Abstract
OprF, the major outer membrane protein of Pseudomonas aeruginosa, is multifunctional in that it can act as a nonspecific porin, plays a role in the maintenance of cell shape, and is required for growth in a low-osmolarity environment. The latter two structural roles of OprF, and OprF’s association with the peptidoglycan, have been proposed to be localized in the carboxy terminus of the protein, based on this region’s similarity to members of the OmpA family of proteins. To determine if this is correct, we constructed a series of C-terminally truncated OprF derivatives and examined their effects on P. aeruginosa cell length and growth in low-osmolarity medium. While the C terminus of OprF was required for wild-type cell length and growth in low-osmolarity medium, expression of the N terminus (first 163 amino acids [aa]) also influenced these phenotypes (compared with OprF deficiency). The first 154 to 164 aa of OprF seemed required for stable protein expression, consistent with the existence of a β-barrel domain in the N terminus of OprF. Greater than 215 aa of the protein were required for strong peptidoglycan association, confirming that residues in the C-terminal end of OprF are required for peptidoglycan binding. OprF deficiency did not affect the in vivo growth of an OprF-deficient strain in a mouse chamber model. Collectively, these data suggest that the C terminus of OprF plays a role in cell length, growth of P. aeruginosa in low-osmolarity media (but not in vivo), and peptidoglycan association, while the N terminus has an influence on the first two characteristics and is additionally important for stable protein expression.
Pseudomonas aeruginosa is an opportunistic pathogen that can cause severe infections in cystic fibrosis patients, burn victims, and patients with a weakened immune system (3). P. aeruginosa is of particular concern because of its intrinsic resistance to antibiotics, a property due largely to the low permeability of its outer membrane, coupled with other resistance mechanisms such as periplasmic β-lactamase or efflux, which benefit from the slow diffusion of such antibiotics into the cell (5, 27). Clinical isolates of P. aeruginosa which are multiply antibiotic resistant and deficient in the major outer membrane protein OprF have been obtained (6, 31). OprF has been shown to function as a nonspecific porin (47). However, OprF seems to be multifunctional since it is noncovalently linked to peptidoglycan and involved in the maintenance of cell shape and growth in low-osmolarity environments (15, 18, 26, 47).
OprF is thought to be structurally similar to Escherichia coli OmpA and other related members of the OmpA family of proteins (48). Its structure has been proposed to comprise three domains: an N-terminal domain (first ∼160 amino acids [aa]) containing eight antiparallel β sheets that are proposed to form a β-barrel structure; a loop or hinge region (161 to 209 aa) containing a poly-proline-alanine repeat region and two disulfide bonds; and a C-terminal domain (210 to 326 aa) which is highly conserved with the corresponding domains of other OmpA family proteins. However, controversy surrounds the proposed structure of OprF. Predictions for the structure of the C-terminal domain have ranged from that of a relatively globular domain which is located exclusively in the periplasm (the most prevalent model for OmpA) to a domain containing a significant proportion of anti-parallel β sheets that are associated with the outer membrane (42, 44, 45). There is a need to verify experimentally some of the theories regarding OprF structure, particularly those based on sequence analysis and extrapolation of studies of other OmpA family proteins. For example, it has been proposed that the C-terminal domain of OprF may contain the residues involved in peptidoglycan binding, since this region shows significant sequence similarity with other proteins that are peptidoglycan associated (9, 17); however, this has not yet been confirmed experimentally. The N terminus of OprF is thought to form a β-barrel structure similar to that proposed for the well-studied E. coli OmpA protein; however, this N-terminal region has no significant sequence similarity to any known protein sequence, including OmpA (with the exception of the equivalent region of OprF found in other Pseudomonas species). Studies such as the mapping of OprF surface epitopes and genetically inserted malarial epitopes (23, 44) have been useful in proposing the topology of this region; however, more experimental data are needed to support these structural predictions for OprF. Our understanding of the relationship between the structure of OprF and its function is also still quite poor.
To further elucidate the structure of OprF and its relationship to function, we have now constructed and characterized a series of C-terminally truncated OprF derivatives. We have examined the effects of truncating OprF on such phenotypes as cell shape and growth in low-osmolarity medium and provide evidence consistent with the existence of a β-barrel domain in the N terminus of OprF and of a peptidoglycan-associated domain in the C terminus of the protein. A new OprF-deficient strain was constructed to confirm the phenotypes associated with OprF deficiency, and we also examined the growth of an OprF-deficient P. aeruginosa strain in a mouse chamber model to see if this strain had any growth disadvantage in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids and in vitro growth conditions.
P. aeruginosa H103 (PAO1 prototroph) is our wild-type reference strain (17), and P. aeruginosa H636 is an OprF-deficient mutant of P. aeruginosa H103 previously constructed by Ω mutagenesis (47). P. aeruginosa M-2 is a mouse-pathogenic strain that was originally isolated from the gastrointestinal tract of normal mice (40) and used as the parent strain for construction of a second Ω-mutagenized OprF-deficient strain (M-2F− [this work]). E. coli DH5α [F− φ80dlacZΔM15 endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 Δ(lacZYA-argF)U169] was used for all basic cloning experiments, and E. coli C441 [(pro r− m+)::RP4-2-Tc::Mu-Km::Tn7] was the donor helper strain in biparental mating experiments for the construction of M-2F−. The plasmids used in this study are listed in Table 1. All medium components were from Difco Laboratories (Detroit, Mich.). E. coli strains were grown on Luria-Bertani medium (LB; 1% Bacto Tryptone, 0.5% yeast extract, 1% NaCl [pH 7.0]), and P. aeruginosa strains were grown on Mueller-Hinton medium or LB high-salt medium (LBHS; 1% tryptone, 0.5% yeast extract, 200 mM NaCl [pH 7.0]). The low-osmolarity medium used in growth experiments was either PP2 (1% Protease Peptone no. 2) or salt-free LB (1% tryptone, 0.5% yeast extract [pH 7.0]). Media were solidified with 2% Bacto Agar. Antimicrobial agents were used at the following concentrations for E. coli: tetracycline at 25 μg/ml, ampicillin at 75 or 100 μg/ml, and kanamycin at 25 μg/ml. For P. aeruginosa, the following concentrations were used: tetracycline at 200 μg/ml, kanamycin at 250 μg/ml, streptomycin at 500 μg/ml, and carbenicillin at 300 μg/ml.
TABLE 1.
