Abstract
CheY serves as a structural prototype for the response regulator proteins of two-component regulatory systems. Functional roles have previously been defined for four of the five highly conserved residues that form the response regulator active site, the exception being the hydroxy amino acid which corresponds to Thr87 in CheY. To investigate the contribution of Thr87 to signaling, we characterized, genetically and biochemically, several cheY mutants with amino acid substitutions at this position. The hydroxyl group appears to be necessary for effective chemotaxis, as a Thr→Ser substitution was the only one of six tested which retained a Che+ swarm phenotype. Although nonchemotactic, cheY mutants with amino acid substitutions T87A and T87C could generate clockwise flagellar rotation either in the absence of CheZ, a protein that stimulates dephosphorylation of CheY, or when paired with a second site-activating mutation, Asp13→Lys, demonstrating that a hydroxy amino acid at position 87 is not essential for activation of the flagellar switch. All purified mutant proteins examined phosphorylated efficiently from the CheA kinase in vitro but were impaired in autodephosphorylation. Thus, the mutant CheY proteins are phosphorylated to a greater degree than wild-type CheY yet support less clockwise flagellar rotation. The data imply that Thr87 is important for generating and/or stabilizing the phosphorylation-induced conformational change in CheY. Furthermore, the various position 87 substitutions differentially affected several properties of the mutant proteins. The chemotaxis and autodephosphorylation defects were tightly linked, suggesting common structural elements, whereas the effects on self-catalyzed and CheZ-mediated dephosphorylation of CheY were uncorrelated, suggesting different structural requirements for the two dephosphorylation reactions.
The signal transduction pathway which governs chemotaxis in Escherichia coli and Salmonella is a well-characterized example of the widely utilized two-component regulatory system (9, 18, 26). The sensor kinase in this pathway, CheA, autophosphorylates at a rate commensurate with the ligand binding status of upstream chemoreceptors. The phosphoryl group is then transferred from CheA to the response regulator CheY, which can also phosphorylate using small-molecule phosphodonors such as acetyl phosphate. Phosphorylated CheY, CheY-P, interacts with flagellar switch proteins to change the direction of flagellar rotation from a default counterclockwise (CCW) to clockwise (CW), altering the swimming mode of the cell. CheY-P can dephosphorylate by a self-catalyzed mechanism or at a much greater rate with the assistance of another chemotaxis protein, CheZ.
CheY has been studied extensively in an attempt to understand the phosphorylation and concomitant activation of response regulators in general. This protein has been used to elucidate the functional roles of active-site residues that are conserved among response regulators. Of five highly conserved active-site residues, four have had functions ascribed to them: Asp57 is the site of phosphorylation (20), Asp12 and Asp13 are involved in coordination of a Mg2+ ion that is essential for phosphorylation and dephosphorylation to occur (24), and Lys109 is involved in the phosphorylation-induced conformational change (16). The function of Thr87, which is also highly conserved within this family of proteins, has not been established.
The position corresponding to Thr87 in CheY is occupied by a hydroxy amino acid (threonine or serine) in virtually all known response regulators (30). The strikingly high degree of conservation at this position, as well as its proximity to the site of phosphorylation in the three-dimensional structure of CheY (17, 21, 24, 25, 31), has prompted speculation about the function of this residue. It has been reported that Thr87 participates in an extensive hydrogen bonding network which may be critical to defining the structure of the active site (31). This observation, in part, has led to the candidacy of Thr87 as a putative proton donor in the CheY phosphorylation reaction (27) or as a participant in the hydrolysis of the acyl phosphate (20). Thr87 has also been proposed as an alternate site of phosphorylation (31) for a mutant CheY molecule in which the primary site of phosphorylation, Asp57, has been rendered nonphosphorylatable by amino acid substitution (4). Recent investigations have ruled out the potential proton donor (23), hydrolysis (7), and phosphorylation site (1) roles. A Thr87→Ile substitution, generated by random mutagenesis and identified by virtue of being nonchemotactic in vivo, was found to be phosphorylatable in vitro, suggesting that the hydroxyl group may be necessary for one or more postphosphorylation signaling events (7).
We report here a comprehensive investigation of the role of Thr87. We have generated and characterized, both genetically and biochemically, six mutants with amino acid substitutions at position 87 in an endeavor to better understand the function of this highly conserved hydroxyl residue. Our results confirm that while Thr87 contributes to an active-site conformation that allows optimal phosphorylation and autodephosphorylation, the residue is not necessary for those catalytic activities. The most important role of Thr87 may be to stabilize the activated conformation of CheY-P.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli KO641 recA carries Δ(cheY)6021 (4). RP5231 carries Δ(cheY-cheZ)4313 (14) and was a gift from J. S. Parkinson (University of Utah).
The ptrp cheYZ oripBR322 (ori from pBR322) plasmid pRBB40 has been described previously (4). The ptrp cheY oripBR322 plasmid pRBB38 was made from pRL22ΔZ (19) by filling in the cohesive ends at the HindIII site, religating to create a NheI site, and converting the NdeI sites at nucleotides 180 and 1216 to BamHI and HindIII sites, respectively, by ligation of synthetic linkers.
