SUMMARY
MicroRNAs (miRNAs) are conserved, short, non-coding RNAs that play a crucial role in the post-transcriptional regulation of gene expression. They have been implicated in the pathogenesis of cancer and neurological, cardiovascular, and autoimmune diseases. Several recent studies have suggested that miRNAs are key players in regulating the differentiation, maturation, and activation of immune cells, thereby influencing the host immune response to infection. The resultant upregulation or downregulation of miRNAs from infection influences the protein expression of genes responsible for the immune response and can determine the risk of disease progression. Recently, miRNAs have been explored as diagnostic biomarkers and therapeutic targets in various infectious diseases. This review summarizes our current understanding of the role of miRNAs during viral, fungal, bacterial, and parasitic infections from a clinical perspective, including critical functional mechanisms and implications for their potential use as biomarkers and therapeutic targets.
KEYWORDS: microRNA, viral infection, fungal infection, bacterial infection, parasite infection, biomarker
INTRODUCTION
MicroRNAs (miRNAs) are evolutionally conserved short (18–25 nucleotides) non-coding RNAs that play vital roles in post-transcriptional regulation of gene expression. Since their discovery in 1993, over 1,900 miRNAs have been identified in the human genome and are thought to maintain homeostasis by regulating over 60% of human protein-coding genes (1, 2). miRNA synthesis begins in the nucleus with the transcription of miRNA genes by RNA polymerase II, producing ~80-bp-long primary miRNA (pri-miRNA) with a hairpin structure (3). pri-mRNA is processed into precursor miRNAs by two endonucleases, first in the nucleus by Drosha, and then further in the cytoplasm by Dicer, to form duplex intermediates of ~22 nucleotides (4, 5). After duplex unwinding, the passenger strand is degraded owing to a lack of biological function, and the remaining stabilized strand acts as a matured miRNA. The matured miRNA forms the microRNA-induced silencing complex (miRISC) together with the Argonaute protein and several other proteins (6). Assembly of the miRISC allows for sequence-specific targeting of mRNA 3′ untranslated regions, leading to reduced translation (6–8).
The discovery of miRNAs has brought another level of complexity to our understanding of gene regulation. miRNAs have been shown to play a critical role in maintaining various biological processes, such as immune cell development, differentiation, activation, proliferation, metabolism, apoptosis, and autophagy (9–15). Dysregulation of miRNA expression has been extensively studied in cancer, neurological and cardiovascular diseases, obesity, and autoimmune diseases (15–18). Additionally, some studies have suggested that miRNAs are key players in regulating the maturation and differentiation of immune cells, which ultimately influence both the innate and adaptive immune responses (19, 20). Some studies have explored the role of human miRNAs during the host immune response to viral, fungal, bacterial, and parasitic infections. For example, toll-like receptors (TLRs), a class of pattern recognition receptors (PRRs) that play an essential role in the innate immune response, have been shown to influence miRNA expression (21). Cell surface TLRs can induce miR-146 after a challenge with lipopolysaccharide (LPS), a common bacterial endotoxin, in a monocytic cell line (22). In this report, upregulated miR-146 downregulated cellular LPS sensitivity via negative feedback and prevented excessive inflammation caused by activation of the LPS-TLR pathway (22). In the adaptive immune response, miRNAs such as miR-155 can affect the generation of anti-viral antibodies and CD4+ helper T and CD8+ cytotoxic T-cell responses in viral infections (23). Thus, it is conceivable that miRNAs play an essential role in infectious diseases. This presents a significant opportunity for novel diagnostics and therapeutics in infectious diseases, particularly infections that pose challenges in timely diagnoses, such as invasive aspergillosis or BK virus (BKV) nephropathy. miRNA profiling has the potential to bridge these gaps as a diagnostic marker for some infectious diseases, similar to studies on oncotherapeutics (24).
To our knowledge, only a few reviews have comprehensively focused on the functions and characteristics of miRNAs in viral, fungal, bacterial, and parasitic infection. Here, we provide an overview of the roles of miRNAs in these infectious diseases. We also discuss the possibility of miRNAs as diagnostic tools for infectious diseases and their potential as new therapeutic targets for gene therapies utilizing miRNA mimics or inhibitors (25). It would be difficult to discuss miRNAs associated with viral, fungal, bacterial, and parasitic infections in a single review. This review focuses on hepatitis viruses [hepatitis C virus (HCV) and hepatitis B virus (HBV)], herpes viruses [herpes simplex virus (HSV), Epstein-Barr virus (EBV), and cytomegalovirus (CMV)], respiratory syncytial virus (RSV), BKV, Aspergillus spp., Candida spp., Staphylococcus aureus, Pseudomonas aeruginosa, Plasmodium falciparum, and Schistosoma japonicum. These pathogens are important from a clinical point of view because of their epidemiological predominancy, high morbidity, and mortality particularly in immunocompromised patients, including solid organ and hematopoietic stem cell transplant recipients, limited treatment options, or lack of efficient diagnostic tools (26–39). For each pathogen, we present miRNAs examined in clinical studies, common mechanisms of action, and potential for use as biomarkers.
miRNA AND INFECTION
miRNAs play an essential role when hosts are infected (40, 41) and can promote or inhibit infectious disease progression by regulating the immune system. Importantly, miRNAs can regulate the proliferation of causative organisms, such as the replication of viruses, and do not directly regulate the immune response (41). For example, some miRNAs regulate pathogen entry into a target cell by downregulating the expression of cell membrane receptor proteins (42, 43). Therefore, modulation of miRNA function could be applied as a therapeutic option. In addition, many miRNAs have been implicated in infectious processes whose functions still need to be well investigated; these have untapped potential for use as biomarkers for infection diagnosis or disease progression (44, 45).
Regarding host-encoded miRNAs, some miRNAs in tissues and experimental cell lines are upregulated or downregulated in patients with viral infections compared to those in control populations (42, 46–52). The reasons why these dysregulations occur have yet to be fully investigated for each miRNA. However, cascade responses caused by pathogenic components and crosstalk between pathogens and PRRs are considered to be associated with several host-encoded miRNAs (48, 53). For instance, miR-21 expression is activated during HCV infection through two signaling pathways: the PKCε/JNK/c-Jun pathway and the PKCα/ERK/c-Fos pathway (48). These pathways are stimulated by HCV viral components NS5A and NS3/4A complex, respectively. Thus, HCV viral proteins stimulate miR-21 expression in hepatocytes (48). In addition, viral attachment to PRRs of target cells can lead to upregulation or downregulation of each miRNA during viral infections. These kinds of mechanisms which cause dysregulation of miRNAs during viral infections are also likely to occur during fungal, bacterial, and parasitic infections. For example, changes in miRNA expression occur after Aspergillus fumigatus binds to PRRs such as TLRs and dectin-1 (45, 47). Subsequent to the changes in miRNA expression during infection, upregulated miRNAs inhibit expression of the target genes by post-transcriptional regulation of gene expression induced by the miRNAs (6, 8). In contrast, downregulated miRNAs promote expression of the target genes (6, 8). These fundamental roles of miRNAs are essential for understanding the relationships between infectious pathogens and miRNAs described in the following sections.
VIRAL INFECTION
Currently, there is a lack of highly effective active anti-viral agents against viruses such as EBV, RSV, and BKV. As a result, these viruses can cause serious illnesses, particularly in immunocompromised patients, including solid organ and hematopoietic stem cell transplant recipients (27–29, 54), and drug discovery remains challenging. For example, no anti-viral agents can treat and prevent post-transplant lymphoproliferative disease caused by EBV, a disease with high mortality (27, 55). In addition, anti-viral resistance is a concern in many viral infections, including HCV, HBV, HSV, and CMV, despite the development of effective anti-virals (26, 36–39). This is especially pertinent for resistant CMV, which can have single or multiple resistant mutations against available anti-CMV agents such as (val)ganciclovir, foscarnet, cidofovir, letermovir, and maribavir (36, 37). Therefore, new therapeutic targets for these viruses are required. Recently, a clinical study showed that a miRNA can be utilized as a therapeutic target during viral infection (56). This indicates that investigating miRNAs may help identify new therapeutic targets. The following section examines host-encoded miRNAs that are associated with HCV, HBV, HSV, EBV, CMV, RSV, and BKV infections. In addition, viral-encoded miRNAs and their influence on the pathogenesis of infectious diseases are reviewed.
Function and characteristics of host-encoded miRNAs during viral infections
Some of the most relevant host-encoded miRNAs associated with viral replication, viral entry into target cells, and immune functions by repression of the target genes are presented in Fig. 1 and Tables 1 to 4. The target genes of host-encoded miRNAs are generally the host genes. However, host-encoded miRNAs can also target viral genes such as miR-122 and miR-125, which target the S genes of HCV and HBV (Table 1) (57, 58). Herein, hepatitis, herpes and atypical viruses are discussed independently.
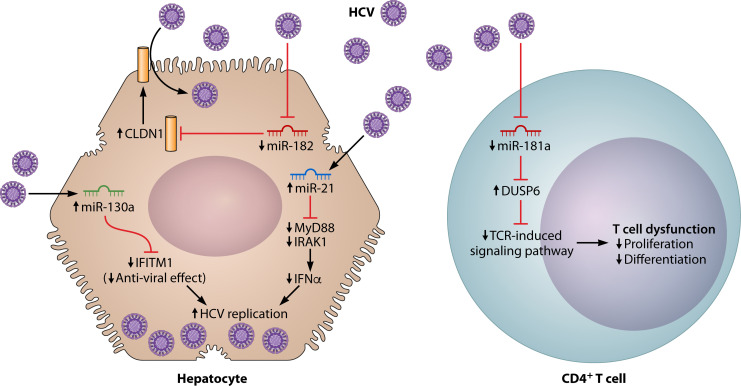
Fig 1.
Functions of host-encoded miRNA during HCV infections. Illustrated here are examples of functional pathways of host-encoded miRNAs. Examples were chosen if they met the following criteria: (i) miRNA expression was compared to control populations or status without the infection; (ii) the target genes of the miRNA were identified in the study; (iii) the functional mechanisms reported had sufficient data; and (iv) the experiments were in human cell lines. In the left, miR-21 and miR-130a are upregulated and miR-182 is downregulated during HCV infection in a hepatocyte. In the right, miR-181a is downregulated during HCV infection in a CD4+ T cell. Abbreviations: CLDN1, claudin-1; DUSP6, dual specific phosphatase 6; IFITM1, interferon-induced transmembrane 1; IFN, interferon; IRAK1, interleukin-1 receptor-associated kinase; MyD88, myeloid differentiation factor 88; TCR, T-cell receptor.
TABLE 1.
Functions and characteristics of microRNA in HBV and HCV infectiona
| Type of miRNAs | Target gene/pathway | Study type | Functions and characteristics of miRNA |
|---|---|---|---|
| Host-encoded microRNA in HCV infection | |||
| miR-122 | |||
| Target sites (S1 and S2) of HCV RNA | Human in vitro study (57, 59) (Huh-7 cells, Hep 3B, and Huh-7.5 cells) | Stimulates viral replication | |
| OCLN | Human in vitro study (60) (Huh-7.5 cells) | Overexpression decreases HCV entry into hepatocytes. | |
| Not reported | Clinical study (61) (liver and blood samples) | Predicts therapeutic response to PEG-IFN/RBV + differentiation between healthy controls and chronic hepatitis C | |
| Not reported | Clinical study (56) (randomized, double-blind, placebo-controlled study) (blood samples) | miR122 inhibitor usage resulted in the reduction of HCV RNA without resistance. | |
| miR-155 | |||
| TLR3-dependent anti-viral pathway | Animal in vitro and human in vitro study (62) [liver (mice and humans) and blood (human) samples] |
Increases hepatic expression in chronic HCV, decreases HCV viral load | |
| T-bet/Tim3 pathway | human in vitro study (63) [PBMCs (from patients and controls)] |
miR-155 is downregulated and inhibits IFN-γ production in NK cells in chronic HCV infection. | |
| Not reported | clinical study (64) (blood samples) | Negative correlation with HCV Viral loads | |
| miR-146 | |||
| (miR-146b-5p) | NF-κB signaling pathway | Human in vitro study (65) PBMCs (from patients and controls) |
Suppression could induce an impaired immune response in monocytes and T cells. |
| (miR-146a) | Not reported | Human in vitro study (66) PBMCs (from patients and controls) |
Regulates the immune responses correlate with cholesterol metabolism |
| miR-21 | |||
| MyD88 and IRAK1 | Human in vitro study (48) (Huh-7 and HEK293 cells) | Upregulated during HCV infection, increases HCV production by suppressing IFN-α production | |
| miR-130a | |||
| IFITM 1 | Human in vitro study (67) (liver samples and Huh-7 cells) | Upregulated during HCV infection, increases HCV replication | |
| ATG5 | Human in vitro study (68) (Huh-7.5.1 cells) | Inhibits HCV replication | |
| miR-27a | |||
| RXRα and ABCA1 | Human in vitro study (69) (Huh-7.5 cells) | Inhibits HCV replication | |
| miR-181a | |||
| DUSP6 | Human in vitro study (51) [PBMCs (from patients and controls) and Huh-7 cells] |
Downregulated during HCV infection in CD4+ cells, resulting in their dysfunction | |
| miR-182 | |||
| CLDN1 | Human in vitro study (42) (liver samples and Huh-7 cells) | Downregulated in HCV-infected patients, inhibits HCV entry into a target cell | |
| miR-200c | |||
| OCLN | Human in vitro study (43) (liver samples and Huf7 cells) | Downregulated in HCV-infected patients, inhibits HCV entry into a target cell | |
| miR-215 | |||
| TRIM | Human in vitro study (70) (Con1b and Hub-7.5.1 cells) |
Promotes HCV replication | |
| miR-373 | |||
| IRF5 | Human in vitro study (71) (liver samples and Huh-7.5 and 293T cells) | Upregulated in HCV infection Inhibits replication of HCV | |
| miR-196 | |||
| Bach1 | Human in vitro study (72) (9–13 cells and Huh-7.5 cells) | Inhibits expression of HCV | |
| Host-encoded microRNA in HBV infection | |||
| miR-122 | |||
| Not reported | Human in vitro study (73) (liver samples and Huh-7 and HspG2 cells) | Upregulated in HBV-infected patients Inhibits HBV replication | |
| The core protein and polymerase of HBV | Human in vitro study (74) (HepG2 and HepG2.2.15) | Inhibits HBV replication | |
| miR-125 | |||
| (miR-125b-5p) | LIN28B | Human in vitro study (75) (Con 1, HepG2, Huh-7, and HepG2.2.15) | Overexpression promotes HBV replication. |
| (miR-125a-5p) | HBV S gene | Human in vitro study (58) (HepG2 cells and Huh-7) | Interferes with the viral translation and downregulates expression of the surface antigen |
| (miR-125a-5p) | Not reported | Clinical study (76) (liver and blood samples) | Associated with severe disease progression (OR = 4.21 for histological activity index of >6 and OR = 3.12, for fibrosis score >2) |
| miR-17–92 | |||
| HBV genome | human in vitro study (77) (HepAD38 cells) | Inhibition of miR-20a and miR-92a-1, both members of the miR-17–92 miRNA cluster, promotes HBV replication. | |
| miR-122 and miR-130a | |||
| Not reported | Clinical study (78) (blood samples) | Can differentiate occult HBV infection from healthy controls, asymptomatic surface antigen of HBV carriers, and chronic hepatitis B (>0.87°F area under the curve) | |
| miR-141 | |||
| Sirt1 | Human in vitro study (79) (HeLa/GFP-LC3, HepG2.2.15, and Huh-7 cells) |
Inhibits HBV expression and replication | |
| miR-146a-5p | |||
| XIAP | Human in vitro study (49) (blood samples and THLE-2 cells) | miR-146a-5p is upregulated in patients with chronic hepatitis B and HBV-expressing. hepatocytes. miR-146a-5p promotes HBV replication. | |
| miR-185–5p | |||
| ELK1 | Human in vitro study (80) (Huh-7 and HepG2.2.15) | Inhibits HBV gene expression and replication | |
| miR-210 | |||
| Not reported | Clinical study (81) (liver and blood samples) | Serum levels correlated with HBV DNA and hemoglobin antigen | |
| miR-548 | |||
| IFN-λ1 | Human in vitro study (82) (blood samples, HepG2, and RD cells) |
Downregulated in infected cells, it downregulates host anti-viral response. | |
| (miR-548ah) | HDAC4 | Animal and human in vitro study (83) (C57BL/6 mice, liver samples (from patients), HepG2.2.15, Huh-7, and 293T cells) | Promotes replication and expression of HBV |
| miR-802 | |||
| SMARCE1 | Human in vitro study (84) (liver samples, HepG2.2.15, and HepG2) | Upregulated in the HBV-associated hepatocellular carcinoma tissues; overexpression promotes HBV replication | |
| miR-1231 | |||
| HBV core gene | Animal and human in vitro study (85) PXB mice, liver (human and mice) and blood samples and HepG2 |
Upregulated in HBV infection Inhibits HBV replication | |
| miR-372/373 | |||
| NFІB | Human in vitro study (86) (liver and blood samples, HepG2, and HepG2.2.15 cells) | Upregulated in infected liver tissues and human hepatic cell line Promote HBV expression | |
ABCA1, ATP-binding cassette subfamily A member 1; ATG5, autophagy-related gene5; CLDN1, claudin-1; DUSP6, dual specific phosphatase 6; ELK1, ETS like-1; HDAC4, histone deacetylase 4; IFN, interferon; IRAK1, interleukin-1 receptor-associated kinase; IRF5; interferon regulatory factor 5; MyD88, myeloid differentiation factor 88; NFІB, nuclear factor І/B; OCLN, occludin; OR, odds ratio; PBMC, peripheral blood mononuclear cell; PEG-IFN/RBV, peginterferon/ribavirin; RXRα, retinoid X receptor alpha; SMARCE1, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily E, member 1.; Tim3, T-cell immunoglobulin and mucin domain proteins 3; TLR, toll-like receptor; TRIM, tripartile motif; XIAP, X-linked inhibitor of apoptosis.
TABLE 2.
