Abstract
The transcription factor Spo0A of Bacillus subtilis has the unique ability to activate transcription from promoters that require different forms of RNA polymerase holoenzyme. One class of Spo0A-activated promoter, which includes spoIIEp, is recognized by RNA polymerase associated with the primary sigma factor, sigma A (ςA); the second, which includes spoIIAp, is recognized by RNA polymerase associated with an early-sporulation sigma factor, sigma H (ςH). Evidence suggests that Spo0A probably interacts directly with RNA polymerase to activate transcription from these promoters. To identify residues of Spo0A that may be involved in transcriptional activation, we used PCR mutagenesis of the entire spo0A gene and designed a screen using two distinguishable reporter fusions, spoIIE-gus and spoIIA-lacZ. Here we report the identification and characterization of five mutants of Spo0A that are specifically defective in activation of ςA-dependent promoters while maintaining activation of ςH-dependent promoters. These five mutants identify a 14-amino-acid segment of Spo0A, from residue 227 to residue 240, that is required for transcriptional activation of ςA-dependent promoters. This region may define a surface or domain of Spo0A that makes direct contacts with ςA-associated holoenzyme.
The phosphorylation-activated transcription factor Spo0A of Bacillus subtilis is a member of the response regulator family of two-component signal transduction proteins that regulates the initiation of sporulation (1, 27, 34). Under conditions of nutrient limitation, extracellular and intracellular signals are processed through a complex signal transduction pathway that controls the phosphorylation state of Spo0A (14, 16, 17). Phosphorylation of Spo0A increases its affinity for a 7-bp consensus DNA sequence, 5′-TGTCGAA-3′, referred to as the 0A box (5, 35). Spo0A binding then serves either to repress transcription of genes such as abrB, which encodes a regulator protein required for the transition into stationary phase (35), or to activate transcription of various sporulation-specific genes, such as spoIIA (9, 37), spoIIE (41), and spoIIG (32). Although much has been learned about the signal transduction network that controls the phosphorylation state of Spo0A, little is known about the mechanism by which Spo0A stimulates transcription from promoters under its control.
Spo0A is unique in its ability to activate transcription from promoters that require different forms of RNA polymerase (RNAP) holoenzyme for transcription. The spoIIA promoter is Spo0A dependent and is recognized by RNAP associated with sigma H (ςH) (38, 39), an early-sporulation sigma factor (11). The spoIIE and spoIIG promoters are also Spo0A dependent but are recognized by RNAP associated with sigma A (ςA) (5, 31, 41), the primary sigma factor of B. subtilis. Genetic and biochemical evidence indicates that Spo0A binds to these promoters at multiple sites (5, 31, 41) and that, upon phosphorylation, it binds with increased affinity to certain sites to activate transcription (5).
In Escherichia coli, two major classes of transcriptional activators have been identified: the class I and class II activators (10, 15, 18). Class I activators are characterized by DNA binding sites upstream of the −35 region of the promoter (18). Evidence suggests that class I activators make direct contacts to the alpha subunit of RNAP to activate transcription (18). In contrast, class II activators promote transcription by binding at or near the −35 region of the promoter and appear to make direct contact with the sigma subunit of RNAP (18, 21, 25). The promoters, spoIIAp, spoIIEp, and spoIIGp, positively regulated by Spo0A have been characterized in detail, and each contains a Spo0A-binding site in its −35 region (5, 31, 32, 37, 41); in some cases, these binding sites have been demonstrated to be of functional importance in vitro and/or in vivo (5, 12, 31, 32, 37, 39, 41). Thus, there was an expectation that Spo0A might conform to the pattern observed for class II activators and that its mechanism of action might involve direct interaction with the sigma subunit of RNAP holoenzyme. This inference was supported by the identification of mutations in both the ςA and ςH factors of B. subtilis that impair expression of Spo0A-dependent promoters but not of Spo0A-independent promoters (4, 33).
