ABSTRACT
Carbapenemase-producing Enterobacterales have emerged in the last decades as one of the main threats in modern medicine. Enterobacter is a genus increasingly recognized as a key player in carbapenemase diffusion besides Escherichia coli and Klebsiella pneumoniae, but information regarding Citrobacter spp. remained scarce. A collection of 803 isolates of Citrobacter spp. recovered from the French National Center for carbapenem resistance over a 2-year period during 2019–2020 was analyzed. A total of 15 different carbapenemases were identified of which OXA-48 followed by NDM-1 and OXA-181 represented the main enzymes. Phylogenetic analysis of this collection revealed that Citrobacter freundii was the main species among which three main clones are circulating in France, being ST8, ST22, and ST91. ST22 and ST8 were distributed in all regions whereas ST91 demonstrated a regional spread. Analysis of the species diversity revealed that among the C. freundii complex, Citrobacter portucalensis and Citrobacter braakii were also disseminated. We revealed the dissemination of a clone carrying both blaDHA-15 (β-lactamase-encoding gene rarely reported in Enterobacterales) and blaOXA-48 in the north of France and allowed to identify an outbreak not previously investigated. This study analyzed the largest collection of carbapenemase-producing Citrobacter spp. and allowed deciphering of molecular epidemiology of Citrobacter spp. in France.
IMPORTANCE
The emergence of carbapenemase producers in Enterobacterales mostly involves Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae complex species. However, in France, we observed the emergence and the rapid dissemination of carbapenemase in Citrobacter spp. In this study, we demonstrated that a wide variety of carbapenemases is produced by many different species of Citrobacter spp. However, we clearly identify three high-risk clones of Citrobacter freundii, ST8, ST22, and ST91 that drive the spread of carbapenemase in France. This epidemiological study paves the way of further analysis that would aim to identify the virulence factors involved in this pellicular ability of these three clones to disseminate at the hospital.
KEYWORDS: Citrobacter, carbapenem, antibiotic, resistance, MLST, core genome
INTRODUCTION
Citrobacter spp. are facultative anaerobic Gram-negative rod-shaped bacteria belonging to Enterobacterales initially named according to their utilization of citrate as carbon source (1, 2). Citrobacter species are found in soil, water, and food and are commensal of the digestive tract (3). They are opportunistic pathogens able to induce urinary tract infections, neonatal sepsis, meningitis inducing brain abscess, respiratory tract infections, gastrointestinal infections, and central nervous system infections, in neonates and in immunocompromised patients (4).
Citrobacter freundii was initially named Bacterium freundii in 1928. In 1932, it was reclassified into the genus Citrobacter that contains now 16 named subspecies: C. freundii, Citrobacter braakii, Citrobacter gillenii, Citrobacter murliniae, Citrobacter rodentium, Citrobacter sedlakii, Citrobacter werkmanii, Citrobacter youngae, Citrobacter koseri (formerly known Citrobacter diversus), Citrobacter farmeri, Citrobacter amalonaticus, Citrobacter pasteurii, Citrobacter europaeus, Citrobacter portucalensis, Citrobacter cronae, and Citrobacter telavivensis (https://lpsn.dsmz.de/genus/citrobacter).
Inside this genus, C. koseri is genetically distant from C. freundii and the other subspecies (5). Moreover, C. koseri belongs to the group II Enterobacterales, as it produces a class A chromosomally encoded penicillinase that confers resistance to amino-, carboxy-, and ureido-penicillins (amoxicillin) and is inhibited by clavulanate or tazobactam (6). C. freundii and the other Citrobacter spp. belong to the group III Enterobacterales as they possess an Ambler class C chromosomally encoded cephalosporinase (blaCMY-like) conferring an intrinsic resistance to aminopenicillins, to classical β-lactamase inhibitors such as clavulanate or tazobactam, and first-generation cephalosporins (7). Overexpression of blaCMY-like genes may lead to resistance to expanded-spectrum cephalosporins and even carbapenems, if associated with decreased permeability (7).
