ABSTRACT
This study compared the impact of sequence type 5 (ST5) and ST764, on the efficacy of vancomycin treatment in methicillin-resistant Staphylococcus aureus (MRSA) infections. From July 2012 to June 2020, a prospective observational study was conducted in five hospitals in China, enrolling 90 patients with MRSA infections, including 44 patients with ST5-MRSA and 46 patients with ST764-MRSA infections. All strains were subjected to minimal inhibitory concentration (MIC) determination, biofilm expression, heterogeneous vancomycin-intermediate S. aureus (hVISA) screening, and whole-genome sequencing. Vancomycin therapeutic drug monitoring was conducted, and the 24-hour area under the curve (AUC0-24) and AUC0-24/MIC values were calculated. ST5-MRSA shows elevated hVISA/VISA presence, increased biofilm formation, and higher presence of virulence genes like tst, sec, and sel. ST764-MRSA expresses seb, aiding infection in elderly patients in the community. Additionally, patients with ST764 infections exhibit higher vancomycin AUC0-24/MIC values, and fewer tracheotomies compared to ST5. Clinical signs and symptoms improvement were observed in 27 (61.4%) and 33 (71.7%) patients in the ST5 and ST764 groups, respectively (P = 0.372). On the other hand, 28 (63.6%) and 39 (84.8%) patients had laboratory-confirmed bacterial eradication in two groups, respectively (P = 0.029). Multivariate analysis showed that the virulence genes, such as the tst gene, were a risk factor for bacterial persistence (adjusted odds ratio, 4.509; 95% confidence interval, 1.216 to 16.724; P = 0.024). Our study showed that vancomycin was less effective in treating ST5-MRSA infection compared to ST764-MRSA infection, in part because ST5-MRSA is healthcare-associated MRSA while ST764-MRSA carries genetic characteristics of community-associated MRSA.
IMPORTANCE
Methicillin-resistant Staphylococcus aureus (MRSA) is a bacterium that is resistant to multiple drugs and can cause serious infections. In recent years, one of the most widespread strains of MRSA worldwide has been the clonal complex 5 (CC5) type. Sequence type 5 (ST5) and ST764 are two prevalent CC5 strains. Although ST5 and ST764 are genotypically identical, ST764 is classified as a hybrid variant of ST5 with characteristics of community-associated MRSA (CA-MRSA). In contrast to ST5, ST764 lacks the tst and sec genes but carries the staphylococcal enterotoxin B (seb) gene. Vancomycin is commonly used as the first-line treatment for MRSA infections. However, it is currently unclear whether the genetic differences between the ST5 and ST764 strains have any impact on the efficacy of vancomycin in treating MRSA infections. We conducted a prospective observational study comparing the efficacy of vancomycin against ST5-MRSA and ST764-MRSA in five hospitals in China. There were significant differences in bacteriological efficacy between the two groups, with virulence genes, such as the tst gene, being a risk factor for bacterial persistence (adjusted odds ratio, 4.509; 95% confidence interval, 1.216 to 16.724; P = 0.024). In the future, it may be necessary to consider personalized vancomycin treatment strategies based on the genetic characteristics of MRSA isolates.
KEYWORDS: ST5, ST764, methicillin-resistant Staphylococcus aureus, vancomycin, efficacy
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is a multidrug-resistant bacterium that causes serious infections, including bloodstream infections, pneumonia, skin and soft tissue infections, and infective endocarditis, with high morbidity and mortality. Additionally, MRSA can lead to prolonged hospitalization, putting pressure on healthcare systems (1–3).
Among prevalent MRSA strains, the clonal complex 5 (CC5) type is one of the most widely spreading strains worldwide in recent years (4, 5). As the predominant subtype of the CC5, sequence type 5 (ST5) mainly originated from New York/Japan (4) and is currently one of the major MRSA endemic strains in East China (6, 7). The ST5 clone is a prevalent strain associated with nosocomial infections (4) and is characterized by the presence of staphylococcal cassette chromosome mec (SCCmec) type II and positive for toxin-type toxin 1 (tst) and enterotoxin C (sec) genes, as well as resistance to multiple antibiotics (8). ST764 also belongs to the CC5 family and was first identified in Japan. It has since been widely disseminated in both hospital and community settings (8–10). Recent reports have shown an increasing detection rate of ST764 in China, and it is gradually becoming the dominant strain in various clinical infections, surpassing ST5 (11–13). ST764 clone is a variant of the ST5 lineage. Despite being a healthcare-associated MRSA (HA-MRSA) clone, it is a hybrid variant with community-associated MRSA (CA-MRSA) features (8). ST764 has acquired two new mobile genetic elements during evolution: the arginine catabolism mobile element II (ACME-II) and SaPInn54. This acquisition resulted in a virulence profile similar to that of CA-MRSA, as these elements carry genes such as ACME arcA and the staphylococcal enterotoxin B gene (seb) (8, 14). Notably, ST764-MRSA also exhibited enhanced expression of cytolytic peptide genes. However, it was observed that ST764-MRSA lacked Panton-Valentine leukocidin, a common virulence factor found in CA-MRSA strains.
Vancomycin is the primary treatment option for MRSA infections, given its strong antibacterial activity against this pathogen (15, 16). Despite having similar genotypes, ST5-MRSA and ST764-MRSA differ in their genetic identity. It is unclear whether this difference affects the effectiveness of vancomycin treatment.
The aim of this study is to conduct a comprehensive comparison of the clinical characteristics, vancomycin pharmacokinetic/pharmacodynamic (PK/PD) indices, and strain genotype characteristics of ST5 and ST764 strains isolated from patients with MRSA infections. The findings of this study will provide insights into the potential impact of vancomycin treatment on the clinical and bacterial efficacy of infections caused by these strains.