If Spo0A does stimulate transcription through direct interaction with ς, it is interesting to consider whether Spo0A contacts ςA and ςH in the same way. We have addressed this question in the present work by asking whether it is possible to isolate mutants of Spo0A that show sigma-specific defects. We report the characterization of five such mutants. In each case, the mutants show a drastic reduction in the ability to stimulate transcription of ςA-dependent promoters while retaining nearly wild-type ability to stimulate ςH-dependent promoters. Interestingly, the five mutations that cause this phenotype are clustered in a 14-amino acid (aa) segment of the protein. We speculate that this segment may represent a surface or domain of Spo0A that interacts directly with the ς subunit of ςA-associated RNAP holoenzyme. No representatives of the reciprocal class of mutant, in which Spo0A-dependent promoters that utilize ςH-associated holoenzyme were specifically affected, were detected. This may indicate mechanistic differences in the way the two holoenzyme forms are influenced by Spo0A, although other explanations are considered.
MATERIALS AND METHODS
Bacterial strains, culture media, genetic techniques, and in vitro manipulation of DNA.
Bacterial strains used in this work are listed in Table 1. Routine microbiological procedures and enzymatic manipulations of DNA were carried out by standard methods (2, 13). The concentrations of antibiotics used for selection on Luria-Bertani (LB) or Difco sporulation medium (DSM) agar and in culture were 5 μg/ml for chloramphenicol, 3 μg/ml for neomycin, 100 μg/ml for spectinomycin, and 100 μg/ml for ampicillin. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronide) (both from U.S. Biological) in dimethyl sulfoxide were used at a final concentration of 75 μg/ml in indicator agar.
TABLE 1.
Bacterial strains used in this work
| Strain | Genotype or description | Source or reference |
|---|---|---|
| E. coli MM294 | endA1 hsdR17 supE44 thi-1 | 3 |
| B. subtilis | ||
| PY79 | Prototroph | 43 |
| JKH63 | PY79 spoIIE::pJKH9 | This work |
| JKH68 | PY79 spoIIA::pPP81 | This work |
| JKH72 | PY79 spoIIA::pPP81 spoIIE::pJKH9 | This work |
| JKH73 | JKH72 spo0A::pSPC101-0A | This work |
| JKH74 | JKH72 spo0A::pSPC101-G227R | This work |
| JKH75 | JKH72 spo0A::pSPC101 | This work |
| JKH104 | JKH72 spo0A::pSPC101-S233P | This work |
| JKH105 | JKH72 spo0A::pSPC101-F236S | This work |
| JKH106 | JKH72 spo0A::pSPC101-V240A | This work |
| JKH107 | JKH72 spo0A::pSPC101-V240G K265R | This work |
| JKH111 | PY79 spoIIG::IIG-lacZ spo0A::pSPC101-0A | This work |
| JKH112 | PY79 spoIIG::IIG-lacZ spo0A::pSPC101 | This work |
| JKH114 | PY79 spoIIG::IIG-lacZ spo0A::pSPC101-G227R | This work |
| JKH115 | PY79 spoIIG::IIG-lacZ spo0A::pSPC101-F236S | This work |
| JKH116 | PY79 spoIIG::IIG-lacZ spo0A::pSPC101-V240A | This work |
| JKH117 | PY79 spoIIG::IIG-lacZ spo0A::pSPC101-S233P | This work |
| JKH118 | PY79 spoIIG::IIG-lacZ spo0A::pSPC101-V240G K265R | This work |
Construction of a B. subtilis screening strain containing two promoter fusions.
