Abstract
Saccharomyces cerevisiae glutamate synthase (GOGAT) is an oligomeric enzyme composed of three 199-kDa identical subunits encoded by GLT1. In this work, we analyzed GLT1 transcriptional regulation. GLT1-lacZ fusions were prepared and GLT1 expression was determined in a GDH1 wild-type strain and in a gdh1 mutant derivative grown in the presence of various nitrogen sources. Null mutants impaired in GCN4, GLN3, GAT1/NIL1, or UGA43/DAL80 were transformed with a GLT1-lacZ fusion to determine whether the above-mentioned transcriptional factors had a role in GLT1 expression. A collection of increasingly larger 5′ deletion derivatives of the GLT1 promoter was constructed to identify DNA sequences that could be involved in GLT1 transcriptional regulation. The effect of the lack of GCN4, GLN3, or GAT1/NIL1 was also tested in the pertinent 5′ deletion derivatives. Our results indicate that (i) GLT1 expression is negatively modulated by glutamate-mediated repression and positively regulated by Gln3p- and Gcn4p-dependent transcriptional activation; (ii) two cis-acting elements, a CGGN15CCG palindrome and an imperfect poly(dA-dT), are present and could play a role in GLT1 transcriptional activation; and (iii) GLT1 expression is moderately regulated by GCN4 under amino acid deprivation. Our results suggest that in a wild-type strain grown on ammonium, GOGAT constitutes an ancillary pathway for glutamate biosynthesis.
The existence of two pathways for glutamate biosynthesis has been demonstrated in a variety of organisms. In one pathway, NADP+-dependent glutamate dehydrogenase (NADP+-GDH; EC 1.4.1.4) catalyzes the reductive amination of 2-oxoglutarate to form glutamate (24). The existence of an alternative pathway for the net biosynthesis of glutamate was demonstrated by Tempest et al. (45). In this pathway, glutamate is aminated to form glutamine by glutamine synthetase (GS; EC 1.4.1.13), the amide group of which is then transferred reductively to 2-oxoglutarate by glutamate synthase (GOGAT; EC 1.4.1.13), resulting in the net conversion of ammonium and 2-oxoglutarate to glutamate. The GS-GOGAT pathway has been found in several microorganisms (8, 25, 30, 32, 40) and in higher plants (32). In Saccharomyces cerevisiae, besides the NADP+-GDH1 encoded by GDH1 and GOGAT encoded by GLT1 (18, 24, 33), there is a third route for glutamate biosynthesis, constituted by a NADP+-GDH1 isozyme (NADP+-GDH3), encoded by GDH3 (2). Thus, in this microorganism, mutations inactivating GDH1, GLT1, and GDH3 are needed in order to attain full glutamate auxotrophy (2).
The presence of multiple pathways for glutamate biosynthesis in several microorganisms has stimulated discussion on the need for several routes for the biosynthesis of the same end product. Since the demonstration of the existence of GOGAT as an alternative pathway for glutamate biosynthesis (45), it was proposed that the role of the GS-GOGAT pathway would be that of ammonium assimilation and glutamate biosynthesis under ammonium limitation (45). In fact, it has been shown that for Klebsiella aerogenes this was the case (40). However, in other microorganisms (18, 28, 40), NADP+-GDH is used to incorporate ammonia during either nitrogen limitation or nitrogen excess. Thus, the initial hypothesis suggesting the differential utilization of NADP+-GDH and GS-GOGAT pathways under excess or limiting ammonia does not hold for most of the microorganisms so far studied. Since in most cases NADP+-GDH seems to be the main pathway for glutamate biosynthesis, the role of GOGAT remains unclear. Physiological studies have been performed with either wild-type or mutant strains impaired in GOGAT or in NADP+-GDH activity. In some cases, this approach has allowed the proposal of different roles for GOGAT in different microorganisms (3, 20, 23, 25, 28, 46). Few studies have been done to investigate the regulation of GOGAT-encoding genes; such studies could also provide information on whether this enzyme is involved in glutamate biosynthesis. In the case of Escherichia coli, it has been found that the two structural genes (gltB and gltD) coding for the two E. coli GOGAT subunits form an operon, with a third regulatory gene, gltF (9), involved in the glutamate-mediated repression of the gltBDF operon (10). In addition, the E. coli gltBDF operon appears to be transcriptionally regulated by the leucine-responsive regulatory protein (Lrp) (16). In Bacillus subtilis, GOGAT gene expression (gltA and gltB) is dependent on a positive regulator (gltC) that is itself transcribed from a divergent but overlapping promoter site (6). It has been postulated that the product of gltC is a positive transcription factor that acts at the gltA promoter to stimulate transcription under conditions of limiting glutamate (7).
During the last years, our group has been interested in defining and understanding the role of each of the pathways involved in glutamate biosynthesis in S. cerevisiae (2, 17, 18). Since in this yeast there are three pathways for glutamate biosynthesis and the precise function of each has not been established, we decided to initiate our study by examining GLT1 transcriptional regulation in S. cerevisiae.
In this work, we prepared GLT1-lacZ fusions which allowed the study of GLT1 expression in GDH1 and gdh1 strains in the presence of various nitrogen sources. A collection of 5′ deletion derivatives of the GLT1 promoter was prepared in order to determine the DNA sequences that could be involved in transcriptional regulation. We also studied the role of three transcriptional activators (Gcn4p, Gln3p, and Gat1p/Nil1p) (5, 11, 22, 34, 41) and of a repressor protein (Uga43p/Dal80p) (13, 15) in GLT1 expression; all of these proteins have been shown to be involved in regulation of expression of genes coding for enzymes of amino acid biosynthesis or of nitrogen catabolism.
