Abstract
Maleylacetate reductases (EC 1.3.1.32) have been shown to contribute not only to the bacterial catabolism of some usual aromatic compounds like quinol or resorcinol but also to the degradation of aromatic compounds carrying unusual substituents, such as halogen atoms or nitro groups. Genes coding for maleylacetate reductases so far have been analyzed mainly in chloroaromatic compound-utilizing proteobacteria, in which they were found to belong to specialized gene clusters for the turnover of chlorocatechols or 5-chlorohydroxyquinol. We have now cloned the gene macA, which codes for one of apparently (at least) two maleylacetate reductases in the gram-positive, chlorophenol-degrading strain Rhodococcus opacus 1CP. Sequencing of macA showed the gene product to be relatively distantly related to its proteobacterial counterparts (ca. 42 to 44% identical positions). Nevertheless, like the known enzymes from proteobacteria, the cloned Rhodococcus maleylacetate reductase was able to convert 2-chloromaleylacetate, an intermediate in the degradation of dichloroaromatic compounds, relatively fast and with reductive dehalogenation to maleylacetate. Among the genes ca. 3 kb up- and downstream of macA, none was found to code for an intradiol dioxygenase, a cycloisomerase, or a dienelactone hydrolase. Instead, the only gene which is likely to be cotranscribed with macA encodes a protein of the short-chain dehydrogenase/reductase family. Thus, the R. opacus maleylacetate reductase gene macA clearly is not part of a specialized chlorocatechol gene cluster.
Maleylacetate reductases (EC 1.3.1.32) have long been known to be involved in the degradation of chloroaromatic compounds via chlorocatechols as intermediates (10, 31). By reduction of a carbon-carbon double bond they form 3-oxoadipate, a metabolite also of catechol catabolism, and thus compensate for the different oxidation states of chlorinated and nonchlorinated compounds. 2-Chloromaleylacetate, which is formed during turnover of several dichlorocatechols, is initially reductively dechlorinated and then reduced to 3-oxoadipate in a second reaction (22, 47).
Corresponding to the biochemical function in chlorocatechol degradation, the following maleylacetate reductase genes have been shown to be associated with dioxygenase, cycloisomerase, and dienelactone hydrolase genes as components of specialized chlorocatechol catabolic operons: tfdF and tfdFII on pJP4 from the 2,4-dichlorophenoxyacetate-utilizing strain Ralstonia eutropha (Alcaligenes eutrophus) JMP134 (29, 33, 37, 44), tcbF on pP51 from the 1,2,4-trichlorobenzene-degrading strain Pseudomonas sp. strain P51 (45), and clcE from the 3-chlorobenzoate catabolizing strains Pseudomonas sp. strain B13 and Pseudomonas putida AC866(pAC27) (15, 20, 21). Catechol degradation, in contrast, does not require a maleylacetate reductase activity, and corresponding genes do not belong to the known catechol operons. Thus, while at least two of the chlorocatechol catabolic enzymes, i.e., the dioxygenases and cycloisomerases, appear to have been recruited from catechol catabolism, maleylacetate reductase genes must have had a different origin and original function (34).
The postulated original function of the maleylacetate reductases is still under discussion. In bacteria, these enzymes have been shown to play a role, for example, in quinol, resorcinol, and 2,4-dihydroxybenzoate degradation (6, 25, 41). Other aromatic growth substrates involving the action of maleylacetate reductase are more exotic, since they carry a fluorine substituent (35), a sulfo group (14), a nitro group (18, 40), or several chlorine substituents (8, 26, 48). Maleylacetate reductase genes have been shown to be part of a specialized gene cluster for 2,4,5-trichlorophenoxyacetate degradation (8, 9) and of a gene cluster for hydroxyquinol conversion which contributes to 4-nitrophenol turnover (4).
The chlorocatechol pathway of the chlorophenol-utilizing strain Rhodococcus opacus (erythropolis) 1CP obviously evolved functionally convergent to the corresponding pathway in the proteobacteria mentioned above (13, 39). Thus, it is not surprising that the chlorocatechol gene cluster of strain 1CP is organized differently from the corresponding proteobacterial operons; in fact, its characterization showed that it does not comprise a maleylacetate reductase gene (13). Thus, the nature of the gene cluster(s) encoding a maleylacetate reductase in R. opacus remained to be elucidated. Such gene clusters could complement otherwise incomplete pathways, and they might also have provided the source from which the maleylacetate reductase gene was recruited during evolution of dedicated pathways, such as the proteobacterial chlorocatechol catabolic route.
(Some of the results presented here have previously been reported in a preliminary communication [38].)
