SUMMARY
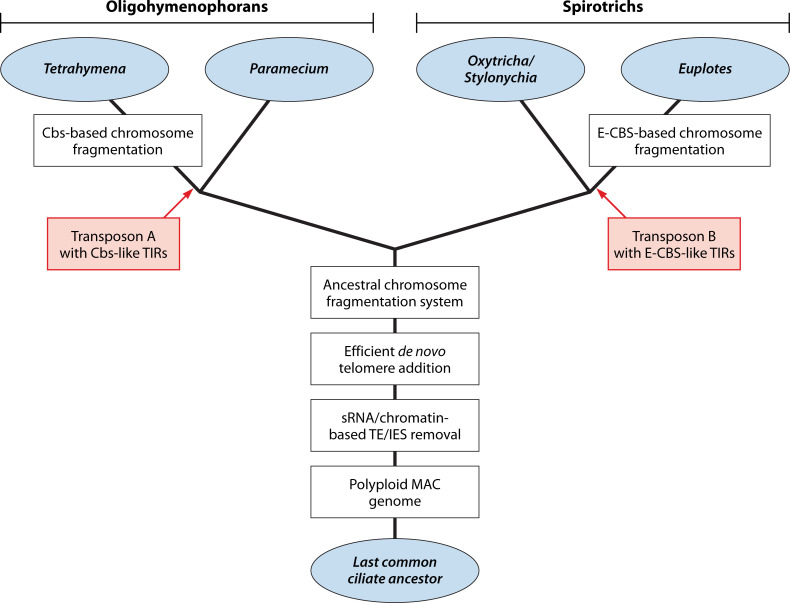
Ciliated protozoa undergo large-scale developmental rearrangement of their somatic genomes when forming a new transcriptionally active macronucleus during conjugation. This process includes the fragmentation of chromosomes derived from the germline, coupled with the efficient healing of the broken ends by de novo telomere addition. Here, we review what is known of developmental chromosome fragmentation in ciliates that have been well-studied at the molecular level (Tetrahymena, Paramecium, Euplotes, Stylonychia, and Oxytricha). These organisms differ substantially in the fidelity and precision of their fragmentation systems, as well as in the presence or absence of well-defined sequence elements that direct excision, suggesting that chromosome fragmentation systems have evolved multiple times and/or have been significantly altered during ciliate evolution. We propose a two-stage model for the evolution of the current ciliate systems, with both stages involving repetitive or transposable elements in the genome. The ancestral form of chromosome fragmentation is proposed to have been derived from the ciliate small RNA/chromatin modification process that removes transposons and other repetitive elements from the macronuclear genome during development. The evolution of this ancestral system is suggested to have potentiated its replacement in some ciliate lineages by subsequent fragmentation systems derived from mobile genetic elements.
KEYWORDS: chromosome fragmentation, transposons, ciliated protozoa, IESs, telomeres, Tetrahymena, Paramecium, spirotrich
INTRODUCTION: CHROMOSOME BREAKAGE IN EUKARYOTES
Doublestranded breaks in DNA occur frequently and can be the result of exogenous factors such as ionizing radiation or reactive oxygen species, or endogenous processes such as DNA replication or the excision of transposable elements (TEs). Such breaks in the DNA are potentially dangerous and/or life threatening, as they can lead to lethal mutations or rearrangements that disrupt the structure of chromosomes. As a result, most organisms have elaborated at least two systems, non-homologous end joining (NHEJ) and homologous recombination repair [reviewed in reference (1)], to mend broken chromosomes and minimize the negative effects of breakage. In rare instances, a double-stranded break in the DNA also may be healed by the de novo addition of telomere repeat sequences [e.g., reference (2)]. As is the case for the natural telomeres of chromosomes, this de novo telomere formation allows for the complete replication and structural integrity of the novel chromosome end.
In contrast to most eukaryotes, a small number of organisms have been identified where chromosome breakage followed by de novo telomere addition is part of the normal life cycle. These include the chromatin diminution process of some parasitic nematodes in the Ascaris genus [reviewed in reference (3)] as well as the unicellular ciliated protozoa (phylum Ciliophora), the latter of which is the focus of the current review. In these instances, chromosomes are broken at specific sites within the genome during a defined time in development, and the resulting ends are stabilized by the efficient de novo addition of telomeres. Such wholesale changes in chromosome structure can be tolerated because they occur only in somatic cells or in nuclei that are analogs of a somatic cell. That is, programmed chromosome breakage coupled with de novo telomere addition does not occur in the germline genome so that unaltered, intact chromosomes still can be transmitted to subsequent generations.
It is of interest to understand how these unusual systems have evolved, as well as the molecular mechanisms that mediate these processes, as this may provide novel means of enhancing chromosome repair in vivo and gene/chromosome engineering in vitro. In this article, we review key findings on developmentally programmed chromosome fragmentation and de novo telomere formation in ciliated protozoa. The ciliates are a diverse group of unicellular eukaryotes that diverged from other eukaryotes over a billion years ago (4). Developmental chromosome fragmentation is common within the group [e.g., references (5–8)], but a great deal of diversity exists, including extreme cases where essentially every gene comes to reside on an individual chromosome following fragmentation. We will focus on ciliates where some molecular details of the fragmentation process have been deduced: the oligotrich ciliates (class Oligohymenophorea) Tetrahymena and Paramecium, which undergo moderate levels of chromosome fragmentation, and members of the spirotrich group (class Spirotrichea; Euplotes, Oxytricha, and Stylonychia), which extensively fragment their genomes. In addition to highlighting shared and unique features of chromosome fragmentation among these ciliates, we will note key unanswered questions and propose model(s) for how fragmentation may have originated and evolved with the ciliate group.
OVERVIEW OF CILIATE NUCLEAR ORGANIZATION AND REORGANIZATION
Nuclear dimorphism
The most distinctive feature of the phylum Ciliophora, and the one that allows for extensive and irreversible modifications to chromosomes such as chromosome fragmentation, is nuclear dimorphism (Fig. 1). Specifically, each ciliate cell contains two types of nuclei that serve distinct functions. The smaller nucleus, the micronucleus (MIC), is diploid, contains large chromosomes with many genes, and is in most respects a typical eukaryotic cell nucleus. That is, the large chromosomes are organized as chromatin, the genome is replicated during the cell cycle, and chromosomes are distributed to daughter cells by mitosis. The MIC is transcriptionally inert during asexual multiplication and plays a major role during sexual reproduction, essentially serving as a “germline nucleus.”
Fig 1.
Nuclear dimorphism in ciliated protozoa. Idealized ciliate cells are shown at the top containing a MIC and a macronucleus (MAC). Both nuclei replicate and divide during asexual or vegetative reproduction. In response to starvation, cells of different mating types pair, the MIC undergoes meiosis, and exchanged haploid meiotic products fuse to form a diploid zygotic nucleus. The zygotic nucleus divides, with one product becoming the MIC, while the other product undergoes developmental genome rearrangement to form a new MAC. The old MAC (hatched circle) is degraded.
In contrast, the large macronucleus (MAC) contains subchromosomal-sized DNA molecules and is invariably polyploid. In extreme cases, the MAC chromosomes of some ciliate species are present in tens of thousands of copies, each of which contains almost exclusively a single gene. MACs typically replicate their genomes during each cell cycle but divide by a process termed amitosis because there is no spindle apparatus to ensure that the two copies of a given chromosome are each distributed to a different daughter cell. These unusual features of the MAC can be tolerated while still maintaining genetic stability because the MAC is transitory. That is, the MAC is differentiated anew during sexual reproduction from a copy of the MIC and then destroyed during the subsequent round of sexual reproduction (Fig. 1).
Mating and macronuclear development
Mating and MAC development are complex processes that occur over periods of at least ~100 hours in some species. Figure 1 provides a generic view of sexual reproduction, summarizing the key common steps shared among ciliates, although details can differ substantially among the various ciliate groups. In response to starvation, cells become mating competent and partially fuse to form mating pairs. The MICs in the paired cells then undergo meiosis, and a haploid meiotic nucleus (migratory pronucleus) is reciprocally exchanged between the two members of the pair. The exchanged haploid nuclei next fuse with one of the resident haploid nuclei (the stationary pronucleus) to generate a diploid zygotic nucleus. This zygotic nucleus replicates its genome and divides generally twice to form the precursor(s) of the MICs and MACs for subsequent asexual reproduction.
The mitotic products of the zygotic nucleus that are destined to form the new MAC then begin a complex series of changes. MAC development typically begins with a number of cycles of DNA replication in the absence of nuclear division (endoreplication). The amount of endoreplication varies between species, but in some groups (e.g., the spirotrichs), visible polytene chromosomes are produced [e.g., reference (9)]. During this initial stage of MAC DNA replication, two forms of DNA rearrangement occur (Fig. 2). First, internal segments of the chromosomes are excised and eliminated, while the flanking sequences are rejoined. Second, the chromosomes are fragmented at specific sites, and de novo telomerization occurs at the resulting ends. Following the reorganization of the genome, development concludes with additional cycles of DNA replication that result in the mature, polyploid MAC genome. In conjunction with the development of a new MAC, the old MAC is destroyed or lost.
Fig 2.
Two forms of DNA rearrangement occur in the developing MAC. A segment of MIC DNA is shown at the top that includes two segments of DNA that will form MAC chromosomes (green rectangles; C1 and C2). MAC development begins with multiple rounds of DNA replication without karyokinesis or cell division. Two types of DNA rearrangement then occur: (i) internal eliminated sequences (IESs; red rectangles) and other repetitive elements are excised from internal regions of the chromosomes, and (ii) the chromosomes are fragmented and telomeric repeats (“T”) are synthesized de novo onto the DNA ends. Following rearrangement, additional cycles of DNA replication give rise to the mature polyploid MAC.
Internal DNA elimination
Much research in recent decades has focused on the internal DNA elimination processes in developing MACs of a select number of ciliate species (Tetrahymena, Paramecium, Euplotes, and Oxytricha/Stylonychia). It has provided key insights into the nature of the eliminated sequences, the proteins involved in their excision, and the possible evolutionary origin of the process. This area has been the subject of a number of recent reviews (10–14), and we will only briefly summarize some of the key findings.
Developmentally programmed elimination of internal sequences during differentiation of the Tetrahymena MAC, followed by rejoining of the flanking DNA, was first documented in 1984 (15, 16). The developmentally excised DNA sequences were generically termed “internal eliminated sequences” (IESs) by analogy to developmentally eliminated sequences reported in the same year in Oxytricha nova (17). Subsequently, comparison of MIC and MAC genome assemblies revealed that ~10,000 IESs, accounting for ~34% of the MIC genome, are eliminated during MAC development in Tetrahymena (18). Two types of IESs have been characterized in the Tetrahymena thermophila MIC genome. Type I IESs constitute the overwhelming majority of them. They are relatively long (85% are in the 1–10 kb range), and nearly half of them overlap germline TEs, mostly DNA transposons. During new MAC differentiation, IES DNA is excised by a domesticated PiggyBac transposase, Tpb2p (19, 20), and the two flanking chromosome ends are joined through NHEJ (21). Type I IESs are located in non-coding regions, and they are excised imprecisely. Tetrahymena Type I IESs are generally flanked in the MIC by pairs of inverted DNA repeats, called “flanking regulatory sequences” (FRS) (22), which are typically located within ~70 bp of each IES end. The existence of at least six different major types of FRSs has been proposed.
In contrast, only 12 Type II IESs exist in the T. thermophila MIC genome. They all include identical 12-bp inverted repeats at each end, which are also excised during MAC differentiation. Type II IESs all occur within exons in protein-coding genes, from which they are precisely excised by the Tpb1p- and Tpb6p-domesticated PiggyBac transposases (18, 23, 24). The Tetrahymena Type II IESs are thought to have arisen from a recent invasion of a member of the PiggyBac transposon family.
The existence of Paramecium IESs was first reported in 1992 in Paramecium tetraurelia (25) and, subsequently, in other members of the P. aurelia group and in P. caudatum (26). In Paramecium, the term “IES” has been restricted to MIC-specific internal sequences (~45,000 in P. tetraurelia) that are precisely eliminated between two conserved 5′-TA-3′ repeats (27) and represent only ~10% of eliminated DNA (26, 28, 29). Paramecium IESs are mostly short (93% are <150 bp) and scattered along the genome, including inside genes. The excision machinery, also composed of domesticated PiggyBac transposases [PiggyMac (Pgm) and its Pgm-like partners], cleaves IES boundaries around the conserved terminal TA repeats (TAs) (30, 31). A hallmark of Paramecium IES excision is the tight coupling between DNA cleavage and subsequent NHEJ-mediated precise double-stranded break repair (32, 33). Indeed, activation of Pgm endonuclease activity requires the presence of NHEJ factors, which would ensure that flanking DNA ends are joined right after cleavage. A weak consensus resembling the termini of Tc1/mariner TEs defines a terminal inverted repeat (TIR) at IES ends (34): this was the first indication that Paramecium IESs are related to TEs (35). Further evidence stemmed from the identification of Tc1/mariner elements among the largest IESs (28) and the finding that several multicopy IESs have recently transposed within Paramecium genomes (26). Besides IESs, ~30% of MIC DNA, including different TE families (both DNA and RNA transposons) and other repeats (28, 29), is eliminated imprecisely, leading to heterogeneous chromosomal deletions (36). Of note, these imprecisely eliminated internal Paramecium MIC DNA sequences are not referred to as IESs because rejoining of flanking DNA does not uniformly occur in conjunction with their excision (see section “Chromosome fragmentation in Paramecium” below). Imprecise DNA elimination is also Pgm dependent (30), except for 3 Mbp that are currently under investigation (29). Whether Pgm plays a direct role, by cutting DNA within or around eliminated regions, or is involved indirectly (e.g., IES excision may be required to activate imprecise elimination) remains to be established.
Similar to Paramecium, Euplotes IESs (estimate number ~100,000) are mostly excised precisely between two 5′-TA-3′ direct repeats, also called “pointers,” a single copy of which is retained at the excision junction (37). Two types of IESs have been distinguished: “short IESs” (<~600 bp) and longer Tc1/mariner-related “transposon IESs” called Tec elements (38–40). A consensus sequence found at the ends of short IESs is similar to the TIRs of Tec elements, which again suggests that the two classes of IESs are evolutionary linked (37). Furthermore, molecular analysis of excision products showed that at least some Tec copies are excised precisely, most likely through the same mechanism as short IESs (41–43). Another elimination pathway, perhaps associated with DNA degradation, was proposed for Tec elements from non-MAC-destined regions (44). The enzymatic machinery involved in DNA elimination in Euplotes, however, has not been identified.
More than 100,000 IESs were also reported in Oxytricha, in which they separate fragments of MAC-destined sequences (MDSs). One particular feature of this ciliate is that ~20% of its genes are scrambled in the MIC and their MDSs must be re-ordered during macronuclear development to form functional MAC chromosomes (45). IESs from non-scrambled genes (median size 180 bp) are excised precisely between directly repeated pointers that vary in length and sequence from one IES to the next, with 5′-TA-3′ and 5′-ANT-3′ representing the majority of 2 and 3 bp pointers, respectively (17, 45). The situation is more complex for scrambled genes, in which the pointers flanking each IES are not direct repeats (46): the rearrangement of scrambled MDSs involves distant identical pointers, which are generally longer and more GC-rich (45). Multicopy TEs (both DNA and RNA transposons) are also found in the MIC genome. The largest class consists of three families of Tc1/mariner-related elements (TBE1, TBE2, and TBE3) that frequently interrupt genes or their immediately surrounding sequences. Similar to IESs, TBEs are flanked by direct repeats (5′-ANT-3′ for TBE1) that they use as excision pointers (47). A mechanistic link between Oxytricha IESs and TBEs was further supported by the demonstration that genome rearrangement requires the expression of transposases from all three TBE families (48).
Epigenetic control of DNA elimination by non-coding RNAs and heterochromatin
Studies performed in a few ciliate species have uncovered the role of different types of non-coding RNAs in the control of developmentally programmed internal DNA elimination [reviewed in reference (49)]. Two pathways, one specifying the sequences to be eliminated from the new MAC and the other specifying those to be maintained in the new MAC, have been reported in different ciliate groups.
First, the “genome scanning” model, initially proposed for Tetrahymena IESs (50), involves the participation of non-coding RNAs called scanRNAs (scnRNAs) in specifying the sequences that will be eliminated from the new developing MAC (Fig. 3). These short scnRNAs (28 nt in Tetrahymena, 25 nt in Paramecium) are specifically produced during meiosis through the cleavage of MIC transcripts by specialized Dicer-like proteins (51, 52). Once loaded onto development-specific Piwi proteins (53, 54), scnRNAs interact with old MAC transcripts through homology-dependent pairing, which mediates a genome-wide comparison of parental MIC and MAC genomes (55, 56). As a result of the molecular subtraction of scnRNAs homologous to old MAC sequences, scnRNAs corresponding to MIC-restricted DNA are directed to their homologous sequences in the new developing MAC through pairing to nascent transcripts (55, 57). Recent work in Paramecium established that scnRNA-guided Piwi proteins recruit the Paramecium PRC2 Polycomb complex (58, 59), which induces the trimethylation of histone H3 on lysines K9 and K27 (60, 61). In Tetrahymena, PRC2-dependent H3K9me3 and H3K27me3 heterochromatin marks also drive the recognition of MIC-specific sequences by the DNA elimination machinery, in a Piwi-dependent manner (62, 63). The genome scanning model fully accounts for the observed trans-generational epigenetic inheritance of DNA elimination patterns in Tetrahymena. In Paramecium, it is supported by the observation that imprecise elimination of repeated DNA and excision of ~70% of IESs are sensitive to depletion of PRC2 subunits (29, 59, 61).
Fig 3.
Genome-scanning model for epigenetic control of DNA elimination in Paramecium. Maternal MAC transcripts (wavy blue lines in top and middle panels) are produced constitutively from the whole rearranged genome. During meiosis, the MIC genome is transcribed on both strands, yielding double-stranded RNAs (dsRNA) that cover the entire non-rearranged genome. After dsRNA processing by Dicer-like proteins Dcl2 and Dcl3, 25-nt scnRNAs (wavy black and red lines, middle panel) are loaded onto Piwi proteins Ptiwi01 and Ptiwi09. Following comparison with maternal MAC transcripts, the scnRNA population is enriched in MIC-specific scnRNAs (wavy red lines in middle and bottom panels, red boxes and double-headed arrow represent IESs and an imprecisely eliminated TE, respectively). The latter scnRNAs are imported into the new developing MACs, where they direct Ptiwi01/09 to homologous sequences by pairing to nascent transcripts (in blue, bottom panel). Ptiwi01/09 recruits the PRC2 polycomb complex, which contains the Ezl1 histone methyl transferase responsible for H3K9 and H3K27 trimethylation (represented by orange and white circles). RNA-mediated heterochromatin formation is thought to drive the elimination complex to MIC-specific DNA. See text for references.
Second, in the spirotrichous ciliate Oxytricha, Piwi-associated small piRNAs (~27 nt) are also produced specifically during conjugation (64, 65), but, in contrast to Paramecium and Tetrahymena scnRNAs, Oxytricha piRNAs appear to be produced from the old MAC (instead of the MIC) and cover the whole rearranged somatic genome. They were shown to guide the retention of MDSs in the new MAC, protecting them from the massive elimination that targets ~95% of the germline genome. Even though the mechanistic details remain to be investigated, both cytosine methylation and hydroxymethylation were proposed to mark eliminated DNA genome-wide (66). Long transcripts from the old MAC were also detected in Oxytricha and appear to act as RNA templates for genome rearrangement and gene unscrambling (see Fig. 9 in section “A potential role for RNA in directing chromosome fragmentation in Oxytricha and Stylonychia”) in the new MAC (67).
CHROMOSOME FRAGMENTATION
The second major type of genome rearrangement during ciliate MAC development, chromosome fragmentation coupled with de novo telomere addition, has received less attention in recent years. However, there have been many key findings concerning this process in a select number of ciliate species. The main goals of this review are to summarize our current knowledge of this process focusing on some of the key model ciliates, identify gaps in our knowledge, and speculate on the origin and function of developmental chromosome fragmentation.