Our results indicate that first, under conditions of glutamate excess, GLT1 expression is governed by both glutamate-mediated repression and Gln3p- and Gcn4p-mediated activation; second, under derepressive conditions, GLT1 expression could be positively regulated by a Zn2-Cys6 binuclear cluster activator, by Gcn4p and Gln3p, and by an imperfect poly(dA-dT) promoter element; and third, under amino acid deprivation, GLT1 expression is moderately regulated by Gcn4p.
MATERIALS AND METHODS
Strains.
Table 1 describes the characteristics of the strains used in this study. Null mutants impaired in GCN4, GLN3, or GAT1 were derived from strain CLA1 by gene replacement using the 3.7-kb BstII-MluI restriction fragment of pM214 (21), AatII-digested pPM62 (34), or plasmid pRR336 previously digested with XbaI-EcoRI (11), thus obtaining CLA100, CLA101, and CLA102. MAR1 was obtained by GDH1 gene disruption with pLV3 linearized with BglII (2).
TABLE 1.
S. cerevisiae strains
| Strain | Genotype | Reference |
|---|---|---|
| CLA1 | MATα GDH1 GDH3 GLT1 ura3 leu2 | 2 |
| CLA1-0 | MATα GDH1 GDH3 GLT1 ura3 leu2/YEp363 (2μm LEU2) | This study |
| CLA1-1 | MATα GDH1 GDH3 GLT1 ura3 leu2/pLOU1(GLT1-lacZ 2μm LEU2) | This study |
| CLA-100 | MATα GDH1 GDH3 GLT1 gcn4Δ::URA3 leu2/pLOU1(GLT1-lacZ 2μm LEU2) | This study |
| CLA-101 | MATα GDH1 GDH3 GLT1 gln3Δ::URA3 leu2/pLOU1(GLT1-lacZ 2μm LEU2) | This study |
| CLA-102 | MATα GDH1 GDH3 GLT1 gat1Δ::URA3 leu2/pLOU1(GLT1-lacZ 2μm LEU2) | This study |
| MAR1 | gdh1Δ::URA3 GDH3 GLT1 leu2 | This study |
| MAR1-0 | gdh1Δ::URA3 GDH3 GLT1 leu2/YEp363 (2μm LEU2) | This study |
| MAR1-1 | gdh1Δ::URA3 GDH3 GLT1 leu2/pLOU1 (GLT1-lacZ 2μm LEU2) | This study |
| 27034b | MATα GDH1 GDH3 GLT1 UGA43 ura3 leu 2/pSIM1 (GLT1-lacZ 2μm URA3) | 13 |
| 30078c | MATα GDH1 GDH3 GLT1 uga43Δ ura3/pSIM1 (GLT1-lacZ 2μm URA3) | 13 |
Growth conditions.
Strains were routinely grown on minimal medium (MM) containing salts, trace elements, and vitamins following the formula of yeast nitrogen base (Difco). Filter-sterilized glucose (2%) was used as the carbon source, and 0.2% (NH4)2SO4 or 0.1% glutamate, glutamine, asparagine, or proline was used as the nitrogen source. Amino acids needed to satisfy auxotrophic requirements were added at 0.01% (wt/vol). Cells were incubated at 30°C with shaking (250 rpm). For amino acid deprivation experiments, CLA1/pLOU1 or its gcn4Δ/pLOU1 derivative was inoculated into 10 ml of YPD, incubated at 30°C with shaking for 6 h, washed twice, and resuspended in MM. An aliquot was inoculated into 100 ml of MM to give at optical density at 600 nm (OD600) of 0.05. This culture was incubated at 30°C with shaking for 6 h, harvested, resuspended in 10 ml of MM, and inoculated into 100 ml of MM to give an OD600 of 0.2 and into 100 ml of MM–10 mM 3-aminotriazole (3-AT) to give an OD600 of 0.5. After 6 h of incubation at 30°C with shaking (250 rpm), cultures were centrifuged and used for β-galactosidase (β-Gal) determinations.
Determination of GOGAT and β-Gal activities.
Yeast total extracts were prepared from cultures inoculated at an OD600 of 0.05 and harvested at an OD600 of between 0.8 and 1.0. Cells were washed twice with H2O and once with the corresponding extraction buffer (12, 37). The pellet was stored at −20°C until used. Soluble extracts were prepared by suspending whole cells in their corresponding extraction buffer and grinding them with glass beads in a Vortex mixer. Yeast GOGAT (EC 1.4.7.1) activity was determined by the method described by Cogoni et al. (12). Specific activity was expressed as nanomoles of NADH oxidized per minute per milligram of protein. β-Gal activities were determined by the method described by Rose and Botstein (37). β-Gal specific activity was expressed as nanomoles of o-nitrophenol produced per minute per milligram of protein. Protein was measured by the method of Lowry et al. (29), with bovine serum albumin as a standard.
Construction of lacZ fusions.
Plasmid Yc14, previously described and sequenced (12, 17), contains 2 kb of the GLT1 coding sequence, the full GLT1 promoter and 30 bp of the UGA3 coding sequence. Yc14 DNA was digested with EcoRI and used as template for PCR amplification. Deoxyoligonucleotide F1 contained a BamHI site and 18 bp of the UGA3 coding region (5′-CGCGCGGGATCCCAATTTCAGCTTCTCCAC-3′). Deoxyoligonucleotide R1 contained a SalI site, 8 bp upstream the GLT1 coding region, and 3 bp downstream the GLT1 promoter region (5′-GCGCGCGGTCGACACTGGCATGCT-3′). Deoxyoligonucleotides F1 and R1 were used to amplify the complete GLT1 promoter. To obtain a 5′ GLT1 promoter deletion series, the pertinent forward deoxyoligonucleotides were designed based on the GLT1 promoter sequence. Deoxyoligonucleotide R1 was also used to amplify the full promoter and the 18 individual deletions. The entire family of PCR products was fused in frame to the E. coli lacZ gene of YEp363 (2μm LEU2) (35), generating 19 fusion plasmids, pLOU1 to pLOU19. The PCR product carrying the full GLT1 promoter was also fused in frame to the E. coli lacZ gene of YEp353 (2μm URA3) (35), generating plasmid pSIM1. All fusion plasmids were sequenced with an automated Applied Biosystems 373 DNA sequencer (W. M. Keck Foundation, Yale University).
