Abstract
Background
Transforming waste and nonfood materials into bulk biofuels and chemicals represents a major stride in creating a sustainable bioindustry to optimize the use of resources while reducing environmental footprint. However, despite these advancements, the production of high-value natural products often continues to depend on the use of first-generation substrates, underscoring the intricate processes and specific requirements of their biosyntheses. This is also true for Streptomyces lividans, a renowned host organism celebrated for its capacity to produce a wide array of natural products, which is attributed to its genetic versatility and potent secondary metabolic activity. Given this context, it becomes imperative to assess and optimize this microorganism for the synthesis of natural products specifically from waste and nonfood substrates.
Results
We metabolically engineered S. lividans to heterologously produce the ribosomally synthesized and posttranslationally modified peptide bottromycin, as well as the polyketide pamamycin. The modified strains successfully produced these compounds using waste and nonfood model substrates such as protocatechuate (derived from lignin), 4-hydroxybenzoate (sourced from plastic waste), and mannitol (from seaweed). Comprehensive transcriptomic and metabolomic analyses offered insights into how these substrates influenced the cellular metabolism of S. lividans. In terms of production efficiency, S. lividans showed remarkable tolerance, especially in a fed-batch process using a mineral medium containing the toxic aromatic 4-hydroxybenzoate, which led to enhanced and highly selective bottromycin production. Additionally, the strain generated a unique spectrum of pamamycins when cultured in mannitol-rich seaweed extract with no additional nutrients.
Conclusion
Our study showcases the successful production of high-value natural products based on the use of varied waste and nonfood raw materials, circumventing the reliance on costly, food-competing resources. S. lividans exhibited remarkable adaptability and resilience when grown on these diverse substrates. When cultured on aromatic compounds, it displayed a distinct array of intracellular CoA esters, presenting promising avenues for polyketide production. Future research could be focused on enhancing S. lividans substrate utilization pathways to process the intricate mixtures commonly found in waste and nonfood sources more efficiently.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-023-02266-0.
Keywords: Streptomyces lividans, Renewable feedstock, Lignin, Polystyrene, Seaweed, 4-hydroxybenzoate, Protocatechuate, Mannitol, Ethylmalonyl-CoA pathway, Bottromycin, Pamamycin, Natural product
Background
Natural products have long been celebrated for their vast therapeutic and commercial potential, and numerous pharmaceuticals, cosmetics, and other beneficial compounds are derived from them. Today, these products are often synthesized by microbes from agricultural feedstocks [1, 2], which demand extensive land use that competes with food production. Typical food-competing raw materials used today are glucose, starch, protein-rich meals from soybean and casein hydrolysate, yeast extract, and different peptones [3–6]. As the world grapples with burgeoning populations and diminishing availability of arable land, a reliance on land-intensive, food-based raw materials for natural product synthesis becomes increasingly unsustainable, and a shift towards more eco-conscious alternatives is needed [7, 8]. Recent prominent studies have shown that petroleum-based polystyrene waste, exhibiting a recovery rate of less than 1% due to recalcitrance [9], underutilized lignin side streams from the pulp and paper industry [10] and (often simply disposed) seaweed residuals from ocean farming [11–13] can be converted into bulk biofuels and chemicals [14–16]. This underscores the potential to use these third-generation renewables for natural product synthesis.
Streptomyces lividans is a prominent filamentous bacterium in the field of natural product synthesis due to its remarkable genetic manipulability and robust secondary metabolic pathways [17]. For instance, S. lividans TK24 has been instrumental in the derivation of heterologous producers for tunicamycin, griseorhodin, and deoxycoformycin [18] and is regarded as a promising nonmodel bacterial chassis for secondary metabolite production [19]. The recently created derivatives S. lividans ΔYA8 and ΔYA9 stand out in terms of their unique genetic makeup; they lack native pathways for natural product synthesis, making them ideal blank slates for the heterologous expression of exogenous biosynthetic pathways [18]. As is typical in the field, previously, production was reliant on first-generation substrates such as glucose, starch, yeast extract, and soytone [20, 21].
In terms of the valorisation of next-generation raw materials, the microbe has a broad pathway repertoire available in its genome [22]. In the context of the use of plastic waste- and lignin-based aromatic monomers, S. lividans possesses the β-ketoadipate pathway, a central route for aromatic catabolism, but it has not been studied experimentally for growth on these substrates. On the other hand, although not specifically tested for S. lividans before, mannitol, a major sugar in different seaweed hydrolysates [16], appears to be a generally well-accepted substrate for Streptomyces [23–26].
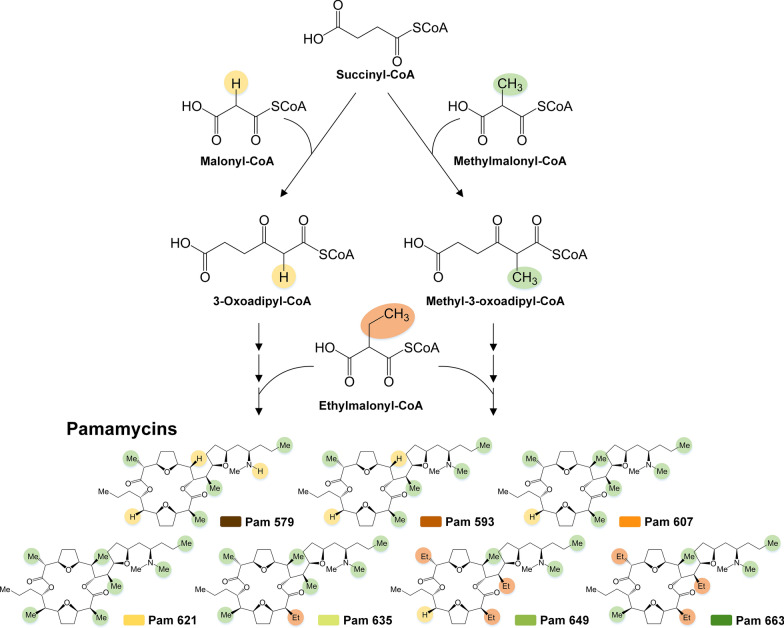
Therefore, in this work, we harnessed the capabilities of S. lividans to produce secondary metabolites from waste- and nonfood-based monomers. On the raw material side, we selected mannitol, a major sugar from seaweed hydrolysates [16, 27, 28], protocatechuate and 4-hydroxybenzoate, representing lignin hydrolysates [29–32], whereby 4-hydroxybenzoate exhibited additional relevance as an intermediate accessible from processed polystyrene waste [14]. On the product side, we selected the ribosomally synthesized and posttranslationally modified peptide bottromycin and the polyketide family of pamamycins, representing two major classes of natural products. Bottromycin exhibits antimicrobial activity against gram-positive pathogens [33, 34]. Its biosynthesis requires the supply of 14 amino acids to form the 7 different core and 36 follower peptides involved [35]. Pamamycins are macrolide polyketides with pronounced anti-insecticidal activity [36, 37] and are synthetized from CoA-ester precursors, including succinyl-CoA, malonyl-CoA, methylmalonyl-CoA, and ethylmalonyl-CoA [38, 39] (Fig. 1). After genomic insertion of the 18 kb bottromycin gene cluster and the 25 kb pamamycin gene cluster in S. lividans ΔYA8, production in the created heterologous producers was studied in batch cultures. Subsequently, comparative transcriptomics and metabolomics analysis of recombinant S. lividans provided a comprehensive systems view of the different waste substrates during strain growth. Finally, we demonstrated the production of bottromycin in a fed-batch process and carried out pamamycin synthesis from a hydrolysate of brown seaweed through Himanthalia elongata, both involving lean mineral media with exclusively next-generation carbon sources and minimized raw material pretreatment.
Fig. 1.
Microbial synthesis of different pamamycin derivatives. Pamamycin is biosynthetically synthesized from 4 different CoA thioesters, namely, succinyl-CoA, malonyl-CoA, methylmalonyl-CoA, and ethylmalonyl-CoA [38]. The promiscuous use of malonyl-CoA, methylmalonyl-CoA, and ethylmalonyl-CoA during the synthetic process leads to the incorporation of different side chains and the creation of pamamycin derivatives of different molecular weights, typically in the range between 579 and 663 Da
Results
The genome-minimized derivative S. lividans ΔYA8-DG2 produces bottromycin A2 from waste- and nonfood-based monomers
In the first step, we verified the principal capability of S. lividans to use sustainable monomers. We incubated S. lividans TK24 on solid minimal media that contained mannitol, protocatechuate or 4-hydroxybenzoate as the sole source of carbon. The strain grew well on all substrates, providing important proof of principle (Additional file 1: Fig. S1).
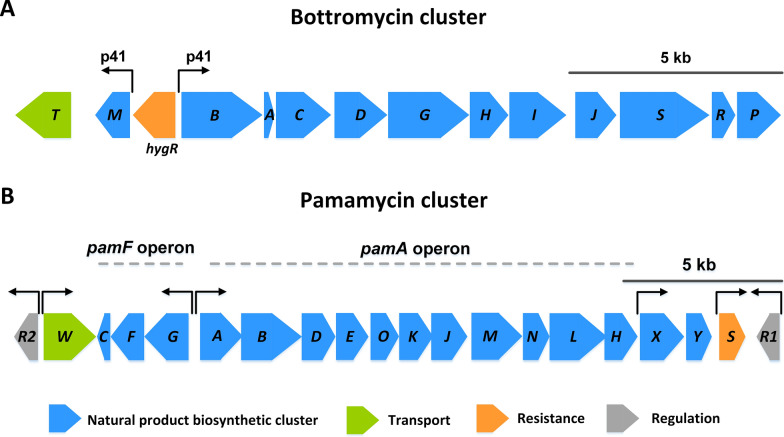
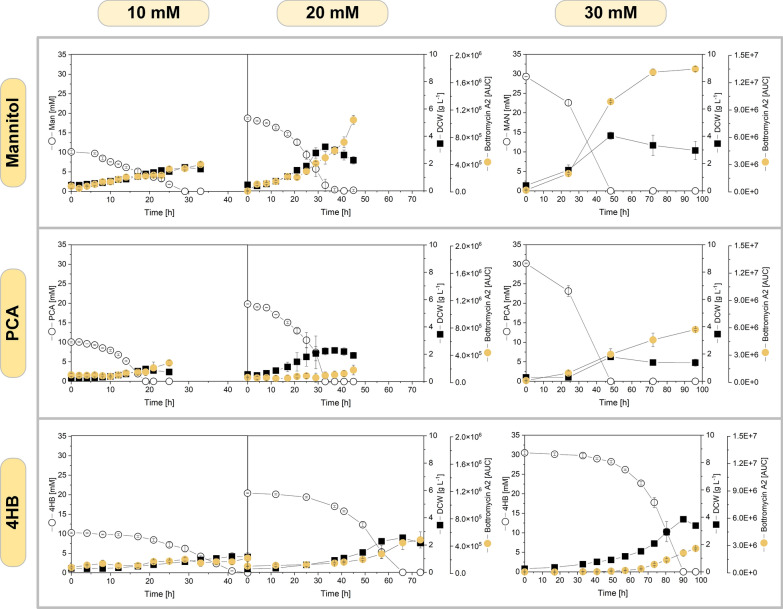
To selectively produce the heterologous natural products of interest, we utilized S. lividans ΔYA8, which lacks eight native biosynthesis-related gene clusters [18], as the chassis strain. It was transformed using conjugation with the cosmid DG2-km-P41hyg, which contains the bottromycin gene cluster regulated by a bidirectional pair of synthetic P41 promoters [20]. The cluster was integrated at the attB site (locus tag, SLIV19310) with the Int-phiC31 recombinase (Fig. 2A). After confirming the modification through PCR and sequencing, we named the modified strain S. lividans ΔYA8-DG2. For production assessment in shake flask cultures, we maintained the minimal nutrient composition used in preliminary growth experiments. Using mannitol concentrations of 10, 20, and 30 mM as the sole carbon source, S. lividans ΔYA8-DG2 successfully synthesized bottromycin (Fig. 3A–C). Cells grew without a lag phase, and higher biomass production correlated with increased mannitol concentrations. The 30 mM mannitol cultures achieved peak bottromycin levels. The mass spectrometric analysis revealed that methylated bottromycin A2 was formed in addition to bottromycin A2 as the major derivative (Additional file 2).
Fig. 2.
Genetic cluster architecture for the synthesis of bottromycin (A) and pamamycin (B) in recombinant S. lividans ΔYA8. The heterologous host S. lividans ΔYA8-DG2 contains a genomic copy of the 18 kb bottromycin biosynthesis-related cluster from Streptomyces sp. BC16019 [20]. The cluster is expressed under the control of synthetic P41 promoters, whereby hygR displays a resistance marker introduced during the cloning process. The heterologous host S. lividans ΔYA8-R2 contains a genomic copy of the 25 kb pamamycin biosynthesis-related cluster from Streptomyces alboniger DSMZ 40043 [38]. The cluster is expressed under the control of native promoters
Fig. 3.
Impact of the carbon source on the growth and bottromycin A2 production of S. lividans ΔYA8-DG2. The recombinant strain was grown in minimal medium containing different levels of mannitol (A–C), protocatechuate (PCA) (D–F), and 4-hydroxybenzoate (4HB) (G–I). The data represent mean values and standard deviations from three biological replicates (n = 3). A minor fraction of the methylated derivative was formed in addition to bottromycin A2 (Additional file 2)
Furthermore, we explored the production capabilities of S. lividans ΔYA8-DG2 grown on protocatechuate (Fig. 3D–F) and 4-hydroxybenzoate (Fig. 3G–I) with starting substrate concentrations up to 30 mM. Remarkably, the strain efficiently processed these toxic aromatic compounds. At 10 mM and 20 mM, protocatechuate was consumed even faster than mannitol. The strain utilized the maximum aromatic level of 30 mM, although this led to initial growth lag and extended cultivation periods on 4-hydroxybenzoate. The resulting biomass was lower than that with mannitol, potentially due to the diminished energy yield from aromatics compared to sugars [40, 41]. Nonetheless, bottromycin A2 was consistently produced across all conditions. This robust production from both aromatic sources and mannitol underscores the promise of S. lividans in converting next-generation substrates to bottromycin A2, along with its notable resilience to toxic raw materials. In all cases, production began early and continued steadily regardless of the substrate. This consistent production seemed to be attributed to the synthetic promoter governing bottromycin gene cluster expression, irrespective of the growth phase [20, 21]. Interestingly, the use of 30 mM mannitol resulted in increased synthesis of methylated bottromycin A2 derivatives (24%) as compared to the culture on 30 mM protocatechuate (11%) and 4-hydroxybenzoate (3%). (Additional file 2). Small amounts of methylated bottromycin A2 were also detected in cultures on 10 mM and 20 mM substrate (data not shown).
Enhanced production of bottromycin A2 from 4-hydroxybenzoate in a fed-batch process
Owing to the high robustness of S. lividans ΔYA8-DG2, we tested this heterologous host in a fed-batch process by repeatedly feeding it 4-hydroxybenzoate, the most toxic of the selected substrates. The batch phase was initiated with a concentration of 30 mM 4-hydroxybenzoate, as illustrated in Fig. 4A. We consistently monitored the metabolization of 4-hydroxybenzoate in real time. Upon substrate depletion after 64 h, we reintroduced the compound, raising the concentration back to 30 mM from a concentrated stock. Interestingly, the microorganism consumed this additional amount at a faster rate compared to the initial supply, and it consistently produced bottromycin. It even managed to fully metabolize a third addition of 4-hydroxybenzoate. In the end, the bottromycin A2 levels surged to almost triple of that in the batch process. Low amounts of methylated bottromycin A2 (3% of the amount of the main derivative) was formed in addition (Additional file 2). This showcases the capability of S. lividans to upgrade aromatics in a fed-batch process for optimized production. Following this process, we extracted the bottromycin A2 mixture from the culture broth. Analysis of the resultant ethyl acetate extract using LC‒MS/MS revealed that our desired product was identical to a commercial bottromycin standard (Fig. 4B, C).
Fig. 4.