The cheYD13K allele has been described previously (4). The cheYT87A, cheYT87C, cheYT87E, cheYT87K, and cheYT87S alleles were created by the site-specific mutagenesis method (13) using dut ung and with M13 cheYZ as a template (4). The cheYT87I allele (7) was a gift of P. Matsumura (University of Illinois, Chicago). Mutant cheY alleles were moved between various vectors and combined to form double mutants by standard techniques.
Phenotype characterization.
Motility agar (1% tryptone, 0.5% NaCl, 0.3% Bacto Agar) plates were stabbed with isolated colonies of KO641 recA/pRBB40 carrying wild-type or mutant cheY genes. Plates were incubated at 30°C for ∼12 h. Diameters of resultant swarms were measured periodically to determine relative swarm rates, where indicated. In order to assess the activity of mutant CheY proteins, the rotational bias of the cells was examined by the tethered-cell assay as previously described (5).
Immunoblots.
Immunoblot analysis was used to assess the relative concentrations of CheY in the strains used in genetic analysis and was performed essentially as described previously (22). Strains containing the pRBB40 plasmid (mutant or wild type) in the KO641 recA background were grown in tryptone broth to an optical density at 600 nm (OD600) between 0.4 and 0.7 at 30°C, conditions identical to those used for the tethering experiments. Cell extracts from 1.0 ml of the cultures were prepared as described previously (22), and portions of the extracts were electrophoresed on a sodium dodecyl sulfate (SDS)–15% polyacrylamide gel. Sample volumes loaded onto the gel were adjusted for slight variations in cell density before the cells were harvested. For the immunoblots, the primary antibody was either purified anti-CheY monoclonal antibody (gift of Birgit Scharf, Harvard University) or affinity-purified polyclonal anti-CheY (gift of Philip Matsamura, University of Illinois, Chicago) and the secondary antibody was peroxidase-conjugated goat anti-mouse immunoglobulin G (Sigma). Detection was by Pierce Supersignal Chemiluminescent system according to the manufacturer’s instructions. The relative intensities of the bands were assessed by eye.
Protein purification.
CheA and CheZ were purified as previously described (8). CheY (wild type and mutant) was purified by a modified version of the previously described method (8). One liter of Luria-Bertani medium LB plus ampicillin (100 μg/ml) was inoculated with 5 ml of a saturated overnight culture of KO641 recA/pRBB40 carrying the cheY mutation of interest. The culture was grown at 37°C with shaking until the OD600 was ≈1, then 3-β-indoleacrylic acid was added to a concentration of 100 μg/ml, and the culture was incubated an additional 16 to 20 h. Cells were harvested by centrifugation for 15 min at 4,000 × g, and the pellet was resuspended in ∼20 ml of TEG (50 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 10% [vol/vol] glycerol). The cells were lysed by sonication and subjected to ultracentrifugation for 1 h at 90,000 × g to remove membranes and cellular debris. The cell lysate was applied to a 10-ml Affi-Gel Blue (Bio-Rad) column equilibrated with TEG and eluted by using a step gradient of 1 M NaCl in TEG. The CheY-containing fractions were pooled, dialyzed against TEG, and applied to a 10-ml DE-52 column equilibrated with TEG. This column was washed extensively with TEG, as some mutant CheY proteins bleed out in washes, and eluted with 0.1 M NaCl in TEG. CheY-containing fractions were pooled, concentrated in Centricon-10 ultrafiltration units, and loaded onto a Superdex-75 fast protein liquid chromatography gel filtration column (Pharmacia). Fractions containing CheY were pooled, concentrated, and stored at −20°C.
Phosphorylation assays.
Steady-state phosphorylation of CheY by CheA was measured in an assay previously described (4). In a 10-μl reaction mixture, 14 pmol of CheA, 70 pmol of CheY, and 14 pmol of CheZ (where indicated) were incubated in a solution containing 50 mM KCl, 5 mM MgCl2, and 50 mM Tris-HCl, pH 7.5. Phosphorylation reactions were initiated by addition of [γ-32P]ATP to a final concentration of 0.3 mM ATP. The reactions were stopped after 10 min by addition of 2× SDS sample buffer (0.125 M Tris-Cl [pH 6.8], 4% SDS, 20% glycerol, 10% 2-mercaptoethanol). The proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 20% polyacrylamide gels, such that Pi and ATP no longer remained on the gels. The gels were dried and exposed to film for autoradiography or a phosphor screen for phosphorimaging.
[32P]CheA-P was used to phosphorylate CheY in an assay which allows comparison of dephosphorylation rates of wild-type and mutant CheY proteins. First, CheA was phosphorylated with [γ-32P]ATP, and radioactive CheA-P was purified essentially as described previously (8). To initiate phosphotransfer, 40 pmol of [32P]CheA-P and 180 pmol of CheY were combined in a solution containing 10 mM MgCl2, 25 mM Tris-HCl, pH 7.5, in a total reaction volume of 45 μl. These reactions were incubated at ambient temperature, and 5-μl aliquots were removed at indicated time points to 5 μl of 2× SDS sample buffer. The reaction products were separated by SDS-PAGE on 15% polyacrylamide gels (Pi was not electrophoresed off). The gels were dried and exposed to a phosphor screen for phosphorimaging and quantitation. Under these conditions, phosphotransfer from CheA-P to CheY occurs before removal of the first time point and the time course of loss of Pi from CheY-P can be monitored without contribution from phosphotransfer. First-order rate constants (k) describing autodephosphorylation were determined as described previously (23).