Yeast transformation.
S. cerevisiae was transformed by the method described by Ito et al. (26). To generate null derivatives, transformants were selected for uracil prototrophy on MM supplemented with auxotrophic requirements as needed. Pertinent strains were transformed with the lacZ fusion plasmids or, when appropriate, with YEp363. Transformants were selected for either leucine or uracil prototrophy on MM supplemented with auxotrophic requirements as needed.
Primer extension RNA analysis.
Primer extension reactions were performed by standard procedures (38). To determine chromosomal GLT1 transcription initiation sites, total RNA was isolated from strain CLA1 grown on MM with 0.2% (NH4)2SO4 as the nitrogen source. A deoxyoligonucleotide containing the first 21 nucleotides of the GLT1 coding region was prepared and used in the primer extension reactions. The transcription initiation sites present in the different lacZ fusion constructs were also determined. Primer extension reactions were carried out with total RNA extracted from the pertinent strains grown on MM with 0.2% (NH4)2SO4 as the nitrogen source and a deoxyoligonucleotide containing 23 nucleotides of the lacZ coding region.
RESULTS AND DISCUSSION
Sequence analysis of GLT1 promoter region and determination of transcription initiation sites.
As stated in the introduction, the role of GOGAT in glutamate biosynthesis and its regulation have not been studied in yeast. To address this matter, we analyzed the nucleotide sequence located upstream of the GLT1 coding region, which was contained in the previously reported plasmid Yc14 (12). As Fig. 1 shows, GLT1 was located in opposite orientation, next to the UGA3 gene, which codes for a transcriptional activator of the genes involved in γ-aminobutyrate catabolism (1). Intragenic sequences may act as sites for trans-acting regulatory elements of either of the two divergent genes, GLT1 and UGA3. Such sequences could face or partly overlap sites with the opposite orientation in the complementary DNA strand that regulate the alternative divergent gene. One could expect that simple occupancy of either sequence by its cognate high-affinity regulator may interfere with regulation of the alternative divergent gene. The study of UGA3 expression may help define this matter.
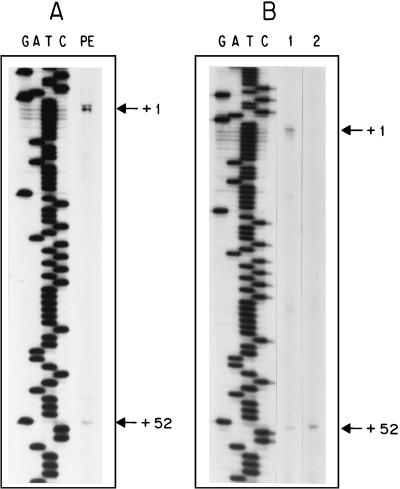
FIG. 1.
GLT1 promoter sequence. Putative Gcn4p (GCN4), Gln3p, and Gat1p binding sites (GATA), CGGN15CCG palindrome, and poly(dA-dT) regions are boxed and numbered starting from the most 5′. Two putative TATA boxes (TATA 1 and TATA 2) as well as the two transcription initiation sites, at positions +1 and +52, are indicated. The 714-bp fragment shown includes a 30-bp sequence of the UGA3 coding region. BamHI and SalI sites were added and used to clone this fragment into the 2μm LEU2 lacZ vector YEp363, generating plasmid pLOU1.
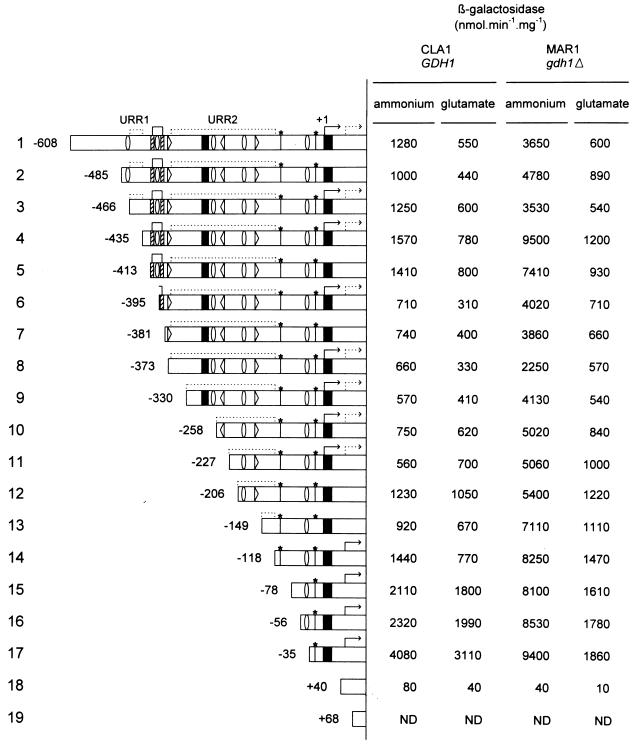
Most of the genes encoding amino acid biosynthetic enzymes in S. cerevisiae are subject to a cross-pathway regulatory system known as the general amino acid control that stimulates their expression under conditions of amino acid starvation. Gcn4p is the direct positive regulator of gene expression in this system (22). Examination of the GLT1 promoter revealed a canonical Gcn4p binding site ATGACTC [GCN4(1)] (Fig. 1) located between positions −477 and −466. The GLT1 promoter also carries four noncanonical binding sites (Fig. 1) with low affinity for Gcn4p (31, 44): TGCGTA from positions −399 to −393 [GCN4(2)], TTAGTCAT from −267 to −260 [GCN4(3)], ATTAATCA from −193 to −186 [GCN4(4)], and GTGATTAAC from −43 to −35 [GCN4(5)].