Carbapenem-hydrolyzing class D β-lactamase (CHDL) OXA-48 and to a lesser extent the class B metallo-β-lactamase NDM-1 are the most frequently carbapenemase encountered in carbapenemase-producing Enterobacterales (CPE) isolated in France (8, 9).
The aim of this study was to characterize the epidemiology of carbapenemase-producing Citrobacter spp. received at the French National Reference Center (F-NRC) for CPEs in 2019–2020.
MATERIALS AND METHODS
Strain collection
A total of 1,121 carbapenem non-susceptible Citrobacter spp. were received at the F-NRC for CPEs between January 1st, 2019, and December 31st, 2020. Clinical isolates were firstly identified by MALDI-TOF mass spectrometry (MALDI Biotyper, Wissembourg, France).
Antimicrobial susceptibility testing and carbapenemase detection
Antimicrobial susceptibility testing was performed using the disc diffusion method on Mueller-Hinton (MH) agar (Bio-Rad, Marnes-La-Coquette, France), and interpreted according to EUCAST guidelines (https://eucast.org/clinical_breakpoints/).
The carbapenemase activity was assessed using the updated Carba NP test as described (10), and the type of carbapenemase was identified by the lateral flow immunoassay (LFIA) NG-Test Carba5 (NG Biotech, Guipry, France) (11).
Whole-genome sequencing and bioinformatics
Whole-genome sequencing was performed using Illumina’s NextSeq 500 as previously described (12). De novo assembly was performed using CLC Genomics Workbench v12.0 (Qiagen, Les Ulis, France). Acquired resistance gene and multilocus sequence typing (MLST) were performed using resfinder 4.1 and MLST 2.0 tools on the Center for Genomic Epidemiology server (http://www.genomicepidemiology.org/). Single nucleotide polymorphism (SNP) analysis and phylogeny were performed as previously described (13).
RESULTS
Whole-genome phylogeny
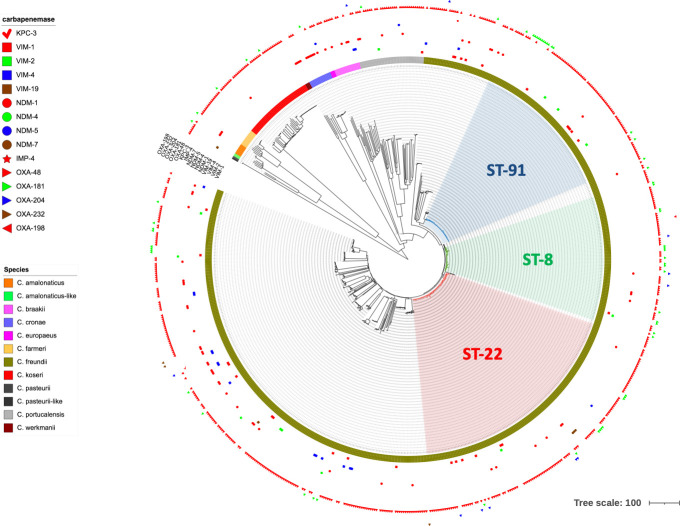
To assess the genetic diversity of Citrobacter spp., a phylogenetic tree was constructed based on the whole genome of the 803 carbapenemase-producing isolates (Fig. 1). As expected, it revealed that the main species corresponded to C. freundii sensu stricto. The main species, apart from C. freundii, correspond to C. koseri (n = 46) followed by C. portucalensis (n = 42), C. braakii (n = 17), and C. cronae (n = 15) (Fig. 1).
Fig 1.
Global phylogeny of carbapenemase-producing Citrobacter spp. isolated in France in 2019 and 2020.
However, WGS results revealed that several isolates sent to the F-NRC were incorrectly identified as C. freundii using MALDI-TOF, including isolates of C. europaeus, C. portucalensis, C. cronae, and C. werkmanii (Fig. 1) This is likely due the inability of MALDI-TOF to distinguish between these C. freundii complex species. The more distant species, and correctly identified by MALDI-TOF, included C. koseri, C. farmeri, C. amalonaticus, and C. pasteurii. Among the two C. pasteurii identified, one was very distant and could be considered as C. pasteurii-like. Further characterizations are needed to decipher if it truly belongs to C. pasteurii or to a novel species.