Plasmids used in this study
| Name | Characteristics and/or origina | Reference |
|---|---|---|
| pUCP19 | pUC19 + 1.8-kb P. aeruginosa stabilizing fragment | 36 |
| pER | pUCP19 + 2-kb fragment coding for Tcr | This study |
| pRW5 | pUCP19 + 1.47-kb HindIII-EcoRI fragment from pRW3 (44) coding for full-length OprF | 43a |
| pER326t | pRW5 + 2-kb fragment coding for Tcr | This study |
| pER290t | Linker insertion mutagenesis of pRW311 (44) so that it encodes only the N-terminal 290 aa of OprF + 2-kb fragment coding for Tcr | This study |
| pER215t | Linker insertion mutagenesis of pRW310 (44) so that it encodes only the N-terminal 215 aa of OprF + 2-kb fragment coding for Tcr | This study |
| pER213t | Linker insertion mutagenesis of pRW309 (44) so that it encodes only the N-terminal 213 aa of OprF + 2-kb fragment coding for Tcr | This study |
| pER188t | Linker insertion mutagenesis of pRW307 (44) so that it encodes only the N-terminal 188 aa of OprF + 2-kb fragment coding for Tcr | This study |
| pER170-26t | pRW5 with its KpnI fragment replaced by the 0.7-kb KpnI fragment from pWW5 (47), + 2-kb fragment coding for Tcr; encodes the first 170 and last 26 aa of OprF | This study |
| pER163t | pER + 0.6-kb HindIII-EcoRI fragment PCR amplified from pRW5 so that it contains a translational stop signal following aa 163 of OprF | This study |
| pFB154 | pUCP19 + 0.6-kb HindIII-EcoRI fragment PCR amplified from pRW5 so that it contains a translational stop signal following aa 154 of OprF | This study |
| pER102t | pRW5 deleted in the 1.1-kb KpnI fragment coding for the C terminus of OprF, + the 2-kb fragment coding for Tcr; encodes the first 102 aa of OprF | This study |
See Materials and Methods for details regarding how the plasmids were constructed.
DNA procedures.
Most common DNA procedures were performed as described by Sambrook et al. (34). Plasmid DNA was routinely isolated by the alkaline lysis method of Birnboim and Doly (2), with further purification by polyethylene glycol precipitation when the DNA was to be used in sequencing reactions (as described in the manual for the Taq DyeDeoxy terminator cycle sequencing kit from Applied Biosystems Incorporated [ABI]). DNA was sequenced with an ABI model 373A automated sequencing system as instructed by the manufacturer, and oligonucleotides were synthesized on an ABI model 392 DNA/RNA synthesizer as described by the manufacturer. Electroporation of P. aeruginosa was done as described by Farinha and Kropinski (10) except that high-salt medium containing 50 mM MgCl2 was used for the recovery period after electroporation. Biparental mating was performed essentially as described by Goldberg and Ohman (14), and P. aeruginosa M-2 was subjected to Ω mutagenesis exactly as described by Woodruff and Hancock (47). For the construction of OprF truncation mutants by PCR, either Vent or Taq DNA polymerase (Fisher Scientific) was used under standard conditions suggested by the manufacturer. PCR was performed in an MJ Research thermal cycler with the following thermal profile repeated 30 times: 95°C for 15 s, 55°C for 30 s, and 72°C for 90 s.
Protein and immunological procedures.
Outer membranes were prepared by the one-step sucrose gradient method of Hancock and Carey (17), and peptidoglycan-associated proteins were prepared by the method of Hancock et al. (18). A modified Lowry assay was used to determine protein concentrations (30). Proteins were prepared for electrophoresis by solubilization in 2% (wt/vol) sodium dodecyl sulfate (SDS) with or without the addition of 2% (wt/vol) 2-mercaptoethanol. Some samples prepared from whole-cell lysates were sonicated three times for 15 s in a sonicating water bath. All samples were heated to 100°C for 10 min immediately before electrophoresis. Heat-treated samples were boiled for a minimum of 30 min in SDS or treated with 5% (wt/vol) trichloroacetic acid (TCA) for 30 min on ice (17). Proteins were separated by electrophoresis in 11, 12, or 14% polyacrylamide gels as previously described (17). The gels were stained with Coomassie brilliant blue R250 (Bio-Rad) for visualization of proteins. OprF-specific monoclonal antibodies were described previously (25, 29, 32). Fast protein liquid chromatography-purified OprF was used to make the polyclonal rabbit and polyclonal mouse sera (32). Colony immunoblotting and Western immunoblotting were performed as described previously (24, 25).
Construction of truncated versions of OprF.
All plasmids constructed in this study were based on pUCP19 and a HindIII/EcoRI insert containing a weakened oprF promoter and a portion of the oprF gene. The weakened oprF promoter permitted wild-type levels of expression of OprF from a multicopy plasmid (reference 44 and this work). Full-length OprF was encoded on plasmid pRW5 (Table 1).
The plasmids encoding truncated versions of OprF (Table 1) were constructed by using the following three basic approaches. First, a KpnI fragment, coding for aa 103 to 170 and 301 to 326 of OprF from pWW5 (47), was used to replace the wild-type KpnI fragment from pRW5, resulting in plasmid pER170-26 (the plasmid number refers to the remaining OprF amino acids, i.e. the N-terminal 170 and C-terminal 26 aa). In a similar manner, plasmid pER102 (encoding the N-terminal 102 aa) was constructed by deleting the 1.1-kb KpnI fragment coding for the C-terminal two-thirds of OprF from plasmid pRW5.
The second approach involved inserting an adapter oligonucleotide, encoding stop codons in all three reading frames, into unique PstI sites present in a set of plasmids encoding linker insertion mutants of OprF (44). Plasmids constructed this way were designated pER188, pER213, pER215, and pER290. This method resulted in the addition of the following amino acids to the C terminus of the truncated protein: DNV (for pER188), VQLDVK (for pER213), LDV (for pER215), and VNAVGY (for pER290).
