Abstract
Mutants of Escherichia coli unable to synthesize a functional pyruvate formate-lyase (PFL) are severely impaired in their capacity to grow by glucose fermentation. In a functional complementation assay designed to isolate the pfl gene from Clostridium butyricum, we fortuitously identified a gene that did not encode a PFL but nonetheless was able to complement the phenotypic defects caused by an E. coli pfl mutation. The clostridial gene encoded a basic 14.5-kDa protein (TcbC) which, based on amino acid similarity and analysis of immediately adjacent DNA sequences, was part of a transposase exhibiting extensive similarity to the product of the site-specific transposon Tn554 from Staphylococcus aureus. Our studies revealed that the clostridial TcbC protein activated the transcription of the E. coli tdcABCDEFG operon, which encodes an anaerobic l-threonine-degradative pathway. Normally, anaerobic synthesis of the pathway is optimal when E. coli grows in the absence of catabolite-repressing sugars and in the presence of l-threonine. Although anaerobic control of pathway synthesis was maintained, TcbC alleviated glucose repression. One of the products encoded by the tdc operon, TdcE, has recently been shown to be a 2-keto acid formate-lyase (C. Heßlinger, S. A. Fairhurst, and G. Sawers, Mol. Microbiol. 27:477–492, 1998) that can accept pyruvate as an enzyme substrate. Here we show that TdcE is directly responsible for the restoration of fermentative growth to pfl mutants.
Pyruvate formate-lyase (PFL) is a glycyl radical enzyme that catalyzes the nonoxidative dissimilation of pyruvate to acetyl coenzyme A (acetyl-CoA) and formate when Escherichia coli grows anaerobically (for a review, see reference 15). The 170-kDa homodimeric PFL enzyme is interconverted between inactive and active forms. Activation of PFL to the radical-bearing species occurs only anaerobically and is catalyzed by an iron-sulfur protein called PFL-activating enzyme. Apart from inactive PFL, the other substrates in the reaction are S-adenosylmethionine and dihydroflavodoxin.
The free radical in PFL is located directly on the polypeptide backbone at Gly-734 (37). Consequently, the active enzyme species is extremely susceptible to dioxygen (37). Exposure to dioxygen results in irreversible inactivation through specific scission of the C-terminal portion of the polypeptide chain between Ser-733 and Gly-734 (37). This scission event results in the appearance of 82- and 3-kDa fragmentation products which can be readily identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Since only one radical is present per homodimer (15, 37), oxygenolytic cleavage of the polypeptide results in the appearance of a characteristic doublet that can be used as a diagnostic tool to identify the existence of active PFL molecules in the anaerobic cell. Only the full-length polypeptide is observed on Western blots of extracts derived from mutants unable to synthesize a functional PFL-activating enzyme (11, 15, 37).
To prevent irreversible damage to the enzyme when E. coli shifts from an anaerobic to an aerobic growth regimen, conversion of the active PFL enzyme back to the inactive, oxygen-stable form occurs. This reaction is catalyzed by the trifunctional AdhE enzyme (16).
A recent study has identified a second enzyme in E. coli with PFL activity (11). This enzyme, TdcE, is encoded by part of the multicistronic tdcABCDEFG operon, whose products form an anaerobic pathway that degrades l-threonine and l-serine to propionate and acetate, respectively, with concomitant generation of ATP (9, 11, 13). TdcE functions as a 2-keto acid formate-lyase, converting 2-ketobutyrate to propionyl-CoA and formate or pyruvate to acetyl-CoA and formate. Like PFL, TdcE is a glycyl radical enzyme, and the proteins have 82% amino identity (11). Moreover, introduction of the protein-based radical into TdcE is catalyzed by PFL-activating enzyme.
Expression of the tdc operon is very complex, being affected by at least five transcription factors (6, 8, 10, 41). Induction of operon expression occurs anaerobically and in the absence of catabolite-repressing sugars, such as glucose. The global transcription factor cyclic AMP (cAMP) receptor protein (CRP) provides the principal control of operon expression, with the LysR-like TdcA protein acting as an upstream regulator, possibly responding to l-threonine levels in the growth medium (8).
PFL enzyme activity has also been detected in a number of anaerobes, including Clostridium butyricum and Clostridium pasteurianum (35, 38). In contrast to the catabolic function PFL assumes in the enterobacteria, it has been proposed that PFL has an anabolic function in C. butyricum, providing formate for C1 metabolism (35). To understand the physiological function of the PFL protein of C. butyricum, we decided to attempt to isolate the corresponding pfl gene. During these studies, we serendipitously discovered a gene from C. butyricum whose product (TcbC) induced the synthesis of the TdcE protein in E. coli. This paper describes the identification and characterization of the TcbC protein.
MATERIALS AND METHODS
Strains and growth conditions.