A spoIIA-lacZ promoter fusion strain was generated by transformation of B. subtilis PY79 to chloramphenicol resistance by integration of the vector pPP81 (30) into the B. subtilis chromosome. This strain was then transformed to neomycin resistance with chromosomal DNA isolated from a spoIIE-gus promoter fusion strain. The spoIIE-gus fusion strain was constructed by integration of the clone pJKH9 into the B. subtilis chromosome. This clone was generated by first cloning a 275-bp HindIII-BamHI fragment containing the spoIIE promoter from pGV49 (12) into the pBluescript polylinker (Stratagene), then subsequently cloning a 290-bp SalI-BamHI fragment containing the promoter into the vector pMLK117 (19). The pMLK117 vector contained a promoterless copy of the gus gene, a neomycin resistance marker selectable in a single copy in B. subtilis, unique sites for the cloning of promoters upstream of the gus gene, and an origin of replication and a selectable marker functional in E. coli. The resulting screening strain, JKH72, carried a spoIIA-lacZ transcriptional fusion and a spoIIE-gus transcriptional fusion and was chloramphenicol resistant, neomycin resistant, and sporulation proficient (Spo+) due to restoration of intact copies of the two fused genes, spoIIA and spoIIE.
Construction of the pSPC101 integrational vector.
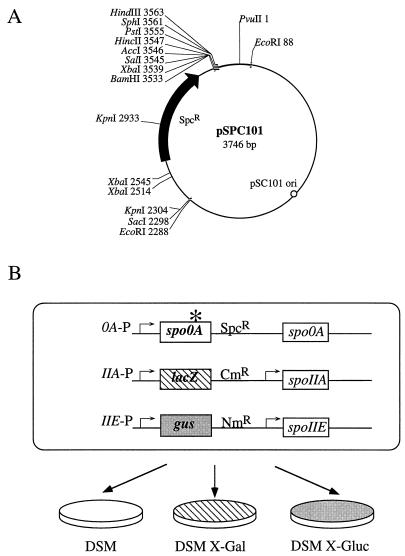
A 1.2-kb blunt-ended fragment carrying the spectinomycin resistance gene from Enterococcus faecalis (22) was cloned by ligation into the SmaI site of pUC19 (40). A 2.2-kb EcoRI fragment from pJRS233 (29) containing the pSC101 origin of replication was then cloned into the EcoRI site of the pUC plasmid carrying the spectinomycin resistance marker. The resulting 6.1-kb plasmid was digested with PvuII, resulting in two fragments of 3.7 and 2.4 kb. The 3.7-kb fragment was then gel purified and recircularized to form the pSPC101 integrational vector. The key features of the vector are the pSC101 origin of replication functional in E. coli, the spectinomycin resistance marker from E. faecalis selectable in both E. coli and B. subtilis, and a portion of the pUC19 polylinker containing unique cloning sites (Fig. 1A). The use of this vector decreased the possibility of homologous recombination into already existing pUC vector sequences in the chromosome of the screening strain.
FIG. 1.
Key components of the mutant screen. (A) Restriction map of the integrational vector pSPC101 used to introduce mutagenized spo0A into the screening strain. SpcR, spectinomycin resistance; pSC101 ori, origin of replication functional in E. coli. (B) Schematic representation of the screen. The screening strain JKH72 contains a spoIIA-lacZ fusion as a reporter of Spo0A-dependent, ςH-dependent transcription activity and a spoIIE-gus fusion as a reporter of Spo0A-dependent, ςA-dependent transcription activity. Transcription activation of these two reporter fusions is screened on X-Gal and X-Gluc indicator agars. The sporulation phenotype is screened on DSM agar. See Materials and Methods for a detailed description of the screen.
Construction of isogenic spo0A wild-type (JKH73) and null (JKH75) control strains.