MLST analysis
MLST analysis evidenced three predominant clones, being ST8, ST22, and ST91, all from C. freundii stricto senso (Fig. 1). These sequence types (STs) represent 40.0% of total isolates with 78, 145, and 98 isolates belonging to ST8, ST22, and ST91, respectively (Fig. 1).
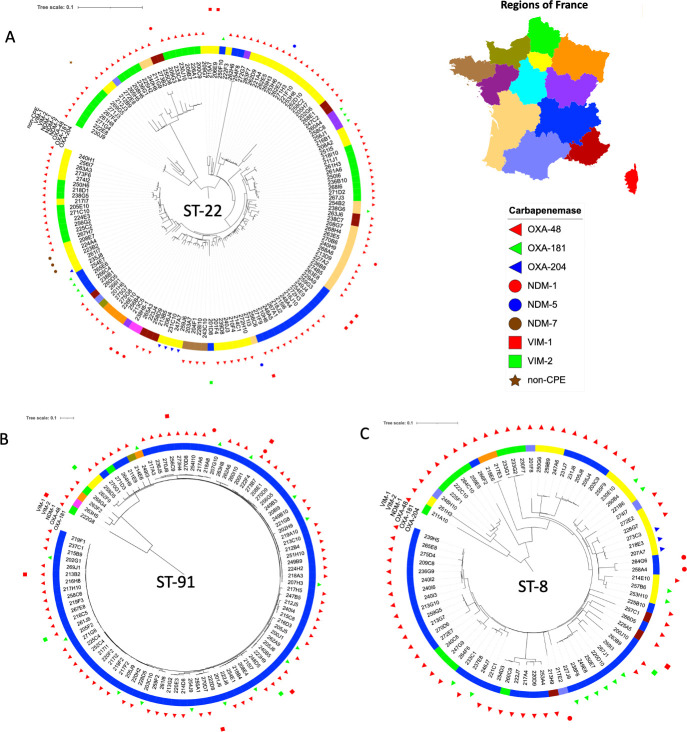
C. freundii isolates of ST22 were the most prevalent (Fig. 1). They were mostly recovered from three regions being Paris area (Ile-de-France, n = 45), north-east of France (Hauts-de-France, n = 22), and south-east (Auvergne-Rhone-Alpes, n = 22). However, isolates belonging to ST22 were also from French island such as Réunion Island (Indian Ocean) (Fig. 2). The in-depth analysis of the phylogenetic tree of ST22 revealed a wide genetic diversity inside this ST. Indeed, the size of the branches demonstrated a broad genetic diversity (Fig. 2A). As an example, despite being part of the ST22, some isolates seemed more distantly related (e.g., 262H6 and 254F8 isolates). This genetic diversity might also be highlighted by the presence in the same branch of clearly unrelated isolates recovered from different regions. Inside this genetic diversity, few outbreaks can be evidenced, such as 9 OXA-48-producing isolates recovered from an outbreak in Hauts-De-France or 14 clonally related strains in the region of Lyon (highlighted in red in Fig. 2A). Regarding this last outbreak which involved 14 isolates in Lyon, it is worthy to note that 3 isolates produced VIM-1, 10 isolates produced OXA-48, and the remaining isolate co-produced both carbapenemases. It suggests a wide spread of this particular clone that independently or successively acquired VIM-1 and OXA-48 carbapenemases in a restricted area (same hospital).
Fig 2.
Phylogeny of the three main clones of C. freundii, ST22 (panel A), ST8 (panel B), and ST91 (panel C).
The second most prevalent cluster corresponds to ST91 (98 isolates). Of note, 90.8% (89/98) of the ST91 isolates were recovered from the same region (Provence-Alpes-Côte d’Azur) strongly indicating a wide dissemination in this particular area (Fig. 2B). Noticeably, although the blaOXA-48 gene is the main carbapenemase gene carried by this clone, 15 blaOXA-181-carrying isolates were also identified. Two isolates carried the carbapenemase-encoding blaVIM-2 gene. In these two strains, blaVIM-2 was carried on a plasmid previously identified in Pseudomonas putida (GenBank accession number MK047608). In ST91, one isolate produced three carbapenemases being OXA-48, NDM-1, and VIM-1 (14).