As none of the linker mutants described above had insertions near the prospective hinge (proline-rich) region of the protein, PCR was used to produce proteins truncated in this region. The primers were designed to introduce a stop codon after a selected amino acid, followed by an EcoRI site to permit cloning (a HindIII site was present on the other primer). The resulting HindIII-EcoRI fragments were cloned into pUCP19. Selection was initially made by colony immunoblotting, selecting for colonies that were reactive with MA7-1. Two mutants, pER158 and pER163, were constructed in this way, but after sequencing it was shown that pER158 contained several unintended sequence rearrangements. The insert in pER163 was fully sequenced to ensure that it was free from errors. No other truncations that expressed fewer than 163 aa and were free from sequence rearrangements could be obtained by this method, which involved screening by colony immunoblotting. However, by screening only for the size of the DNA insert, plasmid pFB154 was constructed successfully by using this PCR method and found to be free of errors.
The addition of carbenicillin to growth media was required to maintain pUCP19-based plasmids in P. aeruginosa. However, the resulting bacterial filamentation compromised the later study of cell length. To solve this problem, a 2.0-kb fragment coding for tetracycline resistance from pHP45Ω-Tc (11) was inserted into the EcoRI site of selected plasmids. The control plasmid, pUCP19, with the added tetracycline resistance cartridge was named pER. The plasmid encoding the entire 326 aa of OprF was named pER326t. Other plasmids had a “t” added to their names to differentiate them from the initial constructs (Table 1).
Cell length measurements.
Bacteria were grown in high-osmolarity medium with 200 μg of tetracycline per ml to mid-logarithmic phase (75 Klett units) or early logarithmic phase (40 Klett units). A sample was dried on a microscope slide, heat fixed, and stained with crystal violet. For image analysis, cells were viewed by oil immersion with a Zeiss Universal microscope at a magnification of ×100. Images were digitized and analyzed with a SEM-IPS image analysis system (Kronton). For microscopy, cells were viewed at a magnification of ×1,000 by oil immersion, using a Zeiss IIIRS microscope fitted with phase rings and connected to a television monitor. Measurements were performed in a single-blinded fashion by Susan Farmer. At least 100 cells of each strain from three different experiments were measured, and the results were analyzed by Student’s t test using the Bonferonni correction.
In vitro growth studies.
Strains of P. aeruginosa with wild-type or mutant OprF were assessed for growth in low-osmolarity medium. Overnight cultures grown in high-salt medium were diluted 1:100 into high- and low-salt media, with or without the addition of antibiotics. Cultures were grown in a shaking water bath at 37°C and 160 to 180 rpm. Cell density was determined by using a Klett-Summerson photometer with a green filter.
In vivo growth studies.
The in vivo chamber model developed by Day et al. (7) was used for growth of P. aeruginosa in mice. Briefly, a Millipore filter (0.2-μm pore size) was glued on one end of the chamber made from a 1-cm section of a 1-ml plastic syringe barrel. Bacteria resuspended in 0.9% saline at approximately 104 cells per ml, as assessed by total count in a Petroff-Hausser bacteria counter, were added to the chambers, and a second filter was attached to the open end. Four chambers were surgically implanted in the peritoneal cavity of B6D2 F1 mice. The chambers were removed from the mice after 4, 8, 16, 20, 24, or 48 h after implantation and plated for a viable count.
RESULTS
Confirmation of the structural roles of OprF.
OprF-deficient strains constructed by chemical mutagenesis have been shown to have a high frequency of reversion (26) and exhibit phenotypic variation (16). Phenotypic variation has also been observed in comparison of OprF-deficient strains insertionally mutagenized by Tn1 mutagenesis or Ω mutagenesis (47), though the Ω-mutagenized strain, named H636, seemed to contain traits most common to all mutants and most varied in phenotype from the parent wild type. Due to this observed phenotypic variation of OprF-deficient strains, and to ensure that the phenotype associated with H636 was not limited to this mutagenized strain, another strain was selected for mutagenesis. This strain, strain M-2, had previously been used in mouse pathogenicity studies (40) and phagocytosis studies (1, 23, 38, 39).
The resulting strain, named M-2F−, was shown by Southern hybridization to contain only one copy of the Ω-cartridge insertion in the chromosome, which was located within the oprF gene (which does not contain a downstream cotranscribed gene [data not shown]). M2F− was found to have phenotypes similar to those of the OprF-deficient strain H636: little to no growth in low-osmolarity medium and a significantly shortened cell length compared with its parent wild type (see below). This similarity in phenotypes between the two constructed OprF− strains, H636 and M-2F−, suggests that these are the true phenotypes associated with OprF deficiency.
OprF-deficient P. aeruginosa has no major growth disadvantage in vivo.
Despite the conditional growth defects of OprF-deficient strains, such strains have been observed in clinical situations (6, 31). Therefore, we used a mouse peritoneal chamber model to determine if the OprF-deficient strain H636 had a growth disadvantage in vivo relative to its parent wild-type strain, H103. This model is useful because it permits the growth of more than one strain of bacteria per animal, in separate chambers, thus reducing the animal-to-animal variation. After a lag of 4 h for H103 and between 4 and 8 h for H636, the strains grew with similar doubling times of approximately 40 and 53 min, respectively, reaching stationary phase after about 20 h. These data suggest that OprF-deficient strains do not have a major growth disadvantage in vivo.
Expression of truncated OprF derivatives.
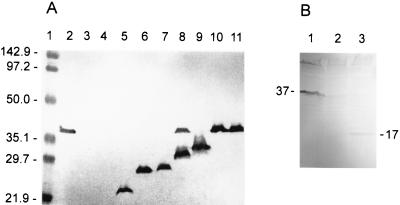
C-terminal truncation of most β-barrel porins results in destabilization of the protein, such that little or no protein is observed (8). We therefore investigated the effect of truncating OprF on expression of the protein. Selected plasmids encoding full-length or truncated OprF (described in Materials and Methods) were electroporated into P. aeruginosa H636, the OprF-deficient strain. As a control, plasmid pER was electroporated into the parent wild-type strain, P. aeruginosa H103. A Western blot of whole-cell lysates of the resulting strains is shown in Fig. 1A. The antibody used was MA7-1, which binds to a linear epitope localized to aa 55 to 62 (12). The results showed that the weakened promoter (see Materials and Methods) allowed good expression of the cloned full-length OprF and of truncated-OprF mutants of 163 aa or larger in an OprF-deficient strain of P. aeruginosa. The relative mobility of these proteins corresponded approximately with the number of amino acids encoded by the plasmid, with specific exceptions: unexpectedly, two bands reactive with MA7-1 were seen with strain H636/pER213t (Fig. 1A, lane 8). One migrated the distance expected for the 213-aa truncated protein, but the other appeared to have the same molecular weight as the native protein. In some experiments, H636/pER290t also appeared to have the same molecular weight as the native protein (Fig. 1A, lane 10). This result suggested either that suppression of the stop codon in these constructs had occurred or that oprF sequences on these plasmids were able to recombine into the chromosome (although it appeared that the plasmid was also retained in the case of H636/pER213t). This apparent recombination-stop codon suppression was not always observed with these constructs and was rarely observed with the other plasmids. However, cultures containing these constructs were monitored closely to ensure that this did not occur during subsequent experiments.
