ABSTRACT
The World Health Organization recently published the first list of priority fungal pathogens highlighting multiple Candida species, including Candida glabrata, Candida albicans, and Candida auris. However, prior studies in these pathogens have been mainly limited to the use of two drug resistance cassettes, NatMX and HphMX, limiting genetic manipulation capabilities in prototrophic laboratory strains and clinical isolates. In this study, we expanded the toolkit for C. glabrata, C. auris, and C. albicans to include KanMX and BleMX when coupled with an in vitro assembled CRISPR-Cas9 ribonucleoprotein (RNP)-based system. Repurposing these drug resistance cassettes for Candida, we were able to make single gene deletions, sequential and simultaneous double gene deletions, epitope tags, and rescue constructs. We applied these drug resistance cassettes to interrogate the ergosterol pathway, a critical pathway for both the azole and polyene antifungal drug classes. Using our approach, we determined for the first time that the deletion of ERG3 in C. glabrata, C. auris, and C. albicans prototrophic strains results in azole drug resistance, which further supports the conservation of the Erg3-dependent toxic sterol model. Furthermore, we show that an ERG5 deletion in C. glabrata is azole susceptible at subinhibitory concentrations, suggesting that Erg5 could act as an azole buffer for Erg11. Finally, we identified a synthetic growth defect when both ERG3 and ERG5 are deleted in C. glabrata, which suggests the possibility of another toxic sterol impacting growth. Overall, we have expanded the genetic tools available to interrogate complex pathways in prototrophic strains and clinical isolates.
IMPORTANCE
The increasing problem of drug resistance and emerging pathogens is an urgent global health problem that necessitates the development and expansion of tools for studying fungal drug resistance and pathogenesis. Prior studies in Candida glabrata, Candida auris, and Candida albicans have been mainly limited to the use of NatMX/SAT1 and HphMX/CaHyg for genetic manipulation in prototrophic strains and clinical isolates. In this study, we demonstrated that NatMX/SAT1, HphMX, KanMX, and/or BleMX drug resistance cassettes when coupled with a CRISPR-ribonucleoprotein (RNP)-based system can be efficiently utilized for deleting or modifying genes in the ergosterol pathway of C. glabrata, C. auris, and C. albicans. Moreover, the utility of these tools has provided new insights into ERG genes and their relationship to azole resistance in Candida. Overall, we have expanded the toolkit for Candida pathogens to increase the versatility of genetically modifying complex pathways involved in drug resistance and pathogenesis.
KEYWORDS: CRISPR-RNP, Candida auris, Candida glabrata, Candida albicans, ERG5 and ERG3, antifungal drug resistance, KanMX and BleMX, ergosterol pathway, toxic sterols, azole antifungal drugs
INTRODUCTION
Fungal infections pose a significant public health concern, with over a billion superficial infections and 1.5 million deaths, mostly from invasive infections, occurring annually worldwide (1, 2). Candida species are responsible for roughly 40%–70% of invasive fungal infections (1–3), and several species are classified as “high priority fungal pathogens” by the World Health Organization (WHO) for study, including Candida glabrata, Candida albicans, and Candida auris. Infections can range from superficial to life-threatening, with invasive candidiasis leading to a mortality rate of 20%–60% (4, 5). Currently, there are three FDA-approved major antifungals clinically used for the treatment of systemic fungal infections: azoles, echinocandins, and polyenes (6–8). However, antifungal drug resistance has become a significant concern, highlighted by the increase in clinically acquired drug resistance in C. albicans and C. glabrata and the recent emergence of a multi-drug-resistant pathogen, C. auris (8, 9).
Two of these drug classes, azole and polyenes, target the ergosterol pathway, an essential but complex biological pathway that is largely conserved across yeast and fungi (10–13). Most of our understanding of the ergosterol pathway has been derived from studies in auxotrophic Saccharomyces cerevisiae and C. albicans strains (13–15), whereas in comparison, our understanding of this pathway in C. glabrata and C. auris has been limited. These genetic and biochemical studies have proposed a model where azoles prevent ergosterol biosynthesis by directly inhibiting Erg11, the lanosterol 14-α-demethylase. Consequently, azole inhibition results in the production of a toxic sterol intermediate, 14α-methyl-3,6-diol, generated by the sterol Δ5,6-desaturase, Erg3 (14). Concurring with this model is the observation that deletion of ERG3 in S. cerevisiae and C. albicans prevents the formation of the 14α-methyl-3,6-diol toxic sterol, resulting in azole resistance (14, 15). However, it has been reported and widely accepted in the field that deletion of ERG3 in C. glabrata is not azole resistant but in contrast azole susceptible (16–18). Contrary to this observation, clinical isolates of C. glabrata and micro-evolved ERG3 mutations can result in azole resistance, thereby providing an unexplained role for ERG3 in C. glabrata (19, 20). Currently, it is unknown if an ERG3 deletion results in azole resistance in C. auris. However, to further characterize the ergosterol pathway in prototrophic strains of C. glabrata, C. auris, and C. albicans, additional genetic tools are needed.
Currently, the C. glabrata, C. auris, and C. albicans toolkit has been mainly limited to the use of nourseothricin (NatMX/SAT1) or hygromycin B (HphMX/CaHyg). In contrast, drug resistance cassettes such as KanMX and BleMX are either rarely or not used in Candida but are commonly used in Saccharomyces cerevisiae. One major reason for this limitation is that Candida species have developed resistance to antibiotics, including kanamycin/geneticin (G418) and phleomycin/zeocin, which results in high background growth and prevents the ability to identify colonies containing KanMX and BleMX cassettes. The ability to repurpose and efficiently use these dominant selection markers in Candida would significantly enhance the flexibility for genetic manipulation of the ergosterol and/or other complex pathways when using prototrophic strains and clinical isolates.
In this study, we demonstrate that multiple drug resistance markers can be effectively used in Candida species when coupled with an in vitro assembled CRISPR-Cas9 ribonucleoprotein (RNP)-based system. Using this approach, we were able to efficiently make erg3 deletions when using NatMX, HphMX, KanMX, or BleMX cassettes for C. glabrata, while KanMX and BleMX were used to make erg3 deletions in C. auris. In addition, endogenous epitope tagging of ERG3 and ERG11 in C. glabrata was done using a 3×HA-KanMX cassette. Subsequently, the ERG3 epitope-tagged gene was used to generate an ERG3-3×HA-KanMX cassette to functionally complement erg3Δ phenotypes. In addition, double gene deletions (erg3Δerg5Δ) were sequentially generated using NatMX and HphMX cassettes in C. glabrata. Finally, the ability to make simultaneous deletions of C. albicans alleles with a combinatorial drug selection approach using CaKanMX and SAT1 or BleMX and SAT1 cassettes was also demonstrated.
Our approach allowed us to not only enhance the Candida toolbox but also provide the genetic capability to efficiently manipulate the ergosterol pathway using prototrophic strains. Using these tools, we showed for the first time that erg3Δ strains are azole resistant in prototrophic C. glabrata strains, which provides support that the toxic sterol model is conserved in C. glabrata. In addition, prototrophic C. auris and C. albicans erg3Δ strains also showed an azole-resistant phenotype, demonstrating that the function of Erg3 is broadly conserved across all three pathogens. We also determined that an erg5Δ strain was susceptible to subinhibitory concentrations of azoles and suggested that Erg5 plays a role in azole buffering in C. glabrata. Finally, a synthetic growth defect was observed for a C. glabrata erg3Δerg5Δ double deletion strain, and we propose that this growth inhibition is caused by another toxic sterol. Overall, we have identified new insights involving the ergosterol pathway, and we anticipate that this expanded toolkit will provide the field the capacity to interrogate other complex pathways.