To assess the sensitivities of the various mutant CheY proteins to CheZ, rates of dephosphorylation of CheY-P were measured as described above but in the presence of various amounts of CheZ (0, 12, 45, or 360 pmol). First-order rate constants for dephosphorylation of CheY-P were plotted versus CheZ concentration. The slope of the resultant line was used as a measure of the CheZ sensitivity and compared to that of wild-type CheY.
RESULTS
Effects of CheY Thr87 substitution mutants on chemotaxis.
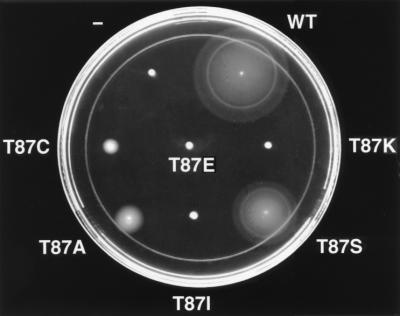
To ascertain the functional significance of the hydroxy amino acid at position 87, we characterized mutants in which Thr87 was replaced by amino acid side chains spanning the spectrum of possible chemical properties. The chosen substitutions were serine (conserved hydroxyl), cysteine (sulfhydryl), alanine (small nonpolar), glutamate (negative charge), lysine (positive charge), or the previously characterized isoleucine (hydrophobic). These mutants were first assayed for chemotactic ability by the swarm agar assay (Fig. 1). Cells carrying cheYT87S displayed chemotactic activity, as evidenced by their ability to form rings demarcating gradients of aspartate and serine, and swarmed at a rate approximately 60% of the rate of cells carrying wild-type cheY in the same strain background. The observation that this mutant supports successful, albeit somewhat slower, chemotaxis is not surprising, as a significant fraction of known response regulators have serine at this position (30). All of the other substitutions examined abolished chemotaxis, but the swarm phenotypes fell into two classes: mutants carrying a glutamate, lysine, or isoleucine at position 87 displayed tight nonchemotactic swarms, whereas those with an alanine or cysteine substitution were able to generate larger nonchemotactic swarms with diffuse edges. This behavior indicates that, although the cheYT87A and cheYT87C mutants cannot successfully follow a chemoattractant gradient, they may be capable of achieving some CW flagellar rotation (33).
FIG. 1.
Ability of CheY Thr87 mutants to display chemotaxis through soft agar. Semisolid agar was stabbed with E. coli KO641 recA containing no plasmid (−) or plasmid pRBB40 encoding wild-type cheY (WT) or cheY mutants with any of the indicated substitutions at position 87. The plate was incubated at 30°C for 12 h.
Ability of mutant CheY proteins to generate CW rotation.
The ability of these mutants to generate CW flagellar rotation was analyzed by the tethering assay (Fig. 2, column A). Consistent with its Che+ swarm phenotype, the cheYT87S mutant displayed frequent reversals, like the wild type. Despite yielding two distinct swarm phenotypes, each of the other mutants displayed exclusively or predominantly CCW rotation, suggesting a loss of cheY function. The apparent discrepancy between the two assays may indicate that the alanine and cysteine mutants are capable of some low level of signaling activity, resulting in only infrequent reversals. These rare reversal events may allow cells to swim out from the point of inoculation over the course of a 12-h swarm assay but may not be detected in the tethering assay, which employs a comparatively short period of analysis.
FIG. 2.
Effect of substitution at Thr87 on rotational behavior. E. coli strains harboring plasmids that encode wild-type (WT) or mutant cheY were grown in tryptone broth at 30°C and tethered to glass coverslips with antiflagellar antibodies (5). For each sample, 20 cells were examined for 20 s/cell, and rotational behavior was recorded in one of seven categories from exclusively CCW (CCW) to exclusively CW (CW). The results are depicted as histograms. Columns A to C refer to strain and plasmid background as follows: (A) KO641 recA (ΔcheY) carrying pRBB40 encoding CheY with the indicated amino acid at position 87 as well as CheZ; (B) KO641 recA carrying pRBB40 encoding CheY with both an Asp→Lys substitution at position 13 and the indicated amino acid at position 87 as well as CheZ; (C) RP5231 (ΔcheY-cheZ) carrying pRBB38 encoding CheY alone with the indicated amino acid at position 87. ND, not done.
