Abstract
Salmonella typhimurium apeR mutations lead to overproduction of an outer membrane-associated N-acetyl phenylalanine β-naphthyl ester-cleaving esterase that is encoded by the apeE gene (P. Collin-Osdoby and C. G. Miller, Mol. Gen. Genet. 243:674–680, 1994). This paper reports the cloning and nucleotide sequencing of the S. typhimurium apeE gene as well as some properties of the esterase that it encodes. The predicted product of apeE is a 69.9-kDa protein which is processed to a 67-kDa species by removal of a signal peptide. The predicted amino acid sequence of ApeE indicates that it is a member of the GDSL family of serine esterases/lipases. It is most similar to a lipase excreted by the entomopathogenic bacterium Photorhabdus luminescens. The Salmonella esterase catalyzes the hydrolysis of a variety of fatty acid naphthyl esters and of C6 to C16 fatty acid p-nitrophenyl esters but will not hydrolyze peptide bonds. A rapid diagnostic test reported to be useful in distinguishing Salmonella spp. from related organisms makes use of the ability of Salmonella to hydrolyze the chromogenic ester substrate methyl umbelliferyl caprylate. We report that the apeE gene product is the enzyme in Salmonella uniquely responsible for the hydrolysis of this substrate. Southern blot analysis indicates that Escherichia coli K-12 does not contain a close analog of apeE, and it appears that the apeE gene is contained in a region of DNA present in Salmonella but not in E. coli.
Mutations at the apeA locus of Salmonella typhimurium lead to loss of a periplasmic enzyme originally identified by its ability to hydrolyze the chromogenic substrate N-acetyl phenylalanine β-naphthyl ester (NAPNE) (17). Mutants with a reduced capacity for NAPNE hydrolysis are easily isolated by using this substrate to detect activity in situ in bacterial colonies growing on an agar surface. NAPNE is a good substrate for chymotrypsin, and ApeA was originally thought to be a protease, protease I (19, 20). Recent work indicates that the Escherichia coli apeA product is a thioesterase (4), and the gene is now designated tesA.
We have previously described the isolation of pseudorevertants of S. typhimurium apeA mutations that lead to restoration of the ability to hydrolyze NAPNE (6). These mutants overproduce a membrane-associated enzyme which is distinct from ApeA and all other known NAPNE hydrolases (12). The mutations affect a locus, apeR, that appears to encode a negative regulator of the transcription of the membrane hydrolase which is thought to be the product of the apeE gene (6). To further characterize this membrane hydrolase, we report the cloning and nucleotide sequence of the apeE gene and further characterization of the enzymatic activity of its product, ApeE.
MATERIALS AND METHODS
Bacterial strains.
Table 1 lists the S. typhimurium LT2 strains used in this project. Other strains used were Photorhabdus luminescens K122 (obtained from Barbara Dowds, St. Patrick’s College, County Kildare, Ireland), Pseudomonas aeruginosa K (obtained from David Nunn, University of Illinois), and derivatives of E. coli K-12.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid |
Genotype or description |
|---|---|
| Strains | |
| TN445 | apeA42 |
| TN478 | apeA42 apeR1 |
| TN925 | apeA42 apeR1 apeE::Tn5 |
| TN1379 | leuBCD485 |
| Plasmids | |
| pCM342 | 10-kb fragment inserted into BamHI site of pBR328 |
| pCM343 | 3.3-kb Sau3A fragment of pCM342 inserted into BamHI site of pBR328 |
Media and growth conditions.
S. typhimurium and E. coli strains were routinely grown at 37°C in Luria broth medium (Gibco BRL) aerated by shaking. P. luminescens was grown under the same conditions but at 28°C. Antibiotics were added as indicated in the following concentrations: ampicillin, 50 mg/ml; chloramphenicol, 15 mg/ml; and tetracycline, 25 mg/ml (18). Minimal (E min) soft agar overlays were prepared from E medium (30) with 0.8% Difco agar.
Isolation of plasmids carrying apeE.
Plasmids containing 8- to 15-kb fragments generated by Sau3A partial digestion of DNA from S. typhimurium TN1379 and inserted into the BamHI site of pBR328 were transformed into strain TN2540. To screen this library, a P22HT 12/4 int-3 lysate was made on the library and used to transduce TN445, an apeA apeE+ apeR+ strain, with selection for chloramphenicol resistance. These transductants were screened for NAPNE-hydrolyzing activity by overlaying the transduction plate with 2.5 ml of E min soft agar; after solidification, 10 ml of a mixture containing 0.1 M phosphate buffer (pH 6.8), 10 mg of Fast Garnet GBC (Sigma), and 2 mg of NAPNE in N,N-dimethylformamide (final concentration is 10%) was poured over the plate, and the plate was incubated at room temperature for about 1 min. The plates were then washed with sterile saline, and any NAPNE-staining clones were picked and restreaked.
