SUMMARY
The characterization of wild-type minimum inhibitory concentration (MIC) and zone diameter distributions with the setting of epidemiological cut-off values (ECOFFs or ECVs) provides a reference for the otherwise relative MIC values in the international system for antimicrobial susceptibility testing. Distributions of MIC values for a species and an agent follow a log-normal distribution, which in the absence of resistance mechanisms is monomodal and designated wild type (WT). The upper end of the WT distribution, the ECOFF, can be identified with statistical methods. In the presence of phenotypically detectable resistance, the distribution has at least one more mode (the non-WT), but despite this, the WT is most often identifiable using the same methods. The ECOFF provides the most sensitive measure of resistance development in a species against an agent. The WT and non-WT modes are independent of the organism´s response to treatment, but when the European Committee on Antimicrobial Susceptibility Testing (EUCAST) determines the clinical breakpoints, the committee avoids breakpoints that split WT distributions of target species. This is to avoid the poorer reproducibility of susceptibility categorization when breakpoints split major populations but also because the EUCAST has failed to identify different clinical outcomes for isolates with different MIC values inside the wild-type distribution. In laboratory practice, the ECOFF is used to screen for and exclude resistance and allows the comparison of resistance between systems with different breakpoints from different breakpoint organizations, breakpoints evolving over time, and different breakpoints between human and animal medicine. The EUCAST actively encourages colleagues to question MIC distributions as presented on the website (https://www.eucast.org/mic_and_zone_distributions_and_ecoffs) and to contribute MIC and inhibition zone diameter data.
KEYWORDS: ECOFF, epidemiological cut-off value, antimicrobial susceptibility testing, AST, phenotypic wild-type distribution, ECV
INTRODUCTION
In the 1990s, the Swedish Reference Group of Antibiotics (SRGA) began systematically characterizing species-specific minimum inhibitory concentration (MIC) distributions of isolates lacking phenotypically detectable resistance mechanisms. The distributions were used as a reference for the discussion and setting of clinical breakpoints (1). They summarized published MIC data and data generated for the committee by the methodology section of the reference group (SRGA-M).
In conjunction with re-starting the European Committee on Antimicrobial Susceptibility Testing (EUCAST) by the European Society for Clinical Microbiology and Infectious Diseasesin 2001, the EUCAST decided to systematically collect large numbers of international MIC distributions (2). Soon after, web-based software was developed for the collection and presentation of the distributions as MIC and zone diameter histograms with epidemiological cut-off values (ECOFFs), and since then, distributions have become freely available through the EUCAST (http://mic.eucast.org). By studying the available data, the Committee realized that, provided a methodology calibrated to the classical broth microdilution method was employed, what was considered, and later defined as the part of the distribution where isolates lacked phenotypically detectable resistance, was the same irrespective of when in time or where isolates were collected (hospital vs general population, geography, disease, and animal species). This knowledge increased the usefulness of gathering, evaluating, and displaying the distributions. Committee members agreed that “it would be difficult to define the abnormal (resistance by breakpoints) without first having agreed on the normal” (3).
PHENOTYPIC EXPRESSION OF ANTIMICROBIAL SENSITIVITY IN BACTERIA AND FUNGI—THE MINIMUM INHIBITORY CONCENTRATION
There is a tendency to consider the MIC as an absolute value on which mathematical and clinical predictions can be based. However, the MIC is a relative value, which like all assay outputs is not only subject to random variation but also to systematic variation since the value will depend significantly on the level of standardization of every detail of methodology (the medium used, inoculum, pH, cation content, incubation time and temperature, and endpoint reading). In 2005, a reference broth microdilution method for the determination of MICs for rapidly growing aerobic bacteria was agreed through the International Organization for Standardization (ISO) (4). As a result, MIC distributions as collected and displayed could be compared and curated against the reference. With the help of an initiative by the EUCAST, an international group of scientists developed the criteria for how to generate and amalgamate agreed MIC distributions and how to estimate the upper end of the wild type of each distribution, defined as the epidemiological cut-off value, abbreviated “ECOFF” by the EUCAST and later “ECV” by the Clinical and Laboratory Standards Institute (CLSI) (5, 6). However, MIC values are still relative, and for most situations, the reproducibility of any single value is significantly lower than for most other laboratory assays. When performed in one laboratory, by the same staff, using the same material from one agreed manufacturer on each occasion, most MIC values can be reproduced to a given value plus or minus one twofold dilution. If, for a given isolate and agent, 1 mg/L is the most commonly obtained value, then 15–25% of the values will, even under the best of circumstances, distribute over 0.5, 1, and 2 mg/L, with the occasional values of 0.25 and 4 mg/L. Under less stringent circumstances, the distribution will become wider, and what appears to be random variation will become a combination of random and systematic variation, where the latter is attributable to the variation produced by an alternative manufacturer of the medium, a discrete change in pH or cation content, an alternative incubation time, a different laboratory, and/or other factors.
WILD-TYPE MIC DISTRIBUTIONS
Contrary to what might be expected for a single species with a single antimicrobial agent, observed wild-type MICs are not a single value, but rather a range of values that follow a log-normal distribution. The range of MICs for a single species/agent combination is due to a combination of technical assay variation and biological variation (7). Studies using disk diffusion have shown that the relative contribution of technical and biological variation differs between species/agent combinations (8). In general, the contribution from technical variation is greater. Technical variation is both intra- and interlaboratory, with the variation between laboratories routinely observed even in well-controlled quality control studies using the same reagents (9). When viewing MIC distributions, such as those displayed in the EUCAST database, importantly all sources of variation are included in the putative wild-type part of the distribution and the calculation of the ECOFF, thereby increasing the general representativeness and utility of the distribution and the ECOFF.