Functions and characteristics of microRNA in herpes viral infectiona
| Type of miRNAs | Target gene/pathway | Study type | Functions and characteristics of miRNA |
|---|---|---|---|
| Host-encoded microRNA in HSV infection | |||
| miR-101 | |||
| ATP5B | Human in vitro study (87) (HeLa cells) | Inhibits HSV-1 replication | |
| miR-23a | |||
| IRF1 | Human in vitro study (88) (HeLa cells) |
Promotes HSV replication | |
| miR-138 | |||
| ICP0 (viral gene) | Animal in vitro and in vivo studies and human in vitro study (89) [CD1 mice, Vero (monkey), 293T, HFF, and neuro-2A (mice) cells] |
Suppresses HSV-1 lytic gene expression | |
| miR-373 | |||
| IRF1 | Human in vitro study (50) (blood samples and HeLa cells) |
Upregulated by HSV-1 infection in HeLa cells and patients with herpetic gingivostomatitis, promotes HSV1 replication | |
| miR-649 | |||
| MALT1 | Human in vitro study (52) (HeLa cells) | Downregulated after HSV-1 infection, promotes HSV-1 replication. | |
| Viral-encoded microRNA in HSV infection | |||
| HSV-1 miR-H6 | |||
| ICP4 | Human in vitro study (90) (human limbal cells) |
Inhibits HSV-1 replication and IL-6 expression in human corneal epithelial cells | |
| Host-encoded microRNA in EBV infection | |||
| miR-155 | |||
| SMAD1, SMAD5, HIVEP2, CEBPB, RUNX2, and MYO10 | Human in vitro study (91) (Mutu І and A549 cells) |
Inhibits BMP-mediated EBV reactivation. | |
| miR-429 | |||
| ZEB1 | Human in vitro study (92) (Rael, Akata, AGS-EBV, and AGS cells) |
Can break latency of EBV | |
| Viral-encoded microRNA in EBV infection | |||
| miR-BART1 | |||
| IL-12B | Human in vitro study (93) (MNCs/PBMCs, Raji,DG-75,and HEK293-based EBV producer cells) |
Inhibits anti-viral CD4+ T cells | |
| (miR-BART1-3p and 5 p) | IFI30 | Human in vitro study (94) (PBMCs and adenoids, Raji and HEK293-based EBV producer cells) |
Reduces immune surveillance by virus-specific T cells |
| miR-BART15 | |||
| NLRP3 | Human in vitro study (95) (CD14+VEand 293T cells) | Inhibits inflammasome activation in non-infected cells by transfer of the miRNA through exosomes. | |
| miR-BART16 | |||
| CREBBP | Human in vitro study (96) (EBV+BL Jijoye, HEK293T, SNU-719, Mutu-1-cl3, and Mutu1 cells) |
Facilitates the establishment of latent EBV infection and promotes viral replication | |
| miR-BART17 | |||
| TAP2 | Human in vitro study (94) (PBMCs and adenoids and Raji, HEK293-based EBV producer cells) |
Reduces immune surveillance by virus-specific T cells | |
| miR-BART6-3p | |||
| RIG-1 | Human in vitro study (97) (HK-1, C666-1, BJAB, and B95.8cells) |
miR-BART6-3p inhibits the EBV-triggered IFN-β response and facilitates EBV infection. | |
| miR-BART18-5p | |||
| MAP 3K2 | Human in vitro study (98) (BL2, Akata2A8, Akata2A8.1, BJAB, B95-8, and B95.8-LCL cells) |
Inhibits EBV replication in B cells | |
| miR-BART22 | |||
| IL-12B | Human in vitro study (93) (PBMCs and adenoids, Raji, DG-75, and HEK293-based EBV producer cells) |
Inhibits anti-viral CD4+ T cells | |
| miR-BART10 | |||
| IL-12B | Human in vitro study (93) (PBMCs and adenoids, Raji, DG-75, and HEK293-based EBV producer cells) |
Inhibits anti-viral CD4+ T cells | |
| miR-BART2 | |||
| IL-12B | Human in vitro study (93) (PBMCs and adenoids, Raji, DG-75, HEK293- based EBV producer cells) |
Inhibits anti-viral CD4+ T cells | |
| (miR-BART2-5p) | MICB | Human in vitro study (99) (293T, RKO, HeLa, 721.221, and BCBL1 cells) | Promotes escape recognition by NK cells |
| (miR-BART2-5p) | CTSB and LGMN | Human in vitro study (94) (PBMCs and adenoids and Raji, HEK293-based EBV producer cells) |
Reduces immune surveillance by virus-specific T cells. |
| miR-BHRF1-2 | |||
| IL-12B | Human in vitro study (93) (PBMCs and adenoids, Raji, DG-75, HEK293-based EBV producer cells) |
Inhibits anti-viral CD4+ T cells | |
| CTSB and IL-12B | Human in vitro study (94) (PBMCs and adenoids and Raji, HEK293-based EBV producer cells) |
Reduces immune surveillance by virus-specific T cells | |
| (miR-BHRF1-2-5p) | IL1R1 | Human in vitro study (100) (LCLs and BJAB cells) |
Inhibits IL-1 cytokine signaling |
| (miR-BHRF1-2-5p) | GRB2 | Human in vitro study (101) (LCLs, BJAB, DLBCLs, and Mutu I cells) |
Inhibition of miR-BHRF1-2-5p enhances BCR (B-cell receptor)-mediated EBV reactivation. |
| Host-encoded microRNA in CMV infection | |||
| miR-155 | |||
| Not reported | Clinical study (102) (blood samples) | Able to discriminate patients with CMV infection from patients without it in a kidney transplant setting | |
| miR-200 | |||
| UL122 (viral gene) | Human in vitro study (103) (Kasumi-3, fibroblast, and CD34+ hematopoietic projenitor cells) |
Maintains CMV virus latency | |
| miR-100 and miR-101 | |||
| mTOR | Human in vitro study (104) (MRC-5, HeLa, and 293T cells) |
Downregulated after CMV infection, inhibits CMV replication | |
| miR-221 | |||
| SOCS1 | Human in vitro and animal (mice) in vivo study (105) (neural precursor cells and C57BL/6 mice) |
Upregulated in CMV infection, inhibits CMV replication | |
| miR-183 and miR-210 | |||
| Not reported | Clinical study (44) (blood samples) | The plasma levels of miR-183–5p and miR-210–3p can discriminate congenital CMV from healthy control infants. | |
| Viral-encoded microRNA in CMV infection | |||
| miR-UL112 | |||
| IL-32 | Human in vitro study (106) (blood samples and HEK293 and MRC-5 cells) |
Reduces IL-32 expression during human CMV infection | |
| MICB | Human in vitro study (107) (HFF cells and HeLa cells) |
Inhibits NK cell cytotoxicity | |
| (miR-UL112-1) | VAMP3, RAB5C, RAB11A, and SNAP23 |
Human in vitro study (108) (HEK293T, NHDF, and HeLa cells) |
miR-UL112-1 inhibits pro-inflammatory cytokine secretion and facilitates formation of the VAC for efficient infectious virus production |
| (miR-UL112-3p) | ATG 5 | Human in vitro study (109) (HFFs, HEK293T, and HeLa cells) |
Inhibits autophagy |
| (miR-UL112-3p) | IKKα and IKKβ | Human in vitro study (110) (NHDF, HeLa, hAEC, THP-1, and 293T cells) |
Inhibits the processing and presentation of the human CMV pp65 peptide to CD8+ T cells |
| (miR-UL112-3p) | TLR2 | Human in vitro study (111) (HEK293T, NHDF, and THP-1 cells) |
Reduces IL-32 expression during human CMV infection |
| (miR-UL112-5p) | ERAP1 | Human in vitro study (112) (HEK293T cells) |
Inhibits the processing and presentation of the human CMV pp65 peptide to CD8+ T cells |
| miR-UL148D | |||
| ACVR1B | Human in vitro study (113) (PBMCs and CD34+ hematopoietic progenitor cells from healthy donors, HFFF2 fibroblast, and KG-1 cells) |
Inhibits IL-6 production | |
| IER5 | Human and animal (murine) in vitro study (114) (Kasumi-3, HFF, and murine AFT024 cells) |
Facilitates latent CMV infection | |
| RANTES | Human in vitro study (115) (HEK293T and HFF cells) |
Reduces RANTES | |
| miR-US25 | |||
| (miR-US25-1-5p) | CD147 | Human in vitro study (116) (HFF cells, U251 cells and HEK293 cells) |
Inhibits Innate immune response |
| (miR-US25-2-3p) | eIF4A1 | Human in vitro study (117) (MRC-5 cells) |
Overexpression inhibits CMV replication |
| miR-US5 | |||
| IKKα and IKKβ | Human in vitro study (110) (NHDF, HeLa, hAEC, THP-1, and 293T cells) |
Inhibits pro-inflammatory cytokine production | |
| (miR-US5-1 and −2) | VAMP3, RAB5C, RAB11A, SNAP23, and CDC42 | Human in vitro study (108) (HEK293T, NHDF, and HeLa cells) |
Inhibits pro-inflammatory cytokine production and facilitates formation of the VAC for efficient infectious virus production |
| miR-US22-5p | |||
| ATG 5 | Human in vitro study (109) (HFFs, HEK293T, and HeLa cells) |
Inhibits autophagy | |
| miR-US29-5p | |||
| ATG 5 | Human in vitro study (109) (HFFs, HEK293T, and HeLa cells) |
Inhibits autophagy | |
| miR-UL22A-5p | |||
| C-MYC | Clinical study (118) (blood samples) |
Detection at baseline independently predicted the recurrence of CMV viremia upon discontinuation of anti-viral therapy among solid organ transplant recipients who diagnosed with CMV diseases (OR = 3.024, P = 0.007). | |
| miR-UL148D, miR-US25-1-5p, and miR-US5-1 | |||
| Not reported | clinical study (119) (Blood samples) |
Plasma concentrations can discriminate pregnant women with adverse pregnancy outcomes associated with CMV from normal controls. (miR-US25-1-5p presented the largest area under the ROC curve (0.735), with a sensitivity of 68% and specificity of 71%.) | |
ACVR1B, activin A receptor 1B; ATG5, autophagy-related gene5; ATP5B, ATP synthase subunit beta; BMP, bone morphogenetic protein; CEBPB, CCAAT/enhancer-binding protein beta; CREBBP, CREB-binding protein; CTSB, cathepsin B; eIF4A1, eukaryotic translation initiation factor 4A1; ERAP1, endoplasmic reticulum aminopeptidase 1; GRB2, growth factor receptor-bound protein 2; HIVEP2; human immunodeficiency virus type 1 enhancer-binding protein 2, ICP; infected cell polypeptide; IER5, immediate early response gene5; IFN, interferon; IFI30, IFN-γ-regulated thiol reductase; IKK, IκB kinase; IL, interleukin; IL1R1, IL-1 receptor 1; IRF1, interferon regulatory factor 1; LGMN, legumain; MALT1, mucosa associated lymphoma translation gene 1; MAP3K2, MAP kinase kinase kinase 2; MICB, major histocompatibility complex class I-related chain B; MRC-5, Medical Research Council cell strain 5; mTOR, mammalian target of rapamycin; MYO10, myosin Ⅹ; NK, natural killer; NLRP3, NLR family pyrin domain containing 3; PBMC, peripheral blood mononuclear cell; RANTES, regulated on activation normal T-cell expressed and secreted; RIG-1, retinoic acid includible gene 1; ROC, receiver operating characteristic; RUNX2, runt-related transcription factor 2; SMAD1, Drosophila mother against decapentaplegic 1; SMAD5, Drosophila mother against decapentaplegic 5; SNAP23, synaptosome associated protein 23; SOCS1, suppressor of cytokine signaling-1; TAP2, transporter2; TLR, toll-like receptor; VAC, virion.
TABLE 3.
Functions and characteristics of microRNA in RSV virus infectiona
| Type of miRNAs | Target gene/pathway | Study type | Functions and characteristics of miRNA |
|---|---|---|---|
| Host-encoded microRNA in RSV infection | |||
| miR-24, miR-124, and miR-744 | |||
| p38 MARK signaling pathway | Human in vitro study (120) (ATCC, MDCK, and Hep-2 cells) | Inhibits RSV infection | |
| miR-221 | |||
| NGF-TrkA axis | Human in vitro study (121) (human bronchial epithelial cells from donors) | Inhibits RSV replication | |
| miR-34b/c-5p | |||
| Not reported | Human in vitro study (122) (HBE cells) | Downregulation of miR-34b/c-5p expression in airway epithelial cells in RSV infection is associated with increasing of mucus secretion. | |
| miR155 | |||
| SOCS1 | Human in vitro and animal (mice) in vivo study (123) [Hep-2, A549, and HPAEpi cells, and MEFs (murine cells), and C57BL/6 mice] |
Inhibits RSV replication | |
| miR-125a and miR-429 | |||
| Not reported | Clinical study (124) (nasal mucosa cytology specimens) | Discrimination between mild and severe RSV infection | |
p38 MARK, P38 mitogen-activated protein kinase; NGF-TrkA, nerve growth factor-tropomyosin-related kinase A; SOCS1, suppressor of cytokine signaling-1.
TABLE 4.
Functions and characteristics of microRNA in BK virus infectiona
| Type of miRNAs | Target gene/pathway | Study type | Functions and characteristics of miRNA |
|---|---|---|---|
| Host-encoded microRNA in BKV infection | |||
| miR-10 and miR-30 | |||
| Not reported | Human in vitro study (125) (RPTEC cells) | Controlled the expression of genes involved in evading the host immune response, nurturing the growth of BKV-infected cells, preventing apoptosis, induction of inflammation, and development of fibrosis | |
| Viral-encoded microRNA in BKV infection | |||
| BKV-miR-B1 | |||
| Not reported | Human in vitro study (125) (RPTEC cells) | bkb-miR-B1-5p and bkv-miR-B1-3p showed a 1,000-fold increase in human tubular epithelium during BKV infection. | |
| Not reported | Clinical study (126) (blood and urine samples) |
bkv-miR-B1-3p and bkv-miR-B1-5p levels in the urine might be able to detect viral replication (plasma BKV DNA) in kidney transplant recipients (the AUCs were 0.7928 and 0.7091, respectively). | |
| Not reported | Clinical study (127) (blood, urine, and cerebrospinal fluid samples) |
bkv-miR-B1-3p and bkv-miR-5p in human plasma, urine, and cerebrospinal fluid might be able to detect suspected or severe BKV disease. | |
| (bkv-miR-B1-5p) | Not reported | Clinical study (128) (blood samples) | Biomarker for BKV nephropathy and infection (sensitivity of 100.0% and specificity of 94.9%; the AUC was 0.97) |
| (bkv-miR-B1-5p) | Not reported | Clinical study (129) (blood and urine samples) | The urinary exosomal microRNA levels of bkv-miR-B1-5p and bkv-miR-B1-5p/miR-16 detect biopsy-proven BKVN. [The AUC was 0.989 for bkv-miR-B1-5p, 0.985 for bkv-miR-B1-5p/miR-16. The cut-off values for bkv-miR-B1-5p and bkv-miR-B1-5p/miR-16 were 5.9 log10 copies/mL (sensitivity, 100%; specificity, 98.5%) and 1.2 log10 copies/mL (sensitivity, 100%; specificity, 98.5%), respectively.] |
AUC, area under the curve.
Host-encoded miRNAs during HCV and HBV infection
When a host is first infected, changes in miRNA expression occur (Fig. 1). We present examples of well-investigated molecular pathways involving human miRNAs in Fig. 1. These data were primarily obtained from in vitro studies.
Studies using human samples and/or human cell lines
In this section, we focus on miRNAs associated with HCV and HBV infection, which are mainly in studies using human samples and/or human cell lines. During HCV infection, miR-21 and miR-130a are upregulated in hepatocytes (48, 67). In contrast, miR-181a and miR-182 are downregulated in a CD4+ T cell and a hepatocyte, respectively, during HCV infection (Fig. 1) (42, 51). Upregulated miRNAs generally repress the expression of their target genes (6, 8). For example, myeloid differentiation factor 88 and interleukin-1 receptor-associated kinase (IRAK1), which are the target genes of miR-21 in hepatocytes and associated with interferon-α (IFN-α) production, are repressed by upregulated miR-21 during HCV infection (Fig. 1) (48). As a result, the output of IFN-α decreases, which permits viral replication of HCV (48). In addition, upregulation of miR-130a during HCV infection represses interferon-induced transmembrane 1 (IFITM1), which is a target gene of miR-130a, in hepatocytes (67). It promotes HCV replication by suppressing the anti-viral effect of IFITM1, which may be associated with the enhancement of neutralizing antibody and inhibition of cell entry of HCV (67, 130).
Similarly, downregulated miRNAs generally lead to overexpression of target genes. For instance, miR-181a is downregulated in CD4+ T cells during HCV infection (51). The downregulation leads to the overexpression of dual specific phosphatase 6 (DUSP6), which is the target gene of miR-181a. DUSP6 overexpression inhibits the proliferation and differentiation of T cells through T-cell receptor-induced signaling pathways. As a result, impairment of the global CD4+ T-cell response (as these T cells were not confirmed to be HCV specific) can occur during HCV infection but through a downregulation mechanism (51). Therefore, reconstitution of miR-181a may restore the impaired immune response. Similarly, downregulation of miR-182 in hepatocytes during HCV infection leads to overexpression of the target gene claudin-1 (CLDN1) (42). Overexpression of CLDN1 in hepatocytes enhances HCV endocytosis (Fig. 1) (42). In summary, these miRNAs are involved in the pathogenesis of HCV in terms of HCV replication, cell entry of HCV, and attenuation of immune response. Hence, these miRNAs may serve as important therapeutic targets.
As described earlier, the primary action of miRNAs to repress gene expression is to bind a target sequence on the resultant mRNA and inhibiting its translation (6, 8). However, there are some exceptions. The most prominent exception is hepatocyte expression of miR-122 during HCV infection (131). miR-122 binds to two closely spaced target sites (S1 and S2) in the highly conserved 5′ untranslated region of the HCV genome, forming an oligomeric miR-122-HCV complex that protects against nucleolytic degradation or host innate immune responses (131–133). Here, miR-122 does not silence mRNA expression but stabilizes the pathogen genome and incites virus replication. This mechanism is clinically significant because the administration of miravirsen, a miR-122 inhibitor developed for patients with chronic HCV genotype 1 infection, showed a prolonged dose-dependent reduction in HCV RNA levels without evidence of resistance in a phase 2 clinical trial (56).
In HBV infection, upregulated miR-146a-5p in hepatocytes represses inflammatory genes [X-linked inhibitor of apoptosis (XIAP)] in the hepatocytes (49). miR-146a-5p overexpression promotes HBV replication through the autophagy pathway, which is mediated by XIAP in an in vitro model of HBV infection (49). Additionally, miR-802 is upregulated in hepatocytes in HBV infection (84). Upregulated miR-802 promotes HBV DNA replication and surface antigen of HBV (HBsAg) and HBeAg expression through the inhibition of SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily E, member 1 (SMARCE1), which is the target gene of miR-802, although the detailed mechanism of SMARCE1 in the expression and replication of HBV remains largely unknown (84, 134). In this context, miR-146a-5p and miR-802 may be potential therapeutic targets for HBV infection.
During HBV infection, some host-encoded miRNAs target genes of HBV. For instance, the HBV genomic segment 3037–3065, which encodes amino acid residues 244–252 of polymerase and 64–72 of HBsAg, is a direct target of miR-125a-5p (58). miR-125a-5p, which is expressed in the human liver, can downregulate the expression of HBV S gene via post-transcriptional regulation, thus reducing the amount of HBsAg (58). The downregulation of HBsAg probably inhibits HBV replication (135). Thus, delivery of synthetic miR-125a-5p mimics may be a therapeutic option for HBV infection.
In addition to mechanistic studies, host-encoded miRNAs have been investigated as biomarkers during viral infection. For instance, miR-125a-5p levels in liver tissue have been investigated as predictors of disease progression in HBV infection (76). More precisely, miR-125a-5p level in the liver tissue was able to predict histological activity index of >6 [odds ratio (OR) = 4.2; 95% confidence interval (CI), 1.1–16.4] and fibrosis score of>2 (OR = 3.1; 95% CI, 1.2–8.3) (76) in patients with chronic hepatitis B who were naïve to nucleoside analogs and interferon therapy.
Moreover, studies of other miRNAs associated with HCV and HBV infection, which have information of target genes and/or are investigated using human samples, are listed in Table 1. Apart from all the miRNAs written in the text and/or Table 1, well-designed functional studies were also performed by using human cell lines for miR-27, miR-130, and miR-491 in HCV infection, although they lacked detailed information of target genes (136–138).
Animal studies
In an animal study of chimpanzees, effectiveness of miravirsen (a miR-122 inhibitor) in HCV infection was demonstrated (139).
Host-encoded miRNAs during HSV, EBV, and CMV infection
Studies using human samples and/or human cell lines
In this section, we focus on miRNAs associated with HSV, EBV, and CMV infection, which are mainly in studies using human samples and/or human cell line. In HSV infection, miR-373 is upregulated in a HeLa cell line in HSV (50). In contrast, miR-649 is downregulated in a HeLa cell in HSV infection (52). Upregulated miR-373 represses interferon regulatory factor 1 (IRF1) in HeLa cells. Owing to the inhibition of IRF1, upregulated miR-373 facilitates HSV replication by suppressing the type 1 interferon response during HSV infection (50). Similarly, downregulation of miR-649 in HeLa cells during HSV infection leads to overexpression of the target gene mucosa-associated lymphoma translation gene 1 (MALT1) (52). MALT1 overexpression inhibited HSV replication in a HeLa cell through activation of NF-κβ (52). Hence, miR-373 and miR-649 may be potential therapeutic targets in HSV infection.
EBV commonly infects with B cells and epithelial cells in vivo (140). In EBV-infected cells, some host-encoded miRNAs are associated with the pathogenesis of EBV infection by escaping the host immune systems and maintaining EBV in its latent phase (141). For example, miR-155 is induced by the oncogeneic latency gene expression program of EBV in B cells (91). miR-155 inhibits bone morphogenetic protein (BMP)-mediated lytic EBV reactivation in an EBV-positive B-cell line by suppressing multiple predicted target genes: Drosophila mother against decapentaplegic 1, Drosophila mother against decapentaplegic 5, human immunodeficiency virus type 1 enhancer-binding protein 2, CCAAT/enhancer-binding protein beta, runt-related transcription factor 2, and myosin X, all of which comprise the BMP signaling cascade (91). These results suggest that miR-155 can maintain EBV-infected B cells latent by suppressing the BMP signaling pathway conferring a survival advantage to EBV (91). Additionally, miR-429, which belongs to the miR-200 family, is associated with EBV reactivation in EBV-infected cells (92). Expression of miR-429 breaks the latency of EBV in both EBV-infected human epithelial and human B cells in vitro through inhibition of ZEB1 (92). ZEB1 is a direct target of the miR-200 family and inhibits EBV reactivation (142–144). Hence, further understanding how host-encoded miRNAs involve a switching mechanism from latent to lytic reactivation of EBV might be helpful to develop a novel therapeutic strategy against EBV infection.
miR-100 and miR-101 are downregulated during CMV infection, although levels of most miRNAs do not change markedly in Medical Research Council cell strain 5 cells (104). These two miRNAs can inhibit CMV replication by repressing the mammalian target of rapamycin kinase (mTOR) pathway, including mTOR and raptor (104). mTOR and raptor are components of the mTOR protein translation initiation regulatory pathway, which is important for CMV replication under some conditions (145). mTOR is a predicted target of both miR-100 and miR-101 (104). In contrast, raptor is a predicted target of miR-100 (104). MiR-100 and miR-101, either alone or in combination, reduce the amount of infectious CMV in vitro (104). Thus, downregulation of miR-100 and miR-101 induced by CMV can assist its replication (104). In contrast, miR-221 is upregulated in human neural precursor cells in CMV infection (105). Subsequently, upregulated miR-221 suppresses replication of CMV by targeting suppressor of cytokine signal 1 (SOCS1) since the SOCS1 downregulation induced by miR-221 leads to type 1 interferon (IFN-α and IFN-β) production in CMV-infected cells (105). In this context, miR-100, miR-101, and miR-221 may be therapeutic targets of CMV.