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
R. opacus 1CP, previously assigned to the species R. erythropolis, has been reported to utilize 4-chlorophenol and 2,4-dichlorophenol as sole sources of carbon and energy (17, 43). Cells for enzyme purification were grown with 4-chlorophenol as described previously (39). Escherichia coli DH5α (32) was obtained from GIBCO BRL, and E. coli BL21(DE3)(pLysS) (32, 42) was obtained from Novagen. E. coli strains were grown aerobically in baffled Erlenmeyer flasks with constant shaking (170 rpm) at 37°C in 2× YT medium (32), if appropriate, with ampicillin at a final concentration of 100 μg/ml. The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Source (reference) |
|---|---|---|
| pBluescript II SK(+) | Phagemid derived from pUC19, Plac, lacZ, Apr, f1(+) origin, ColE1 origin | Stratagene (1) |
| pET11a* | Derivative of pET11a with additional cloning sites (KpnI, XhoI, SalI), PT7lac | J. Altenbuchner, Stuttgart, Germany (42) |
| pMARRE0 | Apr; 0.982-kb PCR product of primers NH2-3 and HOM in the EcoRV site of pBluescript II SK(+) | This study |
| pMARRE1 | Apr; pBluescript II SK(+) containing a 5,657-bp KpnI fragment of R. opacus 1CP DNA | This study |
| pMARRE2 | Apr; pBluescript II SK(+) containing a ca. 7-kb XhoI fragment of R. opacus 1CP DNA | This study |
| pMARRE3 | Apr; pBluescript II SK(+) containing a 4,628-bp PstI fragment of R. opacus 1CP DNA | This study |
| pMARRE5 | Apr; 2,079 bp from pMARRE1 on EcoRI-HindIII fragment in pBluescript II SK(+) | This study |
| pMARRE10 | Apr; PCR-amplified macA in NdeI and BamHI site of pET11a* | This study |
Abbreviation: Apr, ampicillin resistance.
DNA manipulations, cloning procedure, and sequence analysis.
Genomic DNA from R. opacus 1CP was prepared as described by Eulberg et al. (11). Plasmid DNA from E. coli was isolated with a Pharmacia Flexiprep kit or by a modified boiling method (30). Other general genetic methods were performed by standard procedures (3, 32), usually as described previously (11).
Primer NH2-3, 5′-(A/C)G(G/C)GT(G/C)GT(G/C)TTCGG(G/C)GC(G/C)GG-3′, was custom synthesized corresponding to the amino acid sequence RVVFGAG, which is conserved close to the N termini of TfdF and of the previously purified maleylacetate reductase (TfdFII) from R. eutropha JMP134 (37). Primer HOM, 5′-GG(A/T)CG(G/C)GG(A/G)TT(G/C)GG(A/G)TA(C/T)TG-3′, was designed to fit to a conserved region of the maleylacetate reductase genes tfdF and tcbF (positions 976 to 995 and 973 to 992, respectively). PCR mixtures contained, in a total volume of 50 μl, 50 pmol of each primer, 0.1 μg of genomic template DNA, 100 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, 0.3 U of Eurogentech Goldstar polymerase, and the corresponding buffer. PCR was performed with 30 s of denaturing (94°C), 30 s of annealing (45°C), and 60 s of polymerization (72°C) for 33 cycles, with an additional 3 min of denaturing prior to the first cycle (polymerase addition after 2 min) (7) and 15 min of polymerization after the last cycle.
Correctly sized amplification products (ca. 980 bp) were ligated into the T-tailed EcoRV site of pBluescript II SK(+), yielding pMARRE0. After digoxigenin labeling, the cloned PCR product served to identify hybridizing bands on a Southern blot (Amersham Hybond N+ nylon membrane) with XhoI-, PstI-, and KpnI-digested genomic DNA from R. opacus. DNA from corresponding regions on a second gel was eluted and ligated into pBluescript II SK(+). The labeled insert of pMARRE0 was used again to identify by colony hybridization those E. coli DH5α clones which contained the recombinant plasmids pMARRE1, -2, and -3.
The nucleotide sequences of both strands of the inserts of pMARRE1 and -3 were determined and analyzed as described previously (11). In addition to FASTA, the BLAST programs (2, 16) were used under default conditions to screen databases.
Overproduction of the maleylacetate reductase MacA in E. coli.
To detect the polypeptide encoded by the predicted maleylacetate reductase gene, the T7lac expression system of Studier et al. (42) was used. For overexpression of maleylacetate reductase, the macA gene was PCR amplified from pMARRE5 by using primer PND (5′-GATCATATGGCTGAGCGCTCCTCCGAG-3′), with an NdeI site (underlined) and the start of macA (in boldface), as well as primer PBA (5′-GTGGATCCCTATTCGGGCGGGCGATTG-3′) with a BamHI site (underlined) and 19 bases (in boldface) corresponding to the end of macA. The PCR mixture was as described above except that different primers and template (pMARRE5) and 0.5 U of polymerase were used. The PCR program differed from the one given above in that (i) annealing conditions were 45 s at 50°C and (ii) only 22 cycles were performed. The resulting PCR product was digested with NdeI and BamHI and inserted into pET11a*, digested with the same enzymes, to yield plasmid pMARRE10, which contained the intact macA gene under the control of the T7lac promoter of pET11a*.