All strains used in this work are listed in Table 1. Strains were grown in either Luria-Bertani (LB) medium, TYEP-buffered rich medium (TYEP contains, per liter, 10 g of Bacto Tryptone, 5 g of yeast extract, and 100 mM potassium phosphate [pH 6.5]), or the minimal medium described previously (11). Glucose was added to a final concentration of 20 mM in all anaerobic cultures. Aerobic cultures were grown in vigorously shaking conical flasks filled to a maximum of 10% of their volumes, while anaerobic growth of cultures occurred either in 200- or 500-ml stoppered bottles filled to the top or in crimp-sealed serum bottles filled with an atmosphere of dinitrogen and dihydrogen (99:1) according to the method of Balch and Wolfe (2). Monitoring of the specific incorporation of [35S]methionine into polypeptides with the T7 polymerase/promoter system (34) was performed according to the method of Leinfelder et al. (20). C. butyricum was cultivated under strict anaerobic conditions in serum bottles containing 20 ml of thioglycolate bouillon (Merck). Antibiotics were used at the following final concentrations: ampicillin, 50 mg liter−1; chloramphenicol, 15 mg liter−1; and tetracycline, 15 mg liter−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac)U169 ptsF25 deoC1 relA1 flbB530 rpsL150 λ− | 5 |
| FM420 | Like MC4100 but recA56 | 43 |
| RM202 | Like MC4100 but Δpfl-25 Ω(pfl::cat pACYC184) Δ(srl-recA)306::Tn10 | 32 |
| RM221 | Like MC4100 but Δ(pfl-act) Δ(srl-recA)306::Tn10 | 11 |
| RM226 | Like MC4100 but Δpfl-25 Ω(pfl::cat pACYC184) ΔtdcE | 11 |
| 234M11 | Like MC4100 but Δact Ω(act::cat pACYC184) Δ(srl-recA)306::Tn10 | 31 |
| JM109 | Δ(lac-proAB) recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 | 42 |
| W3110 | F− IN(rrnD-rrnE)1 rph-1 λ− | 18 |
| W3110pfl | Like W3110 but Δpfl-25 Ω(pfl::cat pACYC184) | This work |
| K38 | phoA tonA ampF | 22 |
| C. butyricum DSM-552 | Type strain | German Culture Collection, Braunschweig, Germany |
| Plasmids | ||
| pBR322 | Tetr Ampr | 4 |
| pT7-4 | Ampr, T7Φ10 | 34 |
| pT7-5 | Ampr, T7Φ10 | 34 |
| pT7-6 | Ampr, T7Φ10 | 34 |
| pGP1-2 | Kmr, T7 gene 1 (RNA polymerase) | 34 |
| pMU10 | Like pBR322, tcbA′ tcbB+ tcbC+ | This work |
| pMU1011 | Like pBR322, tcbC+ | This work |
| pMU1012 | Like pMU1011 but mutated in tcbC | This work |
| pMU1013 | Like pMU1011 but mutated in tcbC | This work |
| pT7-1011 | Like pT7-5 but tcbC+ | This work |
| pT7-1014 | Like pT7-1011 but with insert in the opposite orientation relative to the T7Φ10 promoter | This work |
Construction of a C. butyricum chromosomal DNA library.
Chromosomal DNA was prepared as described by Ausubel et al. (1). Purified DNA (10 μg) was partially digested with Sau3A, and DNA fragments between 3 and 8 kb in size were isolated after agarose gel electrophoresis and ligated into BamHI-digested pBR322 (29). The ligated DNA mixture was used to transform E. coli JM109. Approximately 10,000 ampicillin-resistant colonies were washed from the agar plates with 10 ml of LB medium and transferred to a sterile 250-ml flask. An additional 10 ml of LB medium was added, and the culture was incubated aerobically for 2 h at 37°C. Plasmid DNA was isolated and after treatment with RNase was resuspended in 1 ml of Tris-EDTA buffer (29). Transformation of 2 μl (50 to 100 ng) of this plasmid DNA into strain RM202 yielded approximately 50,000 ampicillin-resistant colonies.
Screening for clones capable of complementing an E. coli pfl mutant.
Strain RM202 (ΔfocA-pfl) was transformed with an aliquot of the C. butyricum chromosomal DNA gene library and plated on LB agar containing 50 μg of ampicillin ml−1. Plates were incubated anaerobically at 37°C for 24 h. Each plate was overlaid with a mixture held at 45°C and containing 20 mM sodium pyruvate, 5 mg of benzyl viologen (BV) ml−1, and 25 mM potassium phosphate (pH 7.0). Molten agarose (0.4%, wt/vol) was included in the mixture to solidify the overlay (23). Colonies which were unable to synthesize active PFL were small and remained colorless, while the wild-type strain produced large colonies that became dark violet after being overlaid.
DNA manipulations.
Work with recombinant DNA was carried out according to the methods of Sambrook et al. (29).
Analysis and subcloning of the DNA insert from pMU10.
The 3,617-bp DNA insert of plasmid pMU10, which derived from the chromosome of C. butyricum, was sequenced completely on both strands (30). Appropriately designed oligonucleotides were used to complete the sequence. Plasmid pMU1011 was created by deleting the 3.09-kb EcoRI fragment from pMU10 and religating the remaining vector DNA. pMU1012 was created in a similar manner by deleting the 3.74-kb HindIII fragment from pMU10 and religating the residual vector DNA. Plasmid pMU1013 was constructed by digesting pMU1012 with HindIII, filling in the protruding 5′ ends with Klenow polymerase and deoxynucleoside triphosphates, and religating the vector DNA. Plasmid pT7-1011 was constructed by cloning the 900-bp EcoRI-BamHI fragment from pMU10 into EcoRI-BamHI-digested expression vector pT7-5. The same insert was also cloned in the inverse orientation with respect to the T7Φ10 promoter of pT7-6, yielding pT7-1014.
Preparation of cell extracts and determination of PFL enzyme activity.
All steps were performed at 4°C unless otherwise indicated. Desalted crude extracts from approximately 0.5 to 1.0 g (wet weight) of cells were prepared as described by Kaiser and Sawers (14). PFL enzyme activity was determined as described by Knappe and Blaschkowski (17) with [14C]formate (Amersham) (specific radioactivity, 1.48 to 2.22 GBq mmol−1). This assay measures the acetyl-CoA-dependent conversion of formate to pyruvate. Both the activating reaction and the conversion of formate to pyruvate were performed at 30°C in an anaerobic chamber. Assays were performed by mixing extracts (150 μg of protein) prepared from strains that were phenotypically PFL+ ACT− and PFL− ACT+ (ACT is the PFL-activating enzyme). Measurement of 2-ketobutyrate formate-lyase in these extract mixtures was performed according to the method of Heßlinger et al. (11). Assays were found to be reproducible to within 10% of the mean, and activity was linear with respect to time (up to 60 min) and protein concentration.
l-Threonine deaminase assays.
l-Threonine deaminase was assayed in crude extracts by a coupled optical assay, essentially as described by Phillips and Wood (27). Cell pellets (0.5 g) were resuspended in 10 mM potassium phosphate buffer (pH 8.0) containing 1 mM dithiothreitol, 1 mM Pefabloc (Pentapharm AG, Basel, Switzerland), and 10 μg of DNase/ml and were broken by two passages through a French press at 16,000 lb/in2 (110 MPa). After clarification of the supernatant by centrifugation at 15,000 × g for 20 min, threonine deaminase activity was determined in an assay mixture containing 50 mM potassium phosphate buffer (pH 8.0), 5 mM dithiothreitol, 0.4 mM NADH, and 25 μg of rabbit muscle l-lactate dehydrogenase (Sigma) in a final volume of 0.18 ml. The reaction was started by adding 20 μl of l-threonine to a final concentration of 20 mM. Assays were performed at 22°C. Specific activity is given as micromoles of NADH oxidized per minute per milligram of protein.