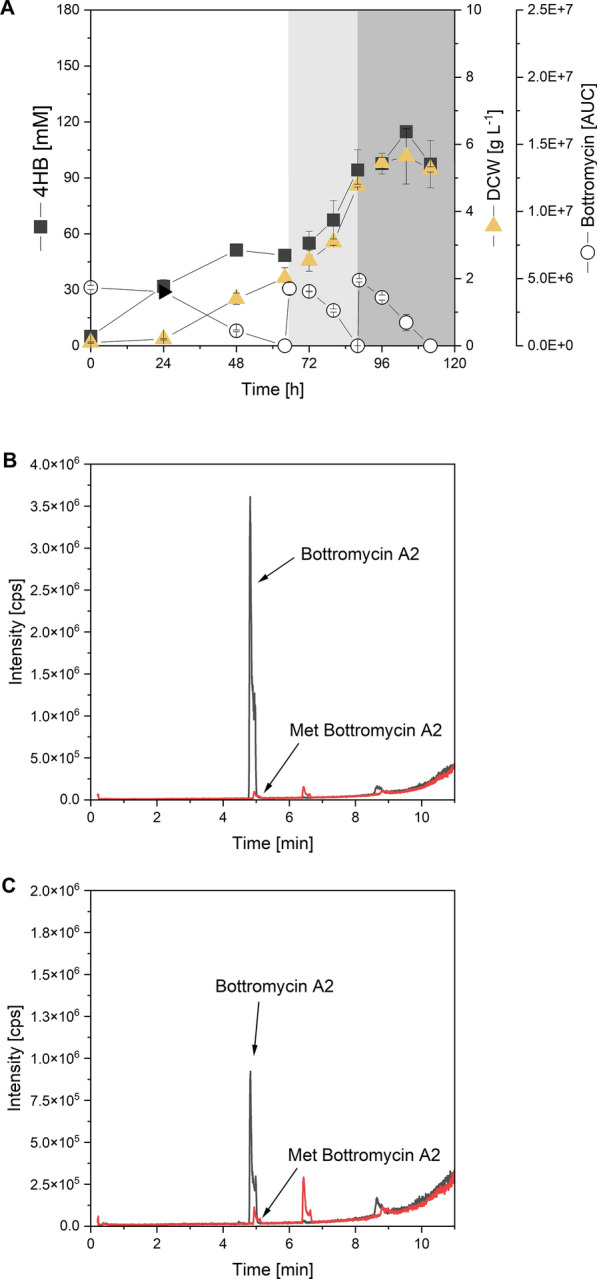
Production of bottromycin A2 from 4-hydroxybenzoate (4HB) in S. lividans ΔYA8-DG2 using a fed-batch process with repeated substrate feeding. The data represent the mean values and standard deviations of three biological replicates (n = 3) (A). At the end of the process, the product was extracted from the culture broth using ethyl acetate and analysed for the presence of bottromycin A2 [M + H]+ = 823.453) and methylated bottromycin A2 [M + H]+ = 837.453) using LC–MS (B). A commercial bottromycin standard was analysed for comparison (C). As shown, a minor fraction of the methylated derivative was formed in addition to bottromycin A2
The overall metabolism of S. lividans exhibited pronounced adaptation in response to different substrates, resulting in varied metabolic gene expression
We subsequently explored the metabolic intricacies of S. lividans ΔYA8-DG2 at the systems level by employing transcriptomic studies. For this investigation, cultures were initiated with three different substrates: 10 mM mannitol, protocatechuate, or 4-hydroxybenzoate. We then conducted RNA sequencing. At the 12-h mark, when the cells were sampled for RNA extraction, they demonstrated active growth, carbon consumption, and bottromycin production across all conditions (Fig. 3). The RNA sequencing results showed high statistical quality, as confirmed by PCA and sample distance analysis (Additional file 1: Fig. S2, S3). Notably, each substrate induced a distinct metabolic state at the global gene expression level, with each set of samples clustering separately. With protocatechuate, there was a significant alteration in the expression of 387 genes (which accounts for 5.2% of the total 7497 encoded genes) compared to the mannitol-based culture. Meanwhile, cultures on 4-hydroxybenzoate exhibited changes in the expression of 1786 genes (23.8%) relative to the mannitol culture. Overall, 1373 gene expression variations (18.3%) were observed between the two aromatic substrates (adjusted p value < 0.05, log2 fold change ≥ 1), including genes linked to energy and carbon core metabolism (see Additional files 3, 4).
The genes with the most distinct expression variations were closely related to central carbon catabolism (Tables 1, 2). This suggests that the microbe fundamentally altered a critical segment of its metabolism to adapt to the carbon source. Moreover, other genes with pronounced changes were responsible for encoding different metabolic process enzymes, regulators, and some proteins of unspecified function. Specifically, when grown on protocatechuate, the S. lividans ΔYA8-DG2 strain showed significant activation of genes associated with TCA cycle enzymes, genes involved in the transformation of phosphorylated intermediates with six and three carbons, belonging to glycolysis and gluconeogenesis, and a series of genes central to aromatic metabolism, including those responsible for the protocatechuate degradation pathway. These include enzymes such as protocatechuate 3,4-dioxygenase, 4-carboxymuconolactone decarboxylase, and 3-carboxy-cis, cis-muconate cycloisomerase (Table 3). Enzymes of the ethyl-malonyl CoA pathway, a signature pathway among various Streptomyces species [42], were also upregulated (Table 4).
Table 1.
Impact of the growth substrate on global gene expression in S. lividans ΔYA8-DG2. The data represent the 40 most strongly upregulated genes during growth on protocatechuate compared to mannitol (p < 0.05, log2fold change > 2.0). The encoded enzymes of the TCA cycle ( ), the EMP pathway, gluconeogenesis (
), the EMP pathway, gluconeogenesis ( ), aromatic catabolism (
), aromatic catabolism ( ), and the ethylmalonyl-CoA pathway (
), and the ethylmalonyl-CoA pathway ( ). Further genes encode other enzymes (M) and regulators (R), as well as proteins of unknown function (U). The samples were taken from 10 mM cultures after 12 h of incubation (Fig. 2). n = 3
). Further genes encode other enzymes (M) and regulators (R), as well as proteins of unknown function (U). The samples were taken from 10 mM cultures after 12 h of incubation (Fig. 2). n = 3
| Function | Annotation | Gene | PCA | 4HB |
|---|---|---|---|---|
| R | Two component histidine kinase | SLYA8N_18305 | 10.2 | 6.3 |
| Cytochrome B subunit | SLYA8N_33310 | 8.7 | 8.5 | |
| M | Glycosyl transferase | SLYA8N_18300 | 8.1 | 3.4 |
| M | ABC transporter, ATP-binding protein | SLYA8N_13690 | 8.0 | 1.1* |
| Succinate dehydrogenase/fumarate reductase iron-sulfur subunit | SLYA8N_33320 | 7.6 | 8.5 | |
| Hypothetical protein | SLYA8N_33325 | 7.4 | 8.3 | |
| M | Protease | SLYA8N_17590 | 7.3 | 0.2* |
| Succinate dehydrogenase flavoprotein subunit | SLYA8N_33315 | 7.1 | 7.6 | |
| Benzaldehyde dehydrogenase | SLYA8N_02130 | 6.7 | 8.4 | |
| R | Transcriptional regulatory protein | SLYA8N_18310 | 6.4 | 2.8 |
| 4-Carboxymuconolactone decarboxylase | SLYA8N_05020 | 6.4 | 6.2 | |
| Crotonyl-CoA reductase | SLYA8N_06060 | 6.3 | 3.6 | |
| β-Ketoadipyl-CoA thiolase | SLYA8N_05000 | 6.1 | 6.2 | |
| U | Integral membrane protein | SLYA8N_13695 | 6.0 | 0.3* |
| Succinyl-CoA:3-ketoacid coenzyme A transferase subunit A | SLYA8N_04990 | 6.0 | 6.3 | |
| U | Hypothetical protein | SLYA8N_18295 | 6.0 | 0.5* |
| 3-Carboxy-cis, cis-muconate cycloisomerase | SLYA8N_05015 | 5.9 | 5.8 | |
| Succinyl-CoA:3-ketoacid coenzyme A transferase subunit B | SLYA8N_04995 | 5.9 | 6.2 | |
| Protocatechuate 3,4-dioxygenase subunit B | SLYA8N_05005 | 5.8 | 5.0 | |
| Pyruvate phosphate dikinase | SLYA8N_25215 | 5.7 | 6.1 | |
| Transmembrane transporter | SLYA8N_02135 | 5.7 | 7.3 | |
| Protocatechuate 3,4-dioxygenase subunit A | SLYA8N_05010 | 5.7 | 5.3 | |
| R | Response regulator | SLYA8N_17595 | 5.5 | − 0.1* |
| Benzoylformate decarboxylase | SLYA8N_02125 | 5.4 | 6.6 | |
| M | Transport integral membrane protein | SLYA8N_13895 | 4.8 | 1.5 |
| β-Ketoadipyl-CoA thiolase | SLYA8N_04415 | 4.5 | 3.8 | |
| M | Alcohol dehydrogenase | SLYA8N_29215 | 4.3 | 3.8 |
| Transcriptional regulator | SLYA8N_04985 | 4.3 | 4.3 | |
| Protein MeaA | SLYA8N_06065 | 4.3 | 3.4 | |
| M | Long-chain-fatty-acid-CoA ligase | SLYA8N_04410 | 4.2 | 3.6 |
| M | Aldehyde dehydrogenase | SLYA8N_29210 | 4.0 | 3.5 |
| M | Exopolysaccharide phosphotransferase | SLYA8N_08030 | 3.9 | 2.0 |
| Oxidoreductase | SLYA8N_06050 | 3.9 | 3.3 | |
| U | Hypothetical protein | SLYA8N_34285 | 3.8 | 3.7 |
| M | Acyl-CoA dehydrogenase | SLYA8N_29235 | 3.7 | 3.5 |
| M | Hypothetical protein | SLYA8N_32305 | 3.6 | 6.1 |
| M | Iron-sulfur oxidoreductase subunit beta | SLYA8N_05120 | 3.5 | 1.9 |
| M | Membrane protein | SLYA8N_03150 | 3.4 | 2.4 |
| Phosphoenolpyruvate carboxykinase | SLYA8N_13500 | 3.3 | 3.3 | |
| p-Hydroxybenzoate hydroxylase | SLYA8N_22275 | 3.2 | 9.0 |
* Genes not significantly affected (Benjamini‒Hochberg, FDR > 0.05) but included for completeness
Table 2.
Impact of the growth substrate on global gene expression in S. lividans ΔYA8-DG2. The data represent the 40 most strongly unregulated genes during growth on protocatechuate compared to mannitol (p < 0.05, log2fold change > 2.0). The encoded enzymes of the EMP pathway are linked to glycolysis and gluconeogenesis ( ), mannitol catabolism (
), mannitol catabolism ( ), and pigment biosynthesis (
), and pigment biosynthesis ( ). Further genes encode other enzymes (M) and regulators (R), as well as proteins of unknown function (U). The samples were taken from 10 mM cultures after 12 h of incubation (Fig. 3). n = 3
). Further genes encode other enzymes (M) and regulators (R), as well as proteins of unknown function (U). The samples were taken from 10 mM cultures after 12 h of incubation (Fig. 3). n = 3
| Function | Annotation | Gene | PCA | 4HB |
|---|---|---|---|---|
| M | Monooxygenase | SLYA8N_19810 | − 5.7 | − 0.9* |
| M | Heavy metal reductase | SLYA8N_19790 | − 5.5 | − 0.7 |
| M | Arsenite resistance protein ArsB | SLYA8N_19800 | − 5.2 | − 2.0 |
| R | Transcriptional regulator | SLYA8N_19795 | − 5.0 | − 2.4 |
| Substrate binding protein, smoE | SLYA8N_28225 | − 4.6 | − 5.3 | |
| Integral membrane sugar transport protein, smoF | SLYA8N_28220 | − 4.6 | − 5.7 | |
| Hypothetical protein | SLYA8N_28205 | − 4.6 | − 5.0 | |
| Transcriptional regulator, smoR | SLYA8N_28230 | − 4.5 | − 4.2 | |
| DNA-binding protein | SLYA8N_19785 | − 4.4 | − 0.6 | |
| Integral membrane sugar transporter, smoG | SLYA8N_28215 | − 4.4 | − 4.3 | |
| Zinc-binding dehydrogenase, smoD | SLYA8N_28210 | − 4.3 | − 3.3 | |
| U | Hypothetical protein | SLYA8N_36995 | − 4.2 | − 5.2 |
| U | Hypothetical protein | SLYA8N_36990 | − 4.1 | − 5.3 |
| U | Hypothetical protein | SLYA8N_36975 | − 4.1 | − 5.3 |
| M | Oxidoreductase | SLYA8N_36985 | − 4.0 | − 5.0 |
| M | Methyltransferase | SLYA8N_36945 | − 4.0 | − 5.6 |
| M | Endoglucanase | SLYA8N_01130 | − 3.9 | − 3.0 |
| M | Methylesterase | SLYA8N_36955 | − 3.9 | − 5.5 |
| Enolase 2 | SLYA8N_01125 | − 3.8 | − 3.4 | |
| Deoxyribodipyrimidine photo-lyase | SLYA8N_36980 | − 3.7 | − 5.3 | |
| Phytoene dehydrogenase | SLYA8N_36965 | − 3.7 | − 4.9 | |
| Geranylgeranyl pyrophosphate synthase | SLYA8N_36970 | − 3.7 | − 4.2 | |
| Lycopene cyclase | SLYA8N_36940 | − 3.7 | − 4.7 | |
| Lipoprotein | SLYA8N_36920 | − 3.5 | − 3.7 | |
| Fructose-specific permease | SLYA8N_21710 | − 3.5 | − 2.4 | |
| M | Secreted protein | SLYA8N_01030 | − 3.4 | − 0.4* |
| DeoR family transcriptional regulator | SLYA8N_21700 | − 3.4 | − 2.4 | |
| Phytoene synthase | SLYA8N_36960 | − 3.3 | − 5.2 | |
| M | Sigma factor | SLYA8N_36925 | − 3.2 | − 1.4 |
| Glyceraldehyde-3-phosphate dehydrogenase | SLYA8N_01755 | − 3.2 | − 2.6 | |
| M | Dehydrogenase | SLYA8N_36950 | − 3.2 | − 4.8 |
| M | Integral membrane lysyl-tRNA synthetase | SLYA8N_20785 | − 3.2 | − 1.5 |
| 1-Phosphofructokinase | SLYA8N_21705 | − 3.1 | − 2.8 | |
| U | Hypothetical protein | SLYA8N_36915 | − 2.8 | − 3.8 |
| M | Secreted protein | SLYA8N_19820 | − 2.6 | 0.1 |
| Fructokinase | SLYA8N_27935 | − 2.6 | − 1.8 | |
| U | Hypothetical protein | SLYA8N_20790 | − 2.5 | − 0.9 |
| M | Neutral zinc metalloprotease | SLYA8N_11275 | − 2.5 | 0.3* |
| M | Lipoprotein | SLYA8N_17715 | − 2.4 | − 2.9 |
| R | MarR family regulatory protein | SLYA8N_01120 | − 2.4 | − 2.6 |
* Genes not significant (Benjamini‒Hochberg FDR > 0.05) but included for completeness
Table 3.