RESULTS
Patient clinical features
In total, 90 patients with MRSA infections were included in this analysis, 44 (48.9%) patients were identified as ST5-MRSA, and 46 (51.1%) patients were ST764-MRSA (Fig. 1). Clinical features of these diagnosed MRSA-infected patients were thoroughly compared between ST5 and ST764 groups (Table 1). A significant difference in age was observed between the two groups, e.g., patients in the ST764-MRSA group were older than those in the ST5-MRSA group (P < 0.001), while no significant differences in gender, weight, body mass index (BMI), admission to the intensive care unit (ICU), or days spent in the ICU were identified. The majority of ST5-MRSA patients (88.6%) originated from tertiary care hospitals, whereas only 26.1% of ST764-MRSA patients exhibited the same origin. In terms of underlying disease, patients in the ST764-MRSA group showed a higher rate of cardiovascular diseases (45.7% vs 22.7%, P = 0.027) and strokes (56.5% vs 15.9%, P < 0.001). In addition, a higher proportion of patients with ST5-MRSA infection underwent more tracheotomy procedures (43.2% vs 10.9%, P = 0.001). Regarding the infection type, pulmonary infections (69 patients, 76.7%) and bloodstream infections (11 patients, 12.2%) accounted for the majority of MRSA infections. ST5-MRSA bloodstream infections were twice as common as ST764, although the difference was not statistically significant (15.9% vs 8.7%, P = 0.348).
Fig 1.
Flowchart of patient enrollment and assessment in this study.
TABLE 1.
Demographics and clinical features of infections caused by ST5-MRSA vs ST764-MRSA isolates
| Characteristica | Total (n = 90) | ST5b (n = 44) |
ST764 (n = 46) |
P valuec |
|---|---|---|---|---|
| Demographic | ||||
| Age, years | 69 (51, 80) | 61 (42, 72) | 77 (64, 84) | <0.001 |
| Gender, male | 63 (70.0%) | 29 (65.9%) | 34 (73.9%) | 0.492 |
| Weight, kg | 60 (55, 70) | 65 (55, 72) | 60 (55, 70) | 0.126 |
| BMI, kg/m2 | 22.0 (20.0, 24.0) | 22.8 (20.3, 24.0) | 21.8 (19.0, 24.0) | 0.190 |
| ICU | ||||
| ICU admission | 53 (58.9%) | 22 (50.0%) | 31 (67.4%) | 0.133 |
| ICU stay days | 11(0, 30) | 0 (0, 28.5) | 18 (0, 31) | 0.206 |
| Occurred in tertiary care hospitals | 51 (56.7%) | 39 (88.6%) | 12 (26.1%) | <0.001 |
| Underlying disease | ||||
| Cardiovascular diseases | 31 (34.4%) | 10 (22.7%) | 21 (45.7%) | 0.027 |
| Diabetes | 14 (15.6%) | 5 (11.4%) | 9 (19.6%) | 0.386 |
| Strokes | 33 (36.7%) | 7 (15.9%) | 26 (56.5%) | <0.001 |
| COPD | 3 (3.3%) | 1 (2.3%) | 2 (4.3%) | >0.999 |
| Autoimmune diseases | 1 (1.1%) | 1 (2.3%) | 0 (0%) | 0.489 |
| Trauma | 11 (12.2%) | 3 (6.8%) | 8 (17.4%) | 0.198 |
| Solid tumor | 17 (18.9%) | 10 (22.7%) | 7 (15.2%) | 0.426 |
| Surgery | 39 (43.3%) | 23 (52.3%) | 16 (34.8%) | 0.136 |
| Implant | ||||
| Venous catheter | 64 (71.1%) | 29 (65.9%) | 35 (76.1%) | 0.355 |
| Endotracheal intubation | 30 (33.3%) | 19 (43.2%) | 11 (23.9%) | 0.074 |
| Tracheotomy | 24 (26.7%) | 19 (43.2%) | 5 (10.9%) | 0.001 |
| Urinary catheter | 54 (60.0%) | 30 (68.2%) | 24 (52.2%) | 0.137 |
| Drainage tube | 22 (24.4%) | 11 (25.0%) | 11 (23.9%) | >0.999 |
| Infection site | ||||
| Bloodstream infection | 11 (12.2%) | 7 (15.9%) | 4 (8.7%) | 0.348 |
| Pulmonary infection | 69 (76.7%) | 32 (72.7%) | 37 (80.4%) | 0.459 |
| Central nervous system infections | 3 (3.3%) | 3 (6.8%) | 0 (0%) | 0.113 |
| Skin and soft tissue infections | 5 (5.6%) | 4 (9.1%) | 1 (2.2%) | 0.198 |
| Urinary tract infection | 4 (4.4%) | 1 (2.3%) | 3 (6.5%) | 0.617 |
| Other infection | 6 (6.7%) | 3 (6.8%) | 3 (6.5%) | >0.999 |
| Treatment | ||||
| Vancomycin daily dose, g | 1.8 (1.2, 2.0) | 2 (1.6, 2.1) | 1.4 (1.0, 2.0) | <0.001 |
| Vancomycin duration | 13 (10,18) | 13 (10,17) | 12 (9,20) | 0.399 |
| Combined with β-lactams | 58 (64.4%) | 30 (68.2%) | 28 (60.9%) | 0.514 |
| Combined with rifampin | 7 (7.8%) | 3 (6.8%) | 4 (8.7%) | >0.999 |
| Combined with quinolones | 4 (4.4%) | 3 (6.8%) | 1 (2.2%) | 0.355 |
| Combined with other antibiotics | 25 (27.8%) | 11 (25.0%) | 14 (30.4%) | 0.641 |
ST, sequence type; BMI, body mass index; ICU, intensive care unit; COPD, chronic obstructive pulmonary disease.
Continuous variables were expressed as median (interquartile range) and categorical variables were summarized as the number of cases (%) in the table.
P values< 0.05 are shown in bold.
Vancomycin therapeutic drug monitoring (TDM) and PK/PD analysis
Vancomycin TDM and PK/PD analysis were performed for all patients to enable personalized therapy. As for vancomycin treatment, the daily doses were higher in the ST5-MRSA group than those in the ST764 group (2 g vs 1.4 g, P < 0.001), with comparable vancomycin in combination with other antibiotics between the two groups. After the implementation of TDM (Table 2), patients with ST764-MRSA infection had higher trough concentration (Cmin) (14.37 mg/L vs 10.09 mg/L, P = 0.011) but comparable peak concentration (Cmax) compared to ST5. The 24-hour area under the curve (AUC0-24) was also higher in ST764 but not significantly different from ST5. Interestingly, the AUC0-24-to-minimal inhibitory concentration ratio (AUC0-24/MIC) is significantly higher in the ST764 group (732 vs 500, P < 0.001).