In CMV infection, the utility of several miRNAs as biomarkers of CMV infection has been reported. For instance, plasma levels of miR-183–5p and miR-210–3p have been investigated as predictors of congenital CMV infection (44). According to this study, plasma levels of miR-183–5p and miR-210–3p were significantly higher in infants with congenital CMV infection than in controls (44).
Studies of other miRNAs associated with HSV, EBV, and CMV infection, which have information regarding target genes and/or are investigated using human samples, are listed in Table 2. Apart from all the miRNAs described here and/or Table 2, well-designed functional studies were also performed in human cell lines for miR-101–1 in HSV infection and miR-146a and miR-155 in EBV infection, although they lack detailed information of target genes (146–148).
Animal studies
In a mouse model, miR-132 and miR-155 were associated with HSV ocular infection (149, 150). In addition, miR-96, miR-141, miR-183, and miR-200 may be associated with the entry of HSV to target cells in a mouse model (151). Furthermore, miR-592, miR-1245b-5p, miR-150, miR-342–5p, miR-1245b-3p, and miR-124 were incorporated into regulation of the TLR pathway during HSV-2 infection in a guinea pig model (152). In CMV infection, function of miR-199a/214 cluster was investigated in a mouse cell line (153).
Host-encoded miRNAs during RSV infection
Although the number of host-encoded miRNAs which have been investigated in RSV infection is fewer than those in the hepatitis and herpes viruses listed in this review, some miRNAs contribute to pathogenesis of these infections (Table 3). These miRNAs have been investigated in studies using human clinical samples and/or human cell lines (Table 3). For example, miR-221 is downregulated in human bronchial epithelial cells during RSV infection (121). This leads to upregulation of the nerve growth factor-tropomyosin-related kinase A axis, which might be caused by upregulated relevant genes that are predicted targets of miR-221 (121). Upregulation of this axis takes advantage of RSV replication by inhibiting apoptotic death of infected cells (121). These findings suggest that miR-221 may be a therapeutic target for RSV infection. In fact, miR-221 transfection in human bronchial epithelial cells reduces RSV replication and infectivity (121).
Host-encoded miRNAs related to RSV infection are summarized in Table 3.
Host-encoded miRNAs during BKV infection
Some miRNAs contribute to pathogenesis of BKV infections and have been investigated in studies using human samples and/or human cell lines. We summarize these in Table 4. In BKV infection, miR-10b and miR-30a are downregulated in normal human renal tubule epithelial cells infected with BKV (125). In addition, these two miRNAs are significantly decreased in specimens obtained from kidney transplant recipients with BKV nephropathy compared to those with a functional graft (125). Moreover, upregulation or downregulation of some genes [e.g., interleukin (IL)-6, IL-8, and tumor protein p53 inducible protein], which are predicted to be direct or indirect targets of miR-10b and miR-30a, may be induced by BKV nephropathy (125). These responses lead to evasion of the host immune response, nurturing the growth of BKV-infected cells, preventing apoptosis, induction of inflammation, and development of fibrosis (125), permitting BKV survival in the tissues. Thus, these miRNAs may be used as therapeutic targets (125).
Function and characteristics of viral-encoded miRNAs during viral infections
miRNAs are also encoded by several types of viruses that have dsDNA genomes, including adenoviruses, herpesviruses, and polyomaviruses (154). Thus far, viral-encoded miRNAs have not been identified in papillomaviruses or poxviruses (154). No viral-encoded miRNAs have been reported in RNA viruses (154).
Viral-encoded miRNAs are incorporated into the miRISC, and this miRNA complex interacts with the 3′ untranslated region of the host and viral-encoded mRNAs. This suppresses the expression of target genes either via translational repression or mRNA degradation (155). Remarkably, the functions of these viral-encoded miRNAs are similar to those of the host-encoded miRNAs as described above (141, 156). These viral-encoded miRNAs usually permit viral persistence and/or promote self-proliferation (141). In particular, viral-encoded miRNAs, which EBV and CMV encode, play an important role in achieving lifelong latency by evading the host immune systems (156, 157). Moreover, some evidence suggests that some viral-encoded miRNAs are active during latency, whereas others are more important during the productive replication phase (155). However, the detailed mechanisms underlying these different expression patterns in each phase remain unclear (155).
Viral miRNAs encoded by HSV, EBV, CMV, and BKV are described in Tables 2 and 4. These viral-encoded miRNAs inhibit target genes within the virus and its host cell (154). The functions of EBV- and CMV-encoded miRNAs have been investigated well in these four viruses.
A pertinent example of immune-related functional changes associated with viral-encoded miRNAs has been observed in B cells during acute EBV infection. EBV-encoded miRNAs have been shown in vitro to inhibit the adaptive and innate immune systems of the host efficiently. miR-BART-1, miR-BART-2, and miR-BHRF1 were shown to suppress the human IL-12B gene expression (93). B cells are the principal cell target of EBV (158). As a result, IL-12 secretion from the infected B cells was decreased. Ultimately, this inhibited Th1 cell differentiation (93). Thus, these three miRNAs can inhibit the adaptive immunity of the host during EBV infection. In addition, miR-BART-2–5p represses the major histocompatibility complex class I-related chain B (MICB) gene in B cells after primary EBV infection. MICB is a receptor of natural killer (NK) cells. As a result, recognition of the infected B cells by NK cells is lost due to decreased cell surface expression of MICB protein on the infected B cells (99). Interestingly, one of the target genes of miR-UL112, which is a CMV-encoded miRNA, is also MICB gene in multiple human cell lines (107). miR-UL112 interferes with the recognition of infected human cells by NK cells by inhibiting the expression of MICBs during CMV infection (107). The other CMV-encoded miRNAs that inhibit host immune defense are presented in Table 2. In this context, inhibition of these viral-encoded miRNAs may be a therapeutic target for these viral infections.
In addition to therapeutic targets, some viral-encoded miRNAs may be useful as biomarkers. For example, in a study in which profiles of viral-encoded miRNA expression were analyzed in samples from a cohort of solid organ transplant patients with CMV disease, the identification of hcmv-miR-UL22A-5p at baseline independently predicted the recurrence of CMV viremia upon discontinuation of anti-viral therapy (OR = 3.024; 95% CI, 1.35–6.8) (118). Additionally, in a study analyzing kidney transplant patients with BKV DNAemia following transplantation, blood BKV-miR-B1-5p detection (cycle threshold value of bkv-miR-B1-5p was 31.9) provided a sensitivity of 100% and a specificity of 94.9% for the diagnosis of biopsy-proven BK virus nephritis. In this study, the area under the curve (AUC) of the receiver operating characteristic (ROC) analysis was 0.97 (128). This miRNA may be an excellent tool for identifying patients at risk of BK virus nephritis, which is especially important because of the severe lack of accurate diagnostic markers for BK nephropathy (159, 160). We describe other candidate viral-encoded miRNAs as diagnostic biomarkers in Tables 2 and 4.
FUNGAL INFECTION
Over the last decade, as invasive fungal infections have risen in immunocompromised populations due to HIV, transplantation, and cancer, there has been renewed focus on reinventing anti-fungal therapy. As only a few classes of anti-fungal drugs are available, the emergence of resistance to single-drug classes and now multi-drug resistance significantly hampers patient management (161). In particular, Candida auris and cryptic species of Aspergillus are emerging concerns because of their multi-drug-resistant nature (32, 33, 162–164). Thus, newer therapeutic agents for highly resistant fungal infections are urgently required. In this context, miRNAs could become a treatment option, although the role of host miRNAs during fungal infections has not been extensively studied in humans. Furthermore, miRNAs may serve as useful diagnostic markers. The laboratory diagnosis of fungal infections such as invasive aspergillosis is often difficult due to the low sensitivity of culture and biomarkers, and consensus criteria have been used to tackle such limitations (165, 166). However, these criteria are not perfect, despite recent revisions (166). In this review, we focused on Aspergillus and Candida infections, which are the most common fungal infections.
Studies using human samples and/or human cell lines
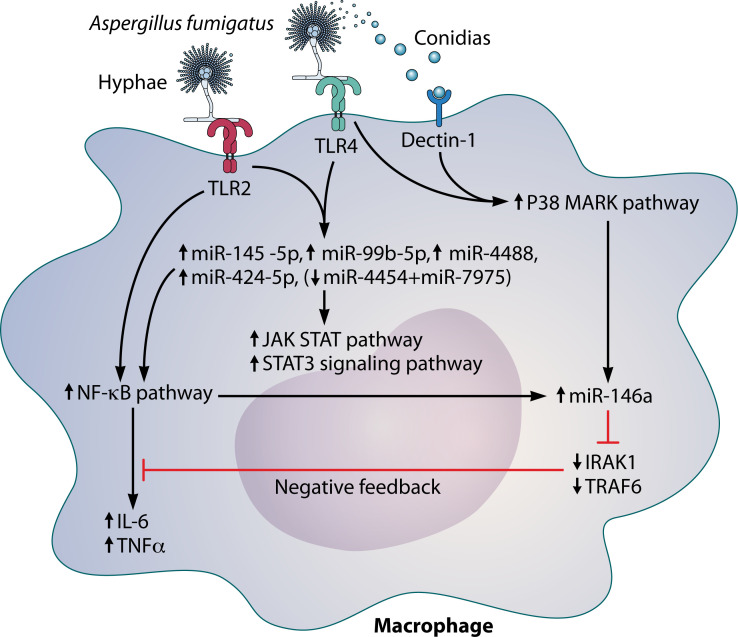
Aspergillus fumigatus is one of the most ubiquitous airborne fungi that causes invasive aspergillosis in immunocompromised patients. Several miRNAs have been implicated in Aspergillus infection and have varying roles. One interesting role of miRNAs is that miRNAs can drive a negative feedback loop in inflammatory response. In vitro, one study demonstrated that A. fumigatus induces activation of the NF-κB and p38 mitogen-activated protein kinase pathways, mediated by TLR2, TLR4 and dectin-1, which contributed to miR-146a upregulation in THP-1 macrophage-like cells (Fig. 2) (47). Upregulation of miR-146a inhibited IRAK1 and tumor necrosis factor receptor-associated factor 6, the target genes of miR-146a, and the inhibition of these genes then led to suppression of the NF-κB pathway. The observed increase in IL-6 and tumor necrosis factor alpha (TNF-α) in the cells was later decreased via a negative feedback loop (Fig. 2) (47). While this study provided evidence that the pro-inflammatory cytokines IL-6 and TNF-α play a central role in regulating the inflammatory response to A. fumigatus, a plausible reason for this mechanism of regulation remains unknown (167). Our group has previously shown that naïve T cells from patients with invasive aspergillosis, but not those with mucormycosis, exhibit reduced responsiveness to IL-6; whether impaired IL-6 responsiveness and downstream Th17 responses observed in T cells from patients with Aspergillus infection are mediated through miRNA upregulation requires further study (168). The function of miR-146a requires further investigation because of its potential value as a therapeutic target for invasive aspergillosis. Interestingly, miR-146a was upregulated in THP-1 cells (a human leukemia monocytic cell line) through the dectin-1 signaling pathway after exposure to Candida albicans (169). miR-146a, in turn, decreased the production of dectin-1-induced cytokines (such as IL-6 and TNF-α) in THP-1 cells (169). This function is quite similar to that observed after exposure to A. fumigatus. In contrast, the expression of miRNA-146a was downregulated in macrophages infected with Candida glabrata (170). This suggests that the mechanisms associated with miR-146a expression differ between other fungal species. In this context, the association between miR-146a and fungal infections has been relatively well investigated in human cell lines. However, the function of miRNAs other than miR-146a has yet to be well investigated during Aspergillus and Candida infection. We describe other functional miRNAs implicated in fungal infections that have been evaluated in studies using human samples and human cell lines in Table 5.
Fig 2.
Regulation pathway and functions of miR-146a and its related pathways during Aspergillus fumigatus infection in a macrophage cell line. Abbreviations: IL-6, interleukin 6; IRAK1, interleukin-1 receptor-associated kinase; NF-κB, nuclear factor-kappa B; p38 MARK, p38 mitogen-activated protein kinase; TLR, toll-like receptor; TNF-α, tumor necrosis factor alpha; TRAF6, TNF receptor-associated factor 6.
TABLE 5.
Functions and characteristics of microRNA in fungal infectionsa
| Type of miRNAs | Target gene/pathway | Study type | Functions and characteristics of miRNA |
|---|---|---|---|
| microRNA in Candida infection | |||
| miR-129–5p, miR-132–3p, miR-212–3p, and miR-212–5p | |||
| Not reported | Human in vitro study (171) (dendritic cells from healthy volunteers) | These are induced by Candida albicans in dendritic cells. | |
| miR-30–5p, miR-146a, and miR-210–3p | |||
| NF-κB signaling pathway | Human in vitro study (169) (THP-1 cells) | These expression levels are increased in human THP-1 cells after exposure of Candida albicans. Overexpression of miR146a significantly suppresses the production of IL-6 and TNF-α. | |
| miR-146a | |||
| Not reported | Human in vitro study (170) (PBMCs) | The expression of miR-146a is downregulated in infected macrophages with Candida glabrata. The downregulated miR-146a might reduce pro-inflammatory cytokine production. | |
| microRNA in Aspergillus infection | |||
| miR-145–5p, miR-424–5p, miR99b-5p, miR-4488, and (miR-4454 + miR-7975) | JAK STAT and NF-κB signaling pathway | Clinical study (45) (BALF samples) | In the lung transplant recipients, a total of five miRNAs are found to be specific to invasive aspergillosis, including four (miR-145–5p, miR-424–5p, miR-99b-5p, and miR-4488) that were upregulated and the pair (miR-4454 + miR-7975) that was downregulated in the invasive aspergillosis group versus controls. |
| miR-21–5p, miR-26b-5p miR-142–3p, and miR-142–5p | |||
| Not reported | Clinical study (172) (blood sample) | These show significant overexpression due to invasive aspergillosis in hemato-oncology patients with profound neutropenia. | |
| miR-191, miR-106, miR16-2, miR-26, miR-15, miR-20, miR-106, and miR-17 | |||
| Not reported | Clinical study (173) (blood sample) | 8 miRNAs (miR-191–5p, miR-106b-5p, miR-16-2-3p, miR-26a-5p, miR-15a-5p, miR-20a-5p, miR-106a-5p, and miR-17–5p) measured by quantitative RT-PCR had high discriminatory power (AUC >0.98 in ROC analysis), which could distinguish proven/probable IA from possible IA. | |
| miR-129–5p, miR-132–3p, miR-132–5p, miR-212–3p, and miR-212–5p | |||
| Not reported | Human in vitro study (171) (PBMCs) | These are induced by Aspergillus fumigatus in dendritic cells. | |
| miR-132 | |||
| Not reported | Human in vitro study (174) (PBMCs) | miR-132 is induced by Aspergillus fumigatus but not by lipopolysaccharide in human monocytes and dendritic cells. | |
| miR-146a | |||
| IRAK1 and TRAF6 | Human in vitro study (47) (THP-1 cells) |
Downregulates the level of TNF-α and IL-6 through the NF-κB signaling pathway in THP-1 macrophage-like cells challenged with A. fumigatus. | |
AUC, area under the curve; BALF, bronchoalveolar lavage fluid; IL, interleukin; IRAK1, interleukin-1 receptor-associated kinase; NF-κB, nuclear factor-kappa B; PBMC, peripheral blood mononuclear cell; ROC, receiver operating characteristic; TNF-α, tumor necrosis factor alpha; TRAF6, tumor necrosis factor receptor‑associated factor 6.
Regarding the application of miRNAs as biomarkers for the diagnosis of fungal infection, there are some clinical studies in which miRNAs were used as diagnostic markers for invasive aspergillosis. Gohir et al. reported that six miRNAs from bronchoalveolar lavage samples of lung transplant recipients were specific for the diagnosis of invasive aspergillosis, including four (miR-145–5p, miR-424–5p, miR-99b-5p, and miR-4488) that were upregulated and a pair (miR-4454 + miR-7975) that was downregulated. These six were found in patients with invasive aspergillosis but not in controls, who did not develop chronic lung allograft dysfunction or Aspergillus colonization after lung transplantation (45). This study reported that signaling from TLR2 and TLR4 might regulate these miRNAs, with downstream effects on the JAK-STAT and NF-κB pathways, both central to the intracellular immune response (Fig. 2). However, the detailed mechanisms and the consequences of this regulation remain unclear. Moreover, miR-142–3p, miR-142–5p, miR-26b-5p, and miR-21–5p in peripheral blood specimens showed significant overexpression associated with invasive aspergillosis in hemato-oncology patients with profound neutropenia (172), suggesting a potential role for these miRNAs as diagnostic biomarkers (Table 5). Some miRNAs might be helpful in Candida infection as diagnostic markers (Table 5); however, most have not been investigated well in clinical settings.
Animal studies
Several miRNAs have been investigated in animal cell lines or animal in vivo studies. For instance, miR-125a, miR-146, miR-155, and miR-455 are upregulated by heat-killed C. albicans in macrophages of mice (175). miR-29a-3p and miR-23b-3p are downregulated during Aspergillus exposure, and they regulate genes involved in innate responses to viable A. fumigatus in a mouse model (176).
BACTERIAL INFECTION
In the following section, we focus on miRNAs during Staphylococcus aureus and Pseudomonas aeruginosa infection because these two pathogens often cause complicated and life-threatening infections (30, 31). Furthermore, drug resistance such as methicillin-resistant Staphylococcus aureus and multi-drug-resistant P. aeruginosa make approaches to therapy very difficult. Therefore, investigating miRNAs during these infections is essential for developing new therapeutic options, in addition to new anti-microbial agents. An overview of functions and characteristics of miRNAs implicated in these infections and their target genes is provided in Tables 6 . In S. aureus and P. aeruginosa infections, most studies investigating miRNAs include animal experiments with or without human cellular experiments. Thus, we discuss the miRNA identified in both types of studies in this section and in Table 6.
TABLE 6.