Impact of the carbon source on the expression of genes encoding substrate uptake and degradation pathways in S. lividans ΔYA8-DG2. The strain was grown on minimal medium with 10 mM mannitol, protocatechuate (PCA), or 4-hydroxybenzoate (4HB) as the sole carbon source. Samples were taken from the cultures after 12 h (Fig. 3). The expression levels are normalized to the mannitol-based culture and are given as log2-fold change. The significance level (Benjamini‒Hochberg, FDR) was set to < 0.05, n = 3
| Gene | Annotation | PCA | 4HB |
|---|---|---|---|
| Mannitol uptake | |||
| SLYA8N_09400 | Phosphocarrier protein HPr | − 1.48 | − 0.87 |
| SLYA8N_17190 | Trehalose import ATP-binding protein SugC | − 1.43 | − 1.13 |
| SLYA8N_21700 | DeoR family transcriptional regulator | − 3.40 | − 2.42 |
| SLYA8N_21705 | 1-Phosphofructokinase | − 3.07 | − 2.75 |
| SLYA8N_21710 | Fructose-specific permease | − 3.47 | − 2.43 |
| SLYA8N_28205 | Hypothetical protein | − 4.56 | − 5.01 |
| SLYA8N_28210 | Zinc-binding dehydrogenase | − 4.28 | − 3.31 |
| SLYA8N_28215 | Integral membrane sugar transporter | − 4.35 | − 4.28 |
| SLYA8N_28220 | Integral membrane sugar transport protein | − 4.57 | − 5.71 |
| SLYA8N_28225 | Substrate binding protein | − 4.61 | − 5.28 |
| SLYA8N_28230 | Transcriptional regulator deoR-type | − 4.54 | − 4.20 |
| Aromatic uptake and degradation | |||
| SLYA8N_02125 | Benzoylformate decarboxylase | 5.35 | 6.61 |
| SLYA8N_02130 | Benzaldehyde dehydrogenase [NAD( +)] | 6.69 | 8.36 |
| SLYA8N_02135 | Transmembrane transporter, Aromatic acid:H+ symporter | 5.66 | 7.30 |
| SLYA8N_04990 | Succinyl-CoA:3-ketoacid coenzyme A transferase subunit A | 6.02 | 6.29 |
| SLYA8N_04995 | Succinyl-CoA:3-ketoacid coenzyme A transferase subunit B | 5.89 | 6.22 |
| SLYA8N_05000 | β-Ketoadipyl-CoA thiolase | 6.13 | 6.17 |
| SLYA8N_05005 | Protocatechuate 3,4-dioxygenase beta chain | 5.78 | 5.04 |
| SLYA8N_05010 | Protocatechuate 3,4-dioxygenase alpha subunit | 5.65 | 5.28 |
| SLYA8N_05015 | 3-Carboxy-cis, cis-muconate cycloisomerase | 5.94 | 5.81 |
| SLYA8N_05020 | 4-Carboxymuconolactone decarboxylase | 6.42 | 6.19 |
| SLYA8N_22275 | p-Hydroxybenzoate hydroxylase | 3.23 | 9.01 |
Table 4.
Expression levels of genes involved in gluconeogenesis, the phosphoenolpyruvate-pyruvate-oxaloacetate node and the ethylmalonyl-CoA pathway (log2 fold change). The strain was grown on minimal medium with 10 mM mannitol, protocatechuate (PCA), or 4-hydroxybenzoate (4HB) as the sole carbon source. Samples were taken from the cultures after 12 h (Fig. 3). The expression levels are normalized to the mannitol-based culture and are given as log2-fold change. The significance level (Benjamini‒Hochberg, FDR) was set to < 0.05, n = 3
| Gene | Annotation | PCA | 4HB |
|---|---|---|---|
| Gluconeogenesis | |||
| SLYA8N_13150 | Fructose-1,6-bisphosphatase | 2.06 | 2.98 |
| SLYA8N_13500 | Phosphoenolpyruvate carboxykinase [GTP] | 3.34 | 3.28 |
| SLYA8N_35210 | Pyruvate carboxylase | 1.12 | 0.46 |
| Phosphoenolpyruvate-pyruvate-oxaloacetate node | |||
| SLYA8N_11390 | Pyruvate kinase | − 1.28 | − 1.05 |
| SLYA8N_12060 | Putative NAD-dependent malic enzyme | 1.14 | 2.06 |
| SLYA8N_22045 | Phosphoenolpyruvate carboxylase | − 0.90 | − 3.63 |
| SLYA8N_22940 | NAD-dependent malic enzyme | 0.16* | 0.82 |
| SLYA8N_25215 | Pyruvate phosphate dikinase | 5.70 | 6.14 |
| SLYA8N_27640 | Pyruvate kinase | − 1.03 | − 1.57 |
| Ethylmalonyl-CoA pathway | |||
| SLYA8N_04575 | Fatty oxidation protein | 0.21* | − 0.31* |
| SLYA8N_04850 | Fatty acid oxidative multifunctional enzyme | − 0.52 | − 1.04 |
| SLYA8N_04855 | Putative acyltransferase | − 0.33* | − 1.27 |
| SLYA8N_06050 | Oxidoreductase | 3.87 | 3.31 |
| SLYA8N_06055 | Transcriptional regulator | 0.41* | − 0.41* |
| SLYA8N_06060 | Crotonyl-CoA reductase | 6.28 | 3.55 |
| SLYA8N_06065 | Protein MeaA | 4.29 | 3.42 |
| SLYA8N_06070 | Citrate lyase | 2.89 | 2.24 |
| SLYA8N_06075 | Hypothetical protein | 2.98 | 2.68 |
| SLYA8N_06080 | Acyl-CoA dehydrogenase | 1.58 | 1.15 |
| SLYA8N_06790 | Acetyl-/propionyl-coenzyme A carboxylase alpha chain | 0.74 | − 0.61 |
| SLYA8N_11430 | Isobutyryl-CoA mutase A | 0.30* | 1.42 |
| SLYA8N_11500 | Putative acetyl-CoA acetyltransferase | 0.65 | 1.47 |
| SLYA8N_11505 | Hypothetical protein | 1.03 | 0.57 |
| SLYA8N_11570 | 3-Hydroxybutyryl-CoA dehydrogenase | 0.58 | 1.29 |
| SLYA8N_13745 | Propionyl-CoA carboxylase beta chain | 0.80 | − 0.56 |
| SLYA8N_13770 | Acetyl-/propionyl-coenzyme A carboxylase alpha chain | 0.58 | − 0.60 |
| SLYA8N_14030 | Methylmalonyl-CoA mutase | 0.51 | 1.30 |
| SLYA8N_14360 | Isobutyryl-CoA mutase small subunit | 0.42 | 0.34 |
| SLYA8N_23780 | Putative acyl-CoA dehydrogenase | 1.24 | 1.57 |
| SLYA8N_23785 | Hydroxymethylglutaryl-CoA lyase | 1.09 | 1.61 |
| SLYA8N_23790 | Acetyl-/propionyl-coenzyme A carboxylase alpha chain | 0.99 | 1.26 |
| SLYA8N_23795 | Methylcrotonoyl-CoA carboxylase beta chain | 1.35 | 1.51 |
* Genes not significant (Benjamini‒Hochberg FDR > 0.05) but included for completeness
Conversely, downregulation was noted in a six-gene operon in protocatechuate-fed cells. This operon seems to produce ABC transporter components for mannitol intake based on BLASTN comparison with the genome of the highly related strain S. coelicolor A3(2) (Table 5), which corroborated past studies [43]. Additionally, we found reduced expression in genes linked to the fructose phosphotransferase system (PTS). This suggests that mannitol, once it is oxidized in the cytoplasm to fructose, is phosphorylated to fructose 1-phosphate by the fructose PTS. This metabolic mechanism mirrors that in the related actinobacterium Corynebacterium glutamicum, as outlined in previous studies [16, 44, 45].
Table 5.
BLAST analysis of a six-gene operon upregulated during growth of S. lividans ΔYA8-DG2 on mannitol against the genome of S. coelicolor A3(2). The analysis revealed that the operon encodes a sugar ABC transporter that exhibits similarity to mannitol transporters [43]
| Gene | Annotation | Reference | Coverage/Identity (%) |
|---|---|---|---|
| SLYA8N_28205 | Hypothetical protein | SCO1902 | 100/99.0 |
| SLYA8N_28210 | Zinc-binding dehydrogenase, smoD | SCO1901 | 100/99.6 |
| SLYA8N_28215 | Membrane sugar transporter, smoG | SCO1900 | 100/99.8 |
| SLYA8N_28220 | Membrane sugar transport protein, smoF | SCO1899 | 100/100 |
| SLYA8N_28225 | Substrate binding protein, smoE | SCO1898 | 100/99.3 |
| SLYA8N_28230 | Transcriptional regulator deoR-type, smoR | SCO1897 | 100/99.7 |
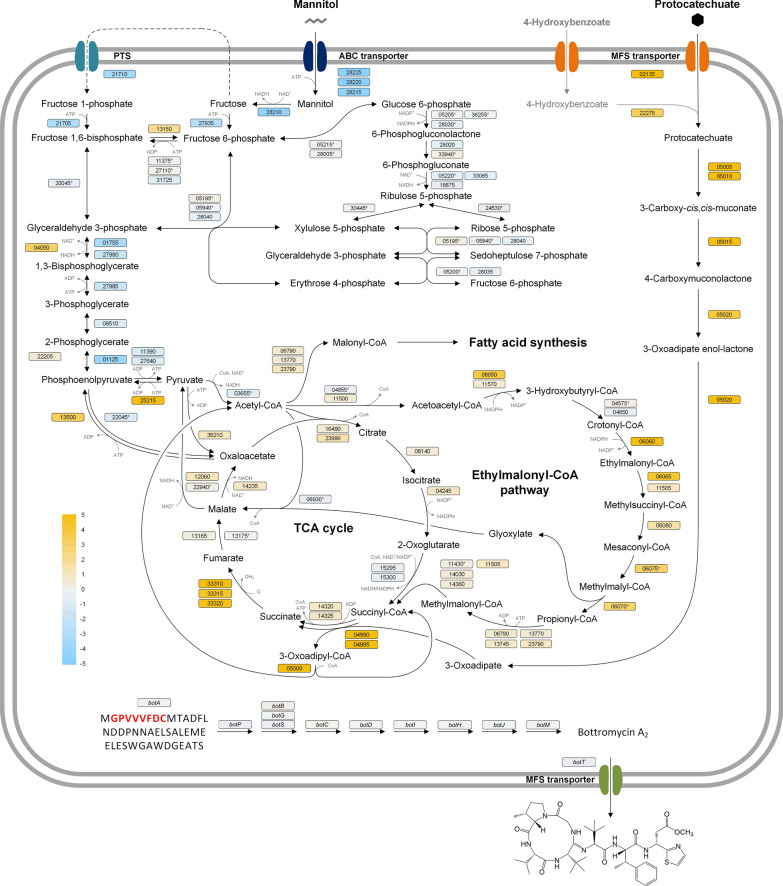
In terms of metabolic adjustment, the use of mannitol or protocatechuate as the growth substrate corresponded to the activation of their specific uptake systems, deactivating alternative pathways (Fig. 5). This trend was also noted in subsequent reactions related to further substrate metabolism. For cells grown on protocatechuate, a comprehensive catabolic pathway was observed, converting the aromatic substrate to 3-oxoadipyl-CoA and eventually yielding central intermediates such as succinyl-CoA and acetyl-CoA. This influx of succinyl-CoA into the TCA cycle led to the heightened activity of succinate dehydrogenase, pushing carbon towards gluconeogenesis. This is supported by the upregulation of several enzymes, including phosphoenolpyruvate carboxykinase, malic enzyme, and pyruvate phosphate dikinase, among others. In contrast, anaplerotic counterparts such as pyruvate carboxylase were downregulated. The generated acetyl-CoA was channelled into the ethylmalonyl pathway. Notably, when grown on protocatechuate, there was a significant increase in the activity of crotonyl-CoA reductase and ethylmalonyl-CoA mutase, pointing to potential shifts in CoA thioester metabolism (Fig. 5). Finally, for genes associated with amino acid biosynthesis, which is crucial for both general cellular construction and bottromycin formation, gene expression remained largely consistent. This suggests that the cells adeptly managed changes within the core central carbon metabolism (see Additional file 3).
Fig. 5.
Transcriptome profiling of bottromycin-producing S. lividans ΔYA8-DG2 during growth on mannitol and protocatechuate (10 mM). The data display differences in the expression of genes encoding central carbon pathways between mannitol- and protocatechuate-cultured cells after 12 h. The differences are highlighted by colour (blue, downregulated on protocatechuate; yellow, upregulated on protocatechuate). The metabolic network was assembled from the KEGG database, including the following manual curation. The genes encoding the mannitol uptake system were identified in this work (Table 4). Structurally, the uptake system matches that from C. glutamicum [16, 103]. The cofactor specificity of phosphoenolpyruvate carboxykinase (PEPCk) and pyruvate carboxylase (PC) was inferred from the corresponding enzymes of S. coelicolor M145 [104]. The cofactor specificity of succinate dehydrogenase was taken from E. coli [105]. All other cofactors refer to S. lividans TK24 [63]. The genes of the ethylmalonyl-CoA pathway were inferred from BLAST analysis (Additional file 1: Table S1). The core peptide of bottromycin is marked in red. + Putative genes, *Genes not significant (Benjamini‒Hochberg FDR > 0.05) but included for completeness. The data represent mean values and standard deviations from three biological replicates (n = 3)
Efficient bottromycin production from different substrate monomers is driven by stable expression of the biosynthesis-related gene cluster and stable intracellular precursor availability
For genes responsible for bottromycin biosynthesis, no major changes were detected, aligning with the observation that the type of substrate had minimal influence on bottromycin production. In-depth analysis of individual RNA sequence reads demonstrated that the expression patterns within the bottromycin cluster remained consistent regardless of the carbon source employed, as visually depicted in Fig. 2B–D. This underscores the efficacy of the synthetic P41 promoter used to guide their stable expression.
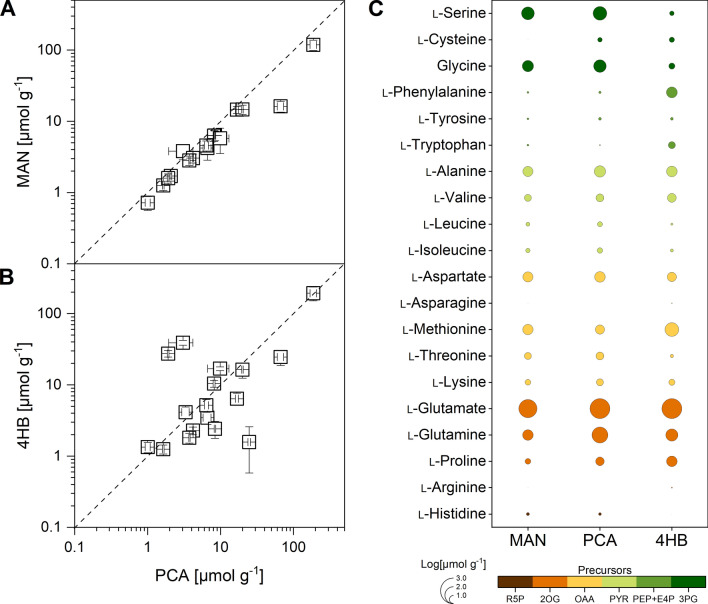
Our curiosity led us to investigate the strain's behaviour with respect to intracellular amino acid availability. For this purpose, we sampled cultures grown on each of the three substrates after 12 h for metabolomic analysis, aligning with the transcriptomics sampling time point. Intriguingly, the intracellular amino acid pools remained largely unaffected (Fig. 6), with only a few exceptions. Growth on aromatic compounds resulted in slightly elevated levels of l-glutamate, l-glutamine, and l-proline, originating from the 2-oxoglutarate node in the TCA cycle. This response appeared to be correlated with the downregulation of 2-oxoglutarate synthase, ultimately promoting the conversion of this intermediate into the corresponding amino acids. Notably, bottromycin biosynthesis requires the use of 14 distinct amino acids. Clearly, S. lividans ΔYA8-DG2 efficiently provides these essential building blocks in a minimal medium, irrespective of whether it is supplemented with mannitol or aromatics.
Fig. 6.
Intracellular amino acid levels of S. lividans ΔYA8-DG2 on different substrates. The minimal medium used contained 10 mM mannitol, protocatechuate, or 4-hydroxybenzoate. The data reflect absolute concentrations after 12 h. In addition to the overall comparison between the substrates (A, B), the data reveal substrate pathway-specific differences related to the biosynthetic origin of the amino acids (C). For the latter, the diameter of the circles reflects log-scaled concentrations. The data represent mean values and standard deviations from three biological replicates (n = 3) and analytical duplicates
The supplementation of aromatic compounds triggers the activation of the ethylmalonyl-CoA pathway in S. lividans
As depicted earlier, cells grown on aromatics displayed a notable upregulation of the ethylmalonyl pathway (Fig. 5), a pathway with a wide array of CoA-based intermediates. To gain a deeper understanding of the substrate’s effects and their metabolic repercussions, we opted to quantitatively assess intracellular CoA esters in S. lividans ΔYA8-DG2 cultured with 10 mM mannitol, protocatechuate, and 4-hydroxybenzoate. These samples were collected during the early growth and production phase at the 12-h mark.