E. coli BL21(DE3)(pLysS) (42) was used as the host strain for inducible overexpression of maleylacetate reductase. The strain containing the recombinant plasmid pMARRE10 was grown at 30°C in shaken 3-liter Erlenmeyer flasks containing 1 liter of 2× YT medium supplemented with 100 μg of ampicillin per ml, 2.5 mM betaine, and 660 mM sorbitol. For induction of macA, 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the cultures at an optical density at 600 nm of 1, and the cells were harvested after 7 h of incubation at 30°C. Cell pellets were washed with 50 mM BisTris [bis(2-hydroxyethyl)imino-tris(hydroxymethyl)methane]-HCl (pH 6.5)–1 mM dithiothreitol (DTT) and then stored at −20°C until required.
Preparation of cell extracts.
Cells of E. coli BL21(DE3)(pLysS)(pMARRE10) were thawed on ice, resuspended in 50 mM BisTris-HCl (pH 6.5)–1 mM DTT, and disrupted by passage through an Aminco French pressure cell (37). Cells of R. opacus 1CP were resuspended in the same buffer and disintegrated by grinding with glass beads (11). In both cases, clear extracts were obtained by ultracentrifugation plus filtration (11).
Enzyme assays and estimation of protein concentrations.
Maleylacetate reductase activities were determined by monitoring NADH oxidation in the presence of maleylacetate as described by Schlömann et al. (35). Kinetic experiments (in the presence of 0.2 mM NADH as cosubstrate) were also performed with 2-chloromaleylacetate. Protein concentrations were determined by the method of Bradford (5), with bovine serum albumin as the standard. Maleylacetate and 2-chloromaleylacetate were prepared by alkaline hydrolysis from cis-dienelactone (cis-4-carboxymethylenebut-2-en-4-olide) or its 2-chlorosubstituted analog, which were kindly provided by S. Kaschabek and W. Reineke (Wuppertal, Germany).
Purification of the maleylacetate reductase MacA from an overexpressing E. coli clone.
Purification was performed at room temperature by using a fast protein liquid chromatography system as well as chromatography media from Pharmacia and 50 mM BisTris (pH 6.5)–1 mM DTT as the basal buffer for all steps. The initial anion-exchange chromatography of the cell extract (volume, 37 ml; protein, 2,040 mg; total activity, 1,470 U) was performed in two parallel runs on a Q Sepharose High Performance HR16/10 column with a linear NaCl gradient of 0.1 to 0.8 M over 300 ml (elution at ca. 0.3 M NaCl). From the following Red Sepharose CL-6B column (bed volume, 85 ml), proteins were eluted by first washing with buffer and by then applying an NaCl step of 0 to 2 M NaCl over 1 ml, which yielded the maleylacetate reductase at the high salt concentration. After concentration of the preparation by ultrafiltration (Amicon PM 10 filters) and addition of NaCl to 3 M (final concentration), the preparation was subjected to two parallel runs of a phenyl-Superose HR 10/10 chromatography with a gradient of 3 to 0.4 M NaCl over 65 ml (elution at ca. 1.7 M NaCl) and to concentration and partial salt removal by ultrafiltration. During the subsequent Mono Q HR 5/5 chromatography (three parallel runs) with an NaCl gradient of 0 to 0.4 M over 92 ml, the maleylacetate reductase eluted at ca. 0.3 M NaCl. In the next step, the maleylacetate reductase pool was chromatographed in four parallel runs on a Blue Sepharose CL-6B HR 10/10 column with an NaCl gradient of 0 to 2 M NaCl over 35 ml (elution at ca. 1 M NaCl). A concentrated and partially desalted preparation was further purified by using a weak anion exchanger (Mono P HR 5/5; three parallel runs) with a gradient of 0 to 0.7 M NaCl over 50 ml (elution at ca. 0.3 M NaCl). The final purification step was another Mono Q HR 5/5 run with an NaCl gradient from 0.2 to 0.7 M over 60 ml. The maleylacetate reductase preparation was concentrated, and 50% (vol/vol) glycerol was added for stabilization.
Partial purification of a maleylacetate reductase induced in 4-chlorophenol-grown cells of R. opacus 1CP.
The purification made use of some of the same chromatographic media and the same buffer solution (50 mM BisTris [pH 6.5]–1 mM DTT) as for the procedure outlined above. A cell extract (volume, 600 ml; protein, 1,650 mg; total activity, 900 U; specific activity, 0.55 U/mg) which had been obtained from 77 g (wet weight) of cells was initially applied to a Q Sepharose High Performance HR16/10 column. Elution with a gradient of 0 to 0.5 M NaCl over 200 ml resulted in two activity peaks. Corresponding pools were further purified in separate runs on a Blue Sepharose CL-6B column (XK 26/20; bed volume, 50 ml) by using a gradient of 0 to 1.5 M NaCl over 300 ml for elution. Active fractions originating from the first peak on Q Sepharose High Performance chromatography appeared to contain relatively pure protein (protein, 0.03 mg; total activity, 6.5 U; specific activity, 217 U/mg) and thus were concentrated and analyzed by electrophoresis.
