Abstract
virF is the master regulator which activates the virulence determinant genes of Shigella spp. such as ipaBCD and virG. We previously reported that expression of virF itself is regulated in a pH-dependent manner and that cpxA, a sensor of a two-component regulatory system, is involved in this regulation (S. Nakayama and H. Watanabe, J. Bacteriol. 177:5062–5069, 1995). Disruption of cpxR, which has been thought to be the cognate response regulator of cpxA (J. Dong, S. Iuchi, H.-S. Kwan, Z. Lue, and E. C. C. Lin, Gene 136:227–230, 1993), abolished virF expression almost completely. Purified CpxR bound directly to the upstream region of virF. Binding capacity was enhanced when CpxR was phosphorylated by coincubation with acetyl phosphate in vitro. Furthermore, we observed that phosphorylated CpxR could activate virF transcription in vitro. These results clearly indicated that CpxR was an essential activator for virF expression and strongly suggested that the binding of phosphorylated CpxR to the target site upstream of the virF gene induced a direct activation of virF transcription.
Studies of the virulence factors of pathogenic bacteria have revealed many genes encoding virulence effectors. They have often been shown to be subject to tight coordinate regulation dependent on many environmental conditions (for reviews, see references 7 and 16). This tight regulation is accomplished by well-suited cascades in which many regulators participate. These cascades are divided into many steps, implying the necessity for accurate checks of the environmental conditions that affect the expression of virulence effectors. This is also true of the regulatory circuit for virulence expression of Shigella spp., which are the causative agents of bacillary dysentery. virF, the master activator for Shigella virulence (1, 12), is contained in the “virulence plasmid” harbored by these bacteria. This plasmid also contains ipaBCD genes, whose products are the direct apparatuses for host cell invasion (17), and genes required for bacterial movement in the host cells such as virG (14). These genes produce the direct effectors for the virulence of Shigella and are positively regulated by virF. VirF induces the transcription of the second activator, invE (virB, ipaR), which is also contained by the plasmid (1, 33). InvE, in turn, induces the transcription of the ipaBCD operon (1, 33). VirF is believed to activate virG induction directly (26). In all, a fine-tuned regulatory cascade starting at virF is established (for reviews, see references 10, 22, and 27).
Research on the environment-dependent regulation of this cascade revealed some important knowledge about the effects of temperature (32), osmolarity (3), and pH (19) on the control of the cascade. Among these effects, the temperature-dependent repression of invE has been investigated most intensively. H-NS, a chromosome-encoded histone-like protein, is required for the apparent repression of invE transcription at 30°C (31). It is believed that although VirF activates the transcription of invE, H-NS interferes with this activation at 30°C and this interference is released at 37°C (31). However, there has been little information about the environmental condition which affects virF expression and the mechanism that produces the effect. We have noticed that the expression of virF itself is also regulated on the basis of the environmental condition and accordingly that Shigella spp. require that the proper conditions for virulence expression be present at the virF gene expression step. Our work showed us that there exists a further upstream regulatory locus that controls virF expression, and we previously reported that chromosome-encoded cpxA, a signal sensor of a two-component regulatory system, is involved in the pH-dependent transcriptional regulation of virF (19). However, the precise mechanism by which cpxA regulates or modulates the expression level of virF and the physiological significance of the regulation are unclear. Generally, a signal sensor functions in combination with a cognately paired response regulator in order to regulate the expression of target genes (for reviews, see references 2, 9, and 29). Therefore, our previous result implied the existence of a response regulator which is paired with cpxA and which regulates virF expression. In order to obtain some information on this issue, we constructed a disrupted mutant version of cpxR, which has been hypothesized to be the cognate response regulator of cpxA in this two-component system (8), and characterized it. In consequence, we revealed that CpxR is a direct activator essential for virF expression.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli K-12 MC1061 (28) was used as the wild-type strain. TB1 (11) was the host for overproduction of the MalE-CpxR fusion protein. pHW848, containing a virF′-′lacZ translational fusion gene, a reporter of virF expression level, was described previously (19). pOK101, which expresses cpxA under the control of the lac promoter (24), was a kind gift from P. M. Silverman.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Genotype or description | Source (reference) |
|---|---|---|
| E. coli K-12 | ||
| MC1061 | hsdR hsdM+ araD139 Δ(ara-leu)7679 Δlac(IPOZYA) galU galK rpsL | E. W. Nester (28) |
| MS8 | MC1061 rnh::cat | H. Ohmori (20) |
| SN1216 | MC1061 cpxR::Kmr | This study |
| SN1044 | MC1061 cpxA146::Tn10 | S. Nakayama (19) |
| TB1 | ara Δ(lac-proAB) rpsL (φ80 lacZΔM15) hsdR | T. C. Johnston (11) |
| Plasmids | ||
| pHW848 | virF′-′lacZ fusion gene cloned into pHSG595, a pSC101 replicon vector | H. Watanabe (19) |
| pACYC177 | p15A replicon cloning vector | A. C. Y. Chang (5) |
| pSN1018 | 2,499-bp BglII-StuI fragment consisting almost entirely of cpxR-cpxA operon cloned into BamHI-SmaI site of pACYC177 | This study |
| pOK101 | 1,508-bp DraI-StuI fragment consisting almost entirely of cpxA reading frame cloned into expression vector pINIIIA3 | P. Silverman (24) |
| pKH5002 | Cloning vector for gene disruption | H. Ohmori (20) |
| pSN1216K+ | 3-kb EcoRI fragment containing cpxR gene cloned into pKH5002 followed by cpxR reading frame disrupted by insertion of 1.4-kb Kmr cassette | This study |
| pMALTM-c2 | MalE fusion vector | Commercial product of New England Biolabs |
| pMAL-CpxR14 | cpxR reading frame fused in frame to malE gene on pMALTM-c2 | This study |
| pHSG397 | pMB1 replicon cloning vector | T. Hashimoto (30) |
| pSN600-T | 547-bp HpaI-ClaI fragment containing a part of virF gene cloned into pHSG397 followed by insertion of rrnB T1 terminator at the ClaI site | This study |
Media and buffers.