In addition to the previously investigated CoA-esters [46, 47], our particular interest lies in evaluating the pool of 3-oxoadipyl-CoA, a pivotal intermediate within the aromatic degradation pathway, which has not been assessed before (Fig. 5). Since this metabolite was not commercially available as a reference standard, we explored its enzymatic synthesis. In brief, we cloned and expressed two proteins from Pseudomonas putida KT2440 [48], namely, 3-oxoadipate CoA-transferase subunit A (PcaI) and 3-oxoadipate CoA-transferase subunit B (PcaJ), in Escherichia coli. These efforts yielded a cell extract exhibiting sufficient enzymatic activity to convert the compounds 3-oxoadipate and succinyl-CoA into 3-oxoadipyl-CoA and succinate. The synthesis process yielded 3-oxoadipyl-CoA at 40% of its theoretical maximum yield based on the components used, and this was confirmed through absorption analysis at 260 nm. Following purification, this intermediate was employed to fine-tune the LC‒MS/MS parameters, ensuring high-sensitivity detection. Furthermore, it served as an external standard for the purpose of absolute quantification.
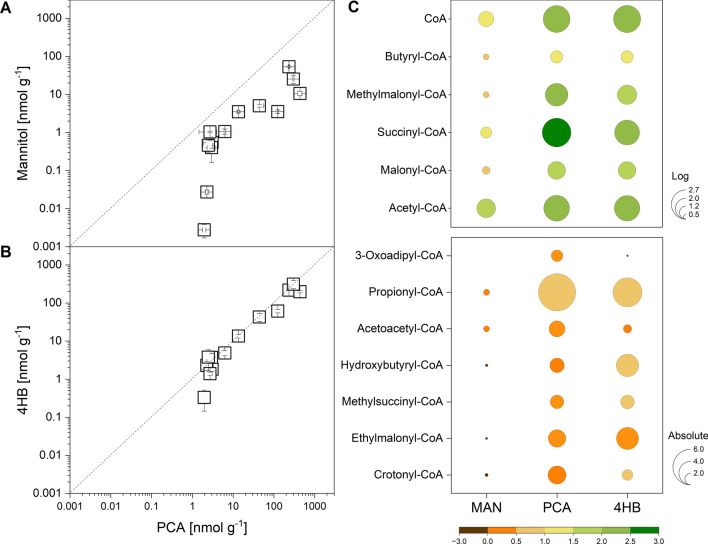
The metabolomics analysis revealed a profound influence of the carbon source on the intracellular composition of CoA esters (Fig. 7). Cells cultivated with mannitol predominantly harboured the "high-abundance" CoA-ester metabolites commonly found in bacteria [46], such as acetyl-CoA, succinyl-CoA, malonyl-CoA, methylmalonyl-CoA, and free CoA, while only minute quantities of other CoA-esters were observed. In stark contrast, cells grown on protocatechuate exhibited substantial deviations. Notably, utilization of the aromatic substrate led to significantly elevated levels of intermediates from the ethylmalonyl-CoA pathway. These included hydroxy-butyryl-CoA, ethylmalonyl-CoA, crotonyl-CoA, butyryl-CoA, methylsuccinyl-CoA, propionyl-CoA, and acetoacetyl-CoA, arranged in order they occur in the pathway. Surprisingly, we detected a notably higher abundance of 3-oxoadipyl-CoA, the first CoA-activated intermediate of protocatechuate degradation. Its pool size was approximately 700-fold higher than that of mannitol-grown cells.
Fig. 7.
Intracellular CoA-thioester levels of S. lividans ΔYA8-DG2. The minimal medium used contained 10 mM mannitol, protocatechuate, or 4-hydroxybenzoate. The data reflect absolute concentrations after 12 h during cell growth on protocatechuate (A) and 4-hydroxybenzoate (B) compared to mannitol. The size and colour of the circles reflect log-scaled concentrations for high (C) and low abundance CoA thioesters (D). The data represent mean values and standard deviations from three biological replicates (n = 3) and analytical duplicates
In total, S. lividans displayed a CoA-ester pool of 105 nmol gDCW−1 when cultured with mannitol. However, this pool expanded dramatically to 1173 nmol gDCW−1, an 11-fold increase, when protocatechuate was employed as the carbon source. An intriguing pattern emerged with the use of 4-hydroxybenzoate. While cells showed a similar trend of the increased abundance of intermediates from the ethylmalonyl-CoA pathway, the presence of 3-oxoadipyl-CoA was significantly lower, approximately sixfold less than in the protocatechuate-grown cells. This disparity might be attributed to differences in the uptake rates of these aromatic compounds. Protocatechuate (10 mM) was depleted at a notably faster rate (Fig. 2D) than 4-hydroxybenzoate (Fig. 2G), potentially leading to a more efficient accumulation of the 3-oxoadipyl-CoA pool. This observation aligns with the slightly lower total CoA-ester pool detected in cells cultivated with 4-hydroxybenzoate, which amounted to 866 nmol gDCW−1. It should be noted, however, that the 4-hydroxybenzoate-grown cells still contained 119-fold more 3-oxoadipyl-CoA than cells cultured on mannitol.
The use of aromatic carbon sources impacts the spectrum of pamamycin derivatives in S. lividans ΔYA8-R2
Inspired by the observed differences in CoA-thioester abundance (Fig. 7), we decided to investigate the production of polyketides synthetized from these precursors on the different substrates. Pamamycin appeared to be a promising candidate, given its broad spectrum of derivatives that are generated based on the availability of malonyl-CoA, methylmalonyl-CoA, and ethylmalonyl-CoA [39, 49]. The pathway proceeds through 3-oxoadipyl-CoA, a key intermediate in the primary metabolism of the degradation of aromatic compounds [38]. In pamamycin production, the R2 cosmid, housing the pamamycin biosynthesis-related gene cluster under control of its native promoters [38], was integrated into the genome of S. lividans ΔYA8 at the attB site, bearing the locus tag SLIV19310 (Fig. 2B). This genetic manipulation yielded the engineered strain designated S. lividans ΔYA8-R2. Verification of the desired mutation was confirmed through PCR analysis and subsequent sequencing.
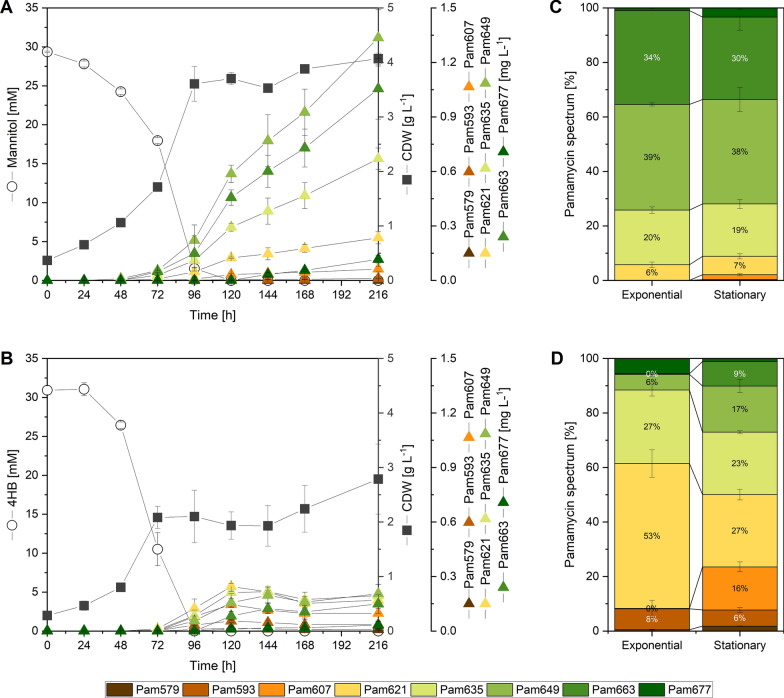
This mutant strain exhibited the ability to produce pamamycins when cultivated on two different substrates, namely, mannitol (30 mM) and 4-hydroxybenzoate (30 mM), as evidenced in Fig. 8A and B, respectively. Intriguingly, the production performance displayed notable disparities depending on the carbon sources utilized. On mannitol, the cells exhibited robust growth from an early stage. When grown on 4-hydroxybenzoate, the cells showed an initial lag phase lasting nearly a day due to the compound's inherent toxicity. However, once the cells were past this phase, they rapidly consumed the substrate and accumulated pamamycin. Subsequently, both cultures entered the stationary phase, where cells grown on mannitol continued to accumulate pamamycins, ultimately reaching a total concentration of 3.3 mg L−1 after 216 h. In contrast, the 4-hydroxybenzoate cultures did not exhibit further significant production of the natural product, resulting in a final titre of 0.9 mg L−1. Notably, the carbon substrate used had a substantial influence on the pamamycin spectrum. On mannitol, the cells generated a substantial fraction of heavy pamamycins, i.e., Pam 649 and Pam 663 (Fig. 8C). Conversely, cultures supplemented with 4-hydroxybenzoate predominantly produced elevated amounts of lighter derivatives such as Pam 621 and Pam 635, accounting for 80% of the total pamamycins (Fig. 8D). Furthermore, during the stationary phase, an intriguing shift in the spectrum of pamamycins was observed.
Fig. 8.
Impact of the carbon source on growth and pamamycin production in S. lividans ΔYA8-R2. The recombinant strain was grown in minimal medium containing either 30 mM mannitol (A) or 30 mM 4-hydroxybenzoate (4HB) (B). In addition, the spectrum of pamamycins formed is given for the mid-exponential (72 h) and stationary phases (216 h) on mannitol (C) and 4HB (D). The data represent mean values and standard deviations from three biological replicates (n = 3)
Demonstration of pamamycin production from a hydrolysate of the brown seaweed Himanthalia elongata using metabolically engineered S. lividans ΔYA8-R2
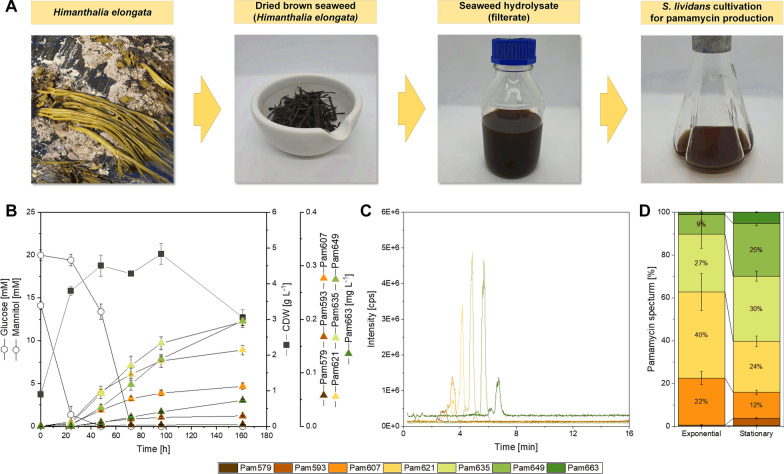
Finally, we aimed to showcase pamamycin production from a seaweed hydrolysate by employing the metabolically engineered strain S. lividans ΔYA8-R2. For this purpose, we selected the brown seaweed Himanthalia elongata, which exhibits commercial potential, as our primary raw material. The dried seaweed was meticulously processed, including grinding, followed by a gentle enzymatic treatment under slightly acidic conditions (pH 5.5). This process resulted in the generation of an aqueous hydrolysate enriched in mannitol (20 mM) and glucose (14 mM), both of which were byproducts of the digestion process (Fig. 9A). After neutralization with a buffer, this hydrolysate served as our sole source for pamamycin production, without any additional supplementation. Upon inoculating the liquid hydrolysate with strain ΔYA8-R2, we observed a sequential utilization of carbon sources. Initially, glucose was consumed, leading to an increase in biomass concentration to 4 g L−1, while total pamamycin production reached 0.02 mg L−1 (Fig. 9B). Subsequently, during the mannitol phase, cell growth continued to occur, accompanied by accelerated pamamycin synthesis, ultimately achieving a titre of 0.7 mg L−1. Notably, these two distinct culture stages resulted in variations in the pamamycin spectrum. In summary, our endeavour proved successful in demonstrating the feasibility of utilizing seaweed hydrolysate as a viable raw material for pamamycin production, aligning with increased sustainability.
Fig. 9.
Pamamycin production from a seaweed hydrolysate using S. lividans ΔYA8-R2. The hydrolysate was prepared from dried Himanthalia elongata [106] (credit for the photo of H. elongate: Willem VK, Saxifraga) (A). The recombinant strain was cultivated on plain seaweed hydrolysate after pH was adjusted to 7.0 (B). Pamamycins in the culture broth were analysed by LC–MS/MS (C). In addition, the spectra of pamamycin derivatives during the mid-exponential phase (24 h) and the stationary phase (96 h) are shown. The data represent mean values and standard deviations from three biological replicates (n = 3)
Discussion
Streptomyces lividans emerges as a robust cell factory for natural products from sustainable resources
The fermentative production of natural products has traditionally been reliant on first-generation carbon sources such as glucose, sucrose, starch, yeast extract, peptone, and soybean meal. In this work, advancements were made beyond this level. As shown, the metabolically engineered S. lividans strains successfully produced the antibiotic bottromycin and the insecticide pamamycin from alternative substrates such as 4-hydroxybenzoate, protocatechuate, and mannitol (Figs. 2, 3, 8). Additionally, we showcased compound production from a seaweed hydrolysate rich in mannitol and established the resilience of S. lividans during sequential 4-hydroxybenzoate feeding, even though this compound is known to be inhibitory to microbial cells [50]. The three substrates highlighted are gaining traction as sustainable inputs sourced from prominent waste and byproducts. This includes polystyrene waste [14], lignin from biorefineries and the pulp and paper sector [10], and residues from seaweed-based food production and high-value ingredient extraction [11]. For instance, 4-hydroxybenzoate is a significant component in poplars and willows, constituting up to 10% of the lignin fraction [51, 52].
Our findings are a promising step in transitioning microbial natural product production to more eco-friendly raw materials. Economically, there is potential value in this shift. In the industrial sphere, antibiotics can cost as little as 20 US dollars per kilogram. This places them in the specialty chemical category, for which economic viability is heavily influenced by raw material costs. Leveraging affordable byproducts instead of pricier conventional ingredients could enhance production cost-effectiveness. It is worth noting that industrial raw materials, such as those derived from lignin, might bring their own sets of impurities, possibly affecting downstream processing [53]. To achieve even broader use, it seems important to extend the capacity of S. lividans to catabolize aromatics. Beyond the two monomers studied here, lignin-derived hydrolysates typically contain a crude mixture of different aromatics such as catechol, phenol, cresols [54], guaiacol [55] and hydroxy-cinnamic acids such as ferulate, caffeate, p-coumarate, and vanillate [41]. These compounds cannot be naturally metabolized by S. lividans, suggesting the implementation of corresponding routes from other hosts, as shown previously for other bacteria [10].
The inherent strain metabolic flexibility and the use of a strong synthetic promotor enable stable compound production from different substrates
Previous research on S. lividans TK24 has primarily been centred around the synthesis of natural products and enzymes. However, its potential in substrate utilization remains relatively unexplored, and this includes the use of mannitol. For mannitol utilization, studies on the closely related S. coelicolor A3(2) strain yielded inconsistent results [43]. While genome analysis of S. coelicolor A3(2) hinted at an ABC-type membrane protein designed for sugar alcohols (potentially including mannitol), the corresponding smo operon was not triggered by introducing mannitol to a culture medium. A six-gene operon induced in S. lividans TK24 during growth on mannitol exhibited high sequence similarity based on BLASTN analysis against the genome of S. coelicolor A3(2) (Table 5). Thus, we infer that these genes constitute the mannitol absorption system in both S. lividans TK24 and S. coelicolor A3(2). Conversely, when protocatechuate and 4-hydroxybenzoate were introduced, the respective catabolic pathways were triggered [10].
Intriguingly, S. lividans TK24 cells showcased comparable levels of intracellular amino acids during growth on both an aromatic compound and a sugar alcohol (Fig. 6), even without notable changes in the transcription of amino acid biosynthetic pathways (Additional file 3). This suggests that simply adjusting the high-flux carbon core pathways was sufficient to enable the effective utilization of these distinct substrates. In context, Bacillus subtilis primarily modifies its central metabolism to maintain metabolic equilibrium under salt stress [56]. In conclusion, this inherent stability across varied environments, coupled with the adoption of a synthetic promoter for consistent cluster expression (Fig. 2A), seems pivotal for the remarkable production performance observed.