IDENTIFYING THE WILD TYPE BY SELECTING EPIDEMIOLOGICAL CUT-OFF VALUES
Because wild-type isolates of a species show a range of values in MIC assays, it is necessary to select an MIC cut-off value that will provide confidence that values at or below that cut-off are without phenotypically detectable resistance mechanisms. By definition, this will exclude or minimize the presence of non-wild-type isolates at MICs below the ECOFF. This is the basis of the concept of the epidemiological cut-off value introduced by the EUCAST in 2003 (2). To favor the participation of many investigators and materials, to make generalized ECOFFs possible, and to increase the representativity of the distributions and ECOFF values, the EUCAST introduced the rules of only defining a wild-type distribution and the ECOFF provided at least five contributions from different sources agreed.
The EUCAST, and later CLSI, have formally defined the ECOFF. The definitions are similar but not identical. The EUCAST definition is “For a given microbial species and antimicrobial agent, the epidemiological cut-off value (ECOFF) is the highest MIC for organisms devoid of phenotypically detectable, acquired resistance mechanisms. It defines the upper end of the wild-type MIC distribution and is typically written as X mg/L, while the wild type is written as ≤X mg/L and the non-wild type as >X mg/L.” The CLSI definition is “the minimal inhibitory concentration (MIC) or zone diameter value that separates microbial populations into those with and without acquired and/or mutational resistance based on their phenotypes (wild type or non-wild type). The ECV defines the upper limit of susceptibility for the wild-type population of isolates.”
A wild-type MIC distribution for an agent and a species consists of all MIC values at or below the ECOFF. The important features are that they are species-specific and will be the same irrespective of the source of the isolates, the time period when isolates were collected, or their geographic origin.
HOW EPIDEMIOLOGICAL CUT-OFF VALUES ARE DETERMINED
There is, to date, no international standard method for selecting ECOFFs, but there are some basic properties of individual distributions that are widely accepted:
Isolates should be identified at the species level.
The MIC values should be based on the traditional twofold dilution series—0.125, 0.25, 0.5, 1, 2, 4, 8, etc. This facilitates analysis. Testing techniques involving concentrations between the traditional twofold values should be rounded up to the next twofold value, e.g., a measured MIC of 3 mg/L should be rounded up to 4 mg/L.
The dilution series should ideally include all concentrations in the putative wild type. Series that are truncated within either end of the putative wild type will distort the analysis and must be excluded.
Apart from these features, there are some differences in the approach from different investigators and organizations. When reviewing ECOFFs, it is important to be aware of these differences in the approach. The two best described methods for selecting ECOFFs are those of EUCAST and CLSI. The former codifies its approach in the EUCAST Standard Operating Procedure SOP 10.2 (5). The latter describes its approach in the M23 and M57 standards (6, 10), Development of In Vitro Susceptibility Testing Criteria and Quality Control Parameters (6) and Principles and Procedures for the Development of Epidemiological Cutoff Values for Antifungal Susceptibility Testing, respectively (10). The approaches are similar but differ in some important ways, where the EUCAST approach is more prescriptive about the analysis (Table 1).
TABLE 1.
A comparison of EUCAST- and CLSI-published approaches to ECOFF setting
| Feature | EUCASTa | CLSI M23 and M57b | Notes |
|---|---|---|---|
| Isolate identification | To species level (or species complex if members of a complex cannot be distinguished by MALDI-TOF | To species level only | |
| Methods used to determine MICs | ISO 20776–1 (4) and methods calibrated to it (which includes both EUCAST’s MIC method and CLSI’s M7). Antifungal methods Antimycobacterial reference method |
M7, M11, M27, M38, M44, M45, M51, and VET05 | CLSI does not have a reference method for Mycobacterium tuberculosis that generates MIC distributions |
| Dilution series | Twofold dilution series based on 0.5, 1, 2, 4, etc. | Not specified but generally defaults to standard twofold dilution series | |
| Minimum number of independent distributions required | 5 for a formal ECOFF and 3 or 4 for a tentative ECOFF (TECOFF) | 3 | |
| Minimum number of datapoints in the putative wild type of each individual distribution | 15 | None, but a total of 100 datapoints for pooled distributions | |
| Other acceptance criteria for distributions | Data not truncated (≤ or >) inside the putative wild type. Mode of the putative wild type is within 2 twofold dilutions of the most common mode. |
Data not truncated (≤ or >) inside the putative wild type. Mode of the putative wild type is within one or two twofold dilutions of the most common mode. |
|
| Analytical methods | Iterative statistical method on individual acceptable distributions, with the (T)ECOFF set as the geometric mean of the individual cut-offs, followed by visual predictive check | Iterative statistical method on pooled distribution data | For CLSI, if one laboratory provides more than 50% of the data points, weighting the data before pooling and analysis should be considered |
| Iterative statistical method cut-off percentage | ≥99% | ≥97.5% | |
| Publication(s) | EUCAST MIC distribution website (https://www.eucast.org/mic_and_zone_distributions_and_ecoffs) | M100, M57S |
EUCAST reference methods can be found at https://www.eucast.org/.
CLSI reference methods can be found at https://clsi.org/standards/products/microbiology/.
The EUCAST accepts distributions whose MICs have been generated by a reference MIC method, or a method calibrated to the reference method, provided that the distributions fulfill the acceptance criteria. The CLSI accepts distributions whose MICs have been generated by reference methods only, a range of which is described in CLSI standard M23, but does not require distributions from several investigators or a defined minimum number of distributions (6).
For the EUCAST, the reference method must be the one described by the International Organization for Standardization (ISO), namely, ISO 20776-1:2019 for rapidly growing aerobic bacteria (4) and ISO 16256:2021 for yeasts (11). For mycobacteria, there is currently no ISO reference method. Instead, the EUCAST has defined its own reference broth microdilution method for Mycobacterium tuberculosis (12). For antifungal testing, there are some important differences between the EUCAST and CLSI that can result in different MICs on the same isolate. For fungi, ECOFFs should only be set for data generated by the EUCAST methodology alone or CLSI alone (10). In cases, where MIC distributions are presented with or without ECOFFs and an accepted reference method is not yet available, it is important that the testing method should be clearly stated and that results obtained with quality control strains of related or similar species are presented.