TABLE 2.
Correlation between vancomycin pharmacokinetics and pharmacokinetic/pharmacodynamic indices of infections caused by ST5-MRSA vs ST764-MRSA isolates
| Characteristica | Total (n = 90) | ST5c (n = 44) |
ST764 (n = 46) |
P valued |
|---|---|---|---|---|
| PKb | ||||
| Cmin, mg/L | 11.18 (7.53,16.70) | 10.09 (5.03,13.72) | 14.37 (8.36, 19.61) | 0.011 |
| Cmax, mg/L | 26.31 (19.73, 32.34) | 25.78 (20.68, 30.02) | 27.94 (19.63,34.40) | 0.522 |
| AUC0-24, mg·h/L | 421 (327, 570) | 398 (329, 482) | 456 (322, 612) | 0.338 |
| PK/PD | ||||
| AUC0-24/MIC | 625 (415, 912) | 500 (365, 659) | 732 (505,1176) | <0.001 |
PK, pharmacokinetic; PK/PD, pharmacokinetic/pharmacodynamic; Cmin, trough concentration; Cmax, peak concentration; AUC0-24, 24-hour area under the curve; AUC0-24/MIC, 24-hour area under the curve over minimal inhibitory concentration.
All the PK parameters were calculated as the weighted average values.
Continuous variables were expressed as median (interquartile range) in the table.
P values< 0.05 are shown in bold.
Phenotypes and molecular characterization of MRSA strains
To assess the distinctions between ST5 and ST764 bacterial phenotypes, we conducted vancomycin MIC assay, heterogeneous vancomycin-intermediate S. aureus (hVISA) screening, and biofilm assessment on all the clinical strains. ST764-MRSA and ST5-MRSA strains showed the same MIC90 value but lower vancomycin MIC50 than that in the ST5-MRSA strain (0.5 mg/L vs 1 mg/L) (Fig. 2A). Similarly, ST5-MRSA strains had a higher detection rate of VISA and hVISA (86.4% vs 26.1%, P<0.001) (Fig. 2B) and increased expression of biofilm (OD570nm: 0.491 vs 0.304, P < 0.001) in comparison to the ST764 strains (Fig. 2C).
Fig 2.
Vancomycin minimum inhibitory concentration (MIC) distribution (A), hVISA/VISA detection rate (B), and biofilm expression (C) of ST5-MRSA and ST764-MRSA.
Furthermore, molecular typing of ST5-MRSA and ST764-MRSA was also investigated to fully explore their distinctions and potential implications for vancomycin treatment and corresponding clinical outcomes (Table 3). Both ST5-MRSA and ST764-MRSA isolates were typed as SCCmecII and accessory gene regulator II (agrII). In terms of the S. aureus protein A (spa) typing, ST764-MRSA are highly clustered in t002 (93.5%), while ST5-MRSA were almost equally scattered among t311 (14/44, 31.8%), t2460 (14/40, 31.8%), and t002 (13, 29.5%).
TABLE 3.
Genotypes of ST5-MRSA and ST764-MRSA isolates
| Characteristica | Total (n = 90) | ST5b (n = 44) |
ST764 (n = 46) |
P valuec |
|---|---|---|---|---|
| Molecular typing | ||||
| SCCmecII | 90 (100%) | 44 (100%) | 46 (100%) | NA |
| agrII | 90 (100%) | 44 (100%) | 46 (100%) | NA |
| spa-t311 | 14 (15.6%) | 14 (31.8%) | 0 (0%) | <0.001 |
| spa-t002 | 56 (62.2%) | 13 (29.5%) | 43 (93.5%) | <0.001 |
| Adhesion and toxins | ||||
| clfA | 74 (82.2%) | 34 (77.3%) | 40 (87.0%) | 0.277 |
| clfB | 75 (83.3%) | 35 (79.5%) | 40 (87.0%) | 0.405 |
| fnbA | 83 (92.2%) | 38 (86.4%) | 45 (97.8%) | 0.056 |
| fnbB | 53 (58.9%) | 24 (54.5%) | 29 (63.0%) | 0.521 |
| sdrC | 79 (87.8%) | 38 (86.4%) | 41 (89.1%) | 0.755 |
| sdrD | 88 (97.8%) | 42 (95.5%) | 46 (100%) | 0.236 |
| sdrE | 81 (90.0%) | 36 (81.8%) | 45 (97.8%) | 0.014 |
| cna | 0 (0%) | 0 (0%) | 0 (0%) | NA |
| ebpS | 90 (100%) | 44 (100%) | 46 (100%) | NA |
| lukS-PV | 0 (0%) | 0 (0%) | 0 (0%) | NA |
| lukF-PV | 90 (100%) | 44 (100%) | 46 (100%) | NA |
| hla | 90 (100%) | 44 (100%) | 46 (100%) | NA |
| hlb | 90 (100%) | 44 (100%) | 46 (100%) | NA |
| hld | 90 (100%) | 44 (100%) | 46 (100%) | NA |
| tst | 39 (43.3%) | 39 (88.6%) | 0 (0%) | <0.001 |
| sea | 11 (12.2%) | 11 (25.0%) | 0 (0%) | <0.001 |
| seb | 43 (47.8%) | 0 (0%) | 43 (93.5%) | <0.001 |
| sec | 39 (43.3%) | 39 (88.6%) | 0 (0%) | <0.001 |
| seh | 0 (0%) | 0 (0%) | 0 (0%) | NA |
| sek | 90 (100%) | 44 (100%) | 46 (100%) | NA |
| sel | 39 (43.3%) | 39 (88.6%) | 0 (0%) | <0.001 |
| eta | 0 (0%) | 0 (0%) | 0 (0%) | NA |
| etb | 0 (0%) | 0 (0%) | 0 (0%) | NA |
| icaA | 88 (97.8%) | 42 (95.5%) | 46 (100%) | 0.236 |
| v8 | 90 (100%) | 44 (100%) | 46 (100%) | NA |
| ACME | ||||
| arcA | 90 (100%) | 44 (100%) | 46 (100%) | NA |
| opp-3c | 0 (0%) | 0 (0%) | 0 (0%) | NA |
ST, sequence type; SCCmec, staphylococcal cassette chromosome mec; agr, accessory gene regulator; spa, staphylococcal protein A; ACME, arginine catabolic mobile element; NA, not applicable.