SDS-polyacrylamide gel electrophoresis and protein sequencing.
Discontinuous sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and staining of gels were performed basically as described previously (11, 37). If bands were to be analyzed by protein sequencing, they were cut from Western blots (for details, see reference 12) and subjected to automated sequencing (Applied Biosystems model 473A).
Investigation of 2-chloromaleylacetate turnover.
The conversion of 2-chloromaleylacetate and the formation of a product from it were investigated in a reaction mixture (100 μl, 37°C) containing 50 mM Tris-HCl (pH 7.5), 0.8 mM NADH, 0.8 mM 2-chloromaleylacetate, and 3.5 U of maleylacetate reductase (MacA) purified from E. coli. After 5 min, an additional 0.8 mM NADH was added. In samples taken from this mixture after 1, 3, 5, 7, and 10 min, the reaction was stopped by addition of equal volumes of solvent for high-performance liquid chromatography (HPLC) and subsequent centrifugation. The substrate consumption and product formation were analyzed by reversed-phase HPLC under conditions described previously (37). The NADH amounts used for the reduction of maleylacetate and 2-chloromaleylacetate, respectively, were determined by recording the absorption at 340 nm over 30 min of a reaction mixture (1 ml, 37°C) containing 50 mM Tris-HCl (pH 7.5), 0.2 mM NADH, 0.08 mM maleylacetate or 2-chloromaleylacetate, and 3.5 U of maleylacetate reductase (MacA) purified from E. coli.
Nucleotide sequence accession number.
The sequence data reported in this article will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AF030176.
RESULTS AND DISCUSSION
The cloned maleylacetate reductase MacA is distantly related to its proteobacterial counterparts but an efficient catalyst of 2-chloromaleylacetate conversion.
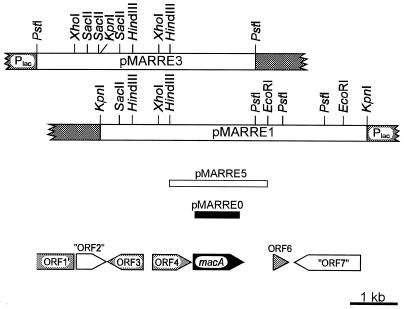
With primers NH2-3 and HOM, which were designed assuming a considerable sequence similarity between proteobacterial and rhodococcal maleylacetate reductases, it was possible to amplify, from genomic DNA of R. opacus 1CP as template, a fragment with the expected size of ca. 980 bp. Labeling of this fragment and its use as a probe allowed the isolation of three recombinant plasmids, pMARRE1, pMARRE2, and pMARRE3 (Table 1; Fig. 1). Sequence analysis of the inserts of pMARRE1 and -3 revealed four complete open reading frames, designated macA, ORF3, ORF4, and ORF6, as well as one incomplete open reading frame, designated ORF1′ (Fig. 1). In addition, we detected two regions with homology to entries in the database which comprised parts of different open reading frames and which were thus designated “ORF2” and “ORF7,” respectively.
FIG. 1.
Restriction and genetic map of the inserts of the original plasmids pMARRE1 and pMARRE3 carrying R. opacus 1CP DNA in the multiple cloning site of pBluescript II SK(+) (vector segments are indicated by shading). The insert of subclone pMARRE5 is shown, as is the PCR product (black) cloned to give pMARRE0, which was used as a digoxigenin-labeled probe for cloning. The positions and orientations of open reading frames are indicated by arrows below the restriction maps. The insert of pMARRE2 (not shown) extends from the XhoI site of pMARRE1 ca. 7 kb to the right.
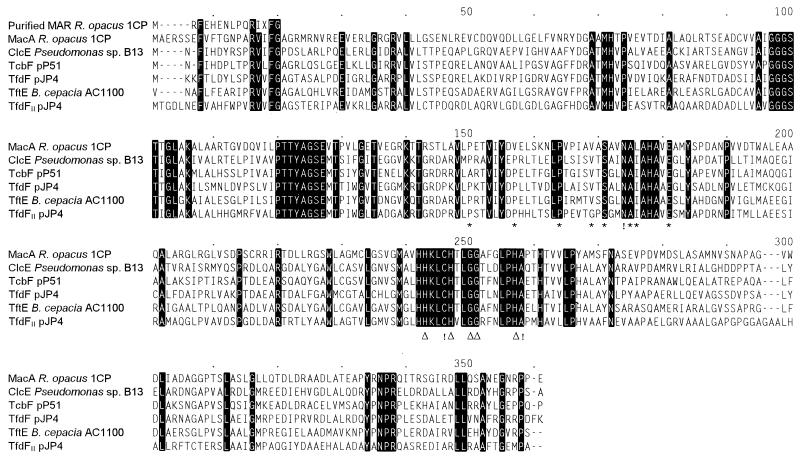
The 1,074-bp maleylacetate reductase gene macA (for maleylacetate conversion) starts at position 3311 of our sequence, is preceded by a putative ribosome binding site, and encodes a protein of 357 amino acids with a calculated molecular mass of 37,798 Da, which is similar to the molecular masses of previously purified maleylacetate reductases (23, 37). The predicted gene product shows significant similarities with known maleylacetate reductases: in the alignment in Fig. 2, MacA shares 43.5% identical positions with ClcE of Pseudomonas sp. strain B13, 42.6% with TcbF of pP51, 42.1% with TfdF of pJP4, 42.6% with TfdFII of pJP4, and 44.3% with TftE of Burkholderia cepacia AC1100. With each other, the known proteobacterial maleylacetate reductases share somewhat higher ratios of identical positions (45.8 to 59.4%). Corresponding to the homology of maleylacetate reductases to type III (iron-containing) alcohol dehydrogenases (46), signature sequences of this protein family could also be detected in MacA of R. opacus 1CP (Fig. 2). However, the sequence similarity to various type III alcohol dehydrogenases tested, i.e., less than 30% identical residues, was considerably lower than to the other maleylacetate reductases.