RESULTS
Short homology regions can be used for gene replacement in C. glabrata when coupled with CRISPR-RNP
CRISPR-mediated or non-CRISPR-based methods generally rely on large flanking homology regions ranging from 500 to 1,000 base pairs (bp) for efficient gene replacement in Candida glabrata (21, 22). Often, steps to generate long flanking regions are time-consuming and tedious using either cloning or multi-step fusion PCR approaches. The initial CRISPR-Cas9 RNP system developed for Candida species, including C. glabrata, utilized long homology regions ranging from 500 to 1,000 bp (21). However, it has been reported for C. glabrata that flanking homology regions ranging from 20 to 200 bp can be used for gene insertions resulting in gene disruption, albeit with the aid of a CRISPR-Cas9 plasmid-based system in an auxotrophic strain (23). To determine if short homology regions (HRs) flanking drug resistance cassettes were efficient in making gene deletions in C. glabrata using a CRISPR-Cas9 RNP method, we PCR amplified drug resistance cassettes using oligonucleotides (IDT Ultramers) of ~130–150 bp of homology to the ADE2 gene. ADE2 was selected due to its red pigment phenotype when the ADE2 gene is disrupted, which allows for an unbiased determination of gene replacement efficiency (24–26). Using the pAG25 NatMX and pAG32 HphMX plasmids (Fig. 1A and B) (27), we deleted the ADE2 open reading frame and counted the proportion of white and red colonies (Fig. 1C). With the addition of a CRISPR-Cas9 RNP containing two gRNAs and 130–150 bp of flanking homology, we observed a fivefold increase in the proportion of red colonies compared to the cassette alone (Fig. 1D). Similarly, we observed a fivefold increase in the proportion of red colonies using hygromycin B (HphMX) when using CRISPR-Cas9 RNP (Fig. 1E). With this efficiency, we determined that 500–1,000-bp homology regions are not required for efficient gene replacement in C. glabrata. To determine the lower limit of homology, we also tested 60-bp HRs and observed a 35% decrease in efficiency when compared to 130-bp HRs (Fig. S1A; Fig. 1D). Based on this comparison, we selected 130–150-bp HRs as the standard length for our CRISPR-RNP-mediated genetic manipulation. Altogether, these data suggest that short HRs are efficient in generating gene deletions in C. glabrata using NatMX and HphMX when coupled to CRISPR-RNP.
Fig 1.
Homology regions of 130–150-bp efficiently generate ADE2 deletions in C. glabrata using NatMX and HphMX when coupled with CRISPR-Cas9 RNP. (A) Schematic of the pAG25 NatMX plasmid. P1 and P2 indicate the locations of amplification sequences. (B) Schematic of the pAG32 HphMX plasmid. P1 and P2 indicate the locations of amplification sequences. (C) Representative transformation plate for ADE2 deletion using NatMX with and without the addition of CRISPR-RNP. (D) Total number of positive transformants using NatMX with and without the addition of CRISPR-RNP. Numbers represent the summation across three separate transformations. (E) Total number of positive transformants using HphMX with and without the addition of CRISPR-RNP. Numbers represent the summation across three separate transformations.
Deletion of ERG3 results in an azole drug-resistant phenotype in C. glabrata
We next applied our optimized approach to investigate the ergosterol pathway, a critical biosynthesis pathway targeted by both the azole and polyene antifungal drug classes. Azole drugs inhibit Erg11, lanosterol 14-α-demethylase, to block ergosterol biosynthesis, which leads to accumulation of an Erg3-dependent toxic sterol 14α-methyl-3,6-diol and growth inhibition (14, 28, 29) (see Fig. S2). While ERG3 is known to have an azole-resistant phenotype when deleted or mutated in S. cerevisiae or C. albicans (14, 15, 30), C. glabrata auxotrophic strains deleted for ERG3 show fluconazole susceptibility (16, 17). In contrast, micro-evolved ERG3 mutations and clinical isolates show resistance to fluconazole (19, 20, 29). To address this unexplained contradiction, we used our optimized CRISPR-RNP method to make erg3Δ strains in the Cg2001 background strain using pAG25-NatMX and pAG32-HphMX as templates (Fig. 1A and B) and reported replacement efficiencies similar to those observed when targeting ADE2 (Fig. S1B). In addition, the use of 60-bp HRs for generating ERG3 deletions also showed 79% decreased efficiency when compared to 130-bp HRs (Fig. S1C).
After the erg3Δ strains were confirmed by PCR, we performed spot assays to confirm and compare their phenotypes with and without 64 µg/mL fluconazole. Both erg3Δ strains demonstrate a slow growth phenotype but also a clear increased resistance to fluconazole, in contrast to the previously published erg3Δ phenotypes in auxotrophic C. glabrata strains (Fig. 2A). To test whether this phenotype was strain specific, we deleted ERG3 in the BG2 strain with CRISPR-RNP and performed spot assays with and without 64 µg/mL fluconazole and observed a similar azole-resistant phenotype to the Cg2001 erg3Δ strain (Fig. 2B). To quantify this difference, we performed liquid growth assays in both strains. We selected 64 µg/mL fluconazole as it led to a significant growth delay in both WT strains (Fig. S3A). Under untreated conditions, we observed minor differences in both doubling time and growth delays when comparing each erg3Δ strain with their parent WT strain (Fig. 2C and D; Fig. S3B). However, under fluconazole treatment, both erg3Δ strains had a significantly shorter growth lag compared to their respective WT, corroborating the plate-based assay results (Fig. 2C and D; Fig. S3C). Altogether, our data show that azole resistance does occur in prototrophic Cg2001 and BG2 strains when deleted for ERG3, suggesting that the function of Erg3 under azole treatment is conserved in C. glabrata.
Fig 2.
Deletion of ERG3 results in an azole drug-resistant phenotype in C. glabrata. (A and B) Fivefold serial dilution spot assays with and without 64 µg/mL fluconazole (FLZ) in SC (A) and YPD (B) media. Indicated deletion strains were generated using CRISPR-Cas9 RNP. Images were captured at 48 hours. (C and D) Liquid growth assays of the indicated strains in YPD over 50 hours with and without 64 µg/mL FLZ, respectively. Growth curves represent the average of three biological replicates per strain.
ERG3 and ERG5 deletions alter growth and azole drug susceptibility in C. glabrata
Similar to Erg11 (CYP51), Erg5 is also a known cytochrome P-450 (CYP61), which can be inhibited by azole drugs in vitro when using purified Erg5 protein from S. cerevisiae and C. glabrata at similar affinities to Erg11 (31–33). In addition, Candida albicans, Neurospora crassa, and Fusarium verticillioides fungal species deleted for ERG5 show susceptibility to azoles, which has led to a hypothesis that Erg5 may serve as an azole buffer for Erg11 in these fungal species (34, 35). To determine the role of Erg5 in growth and azole susceptibility in C. glabrata, we generated an erg5Δ strain using our CRISPR-RNP approach. The erg5Δ strain grew similar to WT but showed susceptibility to 32 µg/mL fluconazole (Fig. 3A), which may be a consequence of more fluconazole inhibiting Erg11, indicating a protective buffering role for Erg5. Alternatively, under subinhibitory concentrations of azoles, an erg5Δ strain would produce Ergosta 5,7 dienol and/or 14α-methyl-3,6-diol, and both could be acting as growth inhibitory sterols (Fig. S2).
Fig 3.
Double deletion of ERG3 and ERG5 results in a synthetic growth defect in C. glabrata. (A) Fivefold serial dilution spot assays with 0, 32, and 64 µg/mL fluconazole (FLZ) in SC media. Indicated deletion strains were generated using CRISPR-Cas9 RNP. (B and C) Expression of the indicated genes was determined by qRT-PCR analysis of mid-log phase cells in SC media. Data were normalized to RDN18 mRNA levels and are the average of three biological replicates with three technical replicates each. Error bars represent the standard deviation. *P-value < 0.05. n.e., not expressed. Statistical analysis for qRT-PCR analysis was done using GraphPad PRISM using an unpaired two-tailed Student’s t-test. (D and E) Liquid growth assays of the indicated strains over 50 hours with and without 20 µg/mL ergosterol in SC media. Growth curves represent the average of three biological replicates per strain.