Luria-Bertani (LB) broth (18) containing 0.1 M sodium phosphate buffer (pH 6.0 or 7.4) was used for bacterial growth. When cultures were grown for plasmid or chromosome DNA preparation, simple unbuffered LB broth was used. Antibiotics were added to the media when necessary, and concentrations (in micrograms per milliliter) were as follows: ampicillin, 100; chloramphenicol, 10; kanamycin, 40; streptomycin, 100. Phosphate-buffered saline (0.8% NaCl, 0.02% KCl, 0.3 mM Na2HPO4, 0.15 mM KH2PO4) was used for bacterial dilution. During the course of protein purification, a protein column buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA) was used for the bacterial suspension step. For the dilution of protein samples, we used a protein elution buffer consisting of 50 mM Tris and 380 mM glycine, which is the same as 0.2× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis running buffer without SDS.
Preparation and manipulation of DNA.
The preparation and manipulation of chromosome and plasmid DNA were carried out essentially as described by Maniatis et al. (15).
Plasmid construction.
pSN1018 was constructed by recloning a 2,499-bp BglII-StuI fragment of pSN0612 (19), which contains almost nothing but the cpxR-cpxA operon (8), into the BamHI-SmaI sites of pACYC177 (5). The orientation of transcription of the Kmr gene from the vector side is opposite to that of the cpxR-cpxA operon in pSN1018. pSN1216K+, a plasmid for the disruption of cpxR, was constructed as follows. A 3-kb EcoRI fragment of pSN0612 containing the cpxR gene was recloned into suicide vector pKH5002 (20), whose replication is sensitive to RNase H. The resultant plasmid was digested by XhoI, the recognition site of which lies between nucleotides (nt) 395 (C) and 400 (G), as shown in Fig. 2 of reference 8 and which is unique in this construct, and the digested site was filled in by Klenow fragments of E. coli DNA polymerase I. A 1.4-kb Kmr cassette fragment derived from Tn903 (21) was ligated into the filled site. The whole construction procedure was performed in MS8 (20), an RNase H-deficient strain. pMAL-CpxR14, an overproducer of the MalE-CpxR fusion protein was constructed as follows. The DNA fragment precisely corresponding to the reading frame of cpxR starting at the initiation codon and ending at the termination codon was synthesized by PCR. The prepared fragment was trimmed by Klenow fragments of E. coli DNA polymerase I and ligated into the XmnI site of pMALTM-c2 (New England Biolabs, Inc., Beverly, Mass.). As the XmnI cutting site is located at the precise position corresponding to the cleavage site of sequence-specific endopeptidase factor Xa (carboxyl-terminal side of I-E-G-R), this construction was expected to enable us to obtain CpxR without any N-terminal tag after factor Xa digestion of the fusion protein. After construction, the complete nucleotide sequence of the cpxR portion of this plasmid was determined in order to check for the precise PCR amplification of cpxR as well as for the precise fusion to malE.
pSN600-T, a template for the in vitro transcription of virF, was constructed by cloning a 547-bp HpaI-ClaI fragment starting at nt −369 and ending at nt +178 (where the transcription start site of virF [19] is at nt +1) into the HincII-ClaI sites of pHSG397 (30), followed by introduction of the 44-bp synthetic rrnB T1 transcriptional terminator fragment (4) at the ClaI site. Suitably oriented integration of the terminator sequence was confirmed by nucleotide sequence analysis. The transcriptional orientation of the lac promoter from the vector is opposite to that of virF in this construct.
Southern hybridization analysis.
Chromosome DNAs were prepared from MC1061 and SN1216. DNA (10 μg) from each strain was digested with EcoRI and run on a 0.7% agarose gel containing TAE buffer. The gel was depurinated with 250 mM HCl, denatured with 1.5 M NaCl–0.5 M NaOH, and renatured with 1.5 M NaCl–0.5 M Tris-HCl, pH 7.5. Then, DNA was blotted onto a nylon membrane (Hybond-N+; Amersham, Buckinghamshire, United Kingdom) with 20× SSC (0.3 M sodium citrate plus 3 M NaCl, pH 7.0) by a capillary blotting method. After fixation of DNA by UV linker (Funakoshi Co., Tokyo, Japan), the blot was subjected to hybridization. Hybridization was carried out with a nonradioactive labeling kit (ECL direct nucleic acid labeling and detection systems; Amersham). A 3-kb EcoRI fragment containing the cpxR gene was purified from pSN0612 (19), labeled with horseradish peroxidase as directed by the manufacturer’s manual, and used as a probe. Hybridization, washing, and detection of signals were also performed in accordance with the manual and were followed by autoradiography at room temperature, usually for 10 min.
Overproduction of MalE-CpxR fusion protein and purification of CpxR.
TB1 harboring pMAL-CpxR14 was cultured to an optical density at 600 nm (OD600) of approximately 0.5. After the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.3 mM to induce expression of the cloned fusion gene, culturing proceeded until an OD600 of 1.0 was reached. Cells were harvested by centrifugation, suspended in the protein column buffer described above, frozen once at −20°C, and then thawed. After adequate sonication, soluble crude extract was obtained by centrifugation. The extract was loaded onto an amylose resin to which the maltose binding domain of the fusion protein binds. The resin was washed with 10 volumes of protein column buffer, and the fusion protein was eluted with a protein column buffer containing 10 mM maltose. The sample was digested with factor Xa for 2 days at room temperature after the addition of SDS to a final concentration of 0.025% for a partial denaturation of the sample, which was required for digestion in this case. After adequate dialysis against the protein column buffer, the digested sample was loaded again onto an amylose resin to trap the portion of maltose-binding protein in the sample, and the flowthrough fraction was collected. The fractionated sample was concentrated adequately by ultrafiltration, divided into small aliquots, and stored at −70°C.
