Abstract
Neutrophil extracellular traps (NETs) not only counteract bacterial and fungal pathogens but can also promote thrombosis, autoimmunity, and sterile inflammation. The presence of citrullinated histones, generated by the peptidylarginine deiminase 4 (PAD4), is synonymous with NETosis and is considered independent of apoptosis. Mitochondrial- and death receptor–mediated apoptosis promote gasdermin E (GSDME)–dependent calcium mobilization and membrane permeabilization leading to histone H3 citrullination (H3Cit), nuclear DNA extrusion, and cytoplast formation. H3Cit is concentrated at the promoter in bone marrow neutrophils and redistributes in a coordinated process from promoter to intergenic and intronic regions during apoptosis. Loss of GSDME prevents nuclear and plasma membrane disruption of apoptotic neutrophils but prolongs early apoptosis-induced cellular changes to the chromatin and cytoplasmic granules. Apoptotic signaling engages PAD4 in neutrophils, establishing a cellular state that is primed for NETosis, but that occurs only upon membrane disruption by GSDME, thereby redefining the end of life for neutrophils.
NETosis and cytoplast formation is the normal fate of apoptotic neutrophils.
INTRODUCTION
Mixed lineage kinase domain-like (MLKL) and gasdermin D (GSDMD) are two non-apoptotic cell death effector proteins that, in neutrophils, also control neutrophil extracellular trap (NET) formation (1–3). In response to MLKL phosphorylation (pMLKL) and activation, necroptotic neutrophils initiate an intricate sequence of signaling events promoting phosphatidylserine exposure on the plasma membrane, NADPH (reduced form of nicotinamide adenine dinucleotide phosphate) oxidase–independent reactive oxygen species production, loss of cytoplasmic granules, and breakdown of the nuclear membrane (1). Peptidylarginine deiminase 4 (PAD4) is hypothesized to neutralize charge on histones to facilitate chromatin decondensation (1, 4). However, genetic deficiency in Padi4 fails to prevent chromatin decondensation but does prevent MLKL-induced NET formation, highlighting a more complex role for PAD4 in the extrusion of nuclear DNA (1, 4).
Existing models of PAD4 function predict that histones are targeted randomly for citrullination during NET formation to alter the charge structure of chromatin. Using a method for visualizing chromatin accessibility, transposase-accessible chromatin with visualization, lamina-associated domains acting as initiation sites for NET formation form at the nuclear periphery of phorbol myristate acetate (PMA)–treated neutrophils. This is suggested to aid in the disassembly of chromatin into mononucleosomes, followed by PAD4-controlled histone citrullination and disassembly of mononucleosomes into free DNA and histones (5). The role of PAD4 in spatial control of histone citrullination at specific genomic loci following induction of regulated cell death and NETosis has not been explored.
While NET formation is a primary outcome of PAD4 activation, concomitant cytoplast formation is the corresponding biological outcome of nuclear DNA extrusion to generate neutrophils lacking nuclear DNA. In the asthmatic lung, PAD4-dependent extrusion of DNA from neutrophils to generate a cytoplast promotes T helper cell 17 responses and amplifies neutrophil recruitment to lung tissues supporting roles of cytoplasts during neutrophilic inflammatory responses. However, the dominant pathways contributing to cytoplast formation in this context remains unclear (6). Diverse stimuli have been proposed to drive NETosis and cytoplast formation but are assumed distinct from apoptosis (7). Our data reveal PAD4 activation and DNA extrusion downstream of GSDME activation as a central target of apoptosis, the default cell death program regulating neutrophil life span (8–10).
RESULTS
Apoptosis activates PAD4
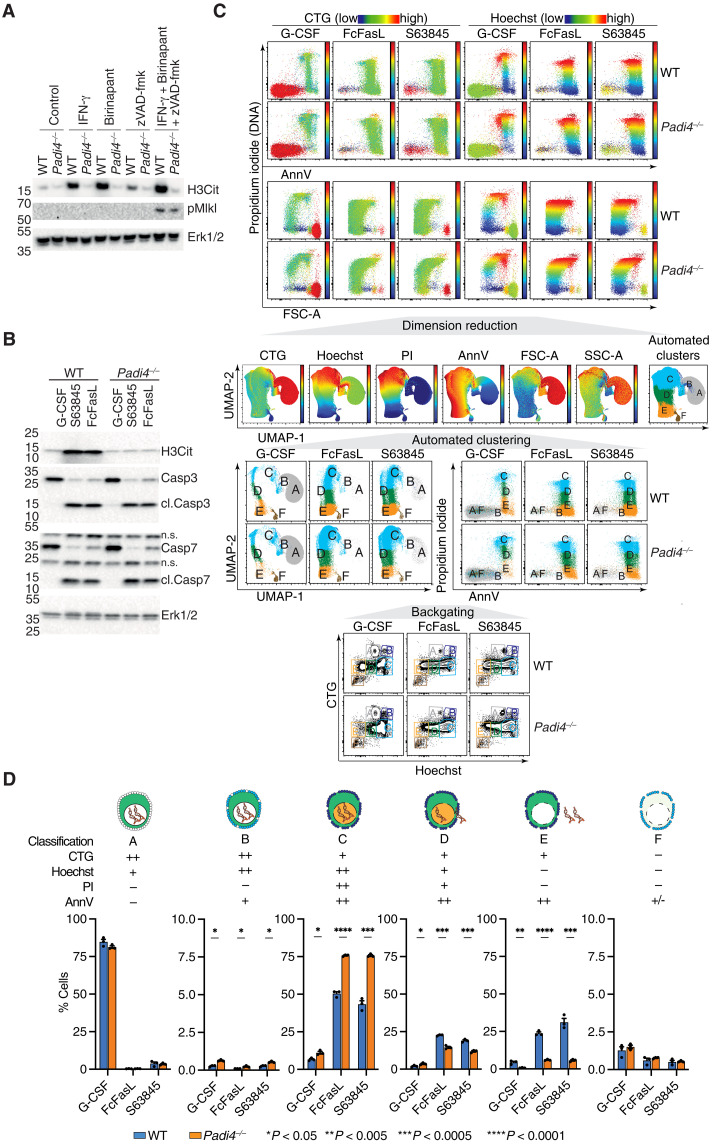
Necroptotic cell death signaling is a trigger for PAD4 activation and generation of histone H3 citrullination (H3Cit) and can be blocked by deficiencies in receptor-interacting protein kinase 1 kinase activity, receptor-interacting protein kinase 3 (RIPK3), and MLKL (1). To investigate the effects of other forms of cell death, including mitochondrial apoptosis and tumor necrosis factor (TNF)- or Fas ligand (FasL)-mediated apoptosis, we treated mouse wild-type (WT) and Padi4−/− bone marrow neutrophils with interferon-γ (IFN-γ) + birinapant (which induces TNF production and triggers apoptosis), IFN-γ + birinapant + z-VAD-fmk (IBZ, which also induces TNF production by neutrophils and engages RIPK3/MLKL], FcFasL (Fas-mediated apoptosis), Q-VD-OPh, S63845 [(myeloid cell leukemia sequence 1 (Mcl-1) antagonist and inducer of BAK/BAX-dependent mitochondrial apoptosis), growth factor withdrawal (dimethyl sulfoxide, mitochondrial apoptosis), or the prosurvival cytokines granulocyte colony-stimulating factor (G-CSF) and IFN-γ as controls (1, 10). Consistent with previous observations, the necroptotic stimulus IBZ induced H3Cit generation in WT but not Padi4−/− neutrophils (Fig. 1A). We also hypothesized that H3Cit generation would be restricted to non-apoptotic stimuli, but, unexpectedly, high levels of H3Cit were detected in response to all apoptotic triggers: birinapant, FcFasL, S63845, and growth factor withdrawal (Fig. 1, A and B). The G-CSF- and IFN-γ–treated samples exhibited low levels of H3Cit consistent with the actions of these prosurvival growth factors, Padi4−/− neutrophils, and the basal H3Cit levels seen in freshly purified cells. These biochemical data reveal that PAD4-dependent H3Cit is induced not only by classical non-apoptotic NET stimuli but also by apoptotic stimuli.
