Abstract
An open reading frame in the genomic database of Mycobacterium tuberculosis H37Rv was identified as having homology with an outer membrane protein. We found that the gene specified a protein belonging to the OmpA family, which includes some porins of gram-negative organisms. The gene was amplified by PCR and cloned into Escherichia coli. Overexpression of the gene was toxic to the host, but limited amounts could be purified from cells before growth ceased. A truncated gene devoid of the code for a presumed signal sequence was well expressed, but the protein had no pore-forming activity in the liposome swelling assay. However, the intact protein, OmpATb, behaved as a porin of low specific activity, with a pore diameter of 1.4 to 1.8 nm, and was also active in planar lipid bilayers, showing a single-channel conductance of 700 pS. The protein had a molecular mass of about 38 kDa in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A polyclonal rabbit antiserum raised to the truncated protein recognized a protein of similar molecular mass in detergent extracts of broken M. tuberculosis cells. Reverse transcription-PCR confirmed that the gene for OmpATb was expressed in M. tuberculosis cells growing in culture. Comparison of the purified protein with that in the detergent-extracted preparation using liposomes and planar lipid bilayers showed that the two materials had similar pore-forming properties. OmpATb is different from either of the mycobacterial porins described so far. This is the first report of a porin-like molecule from M. tuberculosis; the porin is likely to be important in controlling the access of hydrophilic molecules to the bacterial cell.
Mycobacteria are only remotely related to gram-negative bacteria, but the two groups share the property of forming in their envelopes an impermeable outer layer that is distinct from the plasma membrane. The layers are chemically quite different; that in gram-negative bacteria is a free-standing bilayer of phospholipid and a characteristic lipopolysaccharide (16), whereas the barrier in mycobacteria consists of a monolayer of very-long-chain fatty acids (mycolic acids) covalently linked to the rest of the bacterial wall (6). The mycobacterial barrier certainly includes other lipids, and evidence that these are arranged to form a bilayer with the mycolic acids has been presented (9). The existence of a permeability barrier remote from sources of energy to drive transport processes poses a problem to the bacterial cell with regard to obtaining a supply of nutrients and disposing of waste products. Gram-negative bacteria make use of specialized nonspecific pore-forming proteins (porins) to facilitate the passage of small hydrophilic molecules through their outer membranes (13). Mycobacterium chelonae was found to have very restricted permeability to a series of cephalosporins, and the properties of the permeation process were consistent with the presence of porin-like water-filled pores (8). The envelopes of two species of mycobacteria, M. chelonae and Mycobacterium smegmatis, are now known to contain proteins with typical porin-like properties (11, 29, 30). It is important to understand the properties of these pores in the outer permeability barrier in mycobacteria, since they presumably control the access of hydrophilic antibacterial substances to the cell. However, both of the species studied so far belong to the fast-growing group of mycobacteria, whereas the major mycobacterial pathogens, including Mycobacterium tuberculosis (which is the seventh most prevalent cause of death globally [12]), are slow growers. The slow-growing species are biologically distinct from the rapid growers, and their porins are not necessarily similar.
An open reading frame which had homology to an outer membrane protein of gram-negative bacteria was reported in the annotated database of the genome of M. tuberculosis H37Rv, sequenced at the Sanger Centre, Cambridge, United Kingdom (13). We have expressed the protein specified by this gene in Escherichia coli and have shown that it has pore-forming properties typical of porins. The gene is expressed in M. tuberculosis growing in culture. The protein seems to be distinct from the mycobacterial porins already described, and it belongs to the OmpA family of outer membrane proteins; we suggest the name OmpATb for this protein. An abstract referring to the behavior of OmpATb in planar lipid bilayers has been published (10).
MATERIALS AND METHODS
Bacterial strains and vectors.
M. tuberculosis H37Rv NCTC 7416 was obtained from the National Collection of Type Cultures, London, United Kingdom. E. coli DH5α (Clontech Laboratories, Inc., Palo Alto, Calif.) and E. coli BL21(DE3)pLysS (Novagen, R & D Systems Europe Ltd., Abingdon, Oxford, United Kingdom) were used for maintenance of plasmids and expression of foreign proteins, respectively. pET-15b (Novagen) was used as an expression vector in E. coli; pRS1 and pRS2 were derivatives of pET-15b bearing intact and truncated versions, respectively, of the mycobacterial gene being studied here.
Analysis of open reading frame MTCY31.27.
The SwissProt database of the National Center for Biotechnology Information World-Wide Web site was searched with BLASTP for proteins exhibiting homology to the translated DNA sequence of M. tuberculosis open reading frame MTCY31.27 (the accession number for the whole cosmid, MTCY31, is Z73101, while the SwissProt accession number for MTCY31.27 is Q10557). The OmpA-like regions of the 18 proteins with the highest degrees of homology were then matched with the OmpA-like region of the predicted M. tuberculosis protein (residues 258 to 301) by using PileUp from the Genetics Computer Group (GCG) suite of programs. The SwissProt accession numbers of the 18 proteins are shown in Fig. 1.
FIG. 1.
Sequence homologies in OmpA-like regions. Amino acid sequences (44 residues) of the distinctive OmpA-like regions of 18 proteins belonging to the OmpA family were compared to the homologous region (residues 258 to 301) near the C terminus of the predicted sequence of the gene specifying OmpATb of M. tuberculosis, using the program PileUp from the GCG suite. The left-hand column lists the SwissProt accession numbers for the proteins compared. Amino acids in boldfaced type are shared with OmpATb; residues underlined in the mycobacterial sequence are conserved throughout the series of sequences. Dots indicate gaps introduced by the program to maximize matching.
Amplification and cloning of MTCY31.27 and its truncated version.