To determine if Ergosta 5,7 dienol is contributing to azole susceptibility, we generated an erg3Δerg5Δ double deletion strain in a previously constructed erg3Δ background. Based on known genetic and biochemical data (16, 36), the erg3Δerg5Δ double deletion would prevent the production of Ergosta 5,7 dienol (Fig. S2). While the pAG25 NatMX and pAG32 HphMX drug cassettes are effective for use in single deletions, it can be difficult to generate double gene deletions with drug cassettes when using non-CRISPR methods and short homology regions. This is particularly an issue when drug cassettes share similar flanking sequences, such as the AgTEF1 promoter and the AgTEF1 terminator (Fig. 1A and B). In this case, any subsequent gene deletion attempts could replace the drug cassette of the initially deleted gene, leading to drug cassette swapping. To circumvent this issue, we used our CRISPR-RNP approach and generated erg3Δerg5Δ strains using HphMX and NatMX resistance cassettes, which share similar promoter and terminator sequences. Despite the large AgTEF1 homology regions present, we were able to efficiently generate erg3Δerg5Δ strains with the aid of CRISPR-Cas9-RNP (Fig. S1B). For strain confirmation, we performed qRT-PCR and detected no ERG3 transcript in each strain lacking ERG3 and no ERG5 transcript in each strain lacking ERG5 (Fig. 3B and C). Interestingly, we see a trend of decreased ERG3 expression in the erg5Δ strain and increased expression of ERG5 in the erg3Δ strain (Fig. 3B and C; Table S5). Although not statistically significant, this observation is consistent with what is observed in S. cerevisiae (37, 38). After confirming gene deletions by PCR and qRT-PCR analysis, spot assays were performed with and without fluconazole. Interestingly, all erg3Δerg5Δ strains suppressed azole susceptibility of erg5Δ, suggesting that 14α-methyl-3,6-diol and not Ergosta 5,7 dienol is contributing to erg5Δ’s susceptibility to azoles. Overall, our data support the hypothesis that azole susceptibility of erg5Δ with subinhibitory concentrations of azoles is a consequence of losing Erg5’s azole buffering effect, thereby allowing more azoles to inhibit Erg11.
To our surprise, all erg3Δerg5Δ strains display a synthetic growth defect, more than what was observed in the single erg3Δ and erg5Δ strains (Fig. 3A). We suspect that growth inhibition is due to the buildup of Ergosta 7-enol (Fig. S2). Despite this significant growth defect under untreated conditions, erg3Δerg5Δ strains grew on fluconazole-containing plates similar to an erg3Δ strain, which further supports the hypothesis that Ergosta 7-enol and 14α-methyl-3,6-diol are acting as growth inhibitory toxic sterols in C. glabrata.
In addition to plate-based growth assays, we performed liquid growth assays to quantify changes in growth. The erg5Δ strain had a doubling time and lag phase similar to WT. The erg3Δ strain had a growth defect with a twofold longer doubling time and lag phase compared to WT (Fig. 3D; Fig. S4). The erg3Δerg5Δ strain had a synthetic growth defect with a 3.5-fold increase in both doubling time and lag phase compared to WT, as well as a decrease in OD600 saturation (Fig. 3D; Fig. S4). We hypothesized that these defects were due to altered sterol content in the erg3Δ and erg3Δerg5Δ strains, so we tested if supplementation with 20 µg/mL ergosterol could rescue these phenotypes. Strikingly, ergosterol supplementation led to a near complete rescue of the growth delays in each ERG deletion strain as well as a complete rescue of the saturation defect in erg3Δerg5Δ strains (Fig. 3E). Additionally, erg3Δ strains grew better in YPD vs SC media, which we suspect is due to the presence of ergosterol in YPD but not in SC media (Fig. 2C vs 3D), further supporting that the growth defects of ERG deletion strains are likely due to altered sterol content.
Overall, our genetic studies have identified a potential azole buffering effect for Erg11 contributed by Erg5. In addition, when both ERG3 and ERG5 are deleted, we observe a synthetic growth defect that is likely caused by the buildup of Ergosta 7-enol, suggesting that this sterol is acting as a toxic sterol that inhibits growth. Additional biochemical and genetic studies will be needed to fully understand the observed ERG gene deletion phenotypes.
BleMX and KanMX can be used efficiently to make ADE2 and ERG3 deletions in C. glabrata when using CRISPR-RNP, where ERG3 deletion results in azole drug resistance
To further investigate the ergosterol pathway in prototrophic strains, additional drug resistance markers are needed. Since our CRISPR-RNP system is efficient at generating single and double deletions in C. glabrata using NatMX and HphMX, we then tested whether this system was effective for using other drug resistance cassettes typically not used in C. glabrata. We first tested BleMX, which confers resistance to zeocin, as the use of BleMX has been reported once in C. glabrata using a non-CRISPR transformation method, albeit at extremely low efficiency (<1%) (39). To first determine whether the CRISPR-Cas9 RNP system effectively generates gene deletions using BleMX, we deleted the entire open reading frame of ADE2 using pCY3090-07 as a template (Fig. 4A) (40). When comparing the proportion of red colonies with and without the addition of CRISPR-Cas9, a five- to sixfold increase in efficiency was observed when using CRISPR (Fig. 4B).
Fig 4.
BleMX and KanMX can be used as efficient drug resistance cassettes in C. glabrata when coupled with CRISPR-Cas9 RNP. (A) Schematic of the pCY3090-07 plasmid. P1 and P2 indicate the locations of amplification primer sequences. (B) Total number of positive transformants using BleMX with and without the addition of CRISPR-Cas9 RNP. Numbers are the summation across three separate transformations. (C) Schematic of the pUG6 plasmid. P1 and P2 indicate the locations of amplification primer sequences. (D) Total number of positive transformants using KanMX with and without the addition of CRISPR-RNP. Numbers are the summation across three separate transformations. (E) Fivefold serial dilution spot assays of indicated strains with and without 64 µg/mL fluconazole (FLZ) in SC media. Images were captured at 48 hours.
Next, we tested whether this system permitted efficient use of KanMX as a drug resistance cassette in C. glabrata. Although KanMX is routinely used in S. cerevisiae, KanMX has not been successfully utilized for genetic manipulations in C. glabrata, with the exception of one study (41). Effective use of KanMX would allow for direct repurposing of many S. cerevisiae tagging and deletion KanMX cassettes for C. glabrata. To test this, we deleted ADE2 using a KanMX drug resistance cassette amplified from pUG6 (Fig. 4C) (42). With the addition of CRISPR-RNP, we observed a 55-fold increase in efficiency, suggesting that CRISPR-RNP is required for the use of KanMX in C. glabrata (Fig. 4D).
We also deleted ERG3 with KanMX or BleMX using CRISPR-RNP with similar efficiencies to ADE2 (Fig. S1B). By spot assays, we observed an azole-resistant phenotype similar to the other constructed erg3Δ strains (Fig. 4E). These data demonstrate that KanMX and BleMX can be used as effective drug resistance cassettes in C. glabrata when coupled with CRISPR.
The KanMX drug resistance cassette can be repurposed for generating endogenous epitope-tagged proteins in C. glabrata
Because our data indicate that KanMX is a suitable drug resistance cassette for gene deletions in C. glabrata, we wanted to determine if KanMX could be used for endogenous epitope tagging of ERG3 and ERG11 using the C-terminal 3×HA-KanMX plasmid (pFA6a/PYM1) designed for Schizosaccharomyces pombe but also used in S. cerevisiae (Fig. 5A) (43, 44). ERG3 and ERG11 tags were generated using CRISPR-RNP and G418 selection. After PCR confirmation, tagged strains were grown with and without 64 µg/mL fluconazole in SC media and collected at mid-log phase for immunoblotting using anti-HA (12CA5). Histone H3 was used as a loading control. Our data indicate that Erg3 and Erg11 proteins are expressed under untreated conditions and induced under fluconazole treatment (Fig. 5B and C), which is consistent with transcript analysis from previous studies (45, 46). To confirm that the epitope tag does not alter function, we performed spot assays with and without 64 µg/mL fluconazole, using an erg3Δ strain as a control. All epitope-tagged Erg3-3×HA and Erg11-3×HA strains grow similar to WT under both untreated and fluconazole treatment (Fig. 5D and E). Altogether, these data suggest that KanMX epitope tagging constructs used in S. pombe and S. cerevisiae can be repurposed in C. glabrata and that C-terminal tagging of Erg11 and Erg3 does not alter their function.