Fig. 1. PAD4 is activated by apoptotic and necroptotic stimuli and controls cytoplast formation.
(A and B) Protein expression levels of H3Cit, pMLKL, (cl.) caspase 3, and Erk1/2 in freshly isolated WT and Padi4−/− BM neutrophils stimulated with apoptotic and necroptotic stimuli. (C) Flow cytometry evaluation of cell size (FSC-A) and granularity (SSC-A), propidium iodide (PI), annexin V (AnnV), CellTracker Green (CTG), and Hoechst in WT and Padi4−/− neutrophils stimulated as indicated. (D) Statistical analysis of the proportion of each cell cluster identified by dimension reduction (UMAP) and automated clustering (FLOWSOM) from (C). Means ± SEM, n = 3 in each stimuli group, *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001, WT versus Padi4−/−, unpaired t test. n.s., not significant.
PAD4 controls cytoplast formation
The generation of H3Cit in response to apoptotic stimuli suggested that PAD4 may also regulate the extrusion of nuclear DNA in apoptotic neutrophils in a process similar to GSDMD- and MLKL-mediated NET formation. To test this, we designed a flow cytometry panel to simultaneously monitor neutrophils for phosphatidylserine externalization [annexin V (AnnV)], membrane permeability [propidium iodide (PI)], loss of cytoplasmic proteins [CellTracker Green (CTG)], and nuclear DNA (Hoechst). Dimension reduction using Uniform Manifold Approximation and Projection (UMAP) in combination of flow self-organizing maps (FLOWSOM) automated clustering (11) identified six unique cell clusters (A to F) based on the colocalization and intensity of the four markers, as well as the cell size (FSC-A) and granularity (SSC-A) (Fig. 1C). These six clusters vary according to cell death stimuli and correspond to distinct transitional cell death states that reflect changes in DNA content and membrane integrity: cluster A is viable; cluster B is apoptotic with exposed phosphatidylserine (PS) but an intact plasma membrane; cluster C represent a compromised plasma membrane staining with PI that would otherwise be classified as dead; cluster D indicates a partial loss of DNA from cells; and clusters E and F are consistent with a cytoplast lacking DNA (Fig. 1C). Using this gating strategy, we then quantified these transitional cell death states in WT and Padi4−/− neutrophils. Overall we observed increased numbers of AnnV+PI+ neutrophils (clusters B and C) in Padi4−/− neutrophils, suggesting an increase in cell death induced by PAD4 deficiency, or, alternatively, a failure to extrude nuclear DNA from dead neutrophils to form PI− cytoplasts (Fig. 1D). Detailed analysis of these neutrophil subsets indicated that the latter was occurring. We observed an accumulation of the early-stage transitional cell death state cluster C accompanied by a decreased accumulation of clusters D and E indicating a failure to transition toward nuclear DNA-free cytoplasts (Fig. 1D). These data are consistent with a role for PAD4 in priming neutrophils for DNA extrusion.
Apoptotic neutrophils engage PAD4 to externalize DNA
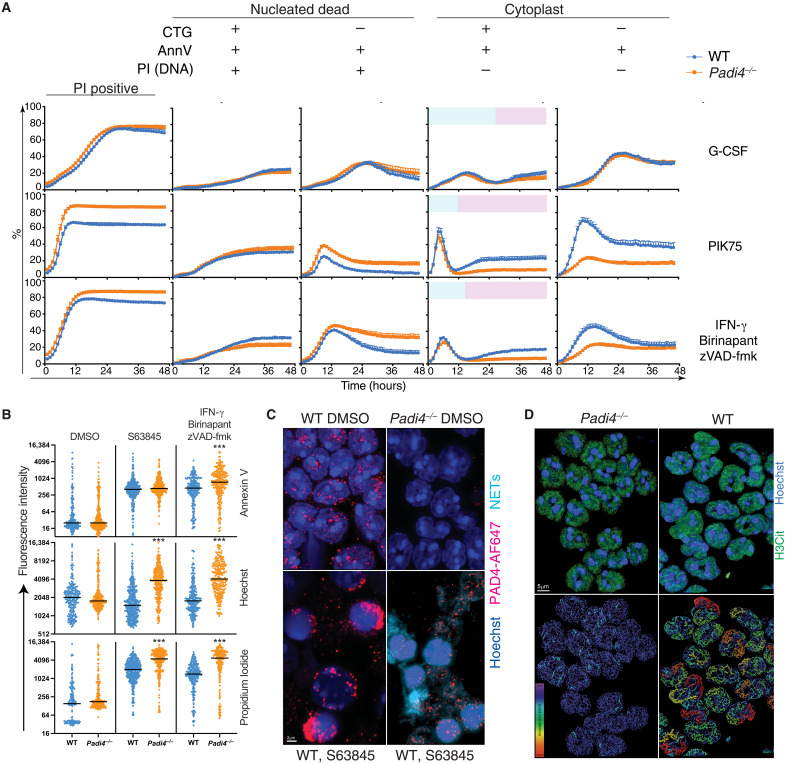
Flow cytometry data suggested that dead Padi4−/− neutrophils were failing to eject DNA to form cytoplasts (Fig. 1, B to D) while also indicating that this cell death transition occurs in a cellular state that is normally considered nonviable because of plasma membrane disruption. To investigate these cell death states, we used live-cell imaging coupled to the StarDist image segmentation deep learning module to study kinetic changes in WT and Padi4−/− neutrophils treated with PIK75 (mitochondrial apoptosis), IBZ (MLKL-dependent necroptosis), or G-CSF as a control. Tracking individual transition states revealed a bimodal CTG+AnnV+PI− population consistent with an early peak of apoptotic neutrophils (blue) and a later generation and accumulation of PI-negative cytoplasts (pink) that were dependent on PAD4 (Fig. 2A). The reduction in frequency of CTG+AnnV+PI− dying cells was accompanied by an increase in frequency of CTG+AnnV+PI+ cells as neutrophils transition to a dead state (Fig. 2A and table S1). Increased DNA retention was also evident by tracking quantitative fluorescence intensity of individual neutrophils using confocal microscopy in response to apoptotic and necroptotic stimuli (Fig. 2B). Both Hoechst and PI fluorescence intensity was increased in Padi4−/− neutrophils (Fig. 2B), consistent with increased DNA retention in the cell as seen by flow cytometry and live-cell imaging (Figs. 1 and 2A). These data support a role for PAD4 in a continuum of DNA externalization during apoptosis.
Fig. 2. PAD4 mediates cytoplast formation after disruption of plasma membrane integrity.
(A) Kinetic changes in WT and Padi4−/− neutrophil viability in 48-hour live-cell imaging in the presence of G-CSF (100 ng/ml), 1 μM PIK75, or combinations of 2 μM BPT, 10 μM zVAD-fmk, and IFN-γ (100 ng/ml). Data represent mean and SEM of independent biological samples from five WT and seven Padi4−/− mice. Blue color in column 4 represents dying neutrophils. Pink color in column 4 represents dead neutrophils lacking a nucleus (cytoplast). (B) Quantitative fluorescence intensity of WT and Padi4−/− neutrophils at 12 hours classified according to staining with Hoechst, CTG, PI, and AnnV. ***P < 0.0001, WT versus Padi4−/−. (C) Fluorescence microscopy showing externalized DNA from neutrophils treated with S63845. Neutrophils were stained with Hoechst and a PAD4 antibody coupled to AF647. A 3D nuclear mask was generated to identify DNA outside the nuclear outline to separate intranuclear (dark blue) from extranuclear (aqua) DNA (NETs). (D) Super-resolution microscopy of H3Cit distribution in WT and Padi4−/− neutrophils indicating the difference in intensity and distribution of signal. A bounding box metric indicates the continuity of H3Cit signal by assessing the length of the longest principal axis inside a rectangular cuboid object and whose local axes are aligned along the principal axis of the object. Data are representative of three independent experiments.