Genomic DNA of M. tuberculosis H37Rv was kindly supplied by Gili Bachrach. Open reading frame MTCY31.27 was amplified from this DNA with the following primers: 1, 5′-AGGGAGTCATATGGTGGCTTCTAAGGCGGGTTTG-3′; and 2, 5′-GAAGGATCCCCCCCAGGAACGCCAGCAGGTA-3′. For amplification of the truncated gene, lacking the code for the presumed amino-terminal signal sequence, an alternative primer, 1a (5′-AGAGAGTCATATGTTCGAGCGGCCCCAGTCC-3′), replaced primer 1. Primers 1 and 1a both contained an NdeI restriction site; primer 2 had a BamHI site. PCR was performed with the Expand High Fidelity PCR system (Boehringer Mannheim Ltd., Lewes, E. Sussex, United Kingdom), using the 1.5 mM MgCl2 Expand buffer supplied with the kit. Annealing temperatures were 58 and 63°C for the full-length gene and the truncated gene, respectively. The products were separated on a 1% agarose gel, isolated with a QIAquick gel extraction kit (Qiagen Ltd., Crawley, W. Sussex, United Kingdom), and digested with restriction endonucleases NdeI and BamHI (Boehringer Mannheim); a corresponding digestion was also applied to plasmid pET-15b. Digested products were isolated as described above and then ligated with T4 DNA ligase (21) to obtain plasmids pRS1 and pRS2, with complete and truncated genes inserted, respectively.
The nucleotide sequence of the gene inserted into plasmid pRS2 was obtained with an ABI Prism 377 machine, using an ABI Prism dye terminator cycle sequencing kit (Perkin-Elmer, Foster City, Calif.) according to the manufacturer’s instructions.
Expression and purification of OmpATb.
Competent cells of E. coli BL21(DE3)pLysS were transformed by the heat shock method for 2 min at 42°C with 100 ng of pRS1 or pRS2 and then plated onto L agar supplemented with ampicillin (200 μg/ml), methicillin (200 μg/ml), and chloramphenicol (34 μg/ml). Single colonies were inoculated into 5 ml of L broth containing the same antibiotics. After incubation for 10 h at 37°C with shaking, the individual cultures were transferred to three 1-liter flasks, each containing 200 ml of the same medium. Incubation was continued at 37°C with shaking until the optical density (OD) at 600 nm was 0.6; then isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the medium was also supplemented with additional ampicillin and methicillin (40 mg of each per flask). After incubation for a further 3 h, the cells were harvested by centrifugation (5,000 × g, 20 min), resuspended in 5 ml of buffer A (10 mM imidazole, 1 M NaCl, 50 mM Tris-HCl [pH 8]), and frozen. To obtain satisfactory yields of nontruncated protein (plasmid pRS1), the procedure was subsequently modified as follows: (i) ampicillin and methicillin were replaced by carbenicillin, (ii) an additional 40 mg of carbenicillin was added to each flask 2 h after inoculation, and (iii) the cells were harvested 1 h after induction with IPTG. (Note that during the course of these experiments, methicillin ceased to be available for laboratory use in the United Kingdom and hence was replaced by carbenicillin in our later experiments.)
Frozen E. coli cells (from 600-ml cultures) in which truncated OmpATb had been expressed were thawed in a water bath at room temperature, treated with 15 μg of RNase A (Sigma Chemical Company, Poole, Dorset, United Kingdom) and 1,000 U of DNase (Promega), and incubated for 30 min. The mixture was centrifuged at 10,000 × g for 20 min, and the supernatant was separated. Truncated OmpATb was found in the supernatant. Cells expressing intact OmpATb were similarly processed, but the protein was extracted from the sediment with 1.4 ml of 1% Zwittergent 3-12 (Boehringer Mannheim) in 20 mM Tris-HCl, pH 8, for 1 h at room temperature (ca. 20°C). The extract was centrifuged at 12,000 × g for 20 min, and the supernatant was collected. Solutions containing truncated or nontruncated OmpATb were added to 0.1 ml or 1 ml, respectively, of nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen), previously equilibrated with buffer A, and stirred on ice for 1 h. The resin was washed by centrifugation, first with buffer A and then with buffer B (same composition as buffer A but at pH 6), in each case until the A280 of the supernatant was <0.01. Bound protein was then eluted with buffer C (0.5 M imidazole in 50 mM sodium phosphate, pH 6). For purification of intact OmpATb, all three buffers were supplemented with 1% Zwittergent 3-12. Protein concentrations were measured with bicinchoninic acid reagent [Peirce & Warriner (UK) Ltd., Chester, United Kingdom]. When necessary, His6 and the 7-residue peptide following this tag were removed from purified recombinant proteins by utilizing the thrombin site built into pET-15b; protein (0.1 mg) in 20 mM Tris-HCl (pH 8.4)–150 mM NaCl–2.5 mM CaCl2 was treated with 1 U of thrombin (Sigma) for 4 h at room temperature. The reaction was stopped with phenylmethylsulfonyl fluoride (PMSF) at a final concentration of 2 mM. The released polyhistidine tag was removed with 0.1 ml of Ni-NTA resin equilibrated with buffer A.
Large-scale production of OmpATb.
For large-scale preparation of intact OmpATb, 5 ml of L broth containing 200 μg of carbenicillin and 34 μg of chloramphenicol per ml was inoculated with E. coli BL21(DE3)pLysS carrying plasmid pRS1 and grown for 6 h at 37°C with shaking. This culture was used as the inoculum for 600 ml of L broth with the same concentrations of antibiotics. The resulting culture was incubated under the same conditions, with further carbenicillin (120 mg) added each hour, until the OD at 600 nm reached 0.6 after 5 h. The culture was kept at 4°C overnight and then used as the inoculum for 40 liters of L broth containing 100 μg of ampicillin and 34 μg of chloramphenicol per ml. The fermentor was stirred at 250 rpm, and further 4-g amounts of ampicillin were added every hour. When the OD at 600 nm reached 0.6 (3.5 h), expression of OmpATb was induced with IPTG at a final concentration of 0.2 mM. The bacteria (115 g [wet weight]) were harvested after 1 h and stored as frozen packed cells at −70°C.