Gel shift assay.
DNA fragments consisting of nt −103 to +110 (probe C) and −37 to +110 (probe D), with the transcription start site of virF as nt +1 (19), were synthesized by PCR. The 3′ termini of 4 pmol of each probe were labeled with 1 nmol of digoxigenin (DIG)–11-ddUTP (Boehringer GmbH, Mannheim, Germany) by 1 U of terminal deoxynucleotidyl transferase at 37°C for 30 min in 200 mM potassium cacodylate–25 mM Tris-HCl (pH 6.6)–0.25 mg of bovine serum albumin per ml–5 mM CoCl2. After the labeling reaction, the probes were precipitated by ethanol (EtOH), suspended in H2O, and stored at −20°C. Eight femtomoles of each DIG-labeled probe was incubated with the CpxR sample at 25°C for 30 min in 20 mM HEPES (pH 7.6)–1 mM EDTA–10 mM (NH4)2SO4–1 mM dithiothreitol (DTT)–0.2% (wt/vol) Tween 20–30 mM KCl–1 μg of poly(dI-dC) per 20 μl–0.1 μg of poly-l-lysine per 20 μl, in a final volume of 20 μl. When phosphorylated CpxR sample was used, 30 pmol of CpxR was phosphorylated by coincubation with acetyl phosphate at 37°C for 30 min in 50 mM Tris-HCl (pH 7.6)–5 mM MgCl2–1 mM DTT–50 mM acetyl phosphate prior to incubation with the DNA probes; this is essentially the same method as that described previously (13). After the addition of 5 μl of 0.25× Tris-borate-EDTA (TBE) containing 40% glycerol, incubated samples were run on a 6% polyacrylamide gel (79:1) in 0.25× TBE, followed by electroblotting onto a nylon membrane (Hybond-N+; Amersham) and fixation by UV cross-linking. Detection of DNA fragments by anti-DIG Fab fragment–alkaline phosphatase conjugate (Boehringer GmbH) and substrate CSPD (Tropix Inc., Bedford, Mass.) was performed as directed by the manual from Boehringer GmbH.
In vitro transcription of virF.
pSN600-T or pHSG397 (0.5 pmol) was preincubated with CpxR sample in 40 mM Tris-HCl (pH 7.9)–10 mM MgCl2–0.1 mM EDTA–150 mM KCl–5% glycerol–2 mM DTT at 37°C for 5 min, followed by the addition of 1 U of E. coli RNA polymerase (Pharmacia Biotech, Uppsala, Sweden). The phosphorylation of the CpxR sample was as described above. After further incubation at 37°C for 5 min, transcription was started by the addition of 400 μM ATP–400 μM GTP–200 μM CTP–100 μM UTP–0.1 μM [α-32P]UTP (3,000 Ci/mmol). The final volume of the reaction mixture was adjusted to 50 μl, and transcription proceeded at 37°C for 30 min. The reaction was stopped by the addition of EDTA and yeast tRNA to final concentrations of 40 mM and 0.1 mg/ml, respectively. The sample was extracted with phenol-CHCl3, precipitated by EtOH, and resuspended in 7.5 μl of H2O. The same volume of 10 mM Tris-HCl (pH 7.5)–10 mM EDTA–0.25% xylenecyanol–0.25% bromophenol blue–95% formamide was added to the synthesized RNA. The sample was heated at 95°C for 3 min and quickly chilled on ice. The denatured RNA was analyzed by electrophoresis on an 8% polyacrylamide gel (19:1) containing 8.3 M urea in 1× TBE, followed by autoradiography. For a molecular weight marker, 5′ termini of the HinfI digest of pBR322 DNA were labeled with [γ-32P]ATP (6,000 Ci/mmol), denatured as described above, and loaded on the same gel.
β-Galactosidase assay.
β-Galactosidase activity was determined essentially as described by Miller (18). Bacterial cultures were grown with shaking at 37°C in 1.5 ml of LB medium containing 0.1 M sodium phosphate buffer (pH 6.0 or 7.4) to an OD600 of approximately 0.4. The cells were harvested and suspended to 1.5 ml of Z buffer (18). After measurement of OD600 and adequate dilution with Z buffer, the samples were used for the assays. Activities were expressed in Miller units. All assays were performed at least four times, and errors in the results obtained were within ± 10% of the averages.
Enzymes, reagents, and radioactive materials.
Restriction enzymes were obtained from Takara Shuzo Co., Kyoto, Japan. Terminal nucleotidyl transferase, DIG–11-ddUTP, anti-DIG Fab–alkaline phosphatase conjugate, and CSPD were from Boehringer GmbH. pMALTM-c2 vector, amylose resin, and factor Xa were from New England Biolabs, Inc. E. coli RNA polymerase was obtained from Pharmacia Biotech. [α-32P]UTP (3,000 Ci/mmol) and [γ-32P]ATP (6,000 Ci/mmol) were purchased from DuPont/NEN Research Products, Boston, Mass.
RESULTS
Disruption of cpxR gene and examination of the effect.