Chromosome fragmentation in Tetrahymena
Chromosome composition of the Tetrahymena thermophila MIC and MAC genomes
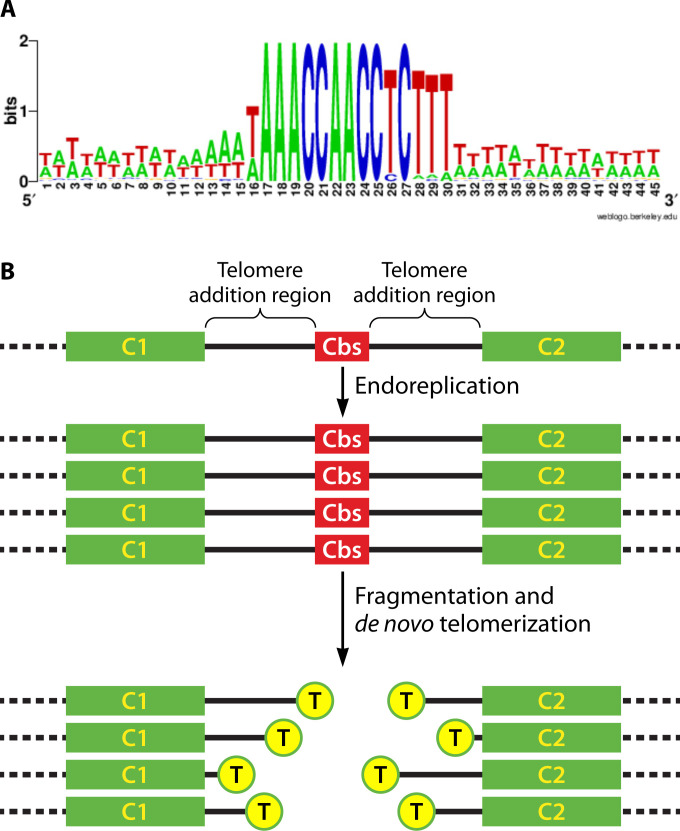
The 157-Mb T. thermophila MIC genome (18) is diploid in vegetative cells and arranged as five pairs of chromosomes of roughly similar size. A number of early studies provided evidence that MAC chromosomes were considerably smaller in size and much more numerous. These studies include the analysis of MAC DNA by sedimentation analysis (68) and pulse-field gel electrophoresis (69, 70), as well as analyzing the MAC telomere content (71). Ultimately, sequencing of the complete T. thermophila MIC and MAC genomes (18, 72) indicated that sexually mature cells contain 181 MAC chromosomes, which are generated during new MAC differentiation by programmed, site-specific fragmentation of the 5 MIC chromosomes. Fragmentation occurs at every copy of a highly conserved, 15-bp “chromosome breakage sequence” (Cbs, described in detail in “The Tetrahymena chromosome breakage sequence”) in the MIC genome, and telomeres are then added de novo to the broken ends.
For decades, it was accepted that essentially all the MAC chromosomes are maintained at an average G1 ploidy of ~45. This was based on genetic observations, i.e., the rate of segregation of assortant cells pure for either allele starting from cells with originally heterozygous MACs (73). However, using “droplet digital PCR” on individual T. thermophila cells, Zhou et al. (74) have concluded that the G1 ploidy of the non-rDNA chromosomes is ~90, instead of ~45. While the two types of observations remain to be reconciled, the resolution of this contradiction is not relevant to programmed chromosome breakage. One significant exception to the above ploidy level is the MAC rDNA minichromosome; it is the smallest chromosome (21 kb) and is maintained at ~9,000 G1 copies (75). MAC chromosomes, other than the rDNA, average 562 kb in length, and range between 37 kb and 3.2 Mb, based on improved MAC sequence assemblies (76).
In addition to the maintained T. thermophila chromosomes in the MAC genome, some MAC chromosomes are generated during MAC differentiation that are lost within the first few fissions after conjugation, as reported first by Cassidy-Hanley et al. (77), and later based on a comparison of MIC and MAC whole-genome assemblies (18, 78). These chromosomes are referred to as “non-maintained chromosomes” (NMCs). Thirty-three NMCs were identified by the Tetrahymena MIC sequencing project. NMCs are relatively small, ranging in predicted length between a few nucleotides and 84 kb. Some NMCs contain predicted genes (18); indeed the function of at least one of these genes during early development stages has been shown to be absolutely required for viability (79).
The failure of NMCs to be maintained is attributed to a putative lack of fully functional origins of replication. This is a reasonable hypothesis, considering that the spacing between chromosome replication origins in the T. thermophila MAC genome averages about 15 kb (80). The identification of NMCs requires a comparison of MIC and MAC genome sequences, so the occurrence of NMCs in species other than T. thermophila, although likely, will remain unknown until the MACs and MICs of related species are sequenced. The possible origin of NMCs is discussed later in the evolutionary context of MIC Cbs duplication and translocation.
The Tetrahymena chromosome breakage sequence
A pioneering series of studies of the Tetrahymena rDNA minichromosome by Meng-Chao Yao and members of his research group led to the discovery and characterization of the Tetrahymena chromosome breakage sequence (Fig. 4) and to the eventual understanding of its role in site-specific fragmentation of the five Tetrahymena MIC chromosomes into around 200 MAC chromosomes and non-maintained chromosomes.
Fig 4.
The Tetrahymena Cbs. (A) DNA sequence logo showing sequence conservation among the 225 Cbss in the T. thermophila MIC genome. The Cbs AC-rich strand is illustrated. The Cbs, comprising the central 15 nucleotides (positions 16–30), is shown along with 15 adjacent nucleotide positions on each side of the Cbs. Only Cbs positions 1, 11, 13, 14, and 15 show any diversity [figure taken from reference (18)]. (B) Schematic of chromosome breakage and telomere addition events in Tetrahymena. A segment of a MIC chromosome containing a Cbs and two flanking MAC chromosome precursors (“C1” and “C2”) is shown at the top. During MAC development, this MIC chromosome region is first endoreplicated and then fragmented at or near the Cbs. Telomeric repeats (5′-CCCCAA-3′/3′-GGGGTT-5′; yellow “T” circles) are added to the ends at various positions within the TAR. Note that the Cbs DNA is lost along with a variable length of sequence on either side of the Cbs.
Sequencing of the entire 21 kb MAC rDNA minichromosome, and of the MIC DNA segment that encodes the MAC rDNA and its flanking regions, led to the discovery of four copies of a highly conserved 15-bp sequence flanking the rDNA minichromosome in the MIC, which are absent from the MAC sequence (81). Three tandem copies of the sequence 5′-TAAACCAACCTCTTT-3′ were found on the 5′ side, and one copy was found on the 3′ side of the MIC rDNA locus. A subsequent analysis found that the 15-bp sequence element was associated with other sites of chromosomal breakage (82). In one case studied in detail, the 15-bp conserved sequence was found to reside within a 54-bp segment of MIC DNA absent from the MAC, such that the ends of two MAC chromosomes were formed from the immediately flanking sequences (82). Furthermore, a naturally occurring “Cbs-like” DNA segment differing at one nucleotide position was found not to be associated with chromosome breakage during MAC differentiation (82).
Direct experimental evidence showed that the Cbs causes chromosome fragmentation in vivo during MAC differentiation (83, 84). Deletion of the three copies of the 15-bp repeats on the 5′ side of the MIC rDNA, or the single copy on the 3′ side, abolished chromosome fragmentation on the respective side of the MIC rDNA in the developing MAC. Single-base pair substitutions at several Cbs positions abolished developmental chromosome fragmentation (83). Moreover, the experimental in vivo insertion of a Cbs within MIC chromosome sites where it previously did not occur induces chromosome fragmentation at that site. These studies led to the conclusion that the Cbs is a necessary and sufficient signal for directing developmental chromosome fragmentation (84).
Subsequent studies characterized additional Cbss in T. thermophila, but it was not until its MIC and MAC whole-genome sequences were obtained and compared that all the Cbss in the MIC genome and their flanking sequences could be analyzed. The 5 MIC chromosomes contain a total of 225 Cbss, including all those that generate the 181 maintained and the 33 non-maintained MAC chromosomes (18). The consensus sequence of all the Cbss in the MIC genome is 5′-tAAACCAACCtCttt-3′ in the “AC-rich” strand, where capital letters represent the nucleotide positions that are absolutely conserved in every one of the 225 Cbss; thus, all the As and Cs in the consensus sequence are universally conserved nucleotides (Fig. 4A). Of the 225 Cbss in the T. thermophila MIC genome, 109 or roughly one half (48%) have the entire, exact consensus sequence: 5′-TAAACCAACCTCTTT-3′. Another 93 Cbss (41%) differ from the consensus sequence at a single position among the 5 variable nucleotide positions. The remaining 23 Cbss (10%) differ from the 15-bp consensus by only 2 nucleotides. Thus, no T. thermophila Cbs differs from the consensus sequence by more than two nucleotides. Although an initial transformation-based study suggested that some deviations from the 15-bp Cbs consensus might have a decreased efficiency in chromosome fragmentation (83), there is as yet no indication that any of the natural Cbs variants have a reduced efficiency of chromosome fragmentation during macronuclear development.
Interestingly, the Cbs shares two sequence features with the Tetrahymena telomere sequence, which consists of repeats of the hexanucleotide 5′-CCCCAA-3′: (i) Both the Cbs and the telomeres have clear C-rich and G-rich DNA strands, and (ii) the Cbs C-rich strand includes an absolutely conserved “CCAACC” hexanucleotide, which is contained within the poly-CCCCAA telomere sequence. The significance of these similarities (if any) remains unclear. Certainly, eukaryotic telomere sequences are far more ancient than the Tetrahymena Cbs.
De novo telomere addition begins at a variable distance away from the Cbs end. The shortest reported distance is 4 bp, and the longest is 30 bp from the end of the Cbs in 90% of the cases investigated (85). Overall, the results to date have led to a model in which the Cbs directs a double-stranded cut(s) in the DNA, and the two ends are trimmed to various extents by a nuclease prior to the addition of telomeres within a “telomere addition region” (TAR; Fig. 4B). The nature and location of the Cbs-induced break are not known. The results also imply that chromosome breakage at the Cbs in some way modifies or tags the resulting ends such that they are recognized by the de novo telomere addition machinery—rather than by the end-rejoining machinery—but there is currently no information on how this might be accomplished.
Cbss are remarkably conserved in the MIC genomes of T. thermophila and other Tetrahymenine species and could be at least 300 million years old
The T. thermophila Cbs sequence shows a remarkable degree of conservation not only in other Tetrahymena species but also in species of closely related genera. The Cbs has been reported in the following Tetrahymena species: T. malaccensis, T. elliotti, T. borealis, T. hegewischi, and T. caudata (18, 86). Coyne and Yao (86) also showed sequence conservation among rDNA-flanking Cbss in Glaucoma chattoni and Colpidium campylum, two genera closely related to Tetrahymena, implying that the Cbs predates the origin of the Tetrahymena genus. All but one of the Cbs variants found in these other species are also among the 225 Cbss in T. thermophila (Table S1), which further illustrates in detail the remarkable degree of Cbs sequence conservation among these species. Only 1.4 mutations have been fixed per Cbs variable site since the divergence of T. thermophila and T. pigmentosa (18), which are estimated to have diverged a little over 100 Mya (87). Based on the estimated age of radiation of the Tetrahymena species (87), the Cbs sequence, as it is found today, could have already existed in Tetrahymenine species as far back as ~300 million years ago.
There also appears to be a strong conservation of the location of Cbss. Nine consecutive Cbss in a randomly chosen MIC chromosome segment in the T. thermophila genome were identified, based on the conservation of genes flanking those sites, in the MIC genomes of three other Tetrahymena species: malaccensis, borealis, and pigmentosa (18). Among the total of 36 chromosome breakage sites investigated, only one site is missing in T. borealis, and a novel one occurs in T. malaccensis. Thus, most of the Cbs sites have been conserved over an estimated period of at least 100 million years; gains and losses of Cbs sites that affect the lengths of the MAC chromosomes have occurred but are rare. Interestingly, among the 36 chromosome breakage sites examined in the four Tetrahymena species, there were four chromosome breakage sites at which tandemly repeated Cbs duplications had occurred. The duplications are likely of recent origin because in every case the two duplicate Cbss have identical 15 bp sequences.
T. thermophila Cbs clades provide evidence for evolutionary Cbs duplication and translocation/transposition
Genome-wide cross alignments of all T. thermophila Cbss and 200 bp of flanking MIC sequence on each side (18) showed that 49 Cbss (nearly a quarter of the 225 Cbss in the genome) cluster into 15 sequence groups, called “Cbs clades” (Table 1). Each Cbs clade is presumed to have been generated by successive duplications of an ancestral Cbs and some flanking sequence. The length of conserved Cbs-flanking DNA sequence among different Cbs clades varies between 45 bp and 17.5 kb (Table 1). Two types of events are proposed to have led to the generation of existing Cbs clades: local tandem duplication and long-range duplication/translocation.
TABLE 1.
Fifteen T. thermophila Cbs cladesa
| Cbs clade name (# members)b,d | Cbs chromosome locationsc,e | Duplication repeat unit |
|---|---|---|
| 1L-16 (2) | 1L-16; 4R-24 | 45 bp |
| 1L-17 (4) | 1L-17, 18, 19; 4R-25 | 45 bp |
| 5R-5 (2) | 5R-5, 6 | 53 bp |
| 5R-14 (2) | 5R-14, 15 | 84 bp |
| 1L-1 (5)* | 1L-1, 2, 3, 4, 5 | 144 bp |
| 1R-35 (2) | 1R-35,36 | 469 bp |
| 1L-28 (2) | 1L-28, 29 | 530 bp |
| 2R-1 (2)* | 2R-1, 2 | 605 bp |
| 1R-37 (2) | 1R-37, 38 | 796 bp |
| 3L-4 (3)* | 3L-4; 4 L-2, 3 | 3.8 Kb |
| 1R-1 (8)* | 1R-1, 2, 3, 4, 5, 6, 7; 2L-2 | 13.6 Kb |
| 5L-9 (3) | 5L-9, 10, 11 | 10.4 kb |
| 4R-3 (6)* | 4R-3, 4, 5, 6, 7; 4R-38 | 17.5 Kb |
| 1L-20 (2) | 1L-20; 3L-14 | Not reported |
| 3L-3 (2)* | 3L-3; 3L-29 | Not reported |
Data were taken from reference (18).
The 15 MIC Cbs clades are listed in order of increasing repeat unit length.
Cbs location includes chromosome arm (e.g., “1L” = left arm of MIC chromosome 1) followed by the consecutive Cbs number along the chromosome arm (e.g., “2” or “28”); thus, XL-1 and XR-1 are the nearest Cbss flanking the centromere of MIC chromosome X.
The six clades that include at least one Cbs located within the first three Cbss closest to a centromere have an asterisk next to their names in the first column.
The seven clades that include long-distance Cbs translocation/transposition—within or between different MIC chromosomes—are underlined.
The members of local tandem Cbs duplication clades are concentrated within centromere-adjacent regions of MIC chromosomes (see Table 1). Hamilton et al. (18) have suggested that local tandem Cbs duplications resulted from unequal crossing-over, based on several features. These Cbss are consecutively arrayed (i.e., contain no interspersed Cbss from another clade), and, with one exception, all Cbss in a tandem array are in the same orientation. In addition, the tandem arrays are concentrated in pericentromeric MIC regions, known to be enriched in functional and decayed transposons and repetitive elements (18). These MIC chromosome regions also show an increased frequency of unequal crossing-over compared to the rest of the MIC genome (87). Thus, a Cbs that became imbedded near pericentromeric transposons or other repetitive sequences would be surrounded by numerous homologous sequences that could facilitate unequal crossing-over, which in turn would lead to a Cbs duplication. Further unequal crossing-over fostered by the duplicate Cbs copies would then be sufficient to quickly extend the length of the duplication array near the MIC chromosome centromere. Moreover, the proximity of a Cbs to pericentromeric transposable elements could also potentiate the association of a Cbs clade member with a mobile element, thus providing an explanation for the “translocated/transposed” Cbs clade members (e.g., in different MIC chromosomes or at great distance within the same MIC chromosome).
A T. thermophila MIC Cbs duplication that leaves at least ~40 bp of intervening sequence but does not include a MAC replication origin is predicted to generate a non-maintained chromosome. Interestingly, among the total of 33 predicted NMCs identified in the T. thermophila MIC genome (18), 24 (73%) are flanked in the MIC by two consecutive members of the same Cbs clade. One additional NMC is flanked by two Cbss belonging to different Cbs clades, and another one is flanked by a Cbs clade member on one side. Only seven NMCs are not flanked by a Cbs clade member in the MIC.
Finally, the events resulting in the currently observed Cbs duplications (local or long-range translocations/duplications) must have occurred relatively recently in evolutionary time, given that the Cbs-flanking sequences have not yet had enough time to mutate to the point that their sequence similarity would have deteriorated beyond recognition.
Biochemical machinery specifically required for Cbs-dependent chromosome breakage
Functional chromosome fragmentation in the developing MAC requires a DNA double-stranded break(s) at the Cbs, putatively generated by a “Cbs endonuclease,” followed by the de novo addition of telomeres to the broken DNA ends. Two proteins have been reported to be specifically required for Cbs-dependent chromosome breakage: the “protection of telomere 2 protein” (Pot2p) (88) and the p68 DEAD box DRH1 RNA helicase (89) (see Table S2). The length and strong sequence conservation of the Tetrahymena Cbs would appear to make it a strong candidate for identifying a protein(s) involved in chromosome fragmentation. Nonetheless, there has been no reported success in either identifying a protein(s) that physically interacts directly with the Cbs or in detecting, in vitro, the catalytic activity required for the Cbs-dependent breakage of the phosphodiester bonds in DNA.
Remarkably, among all the 16 Tetrahymena proteins so far shown experimentally to be required for the excision of thousands of MIC-limited IESs in a newly developing MAC, all but one (apparently not yet tested) are also required for Cbs-dependent chromosome breakage (89) (see Table S2). To explain this observation, those authors have suggested the possibility that a developmental checkpoint exists, such that the successful completion of global IES excision generates a signal that allows genome-wide Cbs-dependent chromosome breakage to proceed. It is also possible that the expression of one or more genes involved in chromosome fragmentation and/or subsequent steps in MAC development is dependent on IES excision. The existence and nature of such a hypothetical checkpoint remain to be addressed experimentally.
In summary, although a good deal of information exists about the Cbs sequence and the products of chromosome fragmentation in Tetrahymena, much remains to be learned about the biochemistry of Cbs-dependent programmed chromosome breakage.
Chromosome fragmentation in Paramecium
Sizes and numbers of MIC and MAC chromosomes in Paramecium
Early microscopic observations of meiotic MICs during metaphase I uncovered 41 to 45 chromosome pairs in P. tetraurelia (90). Similar numbers were obtained for other species of the P. aurelia group, with a haploid count of 37 to 60 MIC chromosomes (91). These chromosome numbers are probably an underestimate because short chromosomes may have been missed due to technical limitations. Recent progress in vegetative MIC purification by cytometry and assembly of the MIC genome from Illumina sequencing (26, 29, 92) have provided an estimate of 108–150 Mbp for the P. tetraurelia MIC genome. This would give a predicted average MIC chromosome size of 2.5–3 Mbp if 60 chromosomes are present. Of note, the organization of MIC chromosomes is still under investigation. In particular, MIC centromeres (eliminated during MAC development) have not been characterized at the molecular level (93), and whether MIC telomeres are retained on MAC chromosomes is under investigation.
Based on whole-genome sequencing data, the size of the P. tetraurelia haploid MAC genome is 72 Mbp (94), with a nuclear ploidy of 1,000 to 1,600 n (95). MAC scaffolds were assembled following whole-genome sequencing of total MAC DNA from young vegetative cells (~10 fissions post autogamy) (94, 96). A majority of the 188 largest MAC scaffolds carry telomere repeats at both ends, as expected for full-length chromosomes (94), and their lengths range between 50 kbp and 1 Mbp, in agreement with earlier pulse-field gel electrophoresis analyses (97–99). The large number of complete MAC scaffolds, together with their shorter length compared with MIC chromosomes, is consistent with programmed chromosome fragmentation taking place during MAC development. Based on the current estimates, one to two fragmentation sites at least may be present on each MIC chromosome. Note that the scaffolds represent a consensus sequence for a larger number of MAC chromosomes owing to heterogeneity generated by DNA elimination, as will be described below.