The remaining main clone, ST8, has been identified in different regions of France and associated with different carbapenemase (Fig. 2C). It might indicate a wide spread of this clone rather than a recent dissemination as observed for ST91.
Antimicrobial susceptibility and carbapenemase production
Among the 1,121 carbapenem non-susceptible Citrobacter spp. received at the F-NRC for CPEs over the study period, 1,023 isolates produced a carbapenemase as revealed by Carba NP. The OXA-48-like carbapenemases were the most frequent, followed by NDM, VIM, KPC, and IMP as revealed by LFIA and further confirmed by WGS (Table 1).
TABLE 1.
Distribution of carbapenemases by Citrobacter species
| Species | n | Carbapenemases (n = 791) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VIM-1 | VIM-2 | VIM-4 | VIM-19 | NDM-1 | NDM-4 | NDM-5 | NDM-7 | IMP-4 | KPC-3 | OXA-48 | OXA-181 | OXA-204 | OXA-232 | OXA-198 | ||
| C. freundii | 643 | 28 | 8 | 16 | 1 | 48 | 3 | 1 | 3 | 1 | 1 | 493 | 56 | 10 | 3 | 1 |
| C. amalonaticus | 9 | 3 | 5 | 1 | ||||||||||||
| C. braakii | 17 | 2 | 4 | 11 | ||||||||||||
| C. cronae | 15 | 9 | 1 | 5 | 1 | |||||||||||
| C. europaeus | 3 | 1 | 2 | |||||||||||||
| C. farmeri | 11 | 10 | 1 | |||||||||||||
| C. koseri | 46 | 1 | 42 | 3 | ||||||||||||
| C. pasteurii-like | 2 | 1 | 2 | |||||||||||||
| C. portucalensis | 42 | 1 | 1 | 4 | 1 | 35 | 1 | 1 | ||||||||
| C. werkmanii | 3 | 2 | 1 | |||||||||||||
| Total | 791 | 29 | 9 | 18 | 2 | 72 | 3 | 3 | 3 | 1 | 1 | 606 | 63 | 11 | 3 | 1 |
Susceptibility testing was performed on the 1,121 carbapenem non-susceptible Citrobacter spp. isolates. All isolates were resistant to ticarcillin and ticarcillin/clavulanate combination, 99.4% were resistant to piperacillin-tazobactam combination, and 97.9% were resistant to temocillin. Resistance rates to cefotaxime, aztreonam, and cefepime were of 86.8%, 72.2%, and 64.7%, respectively. Among these isolates, 91.2% were resistant to ertapenem whereas 11.4% and 13.2% were resistant to meropenem and imipenem, respectively (Table 2). Regarding non-β-lactams, resistance rates to aminoglycosides were of 21.4% for amikacin and 69.6% for gentamicin. Resistance rates to fluoroquinolones were of 81.0% and 73.5% for ciprofloxacin and levofloxacin, respectively (Table 2).
TABLE 2.
Resistance profiles of Citrobacter spp. included in this study
| Molecules | Resistance rates (%) | ||
|---|---|---|---|
| β-Lactams | Ra | Ia | Sa |
| Ticarcillin | 100 | N/Ab | 0.0 |
| Ticarcillin-clavulanate | 100 | N/A | 0.0 |
| Piperacillin-tazobactam | 99.4 | N/A | 0.6 |
| Temocillin | 97.9 | 2.1 | 0.0 |
| Mecillinam | 32.1 | N/A | 67.9 |
| Cefotaxime | 86.8 | 2.6 | 10.6 |
| Ceftazidime | 85.2 | 1.8 | 13.0 |
| Ceftazidime-avibactam | 17.4 | N/A | 82.6 |
| Ertapenem | 91.2 | N/A | 8.8 |
| Meropenem | 11.4 | 22.0 | 66.6 |
| Imipenem | 13.2 | 14.8 | 72.0 |
| Aminoglycosides | |||
| Amikacin | 21.4 | N/A | 78.6 |
| Gentamicin | 69.6 | N/A | 30.4 |
| Fluoroquinolones | |||
| Ciprofloxacin | 81.0 | 4.8 | 14.2 |
| Levofloxacin | 73.5 | 5.2 | 21.3 |
| Others | |||
| Tigecycline | 7.6 | N/A | 92.4 |
| Sulfamethoxazole/trimethoprim | 65.8 | 1.1 | 33.1 |
| Fosfomycin | 9.3 | N/A | 90.7 |
S, susceptible; I, susceptible at high exposure; R, resistant.