Functions and characteristics of microRNA in bacterial infectionsa
| Type of miRNAs | Target gene/pathway | Study type | Function and characteristics of miRNA |
|---|---|---|---|
| microRNA in Staphylococcus aureus infection | |||
| miR-15b-5p | |||
| IKBKB and WEE1 | Animal in vivo and human in vitro study (177) (porcine wound infection model, HaCaT cells, and skin samples from patients) |
Responsible for defective DNA repair, may delay healing of wound in diabetic foot | |
| miR-24 | |||
| Chitinase3-like1 | Animal in vitro and human in vitro study (178) [MC3T3-E1 cells (mouse cell line) and Blood from patients] |
Downregulated in blood of osteomyelitis, protective against worsening of osteomyelitis | |
| miR-128 | |||
| MyD88 | Animal in vivo and in vitro study (179) (BALB/C mice and RAW264.7 macrophages (mouse cell line)] |
Increase expression in S. aureus infection. reduces the secretion of IL-6, IL-1β, and TNF-α | |
| miR-142 | |||
| Not reported | Animal in vivo study (180) (mouse skin wound model) |
Necessary for S. aureus clearance at skin wound sites | |
| miR-155 | |||
| IL-17 pathway | Animal in vivo and human in vitro study (181) (BAL from patients and C57BL/6 mice) |
Treatment with miR-155 antagomir improves lung bacterial clearance by 4.2-fold compared with control. | |
| miR-223 | |||
| IL-6 and Pclo | Animal in vivo and human in vitro study(46) (mouse wound model, skin sample from patients, and HL-60 cells) |
miR-223 is the most highly expressed during the inflammatory phase at wound sites. miR-223 antisense oligodeoxynucleotides in S. aureus-infected wild-type mice's wounds markedly improved the healing of them. | |
| microRNA in Pseudomonas aeruginosa infection | |||
| miR-183/96/182 cluster | |||
| Not reported | Animal in vivo and human in vitro study (182) (129 S2/BL6-mixed mice, human corneas sample, and human PBMCs) |
Inactivation of the miR-183/96/182 cluster decreases the severity of P. aeruginosa-induced keratitis because it decreases expression of pro-inflammatory neuropeptides in the cornea. | |
| DAP12 and Nox2 | Animal in vivo and in vitro study (183) [129 S2/BL6-mixed mice and RAW64.7 cells (mice cell line)] |
Knockdown of miR-183/96/182 cluster results in decreased production of multiple pro-inflammatory cytokines in response to P. aeruginosa or lipopolysaccharide treatment in macrophage-like RAW264.7 cells. | |
| miR-155 | |||
| Not reported | Animal in vivo and human in vitro study (184) (C57BL/6 mice and corneal samples) |
Upregulated during infection, may enhance keratitis | |
| miR-301b | |||
| c-Myb | Animal in vivo and in vitro study (185) [C57BL/6J mice, MLE-12 cells (mice), and MH-S cells (mice)] |
Suppression of miR-301b elevates levels of neutrophil infiltration, thereby alleviating symptoms caused by P. aeruginosa infection in mice. | |
| miR-302/367 cluster | |||
| NF-κB | Animal in vivo and in vitro study (186) [C57BL/6J mice, MLE-12 cells (mice), and MH-S cells (mice)] |
Upregulated after P. aeruginosa infection. | |
DAP12, DNAX activation protein of 12kDa; IKBKB, inhibitor of nuclear factor kappa B kinase subunit beta; IL, interleukin; MyD88, myeloid differentiation primary response 88; NF-κB, nuclear factor-kappa B; Nox2, NADPH oxidase 2; PBMC, peripheral blood mononuclear cell; Pclo, presynaptic cytomatrix protein; TNF-α, tumor necrosis factor alpha.
miR-15b-5p, miR-24, miR-155, and miR-223 have been associated with diabetic foot ulcers, osteomyelitis, pneumonia, and wounds caused by S. aureus (46, 177, 178, 181). These studies were primarily performed by using animal cell lines and/or animal models (46, 177, 178, 181). Interestingly, some of these miRNAs have been investigated for therapeutic applications in animal models. For instance, miR-223 was highly expressed during the inflammatory phase of S. aureus-infected wound sites in mice (46). A known target of miR-223 is IL-6, so the secretion of IL-6 from neutrophils was potentially decreased by the high expression of miR-223 at the wound sites. Support for the important role of miR-223 is illustrated by the delayed healing of the infected sites, as neutrophil-derived IL-6 was positively linked to S. aureus clearance (187). Furthermore, topical miR-223 antisense oligodeoxynucleotides (ODNs) in S. aureus-infected wild-type wounds markedly improved wound healing compared to that in control wounds (wild-type wounds without miR-223 antisense ODNs) in an animal experiment (46). This result suggests that miR-223 antisense ODNs could be utilized for wounds associated with S. aureus infection in a clinical setting.
Regarding P. aeruginosa, miR-155 and a miR-183/96/182 cluster were associated with the pathogenesis of pseudomonal keratitis (182, 184). According to previous studies (182, 184), the severity of the keratitis may be decreased if these miRNAs are inactivated. Therefore, these miRNAs could be considered as new therapeutic targets. In addition, the miR-183/96/182 cluster and the miR-302/367 cluster have been found to influence macrophage function and subsequent bacterial clearance during P. aeruginosa infection (183, 186). In other words, some miRNAs were directly associated with innate immune responses during P. aeruginosa infection. Furthermore, miR-302/367 cluster expression was significantly increased after P. aeruginosa respiratory infection in a mouse model (186).
Additionally, NF-κB, which inhibits mitophagy in macrophages, was identified as a target gene of the miR-302/367 cluster in the same study (186). Mitophagy, the selective autophagy to eliminate damaged mitochondria, is a highly conserved cellular self-digestion and catabolism critical for maintaining cellular homeostasis (188). Various microbial components modulate this process, thereby affecting the innate immune response to infection (189). In this study, ongoing mitophagy was associated with the clearance of P. aeruginosa in the macrophages (186, 190). Therefore, overexpression of the miR-302/367 cluster promotes the clearance of P. aeruginosa in macrophages through mitophagy by inhibiting the target gene (NF-κB) expression (186). Although details of how P. aeruginosa accesses the mitochondria and impacts mitophagy induction require further investigation, it is apt to consider both miRNAs and host mitophagy as potential targets for therapy against P. aeruginosa-associated infection.
PARASITIC INFECTION
The World Health Organization reported 241 million cases and 627 thousand deaths from malaria in 2020 (191). Analogously, it is estimated that at least 230 million people are infected with schistosomes, the pathogen responsible for schistosomiasis (192). Among the parasites that cause malaria or schistosomiasis, P. falciparum and S. japonicum cause the most severe clinical syndromes and/or pathogenicity, respectively (34, 35). To date, effective vaccines against malaria and schistosomiasis have not been established, limiting the prevention of these infections (192, 193). Furthermore, drug resistance to therapeutic agents, including artemisinin-resistant Plasmodium, is a serious concern (193). Regarding schistosomiasis, there is no clear evidence of praziquantel resistance yet, even after its extensive use in many endemic countries. However, such resistance can be experimentally induced; thus, the threat of emerging resistance caused by mass monotherapy remains (192). Hence, investigating miRNAs associated with these parasitic infections is necessary to identify new therapeutic options. Investigations of miRNAs in malaria and schistosomiasis have primarily focused on P. falciparum and S. japonicum (194), including animal studies and/or human in vitro studies (Tables 7 and 8). In this review, host-encoded miRNAs that are associated with Plasmodium falciparum and Schistosoma japonicum were examined. In addition, influence of parasite-encoded miRNAs on the pathogenesis of parasitic diseases is reviewed. miRNAs associated with strongyloidiasis were excluded from this review due to the limited data available.
TABLE 7.
Functions and characteristics of microRNA in malaria caused by Plasmodium falciparuma
| Type of miRNAs | Target gene/pathway | Study type | Functions and characteristics of miRNA |
|---|---|---|---|
| Host-encoded microRNA in P. falciparum infection | |||
| miR-146a | |||
| Not reported | Clinical study (195) (blood sample) | May be associated with protection in pregnant women | |
| Not reported | Clinical study (196) (blood sample) | No major role in the development of cerebral malaria | |
| miR-150 | |||
| Not reported | Clinical study (197) (blood sample) | Differentiation between fatal and non-fatal malaria | |
| miR-150–3p and miR-197–5p | |||
| pfApricon (gene of malaria) | Human in vitro study (198) (human erythrocytes and HEK293T cells) | Inhibits virulence of P. falciparum | |
| miR-155 | |||
| Not reported | Human in vitro and animal (mice) in vivo study (199) (human blood sample) |
Can reduce vascular leak in cerebral malaria | |
| miR-223 | |||
| Not reported | Human in vitro study (200) (human RBCs) |
Negatively regulates P. falciparum along with miR-451 and let-7i in vitro | |
| miR-451 and let-7i | |||
| mRNA of P. falciparum (not specified) | Human in vitro study (200) (human RBCs) | Associated with host protection | |
| miR-451 | |||
| Var gene of malaria | Human in vitro and animal (mice) in vivo study (201) |
Inhibits the parasite virulence factor pfEMP1 | |
| miR-4497 | |||
| Not reported | Clinical study (202) (blood sample) | Discriminate severe malaria from uncomplicated malaria | |
| let-7a-5p | |||
| Not reported | Clinical study (203) (blood sample) | Discrimination between healthy control and P. falciparum infection | |
Studies in mouse models of cerebral malaria using Plasmodium berghei ANKA, which causes cerebral malaria in mice, are included in this table because the cerebral malaria is a significant complication caused by P. falciparum in human. Abbreviations: pfEMP1, P. falciparum erythrocyte membrane protein 1.
TABLE 8.
Functions and characteristics of microRNA in the schistosomiasis caused by Schistosoma japonicuma
| Type of miRNAs | Target gene/pathway | Study type | Functions and characteristics of miRNA |
|---|---|---|---|
| Host-encoded microRNA in S. japonicum infection | |||
| miR-21 | |||
| SMAD7 | Animal (mice) in vitro and in vivo study (204) (mouse hepatic stellate cells, hepatocytes, and Kupper cells) |
Upregulated, pro-fibrotic | |
| miR-92a-2–5p | |||
| TLR2 | Animal (mice) and human in vitro and animal (mice) in vivo study (205) (NIH/3 T3 embryonic fibroblasts and 293T cells) |
Anti-fibrotic | |
| miR-96 | |||
| SMAD7 | Animal (mice and rats) and human in vitro and animal (mice) in vivo study (206) (HSC-T6, HEK293 and mouse primary hepatic stellate cells) |
Upregulated, pro-fibrotic | |
| miR-142 | |||
| WASL | Animal (mice) in vivo study (207) |
Suppresses infection | |
| miR-146b | |||
| STAT1 | Animal (mice) in vivo study (208) |
Upregulated, inhibits differentiation of macrophages to M1 cells | |
| miR-148a | |||
| PTEN | Animal (mice) in vitro and in vivo study (209) (RAW264.7cells and NCTC clone 1469 cells) |
Regulates cytokine production in macrophages | |
| miR-155 | |||
| FOXO3a | Human in vitro study (210) (LX-2 cells) |
Anti-fibrotic | |
| miR-182–5p | |||
| TTP | Animal (mice) in vivo and human and animal (mice) in vitro study (211) (HL7702, AML12 and U937 cells) | Decreased in mice liver, pro-fibrotic | |
| miR-203–3p | |||
| IL-33 | Animal (mice) in vitro and in vivo study (212) (mouse HSCs, hepatocytes, and Kupper cells) |
Downregulated, anti-fibrotic | |
| miR-351 | |||
| VDR | Animal (mice) in vitro and in vivo study (213) (mouse HSCs, hepatocytes, and Kupper cells) |
Pro-fibrotic, initial downregulation during infection followed by upregulation | |
| miR-454 | |||
| SMAD4 | Human in vitro and animal (mice) in vivo study (214) (LX-2 cells) |
Anti-fibrotic | |
| miR-92a-3p, miR-146a-5p, and miR-532–5p | |||
| Not reported | Clinical study (215) | Differentiation between no fibrosis and fibrosis grade | |
| miR-146a-5p, miR-150–5p, Let7a-5p, and Let-7d-5p | |||
| Not reported | Cinical study (216) | Differentiation between fibrotic grades | |
| Parasite-encoded microRNA in S. japonicum infection | |||
| Sja-miR-1 | |||
| SFRP1 | animal (mice) in vivo and human and animal (mice) in vitro study (217) (LX-2 cells and mouse primary HSCs) |
Anti-fibrotic | |
| Sja-miR-71a | |||
| SEMA4D | Animal (mice) in vivo and human in vitro study (218) (LX-2 cells) |
Anti-fibrotic | |
| Sja-miR-124 | |||
| sjDDX1 and sjPOLE2 | Animal (mice) in vivo study (219) | Reduces hepatic egg number Decreases inflammatory cell infiltration | |
| Sja-miR-125b and Sja-bantam | |||
| PROS1, CLMP, and FAM212B | Animal (mice) in vitro and animal (mice and rbits) in vivo study (220) (RAW264.7cells and NCTC clone 1,469 cells) | Increases macrophage proliferation and TNF-α production | |
| Sja-miR-2162 | |||
| TGFBR3 | Human and animal (rats) in vitro and animal (mice) in vivo study (221) (LX-2 cells and HSC-T6 cells) |
Pro-fibrotic | |
CLMP, CXADR-like membrane protein; FAM212B, family with sequence similarity 212 member B; FOXO, forkhead box O; IL, interleukin; PROS, protein S; PTEN, phosphatase and tensin homolog; SFRP1, secretion of frizzled-related protein 1; sjDDX1, S. japonicum DEAD-box ATP-dependent RNA helicase 1; sjPOLE2, S. japonicum DNA polymerase 2 subunit 2; SMAD, Drosophila mother against decapentaplegic; STAT1, signal transducer and activation of transcription 1; TGFBR3, transforming growth factor beta regulator 3; TLR, toll-like receptor. TTP, tristetraprolin; VDR, vitamin D receptor; WASL, Wiskott-Aldrich syndrome protein.
Function and characteristics of host-encoded miRNAs during parasitic infection
Some host-encoded miRNAs are associated with pathogenesis of P. falciparum and S. japonicum infection. Herein, we discuss host-encoded miRNAs associated with P. falciparum and S. japonicum infection separately because they have different characteristics in terms of target cells and functional mechanisms.
Host-encoded miRNAs during P. falciparum infection
Studies using human samples and/or human cell lines with or without an animal experiment
miRNAs investigated in malaria infections caused by P. falciparum are mostly intraerythrocytic (see Table 7). Approximately 100 human (host-encoded) miRNAs have been identified in matured erythrocytes, which is of particular importance in that these cells lack a nucleus and transcription/translation machinery (222). Thus, the miRNAs found in matured erythrocytes primarily act on mRNA synthesis elsewhere (i.e., within the parasite). When merozoites of P. falciparum invade erythrocytes, genetic material between the host erythrocytes and Plasmodium species is exchanged (223). Infection allows the transfer of human miRNAs and Argonaute proteins, which are needed to form the miRISC, from the erythrocyte into the parasite cytoplasm (201, 223). As a result, some of these host-encoded miRNAs can bind the mRNAs of P. falciparum, inhibiting the translation of P. falciparum genes (224). This process has been observed in a previous in vitro study, where a subset of erythrocyte miRNAs, miR-451 and let-7, translocated into the parasite and negatively regulated P. falciparum (200). Moreover, these miRNAs are integrated into essential parasite mRNAs and via impairment of ribosomal loading, resulting in translational inhibition (200). In a more recent study, miR-451 was reported to repress the P. falciparum erythrocyte membrane protein 1 (pfEMP1), an important virulence factor produced by P. falciparum, by inhibiting the var gene encoding pfEMP1 (201). These miRNAs may confer innate resistance to malaria (200, 201).
Interestingly, miR-451 and let-7i were highly enriched in hemoglobin AS (sickle cell trait) erythrocytes and HbSS (sickle cell disease) erythrocytes, compared to HbAA (normal) erythrocytes (200, 225). Thus, the enrichment of these miRNAs in HbAS and HbSS erythrocytes might explain why those with sickle cell disease are resistant to malaria (200). In this context, miR-451 and let-7i could be utilized as therapeutic targets for P. falciparum. In addition, two erythrocytic miRNAs, miR-150–3p and miR-197–5p, have been reported to inhibit P. falciparum growth and invasion by targeting the apicortin gene (198).
Interestingly, some miRNAs may play an essential role in the pathogenesis of cerebral malaria. Cerebral malaria is the most common and severe neuropathological manifestation of malaria caused by P. falciparum in humans (224). In an in vivo experimental model of cerebral malaria (mice infected with Plasmodium berghei), survival was significantly improved in miR-155−/− mice compared to that in wild-type littermate mice (199). The improved survival was associated with preservation of blood-brain barrier integrity and reduced endothelial cell activation (199).
To supplement this evidence, pre-treatment with an antagomir of miR-155 reduced vascular leakage in an ex vivo endothelial microvessel model of cerebral malaria (199). Therefore, miR-155 may be an important therapeutic target to prevent cerebral malaria, although a target gene of miR-155 has not yet been identified. We report additional miRNAs associated with P. falciparum infection in studies using clinical samples and cell lines with or without animal experiments in Table 7.
Several miRNAs have been clinically explored for their application as biomarkers for the diagnosis or evaluation of parasitic infection (Table 7). For example, let-7a-5p was significantly upregulated in the blood of P. falciparum-infected patients compared to uninfected patients (control) (P = 0.01), with the area under the ROC curve equal to 0.82 (P = 0.003) (203). Thus, let-7a-5p could help identify P. falciparum-infected patients, although its sensitivity and specificity warrant confirmation (203). Similarly, plasma concentrations of miR-150 were higher in adult cases of fatal cerebral malaria than those of non-fatal cerebral malaria (median relative expression level 25.4 versus 8.5, P = 0.003) (197). Thus, miR-150 has the potential to discriminate between fatal and non-fatal adult cerebral malaria, and it should be explored further (197).
Animal and bioinformatic studies
In a mouse model experiment and bioinformatic analysis, miR-19a-3p, miR-19b-3p, miR-142–3p, and miR-223–3p were found to be involved in several pathways relevant to cerebral malaria (226). In addition, miR-451a and miR-223–3p may make the most notable contribution to the pathogenesis of P. falciparum based on a bioinformatic analysis (227).
Host-encoded miRNAs during S. japonicum
Most studies investigating miRNAs in S. japonicum infection include an animal experiment with or without a human cellular experiment. Thus, we discuss miRNAs identified in both types of studies in this section and in Table 8. In schistosomiasis caused by S. japonicum, many eggs are permanently lodged in the intestine or liver (192), providing an antigenic stimulus for the immune response, which leads to granuloma formation and hepatic fibrosis (35, 192). Hepatic fibrosis is associated with high morbidity and mortality in schistosomiasis (228), and many miRNAs have been explored in this setting (Table 8). For instance, miR-96 and miR-351 have been recognized as pro-fibrotic miRNAs (206, 213). They were found to be upregulated in hepatic stellate cells during the chronic progressive phase of the schistosomiasis, leading to repression of Drosophila mother against decapentaplegic 7 (SMAD7) and the vitamin D receptor (VDR), which were their direct targets, respectively (206, 213). Repression of these proteins promoted hepatic stellate cell activation and the release of IL-13 and the transforming growth factor beta 1 (TGF-β1). SMAD7 and VDR are antagonists of the TGF-β1/SMAD pathway, and activating these proteins normally inhibits pro-fibrosis responses (35, 206, 213).
An anti-fibrotic miRNA, miR-203–3p, has also been identified. miR-203–3p has been found to target IL-33 directly in hepatic stellate cells during a murine model of S. japonicum infection (212). Lowered levels of miR-203–3p lead to increased expression of IL-33 from hepatic stellate cells, which promotes the expansion and IL-13 production of hepatic group 2 innate lymphoid cells (ILC2s) (212). As a result, hepatic fibrosis pathology progressed through the activation of hepatic stellate cells by IL-13 from ILC2s (212). Elevation of miR-203–3p in the liver through recombinant adeno-associated virus 8 infections was found to protect mice from lethal infection (212). In this context, inhibition of the pro-fibrosis miRNAs and/or induction of anti-fibrosis miRNAs could be explored for the prevention of hepatic fibrosis caused by S. japonicum. We describe other miRNAs that are not directly associated with fibrosis but are associated with S. japonicum infection in Table 8.
Several miRNAs have been explored clinically regarding the application of miRNAs as biomarkers for diagnosis or evaluation of S. japonicum infection (Table 8). For instance, miR-146a-5p and miR-150–5p isolated from the blood could moderately distinguish mild hepatic fibrosis (up to grade I) from severe (grades II and III), with AUC of 0.66, P = 0.0005 and 0.68, and P < 0.0001, respectively, in schistosomiasis caused by S. japonicum (216).
Function and characteristics of parasite-encoded miRNAs during parasitic infection
miRNAs are encoded by almost all eukaryotic parasites, including Schistosoma spp., Clonorchis species, Fasciola species, and Echinococcus species (229). However, neither miRNAs, Dicer, nor Argonaute proteins are encoded in the genome of Plasmodium falciparum (222, 230, 231). Parasite-encoded miRNAs can suppress host and parasite-encoded mRNA (229). These characteristics are similar to those of viral-encoded miRNAs, described in the viral section of this review.