Subcloning.
Subclones of pCM342 were obtained by partial digestion of the plasmid with Sau3A, ligation into the BamHI site of pBR328, and electroporation into E. coli DH5α. The transformants were stained for activity as described above and restreaked. Although DH5α is apeA+, it is easy to distinguish the dark red color of a colony containing an apeE plasmid from the pink color of the parental colony. The presence of an apeE plasmid in the red-staining strains was confirmed by staining sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gels for NAPNE activity after renaturation (see below).
DNA sequencing.
DNA sequencing was carried out with a Sequenase version 2.0 DNA sequencing kit (U.S. Biochemicals) according to manufacturer’s instructions. Compressed areas were resolved by automated cycle sequencing by the University of Illinois Biotechnology Center. The DNA was completely sequenced in both strands. Sequence alignment and analysis were carried out with the Wisconsin sequence analysis package (Genetics Computer Group [GCG]) and the Lasergene software DNAstar.
Southern hybridization.
DNA fragments were transferred to Immobilon N (Millipore) according to the manufacturer’s instructions, using a TransVac Blot apparatus (Hoefer) with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) as the transfer buffer. The DNA was cross-linked to the membrane by UV irradiation as specified by the instructions for the UV cross-linker apparatus.
Hybridization probes were generated by PCR using primers synthesized by the University of Illinois Biotechnology Center. The 2.1-kb apeE-specific probe was constructed by using primers apee3 (5′GCTCCATTATCTGCCTGT3′; bases 813 to 830) and apee17 (5′TAGCTCCAACTGACGAAAT3′; bases 2913 to 2931). The 429-bp probe specific for sequence upstream from apeE was constructed by first generating a 1.1-kb PCR product by using primers BamHIccw (5′CGAATTCATGCGTCCGGCGTAGA3′; located in vector sequence) and apee18 (5′CCGTTTCCGCCGACCCAGTGA3′; bases 1126 to 1106). This product was then cut with SmaI, and the 429-bp fragment (corresponding to bases 1 to 429) was purified by using a Geneclean II kit (Bio 101) according to manufacturer’s instructions. Probes were labeled by random priming according to manufacturer’s directions, using the Multiprime DNA labeling system (Amersham). Hybridization was carried out by using standard procedures (16) with 4× SSC in the prehybridization and hybridization solutions. Probes were hybridized overnight at 50°C and washed as follows: 1× SSC–1% SDS at room temperature for 1 min, 1× SSC–1% SDS at 50°C for 1 h, 0.5× SSC–1% SDS at 50°C for 1 h, and 0.1× SSC–0.2% SDS at 50°C for 1 h.
Preparation of cell extracts.
Bacteria were grown to an optical density at 600 nm of 0.6 to 0.8 and harvested by low-speed centrifugation at 4°C for 10 min. The cell pellets were washed twice with 50 mM Tris-HCl (pH 7.5) (Tris buffer) and resuspended in 1/50 the original culture volume of Tris buffer. The cells were disrupted by sonication (Branson Sonifier 250, microtip) for 1 min and spun at 40,000 × g for 40 min at 4°C. The supernatant was kept as crude cell extract and stored at −70°C. SDS membrane extracts were prepared by resuspension of the pellet in 1/50 the original culture volume of 3% SDS in 50 mM phosphate buffer (pH 8.0), incubation at 100°C for 10 min, and centrifugation at 40,000 × g for 40 min at room temperature. The supernatant was removed and designated SDS-soluble membrane extract. Triton X-100 membrane extracts were prepared by extracting the pellet with 1/50 the original culture volume of 2% Triton X-100 in 50 mM phosphate buffer (pH 8.0) (phosphate buffer). After a 30-min incubation at room temperature, the extract was diluted in half by addition of an equal volume of phosphate buffer and spun at 40,000 × g at 4°C for 40 min. The supernatant was designated 1% Triton-soluble membrane extract and was used for all enzyme assays unless indicated otherwise. This procedure extracts about 25 to 35% of the NAPNE-hydrolyzing activity present in membranes of apeR mutant strains. Extracts of membranes prepared from strains carrying apeE mutations contained no esterase activity.
Enzyme assays.