N/A, not available.
Fosfomycin, tigecycline, and ceftazidime/avibactam remained in the susceptibility range for 90.7%, 92.4%, and 82.6%, respectively. Diameter distribution of ceftazidime/avibactam revealed a bimodal distribution with a highly resistant population likely indicating a production of metal-β-lactamase.
Resistome analysis
Of the 1,023 isolates that produced a carbapenemase, 803 non-duplicate isolates were sequenced. Analysis of their resistomes revealed that the main carbapenemase gene found in these isolates corresponded to blaOXA-48 (n = 606), followed by blaNDM-1 (n = 72) and blaOXA-181 (n = 63) (Table 1). Fifteen different carbapenemase genes were identified during this study including four blaVIM-like variants [blaVIM-1 (n = 29), blaVIM-2 (n = 9), blaVIM-4 (n = 18), and blaVIM-19 (n = 2)], four blaNDM-like variants [blaNDM-1 (n = 72), blaNDM-4 (n = 3), blaNDM-5 (n = 3), and blaNDM-7 (n = 3)], blaIMP-4 (n = 1), blaKPC-3 (n = 1), four blaOXA-48-like variants [blaOXA-48 (n = 606), blaOXA-181 (n = 63), blaOXA-204 (n = 11), and blaOXA-232 (n = 3)], and blaOXA-198 (n = 1).
Besides carbapenemase genes, numerous broad-spectrum β-lactamase-encoding genes were identified including ESBL-encoding genes such as eight different blaCTX-M alleles (blaCTX-M-1, blaCTX-M-3, blaCTX-M-9, blaCTX-M-14, blaCTX-M-15, blaCTX-M-32, blaCTX-M-55, and blaCTX-M-232), blaGES-1, blaGES-7, blaVEB-1, blaDHA-1-like, or plasmid-encoded cephalosporinase genes such as blaDHA-1 or blaDHA-15.
By contrast with blaDHA-1 gene identified in multiple STs, blaDHA-15 gene have only been identified in ST500. Epidemiological data revealed that all these ST500 C. freundii isolates were from the same city and thus revealed a loco-regional spread of a single clone. As described in Morganella, the natural progenitor of blaDHA-like genes, blaDHA-15 is associated to its transcriptional regulator LysR. Analysis of the genetic of the blaDHA-15 environment revealed the presence of a quinolone resistance protein qnrB4 close to this gene.
Among ESBLs identified in this collection, blaGES-1 was present in 10 isolates including 8 C. freundii, 1 C. koseri, and 1 C. amalonaticus. This gene was identified in four different STs in the C. freundii complex being ST116 (n = 4), ST216 (n = 2), ST22 (n = 1), and ST118 (n = 1). Noticeably, the extended-spectrum class D blaOXA-35-like was identified in 22 isolates including 20 C. freundii and 2 C. portucalensis. Analysis of the genetic background identified a wide genetic diversity of STs (n = 10) carrying this gene.
Aminoglycoside resistance genes conferring resistance to gentamicin and amikacin were found in 69.6% and 21.4%, respectively, of the isolates that were resistant to these molecules. Among isolates resistant to both molecules, the 16S RNA methyltransferase-encoding gene armA was identified in 19 isolates (14 C. freundii, 2 C. portucalensis, 2 C. cronae, and 1 C. europaeus). Two other 16S RNA methyltransferase-encoding genes were identified, rmtB1 and rmtC in 1 C. freundii and 16 isolates, respectively (11 C. freundii, 2 C. braakii, 2 C. cronae, and 1 C. werkmanii). Noticeably, two isolates harbored a variant of the tet(X5) gene. These two isolates correspond to a C. portucalensis ST421 and to a C. freundii ST22.