-
The EUCAST also accepts distributions when the method is successfully calibrated to a reference method. Some examples include the following:
commercial and custom-made products (MIC trays) that conform to the reference method and have passed quality control assessment.
gradient diffusion tests where there is evidence that results conform to results obtained with the reference method; importantly, several of the gradient test methods produce biased results, as noted on the EUCAST website (https://www.eucast.org/mic_and_zone_distributions_and_ecoffs).
To be included in the analysis, a distribution must have a putative wild-type mode at or within one twofold dilution of the most common mode of all acceptable distributions.
For the EUCAST, ECOFFs will only be published when at least five acceptable distributions are available. Distributions accepted for aggregation should have at least 15 values in the putative wild-type mode; this is to ensure that there is an agreed identifiable wild-type mode. If three or four acceptable distributions are available, a tentative ECOFF (TECOFF) may be selected and presented, in the hope that with time more distributions will be available to allow the setting of an ECOFF.
Details are provided about the analytical technique used to select an ECOFF. The current EUCAST technique has evolved to provide more utility and confidence in the published ECOFF and includes a confidence interval for the selected ECOFF.
CALCULATION OF ECOFFS
Several analytical techniques have been applied to selecting ECOFFs over the years (13, 14, 15,16,17, 18, 19). The EUCAST uses iterative statistical method designed to fit a log-normal distribution curve to the putative wild type (15). In brief, the method seeks the best fit of a log-normal (log-Gaussian) distribution curve to the putative wild type using increasing subsets of the data commencing one two-fold dilution above the lowest mode in the MIC distribution. Because the log-normal distribution has no upper bound, the user must select a percentage to define an ECOFF that captures at least that percentage of the modeled wild type. Different percentages can be chosen for different purposes. If the aim is to avoid capturing non-wild types at or below the ECOFF, then a lower percentage should be chosen, typically 95.0 or 97.5%. If the aim is to ensure a maximum capture of the wild type and some overlap with non-wild-type isolates is acceptable, then a higher percentage is chosen, commonly 99.9%. The algorithm for this curve-fitting method is implemented in the freely available spreadsheet called ECOFFinder (https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/MIC_distributions/ECOFFinder_XL_2010_v2.1_web_version.xlsm). The spreadsheet offers the user a choice of ECOFFs based on the preferred cut-off percentage. Two examples of ECOFFinder output are shown in Fig. S1.
In the EUCAST database, the ECOFF that is displayed is calculated as follows, working on a log2 scale: (i) the 99.9% cutoff is calculated for each individual distribution accepted in the evaluation process, (ii) the mean and standard deviation of all these cut-offs are calculated, (iii) the mean is converted back to the arithmetic value and rounded up to the next highest twofold dilution, which is the displayed ECOFF, (iv) the standard deviation is used to calculate the confidence interval (CI), with the CI values converted to their arithmetic values and then rounded down and up, respectively, to show the lower and upper ends of the CI, (v) a visual predictive check similar to that used in pharmacokinetic modeling is made to ensure that the calculated ECOFF correctly modeled the graphical wild-type distribution and adjustments made if necessary within the CI. A worked example of how individual distributions are vetted and the procedure for ECOFF calculation are shown in Fig. S2 and S3; Table S1.
The advantage of this EUCAST approach to setting ECOFFs is that it includes the unavoidable variation observed in any laboratory practice, both intra-laboratory and inter-laboratory variation, as well as any variation between the reference method and methods calibrated to the reference method. It is based on the combined effort of many colleagues working independently of each other. This makes the wild-type distributions, as presented on the EUCAST website (http://mic.eucast.org), representative of the best effort of scientists wishing to critically evaluate and acquire good laboratory material and techniques.
WHY THE DEFINITION OF THE WILD-TYPE MIC DISTRIBUTION AND ECOFF ARE BASED PURELY ON PHENOTYPIC EXPRESSION
The correlation between phenotypically and genotypically characterized wild-type and resistant populations has been extensively investigated and found to be excellent for almost all agents and resistance mechanisms (20). In some circumstances, the sensitivity of the phenotypic characterization can be increased by using a surrogate molecule, as exemplified by pefloxacin instead of ciprofloxacin for the detection of fluoroquinolone resistance in Salmonella spp. (21), oxacillin as a sensitive substitute for other beta-lactam agents for the detection of all types of beta-lactam resistance mechanisms in Streptococcus pneumoniae (as recommended by the CLSI and EUCAST for more than 20 years), and the use of benzylpenicillin in Haemophilus influenzae to detect all varieties of beta-lactam resistance mechanisms (22). Using the most sensitive phenotypic method to fine-tune the ECOFF further improves the correlation between phenotypic and genotypic expression when resistance does occur.
ECOFFS DO NOT CHANGE OVER TIME
Once an ECOFF has been established using data from many investigators from many different professional fields, there is rarely a need to change ECOFF values. However, the background material on which ECOFFs are determined varies in size and representativeness from a few hundred MIC values in three or four distributions (for a TECOFF) to >75,000 MIC-values in 5–>25 distributions (for an ECOFF). With an increasing number of distributions and datapoints, there is a need for review and sometimes revision of values. An ECOFF presented in tables on the website against a white background has not been changed over the last 12 months. A yellow background indicates a change over the last month and a light blue background over the last year. The increasing robustness of an ECOFF with added data and a subsequent change may sometimes create problems (23).
THE EUCAST MIC DISTRIBUTION DATABASE
The EUCAST database is a large collection of MIC distributions gathered over many years from a wide range of human, veterinary, and environmental sources, holding more than 30,000 phenotypically characterized MIC and zone diameter distributions (https://mic.eucast.org/). Distributions and ECOFFs are available for many microorganisms, including bacteria, mycobacteria, and fungi. They provide both numeric and graphic views of aggregated MIC distributions and ECOFFs. At present, there are no other similar open databases. Since 2003, it has been freely accessible, which was considered a critical property because it allows users to consult, question, and use the database for a range of laboratory and clinical purposes, described as follows. The software and its upkeep are financed by the European Society for Clinical Microbial and Infectious Disease. As noted previously, the wild-type distributions and ECOFFs have been developed using SOP 10.2 criteria and analytical techniques (5). The entire database was systematically curated during 2021–2023, including reviewing all distributions for acceptability and estimation/re-estimation of all (T)ECOFFs. ECOFFs when presented in tables openly display dates for any change.