PAD4 controls the H3Cit epigenome
To ascertain the impact of PAD4 enzymatic activity on H3Cit in resting neutrophils, we performed airyscan confocal microscopy and lattice structured illumination microscopy (SIM) microscopy of H3Cit distribution. PAD4 protein was found in punctate structures in the nucleus of resting bone marrow neutrophils, which then polarized during apoptosis, and continued to associate with extracellular DNA of NETs released from apoptotic neutrophils (Fig. 2C). WT neutrophils displayed extended “filamentous” tracts of H3Cit staining compared to punctate disrupted H3Cit staining in Padi4−/− neutrophils (Fig. 2D and movie S1). These data indicate a partial role for PAD4 in creating the H3Cit epigenome in the nucleus during neutrophil development. These data support a role for PAD4 in neutrophil morphological changes after loss of plasma membrane integrity but not in the early stages of apoptosis.
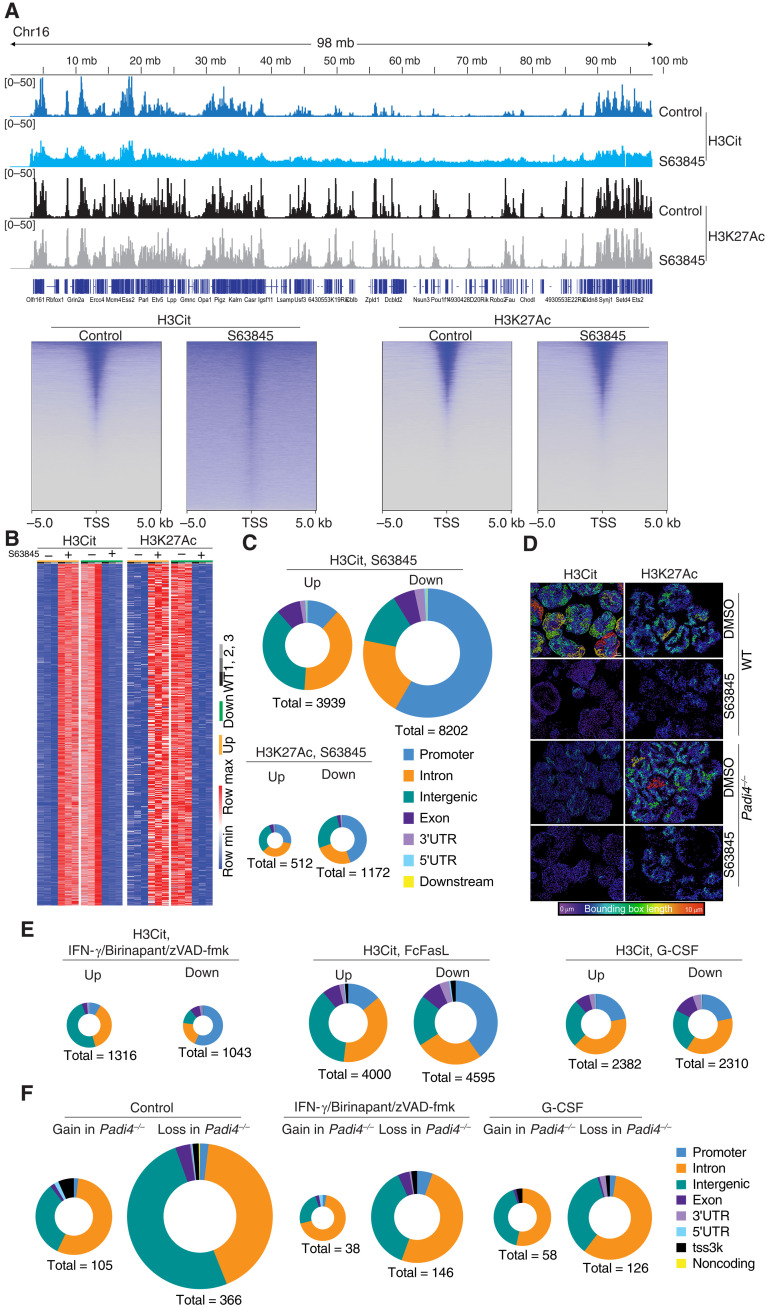
To investigate whether PAD4 randomly citrullinates histones or can specifically target genomic regions for histone citrullination and DNA externalization, we performed epigenomic profiling of H3Cit and H3K27Ac histone modifications using CUT&Tag (12). H3Cit was enriched at gene dense regions, and in particular promoters, at steady state (Fig. 3A). Upon stimulation with S63845 to induce apoptosis, >3000 consensus peaks were differentially citrullinated based on our genome-wide comparison of H3Cit peak intensity using DiffBind (Fig. 3B) (13). These changes were preferentially localized to specific types of genomic regions. In response to S63845, H3Cit accumulated at intronic and intergenic regions and decreased at promoter regions (Fig. 3, A and C). No such effect was observed on H3K27Ac in the same cells. Airyscan enhanced resolution microscopy (Fig. 3D) was complimented with lattice lightsheet structured illumination super-resolution microscopy of the same images (fig. S1) to demonstrate that induction of apoptosis reduced the filamentous H3Cit distribution in the nucleus, coinciding with a near uniform redistribution of H3Cit across the chromatin (Fig. 3D and fig. S1). The dynamic redistribution of H3Cit from promoter elements to intergenic and intronic regions was also evident in neutrophils treated with FcFasL or IBZ (Fig. 3E). These analyses suggest that PAD4 may target enzymatic activity to intergenic and intronic spaces via unknown mechanisms to prime the entire genome for DNA externalization. To understand how redistribution of H3Cit may influence chromatin accessibility more broadly, we used transposase-accessible chromatin with sequencing (ATAC-seq) on WT and Padi4−/− neutrophils. Substantial changes in chromatin accessibility were observed in Padi4−/− neutrophils, with the majority being a loss of signal at intergenic and intronic regions, both in steady state and following induction of regulated cell death compared to WT controls (Fig. 3F). These data demonstrate that PAD4 does influence chromatin accessibility at steady-state and during apoptosis, likely in combination with other proteins, such as PAD2.
Fig. 3. Cell death–triggered epigenetic changes in dying neutrophils.
(A) Top: Genome tracks of H3Cit and H3K27Ac of WT and Padi4−/− control neutrophils or treated with S63845 and analyzed using CUT&Tag. Data representative to three biological replicates. Bottom: Heatmap representation of WT neutrophil H3Cit and H3K27Ac at the TSS following S63845 treatment for 2.5 hours. (B) Heatmap representation of unique differential H3Cit or H3K27Ac peaks at specific genomic loci using CUT&Tag in independent samples of neutrophils treated with S63845 for 2.5 hours or controls. (C) Genomic feature analysis of H3Cit and H3K27Ac differential peaks in WT neutrophils treated with S63845 and analyzed by CUT&Tag. (D) Super-resolution microscopy of H3Cit and H3K27Ac distribution in WT and Padi4−/− neutrophils treated with S63845 and controls. A bounding box metric indicates the continuity of signal. (E) Genomic feature analysis of H3Cit CUT&Tag signal in WT neutrophils treated with S63845, IFN-γ + birinapant + zVAD-fmk, FcFasL, or G-CSF. (F) ATAC-seq evaluation of differential chromatin accessibility in freshly isolated WT and Padi4−/− control neutrophils or treated with G-CSF or IFN-γ + birinapant + zVAD-fmk. 3′UTR, 3′ untranslated region.