FIG. 2.
Multiple sequence alignment of various maleylacetate reductases as calculated by the CLUSTAL program. Amino acids identical in all sequences are highlighted. Numbers above the sequences refer to positions in the alignment, not in a single sequence. Asterisks indicate one pattern typical for type III (iron-containing) alcohol dehydrogenases, [AP]-x(6)-[STAP]-x(5,6)-P-x(4)-[AIV]-x-[GST]-x(2)-D-[AI]-[LIVM]-x(4)-E (Prosite entry PDOC00059; last update, November 1995). A second pattern (Δ) typical for this enzyme family could be important for binding ferrous iron: H-x(2)-[SAE]-[HY]-x(2)-[SGA](2)-x(5)-H-G. Interesting differences from these patterns are marked by exclamation marks. Accession numbers and references for the published sequences (order as in the alignment): AF019038 (21), M57629 (45), M35097 (29), U19883 (8), U16782 (44). MAR, maleylacetate reductase.
After subcloning of macA into the expression vector pET11a*, yielding pMARRE10, induction by IPTG resulted in an extract with a specific maleylacetate reductase activity of 0.72 U/mg. By comparison, the same strain, but with just the vector, under inducing conditions showed an activity of less than 0.02 U/mg. Thus, although according to SDS-electrophoretic analysis most of the MacA protein occurred in an insoluble form (even after some optimization of the inducing conditions), this activity proves that a functional gene had been cloned.
Cell extracts prepared from the overexpressing E. coli cells after induction with IPTG were used for the purification of MacA. The enzyme turned out to be very difficult to purify (the Red and the Blue Sepharose steps being especially detrimental), and in eight steps only 13-fold purification with a 0.07% yield was achieved. The final preparation on an SDS-polyacrylamide gel showed one major band of ca. 38 kDa, in agreement with the mass predicted for MacA.
The purified MacA preparation converted maleylacetate and 2-chloromaleylacetate with similar apparent Km values (100 and 120 μM, respectively) and with similar kcat values (1,210 and 718 min−1, respectively). Since the MacA preparation might have contained inactivated protein, the absolute kinetic constants of a fully active protein might differ from those determined. However, the relative values for the two substrates are less likely to be influenced by inactivation. During conversion of 2-chloromaleylacetate, maleylacetate was detected by HPLC as an intermediate. In addition, the NADH consumption with 2-chloromaleylacetate was twice as high as with maleylacetate. These results showed that MacA of R. opacus, like the maleylacetate reductases of Pseudomonas sp. strain B13 (ClcE) (22), Pseudomonas aeruginosa RHO1 (28), and R. eutropha JMP134 (TfdFII) (37), is able not only to convert 2-chloromaleylacetate with a relatively high turnover rate and affinity but also to bring about a reductive dehalogenation of 2-chloromaleylacetate to maleylacetate. Thus, it appears that the dehalogenation is catalyzed by an enzyme which is (i) relatively distant from other maleylacetate reductases and (ii) apparently not part of a gene cluster dedicated to chlorocatechol conversion (see below). These findings support the hypothesis that no special adaptation of maleylacetate reductases is necessary to effect dehalogenation, but that halide elimination either is the result of hydride addition in the normal reaction mechanism (24), which is a reaction gratuitously catalyzed by the enzyme, or is not enzyme catalyzed at all but instead is the result of a spontaneous decomposition of the initial reaction product.
Indications for the occurrence of different maleylacetate reductases in R. opacus 1CP.