The software provides functions for collecting and uploading contributions of MICs and inhibition zone diameters, for curation of individual and aggregated distributions, and for the display of agreed and aggregated distributions and ECOFFs.
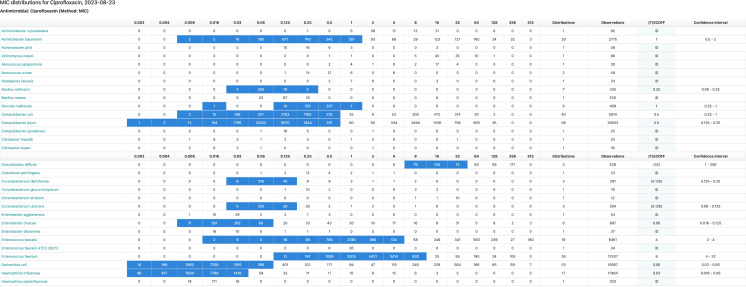
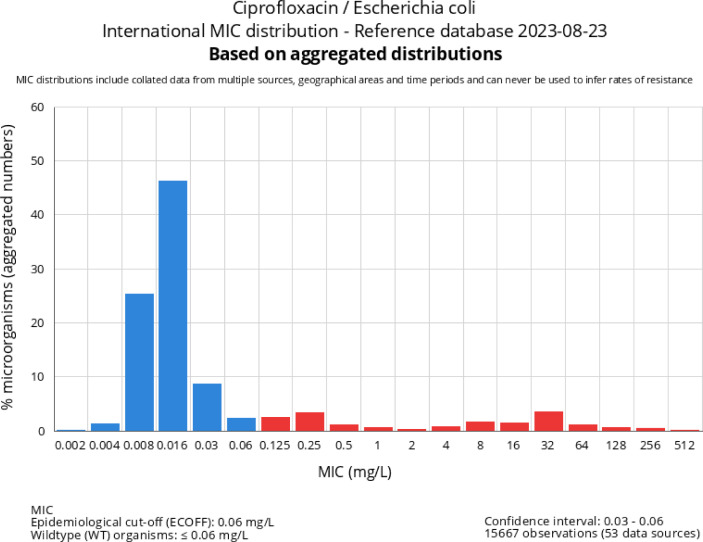
The distributions collected and displayed by the EUCAST are from a wide variety of sources primarily in the human and veterinary environments. These include scientific publications where quality control data could be accessed, breakpoint committee material, contributions from individual researchers, programs for the surveillance of antimicrobial resistance in humans and animals, EUCAST breakpoint development projects [some examples being the organizing of the MIC and zone diameter distributions for Burkholderia pseudomallei (24), Corynebacterium diphtheriae, and C. ulcerans (to be published) and five important Vibrio species (submitted for publication)], and also many pharmaceutical company development programs. Although all distributions are imported to the database, only the MIC distributions that fulfil a set of basic conditions have been accepted for aggregation and display (see introductory notes on the EUCAST MIC distribution website (http://mic.eucast.org) and the criteria published in EUCAST SOP10.2 (5). An aggregated distribution may consist of data from hundred different sources and as many as 90,000 MIC values (e.g., the aggregated distributions of cefuroxime MICs for Escherichia coli and of vancomycin MICs for Staphylococcus aureus). The distributions are presented as tables with raw data (Fig. 1) and as histograms (Fig. 2). A good example of how 72 Escherichia coli cefotaxime distributions are curated is available in Fig. S2 and S3 in a freely accessible reference (3).
Fig 1.
The table of data obtained when searching for “ciprofloxacin” in the EUCAST database (http://mic.eucast.org; the direct link to the specific table above is https://mic.eucast.org/search/?search%5Bmethod%5D=mic&search%5Bantibiotic%5D=60&search%5Bspecies%5D=-1&search%5Bdisk_content%5D=-1&search%5Blimit%5D=50). When entering the name of an agent (rather than that of a species), a list of species is shown, and behind each of these, there is a distribution available. The actual numbers of isolates for each concentration are shown together with the total number of distributions and isolates, the (T)ECOFF, and the confidence intervals. A background color of light blue in the ECOFF column signals a change of values over the past 12 months (a yellow background over the past month). Fly-overs will explain. The software automatically avoids duplicates. (© MIC EUCAST 2023. These data have been produced in part under ECDC service contracts and made available by EUCAST at no cost to the user and can be accessed on the EUCAST website www.eucast.org. The views and opinions expressed are those of EUCAST at a given point in time. EUCAST recommendations are frequently updated and the latest versions are available at www.eucast.org.)
Fig 2.
Graph that is shown on the MIC database website when choosing “Escherichia coli” in Fig. 1. The graph displays the number of distributions included, the number of MIC values included, the mean ECOFF of individual distributions, and the confidence interval for the ECOFF. (© MIC EUCAST 2023. These data have been produced in part under ECDC service contracts and made available by EUCAST at no cost to the user and can be accessed on the EUCAST website www.eucast.org. The views and opinions expressed are those of EUCAST at a given point in time. EUCAST recommendations are frequently updated and the latest versions are available at www.eucast.org.)
There is worldwide interest in the EUCAST MIC distribution database and ECOFFs. It is recognized and referred to by the World Health Organization, the World Organization for Animal Health, the European Centre for Disease Prevention and Control, the European Food Safety Authority, and the Codex Alimentarius Commission as a very important component of international surveillance of antimicrobial resistance and by other breakpoint setting organizations. The practice of defining wild-type distributions and determining ECOFFs has been adopted by colleagues in the antifungal and antimycobacterial fields, and many authors refer to EUCAST MIC distributions in scientific papers and talks (25, 26).