Frozen bacteria (34 g) were placed in 45 ml of buffer A containing 1 mM PMSF and sonicated six times, for 30 s each, in an ice bath at 90 W with a 12.7-mm probe (Soniprobe; Dawe Instruments Ltd., London, United Kingdom), with 1-min intervals between sonications for cooling. The sonicate was centrifuged for 20 min at 12,000 × g, and the supernatant was discarded. The pellet was resuspended in 40 ml of buffer A containing 1 mM PMSF and 1% Zwittergent 3-12 and incubated for 1 h at room temperature. The mixture was centrifuged (10,000 × g, 10 min), and the supernatant was combined with 2 ml of Ni-NTA resin and stirred for 1 h at room temperature. The resin was collected by centrifugation (5,000 × g for 5 min), washed five times with buffer A containing 1% Zwittergent 3-12, and then washed with buffer B containing 1% Zwittergent 3-12 until the A280 of the washings was <0.05. Bound protein was then eluted with buffer B containing 1% Zwittergent 3-12 and 250 mM imidazole; seven 1-ml elutions were done, and the material in the final elution (see Fig. 3) was used for experiments.
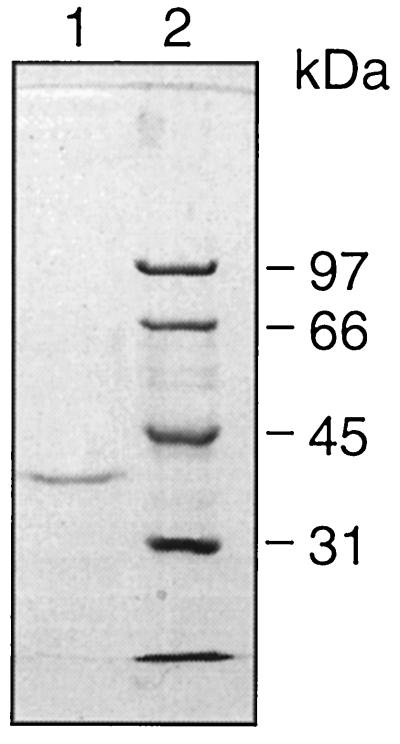
FIG. 3.
SDS-PAGE of purified intact OmpATb. Protein purified with Ni-NTA resin was separated on an SDS–10% polyacrylamide gel along with unstained molecular mass standards (whose positions are indicated on the right). Lanes: 1, purified intact OmpATb; 2, standards. The gel was scanned with a Leaf Lumina camera, and the image was processed on an Apple computer, using Adobe Photoshop version 3 and Macromedia Freehand version 5.5.
Some batches of OmpATb were additionally purified by gel filtration through a Superdex P-75 (Pharmacia) column in 0.5% Zwittergent 3-10 in 25 mM Tris-HCl (pH 8)–50 mM NaCl–1 mM EDTA–1 mM dithiothreitol. Fractions containing protein of the expected molecular mass, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were pooled and desalted with Vivaspin concentrators (10,000-Da molecular mass cutoff; Vivascience Limited, Binbrook, Lincoln, United Kingdom). The concentrated protein was desalted and buffer exchanged, using a PD 10 column (Pharmacia) equilibrated with 0.8% n-octyl-β-d-glucopyranoside (Alexis Corp. Ltd., Bingham, Nottingham, United Kingdom) in 25 mM Tris-HCl (pH 8)–50 mM NaCl–1 mM EDTA–1 mM dithiothreitol.
Production of polyclonal antiserum to truncated OmpATb.
A polyclonal antiserum to truncated OmpATb was prepared by Murex Biotech, Dartford, Kent, United Kingdom, as follows. Two Murex lop rabbits were immunized subcutaneously with 0.2 ml of recombinant protein (0.5 mg/ml in phosphate-buffered saline) in Freund incomplete adjuvant (1:1 [vol/vol]) at each of six sites. Similar booster injections were given at 14 and 28 days. Test bleeds were performed 7 days after each immunization, and the sera were checked for reactivity with OmpATb. Four weeks after the second booster immunization, the animals were bled out and sera were prepared.
Liposome swelling assay.
The assay followed the published method (14) with slight modifications. Protein samples were desalted by subjecting them to three passes through Bio-Spin 6 columns (Bio-Rad, Hemel Hempstead, Hertfordshire, United Kingdom) in accordance with the manufacturer’s instructions. Liposomes were prepared with 2.6 μmol of phosphatidylcholine (Sigma type XVI-E from fresh egg yolk) and 0.4 μmol of dicetyl phosphate (Sigma). Up to 25 μg of OmpATb was used to prepare proteoliposomes; the lipid-protein mixture was dried by rotary evaporation at 40°C. OD changes in liposome suspensions were measured with a UV-2 spectrophotometer (Thermo Unicam, Cambridge, United Kingdom); recorded data were converted to comma-separated value files by using Convert (Thermo Unicam) and then analyzed with SigmaPlot 3.0 (SPSS ASC GmbH, Erkrath, Germany). The swelling rate was calculated as the change in the OD at 450 nm over a 30-s interval; the time was chosen as a compromise between a very short time period, which would be affected by timing uncertainties, and a longer time interval, during which the swelling rate would be affected by concentration changes inside the liposomes.
Lipid bilayer experiments.
Channel-forming activities of recombinant OmpATb and detergent extracts of M. tuberculosis were examined in planar bilayer membranes. Bilayers were formed from soybean lecithin type II-S (Sigma) by the technique of Schindler (22). The buffer used on both sides of the bilayer was 10 mM HEPES, pH 7.4, containing 1 M NaCl and 10 mM CaCl2. Using Ag/AgCl electrodes, the membrane current under the voltage clamp was measured with a bilayer amplifier (HAMK2TC; R.A.P. Montgomery, London, United Kingdom), filtered with a low-pass filter (VBF/3; Kemo Ltd., Beckenham, Kent, United Kingdom) set at 10 kHz, and digitized by a personal computer equipped with a CED 1401 Plus interface (Cambridge Electronic Design, Cambridge, United Kingdom). The data were analyzed with patch clamp software (PAT V1.6; J. Dempster, University of Strathclyde, Glasgow, United Kingdom).
Calculation of pore size.
The size of the pore formed by OmpATb was estimated by the method of Nikaido and Rosenberg (15). Theoretical curves were calculated for pore radii of from 0.6 to 1.4 nm and the following molecules: glycine, glycerol, arabinose, glucose, sucrose, and raffinose. Free-diffusion coefficients in water were obtained from reference 32 or by calculation from the hydrodynamic radii with the Stokes-Einstein equation (1); radii were obtained from references 15 and 24 or, in the case of glycine, by calculation. The experimental curve was obtained from the liposome swelling assay, using the same set of molecules (except glycerol, which diffuses significantly into liposomes even in the absence of porin-like protein, and raffinose, which diffuses too slowly to be measured). Experimental diffusion rates were means of values obtained in at least three experiments.