Apoptotic signaling alters the neutrophil citrullinome
PAD4 can citrullinate histones to increase chromatin decondensation; however, other protein targets may also aid in DNA externalization. To investigate PAD4 targets, we engaged the apoptotic signaling pathway and then used biotin-tagged phenylglyoxal (biotin-PG) to chemically modify citrullinated proteins and then enrich these proteins on streptavidin agarose. Mass spectrometry identified a range of proteins including histones and other nuclear-associated proteins, many of which were shared between WT and Padi4−/− neutrophils (tables S2 to S5). The presence of citrullinated histones in resting bone marrow neutrophils is consistent with the abundance of H3Cit identified at the promoters of WT and Padi4−/− neutrophils as assessed using CUT&Tag and super-resolution microscopy (Figs. 2 and 3, movie S1, and tables S2 to S5). Other citrullinated proteins identified in apoptotic neutrophils were associated with pathways including neutrophil degranulation and Rho guanosine triphosphatases. PAD4 was also detected in WT samples, indicating that it is also citrullinated in both resting and apoptotic bone marrow neutrophils.
GSDME triggers PAD4 activation
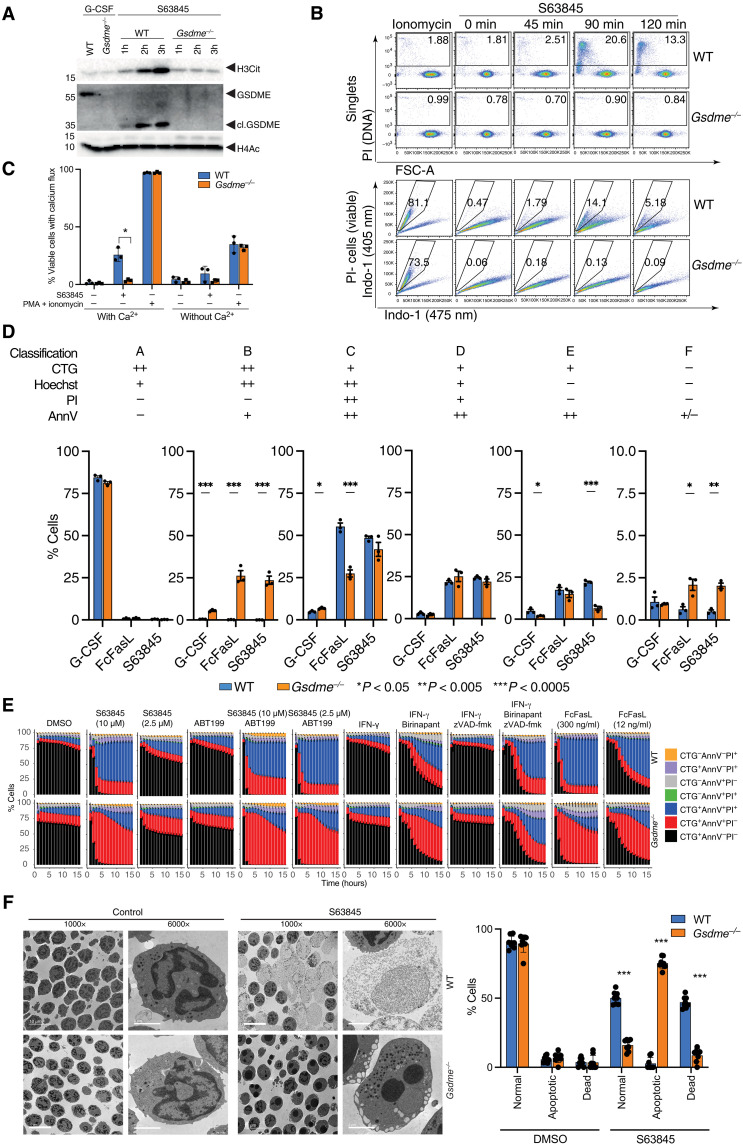
GSDME is a pore-forming protein that is activated by apoptotic caspases and disrupts the integrity of membranes in many cell types, including neutrophils (14). To understand the stage of cell death controlled by GSDME in neutrophils, we examined H3Cit accumulation in neutrophils treated with S63845 (Fig. 4A). WT neutrophils generated a robust induction of H3Cit corresponding with cleaved caspase 3 and a 33-kDa cleaved GSDME. Gsdme−/− neutrophils failed to induce H3Cit in response to S63845, indicating a critical role for GSDME in PAD4 activation. PAD4 is activated by calcium mobilization, and viable Gsdme−/− neutrophils treated with S63845 failed to trigger a calcium flux (Fig. 4B). To investigate the possible sources of calcium signaling during neutrophil apoptosis, we also incubated neutrophils in media lacking calcium (Fig. 4C). A calcium deficiency in the media impaired the calcium flux of apoptotic neutrophils and also reduced calcium flux of neutrophils treated with PMA and ionomycin. Calcium likely enters apoptotic neutrophils through the GSDME pore, but intracellular calcium stores also appear to contribute to the calcium flux of WT apoptotic neutrophils.
Fig. 4. Mcl-1 antagonism triggers GSDME cleavage leading to Ca2+ influx, PAD4 activation, and H3Cit accumulation.
(A) Protein expression levels of H3Cit, (cl.) GSDME, (cl.) caspase 3, and H4Ac in freshly isolated WT and Gsdme−/− BM neutrophils or treated with S63845 and G-CSF. (B) Top: Flow cytometry evaluation of PI+ cells in single WT and Gsdme−/− neutrophils treated with S63845 for 0, 45, 90, and 120 min. Frequency of PI+ cells are indicated on each plot. Bottom: Ca2+ influx is determined in PI-(viable) neutrophils by switch of Indo-1 (475 nm)–positive to Indo-1 (405 nm)–positive staining. Frequency of Indo-1 (405 nm) calcium fluxing cells is indicated on each plot. (C) Calcium flux in neutrophils treated with S63845 for 90 min or PMA + ionomycin for 30 s in the presence or absence of 1.8 mM extracellular calcium in Dulbecco’s modified Eagle’s medium (DMEM). (D) Statistical analysis of automated cell clusters identified by flow cytometry measurement of cell size and granularity, PI, AnnV, CTG, and Hoechst in WT and Gsdme−/− neutrophils treated as indicated. Means ± SEM, n = 3 independent biological replicates, significance is determined by unpaired t tests, *P < 0.05, **P < 0.005, ***P < 0.0005. (E) Kinetic changes in WT and Gsdme−/− neutrophil viability by live-cell imaging and automated image analysis in the presence of the indicated stimuli. (F) Morphology of WT and Gsdme−/− neutrophils after treatment with S63845 for 2.5 hours. ×1000 or ×6000 magnification by electron microscopy.
To identify the transitional state controlled by GSDME, and to understand if it corresponds to the cellular defects seen in the absence of PAD4, we used the same flow cytometry method (as in Fig. 1) to demonstrate an equivalent accumulation of early-stage and intact apoptotic cells in cluster B and a corresponding loss of later-stage apoptotic cells in clusters C&D in Gsdme−/− neutrophils (Fig. 4D). The dominant effect observed was an accumulation of apoptotic AnnV+PI− neutrophils in cluster B, indicating that Gsdme−/− neutrophils can enter an apoptotic state but cannot transition to a PI-positive state. Consistent with flow cytometry data, live-cell imaging demonstrated a consistent accumulation of AnnV+PI− cells that failed to transition to AnnV+PI+ cells when treated with S63845 alone or in combination with the Bcl-2 antagonist ABT-199 (Fig. 4E). Other types of apoptotic cell death were similarly affected by the loss of GSDME, including apoptotic cell death triggered by FcFasL or the SMAC mimetic birinapant. Loss of GSDME was not a neutral event in apoptotic neutrophils resulting in an abnormal accumulation of apoptotic cellular structures including condensed nuclear material inside an intact nuclear membrane, large cytoplasmic vacuoles, and a polarization of cytoplasmic granules (Fig. 4F). These data suggest that caspase cleavage of GSDME, leading to pore formation in the plasma membrane or other intracellular compartments, facilitates calcium accumulation in the cytoplasm and PAD4 activation.
