Abstract
Pathogens generate ubiquitous selective pressures and host-pathogen interactions alter social behaviors in many animals1–4. However, very little is known about the neuronal mechanisms underlying pathogen-induced changes in social behavior. Here we show that in adult Caenorhabditis elegans hermaphrodites, exposure to a bacterial pathogen (Pseudomonas aeruginosa) modulates sensory responses to pheromones by inducing the expression of a chemoreceptor STR-44 to promote mating. Under standard conditions, C. elegans hermaphrodites avoid a mixture of ascaroside pheromones to facilitate dispersal5–13. We find that exposure to the pathogenic Pseudomonas bacteria enables pheromone responses in AWA sensory neurons, which mediate attractive chemotaxis, to suppress the avoidance. Pathogen exposure induces str-44 expression in AWA, a process regulated by a transcription factor zip-5 that also displays a pathogen-induced increase in AWA expression. STR-44 acts as a pheromone receptor and its function in AWA is required for pathogen-induced AWA pheromone response and suppression of pheromone avoidance. Furthermore, we show that C. elegans hermaphrodites, which reproduce mainly through self-fertilization, increase the rate of mating with males after pathogen exposure and this increase requires str-44 in AWA. Thus, our results discover a causal mechanism for pathogen-induced social behavior plasticity, which can promote genetic diversity and facilitate adaptation of the host animals.
Exposure to pathogens and parasites alters social behavior, such as mating, aggregation, aggression, and communication, in many animals, including humans. These behavioral modulations immediately impact the resistance and reproduction of individual animals and shape the adaptation and evolution of animal species in the long run1–4. However, the pathway from pathogen exposure to the social behavior of the host animals is largely unknown. As a free-living worm that feeds on bacteria, C. elegans encounters various pathogenic and nonpathogenic bacteria in its habitats and provides an opportunity to study how host-pathogen interactions regulate the activity and behavioral output of the nervous system14.
Pathogen exposure alters pheromone avoidance
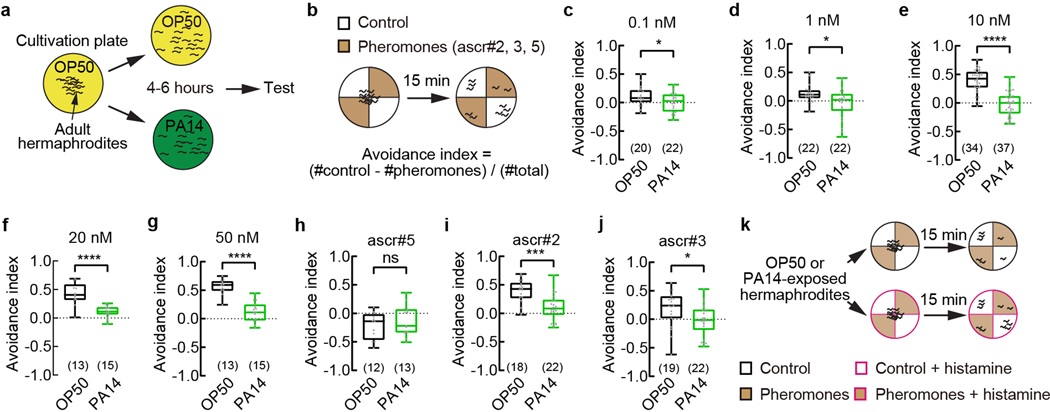
C. elegans hermaphrodites secrete a blend of small-molecule pheromones, including several ascarosides that repel hermaphrodites, attract males, or signal population density5–13,15–21. Under the standard conditions wild-type adult hermaphrodites cultivated on benign bacteria strain Escherichia Coli OP50 avoid the mixture of three ascarosides, ascr#2 (asc-C6-MK; C6), ascr#3 (asc-ΔC9; C9), and ascr#5 (asc-ωC3; C3), at nanomolar to micromolar concentrations5–13. Surprisingly, we found that after feeding on a gram-negative pathogenic bacteria strain Pseudomonas aeruginosa PA14 for 4–6 hours, the adult hermaphrodites avoided the pheromones much less and sometimes were even attracted to them (Fig. 1a, Extended Data Fig. 1a,b). The difference between the avoidance of the ascarosides in OP50-exposed hermaphrodites and in PA14-exposed hermaphrodites indicated the modulation of the pheromone response (Fig. 1a). Exposure to PA14 suppresses the avoidance of the pheromone mixture over a wide range of concentrations and the avoidance of ascr#2 and ascr#3 individually (Extended Data Fig. 1c–j). In contrast, starvation or food scarcity enhanced the avoidance of ascr#3 in adult hermaphrodites and induced the avoidance of another ascaroside, osas#9 9,11,22. Ingestion of PA14 leads to intestinal infection and death of the worms in a few days23. Thus, our results identified a type of social behavioral plasticity induced by exposure to pathogenic bacteria to suppress pheromone avoidance in C. elegans hermaphrodites.
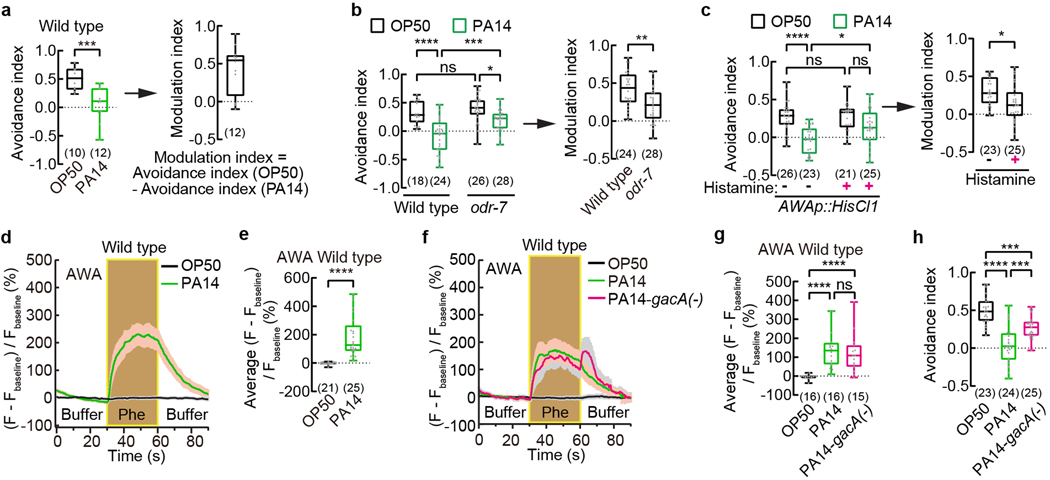
Fig. 1. Exposure to pathogenic Pseudomonas strain PA14 suppresses pheromone avoidance in hermaphrodites and induces pheromone response in AWA.
a - c, Avoidance of ascaroside pheromones when exposed to E. coli OP50 or P. aeruginosa PA14 (avoidance index) and modulation of pheromone response by PA14 (modulation index) in wild-type (a) and odr-7 mutant (b) adult C. elegans hermaphrodites, and in transgenic adult hermaphrodites expressing HisCl1 in AWA (c). Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance. Positive modulation index, suppression of avoidance by PA14.
d – g, Traces of GCaMP6 signals in AWA neurons of wild-type adult hermaphrodites in response to ascaroside pheromones when exposed to different bacteria (d,f) and quantitation of average GCaMP6 signals during pheromone stimulation in the previous panel (e,g). Phe, Pheromone mixture of ascr#2,3,5 (1 μM each). Lines in traces, mean. Shades, s.e.m.. Fbaseline, average GCaMP6 signals in the first 30 seconds.
h, Avoidance of ascaroside pheromones in wild-type adult hermaphrodites exposed to E. coli OP50 or PA14 or PA14-gacA(−). Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance.
Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual assays (a-c,h) or number of individual worms (e,g). Dots, avoidance indexes or modulation indexes of individual assays (a-c,h) or average GCaMP6 signals of individual worms during pheromone stimulation (e,g). P values are derived from two-tailed unpaired t test (avoidance index in a, modulation index in b and c) or Two-way ANOVA with Tukey’s multiple comparisons test (avoidance index in b and c) or two-tailed Mann-Whitney test (e) or Kruskal-Wallis test with Dunn’s multiple comparisons test (g) or One-way ANOVA with Tukey’s multiple comparisons test (h), asterisks indicate significant difference, **** P < 0.0001, *** P < 0.001, ** P < 0.01, * P < 0.05; ns, not significant. P values are shown in Source data.
Pathogens induce AWA pheromone response
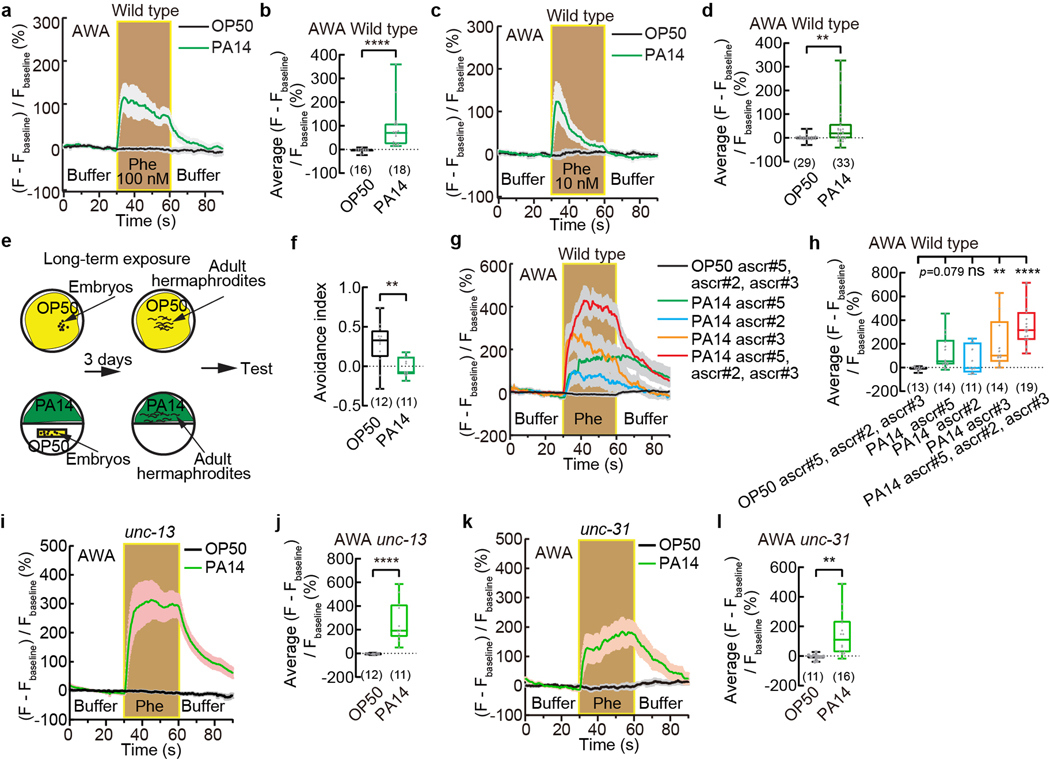
To identify mechanisms underlying pathogen-induced modulation of pheromone response, we first asked which sensory neurons were needed. We found that odr-7(ky4) mutant worms, which were defective in cell-fate determination of AWA olfactory sensory neurons24, avoided the ascaroside pheromones similarly as wild type when exposed to OP50; however, after PA14 exposure odr-7(ky4) mutants avoided the pheromones significantly more than wild type and generated a reduced modulation (Fig. 1b). Inhibiting AWA activity during chemotaxis assay by treating transgenic worms that selectively expressed a histamine-gated chloride channel HisCl1 in AWA25 with histamine does not alter the pheromone response when the transgenic worms are exposed to OP50 but significantly disrupts the pheromone response after PA14 exposure, resulting in a reduced modulation index (Fig. 1c, Extended Data Fig. 1k, Methods). These results together indicate that chemosensory neurons AWA regulate PA14-induced pheromone response.
AWA sensory neurons detect and mediate chemotactic responses to attractive odorants24. Using transgenic worms that selectively expressed a calcium-sensitive fluorescent reporter GCaMP6 in AWA we previously showed that in adult C. elegans hermaphrodites cultivated on E. coli OP50, AWA did not respond to the mixture of ascarosides ascr#2, ascr#3, and ascr#525 (Fig. 1d,e, Methods). However, after exposure to PA14 for 4–6 hours, pheromone stimulation elicited strong calcium transients in AWA that lasted throughout the duration of stimulation over a wide range of pheromone concentrations (Fig. 1d,e, Extended Data Fig. 2a–d). A long-term exposure to the pathogen also suppresses the avoidance of the pheromones and generated strong responses to the pheromone mixture and to ascr#3 alone in AWA (Extended Data Fig. 2e–h). Disrupting synaptic neurotransmission by inactivating unc-1326 or blocking neuropeptide release by inactivating unc-3126 had no effect on the pathogen-induced pheromone response in AWA (Extended Data Fig. 2i–l), suggesting that in PA14-exposed worms, AWA neurons act cell-autonomously to generate the pheromone response. AWA detect and respond to the attractive odorant diacetyl with increased intracellular calcium transients and activation of AWA promotes chemotactic movements towards the odorant27. These findings together with our results show that exposure to the bacterial pathogen PA14 induces the sensory response of AWA to the pheromones to promote attractive chemotaxis and antagonize the avoidance of the pheromones.