Reproducibility and heterogeneity at chromosome fragmentation sites
The pattern of developmentally programmed chromosome fragmentation in P. aurelia is generally reproducible across sexual generations. Different levels of heterogeneity have nevertheless been noted. First, early molecular cloning of MAC chromosome ends revealed the existence of microheterogeneity in the position of the nucleotide to which telomeric repeats are added, with each TAR extending over 500–800 bp (Fig. 5A). MAC telomeres, which are ~200 bp long on average, are composed of a mixture of 5′-T2G4-3′ and 5′-T3G3-3′ repeats (100), both of which are added to free 3′ DNA ends by the same error-prone telomerase using a single RNA template (101, 102). In contrast to Tetrahymena (see section “The Tetrahymena chromosome breakage sequence”) and Euplotes (see section “Identification of the conserved E-CBS near MAC chromosome ends”), no common conserved motif was found in the sequences surrounding telomere addition positions in Paramecium (36, 103–105). At a higher level, macroheterogeneity was also documented in the pattern of chromosome fragmentation (Fig. 5B and C), with the presence of multiple TARs separated by several kilobase pairs from each other (103–105). These two levels of heterogeneity may result from the introduction of multiple chromosome breaks during fragmentation and/or DNA end trimming before telomeres are added.
Fig 5.
Molecular analysis of three Paramecium chromosome fragmentation regions. (A) MIC chromosome (bold line) including the telomere addition region (vertical arrow) downstream of the P. tetraurelia 51G surface antigen gene is shown, along with the corresponding MAC chromosome (thin line). A Tc1/mariner transposon (orange) is adjacent to the TAR on the MIC chromosome. Adapted from reference (28). (B) Multiple TARs in the vicinity of the P. tetraurelia 51A surface antigen gene. Vertical arrows mark the position of the TARs that were mapped upstream and downstream of the 51A gene. Position 1 accounts for 50% of MAC telomeric ends in wild-type strains and overlaps the 3′ end of a minisatellite (blue box). Other as yet uncharacterized germline eliminated sequences were proposed to locate downstream of position 2. The dashed arrow is the major TAR used in the d48 MAC deletion mutant. [See references (103, 104).] (C) Fragmentation region of the P. primaurelia chromosome harboring the 156G surface antigen gene. The three confirmed TARs are marked by vertical arrows. The locus harbors a truncated copy of a Tc1/mariner transposon (orange box) and two minisatellites (blue boxes). [See reference (36).] New MAC chromosomes are represented at the bottom of each panel, with the telomeres shown as hatched boxes to account for the microheterogeneity of telomere addition positions (within 500–800 bp regions). In panel C, internal deletions of germline DNA repeats (minisatellites and transposons) are represented by red dotted lines between brackets on individual chromosomes. The same scale bar applies to all panels.
Chromosome fragmentation in Paramecium is associated with heterogeneous DNA elimination
A more complex view of chromosome fragmentation in Paramecium has emerged from studies of MAC chromosome ends and their corresponding MIC regions in P. primaurelia and P. tetraurelia. In these two species, both fragmented and non-fragmented molecules can originate from the same MIC chromosome (97). Sequence analysis of such alternatively rearranged germline regions (Fig. 5C and 6) revealed that they contain repeated sequences, i.e., TEs (either Tc1/mariner or LINE elements) and/or minisatellites of different lengths and nucleotide composition, all of which are absent from the MAC genome. This led to the hypothesis that chromosome fragmentation is an alternative outcome of imprecise DNA elimination by DNA breakage and rejoining (as noted in section “Internal DNA elimination,” in Paramecium, a distinction is made between “imprecise DNA elimination” and the precise elimination of “IESs” by DNA breakage and rejoining). That is, DNA double-stranded breaks are healed by telomere addition instead of being repaired through end joining [e.g., references (36, 106)]. In support of this hypothesis, all the chromosome fragmentation regions that were previously studied at the molecular level in species from the aurelia group (Fig. 5A and B) coincide with the presence of TEs or minisatellites in their downstream germline sequence.
Fig 6.
Fragmentation of a germline chromosome during development generates multiple somatic chromosomes. The JBrowse image of the MIC scaffold that bears the P. tetraurelia 51G and 51C surface antigen (SAg) genes is shown (top), with tracks for annotations showing genes (107), IESs (28), MAC Illumina read coverage, preliminary TE annotations, curated retro- (LINE) and Tc1/mariner (TIR) transposons (29), and remapped MAC telomere repeats found on individual reads. Note that the arrows representing the orientation of the SAg genes and TEs (TIR and LINE) are not drawn to scale. The true extent of the hypothetical MIC chromosome, beyond the MAC-destined region shown, has not been determined (dotted lines at left and right ends). Sites of fragmentation (light blue shading) are identified from the positions of TEs and remapped MAC telomere repeats combined with the MAC read coverage. The lower part of the figure shows hypothetical MAC chromosomes predicted to result from programmed genome rearrangement. The six molecules carrying the 51C SAg gene (a to f, in black) are compatible with the sizes of the P. tetraurelia MAC chromosomes that were observed on southern blots, following pulsed-field gel electrophoresis and hybridization with a 51C SAg gene probe (108). Note that the region displayed here is homologous to the P. primaurelia region shown in Fig. 5C. (Linda Sperling contributed to the design of this figure.)
In the whole-genome sequence assembly, each MAC scaffold actually represents a population of alternatively rearranged molecules, with co-occurring internal deletions and/or telomeric ends within specific germline regions that likely define internal fragmentation regions [Fig. 6; (109)]. Non-overlapping MAC scaffolds may either originate from distinct MIC chromosomes or from distant parts of the same MIC chromosome separated by a large eliminated germline region (92, 109). Identification of all fragmentation regions genome-wide and their associated imprecisely eliminated sequences awaits the complete assembly of the MIC genome (29).
Whether such a scheme is general for other species of Paramecium is unclear. However, the distantly related species P. bursaria has been shown to also have similar overlapping, or nested, MAC chromosomes, although they are smaller in size than those of the aurelia group species (110). Information on the MIC genome will be needed to determine if they are generated in a similar manner.
Epigenetic control of chromosome fragmentation and experimental induction of heritable variant patterns
Consistent with chromosome fragmentation being associated with imprecise elimination of interstitial DNA, several observations indicate that it is also epigenetically controlled by the old MAC. The first observation is that among the different TARs that surround the P. tetraurelia 51A gene (Fig. 5B), the upstream one is predominantly used in the d48 variant, even though this line carries a fully wild-type MIC genome (103, 105). This alternative fragmentation pattern, which removes the 51A gene from the MAC, is maternally inherited across successive sexual generations. Microinjection of the A gene into the vegetative MAC suppresses the deletion of the A gene and restores the wild-type fragmentation pattern in the following sexual generations (111–113). The rescue is not observed following microinjection of similar paralogous genes, which indicates that it depends on sequence identity or strong similarity between the rescuing transgene and the rescued locus (114). Taken together, these early data fit with the genome scanning model that was originally proposed for internal DNA elimination (Fig. 3) but with de novo telomere addition substituting for chromosome rejoining. Indeed, when the 51A gene is absent from the MAC (e.g., in the d48 line), no homologous maternal transcript is produced. As a consequence, scnRNAs from the wild-type MIC would not be subtracted when compared with old MAC transcripts and can subsequently drive the deletion of the 51A gene in the new MAC. Displacement of the major TAR upstream of 51A, therefore, appears as a consequence of DNA elimination.
Alternative chromosome fragmentation patterns can also be induced experimentally in the sexual offspring of wild-type cells, following RNAi-induced degradation of specific old MAC transcripts. This was first demonstrated for two different subtelomeric regions in P. primaurelia and P. tetraurelia, using either transgene-induced (115, 116) or double-stranded RNA-induced RNAi (115). In each experiment, displacement of the major TAR to an upstream alternative position accompanies the deletion of the gene that was targeted by RNAi. The alternative fragmentation pattern is then inherited across sexual generations. Using the same procedure, heritable somatic deletions could also be induced experimentally in an internal region of a MAC chromosome carrying the trichocyst discharge ND7 gene (115). The resulting internal deletions are heterogeneous and show the same characteristics as those generated upon developmentally programmed imprecise elimination of germline DNA repeats (36), with one or several 5′-TA-3′ dinucleotides at each junction. Interestingly, experimental induction of internal MAC deletions is accompanied by chromosome fragmentation and the detection of new telomeric ends in the vicinity of the deleted ND7 gene (117). These observations strongly suggest that heritable alternative chromosome fragmentation patterns can be triggered experimentally at any genomic region, together with imprecise DNA elimination.
Molecular mechanisms involved in chromosome fragmentation—open questions
As discussed, chromosome fragmentation and heterogeneous internal deletions in Paramecium are most likely alternative products of imprecise DNA elimination. Similar to IES excision, imprecise elimination of TEs and other repeats requires Pgm (30) and the Ku NHEJ factor (33). Significant differences, however, exist between the two DNA elimination processes.
The precision of IES excision can be explained by the conserved position of DNA cleavage sites at the IES-flanking TAs and the obligatory presence of Ku in the cleavage complex allowing for its immediate recruitment to protect and join the two flanking broken ends (118). Although this needs to be examined more closely, telomere addition at cleaved IES ends is likely a rare event in normal conditions, based on the mapping of telomeric reads along the MIC sequence (Fig. 6). One study reported the detection of a minor TAR downstream of an IES in P. primaurelia (119), but no sequence of the corresponding germline region is available, making it difficult to conclude that there is a general mechanistic link between IES excision (i.e., as opposed to imprecise DNA elimination) and chromosome fragmentation.
Imprecise DNA elimination yields heterogeneous internal deletions often involving TA-containing microhomologies (36, 115). Assuming that the Pgm endonuclease cleaves DNA both during IES excision and imprecise DNA elimination, two non-exclusive hypotheses may explain the heterogeneity of the latter [discussed in reference (36)]. Pgm may introduce DSBs at variable TAs on each side of heterochromatin regions with variable boundaries, which may be followed by direct end joining. Alternatively, Pgm may cleave DNA at defined sites, but unprotected broken ends would be trimmed by an exonuclease before they are joined: the AT- richness of eliminated sequences may increase the probability of having self-complementary TA-containing microhomologies at heterogeneous junctions. In either scenario, the finding that chromosome fragmentation takes place as an alternative to end joining suggests that the DSBs that are generated during imprecise elimination are accessible to the telomerase. Further investigation will be needed to unravel whether the recruitment of Paramecium telomerase to broken DNA ends depends on Ku or rather competes with Ku.
An important issue that remains to be addressed is the mechanistic basis for how Paramecium carries out both a precise mode of DNA deletion (IES excision), in which DNA cleavage and end joining are tightly coupled, as well as an imprecise mode associated with chromosome fragmentation (elimination of repeated sequences). A recent genome-wide study has revealed that IES excision takes place well before the bulk of imprecise DNA elimination (120). This delayed elimination kinetics may indicate that different or additional factors contribute to imprecise elimination. Alternatively, DNA elimination linked to chromosome fragmentation may not be directly catalyzed by Pgm and instead use a different, but still Pgm-dependent, pathway.
Chromosome fragmentation in spirotrichs (Euplotes, Oxytricha, and Stylonychia)
Sizes of chromosomes in spirotrich ciliates
Historically, developmental chromosome fragmentation was first inferred in the spirotrich ciliates [class Spirotrichea; (121, 122)], owing in part to the extreme form of chromosome fragmentation that occurs within many of this group’s species. Organisms such as Euplotes, Oxytricha, and Stylonychia all have small MAC chromosomes that predominantly contain single genes. The average size of MAC DNA molecules, often called nanochromosomes, ranges from ~2 to 3 kbp, while the total sizes of MAC genomes range from ~50 to 100 Mb in the various spirotrichs analyzed to date (123–128). MIC genomes have been less well characterized, but the genome sizes typically range from ~500 to >10,000 Mb, and >100 chromosomes have been observed in some species [(45), reviewed in references (129, 130)]. Since genome analyses have identified ~12,000–30,000 different nanochromosomes in the MACs of spirotrich species (123–125, 127, 128), at least this number of chromosome fragmentation sites must exist in their respective MIC genomes. Nonetheless, despite these species sharing MAC nanochromosomes, there appears to be significant diversity in the chromosome fragmentation processes among spirotrichs, similar to the differences observed between the oligohymenophorans Tetrahymena and Paramecium. The differences among spirotrichs are particularly evident when comparing the euplotid (Euplotes) and hypotrich (e.g., Oxytricha and Stylonychia) species.
Chromosome fragmentation in Euplotes
Identification of the conserved E-CBS near MAC chromosome ends
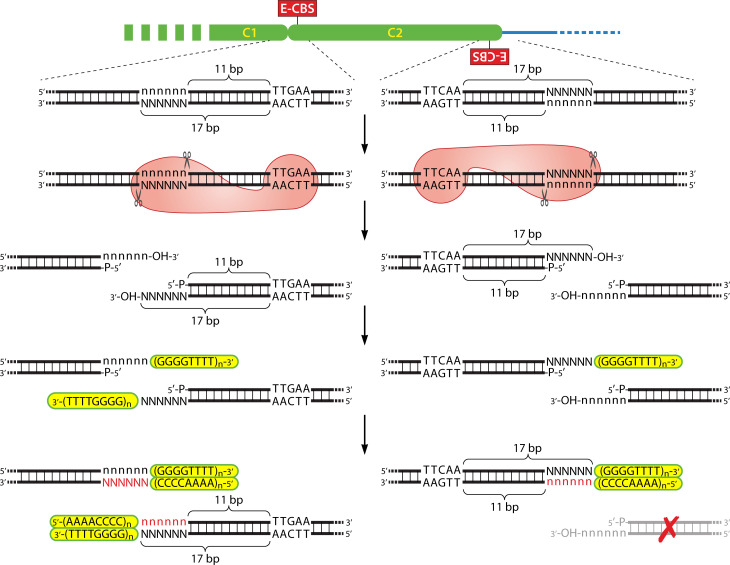
In Euplotes crassus, chromosome fragmentation/telomerization is precise and reproducible to the nucleotide (131, 132). That is, one sees telomeres at exactly the same nucleotide position for the multiple copies of a given nanochromosome generated in a single cell during MAC development, and this holds true for subsequent sexual generations. There is also clear evidence of a conserved sequence element in the vicinity of Euplotes chromosome fragmentation/telomere addition sites. This sequence element was initially observed in global sequence analyses of total MAC DNA of a number of spirotrichs by the Maxam and Gilbert chemical sequencing procedure (133, 134). This sequencing method essentially sequences all chromosome ends simultaneously, and the results indicated that all, or almost all, spirotrich nanochromosomes are capped with short telomeres of a defined length that consist of repeats of 5′-CCCCAAAA-3′/3′-GGGGTTTT-5′. However, in the case of Euplotes aediculatus, an additional conserved 5-bp sequence of 5′-TTGAA-3′ was observed separated by 17 bp from the first telomeric repeat (Fig. 7). Similar sequence analyses, as well as sequencing of individual cloned nanochromosomes and sequencing of MAC genomes, confirm the presence of a 5′-TTGAA-3′ element at the ends of most MAC nanochromosomes in a variety of Euplotes species [e.g., references (124, 125, 128, 135)]. This element appears to be well conserved and has been termed the Euplotes CBS (E-CBS) based on its likely role in chromosome fragmentation. An analysis of 100 nanochromosome ends in E. crassus (136) initially suggested that the 5-bp E-CBS sequence may represent the core of a larger conserved sequence block (5′-HATTGAAaHH-3′; H = A, T, or C), and subsequent MAC genome-level analyses lend support to a larger conserved region (124, 127, 137). However, because the core 5′-TTGAA-3′ is particularly well conserved, for simplicity we will consider it as the “E-CBS” in our further discussion.
Fig 7.
Model of chromosome fragmentation and de novo telomere addition in Euplotes. A segment of MIC DNA is shown at top, which includes two MDSs (green ovals C1 and C2) as well as flanking DNA eliminated during MAC development (thin blue line). An enlargement of the segments of DNA that will be fragmented is shown below the top line, with individual base pairs indicated (N = A, G, C, or T). Note that the right end of MDS “C1” and the left end of MDS “C2” overlap by 6 bp. During MAC development, a fragmentation enzyme (red object) interacts with the E-CBS and cleaves the 5′-strand 11 bases upstream and the 3′-strand 17 bases upstream to generate 6 bp 3′ overhangs. Telomerase then adds telomeric repeats (highlighted in yellow) to the 3′ ends. The 5′ strands of the telomeres are then synthesized, along with the complements of the original six base overhangs (bases in red). Sequences which are not retained as MAC nanochromosomes are degraded, as indicated by the large red “X.”
Positioning of the E-CBS suggests a model for chromosome fragmentation in Euplotes
The E-CBS might be involved in either directing the cellular machinery that fragments the chromosomes or, alternatively, specifying where telomeric repeats will be added. Current data favor the former possibility. First, only ~75% of nanochromosome ends in E. crassus possess a reasonable match to the E-CBS, yet all analyzed ends have a telomere (136). For cases where a nanochromosome end lacks a good match to the E-CBS, analysis of the MIC organization of a few such situations found that they have a strong match to the E-CBS in the flanking, and typically eliminated, MIC DNA. However, when the E-CBS is in flanking MIC DNA, the 5′-TTGAA-3′ is in inverted orientation and separated by 11 bp from the position where a telomere will be added to the nanochromosome (Fig. 7). This difference in orientation and distance of the E-CBS when present in the nanochromosome end versus flanking MIC DNA suggested a model of chromosome fragmentation that explains the “apparent” discrepancy in E-CBS position in a simple and straightforward manner [Fig. 7; (131, 136)]. In the model, the E-CBS serves to position the presumed fragmentation nuclease/complex on the MIC DNA such that it generates a 6-bp staggered cut in the DNA strands, with the resulting 5′ end separated by 11 bp from the E-CBS and the 3′-overhanging end separated by 17 bp from the E-CBS. One or both 3′ ends then serve as the substrate for the addition of telomeric repeats, with the staggered cut subsequently filled in by synthesis of the complementary DNA strand (i.e., the strand containing the 5′ end of the telomere).
Two lines of evidence provide support for fragmentation generating a 6-base 3′ overhang. First, while there is typically some developmentally eliminated spacer DNA separating adjacent precursors of nanochromosomes in the MIC genome of Euplotes crassus, two cases are known in E. crassus where adjacent nanochromosome precursors overlap by 6 bp in the MIC (131, 136). These situations can be explained by a six-base staggered cut in the DNA followed by both ends of the fragmented chromosome undergoing de novo telomere formation as shown in Fig. 7. This would essentially duplicate the 6-bp cut region in both of the resulting nanochromosomes. Second, a ligation-mediated PCR procedure has been used to directly search for the predicted fragmentation intermediates with 6-base 3′ overhangs that have not yet been capped by telomeres (138). Such fragmentation intermediates were observed during MAC development in DNA from E. crassus cells at the period when chromosome fragmentation/telomerization is known to occur but not in the DNA of cells at other stages of MAC development nor in the DNA of asexual cells. Additional analyses indicated that the 5′ end of the staggered cut possesses a phosphate group, while the 3′ end has a hydroxyl group. This represents the only characterized fragmentation intermediate in ciliates and strongly supports the model shown in Fig. 7.
A recent analysis of the Euplotes vannus MAC genome and partial MIC genome (124) both provides support for the above chromosome fragmentation scenario, as well as an indication that it might be more complex. The partial E. vannus MIC genome sequence was obtained from a single E. vannus cell using a whole-genome amplification procedure, followed by high-throughput sequencing and assembly. More than 1,200 chromosome fragmentation sites were obtained. In contrast to E. crassus, where most adjacent nanochromosome precursors are separated by eliminated spacer DNA, the vast majority of adjacent E. vannus nanochromosomes overlapped by up to 10 bp or were separated by less than 10 bp. Significantly, the most frequent situation observed (30%) was a six-bp overlap between adjacent nanochromosomes, which is in agreement with the E. crassus model (Fig. 7). Most of the other junction arrangements are unexpected, particularly since E. crassus and E. vannus are extremely closely related [e.g., reference (139)], and do not fit well with the model of a six-base staggered cut. The results clearly bear further investigation and validation, particularly given that they were based on a specialized high-throughput sequencing procedure.
More generally, multiple MAC genomes of Euplotes species have been obtained but have not been fully analyzed in regard to chromosome fragmentation. In addition to further refining the E-CBS sequence, additional questions that could be addressed include the fidelity of fragmentation/telomerization, as well as how frequently the E-CBS is found at the ends of nanochromosomes and whether its position relative to the site of telomere addition is invariant.