Fig 5.
C-terminal 3×HA tagging of ERG3 and ERG11 does not alter azole susceptibility in C. glabrata. (A) Schematic of the pFA6-3HA-KanMX plasmid. P1 and P2 indicate the locations of amplification primer sequences. (B and C) Indicated strains were either untreated (−) or treated (+) with 64 µg/mL fluconazole (FLZ) for 3 hours. Whole cell extracts were isolated and immunoblotted against an anti-HA antibody for the detection of Erg3 or Erg11. Histone H3 was used as a loading control. Three independent clones were represented for Erg3-3×HA and Erg11-3×HA. (D and E) Fivefold serial dilution spot assays in SC media of indicated strains with 0, 16, and 64 µg/mL FLZ, respectively. Three independent clones were represented for Erg3-3×HA and Erg11-3×HA. Images were captured at 48 hours.
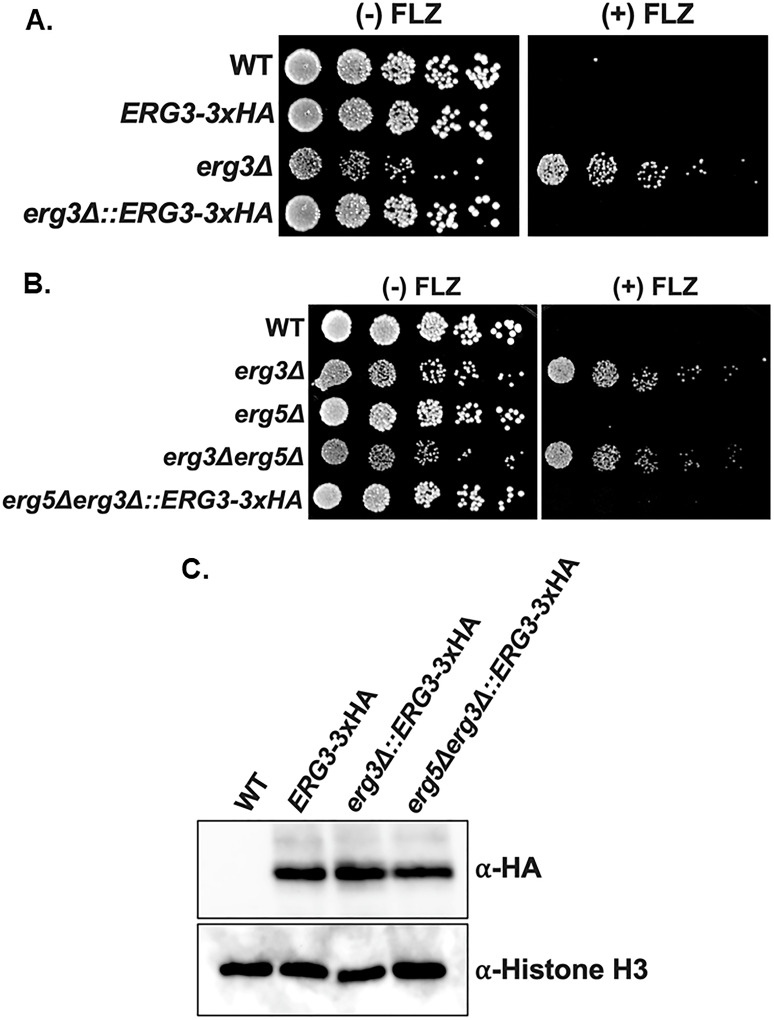
ERG3-3×HA-KanMX complements erg3Δ and erg3Δerg5Δ phenotypes
Because C-terminal tagging of ERG3 did not disrupt function (Fig. 5D), we used the ERG3-3×HA-KanMX strain as a genomic template to generate a replacement cassette to complement the erg3Δ and erg3Δerg5Δ strain phenotypes. The ERG3-3×HA-KanMX cassette was PCR amplified, and CRISPR-RNP was targeted to the NatMX cassette that was used to delete ERG3. Re-introducing ERG3 at its endogenous locus in the erg3Δ strain restored WT growth and azole susceptibility (Fig. 6A). In addition, re-introduction of ERG3 at its endogenous locus in the erg3Δerg5Δ strain reverts erg3Δerg5Δ growth back to erg5Δ and restores azole susceptibility (Fig. 6B). We were able to confirm that Erg3 is expressed by Western blotting (Fig. 6C). Altogether, these data support that the azole-resistant phenotype observed in our erg3Δ strains can be suppressed when complemented with Erg3. These complementation studies indicate that Erg3 was solely responsible for the erg3Δ azole-resistant phenotype. In addition, restoring Erg3 in erg3Δ and erg3Δerg5Δ strains rescues azole susceptibility, which is a consequence of Erg3’s ability to produce the growth inhibitory toxic sterol 14α-methyl-3,6-diol (Fig. S2). Furthermore, complementation of erg3Δerg5Δ also suppressed the growth defect, further supporting that Ergosta-7-enol is another potential growth inhibitory toxic sterol. Because of our demonstrated efficient use of KanMX and BleMX, additional genetic studies can be done to further address these discoveries in prototrophic strains.
Fig 6.
Complementation at ERG3’s endogenous locus with ERG3-3×HA-KanMX rescues erg3Δ and erg3Δerg5Δ phenotypes. (A and B) Fivefold serial dilution spot assays of indicated strains with 0 and 64 µg/mL fluconazole (FLZ) in SC media. Images were captured at 48 hours. (C) Whole cell extracts were isolated and immunoblotted against an anti-HA antibody for the detection of Erg3 in complemented strains. Histone H3 was used as a loading control, and ERG3-3×HA was used as a positive control.
BleMX and KanMX can be used efficiently to make ADE2 and ERG3 deletions in C. auris when using CRISPR-RNP, where ERG3 deletion results in azole drug resistance
Next, we tested whether we could use BleMX and KanMX as drug resistance cassettes in the emerging pathogen, C. auris, since previous studies in C. auris have been limited to using SAT1 and CaHyg as drug resistance cassettes (47, 48). We first generated a codon-optimized BleMX for use in CTG clade species and named the plasmid pCdOpt-BMX (Fig. 7A). Using this codon-optimized BleMX plasmid as a template, we deleted ADE2 in C. auris AR0387 using the CRISPR-RNP method and reported 65% efficiency compared to 0% replacement without CRISPR (Fig. 7B). Next, we used pSFS2A-CaKan codon-optimized KanMX as a template and again targeted ADE2 (Fig. 7C). Using this codon-optimized KanMX, we report 29% efficiency with CRISPR compared to 0% without (Fig. 7D).
Fig 7.
BleMX and KanMX can be used as drug resistance cassettes in C. auris when coupled with CRISPR-Cas9-RNP. (A) Schematic of the pCdOpt-BMX plasmid. P1 and P2 indicate the locations of amplification primer sequences. (B) Total number of positive transformants using BleMX with and without the addition of CRISPR-Cas9 RNP. (C) Schematic of the pSFS2A-CaKan plasmid. P1 and P2 indicate the locations of amplification primer sequences. (D) Total number of positive transformants using KanMX with and without the addition of CRISPR-Cas9 RNP. (E) Fivefold serial dilution spot assays of indicated C. auris strains with and without 64 µg/mL fluconazole (FLZ) in SC media. Three independent clones were represented for each erg3Δ strain deleted with BleMX or KanMX. Images were captured at 48 hours.
Because erg3Δ phenotypes have not been reported in C. auris, we deleted ERG3 with both BleMX and KanMX. After PCR confirmation, we performed spot assays with and without 64 µg/mL fluconazole. Similar to the C. glabrata erg3Δ strains, we observed an azole-resistant phenotype across all clones tested (Fig. 7E). Altogether, these data demonstrate that BleMX and KanMX can be used for efficient gene replacement in C. auris when coupled with CRISPR. This is the first report that erg3Δ strains are azole resistant in C. auris, demonstrating the conservation of Erg3 function in C. auris and providing support for Erg3-dependent production of the toxic 14α-methyl-3,6-diol sterol.