One pertinent example in schistosomiasis caused by S. japonicum is the promotion of hepatic fibrosis by parasite-encoded miRNAs sja-miR-1 and sja-miR-2162, which are found in hepatic stellate cells (217, 221). Sja-miR-1 represses the secretion of frizzled-related protein 1 directly in hepatic stellate cells (217), which leads to the activation of wingless and INT-1/β-catenin, a pro-fibrosis pathway (232, 233). Additionally, sja-miR-2162 directly inhibited transforming growth factor beta regulator 3 in hepatic stellate cells, providing negative feedback to allow for increased TGF-β signaling, which also promoted hepatic fibrosis (221). Thus, these miRNAs may be therapeutic targets. Inhibition of these miRNAs alleviated hepatic fibrosis caused by S. japonicum in a mouse model (217, 221). Moreover, sja-miR-124 might play an important role in this infection, but through targeting a gene in S. japonicum itself. Overexpression of sja-miR-124–3p in infected mice was found to reduce hepatic egg number, produce smaller egg liver granulomas, and diminish inflammatory cell infiltration (219). The target genes of sja-miR-124–3p are S. japonicum DEAD-box ATP-dependent RNA helicase 1 (sjDDX1) and DNA polymerase 2 subunit 2 (219). In addition, RNA interference-mediated sjDDX1 silencing in mice resulted in 24.6% worm reduction rate and 18.4% egg reduction rate (219). This reduction in egg number is significant as schistosome eggs induce the pathology caused by schistosomiasis (192). These findings suggest that sja-miR-124–3p can decrease the hepatic damage caused by S. japonicum.
We present other S. japonicum-encoded miRNAs with therapeutic potential in Table 8.
FUTURE DIRECTIONS OF miRNA
Although the human genome encodes 25,000 genes, it is estimated that only 600 disease-modifying protein drug targets exist (234), and only some proteins can be targeted or modulated by a drug molecule. Therefore, the focus of target selection has shifted to other macromolecules, such as RNAs (25). Recently, RNA-targeted therapy has been developed rapidly (25, 235). One of the miRNA modulation strategies is using anti-sense oligonucleotide agents such as miRNA mimics and miRNA inhibitors (25). miRNA mimics are double-stranded RNA molecules that imitate endogenous miRNA duplexes (236) and induce gene silencing in the same manner as miRNAs, thus resulting in the downregulation of the target protein (25). Furthermore, antimir oligonucleotides, which belong to miRNA inhibitors, are antisense-like oligonucleotide analogs of various types perfectly complementary to the mature miRNA guide strand. miRNA inhibitors (237), including antimir oligonucleotides, induce selective upregulation of one protein population by binding to and inhibiting the target miRNA (25). These techniques have been applied to the production of miRNA drugs, which is a term to define drugs targeting miRNAs. To date, 10 miRNA drugs for various diseases, including infectious diseases, have been in clinical trials (238). A major concern is that an miRNA has the potential to bind to off-target genes, as one miRNA generally has multiple target genes (238–240). Regarding the 10 miRNA drugs in clinical trials, the target genes of each miRNA drug ranged from 30 to 250 (238). Because of its toxicity, no miRNA drug has been used in phase 3 clinical trials and has been approved for clinical usage (238). Thus, the developing of drugs targeting miRNAs may be complex because miRNAs regulate many genes other than the main target gene. The development of target cell-specific and/or target gene-specific miRNA drugs may be required to minimize these concerns.
miRNAs have also been evaluated for their potential as biomarkers of infection, specifically for diagnostic purposes. Both host-encoded and viral-encoded miRNAs have been evaluated in clinical studies (44, 45, 102, 118, 128). Only 4 of the 19 studies (Tables 1 to 8) which evaluated miRNAs as biomarkers reported sensitivity and specificity (61, 119, 128, 129). Ideally, multiple large clinical cohort studies in different patient populations are required to assess the robust sensitivity and specificity in diagnosing a specific infectious disease. However, none of the four studies were large. Thus, sensitivity and specificity which are reliable in specific clinical settings, have yet to be reported in most of these previous studies.
Moreover, to our knowledge, a standard assay to measure an miRNA as a biomarker, including commercially available ones, has yet to be established. The data from the studies reviewed in this paper are mainly obtained from assays developed de novo by a research institution. In this context, validation of these or new standardized assays in large multi-center cohort studies and/or prospective studies is needed to verify the effectiveness of these miRNAs as biomarkers.
To explore the use of miRNAs as potential biomarkers of disease, it is necessary to establish robust detection methods. Northern blots are the gold standard for detecting miRNAs. However, this method is time-consuming and cannot be used to detect trace amounts of miRNAs. Over the past decade, commonly used technologies such as next-generation sequencing (NGS), microarrays, and quantitative reverse transcription PCR (RT-qPCR), along with emerging technologies such as NanoString nCounter Platform, have been used to detect and quantify miRNAs (241, 242). NGS is widely used for both the identification and quantification of miRNAs, and it allows for detection of miRNAs present in very low quantities. However, it has the disadvantage of being relatively expensive (243). Microarrays are one of the most commonly used high-throughput method for detection of miRNAs. Microarrays detect miRNAs through hybridization of the miRNAs to the target probe. They are used to identify known miRNAs and have limited potential for identifying new miRNAs (244). In clinical laboratories, the most frequently used method for detection of specific miRNAs is RT-qPCR because equipment required to run the assays is widely available, and a wide variety of commercial kits exist for detection of miRNA using RT-qPCR (245). The NanoString nCounter system is a new technology that enables direct quantification of individual miRNAs without the need for amplification or reverse transcription (246). The ability to detect and quantify miRNAs without amplification allows it to provide absolute quantification, which reduce potential bias (247). However, this method requires specialized equipment which is not widely available.
Adding further complexity to the practical utility of using miRNA as a biomarker is the availability of other nucleic acid-based molecular targets. Direct comparisons between miRNAs and other established biomarkers, such as microbial cell-free DNA (mcfDNA) obtained from the blood or urine of patients, have yet to be investigated. While mcfDNA can be used to identify causative pathogens at the species level (through sequencing) (248), miRNAs cannot. However, mcfDNA may have less utility in cases of infection by RNA viruses that lack DNA (248). In these contexts, miRNAs may be able to supplement the knowledge gained from mcfDNA testing by providing additional information about the infection severity or treatment response.
CONCLUDING REMARKS
In summary, the role of miRNAs in infectious diseases is emerging rapidly. The miRNAs reviewed are related to immune function and have diagnostic, prognostic, and therapeutic potentials. Their diagnostic potential is largely untapped in infectious diseases where the current molecular diagnostic tool is not perfected or requires a constellation of other clinical criteria, such as invasive mold infections. Although a few miRNAs have been developed and translated into clinical trials, only one miRNA has been evaluated in infectious disease (miravirsen for HCV) (56). Since miRNAs are extensively incorporated into the pathogenesis of infections and immune response, further studies elucidating the mechanistic functions of miRNAs will permit the development of additional targeted therapeutics. Nevertheless, we are only at the beginning of discovering the full potential of miRNA diagnostics and therapeutics in infectious disease.
ACKNOWLEDGMENTS
The authors declare no conflicts of interest related to this study. The authors alone are responsible for the content and writing of this article.
S.H. conceived ideas for this review. M.K. and S.K. wrote the primary draft of the manuscript. W.G., J.F.C., and S.H. revised the manuscript. All the authors approved the final version of the manuscript.
Biographies

Muneyoshi Kimura is a research fellow in the department of Transplant Infectious Diseases, Ajmera Transplant Program, University Health Network, Toronto, Canada. He also works as an infectious diseases physician in Toranomon Hospital, Tokyo, Japan. He has published ten research articles in infectious diseases and his research interest is infectious diseases in immunocompromised patients, particularly invasive fungal infection of allogeneic hematopoietic stem cell transplant recipients.

Sagar Kothari has a passion for infectious diseases and new therapeutics. He studied at the University of Toronto, Canada, where he specialized in immunology and pharmacology, and has conducted research in a breadth of opportunistic infections while at the University Health Network's Ajmera Transplant Centre. He has also had the privilege of working on several clinical trials for new antifungals. Sagar is now starting medical school, with the goal of becoming an infectious disease physician and scientist.

Wajiha Gohir completed her Masters at McMaster University in the department of Biochemistry and Biomedical Sciences, where she investigated role of gut microbiota during pregnancy. At University Health Network, she joined the Transplant Infectious Disease department. Her work focused on understanding the role of STAT3 in invasive aspergillosis and expression of miRNAs in lung transplant recipients.

Jose F. Camargo is a Transplant Infectious Diseases specialist with 10 years of experience. He completed a post-doctoral research fellowship in immunogenetics at the University of Texas Health Science Center at San Antonio; and underwent Internal Medicine training at Jacobi Medical Center/Albert Einstein College of Medicine in New York; followed by an Infectious Diseases fellowship at Westchester Medical Center/New York Medical College, and a Transplant Infectious Diseases fellowship at the University of Toronto. Currently, he is an Associate Professor of Medicine, Immunocompromised Host Section, Division of Infectious Diseases at University of Miami, Miller School of Medicine. He has served as ad-hoc reviewer for Plos One, American Journal of Transplantation, Transplant Infectious Diseases, Clinical Transplantation, Lancet Infectious Diseases, among others; and has several publications in prestigious journals including Nature Medicine, Nature Immunology and Blood. His research interests include CMV and Aspergillus infections, vaccine immunogenicity, and cell-mediated immunity.

Shahid Husain is an internationally recognized expert in the field of Transplant Infectious Diseases, and is an active contributor in the development of guidelines for the American Society of Transplantation and the Infectious Diseases Society of America. Dr. Husain’s research has focused on the risk factors and outcomes of infections in immunocompromised hosts. His funded clinical and translational research is directed towards the evaluation of diagnostic tests for early diagnosis of invasive aspergillosis; especially novel techniques such as droplet digital PCR and biomarkers such as miRNA. He has been funded through the National Institutes of Health, Canadian Institutes of Health Research and the Center for Disease Control. To date, he has published more than 200 peer-reviewed publications and has authored more than seven book chapters. Currently, he serves as the Director of Research for the Ajmera Transplant Centre at Toronto General Hospital, University Health Network.
Contributor Information
Shahid Husain, Email: shahid.husain@uhn.ca.
Graeme N. Forrest, Rush University, Chicago, Illinois, USA
REFERENCES
- 1. Friedman RC, Farh KK-H, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105. doi: 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to Lin-14. Cell 75:843–854. doi: 10.1016/0092-8674(93)90529-y [DOI] [PubMed] [Google Scholar]
- 3. Creugny A, Fender A, Pfeffer S. 2018. Regulation of primary microRNA processing. FEBS Lett 592:1980–1996. doi: 10.1002/1873-3468.13067 [DOI] [PubMed] [Google Scholar]
- 4. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419. doi: 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- 5. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436:740–744. doi: 10.1038/nature03868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fabian MR, Sonenberg N. 2012. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19:586–593. doi: 10.1038/nsmb.2296 [DOI] [PubMed] [Google Scholar]
- 7. Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. 2008. MicroRNAs: new regulators of immune cell development and function. Nat Immunol 9:839–845. doi: 10.1038/ni.f.209 [DOI] [PubMed] [Google Scholar]
- 8. Huntzinger E, Izaurralde E. 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12:99–110. doi: 10.1038/nrg2936 [DOI] [PubMed] [Google Scholar]
- 9. Hwang H-W, Mendell JT. 2006. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 94:776–780. doi: 10.1038/sj.bjc.6603023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. 2007. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083 [DOI] [PubMed] [Google Scholar]
- 11. Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, Dangwal S, Kumarswamy R, Bang C, Holzmann A, Remke J, Caprio M, Jentzsch C, Engelhardt S, Geisendorf S, Glas C, Hofmann TG, Nessling M, Richter K, Schiffer M, Carrier L, Napp LC, Bauersachs J, Chowdhury K, Thum T. 2012. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 3:1078. doi: 10.1038/ncomms2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng AM, Byrom MW, Shelton J, Ford LP. 2005. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 33:1290–1297. doi: 10.1093/nar/gki200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu C-G, Kipps TJ, Negrini M, Croce CM. 2005. miR-15 and miR-16 induce apoptosis by targeting Bcl2. Proc Natl Acad Sci U S A 102:13944–13949. doi: 10.1073/pnas.0506654102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jordan SD, Krüger M, Willmes DM, Redemann N, Wunderlich FT, Brönneke HS, Merkwirth C, Kashkar H, Olkkonen VM, Böttger T, Braun T, Seibler J, Brüning JC. 2011. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol 13:434–446. doi: 10.1038/ncb2211 [DOI] [PubMed] [Google Scholar]
- 15. Wilfred BR, Wang WX, Nelson PT. 2007. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab 91:209–217. doi: 10.1016/j.ymgme.2007.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou S-S, Jin J-P, Wang J-Q, Zhang Z-G, Freedman JH, Zheng Y, Cai L. 2018. miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol Sin 39:1073–1084. doi: 10.1038/aps.2018.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen C, Zhou Y, Wang J, Yan Y, Peng L, Qiu W. 2018. Dysregulated microRNA involvement in multiple sclerosis by induction of T helper 17 cell differentiation. Front Immunol 9:1256. doi: 10.3389/fimmu.2018.01256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riancho J, Vázquez-Higuera JL, Pozueta A, Lage C, Kazimierczak M, Bravo M, Calero M, Gonalezález A, Rodríguez E, Lleó A, Sánchez-Juan P. 2017. MicroRNA profile in patients with Alzheimer's disease: analysis of miR-9-5p and miR-598 in raw and exosome enriched cerebrospinal fluid samples. J Alzheimers Dis 57:483–491. doi: 10.3233/JAD-161179 [DOI] [PubMed] [Google Scholar]
- 19. Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. 2005. Aberrant T cell differentiation in the absence of dicer. J Exp Med 202:261–269. doi: 10.1084/jem.20050678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen C-Z, Li L, Lodish HF, Bartel DP. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86. doi: 10.1126/science.1091903 [DOI] [PubMed] [Google Scholar]
- 21. Kawai T, Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650. doi: 10.1016/j.immuni.2011.05.006 [DOI] [PubMed] [Google Scholar]
- 22. Staedel C, Darfeuille F. 2013. MicroRNAs and bacterial infection. Cell Microbiol 15:1496–1507. doi: 10.1111/cmi.12159 [DOI] [PubMed] [Google Scholar]
- 23. Jafarzadeh A, Naseri A, Shojaie L, Nemati M, Jafarzadeh S, Bannazadeh Baghi H, Hamblin MR, Akhlagh SA, Mirzaei H. 2021. MicroRNA-155 and antiviral immune responses. Int Immunopharmacol 101:108188. doi: 10.1016/j.intimp.2021.108188 [DOI] [PubMed] [Google Scholar]
- 24. Lee YS, Dutta A. 2009. MicroRNAs in cancer. Annu Rev Pathol 4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmidt MF. 2014. Drug target miRNAs: chances and challenges. Trends Biotechnol 32:578–585. doi: 10.1016/j.tibtech.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 26. Piret J, Boivin G. 2011. Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management. Antimicrob Agents Chemother 55:459–472. doi: 10.1128/AAC.00615-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Al Hamed R, Bazarbachi AH, Mohty M. 2020. Epstein-Barr virus-related post-transplant lymphoproliferative disease (EBV-PTLD) in the setting of allogeneic stem cell transplantation: a comprehensive review from pathogenesis to forthcoming treatment modalities. Bone Marrow Transplant 55:25–39. doi: 10.1038/s41409-019-0548-7 [DOI] [PubMed] [Google Scholar]
- 28. Ambalathingal GR, Francis RS, Smyth MJ, Smith C, Khanna R. 2017. BK Polyomavirus: clinical aspects, immune regulation, and emerging therapies. Clin Microbiol Rev 30:503–528. doi: 10.1128/CMR.00074-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Griffiths C, Drews SJ, Marchant DJ. 2017. Respiratory syncytial virus: infection, detection, and new options for prevention and treatment. Clin Microbiol Rev 30:277–319. doi: 10.1128/CMR.00010-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Husain S, Camargo JF. 2019. Invasive aspergillosis in solid-organ transplant recipients: guidelines from the American society of transplantation infectious diseases community of practice. Clin Transplant 33:e13544. doi: 10.1111/ctr.13544 [DOI] [PubMed] [Google Scholar]
- 33. Spivak ES, Hanson KE. 2018. Candida auris: an emerging fungal pathogen. J Clin Microbiol 56:e01588-17. doi: 10.1128/JCM.01588-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Greenwood BM, Bojang K, Whitty CJM, Targett GAT. 2005. Malaria. The Lancet 365:1487–1498. doi: 10.1016/S0140-6736(05)66420-3 [DOI] [PubMed] [Google Scholar]
- 35. Chen Q, Zhang J, Zheng T, Chen H, Nie H, Zheng B, Gong Q. 2019. The role of microRNAs in the pathogenesis, grading and treatment of hepatic fibrosis in schistosomiasis. Parasit Vectors 12:611. doi: 10.1186/s13071-019-3866-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yong MK, Shigle TL, Kim Y-J, Carpenter PA, Chemaly RF, Papanicolaou GA. 2021. American society for transplantation and cellular therapy series: #4 - cytomegalovirus treatment and management of resistant or refractory infections after hematopoietic cell transplantation. Transplant Cell Ther 27:957–967. doi: 10.1016/j.jtct.2021.09.010 [DOI] [PubMed] [Google Scholar]