Chromosome fragmentation and alternative fragmentation in Oxytricha and Stylonychia
Microheterogeneity and macroheterogeneity of chromosome fragmentation in Oxytricha and Stylonychia
Species of Oxytricha and Stylonychia display key differences in chromosome fragmentation compared to Euplotes despite belonging to a related sister group within the spirotrichs. First, in contrast to the “precision-to-the-base” chromosome fragmentation/telomerization process seen in E. crassus, both Oxytricha and Stylonychia display macroheterogeneity and microheterogeneity in the positions of telomere formation.
The macroheterogeneity is the result of alternative use of chromosome fragmentation sites, in which the multiple copies of one region of the genome in the developing MACs are processed into two or more nanochromosomes (Fig. 8). Alternative fragmentation was first documented in Oxytricha (140–142), and subsequent large-scale MAC genome sequence analyses of Oxytricha trifallax (127) and Stylonychia lemnae (123) have provided data on the ubiquity of this phenomenon. In O. trifallax, at least 10% of the MAC chromosomes were found to be members of an alternatively processed family (127). The most common situations observed (74%) were two-gene nanochromosome families, where a shorter nanochromosome exists along with a second family member or “isoform” that is extended at one end (e.g., Fig. 8). Such situations can arise through the alternative use/disuse of a single fragmentation site during MAC development and often generate nanochromosomes containing two or more coding regions. More complicated alternative processing families have also been observed, including a situation involving 14 nanochromosome isoforms that arise through the use of 15 chromosome fragmentation sites encompassing eight coding regions (127).
Fig 8.
Chromosome fragmentation in Oxytricha and Stylonychia. Alternative processing and microheterogeneity of chromosome fragmentation/telomerization are illustrated for a segment of MIC DNA containing two coding regions (green ovals C1 and C2) and three chromosome fragmentation/telomerization regions (F/T), including one that is used in an alternative manner (Alt. F/T). Endoreplication during MAC development generates multiple DNA strands that can be alternatively fragmented to yield a two-nanochromosome family. Chromosome fragmentation and/or de novo telomere addition occurs with variability at all ends, leading to microheterogeneity in the position of telomeres (“T”).
Comparison of the S. lemnae and O. trifallax MAC genomes provides indications that chromosome fragmentation sites display evolutionary change. Specifically, only about half of the two-gene MAC contigs in S. lemnae are found in O. trifallax. Moreover, only about two-thirds of the alternative fragmentation sites that are involved in generating some of these two gene nanochromosomes are conserved between the two species. Finally, a more recent analysis of the O. trifallax MAC genome that incorporated PacBio long sequencing reads into the genome assembly (143) suggests an even higher frequency of alternative fragmentation, with 28% of full-length nanochromosomes having at least one additional alternatively processed isoform.
The function, if any, of alternative fragmentation is not clear. It has been suggested that it could serve to place a gene under different transcriptional controls. That is, the two or more members of an alternatively processed nanochromosome family could differ in regard to the presence of enhancers or other transcriptional control elements (141). Secondly, for cases where alternatively processed chromosomes contain multiple coding functions, it might allow for coordinate expression of gene products from some members of the family. Alternatively, the relative proportions of the members of an alternatively processed nanochromosome family may aid in optimizing the cellular levels of their corresponding expressed proteins (144). Finally, it is quite possible that alternative fragmentation does not have a function per se but is instead the result of neutral, or near-neutral, genetic changes during evolution that impair the recognition of some chromosome fragmentation sites or that create new sites that function suboptimally.
In addition to the macroheterogeneity produced by alternative chromosome fragmentation, microheterogeneity is frequently seen in the base position where the telomere is added for the numerous copies of a given nanochromosome generated in a single cell during MAC development. In the case of chromosome fragmentation sites not subject to alternative processing, there is a minimal amount of microheterogeneity. A single telomere addition site is often used for a given MAC chromosome end of this type, but some of these MAC chromosome ends display a few different sites of telomere addition spanning a region of up to ~50 bp (45, 127, 131, 141, 145). In contrast, multiple TASs are more common for alternatively used chromosome fragmentation sites, and the region of telomere addition may span 250 bp (45, 127, 141, 145).
Finally, similar to Euplotes crassus, adjacent precursors of MAC DNA ends may overlap by a small number of base pairs in the MIC chromosome (45, 142). Indeed, this situation is quite common in Oxytricha trifallax, where almost one-half of MAC precursors overlap by 1–10 bp, but overlaps of up to 34 bp were observed (45). This again suggests that chromosome fragmentation may involve staggered cuts in the DNA.
Sequence features adjacent to Oxytricha and Stylonychia sites of chromosome fragmentation
To date, conserved sequences analogous to the Tetrahymena Cbs or Euplotes E-CBS have not been defined in stichotrichs such as Oxytricha and Stylonychia. The general AT-richness of the ends of the nanochromosomes and their flanking regions in the MIC, coupled with the observed microheterogeneity in the positions where telomeres are added, provide a particularly challenging situation for defining a conserved sequence element. There are, however, indications that sequences near the ends of the nanochromosome precursors are necessary for proper developmental fragmentation/telomerization, as well as some intriguing base composition features for subtelomeric DNA.
First, Jonsson et al. (146, 147) have used transformation to carry out a deletion analysis of a construct with a segment of S. lemnae MIC DNA containing nanochromosome precursors. They found that no particular flanking MIC DNA segment was required for proper fragmentation and telomerization of a nanochromosome precursor but that deletion of the first 70 bp of subtelomeric sequence at one end of the nanochromosome precursor and a region ~250–350 bp inward from the other end resulted in a failure to undergo fragmentation/telomerization. Both of these regions were found to contain the core E-CBS motif, and mutating the motif that exists in close proximity to the 3′ end of the nanochromosome precursor abolished proper processing. The 5′-TTGAA-3′ motif can be found within the first few hundred subtelomeric bases of many S. lemnae nanochromosomes. However, because the motif is AT-rich, which is also a characteristic of the subtelomeric regions and because it is not at a defined position relative to the telomere, its significance in directing chromosome fragmentation remains unclear.
Other analyses have identified unusual global sequence features of the subtelomeric regions of stichotrich nanochromosomes that could potentially play a role in specifying fragmentation and/or telomerization. Prescott and Dizick (148) initially noted a purine versus pyrimidine strand asymmetry within the first 50 bp of the subtelomeric regions of nanochromosomes in Oxytricha and Stylonychia. Specifically, the bases adjacent to the 5′ ends of the telomeres have an abundance of purines, and there were also indications of some periodicity in purine richness. Intriguingly, these features were not seen in the ends of Euplotes nanochromosomes. Subsequent studies examining a larger sample of sequenced O. trifallax nanochromosomes (149), as well as the analyses of the nanochromosomes obtained in the MAC genome sequencing projects of S. lemnae (123) and Oxytricha trifallax (127), have supported the purine richness of the first 50 bp of the subtelomeric regions of 5′ ends. These studies also show a 10-bp periodicity in the AG-richness of the subtelomeric regions. These sequence features have been suggested to either aid in the binding of proteins that foster or carry out chromosome fragmentation or perhaps alter chromatin structure as a way of identifying regions targeted for fragmentation and telomere addition.
A potential role for RNA in directing chromosome fragmentation in Oxytricha and Stylonychia
As noted in section “Epigenetic control of DNA elimination by non-coding RNAs and heterochromatin” above, small RNAs play a role in IES removal during ciliate MAC development, and research has suggested that development-specific RNA molecules also may be involved in chromosome fragmentation and/or telomere addition in stichotrichs. Multiple types of RNAs appear to be involved in MAC development in Oxytricha and Stylonychia [reviewed in references (12, 14)]. First, in contrast to Paramecium and Tetrahymena, small RNAs (piRNAs) are generated from the parental MAC and are thought to migrate to the developing MAC where they interact with homologous DNA sequences “tagging” them for retention during development. Second, nanochromosome-length RNAs that include telomeric repeats are also synthesized in the parental MAC and subsequently play a role in “templating” the rearrangement of nanochromosome precursors, particularly the complex unscrambling of MDSs necessary to form many stichotrich nanochromosomes [(67); Fig. 9]. These nanochromosome-length transcripts, which derive from both DNA strands and appear to be synthesized by RNA polymerase II (150), may also have a role in chromosome fragmentation and/or telomerization. In both O. trifallax (151) and S. lemnae (152), injection of RNAs representing full-length nanochromosomes with telomeres into cells undergoing development resulted in a 2- to 12-fold increase in the corresponding nanochromosome copy number in offspring. This increase in copy number persists in subsequent generations (151, 153) indicating that a transgenerational epigenetic mechanism is involved. Conversely, RNA interference was used with the intention of targeting the endogenous full-length transcripts, and this resulted in decreased copy number of the cognate nanochromosomes in offspring. Two models have been suggested to explain the increase in nanochromosome copy number following developmental injection of full-length RNAs. First, the full-length RNAs may increase the number of DNA strands that are correctly fragmented in the developing nucleus, providing additional copies for amplification during the subsequent stages of MAC development. Second, an increase in the nanochromosome RNA may lead to more efficient (i.e., earlier) processing of the nanochromosome precursors, giving them a head start in the amplification process. Finally, more recent studies (153), discussed in greater detail below, have provided some evidence that the telomeric repeats in the nanochromosomal RNAs may serve as the templates for de novo telomere formation.
Fig 9.
Telomere-to-telomere RNA aids in unscrambling and telomerization. A MAC DNA molecule formed from three MDSs (green rectangles) in the MIC (1, 2, and 3), two of which are scrambled (2 and 3), produces a telomere-to-telomere transcript (red line) in the maternal MAC. The transcript is transferred to the developing MAC where it provides a template for unscrambling the MDSs and for IES (orange lines) removal. The RNA may also play a role in defining chromosome fragmentation and providing a template for the de novo synthesis of telomeres (T).
DE NOVO TELOMERE ADDITION
Developmental chromosome fragmentation is almost certainly dependent on an efficient de novo telomere addition process, as telomeres would both prevent deleterious DNA rearrangements involving the newly generated ends, as well as allow for the complete replication of MAC chromosome ends during subsequent cycles of asexual reproduction. As in most other organisms, ciliate telomeres [reviewed in reference (154)] consist of short sequence repeats with a pronounced G-C base strand bias. In Tetrahymena, the telomere repeat unit is 5′-CCCCAA-3′/3′-GGGGTT-5′, while in Paramecium, the repeats are 5′-CCC[C/A]AA-3′/3’GGG[G/T]TT-5′. The repeats in spirotrichs are 5′-CCCCAAAA-3′/3′-GGGGTTTT-5′.
The enzyme telomerase [reviewed in references (154, 155)] is responsible for synthesizing telomere repeats onto the ends of existing telomeres, thereby maintaining chromosome and telomere length during asexual reproduction. Telomerase is a multi-subunit complex, with two core catalytic subunits: a reverse transcriptase-like protein subunit called TERT (telomerase reverse transcriptase) and an RNA component termed TERC (telomerase RNA component). Telomerase RNAs have a region complementary to the telomeric repeat sequence that binds to existing repeats at the 3′ ends (i.e., the G-rich strand) of telomeres and then provides the template for the synthesis of new repeats.
In almost all cases, the DNA ends that arise as the result of developmental chromosome fragmentation lack pre-existing telomeric repeats or even partial repeats [the Tetrahymena ribosomal DNA is one notable exception; (156)]. As a result, telomeric repeats must be added de novo to the termini of nascent MAC chromosomes during the development of a new MAC. Given that telomerase typically requires pre-existing telomeric repeats to interact with the RNA component of telomerase, de novo telomere addition by telomerase to the newly fragmented ends arising during MAC development would seem to pose a problem. As with chromosome fragmentation, current data indicate that different ciliate groups may have arrived at different solutions for de novo telomere formation, but the results provide indications that some form of telomerase is involved.
De novo telomere addition in Paramecium
Telomerase and de novo telomere formation have received little attention in Paramecium, but key experiments strongly suggest that de novo telomere addition might be performed by the same telomerase that functions during vegetative growth. Studies involving the introduction/transformation of exogenous DNA into vegetative Paramecium have demonstrated that essentially any introduced DNA can be converted into a linear molecule capped by telomeres and maintained as an autonomously replicating mini-chromosome [e.g., references (157–159)]. This suggests that the same telomerase present in asexually reproducing cells might well be sufficient for de novo telomere addition to the DNA ends generated during MAC development. In fact, sequencing of the MAC genome of Paramecium has revealed only one TERT gene (102, 160). It remains possible that some modification or enhancement of the vegetative telomerase activity might be necessary to efficiently deal with the abundance of newly fragmented ends generated in the developing MAC or that fragmentation somehow “marks” newly generated ends in a way that fosters association with telomerase. Transcripts from the TERT gene do increase from two- to threefold during sexual reproduction (107), but the levels of TERT protein, as well as telomerase RNA, have not been evaluated.
De novo telomere addition in Tetrahymena
Perhaps the strongest evidence that components of the vegetative telomerase play a role in the formation of new telomeres comes from work on Tetrahymena. Yu and Blackburn (161) introduced a modified TERC gene into vegetative cells that was designed to template the addition of mutant telomeric repeats (e.g., G5T2 repeats) as opposed to the standard telomeric repeats (G4T2 repeats). When such cells were mated, the resulting offspring contained MAC chromosomes with the variant telomere sequence at, or near, the sites of de novo telomeric repeat addition, providing a clear indication that the telomerase RNA present in vegetative cells prior to mating was also present in the developing MAC where it contributed to the de novo telomeric repeat addition process.
In vitro studies also have provided evidence for the involvement of the telomerase protein complex in de novo telomere formation. Wang and Blackburn (162, 163) utilized partially purified telomerase preparations made from either vegetative cells or cells undergoing MAC development and were able to identify reaction conditions that allowed efficient addition of telomeric repeats onto single-stranded oligonucleotides that represent the ends of natural sites of chromosome fragmentation (i.e., ends lacking any similarity to a telomeric repeat). Double-stranded oligonucleotides could also serve as a substrate for de novo telomere addition, but only if they had a single-stranded 3′ overhang of at least 20 bases and a 3′-OH group. The in vivo substrate for de novo telomere addition in Tetrahymena has not been identified but presumably is similar in structure. In regard to telomerase, no biochemical differences were detected in comparing the vegetative and developing cell telomerase complexes using a number of methods (163), suggesting the absence of development-specific factors that modify its activity.
Intriguingly, the Cbs element appears to play a role in specifying de novo telomere formation. In studies involving transformation of constructs with a Cbs into cells undergoing MAC development, the DNA is both fragmented and undergoes de novo telomere repeat addition (83–85). In contrast, de novo telomere addition did not occur when a construct lacking the Cbs was linearized using a restriction enzyme and introduced into cells (85). Thus, in contrast to Paramecium, a double-stranded break per se does not appear to be sufficient for telomere addition, leading to the conclusion that the Cbs is also a necessary and sufficient signal for de novo telomere addition in the Tetrahymena thermophila developing MAC. How this might be accomplished mechanistically remains unclear, given that the in vivo sites of de novo telomere addition are at a distance from the Cbs (see Fig. 4) such that the Cbs is likely not present on the same DNA strand at the time of telomere synthesis. It is possible that the Cbs is responsible not only for specifying fragmentation but also is responsible for loading a factor onto the flanking DNA that persists following fragmentation and marks the ends for de novo telomere synthesis.
De novo telomere addition in Euplotes
In spirotrichs, the telomeres in asexually reproducing cells are unusual, in that they are quite short and of uniform length. In Euplotes species, telomeres contain a 28-bp double-stranded region of repeats (5′-CCCCAAAA-3′; C4A4 repeats) and a terminal 3′ overhang of 14 bp as illustrated below:
5′-CCCCAAAACCCCAAAACCCCAAAACCCC…3′
3′-GGTTTTGGGGTTTTGGGGTTTTGGGGTTTTGGGGTTTTGGGG…5′
In contrast, the telomeres found on the nanochromosomes in the Euplotes developing MAC during the period immediately following chromosome fragmentation are longer (164, 165) and more variable in size (166), with the most common length of the double-stranded region being 84 bp and a single-stranded 3′ tail of about 10 bp. These long or extended telomeres persist until the late stages of MAC development when they are shortened to the size seen in the mature, vegetative MAC (167). The shortening of the over-sized telomeres occurs even when DNA replication is inhibited, arguing that it represents an active trimming process as opposed to simply loss of telomeric repeats during DNA replication. The function, if any, of these longer telomeres is unknown.
It is likely that the same telomerase RNA that is used in vegetative cells is employed in the de novo addition of telomeres. Price et al. (168) have found that the vegetative-type telomerase RNA is also found in the developing MAC but at levels 12–13 times higher than that seen in vegetative cells. These higher levels presumably are required to efficiently add telomeres to the many DNA ends generated by programmed chromosome fragmentation.
There are indications that a specialized form of telomerase is utilized during Euplotes MAC development (169, 170). Protein extracts from cells undergoing MAC development were found to catalyze the addition of telomeric repeats onto the 3′ ends of single-stranded DNA oligonucleotide substrates lacking telomeric repeats. Similar extracts from vegetative cells could only add telomeres onto substrates that terminate with telomeric repeats. The results of these studies also indicate that telomerase exists in a larger ribonucleoprotein complex in developing cells as compared to vegetative cells, leading to the suggestion that specific factors must associate with the telomerase complex to convert it to a form capable of catalyzing telomeres de novo.
One such change may be in the TERT component itself. Karamysheva et al. (171) found that E. crassus contains at least three TERT genes in its MIC genome. EcTERT-1 and EcTERT-3 are also contained on MAC chromosomes and are expressed in vegetative and developing cells. In contrast, EcTERT-2 is not present in the MAC and is expressed only during MAC development beginning during the period of chromosome fragmentation. EcTert-2 differs significantly in its predicted sequence from the other two EcTERT proteins, which led to the suggestion that it might be the key to the developmental change in telomerase’s de novo telomere addition activity (171).
Finally, the presence of an E-CBS adjacent to a fragmented chromosome end does not appear to be a requirement for de novo telomere addition. As discussed in section “Positioning of the E-CBS suggests a model for chromosome fragmentation in Euplotes,” in the MIC genome, the E-CBS can reside either near the end within the precursor of a MAC chromosome or in the flanking MIC DNA (see Fig. 7). That is, there are MAC chromosome ends that undergo de novo telomere addition even when they lack the E-CBS. Indeed, Mollenbeck and Klobutcher (132) observed that telomeric repeats are added to the ends of the flanking MIC DNA that are subsequently eliminated later in MAC development. This result suggests that telomerase acts on all ends generated by chromosome fragmentation and that addition of a telomere alone is not sufficient for stabilization and subsequent retention of a DNA fragment. It was suggested instead that retained versus eliminated sequences might be discriminated by differences in chromatin structure, as such differences during development have been well documented in both Tetrahymena and Paramecium (see section “Internal DNA elimination”) and are likely to exist in Euplotes as well (172–174).
A possible role for long RNAs in templating de novo telomere addition in stichotrichs
As discussed in section “A potential role for RNA in directing chromosome fragmentation in Oxytricha and Stylonychia,” nanochromosome-length RNAs are thought to be produced by the old MAC during its development in Oxytricha and Stylonychia, and then transferred to the developing MAC where they serve as a guide for both unscrambling and proper chromosome fragmentation (Fig. 9). Injection of synthetic nanochromosome-length RNAs, including terminal telomeric repeats, into cells increases the copy number of the corresponding MAC DNA molecule (151, 152), presumably by enhancing the efficiency of fragmentation. In contrast, injection of nanochromosome-length RNAs that lack telomeric repeats at their ends, or which had inverted telomeric repeats, had no effect on copy number, leading to the suggestion that the long RNAs with telomeres might be needed for de novo telomere addition, perhaps by providing a template for repeat synthesis (153). To test this hypothesis, cells were injected with nanochromosome-length RNAs in which two adenine residues in the telomeric repeat were replaced with thymidines (153). For one of the genes tested, increased copy numbers of the corresponding gene were seen, and a small fraction of injected cells had incorporated the altered bases into their telomeres. The results, however, were complex, as many of the injected cells had telomeres that were abnormal in other ways or alterations in the subtelomeric regions of the MAC chromosome. Thus, the templating of telomeric repeats by nanochromosome-length RNAs is an intriguing possibility in stichotrich ciliates but one that requires additional investigation.