FIG. 1.
(A) Western blot of whole-cell lysates of P. aeruginosa H636 expressing wild-type and truncated OprF proteins encoded on plasmids. The monoclonal antibody used was MA7-1. Plasmid numbers reflect the number of amino acids of OprF that it encodes; e.g., pER102t encodes the first 102 aa of OprF. Lane 1, molecular weight markers; lane 2, H103/pER (positive control); lane 3, H636/pER (negative control); lane 4, H636/pER102t; lane 5, H636/pER163t; lane 6, H636/pER170-26t; lane 7, H636/pER188t; lane 8, H636/pER213t; lane 9, H636/pER215t; lane 10, H636/pER290t; lane 11, H636/pER326t. (B) Western blot of whole-cell lysates of E. coli DH5α containing plasmids encoding wild-type OprF or a truncated product. Product sizes are indicated beside relevant bands (apparent molecular weight in thousands). Lane 1, DH5α/pRW5 (full-length OprF); lane 2, DH5α/pUCP19 (negative control); lane 3, DH5α/pFB154 (the first 154 aa of OprF).
Plasmid pER102t (Fig. 1A, lane 4) did not produce a protein detectable by MA7-1 in P. aeruginosa H636, indicating either that the protein had not been synthesized or that it had been degraded. This plasmid also did not produce a protein detectable by antibody MA7-1 when in an E. coli DH5α background. As mentioned in Materials and Methods, all plasmids encoding fewer than 163 aa of OprF (for example, pER158) had significant DNA arrangements if the constructed plasmid clone was screened for by colony immunoblotting using antibody MA7-1. This suggested to us that deletion to below 163 aa resulted in an unexpressed, degraded, or lethal protein. To further study this, plasmid pFB154 was constructed by screening purely for the correct DNA insert. This plasmid did express in E. coli DH5α a protein which reacted with antibody MA7-1 and corresponded in size to the expected 154-aa product. However, the amount of this product expressed from pFB154 (detected by Western blot analysis of whole-cell lysates) was markedly reduced, and sometimes undetected, compared with levels of expression normally observed for the full-length OprF protein or selected truncated OprF products (Fig. 1B). The cells containing pFB154 grew at a rate comparable to that of cells containing the other plasmids, and pFB154 plasmid DNA extracted from cells after protein expression was of an amount comparable to that of extractions of other OprF-expressing plasmids, suggesting that the reduced amount of protein product from pFB154 was not due to a lethal effect. A proportion of the product of pFB154 may therefore be degraded.
In general, these results suggest that approximately 154 to 163 aa of OprF are necessary for production of a stable protein.
Heat and 2-mercaptoethanol modifiability of OprF truncation derivatives.
The property of heat modifiability refers to a decrease in the mobility of OprF on SDS-polyacrylamide gel electrophoresis (PAGE) when denatured by boiling in SDS or treatment with TCA. When subjected to such treatments, the recombinant OprF encoded on pER326t and pER290t, and the OprF from the wild-type strain H103, were all normally heat modifiable (Table 2). The truncated versions of OprF encoded on pER163t, pER170-26t, pER188t, and pER215t, however, largely ran with an unaltered mobility, except for a portion of the sample which had an increased mobility when pretreated with TCA or boiled in SDS (Table 2). Such modifiability has been observed with truncated derivatives of OmpA (33). Although the explanation for such behavior is unclear, the results are at least inconsistent with heat-TCA-induced cleavage of the protein, since the change in mobility was similar for all mutant proteins, and no common fragment was observed. The results suggest that a structural rearrangement of OprF was occurring, which may be a reflection of the treatment agents (both TCA and SDS are noted for inducing structure formation in peptides [37, 41, 43]). These results in general indicate that between 215 and 290 aa are required for the protein to be normally heat modifiable.
TABLE 2.
Characteristics of truncated-OprF mutants expressed in P. aeruginosa
| Strain/plasmid | Apparent mol wta (103) | Apparent mol wt after heatinga (103) | 2-ME modifiableb |
|---|---|---|---|
| H103/pER | 37 | 41.5 | + |
| H636/pER | − | ||
| H636/pER163t | 23.5 | 20 | − |
| H636/pER188t | 27 | 23.5 | + |
| H636/pER170-26t | 28 | 25.5 | − |
| H636/pER215t | 33 | 28.5 | + |
| H636/pER290t | 35 | 39 | + |
| H636/pER326 | 37 | 41.5 | + |
As assessed by Western blotting.
+, increase in apparent molecular weight after treatment with 2-mercaptoethanol (2-ME); −, no increase.
These truncated-OprF mutant strains were also analyzed for modifiability with 2-mercaptoethanol. OprF contains four cysteines which can form disulfide bridges, and upon treatment with 2-mercaptoethanol, OprF runs in a less compact form, with reduced mobility on SDS-PAGE. This behavior was exhibited by full-length OprF from strain H636/pER326t and by truncated OprF encoded on plasmids pER290t, pER215t, and pER188t (the last of which contains only one pair of cysteines), indicating that the cysteines encoded on these plasmids had formed disulfide bonds (Table 2). The remaining truncated OprF products did not contain cysteines, and their apparent molecular weights were not affected by the addition of 2-mercaptoethanol (Table 2).
Outer membrane and peptidoglycan association of OprF mutants in P. aeruginosa.
Outer membrane preparations were prepared from H103/pER, H636/pER, and H636/pER326t and subjected to SDS-PAGE and Coomassie blue staining. By visual comparison of the samples, it appeared that the cloned OprF was produced at approximately the same levels as the native OprF. Outer membrane preparations were also obtained from cultures of H636 expressing selected truncated OprF proteins (encoded on plasmids pER290t, pER215t, pER188t, pER170-26t, and pER163t). According to a Western blot of these samples probed with antibody MA7-1, all truncated and full-length OprF proteins were associated with the outer membrane. This result suggests that the N-terminal 163 aa of OprF are sufficient for outer membrane association in P. aeruginosa.