CD spectra of recombinant proteins.
Circular dichroism (CD) spectra were recorded on a J-600 spectropolarimeter [JASCO (UK) Ltd., Great Dunmow, Essex, United Kingdom) at 20°C in a solution consisting of 0.8% n-octyl-β-d-glucopyranoside in 25 mM Tris-HCl (pH 8)–50 mM NaCl–1 mM EDTA–1 mM dithiothreitol. Plotted curves were averages of 10 baseline-corrected scans and were drawn in terms of molar ellipticity, calculated by using a mean residue weight of 103.27 Da for intact OmpATb or 103.31 Da for the truncated protein. (Δɛ values can be calculated by dividing the molar ellipticity values by 3,298.)
Preparation of porin-containing extracts from M. tuberculosis.
M. tuberculosis H37Rv was grown at 37°C with shaking in 200 ml of Dubos broth-Dubos albumin (25:1; Difco Laboratories Ltd., West Molesey, Surrey, United Kingdom) in a 500-ml flask until the OD at 600 nm reached 0.6. The bacteria were harvested by centrifugation at 12,000 × g for 20 min, washed in 5 ml of phosphate-buffered saline (pH 8), and resuspended in 0.5 ml of a solution containing 1% Zwittergent 3-12, 40 mM disodium EDTA, and 20 mM Tris-HCl (pH 8); PMSF was added to a final concentration of 2 mM. Cells were lysed by adding 0.5 volume of glass beads (159- to 212-μm diameter; Sigma) and shaking in a Mini Bead Beater (Biospec Products, Bartlesville, Okla.) at 3,800 rpm three times for 1 min each with 1-min intervals on ice between the treatments. The lysate was incubated for 1 h and centrifuged, and the supernatant was collected.
The presence of OmpATb in extracts was demonstrated by immunoblotting with the above-described rabbit antiserum to the recombinant protein. Approximate amounts were determined by enzyme-linked immunosorbent assay, using plates coated with a range of quantities of recombinant OmpATb and of M. tuberculosis extract. Sixteen replicates were made of each level of protein.
RT-PCR of mRNA from M. tuberculosis.
RNA was extracted from 150 ml of an OD 0.8 culture of M. tuberculosis cells by using an RNeasy kit (Qiagen). The protocol given for the extraction of total RNA from the yeast Saccharomyces cerevisiae (RNeasy Mini handbook) was modified as follows: in step 1, centrifugation was extended to 30 min; in step 3, 1.1 ml of RLT buffer was used; in step 4, a RiboLyser (Hybaid Ltd., Teddington, Middlesex, United Kingdom) was used to disrupt the cells (speed 6, 20 s); and in step 5, centrifugation was extended to 20 min. Extracted RNA was treated with 8 μl of RNAguard human placental RNase inhibitor (Pharmacia). One-fourth of the RNA prepared was treated twice with 2 μl (20 U) of RNase-free DNase I (Boehringer Mannheim) in accordance with the manufacturer’s instructions. After each treatment, the RNA was cleaned by using an RNeasy kit according to the manufacturer’s instructions. The reverse transcription (RT) reaction was performed by the method of Papavinasasundaram et al. (17). In the PCR, a 3-μl sample of the RT reaction product was used as a template in Expand HF buffer containing 15 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 300 nM each primer, and 2.6 U of Expand High Fidelity PCR system enzyme mix (Boehringer Mannheim). Since the occurrence of technical difficulties in the amplification of the long cDNA of the complete protein was expected, a fragment truncated at the C-terminal end of the protein was amplified. A new primer (primer 3) was designed as follows: 5′-ATTGGATCCTTATGGCGGTGCCTGGCCCCGTAACCT-3′. This primer was used, together with primer 1, to amplify a fragment of 600 bp.
RESULTS
The open reading frame MTCY31.27 was marked by the annotators of the M. tuberculosis genome at the Sanger Centre as having homology with an outer membrane protein of E. coli (3). Our BLASTP search of the SwissProt database at the National Center for Biotechnology Information Web site identified 26 proteins homologous to the translated mycobacterial DNA sequence with scores exceeding 116 and probabilities of <10−7; of these, 24 were annotated as being members of the OmpA family of outer membrane proteins. The OmpA family is distinguished by the presence of a conserved OmpA-like region at the carboxy-terminal end of the protein, which is believed to extend into the periplasm of the bacterial cell (27). The OmpA-like regions of 18 proteins exhibiting the highest degrees of homology to the protein specified by MTCY31.27 were compared with the corresponding region in the predicted mycobacterial protein (Fig. 1). Three-fourths of the residues in the mycobacterial protein were shared with at least one of the other proteins; nearly half were common to at least one-half of the other members of the OmpA family, and 10 of 44 residues were conserved in all of the members. It seemed clear that the mycobacterial protein was a member of the OmpA family of proteins.
Using the primers described in Materials and Methods, the open reading frame was successfully amplified and cloned into E. coli. However, an attempt to express the gene caused cessation of growth of the bacteria about 1 h after induction, followed by a slow decline of the OD. This event was only slightly delayed when a low concentration of inducer (0.1 mM) was used. E. coli containing the vector alone, or the vector carrying lexA of M. tuberculosis, continued to grow after induction, with only a minor inflection in the curves. We concluded that the mycobacterial OmpA-like protein was toxic to E. coli, probably because it became inserted into one of the membranes of the host. Such toxicity has been reported previously (18, 23); in those cases, toxicity was circumvented by truncating the genes to remove the signal sequences responsible for targeting the proteins to the bacterial membrane, although this resulted in the formation of inclusion bodies and problems with the renaturing of the expressed proteins. Examination of a hydrophobicity plot calculated for the predicted amino acid sequence of the M. tuberculosis open reading frame showed that there was a possible signal sequence at the amino terminus, characterized by a hydrophilic segment followed by a hydrophobic region, centered around residues 18 and 39, respectively. The putative signal sequence conformed with the general rules formulated for identifying such sequences (31, 33), although it was considerably longer than the authentic signal sequence of E. coli OmpA. A new primer was designed to amplify a truncated gene lacking the bases specifying the possible signal sequence. A truncated protein was successfully overexpressed in E. coli and purified. The protein was soluble without the use of surfactants but lacked any porin-like activity in the liposome swelling assay (results not shown), although a complete nucleotide sequence showed that the correct gene had been amplified and cloned.