Hydrolysis of NAPNE was monitored spectrophotometrically essentially as described previously (20). The effects of inhibitors on hydrolysis rates were determined by mixing the inhibitor and enzyme, incubating the mixture for 30 min at room temperature, adding substrate, and monitoring the hydrolysis rate.
Gel electrophoresis.
Six percent nondenaturing polyacrylamide 0.75-mm slab gels were run at 4°C according to the method of Davis (7) except that the stacking gel was omitted. The gels were run at 100 V until the tracking dye entered the gel and then at 250 V until the dye reached the end of the gel. The gels were soaked in deionized water for 30 min and then placed in 100 ml of NAPNE stain solution (90 ml of 0.1 M phosphate buffer [pH 6.8] with 10 mg of Fast Garnet GBC [Sigma] and 10 ml of NAPNE solution [0.2 mg/ml in N,N-dimethylformamide]). The gel was soaked in stain for 5 to 20 min until bands were apparent. The effects of inhibitors (except diisopropylfluorophospate [DFP]) on NAPNE hydrolysis after gel electrophoresis were determined by soaking the gel in the inhibitor solution for 30 min at room temperature and then staining as described above. For DFP, the enzyme was incubated with the inhibitor for 60 min at room temperature and the resulting gel was stained for activity.
Gels were subjected to SDS-PAGE according to the method of Laemmli (15), with the following modifications. Samples were suspended in SDS loading buffer lacking a reducing agent and heated to 55°C for 2 min before loading. After electrophoresis, gels were renatured by soaking in a solution of 1% glycerol–1% Triton X-100–50 mM Tris (pH 7.5) for 30 min. The gels were then stained as described above for nondenaturing gels.
Tris-Tricine SDS-polyacrylamide gels were run according to the method of Schagger and von Jagow (23). Samples were mixed with SDS sample buffer and incubated at 100°C for 10 min before loading. These gels were then stained with Coomassie blue.
Electroblotting.
SDS-polyacrylamide gels were transferred to a ProBlot membrane (Applied Biosystems) with an Electroblot apparatus (Trans Blot) according to manufacturer’s instructions, using 10 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS; pH 11.0)–10% methanol as a transfer buffer.
N-terminal sequencing.
N-terminal sequence was determined by the University of Illinois Biotechnology Center, by using an Applied Biosystems (Perkin-Elmer) model 477A Protein/Peptide Sequencer with a model 120A on-line phenylthiodydantoin analyzer.
Nucleotide sequence accession number.
The DNA sequence presented here is accessible from the GenBank database under accession no. AF047014.
RESULTS AND DISCUSSION
Cloning of the apeE locus.
To isolate clones carrying the apeE gene, plasmids from a pBR328 chromosomal DNA library prepared from strain TN1379 (apeA+ apeE+ apeR+) were transduced into TN445 (apeA apeE+ apeR+) and TN925 (apeA apeE apeR), and the resulting colonies were screened to identify those able to hydrolyze NAPNE. All NAPNE-hydrolyzing colonies were tested by using nondenaturing PAGE followed by NAPNE staining to determine whether they contained ApeA (protease I) or ApeE (see Materials and Methods). A single isolate containing ApeE was identified. This isolate contained plasmid pCM342, which was characterized further. This plasmid was found in the TN445 (apeR+) background, and no plasmids carrying apeE were found in the apeR mutant strain. Subsequent experiments indicated that transfer of pCM342 to an apeR background produced small, slow-growing colonies, suggesting that this level of overproduction of ApeE is toxic. Restriction mapping showed that pCM342 contained an 8.3-kb insert. Sau3A partial digests of pCM342 were generated, cloned into pBR328, and transformed into E. coli DH5α. Screening these transformants for elevated NAPNE-hydrolyzing activity led to the isolation of pCM343, a plasmid with a 3.3-kb insert.
Nucleotide sequence of apeE.
The entire insert DNA in pCM343 was sequenced. An additional 281 bp of the pCM342 insert immediately adjacent to this sequence was also determined, for a total of 3,536 bp. The sequence contained an open reading frame (ORF) consistent with the size predicted for apeE, based on SDS-PAGE of the ApeE enzymatic activity (approximately 60 kDa [5]). This ORF (bp 759 to 2729) predicts a 69.9-kDa protein. The predicted N-terminal amino acid sequence (positions 1 to 25) resembles a signal peptide, and since ApeE is a membrane-associated activity, we expected that the Ala-X-Ala sequence at amino acids 23 to 25 might serve as a signal peptidase cleavage site. This prediction was confirmed by N-terminal sequence analysis of the mature protein, which showed that it carries an N-terminal amino acid sequence beginning with amino acid 26. The predicted molecular mass of the mature protein (67.3 kDa) is somewhat larger than that estimated from SDS-PAGE (60 kDa). The C-terminal region of the protein conforms strikingly to the pattern noted for other outer membrane proteins (25) which contain hydrophobic amino acids at positions 1, 3, 5, 7, and 9 from their C termini. The C-terminal amino acid of ApeE is phenylalanine, and hydrophobic amino acids are located at positions 3, 5, 7, and 9 from the C terminus.
