Abstract
Thymic epithelial cells (TECs) produce glucocorticoids, which antagonize negative selection of autoreactive thymocytes and promote a competent T cell antigen–specific repertoire. To characterize their source, we generated a knock-in reporter mouse in which endogenous Cyp11b1, the final enzyme in de novo production of active glucocorticoids, was fluorescently tagged with mScarlet. Here, we find that Cyp11b1 is expressed in medullary TECs (mTECs) but not cortical TECs or other cells in the thymus. A distinct characteristic of mTECs is the presence of Aire, a transcription factor that drives expression of tissue-restricted antigens (TRAs) important for establishing immune tolerance. Cyp11b1 expression was highest in Aire+ mTECs, lower in post-Aire mTECs, and absent in mTECs of Aire-deficient mice. Transcriptomic analyses found that multiple enzymatic biosynthetic pathways are expressed specifically in mTECs and are also Aire dependent. In particular, we found that the thymus expresses messenger RNA for enzymes that catalyze production of many bioactive steroids and that glucocorticoids and sex steroids were secreted by cultured thymi. Expression of the transcripts for these genes and production of their final steroid products were markedly reduced in the absence of Aire. Thus, in addition to its well-established role in inducing TRAs that promote negative selection, Aire has an additional and contrary function of inducing glucocorticoids that antagonize negative selection, which together may expand and enhance the TCR repertoire. Furthermore, because Aire drives expression of multiple enzymes responsible for production of other non–gene-encoded bioactive molecules, it might have yet other roles in thymus development and function.
INTRODUCTION
Glucocorticoids are cholesterol-derived steroid hormones that circulate in the blood to act on cells and tissues throughout the body and have pleiotropic functions in organismal development and the control of metabolism, immunity, and stress responses (1). De novo biosynthesis of glucocorticoids occurs via a cascade of cytochrome P450 and dehydrogenase enzymes that concludes with the final and critical enzyme Cyp11b1. Historically, the sole source of systemic glucocorticoids was thought to be the adrenals (2). However, in 1994, thymic epithelial cells (TECs) were identified as cells capable of synthesizing glucocorticoids (3). This prompted an investigation of how locally produced glucocorticoids, acting in a paracrine manner, might affect thymocyte development. Thymocytes undergo a process of random T cell antigen receptor (TCR) recombination that generates a huge repertoire of specificities that allow the adaptive immune system to recognize and respond to almost any possible pathogen. However, strongly autoreactive or nonfunctional TCRs must be purged to ensure self-tolerance and effective T cell responses. This antigen-specific selection occurs when thymocytes with subthreshold TCR signaling in response to self-peptide:major histocompatibility complex (MHC) die by neglect, those with moderate signaling survive (positive selection), and those with strong signaling die by apoptosis (negative selection). This process begins in the thymic cortex, where CD4+8+ [double positive (DP)] thymocytes are tested against ubiquitous self-peptides and specific thymoproteasome-, cathepsin L–, and thymus-specific serine protease (TSSP)–generated peptides presented by cortical thymic epithelial cells (cTECs), which appear to function primarily in positive selection (4–9). Antigen-signaled DP thymocytes up-regulate TCR levels (DP TCRhi), migrate to the thymic medulla, and become CD4+CD8− or CD4−CD8+ [single positive (SP)] thymocytes. Ectopic expression of tissue-restricted antigens (TRAs) by thymic medullary epithelial cells (mTECs) ensures negative selection of cells bearing TCRs with high affinity for self (10, 11). The surviving SP thymocytes mature, exit the thymus, and comprise the peripheral T cell repertoire.
The thymic stroma expresses the full suite of enzymes needed to synthesize glucocorticoids de novo from cholesterol (3, 12, 13). The in vitro finding that glucocorticoids inhibit TCR-induced apoptosis led to the hypothesis that they might antagonize negative selection and allow the survival of positively selected cells that otherwise would die (14, 15). This was confirmed by generating mice with Foxn1-Cre–mediated conditional deletion of Cyp11b1. The resulting TEC-specific Cyp11b1 knockout mice had an altered T cell antigen–specific repertoire that resulted in diminished T cell responses to peptide antigens and alloantigen in vitro and impaired immunity to viral infections in vivo (16). Epithelial-derived glucocorticoids do not signal throughout the thymus, as might be expected for lipophilic steroid molecules, but instead are targeted specifically to antigen-signaled DP TCRhi thymocytes (17). Thus, targeted glucocorticoid synthesis and signaling within the thymus antagonizes TCR signaling to promote thymocyte survival and strengthen the TCR repertoire (18). Glucocorticoid synthesis by thymocytes has also been reported (19, 20), although this was not corroborated by others (21, 22), and thymocyte-specific Cyp11b1 deletion had no effect on thymocyte development or T cell responses (16).
Although the impact of de novo glucocorticoid synthesis on antigen-specific thymocyte development is clear, it is unknown whether synthesis occurs in cTECs, thus altering positive selection by distinct cTEC-generated antigens, or in mTECs, altering negative selection by Aire- and Fezf2-driven TRAs. Because the glucocorticoid-signaled DP TCRhi stage is a very brief period in thymocyte development and occurs at a transitional point when cells traffic from the cortex to the medulla, either thymus compartment, or both, could be the site of glucocorticoid synthesis. However, synthesis in the cortex versus medulla would result in glucocorticoid-promoted tolerance to a very different set of antigens. In the cortex, this would increase the range of positively selected TCRs, increasing the diversity of TCRs that progress to the medulla, while leaving the signaling thresholds for negative and agonist selection unaltered. In the medulla, this would instead increase the degree of TCR self-reactivity destined for inclusion in the mature repertoire and perhaps also shift signaling thresholds for agonist selection. At a more fundamental level, targeted glucocorticoid synthesis and signaling are a distinct mechanism of steroid signaling, and it is unclear how such targeted glucocorticoid synthesis can occur, because glucocorticoids, like other steroids, diffuse freely across membranes and are not obviously stored in vesicles for targeted release [but see (23)].
This study aimed to identify which TEC subsets, and potentially other cells in the thymus, synthesize glucocorticoids and how this synthesis is regulated. We found that de novo glucocorticoid synthesis is specific to mTECs and requires Aire. Unexpectedly, a large number of enzymes involved in the synthesis of many steroids and other biologically active compounds was also found to be Aire dependent, indicating that Aire has important roles in addition to driving the expression of TRAs.
RESULTS
A Cyp11b1 KI reporter mouse allows detection of glucocorticoid-synthetic tissues
Glucocorticoids are synthesized de novo from cholesterol via the stepwise activity of a transporter (StAR) and four enzymes (Cyp11a1, 3β-HSD, Cyp21a1, and Cyp11b1) (Fig. 1A). Cyp11b1 is required solely for the biosynthesis of glucocorticoids, whereas upstream enzymes also function in the biosynthesis of progestins, androgens, estrogens, and mineralocorticoids (24). Furthermore, Cyp11b1 is responsible for conversion of inactive precursors (11-deoxycorticosterone in mice and 11-deoxycortisol in humans) to active glucocorticoids (corticosterone in mice and cortisol in humans). Cyp11b1 is thus the definitive mammalian glucocorticoid-synthetic enzyme (24). Mice have more than 100 protein-coding CYP (cytochrome P450) family genes (25), and many of these are highly conserved, complicating immunodetection of Cyp11b1. We tested a panel of anti-Cyp11b1 antibodies using human embryonic kidney (HEK) 293T cells overexpressing a green fluorescent protein (GFP)–tagged or a Myc-FLAG–tagged mouse Cyp11b1 fusion protein. Using flow cytometry, only one antibody gave a Cyp11b1-specific signal, and this was very weak, with a ~30-fold range in Cyp11b1GFP fluorescence corresponding to a twofold range in anti-Cyp11b1 fluorescence (fig. S1A). Western blotting was also unsuccessful, with none of the antibodies giving Cyp11b1-specific signals (fig. S1B). Anti-Cyp11b1 staining also gave equivalent fluorescence in adrenal cells from control and Cyp11b1-deficient mice, as detected by flow cytometry (fig. S1C). These results were not due to protein inaccessibility, because an overexpressed Cyp11b1Myc-FLAG fusion protein was readily detected by anti-FLAG staining using immunoblotting (fig. S1B) and flow cytometry (fig. S1D). Mass spectrometric analyses of purified cTEC and mTEC also failed to detect Cyp11b1 protein (26).
Fig. 1. Cyp11b1mScarlet KI reporter mice indicate that mTECs are the source of de novo synthesized glucocorticoids in the thymus.