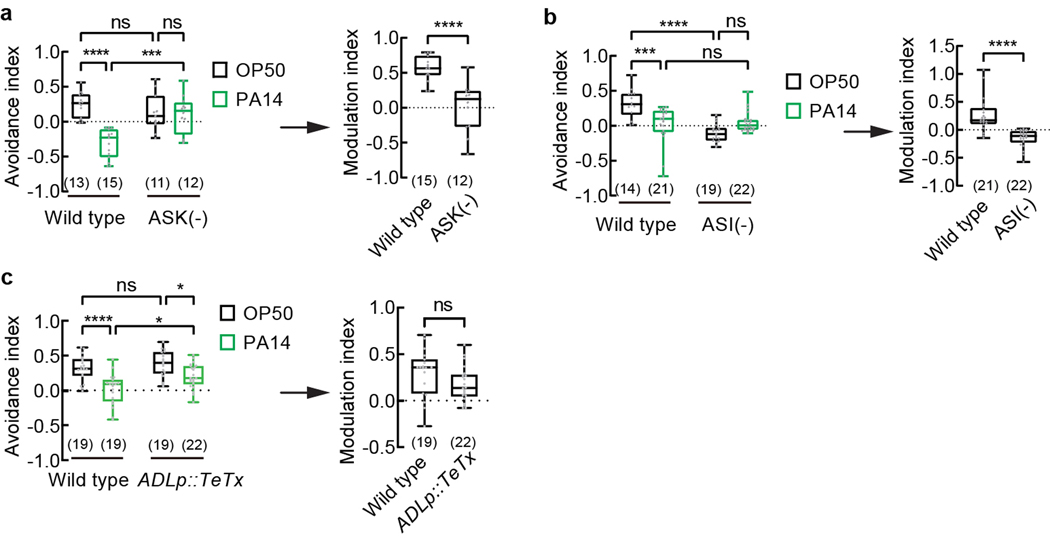
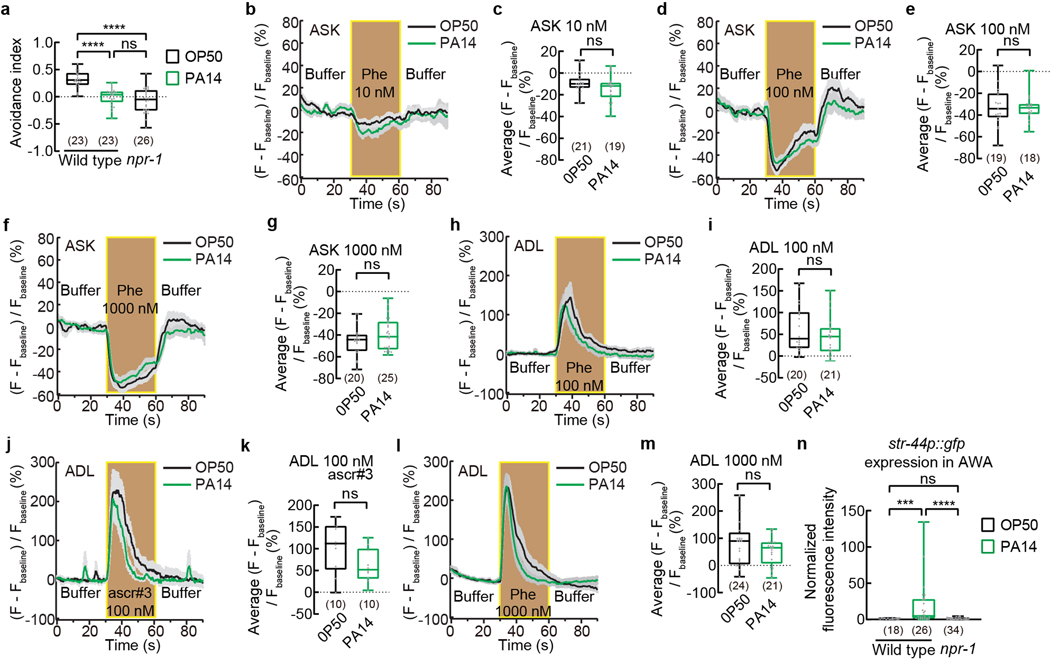
Previous studies showed that the chemosensory neurons ASK responded to the mixture of ascr#2, ascr#3, and ascr#5 and regulated pheromone attraction or avoidance in adult hermaphrodites of various genetic backgrounds7,8,10,13,21. We found that genetic ablation of ASK did not significantly alter the avoidance of the pheromones in OP50-exposed adult hermaphrodites, consistent with previous results8, but disrupted PA14-induced suppression of pheromone avoidance (Extended Data Fig. 3a), implicating ASK in pathogen-induced pheromone response. The chemosensory neurons ASI inhibit dauer formation via the function of several pheromone receptors and inhibit the attraction of adult hermaphrodites to hermaphrodite-secreted pheromones5,12,13,18,20,21,28. Consistently, we found that ablating ASI strongly suppressed the avoidance of the pheromones in OP50-exposed hermaphrodites (Extended Data Fig. 3b), which made it difficult to conclude on the effect of pathogen exposure on ASI-ablated worms. The sensory neurons ADL regulated the acute reversal response to ascr#3 in hermaphrodites8,11. Blocking neurotransmission of ADL did not disrupt the chemotactic response to the pheromone mixture in worms exposed to either OP50 or PA14 (Extended Data Fig. 3c), consistent with a relatively selective response of ADL to ascr#38.
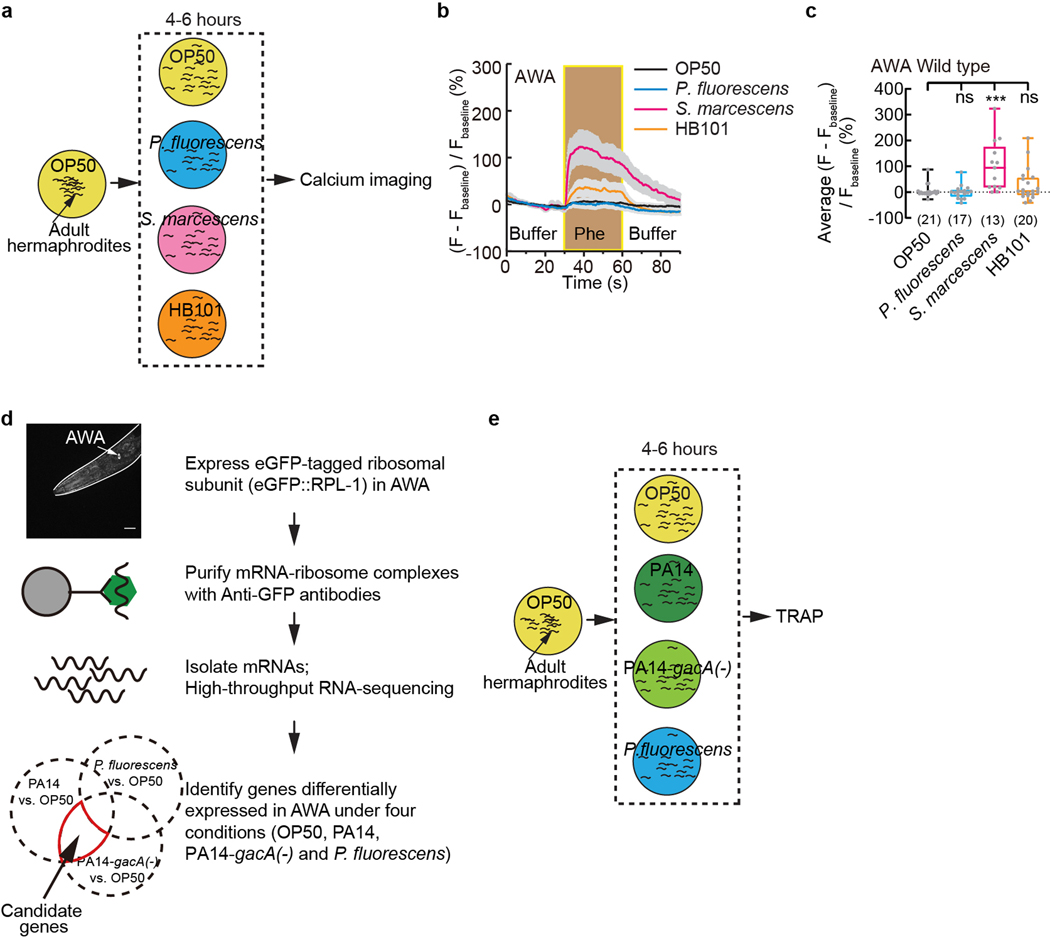
By testing several other bacterial strains, we found that another benign E. coli strain HB101 or a nonvirulent Pseudomonas fluorescens bacteria strain ATCC13525 did not induce a pheromone response in AWA; but a pathogenic Serratia marcescens bacteria strain ATCC1388029 significantly induced the response (Extended Data Fig. 4a–c). In addition, we found that an experimentally generated mutant strain of PA14 that showed reduced virulence, PA14-gacA(−)30, induced the pheromone response in AWA and suppressed the pheromone avoidance in hermaphrodites (Fig. 1f,g). gacA is required for the full pathogenicity of PA14 and inactivating gacA in PA14 does not completely abolish the virulence of PA1430. These results suggest a correlation between the pathogenicity of a bacterial strain and its ability to induce a pheromone response in AWA.
PA14 induces str-44 expression in AWA
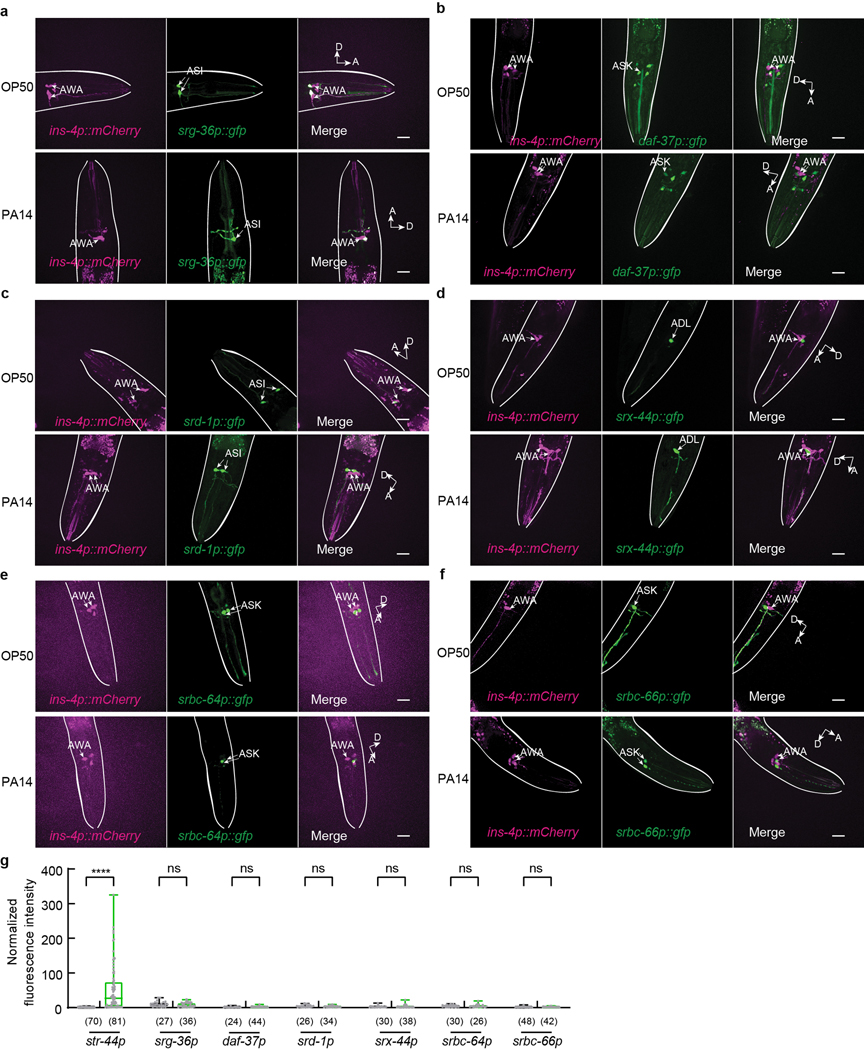
To characterize how pathogen exposure enables the pheromone response in AWA neurons, we examined the expression of previously identified pheromone receptors. Two serpentine receptor class g chemoreceptors that are expressed in sensory neurons ASI, srg-36 and srg-37, function as receptors for ascr#5 to regulate dauer formation20. We found that exposure to PA14 did not induce the expression of a transcriptional reporter srg-36p::gfp in AWA (Extended Data Fig. 5a,g). daf-37, a G-protein coupled pheromone receptor for ascr#2, acts in ASI to regulate dauer formation and in the sensory neurons ASK to mediate the avoidance of ascr#2 in adult hermaphrodites10,21. Exposure to PA14 also did not induce daf-37 expression in AWA (Extended Data Fig. 5b,g). srd-1 is expressed in AWA in C. elegans males to regulate attraction to hermaphrodite-conditioned media. The expression of srd-1 in hermaphrodites is restricted to ASI31 and is not induced in AWA in PA14-exposed hermaphrodites (Extended Data Fig. 5c,g). In addition, we found that expression of srx-44, a chemoreceptor that regulated responses to a dauer-inducing pheromone icas#916,17, and the expression of srbc-64 and srbc-66, two G-protein coupled receptors (GPCRs) expressed in ASK to mediate dauer formation in response to ascr#1, ascr#2, ascr#3, and ascr#519, were also not induced in AWA after exposure to PA14 (Extended Data Fig. 5d–g). Together, these results suggest that none of these characterized chemoreceptors regulates pathogen-induced AWA response to the mixture of ascr#2, ascr#3, and ascr#5 in adult hermaphrodites.
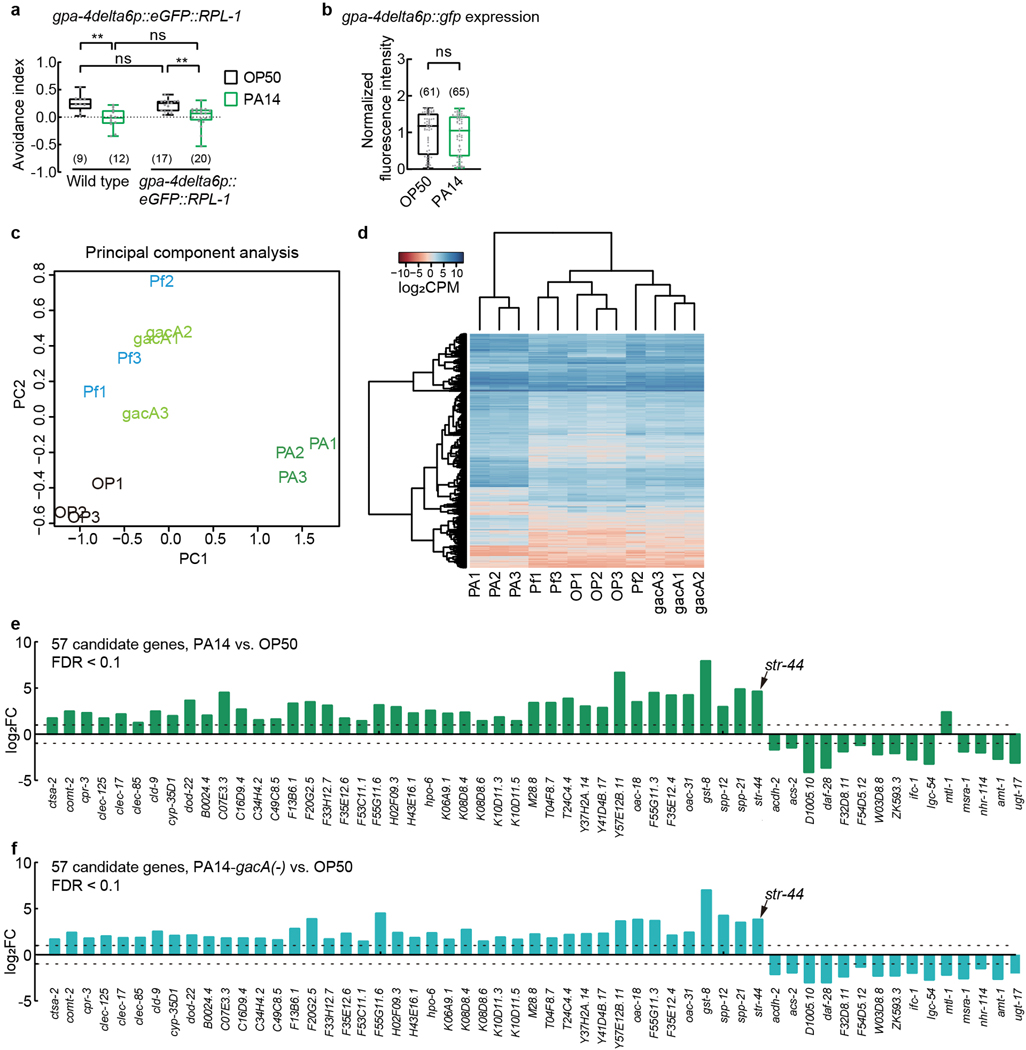
Because previously identified receptors for ascarosides are chemoreceptors16,17,19–22,31, we reasoned that exposure to either PA14 or PA14-gacA(−) (Fig. 1f–h) would induce the expression of an unknown chemoreceptor that elicits the pheromone response of AWA to suppress avoidance, but exposure to P. fluorescens would not (Extended Data Fig. 4b,c). Thus, we performed Translating Ribosome Affinity Purification (TRAP)32 using a transgenic strain that selectively expressed in AWA the ribosomal protein large subunit RPL10a, RPL-1 in C. elegans, tagged with a green fluorescent protein (eGFP) using an AWA-specific promoter (Extended Data Fig. 4d). We first confirmed that PA14 exposure suppressed pheromone avoidance in these transgenic hermaphrodites without altering the activity of the promoter (Extended Data Fig. 6a,b). Next, we cultivated the transgenic hermaphrodites on E. coli OP50 until the adult stage, separated the worms into 4 groups and respectively exposed them to E. coli OP50, PA14, PA14-gacA(−), or P. fluorescens. After 4–6 hours, we collected the worms to isolate actively translated messenger RNAs (mRNAs) from ribosomes to generate cDNA libraries (Extended Data Fig. 4e). We used massively parallel sequencing to sequence 12 cDNA libraries generated from 3 independent experiments. We found that 66% of 4583 AWA-expressed genes detected with expression level more than 10 in a previous gene expression study33 were also detected with more than 10 average counts per million in our cDNA libraries generated from OP50-exposed worms, which validated our approach to identify mRNAs expressed in AWA.