The spo0A gene was amplified by PCR from the chromosome of a wild-type strain of B. subtilis. The primers used to amplify the gene were 0AupBamHI (5′-GCAGTAGGATCCATGTAGCAAGGGTGAATCC-3′) and 0A4HindIII (5′-GCAGGAAGCTTCGCCTCCTATTTATCAGCGC-3′), which incorporate BamHI and HindIII restriction sites, respectively, at the ends of the PCR product to facilitate cloning into the pSPC101 integrational vector. This construct was used to transform the screening strain JKH72 by selecting for spectinomycin resistance. Integration at the spo0A locus was confirmed by PCR, and the wild-type sequence was verified by sequencing the allele on both strands with the fmol sequencing kit (Promega). The JKH73 strain was blue both on X-Gal agar after 20 h of incubation at 37°C and on X-Gluc agar after 48 h of incubation at 37°C when screened for expression of the two promoter fusions carried in the strain. This strain was Spo+ when screened on DSM plates after 48 h of incubation at 37°C. Sporulation phenotypes were assessed directly on DSM agar by the formation of brown pigment by Spo+ strains (26). A null allele of spo0A was generated by cloning an internal 420-bp SauIIIA fragment of the spo0A coding region into the BamHI site of pSPC101. The integration of this clone into the chromosomal spo0A locus disrupted the coding region of the spo0A gene, resulting in a null strain, JKH75. Integration at the spo0A locus was confirmed by PCR. JKH75 was white on both X-Gal agar and X-Gluc agar and had a Spo− phenotype when screened on DSM agar.
Mutagenesis of spo0A.
Random mutations were introduced into the spo0A gene by PCR amplification of the coding sequence under conditions previously described (23). The reaction differed from this protocol by the omission of dimethyl sulfoxide and β-mercaptoethanol and the use of either 0.5 mM MnCl2 alone or 0.25 mM MnCl2 and a 5:1 ratio of dGTP to dATP. The primers 0AupBamHI and 0A4HindIII used for the amplification are described above. BamHI and HindIII restriction sites were used to clone the mutagenized products into the pSPC101 vector.
The screen for Spo0A mutants defective in transcription activation.
The spoIIA-lacZ fusion was used as a reporter of Spo0A-dependent, ςH-dependent gene expression, and the spoIIE-gus fusion was used as a reporter of Spo0A-dependent, ςA-dependent gene expression. PCR-mutagenized spo0A genes were cloned into the pSPC101 integrational vector, and this pool of mutagenized spo0A clones (pSPC101-0A*) was then used to transform E. coli MM294 to spectinomycin resistance. Transformants were pooled, and plasmid DNA was isolated. The plasmid DNA was restricted at a unique SacI site and ligated together to form multimers in order to facilitate plasmid integration into the B. subtilis chromosome. JKH72 was transformed by pSPC101-0A* DNA, and integrants were selected by plating to LB or DSM agar containing spectinomycin (Fig. 1B). Expression of the spoIIA-lacZ fusion in strains transformed by pSPC101-0A* was determined by picking transformants to DSM agar containing chloramphenicol, neomycin, spectinomycin, and X-Gal. Expression of the spoIIE-gus fusion was determined by picking the same transformants to DSM agar containing the same antibiotics and X-Gluc. The ability to sporulate was determined by patching to DSM plates containing the appropriate antibiotics. Expression of the promoter fusions was determined by the presence or absence of blue color on the indicator plates after 20 h of incubation at 37°C for X-Gal and 48 h of incubation at 37°C for X-Gluc. Blue color indicated the expression of the fusion product.
Sequencing of mutant spo0A alleles.
The spo0A gene of spo0A225 was amplified from the chromosome of the mutant strain by PCR using primers which amplify from a site in the promoter region of spo0A (0A71R, 5′-TCTTCACTTCTCAGAATACATACGG-3′) and from a site downstream of the gene (0A1190L, 5′-ACAAATGTCCCCAAAACAAAACGCC-3′). The spo0A alleles of the other mutant strains were amplified from chromosomal DNA by PCR using the same upstream primer (0A71R) and a primer which anneals to a site in the integrated plasmid vector (pUC reverse primer 1224, 5′-GCCAGGGTTTTCCCAGTCACGAC-3′). The PCR products were purified from the reaction mixture by using the Wizard PCR purification kit and were directly sequenced by using the fmol sequencing kit (both from Promega). Sequencing reactions were carried out on both strands of the PCR products.