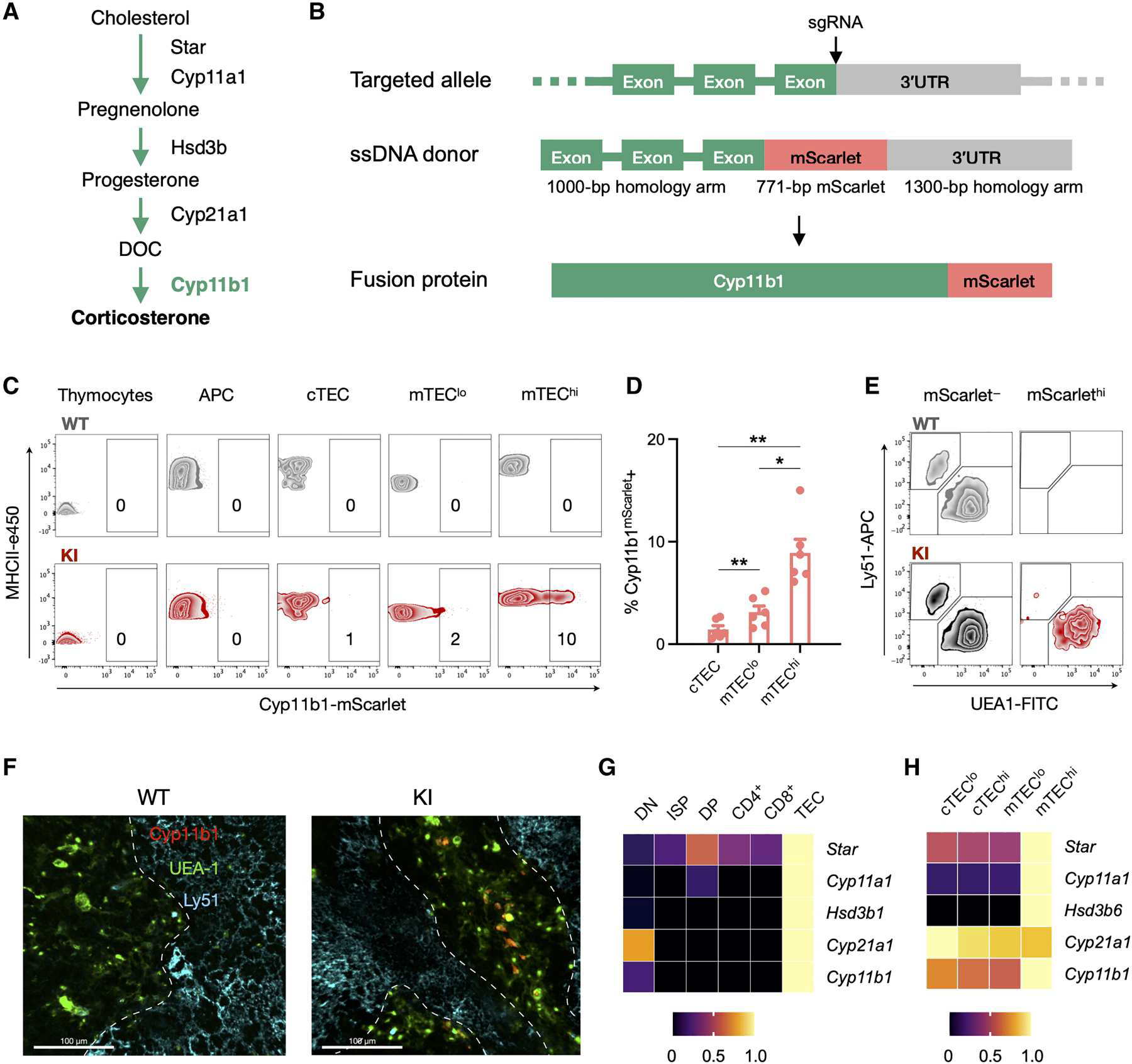
(A) Simplified metabolic pathway for de novo synthesis of glucocorticoids in the mouse. (B) CRISPR-Cas9 targeting strategy to generate KI reporter mice expressing an mScarlet-tagged Cyp11b1 fusion protein (Cyp11b1mScarlet). A guide RNA was selected to cut at the end of the last Cyp11b1 exon to induce homology-directed repair with a single-strand DNA donor containing the mScarlet coding sequence immediately preceding the Cyp11b1 stop codon. (C) Thymi from 2-week-old WT and Cyp11b1mScarlet KI mice were digested, and cells were analyzed by flow cytometry (thymocytes: CD45+EpCAM−MHCII−Ly51−UEA1−; APCs: CD45+EpCAM−MHCII+Ly51−UEA1−; cTEC: CD45− EpCAM+Ly51+UEA1−MHCII+; mTEClo: CD45−EpCAM+Ly51−UEA1+MHCII+; mTEChi: CD45−EpCAM+Ly51−UEA1+MHCIIhi). Contour plots show each subset, and numbers indicate the percentage of cells within the mScarlet+ gate. Data are representative of three independent experiments. (D) Pooled frequencies determined as in (C), presented as mean ± SEM. (C and D) The data are representative of or show data from seven WT and six KI mice, pooled from three independent experiments with 1- to 3-week-old mice. (E) Flow cytometry contour plots of total CD45−EpCAM+MHCII+ TEC isolated from 2-week-old mice were gated for the absence or presence of mScarlet expression, according to fig. S3B (mScarlet− = bottom 70% and mScarlethi = top 5 to 10% of mScarlet fluorescence), and evaluated for expression of Ly51 and UEA-1. (F) Thymi from 1-week-old WT and KI mice were fixed, stained with antibodies against Ly51 (cortex, Alexa Fluor 647), UEA-1 (medulla, FITC), and mScarlet-Atto 565. Confocal images were taken at ×20 magnification, with a 20-μm z-stack. Scale bars, 100 μm. The dashed white lines indicate the corticomedullary junction. Data are representative of two independent experiments, with N = 3 WT and 3 KI mice. (G and H) Gene expression of glucocorticoid biosynthetic enzymes in thymocyte and TEC subsets of the thymus. Bulk RNA-seq data were acquired from the ImmGen database, and each gene is shown relative to its maximum expression level across the analyzed cell types, as indicated by the color scale. Significance between KI subsets in (D) was tested using a mixed linear model with follow-up paired Student’s t tests applying Holm-Šidák correction for multiple comparisons. *P < 0.05 and **P < 0.01. 3′UTR, 3′ untranslated region.
Because antibody staining and mass spectrometry were inadequate for identification of Cyp11b1-expressing cells, we used CRISPR-Cas9 targeting to generate mice expressing an mScarlet-tagged Cyp11b1 fusion protein knocked into the endogenous Cyp11b1 locus [Cyp11b1mScarlet knock-in (KI) mice] (Fig. 1B). Cyp11b1mScarlet alleles were verified by polymerase chain reaction (PCR) (fig. S2A), and mScarlet fluorescence was detectable by microscopy in the adrenal cortex (fig. S2B). Unlike Cyp11b1-deficient mice (16), Cyp11b1mScarlet homozygous mice had normal adrenal mass, spleen mass (fig. S2C), and plasma corticosterone responses to a stressor (fig. S2D), demonstrating that the mScarlet tag did not affect Cyp11b1 function. Flow cytometric analysis of trypsin-digested tissues confirmed that mScarlet-expressing cells were present in the adrenals but were undetectable in brains, livers, and lungs (fig. S2E).
mTECs are the source of de novo glucocorticoid synthesis
To identify the cellular source of thymus glucocorticoids, we used flow cytometry to quantify Cyp11b1mScarlet fluorescence in different thymus cell subsets of the KI mice. Thymic lobes of 1- to 2-week-old were minced and digested, and a portion were enriched for epithelial cells over a Percoll gradient to obtain sufficient TECs for analysis. Consistent with most previous analyses of glucocorticoid production (3, 12, 13, 16, 17) [but see (19, 20) for exceptions], the detection of Cyp11b1mScarlet was restricted to TECs, with no signal detected in thymocytes or CD45+ MHCII+ antigen-presenting cells (APCs) (Fig. 1, C and D). Given that cTECs and mTECs have different functions in thymocyte development, we distinguished these based on the staining of Ly51 and Ulex europaeus agglutinin I (UEA-1), with cTECs being UEA-1−Ly51+ and mTECs being UEA-1+Ly51− (fig. S3, A and B). Cells expressing high levels of Cyp11b1mScarlet (mScarlethi) were nearly all UEA-1+Ly51− mTECs (Fig. 1E and fig. S3C). This same pattern was observed in thymi at embryonic day 18.5 (fig. S3, D and E). Fluorescence microscopy of 2-week-old mouse thymic sections found Cyp11b1mScarlet fluorescence colocalized with UEA-1 in the medulla but not with Ly51 in the cortex (Fig. 1F). These results identify mTECs as the major thymus cell expressing Cyp11b1 and capable of de novo glucocorticoid production.
We undertook transcriptomic analyses of Cyp11b1 and upstream genes involved in steroid biosynthesis. Bulk RNA sequencing (RNA-seq) analysis of various thymus cell subsets, acquired from the ImmGen database (27), showed that the full cascade of genes needed for de novo glucocorticoid synthesis is expressed in TECs and not thymocytes (Fig. 1G). Steroid-efflux transporter genes, which actively export glucocorticoids across cell membranes and may increase or spatially direct secretion, were also expressed by TECs specifically (fig. S3F), consistent with their targeted delivery of paracrine glucocorticoids to TCR-signaled DP thymocytes (16, 17). RNA-seq data from sorted TEC subsets were analyzed to compare expression patterns within mTEC subsets, and glucocorticoid-synthetic enzymes were primarily expressed in mTEChi cells (Fig. 1H). Reverse transcription quantitative PCR (RT-qPCR) showed that whole thymus Cyp11b1 expression was also lost in Rag2−/− mice in which the thymic medulla was underdeveloped because of a lack of thymocyte-derived receptor activator of NF-κB ligand (RANKL), which mTECs depend on for differentiation (fig. S3G) (28). These findings suggest that mTECs express the full enzyme cascade required for de novo glucocorticoid synthesis.
Cyp11b1 is expressed in Aire+ and post-Aire mTEC cells and decreases with age
To further identify characteristics of Cyp11b1-expressing cells, we analyzed single-cell transcriptomic data. Cyp11b1 transcripts were not detectable in datasets obtained from total mouse TECs (29, 30), mouse cTECs (29, 30), or human TECs (31) but were detectable in sorted mouse Pdpn− CD104− mTEClo cells. This cell sample contained Aire-expressing mTECs but was particularly enriched for post-Aire mTECs (32). Although Cyp11b1 transcripts were only detectable in a small number of cells, which may be in part because mTEClo cells were analyzed, these were all located in the same mTEC cell cluster (Fig. 2A). The primary identifying characteristic of this mTEC cluster was expression of Aire (Fig. 2A) and genes involved in antigen presentation (data file S1). Analysis of differentially expressed genes in this cluster compared with all other clusters showed that Aire, Cd40, H2-DMa, H2-DMb1, H2-DMb2, H2-Eb2, H2-M3, H2-Oa, H2-Ob, and H2-Q10 were among the top overexpressed genes. This cluster also had the greatest expression of other genes involved in glucocorticoid synthesis (fig. S4A) and was the only cluster in which all glucocorticoid synthesis genes were expressed (fig. S4B), with the exception of Cyp21a1, which was not detected in any cells. Confocal imaging confirmed that whereas most Aire+ TECs do not express Cyp11b1mScarlet, at least half of the Cyp11b1mScarlet-expressing TECs also express Aire (Fig. 2, B and C). Furthermore, purified mScarlet+ mTECs and mScarlet− mTECs had similar levels of Aire gene expression by RT-qPCR (fig. S4C).
Fig. 2. Cyp11b1 is expressed by Aire+ mTECs and keratin 10+ post-Aire mTECs.