We found that 785 genes were differentially expressed in AWA between PA14-exposed worms and OP50-exposed worms and 103 or 59 genes were respectively differentially expressed in AWA between PA14-gacA(−)-exposed worms or P. fluorescens-exposed worms and OP50-exposed worms (Fig. 2a–c, Supplementary Data 1–3). Principal component analysis and hierarchical clustering based on the expression of these differentially expressed genes separated PA14-exposed worms from the worms under the other three conditions, among which PA14-gacA(−)-exposed worms are more different from OP50-exposed worms than from P. fluorescens-exposed worms (Extended Data Fig. 6c,d). Thus, although exposure to PA14-gacA(−) induces a pheromone response in AWA and suppresses pheromone avoidance, its effect on gene expression is different from exposure to PA14, consistent with the reduced virulence of PA14-gacA(−) bacteria30. The analysis on GO term and KEGG pathways identified enrichment of several signaling processes, including innate immune response, fatty acid metabolism, and stress response in PA14-regulated genes (Supplementary Data 4).
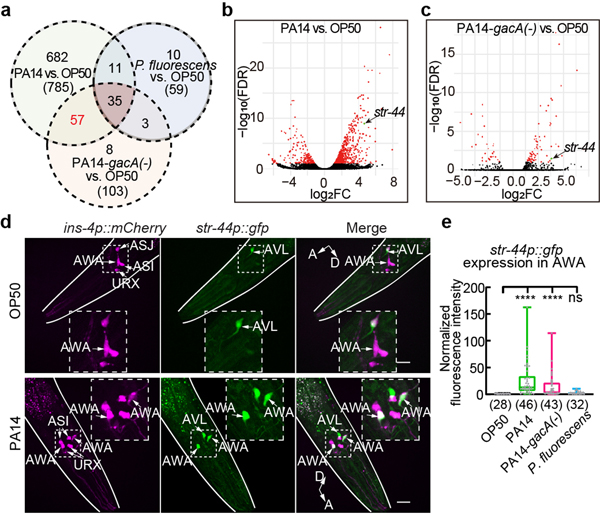
Fig. 2. Pathogenic Pseudomonas PA14 induces expression of str-44 in AWA.
a, Venn diagram showing numbers of genes differentially expressed in AWA between worms exposed to PA14 or PA14-gacA(−) or P. fluorescens and worms exposed to E. coli OP50 (FDR < 0.1). Numbers in parenthesis, total number of genes in each category.
b, c, Volcano plots showing gene expression difference in AWA between PA14-exposed worms and OP50-exposed worms (b) or between PA14-gacA(−)-exposed worms and OP50-exposed worms (c). Genes with FDR < 0.1 are highlighted in red, otherwise highlighted in black; str-44 is highlighted in green. FC, fold change.
d, Sample images for str-44p::gfp expression in adult hermaphrodites exposed to OP50 or PA14 for 4–6 hours. Dashed lines indicate enlarged views. Arrows indicate neuronal cell bodies. Lines outline worm bodies. Scale bar, 20 μm. A, anterior. D, dorsal (points slightly out of page in lower panel).
e, Quantitation of str-44p::gfp signal in AWA in hermaphrodites exposed to different bacteria. Intensity of signals is normalized using average intensity of str-44p::gfp in OP50-exposed worms measured in parallel. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual neurons. Dots, signals of individual neurons. P values are derived from Kruskal-Wallis test with Dunn’s multiple comparisons test, asterisks indicate significant difference, **** P < 0.0001, ns, not significant. P values are shown in Source data.
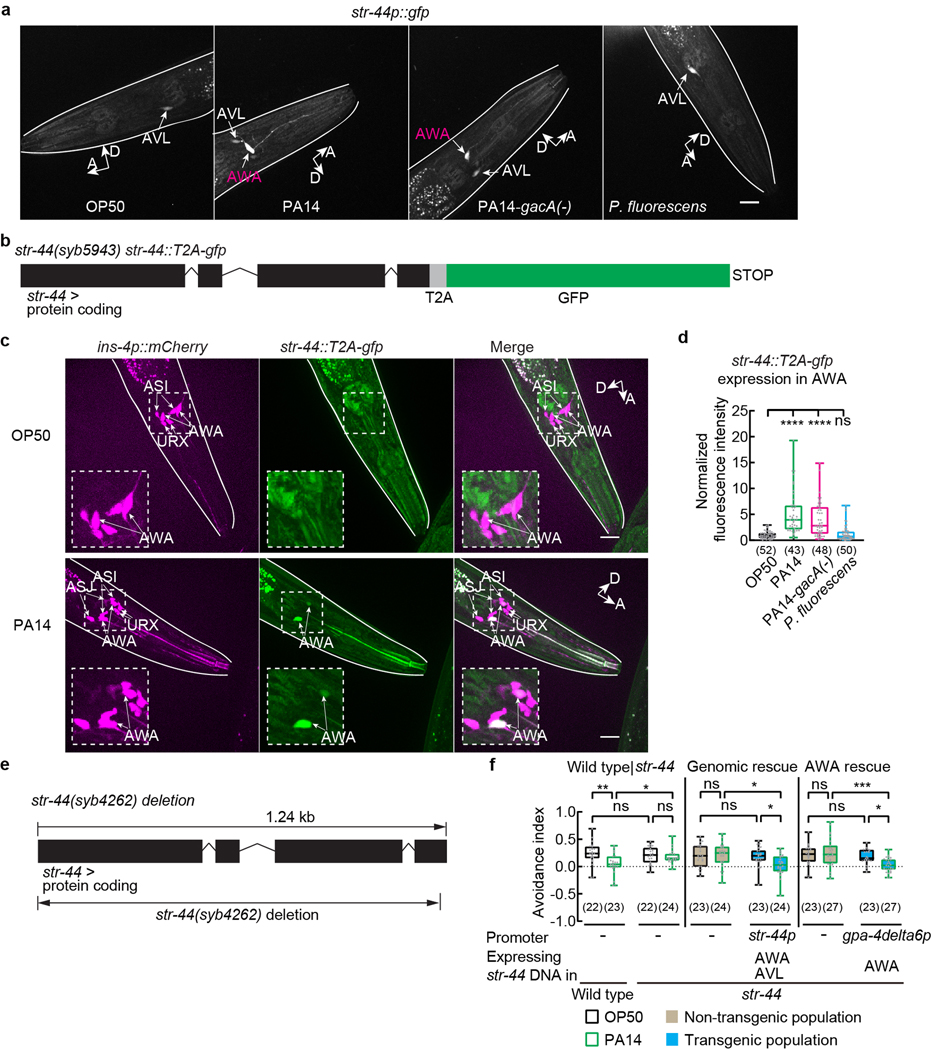
To identify the chemoreceptor that regulates pheromone response in AWA, we examined 57 genes found by excluding 35 genes that showed different expression levels between P. fluorescens-exposed worms and OP50-exposed worms from 92 genes that were differentially regulated in both PA14-exposed worms and PA14-gacA(−)-exposed worms compared with OP50-exposed worms (Fig. 2a–c, Extended Data Fig. 6e,f, Supplementary Data 5). Because previously identified pheromone receptors are G-protein coupled receptors (GPCRs)16,17,19–22,31, we analyzed the single predicted GPCR, str-44, among the 57 candidate genes (Fig. 2a–c, Extended Data Fig. 6e,f). The transcript of str-44 was previously identified in AWA and scarcely in three other pairs of neurons33. We found that a transcriptional reporter str-44p::gfp did not generate a detectable signal in AWA in OP50-exposed adult hermaphrodites. However, exposure to PA14 or PA14-gacA(−) significantly induced expression of str-44p::gfp specifically in AWA, but exposure to P. fluorescens did not (Fig. 2d,e, Extended Data Fig. 7a). We also examined str-44 expression using a CRISPR-generated strain in which a DNA sequence of GFP was inserted into the genomic locus of str-44 and obtained similar results (Extended Data Fig. 7b–d). In addition, using the TRAP-RNAseq results we surveyed the expression of previously identified chemoreceptors for pheromone sensing16,17,19–22,31 and found that none of them showed a consistent increase after exposure to either PA14 or PA14-gacA(−) (Supplementary Data 6). These results together show that exposure to PA14 or PA14-gacA(−) induces the expression of str-44 in AWA of hermaphrodite worms.
STR-44 in AWA regulates pheromone response
STR-44 belongs to the Str chemosensory receptor superfamily that contains an olfactory receptor odr-10 specifically expressed in AWA34. To characterize str-44, we tested a deletion mutant allele, syb4262, generated by CRISPR (Extended Data Fig. 7e). First, we found that the str-44(syb4262) loss-of-function mutant hermaphrodites avoided the ascaroside pheromones similarly as wild type when exposed to OP50. However, the str-44(syb4262) hermaphrodites reduced the pheromone avoidance after PA14 exposure to a significantly less extent than wild type and generated a significantly decreased modulation index (Fig. 3a). Expressing either a wild-type genomic sequence containing the str-44 locus or a wild-type str-44 DNA selectively in AWA rescued the defect of str-44(syb4262) mutants in modulating pheromone avoidance after PA14 exposure (Fig. 3b, Extended Data Fig. 7f). Thus, str-44 acts in AWA to regulate pheromone response in pathogen-exposed worms. Second, by examining the GCaMP6 signal in AWA neurons we found that inactivating str-44 significantly reduced the response of AWA to the ascaroside pheromones after PA14 exposure without affecting AWA response in OP50-exposed hermaphrodites (Fig. 3c,d). The defect was rescued by expressing the wild-type genomic sequence of str-44 or a wild-type str-44 cDNA specifically in AWA (Fig. 3e–h). Together, our results indicate that exposure to the bacterial pathogen P. aeruginosa PA14 induces the expression of str-44 in AWA to activate the response of AWA to the ascarosides and modulate chemotaxis to the pheromones. We note that expressing str-44 in AWA using the cell-specific promoter gpa-4delta6p does not enable AWA response to the pheromones without pathogen exposure (Fig. 3g,h), indicating that other pathogen-induced factors are also required for modulating the pheromone response.
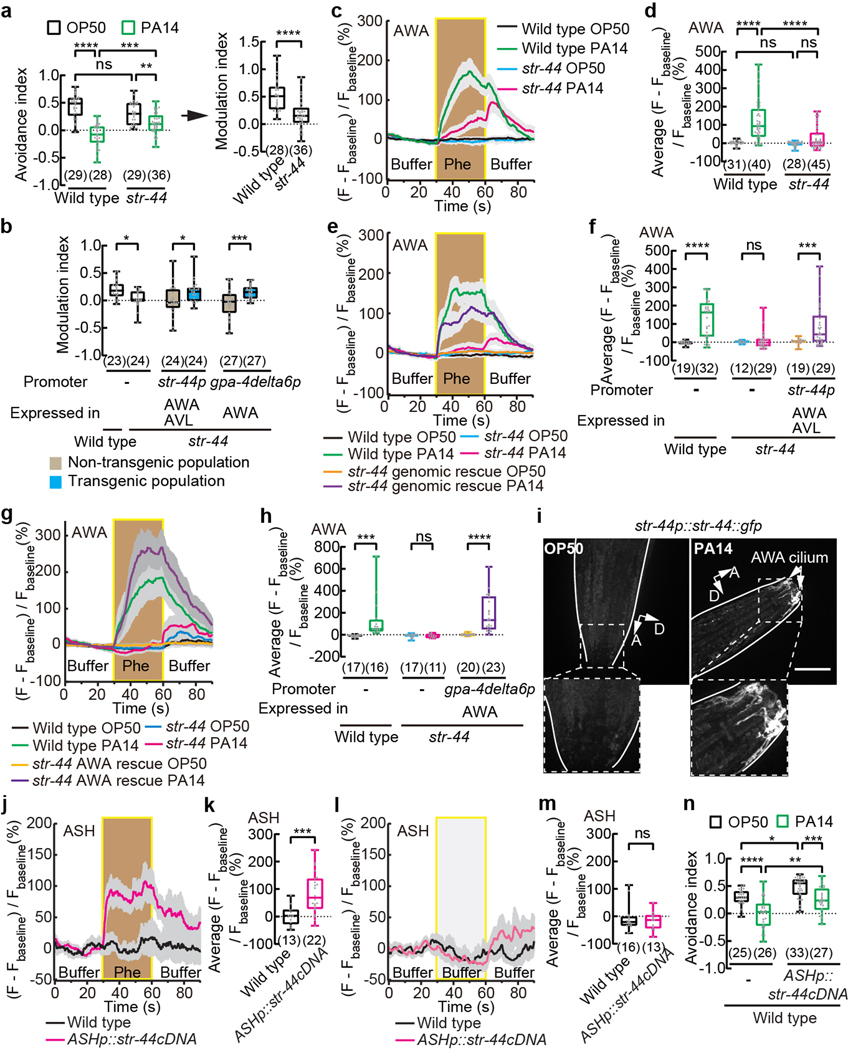
Fig. 3. STR-44 acts in AWA as a pheromone receptor to regulate pheromone response in pathogen-exposed worms.
a, b, n, Avoidance of ascaroside pheromones when exposed to OP50 or PA14 (a,n) and modulation of pheromone avoidance by PA14 exposure (a,b) in wild-type and str-44 mutant hermaphrodites (a, b) or in transgenic hermaphrodites expressing wild-type str-44 DNA using str-44 promoter or AWA-selective promoter gpa-4delta6p in str-44 mutant background (b) or in transgenic hermaphrodites expressing a str-44 cDNA in ASH in wild type (n). Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance. Positive modulation index, suppression of avoidance by PA14.
c - h, j - m, Traces of GCaMP6 signals in AWA neurons in response to ascaroside pheromones in wild-type and str-44 mutant hermaphrodites and in transgenic hermaphrodites expressing a wild-type str-44 DNA using a str-44 promoter or a wild-type str-44 cDNA using an AWA-specific promotor in str-44 mutant background exposed to OP50 or PA14 (c,e,g), or traces of GCaMP6 signals in ASH neurons in response to ascaroside pheromones or buffer control in transgenic hermaphrodites expressing a wild-type str-44 cDNA in ASH and in control hermaphrodites when exposed to OP50 (j,l), and quantitation of average GCaMP6 signals during pheromone stimulation (d,f,h,k) or during the second buffer stimulation (m) in the previous panel. Phe, pheromone mixture of ascr#2,3,5 (1 μM each). Lines in traces, mean. Shades, s.e.m.. Fbaseline, average GCaMP6 signals in the first 30 seconds.
i, Sample images for str-44p::str-44::gfp translational reporter expression in adult hermaphrodites exposed to OP50 or PA14. Dashed lines indicate enlarged views. GFP signals in AWA cilium are shown. Lines outline worm bodies. Scale bar, 20 μm. A, anterior. D, dorsal.
Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual assays (a,b,n) or individual worms (d,f,h,k,m). Dots, avoidance indexes or modulation indexes of individual assays (a,b,n) or average GCaMP6 signals of individual worms during either pheromone stimulation (d,f,h,k) or the second 30-s buffer stimulation (m). P values are derived from Two-way ANOVA with Tukey’s multiple comparisons test [(avoidance index in a),d,n] or two-tailed unpaired t test [(modulation index in a),k] or Two-way ANOVA with Bonferroni’s multiple comparisons test (b,f,h) or two-tailed Mann-Whitney test (m), asterisks indicate significant difference, **** P < 0.0001, *** P < 0.001, ** P < 0.01, * P < 0.05, ns, not significant. P values are shown in Source data.