BleMX and KanMX can be used to make simultaneous ERG3 allele deletions in C. albicans when coupled with SAT1 and CRISPR-RNP
Because our data show that KanMX and BleMX are effective drug resistance cassettes for deleting ERG3 in prototrophic C. glabrata and C. auris strains when coupled to CRISPR-RNP, we next tested if these drug resistance cassettes could be used in C. albicans. Although erg3Δ strains have been generated in auxotrophic C. albicans strains (15), homozygous erg3Δ strains have not been characterized in a prototrophic SC5314 strain or other prototrophic C. albicans isolates. In contrast to C. glabrata and C. auris, C. albicans possesses innate resistance toward G418, hygromycin B, and zeocin even when using high concentrations of these drug compounds, which prevents using them for genetic manipulation. Interestingly, a dual selection approach with both SAT1 and HphMX replacement cassettes coupled with a CRISPR-RNP approach was shown to be effective at generating homozygous double deletions (49). Therefore, we hypothesized that an analogous approach using SAT1 and BleMX or KanMX would work for deleting ERG3.
To test this approach, ERG3 drug resistance cassettes were PCR amplified using both pCdOpt-BMX (BleMX) and pBBS2-SAT1-FLP as templates and simultaneously transformed into SC5314 using our CRISPR-RNP method. When transformants were plated on zeocin alone, we observed no drug selectivity; however, drug selectivity was observed on plates containing both zeocin and nourseothricin or nourseothricin alone (Fig. 8A). From the zeocin and nourseothricin plates, we obtained 16% positive erg3Δ/erg3Δ strains confirmed by PCR (Fig. 8B; Fig. S5A). We then used the same transformation approach but instead with SAT1 and KanMX as replacement templates. Again, we observed no selectivity when plated on G418 plates alone, but drug selectivity was observed on plates containing both G418 and nourseothricin (Fig. 8C). However, we obtained 10% positive erg3Δ/erg3Δ strains, and PCR confirmed the integration of both cassettes (Fig. 8D; Fig. S5B). Interestingly, we were able to get >94% of the SAT1 integration, which allowed us to identify heterozygous ERG3/erg3Δ strains, indicating that both homozygous and heterozygous deletions can be identified from colonies grown on the combined drug plate (Fig. 8B and D; Fig. S5A and B).
Fig 8.
BleMX and KanMX can be used in conjunction with SAT1 for simultaneous homozygous deletions in C. albicans. (A) Representative transformation plated on zeocin alone, nourseothricin alone, and both zeocin and nourseothricin. Images were captured at 48 hours. (B) Percentage of homozygous and heterozygous ERG3 deletions screened by PCR from BleMX+SAT1 double selection plates. (C) Representative transformation plated on G418 alone, nourseothricin alone, and both G418 and nourseothricin. Images were captured at 48 hours. (D) Percentage of homozygous and heterozygous ERG3 deletions screened by PCR from KanMX+SAT1 double selection plates. (E) Fivefold serial dilution spot assays of indicated C. albicans strains with and without 64 µg/mL fluconazole (FLZ) in SC media. Images were captured at 48 hours.
To assess the deletion strains, spot assays were performed with and without 64 µg/mL fluconazole (Fig. 8E). Azole resistance was observed for all erg3Δ/erg3Δ strains, corroborating previous deletions in auxotrophic C. albicans strains (15). Similar to HphMX, our data show that BleMX and KanMX can be used in conjunction with SAT1 for homozygous double deletion engineering in C. albicans. In addition to C. glabrata and C. auris, we have also expanded the tools to genetically manipulate prototrophic C. albicans strains.
DISCUSSION
In this study, we have expanded the Candida toolbox to include KanMX and BleMX when coupled to CRISPR-RNP, which has allowed us to efficiently manipulate the ergosterol pathway. Using these tools, we have established for the first time that deleting ERG3 results in an azole drug-resistant phenotype in C. glabrata, C. auris, and C. albicans prototrophic strains. We also show that a C. glabrata erg5Δ strain is susceptible to azoles, while erg3Δerg5Δ strains show a synthetic growth defect. Overall, we provided new tools for genetic manipulation that allowed us to determine the impact of multiple ERG gene deletions in Candida.
In S. cerevisiae and C. albicans, erg3Δ strains have been well characterized to have an azole drug-resistant phenotype, which is attributed to the failure to make the growth inhibitory toxic sterol, 14α-methyl-3,6-diol (14, 15). In contrast, the prevailing thought in the field is that this does not occur when ERG3 is deleted in C. glabrata because erg3Δ strains have been reported to be susceptible to azole drugs (16, 17). However, this observation has been based mainly on one report where an ERG3 deletion in an auxotrophic C. glabrata strain was reported to be azole susceptible (16). Based on our findings, Erg3’s function is conserved, and deletion of ERG3 leads to azole drug resistance in all prototrophic strains tested. Furthermore, our ERG3 complemented strains generated by our expanded toolkit show that azole drug susceptibility can be restored, indicating that only ERG3 was responsible for the observed phenotypes.
Currently, it is unclear why other groups have observed susceptibility in C. glabrata auxotrophic erg3Δ strains. However, an issue using auxotrophic strains for studying the ergosterol pathway could be an unintentional altered growth phenotype caused by mutations in the amino acid biosynthetic pathway. For example, it has been shown in S. cerevisiae that azole-resistant phenotypes in an erg3Δ strain were contingent on the auxotrophic status of the strain (50). Alternatively, because azoles have been shown to bind to Erg5 and Erg11 with equal affinity and inhibit both enzymes in vitro (31–33), the observed azole susceptibility difference in erg3Δ strains could be caused by differences in Erg5 expression or non-synonymous mutations across C. glabrata strains. These differences could alter the buffering capacity of Erg5 toward azoles and thus change the concentration of azoles that inhibit Erg11. Another possibility could be a paradoxical effect where subinhibitory concentrations of azoles in an erg3Δ strain fail to completely block Erg11, resulting in a potential buildup of another toxic sterol, leading to azole susceptibility. In this case, if Erg5 is effectively inhibited in erg3Δ, this would phenocopy an erg3Δerg5Δ strain, resulting in a buildup in Ergosta-7-enol and growth inhibition (Fig. 3D; Fig. S2). Further investigation will be needed to determine why the reported auxotrophic erg3Δ strains show azole susceptibility and whether this is due to amino acid uptake, Erg5 fluconazole buffering effect, and/or toxic sterol. Nonetheless, we observed broad conservation of erg3Δ phenotypes using prototrophic strains resulting in azole resistance across C. glabrata, C. auris, and C. albicans when treated with growth inhibitory azole concentrations, supporting the notion that Erg3 contributes to the production of the toxic sterol 14α-methyl-3,6-diol in all prototrophic strains tested.
In this study, we show that two additional drug resistance cassettes, KanMX and BleMX, commonly used for S. pombe or S. cerevisiae can be repurposed and used reliably in C. glabrata. Moreover, we show that the codon-optimized KanMX from the pSFS2A-CaKan (KanMX) plasmid (51) and our generated codon-optimized BleMX from the pCdOpt-BMX plasmid can be used for C. auris. In contrast to C. glabrata and C. auris, KanMX or BleMX in C. albicans cannot be used due to this organism’s high tolerance/resistance to the aminoglycoside antibiotic G418 and the glycopeptide-derived antibiotics bleomycin, phleomycin, and zeocin (see Fig. 6). Because the CRISPR-RNP-based system has been used successfully to simultaneously delete both alleles in C. albicans when using SAT1 and HygB (49), we applied a similar approach to use KanMX and BleMX. While we observed lower percentages of homozygous double deletions using this approach, adjuvants such as quinine or molybdate have been shown to suppress background growth of C. albicans when grown on G418 or hygromycin, which allows successful integration of codon-optimized CaKan and CaHygB cassettes (51). The addition of adjuvants and/or the use of integrated/transient expression-based CRISPR systems (26, 52) could further improve the efficiency of simultaneous allele deletions.