In an attempt to delineate the nature of the functional defect introduced by substitution at position 87, we combined those substitutions that resulted in loss of activity (alanine, cysteine, glutamate, lysine, or isoleucine) with a second site-activating mutation. When conserved active-site residue Asp13 is replaced by lysine, the resultant mutant CheY displays phosphorylation-independent CW signaling activity (3, 4). Therefore, double mutants were constructed to carry a lysine at position 13 and each of the aforementioned loss-of-function substitutions at position 87. Mutants with a glutamate, lysine, or isoleucine at position 87 exhibited exclusively CCW flagellar rotation and thus are inactive, despite the presence of a lysine at position 13 (Fig. 2, column B). Double mutants carrying an alanine or cysteine at position 87, however, displayed a significant recovery of CW signaling activity compared to those of the CheYT87X single mutants. This suggests that the hydroxyl group at position 87 is not necessary for directly activating the flagellar switch, as an alanine or cysteine at this position does not preclude the generation of CW rotation favored by the cheYD13K mutation. However, it must also be noted that even those cheYD13K cheYT87X double mutants that displayed reversal were considerably less active than the cheYD13K single mutant.
The lysine at position 13 may mimic phosphorylation by disrupting active-site contacts present in the unphosphorylated wild-type protein and repositioning critical residues to facilitate transition to an active conformation (11). Thr87 may play a role in the propagation of this conformational change, whether it is initiated by phosphorylation or the cheYD13K mutation. Therefore, we hypothesized that the CheYT87X single mutants may not be sufficiently phosphorylated in the cheZ+ background to generate CW rotation with a frequency scorable by tethering. To test this hypothesis, cheYT87X single mutant genes were subcloned onto a plasmid lacking cheZ, the product of which greatly enhances dephosphorylation of CheY-P, and the resultant plasmids were used to transform RP5231, a strain with both cheY and cheZ deleted. As shown in Fig. 2, column C, cells expressing mutant CheY proteins with glutamate, lysine, or isoleucine at position 87 remained CCW in a cheZ mutant background, whereas those with an alanine or cysteine at this site displayed frequent reversal. This clearly demonstrates that the CheYT87A and CheYT87C mutants can adopt a conformation that interacts productively with the flagellar switch to cause CW rotation and that CheZ interferes with this activity. These mutants, however, had impaired activity compared to that of wild-type CheY, which displayed exclusively CW rotation in the same background.
Phosphorylation properties of mutant CheY proteins.
The loss of CW rotation observed for Thr87 mutants might have been due, at least partially, to lower amounts of CheY-P present in these strains. The following scenarios, independently or in combination, would result in a reduced steady-state level of phosphorylated CheY molecules available in a cheZ mutant cell to interact with the flagellar switch: (i) lower amount of CheY due to decreased level of expression or instability of the protein, (ii) increased rate of autodephosphorylation, or (iii) a severely decreased rate of phosphorylation. The possibility that any of the strains containing the amino acid substitutions at position 87 had a reduced CheY concentration (e.g., via enhanced proteolysis) was directly tested by immunoblot analysis which showed that the mutant CheY proteins were present at concentrations similar to that of wild-type CheY (data not shown). Furthermore, all of the CheY proteins that we attempted to purify were recovered in high yield without evidence of degradation.
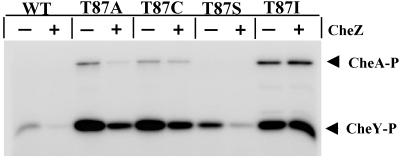
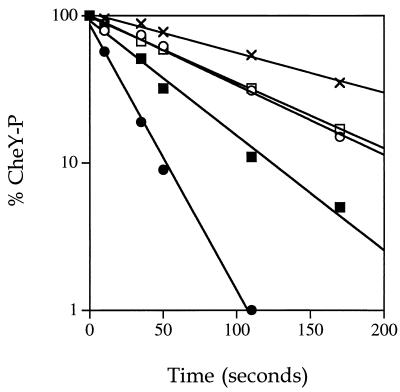
To identify possible perturbations in the phosphorylation and/or dephosphorylation kinetics of mutant CheY proteins relative to wild-type CheY, several phosphorylation assays were carried out on purified CheY proteins. Figure 3 shows the results of an assay that measures steady-state phosphorylation of CheY by CheA in the presence of [γ-32P]ATP. In this experiment, the degree of phosphorylation of CheY reflects the balance between the rates of phosphotransfer from CheA-P and the autodephosphorylation of CheY. For wild-type CheY, there is a relatively low amount of CheY-P present under these conditions (Fig. 3). All of the mutants tested, CheYT87A, CheYT87C, CheYT87I, and CheYT87S, were phosphorylated in this assay. Moreover, there was considerably more mutant than wild-type CheY-P (Fig. 3). This could be due to either an increased rate of phosphotransfer to the mutants or lower rates of autodephosphorylation. Lower rates of autodephosphorylation were directly demonstrated by using an assay in which purified [32P]CheA-P was used to phosphorylate CheY proteins. Under the conditions of this experiment (micromolar protein concentrations and 5:1 ratio of CheY to CheA-P), essentially all of the label was transferred from CheA-P to wild-type CheY by the first time point (10 s). Subsequent time points showed the loss of radioactivity from CheY-P due to the intrinsic autodephosphorylation activity (Fig. 4). Wild-type CheY-P displayed a half-life of ∼14 s (k = 0.049 s−1) at 25°C, which is consistent with published reports (16, 23). CheYT87S-P was found to be slower than the wild type in autodephosphorylation by approximately twofold (k = 0.021 s−1). CheYT87A-P and CheYT87C-P both demonstrated a roughly fourfold reduction in rate (k = 0.012 s−1 for both). CheYT87I-P was approximately six to sevenfold slower than the wild-type protein in our assay (k = 0.0073 s−1), which is in agreement with previous reports for this mutant (7). Therefore, although the degree of impairment varied, all mutants with substitutions at position 87 that were examined were found to have reduced rates of autodephosphorylation. This observation implies that the increased amounts of mutant CheY-P (Fig. 3) can be explained by autodephosphorylation defects in the mutant proteins.