Open questions regarding de novo telomere addition
Results from all the ciliate systems strongly support the notion that at least some components of the vegetative telomerase are essential for de novo telomere formation during MAC development. Indeed, the results from studies on Paramecium and Tetrahymena can be interpreted as indicating that the vegetative cell telomerase may be sufficient for de novo telomere addition. It is hard to rule out the need for other factors that might enhance the efficiency of de novo telomere synthesis, particularly factors that interact with the DNA ends generated by fragmentation. In Euplotes, there are strong indications that at least some of the protein components of telomerase are different during development, and, as discussed above, there may be a role for long, nanochromosome-length RNAs in templating telomeric repeats in ciliates such as Stylonychia. Further biochemical analyses of developmental telomerase are needed to identify if components of the enzyme are altered, added, or replaced. In addition, many of the studies of developmental telomere addition have been carried out in vitro. Additional evidence for the involvement of proposed components of the developmental telomerase in the form of genetic knockout analyses is desirable.
Relative levels of telomerase enzyme components versus components of the DNA repair pathways might also play a role in augmenting de novo telomere formation. Kinzig et al. (175) have found that overexpression of telomerase in human cells increases the frequency of de novo telomere formation at artificially induced chromosome breaks. As noted above, there is evidence that the level of telomerase RNA is significantly increased during development in Euplotes (168), but it is not known if there is a corresponding increase in other components of the enzyme, particularly the telomerase reverse transcriptase component. Telomerase levels in other ciliates also need to be assessed. Conversely, a decrease in components of the non-homologous end joining and homologous recombination repair pathways during MAC development, or downregulation of their activities, might foster de novo telomere formation, and this needs to be assessed.
Another significant question is the nature of the DNA ends to which telomeres are added. To date, Euplotes crassus is the only ciliate where developmentally fragmented DNA intermediates lacking telomeres have been identified (138). One would like to know the nature of the DNA cleavage catalyzed by putative nucleases in other ciliates, as well as the nature of subsequent nuclease processing, if any, of the cleaved ends. Such information on the natural substrate(s) is required to design in vitro systems that reflect the in vivo situation. A related question is whether there are specific proteins that associate with the DNA ends that facilitate recognition by telomerase. This could potentially include components of the chromosome fragmentation machinery. Finally, it will be of interest to determine if the longer telomeres observed during development in Euplotes are also seen in other ciliates. What function the longer telomeres might serve, as well as how they are subsequently processed to the mature length, remain open questions.
DISCUSSION
Evolutionary innovations of the ciliate “way of life”
A number of authors have speculated on factors that may have driven the formation of the ciliate MAC and the fragmentation of chromosomes during its development [e.g., references (6, 176–179)]. In this first section of the Discussion, we seek to summarize these ideas.
Evolutionary selection for bigger ciliate cells
Ciliates are non-photosynthetic unicellular eukaryotes whose cell lengths span a range of approximately two orders of magnitude (~20–~ 2,000 μm) among different species. The majority of species meet their nutritional needs by engulfing and metabolizing other microbial species, including bacteria and other microbial eukaryotes. Accordingly, it has been suggested that a selection for larger cell sizes existed, and ciliates evolved cell volumes that now span approximately six orders of magnitude, with ciliate cells from species in the upper size range visible to the naked eye. Despite the evolution of major size differences among species, the number of distinct genes in ciliate genomes is similar. Likely, this is because ciliate cells are basically built by repetition of structural units, organized around a cilium, that share similar numbers and types of components. Thus, the main evolutionary challenge of evolving ciliate species with larger and larger cells likely was synthesizing greater amounts of structural proteins, needed in stoichiometric amounts, as quickly as possible. This challenge has been answered through a series of striking major evolutionary adaptations, particularly in the MAC.
Evolution of polyploid MACs
Ciliate nuclear dualism may well have been a key step in allowing for increased gene copy number and, thus, increased levels of protein products. The evolution and elaboration of a MAC allowed for a series of genomic changes that would likely be incompatible with a typical eukaryotic nucleus. That is, the development of a MAC allowed for significant experimentation in genome structure to optimize vegetative reproduction, while still maintaining a nucleus (the MIC) that allows for the orderly distribution of the germline genome. Simply increasing the number of nuclei in a cell is one straightforward way of uniformly increasing gene copy number. An early branching group of ciliates, the karyorelictids, appear to have done this by having the “MIC” replicating and dividing multiple times prior to asexual cell division [see reference (180) and references therein]. Some of the daughter nuclei become non-dividing “MACs,” often with a genome size similar to the “MIC,” effectively increasing the number of genomes per cell.
Most other ciliates utilize sexual reproduction as a means of generating a MAC (see Fig. 1), which is also capable of dividing during asexual reproduction. A key MAC innovation in this case was the development of polyploidy to increase the number of gene copies in a cell. MAC ploidy among different currently living ciliate species varies over approximately three orders of magnitude, from tens to tens of thousands, and there is a rough correlation between MAC DNA content/MAC volume/ploidy level and the volume of the cell [e.g., reference (181)].
Evolution of developmentally programmed fragmentation of ciliate MAC chromosomes
Early in ciliate evolution, a MAC generated during the course of sexual reproduction that had a relatively low ploidy level would likely have been able to rely on the standard form of mitosis for distributing chromosomes during subsequent vegetative cell divisions. However, as the number of copies of complete chromosome sets in the MAC increased, it likely would have become increasingly difficult to efficiently disentangle the large number of chromatids and faithfully distribute them to daughter cells using a conventional mitotic mechanism (176, 177). Fragmenting the MAC chromosomes could well have been a common ciliate solution to this problem, which subsequently enabled ciliates to further respond to the selective forces for larger MAC genomes and larger cell size.
Shortening of chromosomes through fragmentation would have required a number of innovations in the MAC. For example, one consequence of the evolution of MAC chromosome fragmentation is that all MAC chromosome fragments, except for the one still containing the centromere, become physically unlinked from a centromere. Thus, conventional mitosis would have been insufficient to distribute the acentric chromosomes to daughter nuclei in an organized manner. However, if the MAC ploidy levels were already sufficiently high, random distribution of the small chromosomes passively at karyokinesis would have been sufficient to give a reasonable probability of each daughter cell receiving at least one copy of each gene, ultimately leading to the amitotic MAC division process. In the absence of copy number control mechanisms, continued divisions would ultimately lead to imbalances or complete loss of some genes, but this also would have provided an additional selection for increasing the overall gene copy number (6, 176, 177).
The current program of MAC development may be an example of “ontogeny recapitulates phylogeny” in regard to providing support for the proposal that the early evolution of the MAC involved some polyploidy prior to the origin of chromosome fragmentation. All characterized ciliates undergo some level of genome endoreplication/polyploidization during MAC development prior to the chromosome fragmentation step. For example, in Tetrahymena, the ploidy of the developing MAC increases to 8C before programmed chromosome breakage begins (182). In the spirotrich species, the level of polytenization prior to chromosome fragmentation is even more dramatic, with many species producing polytene chromosomes similar to those seen in Drosophila salivary glands [reviewed in reference (129)]. For example, Stylonychia lemnae has banded polytene chromosomes in the developing MAC with a ploidy level of 64C (183) and subsequently undergoes additional rounds of DNA replication following fragmentation to give rise to a MAC with thousands of copies of each gene. Similarly, the Paramecium tetraurelia developing MAC is endoreplicated to a level of 64C at the onset of chromosome fragmentation, as indicated by the increased accumulation of new telomeric ends (120).
Other innovations in a MAC with fragmented chromosomes
A number of additional innovations in the MAC were likely needed to allow for developmental chromosome fragmentation, while other features of the current MAC may have evolved as a result of fragmenting the chromosomes. First, a system to stabilize newly generated ends by adding telomeres to them would have been required. As discussed in section “De novo telomere addition,” Paramecium appears to have a system for efficiently adding telomeres to free DNA ends even in vegetative cells, while organisms such as Euplotes appear to have developed modified forms of telomerase that foster de novo telomere addition during MAC development.
Second, some ciliate lineages may have had to evolve modifications to the DNA replication origin mechanism. As we will discuss in greater detail below, organisms such as Tetrahymena, which have relatively large MAC chromosomes, may utilize the same replication origins that function in the MIC. The spirotrich group of ciliates, however, pose a problem in regard to retaining use of the MIC replication origins. Given the small size of the average MAC nanochromosome (~2 kbp), it is hard to envision how each nanochromosome could have maintained a MIC origin of replication, since origins on eukaryotic chromosomes tend to be separated by on the order of 50 kbp on average [see reference (184)]. Thus, as suggested by Herrick (176), it seems likely that some innovation in DNA replication was also required to allow for the high degree of genome fragmentation in spirotrich ciliates. What constitutes an origin of replication for a nanochromosome is unknown, but a number of unusual features of spirotrichs suggest that a specialized mechanism of replication exists. First, DNA replication in spirotrichs is associated with a cytologically visible “replication band” that moves across the MAC, replicating DNA molecules it encounters (185, 186). Second, electron microscopic analyses suggest that DNA replication is initiated near one or both ends of the nanochromosomes (187). Moreover, a telomeric DNA primase activity has been detected (188), leading to a model where the telomeres themselves serve as origins of replication. Perhaps, the extremely short spirotrich telomeres of defined length are somehow relevant in this regard.
Little is known concerning origins of DNA replication in Paramecium, but it is possible that telomeres might also function as origins. As we have discussed, almost any DNA segment introduced into Paramecium cells undergoes de novo telomere addition to generate a mini-chromosome. Since these mini-chromosomes are capable of replication, and it seems unlikely that all such DNA segments would contain an internal functional origin, perhaps, telomeres can also initiate DNA replication in this species. That said, the large size of the natural MAC chromosomes in Paramecium strongly suggests that they also contain internal origins.
Finally, extensive programmed chromosome fragmentation could not have evolved unless all MAC components of the mitosis mechanism (e.g., centromeres, mitotic spindles, etc.) had become useless. This in turn would have made it possible to delete from the developing MAC, without penalty, all silenced transposon-related DNA—centromere DNA included. The ciliate innovation of physically excising IESs/repetitive elements during MAC development, as opposed to simply silencing such elements through small RNA-directed chromatin modification as is typical of other eukaryotes, would have resulted in the loss of the centromeric regions. More broadly, IES/repetitive element excision may also have had other fitness benefits for ciliates. Replicating a highly polyploid genome involves a large energy investment, including the replication of a substantial amount of non-genic DNA. Removal of repetitive DNA sequences during MAC development likely would have shortened S-phase for the vegetative MAC, providing a growth advantage for the cell and a possible opportunity to develop even higher gene copy levels.
Origin(s) of ciliate chromosome fragmentation
Based on our descriptions of the chromosome fragmentation systems in Tetrahymena, Paramecium, Euplotes, Oxytricha, and Stylonychia, major differences have clearly evolved among these ciliates. It seems very possible that chromosome fragmentation systems have originated and/or been modified multiple times within ciliate groups (138, 176, 177). In support of this hypothesis, highly fragmented MAC genomes (e.g., the nanochromosomes of spirotrichs) appear to have arisen independently in three separate ciliate clades (7). The development of fragmentation systems with diverse features would not be surprising, as the evolutionary distances between ciliate groups are large, with the major groups of ciliates having diverged on the order of a billion years ago [e.g., reference (4)]. Even Tetrahymena and Paramecium, which are both Oligohymenophorans, but which have very different fragmentation systems, are estimated to have diverged hundreds of millions of years ago (87).
While the evolutionary pathway(s) leading to developmental chromosome fragmentation in ciliates remains uncertain, we are struck by a number of observations suggesting the involvement of transposable elements in a number of the lineages [e.g., references (18, 83, 138, 189, 190)]. First, the large and well-conserved Tetrahymena Cbs is generally reminiscent of the larger sequence elements that are involved in directing site-specific recombination, including the sequences at the termini of transposons that are recognized by transposases, the sequence elements recognized by the resolvases of replicative transposons, or the sequences recognized by homing endonucleases associated with mobile introns [reviewed in reference (191)]. Second, the observed loss/gain of Cbss during the evolution of Tetrahymenids is suggestive of the movement of transposons in the genome (i.e., insertion and excision). That is, a mobile element carrying the fragmentation signal provides a reasonable explanation for the origin of new fragmentation sites in a genome as well as the loss of such sites. Comparisons of the MAC genomes of Oxytricha and Stylonychia similarly suggest the loss/gain of fragmentation sites (123, 127). Third, the observed association of MIC transposons and repetitive elements with fragmentation sites in Paramecium (see section “Chromosome fragmentation in Paramecium is associated with heterogeneous DNA elimination”) suggests the involvement of these elements in specifying locations of fragmentation. It has also been noted that the combined processes of chromosome fragmentation and de novo telomere addition resemble the target-primed reverse transcription mechanism used for transposition by some non-LTR retrotransposons [see reference (138)]. Finally, as discussed in sections “Internal DNA elimination” and “Epigenetic control of DNA elimination by non-coding RNAs and heterochromatin,” there is strong evidence that one of the other main DNA rearrangement processes of ciliate MAC development, IES excision/splicing, has derived from transposons. The unusual dual nuclear system of ciliates provides some advantages for transposons (192). That is, the primarily transcriptionally silent MIC provides a safe haven for transposable elements, while the development of a MAC presents a means of ridding the functional genome of such elements. Thus, it is conceivable that a transposon(s) might also have been involved in generating a chromosome fragmentation system(s).
Despite the multiple and strong indications of transposon involvement, it remains difficult to propose a single scheme that explains the origin of MAC chromosome fragmentation in all species. As a result, we wish to propose two broad scenarios that we believe are consistent with the varying features of chromosome fragmentation in the species discussed (Fig. 10). In brief, we envision an ancestral fragmentation system in ciliates that is based on the process of ridding the genome of repetitive DNA elements such as transposons and does not rely on well-defined cis-acting sequence elements. Second, we propose that secondary invasions by transposons have occurred that have supplanted the ancestral chromosome fragmentation system, giving rise to systems with well-defined “CBSs” (”CBS” with all capital letters and in parentheses is intended to generically indicate a cis-acting sequence that specifies chromosome fragmentation).
Fig 10.
Summary of proposed two-stage model for the evolution of ciliate chromosome fragmentation systems. See text for details. Relative evolutionary relationships of species are shown, but evolutionary distances are not drawn to scale.
Proposed ancestral mode of chromosome fragmentation
Some ciliates, such as Tetrahymena and Euplotes, have well-conserved “CBS” in the vicinity of fragmentation sites. In contrast, Paramecium, Oxytricha, Stylonychia, and other ciliates have no apparent well-conserved “CBS.” As discussed above (section “Sequence features adjacent to Oxytricha and Stylonychia sites of chromosome fragmentation”), it is still possible that a “CBS” exists in some of these species but that it is short or less well conserved so that its identification has been obscured by the variability in the position of telomere addition. Further analyses of the dynamics of chromosome fragmentation/de novo telomere addition in these species may help in identifying a “CBS” if it exists. For example, if there is nucleolytic degradation of ends prior to telomere addition, then identifying and characterizing the initial site of chromosome cleavage might aid in identifying a nearby conserved sequence element.
It is also possible that these species employ an alternate mode of defining fragmentation sites that does not depend on a well-conserved “CBS.” It has been suggested that chromatin structure might instead be involved in defining chromosome fragmentation/telomere addition sites [e.g., references (61, 132)]. As discussed in section “Epigenetic control of DNA elimination by non-coding RNAs and heterochromatin,” chromatin modifications specified by small RNAs have been documented in a number of ciliates. Indeed, such systems are ubiquitous in eukaryotes (193), suggesting their origin in a common eukaryotic ancestor, and Zufall et al. (194) have suggested that such systems form the basis for the development of numerous genome rearrangement systems in eukaryotes, including ciliates. While chromatin modification is a common method used to silence repetitive elements in genomes, ciliates evolved the ability to rid the MAC genome of such elements by excision and rejoining of flanking sequences.
It is also possible that some specific chromatin modifications specify sites of fragmentation. That is, chromatin modifications might protect sequences destined for the MAC from interacting with a nuclease(s) that carries out fragmentation of the MIC chromosomes such that cleavage occurs only at the boundaries of these protected chromatin domains. Conversely, specific chromatin modifications might designate specific regions for recognition by a fragmentation nuclease. This idea is attractive because the boundaries of chromatin domains are often not precisely defined so that a chromatin-based system could provide an explanation for the variability in the position of telomere addition sites that is seen in some species. That is, variability in the position of the last modified nucleosome would either result in variable sites of chromosome fragmentation or provide the boundary for nucleolytic degradation up to the positions where telomeres are ultimately added.
We suggest that this type of chromatin-based system of chromosome fragmentation evolved in conjunction with, or is derived from, the development of the IES/repetitive element excision system. Specifically, as discussed in Sections “Chromosome fragmentation in Paramecium is associated with heterogeneous DNA elimination” and “Epigenetic control of chromosome fragmentation and experimental induction of heritable variant patterns,” chromosome fragmentation/telomere addition in Paramecium occurs at sites containing interstitial DNA elements (transposons or repetitive elements) that can also be excised with rejoining of the flanking sequences. These elements likely assume a modified chromatin structure via interaction with scnRNAs (58, 59). In this scenario, chromosome fragmentation is simply an alternative outcome for the removal of an interstitial DNA segment where breakage occurs, but there is no rejoining of the chromosome ends (36, 106). These alternative outcomes of element excision are consistent with the nested sets of MAC chromosomes observed in Paramecium (e.g., see Fig. 6).
The notion of something akin to the Paramecium system reflecting the ancestral state is attractive, in that it potentially aids in some of the evolutionary transitions discussed above. First, early on, there may have only been an occasional failure to rejoin ends generated during the removal of repetitive elements. Nonetheless, this failure would have created a selection for an alternative outcome that avoided the possible negative consequences of unrepaired chromosome ends. That is, there would have been a selection to develop or optimize the stabilization of free ends by de novo telomere addition. In turn, once optimized, de novo telomere addition would make it possible for more sites of chromosome breakage to be tolerated, ultimately fostering the development of MACs with smaller chromosomes and higher ploidy levels. As discussed in section ‘Evolutionary innovations of the ciliate “way of life,”’ larger cell size was a likely driver of both fragmented chromosomes and size reduction of the MAC genome via IES/repetitive sequence excision.
Ciliates such as Oxytricha and Stylonychia, where no clear “CBS” has been identified, might represent slight variations on this proposed small RNA/chromatin modification pathway of chromosome fragmentation. Fragmentation is much more frequent in these species, but repetitive sequences are not clearly associated with sites of fragmentation. Nonetheless, the small RNA pathway appears to have diverged in this group, as small RNAs associated with genome rearrangement are produced from the old MAC and correspond to sequences that will be maintained in the MAC [i.e., MDSs; (64, 65)]. Chromatin modification in this instance might serve to roughly define the boundaries of retained sequences, resulting in the observed variability in telomere addition sites.
Finally, it is intriguing that this proposed ciliate ancestral system shares some similarity with the other known group of organisms where developmental chromosome fragmentation has been shown to be coupled with de novo telomere formation. In Ascaris, heterochromatic repetitive DNA sequences are lost from both the internal and terminal regions of chromosomes in conjunction with de novo telomere addition during development (3). The loss of the heterochromatic repetitive sequences, however, occurs only via chromosome fragmentation in Ascaris not via chromosome breakage and rejoining.
Secondary fragmentation systems derived from transposable elements
Other ciliates appear to differ from the proposed ancestral system, in that they utilize well-defined “CBSs” (Tetrahymena and Euplotes) and/or undergo precise chromosome fragmentation/telomerization (Euplotes). We suggest that these organisms represent cases where secondary chromosome fragmentation systems, which derive from transposable elements, have supplanted the proposed ancestral small RNA/chromatin modification system (Fig. 10). More specifically, once a system had been established that efficiently heals chromosome breaks and allows for the replication and distribution of mini-chromosomes, the stage was set for its occasional replacement by an alternative system(s) of fragmenting the chromosomes during MAC development. We note that there is evidence indicating that the current Tetrahymena Cbs-based system is of relatively recent origin. As discussed in section “Chromosome fragmentation in Tetrahymena,” there are five positions within the 15-base Tetrahymena Cbs where sequence variation is tolerated (also see Table S1). However, if one considers the entire repertoire of Cbss, these variable positions in the Cbs do not appear to have reached mutational equilibrium, as would be expected for an ancient system. Thus, it is possible that Oligohymenopherans had an initial fragmentation system similar to the ancestral ciliate system described in the preceding section. In our view, the ancestral system persists in Paramecium but was replaced in the Tetrahymenid lineage by the current Cbs-based system (18).