Our study also successfully demonstrates the repurposing of KanMX-containing plasmids traditionally utilized for making gene deletions or C-terminal epitope tags in S. pombe or S. cerevisiae for use in C. glabrata. While we clearly demonstrate that the endogenous C-terminal 3×HA tagging constructs are suitable for C. glabrata, this approach may not work for all genes, as C-terminal tagging may disrupt the function of the protein. Thus, our approach would also allow for repurposing endogenous N-terminal KanMX tagging constructs designed for S. cerevisiae (53). Additionally, the efficiency of endogenous epitope-tagged proteins using CRISPR allows for more functional and mechanistic studies beyond transcript analysis, as endogenous epitope-tagged proteins have been used sparingly in prototrophic strains and clinical isolates of C. glabrata. This is particularly important since antibodies to endogenous proteins are scarce and costly to make. Finally, we show that endogenous epitope-tagged genes can be used as templates to complement the function of their respective gene deletion.
Overall, our study provides the field with additional ways to efficiently manipulate prototrophic Candida pathogens when using a CRISPR-based approach. Importantly, this approach provides us with further insight into the ergosterol pathway, although additional studies would be needed to address the mechanisms of our new observations. Applying this expanded toolkit in Candida should also enhance our understanding of other complex pathways impacting fungal drug resistance and pathogenesis.
MATERIALS AND METHODS
Yeast strains and plasmids
All strains used are described in Table S1. C. glabrata strains were derived from Cg2001 or BG2 (ATCC 2001). C. albicans strains were derived from SC5314 (54), a gift from William A. Fonzi, Georgetown University. The C. auris AR0387 strain was obtained from the CDC AR Isolate Bank. The pAG25, pAG32, and pUG6 plasmids were obtained from Euroscarf (27, 42). The pFA6a-3HA-KanMX, pCY3090-07, and pSFS2A-CaKan plasmids were obtained from Addgene (40, 43, 51). The pBSS2-SAT1 flipper plasmid was provided to us by P. David Rogers, St. Jude Children’s Research Hospital, with permission from Joachim Morschauser (55). pCdOpt-BMX (BleMX) was synthesized by IDT, where the TEF1p-BleMX-TEF1t sequence was codon optimized for CTG clade Candida species, synthesized, and cloned into the pUCIDT plasmid. The pCdOpt-BMX plasmid can be obtained at Addgene (ID number 203929).
PCR amplification for gene deletion and epitope tagging
All oligonucleotides used are denoted in Table S2. PCR amplification of drug resistance cassettes is as follows: 95°C for 5 minutes, 95°C for 30 seconds, 52°C for 30 seconds, and 72°C for 2–3 minutes for a total of 30 cycles, with a final elongation step at 72°C for 10 minutes. The PCR products were purified from agarose gels.
CRISPR gRNA design and selection
Custom Alt-R CRISPR gRNAs were designed and ordered from Integrated DNA Technologies (Table S3). For each gene deletion, two CRISPR gRNAs were designed close to the 5′ and 3′ open reading frames (ORFs) of the gene of interest. For epitope tagging, one CRISPR gRNA was designed in the 3′ untranslated region (UTR) of the gene of interest. CRISPR gRNAs were selected based on their designated “on-target score” as determined by the CRISPR-Cas9 guide RNA design checker (IDT). Potential gRNAs were screened for off-target events using the CRISPR RGEN Tools Cas OFFinder (http://www.rgenome.net/cas-offinder/). Selected gRNAs required >75 on-target scores as well as 0 potential off-target events with three mismatches or less.
CRISPR-Cas9 RNP system
The CRISPR-Cas9 RNP method was based on Grahl et al. with slight modifications (21). Briefly, Alt-R CRISPR crRNA and tracrRNA were used at a working concentration of 20 µM. The CRISPR-Cas9 crRNAs:tracrRNA hybrid was made by mixing together 1.6 µL of crRNA (8 µM final concentration), 1.6 µL of tracrRNA (8 µM final concentration), and 0.8 µL of RNAse-free water. For gene deletions, two crRNAs, 0.8 µL each, were added at a stoichiometric equivalent to tracrRNA, and for C-terminal tagging, one crRNA, 1.6 µL, was used. The CRISPR-RNP mix was incubated at 95°C for 5 minutes and allowed to cool to room temperature. Three microliters of 4 µM Cas9 (IDT) was added to the mix (final concentration of 1.7 µM) and incubated at room temperature for 5 minutes.
Cell transformation
Twenty-five milliliters of the desired strain was grown to an OD600 of 1.6 to saturation prior to transformation. Cells were resuspended in 10 mL of 1× LiTE Buffer (100 mM LiAc, 10 mM Tris-HCl, and 1 mM EDTA) and shaken at 250 rpm at 30°C for an hour. DTT was added to a final concentration of 100 mM, and cells were incubated at 30°C for an additional 30 minutes. Cells were collected by centrifugation, washed twice with 1 mL of ice-cold water, and washed once with 1 mL of cold sorbitol. Cells were resuspended in 200 µL of cold sorbitol for electroporation.
Electroporation and colony PCR
Twenty microliters of prepared cells, 1–3 µg of drug resistance cassette DNA, CRISPR mix, and RNAse-free water to a final volume of 45 µL was mixed and transferred to a Bio-Rad Gene Pulser cuvette (0.2-cm gap). Cells were pulsed using an Eppendorf Eporator at 1,500 V and immediately resuspended in 1 mL of ice-cold sorbitol. Cells were collected by centrifugation, resuspended in 1 mL of YPD media, and allowed to recover by incubation at 30°C at 250 rpm for 3–24 hours. Recovered cells were resuspended in 100 µL of YPD and plated onto drug-selective media. Nourseothricin (GoldBio) was used at a final concentration of 300 µg/mL for antibiotic selection of the NatMX cassette. Hygromycin B (Cayman) was used at a final concentration of 500 µg/mL. Geneticin (G418, GoldBio) was used at a final concentration of 800 µg/mL. Zeocin (Cayman) was used at a final concentration of 600 µg/mL for C. glabrata and 800 µg/mL for C. auris. For double selection for C. albicans using SAT1 and BleMX or KanMX, plates contained 200 µg/mL nourseothricin and 800 µg/mL zeocin or 800 µg/mL G418 sulfate. Colonies were screened via PCR using primers indicated in Table S2. Three independent clones were used for phenotypic characterizations.
Serial dilution spot assays and liquid growth assays
For serial dilution spot assays, yeast strains were inoculated in SC or YPD media and grown to saturation overnight as previously published (45). Yeast strains were diluted to an OD600 of 0.1 and grown in SC or YPD media to log phase with shaking at 30°C. The indicated strains were spotted in fivefold dilutions starting at an OD600 of 0.01 on untreated SC or YPD plates or plates containing 8, 16, or 64 µg/mL fluconazole (Cayman). For C. glabrata, C. albicans, and C. auris, plates were grown at 30°C for 48 hours prior to imaging. For liquid growth assays, the indicated yeast strains were inoculated in SC or YPD media and grown to saturation overnight. Yeast strains were diluted to an OD600 of 0.1 and grown in SC or YPD media to log phase with shaking at 30°C. The indicated strains were diluted to an OD600 of 0.01 in 100 µL SC or YPD media. Cells were left untreated or treated with 20 µg/mL ergosterol (Alfa Aesar) and grown for 50 hours with shaking at 30°C. The OD600 was determined every 15 minutes using a Bio-Tek Synergy 4 multimode plate reader.
Quantitative real-time PCR analysis
RNA was isolated from cells grown in SC media by standard acid phenol purification as previously described (56). ABM All-In-One 5X RT MasterMix (ABM) was used to generate cDNA. Primers for gene expression analysis are indicated in Table S4. A minimum of three biological replicates, as well as three technical replicates, were performed for each biological replicate using the comparative CT method (2−ΔΔCT) see Table S5.
Cell extract and Western blot analysis
Whole cell extraction and Western blot analysis were performed as previously described (57, 58). The anti-HA (Roche 12CA5, 1:10,000) monoclonal antibody was used as previously described (59). Histone H3 rabbit polyclonal antibody (PRF&L) was used at a 1:100,000 dilution as previously described (60).