Other methods.
Total RNA was isolated from cultures grown to mid-exponential phase, and primer extension analysis was carried out as described by Sawers and Böck (33). Analysis of the tcbC promoter was performed with oligonucleotide Tcb-1 (5′-CCTTTTGTACTTCCAGCCACC-3′), and the sequence ladder was generated with pMU10 DNA. Analysis of the tdc operon promoter was performed with oligonucleotide GS-1 (5′-GCAGCCGAGCCGATAGAACC-3′), and the sequence ladder was generated with pD1 (11). The protein concentration was determined by the method of Lowry et al. (21). SDS-PAGE of proteins was performed as described previously (19), and Western blotting was carried out according to the method of Towbin et al. (36). Anti-E. coli PFL antiserum was diluted 1,500-fold before use, and the antigen-antibody complex was visualized either by using 125I-labelled protein A and autoradiography or with the ECL kit (Amersham) used exactly as recommended by the manufacturer.
Nucleotide sequence accession number.
The nucleotide sequence for the 3,617-bp DNA insert of plasmid pMU10 is available under accession no. Z29084.
RESULTS
Functional complementation of an E. coli pfl deletion mutant.
Mutants unable to synthesize an active PFL grow very poorly in rich medium under fermentative growth conditions (14). Moreover, since these strains do not produce formate, they are incapable of inducing the synthesis of the formate-hydrogen lyase pathway unless formate is supplied exogenously (28). Strains that synthesize a functional formate-hydrogen lyase pathway can be easily and rapidly identified by overlaying colonies of anaerobically cultivated E. coli strains with a solution containing pyruvate or formate and the redox indicator dye BV (23). We used this system to identify plasmids from a C. butyricum chromosomal DNA gene library that could restore both anaerobic growth after 24 h on TYEP plates containing 0.4% (wt/vol) glucose (TGYEP) and a BV+ phenotype to the E. coli Δpfl mutant RM202 (32). Approximately 10,000 transformants were screened, and five dark violet colonies were isolated. However, only one of these clones had the additional desired phenotype of restoring appreciable growth to the pfl mutant under fermentative conditions. Significantly, no complementation was observed in a pfl act double null mutant (RM221), indicating that a PFL-like enzyme was responsible for the phenotype. In liquid minimal medium, MC4100 had a specific growth rate of 1.02 h−1 when grown anaerobically, while the pfl mutant RM202 showed no discernible growth and the complemented mutant had a specific growth rate of 0.18 h−1. The plasmid isolated from the complemented mutant was termed pMU10.
Nucleotide sequence analysis of plasmid pMU10.
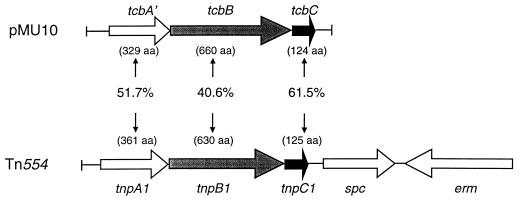
The complete 3,617-bp nucleotide sequence of the pMU10 insert was determined on both strands. The G+C content of the DNA was approximately 28%, which is characteristic for C. butyricum. Analysis of the nucleotide sequence did not reveal an open reading frame with similarity to the sequence encoding PFL. Instead, two complete open reading frames (tcbB and tcbC; transposon from C. butyricum) and one incomplete open reading frame (tcbA′) were identified (Fig. 1). The stop codon of the tcbA gene and the initiation codon of the tcbB gene overlap, while three base pairs separate the stop codon of the tcbB gene from the initiation codon of the tcbC gene. The products of all three genes show high similarity to polypeptides encoded by the Staphylococcus aureus transposon Tn554 (Fig. 1) (24). Southern blotting experiments with the insert from pMU10 revealed a single copy of the tcb genes on the chromosome of C. butyricum (data not shown).
FIG. 1.
Plasmid pMU10 encodes proteins with similarity to proteins encoded by transposon Tn554 from S. aureus. The organization of the genes present on the 3.9-kb insert of pMU10 is shown, together with the percentages of similarity their products exhibit to proteins encoded by transposon Tn554 from S. aureus (24). aa, amino acids.
Restoration of a PFL+ phenotype by pMU10 results from induction of the anaerobic synthesis of the E. coli TdcE enzyme.
The fact that a BV+ phenotype was conferred by pMU10 indicates that the transformants must have recovered the ability to make endogenous formate and strongly suggests that a PFL or PFL-like enzyme is responsible. Furthermore, no complementation by pMU10 was observed in strain RM221 with a deletion of the act gene, confirming this supposition.