The scheme we envision for replacement by a secondary fragmentation system is quite similar to what has been proposed for the evolution of the gene rearrangement process (V(D)J recombination) that produces large numbers of antibodies and T-cell receptor genes in the vertebrate immune system. V(D)J recombination is thought to derive from a “cut and paste” transposon [reviewed in references (195, 196)]. Cut-and-paste transposons are typically flanked by TIRs required for transposition and contain a gene encoding a transposase that initiates transposition by recognizing the TIRs to generate double-stranded breaks that release the transposon from the host chromosome so that it may subsequently integrate into a new site in the genome. V(D)J recombination appears to have arisen from a cut-and-paste element related to the Transib transposons found in vertebrates and the ProtoRAG element in Amphioxus [see references (195, 197)]. The transposase genes of these types of elements were likely domesticated to form the vertebrate RAG1 and RAG2 genes, which specify the proteins that catalyze the assembly of the antibody and T-cell receptor gene segments, while the TIRs of the element have been co-opted as the recombination signal sequences that flank immune gene segments and serve as the cis-acting signals that guide rearrangement.
A situation similar to the origin and establishment of V(D)J recombination can be envisioned for chromosome fragmentation in ciliates (Fig. 11), and the proposed model also shares a number of similarities to what was proposed for the origin of ciliate IESs (35). The initial step is for a cut-and-paste transposon (transposon X) to have invaded the germline (MIC) genome of one or more ciliate ancestors. In ciliates, transposon X would also likely be transcriptionally activated during MAC development such that the transposase protein was produced, leading to breaks at the ends of transposon copies throughout the genome. In eukaryotes, the double-stranded breaks generated during transposon excision are generally repaired either by homologous recombination using the homologous allele as a template or by non-homologous end joining. Typically, unrepaired breaks would lead to a selective force favoring the silencing or suppression of such transposons. However, in a ciliate with the ancestral chromosome fragmentation system in place, this selective force would have been minimized because of the established de novo telomere addition process that mends free DNA ends. In effect, the ancestral fragmentation system paves the way, and lessens the impact, for the invasion of other elements that generate breaks in DNA.
Fig 11.
Transposon-based model for the origin of ”CBS”-based ciliate chromosome fragmentation. The process initiates with the invasion of the MIC genome (blue line with green ovals representing genes/coding regions) by transposon X with terminal inverted repeats (TIRs), which proliferates by transposition. Developmental activation of the transposase would produce breaks at the transposon ends in the genome during MAC development. The transposase gene (hatched rectangle) ultimately is domesticated as the developmental nuclease catalyzing breaks. Further transposition of decayed elements populates the genome with TIRs that are the current ciliate ”CBSs”.
Once such an initial transposon-based chromosome fragmentation system evolved, production of the transposase would be in the host cell’s interest, leading to its separate incorporation into the genome as a domesticated transposase/nuclease (Fig. 11). Other copies of the transposon, either intact or lacking the transposase gene, could continue to proliferate in the genome based on retaining a pair of TIRs but ultimately would be expected to decay leaving only a TIR(s), which are the current “CBSs”.
Mechanistically, cut-and-paste transposons typically require a pair of TIRs to interact with the transposase protein(s) to form a complex, which triggers DNA cleavage to start the transposition process. Formation of such a complex could still be required for fragmentation if pairs of “CBS” at the ends of MAC chromosome precursors need to interact to trigger fragmentation. Alternatively, since the developing MAC has undergone endoreplication prior to chromosome fragmentation, complex formation could also be achieved by interactions of “CBSs” on different chromatids. Lastly, it is possible that the ciliate excision machinery has evolved such that an individual “CBS” is now sufficient to be recognized for efficient DNA cleavage.
In our model, the degree of transposition during the expansion of transposon X influences the degree of chromosome fragmentation and the ultimate size of the MAC chromosomes. In organisms such as Tetrahymena, a modest amount of transposition of the transposon and/or Cbs would have resulted in relatively large MAC chromosomes containing many genes. Transposition into essential genes would clearly not be tolerated, but there also may originally have been additional constraints, including that a pair of Cbss must bracket a MIC origin or replication (176) so that this origin could continue to function in the MAC. The non-maintained chromosomes in Tetrahymena are a likely exception, as they do not contain genes essential for asexual growth, and hence, would not require an origin of replication.
In the spirotrich Euplotes, a mobile E-CBS may have saturated, or essentially saturated, the MIC genome, in the sense that almost every gene ended up being flanked by a pair of E-CBSs, so as to generate its own MAC chromosome during development. As discussed above, such a system likely required the parallel development of novel origins of replication or a novel replication system, as MIC replication origins were too infrequent to provide one for every nanochromosome in this species.
We also suggest that once a “CBS” is established in the genome, it might well propagate itself via a mechanism distinct from the original transposon X. This is suggested by the Tetrahymena Cbs clades (Table 1), in which groups of Cbss are contained within larger conserved repetitive sequence blocks that are preferentially found near centromeres and/or in consecutive arrays along the chromosome. This situation might be explained by Cbss occasionally becoming imbedded within other mobile elements in the genome. It is noteworthy that many Cbs clade members reside near centromeres (Table 1). Eukaryotic centromeres are often littered with transposons, and they could be the location where the initial association of the Cbs with novel mobile elements occurred. Irrespective of what mobile element a “CBS” becomes associated with, it would still be capable of being recognized and cut by the domesticated nuclease to initiate chromosome fragmentation.
The current data also can be interpreted to indicate that, over evolution, the function of some “CBSs” may be impaired by mutation or they may completely lose function. This would give rise to the observed alternative fragmentation seen in some ciliates, as well as the possible loss of some Cbss in tetrahymenids. There would seem to be minimal selection against the loss of a “CBS”, excluding situations where the MAC chromosomes become too large for efficient distribution or where incompatible genes come to reside on the same chromosome.
Finally, there are numerous possible variations on the model. For instance, the relatively large size and strong sequence conservation of the Tetrahymena Cbs have led to suggestions that it may have derived from the recognition site for a homing endonuclease (18, 83, 189, 190). Homing endonucleases have been identified in the Tetrahymena MIC genome. While the recognition sites for homing endonucleases are typically unique sequences and not mobile in a genome, one can envision that such a sequence could proliferate in a genome through becoming associated with a mobile element. The duplicated element sites would then be subject to fragmentation by the homing endonuclease to generate a fragmented MAC genome. As before, it would become advantageous for the homing endonuclease gene to become domesticated within the host genome.
CONCLUSION
Programmed chromosome fragmentation during macronuclear development is likely a shared feature of most, if not all, ciliates. Study of a small number of ciliate species has revealed a great deal of diversity in their chromosome fragmentation processes. This is not entirely surprising, given the ancient origin of ciliates and the large evolutionary distances among the species analyzed.
To explain this astonishing diversity, we have proposed here an evolutionary model in which an initial chromosome fragmentation/de novo telomere addition system arose in the developing MAC as an alternative way to repair chromosome breaks generated by occasional failures of the sRNA and chromatin modification-dependent mechanism that excises transposons and other repetitive elements during macronuclear development. That is, the ancestral chromosome fragmentation system evolved to support the IES/repetitive element excision process. Once established, the ancestral system is proposed to have been replaced in selected lineages by secondary systems derived from mobile elements.
We note that aspects of our model are in some ways similar to a recent proposal by Boscaro and Keeling (178) concerning the evolution of IES excision in ciliates. They propose that IES excision has behaved in an evolutionary ratchet-like manner, in that once it arose it both became essential for the cell and allowed for the subsequent expansion of the system. In the case of chromosome fragmentation, once the proposed ancestral system has been established, it allows for its replacement, and possible expansion, by components of mobile elements that have invaded the MIC genome.
Our model is clearly speculative, owing in part to the relative paucity of data on ciliate chromosome fragmentation, particularly when compared to the IES excision process. Additional details on the mechanism of fragmentation, and particularly the cellular components that carry out chromosome fragmentation, will ultimately provide a better supported and nuanced view of the origin(s) of ciliate chromosome fragmentation. Analyses of additional ciliate species are also needed, in particular, to examine whether the proposed ancestral system may still coexist in some species along with a more recently acquired “CBS”-based system. It will also be of interest to determine if the frequent fragmentation systems (i.e., MAC nanochromosomes) that have independently developed in multiple ciliate lineages (5) arose in a similar manner; in particular, whether there are indications of the involvement of transposable elements in these lineages.
The unique properties of ciliates have led to significant breakthroughs in chromosome and chromatin biology, including catalytic RNA (198) and the initial identification of a histone-modifying enzyme (199) that led to the notion of a “histone code” (200). More within the scope of this review, Greider and Blackburn (201) discovered telomerase in extracts of Tetrahymena mating cells, taking advantage of the massive chromosome fragmentation that takes place during MAC development. It is quite possible that further studies of the chromosome fragmentation/de novo telomere addition process will result in additional surprises. At a minimum, additional work will provide valuable information on the plasticity and evolution of the genomes of early diverging eukaryotes.
ACKNOWLEDGMENTS
We would like to thank Linda Sperling, Eric Meyer, and Glenn Herrick for their insightful comments on the manuscript and/or enlightening discussions. We sincerely apologize to any authors whose work we have failed to cite, as well as to the many others who have influenced our thoughts and proposals but who we have not specifically acknowledged.
Work in the Bétermier laboratory has been supported by intramural funding from the Centre National de la Recherche Scientifique (CNRS) and grants from the Agence Nationale de la Recherche (“PIGGYPACK” ANR-14-CE10-0005, “LaMarque” ANR-18-CE12-0005-02, “CURE” ANR-21-CE12-0019-01) and the Fondation pour la Recherche Médicale (FRM EQU202103012766).
Contributor Information
Lawrence A. Klobutcher, Email: klobutcher@uchc.edu.
Eduardo Orias, Email: orias@lifesci.ucsb.edu.
Melissa Bruckner Lodoen, University of California, Irvine, California, USA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mmbr.00184-22.
Table S1. Known Consensus Cbs and functional variants in T. thermophila and related species Table S2 - Proteins Required for Cbs-dependent Chromosome breakage (CB) and/or IES Excision in Tetrahymena.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Chapman JR, Taylor MRG, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47:497–510. doi: 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- 2. Putnam CD, Pennaneach V, Kolodner RD. 2004. Chromosome healing through terminal deletions generated by de novo telomere additions in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 101:13262–13267. doi: 10.1073/pnas.0405443101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Estrem B, Wang J. 2023. Programmed DNA elimination in the parasitic nematode Ascaris. PLoS Pathog 19:e1011087. doi: 10.1371/journal.ppat.1011087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parfrey LW, Lahr DJG, Knoll AH, Katz LA. 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A 108:13624–13629. doi: 10.1073/pnas.1110633108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McGrath CL, Zufall RA, Katz LA. 2007. Variation in macronuclear genome content of three ciliates with extensive chromosomal fragmentation: a preliminary analysis. J Eukaryot Microbiol 54:242–246. doi: 10.1111/j.1550-7408.2007.00257.x [DOI] [PubMed] [Google Scholar]
- 6. Morgens DW, Lindbergh KM, Adachi M, Radunskaya A, Cavalcanti ARO. 2013. A model for the evolution of extremely fragmented macronuclei in ciliates. PLoS One 8:e64997. doi: 10.1371/journal.pone.0064997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Riley JL, Katz LA. 2001. Widespread distribution of extensive chromosomal fragmentation in ciliates. Mol Biol Evol 18:1372–1377. doi: 10.1093/oxfordjournals.molbev.a003921 [DOI] [PubMed] [Google Scholar]
- 8. Singh M, Seah KBB, Emmerich C, Singh A, Huettel B, Byerly A, Stover NA, Sugiura M, Swart EC. 2021. Macronuclear genome of Blepharisma stoltei 7 strain ATCC 30299. bioRxiv 92 [Google Scholar]
- 9. Ammermann D. 1964. Riesenchromosomen in der makronukleusanlage des ciliaten Stylonychia SPEC. Naturwissenschaften 51:249–249. doi: 10.1007/BF00641378 [DOI] [Google Scholar]
- 10. Betermier M, Duharcourt S. 2014. Programmed rearrangement in ciliates: Paramecium. Microbiol Spectr 2. doi: 10.1128/microbiolspec.MDNA3-0035-2014 [DOI] [PubMed] [Google Scholar]
- 11. Noto T, Mochizuki K. 2017. Whats, hows and whys of programmed DNA elimination in Tetrahymena Open Biol 7:170172. doi: 10.1098/rsob.170172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rzeszutek I, Maurer-Alcalá XX, Nowacki M. 2020. Programmed genome rearrangements in ciliates. Cell Mol Life Sci 77:4615–4629. doi: 10.1007/s00018-020-03555-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yao M-C, Chao J-L, Cheng C-Y. 2014. Programmed genome rearrangements in Tetrahymena. Microbiol Spectr 2. doi: 10.1128/microbiolspec.MDNA3-0012-2014 [DOI] [PubMed] [Google Scholar]
- 14. Yerlici VT, Landweber LF. 2014. Programmed genome rearrangements in the ciliate Oxytricha. Microbiol Spectr 2. doi: 10.1128/microbiolspec.MDNA3-0025-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Callahan RC, Shalke G, Gorovsky MA. 1984. Developmental rearrangements associated with a single type of expressed α-tubulin gene in Tetrahymena. Cell 36:441–445. doi: 10.1016/0092-8674(84)90237-x [DOI] [PubMed] [Google Scholar]
- 16. Yao MC, Choi J, Yokoyama S, Austerberry CF, Yao CH. 1984. DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell 36:433–440. doi: 10.1016/0092-8674(84)90236-8 [DOI] [PubMed] [Google Scholar]
- 17. Klobutcher LA, Jahn CL, Prescott DM. 1984. Internal sequences are eliminated from genes during macronuclear development in the ciliated protozoan Oxytricha nova. Cell 36:1045–1055. doi: 10.1016/0092-8674(84)90054-0 [DOI] [PubMed] [Google Scholar]
- 18. Hamilton EP, Kapusta A, Huvos PE, Bidwell SL, Zafar N, Tang H, Hadjithomas M, Krishnakumar V, Badger JH, Caler EV, Russ C, Zeng Q, Fan L, Levin JZ, Shea T, Young SK, Hegarty R, Daza R, Gujja S, Wortman JR, Birren BW, Nusbaum C, Thomas J, Carey CM, Pritham EJ, Feschotte C, Noto T, Mochizuki K, Papazyan R, Taverna SD, Dear PH, Cassidy-Hanley DM, Xiong J, Miao W, Orias E, Coyne RS. 2016. Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. Elife 5:e19090. doi: 10.7554/eLife.19090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng C-Y, Vogt A, Mochizuki K, Yao M-C. 2010. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol Biol Cell 21:1753–1762. doi: 10.1091/mbc.e09-12-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vogt A, Mochizuki K, Feschotte C. 2013. A domesticated PiggyBac transposase interacts with heterochromatin and catalyzes reproducible DNA elimination in Tetrahymena. PLoS Genet 9:e1004032. doi: 10.1371/journal.pgen.1004032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin I-T, Chao J-L, Yao M-C. 2012. An essential role for the DNA breakage-repair protein Ku80 in programmed DNA rearrangements in Tetrahymena thermophila. Mol Biol Cell 23:2213–2225. doi: 10.1091/mbc.E11-11-0952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin C-Y, Chao J-L, Tsai H-K, Chalker D, Yao M-C. 2019. Setting boundaries for genome-wide heterochromatic DNA deletions through flanking inverted repeats in Tetrahymena thermophila. Nucleic Acids Res 47:5181–5192. doi: 10.1093/nar/gkz209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng C-Y, Young JM, Lin C-Y, Chao J-L, Malik HS, Yao M-C. 2016. The piggyBac Transposon-derived genes TPB1 and TPB6 mediate essential transposon-like excision during the developmental rearrangement of key genes in Tetrahymena thermophila. Genes Dev 30:2724–2736. doi: 10.1101/gad.290460.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fass JN, Joshi NA, Couvillion MT, Bowen J, Gorovsky MA, Hamilton EP, Orias E, Hong K, Coyne RS, Eisen JA, Chalker DL, Lin D, Collins K. 2011. Genome-scale analysis of programmed DNA elimination sites in Tetrahymena thermophila. G3 1:515–522. doi: 10.1534/g3.111.000927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Preer LB, Hamilton G, Preer JR. 1992. Micronuclear DNA from Paramecium tetraurelia: serotype 51 A gene has internally eliminated sequences. J Protozool 39:678–682. doi: 10.1111/j.1550-7408.1992.tb04448.x [DOI] [PubMed] [Google Scholar]
- 26. Sellis D, Guérin F, Arnaiz O, Pett W, Lerat E, Boggetto N, Krenek S, Berendonk T, Couloux A, Aury J-M, Labadie K, Malinsky S, Bhullar S, Meyer E, Sperling L, Duret L, Duharcourt S. 2021. Massive colonization of protein-coding exons by selfish genetic elements in Paramecium germline genomes. PLoS Biol 19:e3001309. doi: 10.1371/journal.pbio.3001309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Steele CJ, Barkocy-Gallagher GA, Preer LB, Preer JR. 1994. Developmentally excised sequences in micronuclear DNA of Paramecium. Proc Natl Acad Sci U S A 91:2255–2259. doi: 10.1073/pnas.91.6.2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arnaiz O, Mathy N, Baudry C, Malinsky S, Aury J-M, Denby Wilkes C, Garnier O, Labadie K, Lauderdale BE, Le Mouël A, Marmignon A, Nowacki M, Poulain J, Prajer M, Wincker P, Meyer E, Duharcourt S, Duret L, Bétermier M, Sperling L. 2012. The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet 8:e1002984. doi: 10.1371/journal.pgen.1002984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guérin F, Arnaiz O, Boggetto N, Denby Wilkes C, Meyer E, Sperling L, Duharcourt S. 2017. Flow cytometry sorting of nuclei enables the first global characterization of Paramecium germline DNA and transposable elements. BMC Genomics 18:327. doi: 10.1186/s12864-017-3713-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baudry C, Malinsky S, Restituito M, Kapusta A, Rosa S, Meyer E, Bétermier M. 2009. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev 23:2478–2483. doi: 10.1101/gad.547309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bischerour J, Bhullar S, Denby Wilkes C, Régnier V, Mathy N, Dubois E, Singh A, Swart E, Arnaiz O, Sperling L, Nowacki M, Bétermier M. 2018. Six domesticated PiggyBac transposases together carry out programmed DNA elimination in Paramecium Elife 7:e37927. doi: 10.7554/eLife.37927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abello A, Régnier V, Arnaiz O, Le Bars R, Bétermier M, Bischerour J. 2020. Functional diversification of Paramecium Ku80 paralogs safeguards genome integrity during precise programmed DNA elimination. PLoS Genet 16:e1008723. doi: 10.1371/journal.pgen.1008723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marmignon A, Bischerour J, Silve A, Fojcik C, Dubois E, Arnaiz O, Kapusta A, Malinsky S, Bétermier M. 2014. Ku-mediated coupling of DNA cleavage and repair during programmed genome rearrangements in the ciliate Paramecium tetraurelia. PLoS Genet 10:e1004552. doi: 10.1371/journal.pgen.1004552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klobutcher LA, Herrick G. 1995. Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes tec transposons. Nucleic Acids Res 23:2006–2013. doi: 10.1093/nar/23.11.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klobutcher LA, Herrick G. 1997. Developmental genome reorganization in ciliated protozoa: the transposon link. Prog Nucleic Acid Res Mol Biol 56:1–62. doi: 10.1016/s0079-6603(08)61001-6 [DOI] [PubMed] [Google Scholar]