All variables were summarized as the number of cases (%) in the table.
P values< 0.05 are shown in bold.
Adhesion factors facilitate the formation of biofilms, enabling MRSA to evade host defenses and impact its pathogenicity. Among the adhesion genes (clfA, clfB, fnbA, fnbB, sdrC, sdrD, sdrE, ebpS), the sdrE gene was significantly different between the two groups (81.8% vs 97.8%, P = 0.014) (Table 3). On the other hand, virulence genes impact infection severity, antibiotic response, and outcomes in CA-MRSA and HA-MRSA. No obvious differences in leukocidin genes (lukS-PV, lukF-PV) and hemolysin genes (hla, hlb, hld) were identified between ST5-MRSA and ST764-MRSA. Notably, ST764 lacked the superantigen genes tst, sec, and sel, whereas 88.6% (39/44) of ST5 strains expressed these genes; sea gene was expressed in 25.0% (11/44) of ST5-MRSA but none in ST764-MRSA (Table 3). On the other way, lacking in ST5, seb genes were identified in 93.5% (43/46) ST764-MRSA.
ACME arcA was detected in all ST5 and ST764 clinical strains, but no opp-3c gene, indicating both ST5-MRSA and ST764-MRSA were ACME-II' type.
Clinical outcomes and risk factors
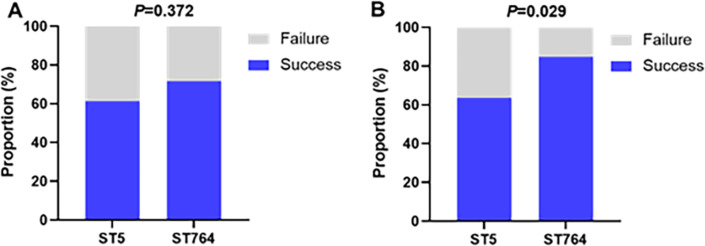
As for the clinical and bacterial efficacy of vancomycin treatment, the two clonal groups also showed different profiles (Fig. 3). The rate of improvement in clinical signs and symptoms was lower in the ST5-MRSA-infected patients compared with ST764, but not significantly different (61.4% vs 71.7%, P = 0.372). Bacterial eradication was also lower in ST5-MRSA patients with statistical significance against ST764 patients (63.6% vs 84.8%, P = 0.029).
Fig 3.
Clinical efficacy (A) and bacteriological efficacy (B) of vancomycin in the treatment of patients with ST5-MRSA and ST764-MRSA infections.
All significant variables identified from patient features, vancomycin PK/PD indices, and MRSA strain phenotype and genotype comparison were included in the correlation analysis (Fig. 4). Cmin was found to be positively correlated with AUC0-24/MIC, and the tst gene was negatively correlated with seb and positively correlated with hVISA/VISA phenotype. The prevalence of the tst gene was 68.0% (34/50) in hVISA/VISA strains and 12.5% (5/40) in VSSA strains. Subsequent multivariate analysis (Table 4) including all selected significant variables showed that the tst toxin was an outstanding risk factor for bacterial persistence with vancomycin [adjusted odds ratio (aOR), 4.509; 95% confidence interval (CI), 1.216 to 16.724, P = 0.024]. The Hosmer-Lemeshow goodness-of-fit test revealed a satisfactory model fit, with a P value of 0.575.
Fig 4.
Correlation analysis of variance variables.
TABLE 4.
Multivariate logistic regression of risk factors associated with bacterial persistence in patients with ST5-/ST764-MRSA infection after vancomycin treatment
| Risk factora | Univariate analysis result OR (95% CI) |
Multivariate analysis result | |
|---|---|---|---|
| aOR (95% CI) | P valueb | ||
| Age, years | 1.009 (0.984, 1.035) | 1.003 (0.970, 1.038) | 0.860 |
| Tracheotomy | 1.700 (0.610, 4.741) | 1.058 (0.323, 3.465) | 0.926 |
| Vancomycin daily dose | 0.910 (0.417, 1.985) | 0.824 (0.296, 2.300) | 0.312 |
| AUC0-24/MIC | 1.000 (0.999, 1.002) | 1.000 (0.999, 1.001) | 0.952 |
| Biofilm | 0.709 (0.329, 1.525) | 1.044 (0.435, 2.507) | 0.923 |
| sdrE | 1.225 (0.236, 6.367) | 1.132 (0.179, 7.159) | 0.895 |
| tst | 3.359 (1.245, 9.066) | 4.509 (1.216, 16.724) | 0.024 |
| sea | 0.614 (0.123, 3.075) | 0.341 (0.061, 1.916) | 0.222 |
ST, sequence type; AUC0-24/MIC, 24-hour area under the curve over minimal inhibitory concentration; OR, odds ratio; CI, confidence interval; aOR, adjusted odds ratio.
P values< 0.05 are shown in bold.
DISCUSSION
This study assessed the efficacy of vancomycin treatment against both ST5-MRSA and ST764-MRSA infections, finding that bacterial eradication was more difficult in the case of ST5-MRSA compared to ST764-MRSA. Between ST5-MRSA and ST764-MRSA infections, clinical characteristics differed in age, hospital setting, stroke, and tracheotomy occurrence for the two patient groups. Patients with ST764-MRSA were older compared to those with ST5-MRSA (77 vs 61 years, P < 0.001), which may be partially attributed to the fact that ST764/SCCmecII-MRSA carries features of CA-MRSA and is therefore more likely to spread, especially among older patients in the community and hospital settings. Baker et al. (17) investigated the risk factors for CA-MRSA infections in two hospitals in New York from 2006 to 2012, and multifactorial analysis revealed age ≥65 years (OR 2–3, 95% CI: 1.2–4.5) as one of the risk factors. In our study, ST5-MRSA mainly originated from tertiary hospitals, while ST764 was sourced from secondary regional hospitals. Previous studies, such as Peng et al. (18), similarly indicated a higher proportion of HA-MRSA in tertiary hospitals, with 660 HA-MRSA and 175 CA-MRSA strains among 835 MRSA isolates. Additionally, patients in the ST5-MRSA-infected group had higher rates of tracheotomy, possibly because these patients were more likely to develop hospital-acquired infections, similarly revealed in a retrospective case-control study that invasive medical procedures, including tracheal intubation and tracheotomy, were risk factors for hospital-acquired pneumonia (P < 0.001) (19).