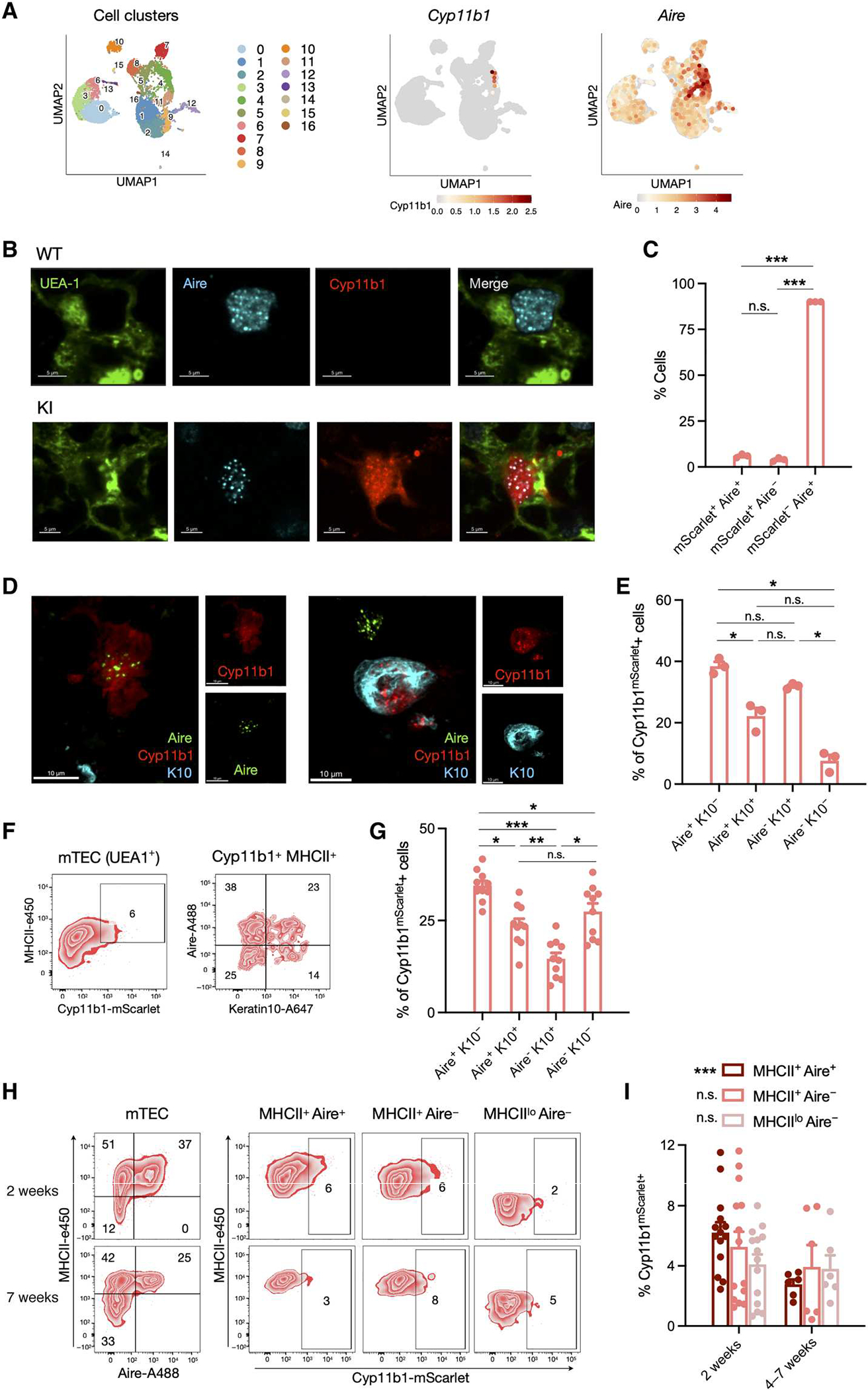
(A) Single-cell transcriptomes from sorted Pdpn− CD104− mTEClo (32) were analyzed for expression of Cyp11b1 and Aire, and the relative expression, indicated by the color scale, was mapped onto a UMAP plot. Numbered populations indicate different cTEC and mTEC cell populations as defined in fig. S4. (B) Thymi from 1-week-old WT and Cyp11b1mScarlet KI mice were fixed, stained for Ly51 (cortex), UEA-1 (medulla), mScarlet-Atto 565, and Aire. Confocal images were taken at ×60 oil immersion magnification. (C) Data from 3936 Aire+ and/or mScarlet+ cells (114 fields from three KI mice) collected as in (B) were analyzed for coexpression of Aire and Cyp11b1mScarlet, shown as mean ± SEM. (D) Thymi from 2-week-old KI mice were fixed and stained for Aire (Alexa Fluor 488), keratin 10 (K10, Alexa Fluor 647), and Cyp11b1mScarlet (Atto 565), and confocal images were taken at ×60 oil immersion magnification. (E) Data from 872 Cyp11b1mScarlet+ cells (114 fields from three KI mice) collected as in (D) were analyzed for expression of Aire and keratin 10. (F) Thymi from 2-week-old KI mice were digested, and cells were analyzed by flow cytometry. Representative contour plots showing mTEC (CD45−EpCAM+UEA1+; left) and Cyp11b1-mScarlet+ MHCIIhi mTEC (right) subsets. (G) Pooled data for samples analyzed as in (F) of 10 KI mice from two independent experiments, presented as mean ± SEM. (H) Thymi collected from 2- and 7-week-old KI mice were digested, and cells were analyzed by flow cytometry. Representative contour plots show Cyp11b1 expression in MHCII+Aire+, MHCII+Aire−, and MHCIIloAire− mTEC (CD45−EpCAM+UEA1+) cell subsets. (I) Pooled data collected and analyzed as in (H), from four independent experiments with 2-week-old (N = 14) and 4- to 7-week-old (N = 6) mice, presented as mean ± SEM. P values were calculated using a mixed linear model analysis (*P < 0.05). Significance in (C), (E), (G), and (I) was assessed using mixed linear models with post hoc paired t tests applying Holm-Šidák corrections for multiple comparisons. *P < 0.05, **P < 0.01, and ***P < 0.001; n.s., not significant.
Aire expression in mTECs decreases as the cells mature. Endstage maturation of a large fraction of post-Aire mTECs resembles that of keratinocytes, with down-regulation of Aire expression and up-regulation of keratin 10 expression, making keratin 10 a suitable marker for late and post-Aire mTECs (33). Confocal imaging showed that Cyp11b1mScarlet was indeed present primarily in early Aire (Aire+ keratin 10−), late Aire (Aire+ keratin 10+), and post-Aire (Aire− keratin 10+) mTECs (Fig. 2, D and E). Flow cytometry analyses similarly found that the majority of Cyp11b1mScarlet was found in Aire+ or keratin 10+ post-Aire cells, with a smaller fraction in Aire− keratin 10− cells (Fig. 2, F and G). Further examination of single-cell RNA-seq data found that Cyp11b1 transcripts were absent from the diverse peripheral tissue “mimetic” mTEC cell subsets that compose the post-Aire compartment (e.g., tuft cells, neuroendocrine cells, and keratinocytes) (fig. S4B) (32). Both confocal microscopy and flow cytometry analyses confirmed that Cyp11b1mScarlet was absent in thymic tuft cells (fig. S4, C to E). A limited number of other post-Aire mTECs may still retain Cyp11b1 protein, but none appears to actively express detectable Cyp11b1 mRNA.
Because thymic glucocorticoid production declines with age (3, 34), we examined thymi of mice at 2 weeks and 4 to 7 weeks. At all ages, Cyp11b1mScarlet was almost exclusively found in mTECs (fig. S5, A to D), although the frequencies of Aire+ mTECs and Cyp11b1mScarlet+ mTECs decreased with age (Fig. 2, H and I). Together, these data suggest that Aire may drive de novo glucocorticoid synthesis in the thymus.
Aire is necessary for mTEC glucocorticoid synthesis
To determine whether Aire is indeed upstream of mTEC glucocorticoid production, we analyzed bulk transcriptomes of wild-type (WT) versus Aire−/− mTECs (35). We also analyzed WT versus Fezf2−/− mTECs (36), because Fezf2 also directs promiscuous expression of TRA genes. Aire deficiency caused a reduction in gene expression of all glucocorticoid synthetic enzymes with the single exception of Cyp21a1, which converts progesterone to deoxycorti-costerone and was reduced only 20%. Of note, Star, Cyp11a1, Hsd3b1, and Cyp11b1 transcripts were nearly absent in Aire−/− mTECs (Fig. 3A). In contrast, Fezf2 deficiency caused a moderate reduction in several of the same enzymes but increases in others (Fig. 3B). More detailed transcriptomic analysis of mTEC subsets (37) similarly found that expression of Cyp11b1, as well as the upstream glucocorticoid-synthesis genes Cyp11a1, Hsd3b1, and Hsd3b6, was found only in Aire+ mTECs and that this expression was lost in mTECs of Aire-deficient mice (Fig. 3C and fig. S6A). Nr5a1 and Nr5a2, which encode steroidogenic factor 1 (SF-1) and liver receptor homolog-1 (LRH-1), respectively, are both capable of driving expression of full glucocorticoid-synthetic programs (38), and expression of Nr5a1 within mTECs was Aire dependent. This raises the possibility that coordinated expression of all glucocorticoid-synthetic enzyme genes within individual cells is due to Aire-driven expression of an upstream regulator that ceases to function in post-Aire mTECs. Gene expression profiles generated by deep sequencing of 174 mTECs (37) allowed detection of most glucocorticoid-synthetic enzymes in one or more cells. Although individual enzyme transcripts were detectable in only 0.6 to 7% of mTECs, we found that glucocorticoid-synthetic enzyme genes were substantially more likely to be coexpressed with one or more other enzyme genes than they were with TRA genes (Fig. 3D). Consistent with this, sorted mTECs with high, intermediate, or no mScarlet fluorescence had high, intermediate, and low expression of multiple steroidogenic enzyme genes, respectively (fig. S6B). These results suggest that there is coexpression of the full steroidogenic enzyme cascade within individual mTECs.
Fig. 3. Aire is required for mTEC glucocorticoid synthesis.
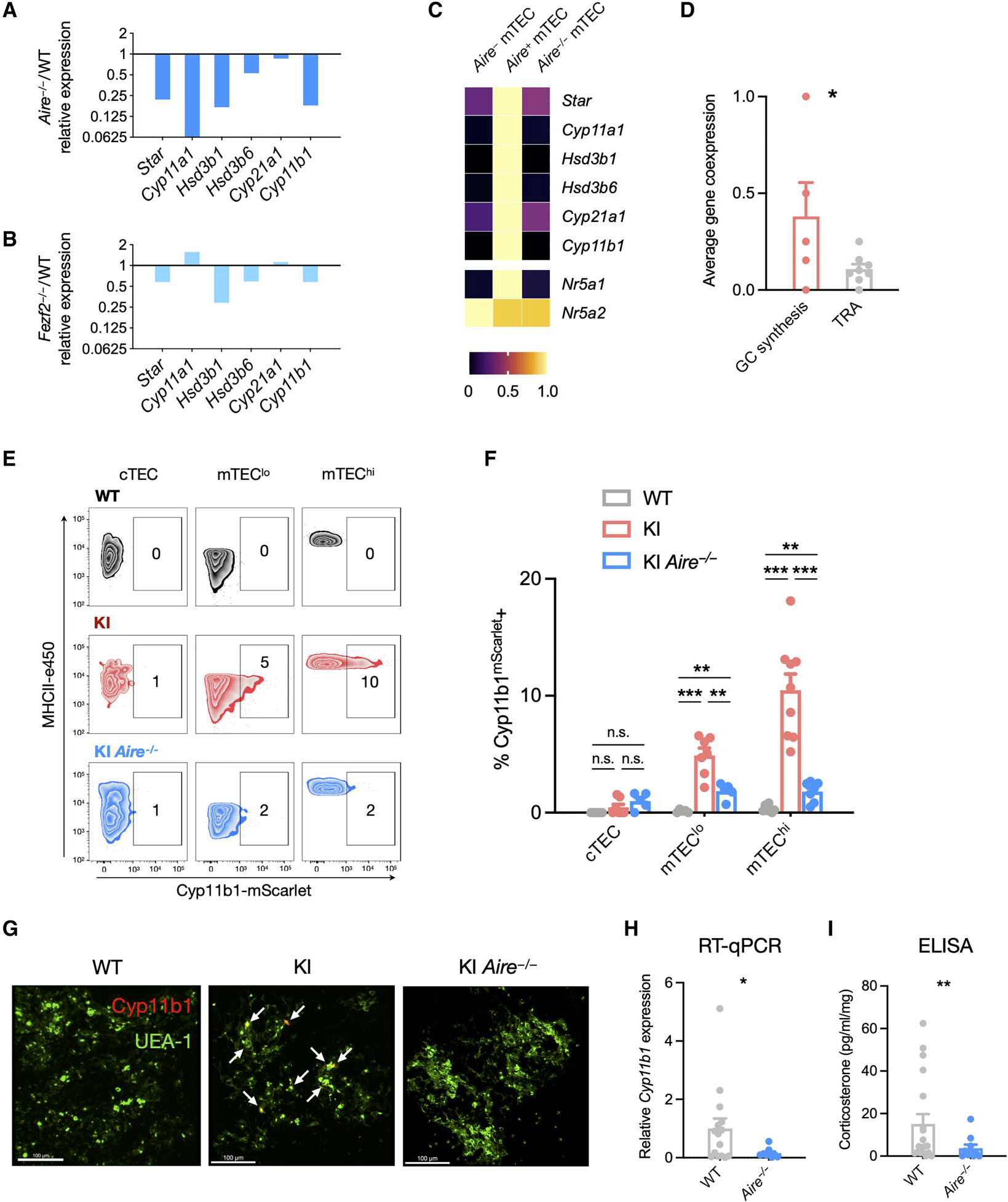
(A) Gene expression of glucocorticoid biosynthetic enzyme genes in mTEC cells of WT and Aire−/− mice. Bulk RNA-seq data from Aire−/− mTEC are shown for each gene relative to WT expression of the same gene. (B) Gene expression of glucocorticoid biosynthetic enzyme genes in mTEC cells of WT and Fezf2−/− mice. Bulk RNA-seq data from Fezf2−/− mTEC are shown for each gene relative to WT expression of the same gene. (C) Expression of glucocorticoid biosynthetic enzyme genes in bulk mTEC subset samples of WT and Aire−/− mice. The expression level of each gene is presented as a proportion of its maximal expression (1.0) across analyzed cell types as indicated by the color scale. (D) Single-cell transcriptomes obtained from deep sequencing of 174 mTECs were analyzed for the coincident expression of glucocorticoid synthesis genes (Star, Cyp11a1, Hsd3b6, Cyp21a1, and Cyp11b1) with each other or with Aire- and Fezf2-dependent TRA genes (Ins1, Spt1, Mup4, S100a8, Resp18, Rabp9, Krt10, and Maoa) in the same cell (see Materials and Methods for calculation details). Data are presented as means ± SEM. (E) Thymi from 2-week-old WT, KI, and KI Aire−/− mice were digested and cells analyzed by flow cytometry. Representative contour plots show each subset, and numbers indicate the percentage of cells within the Cyp11b1mScarlet+ gate. (F) Data collected and analyzed as in (E), pooled from two experiments with 2- and 3-week-old mice, presented as means ± SEM. Data in (E) and (F) are representative or pooled from two independent experiments with N = 7 WT, 9 KI, and 7 KI Aire−/− mice. (G) WT, KI, and KI Aire−/− thymi from 1-week-old mice were fixed, sectioned, and stained for UEA-1 (mTEC, FITC) and Cyp11b1mScarlet (Atto 565). Confocal images were taken at ×60 oil immersion magnification and are representative of two independent experiments. Scale bars, 100 μm. (H) Total RNAwas extracted from whole thymus lobes of WT and Aire−/− mice, and RT-qPCR was used to quantify Cyp11b1 gene expression. Expression was normalized to 18S RNA and is shown relative to WT expression. Data are pooled from four independent experiments, shown as means ± SEM with N = 17 WT and 9 Aire−/− thymi. (I) Thymus lobes were dissected, minced, and cultured for 72 hours in steroid-free medium. Supernatants were collected, and corticosterone was quantified by a commercial ELISA kit. Data are pooled from four independent experiments, with N = 19 WT and 11 Aire−/− thymi. Statistical significance in (D), (H), and (I) was determined by one-tailed t tests and that in (F) was determined by a mixed linear model with post hoc unpaired t tests applying the Holm-Šidák correction for multiple comparisons. *P < 0.05, **P < 0.01, and ***P < 0.001. n.s., not significant.