DISCUSSION
The concept of NETosis has been used broadly to define DNA extrusion events in neutrophils, either as a regulated process during pyroptosis or necroptosis or when externalized citrullinated chromatin is identified in tissue sections, plasma, or in cell cultures (1–3). The use of H3Cit with antibodies is a gold standard biomarker for the identification of NETosis. The extraordinary breadth of stimuli and disease conditions associated with NETosis and H3Cit accumulation has necessitated a reevaluation of the biochemical processes that underpin and link these pathways. A common element for neutrophil bioassays and neutrophilic inflammation is the inevitable induction of regulated cell death. Previous studies revealed critical roles for MLKL-mediated plasma membrane disruption as an essential biochemical step for PAD4 activity and H3Cit accumulation (1). While an important step in the description of a discrete biochemical signaling network controlling NET formation, it did not explain the large number of stimuli that can trigger PAD4 activation independently of the necroptotic pathway. While pyroptotic signaling also engages PAD4 activity and H3Cit, it is yet another non-apoptotic cell death pathway only triggered in unique circumstances in vivo, i.e., the presence of cytoplasmic lipopolysaccharide or PMA-triggered neutrophil elastase release and GSDMD cleavage (2, 3, 15). Our insight into apoptosis and PAD4 activation proposes regulated apoptotic and non-apoptotic cell death as a core set of biochemical pathways controlling DNA externalization from neutrophils.
Epigenetic analyses of neutrophils indicate rapid changes in histone marks during regulated cell death, both at gene-dense regions and in silenced areas of intergenic and intronic spaces. The observed citrullination at promoter regions and subsequent decreases in H3Cit at promoter sites also suggest that PAD2-dependent citrullination of histones, particularly at promoter elements, may help establish how PAD4 redistributes histone marks during regulated cell death. The loss of signal at the promoter region and the gain at intergenic and intronic sites indicate that existing epigenomic marks, including H3Cit itself or H3K27Ac or H4Ac, may also direct PAD4 enzymatic activity to intergenic and intronic elements. Further exploration of this enzymatic activity, the requirement for specific epigenomic marks on the chromatin to direct PAD4 activity, the role of intracellular and extracellular calcium sources for PAD4 activation, and the interdependency of PAD2 and PAD4 epigenetic modification of the genome will likely reveal an intricate network controlling this key neutrophil and innate immune effector function.
These data challenge our conceptual understanding of cell death. The application of membrane-impermeable dyes, including PI, is applied in many applications to signify cell death—a point where regulated cellular processes and biochemical events are not expected to occur. Yet, the description of MLKL-mediated changes in cellular architecture in neutrophils, including chromatin decondensation, degranulation, PAD4 activation, reactive oxygen species production, and regulated DNA extrusion highlighted our lack of understanding of transition states linking MLKL-mediated membrane permeabilization to the final act of a dying cell when no further biochemical activity is possible or physiologically relevant (1). These data indicate roles for GSDME, not in cell death induction, but rather in the transition from apoptotic to membrane permeable—a point occurring in a transitional state before PAD4 establishes control over DNA extrusion in a PI-positive membrane-permeable state. It places a role for GSDME in a similar league to MLKL and GSDMD by activating a series of biochemical events that aid in the regulated extrusion of DNA after the membrane has been permeabilized. A dominant function of GSDME is likely as a factor downstream of apoptotic caspases in PAD4-expressing cells, primarily neutrophils and eosinophils. This last dying act of neutrophils—the life after death—is well recognized to function as not only an antimicrobial event but also as an amplifier of innate immunity and nefariously as a source of autoantigen (16). These insights into pore-forming regulators of regulated DNA extrusion signify NETosis as the default outcome of neutrophil apoptosis, but only if efferocytosis mechanisms fail. These findings reveal targets not only to modulate neutrophil NETosis during infection but also to eliminate chronic autoantigen in autoimmune conditions where histones, nucleic acids, and citrullinated proteins drive disease.
MATERIALS AND METHODS
Mice
The Padi4−/− and Gsdme−/− mouse strains were purchased from JAX Mice. All animal experiments complied with the regulatory standards of and were approved by the Institutional Animal Care and Use Committees at Boston Children’s Hospital and University of California San Diego. Mice were housed under specific pathogen–free conditions with unlimited access to food and water.
Bone marrow neutrophil purification
Murine bone marrow cells were isolated from the femur, tibia, and ossa coxae in Hanks’ balanced salt solution supplemented with 1% fetal bovine serum (FBS) and 15 mM EDTA. Cells were overlayed on 78, 68, and 52% Percoll layers and centrifuged at 400g for 30 min at 4°C. Neutrophils were removed from the 68/78% interface and resuspended in Dulbecco’s modified Eagle’s medium (DMEM)/10% FBS for biochemical assays or phenol red-free DMEM/10% FBS for live-cell imaging viability assays. The purity of neutrophil preparations was greater than 95% as assessed by May-Grünwald Giemsa staining. Electron microscopy of neutrophil preparations was performed with a JOEL 1400Plus transmission electron microscope equipped with Gatan OneView as per published protocols (1). All cell culture media were purchased from Gibco.
Live-cell and confocal imaging
Neutrophils were isolated according to published methods (1). Neutrophils were labeled with 50 nM CTG (Life Technologies) for 15 min at 37°C in serum-free DMEM. A total of 1 × 105 neutrophils were plated in 96-well optical bottom plates (Corning 3904) in recombinant human G-CSF (100 ng/ml; Amgen) or recombinant mouse IFN-γ (100 ng/ml; BioLegend). Neutrophil viability was monitored using AnnV–Alexa Fluor 647 (BioLegend) to detect phosphatidylserine and PI (0.5 μg/ml; Sigma-Aldrich). Cells were treated with 2.5 to 10 μM S63845, 10 μM ABT-199, 2 μM birinapant, 2 μM PIK-75 (Cayman Chemicals), 10 μM zVD-fmk (Cayman Chemicals), 10 μM QVD-OPh (Cayman Chemicals), or FcFasL (300 ng/ml; Adipogen). Confocal experiments in Fig. 2B were performed using a Leica SP8 confocal microscope with lightning deconvolution, and images were segmented with Cellpose. Kinetic imaging was conducted by capturing triplicate fields of view with a 10× [0.45 numerical aperture (NA)] objective every hour using a Nikon Ti2-E microscope with a Qi-2 camera and Nikon Elements 5.02.02 acquisition software using the Jobs module. Cells were maintained at 37°C + 5% CO2 using an Okolab H301 Bold Line stage top incubator. Three non-overlapping 3.125-mm2 zones within each well were imaged for red, green, and far-red fluorescence using a SpectraX light engine (Lumencor) with individual Large-Field-Of-View (LFOV) filter cubes (Semrock) (excitation/emission maxima at 554/609, 470/525, and 618/698 nm, respectively). For some experiments, images from triplicate fields of view were captured at 10× at 37°C + 5% CO2 every hour by the ImageXpress Micro Confocal High-Content Imaging System (Molecular Devices) operating MetaXpress (version 6.2.5) acquisition software. Three non-overlapping 1.96-mm2 zones within each well were imaged for red, green, and far-red fluorescence (excitation/emission at 531/593, 475/536, and 634/692 nm, respectively).
Cell death kinetics were analyzed using a custom macro in the Fiji distribution of ImageJ (17, 18). The macro processed each time point folder that was generated by the Nikon Ti2 imaging system, and time image stacks were constructed of each well field position. The macro segmented each cell population in each frame of the time stacks using custom-scripted code for the Fiji distribution of ImageJ and the deep learning plugin StarDist, and then population percentages were calculated for each time point from the extracted binary images (19). Using these masks, each cell population was defined using Boolean arithmetic. The logic was constructed so that cells were only counted once. Data were logged and percentage populations were calculated at each time point.
Super-resolution imaging
For super-resolution imaging, images were collected with a Zeiss Elyra 7 Lattice SIM super-resolution instrument using a Plan Apochromatic 63× (1.40 NA) oil objective lens with 561- and 642-nm excitation. Z-stack images were collected in Leap-mode with 13 phase positions of the lattice SIM pattern. Images were processed with Zeiss Zen Black SIM2 processing software and analyzed by Imaris software (Bitplane).