Plasmid-encoded colistin resistance genes mcr9.1 (n = 13 isolates) and mcr9.2 (n = 9 isolates) were also identified in different Citrobacter species, suggesting dissemination of these resistance alleles across this genus.
Outbreak of DHA-15 and OXA-48-producing isolates in northern France
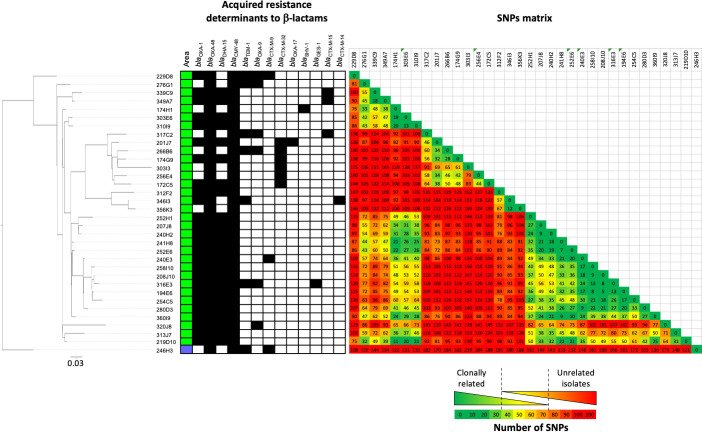
During the analysis of resistomes, some β-lactamase-encoding genes rarely reported in Enterobacterales, but commonly identified in Pseudomonas spp., caught our interest such as blaVIM-2, blaGES-1, and a rare plasmid-encoded cephalosporinase blaDHA-15 gene. The blaDHA-15 gene is present in GenBank database in only two sequences corresponding to the initial description (GenBank accession number NG_049060) and a plasmid-encoded sequence identified in C. freundii (GenBank accession number OW969688). In our collection, DHA-15 was present in nine isolates of C. freundii belonging to ST500. This ST500 was reported only once in PubMLST database (https://pubmlst.org/) in Poland in 2017.
To decipher if these DHA-15-producing ST500 C. freundii corresponded to a clonal diffusion, we collected all DHA-15-producing isolates and all Citrobacter spp. belonging to ST500 from the F-NRC over a 6-year period (2014–2020). A total of 34 C. freundii of ST500 were identified. Among them, 97.1% (33/34) were isolated in the same region in the north of France (Fig. 3). The phylogenetic analysis identified a polyclonal dissemination of several clones producing all the OXA-48 carbapenemase associated with a wide diversity of other β-lactamases (24 DHA-15-, 6 CTX-M-32-, 1 SHV-1-, 1 GES-1-, 3 CTX-M-9-, 3 CTX-M-15-, and 1 CTX-M-14-producing isolates, respectively).
Fig 3.
Genomic and phylogenetic characterization of ST500 Citrobacter freundii.
DISCUSSION
This study characterized the epidemiology of carbapenemase-producing Citrobacter spp. at a genomic level and identified the most prevalent clones that circulated in France in 2019 and 2020. A total of 803 phenotypically characterized isolates were whole-genome sequenced.
Susceptibility testing revealed that among carbapenemase-producing isolates, 8.8%, 66.6%, and 72.0% remained susceptible to ertapenem, meropenem, and imipenem, respectively. Ceftazidime/avibactam combination remained an effective combination except for metallo-β-lactamase producers for which high-level resistance was observed as previously described (15).
Analysis of the resistome revealed that besides the wide dissemination of the blaOXA-48 gene and into a lesser extent blaNDM-1, an unexpected high diversity of other beta-lactamases was identified in this collection. This includes the production of VIM-2 rarely described in Enterobacterales (16) and commonly identified in Pseudomonas spp. (17). Surprisingly, analysis of the genetic context surrounding the blaVIM-2 gene revealed a close genetic context with a plasmid identified in P. aeruginosa suggesting a genetic transfer between these two genus. Another particularity is the presence of the blaGES-1 gene in 10 isolates belonging to different Citrobacter species and the dissemination of the blaDHA-15 gene that seems to be correlated to the spread of C. freundii ST500 and responsible of several outbreaks in the north of France.