β-Galactosidase and β-glucuronidase assays.
β-Galactosidase assays were performed by the fluorometric method of Youngman (42). Control and mutant spo0A strains containing both a spoIIA-lacZ transcriptional fusion and a spoIIE-gus transcriptional fusion were streaked to LB agar containing chloramphenicol, neomycin, and spectinomycin and were incubated overnight at 30°C to produce a very light lawn of growth. Bacteria were washed from the plates with LB medium and were used to directly inoculate 5 ml of LB containing chloramphenicol, neomycin, and spectinomycin to barely detectable turbidity. The 5-ml cultures were allowed to resume growth to mid-log phase and were used to inoculate 30 ml of DSM to a reading of ≤5 Klett units. Bacteria were cultured for assay at 37°C with shaking. At various intervals during growth and sporulation, 0.5-ml samples were collected and frozen in liquid nitrogen. Samples were stored at −70°C until assayed.
β-Glucuronidase assays were performed in an identical manner, except that 0.4 mg of 4-methylumbelliferyl-β-d-glucuronide trihydrate (MUG) substrate (U.S. Biological)/ml specific to β-glucuronidase was used instead of 0.4 mg of 4-methylumbelliferyl-β-d-galactoside (U.S. Biological)/ml. One unit of activity is defined as one picomole of MUG hydrolyzed per milliliter of culture sample per minute, normalized for culture cell density (turbidity). Each sample was assayed for both β-glucuronidase and β-galactosidase activities.
Wild-type and mutant spo0A strains containing a spoIIG-lacZ transcriptional fusion were grown and samples for assay were collected in the manner described above except that the medium used for growth contained chloramphenicol and spectinomycin. β-Galactosidase assays were performed as described above.
Sporulation frequency assay.
B. subtilis strains were grown for 48 h at 37°C on DSM agar containing the appropriate antibiotics. A single colony was resuspended in 0.5 ml of DSM. The number of viable cells was determined by dilution and plating onto LB agar containing the appropriate antibiotics. The number of heat-resistant spores was determined by heating the resuspended cells at 80°C for 20 min and plating appropriate dilutions on selective plates. The sporulation frequency was determined as the percentage of the number of heat-resistant spores compared to the total number of viable cells before heat treatment. The sporulation frequency for each mutant was calculated as the average from three independent assays.
Immunoblot detection of Spo0A proteins.
Polyclonal anti-Spo0A antibodies were raised in rabbits by using heparin agarose-purified Spo0A. Samples (10 to 25 ml) of B. subtilis cultures grown in DSM at 37°C with shaking were collected, and the cells were harvested at various time points. Cell pellets were washed in a buffer previously described (5), quick-frozen in an ethanol-dry ice bath, and stored at −70°C until assayed. Cell pellets were resuspended in 1 ml of buffer (5), and crude extracts were prepared. The cells were lysed at 4°C by two passages through a French pressure cell at 19,000 lb/in2. Total protein was quantitated by using the Bio-Rad Protein microassay procedure as described by the manufacturer. Protein samples (10 and 0.625 μg) were separated by electrophoresis through a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel. Proteins were electroblotted to a PVDF-Plus membrane (Micron Separations, Inc.).
The membrane was blocked by incubation for 1 h at room temperature in Tris-buffered saline–Tween (100 mM Tris · Cl [pH 8.0], 0.9% NaCl, 0.05% Tween) (TBST) containing 3% (wt/vol) bovine serum albumin (BSA). The membrane was subsequently washed three times for 15 min each time in TBST alone and then incubated with 15 ml of TBST containing 1% BSA and a 1:15,000 dilution of rabbit anti-Spo0A antiserum. After incubation for 45 min at room temperature with gentle agitation, the membrane was again washed three times for 15 min each time with TBST alone. The membrane was then incubated with 15 ml of TBST containing 1% BSA and a 1:3,000 dilution of the goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (Promega) for 45 min at room temperature with gentle agitation. After incubation, the membrane was washed three times for 15 min each time with TBST alone and then treated with the Renaissance Chemiluminescent Reagent (DuPont, NEN) according to the manufacturer’s instructions. Treated membranes were immediately exposed to X-ray film for 10 to 30 s.