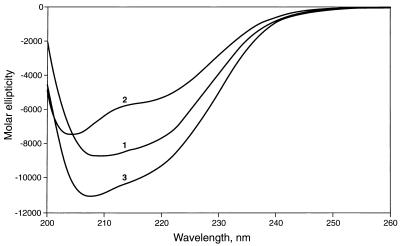
CD spectra of the intact and truncated recombinant proteins are shown in Fig. 2. The plot for the intact protein closely resembled in both shape and magnitude the published plot for OmpA of E. coli (27) and indicated a significant content of α-helix as well as β-sheet. Much of the α-helical structure was lost when the protein was treated with 1% SDS, and the protein became more disordered. The spectrum of the truncated protein indicated that this too contained considerable secondary structure, but with relatively less β-sheet and more α-helix; this may indicate that the first 47 amino acids of the intact protein are partly arranged as a β-sheet.
FIG. 2.
CD spectra of recombinant OmpATb. CD spectra were obtained as described in Materials and Methods. Plot 1, purified intact OmpATb; plot 2, intact OmpATb treated with 1% SDS; plot 3, purified truncated OmpATb.
Assuming that the OmpA-like protein of M. tuberculosis was, in fact, a porin, the inactivity of the truncated protein indicated that expression of the presumed signal sequence was essential for eventual activity. Although the intact protein could not be overexpressed, some small amount must have been present in the induction experiment in order to manifest its toxicity to E. coli. We therefore processed induced E. coli containing the vector with the gene for the intact protein at a time (1 h after induction) just before growth was expected to cease and obtained, after purification with nickel-NTA resin, a single band in SDS-PAGE with an approximate molecular mass, as determined by comparison with markers, of 38 kDa (Fig. 3). (The predicted mass, including the His.Tag fusion, is 35.4 kDa.) The mobility of the recombinant protein on SDS-PAGE was not altered by heat; samples not boiled after addition of sample buffer were indistinguishable from samples treated in the normal way.
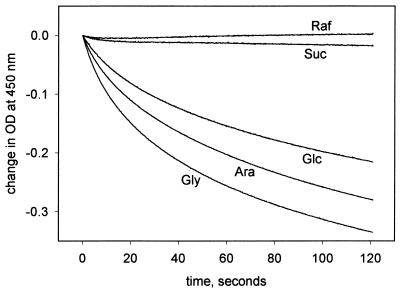
The recombinant protein behaved as a porin in the liposome swelling assay (Fig. 4); its permeability to sucrose was low, and permeability to raffinose could not be detected. The specific activity for arabinose was 6 (in units of ΔOD × 1,000 per minute per microgram of protein), similar to that of OmpA of E. coli (26). We propose the name OmpATb for this protein. Removal of the polyhistidine tag did not alter the behavior of the protein in the liposome swelling assay, and gel-filtered OmpATb also behaved identically. This ruled out the possibility that the activity we had observed was attributable to polyhistidine-tagged signal sequence removed by (natural) processing and conserved throughout the purification with the Ni-NTA column. Protein extracted from E. coli containing the vector alone, without the gene for OmpATb, and purified had no porin-like activity in the liposome swelling assay, confirming that the activity was a property of OmpATb and was not caused by E. coli porin contamination.
FIG. 4.
Porin-like activity of OmpATb. Proteoliposomes prepared with purified intact OmpATb (25 μg of protein; see Materials and Methods) were used in the liposome swelling assay in isotonic solutions of the substances indicated. A decrease in the OD at 450 nm corresponds to swelling of liposomes. Abbreviations: Raf, raffinose; Suc, sucrose; Glc, glucose; Ara, arabinose; Gly, glycine. The slight irregularity of the lines is attributable to instrument noise.
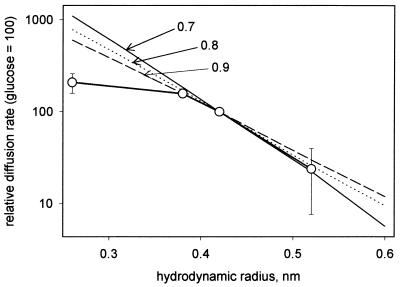
The size of the pore formed in liposomes by OmpATb was estimated from measurements of the rate of swelling in the presence of isotonic concentrations of glycine, arabinose, glucose, and sucrose (Fig. 5). The slope of the experimental line of the relative diffusion rate plotted against the hydrodynamic radius was compared with slopes of theoretical lines calculated for pores with radii of 0.7, 0.8, and 0.9 nm (15). The experimental data did not fall on a straight line: the point for glycine implied a large pore size. However, the data for arabinose, glucose, and sucrose did suggest a linear relationship and indicated a pore diameter of between 1.4 and 1.8 nm.
FIG. 5.
Determination of pore size for OmpATb. The experimental curve obtained with OmpATb in the liposome swelling assay is compared with calculated curves for various possible pore sizes. Data points (○) are means of three measurements.
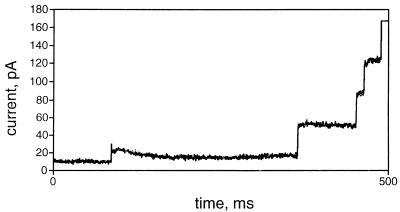
The behavior of recombinant OmpATb in a planar lipid bilayer is shown in Fig. 6. At protein concentrations of about 0.8 μg/ml in the cis compartment of the apparatus, channels incorporated spontaneously at an applied potential difference of 10 mV, visualized as a characteristic “staircase.” A number of different sizes of current steps were seen. The minimum unit was approximately 7 pA (corresponding to a single-channel conductance of 700 pS); the detergent extract of M. tuberculosis also produced pores with a conductance of 700 pS in the membranes (10). Figure 6 also shows steps corresponding to 4 or 5 units being incorporated at once. This behavior resembles that of OmpA of E. coli described by Saint et al. (20); these authors suggest that the protein is partly present in an aggregated form. At protein concentrations of less than 0.1 μg/ml, single channels could be incorporated by applying ±200 mV across the bilayer. The traces showed that the channels were voltage sensitive and closed reversibly in response to a characteristic applied transmembrane potential difference of either polarity (140 to 150 mV) (10). The pores produced by detergent extracts of M. tuberculosis had gating characteristics identical to those of the recombinant protein.