Analysis of the clones circulating in France revealed that three main clones were disseminating carbapenemase being ST8, ST22, and ST91. Despite that these clones are more prevalent, the drivers for their dissemination differ. Indeed, ST91 dissemination corresponded to a clonal expansion mainly within a single region whereas ST8 and ST22 seem to have disseminated in the entire country with a more genetic diversity. In addition, the dissemination of these two STs (ST8 and ST22) is uncorrelated to the production of a given carbapenemase suggesting that these STs can be defined as “high-risk” clones of multidrug resistance. This is the main caveat of this study. We should acknowledge that our collection included only carbapenemase-producing Citrobacter spp. isolates. To really define ST8 and ST22 as a high-risk clone, it will be needed to study antibiotic-susceptible Citrobacter spp. or ESBL producers, too. This additional investigation might help differentiate clones involved in carbapenemase dissemination from those that are intrinsically highly prevalent. Our results pave the way of the analysis of core genomes of these highly prevalent STs to identify features that might be associated with their success.
Finally, as previously observed for other Enterobacterales such as the Enterobacter cloacae complex (18–20), we demonstrated that MALDI-TOF is not precise enough to accurately identify C. freundii complex isolates at the species level. It might be of interest to use our collection of deeply characterized Citrobacter spp. isolates to increase the MALDI-TOF database to see whether specific markers may help separate these species.
Contributor Information
Laurent Dortet, Email: laurent.dortet@aphp.fr.
Paul D. Fey, University of Nebraska Medical Center, Omaha, Nebraska, USA
DATA AVAILABILITY
All genome sequences are stored on the F_NRC webserver. These sequences are available on demand from the corresponding author.
REFERENCES
- 1. Janda JM, Abbott SL, Cheung WK, Hanson DF. 1994. Biochemical identification of citrobacteria in the clinical laboratory. J Clin Microbiol 32:1850–1854. doi: 10.1128/jcm.32.8.1850-1854.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Miki K, Tamura K, Sakazaki R, Kosako Y. 1996. Re-speciation of the original reference strains of serovars in the Citrobacter freundii (Bethesda-Ballerup group) antigenic scheme of West and edwards. Microbiol Immunol 40:915–921. doi: 10.1111/j.1348-0421.1996.tb01160.x [DOI] [PubMed] [Google Scholar]
- 3. Arens S, Verbist L. 1997. Differentiation and susceptibility of Citrobacter isolates from patients in a university hospital. Clin Microbiol Infect 3:53–57. doi: 10.1111/j.1469-0691.1997.tb00251.x [DOI] [PubMed] [Google Scholar]
- 4. Graham DR, Band JD. 1981. Citrobacter diversus brain abscess and meningitis in neonates. JAMA 245:1923–1925. [PubMed] [Google Scholar]
- 5. Yuan C, Yin Z, Wang J, Qian C, Wei Y, Zhang S, Jiang L, Liu B. 2019. Comparative genomic analysis of Citrobacter and key genes essential for the pathogenicity of Citrobacter koseri. Front Microbiol 10:2774. doi: 10.3389/fmicb.2019.02774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Petrella S, Clermont D, Casin I, Jarlier V, Sougakoff W. 2001. Novel class A beta-lactamase Sed-1 from Citrobacter sedlakii: genetic diversity of beta-lactamases within the Citrobacter genus. Antimicrob Agents Chemother 45:2287–2298. doi: 10.1128/AAC.45.8.2287-2298.