To determine whether Cyp11b1 protein expression itself was affected by loss of Aire, we generated Cyp11b1mScarlet Aire−/− mice. Cyp11b1mScarlet expression was indeed lost in Aire-deficient mTECs as detected by flow cytometry (Fig. 3, E and F). The loss of Cyp11b1mScarlet in mTEClo cells of Aire-deficient mice is consistent with these being post-Aire mTECs. Loss of Cyp11b1mScarlet in Aire-deficient thymi was also detected by immunofluorescence microscopy (Fig. 3G and fig. S6C), whereas expression in the adrenals was unaffected (fig. S6D). To test whether Aire is the lynchpin for thymus glucocorticoid synthesis, we quantified total thymus gene expression by RT-qPCR and found that Cyp11b1 mRNA was almost completely lost in Aire−/− thymi (Fig. 3H). Thymus expression of the glucocorticoid-responsive gene Tsc22d3 (encoding Gilz) was also reduced in thymi of Aire−/− mice to a similar extent as in thymi of Cyp11b1-deficient mice (fig. S6E) (16). Correspondingly, Aire−/− thymi produced much less corticosterone in vitro (Fig. 3I), similar to the decrease found in thymi with TEC-specific Cyp11b1 deletion (fig. S6F). In contrast, TEC-specific ablation of Fezf2 had no effect (fig. S6E). The small residual amounts of corticosterone from Aire−/− thymi can be accounted for by metabolic reactivation by thymocytes via Hsd11b1 (22). Together, these data show that Aire expression in mTEChi cells drives glucocorticoid synthesis in the thymus.
Aire drives mTEC biosynthesis of multiple steroids and other hormones
The Aire-dependent enzymes upstream of glucocorticoid production (e.g., Star, Cyp11a1, Hsd3b1, and Hsd3b6) are also involved in the generation of other steroid hormones that mTECs are not known to produce (fig. S7A) (24). To determine whether mTECs are responsible for thymic production of these active products, single-cell mTEC transcriptomic data (32) were analyzed for expression of genes needed for the synthesis of progestins, androgens, estrogens, and mineralocorticoids. The same Aire-expressing cell cluster that expressed the full cascade of glucocorticoid biosynthetic genes also expressed Hsd3b1, Hsd3b6, Hsd17b3, and Cyp19a1, genes that encode critical enzymes in the progestin, androgen, and estrogen pathways (Fig. 4A and figs. S4A and S7A). Cyp11b2, encoding the critical enzyme for mineralocorticoid synthesis, was only detectable in one cell, but this was also in the same cluster. To test whether these pathways were also Aire dependent, we analyzed bulk WT versus Aire−/− mTEC gene expression (37) of the pathways for progestins, corticosteroids, androgens, and estrogens. Most of the genes in each pathway were expressed in an mTEC-specific and Aire-dependent manner (Fig. 4B). To confirm that no other thymus cell subsets were substantial contributors, we next performed RT-qPCR on whole thymus lobes from WT and Aire−/− mice. Expression of the steroid synthetic enzyme genes Hsd3b1, Hsd3b6, Cyp11b1, Cyp11b2, Srd5a2, and Cyp19a1 was almost completely lost in Aire−/− thymi. As a control, we examined the expression of the well-established Aire-dependent TRAs Ins2 and Spt1 and found that they were decreased to a similar extent (Fig. 4C). To determine whether the expression of these transcripts correlated with enzyme activity, we cultured minced thymus lobes for 72 hours and assayed supernatants for the presence of the major bioactive sex steroids progesterone, testosterone, and estradiol. Cultured WT thymi were able to generate all three, whereas Aire−/− thymi exhibited greatly reduced production (Fig. 4D).
Fig. 4. Aire drives mTEC biosynthesis of multiple steroids and other lipid hormones.
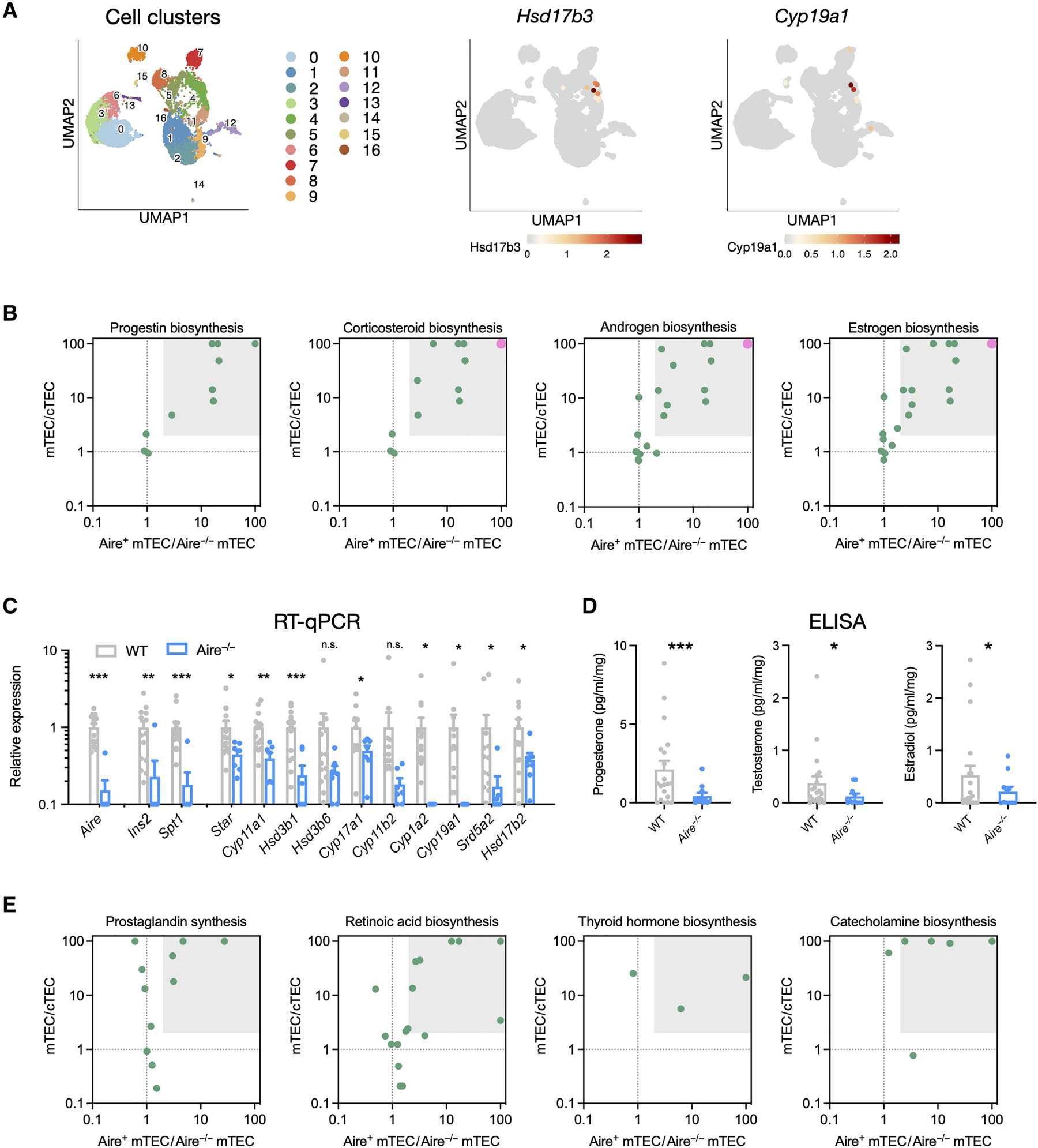
(A) Single-cell RNA-seq analysis of mTEC expression of androgen (Hsd17b3) and estrogen (Cyp19a1) biosynthetic enzymes, with relative expression, as indicated by the color scale, mapped onto a UMAP plot. Numbered populations indicate different cTEC and mTEC cell populations as defined in fig. S4. (B) mTEC specificity and Aire dependence of gene expression in biosynthetic pathways of progestins, corticosteroids (glucocorticoids and mineralocorticoids), androgens, and estrogens. Relative gene expression in bulk Aire+ versus Aire−/− mTEC samples is presented to show Aire-dependent expression, and relative expression in mTECs versus cTECs is presented to show mTEC specificity. Each point represents one gene in the biosynthetic pathway, and the gray box highlights genes with at least twofold higher expression in mTECs versus cTECs and twofold higher expression in Aire+ vs Aire−/− mTEC. A large pink point at the top right represents two or more genes. (C) Total RNAwas extracted from whole thymus lobes of WT and Aire−/− mice, and RT-qPCR was used to quantify gene expression. Ins2 and Spt1 are Aire-dependent TRA genes, and others encode steroidogenic enzymes. Expression was normalized to 18S RNA and is shown relative to WT expression. Data are pooled from four independent experiments, shown as means ± SEM with N = 17 WT and 9 Aire−/− thymi. (D) Thymus lobes were dissected, minced, and cultured for 72 hours in steroid-free medium. Supernatants were collected, and steroids were quantified by commercial ELISA kits. Data were pooled from four independent experiments, with N = 19 WT and 11 Aire−/− thymi. (E) mTEC specificity and Aire dependence of enzyme gene expression in biosynthetic pathways of lipid signaling molecules (prostaglandins, retinoic acid, and thyroid hormone) and amino acid–derived molecules (catecholamines). Relative expression in bulk Aire+ versus Aire−/− mTEC samples is presented to show Aire-dependent expression, and relative expression in mTECs versus cTECs is presented to show mTEC specificity. The gray box highlights genes with at least a twofold expression difference on both axes. Statistical significance in (C) was determined using unpaired t tests with the family-wise error rate set to 0.05 and determined using the correction of Benjamini, Krieger, and Yekutieli and in (D) using one-tailed t tests. *P < 0.05, **P < 0.01, and ***P < 0.001. n.s., not significant.