Since none of the three putative tcb gene products encoded by pMU10 is likely to function as a PFL enzyme, the possibility that one or more of the gene products encoded by pMU10 may induce the synthesis of an E. coli enzyme which can functionally replace PFL was considered. To test this hypothesis, we performed PFL enzyme assays in which [14C]formate was converted to [14C]pyruvate with acetyl-CoA as a substrate. Due to the extreme sensitivity of the radical-bearing species of PFL to oxygen, the activation and assay of PFL and, by implication, TdcE were performed anaerobically by mixing crude extracts of appropriate strains as described in Materials and Methods and by Kaiser and Sawers (14). This test demonstrated that mixing an extract of an act mutant with an extract from a pfl mutant restored a PFL enzyme activity which defined the wild-type level of PFL. No enzyme activity was detectable when the mixture lacked a functional PFL-activating enzyme. Mixing an extract from a pfl mutant and an extract from a pfl act double null mutant carrying pMU10 resulted in a PFL activity that was approximately 8% of that observed in a PFL+ strain. Again, this activity was completely dependent on the presence of a functional PFL-activating enzyme. This low activity is in accord with the partial restoration of anaerobic growth in minimal medium (see above). Specifically, the levels of enzyme activity (in nanomoles of [14C]formate converted per minute per milligram of protein) of extracts of several strains transformed with pMU10 were as follows (PFL and ACT phenotypes are shown in parentheses): 234M11 (PFL+ ACT−) plus RM202 (PFL− ACT+), 610; 234M11 (PFL+ ACT−) plus RM221 (PFL− ACT−), <1; RM202 (PFL− ACT+) plus RM221/pMU10, 42; and RM221 (PFL− ACT−) plus RM221/pMU10, <1.
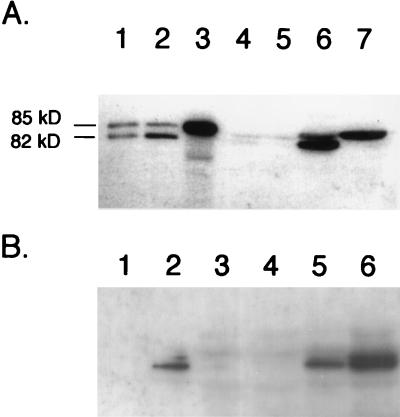
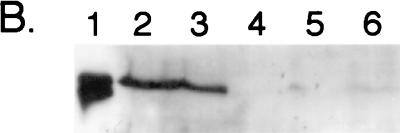
Western blot analysis with anti-PFL antiserum and the crude extracts used in determining PFL enzyme activity revealed that pfl deletion mutants transformed with pMU10 exhibit two strong cross-reacting polypeptides (Fig. 2A, lane 6). Examination of lane 4 in Fig. 2A reveals that cross-reacting polypeptides with similar mobilities were present at very low levels in the control strain. The two polypeptides in extracts of RM202/pMU10 migrated with mobilities slightly different from those of PFL and its specific oxygenolytic fragmentation product (Fig. 2A; compare lanes 1 and 2 with lane 6). Only the slower-migrating polypeptide was observed in an extract of RM221 (pfl act) bearing pMU10 (Fig. 2A, lane 7), which was also observed for PFL in an act mutant (Fig. 2A, lane 3). These findings show a strong correlation with the PFL enzyme activity data and suggest that the new cross-reacting polypeptide is a glycyl radical enzyme, which is converted to the active, radical-bearing species by PFL-activating enzyme.
FIG. 2.
Analysis of polypeptides induced by the presence of plasmid pMU10 and which cross-react with anti-E. coli PFL antibodies. Crude extracts from various E. coli strains were separated in 7.5% (wt/vol) polyacrylamide gels according to the method of Laemmli (19), and after transfer to nitrocellulose membranes, the polypeptides were challenged with anti-E. coli PFL antibodies. (A) Influence of a mutation in the gene encoding PFL-activating enzyme on migration of cross-reacting polypeptides. The positions of the intact PFL polypeptide (85 kDa) and the specific fragmentation product (82 kDa), derived from exposure of the radical form of the enzyme to oxygen, are shown. Lane 1, MC4100 (wild type) grown aerobically (100 μg of protein); lane 2, MC4100 grown anaerobically (7.5 μg of protein); lane 3, 234M11 grown anaerobically (7.5 μg of protein); lane 4, RM202 (Δpfl)/pBR322 grown anaerobically (150 μg of protein); lane 5, RM221 (Δpfl Δact)/pBR322 grown anaerobically (150 μg of protein); lane 6, RM202/pMU10 grown anaerobically (150 μg of protein); lane 7, RM221/pMU10 grown anaerobically (150 μg of protein). (B) Western blot demonstrating that induction of TdcE synthesis by pMU10 occurs anaerobically. Lane 1, MC4100 grown aerobically (7.5 μg of protein); lane 2, MC4100 grown anaerobically (7.5 μg of protein); lane 3, RM202 (Δpfl)/pBR322 grown aerobically (100 μg of protein); lane 4, RM202/pBR322 grown anaerobically (100 μg of protein); lane 5, RM202/pMU10 grown aerobically (100 μg of protein); lane 6, RM202/pMU10 grown anaerobically (100 μg of protein).
Western blotting of extracts from RM202 (Δpfl)/pMU10 grown in the presence and absence of oxygen demonstrated that induction of the new protein occurred primarily during anaerobiosis (Fig. 2B). This result, together with the facts that the PFL enzyme activity of this new enzyme is dependent on PFL-activating enzyme, that it cross-reacts with anti-PFL antibodies, and that its mobility during SDS-PAGE is slightly different from that of PFL, strongly suggests that this polypeptide corresponds to the recently discovered glycyl radical enzyme TdcE, which is a component of the anaerobically induced l-threonine degradation pathway of E. coli (11).
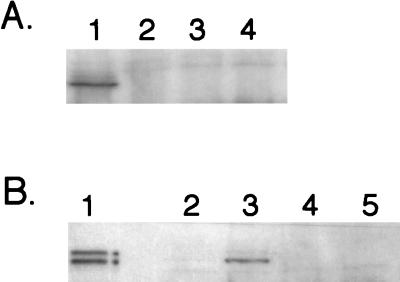
In order to test this hypothesis, we examined the pMU10-induced synthesis of the PFL-like enzyme in strain RM226 (11), which has deletions of both pfl and tdcE. Western blot analysis with anti-PFL antibodies revealed that no cross-reacting polypeptide was induced in extracts of RM226 (Fig. 3A). It is notable that in the extract from RM202/pMU10 more than 50% of the cross-reacting polypeptide was in the fragmented form. We currently have no explanation for this phenomenon.