The GLT1 promoter also contained three GATAA sequences, GATAAG from positions −377 to −372 [GATA(1)], CTTATC (complementary, GATAAG) from −238 to −233 [GATA(2)], and GATAAC from −168 to −164 [GATA(3)] (Fig. 1), which can constitute the cis-acting element, UASNTR (41). UASNTR has been proposed as a binding site for two transcriptional activators, Gln3p and Gat1p/Nil1p (4, 11, 34, 42), which regulate the expression of nitrogen-modulated genes.
Down-regulation of nitrogen-controlled gene expression is accomplished by the action of the GATA family member Dal80p/Uga43p. The Dal80p binding site, URSGATA, consists of a pair of GATA-containing sequences oriented tail to tail or head to tail (15). As can be seen in Fig. 1, the GLT1 promoter harbors two of the above-mentioned GATA sequences oriented tail to tail; these could constitute a Dal80p binding site, although the distance between them (63 bp) is larger than that previously reported (15 to 35 bp) (15).
At least 79 fungal transcription-activating factors containing a Zn2-Cys6 binuclear cluster have been found (39). DNA targets for several members of this family of proteins have two inverted CGG half-sites separated by a spacing characteristic of the particular protein that recognizes it (27). The two inverted CGG half-sites separated by 15 bp present in the GLT1 promoter (Fig. 1) could also constitute a binding site for members of the Zn2-Cys6 binuclear cluster family of proteins.
Many yeast promoters contain homopolymeric (dA-dT) sequences (43). Analysis of the function of these sequences in transcriptional activation has suggested that perfectly homopolymeric sequences function by virtue of their intrinsic structure. For imperfect poly(dA-dT) tracts, it has been proposed that the transcriptional effects might be mediated in part or completely by specific DNA-binding proteins (47). The GLT1 promoter also bears two poly(dA-dT) sequences: one composed of a 16-poly(dA-dT) tract with two imperfections located from positions −292 to −276 [poly(dA-dT)1 in Fig. 1], and another consisting of a 19-poly(dA-dT) tract with two imperfections located from −2 to +17 [poly(dA-dT)2 in Fig. 1].
Primer extension analysis (Fig. 2) defined two transcription initiation sites in GLT1, which are shown in Fig. 1 at positions +1 and +52. The results presented in Fig. 2 indicate that the +1 initiation site is stronger than the +52 site. Two putative TATA boxes differing from the TATAAA canonical sequence were also found (Fig. 1). Either the TATACTA or TATTTA sequence can substitute for TATAAA in transcription initiation (19). Constructions from pLOU14 to −17 were able to initiate transcription only from +52. It is possible that each of these initiation sites together with TATA(1) or TATA(2) can signal transcription under different physiological conditions. If this were the case, the first element would direct transcription regulated by glutamate-mediated repression and by Gcn4p-, Gln3p-, and putative Zn2-Cys6 binuclear cluster-mediated activation. The second element would direct transcription mediated by the poly(dA-dT) element by itself or together with a glutamate-sensitive activator.
FIG. 2.
Primer extension analysis. (A) Assay of transcription initiation sites (lane PE) of the GLT1 gene, carried out with total RNA obtained from the wild-type strain CLA1. (B) Representative results of primer extension analysis carried out with total RNA obtained from strain CLA1 transformed with plasmid pLOU1, -3, -4, -6, -8, -9, -10, or -11 (lane 1) or with pLOU14, -15, -16, or -17 (lane 2). The sequence ladder was produced with the same deoxyoligonucleotide used for the primer extension reaction (described in Materials and Methods).
Regulation of GLT1 expression.
It has been previously observed that mutants impaired in GDH1 display increased GOGAT activity (2), suggesting that GLT1 can be negatively modulated by glutamate and that in a gdh1 mutant, glutamate limitation can result in GLT1 derepression. To determine whether GLT1 expression was regulated by the nature of the nitrogen source, we determined GOGAT and β-Gal activities in a wild-type strain and in a gdh1 mutant. Both strains harbored either plasmid pLOU1, containing the GLT1 promoter fused to the complete β-Gal coding region, or the vector YEp363 (see Materials and Methods). As expected, in the presence of YEp363, no β-Gal activity was detected, and GOGAT activity values were similar to those found in the presence of pLOU1 (Table 2). As Table 2 and Fig. 3 (row 1) show, GOGAT and β-Gal activities were higher in the gdh1 mutant strain grown on ammonium or proline as the sole nitrogen source than in the wild-type strain grown under similar conditions. In the presence of glutamate, glutamine, or asparagine, both GOGAT and β-Gal activities decreased and achieved similar values in extracts obtained from either the wild-type or gdh1 strain (Table 2). These results indicate that GLT1 expression was repressed in the presence of glutamate-rich nitrogen sources.
TABLE 2.
β-Gal and GOGAT specific activities
| Nitrogen source | Sp act (nmol
min−1 mg−1)a
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLA1-0 (GDH1/YEp363) | CLA1-1 (GDH1/pLOU1) | MAR1-0 (gdh1Δ/YEp363) | MAR1-1 (gdh1Δ/pLOU1) | β-Gal in:
|
|||||||||
| CLA100 (GDH1 gcn4Δ/pLOU1) | CLA101 (GDH1 gln3Δ/pLOU1) | CLA102 (GDH1 gat1Δ/pLOU1) | 27034b (GDH1 UGA43/pSIM1) | 30078c (GDH1 uga43Δ/pSIM1) | |||||||||
| β-Gal | GOGAT | β-Gal | GOGAT | β-Gal | GOGAT | β-Gal | GOGAT | ||||||
| Ammonium | ND | 40 | 1,280 | 41 | ND | 71 | 3,650 | 69 | 600 | 230 | 1,150 | 850 | 630 |
| Proline | ND | 40 | 1,580 | 41 | ND | 53 | 2,360 | 55 | 600 | 290 | 1,250 | 3,850 | 4,250 |
| Glutamate | ND | 22 | 550 | 24 | ND | 24 | 600 | 25 | 200 | 270 | 392 | 910 | 750 |
| Glutamine | ND | 22 | 520 | 21 | ND | 27 | 710 | 29 | 200 | 60 | 362 | 410 | 340 |
| Asparagine | ND | 23 | 450 | 25 | ND | 23 | 450 | 24 | 250 | 100 | 308 | 1,340 | 1,000 |
Mean of three independent experiments. Variations were ≤15%. ND, not detected.