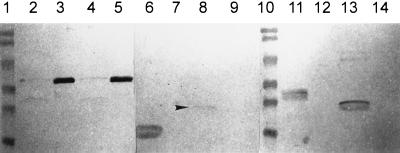
OprF has been shown to be noncovalently associated with the peptidoglycan in strain H103 (17). To determine the regions of OprF involved in peptidoglycan association, Triton-EDTA- and Triton-lysozyme-soluble fractions of outer membrane preparations were prepared (18). The Triton-EDTA-soluble fraction contains proteins that are stabilized in the outer membrane by lipopolysaccharide association with divalent cations like MgCl2. Incubation of the Triton-EDTA-insoluble fraction with lysozyme releases outer membrane proteins that are strongly associated with the peptidoglycan. Full-length OprF from H103/pER and H636/pER326t was found primarily in the Triton-lysozyme-soluble fraction (Fig. 2, lanes 3 and 5). Only a small amount was observed in the Triton-EDTA fraction (Fig. 2, lanes 2 and 4) or the pellet remaining after Triton-lysozyme treatment. All of the truncated proteins tested (encoded on plasmids pER163t, pER170-26t, pER188t, and pER215t) were located in the Triton-EDTA-soluble fraction (Fig. 2, lanes 6, 8, 11, and 13), indicating that they were not strongly associated with the peptidoglycan. These data suggest that more than 215 aa of OprF are required for strong association with the peptidoglycan and confirm that, like the native OprF, the recombinant OprF is peptidoglycan associated.
FIG. 2.
Western blot of outer membrane preparations of P. aeruginosa expressing wild-type or truncated OprF solubilized in Triton-EDTA (lanes 2, 4, 6, 8, 11, and 13) or Triton-lysozyme (lanes 3, 5, 7, 9, 12, and 14). The monoclonal antibody used was MA7-1. Molecular weight markers (lanes 1 and 10), from top to bottom: 112, 84, 53.2, 34.9, 28.7, and 20.5 kDa. Strains used were H103/pER (lanes 2 and 3), H636/pER326t (lanes 4 and 5), H636/pER163t (lanes 6 and 7), H636/pER170-26t (lanes 8 and 9), H636/pER215t (lanes 11 and 12), and H636/pER188t (lanes 13 and 14). The arrowhead indicates the position of the band corresponding to the OprF truncation product of H636/pER170-26t.
Binding of OprF-specific monoclonal antibodies to the truncated OprF mutants.
A series of OprF-specific monoclonal antibodies have been mapped to specific regions or linear epitopes of OprF (12, 23) and were used here for the analysis of the truncated-OprF mutants by colony and Western blot methods. As controls, the strains H636/pER326t and H103/pER were shown to bind all of the monoclonal antibodies tested, while the negative control strain, H636/pER, did not bind any of the antibodies. All of the truncated-OprF mutants studied (containing plasmids pER290t, pER215t, pER188t, pER170-26t, and pER163t) expressed truncated protein which was stable in P. aeruginosa, as shown by their ability to bind antibody MA7-1. Strains H636/pER163t and H636/pER188t did not bind any other antibodies, including MA7-8 and MA4-4, which recognize epitopes that require intact disulfide bonds and have been previously localized between aa 152 and 210 (12). This finding suggests that part or all of these epitopes lie downstream of aa 188 or that the cysteines present in the H636/pER188t product do not form the correct disulfide bond conformation. H636/pER215t bound both MA7-8 and MA4-4, indicating that this truncated protein was not degraded and that the disulfide bond region of this protein had assumed the wild-type conformation. As expected, strain H636/pER290t bound all of the antibodies tested except antibody MA5-8, which binds between amino acid residues 305 and 312 (12). Since seven of the monoclonal antibodies tested recognize conformational epitopes, this finding suggests that the truncated protein folds normally.
Growth of P. aeruginosa expressing truncated OprF derivatives.
OprF is required for the growth of P. aeruginosa in low-osmolarity media (references 16, 26, and 47 and this work). To determine if the entire OprF protein was required for growth under these conditions, the truncated OprF mutants were grown in high- and low-osmolarity media.
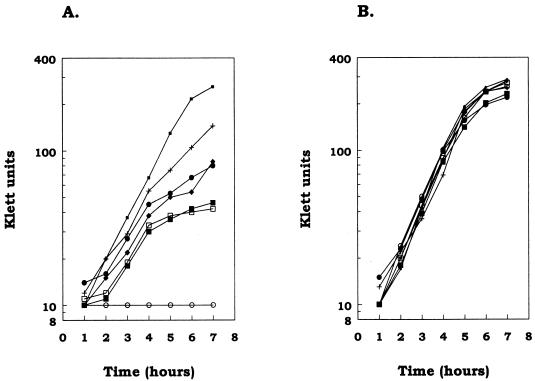
In the initial growth studies, the addition of tetracycline to ensure plasmid maintenance appeared to inhibit the growth of the controls. Since the outer membrane of P. aeruginosa is stabilized by divalent cations, and since tetracycline can act as a divalent-cation chelator (28), MgCl2 was added to the growth media. Figure 3 shows an example of the growth of the truncated OprF strains in low- and high-osmolarity media with the addition of 5 mM MgCl2 and 200 μg of tetracycline per ml. All of the strains tested were able to grow at approximately the same rate in the high-salt medium. In the low-salt medium, H636/pER326t grew at the same rate as the wild-type strain and the OprF-deficient strain H636/pER showed little or no growth during the 7 h tested. In this low-salt medium, the strains with truncated OprF grew at a rate similar to, or slightly less than, the rate for the wild-type strain for about the first 4 h. This initial growth rate lasted from 0 to 4 h in independent experiments. This initial ability to grow at a rate near that of the wild-type strain may have reflected a requirement for adaptation to the low-osmolarity medium. After this initial phase, the remaining strains appeared to fall into two groups. H636/pER290t and H636/pER215t had growth rates less than those of the strains expressing full-length OprF but greater than those of the other strains, with the rate for H636/pER290t being greater than that for strain H636/pER215t. Strains H636/pER163t, H636/pER170-26t, and H636/pER188t always grew at a rate higher than that of the OprF-deficient strain, H636/pER, but lower than that of H636/pER290t or H636/pER215t. Although the growth rate of strain H636/pER163t was higher than those of H636/pER188t and H636/pER170-26t in the experiment presented in Fig. 3, this was not the case in all experiments. One common feature which could differentiate these two groups of strains was that H636/pER290t and H636/pER215t contain a properly folded cysteine disulfide bond region (this work), while H636/pER163t, H636/pER170-26t, and H636/pER188t do not.