FIG. 3.
Western blot demonstrating that synthesis of the polypeptide induced by pMU10 which cross-reacts with anti-PFL antibodies is abolished in a ΔtdcE mutant. Crude extracts were prepared from strains grown anaerobically, and polypeptides were separated in 7.5% (wt/vol) polyacrylamide gels containing SDS. Approximately 100 μg of protein was applied in each lane unless otherwise indicated. (A) Lane 1, RM202 (Δpfl)/pMU10; lane 2, RM202/pBR322; lane 3, RM226 (Δpfl ΔtdcE)/pMU10; lane 4, RM226/pBR322. (B) Lane 1, MC4100 (10 μg of protein); lane 2, RM202/pBR322; lane 3, RM202/pMU10; lane 4, W3110pfl/pBR322; lane 5, W3110pfl/pMU10.
To provide further proof that the polypeptide induced in the presence of pMU10 was indeed the TdcE protein, we analyzed whether it was synthesized in a pfl mutant derivative of W3110. In W3110, the tdcE gene is separated from the rest of the tdc operon by insertion of an IS5 element immediately 5′ of tdcE, which disrupts the operon and prevents tdcE expression (12). The results clearly show that no polypeptide was induced by pMU10 in W3110pfl, confirming that the PFL-like protein induced by pMU10 was the glycyl radical enzyme TdcE.
Finally, we determined 2-ketobutyrate formate-lyase activity (11) in the same mixture of extracts used to determine PFL activity. The activity determined for the extract mixture containing RM202 plus RM221/pMU10 was 51 nmol of NADH formed min−1 mg of protein−1, while no activity was detectable in the absence of PFL-activating enzyme.
The TcbC protein encoded by pMU10 is necessary and sufficient to induce E. coli TdcE synthesis.
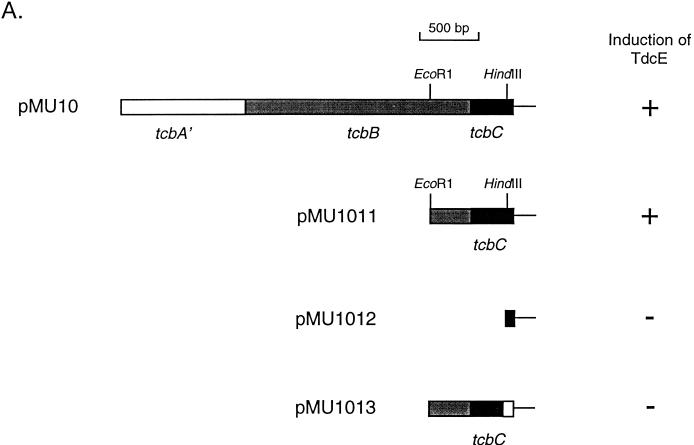
Various subclones of the DNA insert of pMU10 were constructed to determine which of the three tcb genes is responsible for anaerobic induction of E. coli TdcE enzyme synthesis (Fig. 4A). Plasmid pMU1011 has an 897-bp DNA insert that includes a small portion of the tcbB gene and the complete tcbC gene. TdcE was clearly induced in an extract derived from RM221 containing pMU1011 (Fig. 4B), indicating that neither tcbA′ nor tcbB is involved in induction of TdcE synthesis. Removal of 323 bp of the tcbC coding region (pMU1012) abolished induction of TdcE synthesis (Fig. 4B, lane 4). To demonstrate that the TcbC protein and not a portion of the DNA sequence itself on pMU1011 was responsible for enzyme induction, a frameshift mutation was introduced into the tcbC gene by filling in the HindIII restriction site of pMU1011 to create pMU1013 (see Materials and Methods and Fig. 4A). Plasmid pMU1013 was unable to induce TdcE synthesis when transformed into RM221 (Fig. 4B).
FIG. 4.
The tcbC gene of pMU10 induces TdcE synthesis. (A) The subclones derived from pMU10 are shown. The ability and inability of the constructs to induce TdcE enzyme synthesis and to complement an E. coli pfl mutant are indicated by plus and minus signs, respectively. The white box at the end of the tcbC gene in pMU1013 indicates that the gene sequence is out of frame. (B) Western blot showing the ability of the various pMU10 plasmid derivatives to induce anaerobic synthesis of TdcE. Lane 1, MC4100 (10 μg of protein); lane 2, RM221 (Δpfl Δact)/pMU10; lane 3, RM221/pMU1011; lane 4, RM221/pMU1012; lane 5, RM221/pMU1013; lane 6, RM221/pBR322.
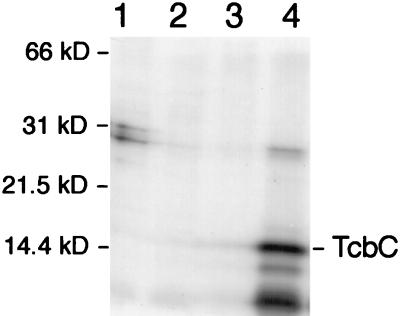
That the C. butyricum tcbC gene does indeed code for a protein product in E. coli was confirmed with the T7 polymerase/promoter system (34). TcbC migrated with a molecular mass of approximately 14.4 kDa, which is in excellent agreement with the molecular weight of 14,527 deduced from the primary sequence (Fig. 5).
FIG. 5.
Analysis of TcbC synthesis with the T7 polymerase/promoter system. Polypeptides were specifically labelled with [35S]methionine as described in Materials and Methods and were separated in a 12.5% (wt/vol) polyacrylamide gel containing SDS. Lane 1, K38/pGP1-2 containing pT7-4; lane 2, K38/pGP1-2 containing pT7-5; lane 3, K38/pGP1-2 containing pT7-6; lane 4, K38/pGP1-2 containing pT7-1011. The molecular masses of the standard proteins used are given.
Heterologous transcription of the tcbC gene in E. coli is independent of the cellular oxygen status.