- 37. Kotton CN, Kumar D, Caliendo AM, Huprikar S, Chou S, Danziger-Isakov L, Humar A, The Transplantation Society International CMV Consensus Group . 2018. The third International consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. transplantation. Transplantation 102:900–931. doi: 10.1097/TP.0000000000002191 [DOI] [PubMed] [Google Scholar]
- 38. Chayama K, Hayes CN. 2015. HCV drug resistance challenges in Japan: the role of pre-existing variants and emerging resistant strains in direct acting antiviral therapy. Viruses 7:5328–5342. doi: 10.3390/v7102876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Song Z-L, Cui Y-J, Zheng W-P, Teng D-H, Zheng H. 2012. Diagnostic and therapeutic progress of multi-drug resistance with anti-HBV nucleos(t)ide analogues. World J Gastroenterol 18:7149–7157. doi: 10.3748/wjg.v18.i48.7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu H, Geng Y, He Q, Li M. 2018. miRNAs regulate immune response and signaling during hepatitis C virus infection. Eur J Med Res 23:19. doi: 10.1186/s40001-018-0317-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cokarić Brdovčak M, Zubković A, Jurak I. 2018. Herpes simplex virus 1 deregulation of host microRNAs. Noncoding RNA 4:36. doi: 10.3390/ncrna4040036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Riad SE, Elhelw DS, Shawer H, El-Ekiaby N, Salah A, Zekri A, Esmat G, Amleh A, Abdelaziz AI. 2018. Disruption of claudin-1 expression by miRNA-182 alters the susceptibility to viral infectivity in HCV cell models. Front Genet 9:93. doi: 10.3389/fgene.2018.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Elhelw DS, Riad SE, Shawer H, El-Ekiaby N, Salah A, Zekri A, Amleh A, Esmat G, Abdelaziz AI. 2017. Ectopic delivery of miR-200C diminishes hepatitis C virus infectivity through transcriptional and translational repression of occludin. Arch Virol 162:3283–3291. doi: 10.1007/s00705-017-3449-3 [DOI] [PubMed] [Google Scholar]
- 44. Kawano Y, Kawada J, Kamiya Y, Suzuki M, Torii Y, Kimura H, Ito Y. 2016. Analysis of circulating human and viral microRNAs in patients with congenital cytomegalovirus infection. J Perinatol 36:1101–1105. doi: 10.1038/jp.2016.157 [DOI] [PubMed] [Google Scholar]
- 45. Gohir W, Klement W, Singer LG, Palmer SM, Mazzulli T, Keshavjee S, Husain S. 2020. Identifying host microRNAs in bronchoalveolar lavage samples from lung transplant recipients infected with Aspergillus. J Heart Lung Transplant 39:1228–1237. doi: 10.1016/j.healun.2020.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Kerckhove M, Tanaka K, Umehara T, Okamoto M, Kanematsu S, Hayashi H, Yano H, Nishiura S, Tooyama S, Matsubayashi Y, Komatsu T, Park S, Okada Y, Takahashi R, Kawano Y, Hanawa T, Iwasaki K, Nozaki T, Torigoe H, Ikematsu K, Suzuki Y, Tanaka K, Martin P, Shimokawa I, Mori R. 2018. Targeting miR-223 in neutrophils enhances the clearance of Staphylococcus aureus in infected wounds. EMBO Mol Med 10. doi: 10.15252/emmm.201809024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tong J, Duan Z, Zeng R, Du L, Xu S, Wang L, Liu Y, Chen Q, Chen X, Li M. 2021. MiR-146A negatively regulates Aspergillus fumigatus-induced TNF-Α and IL-6 secretion in THP-1 macrophages. Mycopathologia 186:341–354. doi: 10.1007/s11046-021-00538-0 [DOI] [PubMed] [Google Scholar]
- 48. Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, Liu Y, Wu J, Rice CM. 2013. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog 9:e1003248. doi: 10.1371/journal.ppat.1003248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fu L, Fu X, Mo J, Li X, Li R, Peng S. 2019. miR-146A-5P enhances hepatitis B virus replication through autophagy to promote aggravation of chronic hepatitis B. IUBMB Life 71:1336–1346. doi: 10.1002/iub.2044 [DOI] [PubMed] [Google Scholar]
- 50. Xie Y, He S, Wang J. 2018. MicroRNA-373 facilitates HSV-1 replication through suppression of type I IFN response by targeting IRF1. Biomed Pharmacother 97:1409–1416. doi: 10.1016/j.biopha.2017.11.071 [DOI] [PubMed] [Google Scholar]
- 51. Li GY, Zhou Y, Ying RS, Shi L, Cheng YQ, Ren JP, Griffin JWD, Jia ZS, Li CF, Moorman JP, Yao ZQ. 2015. Hepatitis C virus-induced reduction in miR-181A impairs Cd4(+) T-cell responses through overexpression of DUSP6. Hepatology 61:1163–1173. doi: 10.1002/hep.27634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y, Dai J, Tang J, Zhou L, Zhou M. 2017. MicroRNA-649 promotes HSV-1 replication by directly targeting MALT1. J Med Virol 89:1069–1079. doi: 10.1002/jmv.24728 [DOI] [PubMed] [Google Scholar]
- 53. Taganov KD, Boldin MP, Chang K-J, Baltimore D. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 103:12481–12486. doi: 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Andrei G, Trompet E, Snoeck R. 2019. Novel Therapeutics for Epstein⁻Barr virus −Novel Therapeutics for Epstein⁻Barr virus. Molecules 24:997. doi: 10.3390/molecules24050997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. AlDabbagh MA, Gitman MR, Kumar D, Humar A, Rotstein C, Husain S. 2017. The role of antiviral prophylaxis for the prevention of Epstein-Barr virus-associated posttransplant lymphoproliferative disease in solid organ transplant recipients: a systematic review. Am J Transplant 17:770–781. doi: 10.1111/ajt.14020 [DOI] [PubMed] [Google Scholar]
- 56. Janssen HLA, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. 2013. Treatment of HCV infection by targeting microRNA. N Engl J Med 368:1685–1694. doi: 10.1056/NEJMoa1209026 [DOI] [PubMed] [Google Scholar]
- 57. Amador-Cañizares Y, Panigrahi M, Huys A, Kunden RD, Adams HM, Schinold MJ, Wilson JA. 2018. MiR-122, small RNA annealing and sequence mutations alter the predicted structure of the hepatitis C virus 5' UTR RNA to stabilize and promote viral RNA accumulation. Nucleic Acids Res 46:9776–9792. doi: 10.1093/nar/gky662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Potenza N, Papa U, Mosca N, Zerbini F, Nobile V, Russo A. 2011. Human microRNA hsa-miR-125A-5P interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res 39:5157–5163. doi: 10.1093/nar/gkr067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cox EM, Sagan SM, Mortimer SAW, Doudna JA, Sarnow P. 2013. Enhancement of hepatitis C viral RNA abundance by precursor miR-122 molecules. RNA 19:1825–1832. doi: 10.1261/rna.040865.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sendi H, Mehrab-Mohseni M, Foureau DM, Ghosh S, Walling TL, Steuerwald N, Zamor PJ, Kaplan KJ, Jacobs C, Ahrens WA, Russo MW, Clemens MG, Schrum LW, Bonkovsky HL. 2015. MiR-122 decreases HCV entry into hepatocytes through binding to the 3' UTR of OCLN mRNA. Liver Int 35:1315–1323. doi: 10.1111/liv.12698 [DOI] [PubMed] [Google Scholar]
- 61. Butt AM, Raja AJ, Siddique S, Khan JS, Shahid M, Tayyab G-U-N, Minhas Z, Umar M, Idrees M, Tong Y. 2016. Parallel expression profiling of hepatic and serum microRNA-122 associated with clinical features and treatment responses in chronic hepatitis C patients. Sci Rep 6:21510. doi: 10.1038/srep21510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jiang M, Broering R, Trippler M, Wu J, Zhang E, Zhang X, Gerken G, Lu M, Schlaak JF. 2014. MicroRNA-155 controls toll-like receptor 3- and hepatitis C virus-induced immune responses in the liver. J Viral Hepat 21:99–110. doi: 10.1111/jvh.12126 [DOI] [PubMed] [Google Scholar]
- 63. Cheng YQ, Ren JP, Zhao J, Wang JM, Zhou Y, Li GY, Moorman JP, Yao ZQ. 2015. MicroRNA-155 regulates interferon-Γ production in natural killer cells via Tim-3 signalling in chronic hepatitis C virus infection. Immunology 145:485–497. doi: 10.1111/imm.12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liao T-L, Hsieh S-L, Chen Y-M, Chen H-H, Liu H-J, Lee H-C, Chen D-Y. 2018. Rituximab may cause increased hepatitis C virus viremia in rheumatoid arthritis patients through declining exosomal microRNA-155. Arthritis Rheumatol 70:1209–1219. doi: 10.1002/art.40495 [DOI] [PubMed] [Google Scholar]
- 65. Kondo Y, Kogure T, Ninomiya M, Fukuda R, Monma N, Ikeo K, Tanaka Y. 2019. The reduction of miR146B-5P in monocytes and T cells could contribute to the immunopathogenesis of hepatitis C virus infection. Sci Rep 9:13393. doi: 10.1038/s41598-019-49706-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sidorkiewicz M, Grek M, Jozwiak B, Krol A, Piekarska A. 2017. The impact of chronic hepatitis C infection on cholesterol metabolism in Pbmcs is associated with microRNA-146A expression. Eur J Clin Microbiol Infect Dis 36:697–702. doi: 10.1007/s10096-016-2851-1 [DOI] [PubMed] [Google Scholar]
- 67. Bhanja Chowdhury J, Shrivastava S, Steele R, Di Bisceglie AM, Ray R, Ray RB. 2012. Hepatitis C virus infection modulates expression of interferon stimulatory gene Ifitm1 by upregulating miR-130A. J Virol 86:10221–10225. doi: 10.1128/JVI.00882-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Duan X, Liu X, Li W, Holmes JA, Kruger AJ, Yang C, Li Y, Xu M, Ye H, Li S, Liao X, Sheng Q, Chen D, Shao T, Cheng Z, Kaj B, Schaefer EA, Li S, Chen L, Lin W, Chung RT. 2019. MicroRNA-130A downregulates HCV replication through an Atg5-dependent autophagy pathway. Cells 8:338. doi: 10.3390/cells8040338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shirasaki T, Honda M, Shimakami T, Horii R, Yamashita T, Sakai Y, Sakai A, Okada H, Watanabe R, Murakami S, Yi M, Lemon SM, Kaneko S. 2013. MicroRNA-27A regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J Virol 87:5270–5286. doi: 10.1128/JVI.03022-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tian H, He Z. 2018. MiR-215 enhances HCV replication by targeting trim22 and inactivating NF-ΚB signaling. Yonsei Med J 59:511–518. doi: 10.3349/ymj.2018.59.4.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gong W, Guo X, Zhang Y. 2018. Depletion of microrna-373 represses the replication of hepatitis C virus via activation of type 1 interferon response by targeting Irf5. Yonsei Med J 59:1181–1189. doi: 10.3349/ymj.2018.59.10.1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hou W, Tian Q, Zheng J, Bonkovsky HL. 2010. MicroRNA-196 represses bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatology 51:1494–1504. doi: 10.1002/hep.23401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ji F, Yang B, Peng X, Ding H, You H, Tien P. 2011. Circulating microRNAs in hepatitis B virus-infected patients. J Viral Hepat 18:e242–51. doi: 10.1111/j.1365-2893.2011.01443.x [DOI] [PubMed] [Google Scholar]
- 74. Chen Y, Shen A, Rider PJ, Yu Y, Wu K, Mu Y, Hao Q, Liu Y, Gong H, Zhu Y, Liu F, Wu J. 2011. A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. FASEB J 25:4511–4521. doi: 10.1096/fj.11-187781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Deng W, Zhang X, Ma Z, Lin Y, Lu M. 2017. MicroRNA-125B-5P mediates post-transcriptional regulation of hepatitis B virus replication via the Lin28B/Let-7 axis. RNA Biol 14:1389–1398. doi: 10.1080/15476286.2017.1293770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Coppola N, Potenza N, Pisaturo M, Mosca N, Tonziello G, Signoriello G, Messina V, Sagnelli C, Russo A, Sagnelli E. 2013. Liver microRNA hsa-miR-125A-5P in HBV chronic infection: correlation with HBV replication and disease progression. PLoS One 8:e65336. doi: 10.1371/journal.pone.0065336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jung YJ, Kim J-W, Park SJ, Min BY, Jang ES, Kim NY, Jeong S-H, Shin CM, Lee SH, Park YS, Hwang J-H, Kim N, Lee DH. 2013. C-Myc-mediated overexpression of miR-17-92 suppresses replication of hepatitis B virus in human hepatoma cells. J Med Virol 85:969–978. doi: 10.1002/jmv.23534 [DOI] [PubMed] [Google Scholar]
- 78. Wang Y, Zhu P, Qiu J, Wang J, Zhu H, Zhu Y, Zhang L, Zhu J, Liu X, Dong C. 2017. Identification and characterization of interferon signaling-related microRNAs in occult hepatitis B virus infection. Clin Epigenetics 9:101. doi: 10.1186/s13148-017-0404-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang Y, Liu Y, Xue J, Yang Z, Shi Y, Shi Y, Lou G, Wu S, Qi J, Liu W, Wang J, Chen Z. 2017. MicroRNA-141 targets sirt1 and inhibits autophagy to reduce HBV replication. Cell Physiol Biochem 41:310–322. doi: 10.1159/000456162 [DOI] [PubMed] [Google Scholar]
- 80. Fan H-X, Feng Y-J, Zhao X-P, He Y-Z, Tang H. 2018. MiR-185-5p suppresses HBV gene expression by targeting Elk1 in hepatoma carcinoma cells. Life Sci 213:9–17. doi: 10.1016/j.lfs.2018.10.016 [DOI] [PubMed] [Google Scholar]
- 81. Yu F, Yang J, Ouyang J, Zheng Y, Chen B, Li G, Lu Z, Dong P, Zheng J. 2015. Serum microRNA-210 levels in different groups of chronic hepatitis B patients. Clin Chim Acta 450:203–209. doi: 10.1016/j.cca.2015.08.022 [DOI] [PubMed] [Google Scholar]
- 82. Li Y, Xie J, Xu X, Wang J, Ao F, Wan Y, Zhu Y. 2013. MicroRNA-548 down-regulates host antiviral response via direct targeting of IFN-Λ1. Protein Cell 4:130–141. doi: 10.1007/s13238-012-2081-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Xing T, Zhu J, Xian J, Li A, Wang X, Wang W, Zhang Q. 2019. miRNA-548Ah promotes the replication and expression of hepatitis B virus by targeting histone deacetylase 4. Life Sci 219:199–208. doi: 10.1016/j.lfs.2018.12.057 [DOI] [PubMed] [Google Scholar]
- 84. Wang Y, Cao J, Zhang S, Sun L, Nan Y, Yao H, Fan J, Zhu LY, Yu L. 2019. MicroRNA-802 induces hepatitis B virus replication and replication through regulating SMARCE1 expression in hepatocellular carcinoma. Cell Death Dis 10:783. doi: 10.1038/s41419-019-1999-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kohno T, Tsuge M, Murakami E, Hiraga N, Abe H, Miki D, Imamura M, Ochi H, Hayes CN, Chayama K. 2014. Human microRNA hsa-miR-1231 suppresses hepatitis B virus replication by targeting core mRNA. J Viral Hepat 21:e89–97. doi: 10.1111/jvh.12240 [DOI] [PubMed] [Google Scholar]
- 86. Guo H, Liu H, Mitchelson K, Rao H, Luo M, Xie L, Sun Y, Zhang L, Lu Y, Liu R, Ren A, Liu S, Zhou S, Zhu J, Zhou Y, Huang A, Wei L, Guo Y, Cheng J. 2011. MicroRNAs-372/373 promote the expression of hepatitis B virus through the targeting of nuclear factor I/B. Hepatology 54:808–819. doi: 10.1002/hep.24441 [DOI] [PubMed] [Google Scholar]
- 87. Zheng S, Li Y, Zhang Y, Li X, Tang H. 2011. MiR-101 regulates HSV-1 replication by targeting Atp5B. Antiviral Res 89:219–226. doi: 10.1016/j.antiviral.2011.01.008 [DOI] [PubMed] [Google Scholar]
- 88. Ru J, Sun H, Fan H, Wang C, Li Y, Liu M, Tang H. 2014. MiR-23A facilitates the replication of HSV-1 through the suppression of interferon regulatory factor 1. PLoS One 9:e114021. doi: 10.1371/journal.pone.0114021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pan D, Flores O, Umbach JL, Pesola JM, Bentley P, Rosato PC, Leib DA, Cullen BR, Coen DM. 2014. A neuron-specific host microRNA targets herpes simplex virus-1 Icp0 expression and promotes latency. Cell Host Microbe 15:446–456. doi: 10.1016/j.chom.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Duan F, Liao J, Huang Q, Nie Y, Wu K. 2012. HSV-1 miR-H6 inhibits HSV-1 replication and IL-6 expression in human corneal epithelial cells in vitro. Clin Dev Immunol 2012:192791. doi: 10.1155/2012/192791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yin Q, Wang X, Fewell C, Cameron J, Zhu H, Baddoo M, Lin Z, Flemington EK. 2010. MicroRNA miR-155 inhibits bone morphogenetic protein (BMP) signaling and BMP-mediated Epstein-Barr virus reactivation. J Virol 84:6318–6327. doi: 10.1128/JVI.00635-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lin Z, Wang X, Fewell C, Cameron J, Yin Q, Flemington EK. 2010. Differential expression of the miR-200 family microRNAs in epithelial and B cells and regulation of Epstein-Barr virus reactivation by the miR-200 family member miR-429. J Virol 84:7892–7897. doi: 10.1128/JVI.00379-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tagawa T, Albanese M, Bouvet M, Moosmann A, Mautner J, Heissmeyer V, Zielinski C, Lutter D, Hoser J, Hastreiter M, Hayes M, Sugden B, Hammerschmidt W. 2016. Epstein-Barr viral miRNAs inhibit antiviral Cd4+ T cell responses targeting IL-12 and peptide processing. J Exp Med 213:2065–2080. doi: 10.1084/jem.20160248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Albanese M, Tagawa T, Bouvet M, Maliqi L, Lutter D, Hoser J, Hastreiter M, Hayes M, Sugden B, Martin L, Moosmann A, Hammerschmidt W. 2016. Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific Cd8+ T cells. Proc Natl Acad Sci U S A 113:E6467–E6475. doi: 10.1073/pnas.1605884113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey A-A, Pich D, McInnes IB, Hammerschmidt W, O’Neill LAJ, Masters SL. 2012. Cutting edge: mir-223 and EBV miR-Bart15 regulate the Nlrp3 inflammasome and IL-1Β production. J Immunol 189:3795–3799. doi: 10.4049/jimmunol.1200312 [DOI] [PubMed] [Google Scholar]
- 96. Hooykaas MJG, van Gent M, Soppe JA, Kruse E, Boer IGJ, van Leenen D, Groot Koerkamp MJA, Holstege FCP, Ressing ME, Wiertz E, Lebbink RJ. 2017. EBV microRNA Bart16 suppresses type I IFN signaling. J Immunol 198:4062–4073. doi: 10.4049/jimmunol.1501605 [DOI] [PubMed] [Google Scholar]
- 97. Lu Y, Qin Z, Wang J, Zheng X, Lu J, Zhang X, Wei L, Peng Q, Zheng Y, Ou C, Ye Q, Xiong W, Li G, Fu Y, Yan Q, Ma J. 2017. Epstein-barr virus miR-Bart6-3p inhibits the RIG-I pathway. J Innate Immun 9:574–586. doi: 10.1159/000479749 [DOI] [PubMed] [Google Scholar]
- 98. Qiu J, Thorley-Lawson DA. 2014. EBV microRNA BART 18-5p targets Map3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proc Natl Acad Sci U S A 111:11157–11162. doi: 10.1073/pnas.1406136111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5:376–385. doi: 10.1016/j.chom.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 100. Skinner CM, Ivanov NS, Barr SA, Chen Y, Skalsky RL. 2017. An epstein-barr virus microrna blocks interleukin-1 (IL-1) signaling by targeting IL-1 receptor 1. J Virol 91:e00530-17. doi: 10.1128/JVI.00530-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chen Y, Fachko D, Ivanov NS, Skinner CM, Skalsky RL, Lin Z. 2019. Epstein-Barr virus microRNAs regulate B cell receptor signal transduction and lytic reactivation. PLoS Pathog 15:e1007535. doi: 10.1371/journal.ppat.1007535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bergallo M, Gambarino S, Montanari P, Daprà V, Rassu M, Galliano I, Ravanini P. 2017. Mir-155 expression is downregulated in kidney transplant patients with human cytomegalovirus infection. Transpl Immunol 43–44:60–63. doi: 10.1016/j.trim.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 103. O’Connor CM, Vanicek J, Murphy EA. 2014. Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J Virol 88:5524–5532. doi: 10.1128/JVI.00481-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE. 2008. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol 82:9065–9074. doi: 10.1128/JVI.00961-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yan B, Ma H, Jiang S, Shi J, Yang Z, Zhu W, Kong C, Chen L, Yan H, Ma C. 2019. microRNA-221 restricts human cytomegalovirus replication via promoting type I IFN production by targeting Socs1/NF-kappaB pathway. Cell Cycle 18:3072–3084. doi: 10.1080/15384101.2019.1667706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Huang Y, Qi Y, Ma Y, He R, Ji Y, Sun Z, Ruan Q. 2013. The expression of interleukin-32 is activated by human cytomegalovirus infection and down regulated by Hcmv-miR-Ul112-1. Virol J 10:51. doi: 10.1186/1743-422X-10-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O. 2007. Host immune system gene targeting by a viral miRNA. Science 317:376–381. doi: 10.1126/science.1140956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hook LM, Grey F, Grabski R, Tirabassi R, Doyle T, Hancock M, Landais I, Jeng S, McWeeney S, Britt W, Nelson JA. 2014. Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 15:363–373. doi: 10.1016/j.chom.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kim S, Seo D, Kim D, Hong Y, Chang H, Baek D, Kim VN, Lee S, Ahn K. 2015. Temporal landscape of microRNA-mediated host-virus crosstalk during productive human cytomegalovirus infection. Cell Host Microbe 17:838–851. doi: 10.1016/j.chom.2015.05.014 [DOI] [PubMed] [Google Scholar]