FIG. 3.
Steady-state phosphorylation properties of CheY proteins with substitutions at position 87. Purified CheA kinase (14 pmol) and wild-type (WT) or mutant CheY protein (70 pmol), in the absence (−) or presence (+) of CheZ (14 pmol), were incubated in the presence of 0.3 mM [γ-32P]ATP as described in Materials and Methods. Reaction products were separated by SDS-PAGE. A phosphorimager scan of a dried gel from one such experiment is shown.
FIG. 4.
Autodephosphorylation rates of wild-type and mutant CheY proteins. CheY proteins were phosphorylated by incubation with purified radiolabelled CheA-P (see Materials and Methods), and aliquots were removed to 2× SDS sample buffer at various time points. The proteins were separated on 15% polyacrylamide gels, and dried gels were analyzed by phosphorimaging. The results are plotted as percent phosphorylated CheY remaining at indicated times. The zero time point represents labelling of CheY after a 10-s incubation with CheA-P. CheY proteins are indicated as follows: wild-type CheY (•), CheYT87S (▪), CheYT87A (□), CheYT87C (○), CheYT87I (×).
The conclusion that diminished rates of autodephosphorylation account for the increased amounts of mutant CheY-P in the steady-state experiment (Fig. 3) is further supported by the presence of [32P]CheA-P in the mutant but not the wild-type samples in the steady-state experiment (Fig. 3). For the mutants, the flow of Pi through the system is retarded due to slowed autodephosphorylation of CheY and subsequent unavailability of free CheY, resulting in a backup of CheA-P. The relative amounts of [32P]CheA-P present at steady state (none detectable in CheYT87S, moderate amounts in CheYT87A and CheYT87C, and a large amount in CheYT87I) correlate exactly with what is expected from the relative autodephosphorylation rates of these proteins (Fig. 4). Taking the phosphorylation experiments together (Fig. 3 and 4), we conclude that the position 87 mutants would be expected to have at least as much CheY-P as the wild-type bacteria. Therefore, the loss of activity observed for these mutant CheY proteins can be attributed to CheY-P species which are impaired in their interaction with the flagellar switch.
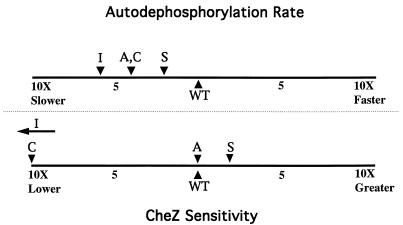
To investigate the sensitivity of the mutant CheY proteins to CheZ, dephosphorylation rates were directly measured in the presence of various amounts of CheZ and the enhancement of dephosphorylation rate relative to autodephosphorylation was quantified as described in Materials and Methods. Interestingly, there appeared to be no correlation between the degree of impairment in autodephosphorylation and the degree of resistance to CheZ (Fig. 5). Although all mutants tested had impaired autodephosphorylation, only two of these displayed reduced sensitivity to CheZ: CheYT87C, which displayed a roughly 10-fold loss of sensitivity, and CheYT87I, which appeared to be CheZ resistant, consistent with previous reports (36). CheZ accelerated the rate of CheYT87A dephosphorylation to a degree similar to that of wild-type CheY. CheYT87S appeared to be slightly more sensitive to CheZ than wild-type CheY, which is in agreement with a published observation (36). Implications of these findings are presented in the Discussion.
FIG. 5.
Comparison of autodephosphorylation rates and degrees of stimulation by CheZ for mutant CheY proteins containing various amino acids at position 87. (Top) Autodephosphorylation rates were derived from the data of Fig. 4 and are plotted in comparison to wild-type CheY. (Bottom) A series of dephosphorylation experiments similar to those displayed in Fig. 4 were done in the presence of different concentrations of CheZ. The magnitudes of rate enhancement achieved per amount of CheZ added are plotted in comparison to wild-type CheY. WT, wild type.
DISCUSSION
Thr87 is not essential for the generation of CW rotation.
In the present study, we have characterized several CheY mutants to further our understanding of the functional role of conserved residue Thr87. A hydroxyl group at this position was found to be critical for chemotaxis, as the only functionally tolerated substitution was Thr→Ser (Fig. 1). Two other mutants which did not support chemotaxis, CheYT87A and CheYT87C, were nevertheless partially active in the absence of CheZ or when paired with a second site-activating mutation, Asp→Lys at position 13 (Fig. 2, columns B and C). Therefore, although the hydroxyl group may be necessary for the environmentally coordinated modulation of CheY activity that permits efficient chemotaxis, it is not strictly essential for the ability to generate CW flagellar rotation.
Thr87 plays a role in signaling by CheY.