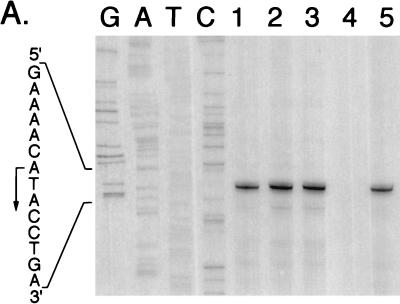
A primer extension analysis of tcbC transcription was performed to determine whether the gene is transcribed from its own promoter and whether transcription is affected by aerobic or anaerobic growth of E. coli. Total RNA was isolated from RM221/pMU10 grown aerobically and from RM221/pMU10, RM221/pMU1011, and RM221/pMU1012 grown anaerobically, and the 5′ end of the tcbC transcript was determined with oligonucleotide Tcb-1, as described in Materials and Methods. Transcription initiated from an adenosine residue 184 bp upstream of the presumptive GTG translation start site of the tcbC gene (Fig. 6). The levels of transcription were similar under aerobic and anaerobic growth conditions. Moreover, while a transcript of the same length as and an intensity similar to those of pMU10 was also observed for pMU1011, no transcript for pMU1012 was observed (Fig. 6A). This result is consistent with these DNA sequences being absent in pMU1012. Analysis of transcription from pMU1013, which carries the frameshift mutation in the tcbC gene, gave the same result as that observed for pMU10 and pMU1011 (Fig. 6A).
FIG. 6.
Primer extension analysis of tcbC gene transcription in the heterologous host E. coli. (A) Total RNA was isolated from E. coli RM221 (Δpfl Δact) containing different mutant derivatives of the pMU10 plasmid. A total of 15 μg of RNA was used for each primer extension experiment. The nucleotide sequence shown was determined with the same oligonucleotide primer with pMU10 as DNA template as that used in the primer extension reactions. Lane 1, RM221/pMU10 grown aerobically; lane 2, RM221/pMU10 grown anaerobically; lane 3, RM221/pMU1011 grown anaerobically; lane 4, RM221/pMU1012 grown anaerobically; lane 5, RM221/pMU1013 grown anaerobically. The arrow indicates the 5′ end of the tcbC mRNA transcript. (B) The location in the DNA sequence of the 5′ end of the tcbC transcript is indicated by the shorter arrow. Putative −10 and −35 RNA polymerase binding sites are underlined, and the first two amino acids of the TcbC protein are shown. The ochre termination codon of the preceding tcbB gene is boxed.
Examination of the DNA sequence upstream of the transcription initiation site of the tcbC gene revealed sequences which match four of the six consensus nucleotides of the E. coli −10 and −35 RNA polymerase recognition sequences (Fig. 6B).
TcbC activates transcription of the tdc operon promoter.
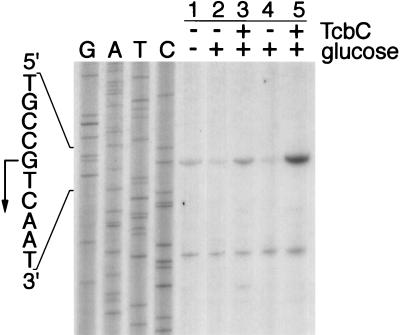
Recent studies have shown that the tdcE gene is cotranscribed with the tdcA, -B, -C, -D, -F, and -G genes (11). Transcript mapping studies were therefore undertaken to determine whether the clostridial TcbC protein activated transcription of the tdc promoter. Expression of the tdc operon is induced anaerobically and is catabolite repressed (41). Primer extension analysis of the transcript generated by wild-type strain FM420 grown in the absence or presence of glucose revealed that transcription levels were lower in the presence of glucose (Fig. 7; compare lanes 1 and 2). The 5′ end of the transcript was three nucleotides downstream of that identified in a previous study (9). It should also be noted that the absence of glucose is not the condition for anaerobic growth under which tdc operon expression is optimal (41).
FIG. 7.
Transcription from the E. coli tdc operon promoter is induced anaerobically by TcbC. Primer extension reactions were carried out with total RNA isolated from anaerobically grown cells. Lane 1, FM420/pBR322 grown in TYEP; lane 2, FM420/pBR322 grown in TGYEP; lane 3, FM420/pMU10 grown in TGYEP; lane 4, RM202/pBR322 grown in TGYEP; lane 5, RM202/pMU10 grown in TGYEP. The absence and presence of the indicated components in the assay are signified by minus and plus signs, respectively. The location of the transcription initiation site of the tdc operon is shown with an arrow. The lower transcript resulted from interaction of the oligonucleotide with an unidentified RNA transcript and was used as a convenient internal control to ensure that equivalent amounts of RNA were used.
Introduction of pMU10 into FM420 resulted in high levels of transcription even in the presence of glucose (Fig. 7, lane 3). Analysis of the pfl mutant RM202 transformed with pBR322 demonstrated that glucose strongly repressed tdc transcription, whereas in the presence of pMU10, transcription from the tdc promoter was no longer repressed by glucose (Fig. 7, lanes 4 and 5). The lower transcript seen in Fig. 7 does not originate from the tdc operon but results from fortuitous hybridization of the oligonucleotide with a distinct mRNA species that is unaffected by the presence of the TcbC protein.
Measurement of the activity of the l-threonine deaminase enzyme, encoded by the tdcB gene (7), revealed that the activity in extracts derived from RM202/pBR322 grown anaerobically in the presence of glucose was 0.06 μmol of NADH oxidized min−1 mg of protein−1, while the activity in RM202/pMU10 grown under the same conditions was 0.64 μmol of NADH oxidized min−1 mg of protein−1. The latter activity is in a range similar to that observed previously in crude extracts of E. coli K-12 strains grown anaerobically in the presence of cAMP (27). These results correlate well with the observed induction of tdc expression by pMU10 (compare lanes 4 and 5 in Fig. 7). The activity of TdcB in RM202/pMU10 grown aerobically was 0.08 μmol of NADH oxidized min−1 mg of protein−1, which confirms that pMU10 induces expression of tdc only anaerobically.