- 36. Le Mouël A, Butler A, Caron F, Meyer E. 2003. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in paramecia. Eukaryot Cell 2:1076–1090. doi: 10.1128/EC.2.5.1076-1090.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jacobs ME, Klobutcher LA. 1996. The long and the short of developmental DNA deletion in Euplotes crassus. J Eukaryot Microbiol 43:442–452. doi: 10.1111/j.1550-7408.1996.tb04503.x [DOI] [PubMed] [Google Scholar]
- 38. Baird SE, Fino GM, Tausta SL, Klobutcher LA. 1989. Micronuclear genome organization in Euplotes crassus: a transposonlike element is removed during macronuclear development. Mol Cell Biol 9:3793–3807. doi: 10.1128/mcb.9.9.3793-3807.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jacobs ME, Sánchez-Blanco A, Katz LA, Klobutcher LA. 2003. Tec3, a new developmentally eliminated DNA element in Euplotes crassus. Eukaryot Cell 2:103–114. doi: 10.1128/EC.2.1.103-114.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krikau MF, Jahn CL. 1991. Tec2, a second transposon-like element demonstrating developmentally programmed excision in Euplotes crassus. Mol Cell Biol 11:4751–4759. doi: 10.1128/mcb.11.9.4751-4759.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jahn CL, Krikau MF, Shyman S. 1989. Developmentally coordinated en masse excision of a highly repetitive element in E. crassus. Cell 59:1009–1018. doi: 10.1016/0092-8674(89)90757-5 [DOI] [PubMed] [Google Scholar]
- 42. Tausta SL, Klobutcher LA. 1989. Detection of circular forms of eliminated DNA during macronuclear development in E. Crassus. Cell 59:1019–1026. doi: 10.1016/0092-8674(89)90758-7 [DOI] [PubMed] [Google Scholar]
- 43. Tausta SL, Turner LR, Buckley LK, Klobutcher LA. 1991. High Fidelity developmental excision of Tec1 transposons and internal eliminated sequences in Euplotes crassus. Nucleic Acids Res 19:3229–3236. doi: 10.1093/nar/19.12.3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frels JS, Tebeau CM, Doktor SZ, Jahn CL. 1996. Differential replication and DNA elimination in the polytene chromosomes of Euplotes crassus. Mol Biol Cell 7:755–768. doi: 10.1091/mbc.7.5.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen X, Bracht JR, Goldman AD, Dolzhenko E, Clay DM, Swart EC, Perlman DH, Doak TG, Stuart A, Amemiya CT, Sebra RP, Landweber LF. 2014. The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell 158:1187–1198. doi: 10.1016/j.cell.2014.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Prescott DM, DuBois ML. 1996. Internal eliminated segments (IESs) of oxytrichidae. J Eukaryot Microbiol 43:432–441. doi: 10.1111/j.1550-7408.1996.tb04502.x [DOI] [PubMed] [Google Scholar]
- 47. Williams K, Doak TG, Herrick G. 1993. Developmental precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J 12:4593–4601. doi: 10.1002/j.1460-2075.1993.tb06148.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. 2009. A functional role for transposases in a large eukaryotic genome. Science 324:935–938. doi: 10.1126/science.1170023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Allen SE, Nowacki M. 2020. Roles of noncoding RNAs in ciliate genome architecture. J Mol Biol 432:4186–4198. doi: 10.1016/j.jmb.2019.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 110:689–699. doi: 10.1016/s0092-8674(02)00909-1 [DOI] [PubMed] [Google Scholar]
- 51. Lepère G, Nowacki M, Serrano V, Gout J-F, Guglielmi G, Duharcourt S, Meyer E. 2009. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res 37:903–915. doi: 10.1093/nar/gkn1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. 2005. Germ line transcripts are processed by a dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol 25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bouhouche K, Gout J-F, Kapusta A, Bétermier M, Meyer E. 2011. Functional specialization of piwi proteins in Paramecium tetraurelia from post-transcriptional gene silencing to genome remodelling. Nucleic Acids Res 39:4249–4264. doi: 10.1093/nar/gkq1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mochizuki K, Gorovsky MA. 2004. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev 18:2068–2073. doi: 10.1101/gad.1219904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KWM, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K. 2008. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev 22:2228–2241. doi: 10.1101/gad.481908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lepère G, Bétermier M, Meyer E, Duharcourt S. 2008. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev 22:1501–1512. doi: 10.1101/gad.473008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maliszewska-Olejniczak K, Gruchota J, Gromadka R, Denby Wilkes C, Arnaiz O, Mathy N, Duharcourt S, Bétermier M, Nowak JK. 2015. TFIIS-dependent non-coding transcription regulates developmental genome rearrangements. PLoS Genet 11:e1005383. doi: 10.1371/journal.pgen.1005383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miró-Pina C, Charmant O, Kawaguchi T, Holoch D, Michaud A, Cohen I, Humbert A, Jaszczyszyn Y, Chevreux G, Del Maestro L, Ait-Si-Ali S, Arnaiz O, Margueron R, Duharcourt S. 2022. Paramecium polycomb repressive complex 2 physically interacts with the small RNA-binding PIWI protein to repress transposable elements. Dev Cell 57:1037–1052. doi: 10.1016/j.devcel.2022.03.014 [DOI] [PubMed] [Google Scholar]
- 59. Wang C, Solberg T, Maurer-Alcalá XX, Swart EC, Gao F, Nowacki M. 2022. A small RNA-guided PRC2 complex eliminates DNA as an extreme form of transposon silencing. Cell Reports 40:111263. doi: 10.1016/j.celrep.2022.111263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Frapporti A, Miró Pina C, Arnaiz O, Holoch D, Kawaguchi T, Humbert A, Eleftheriou E, Lombard B, Loew D, Sperling L, Guitot K, Margueron R, Duharcourt S. 2019. The polycomb protein Ezl1 mediates H3K9 and H3K27 methylation to repress transposable elements in Paramecium. Nat Commun 10:2710. doi: 10.1038/s41467-019-10648-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lhuillier-Akakpo M, Frapporti A, Denby Wilkes C, Matelot M, Vervoort M, Sperling L, Duharcourt S. 2014. Local effect of enhancer of zeste-like reveals cooperation of epigenetic and cis-acting determinants for zygotic genome rearrangements. PLoS Genet 10:e1004665. doi: 10.1371/journal.pgen.1004665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taverna SD, Coyne RS, Allis CD. 2002. Methylation of histone H3 at Lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110:701–711. doi: 10.1016/s0092-8674(02)00941-8 [DOI] [PubMed] [Google Scholar]
- 63. Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. 2007. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev 21:1530–1545. doi: 10.1101/gad.1544207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. 2012. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell 151:1243–1255. doi: 10.1016/j.cell.2012.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zahler AM, Neeb ZT, Lin A, Katzman S. 2012. Mating of the stichotrichous ciliate Oxytricha trifallax induces production of a class of 27 nt small RNAs derived from the parental macronucleus. PLoS One 7:e42371. doi: 10.1371/journal.pone.0042371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bracht JR, Perlman DH, Landweber LF. 2012. Cytosine methylation and hydroxymethylation mark DNA for elimination in Oxytricha trifallax. Genome Biol 13:R99. doi: 10.1186/gb-2012-13-10-r99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. 2008. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature 451:153–158. doi: 10.1038/nature06452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Preer JR, Preer LB. 1979. The size of macronuclear DNA and its relationship to models for maintaining genic balance. J Protozool 26:14–18. doi: 10.1111/j.1550-7408.1979.tb02724.x [DOI] [Google Scholar]
- 69. Altschuler MI, Yao MC. 1985. Macronuclear DNA of Tetrahymena thermophila exists as defined subchromosomal-sized molecules. Nucleic Acids Res 13:5817–5831. doi: 10.1093/nar/13.16.5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Conover RK, Brunk CF. 1986. Macronuclear DNA molecules of Tetrahymena thermophila. Mol Cell Biol 6:900–905. doi: 10.1128/mcb.6.3.900-905.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yao MC, Yao CH. 1981. Repeated hexanucleotide C-C-C-C-A-A is present near free ends of macronuclear DNA of Tetrahymena. Proc Natl Acad Sci U S A 78:7436–7439. doi: 10.1073/pnas.78.12.7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang G, Wang S, Chai X, Zhang J, Yang W, Jiang C, Chen K, Miao W, Xiong J. 2021. A strategy for complete telomere-to-telomere assembly of ciliate macronuclear genome using ultra-high coverage nanopore data. Comput Struct Biotechnol J 19:1928–1932. doi: 10.1016/j.csbj.2021.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nanney DL, Preparata RM. 1979. Genetic evidence concerning the structure of the Tetrahymena thermophila macronucleus . J Protozool 26:2–9. doi: 10.1111/j.1550-7408.1979.tb02722.x [DOI] [Google Scholar]
- 74. Zhou Y, Fu L, Mochizuki K, Xiong J, Miao W, Wang G. 2022. Absolute quantification of chromosome copy numbers in the polyploid macronucleus of Tetrahymena thermophila at the single-cell level. J Eukaryot Microbiol 69:e12907. doi: 10.1111/jeu.12907 [DOI] [PubMed] [Google Scholar]
- 75. Yao MC, Gorovsky MA. 1974. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma 48:1–18. doi: 10.1007/BF00284863 [DOI] [PubMed] [Google Scholar]
- 76. Sheng Y, Duan L, Cheng T, Qiao Y, Stover NA, Gao S. 2020. The completed macronuclear genome of a model ciliate Tetrahymena thermophila and its application in genome scrambling and copy number analyses. Sci China Life Sci 63:1534–1542. doi: 10.1007/s11427-020-1689-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cassidy-Hanley D, Bisharyan Y, Fridman V, Gerber J, Lin C, Orias E, Orias JD, Ryder H, Vong L, Hamilton EP. 2005. Genome-wide characterization of Tetrahymena thermophila chromosome breakage sites. II. Physical and genetic mapping. Genetics 170:1623–1631. doi: 10.1534/genetics.104.031435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lin C-Y, Lin I-T, Yao M-C. 2016. Programmed minichromosome elimination as a mechanism for somatic genome reduction in Tetrahymena thermophila. PLoS Genet 12:e1006403. doi: 10.1371/journal.pgen.1006403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Feng L, Wang G, Hamilton EP, Xiong J, Yan G, Chen K, Chen X, Dui W, Plemens A, Khadr L, Dhanekula A, Juma M, Dang HQ, Kapler GM, Orias E, Miao W, Liu Y. 2017. A germline-limited piggyBac transposase gene is required for precise Excision in Tetrahymena genome rearrangement. Nucleic Acids Res 45:9481–9502. doi: 10.1093/nar/gkx652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gao S, Xiong J, Zhang C, Berquist BR, Yang R, Zhao M, Molascon AJ, Kwiatkowski SY, Yuan D, Qin Z, Wen J, Kapler GM, Andrews PC, Miao W, Liu Y. 2013. Impaired replication elongation in Tetrahymena mutants deficient in histone H3 Lys 27 monomethylation. Genes Dev 27:1662–1679. doi: 10.1101/gad.218966.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yao MC, Zhu SG, Yao CH. 1985. Gene amplification in Tetrahymena thermophila: formation of extrachromosomal palindromic genes coding for rRNA. Mol Cell Biol 5:1260–1267. doi: 10.1128/mcb.5.6.1260-1267.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yao MC, Zheng K, Yao CH. 1987. A conserved nucleotide sequence at the sites of developmentally regulated chromosomal breakage in Tetrahymena. Cell 48:779–788. doi: 10.1016/0092-8674(87)90075-4 [DOI] [PubMed] [Google Scholar]
- 83. Fan Q, Yao MC. 2000. A long stringent sequence signal for programmed chromosome breakage in Tetrahymena thermophila. Nucleic Acids Res 28:895–900. doi: 10.1093/nar/28.4.895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yao MC, Yao CH, Monks B. 1990. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell 63:763–772. doi: 10.1016/0092-8674(90)90142-2 [DOI] [PubMed] [Google Scholar]
- 85. Fan Q, Yao M-C. 1996. New telomere formation coupled with site-specific chromosome breakage in Tetrahymena thermophila. Mol Cell Biol 16:1267–1274. doi: 10.1128/MCB.16.3.1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Coyne RS, Yao MC. 1996. Evolutionary conservation of sequences directing chromosome breakage and rDNA palindrome formation in tetrahymenine ciliates. Genetics 144:1479–1487. doi: 10.1093/genetics/144.4.1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xiong J, Yang W, Chen K, Jiang C, Ma Y, Chai X, Yan G, Wang G, Yuan D, Liu Y, Bidwell SL, Zafar N, Hadjithomas M, Krishnakumar V, Coyne RS, Orias E, Miao W, Malik HS. 2019. Hidden genomic evolution in a morphospecies-the landscape of rapidly evolving genes in Tetrahymena. PLoS Biol 17:e3000294. doi: 10.1371/journal.pbio.3000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cranert S, Heyse S, Linger BR, Lescasse R, Price C. 2014. Tetrahymena Pot2 is a developmentally regulated paralog of Pot1 that localizes to chromosome breakage sites but not to telomeres. Eukaryot Cell 13:1519–1529. doi: 10.1128/EC.00204-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. McDaniel SL, Zweifel E, Harris PKW, Yao M-C, Cole ES, Chalker DL. 2016. DRH1, a p68-related RNA helicase gene, is required for chromosome breakage in Tetrahymena. Biol Open 5:1790–1798. doi: 10.1242/bio.021576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dippell RV. 1954. A preliminary report on the chromosomal constitution of certain variety 4 races of Paramecium aurelia. Suppl Caryologia 6:1109–1111. [Google Scholar]
- 91. Jones K. 1956. Nuclear differentiation in Paramecium Ph. D. thesis, Aberystwyth, University of Wales [Google Scholar]
- 92. Duharcourt S, Sperling L. 2018. The challenges of genome-wide studies in a unicellular eukaryote with two nuclear genomes. Meth Enzymol 612:101–126. doi: 10.1016/bs.mie.2018.08.012 [DOI] [PubMed] [Google Scholar]
- 93. Lhuillier-Akakpo M, Guérin F, Frapporti A, Duharcourt S. 2016. DNA deletion as a mechanism for developmentally programmed centromere loss. Nucleic Acids Res 44:1553–1565. doi: 10.1093/nar/gkv1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Aury J-M, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, Câmara F, Duharcourt S, Guigo R, Gogendeau D, Katinka M, Keller A-M, Kissmehl R, Klotz C, Koll F, Le Mouël A, Lepère G, Malinsky S, Nowacki M, Nowak JK, Plattner H, Poulain J, Ruiz F, Serrano V, Zagulski M, Dessen P, Bétermier M, Weissenbach J, Scarpelli C, Schächter V, Sperling L, Meyer E, Cohen J, Wincker P. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444:171–178. doi: 10.1038/nature05230 [DOI] [PubMed] [Google Scholar]
- 95. Preer JR. 1976. Quantitative predictions of random segregation models of the ciliate macronucleus. Genet Res 27:227–238. doi: 10.1017/s0016672300016426 [DOI] [PubMed] [Google Scholar]
- 96. Gout J-F, Johri P, Arnaiz O, Doak TG, Bhullar S, Couloux A, Guérin F, Malinsky S, Sperling L, Labadie K, Meyer E, Duharcourt S, Lynch M. 2019. Universal trends of post-duplication evolution revealed by the Genomes of 13 Paramecium species sharing an ancestral whole-genome duplication. bioRxiv. doi: 10.1101/573576 [DOI]
- 97. Caron F. 1992. A high degree of macronuclear chromosome polymorphism is generated by variable DNA rearrangements in Paramecium primaurelia during macronuclear differentiation. J Mol Biol 225:661–678. doi: 10.1016/0022-2836(92)90393-x [DOI] [PubMed] [Google Scholar]
- 98. Rautian MS, Potekhin AA. 2002. Electrokaryotypes of macronuclei of several Paramecium species. J Eukaryot Microbiol 49:296–304. doi: 10.1111/j.1550-7408.2002.tb00372.x [DOI] [PubMed] [Google Scholar]
- 99. Zagulski M, Nowak JK, Le Mouël A, Nowacki M, Migdalski A, Gromadka R, Noël B, Blanc I, Dessen P, Wincker P, Keller A-M, Cohen J, Meyer E, Sperling L. 2004. High coding density on the largest Paramecium tetraurelia somatic chromosome. Curr Biol 14:1397–1404. doi: 10.1016/j.cub.2004.07.029 [DOI] [PubMed] [Google Scholar]
- 100. Baroin A, Prat A, Caron F. 1987. Telomeric site position heterogeneity in macronuclear DNA of Paramecium primaurelia. Nucleic Acids Res 15:1717–1728. doi: 10.1093/nar/15.4.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. McCormick-Graham M, Romero DP. 1996. A single telomerase RNA is sufficient for the synthesis of variable telomeric DNA repeats in ciliates of the genus Paramecium. Mol Cell Biol 16:1871–1879. doi: 10.1128/MCB.16.4.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ye AJ, Romero DP. 2002. A unique pause pattern during telomere addition by the error-prone telomerase from the ciliate Paramecium tetraurelia. Gene 294:205–213. doi: 10.1016/s0378-1119(02)00790-4 [DOI] [PubMed] [Google Scholar]
- 103. Amar L, Dubrana K. 2004. Epigenetic control of chromosome breakage at the 5’ end of Paramecium tetraurelia gene A. Eukaryot Cell 3:1136–1146. doi: 10.1128/EC.3.5.1136-1146.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Forney J, Rodkey K. 1992. A repetitive DNA sequence in Paramecium macronuclei is related to the beta subunit of G proteins. Nucleic Acids Res 20:5397–5402. doi: 10.1093/nar/20.20.5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Forney JD, Blackburn EH. 1988. Developmentally controlled telomere addition in wild-type and mutant Paramecia. Mol Cell Biol 8:251–258. doi: 10.1128/mcb.8.1.251-258.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chalker DL, Meyer E, Mochizuki K. 2013. Epigenetics of ciliates. Cold Spring Harb Perspect Biol 5:a017764. doi: 10.1101/cshperspect.a017764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Arnaiz O, Van Dijk E, Bétermier M, Lhuillier-Akakpo M, de Vanssay A, Duharcourt S, Sallet E, Gouzy J, Sperling L. 2017. Improved methods and resources for Paramecium genomics: transcription units, gene annotation and gene expression. BMC Genomics 18:483. doi: 10.1186/s12864-017-3887-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Phan HL, Forney J, Blackburn EH. 1989. Analysis of Paramecium macronuclear DNA using pulsed field GEL electrophoresis. J Protozool 36:402–408. doi: 10.1111/j.1550-7408.1989.tb05535.x [DOI] [PubMed] [Google Scholar]
- 109. Duret L, Cohen J, Jubin C, Dessen P, Goût J-F, Mousset S, Aury J-M, Jaillon O, Noël B, Arnaiz O, Bétermier M, Wincker P, Meyer E, Sperling L. 2008. Analysis of sequence variability in the macronuclear DNA of Paramecium tetraurelia: a somatic view of the germline . Genome Res 18:585–596. doi: 10.1101/gr.074534.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cheng Y-H, Liu C-F, Yu Y-H, Jhou Y-T, Fujishima M, Tsai IJ, Leu J-Y. 2020. Genome plasticity in Paramecium bursaria revealed by population genomics. BMC Biol 18:180. doi: 10.1186/s12915-020-00912-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Jessop-Murray H, Martin LD, Gilley D, Preer JR Jr, Polisky B. 1991. Permanent rescue of a non-mendelian mutation of Paramecium by microinjection of specific DNA sequences. Genetics 129:727–734. doi: 10.1093/genetics/129.3.727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Koizumi S, Kobayashi S. 1989. Microinjection of plasmid DNA encoding the a surface antigen of Paramecium tetraurelia restores the ability to regenerate a wild-type macronucleus. Mol Cell Biol 9:4398–4401. doi: 10.1128/mcb.9.10.4398-4401.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. You Y, Aufderheide K, Morand J, Rodkey K, Forney J. 1991. Macronuclear transformation with specific DNA fragments controls the content of the new macronuclear genome in Paramecium tetraurelia. Mol Cell Biol 11:1133–1137. doi: 10.1128/mcb.11.2.1133-1137.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Scott JM, Mikami K, Leeck CL, Forney JD. 1994. Non-mendelian inheritance of macronuclear mutations is gene specific in Paramecium tetraurelia. Mol Cell Biol 14:2479–2484. doi: 10.1128/mcb.14.4.2479-2484.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Garnier O, Serrano V, Duharcourt S, Meyer E. 2004. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol Cell Biol 24:7370–7379. doi: 10.1128/MCB.24.17.7370-7379.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Meyer E. 1992. Induction of specific macronuclear developmental mutations by microinjection of a cloned telomeric gene in Paramecium primaurelia. Genes Dev 6:211–222. doi: 10.1101/gad.6.2.211 [DOI] [PubMed] [Google Scholar]