STR-44 acts as a pheromone receptor
Next, we tested whether str-44 functioned as a pheromone receptor. First, we found that a str-44 translational fusion reporter str-44p::str-44::gfp did not generate a detectable signal in AWA in OP50-exposed adult hermaphrodites. Exposure to PA14 increased the expression of STR-44::GFP, which was observed in the sensory cilium of AWA, suggesting the function of STR-44 in sensing environmental cues (Fig. 3i). In previous studies, ectopic expression of chemoreceptors srx-43, srg-36 or srg-37 in nociceptive sensory neurons ASH, which normally did not respond to ascaroside pheromones, enabled ASH to respond to the pheromones, demonstrating the function of these chemoreceptors as pheromone receptors16,20. Similarly, we examined transgenic hermaphrodites expressing GCaMP6 in ASH and found that ectopic expression of str-44 generated a robust pheromone response in ASH while ASH without str-44 expression did not respond to the pheromones (Fig. 3j–m). Activation of ASH by strong repellents elicits avoidance35. We found that OP50-exposed transgenic animals that ectopically expressed str-44 in ASH avoided the pheromones significantly more than the control animals that did not express str-44, without interfering with the pathogen-induced suppression of pheromone avoidance (Fig. 3n), further indicating that str-44 expression enables activation of ASH by the pheromones. Together, these results show that STR-44 serves as a receptor or a receptor subunit for the ascaroside pheromones.
zip-5 regulates str-44 and pheromone response
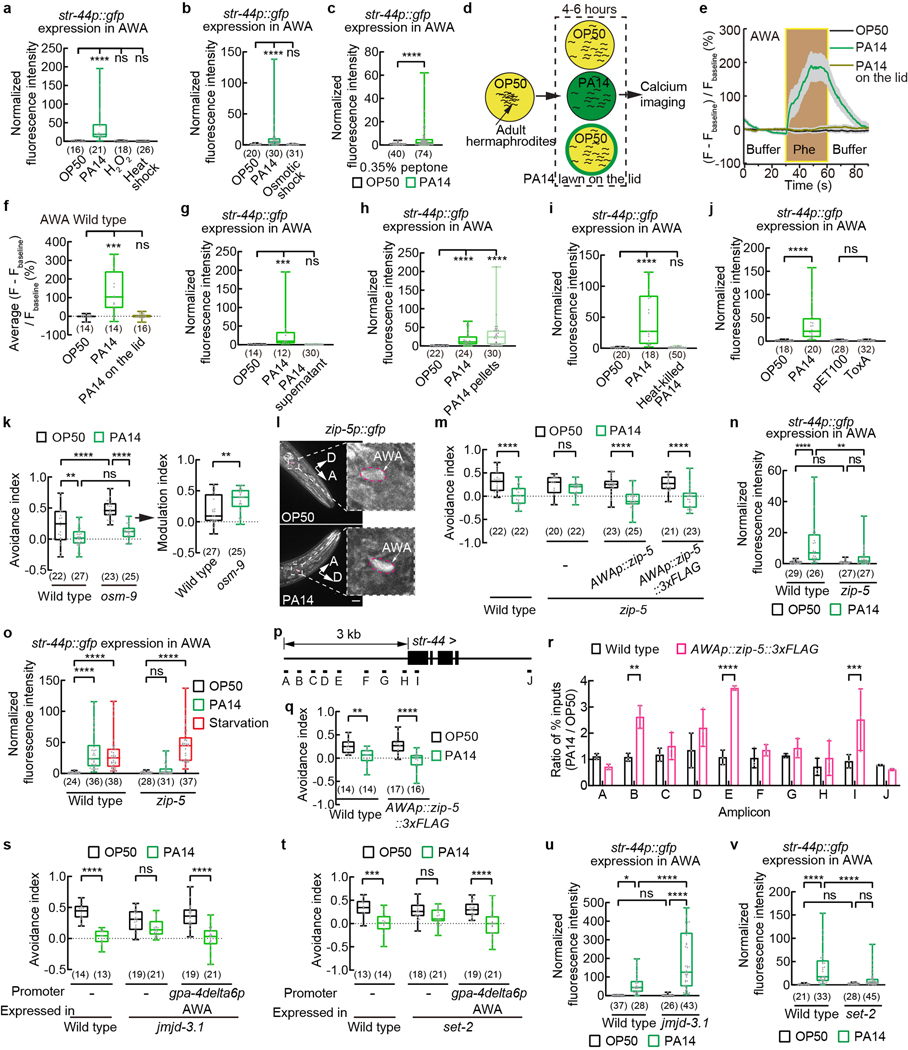
To examine whether the induced AWA expression of str-44 is specific to pathogen exposure, we treated the worms with a heat shock at 32 ⁰C for 8 hours or with hydrogen peroxide for 24 hours36. These stresses did not induce str-44 expression in AWA (Extended Data Fig. 8a). We also treated the worms with an osmotic shock of 300 mOsm for 6 hours37 and did not observe an induction of str-44 expression in AWA (Extended Data Fig. 8b). In contrast, a culturing condition that induces a higher level of virulence in PA1423 generated a robust str-44 expression in AWA (Extended Data Fig. 8c). These results indicate that the pathogen-induced expression of str-44 in AWA is not part of a general response to stresses.
We next asked which biological feature of pathogenic bacteria regulated str-44 expression. First, we found that exposure to PA14 odorants did not induce a pheromone response in AWA (Extended Data Fig. 8d–f). Second, it was shown that C. elegans chemosensory neurons responded to metabolites, such as phenazine-1-carboxamide, that were secreted by PA14 cells38. We found that exposure to the supernatant of a PA14 culture also did not induce str-44 expression; but PA14 cells with the supernatant removed induced robust str-44 expression in AWA (Extended Data Fig. 8g,h). In addition, killing PA14 cells with heat abolished the induction (Extended Data Fig. 8i). We also tested whether the damages of the host generated by PA14 infection induced str-44 expression by exposing the worms to E. coli cells that expressed ToxA39. ToxA encodes Exotoxin A produced by many P. aeruginosa strains to inhibit protein translation, a common target of infection39,40. Exposure to ToxA-expressing E. coli cells resembled the effects of PA14 infection on gene expression and physiological responses39,40, but it did not induce str-44 expression in AWA (Extended Data Fig. 8j). Together, our results suggest that molecules associated with live PA14 cells play a major role in inducing AWA expression of str-44 for pheromone response.
Next, we asked how the worm host regulates str-44 expression in response to pathogen exposure. C. elegans chemosensory neurons use either cGMP-gated (CNG) channels or TRP channels to sense cues41. AWA neurons use the TRPV channels OSM-9 and OCR-2 to mediate their responses to attractive odorants41. The TRPV channels also act in other sensory neurons to regulate avoidance of ascr#3 or predator-released sulfolipids8,11,42. We found that inactivating ocr-2 either with a deletion mutation, ak47, or with a mis-sense mutation, yz5, disrupted the modulation of pheromone response following PA14 exposure (Fig. 4a,b). In contrast, osm-9(ky10) mutant worms containing a loss-of-function mutation generated a robust suppression of pheromone avoidance after PA14 exposure (Extended Data Fig. 8k). Furthermore, loss of ocr-2 in ak47 also abolished PA14-induced str-44 expression in AWA and this defect was rescued by expressing a wild-type ocr-2 gene selectively in AWA (Fig. 4c). These results show that ocr-2 acts in AWA to regulate pathogen-induced expression of str-44.
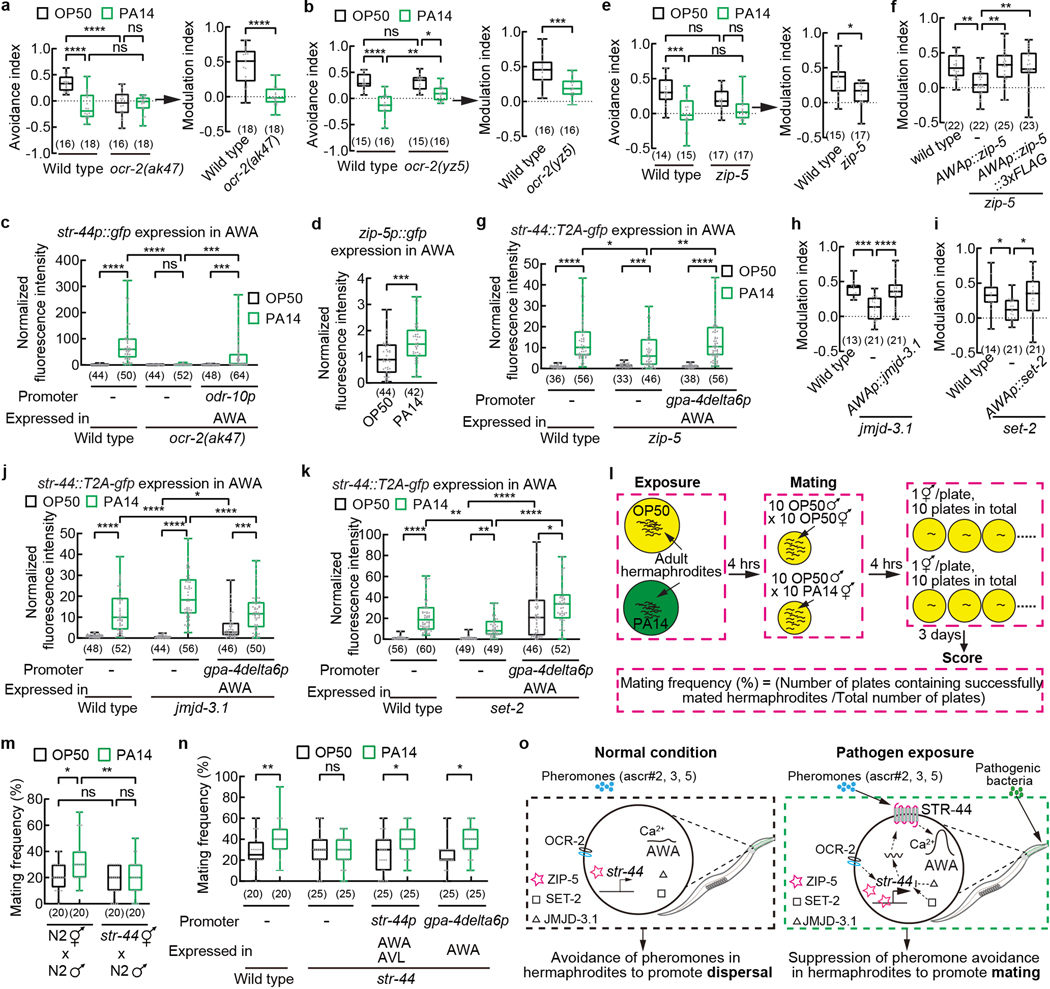
Fig. 4. Pathogen exposure regulates str-44 expression and pheromone response via zip-5 to promote mating.
a, b, e, f, h, i, Avoidance of pheromones in wild-type, ocr-2 (a,b) or zip-5 (e) mutant hermaphrodites exposed to OP50 or PA14 and modulation of pheromone avoidance by PA14 in wild-type, ocr-2 (a,b) or zip-5 (e) or jmjd-3.1 (h) or set-2 (i) mutant hermaphrodites, or in transgenic hermaphrodites expressing specifically in AWA a wild-type zip-5 gene or a wild-type zip-5 gene tagged with a sequence of 3xFLAG in zip-5 mutant background (f), or in transgenic hermaphrodites expressing a wild-type jmjd-3.1 DNA or a wild-type set-2 DNA specifically in AWA in the respective mutant background (h,i). Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance. Positive modulation index, suppression of pheromone avoidance by PA14 exposure.
c, g, j, k, Quantitation of str-44p::gfp fluorescence signals in AWA in wild-type, ocr-2 mutant hermaphrodites and transgenic hermaphrodites expressing a wild-type ocr-2 gene specifically in AWA in ocr-2 mutant background when exposed to OP50 or PA14 (c) or quantitation of AWA expression of str-44 using transgenic animals containing a gfp-tagged str-44 genomic locus in wild-type, zip-5 mutant (g), jmjd-3.1 mutant (j), or set-2 mutant (k) hermaphrodites and in transgenic hermaphrodites expressing a wild-type zip-5 DNA or a wild-type jmjd-3.1 DNA or a wild-type set-2 DNA specifically in AWA in the respective mutant hermaphrodites (g,j,k) when exposed to OP50 or PA14. Intensity of fluorescence signals is normalized using average intensity of str-44p::gfp (c) or str-44::T2A-gfp (g,j,k) in OP50-exposed wild type measured in parallel.
d, Quantitation of zip-5p::gfp signal in AWA when exposed to OP50 or PA14. Intensity of fluorescence signals is normalized using average intensity of zip-5p::gfp in OP50-exposed worms measured in parallel.
l, Cartoon for mating assay.
m, n, Frequency of successful mating for adult wild-type and str-44 mutant hermaphrodites (m,n) or adult transgenic hermaphrodites expressing a wild-type str-44 DNA using str-44 promoter or AWA-selective promoter gpa-4delta6p in str-44 mutant background (n) exposed to OP50 or PA14. All males in mating assays are wild-type raised on OP50.
o, Model for pathogen-induced modulation of pheromone response to promote mating.
Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual assays (a,b,e,f,h,i,m,n) or individual neurons (c,d,g,j,k). Dots, avoidance indexes or modulation indexes of individual chemotaxis assays (a,b,e,f,h,i) or signals of individual neurons (c,d,g,j,k) or frequency of individual mating assays (m,n). P values are derived from Two-way ANOVA with Tukey’s multiple comparisons test [(avoidance index in a,b,e),c,g,j,k,m] or two-tailed unpaired t test [(modulation index in a,b,e), d] or One-way ANOVA with Dunnett’s multiple comparisons test (f,h,i) or Two-way ANOVA with Bonferroni’s multiple comparisons test (n), asterisks indicate significant difference, **** P < 0.0001, *** P < 0.001, ** P < 0.01, * P < 0.05, ns, not significant. P values are shown in Source data.
Next, to further characterize the regulation of str-44 expression, we surveyed the TRAP-RNAseq results for transcription factors. We found that 12 genes encoding transcription factors were differentially expressed between PA14-exposed worms and OP50-exposed worms (Supplementary Data 7). Among them, zip-5 is expressed in neurons, including AWA33. Using a transgene fusion of zip-5 promoter and gfp, we confirmed the expression of zip-5 in AWA and found significantly increased zip-5p::gfp expression in AWA after PA14 exposure (Fig. 4d, Extended Data Fig. 8l). Inactivating zip-5 with a deletion mutation gk646 disrupted PA14-induced suppression of pheromone avoidance (Fig. 4e,f, Extended Data Fig. 8m). It also significantly reduced PA14-induced expression of str-44 in AWA measured using either a genomic locus tagged with gfp (Fig. 4g) or a reporter transgene (Extended Data Fig. 8n). These defects are rescued by expressing a wild-type zip-5 DNA sequence specifically in AWA (Fig. 4f,g, Extended Data Fig. 8m). Together, these results indicate that the transcription factor zip-5 acts in AWA to regulate str-44 expression and pheromone response induced by pathogen exposure. Interestingly, starving the worms for 5 hours also strongly induced the expression of the str-44p::gfp reporter in AWA; however, this regulation did not depend on zip-5 (Extended Data Fig. 8o), indicating that different mechanisms regulate pathogen-induced versus starvation-induced str-44 expression in AWA.
To further analyze the function of ZIP-5, we examined the association of ZIP-5 with str-44 locus using chromatin immunoprecipitation assay combined with quantitative PCR (ChIP-qPCR). We generated transgenic worms that expressed specifically in AWA a fusion transgene of zip-5 and 3xFLAG. We first showed that this transgene was functional because it rescued the defect in pathogen-induced suppression of pheromone avoidance when expressed in the zip-5 mutant worms (Fig. 4f, Extended Data Fig. 8m). We then performed the analysis on several str-44 locus regions and found that compared with wild-type animals that did not express zip-5::3XFLAG, transgenic animals expressing zip-5::3xFLAG in AWA showed significantly increased association between several regions of str-44 locus and immunoprecipitated ZIP-5::3xFLAG after PA14 exposure (Extended Data Fig. 8p–r). Together, our results on zip-5 reveal that pathogen exposure increases AWA expression of zip-5 and promotes the association of ZIP-5 with str-44 sequence, resulting in increased expression of str-44 in AWA.