DISCUSSION
The data presented here corroborate and provide independent support for the findings of Heßlinger et al. (11) that TdcE has a substrate spectrum similar to that of PFL and can partially substitute for PFL in anaerobic catabolism. Indeed, PFL and TdcE have the same activating enzyme. Anaerobic TdcE synthesis was not induced by the TcbC protein (encoded by pMU10) to levels equivalent to those observed for PFL. Our estimates based on densitometry of Western blots indicate that TdcE attained levels of 5 to 10% of those seen for PFL. This is in good agreement with the PFL enzyme activity determined for TdcE, making the assumption that PFL and TdcE have similar Km values for formate in this enzyme assay, and with the fact that complementation of the pfl mutation was only partial. It will be of interest to determine whether, when overproduced to levels similar to those observed with PFL, TdcE can replace PFL completely.
Synthesis of TdcE normally is induced in the absence of both oxygen and catabolite-repressing sugars. E. coli strains harboring TcbC synthesize TdcE even in the presence of glucose, but only under anaerobic conditions. Our studies have shown that TcbC functions at the transcriptional level, inducing expression of the tdcABCDEFG operon without altering the site of transcription initiation.
How is TcbC able to abrogate glucose repression and activate tdc transcription? Although TcbC is encoded by part of a presumptive transposon from C. butyricum, activation of E. coli tdc transcription is unlikely to be the result of a transposition event, since by analogy with Tn554 of S. aureus, transposition of the C. butyricum transposon is expected to require the TcbA and TcbB proteins (24, 25). Neither of these proteins is required for complementation of the pfl mutant. Also, a transposition event would not be expected to affect tdc expression only anaerobically. Furthermore, our transcript mapping data did not indicate any alteration in the location of the start site of the tdc transcript, which might be anticipated if a transposon were to insert in the neighborhood of the promoter. Rather, we believe that TcbC may be functioning directly by interacting with the tdc transcription machinery. As mentioned above, the deduced amino acid sequences of the tcbA, tcbB, and tcbC products are highly similar to the corresponding TnpA, TnpB, and TnpC proteins (24, 25). Insertion of Tn554 occurs with high frequency, is site specific, and is always in one orientation. Mutational analyses have demonstrated that TnpC determines the orientation specificity of the element (3). Although tnpC mutants are still capable of transposition, the frequency is reduced by approximately 2 orders of magnitude compared with the wild-type element. Insertion of Tn554 at its unique chromosomal site, att554, requires a core hexanucleotide recognition sequence for the transposase complex (26). Thus, in S. aureus, it is likely that TnpC may be involved in binding the transposase complex to the att554 recognition sequence. By analogy, TcbC may be a DNA binding protein that fortuitously recognizes a sequence in the tdc operon promoter region with similarity to the C. butyricum transposase att site. This, however, does not provide an explanation of how TcbC may activate transcription. Assuming that TcbC functions in a direct manner (rather than indirectly, for example, by affecting cAMP levels in glucose-grown cells), it is improbable that TcbC simply overrides transcriptional control of the tdc promoter, since anaerobic transcriptional regulation is maintained. Hence, it is anticipated that TcbC-induced transcriptional activation still depends on Fnr and ArcA (6). Since anaerobic regulation of tdc expression is strongly dependent on the cAMP-CRP complex (41), it is conceivable that TcbC is capable of enhancing tdc expression in the presence of glucose either by substituting for cAMP-CRP in transcriptional activation or by improving interaction of the complex with the CRP binding site at the tdc promoter at lower cAMP levels. The former is unlikely, since TcbC has no similarity to CRP at the primary sequence level. However, TcbC is a very basic protein (pI = 10.32), and although it has no obvious DNA binding domain, it nevertheless has some features of histone-like proteins. Expression of tdc has been shown to be influenced both by the topological constraint of the DNA (40) and by integration host factor, which functions in concert with cAMP-CRP (39, 41). It is therefore possible that TcbC facilitates the binding of CRP to the tdc promoter by influencing DNA topology. The consequences of this could be either that the cAMP-CRP complex has a higher affinity for the CRP binding site in the tdc promoter or that the efficiency of transcriptional activation through improved CRP-RNA polymerase interaction is increased. We are currently conducting in vitro studies with purified TcbC to address this possibility.
ACKNOWLEDGMENTS
We thank Ray Dixon for his critical comments on the manuscript and Andrea Sawers for expert technical assistance.