In terms of bacterial susceptibility, the vancomycin MIC90 was 1 mg/L for both ST5 and ST764 strains. However, the MIC50 was 1 mg/L and 0.5 mg/L, respectively. Interestingly, the hVISA/VISA detection rate in ST5-MRSA isolates was significantly higher compared to ST764-MRSA, which has not been reported in previous studies. These findings suggest that ST764-MRSA exhibited greater sensitivity to vancomycin than ST5-MRSA. Biofilm expression of ST5-MRSA isolates was also significantly higher than ST764-MRSA (0.491 vs 0.304, P < 0.001), which is similar to those of Suzuki et al. (20).
The presence of an arginine deaminase system coding for ACME is one of the features of CA-MRSA, which carries arc and opp-3 regions in the classical USA300 (CC8 CA-MRSA) and has a potential role in enhancing the ability of pathogenic bacteria to grow and survive in the host (21). Both ST5-MRSA and ST764-MRSA in this study were ACME-II' type, carrying only the arcA gene and lacking the opp-3c gene. This is in line with Takano et al. (8) who first isolated and characterized seven strains of ST764/SCCmecII/t002, which carried the arcA gene and lacked the opp-3c gene, from community and hospital-acquired patients in Japan; however, the other seven strains of ST5/SCCmecII/t002 did not carry the aforementioned gene. Urushibara et al. (14) and Kawaguchiya et al. (22) found expression of arcA in ST5-MRSA, suggesting that the classical ST5 New York/Japan clone strain may also enhance growth ability in the host by acquiring ACME-II'.
Multivariate analysis suggests that virulence factors, such as the tst gene, may be an important risk factor responsible for differential vancomycin efficacy between ST5-MRSA and ST764-MRSA infections. Although tst, sea, seb, sec, and sel are all superantigen genes, seb is expressed in ST764-MRSA but not in ST5-MRSA. seb is thought to act as an immune evader during staphylococcal infections (23), thereby promoting infection disseminated in the community. For example, Xie et al. (24) found that the seb gene was detected in 52.2% of CA-MRSA isolates, but not in HA-MRSA, speculating that the seb gene may be a marker of CA-MRSA isolates. On the other hand, Ho et al. (25) detected that most of the ST5-SCCmecII isolates carried both sec and tst genes, but not in the ST239-SCCmecIII isolates of HA-MRSA. This suggests that tst and sec may be ST5-specific features, rather than characteristic genes of all HA-MRSA strains. Additionally, the absence of the pvl gene, a characteristic of CA-MRSA, was consistent with the literature (8) in the ST764-MRSA strains.
The difference in efficacy between ST5-MRSA and ST764-MRSA may also be related to the difference in vancomycin trough concentration and AUC0-24/MIC between the two groups. The 2020 vancomycin international consensus guidelines recommend maintaining vancomycin AUC0-24/MIC at 400–600 (15, 16). The 2020 Chinese vancomycin TDM guidelines also state that for adult patients with MRSA infection, vancomycin trough concentrations should be kept at 10–20 mg/L (26). In our analysis, the median vancomycin trough concentration in ST5-MRSA infection patients was 10.09 mg/L and 14.37 mg/L in the ST764-MRSA group. The median AUC0-24 was around 400 mg·h/L in both groups, although AUC0-24/MIC was within the recommended threshold, the differences were significant (500 vs 732, P < 0.001), indicating the MIC values contribute to the parameter difference. Multifactorial analysis showed that the genetic characteristics of ST5-MRSA and ST764-MRSA are risk factors for bacteriological failure after vancomycin treatment, warranting different vancomycin treatment strategies might be recommended for ST5-MRSA and ST764-MRSA.
Limitations of this study include the absence of psmα, hld, or ACME arcA expression measurements in ST764 and ST5. Further investigation of these virulence factors within our study is warranted.
In summary, there were significant differences in the characteristics of patients and pathogens between ST5-MRSA and ST764-MRSA infections, likely due to the fact that ST764-MRSA, while a variant of ST5-MRSA, exhibits features of CA-MRSA. Patients with ST5-MRSA infections had a lower bacterial eradication rate than those with ST764-MRSA infections. Moreover, virulence factors such as tst were found to be the risk factors for treatment failure in ST5 infections. These findings suggest the need for personalized vancomycin treatment strategies based on the genetic characteristics of MRSA isolates. Further studies are needed to confirm these results.
MATERIALS AND METHODS
Study design and bacterial isolates
This observational study was conducted at five hospitals from July 2012 to June 2020, using a database constituted of two prospective multicenter clinical studies that were approved by the Ethics Committee of Huashan Hospital and registered with the China Clinical Trials Registry (ChiCTR-OPC-16007920 and ChiCTR-OPC-17012567).
All clinical strains of S. aureus prior to vancomycin administration were collected and identical strains from the same patient were excluded. For patients diagnosed with pneumonia, eligible sputum specimens were defined as those with low magnification observation of squamous epithelial cells ≤10 and a minimum leukocyte count of ≥25. MIC of oxacillin and vancomycin were determined by agar dilution method at a CHINET microbiology laboratory (27). Oxacillin ≥ 4 mg/L was defined as MRSA.
Genotyping
The MRSA strains were extracted using the TaKaRa MiniBEST Bacteria Genomic DNA Extraction Kit Ver.3.0 (Takara Biomedical Technology Co., Ltd., Beijing, China) and stored at −70°C before sequencing. A 300-bp paired-end library was constructed using the NEXTflex DNA Sequencing Kit (Bio Scientific, AZ, USA), and 2 × 150-bp paired-end sequencing was performed on the Illumina X10 platform (Illumina, San Diego, CA, USA). Genome assembly was conducted using the Velvet 1.0.15 program (28), with various hash lengths and coverage cutoffs. Sequence data of seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, yqiL) were extracted and analyzed for multifocal sequence typing (MLST) according to the pubMLST database (https://pubmlst.org/). Only ST5-MRSA and ST764-MRSA strains were included in this study.