FIG. 6.
Channel-forming activity of recombinant OmpATb in a planar lipid bilayer. Measurements of channel-forming activity were made as described in Materials and Methods. The trace shows the final 500 ms of a 1,300-ms recording, selected to show several increments in the current that passed by the membrane. One channel was open when the trace shown was begun.
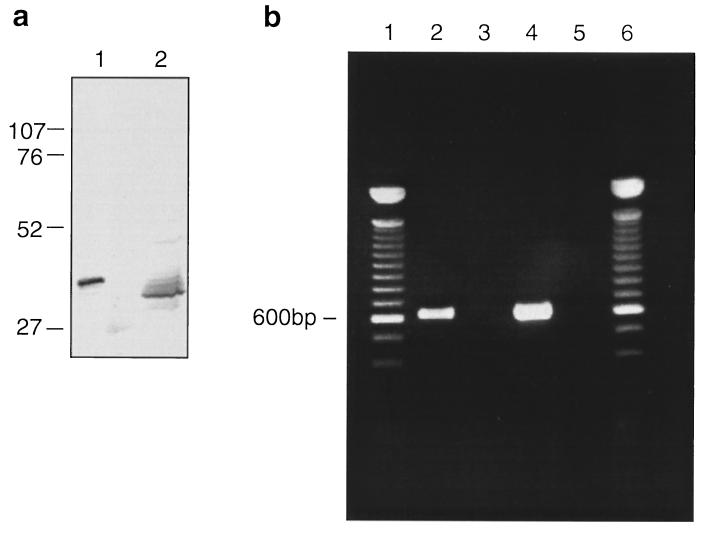
By immunoblotting with a polyclonal rabbit serum raised to purified truncated OmpATb, we were able to detect a band of the appropriate molecular mass in extracts of M. tuberculosis H37Rv (Fig. 7a). The M. tuberculosis protein had a somewhat smaller apparent molecular mass than that of recombinant OmpATb from which the polyhistidine tag had been removed.
FIG. 7.
Expression of the gene for OmpATb in growing M. tuberculosis. Shown are gels scanned with a Leaf Lumina camera; images were processed on an Apple computer, using Adobe Photoshop version 3 and Macromedia Freehand version 5.5. (a) OmpATb and protein extracted from M. tuberculosis H37Rv were separated in SDS–10% polyacrylamide gels, blotted onto Immobilon-P membranes, and then detected with rabbit antiserum to truncated OmpATb. Lanes: 1, purified OmpATb; 2, protein from M. tuberculosis. The positions of marker proteins (not visible on the photograph) are indicated (in kilodaltons). Sample sizes were 50 ng for recombinant OmpATb and 100 μg for M. tuberculosis extract. (b) Gel of RT-PCR product obtained from mRNA of M. tuberculosis as described in Materials and Methods. Lanes 2 to 5 show products of PCRs with primers 1 and 3 and templates as follows: lane 2, reverse-transcribed M. tuberculosis RNA; lane 3, as for lane 2, but reverse transcriptase was omitted from the RT reaction mixture; lane 4, M. tuberculosis DNA; and lane 5, no template. Lanes 1 and 6 contain 100-bp ladder DNA (GIBCO-BRL; Life Technologies Ltd, Paisley, United Kingdom).
RT-PCR analysis of RNA extracted from M. tuberculosis growing in culture, using primers 1 and 3, amplified a fragment of DNA of the expected size (600 bp) (Fig. 7b). This indicated that the gene for OmpATb is expressed in the organism growing in culture.
Enzyme-linked immunosorbent assay determination of the concentration of material in the M. tuberculosis extracts reacting with rabbit anti-OmpATb, presumed to be OmpATb itself since there is no other gene in the genome with close homology, showed that about 0.02% of the total protein was OmpATb. Estimates based on the activity of the extract in the liposome swelling assay were much higher (about 2%), suggesting that a large proportion of the recombinant OmpATb may be in an inactive form.
DISCUSSION
M. tuberculosis displays some general resistance to antibiotics, notably β-lactams, although it does not have the extreme degree of nonspecific resistance found in strains of Mycobacterium avium or M. chelonae. Porin-like proteins from M. chelonae have already been characterized (30), although their sequences are not available. Since, according to present information, the structures of the envelopes of all mycobacteria are similar (6), it was expected that porin analogs would also be present in species in the M. tuberculosis complex. Characterization of such an analog was greatly simplified for us by the publication of the DNA sequence for a candidate porin gene in the M. tuberculosis database.
Our inability to overexpress OmpATb was typical of difficulties in expression of foreign membrane-active proteins which can interfere with the function of the host membrane. A solution to this problem has been to remove the signal sequences which direct the proteins to their intramembrane location (18, 23), although the expressed proteins were then produced in an insoluble form in inclusion bodies and had to be solubilized by denaturation and refolded before their biological activity could be recovered. The gene specifying OmpATb appeared to include a signal sequence. Although this was rather long compared with such sequences in many bacteria, it was very similar in length to known signal sequences of the secreted antigen 85 proteins of M. tuberculosis (5), and the presumed cleavage site had some similarity. Thus, we were fairly confident that we had identified the signal sequence correctly. The truncated protein was readily expressed and was soluble without the use of surfactants, but it lacked activity in the liposome swelling assay. It is clear that the signal sequence (if it is one) is only partly, if at all, removed during the expression of intact OmpATb in E. coli, since our successful purification of active porin depended on the presence of the amino-terminal polyhistidine tag, which would have been removed along with the signal sequence. It is possible that the E. coli proteases do not recognize mycobacterial signals. The putative OmpATb protein identified in the extracts of M. tuberculosis had a lower apparent molecular mass, but the decrement in mass (approximately 2.6 kDa, as determined by measuring the distance between positions on immunoblots) was less than might have been expected if the presumed signal sequence had been removed (4.9 kDa). Additional experiments and considerably more material would be needed to demonstrate processing conclusively. Our experiments show, though, that recombinant OmpATb, with the presumed signal sequence, is active as a porin in the planar lipid bilayer assay.