It has been agreed that the rules for including and excluding MIC distributions and the methods used to set ECOFFs need to be defined and agreed at an international level. The closest to an international agreement is the EUCAST SOP10.2, organized via a group of international experts invited by the EUCAST to help agree to a set of criteria (5). The role of genetics in the setting of ECOFFs has been a topic for much discussion, as has the need for a minimum number of distributions before a general and representative ECOFF can be calculated. The conclusion has been that although there is generally an excellent correlation between a wild-type distribution defined on phenotypic traits alone and one primarily based on the presence and absence of identifiable resistance genes (27), a definition requiring the absence of all possibly relevant resistance genes, expressed or not, encoding for resistance to the agent or group of agents will fail to be practical (5, 28). Some colleagues have insisted on using their own definition, where emphasis has been placed on genetic rather than phenotypic traits (28). In our view, the term ECOFF should be reserved for the joint effort of phenotypically defining a wild-type distribution.
OTHER PUBLISHED “ECOFFS”
There are now many publications defining so-called “ECOFFs” for bacteria and fungi. As noted previously, different acceptance criteria for distributions and different analytical techniques may yield different “ECOFFs,” although most often the difference is only one or two twofold dilutions. The EUCAST promotes their method as the “reference” method for establishing true ECOFFs (5) and stresses the point that attempting to establish a true ECOFF from a single distribution performed in a single laboratory is, by definition, not possible since it will not encompass inter-laboratory variation or the variation provided by differences in MIC (or zone diameters) attributable to differences in the material obtained from different sources/manufacturers and in laboratory skills and techniques. These differences are all embraced by the EUCAST-aggregated distributions from multiple sources. A locally generated cut-off value for a single distribution can be useful but only to the laboratory that generated it and only as long as there is no change in the materials used. The EUCAST is open to discussion with researchers interested in establishing ECOFFs for combinations of agents and species where it has yet not had access to enough quality MIC distributions. For such situations, we have helped establish ECOFFs for several agents (23, 29, 30).
Are differences between wild-type distributions and ECOFFs of one or twofold dilutions important? Yes, because in the EUCAST procedure for setting clinical breakpoints, the agreed ECOFF will influence the discussion of several possible issues related to clinical breakpoints, susceptibility testing, pharmacokinetics and pharmacodynamics, surveillance of antimicrobial resistance, and the comparison of resistance between humans and animals and between animal species (22), as outlined below.
THE APPLICATIONS OF WILD-TYPE MIC DISTRIBUTIONS AND ECOFFS
When determining clinical breakpoints for an agent and a species, it is of major importance to be aware of the wild-type distribution and the ECOFF. Once the wild-type distribution is known and agreed upon, it serves as a reference for deliberations on clinical breakpoints, the detection and surveillance of resistance, and comparison of resistance between humans and animals and between animal species(23).
As a reference. A wild-type distribution agreed by many investigators will serve as a reference of agent activity against a defined species. The robustness of the wild-type and ECOFF characterization will depend on the number of investigators involved and in agreement. For instance, by reviewing the database, it becomes evident that one will not lightly publish an alternative vancomycin MIC distribution or ECOFF for Staphylococcus aureus when the ECOFF of 2 mg/L is based on 91,635 MIC values from 37 different investigators.
-
In the determination of clinical breakpoints: At present, clinical breakpoints (interpretive criteria) are set by the EUCAST, CLSI, and the United States Food and Drug Administration. Of these, only the EUCAST requires that clinical breakpoints should not be set to split wild-type distributions. The two main reasons are (i) EUCAST has not been able to identify a situation where clinical outcome can be related to different MIC values inside a wild-type distribution of a species; (ii) favoring breakpoints outside a wild-type distribution avoids the region where unavoidable assay variation will have the most detrimental effect in the form of poor reliability of a single result. The susceptibility reporting categories of S, I, and R are dichotomous by nature, and if clinical breakpoints are allowed to divide homogenous populations of important target species, the “flip-flop” between S, I, and R will be frequent and random. Thus, for the EUCAST, the process for determining clinical breakpoints requires the identification of the wild-type distributions of all important target species.
For other recognized organizations, wild-type populations are required for consideration, but they have no codified requirements to prevent clinical breakpoints from splitting the wild-type population.
At times, when attempting to set clinical breakpoints, the ECOFF of a target pathogen may be higher than a cutoff determined using pharmacokinetic/pharmacodynamic (PK/PD) analyses, i.e., the PK/PD cutoff splits the wild type. Because ECOFFs are a measure of the phenotype only, ECOFFs are not influenced by this difference; instead, the interpretation is that the dosage regimen of the antimicrobial in question may be suboptimal.
-
To exclude resistance: The ECOFF by definition and nature provides the most sensitive phenotypic measurement with which to exclude resistance. This is of interest for resistance screening purposes and as an initial step in the process of defining certain types of resistance. Indeed, one great value of maintaining skills in phenotypic susceptibility testing is that this is the only way that new resistance mechanisms will be detected. Genotypic methods of resistance detection work only on known resistance mechanisms and their genes.
The following are examples of how ECOFFs can be applied to resistance detection and the development of alternative screening strategies. The most sensitive phenotypic measure for excluding fluoroquinolone resistance in Enterobacterales, primarily Salmonella spp., is using pefloxacin and its ECOFF(21). Likewise, the ECOFFs of oxacillin in Streptococcus pneumoniae and cefoxitin for Staphylococcus aureus (MIC, and even more so, the ECOFF of the zone diameter) have for many years provided a means for the sensitive detection of beta-lactam resistance. By choosing the agent with care, the screen can exclude resistance to a host of related agents (e.g., pefloxacin to exclude all fluoroquinolone resistance mechanisms). The EUCAST guidance document will provide many more examples(31).