Thymocyte development, activation, and selection are processes that integrate signals from multiple non–gene-encoded, enzyme-generated molecules, including steroids (39), retinoids (40), oxysterols (41, 42), catecholamines (43), eicosanoids (44), and aryl hydrocarbon receptor ligands (45). We reasoned that Aire might also drive expression of multiple ligand-generating biosynthetic enzyme cascades, generating other signal molecules that could operate in thymocyte development. Differential Pathway Perturbation analysis (46) was used to compare bulk Aire+ and Aire−/− mTEC transcriptomes (37) and identify biosynthetic pathways that were severely altered in the absence of Aire expression. Multiple pathways that function in the generation of enzyme-generated signaling molecules were altered, including biosynthesis of eicosanoids, retinoic acid, thyroid hormones, and all major classes of steroid hormones (fig. S7B). We further analyzed gene expression in the pathways that have known function in thymocyte development. Specifically, gene expression ratios of mTEC:cTEC and Aire+/Aire−/− mTEC cells were used to identify mTEC-specific and Aire-dependent pathways. Complete expression of metabolic pathways needed to synthesize steroids (androgens, estrogens, corticosteroids, and progestins), prostaglandins, retinoids, thyroid hormone, and catecholamines was mTEC specific and Aire dependent (Fig. 4E). In contrast, expression of the cholesterol biosynthetic pathway, required for basal cell functions, was largely unaltered (fig. S7, C and D). Therefore, Aire not only is responsible for driving synthesis of steroids but also drives mTEC synthesis of a large number of non–gene-encoded biologically active products.
DISCUSSION
Steroids function largely as systemic hormones, with glucocorticoids and mineralocorticoids being secreted by the adrenals and progestins, androgens, and estrogens secreted by the gonads. Glucocorticoids can also be synthesized in peripheral tissues such as the thymus (3, 16), intestine (47), and skin (48). These autonomously generated glucocorticoids act in a paracrine fashion, signaling other cells in the nearby environment. In the gut, Cyp11b1 is expressed in intestinal epithelial cells (47) and is up-regulated in response to immune activation, particularly the proinflammatory cytokine tumor necrosis factor (49). Local induction of and signaling by glucocorticoids result in potent immunosuppression that mitigates the damaging effects of excessive inflammation while avoiding the costs that would be incurred by adrenal glucocorticoid production and systemic metabolic changes and immunosuppression (50). In the skin, Cyp11b1 is expressed in keratinocytes and up-regulated in response to inflammatory signals (51). Keratinocyte Cyp11b1-dependent production of glucocorticoids inhibits spontaneous inflammation and, in particular, reduces psoriatic inflammation and contact hypersensitivity (51), similar to Cyp11b1 function in the intestine. Paracrine glucocorticoid signaling is thus an adaptation that allows spatially targeted glucocorticoid immunosuppression and perhaps other functions, while avoiding the global effects of systemic glucocorticoid elevation.
In the thymus, the first identified source of extra-adrenal glucocorticoid synthesis, initial studies found that steroids were produced by radioresistant stromal cells (3, 13). Blockade of glucocorticoid synthesis or reduction in thymocyte glucocorticoid receptor (GR) levels resulted in the antigen-specific loss of thymocytes that would otherwise be positively selected (52, 53). The importance of glucocorticoids in selection was confirmed with thymocyte-specific knockout of the GR, which resulted in altered antigen-specific selection and impaired peripheral immune responses (39). The source of the glucocorticoids, thymus or adrenal, was addressed using epithelial cell–specific deletion of Cyp11b1. The resulting immune phenotype was similar to that of the thymocyte GR knockout, demonstrating that locally produced, paracrine glucocorticoids were essential to thymocyte selection (16). Because Cyp11b1 deletion was targeted using Cre driven by the Foxn1 promoter, deletion occurred in both cTEC and mTEC, so the cellular source and thymic location of glucocorticoid synthesis remained undetermined. In the present study, a Cyp11b1 reporter mouse was created to address this issue, and it was found that Cyp11b1 is expressed exclusively in mTECs. This specificity was explained by the finding that expression of Cyp11b1, as well as that of upstream enzymes in the glucocorticoid synthetic pathway, was dependent on the transcription factor Aire, which in the thymus is expressed by mTECs only and not by other cell types.
Aire is a central component in the generation of an adaptive immune system and is present across vertebrates from bony fish to mammals (54). The hallmark of adaptive immunity, random generation of distinct antigen receptors by developing lymphocytes, inevitably generates autoreactive specificities that must be purged from the repertoire. Removal of autoreactive thymocytes by negative selection is primarily mediated by self-antigen–bearing mTECs and dendritic cells present in the corticomedullary junction and medulla. mTECs express Aire, which drives ectopic gene expression of about 4000 tissue-restricted genes. In cooperation with other transcription factors, this results in TEC expression of more than 19,000 gene products, the most of any cell type (37). Ectopic expression of tissue-restricted gene products ensures that thymocytes are tested against antigens that are normally expressed only outside of the thymus. Thymocytes overly reactive to antigens expressed at specific sites, such as the eye or pancreas, will thus be eliminated. Mutations in Aire that impair its function result in survival of such autoreactive cells (10), causing systemic autoimmunity in mice and humans, in the latter termed autoimmune polyendocrine syndrome type 1. Rather than acting by binding a specific DNA consensus sequence, Aire is thought to regulate gene expression by a variety of nonspecific activities, including the release of stalled polymerases (55), activation of superenhancers (35), and control of open chromatin (56). Although most work on Aire has focused on TRA as mediators of negative selection, it has also been shown to have another important role in up-regulating expression of chemokines, including XCL1 and CCL5. mTEC expression of XCL1 is important for recruitment of dendritic cells to the thymic medulla and effective generation of thymic T regulatory cells (Tregs). In the absence of Aire-driven XCL1, a lack of Treg results in spontaneous autoimmunity, especially of the lacrimal glands (57). Aire-dependent expression of CCL5 and other ligands of CCR4 and CCR7 promotes normal thymocyte migration into the medulla (58). The production of steroids and other non–gene-encoded molecules found in this report indicate an additional function of Aire. Whereas Aire affects development and survival by promoting negative selection (10, 11), Aire-driven mTEC-synthesized glucocorticoids antagonize signals generated by the TCR, altering a thymocyte’s perceived strength of signaling and increasing the affinity for peptide:MHC that is required to induce negative selection (16, 17). In this way, Aire could be considered to have opposing effects on selection: induction of negative selection against TRAs on one hand and antagonism of negative selection by glucocorticoids on the other. These functions might work in concert to optimize fitness of the TCR repertoire, with TRAs expanding the pool of potentially recognized antigens and glucocorticoids regulating the threshold of TCR affinity for antigen required for survival. Although less studied than glucocorticoids, the other Aire-dependent mTEC-synthesized steroids identified in this report can also affect T cell development and T cell responses: Progestins suppress T cell activation and promote Treg immunosuppression (59, 60), androgens inhibit T helper cell 1 (TH1) (61) while promoting TH2 differentiation and interleukin-10 production (62, 63), and estrogens promote thymocyte maturation (64, 65) and TH1 differentiation (66).
The finding that expression of Cyp11b1 is present in mTECs rather than cTECs also resolves a long-standing question about the spatial coordination of glucocorticoid-TCR antagonism within the thymus. cTEC-mediated positive selection occurs on thymoproteasome-, cathepsin L–, and TSSP-generated self-antigen fragments, which are largely distinct from the self-antigen fragments generated in the periphery by constitutive proteasomes and immunoproteasomes (4–8). The cortex is thus adapted to promote positive selection of a maximally diverse TCR repertoire (8, 67), although it can also be a site of negative selection (68). Glucocorticoid antagonism of TCR signaling in the cortex, therefore, would be expected to also maximize positive selection by reducing the number of negatively selected TCRs in the cortex. In contrast, glucocorticoid production by mTECs suggests that antagonism of TCR signaling largely occurs in the context of TCRs recognizing self-peptides, an environment optimized for negative and agonist selection. This means glucocorticoids increase the autoreactivity of the TCR repertoire, which might be expected to increase tonic TCR signaling and perhaps result in a more reactive or “stronger” TCR repertoire. This is consistent with the reduction in fitness (39) and loss of particular TCR specificities (69) of GR-deficient T cells.
Although studies of Aire have focused on its role in the ectopic generation of peptides, those peptides are themselves derived from gene products that have roles in a wide array of biologic functions. It is perhaps expected, therefore, that the Aire-coordinated expression of enzymes involved in biosynthetic pathways results in the production of mediators that affect the thymocytes in their vicinity. Our analyses of single-cell and bulk transcriptomes suggest that there is coexpression of glucocorticoid synthetic enzyme cascades within individual Aire-expressing cells, which might be mediated by Aire-dependent expression of an upstream transcriptional regulator that promotes steroidogenesis. One limitation of the present study is that we were unable to identify what molecule, if any, performs this orchestrative function. Identification and manipulation of such a factor, in addition to providing a mechanistic understanding of enzyme coexpression, would also allow direct examination of the aggregate effects of mTEC-synthesized steroids—androgens, estrogens, progestins, mineralocorticoids, and glucocorticoids—on thymus development and thymocyte selection. Another limitation is the difficulty in linking Cyp11b1 expression in one mTEC to altered selection outcomes in adjacent thymocytes. Development of steroid sensors and their use in combination with TCR signaling reporters, such as calcium indicators, might be used to spatiotemporally link these processes within the thymus.
The present study focused on steroid hormone production, but other Aire-dependent biosynthetic pathways were identified as well, including those producing other lipid hormones with known functions in thymocyte development. Oxysterols (Ebi2 ligands) promote negative selection via increased motility of SP thymocytes within the medulla (42), multiple eicosanoid species inhibit thymocyte proliferation (70, 71), and retinoids (RAR and RXR ligands) induce apoptosis via induction of Nur77 and Bim expression (72) and enhance glucocorticoid signaling in thymocytes (73). Together, these suggest that steroid generation represents only one example of the antigen-independent functions of Aire in the thymic medulla. Generation of TEC-specific deletions of enzymes in these and other pathways would be useful in exploring their individual and combinatorial functions and thus potentially identify additional roles for Aire in the thymus.
MATERIALS AND METHODS
Study design
The main objectives of this study are to (i) identify the cellular source of thymus glucocorticoid synthesis, (ii) identify upstream molecules controlling synthesis, and (iii) determine whether glucocorticoids are the only lipid signaling molecules produced in the thymus or whether there are others. We generated a KI Cyp11b1 reporter mouse expressing a Cyp11b1-mScarlet fluorescent fusion protein to identify glucocorticoid-synthesizing cells and used a combination of transcriptomic analyses and mutant mouse models to further identify and perturb gene expression programs within cells of interest. Key reagents used in the study can be found in table S1.