This work was supported by the BBSRC via a grant-in-aid to the John Innes Centre and the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1987. [Google Scholar]
- 2.Balch W E, Wolfe R S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol. 1976;32:781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastos M, Murphy E. Transposon Tn554 encodes three products required for transposition. EMBO J. 1988;7:2935–2941. doi: 10.1002/j.1460-2075.1988.tb03152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Cloning and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 5.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chattopadhyay S, Wu Y, Datta P. Involvement of Fnr and ArcA in anaerobic expression of the tdc operon of Escherichia coli. J Bacteriol. 1997;179:4868–4873. doi: 10.1128/jb.179.15.4868-4873.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta P, Goss T J, Omnass J R, Patil R V. Covalent structure of biodegradative threonine dehydratase of Escherichia coli: homology with other dehydratases. Proc Natl Acad Sci USA. 1987;84:393–397. doi: 10.1073/pnas.84.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganduri Y L, Sadda S R, Datta M W, Jambukeswaran R K, Datta P. TdcA, a transcriptional activator of the tdcABC operon of Escherichia coli, is a member of the LysR family of proteins. Mol Gen Genet. 1993;240:395–402. doi: 10.1007/BF00280391. [DOI] [PubMed] [Google Scholar]
- 9.Goss T J, Schweizer H P, Datta P. Molecular characterization of the tdc operon of Escherichia coli K-12. J Bacteriol. 1988;170:5352–5359. doi: 10.1128/jb.170.11.5352-5359.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagewood B T, Ganduri Y L, Datta P. Functional analysis of the tdcABC promoter of Escherichia coli: roles of TdcA and TdcR. J Bacteriol. 1994;176:6214–6220. doi: 10.1128/jb.176.20.6214-6220.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heßlinger C, Fairhurst S A, Sawers G. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades l-threonine to propionate. Mol Microbiol. 1998;27:477–492. doi: 10.1046/j.1365-2958.1998.00696.x. [DOI] [PubMed] [Google Scholar]
- 12.Heßlinger, C., and G. Sawers. The tdcE gene in Escherichia coli strain W3110 is separated from the rest of the tdc operon by insertion of IS5 elements. DNA Sequence, in press. [DOI] [PubMed]
- 13.Hobert E H, Datta P. Synthesis of biodegradative threonine dehydratase in Escherichia coli: role of amino acids, electron acceptors, and certain intermediary metabolites. J Bacteriol. 1983;155:586–592. doi: 10.1128/jb.155.2.586-592.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaiser M, Sawers G. Pyruvate formate-lyase is not essential for nitrate respiration by Escherichia coli. FEMS Microbiol Lett. 1994;117:163–168. doi: 10.1111/j.1574-6968.1994.tb06759.x. [DOI] [PubMed] [Google Scholar]
- 15.Kessler D, Knappe J. Anaerobic dissimilation of pyruvate. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 199–205. [Google Scholar]
- 16.Kessler D, Herth W, Knappe J. Ultrastructure and pyruvate formate-lyase radical quenching property of the multienzymic AdhE protein of Escherichia coli. J Biol Chem. 1992;267:18073–18079. [PubMed] [Google Scholar]
- 17.Knappe J, Blaschkowski H P. Pyruvate formate-lyase from Escherichia coli and its activation system. Methods Enzymol. 1975;41:508–518. doi: 10.1016/s0076-6879(75)41107-7. [DOI] [PubMed] [Google Scholar]
- 18.Kohara Y, Akiyama K, Isono K. The physical map of the whole Escherichia coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Leinfelder W, Forchhammer K, Zinoni F, Sawers G, Mandrand-Berthelot M-A, Böck A. Escherichia coli genes whose products are involved in selenium metabolism. J Bacteriol. 1988;170:540–546. doi: 10.1128/jb.170.2.540-546.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.Lyons L B, Zinder N D. The genetic map of the filamentous phage f1. Virology. 1972;49:45–60. doi: 10.1016/s0042-6822(72)80006-0. [DOI] [PubMed] [Google Scholar]
- 23.Mandrand-Berthelot M-A, Wee M K Y, Haddock B A. An improved method for identification and characterisation of mutants of Escherichia coli deficient in formate dehydrogenase activity. FEMS Microbiol Lett. 1978;4:37–40. [Google Scholar]
- 24.Murphy E, Huwyler L, Bastos M. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 1985;4:3357–3365. doi: 10.1002/j.1460-2075.1985.tb04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy E, Löfdahl S. Transposition of Tn554 does not generate a target duplication. Nature. 1984;307:292–294. doi: 10.1038/307292a0. [DOI] [PubMed] [Google Scholar]
- 26.Murphy E, Reinheimer E, Huwyler L. Mutational analysis of att554, the target of the site-specific transposon Tn554. Plasmid. 1991;26:20–29. doi: 10.1016/0147-619x(91)90033-s. [DOI] [PubMed] [Google Scholar]
- 27.Phillips A T, Wood W A. The mechanism of action of 5′-adenylic acid-activated threonine dehydratase. J Biol Chem. 1965;240:4703–4709. [PubMed] [Google Scholar]
- 28.Rossmann R, Sawers G, Böck A. Mechanism of regulation of the formate-hydrogen lyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol Microbiol. 1991;5:2807–2814. doi: 10.1111/j.1365-2958.1991.tb01989.x. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauter M, Sawers R G. Transcriptional analysis of the gene encoding pyruvate formate-lyase-activating enzyme of Escherichia coli. Mol Microbiol. 1990;4:355–363. doi: 10.1111/j.1365-2958.1990.tb00603.x. [DOI] [PubMed] [Google Scholar]
- 32.Sawers G, Böck A. Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K-12. J Bacteriol. 1988;170:5330–5336. doi: 10.1128/jb.170.11.5330-5336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawers G, Böck A. Novel transcriptional control of the pyruvate formate-lyase gene: upstream regulatory sequences and multiple promoters regulate anaerobic expression. J Bacteriol. 1989;171:2485–2498. doi: 10.1128/jb.171.5.2485-2498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabor S, Richardson C C. A bacteriophage T7 polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thauer R K, Kirschniawy F H, Jungermann K A. Properties and function of the pyruvate formate-lyase reaction in clostridia. Eur J Biochem. 1972;27:282–290. doi: 10.1111/j.1432-1033.1972.tb01837.x. [DOI] [PubMed] [Google Scholar]
- 36.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner A F V, Frey M, Neugebauer F A, Schäfer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci USA. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weidner G, Sawers G. Molecular characterization of the genes encoding pyruvate formate-lyase and its activating enzyme of Clostridium pasteurianum. J Bacteriol. 1996;178:2440–2444. doi: 10.1128/jb.178.8.2440-2444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y, Datta P. Integration host factor is required for positive regulation of the tdc operon of Escherichia coli. J Bacteriol. 1992;174:233–240. doi: 10.1128/jb.174.1.233-240.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y, Datta P. Influence of DNA topology on expression of the tdc operon in Escherichia coli K-12. Mol Gen Genet. 1995;247:764–767. doi: 10.1007/BF00290409. [DOI] [PubMed] [Google Scholar]
- 41.Wu Y, Patil R V, Datta P. Catabolite gene activator protein and integration host factor act in concert to regulate tdc operon expression in Escherichia coli. J Bacteriol. 1992;174:6918–6927. doi: 10.1128/jb.174.21.6918-6927.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 43.Zinoni F, Birkmann A, Stadtman T C, Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci USA. 1986;83:4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]