Aromatic substrates open new avenues for tailor-made product syntheses due to their modulation of the CoA ester spectrum
The metabolism of 4-hydroxybenzoate and protocatechuate resulted in the generation of intermediates of the corresponding protocatechuate 3,4-cleavage pathway, such as 3-oxoadipyl-CoA, succinyl-CoA, and acetyl-CoA. Given the absence of the glyoxylate shunt in S. lividans [57]—a typical mechanism for assimilating two-carbon compounds [58]—we theorized that the assimilation of acetyl-CoA might engage the ethylmalonyl-CoA pathway. This hypothesis was supported by the observed upregulation of key enzymes in the ethylmalonyl-CoA pathway: crotonyl-CoA reductase and ethylmalonyl-CoA mutase (Fig. 5). This was further supported by the increase in specific intracellular pathway intermediates in cells grown on aromatics (Fig. 7). The ethylmalonyl-CoA pathway plays a crucial role in the synthesis of polyketides, a commercially significant class of natural products [59], as it supplies the CoA thioester intermediates essential for building these compounds [60].
The pamamycins studied here are notable polyketides known for their intriguing antibiotic activity [36]. They encompass a variety of derivatives marked by structural variations in their side chains at six distinct positions [37, 38]. All pamamycins incorporate succinyl-CoA as a core starter, but their diverse derivatives stem from the alternative addition of three unique CoA thioesters: malonyl-CoA, methyl-malonyl-CoA, and ethyl-malonyl-CoA [38]. Prior research has demonstrated that the pamamycin spectrum in the related strain S. albus can be tailored by manipulating the intracellular concentrations of these building blocks. This can be achieved, for example, through genetic alterations of CoA ester metabolism [49] or by feeding the organism branched-chain amino acids [25]. Our findings reveal that the substrate selected significantly influences the intracellular CoA ester profile (Fig. 7). Consequently, this enables the production of diverse pamamycin combinations (Fig. 8). Aromatics tend to drive the synthesis of smaller pamamycin derivatives, whereas mannitol promotes the formation of bulkier versions. This suggests that employing aromatics could be a strategic method to fine-tune the pamamycin spectrum. Undoubtedly, 3-oxoadipyl-CoA played a pivotal role in determining the product spectrum. Cell growth on aromatic compounds led to an accumulation of this uncommon CoA-thioester, which was not observed in cells cultured on mannitol (Fig. 7). In cells grown on aromatics, 3-oxoadipyl-CoA had a twofold role. On the one hand, it emerged during the catabolic breakdown of 4-hydroxybenzoate. On the other hand, it acted as an intermediate in pamamycin biosynthesis, stemming from the union of malonyl-CoA and succinyl-CoA. This particular interaction was instrumental in producing lighter pamamycin derivatives [38]. Given the swifter metabolism of protocatechuate compared to 4-hydroxybenzoate and the substantial increase in the 3-oxoadipyl-CoA pool (Fig. 7), it is plausible that using protocatechuate as a substrate could further amplify the selective synthesis of lighter pamamycins. For a more comprehensive understanding, in future studies, the enzymatic synthesis of methyl-3-oxoadipyl-CoA should be explored, paralleling the approach taken for oxoadipyl-CoA in our study, to facilitate its LC‒MS analysis in cell extracts. This derivative plays a significant role in the pamamycin pathways, guiding the formation of heavier product variants (Fig. 1). In addition, the substrates had an influence on the bottromycin spectrum (Additional file 2).
Beyond pamamycins, S. lividans [22] is a well-known heterologous host to produce natural products and proteins, which is attributed to its acceptance of methylated DNA, its low endogenous protease activity [17, 61], the availability of clean strains that have been cleared of their native natural product gene clusters [18], and the development of promoters for tuneable promotion [62]. This positions S. lividans as an optimized host strain for heterologous expression. In this regard, it seems promising to evaluate S. lividans to produce other compounds from, what one might call, sustainable monomers. As an example, a stable supply of amino acids in the different substrates (Fig. 6) seems to be a valuable trait to synthetize heterologous proteins such as cellulase [63] and human proteins [64] in the host.
The synthesis of natural products from sustainable raw materials promises to be an attractive complement to existing biorefineries
Integrating the production of high-value bioactive natural products into existing biorefineries, which have traditionally focused on low-value bulk chemicals, would offer several compelling advantages. By producing both high- and low-value products, biorefineries can diversify their portfolio, enhancing economic stability in a still challenging market [65, 66]. With regard to the valorisation of lignin, the previously demonstrated production of bulk chemicals such as cis, cis-muconic acid [41, 67, 68], β-ketoadipic acid [69, 70], lactate and pyruvate [71], and aromatic monomers such as ferulic acid and p-coumaric acid as antioxidants [72] could be well complemented by high-value natural products. For seaweed biorefineries, natural product synthesis from algal residuals could be linked into cascaded value chains that sequentially extract high-value ingredients and then ferment the resulting hydrolysates and extracts into value-added chemicals [11]. Thus far, the latter addresses biofuels and bulk chemicals such as ethanol [28, 73, 74], butanol [75, 76], lactate [77], and L-lysine [16], offering promising opportunities for additional value creation by natural product synthesis. It is worth mentioning that when demonstrating pamamycin production in our work, we mimicked the sugar- and mineral-rich residual streams that typically accumulate as a side product from the extraction of algal biomass [78]. This increased diversification and economic resilience would help to buffer biorefineries against market fluctuations in the bulk chemical sector and add revenue streams from premium products [79].
Conclusions
High-value natural products, especially those with therapeutic, cosmetic, or specialized industrial applications, have a growing demand. Notably, the global spending on pharmaceuticals expanded by 56% from 2007 to 2017 [80]. By meeting this demand and integrating microbial production of these types of products, biorefineries could place themselves in advantageous position in the market. In this regard, our findings provide an exciting proof-of-principle for expanding this field. However, one should note that the process is far from being industrially implementable yet. Significant research and development efforts are needed to meet the challenges ahead [81, 82]. On the technological side, these factors relate to reducing energy costs during raw material pretreatment, e.g., drying seaweed [11] or efficiently depolymerizing the rigid structure of lignin [83]. With respect to scaling, the obtained hydrolysates are rather dilute and contain only low levels of available carbon-limiting product titres and demand high-volume fermentation equipment [84]. Furthermore, in practice, raw materials from waste and residual streams are crude mixtures of substrates and might contain toxins, exceeding the capabilities of the cell factories used [85]. In addition, the supply of lignin and seaweed used in industries, except for lignin that is derived from pulp and paper plants, are not centralized, and require the implementation of new structures [86]. To address these issues associated with industrial implementation, we need new interdisciplinary value chains [16, 41, 54, 67]. In this regard, a shift towards producing complex natural products from new raw material streams seems promising to drive innovation, potentially leading to the discovery of new processes, products, or applications and opening doors for collaborative research and development [79].
In addition to S. lividans, other Streptomyces and related strains that are potent in forming natural products should also be considered in future studies. Streptomyces are known to grow well on mannitol and glucose, which are found in seaweed hydrolysates [87]. Furthermore, they contain pathways to degrade a range of other sugars potentially contained in algal streams, such as mannose, galactose, and rhamnose [11], suggesting that seaweed biomass could be valorised in a rather straightforward manner. On the other hand, the spectrum of metabolized aromatics differs quite substantially between strains. As an example, Amycolatopsis sp. can degrade benzoate, phenol, and guaiacol [55], while S. albus lacks the β-ketoadipate pathway necessary for aromatics utilization [88]. In this regard, metabolic engineering strategies for the conversion of aromatic mixtures and aromatic-rich waste streams is likely to be rather strain specific.
Material and methods
Microorganisms and plasmids
Streptomyces lividans ΔYA8 was obtained from previous work [18]. DNA of Pseudomonas putida KT2440 was utilized to amplify the two genes pcaI and pcaJ, encoding 3-oxoadipate:succinyl-CoA transferase [48]. Escherichia coli DH5α (Invitrogen, Carlsbad, USA) was employed for plasmid amplification. E. coli ET12567/pUZ8002 was used for conjugal gene transfer [23]. E. coli BL21 (DE3) (Thermo Fisher Scientific, Karlsruhe, Germany) and the plasmid pET-21a (Invitrogen) were used for protein expression. The cosmids DG2-km-P41hyg (bottromycin biosynthesis-related gene cluster under synthetic promoter control) [20] and R2 (pamamycin biosynthesis-related gene cluster under native promoter control) [38] were taken from previous work. All strains were stored in 20% glycerol at −80 °C. All strains and plasmids are listed in Table 6.
Table 6.
Strains and plasmids
| Strains and plasmids | Description | References |
|---|---|---|
| Strains | ||
| E. coli DH5α | Strain for plasmid amplification | Invitrogen |
| E. coli ET12567 | Strain harbouring pUZ8002 for conjugal gene transfer | [23] |
| E. coli BL21 (DE3) | Strain for high-level expression of recombinant proteins | Thermo Fisher Scientific |
| E. coli p21pcaI | Derivative of E. coli BL21 (DE3) harbouring p21pcaI gene | This work |
| E. coli p21pcaJ | Derivative of E. coli BL21 (DE3) harbouring p21pcaJ gene | This work |
| S. lividans ΔYA8 | Derivative of S. lividans TK24, 8 secondary metabolite gene clusters were removed from genomic DNA | [18] |
| S. lividans ΔYA8-DG2 | Derivative of S. lividans ΔYA8 containing bottromycin biosynthetic cluster | This work |
| S. lividans ΔYA8-R2 | Derivative of S. lividans ΔYA8 containing pamamycin biosynthetic cluster | This work |
| Plasmids | ||
| pET21a | Vector for protein overexpression | Invitrogen |
| p21pcaI | Derivative of pET21a plasmid. Protein expression vector for pcaI gene of P. putida KT2440 | This work |
| p21pcaJ | Derivative of pET21a plasmid. Protein expression vector for pcaJ gene of P. putida KT2440 | This work |
| DG2-Km-P41hyg | Derivative of integrative DG2-cosmid with KmR marker and P41 promoter pairs, contains bottromycin biosynthetic cluster | [20] |
| R2 | An Integrative cosmid containing pamamycin biosynthetic gene cluster | [38] |
Media
LB medium (Becton & Dickinson, Heidelberg, Germany) and terrific broth were used to culture E. coli. The terrific broth contained 24 g of yeast extract (Sigma‒Aldrich, Taufkirchen, Germany), 12 g of tryptone (Fluka, Buchs, Switzerland), 5 g of glycerol, 12.5 g of K2HPO4, and 2.3 g of KH2PO4 per litre. Plate cultures of S. lividans were grown on mannitol soy (MS) flour agar containing 20 g of mannitol, 20 g of soy flour (Schoenenberger Hensel, Magstadt, Germany), and 20 g of agar (Becton & Dickinson) per litre. Liquid cultures of S. lividans involved two sequential precultures, followed by the main culture. Tryptic soy broth (30 g L−1, TSB, Sigma‒Aldrich) was used for the first precultivation. The second precultivation and the main cultivation were conducted in minimal medium [46] containing 200 mM potassium phosphate buffer (pH 7.8): 15 g of (NH4)2SO4, 1 g of NaCl, 200 mg of MgSO4·7H2O, 55 mg of CaCl2, 20 mg of FeSO4·7H2O, 2 mg of FeCl3·6H2O, 2 mg of MnSO4·H2O, 0.5 mg of ZnSO4·H2O, 0.2 mg of CuCl2·2H2O, 0.2 mg of Na2B4O7·10H2O, 0.1 mg of (NH4)6Mo7O24·4H2O, 1 mg of riboflavin, 1 mg of nicotinamide, 0.5 mg of thiamine hydrochloride, 0.5 mg of pyridoxine hydrochloride, 0.2 mg of biotin, and 0.1 mg of p-aminobenzoate per litre. In parallel, mannitol, protocatechuate (PCA), and 4-hydroxybenzoate (4HB) were added as the sole carbon sources, as given below. When needed, kanamycin (50 μg mL−1), apramycin (20 μg mL−1), phosphomycin (200 μg mL−1), and ampicillin (100 μg mL−1) were added to filter sterilized stocks.
Genetic engineering
SnapGene software (GSL Biotech LLC, San Diego, USA) was used for strain and primer design. Transformation and conjugation of S. lividans ΔYA8 was based on standard methods [23]. The site-specific integration of biosynthesis-related gene clusters into the chromosome of S. lividans ΔYA8 was carried out using the phiC31 integrase system [20] and site specific primers (Additional file 1: Table S2). In short, the corresponding cosmid was transformed into E. coli DH5α using heat shock, amplified, isolated (QIAprep Spin MiniPrep Kit, Qiagen, Hilden, Germany), and transformed into E. coli ET12567/pUZ8002 by electroporation. The obtained mutant then served as a donor for conjugal transfer. For this purpose, it was mixed with spores of S. lividans ΔYA8, plated on MS agar, and incubated at 30 °C overnight. For the selection of transconjugants, the agar was then overlaid with phosphomycin and selective antibiotics. Afterwards, the plates were further incubated until sporulation. The obtained transformants were verified for correctness of the desired genetic change by PCR (Phire Green Hot Start II PCR Mastermix, Thermo Scientific, Waltham, MA, USA) and by sequencing. Furthermore, we created E. coli mutants to enable the enzymatic synthesis of 3-oxoadipyl-CoA, which is needed as a standard for its intracellular analysis by LC‒MS/MS [46]. For this purpose, we selected the genes pcaI and pcaJ encoding 3-oxoadipate:succinyl-CoA transferase in the genome of P. putida KT 2440 [48]. First, the backbone of the pET-21a expression vector was digested with FastDigest NdeI and HindIII (Thermo Fisher Scientific) according to the manufacturer’s protocol. Then, the two genes of P. putida KT 2440 were separately amplified from genomic DNA (Q5 HotStart Polymerase, New England Biolabs, Frankfurt am Main, Germany). Following amplification, each gene was cloned and inserted into the vector (Gibson Assembly MasterMix, New England Biolabs). Vector overhangs are underscored. Subsequently, the plasmids were transformed into E. coli DH5α, amplified, isolated, and verified by sequencing. E. coli BL21 (DE3) was then transformed with the expression plasmids using heat shock.
Preparation of brown seaweed hydrolysate
To prepare mannitol-rich seaweed hydrolysate, 50 g of dried Himanthalia elongata (PureRaw, Klötze, Germany) was blended into powder, suspended in 500 mL of deionized water, and extracted (121 °C, 18 min) [16]. Celluclast 1.5 L and Viscozyme L (Sigma‒Aldrich, Steinheim, Germany) were added to the mixture at 0.01 g of the enzyme mix per g of dry biomass, followed by pH adjustment to 5.5. The mixture was incubated over 48 h. Afterwards, the hydrolysate was clarified (4500 ×g, 15 min, 4 °C), and the solution pH was adjusted to pH 7.0 (6 M NaOH). The obtained solution was autoclaved prior to further use.
Batch cultivation in shake flasks
Cultivations were conducted in 500 mL baffled shake flasks filled with 50 mL medium and 30 g soda-lime glass beads (5 mm, Sigma‒Aldrich) on an orbital shaker (230 rpm, 28 °C, 75% relative humidity, 5 cm shaking diameter, Multitron, Infors AG, Bottmingen, Switzerland). For bottromycin production, 107 spores of the corresponding producer were inoculated into TSB medium and incubated for 48 h. Then, the cells were harvested (8000 ×g, 2 min, 25 °C) and transferred to the second preculture in minimal medium containing 10 g L−1 mannitol as the carbon source, followed by incubation over 72 h, harvesting, and inoculation of the main culture, which contained either mannitol, PCA or HBA, as specified below. For pamamycin production, 107 spores of the corresponding producer were inoculated into TSB medium and incubated for 24 h. Then, either 10 mM mannitol or 4-HB was added to the preculture for adaptation, and the preculture was further incubated for an additional 24 h. Cells were collected (8000 ×g, 2 min, 25 °C) and inoculated into the main culture in minimal medium containing mannitol or 4HB. Generally, the main cultures were inoculated to a starting optical density (OD600) = 0.5. All cultivations were carried out in triplicate.