A search of GenBank using the BLAST program turned up another protein with strong similarity to ApeE. This protein (LipI; GenBank accession no. P40601) is an extracellular lipase from P. luminescens, an entomopathogenic member of the family Enterobacteriaceae (31). These predicted proteins show 41% amino acid sequence identity and 62% similarity (Fig. 1). The two proteins are approximately the same length (656 [Salmonella] and 645 [Photorhabdus] amino acids for the unprocessed proteins), and regions of identity and strong similarity extend throughout the sequences. An ORF of unknown function located between the trpE and the trpGDC genes of Pseudomonas putida (10) has a product that also shows significant similarity to ApeE. Alignment of the two sequences using the GCG Gap program revealed 49.5% similarity and 29% identity. A number of conserved regions could be identified by aligning ApeE, LipI, and the Pseudomonas ORF product. Although E. coli does not appear to have an apeE gene (see below), it does contain an ORF (YHJY_ECOLI; accession no. P37663) (24) whose product has significant similarity to an approximately 200-amino-acid region at the C terminus of ApeE.
FIG. 1.
Comparison of P. luminescens lipase and S. typhimurium esterase amino acid sequences, generated by the GCG computer program Gap. The top sequence is that of the P. luminescens lipI (GenBank accession no. P40601) product, and the bottom sequence is that of the S. typhimurium apeE product. The row of asterisks above the sequence designates the signal peptide, and ↓ designates the first amino acid of the mature protein. The underlined sequence is the conserved motif characteristic of the GDSL family of esterases. A vertical line indicates sequence identity, a double dot represents very similar residues, and a single dot represents similar residues.
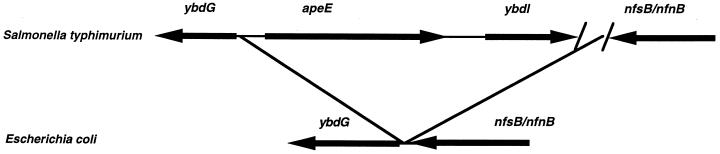
Further analysis of the sequence carried by pCM343 revealed two additional ORFs (Fig. 2). One of these is in the opposite strand and upstream from apeE extending to the end of the region sequenced. This ORF shows strong similarity to ybdG, an uncharacterized ORF in E. coli. This E. coli ORF is located immediately downstream from nfsB/nfnB (encoding a nitroreductase) of E. coli. Salmonella contains a homolog of nfsB/nfnB which is situated at approximately the same place in the Salmonella chromosome as in that of E. coli (32). The apeE gene had been previously mapped by genetic methods to this region of the chromosome (6). The other ORF lies downstream from apeE and in the same strand. This ORF (ybdI) shows significant similarity to the levR gene of Bacillus subtilis, which encodes a transcriptional regulator of the levanase operon of this organism (9). LevR is a member of a family of regulators of ς54-dependent promoters (26). No analog of this gene is found in E. coli. A comparison of the gene order in this region based on the E. coli and Salmonella sequences is shown in Fig. 2. It appears that both apeE and ybdI are part of a segment of DNA present in Salmonella but not in E. coli that is inserted (relative to E. coli) between the Salmonella homolog of ybdG and nfsB/nfsN. The inserted DNA extends at least to the apeE-distal end of the Salmonella DNA segment cloned into pCM342. This indicates that the insertion in Salmonella is at least 3 kb in length. Preliminary experiments in which a primer sequence taken from ybdI and another from the published S. typhimurium nfnB (32) sequence were used to PCR amplify S. typhimurium chromosomal DNA yielded an amplified fragment of approximately 6 kb. This implies that the insertion in S. typhimurium may be nearly 9 kb in length.
FIG. 2.
Comparison of the gene order near apeE in S. typhimurium and E. coli. apeE and ybdI are part of a segment of DNA present in Salmonella but not in E. coli that is believed to be inserted between the Salmonella homolog of E. coli ybdG and nfsB/nfsN.
apeE is not present in E. coli.