ACKNOWLEDGMENTS
We thank Drs. Majid Kazemian and Mark Hall for critical review of our manuscript.
This publication was supported by grants from the National Institute of Allergy and Infectious Diseases of NIH under award numbers T32AI148103 (to J.B.G.) and AI136995 (to S.D.B.). Funding support was also provided by the NIFA 1007570 (to S.D.B) and NSF DBI-2150331 (to M.G.B. and C.V.).
Contributor Information
Scott D. Briggs, Email: sdbriggs@purdue.edu.
Aaron P. Mitchell, University of Georgia, Athens, Georgia, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msphere.00311-23.
Fig. S1 to S5 and Tables S1 to S5.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Bongomin F, Gago S, Oladele RO, Denning DW. 2017. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi (Basel) 3:57. doi: 10.3390/jof3040057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rayens E, Norris KA. 2022. Prevalence and healthcare burden of fungal infections in the United States, 2018. Open Forum Infect Dis 9:ofab593. doi: 10.1093/ofid/ofab593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borjian Boroujeni Z, Shamsaei S, Yarahmadi M, Getso MI, Salimi Khorashad A, Haghighi L, Raissi V, Zareei M, Saleh Mohammadzade A, Moqarabzadeh V, Soleimani A, Raeisi F, Mohseni M, Mohseni MS, Raiesi O. 2021. Distribution of invasive fungal infections: molecular epidemiology, etiology, clinical conditions, diagnosis and risk factors: a 3-year experience with 490 patients under intensive care. Microb Pathog 152:104616. doi: 10.1016/j.micpath.2020.104616 [DOI] [PubMed] [Google Scholar]
- 4. Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. doi: 10.1128/CMR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Webb BJ, Ferraro JP, Rea S, Kaufusi S, Goodman BE, Spalding J. 2018. Epidemiology and clinical features of invasive fungal infection in a US health care network. Open Forum Infect Dis 5:ofy187. doi: 10.1093/ofid/ofy187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodrigues CF, Silva S, Henriques M. 2014. Candida glabrata: a review of its features and resistance. Eur J Clin Microbiol Infect Dis 33:673–688. doi: 10.1007/s10096-013-2009-3 [DOI] [PubMed] [Google Scholar]
- 7. White TC, Holleman S, Dy F, Mirels LF, Stevens DA. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob Agents Chemother 46:1704–1713. doi: 10.1128/AAC.46.6.1704-1713.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rybak JM, Barker KS, Muñoz JF, Parker JE, Ahmad S, Mokaddas E, Abdullah A, Elhagracy RS, Kelly SL, Cuomo CA, Rogers PD. 2022. In vivo emergence of high-level resistance during treatment reveals the first identified mechanism of amphotericin B resistance in Candida auris. Clin Microbiol Infect 28:838–843. doi: 10.1016/j.cmi.2021.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x [DOI] [PubMed] [Google Scholar]
- 10. Johnston EJ, Moses T, Rosser SJ. 2020. The wide-ranging phenotypes of ergosterol biosynthesis mutants, and implications for microbial cell factories. Yeast 37:27–44. doi: 10.1002/yea.3452 [DOI] [PubMed] [Google Scholar]
- 11. Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AGS, Rolley N, Kelly DE, Kelly SL. 2010. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob Agents Chemother 54:4527–4533. doi: 10.1128/AAC.00348-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alcazar-Fuoli L, Mellado E. 2012. Ergosterol biosynthesis in Aspergillus fumigatus: its relevance as an antifungal target and role in antifungal drug resistance. Front Microbiol 3:439. doi: 10.3389/fmicb.2012.00439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jordá T, Puig S. 2020. Regulation of ergosterol biosynthesis in Saccharomyces cerevisiae. Genes (Basel) 11:795. doi: 10.3390/genes11070795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Watson PF, Rose ME, Ellis SW, England H, Kelly SL. 1989. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem Biophys Res Commun 164:1170–1175. doi: 10.1016/0006-291x(89)91792-0 [DOI] [PubMed] [Google Scholar]
- 15. Sanglard D, Ischer F, Parkinson T, Falconer D, Bille J. 2003. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob Agents Chemother 47:2404–2412. doi: 10.1128/AAC.47.8.2404-2412.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geber A, Hitchcock CA, Swartz JE, Pullen FS, Marsden KE, Kwon-Chung KJ, Bennett JE. 1995. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother 39:2708–2717. doi: 10.1128/AAC.39.12.2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhakt P, Raney M, Kaur R. 2022. The SET-domain protein CgSet4 negatively regulates antifungal drug resistance via the ergosterol biosynthesis transcriptional regulator CgUpc2A. J Biol Chem 298:102485. doi: 10.1016/j.jbc.2022.102485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Akins RA. 2005. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med Mycol 43:285–318. doi: 10.1080/13693780500138971 [DOI] [PubMed] [Google Scholar]
- 19. Ksiezopolska E, Schikora-Tamarit MÀ, Beyer R, Nunez-Rodriguez JC, Schüller C, Gabaldón T. 2021. Narrow mutational signatures drive acquisition of multidrug resistance in the fungal pathogen Candida glabrata. Curr Biol 31:5314–5326. doi: 10.1016/j.cub.2021.09.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pais P, Galocha M, Takahashi-Nakaguchi A, Chibana H, Teixeira MC. 2022. Multiple genome analysis of Candida glabrata clinical isolates renders new insights into genetic diversity and drug resistance determinants. Microb Cell 9:174–189. doi: 10.15698/mic2022.11.786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grahl N, Demers EG, Crocker AW, Hogan DA. 2017. Use of RNA-protein complexes for genome editing in non-albicans Candida species. mSphere 2:e00218-17. doi: 10.1128/mSphere.00218-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwarzmüller T, Ma B, Hiller E, Istel F, Tscherner M, Brunke S, Ames L, Firon A, Green B, Cabral V, Marcet-Houben M, Jacobsen ID, Quintin J, Seider K, Frohner I, Glaser W, Jungwirth H, Bachellier-Bassi S, Chauvel M, Zeidler U, Ferrandon D, Gabaldón T, Hube B, d’Enfert C, Rupp S, Cormack B, Haynes K, Kuchler K. 2014. Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog 10:e1004211. doi: 10.1371/journal.ppat.1004211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Enkler L, Richer D, Marchand AL, Ferrandon D, Jossinet F. 2016. Genome engineering in the yeast pathogen Candida glabrata using the CRISPR-Cas9 system. Sci Rep 6:35766. doi: 10.1038/srep35766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edlind TD, Henry KW, Vermitsky JP, Edlind MP, Raj S, Katiyar SK. 2005. Promoter-dependent disruption of genes: simple, rapid, and specific PCR-based method with application to three different yeast. Curr Genet 48:117–125. doi: 10.1007/s00294-005-0008-3 [DOI] [PubMed] [Google Scholar]
- 25. Vyas VK, Bushkin GG, Bernstein DA, Getz MA, Sewastianik M, Barrasa MI, Bartel DP, Fink GR. 2018. New CRISPR mutagenesis strategies reveal variation in repair mechanisms among fungi. mSphere 3:00154–18. doi: 10.1128/mSphere.00154-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Min K, Ichikawa Y, Woolford CA, Mitchell AP, Imperiale MJ. 2016. Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere 1:00130–16. doi: 10.1128/mSphere.00130-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goldstein AL, McCusker JH. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553. doi: [DOI] [PubMed] [Google Scholar]
- 28. Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S. 2002. Molecular basis of resistance to azole antifungals. Trends Mol Med 8:76–81. doi: 10.1016/s1471-4914(02)02280-3 [DOI] [PubMed] [Google Scholar]
- 29. Robbins N, Cowen LE. 2021. Antifungal drug resistance: deciphering the mechanisms governing multidrug resistance in the fungal pathogen Candida glabrata. Curr Biol 31:R1520–R1523. doi: 10.1016/j.cub.2021.09.071 [DOI] [PubMed] [Google Scholar]