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182, doi: 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Emeraud C, Girlich D, Bonnin RA, Jousset AB, Naas T, Dortet L. 2021. Emergence and polyclonal dissemination of OXA-244-producing Escherichia coli, France. Emerg Infect Dis 27:1206–1210. doi: 10.3201/eid2704.204459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patiño-Navarrete R, Rosinski-Chupin I, Cabanel N, Zongo PD, Héry M, Oueslati S, Girlich D, Dortet L, Bonnin RA, Naas T, Glaser P. 2022. Specificities and commonalities of carbapenemase-producing Escherichia coli isolated in France from 2012 to 2015. mSystems 7:e0116921. doi: 10.1128/msystems.01169-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dortet L, Bréchard L, Cuzon G, Poirel L, Nordmann P. 2014. Strategy for rapid detection of carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 58:2441–2445. doi: 10.1128/AAC.01239-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boutal H, Vogel A, Bernabeu S, Devilliers K, Creton E, Cotellon G, Plaisance M, Oueslati S, Dortet L, Jousset A, Simon S, Naas T, Volland H. 2018. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 73:909–915. doi: 10.1093/jac/dkx521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Girlich D, Bonnin RA, Bogaerts P, De Laveleye M, Huang DT, Dortet L, Glaser P, Glupczynski Y, Naas T. 2017. Chromosomal amplification of the blaOXA-58 carbapenemase gene in a proteus mirabilis clinical isolate. Antimicrob Agents Chemother 61:e01697-16. doi: 10.1128/AAC.01697-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bonnin RA, Jousset AB, Chiarelli A, Emeraud C, Glaser P, Naas T, Dortet L. 2020. Emergence of new non-clonal group 258 high-risk clones among Klebsiella pneumoniae carbapenemase-producing K. pneumoniae isolates, France. Emerg Infect Dis 26:1212–1220. doi: 10.3201/eid2606.191517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Biez L, Bonnin RA, Naas T, Dortet L. 2022. Characterization of VIM-1-, NDM-1- and OXA-48-producing Citrobacter freundii in France. J Antimicrob Chemother 77:1200–1202. doi: 10.1093/jac/dkac005 [DOI] [PubMed] [Google Scholar]
- 15. Bonnin RA, Bernabeu S, Emeraud C, Creton E, Vanparis O, Naas T, Jousset AB, Dortet L. 2022. Susceptibility of OXA-48-producing Enterobacterales to imipenem/relebactam, meropenem/vaborbactam and ceftazidime/avibactam. Int J Antimicrob Agents 60:106660. doi: 10.1016/j.ijantimicag.2022.106660 [DOI] [PubMed] [Google Scholar]
- 16. Bonnin RA, Girlich D, Jousset AB, Emeraud C, Creton E, Gauthier L, Jové T, Dortet L, Naas T. 2021. Genomic analysis of VIM-2-producing Enterobacter hormaechei subsp. steigerwaltii. Int J Antimicrob Agents 57:106285. doi: 10.1016/j.ijantimicag.2021.106285 [DOI] [PubMed] [Google Scholar]
- 17. Tenover FC, Nicolau DP, Gill CM. 2022. Carbapenemase-producing Pseudomonas aeruginosa -an emerging challenge. Emerg Microbes Infect 11:811–814. doi: 10.1080/22221751.2022.2048972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emeraud C, Petit C, Gauthier L, Bonnin RA, Naas T, Dortet L. 2022. Emergence of VIM-producing Enterobacter cloacae complex in France between 2015 and 2018. J Antimicrob Chemother 77:944–951. doi: 10.1093/jac/dkab471 [DOI] [PubMed] [Google Scholar]
- 19. Matsumura Y, Peirano G, Devinney R, Bradford PA, Motyl MR, Adams MD, Chen L, Kreiswirth B, Pitout JDD. 2017. Genomic epidemiology of global VIM-producing Enterobacteriaceae. J Antimicrob Chemother 72:2249–2258. doi: 10.1093/jac/dkx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peirano G, Matsumura Y, Adams MD, Bradford P, Motyl M, Chen L, Kreiswirth BN, Pitout JDD. 2018. Genomic epidemiology of global carbapenemase-producing Enterobacter spp., 2008-2014. Emerg Infect Dis 24:1010–1019. doi: 10.3201/eid2406.171648 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All genome sequences are stored on the F_NRC webserver. These sequences are available on demand from the corresponding author.