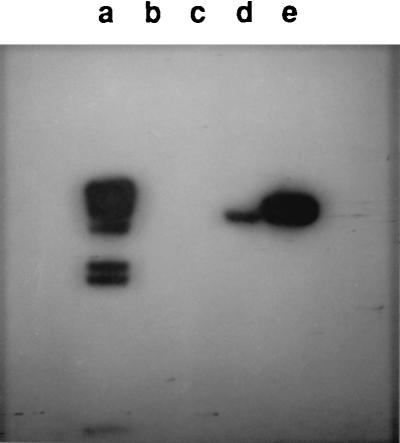
Although mutations that lead to overexpression of ApeE are easily isolated as NAPNE-staining pseudorevertants of S. typhimurium apeA mutations, E. coli apeA mutants do not appear to give rise to NAPNE-staining pseudorevertants (5). This result combined with the observation that apeE might be contained on a DNA segment present in Salmonella but not E. coli suggested that E. coli may not have an apeE homolog. To test this, Southern blot analysis was carried out with a probe containing only apeE coding sequence and a probe carrying the ybdG ORF known to be present in both organisms. The apeE-specific probe hybridized to both Salmonella genomic DNA and DNA from the plasmid pCM343 but not to E. coli DNA (Fig. 3). A probe consisting of the 5′ ORF hybridized to chromosomal DNA from both E. coli and S. typhimurium (data not shown). We conclude that the apeE gene is not present in E. coli K-12. The sequence of the E. coli genome which appeared after this work was complete confirms that this organism does not contain an apeE homolog.
FIG. 3.
Southern blot with apeE probe. DNA was digested with EcoRI, and hybridization was carried out with the apeE-specific probe described in Materials and Methods. Lanes: a, standards; b, E. coli DH5α; c, P. luminescens; d, S. typhimurium; e, pCM343.
Substrate specificity of apeE.
The ApeE activity was originally recognized by its hydrolysis of NAPNE. We have previously reported evidence suggesting that ApeE does not have proteolytic activity despite the apparent specificity of the enzyme for the amino acid residue in the ester substrate (ApeE hydrolyzes the Phe ester but not the corresponding Leu substrate [6]). To learn more about the enzyme’s specificity, we tested the ability of ApeE to hydrolyze a variety of chromogenic substrates (Table 2). Since none of the naphthylamides, nitroanilides, or peptides were hydrolyzed, we tentatively conclude that the enzyme will not cleave amide bonds. Clearly the enzyme is not specific for amino acid esters since naphthyl esters of short-chain fatty acids are rapidly cleaved. Indeed, the best substrates, naphthyl butyrate and naphthyl caproate, are hydrolyzed much more rapidly than the Phe derivatives. Many of the ester substrates that were not hydrolyzed are significantly more polar than those which were. Although naphthyl esters of lauric and palmitic acid were not hydrolyzed, p-nitrophenyl palmitate is a good substrate.
TABLE 2.
Substrate specificity of ApeEa
| Type | Substrates |
|---|---|
| Hydrolyzed | β-Naphthyl esters of N-acetyl, N-benzoyl, and N-benzyloxycarbonyl Phe and of caproic acid; α- and β-naphthyl esters of acetic, propionic, and butyric acids; p-nitrophenyl esters of all straight-chain fatty acids C6 to C16 |
| Not hydrolyzed | β-Naphthyl esters of N-acetyl Leu, N-methyl-N-toluene-p-sulfonyl Lys, and N-acetyl Arg and of lauric and palmitic acids; naphthol AS esters of propionic, phenylpropionic, and benzoic acids; p-nitrophenyl esters of N-acetyl Phe; β-naphthylamides of N-benzoyl Phe, Leu, Trp, and Leu-Phe; p-nitroanilide of N-acetyl Phe; peptidesb PheAsp, AspPhe, PheGly, Phe4, Phe5, and PheGlyGly |
Based on qualitative assays using Triton X-100 membrane extracts as described in Materials and Methods. Membranes from a wild-type strain, an ApeR− strain overproducing ApeE, and an ApeE− strain were compared. In all cases in which hydrolysis was observed, the overproducer strain showed the most rapid color development and the ApeE− strain showed no activity. The most rapidly hydrolyzed compounds were the α- and β-naphthyl esters of butyric and caproic acids. Naphthol AS, 3-hydroxy-2-naphthoic acid anilide.
Peptide hydrolysis was tested after electrophoresis of detergent extracts on nondenaturing polyacrylamide gels.