- 110. Hancock MH, Hook LM, Mitchell J, Nelson JA. 2017. Human cytomegalovirus MicroRNAs miR-Us5-1 and miR-Ul112-3p block proinflammatory cytokine production in response to NF-ΚB-activating factors through direct downregulation of IKKα and IKKβ. mBio 8:e00109-17. doi: 10.1128/mBio.00109-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Landais I, Pelton C, Streblow D, DeFilippis V, McWeeney S, Nelson JA. 2015. Human cytomegalovirus miR-Ul112-3p targets Tlr2 and modulates the Tlr2/Irak1/Nfkappab signaling pathway. PLoS Pathog 11:e1004881. doi: 10.1371/journal.ppat.1004881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Romania P, Cifaldi L, Pignoloni B, Starc N, D’Alicandro V, Melaiu O, Li Pira G, Giorda E, Carrozzo R, Bergvall M, Bergström T, Alfredsson L, Olsson T, Kockum I, Seppälä I, Lehtimäki T, Hurme MA, Hengel H, Santoni A, Cerboni C, Locatelli F, D’Amato M, Fruci D. 2017. Identification of a genetic variation in Erap1 aminopeptidase that prevents human cytomegalovirus miR-Ul112-5p-mediated immunoevasion. Cell Rep 20:846–853. doi: 10.1016/j.celrep.2017.06.084 [DOI] [PubMed] [Google Scholar]
- 113. Lau B, Poole E, Krishna B, Montanuy I, Wills MR, Murphy E, Sinclair J. 2016. Corrigendum: the expression of human cytomegalovirus MicroRNA Mir-Ul148D during latent infection in primary myeloid cells inhibits activin a-triggered secretion of IL-6. Sci Rep 6:33771. doi: 10.1038/srep33771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Pan C, Zhu D, Wang Y, Li L, Li D, Liu F, Zhang C-Y, Zen K, Flemington EK. 2016. Human cytomegalovirus miR-Ul148D facilitates latent viral infection by targeting host cell immediate early response gene 5. PLoS Pathog 12:e1006007. doi: 10.1371/journal.ppat.1006007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kim Y, Lee S, Kim S, Kim D, Ahn J-H, Ahn K, Gao S-J. 2012. Human cytomegalovirus clinical strain-specific microRNA miR-Ul148D targets the human chemokine RANTES during infection. PLoS Pathog 8:e1002577. doi: 10.1371/journal.ppat.1002577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Chen J, Xia S, Yang X, Chen H, Li F, Liu F, Chen Z. 2017. Human cytomegalovirus encoded miR-Us25-1-5p attenuates Cd147/EMMPRIN-mediated early antiviral response. Viruses 9:365. doi: 10.3390/v9120365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Qi M, Qi Y, Ma Y, He R, Ji Y, Sun Z, Ruan Q. 2013. Over-expression of human cytomegalovirus miR-Us25-2-3p downregulates Eif4A1 and inhibits HCMV replication. FEBS Lett 587:2266–2271. doi: 10.1016/j.febslet.2013.05.057 [DOI] [PubMed] [Google Scholar]
- 118. Lisboa LF, Egli A, O’Shea D, Åsberg A, Hartmann A, Rollag H, Pang XL, Tyrrell DL, Kumar D, Humar A. 2015. Hcmv-miR-Ul22A-5P: a biomarker in transplantation with broad impact on host gene expression and potential immunological implications. Am J Transplant 15:1893–1902. doi: 10.1111/ajt.13222 [DOI] [PubMed] [Google Scholar]
- 119. Gao Z, Zhou L, Bai J, Ding M, Liu D, Zheng S, Li Y, Li X, Wang X, Jin M, Shangting H, Qiu C, Wang C, Zhang X, Zhang C, Chen X. 2021. Assessment of HCMV-encoded microRNAs in plasma as potential biomarkers in pregnant women with adverse pregnancy outcomes. Ann Transl Med 9:638–638. doi: 10.21037/atm-20-7354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. McCaskill JL, Ressel S, Alber A, Redford J, Power UF, Schwarze J, Dutia BM, Buck AH. 2017. Broad-spectrum inhibition of respiratory virus infection by microrna mimics targeting P38 MAPK signaling. Mol Ther Nucleic Acids 7:256–266. doi: 10.1016/j.omtn.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Othumpangat S, Walton C, Piedimonte G. 2012. MicroRNA-221 modulates RSV replication in human bronchial epithelium by targeting NGF expression. PLoS One 7:e30030. doi: 10.1371/journal.pone.0030030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Du X, Yang Y, Xiao G, Yang M, Yuan L, Qin L, He R, Wang L, Wu M, Wu S, Feng J, Xiang Y, Qu X, Liu H, Qin X, Liu C. 2020. Respiratory syncytial virus infection-induced mucus secretion by down-regulation of miR-34B/C-5P expression in airway epithelial cells. J Cell Mol Med 24:12694–12705. doi: 10.1111/jcmm.15845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wang S, Ling Y, Yao Y, Zheng G, Chen W. 2020. Luteolin inhibits respiratory syncytial virus replication by regulating the Mir-155/Socs1/Stat1 signaling pathway. Virol J 17:187. doi: 10.1186/s12985-020-01451-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Inchley CS, Sonerud T, Fjærli HO, Nakstad B. 2015. Nasal mucosal microRNA expression in children with respiratory syncytial virus infection. BMC Infect Dis 15:150. doi: 10.1186/s12879-015-0878-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zeng G, Wang Z, Huang Y, Abedin Z, Liu Y, Randhawa P. 2019. Cellular and viral miRNA expression in polyomavirus BK infection. Transpl Infect Dis 21:e13159. doi: 10.1111/tid.13159 [DOI] [PubMed] [Google Scholar]
- 126. Demey B, Descamps V, Presne C, Helle F, Francois C, Duverlie G, Castelain S, Brochot E. 2021. BK polyomavirus micro-Rnas: time course and clinical relevance in kidney transplant recipients. Viruses 13:351. doi: 10.3390/v13020351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pietilä T, Nummi M, Auvinen P, Mannonen L, Auvinen E. 2015. Expression of BKV and JCV encoded microRNA in human cerebrospinal fluid, plasma and urine. J Clin Virol 65:1–5. doi: 10.1016/j.jcv.2015.01.019 [DOI] [PubMed] [Google Scholar]
- 128. Li JYZ, McNicholas K, Yong TY, Rao N, Coates PTH, Higgins GD, Carroll RP, Woodman RJ, Michael MZ, Gleadle JM. 2014. BK virus encoded microRNAs are present in blood of renal transplant recipients with BK viral nephropathy. Am J Transplant 14:1183–1190. doi: 10.1111/ajt.12694 [DOI] [PubMed] [Google Scholar]
- 129. Kim MH, Lee YH, Seo J-W, Moon H, Kim JS, Kim YG, Jeong K-H, Moon J-Y, Lee TW, Ihm C-G, Kim C-D, Park JB, Chung BH, Kim Y-H, Lee S-H. 2017. Urinary exosomal viral microRNA as a marker of BK virus nephropathy in kidney transplant recipients. PLoS One 12:e0190068. doi: 10.1371/journal.pone.0190068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Wrensch F, Ligat G, Heydmann L, Schuster C, Zeisel MB, Pessaux P, Habersetzer F, King BJ, Tarr AW, Ball JK, Winkler M, Pöhlmann S, Keck Z-Y, Foung SKH, Baumert TF. 2019. Interferon-induced transmembrane proteins mediate viral evasion in acute and chronic hepatitis C virus infection. Hepatology 70:1506–1520. doi: 10.1002/hep.30699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581. doi: 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- 132. Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. 2012. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci U S A 109:941–946. doi: 10.1073/pnas.1112263109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Machlin ES, Sarnow P, Sagan SM. 2011. Masking the 5' terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A 108:3193–3198. doi: 10.1073/pnas.1012464108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Pan H, Niu DD, Feng H, Ng LFP, Ren EC, Chen WN. 2007. Cellular transcription modulator Smarce1 binds to HBV core promoter containing naturally occurring deletions and represses viral replication. Biochim Biophys Acta 1772:1075–1084. doi: 10.1016/j.bbadis.2007.06.005 [DOI] [PubMed] [Google Scholar]
- 135. Xiangji L, Feng X, Qingbao C, Weifeng T, Xiaoqing J, Baihe Z, Feng S, Hongyang W, Mengchao W. 2011. Knockdown of HBV surface antigen gene expression by a lentiviral microRNA-based system inhibits HBV replication and HCC growth. J Viral Hepat 18:653–660. doi: 10.1111/j.1365-2893.2010.01346.x [DOI] [PubMed] [Google Scholar]
- 136. Ishida H, Tatsumi T, Hosui A, Nawa T, Kodama T, Shimizu S, Hikita H, Hiramatsu N, Kanto T, Hayashi N, Takehara T. 2011. Alterations in microRNA expression profile in HCV-infected hepatoma cells: involvement of miR-491 in regulation of HCV replication via the Pi3 kinase/AKT pathway. Biochem Biophys Res Commun 412:92–97. doi: 10.1016/j.bbrc.2011.07.049 [DOI] [PubMed] [Google Scholar]
- 137. Li S, Duan X, Li Y, Liu B, McGilvray I, Chen L. 2014. MicroRNA-130A inhibits HCV replication by restoring the innate immune response. J Viral Hepat 21:121–128. doi: 10.1111/jvh.12131 [DOI] [PubMed] [Google Scholar]
- 138. Sakurai F, Hashimoto R, Inoue C, Wakabayashi K, Tsukamoto T, Imaizumi T, Andres TGM, Sakai E, Itsuki K, Sakamoto N, Wakita T, Mizuguchi H. 2019. miR-27B-mediated suppression of aquaporin-11 expression in hepatocytes reduces HCV genomic RNA levels but not viral titers. Virol J 16:58. doi: 10.1186/s12985-019-1160-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327:198–201. doi: 10.1126/science.1178178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Forte E, Luftig MA. 2011. The role of microRNAs in Epstein-Barr virus latency and lytic reactivation. Microbes Infect 13:1156–1167. doi: 10.1016/j.micinf.2011.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Iizasa H, Kim H, Kartika AV, Kanehiro Y, Yoshiyama H. 2020. Role of viral and host microRNAs in immune regulation of Epstein-Barr virus-associated diseases. Front Immunol 11:498. doi: 10.3389/fimmu.2020.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Park S-M, Gaur AB, Lengyel E, Peter ME. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22:894–907. doi: 10.1101/gad.1640608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Feng W, Kraus RJ, Dickerson SJ, Lim HJ, Jones RJ, Yu X, Mertz JE, Kenney SC. 2007. ZEB1 and c-jun levels contribute to the establishment of highly lytic Epstein-Barr virus infection in gastric AGS cells. J Virol 81:10113–10122. doi: 10.1128/JVI.00692-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yu X, Wang Z, Mertz JE. 2007. ZEB1 regulates the latent-lytic switch in infection by Epstein-Barr virus. PLoS Pathog 3:e194. doi: 10.1371/journal.ppat.0030194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. 2006. Human cytomegalovirus infection alters the substrate specificities and rapamycin sensitivities of raptor- and rictor-containing complexes. Proc Natl Acad Sci U S A 103:14182–14187. doi: 10.1073/pnas.0605825103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Godshalk SE, Bhaduri-McIntosh S, Slack FJ. 2008. Epstein-Barr virus-mediated dysregulation of human microRNA expression. Cell Cycle 7:3595–3600. doi: 10.4161/cc.7.22.7120 [DOI] [PubMed] [Google Scholar]
- 147. Sadegh Ehdaei B, Pirouzmand A, Shabani M, Mirzaei A, Moghim S. 2021. Cellular miR-101-1 reduces efficiently the replication of HSV-1 in HeLa cells. Intervirology 64:88–95. doi: 10.1159/000512956 [DOI] [PubMed] [Google Scholar]
- 148. Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. 2010. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J Virol 84:11670–11678. doi: 10.1128/JVI.01248-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Mulik S, Xu J, Reddy PBJ, Rajasagi NK, Gimenez F, Sharma S, Lu PY, Rouse BT. 2012. Role of miR-132 in angiogenesis after ocular infection with herpes simplex virus. Am J Pathol 181:525–534. doi: 10.1016/j.ajpath.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Bhela S, Mulik S, Reddy PBJ, Richardson RL, Gimenez F, Rajasagi NK, Veiga-Parga T, Osmand AP, Rouse BT. 2014. Critical role of microRNA-155 in herpes simplex encephalitis. J Immunol 192:2734–2743. doi: 10.4049/jimmunol.1302326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Majer A, Caligiuri KA, Gale KK, Niu Y, Phillipson CS, Booth TF, Booth SA. 2017. Correction: induction of multiple miR-200/182 members in the brains of mice are associated with acute herpes simplex virus 1 encephalitis. PLoS One 12:e0172815. doi: 10.1371/journal.pone.0172815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Kuang L, Deng Y, Liu X, Zou Z, Mi L. 2017. Differential expression of mRNA and miRNA in guinea pigs following infection with Hsv2V. Exp Ther Med 14:2577–2583. doi: 10.3892/etm.2017.4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Santhakumar D, Forster T, Laqtom NN, Fragkoudis R, Dickinson P, Abreu-Goodger C, Manakov SA, Choudhury NR, Griffiths SJ, Vermeulen A, Enright AJ, Dutia B, Kohl A, Ghazal P, Buck AH. 2010. Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc Natl Acad Sci U S A 107:13830–13835. doi: 10.1073/pnas.1008861107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Dhuruvasan K, Sivasubramanian G, Pellett PE. 2011. Roles of host and viral microRNAs in human cytomegalovirus biology. Virus Res 157:180–192. doi: 10.1016/j.virusres.2010.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Cullen BR. 2009. Viral and cellular messenger RNA targets of viral microRNAs. Nature 457:421–425. doi: 10.1038/nature07757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Albanese M, Tagawa T, Buschle A, Hammerschmidt W. 2017. microRNAs of Epstein-Barr virus control innate and adaptive antiviral immunity. J Virol 91:e01667-16. doi: 10.1128/JVI.01667-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Abdalla AE, Mahjoob MO, Abosalif KOA, Ejaz H, Alameen AAM, Elsaman T. 2020. Human cytomegalovirus-encoded microRNAs: a master regulator of latent infection. Infect Genet Evol 78:104119. doi: 10.1016/j.meegid.2019.104119 [DOI] [PubMed] [Google Scholar]
- 158. Cohen JI. 2000. Epstein-Barr virus infection. N Engl J Med 343:481–492. doi: 10.1056/NEJM200008173430707 [DOI] [PubMed] [Google Scholar]
- 159. Drachenberg CB, Papadimitriou JC, Ramos E. 2006. Histologic versus molecular diagnosis of BK polyomavirus-associated nephropathy: a shifting paradigm? Clin J Am Soc Nephrol 1:374–379. doi: 10.2215/CJN.02021205 [DOI] [PubMed] [Google Scholar]
- 160. Hoffman NG, Cook L, Atienza EE, Limaye AP, Jerome KR. 2008. Marked variability of BK virus load measurement using quantitative real-time PCR among commonly used assays. J Clin Microbiol 46:2671–2680. doi: 10.1128/JCM.00258-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. 2017. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis 17:e383–e392. doi: 10.1016/S1473-3099(17)30316-X [DOI] [PubMed] [Google Scholar]
- 162. Yamamuro R, Kimura M, Asano-Mori Y, Abe M, Nakamura S, Umeyama T, Yamagoe S, Miyazaki Y, Ogura S, Sakoh T, Mitsuki T, Yamaguchi K, Yuasa M, Kaji D, Kageyama K, Nishida A, Taya Y, Ishiwata K, Takagi S, Yamamoto H, Yamamoto G, Uchida N, Wake A, Taniguchi S, Araoka H. 2022. Clinical and microbiological characteristics of proven invasive Aspergillosis due to rare/cryptic species in allogeneic hematopoietic stem cell transplant recipients. Antimicrob Agents Chemother 66:e0163021. doi: 10.1128/AAC.01630-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Mendoza MA, Anderson A, Morris MI, Lekakis L, Simkins J, Prado CE, Martinez OV, Komanduri KV, Camargo JF. 2020. Successful treatment of invasive fungal infection due to highly resistant Aspergillus calidoustus in an allogeneic hematopoietic cell transplant recipient. Mycopathologia 185:399–403. doi: 10.1007/s11046-019-00423-x [DOI] [PubMed] [Google Scholar]
- 164. Camargo JF, Jabr R, Anderson AD, Lekakis L, Diaz-Paez M, Briski LM, Raja M, Morris MI, Komanduri KV, Pereira D. 2022. Successful treatment of disseminated disease due to highly resistant Aspergillus calidoustus with a novel antifungal therapy. Antimicrob Agents Chemother 66:e0220621. doi: 10.1128/aac.02206-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Munoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE. 2008. European organization for R, treatment of cancer/invasive fungal infections cooperative G, national Institute of A, infectious diseases mycoses study group consensus G. 2008. revised definitions of invasive fungal disease from the European organization for research and treatment of cancer/invasive fungal infections cooperative group and the national institute of allergy and infectious diseases mycoses study group (EORTC/MSG) consensus group. Clin infect dis 46. doi: 10.1086/588660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Donnelly JP, Chen SC, Kauffman CA, Steinbach WJ, Baddley JW, Verweij PE, Clancy CJ, Wingard JR, Lockhart SR, Groll AH, Sorrell TC, Bassetti M, Akan H, Alexander BD, Andes D, Azoulay E, Bialek R, Bradsher RW, Bretagne S, Calandra T, Caliendo AM, Castagnola E, Cruciani M, Cuenca-Estrella M, Decker CF, Desai SR, Fisher B, Harrison T, Heussel CP, Jensen HE, Kibbler CC, Kontoyiannis DP, Kullberg B-J, Lagrou K, Lamoth F, Lehrnbecher T, Loeffler J, Lortholary O, Maertens J, Marchetti O, Marr KA, Masur H, Meis JF, Morrisey CO, Nucci M, Ostrosky-Zeichner L, Pagano L, Patterson TF, Perfect JR, Racil Z, Roilides E, Ruhnke M, Prokop CS, Shoham S, Slavin MA, Stevens DA, Thompson GR, Vazquez JA, Viscoli C, Walsh TJ, Warris A, Wheat LJ, White PL, Zaoutis TE, Pappas PG. 2020. Revision and update of the consensus definitions of invasive fungal disease from the European organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin Infect Dis 71:1367–1376. doi: 10.1093/cid/ciz1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Pylkkänen L, Gullstén H, Majuri M-L, Andersson U, Vanhala E, Määttä J, Meklin T, Hirvonen M-R, Alenius H, Savolainen K. 2004. Exposure to Aspergillus fumigatus spores induces chemokine expression in mouse macrophages. Toxicology 200:255–263. doi: 10.1016/j.tox.2004.03.019 [DOI] [PubMed] [Google Scholar]
- 168. Camargo JF, Bhimji A, Kumar D, Kaul R, Pavan R, Schuh A, Seftel M, Lipton JH, Gupta V, Humar A, Husain S. 2015. Impaired T cell responsiveness to interleukin-6 in hematological patients with invasive aspergillosis. PLoS One 10:e0123171. doi: 10.1371/journal.pone.0123171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Du L, Chen X, Duan Z, Liu C, Zeng R, Chen Q, Li M. 2017. MiR-146A negatively regulates dectin-1-induced inflammatory responses. Oncotarget 8:37355–37366. doi: 10.18632/oncotarget.16958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Arghavan B, Sharifi M, Shafiee M, Mohammadi R. 2016. Evaluation of miR-146A expression level in macrophages exposed to Candida glabrata. Curr Med Mycol 2:16–19. doi: 10.18869/acadpub.cmm.2.2.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Dix A, Czakai K, Leonhardt I, Schäferhoff K, Bonin M, Guthke R, Einsele H, Kurzai O, Löffler J, Linde J. 2017. Specific and novel microRNAs are regulated as response to fungal infection in human dendritic cells. Front Microbiol 8:270. doi: 10.3389/fmicb.2017.00270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Tolnai E, Fidler G, Szász R, Rejtő L, Nwozor KO, Biró S, Paholcsek M. 2020. Free circulating mircoRNAs support the diagnosis of invasive aspergillosis in patients with hematologic malignancies and neutropenia. Sci Rep 10:16532. doi: 10.1038/s41598-020-73556-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Fidler G, Szilágyi-Rácz AA, Dávid P, Tolnai E, Rejtő L, Szász R, Póliska S, Biró S, Paholcsek M. 2022. Circulating microRNA sequencing revealed miRNome patterns in hematology and oncology patients aiding the prognosis of invasive aspergillosis. Sci Rep 12:7144. doi: 10.1038/s41598-022-11239-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Das Gupta M, Fliesser M, Springer J, Breitschopf T, Schlossnagel H, Schmitt A-L, Kurzai O, Hünniger K, Einsele H, Löffler J. 2014. Aspergillus fumigatus induces microRNA-132 in human monocytes and dendritic cells. Int J Med Microbiol 304:592–596. doi: 10.1016/j.ijmm.2014.04.005 [DOI] [PubMed] [Google Scholar]