FIG. 3.
β-Gal activities of 5′ deletions of the GLT1 promoter. The GLT1 full promoter and 5′ deletions were cloned into the 2μm LEU2 lacZ vector YEp363, generating plasmids pLOU1 to -19. These plasmids were transformed into either the GDH1 wild-type strain CLA1 or the gdh1 mutant strain MAR1. The 5′ region carried in each plasmid is indicated in rows 1 to 19. β-Gal activity was determined in extracts obtained from cells grown on either 0.2% ammonium sulfate or 0.1% glutamate. ND, not detected. Diagrams depict Gcn4p putative binding sites ( ), palindrome (▨⊓▨), Gln3p putative binding sites (▹, ◃), poly(dA-dT) (▪), putative TATA boxes (*), transcription initiation sites ( , ), and putative URRs.
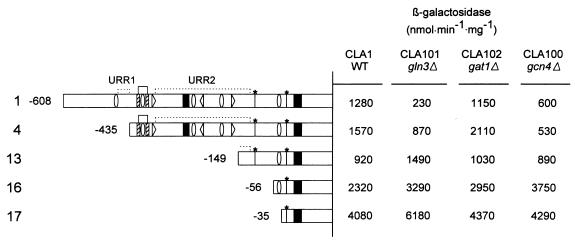
To analyze whether Gln3p, Gat1p/Nil1p, Gcn4p, or Dal80p/Uga43p had a role in GLT1 expression, plasmid pLOU1 was transformed into gcn4Δ, gln3Δ, gat1Δ, and uga43Δ mutant strains, and β-Gal activity was determined (Table 1; Fig. 4, row 1). It was found that with ammonium or proline as the nitrogen source, the lack of Gln3p severely diminished β-Gal activity; impairment of Gcn4p had a slight effect on this activity, while the lack of Gat1p had no effect. Extracts obtained from cultures of the gln3Δ or gcn4Δ mutant showed decreased β-Gal activity compared to extracts obtained from the wild type when either strain was grown on glutamate, glutamine, or asparagine. These results suggested that GLT1 transcription of yeast cells grown in glutamate-rich nitrogen sources was down-regulated by glutamate repression and up-regulated by transcriptional activators Gln3p and Gcn4p. The capacity of Gln3p and Gat1p to activate transcription appears to be nitrogen regulated in such a way that GLN3 stimulates transcription on glutamate and proline and GAT1 does so on ammonium and urea (42). However, neither Gln3p or Gat1p promotes the expression of nitrogen-regulated genes on glutamine (42). Our results indicate (i) that GLT1 is not regulated by GAT1 and (ii) that GLN3 activates GLT1 expression with all nitrogen sources tested, including glutamine. Possibly the GLT1 promoter has a higher affinity for the GLN3 inactive form that has been postulated to be present in glutamine (5). Also, as has been observed in other cases (36), GLN3 may act in combination with the positive activator that should bind the CGG palindrome. Maybe in the case of GLT1 expression, a less active form of GLN3 has an important effect on glutamine when assisted by another activator.
FIG. 4.
Effects of gln3, gat1, and gcn4 null mutations on β-Gal activity in 5′ deletions of the GLT1 promoter. Mutant strains CLA101 (gln3Δ), CLA102 (gat1Δ), and CLA100 (gcn4Δ) harboring plasmids pLOU1, pLOU4, pLOU13, pLOU16, and pLOU17 (lines 1, 4, 13, 16, and 17) were grown on 0.2% ammonium sulfate as the nitrogen source, and β-Gal activity was determined. The reported β-Gal activities are averages of values obtained in three independent experiments. GLT1 promoter regions are represented as in Fig. 3. Variations were <15%.
Isogenic strains carrying the wild-type UGA43/DAL80 gene or the null allele were also transformed with pLOU1. As Table 2 shows, lack of Uga43p/Dal80p did not result in derepressed GLT1 expression in the presence of glutamate, indicating that glutamate-mediated repression was not UGA43/DAL80 dependent. Further experiments will be required to determine the nature of the cis- and trans-acting elements which mediate glutamate repression.
To address if GLT1 expression was regulated during amino acid deprivation by the general amino acid control mediated by Gcn4p (22), β-Gal was determined in extracts from cultures of the wild-type strain and of the gcn4Δ mutant grown in the presence and absence of 3-AT, a competitive inhibitor of His3p. In the presence of this analog, cells become deprived for histidine. β-Gal activity was twofold higher in extracts obtained from the wild-type strain grown in the absence of 3-AT compared to that found in its presence (1,280 versus 2,170 nmol min−1 mg−1). This increment was not observed in the gcn4Δ mutant strain (600 versus 550 nmol min−1 mg−1). These results indicate that GLT1 expression was increased during amino acid deprivation and that this increase was Gcn4p dependent (22). Since glutamate is a precursor in the biosynthesis of most amino acids, the genes coding for the enzymes involved in its biosynthesis would likely have to be responsive to starvation of a number of amino acids. However, our results indicate that GOGAT (GLT1) is not strongly regulated by Gcn4p. The analysis of whether GDH1 or GDH3 transcription responds to amino acid limitation will be very useful to fully understand the pathway(s) through which glutamate biosynthesis could be increased during amino acid starvation. The exact binding site(s) for Gcn4p on GLT1 promoter remains to be determined. However, our 5′ deletion analysis suggests that the canonical GCN4 binding site [GCN4(1)] plays no role in GLT1 GCN4-dependent transcriptional activation, since when it is deleted (pLOU3), GLT1 transcription is not decreased. It is clear that pLOU4-dependent β-Gal activity is decreased in a null gcn4Δ derivative, indicating that the GCN4 binding site [GCN4(2)] could play a more important role than [GCN4(1)] in GCN4-mediated transcriptional activation. It is also possible that the GCN4(3) putative binding site plays a role in GLT1 gene activation together with the poly(dA-dT)(1), since it has been suggested that during gene activation of promoters harboring both a poly(dA-dT) tract and a GCN4 binding site, transcription can be either hindered or promoted through chromatin reorganization (47).