The P. luminescens lipase is able to hydrolyze Tween 80, a water-soluble oleic acid ester of a polyoxyalkylene derivative of sorbitan (31). A plate assay can be used to detect precipitation of liberated water-insoluble fatty acids, the product formed upon hydrolysis of Tween 80. This precipitation is visible as halos around the colonies. Salmonella strains TN1379, TN445, TN478, and TN925 were screened by using this plate assay for their abilities to hydrolyze Tween 80. After overnight incubation at 37°C, no precipitation was detectable with any of the Salmonella strains, but a Pseudomonas aeruginosa strain used as a positive control showed a large zone of precipitation. After overnight incubation at 4°C, however, a faint halo was apparent around S. typhimurium TN478 colonies but none of the other Salmonella strains. Since TN478 is the only strain in the group that overexpresses the ApeE esterase, this result suggests that ApeE cleaves Tween 80 but not as efficiently as the P. luminescens lipase. This could be due to a lower activity against this substrate or to the difference in localization of the two enzymes. The Photorhabdus enzyme is secreted into the culture medium, whereas the Salmonella activity is membrane bound.
The deduced amino acid sequence of ApeE indicates that ApeE is a member of the GDSL family of ester hydrolases (28). This family of enzymes is characterized by an active site Ser residue located in most cases very close to the N terminus within the sequence GDSL (amino acids 33 to 36 in the S. typhimurium sequence of Fig. 1). This sequence differs from the GXSXG sequence found in most esterases, and the GDSL family appears to represent a distinct subfamily of serine hydrolases. The group also displays several other blocks of sequence similarity, including blocks that are thought to contain the Asp and His residues of the serine hydrolase catalytic triad (28). Although all act as esterases/lipases, the family appears to contain activities with distinct and varying specificities. Most members of the family hydrolyze a variety of ester substrates, however, and a full range of substrates has not been tested with all members of the family. It is interesting that the thioesterase product of the tesA (apeA) gene is also a member of this family although it is not similar to ApeE outside the blocks of similarity noted above. An arylesterase of Vibrio mimicus which is quite similar to TesA also contains the GDSL sequence (3). ApeE is most closely related to the lipase produced by P. luminescens and more distantly related to a protein encoded by a Pseudomonas putida ORF (10) and to a lipase/acyltransferase from Aeromonas hydrophila (27). The Aeromonas enzyme not only hydrolyzes soluble esters, neutral lipids, and phospholipids but also acts as a specific acyltransferase (13, 21, 27). Clearly a more detailed characterization of the substrate specificity of ApeE is in order. We do not know whether ApeE is activated by a lipid-water interface as are classical lipases, nor have we characterized its ability to hydrolyze neutral or phospholipid substrates. ApeE clearly does not belong to the class of lipases which require a lipid-water interface for activity (14, 29) since it hydrolyzes soluble esters such as NAPNE and β-naphthyl butyrate at an appreciable rate. The apeE gene designation seems clearly inappropriate since the acetyl-phenylalanine naphthyl ester esterase activity of the gene product almost certainly has no physiological significance. It seems reasonable to delay such a name change, however, until a better understanding of the enzyme’s specificity and function allow assignment of a meaningful mnemonic.
Hydrolysis of MUCAP.
One method for identifying Salmonella spp. in the clinical laboratory involves the use of methylumbelliferyl caprylate (MUCAP), a substrate that fluoresces upon hydrolysis of the ester bond (1, 8, 11, 22). This substrate can be used to distinguish salmonellae from other bacteria, including Escherichia, Enterobacter, Yersinia, and Shigella spp. Colonies of Salmonella strains TN1379, TN445, TN478, and TN925 were tested for their abilities to hydrolyze MUCAP. All apeE+ strains fluoresced, but TN925, an apeE strain lacking the enzyme, did not fluoresce. In addition, E. coli DH5α containing plasmid pCM343 fluoresced, while the same E. coli strain with pBR328, the parent vector for pCM343, did not. These results indicate that the apeE gene product is responsible for the hydrolysis of MUCAP in salmonellae.
Inhibitors of ApeE.