Enhanced confocal imaging was performed with a Zeiss laser scanning confocal microscope 880 Airyscan using a 63× (1.4 NA) objective and the 32-channel GaAsP-PMT area detector. All image stacks (on average of 45 slices) were acquired with Nyquist resolution parameters using a 0.159-μm step size and optimal frame size of 2124 × 2124. All 12-bit images were acquired using the full dynamic intensity range (0 to 4096) that was determined with the population of unstimulated neutrophils. All Airyscan-acquired images were batch-processed using the ideal three-dimensional (3D) defaults settings of the Airyscan processing module, which briefly involves linear deconvolution and pixel reassignment for recentering of the raw data. Images were further processed for maximum intensity projections in either Zen Pro in 2D (Zeiss) or Imaris in 3D (Bitplane) for figures. In addition, 3D images were imported into Imaris and rendered into iso-surfaces based on a threshold and fluorescent intensity range. Thresholds above 400 to 4096 were conservatively deemed relevant based on secondary alone labeled controls and autofluorescence. The rendered 3D surfaces were then displayed as irregular quadrilateral bounding box lengths.
DeathTrackR data visualization
ImageJ datalogs from live-cell imaging analyses were visualized using the R Shiny app DeathTrackR (https://croker.shinyapps.io/project/). The DeathTrackR package was developed using Shiny in R 3.6.3., the R packages Tidyverse, and plotrix for basic data loading and plotting utilities and shinydashboard for module management (20–23).
Flow cytometry
Flow cytometry analysis was performed on neutrophils stained with Cell Tracker Green (200 nM), Hoechst, AnnV-Alexa Fluor 647 (BioLegend) in AnnV staining buffer, and PI (Sigma-Aldrich) using a LSR Fortessa (BD Biosciences) and analyzed with OMIQ. For calcium experiments, purified bone marrow neutrophils were loaded with 2 μM Indo-1 a.m. for 30 min at 37°C. Cells were washed twice in Hanks’ balanced salt solution/1% FBS and resuspended in DMEM/10% FBS without phenol red for assay. Cells were treated with 200 nM PMA + ionomycin (200 ng/ml) or 10 mM S63845 for the indicated times. Cells were washed, stained with PI (1 μg/ml), acquired on an LSRII flow cytometer, and analyzed with FlowJo. Statistical analyses were performed with Prism software (GraphPad). Differences between groups were determined via unpaired t tests.
Immunoblot
Neutrophil proteins were extracted with 4 volumes of methanol and 1 volume of chloroform and then centrifuged at 9000g for 5 min. Proteins at the interface were precipitated with 3 volumes of methanol at 9000g for 10 min, dried, and resuspended in SDS Gel loading buffer. Cell lysates and supernatants were analyzed using antibodies specific to H3Cit (Abcam), GSDME (Abcam), caspase 3 (Cell Signaling), cleaved caspase 3 (Cell Signaling), H4Ac (Abcam, ab177790), or p42/44 mitogen-activated protein kinase (ERK1/2) (Cell Signaling Technology, 137F5). Westerns were developed using enhanced chemiluminescence substrate, visualized on a ChemiDoc, and analyzed with ImageLab Software (Bio-Rad).
Biotin-PG labeling and LC/LC-MS/MS proteomic analysis
Samples were labeled with biotin-PG and prepared according to published methods (24–26). For mass spectrometry, 5 μl of samples was diluted in TNE [50 mM Tris (pH 8.0), 100 mM NaCl, and 1 mM EDTA] buffer. RapiGest SF reagent (Waters Corp.) was added to the mix to a final concentration of 0.1%, and samples were boiled for 5 min. TCEP [tris (2-carboxyethyl) phosphine] was added to 1 mM (final concentration), and the samples were incubated at 37°C for 30 min. Subsequently, the samples were carboxymethylated with iodoacetamide (0.5 mg/ml) for 30 min at 37°C followed by neutralization with 2 mM TCEP (final concentration). Protein samples prepared as above were digested with trypsin (trypsin:protein ratio, 1:50) overnight at 37°C. RapiGest was degraded and removed by treating the samples with 250 mM HCl at 37°C for 1 hour followed by centrifugation at 13,000g for 30 min at 4°C. The soluble fraction was then added to a new tube, and the peptides were extracted and desalted using C18 desalting columns (Thermo Fisher Scientific, PI-87782). Peptides were quantified using bicinchoninic acid assay (BCA) assay, and a total of 1 μg of peptides was injected for liquid chromatography–mass spectrometry analysis. Trypsin-digested peptides were analyzed by ultrahigh pressure liquid chromatography (UPLC) coupled with tandem mass spectroscopy (LC-MS/MS) using nanospray ionization. The nanospray ionization experiments were performed using a TimsTOF 2 pro hybrid mass spectrometer (Bruker) interfaced with nanoscale reversed-phase UPLC (Thermo Dionex UltiMate 3000 RSLC nano System) using a 25-cm, 75-μm inner diameter (ID) glass capillary packed with 1.7-μm C18 (130) BEHTM beads (Waters Corporation). Peptides were eluted from the C18 column into the mass spectrometer using a linear gradient (5 to 80%) of ACN (acetonitrile) at a flow rate of 375 μl/min for 1 hour. The buffers used to create the ACN gradient were as follows: buffer A (98% H2O, 2% ACN, and 0.1% formic acid) and buffer B (100% ACN and 0.1% formic acid). Mass spectrometry analysis was carried out using the Bruker TimsTOF Pro 2. For the TimsTOF Pro 2 settings, the following parameters were adapted, starting from the parallel accumulation-serial fragmentation (PASEF) method for standard proteomics. The values for mobility-dependent collision energy ramping were set to 95 eV at an inversed reduced mobility (1/k0) of 1.6 V s/cm2 and 23 eV at 0.73 V s/cm2. Collision energies were linearly interpolated between these two 1/k0 values and kept constant above or below. No merging of thermal ionization mass spectrometry scans was performed. Target intensity per individual PASEF precursor was set to 20,000. The scan range was set between 0.6 and 1.6 V s/cm2 with a ramp time of 166 ms. Fourteen PASEF MS/MS scans were triggered per cycle (2.57 s) with a maximum of seven precursors per mobilogram. Precursor ions in a mass/charge ratio range between 100 and 1700 with charge states ≥3+ and ≤ 8+ were selected for fragmentation. Active exclusion was enabled for 0.4 min (mass width, 0.015 Th; 1/k0 width, 0.015 V s/cm2). Protein identification and label-free quantification were carried out using Peaks Studio X (Bioinformatics Solutions Inc.)