Batch cultivation on seaweed hydrolysate
First, 90% (v/v) seaweed hydrolysate was mixed with 10% (v/v) 2 M MOPS buffer (pH 7.0). As described above, 107 spores of the pamamycin producer were inoculated into TSB medium and incubated for 48 h. Then, the cells were centrifuged (8000 ×g, 2 min, 25 °C) and inoculated into seaweed hydrolysate medium.
Fed-batch cultivation in shake flasks
First, 107 spores of the bottromycin-producing mutant were inoculated into TSB medium and incubated for 48 h. Then, the cells were collected (8000 ×g, 2 min, 25 °C) and inoculated into minimal medium containing 10 g L−1 mannitol. The second preculture was grown for 72 h and then used to inoculate the main culture containing 30 mM 4HB. When the substrate was completely consumed, a pulse of the substrate was added from a concentrated stock (500 mM, pH 7.0), which increased the 4HB level in the broth to 30 mM.
Quantification of cell concentration
Generally, growth was inferred from optical density (OD) based on photometric measurements at 600 nm. In addition, the dry cell weight (CDW) was measured gravimetrically. For this purpose, cells were harvested (10,000 ×g, 4 °C, 10 min), washed with 15 mL deionized water, freeze-dried, and weighed [39]. Systematic measurements provided a substrate-specific correlation between OD600 and CDW, which allowed us to infer the latter from OD readings: CDW (g L−1) = 0.946 × OD600 (mannitol, 4HB), CDW (g L−1) = 0.474 × OD600 (PCA).
Quantification of substrates
Sugars were analysed by HPLC (1260 Infinity Series, Agilent, Waldbronn, Germany) using a column (NUCLEOGEL SUGAR Pb, 300 × 7.8 mm, Macherey–Nagel, Düren, Germany) at 80 °C as the stationary phase and deionized water as the mobile phase (0.4 mL min−1). Refraction index measurements and external standards were used for quantification. HPLC-based analysis of aromatics (PCA, 4HB) involved separation on a C18 column at 25 °C (Nucleodur C18 Isis, 100 × 3 mm, Macherey–Nagel) with a gradient of 0.025% H3PO4 and acetonitrile (1 mL min−1) [55]. The analytes were detected by UV absorbance at compound-specific wavelengths (210 nm for PCA, 260 nm for 4HB). External standards were used for quantification.
Quantification of intracellular amino acids
Intracellular amino acids were quantified as previously described [89, 90]. In brief, 2 mg of biomass was harvested and vacuum-filtered (cellulose nitrate, 0.2 μm pore size, 47 mm, Sartorius, Göttingen, Germany). The filter with the cells was washed (15 mL 2.5% NaCl, 25 °C) and quickly transferred into 2 mL of a 200 μM α-aminobutyrate solution, followed by extraction in boiling water (15 min, 100 °C). Afterwards, the extract was cooled on ice and clarified from debris (20,000 ×g, 5 min, 4 °C). The supernatant was used for analysis. For this purpose, the amino acids were separated by HPLC on a reversed-phase column (Gemini 5 µm C18 110 Å, 150 × 4.6 mm, Phenomenex) after precolumn derivatization with o-phthaldialdehyde and fluorenylmethyloxycarbonyl chloride [91]. Quantification was based on α-aminobutyrate as an internal standard.
Analysis of bottromycin and pamamycin
In brief, 300 μL of culture broth was mixed with 300 μL of acetone and shaken for 10 min at room temperature (1000 rpm, Thermomixer F1.5; Eppendorf, Wesseling, Germany). Then, 300 μL of ethyl acetate was added, and the mixture was incubated under the same conditions for another 10 min. The organic phase was separated (20,000 ×g, room temperature, 10 min), collected, and evaporated under nitrogen. The obtained extract was dissolved in 300 μL methanol, clarified by centrifugation (20,000 ×g, 4 °C, 10 min) and analysed by LC–ESI–MS (Agilent Infinity 1290; AB Sciex QTrap 6500, Darmstadt, Germany) [20, 21, 46]. Separation of the analytes was conducted using a reversed-phase column (Vision HT C18 HighLoad, 100 × 2 mm, Dr. Maisch, Ammerbuch-Entringen, Germany) operated at 45 °C at a flow rate of 0.55 mL min−1 with the following linear gradient of 0.1% formic acid in deionized water (A) and 0.1% formic acid in acetonitrile (B): 0–11 min, 95–5% A, 5–95% B. Based on LC–ESI–MS analysis [20], bottromycin A2 ([M + H] + = 823.453) was the major derivative formed, while methylated bottromycin A2 [M + H] + = 837.453) accumulated in traces. As a reference, a commercial standard (purity 85%) was obtained from Cayman Chemicals (Ann Arbor, MI, USA). Pamamycins were separated using 8 mM ammonium formate in 92% acetonitrile at a flow rate of 0.3 mL min−1 and analysed by LC–ESI–MS as described previously [39]. The chromatographic and mass spectrometric settings for natural product analysis are described in Additional file 1: Table S3.
Enzymatic synthesis and purification of 3-oxoadipyl-CoA
Escherichia coli BL21 (DE3) harbouring p21pcaI and p21pcaJ, respectively, was plated on LB agar supplemented with the appropriate antibiotics. After overnight incubation at 37 °C, a single colony of each mutant was inoculated in 10 mL of antibiotic-amended LB medium (100 mL shake flask), grown overnight on an orbital shaker (37 °C, 160 rpm) and inoculated into 1 L of terrific broth containing the respective antibiotics (3 L shake flask). When the OD reached 0.7, the cultures were cooled to 23 °C, and protein production was induced by the addition of IPTG to a level of 0.5 mM. The incubation was continued overnight. Afterwards, the cells were harvested (5000 ×g, 4 °C, 45 min) and resuspended in buffer A (50 mM HEPES–KOH, 450 mM NaCl, 15% (v/v) glycerol, pH 7.6) at a ratio of 2:1 (v/w), followed by sonication for cell lysis. Cell debris was removed by centrifugation (55,000 ×g, 4 °C, 45 min). The supernatant was purified using Ni–NTA agarose beads (Protino, Macherey–Nagel, Düren, Germany), dialyzed over a PD-10 column (Cytiva, Freiburg, Germany), and concentrated (Amicon centrifugal filter, Merck, Darmstadt, Germany), and the protein size was verified via SDS‒PAGE (PcaI = 26.6 kDa; PcaJ = 24.7 kDa). The protein concentration was determined spectroscopically at 260 nm using the corresponding molar extinction coefficient [92]. All CoA-thioesters were purified by a 1260 Infinity LC system (Agilent) using a reversed-phase column (Gemini 10 μm NX-C18 110 Å, 100 × 21.2 mm AXIA packed column, Phenomenex, Aschaffenburg, Germany). Succinyl-CoA was obtained using previously established methods [93], wasy purified by a gradient of solvent A (25 mM NH4HCO2, pH 4.2) and solvent B (methanol) at a flow rate of 25 mL min−1: 5–23% B in 15 min, followed by 3 min washing (95% B) and subsequent re-equilibration of the column for 3 min (5% B), and was then used as a substrate. For enzymatic synthesis of 3-oxoadipyl-CoA, 30 mg of succinyl-CoA (8.25 mM final concentration, 1 eq.) was dissolved in 25 mM ammonium formate (pH 4.2) and added to a mixture containing 33 mM 3-oxoadipic acid (4 eq.), 20 mM MgCl2, 100 mM KHCO3 and 100 mM HEPES–KOH (pH 7.5) at a final volume of 4 mL. After brief equilibration at 30 °C, 10 µM of each PcaI and PcaJ was added, and the mixture was further incubated (30 °C, 200 rpm). After 3 h, the assay was quenched with a final concentration of 10% (v/v) formic acid, centrifuged (4500 × g, room temperature, 20 min), and filtered (0.45 µm). The purification of 3-oxoadipyl-CoA was conducted by using solvent A (50 mM NH4CH3CO2, pH 8.1) and solvent B (methanol) at isocratic flow (2.5% of solvent B) for 17 min with a flow rate of 25 mL min−1, followed by 3 min washing at 95% B and 3 min re-equilibration with 2.5% B. Fractions containing either product were pooled, flash-frozen in liquid nitrogen, lyophilized and stored at − 20 °C. The obtained concentrations were determined photometrically, considering the known extinction coefficient of saturated acyl-CoA (ε260 nm = 16.4 mM−1 cm−1) [93, 94].
Analysis of intracellular CoA thioesters
Intracellular CoA thioesters were quantified using a recently established protocol [39, 46] with slight adaptations. In short, cell broth, containing approximately 8 mg CDW, was transferred into a quenching solution (95% acetonitrile with 25 mM formic acid, − 20 °C) at a volume ratio of 1:2, mixed and kept on ice for 10 min. Cell debris was removed (10 min, 4 °C, 10,000 ×g). The supernatant was transferred into 5 ml of supercooled deionized water. The remaining pellet was washed once with supercooled deionized water. Then, the two supernatants were combined, frozen in liquid nitrogen, and lyophilized. The obtained extract was resuspended in 1 mL of resuspension buffer (25 mM ammonium formate, pH 5.6, 2% methanol, 4 °C). Afterwards, the CoA-thioesters were analysed using LC‒ESI‒MS/MS. Analyte separation was conducted on a reversed-phase column (Kinetex 2.6 μ XB-C18 100 Å, 100 × 2.1 mm, Phenomenex) at 40 °C with a gradient of eluent A (50 mM formic acid, adjusted to pH 8.1 with 25% ammonium hydroxide) and eluent B (methanol) at a flow rate of 300 μL min−1. Multiple reaction monitoring (MRM) was used for the detection of CoA thioesters. Absolute quantification of thioesters was performed as described previously [46]. For absolute quantification of 3-oxoadipyl-CoA, a 13C-labelled internal standard was newly obtained from S. lividans cells pregrown on 99% 4-hydroxybenzoic acid-[phenyl-13C6] (Sigma‒Aldrich) [46]. The optimized instrumental settings applied for the analysis of 3-oxoadipyl-CoA were operation in positive ion mode [M + H]+ and specific values for the curtain gas (35 psi), collision gas flow rate (medium), ion spray voltage (4.5 kV), temperature (400 °C), ion source gas (60 psi), entrance potential (10 V), mass of the parent ion (m/z 910.3) and the daughter ion (m/z 403.1), declustering potential (173 V), collision energy (43.2 V) and cell exit potential (24.3 V). The settings for the other CoA esters were taken from previous work [46, 93, 95, 96].
Transcriptomic analysis
Samples were collected from the production process after 12 h (20,000 ×g, 1 min, 4 °C) and immediately frozen in liquid nitrogen. Sample processing, RNA extraction, and RNA sequencing were performed in biological triplicate as described previously [21, 25, 39]. In short, cells (1 mL broth) were collected by centrifugation (20,000 ×g, 4 °C, 1 min) and immediately frozen in liquid nitrogen. RNA was extracted with the Qiagen RNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Residual DNA was removed by digestion with 10 U RNase-free DNase I (Thermo Scientific) for 1 h in the presence of RiboLock RNase inhibitor (Thermo Scientific). After DNA digestion, the RNA was again purified with the same kit. RNA quality was checked by Trinean Xpose (Gentbrugge, Belgium) and the Agilent RNA 6000 Nano Kit on an Agilent 2100 Bioanalyzer (Agilent Technologies, Böblingen, Germany). Ribosomal RNA (rRNA) molecules were removed from the total RNA with the Ribo-Zero rRNA Removal Kit (Illumina, San Diego, USA). The removal of rRNA was checked with the Agilent RNA 6000 Pico Kit on an Agilent 2100 Bioanalyzer (Agilent Technologies). cDNA libraries were prepared with the TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, USA), and the resulting cDNA was paired-end sequenced on an Illumina NextSeq 500 system using a 2 × 75 bp read length. Reads were mapped to the S. lividans TK24 ΔYA8-DG2 genome sequence (CP111182.1) with Bowtie2 using standard settings [97], except the maximal allowed distance for paired reads was increased to 600 bases. For visualization of read alignments and raw read count calculation, ReadXplorer 2.2.3 was used [98]. Using the resulting data, DESeq2 [99] was used to QC the datasets via, for example, calculation of the sample-to-sample distances (Additional file 1: Fig. S3) and PCA (Additional file 1: Fig. S2). In addition, DESeq2 was used to calculate the DGE datasets. The raw datasets (sequenced reads) as well as processed datasets (input matrix & normalized read counts from DESeq2) are available from GEO (GSE246798). For statistical analysis, Student’s t test was carried out, and the data were filtered for genes with a log2-fold change ≥ 1 (p value ≤ 0.05). Hierarchical clustering was conducted using the software package gplots [100, 101]. RNA extraction and sequencing were conducted in biological triplicates.
Data processing and statistical analysis
All results displayed in Figures and Tables are shown as the mean values ± standard deviations (SD). Statistical evaluation of the data was conducted by one-way analysis of variance (ANOVA) [102]. Statistical analyses were performed by using SPSS (version 24.0).
Supplementary Information
Additional file 1: Table S1. BLAST analysis of amino acid sequences of genes responsible for the ethylmalonyl-CoA pathway in S. lividans ΔYA8-DG2 against S. venezuelae ATCC 15439. Table S2. Primers for genetic engineering and sequencing. Table S3. Chromatographic and mass spectrometric settings during LC–ESI–MS analysis. The conditions for the analysis of bottromycins [20] and pamamycins [39] were adapted from previous work. Figure S1. Substrate screening of S. lividans TK24. The minimal plate medium used contained 10 g L−1 mannitol (A), 5 mM protocatechuic acid (B), 5 mM 4-hydroxybenzoic acid (C) as the sole source of carbon, or no carbon source (D). The plates were incubated for 5 days at 28 °C. The different concentrations used were chosen based on the expected toxicity of the aromatics. Figure S2. Quality assessment of RNA sequencing using PCA. The data comprise the global transcriptomes of S. lividans ΔYA8-DG2 grown on mannitol (10 mM), protocatechuate (10 mM), or 4-hydroxybenzoate (10 mM) and sampled after 12 h, n = 3. Figure S3. Sample distance plot. The data originate from RNA sequencing of S. lividans ΔYA8-DG2 grown on mannitol (10 mM), protocatechuate (10 mM), or 4-hydroxybenzoate (10 mM) and sampled after 12 h, n = 3. Figure S4. Volcano plot. The data reflect the differences in gene expression between mannitol- and protocatechuate-grown S. lividans ΔYA8-DG2 sampled after 12 h, n = 3. Figure S5. Volcano plot. The data reflect the differences in gene expression between mannitol- and 4-hydroxybenzoate-grown S. lividans ΔYA8-DG2 sampled after 12 h, n = 3. Figure S6. Correlation between optical density and cell dry weight for S. lividans ΔYA8-DG2. The strain was grown on 10 mM protocatechuate (A) and 30 mM mannitol (B), and parallel measurements at different culture time points were performed, n = 3.
Additional file 2. LC-MS analysis of bottromycin A2 and methylated bottromycin A2.
Additional file 3. Gene expression data reflecting genes linked to amino acid biosynthesis.
Additional file 4. Gene expression data reflecting genes linked to oxidative stress response and energy metabolism.
Acknowledgements
This work is dedicated to the memory of Dr. Judith Becker (2.2. 1981–27.4.2021), our close and cherished colleague at the Institute of Systems Biotechnology, Saarland University, and a true friend.
Author contributions
KS conducted genetic engineering, cultures of the strains, and intracellular and extracellular analytics. PG contributed to CoA-ester synthesis for analytical purposes. WS prepared seaweed extracts. CR and JK performed RNA sequencing and data processing. CW conceived and structured the work. KS and CW wrote the first draft of the manuscript. KS and CW prepared all figures. All authors critically commented, improved, and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. JK, CR, and CW acknowledge financial support from the Federal Ministry of Education and Research, Germany, through grant Explomare (031B0868). CW acknowledges financial support from the Federal Ministry of Education and Research, Germany, through grant ERA CoBioTech MISSION (031B0611) and the German Research Foundation through grant INST 256/418–1.
Availability of data and materials
All dataset(s) supporting the conclusions of this article are included within the article.