Although two of the substitutions at position 87, T87A and T87C, showed some ability to generate CW rotation, this activity could be observed only in the absence of CheZ and was, under all conditions, significantly reduced compared to that of wild-type CheY (Fig. 2, column C). Several experimental observations eliminated the possibility that the mutant phenotypes were due to reduced levels of CheY-P. First, none of the mutant CheY proteins were detected by immunoblotting to be present in the cell at concentrations less than that of wild-type CheY. Second, biochemical characterization of the phosphorylation and dephosphorylation properties of the purified CheY proteins indicate that there should be at least as much CheY-P present in cells encoding mutant CheY as wild-type cells. Therefore, we conclude that the mutant CheY-P proteins are defective in signaling.
Thr87 plays a role in the phosphorylation-induced conformational change of CheY.
It has been proposed that CheY exists in an equilibrium between active and inactive conformations (11). Phosphorylation presumably shifts the equilibrium strongly towards the active conformation, as CheY-P has been reported to display a CW signaling ability roughly 100-fold greater than that of unphosphorylated CheY (2). In addition to activating the flagellar switch, several reactions in which CheY participates may serve as indicators of structural differences between active and inactive CheY. For instance, CheY-P displays greater binding affinity than CheY for FliM or CheZ and reduced affinity for CheA. CheY exhibits a catalytic phosphorylation activity, whereas CheY-P adopts a structure promoting catalytic autodephosphorylation. It is striking that the rank order of autodephosphorylation ability in the CheY Thr87 substitution mutants (Thr > Ser > Ala,Cys > Ile) (Fig. 4) is the same as the rank order of CW signaling ability (Fig. 2). The simplest way to account for this correlation is to postulate that Thr87 affects a common step in CheY function prior to either the autodephosphorylation reaction or the interaction of CheY-P with FliM, specifically achievement of the active conformation following phosphorylation. Furthermore, this linkage strongly suggests common structural requirements for the autodephosphorylation and FliM signaling reactions.
In light of the above, it appears likely that the autodephosphorylation defects seen here for mutants of CheY at position 87 are due to misalignment of pertinent residues in CheY-P. An alternate possibility is that the observed autodephosphorylation defects (Fig. 4) reflect a catalytic rather than structural role for Thr87. However, the increased stability of the phosphoryl group on the mutant CheY proteins is not great enough to conclude that the hydroxyl is necessary for catalytic autodephosphorylation, as noncatalytic hydrolysis of an acyl phosphate is on the order of hours (12). A hydroxy amino acid is present at this position in almost all known response regulators (30), and these proteins display widely different dephosphorylation rates, with half-lives ranging from seconds in the case of CheY to hours in the case of OmpR (10). The possibility that the interaction of a threonine or serine with nonconserved residues in the active sites of these proteins may, in part, help to modulate the rates of dephosphorylation displayed by the response regulators cannot be excluded.
The structural requirements for autodephosphorylation and CheZ-mediated dephosphorylation of CheY-P are different.
There is no correlation between the autodephosphorylation rates of the Thr87 mutants and the degrees of sensitivity to CheZ-mediated dephosphorylation (Fig. 5). For instance, although the rates of autodephosphorylation of CheYT87A-P and CheYT87C-P were indistinguishable, CheZ accelerated CheYT87A-P dephosphorylation to a degree similar to that of wild-type CheY-P, whereas CheYT87C-P displayed a roughly 10-fold loss of sensitivity to CheZ. Similarly, CheYT87S-P showed a twofold decrease in autodephosphorylation rate compared to that of wild-type CheY-P but appeared to be slightly more sensitive to CheZ than wild-type CheY-P, as previously noted by Zhu et al. (36). It may be that the differences in sensitivity to CheZ that we observed are due entirely or in part to variations in CheZ binding affinities. Nonetheless, these data suggest that the structural requirements for autodephosphorylation and CW signaling are distinct from those mediating CheZ sensitivity.
Proposed molecular basis of mutant phenotypes.
The loss of function substitutions can be divided into two classes with regard to in vivo phenotype and chemical properties of the amino acid introduced at position 87. Those substitutions that rendered cells nonchemotactic but able to generate some CW rotation were cysteine and alanine (Fig. 2). Both of these differ from serine or threonine in that they are not hydroxy amino acids. However, both of these have relatively small uncharged side chains that presumably do not significantly disrupt distal structure and are unlikely to disallow steric shifts of other important residues which may be induced by phosphorylation. Therefore, although they are unable to simulate the active contribution made by the hydroxyl group of a serine or threonine, these substitutions may not preclude the repositioning of other amino acids whose phosphorylation-induced movements favor enhanced activity at the flagellar switch. Those substitutions for which CW rotation was not observed under any of the conditions examined were lysine, glutamate, and isoleucine (Fig. 2). Lysine and glutamate have charged hydrophilic side chains, whereas isoleucine is a nonpolar hydrophobic residue. What these amino acids share in common is their relatively large size. Thus, not only are they unable to engage in the same hydroxy-mediated interactions as a threonine, their large side chains may sterically obstruct the phosphorylation-induced shifts of other critical residues necessary for activation. In the case of the CheYT87I mutant, this has been directly demonstrated by X-ray crystallographic analysis (7, 35). In these reports, the isoleucine side chain at position 87 was found to occupy a cavity in the CheY protein that in wild-type CheY may be occupied upon phosphorylation by the rotomeric side chain of Tyr106. Because of the bulk and hydrophobicity of the isoleucine, the Tyr106 side chain is forced to reside exclusively in the outside, solvent-exposed position which, from studies of CheY proteins mutated at that position, negatively correlates with activity at the flagellar switch (34, 35). In wild-type CheY, it has been demonstrated that Thr87 forms a hydrogen bond with the hydroxyl group of Tyr106 through one intervening solvent molecule (35), perhaps stabilizing the internal position of this tyrosine. A simple explanation in light of this hypothesis, then, may be that a lysine or glutamate at position 87 also prevents or impedes the inward shift of Tyr106, and that an alanine or cysteine, while not actively stabilizing the solvent-inaccessible placement of this tyrosine, may nonetheless allow for unhindered rotation and putative interactions with other amino acid side chains that favor an internal position for Tyr106.