The molecular typing results of Takano et al. (8) showed that ST764 and ST5-MRSA were both SCCmecII and agrII. In this study, SCCmec type II (kdpC/B/E, GenBank accession number BA000018.3) and agrII (GenBank accession number AF001782) were used as reference for confirmation. The spa gene of the bacteria was extracted and classified as spa genotypes according to the Ridom SpaServer database (http://www.spaserver.ridom.de/). Virulence and adhesion genes were screened using the Virulence Factor Database (VFDB, http://www.mgc.ac.cn/VFs/main.htm). A total of 25 virulence or adhesion genes, commonly found in S. aureus (29), were included in the analysis. These genes were selected based on the factors that may influence vancomycin efficacy (30). ACME was identified by arcA (GenBank accession number BA000017.4: c2790552-2789317) and opp-3c (GenBank accession number MF346685.1:43104–43871). ACME-II' was defined as carrying only the arcA and not the opp-3c region (8, 21). BLAST software was used to find the similarity and respective lengths between sequences.
hVISA screening
As previously described, we used a modified population analysis area under the curve method (PAP-AUC) to screen for hVISA (31, 32). For classification, strains with ratios greater than 1.3 were categorized as VISA, ratios between 0.9 and 1.3 were classified as hVISA, and strains with ratios less than 0.9 were considered as vancomycin-susceptible S. aureus (VSSA).
Biofilm formation
Biofilm formation was determined by a crystalline violet method (32) and controlled against the ATCC29213 standard strain. Fresh overnight bacteria were selected, adjusted for turbidity to 0.5 McFarland, and shaken overnight at 37°C 180 rpm. Bacterial broth was diluted 1:100 and inoculated into BHI + 1% glucose broth after adding to 96-well Costa plates making three replicate wells per strain. After incubation at 37°C for 24 hours, the wells were washed three times with phosphate-buffered saline [(PBS); pH 7.2]. Methanol was used to stabilize the biofilm, followed by 15 minutes of staining with 1% crystal violet dye. The wells were washed with slow-flowing water until the water was colorless and dried at room temperature. Each well was dissolved by adding 0.2 mL of an aqueous 80% ethanol solution, and the optical density (OD) was measured at 570 nm on a spectrophotometer (model ELX800; BioTek, Winooski, VT, USA).
Patients and clinical data collection
Adult patients (age ≥18 years) with clinical signs, symptoms, laboratory tests, and microbiological culture results diagnosed with ST5-/ST764-MRSA infection and taking vancomycin for ≥5 days were included in the data set. Patients were excluded if they had received any other anti-Gram-positive agents besides vancomycin for ≥24 hours within 72 hours prior to admission, if Gram-positive bacteria colonized, and if they were pregnant or lactating women or if they were taking concomitant aminoglycosides. Patient demographics, ICU admissions, underlying diseases, curative treatment, clinical laboratory tests, bacteriological tests, and vancomycin doses were collected. In this study, we included underlying diseases such as cardiovascular diseases, diabetes, and strokes, as they have the potential to impact the efficacy of anti-infection therapy (32, 33).
Vancomycin TDM and PK/PD analysis
Serum Cmin samples were collected no earlier than 0.5 hours prior to the fifth dose, and Cmax samples were collected 0.5–1 hour after dosing. If the patient’s creatine clearance (CLCr) was less than 30 mL/min, a serum sample was collected at the second dose. Vancomycin serum concentrations were determined by fluorescence polarization immunoassay (FPIA) or chemiluminescent microparticle immunoassay (CMIA) with a linear range of 3 to 100 mg/L.
A previously published method (32) was used to simulate the concentration-time profile of patients and calculate the PK/PD indices of patients after vancomycin dosing. Cmin, Cmax, and AUC0-24 values were calculated as weighted means using the dose and interval of administration, respectively. AUC0-24/MIC was evaluated based on the calculated AUC0-24 and laboratory vancomycin MIC values.
Clinical outcome definitions
Clinical outcomes include clinical efficacy (improvement in clinical signs and symptoms) and bacteriological efficacy (eradication of bacteria). Improvement in clinical signs and symptoms means that the patient’s clinical signs and symptoms of infection and laboratory test results (other than bacteriological tests) have returned to normal or pre-infection status, and vancomycin is no longer required for 7 days since vancomycin discontinuation. The assessment of clinical signs and symptoms may vary depending on the type of infection. For instance, in patients with pneumonia, typical clinical signs and symptoms include fever, cough, sputum production, both dry and moist rales, and observable changes in imaging. Laboratory tests such as white blood cell count, neutrophil count, and serum creatinine values are also considered. Bacterial eradication means that the original pathogen cannot be cultured in 7 days after stopping vancomycin treatment and antibiotics against Gram-positive organisms are no longer required.
Statistical analysis
All statistical analyses were performed using SPSS 19 (SAS Institute, Cary, NC, USA), and figures were plotted using GraphPad Prism software version 8.4.3 (GraphPad Software, LLC, San Diego, CA, USA). Categorical variables were presented in descriptive statistics as number of cases (percent %) and continuous variables as median [interquartile range (IQR)]. Univariate comparisons were performed using Fisher’s exact test for categorical variables and Mann-Whitney U test for continuous variables. P values less than 0.05 were considered statistically significant. All significant variables were included in the correlation analysis using the OriginPro software version 2023 (OriginLab Corporation, OriginLab Corporation, MA, USA). Variables with correlation coefficients below 0.5 and deemed potentially relevant to the treatment outcomes were included in the final multivariate analysis. The adequacy of the model fit was assessed using the Hosmer-Lemeshow goodness-of-fit test, with a P value exceeding 0.05 indicating a satisfactory model fit.
ACKNOWLEDGMENTS
We sincerely thank Hai-Lan Wu, Yu Wang, Yi Li, and Xin Li for their help in the TDM of vancomycin and Ji-Cheng Yu, Yang Yang, Prof. Yan Guo, and Prof. De-Mei Zhu for support with MIC determination.