A shoulder in the elution profile during anion-exchange chromatography of cell extracts had previously indicated that there might be more than one maleylacetate reductase induced in 4-chlorophenol-grown cells of R. opacus 1CP (27). To investigate whether the protein encoded by macA is in fact induced during growth of strain 1CP with 4-chlorophenol, we attempted to purify the maleylacetate reductases present under these conditions. However, the enzyme responsible for the shoulder or second activity peak during anion-exchange chromatography on a Q Sepharose High Performance column appeared to be unstable under the conditions used and could not be isolated. Only the enzyme responsible for the first peak could be further enriched by using a Blue Sepharose column. Active fractions from this chromatography on an SDS-polyacrylamide gel had a major band in a position corresponding to the molecular mass of other maleylacetate reductases. This band after excision from a Western blot was subjected to N-terminal amino acid sequencing and yielded the sequence MRFEHENLPQRIXFG. As shown in Fig. 2, on the one hand this sequence is clearly distinct from the predicted MacA sequence, but on the other hand four of the five amino acids conserved among the other maleylacetate reductases were also present in the partially purified maleylacetate reductase, thus proving that a maleylacetate reductase, not a contaminating protein, had been sequenced. The result that the maleylacetate reductase (partially) purified from 4-chlorophenol-grown cells of R. opacus 1CP has an N-terminal sequence distinct from that of the predicted MacA supports the hypothesis, derived from anion-exchange chromatography, that strain 1CP has at least two different maleylacetate reductases. Whether, however, MacA is in fact the enzyme which gave rise to the second activity peak or shoulder during the chromatography or whether MacA represents yet a third isoenzyme, not induced during growth with 4-chlorophenol, is not clear.
MacA is a maleylacetate reductase not encoded by a major gene cluster specialized on catabolism of chloroaromatic compounds.
Sequence characterization of the cloned fragment (Fig. 1) revealed most upstream of macA an incomplete open reading frame (ORF1′) and a region comprising several partial reading frames (“ORF2”), the products of which have significant sequence similarity to transposition-related proteins of IS117 or of IS1237 and IS402, respectively. ORF3, which might code for a protein homologous to flavin adenine dinucleotide synthetase, appears to be transcribed divergently from macA and thus not part of the same operon. This conclusion may also be drawn for ORF6, since it is relatively distant from macA (666 bp) and seems to code for a transcription factor-related protein, as well as for a downstream region (“ORF7”) which, when translated in different frames, shows sequence similarity to p-aminobenzoate synthases.
Thus, ORF4 (positions 2455 to 3264 of our sequence) is the only one which is likely to be cotranscribed with macA: it is relatively close (46 bp upstream) and apparently transcribed in the same direction. By comparison of ORF4 to database entries, the predicted gene product was shown to share significant similarity with members of the family of short-chain (pyridine nucleotide-linked) dehydrogenases/reductases (19). Among the proteins with relatively high sequence similarity (ca. 28 to 33% identical positions) were 3-oxoacyl-(acyl carrier protein) reductases, tropinone reductases of solanaceous plants, hydroxysteroid dehydrogenases, and cis-1,2-dihydroxy-3,4-cyclohexadiene-1-carboxylate dehydrogenases. The functional divergence of these oxidoreductases as well as their limited sequence similarity to the ORF4 product did not allow any clear-cut prediction with respect to the actual function of the latter. If ORF4 is, in fact, cotranscribed with macA, one might assume its gene product to be an oxidoreductase which either forms or further converts maleylacetate. The pnpD gene of the maleylacetate forming 4-hydroxymuconic semialdehyde dehydrogenase (40) has recently been sequenced from 4-nitrophenol-utilizing Pseudomonas fluorescens ENV2030 and was shown to code for a protein of 494 amino acids with sequence similarity to aldehyde dehydrogenases (4). Since the predicted ORF4 product is only 269 amino acids long and related to different enzymes, it is unlikely to be similar to PnpD.
Whatever the function of the product of ORF4 turns out to be, macA, unlike other known maleylacetate reductase genes, appears not to be part of a specialized gene cluster encoding all enzymes between two major catabolic hubs, like chlorocatechols and 3-oxoadipate. Instead, the presumed ORF4-macA gene cluster could be assumed to code for only a small part of a major pathway or to have some auxiliary function. A similar situation may be found for maleylacetate reductase genes located on proteobacterial chromosomes (36).
ACKNOWLEDGMENTS
We are indebted to H.-J. Knackmuss for providing excellent facilities and for stimulating discussions. We thank D. Eulberg for chromosomal DNA from R. opacus 1CP and S. Lakner for help with some of the experiments. Thanks are due to R. Schmid and H. Atomi, Institut für Technische Biochemie, for making it possible for us to use the automated sequencer and to S. Bürger for performing the sequencing. We are grateful to S. R. Kaschabek and W. Reineke for providing dienelactone and chlorodienelactone and to H. Weber for N-terminal sequencing.