-
For surveillance of resistance development: Clinical breakpoints are in many ways unsuitable for determination and surveillance of resistance rates, especially for agents where toxicology profiles, pharmacokinetic–pharmacodynamic (PK-PD), and clinical evidence of efficacy have allowed clinical breakpoints to be set higher than the ECOFF. This is most often true for many beta-lactams because the safety of higher dosages has allowed breakpoints that are sometimes several dilutions above those of the wild type. For a more sensitive and robust measure of resistance and resistance development, ECOFFs are preferable since
clinical breakpoints change over time, making comparing rates calculated before and after such changes difficult.
clinical breakpoints differ between systems. Breakpoints determined by the EUCAST, CLSI, and FDA “systems” (e.g., procedures for determining breakpoints) often differ and when they change over time, the change is not synchronized. The general trend, with only a few exceptions, is for initial breakpoints to be decreased over time. This is to align with evolving knowledge on resistance mechanisms and levels and their importance for clinical outcome, the broadening knowledge of pharmacokinetics/pharmacodynamics, and sometimes with the change or widening of clinical indications.
surveillance programs, official and privately run, have traditionally merged the “I” and “R” categories as “non-susceptible” and reported frequencies for “non-susceptible” isolates. This has created uncertainty since the definition of “intermediate,” both in the CLSI and EUCAST, was ambiguous and merging “I” and “R” was incorrect. With the fundamental change in the EUCAST definitions of “S”, “I”, and “R” in 2019, merging “I” and “R” became unacceptable for surveillance in Europe, and lumping categories together was abandoned in European surveillance programs, as exemplified by the criteria for participation in EARS-Net, the surveillance program for antimicrobial resistance in blood stream infections from the European Centre for Disease Prevention and Control (ECDC) (32).
For local, national, and international comparisons: Using ECOFFs, and thus wild type versus non-wild type, resistance can be compared irrespective of differences in clinical breakpoints between breakpoint systems and irrespective of the origin of the isolates (human, animal, environment, time period, geography, etc.). The only requirement is that the ECOFFs used for comparison are based on the same published testing methodology.
In lieu of breakpoints: Wild-type isolates of a species cannot be automatically considered susceptible to the agent. Many wild-type populations are either not clinically categorized as “S”, “I,” or “R” or are categorized as “R.” “Susceptible” in the clinical sense includes consideration of drug administration, including pharmacokinetics, pharmacodynamics, and dosing regimens, which together result in drug exposure at the site of infection and its adequacy for effective treatment. There are many examples of where the achievable exposure to an antimicrobial is inadequate for the treatment of wild-type isolates, e.g., benzylpenicillin and Escherichia coli and cephalosporins and aminoglycosides to Enterococcus spp. In other circumstances, the wild-type distribution and ECOFF for a pathogen without clinical breakpoints may be similar to that of related pathogens with clinical breakpoints. If so, the MIC and ECOFF can be cautiously interpreted to predict the clinical outcome. However, formally approved wild-type MIC distributions may be lacking for rarely isolated organisms, and then the EUCAST-published guidance on “What to do when there are no clinical breakpoints” may be of value(33).
In therapeutic drug monitoring: ECOFFs can play a role in therapeutic drug monitoring and dosage adjustment. It has become common practice, mainly in seriously ill patients, to estimate drug exposure in an individual patient and compare that to a single measure of the MIC of the infecting pathogen, which will be used to estimate whether the patient is receiving sufficient exposure (dosage) to ensure that PK-PD targets are reached. The EUCAST has pointed out that a single MIC measurement cannot be relied upon for such purposes due to the intrinsic variation in assays, between assays, and between assay systems. In situations where resistance has been ruled out because the isolate has been shown to belong to the wild type for the agent, the EUCAST promotes the use of the species ECOFF, aiming for a PK-PD target based on an MIC two dilutions higher than the ECOFF. This approach guarantees that assay variation has been accounted for and ensures the highest margin for efficacy should dosage adjustment be required (34, 28.
ECOFFs should not be used for any purpose (i) if they have not been determined by the methods described previously, or (ii) if adequate PK/PD and clinical outcome data are available, which suggest clinical breakpoints that differ from the ECOFF.
In conclusion, the characterization of wild-type MIC and zone diameter distributions with the setting of ECOFFs will provide a reference in a system where the MIC value is otherwise relative. ECOFFs and wild-type distributions are an invaluable resource and have many uses in the fields of determining clinical breakpoints, development of susceptibility testing methods, surveillance of antimicrobial resistance development, comparison of resistance rates between different breakpoint systems, and in the “One Health” concept.
Biographies

Gunnar Kahlmeter is a clinical microbiologist, senior professor, and lecturer of the medical faculties of Lund and Uppsala Universities. He was the head of clinical microbiology in Kronoberg county 1985 – 2016, Kalmar county 1989 – 1996, and Blekinge county 2010 – 2016. He was asked by ESCMID to rejuvenate EUCAST in 2001 and he chaired EUCAST between 2001 – 2012. He was the president of ESCMID 2012 – 14 and has been the advisor to many national and international organizations involved in antimicrobial resistance surveillance, antimicrobial susceptibility testing and external quality control. He recently chaired the WHO AMR STAG committee. He is currently the EUCAST Technical Data Coordinator and webmaster and the head of the EUCAST Development Laboratory, and the Swedish Reference Laboratory for phenotypic susceptibility testing of bacteria.

John Turnidge is an infectious diseases physician and microbiologist, currently holding professorships in the School of Biological Sciences at the University of Adelaide, South Australia. He holds the Document and Technical Support role in the European Committee on Antimicrobial Susceptibility Testing and is a Senior Medical Advisor to the Australian Commission on Safety and Quality in Health Care. He obtained his medical degree from the University of Sydney and subsequently received fellowship degrees from the Royal Australasian College of Physicians and the Royal College of Pathologists of Australasia. He held senior positions in Microbiology and Infectious Diseases in Melbourne and Adelaide, Australia. His current interests include the science of phenotypic antimicrobial susceptibility testing, surveillance of antimicrobial resistance and One Health policy and management of antimicrobial resistance.
Contributor Information
Gunnar Kahlmeter, Email: Gunnar.kahlmeter@kronoberg.se.