Deletion analysis of GLT1 promoter.
A collection of 5′ deletions of increasing size affecting the GLT1 promoter was prepared as described in Material and Methods. When a GDH1 strain harboring pLOU4, which lacks the most 5′ 173 bp of the GLT1 promoter, was grown on ammonium, β-Gal activity was slightly higher than that obtained with the GDH1 strain carrying pLOU1 (Fig. 3, row 4). This increment was more evident when β-Gal was determined in a gdh1 strain carrying pLOU4, which showed β-Gal activity nearly threefold higher than that found in the gdh1 strain carrying pLOU1. These results suggested that pLOU4 had lost a target for negative regulation (upstream repressing region 1 [URR1]) (Fig. 3). Deletions from bp −608 to −413, −608 to −395, −608 to −381, and −608 to −373 (pLOU5 to -8) resulted in decreased β-Gal activity in both GDH1 and gdh1 strains, indicating that this region (−413 to −373) could contain DNA binding sites for transcriptional activators. As Fig. 1 shows, this region contained putative binding sites for Gcn4p, Gln3p, and a Zn2-Cys6 binuclear cluster activator. To determine whether the observed increase in β-Gal activity, conferred by pLOU4, was GCN4, GLN3, or GAT1/NIL1 dependent, pertinent strains were transformed with this plasmid. Increased β-Gal activity was mainly GLN3 and GCN4 dependent (Fig. 4, row 2). Fig. 3 also shows that increased β-Gal activity was still glutamate sensitive, indicating that pLOU4 had retained a cis-acting region able to respond to glutamate. Further deletions (pLOU9 to -14) resulted in a constant increase of β-Gal activity in the gdh1 derivatives but practically no changes in β-Gal activity of the corresponding GDH1 strains. Deletions present in pLOU15 to -17 resulted in a clear increase of β-Gal activity in both GDH1 and gdh1 strains, the highest activity being observed after removal of the first 573 bp (pLOU17). β-Gal activity of constructions pLOU1 to -13 was clearly diminished by the presence of glutamate in the medium; however, when β-Gal was determined in strains harboring constructions pLOU14 to -17, although addition of glutamate to the medium reduced β-Gal activity, the values were severalfold higher than those found with the full promoter in cells grown in the presence of glutamate. These results suggested (i) that a glutamate-responsive negative-acting region (URR2) was localized from positions −373 to −119 and (ii) that the region between −35 and +40 contained a target for a trans-acting positive regulatory element. As Fig. 3 shows, this DNA segment contained a poly(dA-dT) tract, which has been considered a promoter element able to stimulate transcription (47). In the presence of glutamate, transcription conferred by pLOU17 is diminished, although the β-Gal levels determined in this condition are threefold higher than those found under repressive conditions (MAR1/pLOU1 on glutamate). Thus, it is possible that the as yet undetermined activator, which we propose acts in combination with the poly(dA-dT) tract, could be glutamate inactivated. β-Gal activity fostered by pLOU16 and -17 was also found in gln3Δ, gat1Δ, and gcn4Δ null derivatives (Fig. 4), indicating that the poly(dA-dT) tract acted either independently of activators or was assisted by an as yet unrecognized positive regulatory element. This analysis suggested that GLT1 transcriptional regulation depended on the action of both negative regulatory regions (URR1 and URR2) and positive-acting elements. Both URR regions could be targets for glutamate-mediated repression, since when they were removed, GLT1 expression was no longer fully repressed by glutamate. In addition, our results indicate that of the putative cis-acting sites depicted in Fig. 1, the following could have a positive role in GLT1 transcription: (i) the GCN4(2) binding site from positions −396 to −390, (ii) the GLN3 binding site from −377 to −372 [GATA(1)], (iii) the CGG palindromic region located from −412 to −388, and (iv) the poly(dA-dT) tract located from −2 to +17. No β-Gal activity was determined in strains carrying constructions present in pLOU18 and pLOU19, indicating that the promoter fragment contained from +40 to +100 was unable to initiate GLT1 transcription.
In regard to the role of GOGAT in glutamate biosynthesis, our results indicate that (i) under low-glutamate conditions GLT1 transcription is considerably low, which suggests that GOGAT may have an important role in glutamate biosynthesis under conditions where this amino acid becomes limiting; and (ii) GOGAT could constitute an ancillary pathway furnishing low but sustained glutamate production, even in the presence of NADP+-GDH, i.e., in the presence of a relatively high glutamate pool, suggesting that a high intracellular glutamate pool may be needed for optimal growth. Since it has been reported that null GOGAT mutants grow as well as the wild-type strain on ammonium (2), the high glutamate need could be restricted to certain physiological conditions, such as high external osmolality (14), or during sporulation, since in this condition, both carbon and nitrogen are limiting and this could result in glutamate deprivation.