This work was supported by grants from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Alting-Mees M A, Sorge J A, Short J M. pBluescriptII: multifunctional cloning and mapping vectors. Methods Enzymol. 1992;216:483–495. doi: 10.1016/0076-6879(92)16044-k. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 4.Bang S-W, Zylstra G J. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Cloning and sequencing of the hydroquinone 1,2-dioxygenase, γ-hydroxymuconic semialdehyde dehydrogenase, maleylacetate reductase genes from Pseudomonas fluorescens ENV2030, abstr. Q-383; p. 519. [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Chapman P J, Ribbons D W. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J Bacteriol. 1976;125:985–998. doi: 10.1128/jb.125.3.985-998.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Aquila R T, Bechtel L J, Videler J A, Eron J J, Gorczyca P, Kaplan J C. Maximizing sensitivity and specificity of PCR by preamplification heating. Nucleic Acids Res. 1991;19:3749. doi: 10.1093/nar/19.13.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daubaras D L, Hershberger C D, Kitano K, Chakrabarty A M. Sequence analysis of a gene cluster involved in metabolism of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepacia AC1100. Appl Environ Microbiol. 1995;61:1279–1289. doi: 10.1128/aem.61.4.1279-1289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daubaras D L, Saido K, Chakrabarty A M. Purification of hydroxyquinol 1,2-dioxygenase and maleylacetate reductase: the lower pathway of 2,4,5-trichlorophenoxyacetic acid metabolism by Burkholderia cepacia AC1100. Appl Environ Microbiol. 1996;62:4276–4279. doi: 10.1128/aem.62.11.4276-4279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duxbury J M, Tiedje J M, Alexander M, Dawson J E. 2,4-D metabolism: enzymatic conversion of chloromaleylacetic acid to succinic acid. J Agric Food Chem. 1970;18:199–201. doi: 10.1021/jf60168a029. [DOI] [PubMed] [Google Scholar]
- 11.Eulberg D, Golovleva L A, Schlömann M. Characterization of catechol catabolic genes from Rhodococcus erythropolis 1CP. J Bacteriol. 1997;179:370–381. doi: 10.1128/jb.179.2.370-381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eulberg D, Lakner S, Golovleva L A, Schlömann M. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J Bacteriol. 1998;180:1072–1081. doi: 10.1128/jb.180.5.1072-1081.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eulberg D, Kourbatova E M, Golovleva L A, Schlömann M. Evolutionary relationships between chlorocatechol catabolic enzymes from Rhodococcus opacus 1CP and their counterparts in proteobacteria: sequence divergence and functional convergence. J Bacteriol. 1998;180:1082–1094. doi: 10.1128/jb.180.5.1082-1094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feigel B J, Knackmuss H-J. Syntrophic interactions during degradation of 4-aminobenzenesulfonic acid by a two species bacterial culture. Arch Microbiol. 1993;159:124–130. doi: 10.1007/BF00250271. [DOI] [PubMed] [Google Scholar]
- 15.Frantz B, Chakrabarty A M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci USA. 1987;84:4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 17.Gorlatov S N, Maltseva O V, Shevchenko V I, Golovleva L A. Degradation of chlorophenols by a culture of Rhodococcus erythropolis. Mikrobiologiya. 1989;58:802–806. . (Microbiology 58:647–651.) [Google Scholar]
- 18.Jain R K, Dreisbach J H, Spain J C. Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by an Arthrobacter sp. Appl Environ Microbiol. 1994;60:3030–3032. doi: 10.1128/aem.60.8.3030-3032.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jörnvall H, Persson B, Krook M, Atrian S, González-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 20.Kasberg T, Daubaras D L, Chakrabarty A M, Kinzelt D, Reineke W. Evidence that operons tcb, tfd, and clc encode maleylacetate reductase, the fourth enzyme of the modified ortho pathway. J Bacteriol. 1995;177:3885–3889. doi: 10.1128/jb.177.13.3885-3889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasberg T, Seibert V, Schlömann M, Reineke W. Cloning, characterization, and sequence analysis of the clcE gene encoding the maleylacetate reductase of Pseudomonas sp. strain B13. J Bacteriol. 1997;179:3801–3803. doi: 10.1128/jb.179.11.3801-3803.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaschabek S R, Reineke W. Maleylacetate reductase of Pseudomonas sp. strain B13: dechlorination of chloromaleylacetates, metabolites in the degradation of chloroaromatic compounds. Arch Microbiol. 1992;158:412–417. doi: 10.1007/BF00276301. [DOI] [PubMed] [Google Scholar]
- 23.Kaschabek S R, Reineke W. Degradation of chloroaromatics: purification and characterization of maleylacetate reductase from Pseudomonas sp. strain B13. J Bacteriol. 1993;175:6075–6081. doi: 10.1128/jb.175.19.6075-6081.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaschabek S R, Reineke W. Maleylacetate reductase of Pseudomonas sp. strain B13: specificity of substrate conversion and halide elimination. J Bacteriol. 1995;177:320–325. doi: 10.1128/jb.177.2.320-325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larway P, Evans W C. Metabolism of quinol and resorcinol by soil pseudomonads. Biochem J. 1965;95:52. p. [Google Scholar]