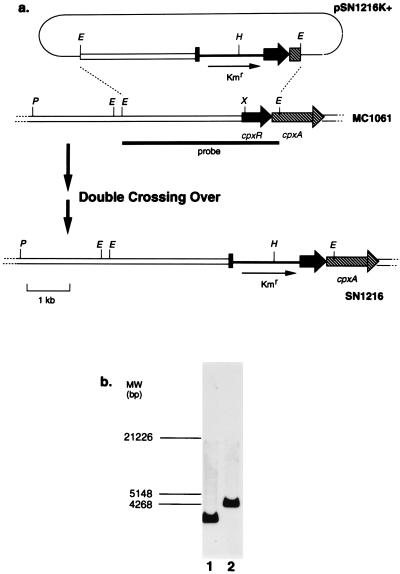
In a previous paper, we reported that the expression level of virF was regulated in a pH-dependent manner in E. coli as well as in Shigella spp. and that cpxA, a signal sensor gene of a two-component signal transduction system encoded on 88 min of the E. coli genome, is involved in this regulation (19). Generally, a signal sensor regulates the expression of the target gene(s) through the cognately paired response regulator by phosphorylation/dephosphorylation. Therefore, our previous observation implied the existence of a response regulator which pairs with the cpxA sensor protein by transmission of a phosphate group. Previously, Dong et al. reported that a response regulator, cpxR, is located just upstream of cpxA (8) and Raivio and Silhavy reported that phosphotransfer events between CpxA and CpxR played an important role in Cpx signal transduction (25). This is supporting evidence that cpxR is the cognately paired regulator gene of cpxA. If this is indeed the case, the inactivation of cpxR must also affect the expression of virF cloned in E. coli. With this idea in mind, we decided to construct a cpxR disruptant strain. For this purpose, we constructed plasmid pSN1216K+, as described in Materials and Methods. This plasmid was introduced into MC1061. pSN1216K+ is a derivative of pKH5002, whose replication is sensitive to RNase H (20). Therefore, we could obtain a strain in which the whole plasmid was integrated into the cpxR region of the chromosome via homologous recombination by selection of both Apr and Kmr transformants. In this strain, both intact and Kmr cassette-disrupted cpxR genes are located contiguously, together with plasmid sequences (single crossover). The plasmid contains a wild-type rpsL gene, which confers Sms on MC1061, which was originally an Smr strain (28). So, subsequent positive selection of both Smr and Kmr colonies from the single-crossover strain resulted in the deletion of the intact cpxR gene as well as the plasmid sequences (double crossover). The procedure of disruption of cpxR is summarized in Fig. 1a. The chromosomal construction of the double-crossover strain was confirmed by Southern hybridization of EcoRI-digested chromosomes of the obtained strain and the parent, MC1061, with a probe consisting of a 3-kb EcoRI fragment containing the cpxR region. In MC1061, a single 3-kb band was detected as expected (lane 1 of Fig. 1b). On the other hand, we observed a single band about 4.5 kb long, instead of the 3-kb band, in the obtained strain (lane 2 of Fig. 1b), which is consistent with the length of the inserted Kmr cassette, 1.4 kb. Thus, we named the strain SN1216 and used it as a cpxR disruptant thereafter.
FIG. 1.
(a) Schematic drawings of the constructs of plasmid pSN1216K+ and of chromosomes around the cpxR-cpxA regions in MC1061 and SN1216. Thick arrows indicate the locations and direction of cpxR and cpxA genes. Thin arrows in pSN1216K+ and SN1216 show the sites and the direction of the inserted Kmr cassette. The location of the 3-kb EcoRI probe used in the Southern hybridization analysis is shown by a thick line under the MC1061 drawing. Abbreviations: E, EcoRI; H, HindIII; P, PstI; X, XhoI. (b) Confirmation of chromosomal constructions of MC1061 and SN1216 by Southern hybridization with the probe described above. Ten micrograms of the chromosome DNA of MC1061 (lane 1) and SN1216 (lane 2) was completely digested by EcoRI and run on a 0.7% agarose gel containing 1× TAE buffer. After the DNA was blotted onto a nylon membrane, hybridization was performed with the horseradish peroxidase-labeled probe, followed by washing and signal detection (see Materials and Methods for the detailed protocol). The positions of DNA molecular size markers in base pairs are indicated on the left.
We examined the expression level of virF in SN1216 at pH 6.0 and 7.4, with pHW848 as a reporter plasmid (19), according to the hypothesis described above. The results are shown in Table 2. The effect of the gene disruption in SN1216 was remarkable. Virtually no detectable expression of virF was observed in this strain at both pHs. As the complementation analyses clearly indicated that cpxR was essential for virF expression (see below), it was revealed that cpxR regulated virF expression, as cpxA did (19). This is strong supporting evidence of cognate pairing between cpxA and cpxR, a signal sensor and a response regulator, respectively, constituting a two-component regulatory system. However, the specific expression pattern of virF in cpxR mutant SN1216 is quite different from that in previously described cpxA mutant SN1044 (19) (Table 2). We discuss this issue below (see Discussion).
TABLE 2.
Effects of cpxR and cpxA mutants on virF expression
| Strain (relevant genotype)a | β-Galactosidase activity (U) at pH
|
|
|---|---|---|
| 6.0 | 7.4 | |
| MC1061 (cpxR+cpxA+) | 308 | 2,840 |
| SN1216 (cpxR::Kmr) | 50 | 50 |
| SN1044 (cpxA146::Tn10) | 3,500 | 1,670 |
The reporter plasmid in each case was pHW848.
Identification of cpxR as a genetic locus essential for virF expression.
In order to distinguish whether the effect of cpxR disruption in SN1216 is indeed the outcome of inactivation of the gene or whether it is due to a polar effect on a downstream gene(s) such as cpxA or an accidental mutation in SN1216, we performed a complementation test. For this purpose, we constructed a plasmid containing the cpxR-cpxA operon only, pSN1018, and compared the effect of it with that of pOK101, which expresses cpxA only under the control of the lac promoter (25). Our effort to clone the cpxR gene only was unsuccessful (see Discussion). These plasmids were introduced separately into SN1216 and used for the complementation test by monitoring virF expression at pH 6.0 and 7.4. In order to standardize the dosage effect of cpxR and cpxA, we compared the expression levels of pHW848 in MC1061 with those in SN1216, which harbors the same effector plasmid. The results of the assays are shown in Table 3. We judged that pSN1018 fully complemented SN1216; MC1061 and SN1216 showed almost the same activities at both pH 6.0 and 7.4 when they harbored pSN1018, although the apparent repression at pH 6.0 became somewhat worse than that in MC1061 without the effector plasmid (Table 3). On the other hand, the inability of SN1216 to express virF was not altered at all by the introduction of pOK101 (Table 3). In all, these results clearly indicated that cpxR was essential for virF expression and that the cpxR-cpxA operon was sufficient for that.