ACKNOWLEDGMENTS
We are grateful to Soledad Moreno, Simón Guzmán, and Marco Martegani for skillful technical assistance, to the Molecular Biology Unit of the Instituto de Fisiología Celular for the synthesis of deoxyoligonucleotides, to Guadalupe Espín and Hiram Olivera for helpful discussions, and to Fernando Bastarrachea for critical review of the manuscript. We also thank Boris Magasanik, Terrance G. Cooper, and Allan Hinnebusch for kindly providing plasmids pPM62, pRR336, and pM214 and Bruno André for providing strains 27034b and 30078c.
This work was supported in part by the Dirección de Asuntos del Personal Académico, Universidad Nacional Autónoma de México (IN204695), by the Programa de Apoyo a las Divisiones de Estudios de Posgrado (030359, 030375, and 030366), by CONACyT (400360-5-2549PN), and by Fondazione Pasteur Cenci Bologneti.
REFERENCES
- 1.André B. The UGA3 gene regulating the GABA catabolic pathway in Saccharomyces cerevisiaecodes for a putative zinc-finger protein acting on RNA amount. Mol Gen Genet. 1990;220:269–276. doi: 10.1007/BF00260493. [DOI] [PubMed] [Google Scholar]
- 2.Avendaño A, DeLuna A, Olivera H, Valenzuela L, González A. GDH3 encodes a glutamate dehydrogenase isozyme, a previously unrecognized route for glutamate biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1997;179:5594–5597. doi: 10.1128/jb.179.17.5594-5597.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barel I, MacDonald D W. Enzyme defects in glutamate-requiring strains of Schizosaccharomyces pombe. FEMS Microbiol Lett. 1993;113:267–272. doi: 10.1111/j.1574-6968.1993.tb06525.x. [DOI] [PubMed] [Google Scholar]
- 4.Blinder D, Magasanik B. Recognition of nitrogen-responsive upstream activation sequences of Saccharomyces cerevisiae by the product of the GLN3gene. J Bacteriol. 1995;177:4190–4193. doi: 10.1128/jb.177.14.4190-4193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blinder D, Coschigano P W, Magasanik B. Interaction of the GATA factor Gln3p with the nitrogen regulator Ure2p in Saccharomyces cerevisiae. J Bacteriol. 1996;178:4734–4736. doi: 10.1128/jb.178.15.4734-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohannon D E, Sonenshein A L. Positive regulation of glutamate biosynthesis in Bacillus subtilis. J Bacteriol. 1989;171:4718–4727. doi: 10.1128/jb.171.9.4718-4727.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohannon D E, Rosenkrantz M S, Sonenshein A L. Regulation of Bacillus subtilisglutamate synthase genes by the nitrogen source. J Bacteriol. 1985;163:957–964. doi: 10.1128/jb.163.3.957-964.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bravo A, Mora J. Ammonium assimilation in Rhizobium phaseoliby glutamine synthetase-glutamate synthase pathway. J Bacteriol. 1988;170:980–984. doi: 10.1128/jb.170.2.980-984.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castaño I, Bastarrachea F, Covarrubias A A. gltBDF operon of Escherichia coli. J Bacteriol. 1988;170:821–827. doi: 10.1128/jb.170.2.821-827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castaño I, Flores N, Valle F, Covarrubias A A, Bolivar F. gltF, a member of the gltBDF operon of Escherichia coli, is involved in nitrogen regulated gene expression. Mol Microbiol. 1992;6:2733–2741. doi: 10.1111/j.1365-2958.1992.tb01450.x. [DOI] [PubMed] [Google Scholar]
- 11.Coffman J A, Rai R, Loprete D M, Cunninham T, Svetlov V, Cooper T G. Cross regulation of four GATA factors that control nitrogen catabolic gene expression in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3416–3429. doi: 10.1128/jb.179.11.3416-3429.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cogoni C, Valenzuela L, González-Halphen D, Olivera H, Macino G, Ballario P, González A. Saccharomyces cerevisiaehas a single glutamate synthase gene coding for a plant-like high-molecular-weight polypeptide. J Bacteriol. 1995;177:792–798. doi: 10.1128/jb.177.3.792-798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coornaert D, Vissers S, Andre B, Grenson M. The UGA43 negative regulatory gene of Saccharomyces cerevisiaecontains both a GATA-1 type zinc finger and a putative leucine zipper. Curr Genet. 1992;21:301–307. doi: 10.1007/BF00351687. [DOI] [PubMed] [Google Scholar]
- 14.Csonka L N, Ikeda T P, Fletcher S A, Kustu S. The accumulation of glutamate is necessary for optimal growth of Salmonella typhimurium in media of high osmolality but not induction of the proUoperon. J Bacteriol. 1994;176:6324–6333. doi: 10.1128/jb.176.20.6324-6333.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham T S, Cooper T G. The Saccharomyces cerevisiae DAL80 repressor protein binds to multiple copies of GATAA-containing sequences (URSGATA) J Bacteriol. 1993;175:5851–5861. doi: 10.1128/jb.175.18.5851-5861.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernsting B R, Denninger J W, Blumenthal R M, Matthews R G. Regulation of the gltBDF operon of Escherichia coli: how is a leucine-insensitive operon regulated by the leucine responsive regulatory protein? J Bacteriol. 1993;175:7160–7169. doi: 10.1128/jb.175.22.7160-7169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filetici P, Martegani M P, Valenzuela L, González A, Ballario P. Sequence of the GLT1 gene from Saccharomyces cerevisiaereveals the domain structure of yeast glutamate synthase. Yeast. 1996;12:1359–1366. doi: 10.1002/(sici)1097-0061(199610)12:13<1359::aid-yea3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 18.Folch J L, Antaramián A, Rodríguez L, Bravo A, Brunner A, González A. Isolation and characterization of a Saccharomyces cerevisiaemutant with impaired glutamate synthase activity. J Bacteriol. 1989;171:6776–6781. doi: 10.1128/jb.171.12.6776-6781.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harbury P A B, Struhl K. Functional distinctions between yeast TATA elements. Mol Cell Biol. 1989;9:5298–5304. doi: 10.1128/mcb.9.12.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helling R B. Why does Escherichia coli have two primary pathways for synthesis of glutamate? J Bacteriol. 1994;176:4664–4668. doi: 10.1128/jb.176.15.4664-4668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinnebusch A G. A hierarchy of trans-acting factors modulated translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinnebusch A G. The general control of amino acid biosynthetic genes in the yeast Saccharomyces cerevisiae. Crit Rev Biochem. 1986;21:277–317. doi: 10.3109/10409238609113614. [DOI] [PubMed] [Google Scholar]