RESULTS
Mutant spo0A225.
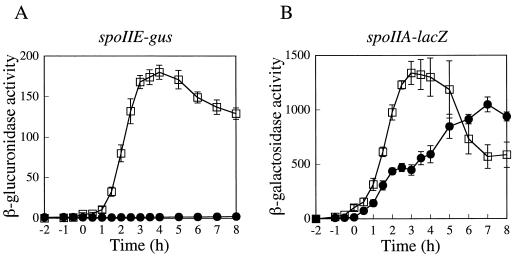
While screening for mutants that maintained expression of the spoIIA operon (which encodes ςF) but failed to activate ςF-controlled gene expression in the forespore, Levin and Losick (24) identified a mutation that mapped within or near the spo0A locus. Strains containing this mutation were strongly impaired in the expression of spoIIE, raising the possibility that the mutant phenotype was the result of an alteration in Spo0A that produced a promoter-specific defect in transcription activation. After confirming by genetic methods that this mutation was within the spo0A coding sequence (data not shown), we sequenced the entire spo0A gene from the mutant strain. The sequence revealed a GC-to-AT base pair substitution that resulted in a glycine-to-arginine change at residue 227 of the mutant Spo0A protein. We refer to the mutant gene as spo0A225 and to the mutant protein as G227R. The strongly differential effect of spo0A225 on transcription from the spoIIE and spoIIA promoters is apparent in the behavior of transcriptional fusions to these two promoters (Fig. 2).
FIG. 2.
Effects of the spo0A225 mutation on transcription from two Spo0A-dependent promoters. (A) spoIIE-gus expression; (B) spoIIA-lacZ expression. Strains were cultured and assayed as described in Materials and Methods. T0 marks the end of exponential growth. Samples were collected at 1/2-h time points from T−2 until T4 and then hourly until T8 unless otherwise indicated. Open squares, wild type (JKH73); filled circles, spo0A225 strain. Data from at least three independent trials were averaged. Error bars, standard error of the mean (SEM). One unit of activity is defined as one picomole of MUG hydrolyzed per milliliter of culture sample per minute, normalized for culture cell density (turbidity).
Identification of additional mutations in spo0A that produce promoter-specific defects.
Available evidence supports a model in which Spo0A activates transcription through direct contact with RNAP (4, 8, 33). The spo0A225 mutation might therefore act by affecting such contacts, possibly by altering the surface or domain of Spo0A that contacts ςA. If so, we reasoned that random mutagenesis within the spo0A coding sequence with a screen for a phenotype similar to that of spo0A225 might identify additional mutations that would help to further define this surface or domain. In addition, we speculated that it might be possible to identify mutants with the reciprocal phenotype, a defect in activation of ςH-dependent promoters but not ςA-dependent promoters under Spo0A control.