TABLE 3.
Complementation of SN1216 by cpxR clone
| Strain (relevant genotype)a | Added plasmid | β-Galactosidase activity (u) at pH
|
|
|---|---|---|---|
| 6.0 | 7.4 | ||
| MC1061 (cpxR+cpxA+) | None | 308 | 2,840 |
| pSN1018 | 700 | 2,530 | |
| pOK101 | 40 | 1,100 | |
| SN1216 (cpxR::Kmr) | None | 50 | 50 |
| pSN1018 | 610 | 2,640 | |
| pOK101 | 50 | 50 | |
The reporter plasmid in each case was pHW848.
Direct binding of CpxR to the virF upstream region.
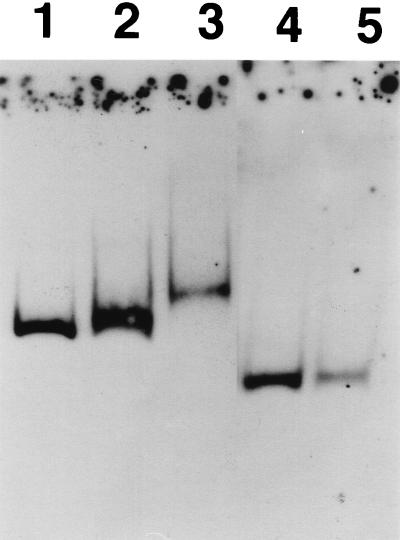
In order to discover the mechanism by which cpxR activates virF expression, we intended to examine whether the activation pathway is direct or not. First, we investigated the capacity of CpxR to bind to the DNA fragment corresponding to the virF upstream region. For this purpose, we established an overproduction and purification system for CpxR by using a MalE fusion vector as described in Materials and Methods. The product prepared in this system was used in the gel shift assay. We used two probes consisting of nt −103 to +110 (probe C) and −37 to +110 (probe D), with the virF transcription start site as nt +1 (19), because in another experiment we observed that the deletion of the upstream region to nt −103 did not affect virF expression, whereas deletion to nt −37 abolished the expression almost completely (unpublished results); nevertheless, the putative promoter for virF (19) is present downstream of the nt −37 site, which implied that there might be a binding site(s) for the activator between those two sites. The results of the assays using the CpxR sample and probe C or D are shown in Fig. 2. Probe C was clearly shifted when incubated with CpxR at a final concentration of 1.8 μM (lane 3 of Fig. 2); however, probe D was not shifted under the same condition (lane 5 of Fig. 2). This result strongly suggested that CpxR had the capacity to bind the virF upstream region between nt −103 and −37. This observation was apparently consistent with the expression patterns of virF in the deletion constructs described above.
FIG. 2.
Gel shift assay. Eight femtomoles of probe C consisting of nt −103 to +110, with the virF transcription start site as nt +1 (lanes 1, 2, and 3), and probe D, nt −37 to +110 (lanes 4 and 5), was generated by PCR, and the 3′ termini were labeled with DIG–11-ddUTP. Probes were incubated at 25°C for 30 min in 20 mM HEPES (pH 7.6)–1 mM EDTA–10 mM (NH4)2SO4–1 mM DTT–0.2% (wt/vol) Tween 20–30 mM KCl–1 μg of poly(dI-dC) per 20 μl–0.1 μg of poly-l-lysine per 20 μl in the absence of CpxR (lanes 1 and 4) or the presence of 0.9 (lane 2) or 1.8 μM (lanes 3 and 5) (final concentrations) CpxR sample in a final reaction volume of 20 μl. After electrophoresis on a 6% polyacrylamide gel containing 0.25× TBE, DNA was blotted onto a nylon membrane, followed by incubation with anti-DIG Fab–alkaline phosphatase conjugate and signal detection (see Materials and Methods for the detailed protocol).
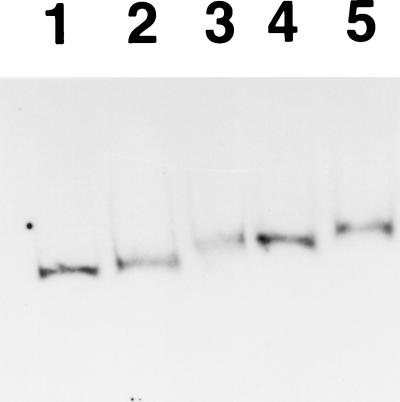
It is generally accepted that a response regulator increases its binding capacity, and consequently its ability to control transcription of the target gene(s), when it is phosphorylated (for reviews, see references 2, 9, and 29). In order to examine whether this is also the case for this system, we tried to prepare the phosphorylated form of CpxR and compare its binding capacity with that of the nonphosphorylated form. The phosphorylation of CpxR in vitro was performed by coincubation with acetyl phosphate essentially as described previously (13). The results of this examination are shown in Fig. 3. In this assay, we observed that incubation with 0.9 μM phosphorylated CpxR clearly shifted probe C, whereas incubation with nonphosphorylated CpxR at the same concentration resulted in little shift (lanes 2 and 4 of Fig. 3), although we could not quantify the extent of the phosphorylation in our system. The position of the shifted band was comparable to that for 1.8 μM nonphosphorylated CpxR (lanes 3 and 4 of Fig. 3). In 1.8 μM CpxR, the mobility of the band decreased more when CpxR was phosphorylated than when it was not phosphorylated (lanes 3 and 5 of Fig. 3). This suggests the existence of more than one binding site with different affinities to CpxR within this probe, although we must determine the specific binding site(s) to draw this conclusion. In all, these series of gel shift assays have indicated that the CpxR product can directly bind to the virF upstream region without the assistance of any other factor and that, as in many response regulators, phosphorylation enhances the binding capacity.
FIG. 3.