- 23.Holmes A R, Collings A, Farnden K J F, Sheperd M G. Ammonium assimilation by Candida albicansand other yeasts: evidence for activity of glutamate synthase. J Gen Microbiol. 1989;135:1423–1430. doi: 10.1099/00221287-135-6-1423. [DOI] [PubMed] [Google Scholar]
- 24.Holzer H, Schneider S. Anreicherung und Trennung einer DPN-spezifischen und einer TPN-spezifischen Glutaminosaure Dehydrogenase aus Hefe. Biochem Z. 1957;329:361–367. [PubMed] [Google Scholar]
- 25.Hummelt G, Mora J. Regulation and function of glutamate synthase in Neurospora crassa. Biochem Biophys Res Commun. 1980;96:1688–1694. doi: 10.1016/0006-291x(80)91368-6. [DOI] [PubMed] [Google Scholar]
- 26.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang S D, Marmorstein R, Harrison S C, Ptashne M. Sequence preferences of GAL4 and PPR1: how a subset of Zn2 Cys6binuclear cluster proteins recognize DNA. Mol Cell Biol. 1996;16:3773–3780. doi: 10.1128/mcb.16.7.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomnitz A, Calderón J, Hernandez G, Mora J. Functional analysis of ammonium assimilation enzymes in Neurospora crassa. J Gen Microbiol. 1987;133:2333–2340. [Google Scholar]
- 29.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 30.Marqués S, Florencio F J, Candau P. Purification and characterization of the ferredoxin-glutamate synthase from the unicellular cyanobacterium Synechoccussp. PCC 6301. Eur J Biochem. 1992;206:69–77. doi: 10.1111/j.1432-1033.1992.tb16902.x. [DOI] [PubMed] [Google Scholar]
- 31.Mavrothalassitis G, Beal G, Papas T S. Defining target sequences of DNA-binding proteins by random selection and PCR: determination of the GCN4binding sequence repertoire. DNA Cell Biol. 1990;9:783–788. doi: 10.1089/dna.1990.9.783. [DOI] [PubMed] [Google Scholar]
- 32.Miflin B J, Lea P J, Wallsgrove R M. The role of glutamine in amonnium assimilation and reassimilation in plants. In: Mora J, Palacios R, editors. Glutamine: metabolism, enzymology and regulation. New York, N.Y: Academic Press, Inc.; 1980. pp. 213–234. [Google Scholar]
- 33.Miller S M, Magasanik B. Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. J Bacteriol. 1990;172:4927–4935. doi: 10.1128/jb.172.9.4927-4935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minehart P L, Magasanik B. Sequence and expression of GLN3, a positive regulatory gene of Saccharomyces cerevisiaeencoding a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1991;11:6216–6228. doi: 10.1128/mcb.11.12.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers A M, Tzagaloff A, Kinney D M, Lusty C J. Yeast shuttle integrative vectors with multiple cloning sites suitable for construction of lacZfusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 36.Rai R, Daugherty J R, Cooper T G. UASNTR functioning in combination with other UAS elements underlies exceptional patterns of nitrogen regulation in Saccharomyces cerevisiae. Yeast. 1995;11:247–260. doi: 10.1002/yea.320110307. [DOI] [PubMed] [Google Scholar]
- 37.Rose M, Botstein D. Construction and use of gene fusions lacZ(β-galactosidase) which are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Schjerling P, Holmberg S. Comparative amino acid sequence analysis of the C6zinc cluster family of transcriptional regulators. Nucleic Acids Res. 1996;24:4599–4607. doi: 10.1093/nar/24.23.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senior P J. Regulation of nitrogen metabolism in Escherichia coli and Klebsiella aerogenes: studies with the continuous-culture technique. J Bacteriol. 1975;123:407–418. doi: 10.1128/jb.123.2.407-418.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanbrough M, Magasanik B. Two transcription factors, Gln3p and Nil1p, use the same GATAAG sites to activate the expression of GAP1 of Saccharomyces cerevisiae. J Bacteriol. 1996;178:2465–2468. doi: 10.1128/jb.178.8.2465-2468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanbrough M, Rowen R D W, Magasanik B. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiaein the expression of nitrogen-regulated genes. Proc Natl Acad Sci USA. 1995;92:9450–9454. doi: 10.1073/pnas.92.21.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci USA. 1985;82:8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavernarakis N, Thireos G. The DNA target sequence influences the dependence of the yeast transcriptional activator GCN4on co-factors. Mol Gen Genet. 1997;253:766–769. doi: 10.1007/s004380050382. [DOI] [PubMed] [Google Scholar]
- 45.Tempest D W, Meers J L, Brown C M. Synthesis of glutamate in Aerobacter aerogenesby hitherto unknown route. Biochem J. 1970;117:405–507. doi: 10.1042/bj1170405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valenzuela L, Guzmán León S, Coria R, Ramírez J, Aranda C, González A. A NADP+-glutamate dehydrogenase mutant of the petit-negative yeast Kluyveromyces lactisuses the glutamine synthase-glutamate synthase pathway for glutamate biosynthesis. Microbiology. 1995;141:2443–2447. doi: 10.1099/13500872-141-10-2443. [DOI] [PubMed] [Google Scholar]
- 47.Vishwanath I, Struhl K. Poly (dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsec DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]