Cleavage under targets and tagmentation (CUT&Tag)
CUT&Tag assays were performed as described previously with the modifications described below (12). A step-by-step protocol is also available at https://protocols.io/view/bench-top-cut-amp-tag-bcuhiwt6. Briefly, 2 × 105 cells were washed in 200 μl of wash buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, 0.5 mM spermidine, and 1× protease inhibitors] by gentle pipetting and then resuspended in 1.5 ml of wash buffer. Ten microliters of concanavalin A–coated magnetic beads (Bangs Laboratories, BP531) were activated and then added to 2 × 105 cells with incubation on an end-over-end rotator at room temperature for 10 min. The supernatant was removed, and bead-bound cells were resuspended in 50 μl of antibody buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, 0.5 mM spermidine, one Roche Complete Protease Inhibitor EDTA-Free tablet, 0.05% digitonin, 0.1% bovine serum albumin, and 2 mM EDTA]. Rabbit polyclonal anti-histone H3 (citrulline R2 + R8 + R17) antibody (Abcam, ab5103) was added to bead-bound cells (1:50 dilution) and incubated on a nutator overnight at 4°C. As a control, histone H3 (acetyl K27) (Abcam, ab4729) was added to bead-bound cells (1:50 dilution) and incubated on a nutator overnight at 4°C. After removing the supernatant in the next morning, the cells were incubated with donkey anti-rabbit immunoglobulin G (IgG) antibody (1:100 dilution, Jackson ImmunoResearch) in 100 μl of antibody buffer on a nutator at room temperature for 1 hour. The control cells were incubated with rabbit anti-mouse IgG antibody (1:100 dilution, Jackson ImmunoResearch). Bead-bound cells were washed twice with 1 ml dig-wash buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, 0.5 mM spermidine, 1× protease inhibitors, and 0.05% digitonin] to remove the unbound antibodies. CUTANA pAG-Tn5 (2.5 μl; EpiCypher) was added into 50 μl of dig-300 buffer [20 mM Hepes (pH 7.5), 300 mM NaCl, 0.5 mM spermidine, one Roche Complete Protease Inhibitor EDTA-Free tablet, and 0.01% digitonin], mixed with the bead-bound cells, and then rotated at room temperature for 1 hour. Bead-bound cells were washed twice in 1 ml of dig-300 buffer to remove unbound pA-Tn5 protein. Next, cells were resuspended in 300 μl of tagmentation buffer [20 mM Hepes (pH 7.5), 300 mM NaCl, 0.5 mM spermidine, 1 Roche Complete Protease Inhibitor EDTA-Free tablet, 0.01% digitonin, and 10 mM MgCl2] and incubated at 37°C for 1 hour. To stop tagmentation and solubilize DNA fragments, 10 μl of 0.5 M EDTA, 3 μl of 10% SDS, and 2.5 μl of Proteinase K (20 mg/ml; Thermo Fisher Scientific) were added to each sample and were further incubated at 55°C for 60 min. To extract the DNA, cells were mixed with 300 μl of phenol:chloroform:isoamyl alcohol (PCI) (Thermo Fisher Scientific), centrifuged at 16,000g for 3 min, and then mixed with 300 μl of chloroform (Sigma-Aldrich). The aqueous layer was added into 300 μl of 100% ethanol, centrifuged at 4°C 16,000g for 10 min, and then rinsed with 1 ml of 100% ethanol. The DNA was dissolved in 30 μl of 1 mM tris-HCl (pH 8) and 0.1 mM EDTA. To generate the sequencing libraries, the mixture was mixed with a universal i5 primer and a uniquely barcoded i7 primer and then amplified with the NEBNext HiFi 2x PCR Master mix (New England Biolabs Inc. (EB), M0541). The libraries were cleaned-up with solid-phase reversible immobilization (SPRI) paramagnetic beads (Beckman Coulter), washed with 80% ethanol, and then dissolved in 25 μl of 10 mM tris-HCl (pH 8). TapeStation D1000 capillary electrophoresis was performed to determine the size distribution and concentration of libraries. Paired-end Illumina sequencing was performed on the barcoded libraries following the manufacturer’s instructions.
Computational analyses of CUT&Tag and ATAC-seq data
Sequencing reads were mapped to mm9 reference genome using BWA v0.7.17 aligner (27). Hotspot peak caller was used to identify peaks of read density (28). Heatmaps of signal densities at promoter regions were generated using deepTools v3.0.2 (29). The DiffBind v3.0.15 Bioconductor package was used to identify differential peaks within the set of consensus peaks among replicates, based on the cutoffs of >2-fold change in peak intensity and false discovery rate < 0.05 (13).
Genotyping
Mice were genotyped with the following primers: Pad4: 5′-AGC TTT GTA AGG GGC ATC CT-3′; 5′-TCA AAG TAC CTG ATG TGT TGA CTG-3′; 5′-TCA TGA CCC CCA ACA CTC A-3′. Gsdme−/−: 5′-CAT GCG AAA AGA AGC TGT CA-3′; 5′-GAT CCT TTC AGG TTG GCA GT-3; 5′-CCA ATC CCA TGC TTC GAC-3′.
GLIMMIX
For live-cell image statistical analysis, the generalized linear mixed model (GLIMMIX) was used. Variables are assigned according to time I, well j, and cell number cij, in a transition state (or states) of interest, which are a subset of some total number dij of cells in the image. A generalized linear mixed model was fit to the outcome Yij = cij/dij, the proportion of cells of interest in well j at time I. Txgi represents the treatment-genotype combination for well j. Then
Interwell correlation between genetic replicates is accounted for by Txgi which specifies a fixed effect for each treatment-genotype combination. Define bi = ci1/di1, a random intercept, and ai, a time slope, for well j, accounting for correlation of images of each well. Then, given ai and bi, it is assumed that dij and Yij are independent and binomial-distributed as bin(dij, πij) with
Precision is increased due to the ability to distinguish individual cells in each image in the live-cell imaging technique: Variance of a binomial outcome varies inversely with the denominator. Underestimation of the SEs due to overdispersion is remedied by use of the classical empirical (“sandwich”) estimator of Cov(β).
Last, we assume that the random effects ai and bi have a bivariate normal distribution with mean [0, 0], covariance , and variances and , so . The model supposes each genetic replicate in a treatment has its own underlying probability of transitioning out of the state(s) of interest that does not vary over the course of the experiment. All data preparation for and inference by GLIMMIX in this paper was performed with SAS 9.4 (SAS Institute, Cary, NC).
Acknowledgments
Funding: This work was supported by National Institutes of Health grant 5RO1HL124209 (B.A.C.), National Institutes of Health grant R01AI155869 (B.A.C. and H.M.H.), National Institutes of Health grant P01HL152958 (B.A.C., H.M.H., and S.D.C.), National Institutes of Health grant R35 GM118112 (P.R.T.), American Asthma Foundation (B.A.C.), Alpha-1 Foundation grant #615533 (M.Ge.), ISF grant #818/18 (M.Ge.), Israel Cancer Association Grant # 20220099 (M.Ge.), Varda and Boaz Dotan Research Center in Hemato-Oncology (M.Ge.), US-BSF grant #2017176 (B.A.C. and M.Ge.), National Institutes of Health grant 1U54HD0902565, Kenneth Rainin Foundation Innovator and Synergy Award (KLJ). National Institutes of Health grant R21AI144877 (KLJ). National Institutes of Health grant R01DK119996 (K.L.J.), National Institutes of Health grant UL 1TR002541, American Heart Association Career Development Award (Y.P.Z.), National Institutes of Health grant P30DK040561 (R.S.), National Institutes of Health grant NINDS P30 NS047101 (J.S.), National Cancer Institute Cancer Center Support grant 3P30CA006516-52 (C.J.H.), National Institutes of Health S10D021831, and UCSD-CMM-EM Core, RRID: SCR_022039, S10OD023527.
Author contributions: Conceptualization: Y.P.Z., M.S., Z.T., K.L.J., and B.A.C. Methodology: Y.P.Z., M.S., Z.T., J.C.L., C.J.N., A.A.C., A.J.S., C.J.H., G.W., K.A., E.G., M.Gh., J.S., M.Ge., W.B.K., S.D.C., K.F.G., E.W., P.R.T., W.P., L.P.W., R.S., K.L.J., and B.A.C. Investigation: Y.P.Z., M.S., Z.T., J.C.L., A.A.C., H.A., A.J.S., C.H., K.A., J.S., P.R.T., S.D.C., L.P.W., R.S., and B.A.C. Visualization: C.J.N., W.P., L.P.W., and R.S. Funding acquisition: Y.P.Z., M.Gh., S.D.C., H.M.H., P.R.T., K.L.J., and B.A.C. Supervision: Y.P.Z., M.S., P.R.T., R.S., K.L.J., and B.A.C. Writing—original draft: Y.P.Z., M.S., K.L.J., and B.A.C. Writing—review and editing: Y.P.Z., M.S., Z.T., A.J.S., C.H., J.S., , M.Gh., P.R.T., L.P.W., R.S., K.L.J., and B.A.C.
