Z. Carbohydrate metabolism in the human colon. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press, Inc.; 1983. pp. 129–146. [Google Scholar]
- 63.Sweeney N J, Laux D C, Cohen P S. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect Immun. 1996;64:3504–3511. doi: 10.1128/iai.64.9.3504-3511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sweeney N J, Klemm P, McCormick B A, Moller-Nielsen E, Utley M, Schembri M A, Laux D C, Cohen P S. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct niche in the streptomycin-treated mouse large intestine. Infect Immun. 1996;64:3497–3503. doi: 10.1128/iai.64.9.3497-3503.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tong S, Porco A, Isturiz T, Conway T. Cloning and molecular characterization of the Escherichia coli gntR, gntK, and gntU genes of GntI, the main system for gluconate metabolism. J Bacteriol. 1996;178:3260–3269. doi: 10.1128/jb.178.11.3260-3269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Truesdell S J, Sims J C, Boerman P A, Seymour J L, Lazarus R A. Pathways for metabolism of keto-aldonic acids in an Erwinia sp. J Bacteriol. 1991;173:6651–6656. doi: 10.1128/jb.173.21.6651-6656.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.VanBogelen R, Olson E R, Wanner B L, Neidhardt F C. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J Bacteriol. 1996;178:4344–4366. doi: 10.1128/jb.178.15.4344-4366.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wadolowski E A, Laux D C, Cohen P S. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect Immun. 1988;56:1030–1035. doi: 10.1128/iai.56.5.1030-1035.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]