OmpATb exhibits appreciable homology to OmpA family porins of gram-negative organisms. This resemblance is largely confined to the so-called OmpA region, toward the carboxy terminus of the protein, which is believed to have a periplasmic location in gram-negative species (19, 27). The mycobacterial protein also has a proline-rich region in a position corresponding to the position of the proline-rich hinge of OmpA of gram-negative bacteria. This suggests that the physical organization of OmpATb may be similar to that of other members of the OmpA family. OmpA was only relatively recently recognized as a porin (20, 25, 26); it apparently exists as a monomer, rather than as a trimer like the classical porins, and has a low specific activity compared with that of classical porins. There is no information yet about whether OmpATb exists as a monomer in the cell envelope, but it too has a low specific activity. A distinctive feature of OmpA of E. coli is the low specific activity of the purified protein in the liposome swelling assay (6 U of ΔOD × 1,000/min/μg of protein, compared with 400 U of ΔOD × 1,000/min/μg of protein for the classical trimeric porin OmpF [26]); OmpATb shares this low activity.
OmpATb is clearly different from the porins of rapid-growing mycobacteria (29, 30); it has a different molecular mass, and its amino-terminal sequence differs from that reported for the porin of M. smegmatis (11). Our attempt to measure its pore diameter was not wholly successful, because the behavior of glycine was anomalous. The method depends on the assumption that the molecules studied behave as spherical entities in water, and it is possible that glycine does not behave in this manner. Using the data points for three carbohydrates gave a value between 1.4 and 1.8 nm, which is slightly smaller than the 2 nm calculated for the pore of the M. chelonae porin (30) and the 2 to 3 nm (11, 29) determined for that of M. smegmatis, although larger than OmpA or the classical OmpF of E. coli (both of which are about 1.1 nm [25], [26]). The M. chelonae porin has a specific activity for arabinose of 27 U of ΔOD × 1,000/min/μg of protein (30), compared with our value of 6 U of ΔOD × 1,000/min/μg of protein for recombinant OmpATb. The single-channel conductance of OmpATb was 700 U of ΔOD × 1,000/min/μg of protein, similar to that of OmpF of E. coli but considerably lower than that of the porin of M. chelonae (1,400 U of ΔOD × 1,000/min/μg of protein) (28) or M. smegmatis (2,900 U of ΔOD × 1,000/min/μg of protein) (29).
The question of whether OmpATb is the sole such molecule present in M. tuberculosis arises. Our approach, starting with a DNA sequence identified on theoretical grounds, cannot give a direct answer. There is no other gene in the M. tuberculosis genome showing homology to OmpATb, and no DNA sequences specifying homologs of classical, trimeric porins of gram-negative species have yet been identified. The similar behaviors of the recombinant protein and the detergent extract of M. tuberculosis in planar lipid bilayers suggest that OmpATb may be the only porin active in the extracts. On the other hand, M. tuberculosis is about 10 times more permeable to cephalosporins than M. chelonae (2, 4), which seems inconsistent with the relative pore sizes and specific activities determined for the porins of the two species. The simplest explanation of this discrepancy is that there are more porin molecules in the envelope of M. tuberculosis than in that of M. chelonae, but an alternative theory is that additional types of porin are present, while the low specific activity may be due to the presence of a high proportion of inactive (perhaps aggregated) protein in our preparations. A peptidoglycan-associated polypeptide of M. tuberculosis which exhibits some amino acid homology to porins of gram-negative bacteria has been described (7); limited amino acid sequences reported for this protein do not appear in OmpATb, so this may represent a second porin type. Resolution of this problem depends on improving yields of purified envelope from M. tuberculosis, so that a thorough search for porins can be made, and developing methods of measuring levels of individual porin species in mycobacterial envelopes.
If porins do, indeed, control the access of small hydrophilic molecules to the mycobacterial cell, as appears to be the case, then the properties of OmpATb (and of other porin types, if present) put a constraint on the size of antimycobacterial drugs which can enter the cells by this route. Molecules much bigger than sucrose are unlikely to be able to pass through the pores. This raises the interesting question of how the large, polar streptomycin molecule reaches the mycobacterial cell and makes it clear that the identification of OmpATb is only a partial solution to the problem of understanding the permeability of the tubercle bacillus.
ACKNOWLEDGMENTS
We thank Gili Bachrach for advice and for supplying M. tuberculosis genomic DNA, Nichola Thomas-Nuttall and Farahnaz Movahedzadeh for providing cell extracts of M. tuberculosis, F. Movahedzadeh for providing pET-15b containing lexA of M. tuberculosis, Steve Martin for preparing CD spectra, and Susana Gonzalez-Rico and Paul Thurman for advice and assistance.
The M. tuberculosis sequencing project at the Sanger Centre is funded by The Wellcome Trust.