Gel shift assay with phosphorylated or nonphosphorylated CpxR. Eight femtomoles of DIG-labeled probe C (see legend to Fig. 2) (lanes 1 to 5) was incubated under the same condition as those for Fig. 2 in the absence of CpxR (lane 1) or the presence of 0.9 (lanes 2 and 4) or 1.8 μM (lanes 3 and 5) (final concentrations) CpxR samples preincubated in 50 mM acetyl phosphate at 37°C for 30 min (lanes 4 and 5) or in phosphorylation buffer only (lanes 2 and 3) (see Materials and Methods for the detailed conditions of the phosphorylation of CpxR). After the electrophoresis, the same protocol as that of Fig. 2 was performed.
In vitro transcription assay of virF.
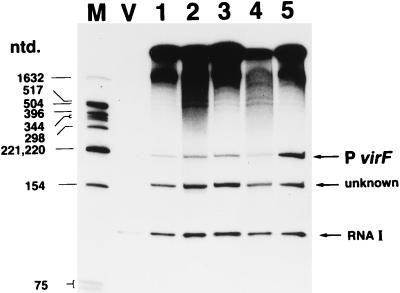
The results in this study raised two possibilities concerning the mechanism of activation of virF by CpxR, i.e., either that CpxR binds to the target site and, in consequence, directly activates transcription of virF or that at the binding step, CpxR releases some repressor of virF that shares the binding site with CpxR, which results in an apparent activation of virF. In order to examine when these mechanisms are operative and, when the former is operative, in order to determine whether the supply of CpxR by itself is sufficient to induce transcription of virF, we performed in vitro transcription assays. The results of the assays are shown in Fig. 4. For this experiment, we constructed template plasmid pSN600-T as described in Materials and Methods. In this construct we could detect a virF transcript as a short band about 220 nt long, which is consistent with the length calculated from the transcription start site of virF in vivo in our previous work (19). At the same time, RNAI of the pMB1 replicon of the vector side, which is 108 nt long and which was a convenient internal control for the assays, was observed. The emergence of a band about 150 nt long was unexpected. At present, we do not know the origin of it. However, preliminary investigation suggested that it started within the reading frame of virF and ran in the same direction as virF, i.e., when we used ClaI-linearized pSN600-T as the template in transcription assays, we could observe two bands with lengths of 170 and 100 nt, which must correspond to the 220- and 150-nt bands, respectively, in the circular template system (data not shown).
FIG. 4.
In vitro transcription assay of virF transcript. pSN600-T (lanes 1 to 5) or pHSG397 (lane V) (0.5 pmol) was preincubated without CpxR (lanes V and 1) or with nonphosphorylated (lanes 2 and 3) or phosphorylated (lanes 4 and 5) CpxR samples in 40 mM Tris-HCl (pH 7.9)–10 mM MgCl2–0.1 mM EDTA–150 mM KCl–5% glycerol–2 mM DTT at 37°C for 5 min, followed by the addition of 1 U of E. coli RNA polymerase (for the phosphorylation of CpxR samples, see the legend to Fig. 3 and Materials and Methods). The final concentrations of CpxR were 0 (lanes V and 1), 0.14 (lanes 2 and 4), and 1.4 μM (lanes 3 and 5). After further incubation at 37°C for 5 min, transcription was started by the addition of nucleoside triphosphates and [α-32P]UTP. The final volume of the reaction mixture was adjusted to 50 μl. After the reaction, the samples were extracted with phenol-CHCl3, precipitated by EtOH, and denatured as described in Materials and Methods. RNA was analyzed by electrophoresis on a urea–denatured 8% polyacrylamide gel containing 1× TBE, followed by autoradiography. The positions of the virF transcript (P virF), internal control RNAI, and a cryptic RNA about 150 nt long of unknown origin are indicated by horizontal arrows on the right. The positions of denatured DNA molecular size markers in nucleotides (ntd.) (lane M) are indicated on the left.
Without a supply of CpxR, the intensity of the band corresponding to the virF transcript was very much weaker than that corresponding to the internal control, RNAI (lane 1 of Fig. 4). The addition of nonphosphorylated CpxR, at least to the final concentration of 1.4 μM, did not alter the intensity of the band much (lanes 2 and 3 of Fig. 4). Supplying phosphorylated CpxR to the final concentration of 0.14 μM also had little effect (lane 4 of Fig. 4). Some might argue that the intensity of the band is weaker than those of bands shown in lanes 2 and 3. This may be due to less-efficient recovery of the transcript by precipitation in this tube (compare the intensities of the bands at the origin of electrophoreses). In the replicated experiments, we could observe neither a positive nor a negative effect of phosphorylation at 0.14 μM CpxR (data not shown). However, when we added the phosphorylated form at the final concentration of 1.4 μM to the reaction system, the clear enhancement of the virF transcript was observed (lane 5 of Fig. 4). Under this condition, the intensity of the band corresponding to virF got much stronger than that of the band corresponding to RNAI. Thus, these results have given us important information. First, the mechanism of induction of virF transcription by CpxR is true activation by CpxR, not a double-negative mechanism such as the release of repressor. Indeed, when no factors other than E. coli RNA polymerase were present, the level of the virF transcript was very low (lane 1 of Fig. 4), and the level was greatly increased by the addition of phosphorylated CpxR (lane 5 of Fig. 4). Such an activation could not have been observed in this system if the mechanism had been a double-negative regulation. Second, CpxR has the capacity to induce transcription of virF alone without any assistance from other factors, although of course we cannot rule out the existence of some factor(s) that modulates this reaction. Third, phosphorylation increases the induction activity of CpxR, at least when the concentration of CpxR is as high as 1.4 μM (compare lanes 3 and 5 of Fig. 4), which is consistent with the enhancement of the binding ability of CpxR by phosphorylation (see above), although we could not quantify the extent of CpxR phosphorylation in our systems. Thus, we concluded that, when phosphorylated, CpxR efficiently bound upstream of virF and, in consequence, directly activated the transcription of virF.