Role of the conserved active-site hydroxy amino acid in other response regulators.
It should be noted that, although the residue corresponding to CheY Thr87 is conserved among response regulators, similar mutations at the analogous site in other members of this superfamily of proteins may yield quite different phenotypes. For instance, a Thr→Ile substitution at position 82 in FixJ (32) or a Thr→Ala substitution at this position in Spo0F (29) renders the FixJ and Spo0F proteins severely impaired in their ability to accept phosphate from the cognate sensor kinases FixL and KinA, respectively, whereas CheYT87I and CheYT87A phosphorylate readily from CheA. It has been reported by Brissette et al. (6) that a Thr83→Ala mutant of E. coli OmpR was activated in the absence of its cognate sensor kinase, EnvZ. Furthermore, this substitution was able to suppress the inactive phenotype of the OmpRD55Q mutant in which the site of phosphorylation was replaced with a glutamine. In contrast, CheYT87A does not exhibit constitutive phosphorylation-independent activity. It has been postulated that, despite structural homology and the high degree of conservation of several key residues among response regulators, each member of this superfamily may utilize a different subset of a global conformational change (15, 28). Moreover, it may be that different response regulators employ similar conformational changes to contrasting ends, and interactions between conserved residues and nonconserved residues in different proteins would be expected to render different consequences. Therefore, perhaps Thr83 of OmpR engages in important interactions with other residues in the unphosphorylated protein that maintain an inactive conformation. Disruption of these putative interactions upon phosphorylation may serve as the activation “switch” of OmpR. An alanine at position 83 may be unable to participate in these critical interactions in unphosphorylated OmpR, allowing the protein to adopt an active state in the absence of the disruptive influence of phosphorylation. If this model is correct, this conserved hydroxy residue may be primarily important for the stabilization of the prephosphorylation conformation in some response regulators and the postphosphorylation conformation in others.
ACKNOWLEDGMENTS
We thank Phil Matsumura and Xiangyang Zhu for strains and communication of results prior to publication, Birgit Scharf and Howard Berg for anti-CheY antibody and advice on its use, and Ruth Silversmith for numerous helpful discussions and assistance with the manuscript. We also thank Michael Eisenbach, Phil Matsumura, Sandy Parkinson, and Rick Stewart for detailed comments on an early version of the manuscript.
This work was supported by Public Health Service grant GM-50860 from the National Institute of General Medical Sciences (to R.B.B.).
REFERENCES
- 1.Appleby, J. L., and R. B. Bourret. Activation of CheY mutant D57N by phosphorylation at an alternate site, Ser56. Submitted for publication. [DOI] [PubMed]
- 2.Barak R, Eisenbach M. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry. 1992;31:1821–1826. doi: 10.1021/bi00121a034. [DOI] [PubMed] [Google Scholar]
- 3.Bourret R B, Drake S K, Chervitz S A, Simon M I, Falke J J. Activation of the phosphosignaling protein CheY. II. Analysis of activated mutants by 19F NMR and protein engineering. J Biol Chem. 1993;268:13089–13096. [PMC free article] [PubMed] [Google Scholar]
- 4.Bourret R B, Hess J F, Simon M I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci USA. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray D, Bourret R B, Simon M I. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol Biol Cell. 1993;4:469–482. doi: 10.1091/mbc.4.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brissette R E, Tsung K, Inouye M. Intramolecular second-site revertants to the phosphorylation site mutation in OmpR, a kinase-dependent transcriptional activator in Escherichia coli. J Bacteriol. 1991;173:3749–3755. doi: 10.1128/jb.173.12.3749-3755.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganguli S, Wang H, Matsumura P, Volz K. Uncoupled phosphorylation and activation in bacterial chemotaxis: the 2.1-Å structure of a threonine to isoleucine mutant at position 87 of CheY. J Biol Chem. 1995;270:17386–17393. [PubMed] [Google Scholar]
- 8.Hess J F, Bourret R B, Simon M I. Phosphorylation assays for proteins of the two-component regulatory system controlling chemotaxis in Escherichia coli. Methods Enzymol. 1991;200:188–204. doi: 10.1016/0076-6879(91)00139-n. [DOI] [PubMed] [Google Scholar]
- 9.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 10.Igo M M, Ninfa A J, Stock J B, Silhavy T J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- 11.Jiang M, Bourret R B, Simon M I, Volz K. Uncoupled phosphorylation and activation in bacterial chemotaxis: the 2.3-Å structure of an aspartate to lysine mutant at position 13 of CheY. J Biol Chem. 1997;272:11850–11855. doi: 10.1074/jbc.272.18.11850. [DOI] [PubMed] [Google Scholar]