- 175. Monk CE, Hutvagner G, Arthur JSC. 2010. Regulation of miRNA transcription in macrophages in response to Candida albicans. PLoS One 5:e13669. doi: 10.1371/journal.pone.0013669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Croston TL, Nayak AP, Lemons AR, Goldsmith WT, Gu JK, Germolec DR, Beezhold DH, Green BJ. 2016. Influence of Aspergillus fumigatus conidia viability on murine pulmonary microRNA and mRNA expression following subchronic inhalation exposure. Clin Exp Allergy 46:1315–1327. doi: 10.1111/cea.12783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Ramirez HA, Pastar I, Jozic I, Stojadinovic O, Stone RC, Ojeh N, Gil J, Davis SC, Kirsner RS, Tomic-Canic M. 2018. Staphylococcus aureus triggers induction of miR-15B-5P to diminish DNA repair and deregulate inflammatory response in diabetic foot ulcers. J Invest Dermatol 138:1187–1196. doi: 10.1016/j.jid.2017.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Jin T, Lu Y, He QX, Wang H, Li BF, Zhu LY, Xu QY. 2015. The role of microRNA, miR-24, and its target Chi3L1 in Osteomyelitis caused by Staphylococcus aureus. J Cell Biochem 116:2804–2813. doi: 10.1002/jcb.25225 [DOI] [PubMed] [Google Scholar]
- 179. Ma X, Guo S, Jiang K, Wang X, Yin N, Yang Y, Zahoor A, Deng G. 2019. Mir-128 mediates negative regulation in Staphylococcus aureus induced inflammation by targeting Myd88. Int Immunopharmacol 70:135–146. doi: 10.1016/j.intimp.2018.11.024 [DOI] [PubMed] [Google Scholar]
- 180. Tanaka K, Kim SE, Yano H, Matsumoto G, Ohuchida R, Ishikura Y, Araki M, Araki K, Park S, Komatsu T, Hayashi H, Ikematsu K, Tanaka K, Hirano A, Martin P, Shimokawa I, Mori R. 2017. Mir-142 is required for Staphylococcus aureus clearance at skin wound sites via small Gtpase-mediated regulation of the neutrophil actin cytoskeleton. J Invest Dermatol 137:931–940. doi: 10.1016/j.jid.2016.11.018 [DOI] [PubMed] [Google Scholar]
- 181. Podsiad A, Standiford TJ, Ballinger MN, Eakin R, Park P, Kunkel SL, Moore BB, Bhan U. 2016. MicroRNA-155 regulates host immune response to postviral bacterial pneumonia via IL-23/IL-17 pathway. Am J Physiol Lung Cell Mol Physiol 310:L465–75. doi: 10.1152/ajplung.00224.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Muraleedharan CK, McClellan SA, Barrett RP, Li C, Montenegro D, Carion T, Berger E, Hazlett LD, Xu S. 2016. Inactivation of the miR-183/96/182 cluster decreases the severity of Pseudomonas aeruginosa-induced keratitis. Invest Ophthalmol Vis Sci 57:1506–1517. doi: 10.1167/iovs.16-19134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Muraleedharan CK, McClellan SA, Ekanayaka SA, Francis R, Zmejkoski A, Hazlett LD, Xu S. 2019. The miR-183/96/182 cluster regulates macrophage functions in response to Pseudomonas Aeruginosa. J Innate Immun 11:347–358. doi: 10.1159/000495472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Yang K, Wu M, Li M, Li D, Peng A, Nie X, Sun M, Wang J, Wu Y, Deng Q, Zhu M, Chen K, Yuan J, Huang X. 2014. miR-155 suppresses bacterial clearance in Pseudomonas aeruginosa–induced keratitis by targeting Rheb. J Infect Dis 210:89–98. doi: 10.1093/infdis/jiu002 [DOI] [PubMed] [Google Scholar]
- 185. Li X, He S, Li R, Zhou X, Zhang S, Yu M, Ye Y, Wang Y, Huang C, Wu M. 2016. Pseudomonas aeruginosa infection augments inflammation through miR-301B repression of C-Myb-mediated immune activation and infiltration. Nat Microbiol 1:16132. doi: 10.1038/nmicrobiol.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Huang T, Pu Q, Zhou C, Lin P, Gao P, Zhang X, Chu Y, Yue B, Wu M. 2020. MicroRNA-302/367 cluster impacts host antimicrobial defense via regulation of mitophagic response against Pseudomonas aeruginosa infection. Front Immunol 11:569173. doi: 10.3389/fimmu.2020.569173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Hruz P, Zinkernagel AS, Jenikova G, Botwin GJ, Hugot J-P, Karin M, Nizet V, Eckmann L. 2009. NOD2 contributes to cutaneous defense against Staphylococcus aureus through alpha-toxin-dependent innate immune activation. Proc Natl Acad Sci U S A 106:12873–12878. doi: 10.1073/pnas.0904958106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Lahiri V, Klionsky DJ. 2017. Phb2/Prohibitin 2: an inner membrane mitophagy receptor. Cell Res 27:311–312. doi: 10.1038/cr.2017.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Cho DH, Kim JK, Jo EK. 2020. Mitophagy and innate immunity in infection. Mol Cells 43:10–22. doi: 10.14348/molcells.2020.2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Kirienko NV, Ausubel FM, Ruvkun G. 2015. Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:1821–1826. doi: 10.1073/pnas.1424954112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. WHO . 2021. World Malaria Report 2021. Available from: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2021
- 192. Colley DG, Bustinduy AL, Secor WE, King CH. 2014. Human schistosomiasis. Lancet 383:2253–2264. doi: 10.1016/S0140-6736(13)61949-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu OA, Dondorp AM. 2014. Malaria. The Lancet 383:723–735. doi: 10.1016/S0140-6736(13)60024-0 [DOI] [PubMed] [Google Scholar]
- 194. Paul S, Ruiz-Manriquez LM, Serrano-Cano FI, Estrada-Meza C, Solorio-Diaz KA, Srivastava A. 2020. Human microRNAs in host-parasite interaction: a review. 3 Biotech 10:510. doi: 10.1007/s13205-020-02498-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. van Loon W, Gai PP, Hamann L, Bedu-Addo G, Mockenhaupt FP. 2019. MiRNA-146A polymorphism increases the odds of malaria in pregnancy. Malar J 18:7. doi: 10.1186/s12936-019-2643-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Wah ST, Hananantachai H, Patarapotikul J, Ohashi J, Naka I, Nuchnoi P. 2019. microRNA-27A and microRNA-146A SNP in cerebral malaria. Mol Genet Genomic Med 7:e00529. doi: 10.1002/mgg3.529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Sahu PK, Hoffmann A, Majhi M, Pattnaik R, Patterson C, Mahanta KC, Mohanty AK, Mohanty RR, Joshi S, Mohanty A, Bage J, Maharana S, Seitz A, Bendszus M, Sullivan SA, Turnbull IW, Dondorp AM, Gupta H, Pirpamer L, Mohanty S, Wassmer SC. 2021. Brain magnetic resonance imaging reveals different courses of disease in pediatric and adult cerebral malaria. Clin Infect Dis 73:e2387–e2396. doi: 10.1093/cid/ciaa1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Chakrabarti M, Garg S, Rajagopal A, Pati S, Singh S. 2020. Targeted repression of Plasmodium apicortin by host microRNA impairs malaria parasite growth and invasion. Dis Model Mech 13:dmm042820. doi: 10.1242/dmm.042820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Barker KR, Lu Z, Kim H, Zheng Y, Chen J, Conroy AL, Hawkes M, Cheng HS, Njock M-S, Fish JE, Harlan JM, López JA, Liles WC, Kain KC. 2017. MiR-155 modifies inflammation, endothelial activation and blood-brain barrier dysfunction in cerebral malaria. Mol Med 23:24–33. doi: 10.2119/molmed.2016.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. LaMonte G, Philip N, Reardon J, Lacsina JR, Majoros W, Chapman L, Thornburg CD, Telen MJ, Ohler U, Nicchitta CV, Haystead T, Chi J-T. 2012. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 12:187–199. doi: 10.1016/j.chom.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Wang Z, Xi J, Hao X, Deng W, Liu J, Wei C, Gao Y, Zhang L, Wang H. 2017. Red blood cells release microparticles containing human argonaute 2 And miRNAs to target genes of Plasmodium falciparum. Emerg Microbes Infect 6:e75. doi: 10.1038/emi.2017.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Gupta H, Rubio M, Sitoe A, Varo R, Cisteró P, Madrid L, Cuamba I, Jimenez A, Martiáñez-Vendrell X, Barrios D, Pantano L, Brimacombe A, Bustamante M, Bassat Q, Mayor A. 2021. Plasma microrna profiling of Plasmodium falciparum biomass and association with severity of malaria disease. Emerg Infect Dis 27:430–442. doi: 10.3201/eid2702.191795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Ketprasit N, Cheng IS, Deutsch F, Tran N, Imwong M, Combes V, Palasuwan D. 2020. The characterization of extracellular vesicles-derived microRNAs in thai malaria patients. Malar J 19:285. doi: 10.1186/s12936-020-03360-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. He X, Xie J, Zhang D, Su Q, Sai X, Bai R, Chen C, Luo X, Gao G, Pan W. 2015. Recombinant adeno-associated virus-mediated inhibition of microRNA-21 protects mice against the lethal schistosome infection by repressing both IL-13 and transforming growth factor beta 1 pathways. Hepatology 61:2008–2017. doi: 10.1002/hep.27671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Zhao Y, Dang Z, Chong S. 2019. Mmu-miR-92A-2-5p targets Tlr2 to relieve Schistosoma japonicum-induced liver fibrosis. Int Immunopharmacol 69:126–135. doi: 10.1016/j.intimp.2019.01.007 [DOI] [PubMed] [Google Scholar]
- 206. Luo X, Zhang D, Xie J, Su Q, He X, Bai R, Gao G, Pan W. 2018. MicroRNA-96 promotes schistosomiasis hepatic fibrosis in mice by suppressing Smad7. Molecular Therapy - Methods & Clinical Development 11:73–82. doi: 10.1016/j.omtm.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Wang L, Zhu Z, Liao Y, Zhang L, Yu Z, Yang R, Wu J, Wu Z, Sun X. 2022. Host liver-derived extracellular vesicles deliver miR-142A-3P induces neutrophil extracellular traps via targeting WASL to block the development of Schistosoma japonicum. Mol Ther 30:2092–2107. doi: 10.1016/j.ymthe.2022.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. He X, Tang R, Sun Y, Wang Y-G, Zhen K-Y, Zhang D-M, Pan W-Q. 2016. Micror-146 blocks the activation of M1 macrophage by targeting signal transducer and activator of transcription 1 in hepatic schistosomiasis. EBioMedicine 13:339–347. doi: 10.1016/j.ebiom.2016.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Giri BR, Cheng G. 2019. Host miR-148 regulates a macrophage-mediated immune response during Schistosoma japonicum infection. Int J Parasitol 49:993–997. doi: 10.1016/j.ijpara.2019.08.002 [DOI] [PubMed] [Google Scholar]
- 210. Zhu D, Yang C, Shen P, Chen L, Chen J, Sun X, Duan L, Zhang L, Zhu J, Duan Y. 2018. Rsjp40 suppresses hepatic stellate cell activation by promoting microRNA-155 expression and inhibiting Stat5 and Foxo3A expression. J Cell Mol Med 22:5486–5493. doi: 10.1111/jcmm.13819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Zhao X, Xia Z, Wang Z, Zhou M, Qiu X, Wang C, Xu T, Fang Q, Ming Z, Dong H. 2022. miR-182-5p attenuates Schistosoma japonicum-induced hepatic fibrosis by targeting tristetraprolin. Acta Biochim Biophys Sin (Shanghai) 54:1421–1430. doi: 10.3724/abbs.2022130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. He X, Xie J, Wang Y, Fan X, Su Q, Sun Y, Lei N, Zhang D, Gao G, Pan W, Loke P. 2018. Down-regulation of microRNA-203-3p initiates type 2 pathology during schistosome infection via elevation of interleukin-33. PLoS Pathog 14:e1006957. doi: 10.1371/journal.ppat.1006957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. He X, Sun Y, Lei N, Fan X, Zhang C, Wang Y, Zheng K, Zhang D, Pan W. 2018. MicroRNA-351 promotes schistosomiasis-induced hepatic fibrosis by targeting the vitamin D receptor. Proc Natl Acad Sci U S A 115:180–185. doi: 10.1073/pnas.1715965115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Zhu D, He X, Duan Y, Chen J, Wang J, Sun X, Qian H, Feng J, Sun W, Xu F, Zhang L. 2014. Expression of microRNA-454 in TGF-Β1-stimulated hepatic stellate cells and in mouse livers infected with Schistosoma japonicum. Parasit Vectors 7:148. doi: 10.1186/1756-3305-7-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Cai P, Mu Y, Olveda RM, Ross AG, Olveda DU, McManus DP. 2020. Serum exosomal miRNAs for grading hepatic fibrosis due to schistosomiasis. Int J Mol Sci 21:3560. doi: 10.3390/ijms21103560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Cai P, Mu Y, Olveda RM, Ross AG, Olveda DU, McManus DP. 2018. Circulating miRNAs as footprints for liver fibrosis grading in schistosomiasis. EBioMedicine 37:334–343. doi: 10.1016/j.ebiom.2018.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Wang Y, Fan X, Lei N, He X, Wang X, Luo X, Zhang D, Pan W. 2020. A microRNA derived from Schistosoma japonicum promotes schistosomiasis hepatic fibrosis by targeting host secreted frizzled-related protein 1. Front Cell Infect Microbiol 10:101. doi: 10.3389/fcimb.2020.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Wang L, Liao Y, Yang R, Yu Z, Zhang L, Zhu Z, Wu X, Shen J, Liu J, Xu L, Wu Z, Sun X. 2020. Sja-miR-71A in Schistosome egg-derived extracellular vesicles suppresses liver fibrosis caused by schistosomiasis via targeting semaphorin 4D. J Extracell Vesicles 9:1785738. doi: 10.1080/20013078.2020.1785738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Zhou X, Hong Y, Shang Z, Abuzeid AMI, Lin J, Li G. 2022. The potential role of microRNA-124-3p in growth, development, and reproduction of Schistosoma japonicum. Front Cell Infect Microbiol 12:862496. doi: 10.3389/fcimb.2022.862496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Liu J, Zhu L, Wang J, Qiu L, Chen Y, Davis RE, Cheng G. 2019. Schistosoma japonicum extracellular vesicle miRNA cargo regulates host macrophage functions facilitating parasitism. PLoS Pathog 15:e1007817. doi: 10.1371/journal.ppat.1007817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. He X, Wang Y, Fan X, Lei N, Tian Y, Zhang D, Pan W. 2020. A schistosome miRNA promotes host hepatic fibrosis by targeting transforming growth factor beta receptor III. J Hepatol 72:519–527. doi: 10.1016/j.jhep.2019.10.029 [DOI] [PubMed] [Google Scholar]
- 222. Xue X, Zhang Q, Huang Y, Feng L, Pan W. 2008. No miRNA were found in Plasmodium and the ones identified in erythrocytes could not be correlated with infection. Malar J 7:47. doi: 10.1186/1475-2875-7-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Kataria P, Surela N, Chaudhary A, Das J. 2022. MirRNA: biological regulator in host-parasite interaction during malaria infection. Int J Environ Res Public Health 19:2395. doi: 10.3390/ijerph19042395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Prabhu SR, Ware AP, Saadi AV. 2021. Erythrocyte miRNA regulators and malarial pathophysiology. Infect Genet Evol 93:105000. doi: 10.1016/j.meegid.2021.105000 [DOI] [PubMed] [Google Scholar]
- 225. Chen S-Y, Wang Y, Telen MJ, Chi J-T. 2008. The genomic analysis of erythrocyte microRNA expression in sickle cell diseases. PLoS One 3:e2360. doi: 10.1371/journal.pone.0002360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Martin-Alonso A, Cohen A, Quispe-Ricalde MA, Foronda P, Benito A, Berzosa P, Valladares B, Grau GE. 2018. Differentially expressed microRNAs in experimental cerebral malaria and their involvement in endocytosis, adherens junctions, FoxO and TGF-Β signalling pathways. Sci Rep 8:11277. doi: 10.1038/s41598-018-29721-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Grinev A, Fokina N, Bogomolov D, Berechikidze I, Lazareva Y. 2021. Prediction of gene expression regulation by human microRNAs in Plasmodium falciparum. Genes Environ 43:22. doi: 10.1186/s41021-021-00198-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Zhong H, Jin Y. 2022. Multifunctional roles of microRNAs in schistosomiasis. Front Microbiol 13:925386. doi: 10.3389/fmicb.2022.925386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. He X, Pan W. 2022. Host-parasite interactions mediated by cross-species microRNAs. Trends Parasitol 38:478–488. doi: 10.1016/j.pt.2022.02.011 [DOI] [PubMed] [Google Scholar]
- 230. Rathjen T, Nicol C, McConkey G, Dalmay T. 2006. Analysis of short Rnas in the malaria parasite and its red blood cell host. FEBS Lett 580:5185–5188. doi: 10.1016/j.febslet.2006.08.063 [DOI] [PubMed] [Google Scholar]
- 231. Hall N, Karras M, Raine JD, Carlton JM, Kooij TWA, Berriman M, Florens L, Janssen CS, Pain A, Christophides GK, James K, Rutherford K, Harris B, Harris D, Churcher C, Quail MA, Ormond D, Doggett J, Trueman HE, Mendoza J, Bidwell SL, Rajandream M-A, Carucci DJ, Yates JR, Kafatos FC, Janse CJ, Barrell B, Turner CMR, Waters AP, Sinden RE. 2005. A comprehensive survey of the plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307:82–86. doi: 10.1126/science.1103717 [DOI] [PubMed] [Google Scholar]
- 232. Myung SJ, Yoon J-H, Gwak G-Y, Kim W, Lee J-H, Kim KM, Shin CS, Jang JJ, Lee S-H, Lee S-M, Lee H-S. 2007. Wnt signaling enhances the activation and survival of human hepatic stellate cells. FEBS Lett 581:2954–2958. doi: 10.1016/j.febslet.2007.05.050 [DOI] [PubMed] [Google Scholar]
- 233. Jiang F, Parsons CJ, Stefanovic B. 2006. Gene expression profile of quiescent and activated rat hepatic stellate cells implicates Wnt signaling pathway in activation. J Hepatol 45:401–409. doi: 10.1016/j.jhep.2006.03.016 [DOI] [PubMed] [Google Scholar]
- 234. Hopkins AL, Groom CR. 2002. The druggable genome. Nat Rev Drug Discov 1:727–730. doi: 10.1038/nrd892 [DOI] [PubMed] [Google Scholar]
- 235. Crooke ST, Witztum JL, Bennett CF, Baker BF. 2018. RNA-targeted therapeutics. Cell Metab 27:714–739. doi: 10.1016/j.cmet.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 236. Schöniger C, Arenz C. 2013. Perspectives in targeting miRNA function. Bioorg Med Chem 21:6115–6118. doi: 10.1016/j.bmc.2013.03.040 [DOI] [PubMed] [Google Scholar]
- 237. Lu TX, Rothenberg ME. 2018. MicroRNA. J Allergy Clin Immunol 141:1202–1207. doi: 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Zhang S, Cheng Z, Wang Y, Han T. 2021. The risks of miRNA therapeutics: In a drug target perspective. Drug Des Devel Ther 15:721–733. doi: 10.2147/DDDT.S288859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. 2004. Human microRNA targets. PLoS Biol 2:e363. doi: 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. 2004. A combined computational-experimental approach predicts human microRNA targets. Genes Dev 18:1165–1178. doi: 10.1101/gad.1184704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. de Planell-Saguer M, Rodicio MC. 2013. Detection methods for microRNAs in clinic practice. Clin Biochem 46:869–878. doi: 10.1016/j.clinbiochem.2013.02.017 [DOI] [PubMed] [Google Scholar]
- 242. Kappel A, Keller A. 2017. miRNA assays in the clinical laboratory: workflow, detection technologies and automation aspects. Clin Chem Lab Med 55:636–647. doi: 10.1515/cclm-2016-0467 [DOI] [PubMed] [Google Scholar]
- 243. Akhtar MM, Micolucci L, Islam MS, Olivieri F, Procopio AD. 2016. Bioinformatic tools for microRNA dissection. Nucleic Acids Res 44:24–44. doi: 10.1093/nar/gkv1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Liu C-G, Calin GA, Volinia S, Croce CM. 2008. MicroRNA expression profiling using microarrays. Nat Protoc 3:563–578. doi: 10.1038/nprot.2008.14 [DOI] [PubMed] [Google Scholar]
- 245. Forero DA, González-Giraldo Y, Castro-Vega LJ, Barreto GE. 2019. qPCR-based methods for expression analysis of miRNAs. Biotechniques 67:192–199. doi: 10.2144/btn-2019-0065 [DOI] [PubMed] [Google Scholar]
- 246. Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26:317–325. doi: 10.1038/nbt1385 [DOI] [PubMed] [Google Scholar]
- 247. Hong LZ, Zhou L, Zou R, Khoo CM, Chew ALS, Chin C-L, Shih S-J. 2021. Systematic evaluation of multiple qPCR platforms, nanostring and miRNA-Seq for microRNA biomarker discovery in human biofluids. Sci Rep 11:4435. doi: 10.1038/s41598-021-83365-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Han D, Li R, Shi J, Tan P, Zhang R, Li J. 2020. Liquid biopsy for infectious diseases: a focus on microbial cell-free DNA sequencing. Theranostics 10:5501–5513. doi: 10.7150/thno.45554 [DOI] [PMC free article] [PubMed] [Google Scholar]