The ability of various inhibitors to inhibit the hydrolysis of NAPNE by ApeE was tested either by spectrophotometric assays or by incubating the inhibitors with nondenaturing gels through which extracts containing the activity had been incubated prior to staining. Spectrophotometric assays showed that none of the following inhibitors had a significant effect (>20%) on ApeE activity: phenylmethylsulfonyl fluoride (3 mM), EDTA (0.1 M), p-chloromercuribenzoate (10 mM), iodoacetamide (1 mM), pepstatin A (0.25 mM), and β-mercaptoethanol (50 mM). In the gel activity stain assay, DFP (1 mM) showed strong inhibition although a faint band of activity could be observed after treatment with the inhibitor. We believe that this result indicates that ApeE is DFP sensitive. The residual activity may be a result of the failure of the inhibitor to fully inactivate the enzyme during the incubation time allowed, or it may represent a small fraction of the enzyme that is not sensitive to the inhibitor (2). Under the same conditions, none of the following had any observable effect: N-tosyl-l-lysine chloromethyl ketone (0.5 mM), N-tosyl-l-phenylalanine chloromethyl ketone (0.5 mM), eserine (1 mM), EDTA (100 mM), bis-p-nitrophenyl phosphate (1 mM), and N-ethylmaleimide (100 mM).
The ApeE esterase can be reactivated after SDS-PAGE.
When a Triton X-100 extract of whole membranes containing ApeE is subjected to SDS-PAGE and the resulting gel is incubated in a renaturation buffer (see Materials and Methods), renatured enzyme is easily detected (data not shown). Using the more sensitive β-naphthyl caproate substrate, activity can be observed after electrophoresis of either boiled or unboiled samples. The electrophoretic mobility of the activity in the unboiled samples is somewhat greater than that observed after boiling, suggesting that the enzyme is not completely unfolded by treatment with SDS at room temperature.
Other enzymatic properties.
When NAPNE was used as a substrate, the enzyme was found to have a pH optimum of approximately 8.0. Attempts to determine kinetic constants for this substrate were limited by its insolubility. No indication of saturation was observed at 0.15 mM NAPNE.
Physiological function of ApeE.
The data that we present provide few clues concerning the physiological function of the ApeE protein. It is clearly not required for growth. It is conceivable that it is involved in the catabolism of fatty acid esters, although it is apparently not regulated by cyclic AMP receptor protein (based on the absence of a cyclic AMP receptor protein binding site in the promoter region). Preliminary experiments indicate that an apeE+ strain but not an apeE mutant utilizes Tween 80 as a sole carbon source (6a). Perhaps elucidation of the nature of the regulatory gene which controls its transcription (apeR) will provide clues concerning ApeE’s physiological role.
ACKNOWLEDGMENT
This work was supported by grant AI10333 from the National Institute for Allergy and Infectious Diseases.
REFERENCES
- 1.Aguirre P M, Cacho J B, Folgueira L, Lopez M, Garcia J, Velasco A C. Rapid fluorescence method for screening Salmonella spp. from enteric differential agars. J Clin Microbiol. 1990;28:148–149. doi: 10.1128/jcm.28.1.148-149.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balls A K, Jensen E F. Stoichiometric inhibition of chymotrypsin. Adv Enzymol. 1952;13:321–343. doi: 10.1002/9780470122587.ch8. [DOI] [PubMed] [Google Scholar]
- 3.Chang R C, Chen J C, Shaw J F. Vibrio mimicus arylesterase has thioesterase and chymotrypsin-like activity. Biochem Biophys Res Commun. 1995;213:475–483. doi: 10.1006/bbrc.1995.2156. [DOI] [PubMed] [Google Scholar]
- 4.Cho H, Cronan J E., Jr “Protease I” of Escherichia coli functions as a thioesterase in vivo. J Bacteriol. 1994;176:1793–1795. doi: 10.1128/jb.176.6.1793-1795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collin-Osdoby P. Studies of a regulated outer membrane esterase from Salmonella typhimurium. Ph.D. thesis. Cleveland, Ohio: Case Western Reserve University; 1982. [Google Scholar]
- 6.Collin-Osdoby P, Miller C G. Mutations affecting a regulated, membrane-associated esterase in Salmonella typhimurium LT2. Mol Gen Genet. 1994;243:674–680. doi: 10.1007/BF00279577. [DOI] [PubMed] [Google Scholar]
- 6a.Conlin, C. A. Unpublished data.