Graeme N. Forrest, Rush University, Chicago, Illinois, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/cmr.00100-22.
Wild type MIC distributions and ECOFFs - the how.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Ringertz S, Olsson-Liljequist B, Kahlmeter G, Kronvall G. 1997. Antimicrobial susceptibility testing in Sweden. II. species-related zone diameter breakpoints to avoid interpretive errors and guard against unrecognized evolution of resistance. Scand J Infect Dis Suppl 105:8–12. [PubMed] [Google Scholar]
- 2. Kahlmeter G, Brown DFJ, Goldstein FW, MacGowan AP, Mouton JW, Osterlund A, Rodloff A, Steinbakk M, Urbaskova P, Vatopoulos A. 2003. European harmonization of MIC breakpoints for antimicrobial susceptibility testing of bacteria. J Antimicrob Chemother 52:145–148. doi: 10.1093/jac/dkg312 [DOI] [PubMed] [Google Scholar]
- 3. Kahlmeter G. 2015. The 2014 garrod lecture: EUCAST - are we heading towards international agreement? J Antimicrob Chemother 70:2427–2439. doi: 10.1093/jac/dkv145 [DOI] [PubMed] [Google Scholar]
- 4. International Organization for Standardization . 2019. Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility devices —part 1: reference methods for testing the in vitro activity of antimicrobial agents against bacteria involved in infectious diseases (ISO 20776-1:2019). Geneva: ISO [Google Scholar]
- 5. European Committee on Antimicrobial Susceptibility Testing – Standard Operating Procedure 10.2 (SOP 10.2) . 2021. MIC distributions and the setting of epidemiological cut-off (ECOFF) values. Available from: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/EUCAST_SOPs/2021/EUCAST_SOP_10.2_MIC_distributions_and_epidemiological_cut-off_value__ECOFF__setting_20211202.pdf
- 6. Clinical and Laboratory Standards Institute . 2023. Development of in vitro susceptibility testing criteria and quality control parameters. In CLSI guideline M23, 6th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7. Mouton JW, Meletiadis J, Voss A, Turnidge J. 2018. Variation of MIC measurements: the contribution of strain and laboratory variability to measurement precision. J Antimicrob Chemother 73:2374–2379. doi: 10.1093/jac/dky232 [DOI] [PubMed] [Google Scholar]
- 8. Hombach M, Ochoa C, Maurer FP, Pfiffner T, Böttger EC, Furrer R. 2016. Relative contribution of biological variation and technical variables to zone diameter variations of disc diffusion susceptibility testing. J Antimicrob Chemother 71:141–151. doi: 10.1093/jac/dkv309 [DOI] [PubMed] [Google Scholar]
- 9. Annis DH, Craig BA. 2005. The effect of interlaboratory variability on antimicrobial susceptibility determination. Diagn Microbiol Infect Dis 53:61–64. doi: 10.1016/j.diagmicrobio.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standards Institute . 2016. Principles and procedures for the development of epidemiological cutoff values for antifungal susceptibility testing. In CLSI guideline M57, 1st. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11. International Organization for Standardization . 2021. Clinical laboratory testing and in vitro diagnostic test systems — broth micro-dilution reference method for testing the in vitro activity of antimicrobial agents against yeast fungi involved in infectious diseases (ISO 16256-1:2021). Geneva: ISO [Google Scholar]
- 12. Schön T, Werngren J, Machado D, Borroni E, Wijkander M, Lina G, Mouton J, Matuschek E, Kahlmeter G, Giske C, Santin M, Cirillo DM, Viveiros M, Cambau E. 2020. Antimicrobial susceptibility testing of Mycobacterium tuberculosis complex isolates – the EUCAST broth Microdilution reference method for MIC determination. Clin Microbiol Infect 26:1488–1492. doi: 10.1016/j.cmi.2020.07.036 [DOI] [PubMed] [Google Scholar]
- 13. Pfaller MA, Diekema DJ, Ghannoum MA, Rex JH, Alexander BD, Andes D, Brown SD, Chaturvedi V, Espinel-Ingroff A, Fowler CL, Johnson EM, Knapp CC, Motyl MR, Ostrosky-Zeichner L, Sheehan DJ, Walsh TJ, Clinical and Laboratory Standards Institute Antifungal Testing Subcommittee . 2009. Wild-type MIC distribution and epidemiological cutoff values for Aspergillus fumigatus and three triazoles as determined by the clinical and laboratory standards Institute broth microdilution methods. J Clin Microbiol 47:3142–3146. doi: 10.1128/JCM.00940-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kronvall G. 2010. Normalized resistance interpretation as a tool for establishing epidemiological MIC susceptibility breakpoints. J Clin Microbiol 48:4445–4452. doi: 10.1128/JCM.01101-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turnidge J, Kahlmeter G, Kronvall G. 2006. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin Microbiol Infect 12:418–425. doi: 10.1111/j.1469-0691.2006.01377.x [DOI] [PubMed] [Google Scholar]
- 16. Meletiadis J, Mavridou E, Melchers WJG, Mouton JW, Verweij PE. 2012. Epidemiological cutoff values for azoles and Aspergillus fumigatus based on a novel mathematical approach incorporating cyp51A sequence analysis. Antimicrob Agents Chemother 56:2524–2529. doi: 10.1128/AAC.05959-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jaspers S, Aerts M, Verbeke G, Beloeil PA. 2014. Estimation of the wild-type minimum inhibitory concentration value distribution. Stat Med 33:289–303. doi: 10.1002/sim.5939 [DOI] [PubMed] [Google Scholar]