FIG. 3.
Growth of P. aeruginosa expressing truncated forms of OprF in salt-free LB plus 200 μg of tetracycline per ml and 5 mM MgCl2 (A) or LBHS plus 200 μg of tetracycline per ml and 5 mM MgCl2 (B). The strains used were H636/pER (○), H636/pER163t (•), H636/pER170/26t (□), H636/188t (▪), H636/pER215t (⧫), H636/pER290t (+), and H636/pER326 (▪). H103 behaved identically to H636/pER326t and was omitted for clarity.
The results from these experiments indicate that strains containing the truncated versions of OprF are able to grow in a low-osmolarity medium, but at a lower growth rate than the wild-type strain. Thus, both the N- and C-terminal regions of OprF contribute to the reconstitution of growth in low-osmolarity medium.
Length of P. aeruginosa cells expressing truncated OprF.
Previous studies have shown that OprF-deficient strains are significantly shorter than their wild-type parent strains, as judged by image analysis (47) and by electron microscopy (16). In this study, cells were grown with 200 μg of tetracycline per ml in LBHS to mid-log phase (50 Klett units), fixed, stained, and measured in a blinded fashion directly from a video monitor connected to a phase-contrast microscope. All of the cells had similar widths but varied in length. Results from one of these experiments are shown in Table 3. The OprF-deficient strain, H636/pER, was 61% of the length of its parent strain, H103/pER. Consistent with this, the newly constructed OprF-deficient strain, M-2F−, was about 70% of the length of the parent strain, M-2. Strain H636 containing plasmid pER163t, pER188t, pER170-26t, pER215t, pER290t, or pER326t was an average of 77, 85, 86, 88, 91 or 97%, respectively, the length of H103/pER. All of the strains expressing truncated OprF were significantly larger than the OprF-deficient strain H636/pER (P ≤ 0.05 in this case and all subsequent comparisons). There was no significant difference between the wild-type strain H103/pER and the strain expressing recombinant full-length OprF, H636/pER326t. There was also no significant difference between H636/pER290t and H636/pER326t. None of the truncated OprF mutants were significantly different from one another, with the exception of H636/pER163t. H636/pER163t appeared to be intermediate in size between OprF-deficient H636/pER and H636 expressing recombinant full-length OprF and was significantly different from both of them. These results indicate that the cloned full-length OprF was able to complement the size defect in the OprF-deficient strain, H636. While truncating OprF did appear to reduce cell length, the cells were never as short as those of the OprF-deficient strain.
TABLE 3.
Lengths of cells of P. aeruginosa expressing full-length or truncated versions of OprF
| Strain/plasmid | Cell lengtha (mean ± SD) |
|---|---|
| H103/pER | 1.57 ± 0.22 |
| H636/pER | 0.96 ± 0.19 |
| H636/pER163t | 1.21 ± 0.23 |
| H636/pER188t | 1.40 ± 0.20 |
| H636/pER170-26t | 1.47 ± 0.21 |
| H636/pER215t | 1.37 ± 0.16 |
| H636/pER290t | 1.41 ± 0.19 |
| H636/pER326t | 1.52 ± 0.20 |
| M-2 | 2.00 ± 0.16 |
| M-2F− | 1.49 ± 0.14 |
At least 100 cells were measured in a blinded fashion, and the mean was normalized to the length of H103/pER as determined by image analysis.
DISCUSSION
Our results provide some important confirmation of theories regarding OprF structure and function. We have demonstrated that residues in the C-terminal domain of OprF are required for strong peptidoglycan association, as previously proposed (9). Expression of our truncated OprF mutants in E. coli or P. aeruginosa suggests that the first 154 to 163 aa of OprF are needed to produce a stable protein. This possibility is in strong agreement with the proposal that the first 160 aa of OprF form a β-barrel structure and suggests that residues critical to proper conformation of this domain are present between aa 154 and 163. The residues between aa 150 and 160 conform to the consensus sequence of the 16th C-terminal β strand of the porin superfamily (21, 22). Studies of other porins, such as E. coli PhoE, suggest that a phenylalanine residue in this terminal strand (equivalent to OprF residue 160) is critical for protein stability and proper folding (8). Our results may thus reflect deletion of such a critical residue. The minimum number of residues required for stable OprF expression correlated with the number required for the expression of OmpA in E. coli (4), even though there is no sequence similarity between OprF and OmpA in this N-terminal region. However, there is growing evidence that porins can adopt a similar β-barrel structure despite having virtually no sequence similarity with one another (35).
Insight into the role of OprF in cell structure and the effect of OprF deficiency was gained through studies of the length and growth in vitro of P. aeruginosa strains expressing truncated OprF derivatives. While all of the truncated OprF mutants examined grew significantly better than the OprF-deficient mutant in a low-osmolarity environment, it seemed that wild-type-length OprF was required for wild-type growth. Likewise for cell length, all of the truncated OprF mutant cells were significantly longer than those of the OprF-deficient mutant, though almost wild-type-length OprF was required for full-length cells. There seemed to be some gradation in both of these properties, with the strain expressing the 163-aa OprF truncation supporting an intermediate growth rate in low-osmolarity medium and intermediate cell length relative to full-length OprF and OprF-deficient strains. Overall, the results suggest that the N-terminal domain of OprF (present in all truncated OprF mutants studied), as well as the C-terminal domain, plays a role in the maintenance of both wild-type cell length and growth in low-osmolarity medium. This has some notable implications; for example, it suggests that the loss of strong association between the peptidoglycan and the outer membrane-anchored OprF protein is not the sole explanation for the change in cell shape observed in OprF-deficient P. aeruginosa (as has been previously alluded to [9, 48]). In fact, our cell length studies suggest that loss of strong peptidoglycan association was no more influential than loss of the N terminus of OprF. For example, the change in cell length between the strain expressing full-length recombinant OprF and the 215-aa truncation (which does not bind to peptidoglycan) is less than the change in cell length between the OprF-deficient strain and cells expressing the 163-aa truncation (which contains the whole proposed β-barrel region). It should be noted that the available methods for assessing peptidoglycan association reveal only strong associations, and so it cannot be discounted that the results are a reflection of a gradually weakening peptidoglycan association as OprF is truncated. However, all of these results seem to reflect a general destabilization of the outer membrane due to truncation or deficiency of OprF, in which both the N terminus and the C terminus cooperatively play a role. The N terminus may act as an additional membrane anchor for the C-terminal regions of the protein which bind peptidoglycan. Possible cooperativity between OprF and other peptidoglycan-associated proteins such as OprL and OprI also needs to be investigated.