- 26.Latus M, Seitz H-J, Eberspächer J, Lingens F. Purification and characterization of hydroxyquinol 1,2-dioxygenase from Azotobacter sp. strain GP1. Appl Environ Microbiol. 1995;61:2453–2460. doi: 10.1128/aem.61.7.2453-2460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maltseva O V, Solyanikova I P, Golovleva L A, Schlömann M, Knackmuss H-J. Dienelactone hydrolase from Rhodococcus erythropolis 1CP: purification and properties. Arch Microbiol. 1994;162:368–374. [Google Scholar]
- 28.Müller D, Schlömann M, Reineke W. Maleylacetate reductases in chloroaromatic-degrading bacteria using the modified ortho pathway: comparison of catalytic properties. J Bacteriol. 1996;178:298–300. doi: 10.1128/jb.178.1.298-300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perkins E J, Gordon M P, Caceres O, Lurquin P F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990;172:2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajeevan M S, Bassett C L. Flexibility in the boiling method of extracting plasmid DNA directly from cell culture. BioTechniques. 1994;16:376–380. [PubMed] [Google Scholar]
- 31.Reineke W. Microbial degradation of halogenated aromatic compounds. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 319–360. [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schell U, Seibert V, Vollmer M, Schlömann M. TfdF—a second, plasmid-encoded maleylacetate reductase of Alcaligenes eutrophus JMP134 (pJP4), abstr. P413. Bioengineering. 1994;10(2):83. [Google Scholar]
- 34.Schlömann M. Evolution of chlorocatechol catabolic pathways. Conclusions to be drawn from comparisons of lactone hydrolases. Biodegradation. 1994;5:301–321. doi: 10.1007/BF00696467. [DOI] [PubMed] [Google Scholar]
- 35.Schlömann M, Schmidt E, Knackmuss H-J. Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J Bacteriol. 1990;172:5112–5118. doi: 10.1128/jb.172.9.5112-5118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seibert V, Schlömann M. Cloning, sequencing and purification of maleylacetate reductase from Alcaligenes eutrophus 335. Biospektrum (special issue) 1996. p. 123. , abstr. PE143. [Google Scholar]
- 37.Seibert V, Stadler-Fritzsche K, Schlömann M. Purification and characterization of maleylacetate reductase from Alcaligenes eutrophus JMP134(pJP4) J Bacteriol. 1993;175:6745–6754. doi: 10.1128/jb.175.21.6745-6754.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seibert V, Golovleva L A, Schlömann M. Cloning of maleylacetate reductase genes from Alcaligenes eutrophus JMP289 and Rhodococcus erythropolis 1CP. Biospektrum (special issue) 1995. p. 57. , abstr. PA013. [Google Scholar]
- 39.Solyanikova I P, Maltseva O V, Vollmer M D, Golovleva L A, Schlömann M. Characterization of muconate and chloromuconate cycloisomerase from Rhodococcus erythropolis 1CP: indications for functionally convergent evolution among bacterial cycloisomerases. J Bacteriol. 1995;177:2821–2826. doi: 10.1128/jb.177.10.2821-2826.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spain J C, Gibson D T. Pathway for biodegradation of p-nitrophenol in a Moraxella sp. Appl Environ Microbiol. 1991;57:812–819. doi: 10.1128/aem.57.3.812-819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stolz A, Knackmuss H-J. Degradation of 2,4-dihydroxybenzoate by Pseudomonas sp. BN9. FEMS Microbiol Lett. 1993;108:219–224. doi: 10.1111/j.1574-6968.1993.tb06102.x. [DOI] [PubMed] [Google Scholar]
- 42.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 43.Tsitko, T. V., G. M. Zaitsev, A. G. Lobanok, and M. S. Salkinoja-Salonen. Tolerance of Rhodococcus opacus strains to aromatic and aliphatic solvents and the role of cellular fatty acids in the adaptation. Submitted for publication.
- 44.van der Meer, J. R. 1996. Direct submission to the databases. Accession no. U16782.
- 45.van der Meer J R, Eggen R I L, Zehnder A J B, de Vos W M. Sequence analysis of the Pseudomonas sp. strain P51 tcb gene cluster, which encodes metabolism of chlorinated catechols: evidence for specialization of catechol 1,2-dioxygenases for chlorinated substrates. J Bacteriol. 1991;173:2425–2434. doi: 10.1128/jb.173.8.2425-2434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Meer J R, de Vos W M, Harayama S, Zehnder A J B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vollmer M D, Stadler-Fritzsche K, Schlömann M. Conversion of 2-chloromaleylacetate in Alcaligenes eutrophus JMP134. Arch Microbiol. 1993;159:182–188. doi: 10.1007/BF00250280. [DOI] [PubMed] [Google Scholar]
- 48.Zaborina O, Latus M, Eberspächer J, Golovleva L A, Lingens F. Purification and characterization of 6-chlorohydroxyquinol 1,2-dioxygenase from Streptomyces rochei 303: comparison with an analogous enzyme from Azotobacter sp. strain GP1. J Bacteriol. 1995;177:229–234. doi: 10.1128/jb.177.1.229-234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]