This study was supported by the National Natural Science Foundation of China (82204467), Research Startup Fund of Huashan Hospital, Fudan University (2021QD033) and Municipal Hospital Emerging Frontier Technology Joint Research Project of Shanghai Shenkang Development Center (SHDC12020106).
Contributor Information
Jing Zhang, Email: zhangj_fudan@aliyun.com.
David S. Perlin, Hackensack Meridian Health Center for Discovery and Innovation, Nutley, New Jersey, USA
REFERENCES
- 1. Wang F-D, Chen Y-Y, Chen T-L, Liu C-Y. 2008. Risk factors and mortality in patients with nosocomial Staphylococcus aureus bacteremia. Am J Infect Control 36:118–122. doi: 10.1016/j.ajic.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 2. David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687. doi: 10.1128/CMR.00081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Self WH, Wunderink RG, Williams DJ, Zhu Y, Anderson EJ, Balk RA, Fakhran SS, Chappell JD, Casimir G, Courtney DM, Trabue C, Waterer GW, Bramley A, Magill S, Jain S, Edwards KM, Grijalva CG. 2016. Staphylococcus aureus community-acquired pneumonia: prevalence, clinical characteristics, and outcomes. Clin Infect Dis 63:300–309. doi: 10.1093/cid/ciw300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lakhundi S, Zhang K. 2018. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 31:e00020-18. doi: 10.1128/CMR.00020-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Y, Sun L, Ba X, Jiang S, Zhuang H, Zhu F, Wang H, Lan P, Shi Q, Wang Z, Chen Y, Shi K, Ji S, Jiang Y, Holmes MA, Yu Y. 2022. Epidemiology, evolution and cryptic susceptibility of methicillin-resistant Staphylococcus aureus in China: a whole-genome-based survey. Clin Microbiol Infect 28:85–92. doi: 10.1016/j.cmi.2021.05.024 [DOI] [PubMed] [Google Scholar]
- 6. Wu D, Wang Z, Wang H, Sun L, Chen Y, Ji S, Shi K, Yu Y. 2018. Predominance of ST5-II-t311 clone among healthcare-associated methicillin-resistant Staphylococcus aureus isolates recovered from Zhejiang, China. Int J Infect Dis 71:107–112. doi: 10.1016/j.ijid.2018.04.798 [DOI] [PubMed] [Google Scholar]
- 7. Dai Y, Liu J, Guo W, Meng H, Huang Q, He L, Gao Q, Lv H, Liu Y, Wang Y, Wang H, Liu Q, Li M. 2019. Decreasing methicillin-resistant Staphylococcus aureus (MRSA) infections is attributable to the disappearance of predominant MRSA ST239 clones, Shanghai, 2008-2017. Emerg Microbes Infect 8:471–478. doi: 10.1080/22221751.2019.1595161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takano T, Hung W-C, Shibuya M, Higuchi W, Iwao Y, Nishiyama A, Reva I, Khokhlova OE, Yabe S, Ozaki K, Takano M, Yamamoto T. 2013. A new local variant (ST764) of the globally disseminated ST5 lineage of hospital-associated methicillin-resistant Staphylococcus aureus (MRSA) carrying the virulence determinants of community-associated MRSA. Antimicrob Agents Chemother 57:1589–1595. doi: 10.1128/AAC.01147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ozaki K, Takano M, Higuchi W, Takano T, Yabe S, Nitahara Y, Nishiyama A, Yamamoto T. 2009. Genotypes, intrafamilial transmission, and virulence potential of nasal methicillin-resistant Staphylococcus aureus from children in the community. J Infect Chemother 15:84–91. doi: 10.1007/s10156-009-0668-x [DOI] [PubMed] [Google Scholar]
- 10. Nakaminami H, Noguchi N, Ito A, Ikeda M, Utsumi K, Maruyama H, Sakamoto H, Senoo M, Takasato Y, Nishinarita S. 2014. Characterization of methicillin-resistant Staphylococcus aureus isolated from tertiary care hospitals in Tokyo, Japan. J Infect Chemother 20:512–515. doi: 10.1016/j.jiac.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 11. Chen S, Jin Y, Lin C, Hao Z, Duan J, Guo Y, Wang S, Hu L, Wang L, Yu F. 2019. Low prevalence of mupirocin resistance among Staphylococcus aureus clinical isolates from a Chinese tertiary hospital. J Med Microbiol 68:201–205. doi: 10.1099/jmm.0.000911 [DOI] [PubMed] [Google Scholar]
- 12. Wang B, Xu Y, Zhao H, Wang X, Rao L, Guo Y, Yi X, Hu L, Chen S, Han L, Zhou J, Xiang G, Hu L, Chen L, Yu F. 2022. Methicillin-resistant Staphylococcus aureus in China: a multicentre longitudinal study and whole-genome sequencing. Emerg Microbes Infect 11:532–542. doi: 10.1080/22221751.2022.2032373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang H, Tian L, Chen T, Chen W, Ge Y, Bi J, Fang Z, Chen M. 2022. Prevalence and WGS-based characteristics of MRSA isolates in hospitals in Shanghai, China. Front Microbiol 13:1002691. doi: 10.3389/fmicb.2022.1002691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Urushibara N, Kawaguchiya M, Onishi M, Mise K, Aung MS, Kobayashi N. 2016. Novel structures and temporal changes of arginine catabolic mobile elements in methicillin-resistant Staphylococcus aureus genotypes ST5-MRSA-II and ST764-MRSA-II in Japan. Antimicrob Agents Chemother 60:3119–3122. doi: 10.1128/AAC.02356-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rybak MJ, Le J, Lodise TP, Levine DP, Bradley JS, Liu C, Mueller BA, Pai MP, Wong-Beringer A, Rotschafer JC, Rodvold KA, Maples HD, Lomaestro B. 2020. Therapeutic monitoring of vancomycin for serious methicillin-resistant Staphylococcus aureus infections: a revised consensus guideline and review by the American Society of Health-System Pharmacists, the Infectious Diseases Society of America, the Pediatric Infectious Diseases Society, and the Society of Infectious Diseases Pharmacists. Clin Infect Dis 71:1361–1364. doi: 10.1093/cid/ciaa303 [DOI] [PubMed] [Google Scholar]