- 12.Koshland D E., Jr Effect of catalysts on the hydrolysis of acetyl phosphate: nucleophilic displacement mechanisms in enzymatic reactions. J Am Chem Soc. 1952;74:2286–2292. [Google Scholar]
- 13.Kunkel T A, Roberts J D, Zakour R. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 14.Liu J D, Parkinson J S. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci USA. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowry D F, Roth A F, Rupert P B, Dahlquist F W, Moy F J, Domaille P J, Matsumura P. Signal transduction in chemotaxis. A propagating conformation change upon phosphorylation of CheY. J Biol Chem. 1994;269:26358–26362. [PubMed] [Google Scholar]
- 16.Lukat G S, Lee B H, Mottonen J M, Stock A M, Stock J B. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem. 1991;266:8348–8354. [PubMed] [Google Scholar]
- 17.Moy F J, Lowry D F, Matsumura P, Dahlquist F W, Krywko J E, Domaille P J. Assignments, secondary structure, global fold and dynamics of chemotaxis Y protein using three- and four-dimensional heteronuclear (13C, 15N) NMR spectroscopy. Biochemistry. 1994;33:10731–10742. doi: 10.1021/bi00201a022. [DOI] [PubMed] [Google Scholar]
- 18.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 19.Roman S J, Meyers M, Volz K, Matsumura P. A chemotactic signaling surface on CheY defined by suppressors of flagellar switch mutations. J Bacteriol. 1992;174:6247–6255. doi: 10.1128/jb.174.19.6247-6255.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanders D A, Gillece-Castro B L, Stock A M, Burlingame A L, Koshland D E., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- 21.Santoro J, Bruix M, Pascual J, Lopez E, Serrano L, Rico M. Three-dimensional structure of chemotactic CheY protein in aqueous solution by nuclear magnetic resonance methods. J Mol Biol. 1995;247:717–725. doi: 10.1006/jmbi.1995.0175. [DOI] [PubMed] [Google Scholar]
- 22.Scharf B E, Fahrner K A, Turner L, Berg H C. Control of direction of flagellar rotation in bacterial chemotaxis. Proc Natl Acad Sci USA. 1998;95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silversmith R E, Appleby J L, Bourret R B. The catalytic mechanism of phosphorylation and dephosphorylation of CheY: kinetic characterization of imidazole-phosphates as phosphodonors and the role of acid catalysis. Biochemistry. 1997;36:14965–14974. doi: 10.1021/bi9715573. [DOI] [PubMed] [Google Scholar]
- 24.Stock A M, Martinez-Hackert E, Rasmussen B F, West A H, Stock J B, Ringe D, Petsko G A. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 25.Stock A M, Mottonen J M, Stock J B, Schutt C E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989;337:745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- 26.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1103–1129. [Google Scholar]
- 27.Stock J B, Surrette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 25–51. [Google Scholar]
- 28.Swanson R V, Alex L A, Simon M I. Histidine and aspartate phosphorylation: two-component regulatory systems and the limits of homology. Trends Biochem Sci. 1994;19:485–490. doi: 10.1016/0968-0004(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 29.Tzeng Y-L, Hoch J A. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J Mol Biol. 1997;272:200–212. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- 30.Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 31.Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-Å resolution. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 32.Weinstein M, Lois A F, Monson E K, Ditta G S, Helinski D R. Isolation of phosphorylation-deficient mutants of the Rhizobium meliloti two-component regulatory protein, FixJ. Mol Microbiol. 1992;6:2041–2049. doi: 10.1111/j.1365-2958.1992.tb01377.x. [DOI] [PubMed] [Google Scholar]
- 33.Wolfe A J, Berg H C. Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA. 1989;86:6973–6977. doi: 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu X, Amsler C D, Volz K, Matsumura P. Tyrosine 106 of CheY plays an important role in chemotaxis signal transduction in Escherichia coli. J Bacteriol. 1996;178:4208–4215. doi: 10.1128/jb.178.14.4208-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X, Rebello J, Matsumura P, Volz K. Crystal structures of CheY mutants Y106W and T87I/Y106W: CheY activation correlates with movement of residue 106. J Biol Chem. 1997;272:5000–5006. doi: 10.1074/jbc.272.8.5000. [DOI] [PubMed] [Google Scholar]
- 36.Zhu X, Volz K, Matsumura P. The CheZ-binding surface of CheY overlaps with the CheA- and FliM-binding surfaces. J Biol Chem. 1997;272:23758–23764. doi: 10.1074/jbc.272.38.23758. [DOI] [PubMed] [Google Scholar]