To identify activation-defective spo0A mutants, we constructed a B. subtilis screening strain, JKH72, containing transcriptional fusions with two distinguishable reporters: a spoIIA-lacZ promoter fusion, used as a reporter of Spo0A-dependent, ςH-dependent transcription, and a spoIIE-gus promoter fusion, used as a reporter of Spo0A-dependent, ςA-dependent transcription. Mutations were generated by PCR amplification of the entire spo0A gene under conditions favoring misincorporation errors, followed by direct cloning of the amplification products into the pSPC101 integrational vector (Fig. 1A). After passage through E. coli MM294, plasmid DNA was pooled and used to transform JKH72. Chromosomal integrations at the spo0A locus were selected on medium containing spectinomycin, then transferred to DSM, DSM X-Gal, and DSM X-Gluc plates containing the appropriate antibiotics for assessment of sporulation phenotypes, spoIIA expression, and spoIIE expression, respectively (Fig. 1B). Because we expected that a defect in transcriptional activation of one of the stage II genes would block sporulation, the primary screen was for a Spo− phenotype. We screened 3,069 transformants and found 72 strains which were phenotypically Spo−, white on X-Gluc, and blue on X-Gal, which we interpreted as indicating a defect in transcriptional activation at the spoIIE promoter. None of the Spo− transformants screened had the reciprocal expression pattern on the two indicator plates. Nineteen strains among the 72 mutant candidates were tested for linkage between the Spo− phenotype and spectinomycin resistance. Linkage would indicate that the phenotype observed was linked to the allele of spo0A present in the mutant strain. Linkage was tested by transformation of the wild-type B. subtilis strain PY79 to Spcr by using chromosomal DNA from candidate Spo− strains, then assessing the number of Spo− transformants produced. Sixteen strains exhibited cotransformation frequencies greater than 90%. Five of these strains exhibiting the strongest phenotypes in our screen were chosen for further study.
Sequencing of mutant spo0A alleles.
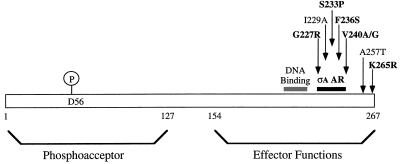
All five independently isolated mutant alleles of spo0A were sequenced through the entire structural gene, and the amino acid substitutions resulting from each mutation were determined. Remarkably, all five strains identified in the screen, like spo0A225, carried mutations within a 14-aa stretch in the C-terminal effector domain of the 267-aa Spo0A protein (Fig. 3). The S233P mutant was isolated as a single-base-pair substitution. As expected, however, considering the intensity of mutagenesis, most of the mutant alleles also contained substitutions at other locations within the spo0A gene. The S233P* mutant (the asterisk denotes a second substitution in Spo0A other than the substitution indicated) was isolated from a mutagenesis independent of that for the S233P single mutant. Each of the F236S* and V240A* mutants also contained a second substitution outside the 14-aa region. To determine whether the substitutions within the 14-aa region were responsible for the mutant phenotype, the F236S and V240A mutations were subcloned into a wild-type copy of spo0A in order to isolate the single substitutions and were reintroduced into JKH72. The subcloned mutants were phenotypically identical to the strains containing the double substitutions in Spo0A (data not shown). One mutant, V240G K265R, contained a second substitution near the 14-aa segment; no attempt was made to resolve whether either substitution alone might cause the mutant phenotype. The sporulation defects caused by the spo0A mutations are quantified in Table 2.
FIG. 3.
Map of the activating region of Spo0A proposed for ςA-dependent transcription. The Spo0A protein consists of two domains. The phosphoacceptor domain is formed by the N-terminal 127 aa, and the effector domain is formed by the C-terminal 113 residues; they are joined by a linker region of 27 aa, as defined by Brown et al. (7). Mutations in Spo0A protein which decrease activation of ςA-dependent promoters under Spo0A control described in this work and elsewhere (8) are mapped. Residues examined in this study are in boldface. The ςA activating region (ςA AR) (solid bar) includes residues from aa 227 to aa 240. The K265 residue may also have some effect on transcription activation of ςA-dependent promoters. Also indicated is position A257, which, when mutated, generates the spo0A9V mutant, which does not activate transcription of ςH-dependent promoters (28). Shaded bar, putative DNA binding domain (aa 194 to 224) proposed for Spo0A based on sequence conservation of this region in spo0A homologs from diverse Bacillus and Clostridium species (7).
TABLE 2.