Mice
WT C57BL/6 mice and FoxN1-Cre-transgenic mice were obtained from the Jackson Laboratory. Aire-deficient and Fezf2flox/flox mice were gifts of Y. Takahama and N. Sestan, respectively (57, 74). Fezf2FoxN1-Cre mice were generated by crossing Fezf2fl/fl and FoxN1-Cre mice. Mice were used between embryonic day 18.5 and 7 weeks of age, depending on the experiment. Embryonic mice were collected from timed pregnancies of 2- to 3-month-old females. Males were added to the cage and removed after detection of a vaginal plug (embryonic day 0.5). We did not detect any sex differences in 3- to 7-week mice and therefore pooled female and male mice for all analyses. Mice were housed on a 12-hour light/12-hour dark cycle, with ad libitum access to standard chow (NIH-31, Teklad). For experiments examining tissue steroid production, mice were collected between 10 a.m. and 3 p.m. (Zeitgeiber time 4 to 9). Sample sizes were selected on the basis of previous studies performed by the laboratory. Mouse genotypes were not randomized, and experimenters were not blinded. All protocols and procedures were approved by the National Cancer Institute (NCI) Animal Care and Use Committee (LICB-008) and followed the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals guidelines.
Cyp11b1mScarlet KI reporter mice
The Cyp11b1mScarlet mice were generated using CRISPR-Cas9 to create a double-strand DNA break immediately upstream of the Cyp11b1 stop codon followed by introduction of the mScarlet coding sequence by homologous recombination. Single-stranded DNA (ssDNA) donor was generated as follows: A gBLOCK DNA fragment encoding 997 base pairs (bp) of mouse genomic sequence directly upstream of the Cyp11b1 stop codon (left homology arm) was synthesized by Integrated DNA Technologies (IDT) and cloned into the pLifeAct-mScarlet-N1 plasmid (Addgene, 85054) immediately upstream of the mScarlet coding sequence using Xho I and Age I restriction sites. A gBLOCK DNA fragment (IDT) encoding 1270 bp of genomic sequence directly downstream of the Cyp11b1 stop codon (right homology arm) was cloned into the pLifeActmScarlet-N1 plasmid immediately downstream of the mScarlet stop codon using Sgr AI and Mfe I restriction sites. Correct assembly was verified by Sanger sequencing, and the fragment containing mScarlet and flanking homology arms was amplified by PCR and converted to ssDNA using the Guide-it Long ssDNA Strandase kit (Takara Bio, 632645). Mouse embryos were injected with Cas9 protein, 2′-modified synthetic short guide RNA (Synthego, AGGCT GTGTGACTAGTTGAC), and donor ssDNA. Founders were screened by PCR genotyping and Sanger sequencing and crossed to WT mice to create the Cyp11b1mScarlet KI reporter mouse line.
Cell culture
HEK 293T cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 25 mM Hepes, and gentamicin (50 mg/ml). Cells were transfected using Lipofectamine 2000 and commercially available pCMV6 plasmids encoding mouse Cyp11b1-GFP (OriGene, MG219439) or Cyp11b1-Myc-FLAG (OriGene, MR219439) fusion proteins. Cells were used for analysis (flow cytometry) 72 hours after transfection.
Flow cytometry
Thymi were dissected into ice-cold phosphate-buffered saline (PBS), and adipose and connective tissue were removed, transferred into 2 ml of basic medium [RPMI 1640 supplemented with 2 mM l-glutamine, 1 mM sodium pyruvate, 25 mM Hepes, 1× nonessential amino acids, and gentamicin (50 mg/ml)], and minced into 1-mm3 pieces. Minced thymi were then transferred into round-bottom 15-ml tubes; supernatants were removed and replaced with 2 ml of digestion buffer [basic medium supplemented with Liberase TM (62.5 mg/ml) and deoxyribonuclease I (20 mg/ml)] and stirred gently at 37°C for 40 min. Tissue fragments were gently aspirated with a wide-bore pipet, dispersed cells were collected into ice-cold tubes, and enzymes were inactivated with 5 mM EDTA. Fresh digestion buffer was added to the remaining thymus fragments, which were stirred at 37°C for an additional 20 min; dispersed cells were combined with the previously collected cells, and enzymes were inactithrough with EDTA and incubated on ice for 15 min. Cells were passed a 100-mm strainer and washed with PBS. Postnatal thymus samples were resuspended in isotonic Percoll (1.115 g/ml), which was overlaid with isotonic Percoll (1.065 g/ml) and an additional layer of PBS. Epithelial cells were enriched by centrifuging at 2700 rpm for 30 min at 4°C with brakes off and collection of the interface between the Percoll and PBS layers. Embryonic thymus cells were not Percoll-separated because of the small number of cells. Cells from all thymi were washed twice with magnetic-activated cell sorting buffer (PBS with 5% FBS and 2 mM EDTA), resuspended in fluorescence-activated cell sorting (FACS) buffer (PBS containing 2% FBS and 0.05% sodium azide), and counted on a hemocytometer using Trypan blue, and about 5 × 106 cells were aliquoted into FACS tubes. Cells were resuspended in 100 ml of surface stain mix for 30 min on ice, washed, stained with fluorophore-conjugated streptavidin or secondary antibodies for 15 min on ice, washed, and, in some cases, fixed and permeabilized for intracellular staining using the eBioscience Foxp3 Transcription Factor staining kit. Intracellular staining was done overnight at 4°C, cells were washed twice with perm/wash buffer, and cells were kept on ice or at 4°C until data acquisition. Data were acquired with a BD LSRFortessa or a BD FACSymphony and analyzed using FlowJo (Tree Star Inc.). Markers used to define cell subsets are described in the text and figure panels, and mTEClo and mTEChi cells were distinguished by expression of MHCII, gated on the bottom 30 to 40% and top 30 to 40% of MHCII fluorescence, respectively.
Immunofluorescence microscopy
Tissues were dissected into ice-cold PBS and fixed by rocking overnight in 4% paraformaldehyde at 4°C. Tissues were then washed with PBS for 10 min at room temperature and cryopreserved by rocking in 10% sucrose in PBS for 1 hour at 4°C, 20% sucrose in PBS for 3 hours at 4°C, and 30% sucrose in PBS overnight at 4°C. Cryopreserved tissues were embedded in optimal cutting temperature compound (Tissue-Tek), frozen on dry ice, and stored overnight at −80°C. Ten- to 20-mm cryosections were obtained at −20°C on a Leica cryostat, slide-mounted, and dried onto slides by incubating at room temperature for 15 to 30 min. Dried tissues were rehydrated in PBS for 5 min at room temperature, fixed in 4% paraformaldehyde for 30 min at room temperature, quenched in 0.1 M glycine in PBS for 10 min at room temperature, washed three times with 0.1% Tween-20 in PBS (PBS-T) for 5 min at room temperature, and blocked with 5% BSA in PBS for 60 min at room temperature. All tissue sections were incubated with Hoechst 33342 (1:1000; Invitrogen) for 5 to 10 min at room temperature in the dark and washed three times with 0.1% Tween-20 in PBS-T. Thymus lobe sections were stained with UEA1–fluorescein isothiocyanate (FITC) (1:300; Vector), Ly51–Alexa Fluor 647 (1:100; BioLegend), Aire-eFluor 660 (1:500), Aire–Alexa Fluor 488 (1:250; Invitrogen), and cytokeratin 10 recombinant rabbit (1:100; Invitrogen) for 1 hour at room temperature; washed three times with PBS-T; stained with secondary anti-rabbit Alexa Fluor 647 (1:1000; Invitrogen) for 1 hour at room temperature; washed three times with PBS-T; stained with mScarlet-Atto 565 (1:200; FluoTag-X2, NanoTag) for 1 hour at room temperature; washed three times in PBS-T; and washed once in 0.5 M NaCl in PBS. All slides were rinsed with PBS and rinsed with water, and coverslips were mounted with ProLong Glass Antifade Mountant (Invitrogen) and kept overnight at 4°C. Images were acquired on a Nikon SoRa Spinning Disk confocal microscope and processed using Imaris 9.6.0 software.
Western blot
Cell samples were boiled in SDS sample buffer for 5 to 10 min, and proteins were separated by SDS–polyacrylamide gel electrophoresis and transferred using a Bio-Rad Trans-Blot Turbo. Blots were blocked with 5% dried milk, incubated overnight with primary antibodies against Cyp11b1 (bs-3898R, Bioss) or β-actin (AC-15), and detected with horseradish peroxidase–conjugated secondary antibodies and enhanced chemiluminescence substrate (SuperSignal West Dura or SuperSignal West Femto, Thermo Fisher Scientific) using a ChemiDoc imaging system (Bio-Rad).
Bulk transcriptome analyses
Data comparing steroidogenic enzyme gene expression in thymocytes and TECs were obtained from the ImmGen database (https://immgen.org) using the Databrowser tool with Gene Skyline ULI RNA-seq and selecting the subpopulations of interest. Data comparing steroidogenic gene expression in TEC subsets were obtained from the ImmGen database using the Databrowser tool with Gene Skyline Microarray and selecting the subpopulations of interest. For each analysis, the highest expression of a particular gene was set equal to 1, and expression of the same gene in other cell types is presented as a proportion of this maximal expression.
Single-cell SMART-seq transcriptomes from 174 isolated mTEC are described in (37). Coexpression was calculated for each of the five glucocorticoid synthetic genes: Star, Cyp11a1, Hsd3b6, Cyp21a1, and Cyp11b1. For a given gene, e.g., Star, the proportion of coexpression was calculated as the number of Star-expressing cells also expressing one or more other steroidogenic genes, divided by the total number of Star-expressing cells. Baseline coexpression was calculated in the same way using the Aire- and Fezf2-dependent TRA genes Ins1, Fabp9, Krt10, Maoa, Mup4, Resp18, S100a8, and Spt1. For a given TRA gene, e.g., Ins1, the proportion of coexpression was calculated as the number of Ins1-expressing cells expressing one of more steroidogenic genes, divided by the total number of Ins1-expressing cells.
Data comparing TEC transcriptomes of WT and Aire−/− or WT and Fezf2−/− mice are described in (35–37). For each analysis, the WT expression of a particular gene was set equal to 1, and expression of the same gene in mutant mouse strains is presented as a proportion of WT expression. Aire-dependent expression of biosynthetic pathways was further analyzed in two complementary ways. First, the complete gene expression (fragments per kilobase per million mapped fragments) data from Aire+ mTECs and Aire−/− mTECs (37) were imported into MetaCyc (metacyc.org) and analyzed by calculating differential perturbation pathway scores (DPPS) in which changes in expression of metabolic pathway genes were used to predict changes in activity of that metabolic pathway (75). A DPPS score was generated using the formula , where RPS1 is the maximum differential expression of genes in one reaction of a pathway with distinct reactions. Second, we generated two gene lists, one containing genes whose expression was greater than threefold higher in Aire+ mTECs than Aire−/− mTECs, and one containing genes whose expression was greater than threefold higher in mTECs than in cTECs. Both lists were entered into Database for Annotation, Visualization and Integrated Discovery (DAVID) (v6.8, david.ncifcrf.gov), and biosynthetic/metabolic pathways independently identified by both gene sets were used for further analysis. All analyses returned androgen, estrogen, and corticosteroid biosynthetic pathways as among the most highly altered pathways.