DISCUSSION
A two-component regulatory system is one of the genetic elements that regulate the expression of the target genes in trans in response to environmental signals (for reviews, see references 2, 9, and 29). We previously reported that virF expression is regulated in a pH-dependent manner and that a sensor of a two-component system is involved in the regulation (19). This led us to the hypothesis that the expression of virF is regulated by a response regulator which is cognately paired with cpxA. In this study, we indicated that response regulator CpxR, whose homolog also exists on chromosomes of Shigella spp. (unpublished data), was an essential activator for virF expression. It is the first example of a chromosome-encoded element indispensable for virF transcription, which has been recognized as the key step in turning on the total virulence cascade of Shigella. Therefore, we can conclude that the very first switch of the virulence cascade is under the control of a chromosome factor.
We also showed that the binding of CpxR to the upstream region of virF transcriptionally activated the expression of virF. Direct binding of CpxR to the target DNA was also reported for degP, yihE, and ppiA by Pogliano et al. (23). They determined binding sites upstream of those genes and concluded that the consensus sequences for the recognition sites for CpxR binding are 5′-GTAAN(6-7)GTAA-3′ and, in some cases, one copy of 5′-GTAA-3′ (23). We reported in this paper that CpxR bound to the virF region between nt −103 and −37. Within this region, sequences 5′-GTAAATAAAGTTAAA-3′ and 5′-TTAC-3′ (GTAA in the opposite strand), which resemble the reported consensus sequences, are present from nt −75 to −61 and nt −49 to −46, respectively. We preliminarily investigated the inhibitory effect of DNA fragments mutated at the predicted binding sites. As a result, we found that fragments with mutations at either of the described regions reduced the inhibitory effect on the shift of probe C compared to the fragment with the wild-type sequence or to the fragment mutated within the region from nt −60 to −50 (data not shown). This may be a supporting evidence that the consensus region within the upstream portion of virF has a function for CpxR binding.
We confirmed that cpxR was the cognate partner of cpxA, the pair constituting a two-component system; a mutation in either gene affected virF expression (Table 2) (19). And, to our knowledge, there have been no cases in which a contiguously encoded sensor and regulator are not cognately paired. Danese et al. have reached the same conclusion through the mechanistic analyses of the modulation of degP expression (6). Furthermore, direct transmission of a phosphate group between a MalE-CpxA fusion protein and a MalE-CpxR fusion protein in vitro was recently demonstrated (25). Given that cpxA and cpxR are the cognate pair of a two-component system, we cannot simply explain the difference between virF expression levels in cpxA and cpxR mutants. In a cpxR mutant, virF is hardly expressed at all (Table 2), whereas in a cpxA mutant, virF expression is never abolished (Table 2) (19). Such a discrepancy between the effects of the mutation of cpxA and cpxR in the regulation of degP was also reported by Danese et al. (6). They attributed it to the direct phosphorylation of CpxR by acetyl phosphate in vivo in the absence of CpxA; indeed, they observed that inactivation of pta and ackA, which are involved in the biosynthesis of acetyl phosphate, in a cpxA mutant almost eliminated that discrepancy (Fig. 4 of reference 6). However, in our preliminary investigation, these genes seemed to play a small role in the in vivo expression of virF in a cpxA mutant (unpublished results). This implied that there might be qualitative and/or quantitative differences in dependency upon cpxR-cpxA of the mechanisms by which virF and degP are regulated. In this context, it may also be noteworthy that the effect of disruption of cpxR on virF expression was quite remarkable, whereas the effect of that on degP expression was drastic only when NlpE, a new kind of lipoprotein, was overproduced artificially (compare Table 2 of this study and Fig. 5 of reference 6).
Thus, we raised two possibilities which could explain the phenotypic discrepancy between cpxA and cpxR mutants. First, although the virF expression level simply corresponds to the phosphorylation level of CpxR as implied from the results of the gel shift assay and in vitro transcription assay (Fig. 3 and 4; also, see above), CpxR is phosphorylated by another phosphate donor(s) besides CpxA and acetyl phosphate. In this case, virF expression in the cpxA mutant could be attributed to the phosphorylation of CpxR by this putative phosphate donor(s). Second, the phosphorylation level of CpxR is not the only factor that determines the expression level of virF in vivo. This is to say that the presence of phosphorylated CpxR is a prerequisite for virF expression but that the expression level of virF is finally determined by some other factor independent of cpxR. This second scenario is rather complex and seems unlikely; however, we cannot exclude it at present. In either case, there must still be another regulatory locus (or loci) besides cpxR-cpxA that controls virF expression. The systematic screening for a secondary mutation that affects the expression is now being undertaken.
As we have failed to clone cpxR without a cpxA region (see above), we cannot conclude that cpxR is sufficient for virF expression. However, cloning of a cpxR homolog from Salmonella spp., whose amino acid sequences showed 97.4% homology with that of E. coli CpxR, was successful even when cpxA homolog region was absent (18a). Introduction of this clone into SN1216 recovered the expression of virF, although the apparent repression at low pH was weak (18a). This may imply that cpxR is sufficient for virF expression, although the strict regulation may require cpxA in addition to cpxR. This observation, together with the results in Tables 2 and 3, implied something important about the role of cpxA in the repression of virF at low pH, i.e., that inactivation of cpxA resulted in a higher expression level of virF at low pH (Table 2). Furthermore, the introduction of the E. coli cpxR cpxA operon or the Salmonella cpxR homolog on a plasmid resulted in inefficient repression of virF both in the wild type and in the cpxR disruptant (Table 3) (18a). Because phosphorylated CpxR is the activator of virF and CpxA is the cognate sensor of CpxR, the most simple interpretation is that CpxA functions as a phosphatase of CpxR at low pH.
In order to approach to this issue, we are planning to examine the relationship between the phosphorylation level of CpxR and expression level of virF in vivo. If these two levels are always directly proportional to each other regardless of genetic background or environmental condition, we could simply expect that specific inhibition of CpxR might be a new method for the control of the virulence of Shigella. Anyway, we believe our results have presented information about the mechanism of the regulatory circuit of virulence expression of Shigella which is important from both the basic and applied points of view.