- 117. Marmignon A. 2014. Ku-mediated coupling of DNA cleavage and repair during programmed genome Rearrangements in the Ciliate Paramecium tetraurelia Thesis, Université Paris Sud - Paris XI, 2, Paris, FR. doi: 10.1371/journal.pgen.1004552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bétermier M, Borde V, de Villartay J-P. 2020. Coupling DNA damage and repair: an essential safeguard during programmed DNA double-strand breaks?. Trends Cell Biol 30:87–96. doi: 10.1016/j.tcb.2019.11.005 [DOI] [PubMed] [Google Scholar]
- 119. Amar L. 1994. Chromosome end formation and internal sequence elimination as alternative genomic rearrangements in the ciliate Paramecium. J Mol Biol 236:421–426. doi: 10.1006/jmbi.1994.1154 [DOI] [PubMed] [Google Scholar]
- 120. Zangarelli C, Arnaiz O, Bourge M, Gorrichon K, Jaszczyszyn Y, Mathy N, Escoriza L, Bétermier M, Régnier V. 2022. Developmental timing of programmed DNA elimination in Paramecium tetraurelia recapitulates germline transposon evolutionary dynamics. Genome res 32:2028–2042. doi: 10.1101/gr.277027.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Prescott DM, Bostock CJ, Murti KG, Lauth MR, Gamow E. 1971. DNA of ciliated protozoa. I. Electron microscopic and sedimentation analyses of macronuclear and micronuclear DNA of Stylonychia mytilus. Chromosoma 34:355–366. doi: 10.1007/BF00326311 [DOI] [Google Scholar]
- 122. Ammermann D. 1971. Morphology and development of the macronuclei of the ciliates Stylonychia mytilus and Euplotes aediculatus. Chromosoma 33:209–238. doi: 10.1007/BF00285634 [DOI] [PubMed] [Google Scholar]
- 123. Aeschlimann SH, Jönsson F, Postberg J, Stover NA, Petera RL, Lipps H-J, Nowacki M, Swart EC. 2014. The draft assembly of the radically organized Stylonychia lemnae macronuclear genome. Genome Biol Evol 6:1707–1723. doi: 10.1093/gbe/evu139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chen X, Jiang Y, Gao F, Zheng W, Krock TJ, Stover NA, Lu C, Katz LA, Song W. 2019. Genome analyses of the new model protist Euplotes vannus focusing on genome rearrangement and resistance to environmental stressors . Mol Ecol Resour 19:1292–1308. doi: 10.1111/1755-0998.13023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Mozzicafreddo M, Pucciarelli S, Swart EC, Piersanti A, Emmerich C, Migliorelli G, Ballarini P, Miceli C. 2021. The macronuclear genome of the Antarctic psychrophilic marine ciliate Euplotes focardii reveals new insights on molecular cold adaptation. Sci Rep 11:18782. doi: 10.1038/s41598-021-98168-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Swanton MT, Heumann JM, Prescott DM. 1980. Gene-sized DNA molecules of the macronuclei in three species of hypotrichs: size distributions and absence of nicks. DNA of ciliated protozoa. VIII. Chromosoma 77:217–227. doi: 10.1007/BF00329546 [DOI] [PubMed] [Google Scholar]
- 127. Swart EC, Bracht JR, Magrini V, Minx P, Chen X, Zhou Y, Khurana JS, Goldman AD, Nowacki M, Schotanus K, Jung S, Fulton RS, Ly A, McGrath S, Haub K, Wiggins JL, Storton D, Matese JC, Parsons L, Chang W-J, Bowen MS, Stover NA, Jones TA, Eddy SR, Herrick GA, Doak TG, Wilson RK, Mardis ER, Landweber LF. 2013. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol 11:e1001473. doi: 10.1371/journal.pbio.1001473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wang R, Xiong J, Wang W, Miao W, Liang A. 2016. High frequency of +1 programmed ribosomal frameshifting in Euplotes octocarinatus. Sci Rep 6:21139. doi: 10.1038/srep21139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Klobutcher LA, Prescott DM. 1986. The special case of the Hypotrichs, p 111–154. In Gall JG (ed), The molecular biology of ciliated protozoa. Academic Press, Inc. [Google Scholar]
- 130. Prescott DM. 1994. The DNA of ciliated protozoa. Microbiol Rev 58:233–267. doi: 10.1128/mr.58.2.233-267.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Baird SE, Klobutcher LA. 1989. Characterization of chromosome fragmentation in two protozoans and identification of a candidate fragmentation sequence in Euplotes crassus. Genes Dev 3:585–597. doi: 10.1101/gad.3.5.585 [DOI] [PubMed] [Google Scholar]
- 132. Möllenbeck M, Klobutcher LA. 2002. De novo telomere addition to spacer sequences prior to their developmental degradation in Euplotes crassus. Nucleic Acids Res 30:523–531. doi: 10.1093/nar/30.2.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Klobutcher LA, Swanton MT, Donini P, Prescott DM. 1981. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3’ terminus. Proc Natl Acad Sci U S A 78:3015–3019. doi: 10.1073/pnas.78.5.3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Maxam AM, Gilbert W. 1977. A new method for sequencing DNA. Proc Natl Acad Sci U S A 74:560–564. doi: 10.1073/pnas.74.2.560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Vinogradov DV, Tsoi OV, Zaika AV, Lobanov AV, Turanov AA, Gladishev VN, Gel’fand MS. 2012. Draft macronucleus genome of Euplotes crassus ciliate. Mol Biol 46:328–333. doi: 10.1134/S0026893312020197 [DOI] [PubMed] [Google Scholar]
- 136. Klobutcher LA, Gygax SE, Podoloff JD, Vermeesch JR, Price CM, Tebeau CM, Jahn CL. 1998. Conserved DNA sequences adjacent to chromosome fragmentation and telomere addition sites in Euplotes crassus. Nucleic Acids Res 26:4230–4240. doi: 10.1093/nar/26.18.4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Jin D, Li C, Chen X, Byerly A, Stover NA, Zhang T, Shao C, Wang Y. 2023. Comparative genome analysis of three euplotid protists provides insights into the evolution of nanochromosomes in unicellular eukaryotic organisms. Mar Life Sci Technol 5:300–315. doi: 10.1007/s42995-023-00175-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Klobutcher LA. 1999. Characterization of in vivo developmental chromosome fragmentation Intermediates in E. crassus. Mol Cell 4:695–704. doi: 10.1016/s1097-2765(00)80380-9 [DOI] [PubMed] [Google Scholar]
- 139. Petroni G, Dini F, Verni F, Rosati G. 2002. A molecular approach to the tangled Intrageneric relationships underlying phylogeny in Euplotes (ciliophora, spirotrichea). Mol Phylogenet Evol 22:118–130. doi: 10.1006/mpev.2001.1030 [DOI] [PubMed] [Google Scholar]
- 140. Cartinhour SW, Herrick GA. 1984. Three different macronuclear DNAs in Oxytricha fallax share a common sequence block. Mol Cell Biol 4:931–938. doi: 10.1128/mcb.4.5.931-938.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Herrick G, Hunter D, Williams K, Kotter K. 1987. Alternative processing during development of a macronuclear chromosome family in Oxytricha fallax. Genes Dev 1:1047–1058. doi: 10.1101/gad.1.10.1047 [DOI] [PubMed] [Google Scholar]
- 142. Klobutcher LA, Huff ME, Gonye GE. 1988. Alternative use of chromosome fragmentation sites in the ciliated protozoan Oxytricha nova . Nucleic Acids Res 16:251–264. doi: 10.1093/nar/16.1.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Lindblad KA, Pathmanathan JS, Moreira S, Bracht JR, Sebra RP, Hutton ER, Landweber LF. 2019. Capture of complete ciliate chromosomes in single sequencing reads reveals widespread chromosome isoforms. BMC Genomics 20:1037. doi: 10.1186/s12864-019-6189-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Seegmiller A, Williams KR, Herrick G. 1997. Two two-gene macronuclear chromosomes of the hypotrichous ciliates Oxytricha fallax and O. trifallax generated by alternative processing of the 81 locus. Dev Genet 20:348–357. doi: [DOI] [PubMed] [Google Scholar]
- 145. Williams KR, Doak TG, Herrick G. 2002. Telomere formation on macronuclear chromosomes of Oxytricha trifallax and O. fallax: alternatively processed regions have multiple telomere addition sites. BMC Genet 3:16. doi: 10.1186/1471-2156-3-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Jönsson F, Wen JP, Fetzer CP, Lipps HJ. 1999. A subtelomeric DNA sequence is required for correct processing of the macronuclear DNA sequences during macronuclear development in the hypotrichous ciliate Stylonychia lemnae. Nucleic Acids Res 27:2832–2841. doi: 10.1093/nar/27.14.2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Jönsson F, Steinbrück G, Lipps HJ. 2001. Both subtelomeric regions are required and sufficient for specific DNA fragmentation during macronuclear development in Stylonychia lemnae. Genome Biol 2:research0005. doi: 10.1186/gb-2001-2-2-research0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Prescott DM, Dizick SJ. 2000. A unique pattern of intrastrand anomalies in base composition of the DNA in hypotrichs. Nucleic Acids Res 28:4679–4688. doi: 10.1093/nar/28.23.4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Cavalcanti ARO, Dunn DM, Weiss R, Herrick G, Landweber LF, Doak TG. 2004. Sequence features of Oxytricha trifallax (class spirotrichea) macronuclear telomeric and subtelomeric sequences. Protist 155:311–322. doi: 10.1078/1434461041844196 [DOI] [PubMed] [Google Scholar]
- 150. Khurana JS, Wang X, Chen X, Perlman DH, Landweber LF. 2014. Transcription-independent functions of an RNA polymerase II subunit, Rpb2, during genome rearrangement in the ciliate, Oxytricha trifallax . Genetics 197:839–849. doi: 10.1534/genetics.114.163279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Nowacki M, Haye JE, Fang W, Vijayan V, Landweber LF. 2010. RNA-mediated epigenetic regulation of DNA copy number. Proc Natl Acad Sci U S A 107:22140–22144. doi: 10.1073/pnas.1012236107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Heyse G, Jönsson F, Chang W-J, Lipps HJ. 2010. RNA-dependent control of gene amplification. Proc Natl Acad Sci U S A 107:22134–22139. doi: 10.1073/pnas.1009284107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Fuhrmann G, Jönsson F, Weil PP, Postberg J, Lipps HJ. 2016. RNA-template dependent De Novo Telomere addition. RNA Biol 13:733–739. doi: 10.1080/15476286.2015.1134414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. de Lange T, Lundblad V, Blackburn E, eds. 2006. Telomeres. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 155. Bryan TM, Cech TR. 1999. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol 11:318–324. doi: 10.1016/S0955-0674(99)80043-X [DOI] [PubMed] [Google Scholar]
- 156. King BO, Yao MC. 1982. Tandemly repeated hexanucleotide at Tetrahymena rDNA free end is generated from a single copy during development. Cell 31:177–182. doi: 10.1016/0092-8674(82)90417-2 [DOI] [PubMed] [Google Scholar]
- 157. Gilley D, Preer JR Jr, Aufderheide KJ, Polisky B. 1988. Autonomous replication and addition of telomerelike sequences to DNA microinjected into Paramecium tetraurelia macronuclei. Mol Cell Biol 8:4765–4772. doi: 10.1128/mcb.8.11.4765-4772.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Kim CS, Preer JR, Polisky B. 1992. Bacteriophage lambda DNA fragments replicate in the Paramecium macronucleus: absence of active copy number control. Dev Genet 13:97–102. doi: 10.1002/dvg.1020130202 [DOI] [PubMed] [Google Scholar]
- 159. Kim CS, Preer JR, Polisky B. 1994. Identification of DNA segments capable of rescuing a non-mendelian mutant in Paramecium. Genetics 136:1325–1328. doi: 10.1093/genetics/136.4.1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Arnaiz O, Meyer E, Sperling L. 2020. Parameciumdb 2019: Integrating genomic data across the genus for functional and evolutionary biology. Nucleic Acids Res 48:D599–D605. doi: 10.1093/nar/gkz948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Yu GL, Blackburn EH. 1991. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell 67:823–832. doi: 10.1016/0092-8674(91)90077-c [DOI] [PubMed] [Google Scholar]
- 162. Wang H, Gilley D, Blackburn EH. 1998. A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. EMBO J 17:1152–1160. doi: 10.1093/emboj/17.4.1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Wang H, Blackburn EH. 1997. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J 16:866–879. doi: 10.1093/emboj/16.4.866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Roth M, Prescott DM. 1985. DNA intermediates and telomere addition during genome reorganization in Euplotes crassus. Cell 41:411–417. doi: 10.1016/s0092-8674(85)80014-3 [DOI] [PubMed] [Google Scholar]
- 165. Tausta SL, Klobutcher LA. 1990. Internal eliminated sequences are removed prior to chromosome fragmentation during development in Euplotes crassus. Nucleic Acids Res 18:845–853. doi: 10.1093/nar/18.4.845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Vermeesch JR, Price CM. 1994. Telomeric DNA sequence and structure following de novo telomere synthesis in Euplotes crassus. Mol. Cell. Biol 14:554–566. doi: 10.1128/MCB.14.1.554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Vermeesch JR, Williams D, Price CM. 1993. Telomere processing in Euplotes. Nucleic Acids Res 21:5366–5371. doi: 10.1093/nar/21.23.5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Price CM, Adams AK, Vermeesch JR. 1994. Accumulation of telomerase RNA and telomere protein transcripts during Telomere synthesis in Euplotes. J Eukaryot Microbiol 41:267–275. doi: 10.1111/j.1550-7408.1994.tb01507.x [DOI] [PubMed] [Google Scholar]
- 169. Bednenko J, Melek M, Shippen DE. 1998. Reiterative dG addition by Euplotes crassus telomerase during extension of non-telomeric DNA. Nucleic Acids Res 26:3998–4004. doi: 10.1093/nar/26.17.3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Bednenko J, Melek M, Greene EC, Shippen DE. 1997. Developmentally regulated initiation of DNA synthesis by telomerase: evidence for factor-assisted de novo telomere formation. EMBO J 16:2507–2518. doi: 10.1093/emboj/16.9.2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Karamysheva Z, Wang L, Shrode T, Bednenko J, Hurley LA, Shippen DE. 2003. Developmentally programmed gene elimination in Euplotes crassus facilitates a switch in the telomerase catalytic subunit. Cell 113:565–576. doi: 10.1016/s0092-8674(03)00363-5 [DOI] [PubMed] [Google Scholar]
- 172. Ghosh S, Klobutcher LA. 2000. A development-specific histone H3 localizes to the developing macronucleus of Euplotes. Genesis 26:179–188. doi: [DOI] [PubMed] [Google Scholar]
- 173. Jahn CL. 1999. Differentiation of chromatin during DNA elimination in Euplotes crassus. Mol Biol Cell 10:4217–4230. doi: 10.1091/mbc.10.12.4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Jahn CL, Ling Z, Tebeau CM, Klobutcher LA. 1997. An unusual histone H3 specific for early macronuclear development in Euplotes crassus. Proc Natl Acad Sci U S A 94:1332–1337. doi: 10.1073/pnas.94.4.1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Kinzig CG, Zakusilo G, Takai KK, de Lange T. 2022. Neotelomere formation by human Telomerase. bioRxiv. doi: 10.1101/2022.10.31.514589 [DOI]
- 176. Herrick G. 1994. Germline-soma relationships in ciliated protozoa: the inception and evolution of nuclear dimorphism in one-celled animals. Semin Dev Biol 5:3–12. doi: 10.1006/sedb.1994.1002 [DOI] [Google Scholar]
- 177. Orias E. 1991. Evolution of amitosis of the ciliate macronucleus: gain of the capacity to divide. J Protozool 38:217–221. doi: 10.1111/j.1550-7408.1991.tb04431.x [DOI] [PubMed] [Google Scholar]
- 178. Boscaro V, Keeling PJ. 2023. How ciliates got their nuclei. Proc Natl Acad Sci U S A 120:e2221818120. doi: 10.1073/pnas.2221818120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Zufall RA, Robinson T, Katz LA. 2005. Evolution of developmentally regulated genome rearrangements in eukaryotes. J Exp Zool B Mol Dev Evol 304:448–455. doi: 10.1002/jez.b.21056 [DOI] [PubMed] [Google Scholar]
- 180. Orias E. 1991. On the evolution of the karyorelict ciliate life cycle: heterophasic ciliates and the origin of ciliate binary fission. Biosystems 25:67–73. doi: 10.1016/0303-2647(91)90013-b [DOI] [PubMed] [Google Scholar]
- 181. Taylor WD, Shuter BJ. 1981. Body size, genome size, and intrinsic rate of increase in ciliated protozoa. Am Nat 118:160–172. doi: 10.1086/283812 [DOI] [Google Scholar]
- 182. Austerberry CF, Allis CD, Yao MC. 1984. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc Natl Acad Sci U S A 81:7383–7387. doi: 10.1073/pnas.81.23.7383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Ammermann D, Steinbrück G, von Berger L, Hennig W. 1974. The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma 45:401–429. doi: 10.1007/BF00283386 [DOI] [PubMed] [Google Scholar]
- 184. Yella VR, Vanaja A, Kulandaivelu U, Kumar A. 2020. Delving into eukaryotic origins of replication using DNA structural features. ACS Omega 5:13601–13611. doi: 10.1021/acsomega.0c00441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Irwin NAT, Pittis AA, Mathur V, Howe LJ, Keeling PJ, Lynn DH, Bourland WA. 2021. The function and evolution of motile DNA replication systems in ciliates. Curr Biol 31:66–76. doi: 10.1016/j.cub.2020.09.077 [DOI] [PubMed] [Google Scholar]
- 186. Olins DE, Olins AL. 2021. DNA replication: an in vivo space-time continuum in the ciliate replication band. Curr Biol 31:R16–R17. doi: 10.1016/j.cub.2020.10.058 [DOI] [PubMed] [Google Scholar]
- 187. Murti KG, Prescott DM. 1983. Replication forms of the gene-sized DNA molecules of hypotrichous ciliates. Mol Cell Biol 3:1562–1566. doi: 10.1128/mcb.3.9.1562-1566.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Zahler AM, Prescott DM. 1989. DNA primase and the replication of the telomeres in Oxytricha nova . Nucl Acids Res 17:6299–6317. doi: 10.1093/nar/17.15.6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Hamilton EP, Williamson S, Dunn S, Merriam V, Lin C, Vong L, Russell-Colantonio J, Orias E. 2006. The highly conserved family of Tetrahymena thermophila chromosome breakage elements contains an invariant 10-base-pair core. Eukaryot Cell 5:771–780. doi: 10.1128/EC.5.4.771-780.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Wuitschick JD, Lindstrom PR, Meyer AE, Karrer KM. 2004. Homing endonucleases encoded by germ line-limited genes in Tetrahymena thermophila have APETELA2 DNA binding domains. Eukaryot Cell 3:685–694. doi: 10.1128/EC.3.3.685-694.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Craig NL, Chandler M, Gellert M, Lambowitz AM, Rice P, Sandmeyer SB, eds. 2015. Mobile DNA III. ASM Press, Washington, DC, USA. doi: 10.1128/9781555819217 [DOI] [Google Scholar]
- 192. Seah BKB, Swart EC. 2023. When cleaning facilitates cluttering - genome editing in ciliates. Trends Genet 39:344–346. doi: 10.1016/j.tig.2023.02.016 [DOI] [PubMed] [Google Scholar]
- 193. Shabalina SA, Koonin EV. 2008. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol 23:578–587. doi: 10.1016/j.tree.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Zufall RA, Robinson T, Katz LA. 2005. Evolution of developmentally regulated genome rearrangements in eukaryotes. J Exp Zool B Mol Dev Evol 304:448–455. doi: 10.1002/jez.b.21056 [DOI] [PubMed] [Google Scholar]
- 195. Fugmann SD. 2010. The origins of the rag genes—from transposition to V(D)J recombination. Semin Immunol 22:10–16. doi: 10.1016/j.smim.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Schatz DG, Ji Y. 2011. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol 11:251–263. doi: 10.1038/nri2941 [DOI] [PubMed] [Google Scholar]
- 197. Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, Zhang Y, Yu W, Pontarotti P, Escriva H, Le Petillon Y, Liu X, Chen S, Schatz DG, Xu A. 2016. Discovery of an active RAG transposon illuminates the origins of V(D)J recombination. Cell 166:102–114. doi: 10.1016/j.cell.2016.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Cech TR, Zaug AJ, Grabowski PJ. 1981. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 27:487–496. doi: 10.1016/0092-8674(81)90390-1 [DOI] [PubMed] [Google Scholar]
- 199. Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843–851. doi: 10.1016/S0092-8674(00)81063-6 [DOI] [PubMed] [Google Scholar]
- 200. Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403:41–45. doi: 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 201. Greider CW, Blackburn EH. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405–413. doi: 10.1016/0092-8674(85)90170-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Known Consensus Cbs and functional variants in T. thermophila and related species Table S2 - Proteins Required for Cbs-dependent Chromosome breakage (CB) and/or IES Excision in Tetrahymena.