Single-cell RNA-seq analyses
Single-cell RNA-seq data of mouse Aire- and post-Aire mTECs (GSM5831744) were downloaded from the Gene Expression Omnibus GSE194253 (32). The corresponding gene matrix, features, barcode, and metadata files were used to create a Seurat Object through the CreateSeuratObject function. Downstream analysis was performed within the NIH Integrated Data Analysis Platform (NIDAP) using R programs developed on the Palantir Foundry Platform (Palantir Technologies). Web-based visualization of the data was done using a ShinyCell (76) application published on the NIH RStudio Connect Server. Cells were preprocessed according to unique molecular identifier (UMI) counts, number of expressed genes, and mitochondrial content; cells having low UMI counts (>500) or low complexity (<0.8 genes/UMI) were filtered from the data, along with cells whose gene or mitochondrial content exceeds 3 absolute deviations above the respective medians. The gene expression data were then normalized using the Seurat sctransform (77) function. Highly variable genes were summarized by principal components analysis, and the first 19 principal components were further projected using uniform manifold approximation and projection (UMAP) (78). Unsupervised clustering was performed using Seurat FindClusters function. We chose a resolution value of 0.7 to separate cell populations because it contained a similar number of clusters as those published in the original analysis of this dataset, and we were interested in obtaining similar cell clusters. Gene expression data of cells from the sctransform scale.data slot were aggregated and rescaled across clusters before visualization using the pheatmap function. Differential gene expression analysis was performed using Model-based Analysis of Single-cell Transcriptomics (MAST) (79). Cell clusters not mapping to signatures from the original analyses were further analyzed using the top differentially expressed genes. These genes were then explored in BioGPS to show tissue-specific expression patterns and identify potential cell types. Correlation analyses were used to identify cluster-specific enrichment of TRA and non-TRA genes following previously published TRA and non-TRA gene sets (37).
Reverse transcription quantitative PCR
Thymi were dissected into ice-cold RNAlater and incubated at 4°C for at least 24 hours. Total RNA was extracted using RNeasy Mini kits (Qiagen), complementary DNA was synthesized using a SuperScript IV First Strand synthesis kit (Thermo Fisher Scientific), and qPCR was performed using PowerUp SYBR Green Master Mix (Applied Biosystems) on a QuantStudio 6 quantitative PCR thermocycler. Gene expression was normalized to 18S RNA, and results are expressed relative to WT thymi.
Ex vivo steroid production
Thymi were dissected into ice-cold PBS, and adipose and connective tissue were removed, transferred into about 5 ml of steroid-free culture medium [RPMI 1640 supplemented with 10% charcoalstripped heat-inactivated FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 25 mM Hepes, 0.1 mM nonessential amino acids, and gentamicin (50 mg/ml)], and minced into 1-mm3 pieces using forceps and scalpel. Fragments were left for at least 10 min to deplete endogenous steroid substrates and then transferred with forceps to 24-well plates containing 400 ml of steroid-free culture medium. Samples were cultured for 72 hours at 37°C in 5% CO2, and then supernatants were collected and immediately assayed or stored at −20°C until assayed for steroids. Steroids were quantified using commercial enzyme-linked immunosorbent assay (ELISA) kits (corticosterone, Arbor Assays, K014; progesterone, Cayman Chemical, 582601; testosterone, Cayman Chemical, 582701; estradiol, Cayman Chemical, 501890) according to the manufacturer’s protocol, with standards diluted in steroid-free culture medium. One outlier subject, with all steroid measurements greater than threefold higher than all other subjects, was omitted from these analyses. Inclusion of this subject did not alter the results of these experiments.
Statistical analyses
Analyses were performed using Excel, GraphPad Prism, and R, using t tests to compare two groups or linear mixed-effects models to compare more than two groups with post hoc pairwise comparison tests as appropriate. Multiple pairwise comparisons applied the Holm-Šidák correction to control for family-wise error rate at 0.05. Error bars indicate SEM unless otherwise stated, and significance was set at P = 0.05.
Supplementary Material
Acknowledgments:
We wish to thank F. Livak, C. C. Li, and S. Siddiqui of the NCI Flow Core Facility for help with flow cytometry; P. R. Mittelstadt, J. E. Cowan, and A. Bhandoola for discussions and technical advice; R. Awasthi and R. Chari for assistance with transgenic mouse generation; Y. Takahama for the Aire−/− mice; and N. Sestan for the Fezf2fl/fl mice. For help with microscopy, we thank M. Kruhlak and L. Lim from the CCR Microscopy Core Facility, R. Weigert and N. Melis from the CCR Intravital Microscopy Core, and A. Jacques. We also thank B. Pandalai for help with Western blots. We are grateful to A. Bhandoola, Y. Takahama, and R. Bosselut for critical review of this manuscript.
Funding:
This research was supported by the Center for Cancer Research, NCI, NIH intramural research program, and a CIHR postdoctoral fellowship to M.D.T.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability:
The Cyp11b1mScarlet KI mice used in this study are available under a material transfer agreement with the NCI, NIH. The transcriptomic analyses performed as part of this study used bulk RNA-seq and single-cell RNA-seq data previously deposited in public databases and available under the following accession numbers: GSE92509 (35), GSE69105 (36), GSE53111 (37), and GSE194252 (32). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Sapolsky RM, Romero LM, Munck AU, How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr. Rev. 21, 55–89 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Miller WL, Steroidogenesis: Unanswered questions. Trends Endocrinol. Metab. 28, 771–793 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Vacchio MS, Papadopoulos V, Ashwell JD, Steroid production in the thymus: Implications for thymocyte selection. J. Exp. Med. 179, 1835–1846 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gommeaux J, Grégoire C, Nguessan P, Richelme M, Malissen M, Guerder S, Malissen B, Carrier A, Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes. Eur. J. Immunol. 39, 956–964 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Honey K, Nakagawa T, Peter C, Rudensky A, Cathepsin L regulates CD4+ T cell selection independently of its effect on invariant chain: A role in the generation of positively selecting peptide ligands. J. Exp. Med. 195, 1349–1358 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K, Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316, 1349–1353 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, Villadangos JA, Ploegh H, Peters C, Rudensky AY, Cathepsin L: Critical role in Ii degradation and CD4 T cell selection in the thymus. Science 280, 450–453 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Ohigashi I, Takahama Y, Thymoproteasome optimizes positive selection of CD8+ T cells without contribution of negative selection. Adv. Immunol. 149, 1–23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki K, Takada K, Ohte Y, Kondo H, Sorimachi H, Tanaka K, Takahama Y, Murata S, Thymoproteasomes produce unique peptide motifs for positive selection of CD8+ T cells. Nat. Commun. 6, 7484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D, Projection of an immunological self shadow [DOI] [PubMed] [Google Scholar]
- 11.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC, Aire regulates negative selection of organ-specific T cells. Nat. Immunol. 4, 350–354 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Lechner O, Wiegers GJ, Oliveira-Dos-Santos AJ, Dietrich H, Recheis H, Waterman M, Boyd R, Wick G, Glucocorticoid production in the murine thymus. Eur. J. Immunol. 30, 337–346 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Pazirandeh A, Xue Y, Rafter I, Sjövall J, Jondal M, Okret S, Paracrine glucocorticoid activity produced by mouse thymic epithelial cells. FASEB J. 13, 893–901 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Zacharchuk CM, Merćep M, Chakraborti PK, Simons SS, Ashwell JD, Programmed T lymphocyte death. Cell activation- and steroid-induced pathways are mutually antagonistic. J. Immunol. 145, 4037–4045 (1990). [PubMed] [Google Scholar]
- 15.Iwata M, Hanaoka S, Sato K, Rescue of thymocytes and T cell hybridomas from glucocorticoid-induced apoptosis by stimulation via the T cell receptor/CD3 complex: A possible in vitro model for positive selection of the T cell repertoire. Eur. J. Immunol. 21, 643–648 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Mittelstadt PR, Taves MD, Ashwell JD, Cutting Edge: De novo glucocorticoid synthesis by thymic epithelial cells regulates antigen-specific thymocyte selection. J. Immunol. 200, 1988–1994 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taves MD, Mittelstadt PR, Presman DM, Hager GL, Ashwell JD, Single-cell resolution and quantitation of targeted glucocorticoid delivery in the thymus. Cell Rep. 26, 3629–3642.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taves MD, Ashwell JD, Glucocorticoids in T cell development, differentiation and function. Nat. Rev. Immunol. 21, 233–243 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Qiao S, Chen L, Okret S, Jondal M, Age-related synthesis of glucocorticoids in thymocytes. Exp. Cell Res. 314, 3027–3035 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Qiao S, Okret S, Jondal M, Thymocyte-synthesized glucocorticoids play a role in thymocyte homeostasis and are down-regulated by adrenocorticotropic hormone. Endocrinology 150, 4163–4169 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Rocamora-Reverte L, Reichardt HM, Villunger A, Wiegers G, T-cell autonomous death induced by regeneration of inert glucocorticoid metabolites. Cell Death Dis. 8, e2948 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taves MD, Plumb AW, Korol AM, Van Der Gugten JG, Holmes DT, Abraham N, Soma KK, Lymphoid organs of neonatal and adult mice preferentially produce active glucocorticoids from metabolites, not precursors. Brain Behav. Immun. 57, 271–281 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Yamanaka N, Marqués G, O’Connor MB, Vesicle-mediated steroid hormone secretion in drosophila melanogaster. Cell 163, 907–919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller WL, Auchus RJ, The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev. 32, 81–151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW, Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 14, 1–18 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Ohigashi I, Tanaka Y, Kondo K, Fujimori S, Kondo H, Palin AC, Hoffmann V, Kozai M, Matsushita Y, Uda S, Motosugi R, Hamazaki J, Kubota H, Murata S, Tanaka K, Katagiri T, Kosako H, Takahama Y, Trans-omics impact of thymoproteasome in cortical thymic epithelial cells. Cell Rep. 29, 2901–2916.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heng TSP, Painter MW; The Immunological Genome Project Consortium, Elpek K, Lukacs-Kornek V, Mauermann N, Turley SJ, Koller D, Kim FS, Wagers AJ, Asinovski N, Davis S, Fassett M, Feuerer M, Gray DHD, Haxhinasto S, Hill JA, Hyatt G, Laplace C, Leatherbee K, Mathis D, Benoist C, Jianu R, Laidlaw DH, Best JA, Knell J, Goldrath AW, Jarjoura J, Sun JC, Zhu Y, Lanier LL, Ergun A, Li Z, Collins JJ, Shinton SA, Hardy RR, Friedline R, Sylvia K, Kang J, The immunological genome project: Networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Hikosaka Y, Nitta T, Ohigashi I, Yano K, Ishimaru N, Hayashi Y, Matsumoto M, Matsuo K, Penninger JM, Takayanagi H, Yokota Y, Yamada H, Yoshikai Y, Inoue J-I, Akiyama T, Takahama Y, The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity 29, 438–450 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Bornstein C, Nevo S, Giladi A, Kadouri N, Pouzolles M, Gerbe F, David E, Machado A, Chuprin A, Tóth B, Goldberg O, Itzkovitz S, Taylor N, Jay P, Zimmermann VS, Abramson J, Amit I, Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 559, 622–626 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Cowan JE, Malin J, Zhao Y, Seedhom MO, Harly C, Ohigashi I, Kelly M, Takahama Y, Yewdell JW, Cam M, Bhandoola A, Myc controls a distinct transcriptional program in fetal thymic epithelial cells that determines thymus growth. Nat. Commun. 