- 7.Davis B J. Disc electrophoresis. II. Method and application to human serum proteins. Ann N Y Acad Sci. 1964;121:404. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 8.Dealler S F, Collins J, James A L. A rapid heat-resistant technique for presumptive identification of Salmonella on desoxycholate-citrate agar. Eur J Clin Microbiol Infect Dis. 1992;11:249–252. doi: 10.1007/BF02098090. [DOI] [PubMed] [Google Scholar]
- 9.Debarbouille M, Martin-Verstraete I, Klier A, Rapoport G. The transcriptional regulator LevR of Bacillus subtilis has domains homologous to both sigma 54- and phosphotransferase system-dependent regulators. Proc Natl Acad Sci USA. 1991;88:2212–2216. doi: 10.1073/pnas.88.6.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Essar D W, Eberly L, Crawford I P. Evolutionary differences in chromosomal locations of four early genes of the tryptophan pathway in fluorescent pseudomonads: DNA sequences and characterization of Pseudomonas putida trpE and trpGDC. J Bacteriol. 1990;172:867–883. doi: 10.1128/jb.172.2.867-883.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freydiere A M, Gille Y. Detection of salmonellae by using Rambach agar and by a C8 esterase spot test. J Clin Microbiol. 1991;29:2357–2359. doi: 10.1128/jcm.29.10.2357-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heiman C, Miller C G. Acylaminoacid esterase mutants of Salmonella typhimurium. Mol Gen Genet. 1978;164:57–62. doi: 10.1007/BF00267599. [DOI] [PubMed] [Google Scholar]
- 13.Hilton S, Buckley J T. Studies on the reaction mechanism of a microbial lipase/acyltransferase using chemical modification and site-directed mutagenesis. J Biol Chem. 1991;266:997–1000. [PubMed] [Google Scholar]
- 14.Kazlauskas R J. Elucidating structure-mechanism relationships in lipases: prospects for predicting and engineering catalytic properties. Trends Biotechnol. 1994;12:464–472. doi: 10.1016/0167-7799(94)90022-1. . (Erratum, 13:195, 1995.) [DOI] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 17.Miller C G, Heiman C, Yen C. Mutants of Salmonella typhimurium deficient in an endoprotease. J Bacteriol. 1976;127:490–497. doi: 10.1128/jb.127.1.490-497.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 19.Pacaud M, Sibilli S, Bras G. Protease I from Escherichia coli. Some physicochemical properties and substrate specificity. Eur J Biochem. 1976;69:141–151. doi: 10.1111/j.1432-1033.1976.tb10867.x. [DOI] [PubMed] [Google Scholar]
- 20.Pacaud M, Uriel J. Isolation and some properties of a proteolytic enzyme from Escherichia coli (protease I) Eur J Biochem. 1971;23:435–442. doi: 10.1111/j.1432-1033.1971.tb01638.x. [DOI] [PubMed] [Google Scholar]
- 21.Robertson D L, Hilton S, Wong K R, Koepke A, Buckley J T. Influence of active site and tyrosine modification on the secretion and activity of the Aeromonas hydrophila lipase/acyltransferase. J Biol Chem. 1994;269:2146–2150. [PubMed] [Google Scholar]
- 22.Ruiz J, Sempere M A, Varela M C, Gomez J. Modification of the methodology of stool culture for Salmonella detection. J Clin Microbiol. 1992;30:525–526. doi: 10.1128/jcm.30.2.525-526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 24.Sofia H J, Burland V, Daniels D L, Plunkett G R, Blattner F R. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. Nucleic Acids Res. 1994;22:2576–2586. doi: 10.1093/nar/22.13.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 26.Stulke J, Martin-Verstraete I, Charrier V, Klier A, Deutscher J, Rapoport G. The HPr protein of the phosphotransferase system links induction and catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol. 1995;177:6928–6936. doi: 10.1128/jb.177.23.6928-6936.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thornton J, Howard S P, Buckley J T. Molecular cloning of a phospholipid-cholesterol acyltransferase from Aeromonas hydrophila. Sequence homologies with lecithin-cholesterol acyltransferase and other lipases. Biochim Biophys Acta. 1988;959:153–159. doi: 10.1016/0005-2760(88)90026-4. [DOI] [PubMed] [Google Scholar]
- 28.Upton C, Buckley J T. A new family of lipolytic enzymes? Trends Biochem Sci. 1995;20:178–179. doi: 10.1016/s0968-0004(00)89002-7. [DOI] [PubMed] [Google Scholar]
- 29.Verger R. Enzyme kinetics of lipolysis. Methods Enzymol. 1980;64:340–392. doi: 10.1016/s0076-6879(80)64016-6. [DOI] [PubMed] [Google Scholar]
- 30.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 31.Wang H, Dowds B C A. Phase variation in Xenorhabdus luminescens: cloning and sequencing of the lipase gene and analysis of its expression in primary and secondary phases of the bacterium. J Bacteriol. 1993;175:1665–1673. doi: 10.1128/jb.175.6.1665-1673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe M, Ishidate M, Jr, Nohmi T. Nucleotide sequence of Salmonella typhimurium nitroreductase gene. Nucleic Acids Res. 1990;18:1059. doi: 10.1093/nar/18.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]