- 30. Bhattacharya S, Esquivel BD, White TC, Lorenz M. 2018. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. mBio 9:e01291-18. doi: 10.1128/mBio.01291-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hitchcock CA, Dickinson K, Brown SB, Evans EG, Adams DJ. 1990. Interaction of azole antifungal antibiotics with cytochrome P-450-dependent 14 alpha-sterol demethylase purified from Candida albicans. Biochem J 266:475–480. doi: 10.1042/bj2660475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kelly SL, Lamb DC, Baldwin BC, Corran AJ, Kelly DE. 1997. Characterization of Saccharomyces cerevisiae CYP61, sterol delta22-desaturase, and inhibition by azole antifungal agents. J Biol Chem 272:9986–9988. doi: 10.1074/jbc.272.15.9986 [DOI] [PubMed] [Google Scholar]
- 33. Lamb DC, Maspahy S, Kelly DE, Manning NJ, Geber A, Bennett JE, Kelly SL. 1999. Purification, reconstitution, and inhibition of cytochrome P-450 sterol delta22-desaturase from the pathogenic fungus Candida glabrata. Antimicrob Agents Chemother 43:1725–1728. doi: 10.1128/AAC.43.7.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun X, Wang W, Wang K, Yu X, Liu J, Zhou F, Xie B, Li S. 2013. Sterol C-22 desaturase ERG5 mediates the sensitivity to antifungal azoles in Neurospora crassa and Fusarium verticillioides. Front Microbiol 4:127. doi: 10.3389/fmicb.2013.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mount HO, Revie NM, Todd RT, Anstett K, Collins C, Costanzo M, Boone C, Robbins N, Selmecki A, Cowen LE. 2018. Global analysis of genetic circuitry and adaptive mechanisms enabling resistance to the azole antifungal drugs. PLoS Genet 14:e1007319. doi: 10.1371/journal.pgen.1007319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu G, Chen Y, Færgeman NJ, Nielsen J. 2017. Elimination of the last reactions in ergosterol biosynthesis alters the resistance of Saccharomyces cerevisiae to multiple stresses. FEMS Yeast Res 17. doi: 10.1093/femsyr/fox063 [DOI] [PubMed] [Google Scholar]
- 37. Arthington-Skaggs BA, Crowell DN, Yang H, Sturley SL, Bard M. 1996. Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the post-squalene portion of the yeast ergosterol pathway. FEBS Lett 392:161–165. doi: 10.1016/0014-5793(96)00807-1 [DOI] [PubMed] [Google Scholar]
- 38. Skaggs BA, Alexander JF, Pierson CA, Schweitzer KS, Chun KT, Koegel C, Barbuch R, Bard M. 1996. Cloning and characterization of the Saccharomyces cerevisiae C-22 sterol desaturase gene, encoding a second cytochrome P-450 involved in ergosterol biosynthesis. Gene 169:105–109. doi: 10.1016/0378-1119(95)00770-9 [DOI] [PubMed] [Google Scholar]
- 39. Alderton AJ, Burr I, Mühlschlegel FA, Tuite MF. 2006. Zeocin resistance as a dominant selective marker for transformation and targeted gene deletions in Candida glabrata. Mycoses 49:445–451. doi: 10.1111/j.1439-0507.2006.01271.x [DOI] [PubMed] [Google Scholar]
- 40. Young CL, Raden DL, Caplan JL, Czymmek KJ, Robinson AS. 2012. Cassette series designed for live-cell imaging of proteins and high-resolution techniques in yeast. Yeast 29:119–136. doi: 10.1002/yea.2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cormack BP, Falkow S. 1999. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 151:979–987. doi: 10.1093/genetics/151.3.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24:2519–2524. doi: 10.1093/nar/24.13.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bähler J, Wu J-Q, Longtine MS, Shah NG, Mckenzie III A, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951. doi: [DOI] [PubMed] [Google Scholar]
- 44. Chandrasekharan MB, Huang F, Chen YC, Sun ZW. 2010. Histone H2B C-terminal helix mediates trans-histone H3K4 methylation independent of H2B ubiquitination. Mol Cell Biol 30:3216–3232. doi: 10.1128/MCB.01008-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baker KM, Hoda S, Saha D, Gregor JB, Georgescu L, Serratore ND, Zhang Y, Cheng L, Lanman NA, Briggs SD. 2022. The Set1 histone H3K4 methyltransferase contributes to azole susceptibility in a species-specific manner by differentially altering the expression of drug efflux pumps and the ergosterol gene pathway. Antimicrob Agents Chemother 66:e02250–21. doi: 10.1128/aac.02250-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vu BG, Thomas GH, Moye-Rowley WS. 2019. Evidence that ergosterol biosynthesis modulates activity of the Pdr1 transcription factor in Candida glabrata. mBio 10:e00934-19. doi: 10.1128/mBio.00934-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim SH, Iyer KR, Pardeshi L, Muñoz JF, Robbins N, Cuomo CA, Wong KH, Cowen LE. 2019. Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. mBio 10:e00346-19. doi: 10.1128/mBio.00346-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rybak JM, Doorley LA, Nishimoto AT, Barker KS, Palmer GE, Rogers PD. 2019. Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris. Antimicrob Agents Chemother (Bethesda) 63. doi: 10.1128/AAC.00057-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu J, Vogel AK, Miao J, Carnahan JA, Lowes DJ, Rybak JM, Peters BM, O’Meara TR. 2022. Rapid hypothesis testing in Candida albicans clinical isolates using a cloning-free, modular, and recyclable system for CRISPR-Cas9 mediated mutant and revertant construction. Microbiol Spectr 10. doi: 10.1128/spectrum.02630-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robbins N, Collins C, Morhayim J, Cowen LE. 2010. Metabolic control of antifungal drug resistance. Fungal Genet Biol 47:81–93. doi: 10.1016/j.fgb.2009.07.004 [DOI] [PubMed] [Google Scholar]
- 51. Park SO, Frazer C, Bennett RJ. 2022. An adjuvant-based approach enables the use of dominant HYG and KAN selectable markers in Candida albicans. mSphere 7:e0034722. doi: 10.1128/msphere.00347-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vyas VK, Barrasa MI, Fink GR. 2015. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv 1:e1500248. doi: 10.1126/sciadv.1500248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y, Serratore ND, Briggs SD. 2017. N-ICE plasmids for generating N-terminal 3× FLAG tagged genes that allow inducible, constitutive or endogenous expression in Saccharomyces cerevisiae. Yeast 34:223–235. doi: 10.1002/yea.3226 [DOI] [PubMed] [Google Scholar]
- 54. Fonzi WA, Irwin MY. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728. doi: 10.1093/genetics/134.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reuss O, Vik A, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021 [DOI] [PubMed] [Google Scholar]
- 56. Baker KM, Hoda S, Saha D, Gregor JB, Georgescu L, Serratore ND, Zhang Y, Cheng L, Lanman NA, Briggs SD. 2022. The Set1 histone H3K4 methyltransferase contributes to azole susceptibility in a species-specific manner by differentially altering the expression of drug efflux pumps and the ergosterol gene pathway. Antimicrob Agents Chemother 66:e0225021. doi: 10.1128/aac.02250-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. 2012. Charge-based interaction conserved within histone H3 lysine 4 (H3K4) methyltransferase complexes is needed for protein stability, histone methylation, and gene expression. J Biol Chem 287:2652–2665. doi: 10.1074/jbc.M111.280867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fingerman IM, Wu CL, Wilson BD, Briggs SD. 2005. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J Biol Chem 280:28761–28765. doi: 10.1074/jbc.C500097200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. South PF, Fingerman IM, Mersman DP, Du HN, Briggs SD. 2010. A conserved interaction between the SDI domain of Bre2 and the Dpy-30 domain of Sdc1 is required for histone methylation and gene expression. J Biol Chem 285:595–607. doi: 10.1074/jbc.M109.042697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Milholland KL, Gregor JB, Hoda S, Píriz-Antúnez S, Dueñas-Santero E, Vu BG, Patel KP, Moye-Rowley WS, Vázquez de Aldana CR, Correa-Bordes J, Briggs SD, Hall MC. 2023. Rapid, efficient auxin-inducible protein degradation in Candida pathogens. mSphere:e0028323. doi: 10.1128/msphere.00283-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5 and Tables S1 to S5.