While OprF deficiency seems to place a significant degree of stress on the cell, our mouse model studies suggest that P. aeruginosa is not adversely affected by an OprF deficiency during growth in vivo. It is possible that P. aeruginosa requires OprF solely for growth and survival in low-osmolarity niches such as water and that the lack of OprF actually confers an advantage to the cell when confronting a barrage of antibiotics and host defenses during human infection. Its function as a porin has led to the proposal that OprF deficiency plays a role in the resistance of P. aeruginosa to antimicrobial therapy in vivo (20). Moreover, OprF deficiency may also be of advantage to P. aeruginosa when infecting humans, since OprF has been shown to function as a significant antigen (13, 19). It might therefore be to the bacterium’s benefit to rid itself of this protein in order to evade the host’s immune response. We suspect that both of these issues might play a role in explaining the occurrence of OprF deficiency in clinical isolates of P. aeruginosa.
ACKNOWLEDGMENTS
We thank Susan Farmer (Department of Microbiology and Immunology, UBC) for assistance with cell length measurement experiments.
This work was financially supported by the Canadian Cystic Fibrosis Foundation (CCFF) and the Medical Research Council of Canada. E.G.R. and F.S.L.B. were the recipients of a CCFF studentship and CCFF fellowship, respectively.
REFERENCES
- 1.Battershill J, Speert D P, Hancock R E W. Use of monoclonal antibodies against protein F of Pseudomonas aeruginosa as opsonins for phagocytosis by macrophages. Infect Immun. 1987;55:2531–2533. doi: 10.1128/iai.55.10.2531-2533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwood L L, Pennington J E. Influence of mucoid coating on clearance of Pseudomonas aeruginosa. Infect Immun. 1981;32:443–448. doi: 10.1128/iai.32.2.443-448.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bremer E, Cole S T, Hindennach I, Henning U, Beck E, Kurz C, Schaller H. Export of a protein into the outer membrane of Escherichia coli K12: stable incorporation of the OmpA protein requires less than 193 amino-terminal amino-acids residues. Eur J Biochem. 1982;122:223–231. doi: 10.1111/j.1432-1033.1982.tb05870.x. [DOI] [PubMed] [Google Scholar]
- 5.Bryan L E. Resistance to antimicrobial agents: the general nature of the problem and the basis of resistance. In: Doggett R, editor. Pseudomonas aeruginosa: clinical manifestations of infection and current therapy. New York, N.Y: Academic Press; 1979. pp. 219–270. [Google Scholar]
- 6.Chamberland S, Malouin F, Rabin H R, Schollaardt T, Parr T R, Jr, Bryan L E. Persistence of Pseudomonas aeruginosa during ciprofloxacin therapy of a cystic fibrosis patient: transient resistance to quinolones and protein F-deficiency. J Antimicrob Chem. 1990;25:995–1010. doi: 10.1093/jac/25.6.995. [DOI] [PubMed] [Google Scholar]
- 7.Day S E J, Vasil K K, Russel R J, Arbuthnott J P. A simple method for the study in vivo of bacteria growth and accompanying host response. J Infect Dis. 1980;2:39–51. doi: 10.1016/s0163-4453(80)91773-9. [DOI] [PubMed] [Google Scholar]
- 8.de Cock H, Struyvé M, Kleerebesem M, van der Krift T, Tommassen J. Role of the carboxy-terminal phenylalanine in the biogenesis of outer membrane protein PhoE of Escherichia coli K-12. J Mol Biol. 1997;269:473–478. doi: 10.1006/jmbi.1997.1069. [DOI] [PubMed] [Google Scholar]
- 9.De Mot R, Vanderleyden J. MicroCorrespondence. Mol Microbiol. 1994;13:379–380. doi: 10.1111/j.1365-2958.1994.tb00431.x. [DOI] [PubMed] [Google Scholar]
- 10.Farinha M A, Kropinski A M. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol Lett. 1990;70:221–226. doi: 10.1111/j.1574-6968.1990.tb13982.x. [DOI] [PubMed] [Google Scholar]
- 11.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 12.Finnen R L, Martin N L, Siehnel R J, Woodruff W A, Rosok M, Hancock R E W. Analysis of the major outer membrane protein OprF from Pseudomonas aeruginosa using truncated derivatives and monoclonal antibodies. J Bacteriol. 1992;174:4977–4985. doi: 10.1128/jb.174.15.4977-4985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilleland H E, Jr, Parker M G, Matthews J M, Berg R D. Use of a purified outer membrane protein F (porin) preparation of Pseudomonas aeruginosa as a protective vaccine in mice. Infect Immun. 1984;44:49–54. doi: 10.1128/iai.44.1.49-54.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg J B, Ohman D E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984;158:1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotoh N, Wakebe H, Nishino T. Ultrastructural aspects of fragility of Pseudomonas aeruginosa outer membrane devoid of protein F. FEMS Microbiol Lett. 1989;59:51–54. doi: 10.1016/0378-1097(89)90457-6. [DOI] [PubMed] [Google Scholar]
- 16.Gotoh N, Wakebe H, Yoshihara E. Role of protein F in maintaining structural integrity of the Pseudomonas aeruginosa outer membrane. J Bacteriol. 1989;171:983–990. doi: 10.1128/jb.171.2.983-990.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock R E W, Carey A M. Outer membrane of Pseudomonas aeruginosa: heat- and 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979;140:902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock R E W, Irwin R T, Costerton J W, Carey A M. The outer membrane of Pseudomonas aeruginosa: peptidoglycan associated proteins. J Bacteriol. 1981;145:628–631. doi: 10.1128/jb.145.1.628-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock R E W, Mutharia L M, Mouat E C A. Immunotherapeutic potential of monoclonal antibodies against Pseudomonas aeruginosa protein F. Eur J Clin Microbiol. 1985;4:224–227. doi: 10.1007/BF02013602. [DOI] [PubMed] [Google Scholar]
- 20.Hancock R E W, Siehnel R, Martin N. Outer membrane proteins of Pseudomonas. Mol Microbiol. 1990;4:1069–1075. doi: 10.1111/j.1365-2958.1990.tb00680.x. [DOI] [PubMed] [Google Scholar]