Declarations
Ethics approval and consent to participate
Not applicable. The manuscript does not contain data collected from humans or animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ögmundarson Ó, Sukumara S, Laurent A, Fantke P. Environmental hotspots of lactic acid production systems. GCB Bioenergy. 2020;12:19–38. doi: 10.1111/gcbb.12652. [DOI] [Google Scholar]
- 2.Drescher A, Kienberger M. A systematic review on waste as sustainable feedstock for bioactive molecules-extraction as isolation technology. Processes. 2022;10:1668. doi: 10.3390/pr10081668. [DOI] [Google Scholar]
- 3.Lichtenthaler FW, Peters S. Carbohydrates as green raw materials for the chemical industry. C R Chim. 2004;7:65–90. doi: 10.1016/j.crci.2004.02.002. [DOI] [Google Scholar]
- 4.Wang Y, Fang X, An F, Wang G, Zhang X. Improvement of antibiotic activity of Xenorhabdus bovienii by medium optimization using response surface methodology. Microb Cell Fact. 2011;10:98. doi: 10.1186/1475-2859-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahsan T, Chen JG, Wu YH, Irfan M. Application of response surface methodology for optimization of medium components for the production of secondary metabolites by Streptomyces diastatochromogenes KX852460. AMB Express. 2017;7:1. doi: 10.1186/s13568-017-0388-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elibol M. Optimization of medium composition for actinorhodin production by Streptomyces coelicolor A3(2) with response surface methodology. Process Biochem. 2004;39:1057–1062. doi: 10.1016/S0032-9592(03)00232-2. [DOI] [Google Scholar]
- 7.Straathof AJJ, Wahl SA, Benjamin KR, Takors R, Wierckx N, Noorman HJ. Grand research challenges for sustainable industrial biotechnology. Trends Biotechnol. 2019;37:1042–1050. doi: 10.1016/j.tibtech.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Jong E, Stichnothe H, Bell G, Jørgensen H. Bio-based chemicals: a 2020 update. In: Jong E, editor. IEA bioenergy, volume task 42. Wageningen: IEA; 2020. [Google Scholar]
- 9.Rahimi A, García JM. Chemical recycling of waste plastics for new materials production. Nat Rev Chem. 2017;1:0046. doi: 10.1038/s41570-017-0046. [DOI] [Google Scholar]
- 10.Becker J, Wittmann C. A field of dreams: Lignin valorization into chemicals, materials, fuels, and health-care products. Biotechnol Adv. 2019;37:107360. doi: 10.1016/j.biotechadv.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Poblete-Castro I, Hoffmann SL, Becker J, Wittmann C. Cascaded valorization of seaweed using microbial cell factories. Curr Opin Biotechnol. 2020;65:102–113. doi: 10.1016/j.copbio.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Yun JH, Archer SD, Price NN. Valorization of waste materials from seaweed industry: an industry survey based biorefinery approach. Rev Aquac. 2023;15:1020–1027. doi: 10.1111/raq.12748. [DOI] [Google Scholar]
- 13.Seghetta M, Tørring D, Bruhn A, Thomsen M. Bioextraction potential of seaweed in Denmark: an instrument for circular nutrient management. Sci Total Environ. 2016;563–564:513–529. doi: 10.1016/j.scitotenv.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan KP, Werner AZ, Ramirez KJ, Ellis LD, Bussard JR, Black BA, Brandner DG, Bratti F, Buss BL, Dong X, et al. Mixed plastics waste valorization through tandem chemical oxidation and biological funneling. Science. 2022;378:207. doi: 10.1126/science.abo4626. [DOI] [PubMed] [Google Scholar]
- 15.Weiland F, Kohlstedt M, Wittmann C. Guiding stars to the field of dreams: metabolically engineered pathways and microbial platforms for a sustainable lignin-based industry. Metab Eng. 2022;71:13–41. doi: 10.1016/j.ymben.2021.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann SL, Kohlstedt M, Jungmann L, Hutter M, Poblete-Castro I, Becker J, Wittmann C. Cascaded valorization of brown seaweed to produce l-lysine and value-added products using Corynebacterium glutamicum streamlined by systems metabolic engineering. Metab Eng. 2021;67:293–307. doi: 10.1016/j.ymben.2021.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Myronovskyi M, Luzhetskyy A. Heterologous production of small molecules in the optimized Streptomyces hosts. Nat Prod Rep. 2019;36:1281–1294. doi: 10.1039/C9NP00023B. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed Y, Rebets Y, Estévez MR, Zapp J, Myronovskyi M, Luzhetskyy A. Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters. Microb Cell Fact. 2020;19:5. doi: 10.1186/s12934-020-1277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang S, Joung C, Kim W, Palsson B, Cho B-K. Recent advances in non-model bacterial chassis construction. Curr Opin Syst Biol. 2023;36:100471. doi: 10.1016/j.coisb.2023.100471. [DOI] [Google Scholar]
- 20.Horbal L, Marques F, Nadmid S, Mendes MV, Luzhetskyy A. Secondary metabolites overproduction through transcriptional gene cluster refactoring. Metab Eng. 2018;49:299–315. doi: 10.1016/j.ymben.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 21.Kuhl M, Rückert C, Gläser L, Beganovic S, Luzhetskyy A, Kalinowski J, Wittmann C. Microparticles enhance the formation of seven major classes of natural products in native and metabolically engineered actinobacteria through accelerated morphological development. Biotechnol Bioeng. 2021;118:3076–3093. doi: 10.1002/bit.27818. [DOI] [PubMed] [Google Scholar]
- 22.Rückert C, Albersmeier A, Busche T, Jaenicke S, Winkler A, Friðjónsson OH, Hreggviðsson GO, Lambert C, Badcock D, Bernaerts K, et al. Complete genome sequence of Streptomyces lividans TK24. J Biotechnol. 2015;199:21–22. doi: 10.1016/j.jbiotec.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. Norwich: The John Innes Foundation; 2000. [Google Scholar]
- 24.Vicente RL, Gullón S, Marín S, Mellado RP. The Three Streptomyces lividans HtrA-like proteases involved in the secretion stress response act in a cooperative manner. PLoS ONE. 2016;11:e0168112. doi: 10.1371/journal.pone.0168112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gläser L, Kuhl M, Stegmüller J, Rückert C, Myronovskyi M, Kalinowski J, Luzhetskyy A, Wittmann C. Superior production of heavy pamamycin derivatives using a bkdR deletion mutant of Streptomyces albus J1074/R2. Microb Cell Fact. 2021;20:111. doi: 10.1186/s12934-021-01602-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beganovic S, Rückert-Reed C, Sucipto H, Shu W, Gläser L, Patschkowski T, Struck B, Kalinowski J, Luzhetskyy A, Wittmann C. Systems biology of industrial oxytetracycline production in Streptomyces rimosus : the secrets of a mutagenized hyperproducer. Microb Cell Fact. 2023;22:222. doi: 10.1186/s12934-023-02215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsson J, Toth GB, Albers E. Biochemical composition of red, green and brown seaweeds on the Swedish west coast. J Appl Phycol. 2020;32:3305–3317. doi: 10.1007/s10811-020-02145-w. [DOI] [Google Scholar]
- 28.Chades T, Scully SM, Ingvadottir EM, Orlygsson J. Fermentation of mannitol extracts from brown macro algae by Thermophilic Clostridia. Front Microbiol. 1931;2018:9. doi: 10.3389/fmicb.2018.01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otsuka Y, Nakamura M, Shigehara K, Sugimura K, Masai E, Ohara S, Katayama Y. Efficient production of 2-pyrone 4,6-dicarboxylic acid as a novel polymer-based material from protocatechuate by microbial function. Appl Microbiol Biotechnol. 2006;71:608–614. doi: 10.1007/s00253-005-0203-7. [DOI] [PubMed] [Google Scholar]
- 30.Niu W, Willett H, Mueller J, He X, Kramer L, Ma B, Guo J. Direct biosynthesis of adipic acid from lignin-derived aromatics using engineered Pseudomonas putida KT2440. Metab Eng. 2020;59:151–161. doi: 10.1016/j.ymben.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Johnson CW, Salvachúa D, Rorrer NA, Black BA, Vardon DR, St John PC, Cleveland NS, Dominick G, Elmore JR, Grundl N, et al. Innovative chemicals and materials from bacterial aromatic catabolic pathways. Joule. 2019;3:1523–1537. doi: 10.1016/j.joule.2019.05.011. [DOI] [Google Scholar]
- 32.Kosa M, Ragauskas AJ. Bioconversion of lignin model compounds with oleaginous Rhodococci. Appl Microbiol Biotechnol. 2012;93:891–900. doi: 10.1007/s00253-011-3743-z. [DOI] [PubMed] [Google Scholar]
- 33.Waisvisz JM, van der Hoeven MG, van Peppen J, Zwennis WCM. Bottromycin. I. A new sulfur-containing antibiotic. J Am Chem Soc. 1957;79:4520–4521. doi: 10.1021/ja01573a072. [DOI] [Google Scholar]
- 34.Kobayashi Y, Ichioka M, Hirose T, Nagai K, Matsumoto A, Matsui H, Hanaki H, Masuma R, Takahashi Y, Ōmura S, Sunazuka T. Bottromycin derivatives: efficient chemical modifications of the ester moiety and evaluation of anti-MRSA and anti-VRE activities. Bioorg Med Chem Lett. 2010;20:6116–6120. doi: 10.1016/j.bmcl.2010.08.037. [DOI] [PubMed] [Google Scholar]
- 35.Franz L, Kazmaier U, Truman AW, Koehnke J. Bottromycins - biosynthesis, synthesis and activity. Nat Prod Rep. 2021;38:1659–1683. doi: 10.1039/D0NP00097C. [DOI] [PubMed] [Google Scholar]
- 36.Kondo S, Yasui K, Natsume M, Katayama M, Marumo S. Isolation, physico-chemical properties and biological activity of pamamycin-607, an aerial mycelium-inducing substance from Streptomyces alboniger. J Antibiot. 1988;41:1196–1204. doi: 10.7164/antibiotics.41.1196. [DOI] [PubMed] [Google Scholar]
- 37.Hanquet G, Salom-Roig X, Lanners S. New insights into the synthesis and biological activity of the Pamamycin Macrodiolides. Chimia. 2016;70:20–28. doi: 10.2533/chimia.2016.20. [DOI] [PubMed] [Google Scholar]
- 38.Rebets Y, Brotz E, Manderscheid N, Tokovenko B, Myronovskyi M, Metz P, Petzke L, Luzhetskyy A. Insights into the pamamycin biosynthesis. Angew Chem Int Ed Engl. 2015;54:2280–2284. doi: 10.1002/anie.201408901. [DOI] [PubMed] [Google Scholar]
- 39.Kuhl M, Glaser L, Rebets Y, Ruckert C, Sarkar N, Hartsch T, Kalinowski J, Luzhetskyy A, Wittmann C. Microparticles globally reprogram Streptomyces albus toward accelerated morphogenesis, streamlined carbon core metabolism, and enhanced production of the antituberculosis polyketide pamamycin. Biotechnol Bioeng. 2020;117:3858–3875. doi: 10.1002/bit.27537. [DOI] [PubMed] [Google Scholar]
- 40.Becker J, Zelder O, Hafner S, Schroder H, Wittmann C. From zero to hero-Design-based systems metabolic engineering of Corynebacterium glutamicum for L-lysine production. Metab Eng. 2011;13:159–168. doi: 10.1016/j.ymben.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 41.Weiland F, Barton N, Kohlstedt M, Becker J, Wittmann C. Systems metabolic engineering upgrades Corynebacterium glutamicum to high-efficiency cis, cis-muconic acid production from lignin-based aromatics. Metab Eng. 2023;75:153–169. doi: 10.1016/j.ymben.2022.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Wang C, Song K, Wen J. Metabolic network model guided engineering ethylmalonyl-CoA pathway to improve ascomycin production in Streptomyces hygroscopicus var. ascomyceticus. Microb Cell Fact. 2017;16:169. doi: 10.1186/s12934-017-0787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertram R, Schlicht M, Mahr K, Nothaft H, Saier MH, Jr, Titgemeyer F. In silico and transcriptional analysis of carbohydrate uptake systems of Streptomyces coelicolor A3(2) J Bacteriol. 2004;186:1362–1373. doi: 10.1128/JB.186.5.1362-1373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiefer P, Heinzle E, Zelder O, Wittmann C. Comparative metabolic flux analysis of lysine-producing Corynebacterium glutamicum cultured on glucose or fructose. Appl Environ Microbiol. 2004;70:229–239. doi: 10.1128/AEM.70.1.229-239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffmann SL, Jungmann L, Schiefelbein S, Peyriga L, Cahoreau E, Portais JC, Becker J, Wittmann C. Lysine production from the sugar alcohol mannitol: design of the cell factory Corynebacterium glutamicum SEA-3 through integrated analysis and engineering of metabolic pathway fluxes. Metab Eng. 2018;47:475–487. doi: 10.1016/j.ymben.2018.04.019. [DOI] [PubMed] [Google Scholar]
- 46.Gläser L, Kuhl M, Jovanovic S, Fritz M, Vogeli B, Erb TJ, Becker J, Wittmann C. A common approach for absolute quantification of short chain CoA thioesters in prokaryotic and eukaryotic microbes. Microb Cell Fact. 2020;19:160. doi: 10.1186/s12934-020-01413-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jovanovic Gasovic S, Dietrich D, Gläser L, Cao P, Kohlstedt M, Wittmann C. Multi-omics view of recombinant Yarrowia lipolytica: Enhanced ketogenic amino acid catabolism increases polyketide-synthase-driven docosahexaenoic production to high selectivity at the gram scale. Metab Eng. 2023;80:45–65. doi: 10.1016/j.ymben.2023.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Parales RE, Harwood CS. Characterization of the genes encoding beta-ketoadipate: succinyl-coenzyme A transferase in Pseudomonas putida. J Bacteriol. 1992;174:4657–4666. doi: 10.1128/jb.174.14.4657-4666.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gummerlich N, Manderscheid N, Rebets Y, Myronovskyi M, Gläser L, Kuhl M, Wittmann C, Luzhetskyy A. Engineering the precursor pool to modulate the production of pamamycins in the heterologous host S. albus J1074. Metab Eng. 2021;67:11–18. doi: 10.1016/j.ymben.2021.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Cho JY, Moon JH, Seong KY, Park KH. Antimicrobial activity of 4-hydroxybenzoic acid and 4-hydroxycinnamic acid isolated and identified from rice hull. Biosci Biotechnol Biochem. 1998;62:2273–2276. doi: 10.1271/bbb.62.2273. [DOI] [PubMed] [Google Scholar]
- 51.Goacher RE, Mottiar Y, Mansfield SD. ToF-SIMS imaging reveals that p-hydroxybenzoate groups specifically decorate the lignin of fibres in the xylem of poplar and willow. Holzforschung. 2021;75:452–462. doi: 10.1515/hf-2020-0130. [DOI] [Google Scholar]
- 52.Smith DC. p-Hydroxybenzoate groups in the lignin of aspen (Populus Tremula) J Chem Soc. 1955 doi: 10.1039/jr9550002347. [DOI] [Google Scholar]
- 53.Rathore AS, Kumar D, Kateja N. Role of raw materials in biopharmaceutical manufacturing: risk analysis and fingerprinting. Curr Opin Biotechnol. 2018;53:99–105. doi: 10.1016/j.copbio.2017.12.022. [DOI] [PubMed] [Google Scholar]
- 54.Kohlstedt M, Starck S, Barton N, Stolzenberger J, Selzer M, Mehlmann K, Schneider R, Pleissner D, Rinkel J, Dickschat JS, et al. From lignin to nylon: cascaded chemical and biochemical conversion using metabolically engineered Pseudomonas putida. Metab Eng. 2018;47:279–293. doi: 10.1016/j.ymben.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 55.Barton N, Horbal L, Starck S, Kohlstedt M, Luzhetskyy A, Wittmann C. Enabling the valorization of guaiacol-based lignin: Integrated chemical and biochemical production of cis, cis-muconic acid using metabolically engineered Amycolatopsis sp ATCC 39116. Metab Eng. 2018;45:200–210. doi: 10.1016/j.ymben.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 56.Kohlstedt M, Sappa PK, Meyer H, Maaß S, Zaprasis A, Hoffmann T, Becker J, Steil L, Hecker M, van Dijl JM, et al. Adaptation of Bacillus subtilis carbon core metabolism to simultaneous nutrient limitation and osmotic challenge: a multi-omics perspective. Environ Microbiol. 2014;16:1898–1917. doi: 10.1111/1462-2920.12438. [DOI] [PubMed] [Google Scholar]