Furthermore, we investigated the role of histone and chromatin modifications. We focused on mutants without a significant developmental or locomotory defect. Inactivating jmjd-3.1 that encoded a H3K27 demethylase43 with a deletion mutation gk384 or inactivating set-2 that encoded a H3K4 methyltransferase44 with a deletion ok952 abolished PA14-induced modulation of pheromone avoidance (Fig. 4h,i, Extended Data Fig. 8s,t). Intriguingly, while inactivating set-2 decreased PA14-induced AWA expression of str-44 in hermaphrodites, inactivating jmjd-3.1 enhanced the induction, analyzed using either a str-44 genomic locus tagged with a gfp sequence or a str-44p::gfp transgene (Fig. 4j,k, Extended Data Fig. 8u,v). Both jmjd-3.1 and set-2 are widely expressed in the nervous system33. Expressing a wild-type jmjd-3.1 DNA or a wild-type set-2 DNA in the AWA neurons of their respective mutant animals rescued the defects in PA14-induced responses (Fig. 4h–k, Extended Data Fig. 8s,t). We note that the rescuing genes also mildly regulate str-44 expression in OP50-exposed worms. These results indicate that the H3K27 demethylase JMJD-3.1 and the H3K4 methyltransferase SET-2 act in AWA to mediate pathogen-induced str-44 expression and pheromone response.
As previously shown, while wild-type hermaphrodites avoided the mixture of ascr#2, ascr#3 and ascr#5, a loss-of-function mutation in the neuropeptide Y receptor NPR-1, npr-1(ad609), suppressed the avoidance7,8,10(Extended Data Fig. 9a). Using transgenic lines that expressed GCaMP reporters in chemosensory neurons ASK or ADL, it has been shown that ASK, which mediate attraction of npr-1 to the pheromone mixture, and ADL, which mediate the avoidance of ascr#3, exhibit stronger and weaker responses compared with the wild type, respectively, in response to the pheromones in npr-1(ad609) hermaphrodites, which contributes to the attraction of npr-1(ad609) worms to the pheromones7,8,10,11. In contrast, exposure of wild-type adult hermaphrodites to PA14 for 4–6 hours did not significantly alter the response of either ASK or ADL to the pheromones, although a long-term exposure reduced the pheromone response in ADL25 (Extended Data Fig. 9b–m). Furthermore, str-44 was not expressed in AWA in OP50-exposed npr-1(ad609) hermaphrodites (Extended Data Fig. 9n). Therefore, although inactivating npr-1 and exposure to pathogenic PA14 bacteria both suppress pheromone avoidance in hermaphrodites, the underlying mechanisms are different.
PA14 exposure promotes mating via str-44
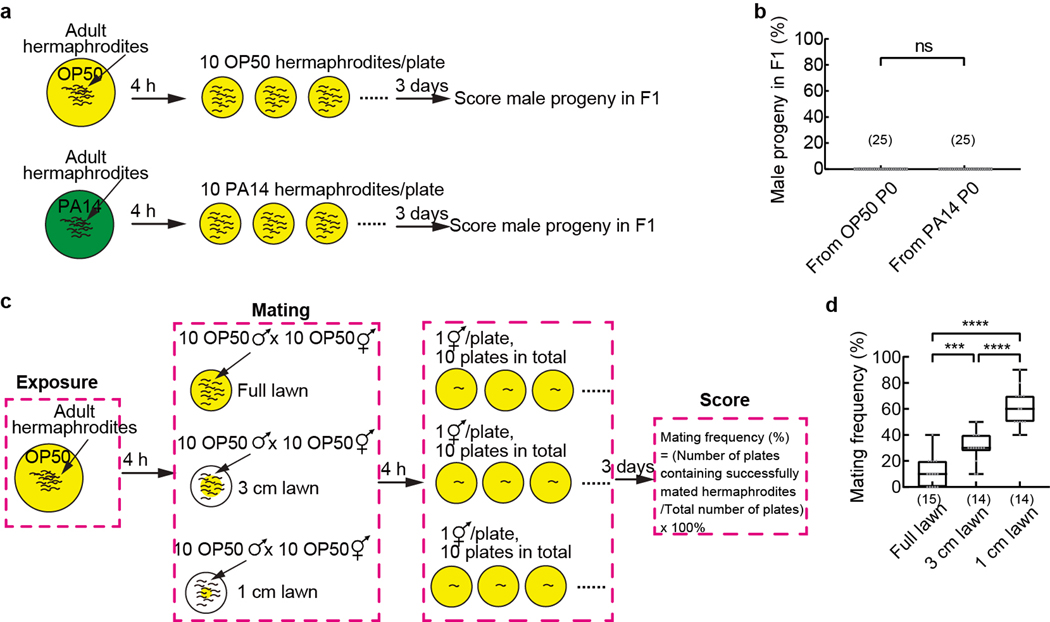
Finally, we addressed the biological significance for pathogen-induced suppression of pheromone avoidance in hermaphrodites. C. elegans hermaphrodites reproduce mainly through self-fertilization45. Meanwhile, the hermaphrodites retain the ability to mate with males. Under normal conditions, adult hermaphrodites avoid the mixture of ascr#2, ascr#3, and ascr#5 at nanomolar to micromolar concentrations to reduce aggregation and facilitate dispersal5–13. We reasoned that suppressing avoidance of the ascarosides in hermaphrodites by pathogen exposure would antagonize dispersal to promote mating with males by increasing the effective density of the hermaphrodites and the chance for males to encounter hermaphrodites in an area. To test this possibility, we placed 10 OP50-exposed adult hermaphrodites or 10 PA14-exposed adult hermaphrodites together with 10 males, which were grown on OP50, on a standard plate and allowed mating for 4 hours (Fig. 4l). We scored the outcome of mating for each hermaphrodite by identifying male progenies (Methods), because mating with males produced 50% males in the mated progeny while self-reproduction produced male progenies at a very low frequency45. We found that PA14-exposed hermaphrodites mated at a significantly higher rate than OP50-exposed hermaphrodites (Fig. 4m). Neither OP50-exposed hermaphrodites nor PA14-exposed hermaphrodites produced any male progeny through selfing under the same conditions (Extended Data Fig. 10a,b). We also showed that decreasing the size of a bacterial lawn used for mating significantly increased the mating rate (Extended Data Fig. 10c,d), supporting that increasing the density of the hermaphrodites promotes mating. Deleting str-44 in hermaphrodites abolished PA14-induced increase in mating frequency without altering mating frequency in OP50-exposed hermaphrodites (Fig. 4m) and this defect was rescued by expressing a wild-type str-44 genomic DNA fragment or a wild-type str-44 DNA selectively in AWA (Fig. 4n). Together, these results indicate that exposure to bacterial pathogen PA14 suppresses pheromone avoidance in hermaphrodites by inducing str-44-mediated pheromone response of AWA to promote mating (Fig. 4o).
Discussion
Here, we show that exposing C. elegans hermaphrodites to a virulent Pseudomonas aeruginosa bacterial pathogen suppresses the avoidance of ascaroside pheromones, which facilitates dispersal5–13, to promote mating. We propose that this pathogen-induced change in pheromone response provides an adaptive value for the host, because compared with selfing, mating with males and outcrossing offer more opportunities to generate novel genotypes through recombination46. Consistently, previous studies on C. elegans and other organisms have found increased mating or recombination rate under the condition of pathogen stress and some of these effects benefit the hosts and facilitate adaptation3,47,48. In addition to mating, exposure to pathogens or parasites alters other behaviors known to be regulated by pheromones49. Our study suggests that modulation of pheromone response in host animals provides a pathway from pathogen stress to social behavior regulation. We further show that the modulation of pheromone response in pathogen-exposed hermaphrodite worms is achieved by inducing the expression of a chemoreceptor str-44 in olfactory sensory neurons AWA. We also show that STR-44 acts as a receptor (or a receptor subunit) for the pheromones. Previously identified ascaroside pheromone receptors in C. elegans are also chemoreceptors16,17,19–22,31. While some pheromone receptors are specific, others are broadly tuned, such as the Drosophila Or7a that responds to an aggregation pheromone as well as food odorants50. We speculate that widely tuned sensory receptors allow animals to integrate complex environmental cues, such as the presence of food and conspecifics, for a coherent behavioral response. Chemoreceptors are encoded as large families in many animals34, which may serve as a collection of sensory receptors that can be rapidly induced or suppressed under novel conditions. We propose that regulation of chemoreceptors for pheromones represents a mechanism to generate social behavioral plasticity that can be deployed under varying environmental conditions.
Methods
Strains
This study uses invertebrate Caenorhabditis elegans. The C. elegans strains were maintained under standard conditions at 20–22°C on Nematode Growth Medium (NGM, 2.5 g/L BactoPeptone, 3 g/L NaCl, 25 mM KPO4 pH 6.0, 1 mM CaCl2, 1 mM MgSO4, 5 mg/L cholesterol) plates containing E. coli OP50 bacterial lawns51. Adult hermaphrodites were used in this study unless otherwise specified. The strains used in this study include: N2 Bristol (Wild type), CX4 odr-7(ky4)X, CX10 osm-9 (ky10)IV, CX4544 ocr-2(ak47)IV, JY243 ocr-2(yz5)IV, VC1392 zip-5(gk646)V, DA609 npr-1(ad609)X, PY7505 oyIs84[gpa-4p::TU#813; gcy-27p::TU#814; gcy-27p::GFP; unc-122p::DsRed], CX12330 Ex[sre-1p::TeTx], PS6025 qrIs2 [sra-9p::mCasp1], KHK225 lskEx218[sre-1p::GCaMP3;unc-122p:dsRed], CX10981 kyEx2866[sra-9p::GCaMP3.1; ofm-1p::gfp], ZC2706 yxEx1393[gpa-4delta6p::GCaMP6; unc-122p::DsRed], ZC3036 jxSi27; yxEx1581[gpa-4delta6p::gfp; unc-122p::gfp], ZC3142 jxSi27; yxEx1628[srg-36p::gfp; unc-122p::gfp], ZC3150 jxSi27; yxEx488[srd-1p::gfp; unc-122p::gfp], ZC3151 jxSi27; yxEx1631[daf-37p::gfp; unc-122p::gfp], ZC3153 jxSi27; yxEx1633[srx-44p::gfp; unc-122p::gfp], ZC3160 yxEx1638[odr-10p::HisCl1; unc-122p::gfp], ZC3172 unc-13(e51)I; yxEx1393, ZC3173 unc-31(e928)IV; yxEx1393, ZC3175 jxSi27[ins-4p::mCherry]; oyEx11[srbc-64p::gfp; unc-122p::dsRed], ZC3176 jxSi27; oyEx1[srbc-66p::gfp; unc-122p::dsRed], ZC3291 yxIs49[gpa-4delta6p::egfp::rpl-1; lin-15(+)], ZC3335 set-2(ok952)III, ZC3337 jmjd-3.1(gk384)X, ZC3403 jxSi27; yxEx1767[str-44p::gfp; unc-122p::gfp], ZC3443 jmjd-3.1(gk384)X; jxSi27; yxEx1767, ZC3463 zip-5(gk646)V; yxEx1767, ZC3467 set-2(ok952)III; jxSi27; yxEx1767, ZC3524 str-44(syb4262)IV; yxEx1393, ZC3527 str-44(syb4262)IV 2xoutcrossed, ZC3535 str-44(syb4262)IV; yxEx1872[str-44p::str-44DNA::3’UTR; unc-122p::gfp], ZC3638 yxEx1952[gpa-4delta6p::zip-5::3xFLAG; unc-122p::gfp], ZC3540 str-44(syb4262)IV; yxEx1877[gpa-4delta6p::str-44DNA; unc-122p::gfp], ZC3565 str-44(syb4262)IV; yxEx1872; yxEx1393, ZC3582 yxEx1903[sra-6p::GCaMP6; unc-122p::gfp], ZC3585 yxEx1906[sra-6p::str-44cDNA; sra-6p::GCaMP6; unc-122p::gfp], ZC3609 str-44(syb4262)IV; yxEx1393; yxEx1930[gpa-4delta6p::str-44cDNA; unc-122p::gfp], ZC3618 ocr-2(ak47)IV; jxSi27; yxEx1767, ZC3653 str-44(syb5943)IV; jxSi27, ZC3660 str-44(syb5943)IV; jmjd-3.1(gk384)X; jxSi27, ZC3663 set-2(ok952)III; str-44(syb5943)IV; jxSi27, ZC3669 str-44(syb5943)IV; zip-5(gk646)V; jxSi27, ZC3624 jmjd-3.1(gk384)X; yxEx1938[gpa-4delta6p::jmjd-3.1DNA; unc-122p::gfp], ZC3628 set-2(ok952)III; yxEx1943[gpa-4delta6p::set-2DNA; unc-122p::gfp], ZC3632 jxSi27; yxEx1947[zip-5p::gfp; unc-122p::gfp], ZC3642 zip-5(gk646)V; yxEx1953[gpa-4delta6p::zip-5::3xFLAG; unc-122p::gfp], ZC3646 zip-5(gk646)V; yxEx1957[gpa-4delta6p::zip-5DNA::unc-54 3’UTR; unc-122p::gfp], ZC3695 zip-5(gk646)V; str-44(syb5943)IV; jxSi27; yxEx1988[gpa-4delta6p::zip-5DNA::unc-54 3’UTR; unc-122p::gfp], ZC3709 jmjd-3.1(gk384)X; str-44(syb5943)IV; jxSi27; yxEx2002[gpa-4delta6p::jmjd-3.1DNA::unc-54 3’UTR; unc-122p::gfp], ZC3712 set-2(ok952)III; str-44(syb5943)IV; jxSi27; yxEx2005[gpa-4delta6p::set-2DNA::unc-54 3’UTR; unc-122p::gfp], ZC3724 ocr-2(ak47)IV; kyEx685[odr-10p::ocr-2; unc-122p::gfp]; yxEx2016[str-44p::gfp; lin-44p::gfp], ZC3444 jxSi27; yxEx1803[str-44p:str-44::GFP; unc-122p::gfp], ZC3639 npr-1(ad609)X; yxEx1767. PHX4262 str-44(syb4262)IV and PHX5943 str-44(syb5943)IV were generated by SunyBiotech and contain CRISPR-Cas9 engineered deletion or insertion of sequences of T2A and GFP, respectively.