REFERENCES
- 1.Atkins P W. Physical chemistry. 5th ed. Oxford, United Kingdom: Oxford University Press; 1994. p. 795. [Google Scholar]
- 2.Chambers H F, Moreau D, Yajko D, Miick C, Wagner C, Hackbarth C, Kocagöz S, Rosenberg E, Hadley W K, Nikaido H. Can penicillins and other β-lactam antibiotics be used to treat tuberculosis? Antimicrob Agents Chemother. 1995;39:2620–2624. doi: 10.1128/aac.39.12.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 4.Connell N D, Nikaido H. Membrane permeability and transport in Mycobacterium tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: ASM Press; 1994. pp. 333–352. [Google Scholar]
- 5.Content J, de la Cuvellerie A, De Wit L, Vincent-Levy-Frébault V, Ooms J, De Bruyn J. The genes coding for the antigen 85 complexes of Mycobacterium tuberculosis and Mycobacterium bovis BCG are members of a gene family: cloning, sequence determination, and genomic organization of the gene coding for antigen 85-C of M. tuberculosis. Infect Immun. 1991;59:3205–3212. doi: 10.1128/iai.59.9.3205-3212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daffé M, Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol. 1998;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 7.Hirschfield G R, McNeil M, Brennan P J. Peptidoglycan-associated polypeptides of Mycobacterium tuberculosis. J Bacteriol. 1990;172:1005–1013. doi: 10.1128/jb.172.2.1005-1013.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarlier V, Nikaido H. Permeability barrier to hydrophilic solutes in Mycobacterium chelonei. J Bacteriol. 1990;172:1418–1423. doi: 10.1128/jb.172.3.1418-1423.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Rosenberg E Y, Nikaido H. Fluidity of the lipid domain of cell wall from Mycobacterium chelonae. Proc Natl Acad Sci USA. 1995;92:11254–11258. doi: 10.1073/pnas.92.24.11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mobasheri H, Senaratne R H, Draper P, Lea E J A. Single channel properties of a porin-like protein from Mycobacterium tuberculosis H37Rv in planar lipid bilayers. Biophys J. 1998;74:A320. doi: 10.1128/jb.180.14.3541-3547.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukhopadhyay S, Basu D, Chakrabarti P. Characterization of a porin from Mycobacterium smegmatis. J Bacteriol. 1997;179:6205–6207. doi: 10.1128/jb.179.19.6205-6207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray C J L, Lopez A D. Mortality by cause for eight regions of the world: Global Burden of Disease Study. Lancet. 1997;349:1269–1276. doi: 10.1016/S0140-6736(96)07493-4. [DOI] [PubMed] [Google Scholar]
- 13.Nikaido H. Transport across the bacterial outer membrane. J Bioenerg Biomembr. 1993;25:581–589. doi: 10.1007/BF00770245. [DOI] [PubMed] [Google Scholar]
- 14.Nikaido H, Nikaido K, Harayama S. Identification and characterization of porins in Pseudomonas aeruginosa. J Biol Chem. 1991;266:770–779. [PubMed] [Google Scholar]
- 15.Nikaido H, Rosenberg E Y. Effect of solute size on diffusion rates through the transmembrane pores of the outer membrane of Escherichia coli. J Gen Physiol. 1981;77:121–135. doi: 10.1085/jgp.77.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papavinasasundaram K G, Movahedzadeh F, Keer J T, Stoker N G, Colston M J, Davis E O. Mycobacterial recA is cotranscribed with a potential regulatory gene called recX. Mol Microbiol. 1997;24:141–153. doi: 10.1046/j.1365-2958.1997.3441697.x. [DOI] [PubMed] [Google Scholar]
- 18.Pullen J K, Liang S-M, Blake M S, Mates S, Tai J Y. Production of Haemophilus influenzae type-b porin in Escherichia coli and its folding into the trimeric form. Gene. 1995;152:85–88. doi: 10.1016/0378-1119(94)00706-x. [DOI] [PubMed] [Google Scholar]
- 19.Ried G, Koebnik R, Hindennach I, Mutschler B, Henning U. Membrane topology and assembly of outer membrane protein OmpA of Escherichia coli K12. Mol Gen Genet. 1994;243:127–135. doi: 10.1007/BF00280309. [DOI] [PubMed] [Google Scholar]
- 20.Saint N, De E, Julien S, Orange N, Molle G. Ionophore properties of OmpA of Escherichia coli. Biochim Biophys Acta. 1992;1145:119–123. doi: 10.1016/0005-2736(93)90388-g. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. pp. 5.60–5.71. [Google Scholar]
- 22.Schindler H. Formation of planar bilayers from artificial or native membrane vesicles. FEBS Lett. 1980;122:77–79. doi: 10.1016/0014-5793(80)80405-4. [DOI] [PubMed] [Google Scholar]
- 23.Schmid B, Krömer M, Schulz G E. Expression of porin from Rhodopseudomonas blastica in Escherichia coli inclusion bodies and folding into exact native structure. FEBS Lett. 1996;381:111–114. doi: 10.1016/0014-5793(96)00080-4. [DOI] [PubMed] [Google Scholar]
- 24.Schultz S G, Solomon A K. Determination of the effective hydrodynamic radii of small molecules by viscometry. J Gen Physiol. 1961;44:1189–1199. doi: 10.1085/jgp.44.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugawara E, Nikaido H. Pore-forming activity of OmpA porin of Escherichia coli. J Biol Chem. 1992;267:2507–2511. [PubMed] [Google Scholar]
- 26.Sugawara E, Nikaido H. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J Biol Chem. 1994;269:17981–17987. [PubMed] [Google Scholar]
- 27.Sugawara E, Steiert M, Rouhani S, Nikaido H. Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J Bacteriol. 1996;178:6067–6069. doi: 10.1128/jb.178.20.6067-6069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trias J, Benz R. Characterization of the channel formed by mycobacterial porin in lipid bilayer membranes. Demonstration of voltage gating and of negative point charges at the channel mouth. J Biol Chem. 1993;268:6234–6240. [PubMed] [Google Scholar]
- 29.Trias J, Benz R. Permeability of the cell wall of Mycobacterium smegmatis. Mol Microbiol. 1994;14:283–290. doi: 10.1111/j.1365-2958.1994.tb01289.x. [DOI] [PubMed] [Google Scholar]
- 30.Trias J, Jarlier V, Benz R. Porins in the cell wall of mycobacteria. Science. 1992;258:1479–1481. doi: 10.1126/science.1279810. [DOI] [PubMed] [Google Scholar]
- 31.von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984;173:243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]
- 32.Weast R C, editor. CRC handbook of chemistry and physics. 70th ed. Boca Raton, Fla: CRC Press, Inc.; 1989. p. F-50. [Google Scholar]
- 33.Wickner W, Driessen A J M, Hartl F-U. The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu Rev Biochem. 1991;60:101–124. doi: 10.1146/annurev.bi.60.070191.000533. [DOI] [PubMed] [Google Scholar]