10, 5498 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bautista JL, Cramer NT, Miller CN, Chavez J, Berrios DI, Byrnes LE, Germino J, Ntranos V, Sneddon JB, Burt TD, Gardner JM, Ye CJ, Anderson MS, Parent AV, Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat. Commun. 12, 1096 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michelson DA, Hase K, Kaisho T, Benoist C, Mathis D, Thymic epithelial cells co-opt lineage-defining transcription factors to eliminate autoreactive T cells. Cell 185, 2542–2558.e18 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Laan M, Bichele R, Kisand K, Scott HS, Peterson P, Post-Aire maturation of thymic medullary epithelial cells involves selective expression of keratinocyte-specific autoantigens. Front. Immunol. 3, 19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taves MD, Plumb AW, Sandkam BA, Ma C, Van Der Gugten JG, Holmes DT, Close DA, Abraham N, Soma KK, Steroid profiling reveals widespread local regulation of glucocorticoid levels during mouse development. Endocrinology 156, 511–522 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Bansal K, Yoshida H, Benoist C, Mathis D, The transcriptional regulator Aire binds to and activates super-enhancers. Nat. Immunol. 18, 263–273 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takaba H, Morishita Y, Tomofuji Y, Danks L, Nitta T, Komatsu N, Kodama T, Takayanagi H, Fezf2 orchestrates a thymic program of self-antigen expression for immune tolerance. Cell 163, 975–987 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Sansom SN, Shikama-Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME, Heger A, Ponting CP, Holländer GA, Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 24, 1918–1931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meinsohn MC, Smith OE, Bertolin K, Murphy BD, The orphan nuclear receptors steroidogenic factor-1 and liver receptor homolog-1: Structure, regulation, and essential roles in mammalian reproduction. Physiol. Rev. 99, 1249–1279 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Mittelstadt PR, Monteiro JP, Ashwell JD, Thymocyte responsiveness to endogenous glucocorticoids is required for immunological fitness. J. Clin. Invest. 122, 2384–2394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szegezdi E, Kiss I, Simon A, Blaskó B, Reichert U, Michel S, Sándor M, Fésüs L, Szondy Z, Ligation of retinoic acid receptor alpha regulates negative selection of thymocytes by inhibiting both DNA binding of nur77 and synthesis of bim. J. Immunol. 170, 3577–3584 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Chan CT, Fenn AM, Harder NK, Mindur JE, Mc Alpine CS, Patel J, Valet C, Rattik S, Iwamoto Y, He S, Anzai A, Kahles F, Poller WC, Janssen H, Wong LP, Fernandez-Hernando C, Koolbergen DR, van der Laan AM, Yvan-Charvet L, Sadreyev RI, Nahrendorf M, Westerterp M, Tall AR, Gustafsson J-A, Swirski FK, Liver X receptors are required for thymic resilience and T cell output. J. Exp. Med. 217, e20200318 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ki S, Thyagarajan HM, Hu Z, Lancaster JN, Ehrlich LIR, EBI2 contributes to the induction of thymic central tolerance in mice by promoting rapid motility of medullary thymocytes. Eur. J. Immunol. 47, 1906–1917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leposavić G, Perisić M, Age-associated remodeling of thymopoiesis: Role for gonadal hormones and catecholamines. Neuroimmunomodulation 15, 290–322 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Lone AM, Taskén K, Proinflammatory and immunoregulatory roles of eicosanoids in T cells. Front. Immunol. 4, 130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nohara K, Pan X, Tsukumo S, Hida A, Ito T, Nagai H, Inouye K, Motohashi H, Yamamoto M, Fujii-Kuriyama Y, Tohyama C, Constitutively active aryl hydrocarbon receptor expressed specifically in T-lineage cells causes thymus involution and suppresses the immunization-induced increase in splenocytes. J. Immunol. 174, 2770–2777 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Caspi R, Billington R, Keseler IM, Kothari A, Krummenacker M, Midford PE, Ong WK, Paley S, Subhraveti P, Karp PD, The MetaCyc database of metabolic pathways and enzymes—A 2019 update. Nucleic Acids Res. 48, D445–D453 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cima I, Corazza N, Dick B, Fuhrer A, Herren S, Jakob S, Ayuni E, Mueller C, Brunner T, Intestinal epithelial cells synthesize glucocorticoids and regulate T cell activation. J. Exp. Med. 200, 1635–1646 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slominski A, Zbytek B, Szczesniewski A, Semak I, Kaminski J, Sweatman T, Wortsman J, CRH stimulation of corticosteroids production in melanocytes is mediated by ACTH. Am. J. Physiol. Endocrinol. Metab. 288, E701–E706 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Noti M, Corazza N, Mueller C, Berger B, Brunner T, TNF suppresses acute intestinal inflammation by inducing local glucocorticoid synthesis. J. Exp. Med. 207, 1057–1066 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taves MD, Gomez-Sanchez CE, Soma KK, Extra-adrenal glucocorticoids and mineralocorticoids: Evidence for local synthesis, regulation, and function. Am. J. Physiol. Endocrinol. Metab. 301, E11–E24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.San Phan T, Schink L, Mann J, Merk VM, Zwicky P, Mundt S, Simon D, Kulms D, Abraham S, Legler DF, Keratinocytes control skin immune homeostasis through de novo–synthesized glucocorticoids. Sci. Adv. 7, eabe0337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tolosa E, King LB, Ashwell JD, Thymocyte glucocorticoid resistance alters positive selection and inhibits autoimmunity and lymphoproliferative disease in MRL-lpr/lpr mice. Immunity 8, 67–76 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Vacchio MS, Lee JYM, Ashwell JD, Thymus-derived glucocorticoids set the thresholds for thymocyte selection by inhibiting TCR-mediated thymocyte activation. J. Immunol. 163, 1327–1333 (1999). [PubMed] [Google Scholar]
- 54.Saltis M, Criscitiello MF, Ohta Y, Keefe M, Trede NS, Goitsuka R, Flajnik MF, Evolutionarily conserved and divergent regions of the autoimmune regulator (Aire) gene: A comparative analysis. Immunogenetics 60, 105–114 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giraud M, Yoshida H, Abramson J, Rahl PB, Young RA, Mathis D, Benoist C, Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 109, 535–540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koh AS, Miller EL, Buenrostro JD, Moskowitz DM, Wang J, Greenleaf WJ, Chang HY, Crabtree GR, Rapid chromatin repression by Aire provides precise control of immune tolerance. Nat. Immunol. 19, 162–172 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lei Y, Ripen AM, Ishimaru N, Ohigashi I, Nagasawa T, Jeker LT, Bösl MR, Holländer GA, Hayashi Y, de Waal Malefyt R, Nitta T, Takahama Y, Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J. Exp. Med. 208, 383–394 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laan M, Kisand K, Kont V, Möll K, Tserel L, Scott HS, Peterson P, Autoimmune regulator deficiency results in decreased expression of CCR4 and CCR7 ligands and in delayed migration of CD4+ thymocytes. J. Immunol. 183, 7682–7691 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hierweger AM, Engler JB, Friese MA, Reichardt HM, Lydon J, DeMayo F, Mittrücker HW, Arck PC, Progesterone modulates the T-cell response via glucocorticoid receptor-dependent pathways. Am. J. Reprod. Immunol. 81, e13084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Solano ME, Kowal MK, O’Rourke GE, Horst AK, Modest K, Plösch T, Barikbin R, Remus CC, Berger RG, Jago C, Ho H, Sass G, Parker VJ, Lydon JP, De Mayo FJ, Hecher K, Karimi K, Arck PC, Progesterone and HMOX-1 promote fetal growth by CD8+ T cell modulation. J. Clin. Invest. 125, 1726–1738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kissick HT, Sanda MG, Dunn LK, Pellegrini KL, On ST, Noel JK, Arredouani MS, Androgens alter T-cell immunity by inhibiting T-helper 1 differentiation. Proc. Natl. Acad. Sci. U.S.A. 111, 9887–9892 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bebo BF, Schuster JC, Vandenbark AA, Offner H, Androgens alter the cytokine profile and reduce encephalitogenicity of myelin-reactive T cells. J. Immunol. 162, 35–40 (1999). [PubMed] [Google Scholar]
- 63.Dalal M, Kim S, Voskuhl RR, Testosterone therapy ameliorates experimental autoimmune encephalomyelitis and induces a T helper 2 bias in the autoantigen-specific T lymphocyte response. J. Immunol. 159, 3–6 (1997). [PubMed] [Google Scholar]
- 64.Staples JE, Gasiewicz TA, Fiore NC, Lubahn DB, Korach KS, Silverstone AE, Estrogen receptor alpha is necessary in thymic development and estradiol-induced thymic alterations. J. Immunol. 163, 4168–4174 (1999). [PubMed] [Google Scholar]
- 65.Erlandsson MC, Ohlsson C, Gustafsson JA, Carlsten H, Role of oestrogen receptors α and β in immune organ development and in oestrogen-mediated effects on thymus. Immunology 103, 17–25 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fox HS, Bond BL, Parslow TG, Estrogen regulates the IFN-γ promoter. J. Immunol. 146, 4362–4367 (1991). [PubMed] [Google Scholar]
- 67.Nakagawa Y, Ohigashi I, Nitta T, Sakata M, Tanaka K, Murata S, Kanagawa O, Takahama Y, Thymic nurse cells provide microenvironment for secondary T cell receptor α rearrangement in cortical thymocytes. Proc. Natl. Acad. Sci. U.S.A. 109, 20572–20577 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCaughtry TM, Baldwin TA, Wilken MS, Hogquist KA, Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J. Exp. Med. 205, 2575–2584 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu FW, Yasutomo K, Goodman GB, McHeyzer-Williams LJ, McHeyzer-Williams MG, Germain RN, Ashwell JD, Thymocyte resistance to glucocorticoids leads to antigen-specific unresponsiveness due to “holes” in the T cell repertoire. Immunity 12, 183–192 (2000). [DOI] [PubMed] [Google Scholar]
- 70.Delebassée S, Gualde N, Effect of arachidonic acid metabolites on thymocyte proliferation. Ann. Inst. Pasteur Immunol. 139, 383–399 (1988). [DOI] [PubMed] [Google Scholar]
- 71.Rotondo D, Earl CR, Laing KJ, Kaimakamis D, Inhibition of cytokine-stimulated thymic lymphocyte proliferation by fatty acids: The role of eicosanoids. Biochim. Biophys. Acta 1223, 185–194 (1994). [DOI] [PubMed] [Google Scholar]
- 72.Kiss B, Tóth K, Sarang Z, Garabuczi É, Szondy Z, Retinoids induce Nur77-dependent apoptosis in mouse thymocytes. Biochim. Biophys. Acta 1853, 660–670 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Tóth K, Sarang Z, Scholtz B, Brázda P, Ghyselinck N, Chambon P, Fésüs L, Szondy Z, Retinoids enhance glucocorticoid-induced apoptosis of T cells by facilitating glucocorticoid receptor-mediated transcription. Cell Death Differ. 18, 783–792 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han W, Kwan KY, Shim S, Lam MMS, Shin Y, Xu X, Zhu Y, Li M, Sestan N, TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc. Natl. Acad. Sci. U.S.A. 108, 3041–3046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karp PD, Midford PE, Billington R, Kothari A, Krummenacker M, Latendresse M, Ong WK, Subhraveti P, Caspi R, Fulcher C, Keseler IM, Paley SM, Pathway Tools version 23.0 update: Software for pathway/genome informatics and systems biology. Brief. Bioinform. 22, 109–126 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ouyang JF, Kamaraj US, Cao EY, Rackham OJL, Gaussian mixture clustering and imputation of microarray data. Bioinformatics 20, 917–923 (2004). [DOI] [PubMed] [Google Scholar]
- 77.Hao Y, Hao S, Andersen-Nissen E, Mauck III WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McInnes L, Healy J, Melville J, UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426 [stat.ML] (18 September 2020). [Google Scholar]
- 79.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Cyp11b1mScarlet KI mice used in this study are available under a material transfer agreement with the NCI, NIH. The transcriptomic analyses performed as part of this study used bulk RNA-seq and single-cell RNA-seq data previously deposited in public databases and available under the following accession numbers: GSE92509 (35), GSE69105 (36), GSE53111 (37), and GSE194252 (32). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.