- 18. Meletiadis J, Curfs-Breuker I, Meis JF, Mouton JW. 2017. In vitro antifungal susceptibility testing of Candida isolates with the EUCAST methodology, a new method for ECOFF determination. Antimicrob Agents Chemother 61:e02372-16. doi: 10.1128/AAC.02372-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. CRyPTIC Consortium . 2022. Epidemiological cut-off values for a 96-well broth microdilution plate for high-throughput research antibiotic susceptibility testing of M. tuberculosis. Eur Respir J 60:2200239. doi: 10.1183/13993003.00239-2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, Grundman H, Hasman H, Holden MTG, Hopkins KL, Iredell J, Kahlmeter G, Köser CU, MacGowan A, Mevius D, Mulvey M, Naas T, Peto T, Rolain JM, Samuelsen Ø, Woodford N. 2017. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST subcommittee. Clin Microbiol Infect 23:2–22. doi: 10.1016/j.cmi.2016.11.012 [DOI] [PubMed] [Google Scholar]
- 21. Skov R, Matuschek E, Sjölund-Karlsson M, Åhman J, Petersen A, Stegger M, Torpdahl M, Kahlmeter G. 2015. Development of a pefloxacin disk diffusion method for detection of fluoroquinolone-resistant Salmonella enterica. J Clin Microbiol 53:3411–3417. doi: 10.1128/JCM.01287-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skaare D, Lia A, Hannisdal A, Tveten Y, Matuschek E, Kahlmeter G, Kristiansen B-E. 2015. Haemophilus influenzae with non-beta-lactamase-mediated beta-lactam resistance: easy to find but hard to categorize. J Clin Microbiol 53:3589–3595. doi: 10.1128/JCM.01630-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Jong A, Thomas V, Klein U, Marion H, Moyaert H, Simjee S, Vallé M. 2013. Pan-European resistance monitoring programmes encompassing food-borne bacteria and target pathogens of food-producing and companion animals. Int J Antimicrob Agents 41:403–409. doi: 10.1016/j.ijantimicag.2012.11.004 [DOI] [PubMed] [Google Scholar]
- 24. Karatuna O, Dance DAB, Matuschek E, Åhman J, Turner P, Hopkins J, Amornchai P, Wuthiekanun V, Cusack T-P, Baird R, Hennessy J, Norton R, Armstrong M, Zange S, Zoeller L, Wahab T, Jacob D, Grunow R, Kahlmeter G. 2021. Burkholderia pseudomallei multi-centre study to establish EUCAST MIC and zone diameter distributions and epidemiological cut-off values. Clin Microbiol Infect 27:736–741. doi: 10.1016/j.cmi.2020.07.001 [DOI] [PubMed] [Google Scholar]
- 25. Arendrup MC, Kahlmeter G, Rodriguez-Tudela JL, Donnelly JP. 2009. Breakpoints for susceptibility testing should not divide wild-type distributions of important target species. Antimicrob Agents Chemother 53:1628–1629. doi: 10.1128/AAC.01624-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Angeby KA, Jureen P, Giske CG, Chryssanthou E, Sturegård E, Nordvall M, Johansson AG, Werngren J, Kahlmeter G, Hoffner SE, Schön T. 2010. Wild-type MIC distributions of four fluoroquinolones active against Mycobacterium tuberculosis in relation to current critical concentrations and available pharmacokinetic and pharmacodynamic data. J Antimicrob Chemother 65:946–952. doi: 10.1093/jac/dkq091 [DOI] [PubMed] [Google Scholar]
- 27. Avershina E, Khezri A, Ahmad R. 2023. Clinical diagnostics of bacterial infections and their resistance to antibiotics-current state and whole genome sequencing implementation perspectives. Antibiotics (Basel) 12:781. doi: 10.3390/antibiotics12040781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kahlmeter G, Turnidge J. 2022. How to: ECOFFs-the why, the how, and the Don’ts of EUCAST epidemiological cutoff values. Clin Microbiol Infect 28:952–954. doi: 10.1016/j.cmi.2022.02.024 [DOI] [PubMed] [Google Scholar]
- 29. Yang Q, Li X, Jia P, Giske C, Kahlmeter G, Turnidge J, Yu Y, Lv Y, Wang M, Sun Z, Lin J, Li Y, Zheng B, Hu F, Guo Y, Chen Z, Li H, Zhang G, Zhang J, Kang W, Duan S, Wang T, Jing R, Xu Y, Chinese Committee on Antimicrobial Susceptibility Testing (ChiCAST) . 2021. Determination of norvancomycin epidemiological cut-off values (ECOFFs) for Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus and Staphylococcus hominis. J Antimicrob Chemother 76:152–159. doi: 10.1093/jac/dkaa414 [DOI] [PubMed] [Google Scholar]
- 30. Li X, Jia P, Zhu Y, Xu Y, Yu Y, Lv Y, Wang M, Sun Z, Lin J, Li Y, Zheng B, Hu F, Guo Y, Chen Z, Li H, Zhang G, Zhang J, Kang W, Duan S, Wang T, Jing R, Yang Q. 2021. Establishment of epidemiological cut-off values for cefoselis, a new fourth-generation cephalosporin, against Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae, Proteus mirabilis and Pseudomonas aeruginosa. J Antimicrob Chemother 76:2593–2599. doi: 10.1093/jac/dkab216 [DOI] [PubMed] [Google Scholar]
- 31. European Committee on Antimicrobial Susceptibility Testing . 2022. EUCAST Phenotypic screening tests to detect / exclude resistance of clinical relevance. Available from: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Guidance_documents/Screening_to_detect_and_exclude_resistance_2022_08_22.pdf
- 32. European Centres for Disease Control and Prevention . 2020. EARS-net-reporting-protocol. Available from: https://www.ecdc.europa.eu/sites/default/files/documents/EARS-Net-reporting-protocol-2020.pdf
- 33. European Committee on Antimicrobial Susceptibility Testing . 2023. EUCAST guidance on ”what to do when there are no breakpoints” Available from: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Guidance_documents/When_there_are_no_breakpoints_20230630_Final.pdf
- 34. Mouton JW, Muller AE, Canton R, Giske CG, Kahlmeter G, Turnidge J. 2018. MIC-based dose adjustment: facts and fables. J Antimicrob Chemother 73:564–568. doi: 10.1093/jac/dkx427 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wild type MIC distributions and ECOFFs - the how.