- 21.Jeanteur D, Lakey J H, Pattus F. The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 22.Jeanteur D, Lakey J H, Pattus F. The porin superfamily: diversity and common features. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. pp. 363–380. [Google Scholar]
- 23.Martin N L, Rawling E G, Wong R S Y, Rosok M, Hancock R E W. Conservation of surface epitopes in Pseudomonas aeruginosa outer membrane porin protein OprF. FEMS Microbiol Lett. 1993;113:261–266. doi: 10.1111/j.1574-6968.1993.tb06524.x. [DOI] [PubMed] [Google Scholar]
- 24.Mutharia L M, Hancock R E W. Surface localization of Pseudomonas aeruginosa outer membrane porin protein using monoclonal antibodies. Infect Immun. 1983;42:1027–1033. doi: 10.1128/iai.42.3.1027-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mutharia L M, Hancock R E W. Characterization of two surface-localized antigenic sites on porin protein F of Pseudomonas aeruginosa. Can J Microbiol. 1985;31:381–390. doi: 10.1139/m85-073. [DOI] [PubMed] [Google Scholar]
- 26.Nicas T I, Hancock R E W. Pseudomonas aeruginosa outer membrane permeability: isolation of a porin F-deficient mutant. J Bacteriol. 1983;153:281–285. doi: 10.1128/jb.153.1.281-285.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–387. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 28.Nikaido H, Thanassi D G. Penetration of lipophilic agents with multiple protonation sites into bacterial cells: tetracyclines and fluoroquinolones as examples. Antimicrob Agents Chemother. 1993;37:1393–1399. doi: 10.1128/aac.37.7.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pennington J E, Small G E, Lostrom M E, Pier G B. Polyclonal and monoclonal antibody therapy for exponential Pseudomonas aeruginosa pneumonia. Infect Immun. 1986;54:239–244. doi: 10.1128/iai.54.1.239-244.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson G L. A simplification of the protein assay method of Lowry et al. which is more generally acceptable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 31.Piddock L J V, Hall M C, Bellido F, Bains M, Hancock R E W. A pleiotropic, posttherapy, enoxacin-resistant mutant of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1057–1061. doi: 10.1128/aac.36.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rawling E G, Martin N L, Hancock R E W. Epitope mapping of the Pseudomonas aeruginosa major outer membrane porin protein OprF. Infect Immun. 1995;63:38–42. doi: 10.1128/iai.63.1.38-42.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ried G, Koebnik R, Hindennach I, Mutschler B, Henning U. Membrane topology and assembly of the outer membrane protein OmpA of Escherichia coli K12. Mol Gen Genet. 1994;243:127–135. doi: 10.1007/BF00280309. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Schulz G E. Bacterial porins: structure and function. Curr Opin Cell Biol. 1993;5:701–707. doi: 10.1016/0955-0674(93)90143-e. [DOI] [PubMed] [Google Scholar]
- 36.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 37.Sivaraman T, Kumar T K S, Jayaraman G, Han C C, Yu C. Characterization of a partially structured state in an all-beta-sheet protein. Biochem J. 1997;321:457–464. doi: 10.1042/bj3210457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Speert D P, Bond M, Woodman R C, Curnutte J T. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J Infect Dis. 1994;170:1524–1531. doi: 10.1093/infdis/170.6.1524. [DOI] [PubMed] [Google Scholar]
- 39.Speert D P, Thorson L. Suppression by human recombinant gamma interferon of in vitro macrophage nonopsonic and opsonic phagocytosis and killing. Infect Immun. 1991;59:1893–1898. doi: 10.1128/iai.59.6.1893-1898.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stieritz D D, Holder I A. Experimental studies of the pathogenesis of infections due to Pseudomonas aeruginosa: description of a burned mouse model. Infect Immun. 1975;131:688–691. doi: 10.1093/infdis/131.6.688. [DOI] [PubMed] [Google Scholar]
- 41.Takenaka O, Takenaka A, Inada Y. Sodium trichloroacetate-induced helical conformation of poly(l-lysine) Biochim Biophys Acta. 1976;439:302–309. doi: 10.1016/0005-2795(76)90065-9. [DOI] [PubMed] [Google Scholar]
- 42.Vogel H, Jahnig F. Models for the structure of outer-membrane proteins of Escherichia coli derived from Raman spectroscopy and prediction methods. J Mol Biol. 1986;190:191–199. doi: 10.1016/0022-2836(86)90292-5. [DOI] [PubMed] [Google Scholar]
- 43.Waterhous D V, Johnson W C. The importance of environment in determining secondary structure in protein. Biochemistry. 1994;33:212–218. doi: 10.1021/bi00174a019. [DOI] [PubMed] [Google Scholar]
- 43a.Wong, R. Unpublished data.
- 44.Wong R S Y, Jost H, Hancock R E W. Linker-insertion mutagenesis of Pseudomonas aeruginosa outer membrane protein OprF. Mol Microbiol. 1993;10:283–292. [PubMed] [Google Scholar]
- 45.Wong R S Y, Wirtz R A, Hancock R E W. Pseudomonas aeruginosa outer membrane protein OprF as an expression vector for foreign epitopes: the effects of positioning and length on the antigenicity of the epitope. Gene. 1995;158:55–60. doi: 10.1016/0378-1119(95)00155-y. [DOI] [PubMed] [Google Scholar]
- 46.Woodruff W A, Parr T R, Jr, Hancock R E W, Hanne L F, Nicas T I, Iglewski B H. Expression in Escherichia coli and function of Pseudomonas aeruginosa outer membrane porin protein F. J Bacteriol. 1986;167:473–479. doi: 10.1128/jb.167.2.473-479.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woodruff W A, Hancock R E W. Construction and characterization of Pseudomonas aeruginosa porin protein F-deficient mutants after in vivo and in vitro mutagenesis of the cloned protein F gene in Escherichia coli. J Bacteriol. 1988;170:2592–2598. doi: 10.1128/jb.170.6.2592-2598.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodruff W A, Hancock R E W. Pseudomonas aeruginosa outer membrane protein F: structural role and relationship to the Escherichia coli OmpA protein. J Bacteriol. 1989;171:3304–3309. doi: 10.1128/jb.171.6.3304-3309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]