ACKNOWLEDGMENTS
We thank Philip M. Silverman who kindly provided us with plasmid pOK101, which was very helpful to us as a CpxA supplier.
This work was supported by grants from the Japan Health Sciences Foundation and the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Adler B, Sasakawa C, Tobe T, Makino S, Komatsu K, Yoshikawa M. A dual transcriptional activation system for the 230kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol Microbiol. 1989;3:627–635. doi: 10.1111/j.1365-2958.1989.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 2.Albright L M, Huala E, Ausubel F M. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu Rev Genet. 1989;23:311–336. doi: 10.1146/annurev.ge.23.120189.001523. [DOI] [PubMed] [Google Scholar]
- 3.Bernardini M L, Fontaine A, Sansonetti P J. The two-component regulatory system OmpR-EnvZ controls the virulence of Shigella flexneri. J Bacteriol. 1990;172:6274–6281. doi: 10.1128/jb.172.11.6274-6281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 5.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danese P N, Snyder W B, Cosma C L, Davis L J B, Silhavy T J. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 7.Di Rita V J, Mekalanos J J. Genetic regulation of bacterial virulence. Annu Rev Genet. 1989;23:455–482. doi: 10.1146/annurev.ge.23.120189.002323. [DOI] [PubMed] [Google Scholar]
- 8.Dong J, Iuchi S, Kwan H-S, Lue Z, Lin E C C. The deduced amino-acid sequence of the cloned cpxR gene suggests the protein is the cognate regulator for the membrane sensor, CpxA, in a two-component signal transduction system of Escherichia coli. Gene. 1993;136:227–230. doi: 10.1016/0378-1119(93)90469-j. [DOI] [PubMed] [Google Scholar]
- 9.Gross R, Arico B, Rappuoli R. Families of bacterial signal-transducing proteins. Mol Microbiol. 1989;3:1661–1667. doi: 10.1111/j.1365-2958.1989.tb00152.x. [DOI] [PubMed] [Google Scholar]
- 10.Hale T L. Genetic basis of virulence in Shigella species. Microbiol Rev. 1991;55:206–224. doi: 10.1128/mr.55.2.206-224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnston T C, Thompson R B, Baldwin T O. Nucleotide sequence of the luxB gene of Vibrio harveyi and the complete amino acid sequence of the beta subunit of bacterial luciferase. J Biol Chem. 1986;261:4805–4811. [PubMed] [Google Scholar]
- 12.Kato J, Ito K, Nakamura A, Watanabe H. Cloning of regions required for contact hemolysis and entry into LLC-MK2 cells from Shigella sonnei form I plasmid: virF is a positive regulator gene for these phenotypes. Infect Immun. 1989;57:1391–1398. doi: 10.1128/iai.57.5.1391-1398.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukat G S, McClerary W R, Stock A M, Stock J B. Phosphorylation of bacterial response regulator proteins by low molecular weight phosphodonors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makino S-I, Sasakawa C, Komatsu K, Kurata T, Yoshikawa M. A genetic determinant required for continuous reinfection of adjacent cells on a large plasmid in Shigella flexneri 2a. Cell. 1986;46:551–555. doi: 10.1016/0092-8674(86)90880-9. [DOI] [PubMed] [Google Scholar]
- 15.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 16.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 18a.Nakayama, S., S. Miyake, and H. Watanabe. Unpublished data.
- 19.Nakayama S, Watanabe H. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J Bacteriol. 1995;177:5062–5069. doi: 10.1128/jb.177.17.5062-5069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohmori H, Saito M, Yasuda T, Nagata T, Fujii T, Wachi M, Nagai K. The pcsA gene is identical to dinD in Escherichia coli. J Bacteriol. 1995;177:156–165. doi: 10.1128/jb.177.1.156-165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 22.Parsot C. Shigella flexneri: genetics of entry and intracellular dissemination in epithelial cells. Curr Top Microbiol Immunol. 1994;192:217–241. doi: 10.1007/978-3-642-78624-2_10. [DOI] [PubMed] [Google Scholar]
- 23.Pogliano J, Lynch A S, Belin D, Lin E C C, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 24.Rainwater S, Silverman P M. The Cpx proteins of Escherichia coli K-12: evidence that cpxA, ecfB, ssd, and eup mutations all identify the same gene. J Bacteriol. 1990;172:2456–2461. doi: 10.1128/jb.172.5.2456-2461.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raivio T L, Silhavy T J. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol. 1997;179:7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai T, Sasakawa C, Yoshikawa M. Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30 kD virF protein. Mol Microbiol. 1988;2:589–597. doi: 10.1111/j.1365-2958.1988.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 27.Sasakawa C, Buysse J M, Watanabe H. The large virulence plasmid of Shigella. Curr Top Microbiol Immunol. 1992;180:21–44. doi: 10.1007/978-3-642-77238-2_2. [DOI] [PubMed] [Google Scholar]
- 28.Stachel S E, An G, Flores C, Nester E W. A Tn3-lacZ transposon for the random generation of β-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol-or-kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 31.Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by VirF and repression by H-NS. J Bacteriol. 1993;175:6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkatesan M, Buysse J M, Kopecko D J. Characterization of invasion plasmid antigen (ipaBCD) genes from Shigella flexneri: DNA sequence analysis and control of gene expression. Proc Natl Acad Sci USA. 1988;85:9317–9321. doi: 10.1073/pnas.85.23.9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe H, Arakawa E, Ito K, Kato J, Nakamura A. Genetic analysis of an invasion region by use of a Tn3-lac transposon and identification of a second positive regulator gene, invE, for cell invasion of Shigella sonnei: significant homology of InvE with ParB of plasmid P1. J Bacteriol. 1990;172:619–629. doi: 10.1128/jb.172.2.619-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]