Transgenes and transgenic animals
To make DNA plasmid gpa-4delta6p::egfp::rpl-1::unc-54 3’UTR, a 1.82 kb DNA sequence upstream of gpa-4 amplified using PCR and primers oTW269 5’GCAGGCTCCGAATTCTAGACCGCTCGCCCTTCGATGATCATT and oTW270 5’GGTCGAATTCTAGAGGCGCGTGTTGAAAAGTGTTCACAAAATGAA and the vector containing the sequence of egfp::rpl-1::unc-54 3’UTR amplified from plasmid rgef-1p::egfp::rpl-1::unc-54 3’UTR52 using PCR and primers oTW275 5’CAATGATCATCGAAGGGCGAGCGGTCTAGAAT TCGGAGCCTGCT and oTW276 5’CTTATTCATTTTGTGAACACTTTTCAACACGCGCCTCTAG AATTCGACCCAG were used to generate gpa-4delta6p::egfp::rpl-1::unc-54 3’UTR by using Gibson assembly (NEB #E2611). To make the DNA plasmid str-44p::gfp, a 3 kb DNA sequence upstream of str-44 was amplified using PCR and primers oTW460 5’AGAAGCGAGCTTCTC AAATCCTTCGAGTCT and oTW455 5’CATTTCGCTCGGAAGTGTGACTAAAATCCC, cloned into the pCR8 Gateway entry vector (Invitrogen #K250020), and recombined with a destination vector converted from pPD95.75 (a gift from Dr. Andrew Fire containing sequence for gfp). To make DNA plasmid str-44p::str-44DNA::3’UTR, a 5 kb DNA sequence containing 3 kb DNA sequence upstream of str-44, str-44 coding sequence and 0.8 kb DNA sequence downstream of str-44 were amplified using PCR and primers oTW460 5’AGAAGCGAGCTTCTCAAATCCTTCGAGTCT and oTW410 5’CCCTTTCAATTCCCTTTTATACCATCGGGAC and cloned into the pCR8 Gateway entry vector (Invitrogen #K250020). To make DNA plasmids srg-36p::gfp, daf-37p::gfp, srx-44p::gfp, and gpa-4delta6p::gfp, a 2.6 kb, 3 kb, 0.53 kb and 1.82 kb DNA sequence upstream of each gene was respectively amplified and cloned into pCR8 Gateway entry vector (Invitrogen #K250020), and recombined with a destination vector converted from pPD95.75 (a gift from Dr. Andrew Fire containing sequence for gfp). To make cDNA plasmids gpa-4delta6p::str-44cDNA::unc-54 3’UTR, and sra-6p::str-44cDNA::unc-54 3’UTR, a 1.82 kb DNA sequence upstream of gpa-4 and a 3.25 kb DNA sequence upstream of sra-6 were respectively amplified and cloned into pCR8 Gateway entry vector (Invitrogen #K250020), and recombined with a destination vector pDEST::str-44cDNA::unc-54 3’UTR by using LR reaction (Invitrogen #11791020). To make DNA plasmids gpa-4delta6p::zip-5DNA::unc-54 3’UTR, gpa-4delta6p::jmjd-3.1DNA::unc-54 3’UTR, and gpa-4delta6p::set-2DNA::unc-54 3’UTR, a 5.27 kb zip-5 DNA sequence was amplified using PCR and primers oTW563 5’CAGGAGGACCCTTGGCTAGCATGAATTGCGCATGTGGAGATC and oTW564 5’ GATATCAATACCATGGTACCTCAATATAATTTATTGCATGTCATATCAAAA TAATCTTG, a 4.4 kb jmjd-3.1 DNA sequence was amplified using PCR and primers oTW567 5’CAGGAGGACCCTTGGCTAGCTTTGTTCCCCAGCATTTACC and oTW568 5’GATATCAA TACCATGGTACCCAGAAGGACACGCGCTTTATA, a 7.56 kb set-2 DNA sequence was amplified using PCR and primers oTW569 5’ CAGGAGGACCCTTGGCTAGCATTTCTACTGTGCATC GGATG and oTW570 5’GATATCAATACCATGGTACCTCCTCCAAGAAAACAATTCGT AAACCTATAGAG. Each of the PCR fragments was used to replace str-44cDNA in gpa-4delta6p::str-44cDNA::unc-54 3’UTR by using Gibson assembly (NEB #2611). To make DNA plasmid gpa-4delta6p::zip-5::3xFLAG, a 78 bp sequence of 3xFLAG (gattataaagacgatgacgataagcgtgact acaaggacgacgacgacaagcgtgattacaaggatgacgatgacaag) was amplified and inserted into gpa-4delta6p::zip-5DNA::unc-54 3’UTR before the stop code of zip-5 gene by using primers oTW575 5’ TCCTTGTAGTCACGCTTATCGTCATCGTCTTTATAATCATATAATTTATTGCATGTCATA and oTW576 5’ CGACGACGACAAGCGTGATTACAAGGATGACGATGACAAGTGAGGTACCA TGGTATTGAT and Q5 Site-Directed Mutagenesis Kit (NEB #E0554S). The DNA plasmid expressing a GCaMP6 reporter53 in AWA and the DNA plasmid expressing the histamine-gated chloride channel HisCl154 in AWA were previously published25. Transgenic animals were generated by injecting transgene(s) (30 ng/μL each) mixed with a co-injection marker unc-122p::gfp or unc-122p::deRed or lin-44p::gfp (10 ng/μL) using standard methods55.
Behavioral assays
Exposure to pathogenic bacteria P. aeruginosa PA14 and benign bacteria E. coli OP50 was performed similarly as previously described56 unless otherwise specified. The worms were cultivated under the standard conditions on E. coli OP50 till the adult stage and then were randomly split into groups to transfer to plates that were respectively prepared by inoculating 10-cm NGM plates with freshly prepared culture (overnight at 26°C) of P. aeruginosa PA14 or E. coli OP50 in NGM medium and incubated at 26°C for 36–48 hours. A PA14 lawn completely covered each PA14 plate57. To grow PA14 lawns under a more virulence-inducing condition, 3.5 g/L BactoPeptone was used for the NGM plates followed by incubation at 37°C for 24 hours and at 25°C for 24 hours23. Adult hermaphrodites were exposed to the bacteria for 4–6 hours (unless otherwise specified) before being tested in chemotaxis assay, calcium imaging or collected for TRAP-RNAseq analysis or analyzed using confocal microscopy. The same procedure was used to expose worms to other bacteria strains. For long-term exposure, embryos were collected and placed on NGM plates containing either OP50 or a large lawn of PA14 with a small lawn of OP50 (Extended Data Fig. 2e) and grown for three days before testing.
Pheromone quadrant assay To examine the response of a population of worms to ascaroside pheromones, we used the quadrant assay7,10 with small modifications. To perform the assay, worms grown on E. coli OP50 under the standard conditions were collected, washed with assay buffer (5 mM KPO4 pH 6.0, 1 mM CaCl2 and 1 mM MgSO4) twice, transferred to plates seeded with OP50 or plates seeded with PA14 or other indicated bacteria strains, and exposed the worms to the bacteria for 4–6 hours before testing. The avoidance of pheromones was assayed on 10-cm chemotaxis plates (1.6 % agar, 5 mM KPO4 pH 6.0, 1 mM CaCl2, 1 mM MgSO4)58 that were divided into four quadrants and spread with the ascaroside mixture (ascr#2, 3 and 5 mixed with 1:1:1 ratio) in 2 quadrants in opposite directions and with control buffer in the other 2 quadrants. Twenty-five μL of 1 μM ascarosides mixture was spread on 2.5 mL agar of each of the two quadrants for pheromones to reach the final concentration of 10 nM for each ascaroside at equilibrium (unless otherwise specified). Twenty-five μL of control buffer with the same amount of ethanol used to solve pheromones was spread on each of the two quadrants for control. Assays were started around 1.5 hours afterwards. After bacteria exposure, approximately 500 worms were washed for three times with chemotaxis buffer (5 mM KPO4 pH 6.0, 1 mM MgSO4, 1 mM CaCl2)58 and around 50 – 200 worms were placed in the center of each assay plate. Assays were allowed for 15 minutes and stopped by freezing the plates at −20°C for 10 minutes and then kept at 4°C. The number of worms in pheromone and control quadrants were counted. Avoidance index for each chemotaxis assay was defined as (The number of worms in control quadrants – the number of worms in pheromone quadrants) / (The total number of worms). A positive avoidance index indicates the avoidance of the pheromones. For each assay on PA14-exposed worms, Modulation index was defined as the difference between the average avoidance index of OP50-exposed worms tested in parallel and the avoidance index of PA14-exposed worms. A positive modulation index indicates suppression of pheromone avoidance by PA14 exposure. Sample sizes were determined based on previous studies7,10. The investigators were not blinded to the condition and genotype of the worms.
Histamine supplementation
1 M histamine-dihydrochloride (Sigma-Aldrich) stock solution was made and used to generate final concentration of 5 mM in NGM plate as previously described54. In the quadrant assays on plates, 50 μL of 1 M histamine stock solution was added into 10 mL medium for each plate to reach a final concentration of 5 mM.
Hermaphrodite mating frequency assay
C. elegans hermaphrodites produced male progenies at a very low frequency (≤ 0.3%) via self-reproduction45,59. The mating frequency assay was performed essentially as described60 with small modifications. For each mating assay replicate, worms were grown on E. coli OP50 under the standard conditions till adult stage and exposed to either E. coli OP50 or P. aeruginosa PA14 for 4 hours as described above. Then, 10 OP50-exposed or 10 PA14-exposed wild-type or mutant or transgenic adult hermaphrodites were transferred to a 6 cm NGM plate containing a full lawn of E. coli OP50 to mate with 10 wild-type adult males grown on E. coli OP50 and allowed to mate for 4 hours at 20–22°C. After mating, each hermaphrodite was transferred onto one NGM plate containing a lawn of E. coli OP50. After three days, the result of mating was quantified by scoring males among F1 progeny on each plate. If > 3 males (typically 3 – 100 males) were identified among F1 progeny of a hermaphrodite, the mating of the hermaphrodite was scored as successful; if < 3 males (but usually 0) were identified among F1 progeny of a hermaphrodite, the mating of the hermaphrodite was scored as failed. “Mating frequency” was defined as the number of hermaphrodites with successful mating divided by the total number of hermaphrodites tested in each assay (which was 10). Each data point in Fig. 4m and 4n indicates mating frequency for one assay of 10 hermaphrodites. For each control assay, 10 OP50-exposed or 10 PA14-exposed wild-type adult hermaphrodites were respectively transferred to a 6 cm NGM plate seeded with E. coli OP50 and number of males among F1 progeny on each plate was scored 3 days later. For mating assays on lawns of different sizes, 10 OP50-exposed hermaphrodites and 10 OP50-exposed males were put on a lawn to mate for 4 hours and the mating outcome was scored for each hermaphrodite three days later. Sample sizes were determined based on previous studies60. The investigators were blinded to the condition and genotype of the worms.
Generation of TRAP-RNAseq Libraries, sequencing and analysis
Translating ribosome affinity purification (TRAP) was performed essentially as described32,52. Synchronized hermaphrodites (ZC3291 yxIs49[gpa-4delta6p::egfp::rpl-1; lin-15(+)]) were grown at 20–22°C on NGM plates containing E. coli OP50 till young adult stage, and then washed and placed onto 15-cm NGM plates that contained a fresh lawn of E. coli OP50 (control condition), or pathogenic bacteria Pseudomonas aeruginosa PA14 (PA14-exposed condition), or Pseudomonas aeruginosa PA14-gacA(−) (PA14-gacA(−)-exposed condition), or Pseudomonas fluorescens (P. fluorescens-exposed condition). The worms were exposed to the bacteria at 20–22°C for 4–6 hours and then collected to make lysates52. Ribosome proteins were precipitated using an anti-GFP antibody (Rockefeller University Cat# Htz-GFP-19F7, RRID:AB_2716736; Cat# Htz-GFP-19C8, RRID:AB_2716737) and messenger RNAs (mRNAs) associated with the ribosome proteins were isolated and purified to make cDNA libraries. In total, 12 cDNA libraries (4 conditions) were generated from 3 independent experiments and sequenced by Illumina NovaSeq. Sample size was determined based on previous studies52. An average of 41 million reads per library were obtained. C. elegans genome version WBcel235.101 was used as reference for transcriptomic mapping using STAR version 2.7.0e. Numbers of uniquely mapped reads were counted using HTSeq version 0.11.2. Differential gene expression was analyzed using edgeR in R version 4.0.261,62. Genes with >=20 reads were filtered as background reference for Gene ontology and KEGG pathways enrichment analysis using functional annotation tools on DAVID Bioinformatics Resources 6.863,64. Volcano plots compare log2-transformed fold change (FC) versus −log10-transformed Benjamini-Hochberg adjusted P-value (FDR). Principal component analysis and hierarchical clustering plot were based on log2CPM (counts per million). The data were normalized using the trimmed mean of the M-values (TMM) method. log2CPM values were calculated using cpm function with prior.count = 2 in edgeR. All the plots were generated in R version 4.0.2. The survey of transcription factors (TFs) was done using C. elegans TFs previously identified with high confidence65.
Calcium imaging
Calcium imaging and analysis were performed as previously described25 using a PDMS microfluidic device that limited movements of the tested worms and stimulated worms with the pheromone mixture solution66. Exposure of young adult hermaphrodites to different bacteria was performed as described above using 10 cm NGM plates containing a fresh lawn of E. coli OP50, or Pseudomonas aeruginosa PA14, or Pseudomonas aeruginosa PA14-gacA(−), or Pseudomonas fluorescens, or Serratia marcescens, or E. coli HB101, or E. coli OP50 with PA14 lawn on the lid for 4–6 hours at 20–22°C before imaging. Fluorescence images were taken at 5 frames per second using a 40X oil immersion objective, a Yokogawa CSU-X1 scanner unit on a confocal Nikon Eclipse Ti-E inverted microscope with a Photometrics CoolSnap EZ camera or an Andor iXon Ultra EMCCD camera and NIS-Elements (version AR 4.13.04). Hermaphrodite worms were stimulated with NGM buffer or pheromone mixture solved in NGM buffer (mixture of ascr#2,3,5 in 1:1:1 ratio with final concentration at 1 μM each or as otherwise described). GCaMP signal in neuronal cell body was quantified using ImageJ. Average fluorescence intensity for the first 30 seconds in each recording was defined as baseline (Fbaseline). Change in fluorescence intensity relative to the baseline, [(F – Fbaseline) / Fbaseline] x 100%, was plotted as a function of time for all traces. Average (F – Fbaseline) / Fbaseline (%) during pheromone stimulation or during the second buffer stimulation in Fig. 3l was calculated for each worm. Sample sizes were determined based on previous studies10,25. The investigators were not blinded to the condition and genotype of the worms.
Chromatin immunoprecipitation (ChIP) and qPCR
Young adult hermaphrodites were exposed to bacteria as described above using 10 cm NGM plates with E. coli OP50 or Pseudomonas aeruginosa PA14 for 4–6 hours at 20–22°C. Chromatin immunoprecipitation was performed essentially as described67 with the following modifications. Sample size was determined based on previous studies67. Worms were collected and washed 3X in phosphate-buffered saline (PBS) with 0.01% Triton X-100 (PBS/Tx). The volume was adjusted to at least 10 times the volume of the pellet. The samples were crosslinked in 1.8% formaldehyde (Sigma F8775) for 22 minutes with a snap freeze-thaw at 6 minutes. After quenching and washing as described, samples were sonicated in FA buffer67 supplemented with 0.1% sodium deoxycholate and 0.35% sarkosyl in a QSonica Q800R3–110 sonicator for 6–8 minutes (50% power, 20s on/40s off), gently mixed, and sonicated for another 6–8 minutes. Following centrifugation (13,000g, 15 min, 4°C), supernatants were aliquoted for immunoprecipitation with 5–10% of the volume saved as Input. Immunoprecipitations were performed with 1–2 μg of anti-Flag antibody (Sigma F1804) and Protein G Dynabeads (Invitrogen 10003D, 7.5 μL/μg antibody) and were washed and eluted as described67. Inputs and eluted IP samples were treated with RNase A and proteinase K, and crosslinks were reversed by overnight incubation at 65°C. DNA was isolated using the QIAquick PCR purification kit (Qiagen 28104). qPCRs were performed with iTaq Universal SYBR Green Supermix (Biorad 1725122) in 10 μL reactions for 40 cycles on the CFX96 Touch Real-Time PCR Detection System. Cq values were determined on CFX Maestro 2.0 Version 5.0.021.0616 using the default single threshold method. Input DNA was used to create a standard curve with a 4-fold dilution series. Each IP Cq value was compared to its corresponding Input Cq values to calculate the % input.
Confocal microscopy
All confocal images were taken and analyzed as previously described25. Briefly, for each image a set of Z-stack images was captured on a confocal Nikon Eclipse Ti-E inverted microscope with a 40x oil-immersion objective (100x oil immersion objective for Fig. 3i), an ANDOR iXon Ultra EMCCD camera with NIS-Elements (version AR 4.13.04) and a maximum intensity projection was generated using NIH ImageJ. Fluorescence intensity for each neuron was obtained by quantifying the average intensity of a ROI containing the neuronal cell body subtracted by average intensity of a background area of the same size. Intensity of fluorescence signals was normalized using average intensity of wild-type or control worms exposed to OP50 or control condition measured in parallel as defined: Normalized fluorescence intensity = Fluorescence intensity / Average fluorescence intensity of OP50-exposed wild-type worms (or specified control worms). Sample sizes were determined based on previous studies25. The investigators were not blinded to the condition and genotype of the worms.