Competing interests: P.R.T. holds several patents for PAD inhibitors and holds an equity interest in Padlock Therapeutics, a wholly owned subsidiary of Bristol Myers Squibb. P.R.T. is a consultant for Related Sciences and holds equity in Danger Bio. P.R.T. and Y. L. (Synthesis and use of novel inhibitors and inactivators of protein arginine deiminases, US Patent 7964636, issued on 21 June 2011, University of South Carolina); P.R.T., K. L. Bicker, and V. Subramanian (Phenylglyoxal probes, US Patent 9,921,225, 2018, issued 20 March 2018, Scripps Research); P.R.T., A. Muth, and V. Subramanian [Inhibitors of protein arginine deiminases (PADs) and methods of preparation and use thereof, US Patent 11,208,386, issued 28 December 2021, Umass Medical School]; P.R.T. and S. Mondal (Inhibitors of protein arginine deiminase 1 and methods of preparation and use thereof, US Patent App. 17/622,200 UMass Medical School). K.L.J. and H.A. are currently employees of Moderna. C.J.H. is currently an employee of Pfizer. M.S. is currently an employee of Noxopharm Ltd. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sequencing data are available at GSE241485.
Supplementary Materials
This PDF file includes:
Fig. S1
Table S1 to S5
Legend for movie S1
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.D’Cruz A. A., Speir M., Bliss-Moreau M., Dietrich S., Wang S., Chen A. A., Gavillet M., Al-Obeidi A., Lawlor K. E., Vince J. E., Kelliher M. A., Hakem R., Pasparakis M., Williams D. A., Ericsson M., Croker B. A., The pseudokinase MLKL activates PAD4-dependent NET formation in necroptotic neutrophils. Sci. Signal. 11, eaao1716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen K. W., Monteleone M., Boucher D., Sollberger G., Ramnath D., Condon N. D., von Pein J. B., Broz P., Sweet M. J., Schroder K., Noncanonical inflammasome signaling elicits gasdermin D-dependent neutrophil extracellular traps. Sci. Immunol. 3, (2018). [DOI] [PubMed] [Google Scholar]
- 3.Sollberger G., Choidas A., Burn G. L., Habenberger P., Di Lucrezia R., Kordes S., Menninger S., Eickhoff J., Nussbaumer P., Klebl B., Kruger R., Herzig A., Zychlinsky A., Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Sci. Immunol. 3, (2018). [DOI] [PubMed] [Google Scholar]
- 4.Li P., Li M., Lindberg M. R., Kennett M. J., Xiong N., Wang Y., PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 207, 1853–1862 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X., Shen Y., Draper W., Buenrostro J. D., Litzenburger U., Cho S. W., Satpathy A. T., Carter A. C., Ghosh R. P., East-Seletsky A., Doudna J. A., Greenleaf W. J., Liphardt J. T., Chang H. Y., ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods 13, 1013–1020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krishnamoorthy N., Douda D. N., Bruggemann T. R., Ricklefs I., Duvall M. G., Abdulnour R. E., Martinod K., Tavares L., Wang X., Cernadas M., Israel E., Mauger D. T., Bleecker E. R., Castro M., Erzurum S. C., Gaston B. M., Jarjour N. N., Wenzel S., Dunican E., Fahy J. V., Irimia D., Wagner D. D., Levy B. D.; National Heart, Lung, and Blood Institute Severe Asthma Research Program-3 Investigators , Neutrophil cytoplasts induce TH17 differentiation and skew inflammation toward neutrophilia in severe asthma. Sci. Immunol. 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenny E. F., Herzig A., Krüger R., Muth A., Mondal S., Thompson P. R., Brinkmann V., von Bernuth H., Zychlinsky A., Diverse stimuli engage different neutrophil extracellular trap pathways. eLife 6, e24437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steimer D. A., Boyd K., Takeuchi O., Fisher J. K., Zambetti G. P., Opferman J. T., Selective roles for antiapoptotic MCL-1 during granulocyte development and macrophage effector function. Blood 113, 2805–2815 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dzhagalov I., St John A., He Y. W., The antiapoptotic protein Mcl-1 is essential for the survival of neutrophils but not macrophages. Blood 109, 1620–1626 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McArthur K., D’Cruz A. A., Segal D., Lackovic K., Wilks A. F., O’Donnell J. A., Nowell C. J., Gerlic M., Huang D. C. S., Burns C. J., Croker B. A., Defining a therapeutic window for kinase inhibitors in leukemia to avoid neutropenia. Oncotarget 8, 57948–57963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quintelier K., Couckuyt A., Emmaneel A., Aerts J., Saeys Y., Van Gassen S., Analyzing high-dimensional cytometry data using FlowSOM. Nat. Protoc. 16, 3775–3801 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Kaya-Okur H. S., Wu S. J., Codomo C. A., Pledger E. S., Bryson T. D., Henikoff J. G., Ahmad K., Henikoff S., CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross-Innes C. S., Stark R., Teschendorff A. E., Holmes K. A., Ali H. R., Dunning M. J., Brown G. D., Gojis O., Ellis I. O., Green A. R., Ali S., Chin S.-F., Palmieri C., Caldas C., Carroll J. S., Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K. W., Demarco B., Ramos S., Heilig R., Goris M., Grayczyk J. P., Assenmacher C.-A., Radaelli E., Joannas L. D., Henao-Mejia J., Tacchini-Cottier F., Brodsky I. E., Broz P., RIPK1 activates distinct gasdermins in macrophages and neutrophils upon pathogen blockade of innate immune signaling. Proc. Natl. Acad. Sci. U.S.A. 118, e2101189118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh C., Li L., Verma A., Reuven A. D., Miao E. A., Bliska J. B., Aachoui Y., Neutrophil inflammasomes sense the subcellular delivery route of translocated bacterial effectors and toxins. Cell Rep. 41, 111688 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y., Zychlinsky A., Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Speir M., Nowell C. J., Chen A. A., O’Donnell J. A., Shamie I., Lakin P. R., D’Cruz A. A., Braun R. O., Babon J. J., Lewis R. S., Bliss-Moreau M., Shlomovitz I., Wang S., Cengia L. H., Stoica A. I., Hakem R., Kelliher M. A., O’Reilly L. A., Patsiouras H., Lawlor K. E., Weller E., Lewis N. E., Roberts A. W., Croker B. A., Ptpn6 inhibits caspase-8- and Ripk3/Mlkl-dependent inflammation. Nat. Immunol. 21, 54–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.U. Schmidt, M. Weigert, C. Broaddus, G. Myers, Cell detection with star-convex polygons in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) (2018), p. 1806.03535v2. [Google Scholar]
- 20.W. Chang, J. Cheng, J. Allaire, Y. Xie, J. McPherson, shiny: Web Application Framework for R. R package version (2016).
- 21.R Core Team, R: A language and environment for statistical computing. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2020). [Google Scholar]
- 22.Wickham H., Francois R., Henry L., K., DPLYR. R package version 1.0.6, (2021). (available at https://CRAN.R-project.org/package=dplyr [Google Scholar]
- 23.Lemon J., Plotrix: A package in the red light district of R. R-News. 6, 8–12 (2006). [Google Scholar]
- 24.Tilvawala R., Nguyen S. H., Maurais A. J., Nemmara V. V., Nagar M., Salinger A. J., Nagpal S., Weerapana E., Thompson P. R., The Rheumatoid arthritis-associated citrullinome. Cell Chem Biol. 25, 691–704.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clancy K. W., Weerapana E., Thompson P. R., Detection and identification of protein citrullination in complex biological systems. Curr. Opin. Chem. Biol. 30, 1–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewallen D. M., Bicker K. L., Subramanian V., Clancy K. W., Slade D. J., Martell J., Dreyton C. J., Sokolove J., Weerapana E., Thompson P. R., Chemical proteomic platform to identify citrullinated proteins. ACS Chem. Biol. 10, 2520–2528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.John S., Sabo P. J., Thurman R. E., Sung M.-H., Biddie S. C., Johnson T. A., Hager G. L., Stamatoyannopoulos J. A., Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43, 264–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1
Table S1 to S5
Legend for movie S1
Movie S1