- 16. Rybak MJ, Le J, Lodise TP, Levine DP, Bradley JS, Liu C, Mueller BA, Pai MP, Wong-Beringer A, Rotschafer JC, Rodvold KA, Maples HD, Lomaestro BM. 2020. Therapeutic monitoring of vancomycin for serious methicillin-resistant Staphylococcus aureus infections: a revised consensus guideline and review by the American Society of Health-System Pharmacists, the Infectious Diseases Society of America, the Pediatric Infectious Diseases Society, and the Society of Infectious Diseases Pharmacists. Am J Health Syst Pharm 77:835–864. doi: 10.1093/ajhp/zxaa036 [DOI] [PubMed] [Google Scholar]
- 17. Baker P, Cohen B, Liu J, Larson E. 2016. Incidence and risk factors for community-associated methicillin-resistant Staphylococcus aureus in New York City, 2006-2012. Epidemiol Infect 144:1014–1017. doi: 10.1017/S095026881500196X [DOI] [PubMed] [Google Scholar]
- 18. Peng H, Liu D, Ma Y, Gao W. 2018. Comparison of community- and healthcare-associated methicillin-resistant Staphylococcus aureus isolates at a Chinese tertiary hospital, 2012-2017. Sci Rep 8:17916. doi: 10.1038/s41598-018-36206-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang H, Fan Y, Li C, Zhang M, Liu W. 2022. A retrospective study on risk factors and disease burden for hospital-acquired pneumonia caused by multi-drug-resistant bacteria in patients with intracranial cerebral hemorrhage. Neurol Sci 43:2461–2467. doi: 10.1007/s10072-021-05721-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suzuki T, Yamamoto T, Kaito C, Miyamoto H, Ohashi Y. 2016. Impact of psm-mec in methicillin-resistant Staphylococcus aureus (ST764) strains isolated from keratitis patients. Microb Drug Resist 22:589–597. doi: 10.1089/mdr.2015.0315 [DOI] [PubMed] [Google Scholar]
- 21. Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 22. Kawaguchiya M, Urushibara N, Ghosh S, Kuwahara O, Morimoto S, Ito M, Kudo K, Kobayashi N. 2013. Genetic diversity of emerging Panton-Valentine leukocidine/arginine catabolic mobile element (ACME)-positive ST8 SCCmec-IVa meticillin-resistant Staphylococcus aureus (MRSA) strains and ACME-positive CC5 (ST5/ST764) MRSA strains in Northern Japan. J Med Microbiol 62:1852–1863. doi: 10.1099/jmm.0.062125-0 [DOI] [PubMed] [Google Scholar]
- 23. Vojtov N, Ross HF, Novick RP. 2002. Global repression of exotoxin synthesis by staphylococcal superantigens. Proc Natl Acad Sci U S A 99:10102–10107. doi: 10.1073/pnas.152152499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xie X, Bao Y, Ouyang N, Dai X, Pan K, Chen B, Deng Y, Wu X, Xu F, Li H, Huang S. 2016. Molecular epidemiology and characteristic of virulence gene of community-acquired and hospital-acquired methicillin-resistant Staphylococcus aureus isolates in Sun Yat-Sen Memorial Hospital, Guangzhou, Southern China. BMC Infect Dis 16:339. doi: 10.1186/s12879-016-1684-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ho C-M, Ho M-W, Lee C-Y, Tien N, Lu J-J. 2012. Clonal spreading of methicillin-resistant SCCmec Staphylococcus aureus with specific spa and dru types in central Taiwan. Eur J Clin Microbiol Infect Dis 31:499–504. doi: 10.1007/s10096-011-1338-3 [DOI] [PubMed] [Google Scholar]
- 26. He N, Su S, Ye Z, Du G, He B, Li D, Liu Y, Yang K, Zhang X, Zhang Y, et al. 2020. Evidence-based guideline for therapeutic drug monitoring of vancomycin: 2020 update by the Division of Therapeutic Drug Monitoring, Chinese Pharmacological Society. Clin Infect Dis 71:S363–S371. doi: 10.1093/cid/ciaa1536 [DOI] [PubMed] [Google Scholar]
- 27. Clinical and Laboratory Standards Institute . 2022. Approved standard; 32th informational supplement. Edited by W P. A.. Performance standards for antimicrobial susceptibility testing [Google Scholar]
- 28. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peacock SJ, Moore CE, Justice A, Kantzanou M, Story L, Mackie K, O’Neill G, Day NPJ. 2002. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect Immun 70:4987–4996. doi: 10.1128/IAI.70.9.4987-4996.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chong YP, Park K-H, Kim ES, Kim M-N, Kim S-H, Lee S-O, Choi S-H, Jeong J-Y, Woo JH, Kim YS. 2015. Clinical and microbiologic analysis of the risk factors for mortality in patients with heterogeneous vancomycin-intermediate Staphylococcus aureus bacteremia. Antimicrob Agents Chemother 59:3541–3547. doi: 10.1128/AAC.04765-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wootton M, Howe RA, Hillman R, Walsh TR, Bennett PM, MacGowan AP. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother 47:399–403. doi: 10.1093/jac/47.4.399 [DOI] [PubMed] [Google Scholar]
- 32. Fan Y-X, Chen M-T, Li N-Y, Liu X-F, Yang M-J, Chen Y-C, Liang X-Y, Wu J-F, Guo B-N, Song S-C, Zhu Y-Q, Zhang F-Y, Hang J-Q, Wu S-B, Shen B, Li H-Y, Wang Q, Luo X-M, Chen Q-G, Zhang H-F, Wang R-L, Shen L-H, Fu F-M, Song X-L, Zhang J. 2022. Sequence type 5 (ST5) as a possible predictor of bacterial persistence in adult patients with methicillin-resistant Staphylococcus aureus pneumonia treated with vancomycin. Microbiol Spectr 10:e0134822. doi: 10.1128/spectrum.01348-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dhainaut J-F, Claessens Y-E, Janes J, Nelson DR. 2005. Underlying disorders and their impact on the host response to infection. Clin Infect Dis 41 Suppl 7:S481–S489. doi: 10.1086/432001 [DOI] [PubMed] [Google Scholar]