Statistics and reproducibility
Boxplots and statistical tests were generated using GraphPad Prism 9, plots for calcium imaging traces were generated using Igor pro 6.12, analysis of confocal microscopy images and calcium imaging recording were done using ImageJ. The types of statistical tests used in each experiment, n numbers, P values, and other related measures are indicated in each figure and the associated legend and reported in Source Data. All data reported in figures are shown in Source Data. In all figures, **** P < 0.0001, *** P < 0.001, ** P < 0.01, * P < 0.05, ns, not significant. Replications were done by testing different worms, different assays, different neurons and by doing experiments on different days. All replication attempts were successful. The n numbers for chemotaxis pheromone assays or mating assays are the numbers of different assays performed on at least two independent days; the n numbers for calcium imaging experiments are the numbers of different worms examined generally on at least two independent days (worms in Extended Data Fig. 9j,k were examined on one day; but the same cue was also included in the test in Extended Data Fig. 9h,i); the n numbers for gene expression analysis using reporters are the numbers of different neurons examined generally on at least two independent days. TRAP–RNA-seq analysis and ChIP–qPCR analysis were performed in three independent experiments. Sample images were replicated generally in more than 10 worms in at least two independent experiments. For each C. elegans strain used in this study, a population of worms was grown together under identical conditions, and the worms were randomly distributed into different conditions. Worms from a population were randomly chosen for behavioral test, calcium imaging experiment and gene expression analysis.
Data Availability.
All data generated or analyzed during this study are included in the manuscript (and its supplementary information data files and the source data files). The statistical tests used in each experiment, n numbers, P values, and other related measures are indicated in each figure and the associated legend and reported in Source Data. All data reported in figures are shown in Source data. The sequencing results (accession#: PRJNA789902) are deposited at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA789902
Extended Data
Extended Data Fig. 1. Pathogen exposure suppresses avoidance of ascaroside pheromones.
a, b, k, Schematics for bacteria exposure (a), chemotaxis assay on ascaroside pheromones (b), and histamine treatment during chemotaxis (k).
c – j, Avoidance of the mixture of ascaroside pheromones ascr#2,3,5 over a range of concentrations (c-g) or avoidance of individual ascarosides (h-j, 10 nM each at equilibrium) in wild-type adult hermaphrodites when exposed to E. coli OP50 or P. aeruginosa PA14. Positive avoidance index, avoidance. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual assays. Dots, avoidance indexes of individual assays. P values are derived from two-tailed unpaired t test, asterisks indicate significant difference, **** P < 0.0001, *** P < 0.001, * P < 0.05, ns, not significant. P values are shown in Source data.
Extended Data Fig. 2. Pathogen exposure induces pheromone response of AWA neurons.
a - d, Traces of GCaMP6 signals in AWA neurons of wild-type adult hermaphrodites in response to pheromones at different concentrations when exposed to OP50 or PA14 (a,c) and quantitation of average GCaMP6 signals in AWA during pheromone stimulation in the previous panel (b,d). Phe, pheromone mixture of ascr#2,3,5. Lines in traces, mean. Shades, s.e.m.. Fbaseline, average GCaMP6 signals in the first 30 seconds. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual worms. Dots, average GCaMP6 signals of individual worms during pheromone stimulation. P values are derived from two-tailed Mann-Whitney test, asterisks indicate significant difference, **** P < 0.0001, ** P < 0.01.
e, Paradigm of a long-term exposure to bacterial pathogen PA14.
f, Avoidance of ascaroside pheromones in wild-type adult hermaphrodites exposed to OP50 or PA14 in a long-term exposure paradigm as shown in (e). Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance. Numbers in parenthesis, number of individual assays. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Dots, avoidance indexes of individual assays. P values are derived from two-tailed unpaired t test, asterisks indicate significant difference, ** P < 0.01.
g, h, Traces of GCaMP6 signals in AWA neurons of wild-type adult hermaphrodites in response to ascaroside pheromones when exposed to OP50 or PA14 in the long-term exposure paradigm (g) and quantitation of average GCaMP6 signals in AWA during pheromone stimulation (h). Phe, pheromones, individual or mixture of ascr#2,3,5 (1 μM each). Lines in traces, mean. Shades, s.e.m.. Fbaseline, average GCaMP6 signals in the first 30 seconds. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual worms. Dots, average GCaMP6 signals of individual worms during pheromone stimulation. P values are derived from One-way ANOVA with Dunnett’s multiple comparisons test, asterisks indicate significant difference, **** P < 0.0001, ** P < 0.01, ns, not significant.
i – l, Traces of GCaMP6 signals in AWA neurons of unc-13(e51) (i) or unc-31(e928) (k) mutant adult hermaphrodites in response to pheromones when exposed to OP50 or PA14 and quantitation of average GCaMP6 signals during pheromone stimulation (j,l) in the previous panel. Phe, Pheromone mixture of ascr#2,3,5 (1 μM each). Lines in traces, mean. Shades, s.e.m.. Fbaseline, average GCaMP6 signals in the first 30 seconds. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual worms. Dots, average GCaMP6 signals of individual worms during pheromone stimulation. P values are derived from two-tailed unpaired t test, asterisks indicate significant difference, **** P < 0.0001, ** P < 0.01. P values are shown in Source data.
Extended Data Fig. 3. Different roles of sensory neurons ASK, ASI and ADL in pheromone response and PA14-induced modulation.
a - c, Avoidance of ascaroside pheromones when exposed to E. coli OP50 or P. aeruginosa PA14 (avoidance index) and modulation of pheromone response by PA14 (modulation index) in wild-type adult hermaphrodites and transgenic adult hermaphrodites in which ASK neurons are genetically ablated (a), or ASI neurons are genetically ablated (b), or neurotransmission of ADL is blocked (c). Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance. Positive modulation index, suppression of avoidance by PA14 exposure. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual assays. Dots, avoidance indexes or modulation indexes of individual assays. P values are derived from Two-way ANOVA with Tukey’s multiple comparisons test (avoidance index in a-c) or two-tailed unpaired t test (modulation index in a and c) or two-tailed Mann-Whitney test (modulation index in b), asterisks indicate significant difference, **** P < 0.0001, *** P < 0.001, * P < 0.05, ns, not significant. P values are shown in Source data.
Extended Data Fig. 4. Exposure to pathogenic Serratia bacteria induces pheromone response of AWA.
a, Cartoon for exposure to different bacteria strains for calcium imaging analysis.
b, c, Traces of GCaMP6 signals in AWA neurons of wild-type adult hermaphrodites in response to ascaroside pheromones after 4–6 hour exposure to different bacteria strains (b) and quantitation of average GCaMP6 signals in AWA during pheromone stimulation (c). Phe, pheromone mixture of ascr#2,3,5 (1 μM each). Lines in traces, mean. Shades, s.e.m.. Fbaseline, average GCaMP6 signals in the first 30 seconds. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual worms. Dots, average GCaMP6 signals of individual worms during pheromone stimulation. P values are derived from Kruskal-Wallis test with Dunn’s multiple comparisons test, asterisks indicate significant difference, *** P < 0.001, ns, not significant. P values are shown in Source data.
d, e, Cartoon for TRAP-RNAseq (d) and for exposure to different bacteria strains for TRAP-RNAseq analysis (e).
Extended Data Fig. 5. Exposure to PA14 does not induce AWA expression of several previously identified receptors for pheromone sensing.
a - g, Sample images (a - f) and quantitation in AWA neurons (g) for srg-36p::gfp, daf-37p::gfp, srd-1p::gfp, srx-44p::gfp, srbc-64p::gfp, and srbc-66p::gfp expression in adult hermaphrodites exposed to OP50 or PA14 for 4–6 hours (reporters are indicated by the name of the promoters in g). Arrows point to cell bodies of several sensory neurons. Lines outline worm bodies. Scale bar, 20 μm. A, anterior. D, dorsal (arrows point out of the page for OP50 condition in c and for PA14 condition in d). Intensity of fluorescence signals is normalized using average intensity of AWA expression of str-44p::gfp in OP50-exposed worms measured in parallel. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual neurons. Dots, signals of individual neurons. P values are derived from Two-way ANOVA with Bonferroni’s multiple comparisons test, asterisks indicate significant difference, **** P < 0.0001, ns, not significant. P values are shown in Source data.
Extended Data Fig. 6. TRAP-RNAseq analysis on AWA neurons in worms exposed to OP50, or PA14, or PA14-gacA(−), or P. fluorescens.
a, Avoidance of ascaroside pheromones in adult transgenic hermaphrodites expressing eGFP::RPL-1 selectively in AWA using gpa-4delta6p promoter exposed to OP50 or PA14 for 4–6 hours. Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, numbers of individual assays. Dots, avoidance indexes of individual assays. P values are derived from Two-way ANOVA with Tukey’s multiple comparisons test, asterisks indicate significant difference, ** P < 0.01, ns, not significant. P values are shown in Source data.
b, Expression of a transcriptional reporter using an AWA-specific promoter (gpa-4delta6p) in adult transgenic hermaphrodites exposed to OP50 or PA14 for 4–6 hours. Intensity of fluorescence signals is normalized using average intensity of AWA expression of gpa-4delta6p::gfp in OP50-exposed worms measured in parallel. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, numbers of individual neurons. Dots, signals of individual neurons. P values are derived from two-tailed Mann-Whitney test, ns, not significant. P values are shown in Source data.
c, d, Principal component analysis (c) and hierarchical clustering of samples (d) based on the expression of 806 genes that were differentially expressed in AWA neurons under 4 conditions (exposure to OP50, or PA14, or PA14-gacA(−), or P. fluorescens). log2CPM (counts per million) values calculated using cpm function in edgeR package represent expression levels. OP1–3, samples 1–3 for OP50-exposed hermaphrodites; PA1–3, samples 1–3 for PA14-exposed hermaphrodites; gacA1–3, samples 1–3 for PA14-gacA(−)-exposed hermaphrodites; Pf1–3, samples 1–3 for P. fluorescens-exposed hermaphrodites.
e, f, Expression levels of 57 genes showing differential expression between PA14-exposed worms and OP50-exposed worms (e, FDR < 0.1) and differential expression between PA14-gacA(−)-exposed worms and OP50-exposed worms (f, FDR < 0.1), but showing no difference between P. fluorescens-exposed worms and OP50-exposed worms. Dashed lines indicate 1 or −1. FC, fold change (in comparison with expression in OP50-exposed worms). P values and FDR are shown in Supplementary Data 5.
Extended Data Fig. 7. Exposure to PA14 or PA14-gacA(−) induces expression of str-44 in AWA and analysis of str-44 function in pheromone response.
a, Sample images of adult transgenic hermaphrodites expressing str-44p::gfp exposed to OP50, PA14, PA14-gacA(−) or P. fluorescens for 4–6 hours. Arrows indicate neuronal cell bodies. Lines outline worm bodies. Scale bar (applicable to images in the same row), 20 μm. A, anterior. D, dorsal.
b, Schematics showing the allele str-44(syb5943) that contains the str-44 genomic locus tagged with sequences of T2A peptide and GFP. Boxes indicate protein coding exons.
c, Sample images of AWA expression of str-44::T2A-gfp in adult str-44(syb5943) hermaphrodites containing the str-44 genomic locus tagged with a gfp sequence when exposed to OP50 or PA14 for 4–6 hours. Dashed lines indicate enlarged views. Arrows indicate neuronal cell bodies. Lines outline worm bodies. Scale bar, 20 μm. A, anterior. D, dorsal (arrows point slightly out of the page).
d, Quantitation of AWA expression of str-44::T2A-gfp in adult str-44(syb5943) hermaphrodites containing the str-44 genomic locus tagged with a gfp sequence when exposed to OP50, PA14, PA14-gacA(−) or P. fluorescens for 4–6 hours. Intensity of fluorescence signals is normalized using average intensity of AWA expression of str-44::T2A-gfp in OP50-exposed worms measured in parallel. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual neurons. Dots, signals of individual neurons. P values are derived from Kruskal-Wallis test with Dunn’s multiple comparisons test, asterisks indicate significant difference, **** P < 0.0001, ns, not significant. P values are shown in Source data.
e, Schematics showing the deletion mutation in the str-44(syb4262) allele.
f, Avoidance index in wild-type and str-44 mutant adult hermaphrodites, and transgenic adult hermaphrodites expressing a wild-type str-44 DNA using either str-44 promoter or AWA-specific promoter gpa-4deta6p in str-44 mutant background exposed to OP50 or PA14. Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual assays. Dots, avoidance indexes of individual assays. P values are derived from Two-way ANOVA with Tukey’s multiple comparisons test, asterisks indicate significant difference, *** P < 0.001, ** P < 0.01, * P < 0.05, ns, not significant. P values are shown in Source data.
Extended Data Fig. 8. AWA expression of str-44 is regulated by biological features associated with live PA14 cells and the function of zip-5, jmjd-3.1 and set-2 in AWA.
a - c, Quantitation of str-44p::gfp signals in AWA in adult hermaphrodites exposed to OP50 or PA14 for 4–6 hours, or to a heat shock at 32⁰C for 8 hours, or to 10 mM hydrogen peroxide for 24 hours (a), or to a osmotic shock at 300 mOsm for 24 hours (b), or to OP50 and PA14 cultivated under conditions that induce a higher level of virulence in PA14 (c, Methods). Intensity of fluorescence signals is normalized using average intensity of str-44p::gfp in OP50-exposed worms measured in parallel.
d, Cartoon for exposure to PA14 odorants by placing a lawn of PA14 on the lid.
e, f, Traces of GCaMP6 signals in AWA neurons of wild-type adult hermaphrodites in response to pheromones after 4 to 6-hour exposure to OP50, or PA14, or PA14 odorants (a lawn of PA14 on the lid) (e) and quantitation of average GCaMP6 signals in AWA during pheromone stimulation (f). Phe, pheromone mixture of ascr#2,3,5 (1 μM each). Lines in traces, mean. Shades, s.e.m.. Fbaseline, average GCaMP6 signals in the first 30 seconds.
g - j, Exposure to supernatant of PA14 culture does not induce AWA expression of str-44p::gfp (g), but exposure to PA14 cells with supernatant removed does (h); and heat-killing of PA14 cells abolishes the induction (i). Exposure to E. coli expressing the exotoxin ToxA of PA14 also does not induce str-44 expression (j). Intensity of fluorescence signals is normalized using average intensity of str-44p::gfp in OP50-exposed worms measured in parallel. pET100, cloning vector for ToxA.
k, m, q, s, t, Avoidance of pheromones in wild-type, osm-9 mutant (k), zip-5 mutant (m), jmjd-3.1 mutant (s), or set-2 mutant (t) adult hermaphrodites, or in transgenic adult hermaphrodites specifically expressing in AWA a wild-type zip-5 DNA or a wild-type zip-5 DNA tagged with a sequence of 3xFLAG in zip-5 mutant background (m), or in transgenic adult hermaphrodites specifically expressing in AWA a wild-type zip-5 DNA tagged with a sequence of 3xFLAG in wild-type background (q), or in transgenic adult hermaphrodites expressing specifically in AWA a wild-type jmjd-3.1 DNA or a wild-type set-2 DNA in the respective mutant background (s,t) when exposed to OP50 or PA14 (avoidance indexes in q were measured during sample collection for ChIP-qPCR assays), and modulation of pheromone avoidance by PA14 (k). Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance. Positive modulation index, suppression of avoidance by PA14 exposure.
l, Sample images of adult transgenic hermaphrodites expressing zip-5p::gfp exposed to OP50 or PA14 for 4–6 hours. Dashed lines indicate enlarged views. Arrows indicate neuronal cell bodies. Scale bar, 20 μm. A, anterior. D, dorsal.
n, o, u, v, Quantitation of str-44p::gfp signals in AWA in wild-type and zip-5 mutant adult hermaphrodites when exposed to OP50 or PA14 for 4–6 hours (n), or in wild-type and zip-5 mutant adult hermaphrodites when exposed to OP50 or PA14 for 4–6 hours or starved for 5 hours (o), or in jmjd-3.1 mutant (u) or set-2 mutant (v) hermaphrodites exposed to OP50 or PA14. Intensity of fluorescence signals is normalized using average intensity of str-44p::gfp in OP50-exposed wild-type worms measured in parallel.
p, Diagram of qPCR amplicon positions on str-44 genomic locus in ChIP assays for association of ZIP-5::3xFLAG with str-44 sequence.
r, Ratio of ChIP signal (% of input) in PA14 versus OP50-exposed animals. n = 3 independent assays, mean ± SD. Dots, ratios of individual assays. P values are derived from Two-way ANOVA with Bonferroni’s multiple comparisons test, asterisks indicate significant difference, **** P < 0.0001, *** P < 0.001, ** P < 0.01. P values are shown in Source data.
Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual neurons (a-c,g-j,n,o,u,v) or individual worms (f) or individual assays (k,m,q,s,t). Dots, signals of individual neurons (a-c,g-j,n,o,u,v) or individual worms (f), or avoidance indexes (k,m,q,s,t) or modulation indexes (k) of individual assays. P values are derived from Kruskal-Wallis test with Dunn’s multiple comparisons test (a,b,f-j) or two-tailed Mann-Whitney test (c) or Two-way ANOVA with Tukey’s multiple comparisons test [(avoidance index in k),n,o,q,u,v] or Two-way ANOVA with Bonferroni’s multiple comparisons test (m,s,t) or two-tailed unpaired t test (modulation index in k), asterisks indicate significant difference, **** P < 0.0001, *** P < 0.001, ** P < 0.01, * P < 0.05, ns, not significant. P values are shown in Source data.
Extended Data Fig. 9. Inactivating npr-1 regulates pheromone response differently from pathogen exposure.
a, Avoidance of pheromones in wild-type and npr-1 mutant adult hermaphrodites. Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance.
b - m, Traces of GCaMP signals in ASK (b,d,f) or ADL (h,j,l) neurons of wild-type adult hermaphrodites in response to ascaroside pheromones and quantitation of average GCaMP signals during pheromone stimulation in the previous panel (c,e,g,i,k,m). Phe, pheromone mixture of ascr#2,3,5 at indicated concentrations (b,d,f,h,l). Lines in traces, mean. Shades, s.e.m.. Fbaseline, average GCaMP signals in the first 30 seconds.
n, Quantitation of str-44p::gfp signals in AWA in wild-type and npr-1 mutant adult hermaphrodites. Intensity of fluorescence signals is normalized using average intensity of str-44p::gfp in OP50-exposed wild-type worms.
Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual assays (a) or individual worms (c,e,g,i,k,m) or individual neurons (n). Dots, Avoidance indexes of individual assays (a) or signals of individual worms (c,e,g,i,k,m) or signals of individual neurons (n). P values are derived from One-way ANOVA with Tukey’s multiple comparisons test (a) or two-tailed unpaired t test (c,g,i,k,m) or two-tailed Mann-Whitney test (e) or Kruskal-Wallis test with Dunn’s multiple comparisons test (n), asterisks indicate significant difference, **** P < 0.0001, *** P < 0.001, ns, not significant. P values are shown in Source data.
Extended Data Fig. 10. Wild-type hermaphrodites exposed to OP50 or PA14 produce male progeny by selfing at a very low frequency, and mating on lawns of different sizes.
a, Cartoon for control experiment.
b, Percentage of male progeny produced by selfing of adult hermaphrodites exposed to OP50 or PA14 for 4 hours. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, numbers of control assays (progeny of 10 hermaphrodites were scored for each assay). Dots, male progeny in F1 (%) of individual assays. Two-tailed Mann-Whitney test, ns, not significant. P values are shown in Source data.
c, Cartoon for mating on lawns of different sizes.
d, Frequency of successful mating for adult wild-type hermaphrodites and males on lawns of different sizes. Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of assays (each contains 10 hermaphrodites with their outcome of mating scored individually). Dots, mating frequency of individual assays. P values are derived from One-way ANOVA with Tukey’s multiple comparisons test, asterisks indicate significant difference, **** P < 0.0001, *** P < 0.001. P values are shown in Source data.
Supplementary Material
Acknowledgements
We thank Caenorhabditis Genetics Center that is funded by NIH Office of Research Infrastructure Programs (P40 OD010440) for strains. We thank Dr. C.I. Bargmann and Dr. P. Sengupta for plasmids and strains, Dr. N. Pierce, Dr. M.-K. Choi, and Zhang lab members for discussion on manuscript. A.L.S. is supported by NSERC (RGPIN-2019-06843). C.L. is supported by an NSERC PGS-D. R.A.B. is supported by NIH (GM118775, R35GM144076). Y.Z. is supported by NIH (NS115484).
Footnotes
Competing interests.
The authors declare no competing interests.
References
- 1.Hemachudha T, Laothamatas J. & Rupprecht CE Human rabies: a disease of complex neuropathogenetic mechanisms and diagnostic challenges. Lancet Neurol 1, 101–109, doi: 10.1016/s1474-4422(02)00041-8 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Keesey IW et al. Pathogenic bacteria enhance dispersal through alteration of Drosophila social communication. Nat Commun 8, 265, doi: 10.1038/s41467-017-00334-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morran LT, Schmidt OG, Gelarden IA, Parrish RC 2nd & Lively CM. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science 333, 216–218, doi: 10.1126/science.1206360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paciencia FMD et al. Mating avoidance in female olive baboons (Papio anubis) infected by Treponema pallidum. Sci Adv 5, eaaw9724, doi: 10.1126/sciadv.aaw9724 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butcher RA, Fujita M, Schroeder FC & Clardy J. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat Chem Biol 3, 420–422, doi: 10.1038/nchembio.2007.3 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Fagan KA et al. A single-neuron chemosensory switch determines the valence of a sexually dimorphic sensory behavior. Curr Biol 28, 902–914 e905, doi: 10.1016/j.cub.2018.02.029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenk LA & de Bono M. Memory of recent oxygen experience switches pheromone valence in Caenorhabditis elegans. Proc Natl Acad Sci U S A 114, 4195–4200, doi: 10.1073/pnas.1618934114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang H. et al. Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron 75, 585–592, doi: 10.1016/j.neuron.2012.06.034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo J. & Portman DS Sex-specific, pdfr-1-dependent modulation of pheromone avoidance by food abundance enables flexibility in C. elegans foraging behavior. Curr Biol 31, 4449–4461 e4444, doi: 10.1016/j.cub.2021.07.069 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macosko EZ et al. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458, 1171–1175, doi: 10.1038/nature07886 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryu L. et al. Feeding state regulates pheromone-mediated avoidance behavior via the insulin signaling pathway in Caenorhabditis elegans. EMBO J 37, e98402. doi: 10.15252/embj.201798402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan J. et al. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature 454, 1115–1118, doi: 10.1038/nature07168 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White JQ & Jorgensen EM Sensation in a single neuron pair represses male behavior in hermaphrodites. Neuron 75, 593–600, doi: 10.1016/j.neuron.2012.03.044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DH & Flavell SW Host-microbe interactions and the behavior of Caenorhabditis elegans. J Neurogenet 34, 500–509, doi: 10.1080/01677063.2020.1802724 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aprison EZ & Ruvinsky I. Counteracting ascarosides act through distinct neurons to determine the sexual identity of C. elegans pheromones. Curr Biol 27, 2589–2599 e2583, doi: 10.1016/j.cub.2017.07.034 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Greene JS et al. Balancing selection shapes density-dependent foraging behaviour. Nature 539, 254–258, doi: 10.1038/nature19848 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene JS, Dobosiewicz M, Butcher RA, McGrath PT & Bargmann CI. Regulatory changes in two chemoreceptor genes contribute to a Caenorhabditis elegans QTL for foraging behavior. eLife 5, e21454. doi: 10.7554/eLife.21454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong PY et al. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature 433, 541–545, doi: 10.1038/nature03201 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Kim K. et al. Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science 326, 994–998, doi: 10.1126/science.1176331 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGrath PT et al. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477, 321–325, doi: 10.1038/nature10378 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park D. et al. Interaction of structure-specific and promiscuous G-protein-coupled receptors mediates small-molecule signaling in Caenorhabditis elegans. Proc Natl Acad Sci U S A 109, 9917–9922, doi: 10.1073/pnas.1202216109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chute CD et al. Co-option of neurotransmitter signaling for inter-organismal communication in C. elegans. Nat Commun 10, 3186, doi: 10.1038/s41467-019-11240-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan MW, Rahme LG, Sternberg JA, Tompkins RG & Ausubel FM Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A 96, 2408–2413, doi: 10.1073/pnas.96.5.2408 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sengupta P, Colbert HA & Bargmann CI The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell 79, 971–980, doi: 10.1016/0092-8674(94)90028-0 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Wu T. et al. Pheromones modulate learning by regulating the balanced signals of two insulin-like peptides. Neuron 104, 1095–1109 e1095, doi: 10.1016/j.neuron.2019.09.006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richmond J. Synaptic function. WormBook. 2005. Dec 30:1–14. doi: 10.1895/wormbook.1.69.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsch J. et al. A circuit for gradient climbing in C. elegans chemotaxis. Cell Rep 12, 1748–1760, doi: 10.1016/j.celrep.2015.08.032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bargmann CI & Horvitz HR Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251, 1243–1246, doi: 10.1126/science.2006412 (1991). [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Lu H. & Bargmann CI Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184, doi: 10.1038/nature04216 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Rahme LG et al. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci U S A 94, 13245–13250, doi: 10.1073/pnas.94.24.13245 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan X. et al. SRD-1 in AWA neurons is the receptor for female volatile sex pheromones in C. elegans males. EMBO Rep 20, e46288. doi: 10.15252/embr.201846288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heiman M. et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748, doi: 10.1016/j.cell.2008.10.028 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor SR et al. Molecular topography of an entire nervous system. Cell 184, 4329–4347 e4323, doi: 10.1016/j.cell.2021.06.023 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson HM & Thomas JH. The putative chemoreceptor families of C. elegans. WormBook. 2006. Jan 6:1–12. doi: 10.1895/wormbook.1.66.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilliard MA et al. In vivo imaging of C. elegans ASH neurons: cellular response and adaptation to chemical repellents. EMBO J 24, 63–72, doi: 10.1038/sj.emboj.7600493 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiffer JA et al. Caenorhabditis elegans processes sensory information to choose between freeloading and self-defense strategies. eLife 9, e56186. doi: 10.7554/eLife.56186 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J, Yang W, Liu H, Hao Y. & Zhang Y. An aversive response to osmotic upshift in Caenorhabditis elegans. eNeuro 4, doi: 10.1523/ENEURO.0282-16.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meisel JD, Panda O, Mahanti P, Schroeder FC & Kim DH Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell 159, 267–280, doi: 10.1016/j.cell.2014.09.011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McEwan DL, Kirienko NV & Ausubel FM Host translational inhibition by Pseudomonas aeruginosa Exotoxin A triggers an immune response in Caenorhabditis elegans. Cell Host Microbe 11, 364–374, doi: 10.1016/j.chom.2012.02.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunbar TL, Yan Z, Balla KM, Smelkinson MG & Troemel ER. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe 11, 375–386, doi: 10.1016/j.chom.2012.02.008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bargmann CI. Chemosensation in C. elegans. WormBook. 2006. Oct 25:1–29. doi: 10.1895/wormbook.1.123.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z. et al. Predator-secreted sulfolipids induce defensive responses in C. elegans. Nat Commun 9, 1128, doi: 10.1038/s41467-018-03333-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agger K. et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734, doi: 10.1038/nature06145 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Xu L. & Strome S. Depletion of a novel SET-domain protein enhances the sterility of mes-3 and mes-4 mutants of Caenorhabditis elegans. Genetics 159, 1019–1029, doi: 10.1093/genetics/159.3.1019 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hodgkin J, Horvitz HR & Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91, 67–94, doi: 10.1093/genetics/91.1.67 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agrawal AF & Lively CM. Parasites and the evolution of self-fertilization. Evolution 55, 869–879, 10.1111/j.0014-3820.2001.tb00604.x (2001). [DOI] [PubMed] [Google Scholar]
- 47.Ebert D, Altermatt F. & Lass S. A short term benefit for outcrossing in a Daphnia metapopulation in relation to parasitism. J R Soc Interface 4, 777–785, doi: 10.1098/rsif.2007.0232 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerstes NA, Berenos C, Schmid-Hempel P. & Wegner KM Antagonistic experimental coevolution with a parasite increases host recombination frequency. BMC Evol Biol 12, 18, doi: 10.1186/1471-2148-12-18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karlson P. & Luscher M. Pheromones’: a new term for a class of biologically active substances. Nature 183, 55–56, doi: 10.1038/183055a0 (1959). [DOI] [PubMed] [Google Scholar]
- 50.Lin CC, Prokop-Prigge KA, Preti G. & Potter CJ. Food odors trigger Drosophila males to deposit a pheromone that guides aggregation and female oviposition decisions. eLife 4, e08688. doi: 10.7554/eLife.08688 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94, doi: 10.1093/genetics/77.1.71 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gracida X. & Calarco JA Cell type-specific transcriptome profiling in C. elegans using the Translating Ribosome Affinity Purification technique. Methods 126, 130–137, doi: 10.1016/j.ymeth.2017.06.023 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Chen TW et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300, doi: 10.1038/nature12354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pokala N, Liu Q, Gordus A. & Bargmann CI Inducible and titratable silencing of Caenorhabditis elegans neurons in vivo with histamine-gated chloride channels. Proc Natl Acad Sci U S A 111, 2770–2775, doi: 10.1073/pnas.1400615111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mello CC, Kramer JM, Stinchcomb D. & Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10, 3959–3970, doi: 10.1002/j.1460-2075.1991.tb04966.x. (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ha HI et al. Functional organization of a neural network for aversive olfactory learning in Caenorhabditis elegans. Neuron 68, 1173–1186, doi: 10.1016/j.neuron.2010.11.025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reddy KC, Andersen EC, Kruglyak L. & Kim DH A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science 323, 382–384, doi: 10.1126/science.1166527 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bargmann CI, Hartwieg E. & Horvitz HR Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74, 515–527, doi: 10.1016/0092-8674(93)80053-h(1993). [DOI] [PubMed] [Google Scholar]
- 59.Hodgkin J. & Doniach T. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics 146, 149–164, doi: 10.1093/genetics/146.1.149 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bahrami AK & Zhang Y. When females produce sperm: genetics of C. elegans hermaphrodite reproductive choice. G3 (Bethesda) 3, 1851–1859, doi: 10.1534/g3.113.007914. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, doi: 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCarthy DJ, Chen Y. & Smyth GK Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297, doi: 10.1093/nar/gks042 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang DW, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57, doi: 10.1038/nprot.2008.211 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T and Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50(W1), W216–221, doi: 10.1093/nar/gkac194 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narasimhan K. et al. Mapping and analysis of Caenorhabditis elegans transcription factor sequence specificities. eLife 4, e06967. doi: 10.7554/eLife.06967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chronis N, Zimmer M. & Bargmann CI Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Methods 4, 727–731, doi: 10.1038/nmeth1075 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Askjaer P, Ercan S & Meister P. Modern techniques for the analysis of chromatin and nuclear organization in C. elegans. WormBook. 2014. Apr 2:1–35. doi: 10.1895/wormbook.1.169.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in the manuscript (and its supplementary information data files and the source data files). The statistical tests used in each experiment, n numbers, P values, and other related measures are indicated in each figure and the associated legend and reported in Source Data. All data reported in figures are shown in Source data. The sequencing results (accession#: PRJNA789902) are deposited at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA789902