- 57.Lewis RA, Laing E, Allenby N, Bucca G, Brenner V, Harrison M, Kierzek AM, Smith CP. Metabolic and evolutionary insights into the closely-related species Streptomyces coelicolor and Streptomyces lividans deduced from high-resolution comparative genomic hybridization. BMC Genomics. 2010;11:682. doi: 10.1186/1471-2164-11-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Renilla S, Bernal V, Fuhrer T, Castaño-Cerezo S, Pastor JM, Iborra JL, Sauer U, Cánovas M. Acetate scavenging activity in Escherichia coli: interplay of acetyl-CoA synthetase and the PEP-glyoxylate cycle in chemostat cultures. Appl Microbiol Biotechnol. 2012;93:2109–2124. doi: 10.1007/s00253-011-3536-4. [DOI] [PubMed] [Google Scholar]
- 59.Alber BE. Biotechnological potential of the ethylmalonyl-CoA pathway. Appl Microbiol Biotechnol. 2011;89:17–25. doi: 10.1007/s00253-010-2873-z. [DOI] [PubMed] [Google Scholar]
- 60.Chan YA, Podevels AM, Kevany BM, Thomas MG. Biosynthesis of polyketide synthase extender units. Nat Prod Rep. 2009;26:90–114. doi: 10.1039/B801658P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Busche T, Tsolis KC, Koepff J, Rebets Y, Rückert C, Hamed MB, Bleidt A, Wiechert W, Lopatniuk M, Yousra A, et al. Multi-omics and targeted approaches to determine the role of cellular proteases in Streptomyces protein secretion. Front Microbiol. 2018;9:1174. doi: 10.3389/fmicb.2018.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao M, Wang SL, Tao XY, Zhao GL, Ren YH, Wang FQ, Wei DZ. Engineering diverse eubacteria promoters for robust gene expression in Streptomyces lividans. J Biotechnol. 2019;289:93–102. doi: 10.1016/j.jbiotec.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 63.Daniels W, Bouvin J, Busche T, Rückert C, Simoens K, Karamanou S, Van Mellaert L, Friðjónsson ÓH, Nicolai B, Economou A, et al. Transcriptomic and fluxomic changes in Streptomyces lividans producing heterologous protein. Microb Cell Fact. 2018;17:198. doi: 10.1186/s12934-018-1040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bender E, Koller KP, Engels JW. Secretory synthesis of human interleukin-2 by Streptomyces lividans. Gene. 1990;86:227–232. doi: 10.1016/0378-1119(90)90283-W. [DOI] [PubMed] [Google Scholar]
- 65.Söderholm P. The green economy transition: the challenges of technological change for sustainability. Sustainable Earth. 2020;3:6. doi: 10.1186/s42055-020-00029-y. [DOI] [Google Scholar]
- 66.Gallezot P. Process options for converting renewable feedstocks to bioproducts. Green Chem. 2007;9:295–302. doi: 10.1039/b615413a. [DOI] [Google Scholar]
- 67.Kohlstedt M, Weimer A, Weiland F, Stolzenberger J, Selzer M, Sanz M, Kramps L, Wittmann C. Biobased PET from lignin using an engineered cis, cis-muconate-producing Pseudomonas putida strain with superior robustness, energy and redox properties. Metab Eng. 2022;72:337–352. doi: 10.1016/j.ymben.2022.05.001. [DOI] [PubMed] [Google Scholar]
- 68.Sonoki T, Takahashi K, Sugita H, Hatamura M, Azuma Y, Sato T, Suzuki S, Kamimura N, Masai E. Glucose-free cis, cis-muconic acid production via new metabolic designs corresponding to the heterogeneity of lignin. ACS Sustain Chem Eng. 2018;6:1256–1264. doi: 10.1021/acssuschemeng.7b03597. [DOI] [Google Scholar]
- 69.Okamura-Abe Y, Abe T, Nishimura K, Kawata Y, Sato-Izawa K, Otsuka Y, Nakamura M, Kajita S, Masai E, Sonoki T, Katayama Y. Beta-ketoadipic acid and muconolactone production from a lignin-related aromatic compound through the protocatechuate 3,4-metabolic pathway. J Biosci Bioeng. 2016;121:652–658. doi: 10.1016/j.jbiosc.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki Y, Otsuka Y, Araki T, Kamimura N, Masai E, Nakamura M, Katayama Y. Lignin valorization through efficient microbial production of β-ketoadipate from industrial black liquor. Biores Technol. 2021;337:125489. doi: 10.1016/j.biortech.2021.125489. [DOI] [PubMed] [Google Scholar]
- 71.Johnson CW, Beckham GT. Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metab Eng. 2015;28:240–247. doi: 10.1016/j.ymben.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 72.Karagoz P, Khiawjan S, Marques MPC, Santzouk S, Bugg TDH, Lye GJ. Pharmaceutical applications of lignin-derived chemicals and lignin-based materials: linking lignin source and processing with clinical indication. Biomass Convers Biorefinery. 2023 doi: 10.1007/s13399-023-03745-5. [DOI] [Google Scholar]
- 73.Sunwoo I, Kwon JE, Jeong GT, Kim SK. Optimization of hyper-thermal acid hydrolysis and enzymatic saccharification of Ascophyllum nodosum for ethanol production with mannitol-adapted yeasts. Bioprocess Biosyst Eng. 2019;42:1255–1262. doi: 10.1007/s00449-019-02123-8. [DOI] [PubMed] [Google Scholar]
- 74.Sasaki Y, Takagi T, Motone K, Shibata T, Kuroda K, Ueda M. Direct bioethanol production from brown macroalgae by co-culture of two engineered Saccharomyces cerevisiae strains. Biosci Biotechnol Biochem. 2018;82:1459–1462. doi: 10.1080/09168451.2018.1467262. [DOI] [PubMed] [Google Scholar]
- 75.Sunwoo IY, Hau NT, Ra CH, Jeong GT, Kim SK. Acetone-butanol-ethanol production from waste seaweed collected from Gwangalli Beach, Busan, Korea, based on pH-controlled and sequential fermentation using two strains. Appl Biochem Biotechnol. 2018;185:1075–1087. doi: 10.1007/s12010-018-2711-9. [DOI] [PubMed] [Google Scholar]
- 76.Hou X, From N, Angelidaki I, Huijgen WJJ, Bjerre AB. Butanol fermentation of the brown seaweed Laminaria digitata by Clostridium beijerinckii DSM-6422. Biores Technol. 2017;238:16–21. doi: 10.1016/j.biortech.2017.04.035. [DOI] [PubMed] [Google Scholar]
- 77.Mazumdar S, Bang J, Oh MK. L-lactate production from seaweed hydrolysate of Laminaria japonica using metabolically engineered Escherichia coli. Appl Biochem Biotechnol. 2014;172:1938–1952. doi: 10.1007/s12010-013-0653-9. [DOI] [PubMed] [Google Scholar]
- 78.Torres MD, Kraan S, Domínguez H. Seaweed biorefinery. Rev Environ Sci Biotechnol. 2019;18:335–388. doi: 10.1007/s11157-019-09496-y. [DOI] [Google Scholar]
- 79.Commission E, Research D-Gf, Innovation. Platt R, Bauen A, Reumerman P, Geier C, Van Ree R, Vural Gursel I, Garcia L, et al. EU biorefinery outlook to 2030 – Studies on support to research and innovation policy in the area of bio-based products and services. Brussels: European Commission; 2021. [Google Scholar]
- 80.Morgan SG, Bathula HS, Moon S. Pricing of pharmaceuticals is becoming a major challenge for health systems. BMJ. 2020;368:l4627. doi: 10.1136/bmj.l4627. [DOI] [PubMed] [Google Scholar]
- 81.van Hal JW, Huijgen WJ, Lopez-Contreras AM. Opportunities and challenges for seaweed in the biobased economy. Trends Biotechnol. 2014;32:231–233. doi: 10.1016/j.tibtech.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 82.Wang H, Pu Y, Ragauskas A, Yang B. From lignin to valuable products-strategies, challenges, and prospects. Bioresour Technol. 2019;271:449–461. doi: 10.1016/j.biortech.2018.09.072. [DOI] [PubMed] [Google Scholar]
- 83.Becker J, Wittmann C. A field of dreams: lignin valorization into chemicals, materials, fuels, and health-care products. Biotechnol Adv. 2019;37:107360. doi: 10.1016/j.biotechadv.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 84.van Duuren J, de Wild PJ, Starck S, Bradtmoller C, Selzer M, Mehlmann K, Schneider R, Kohlstedt M, Poblete-Castro I, Stolzenberger J, et al. Limited life cycle and cost assessment for the bioconversion of lignin-derived aromatics into adipic acid. Biotechnol Bioeng. 2020;117:1381–1393. doi: 10.1002/bit.27299. [DOI] [PubMed] [Google Scholar]
- 85.Weiland F, Kohlstedt M, Wittmann C. Guiding stars to the field of dreams: metabolically engineered pathways and microbial platforms for a sustainable lignin-based industry. Metab Eng. 2021;71:13–41. doi: 10.1016/j.ymben.2021.11.011. [DOI] [PubMed] [Google Scholar]
- 86.Sethupathy S, Murillo Morales G, Gao L, Wang H, Yang B, Jiang J, Sun J, Zhu D. Lignin valorization: status, challenges and opportunities. Bioresour Technol. 2022;347:126696. doi: 10.1016/j.biortech.2022.126696. [DOI] [PubMed] [Google Scholar]
- 87.Sánchez S, Chávez A, Forero A, García-Huante Y, Romero A, Sánchez M, Rocha D, Sánchez B, Ávalos M, Guzmán-Trampe S, et al. Carbon source regulation of antibiotic production. J Antibiot. 2010;63:442–459. doi: 10.1038/ja.2010.78. [DOI] [PubMed] [Google Scholar]
- 88.Zaburannyi N, Rabyk M, Ostash B, Fedorenko V, Luzhetskyy A. Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genomics. 2014;15:1. doi: 10.1186/1471-2164-15-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bolten CJ, Kiefer P, Letisse F, Portais JC, Wittmann C. Sampling for metabolome analysis of microorganisms. Anal Chem. 2007;79:3843–3849. doi: 10.1021/ac0623888. [DOI] [PubMed] [Google Scholar]
- 90.Schwechheimer SK, Becker J, Peyriga L, Portais JC, Sauer D, Müller R, Hoff B, Haefner S, Schröder H, Zelder O, Wittmann C. Improved riboflavin production with Ashbya gossypii from vegetable oil based on 13C metabolic network analysis with combined labeling analysis by GC/MS, LC/MS, 1D, and 2D NMR. Metab Eng. 2018;47:357–373. doi: 10.1016/j.ymben.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 91.Rohles CM, Giesselmann G, Kohlstedt M, Wittmann C, Becker J. Systems metabolic engineering of Corynebacterium glutamicum for the production of the carbon-5 platform chemicals 5-aminovalerate and glutarate. Microb Cell Fact. 2016;15:154. doi: 10.1186/s12934-016-0553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 93.Peter DM, Vögeli B, Cortina NS, Erb TJ. A chemo-enzymatic road map to the synthesis of CoA esters. Molecules. 2016;21:517. doi: 10.3390/molecules21040517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dawson RMC, Elliott DC, Elliott WH, Jones KM. Data for biochemical research. 3. Oxford: Clarendon press; 2002. [Google Scholar]
- 95.Gerlinger PD, Angelidou G, Paczia N, Erb TJ. Ketosynthase engineering enhances activity and shifts specificity towards non-native extender units in type I linear polyketide synthase. bioRxiv. 2023 doi: 10.1101/2023.02.16.528826. [DOI] [Google Scholar]
- 96.Diehl C, Gerlinger PD, Paczia N, Erb TJ. Synthetic anaplerotic modules for the direct synthesis of complex molecules from CO(2) Nat Chem Biol. 2023;19:168–175. doi: 10.1038/s41589-022-01179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hilker R, Stadermann KB, Doppmeier D, Kalinowski J, Stoye J, Straube J, Winnebald J, Goesmann A. ReadXplorer—visualization and analysis of mapped sequences. Bioinformatics. 2014;30:2247–2254. doi: 10.1093/bioinformatics/btu205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Warnes MGR, Bolker B, Bonebakker L, Gentleman R. Package ‘gplots’. Various R Programming Tools for Plotting Data. 2016.
- 101.Team R. Core . R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 102.Tangyu M, Fritz M, Ye L, Aragão Börner R, Morin-Rivron D, Campos-Giménez E, Bolten CJ, Bogicevic B, Wittmann C. Co-cultures of Propionibacterium freudenreichii and Bacillus amyloliquefaciens cooperatively upgrade sunflower seed milk to high levels of vitamin B12 and multiple co-benefits. Microb Cell Fact. 2022;21:48. doi: 10.1186/s12934-022-01773-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Peng X, Okai N, Vertes AA, Inatomi K, Inui M, Yukawa H. Characterization of the mannitol catabolic operon of Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2011;91:1375–1387. doi: 10.1007/s00253-011-3352-x. [DOI] [PubMed] [Google Scholar]
- 104.Llamas-Ramírez R, Takahashi-Iñiguez T, Flores ME. The phosphoenolpyruvate-pyruvate-oxaloacetate node genes and enzymes in Streptomyces coelicolor M-145. Int microb. 2020;23:429–439. doi: 10.1007/s10123-019-00116-x. [DOI] [PubMed] [Google Scholar]
- 105.Cecchini G, Schröder I, Gunsalus RP, Maklashina E. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochem Biophys Acta. 2002;1553:140–157. doi: 10.1016/s0005-2728(01)00238-9. [DOI] [PubMed] [Google Scholar]
- 106.Willem VK. Saxifraga: Himanthalia elongata 5. 2018. http://www.freenatureimageseu/. Accessed 15 July 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. BLAST analysis of amino acid sequences of genes responsible for the ethylmalonyl-CoA pathway in S. lividans ΔYA8-DG2 against S. venezuelae ATCC 15439. Table S2. Primers for genetic engineering and sequencing. Table S3. Chromatographic and mass spectrometric settings during LC–ESI–MS analysis. The conditions for the analysis of bottromycins [20] and pamamycins [39] were adapted from previous work. Figure S1. Substrate screening of S. lividans TK24. The minimal plate medium used contained 10 g L−1 mannitol (A), 5 mM protocatechuic acid (B), 5 mM 4-hydroxybenzoic acid (C) as the sole source of carbon, or no carbon source (D). The plates were incubated for 5 days at 28 °C. The different concentrations used were chosen based on the expected toxicity of the aromatics. Figure S2. Quality assessment of RNA sequencing using PCA. The data comprise the global transcriptomes of S. lividans ΔYA8-DG2 grown on mannitol (10 mM), protocatechuate (10 mM), or 4-hydroxybenzoate (10 mM) and sampled after 12 h, n = 3. Figure S3. Sample distance plot. The data originate from RNA sequencing of S. lividans ΔYA8-DG2 grown on mannitol (10 mM), protocatechuate (10 mM), or 4-hydroxybenzoate (10 mM) and sampled after 12 h, n = 3. Figure S4. Volcano plot. The data reflect the differences in gene expression between mannitol- and protocatechuate-grown S. lividans ΔYA8-DG2 sampled after 12 h, n = 3. Figure S5. Volcano plot. The data reflect the differences in gene expression between mannitol- and 4-hydroxybenzoate-grown S. lividans ΔYA8-DG2 sampled after 12 h, n = 3. Figure S6. Correlation between optical density and cell dry weight for S. lividans ΔYA8-DG2. The strain was grown on 10 mM protocatechuate (A) and 30 mM mannitol (B), and parallel measurements at different culture time points were performed, n = 3.
Additional file 2. LC-MS analysis of bottromycin A2 and methylated bottromycin A2.
Additional file 3. Gene expression data reflecting genes linked to amino acid biosynthesis.
Additional file 4. Gene expression data reflecting genes linked to oxidative stress response and energy metabolism.
Data Availability Statement
All dataset(s) supporting the conclusions of this article are included within the article.