Sporulation frequencies
| Strain | Allele | % Sporulationa |
|---|---|---|
| JKH73 | wtb | 88.0 |
| JKH75 | Nullc | 0.006 |
| JKH74 | G227R | 0.024 |
| JKH104 | S233P | 0.141 |
| JKH105 | F236S | 0.526 |
| JKH106 | V240A | 0.0008 |
| JKH107 | V240G K265R | 0.0002 |
Sporulation frequencies were calculated as described in Materials and Methods.
wt, wild type.
Inactivation of spo0A generated by Campbell integration (spo0A::pSPC101).
spo0A mutants that are differentially defective in activation of ςA-dependent promoters.
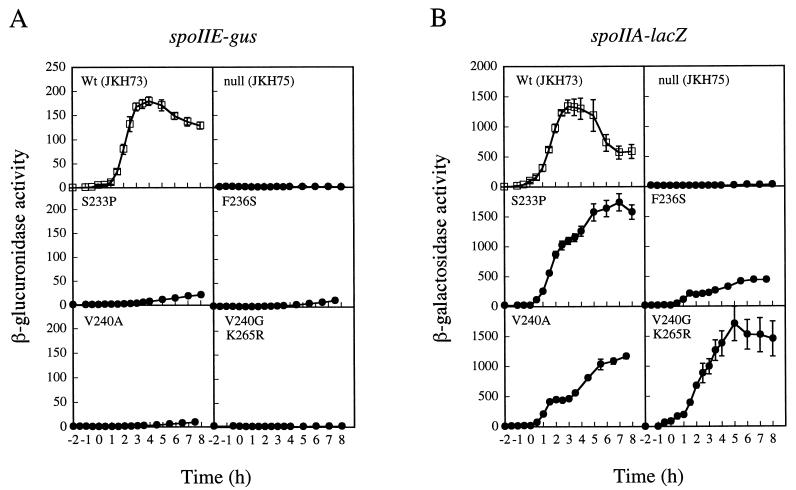
Each of the mutants was tested for its ability to activate transcription from the spoIIA and spoIIE promoters. The two reporter fusions in these strains allowed us to quantitate the level of transcription from both promoters by monitoring β-galactosidase and β-glucuronidase accumulation. All five mutant strains were drastically reduced in their abilities to activate transcription of the ςA-dependent promoter spoIIE (Fig. 2A and 4A) but only slightly or partially impaired at the ςH-dependent promoter spoIIA (Fig. 2B and 4B). At 2 h after the start of sporulation, when the stage II gene products are expressed, spoIIE transcription was limited to 2.5% or less of the wild-type levels in all five mutants. These levels were similar to the activity of the promoter in a Spo0A null strain, JKH75 (<1% of wild type). All five of the mutants tested, however, showed substantial ability to activate transcription from the spoIIA or ςH-dependent promoter. The S233P and V240G K265R mutants activated transcription from the spoIIA promoter only slightly less efficiently than the wild-type Spo0A strain, JKH73. The G227R and V240A mutants each showed approximately a twofold decrease in ability to activate transcription from this promoter. For the mutant with the least ability to activate transcription from the spoIIA promoter, F236S, transcription was approximately 4-fold less than that for the wild-type strain, JKH73, but 10-fold greater than that for the Spo0A null strain, JKH75. Because transcriptional activation at spoIIA is dependent on Spo0A (30, 37, 38), these results suggested that the Spo0A protein in the mutants retained the ability to bind to 0A box sites and activate transcription from the spoIIA promoter.
FIG. 4.
Effects of spo0A mutants on the expression of spoIIE-gus (A) and spoIIA-lacZ (B) transcriptional fusions. Samples were collected and assayed as described in the legend to Fig. 2. In each panel, the filled circles represent the mutant indicated. Open squares represent the wild type (JKH73). Data were averaged from at least three independent trials. Error bars, SEM. One unit of activity is defined as described in the legend to Fig. 2.
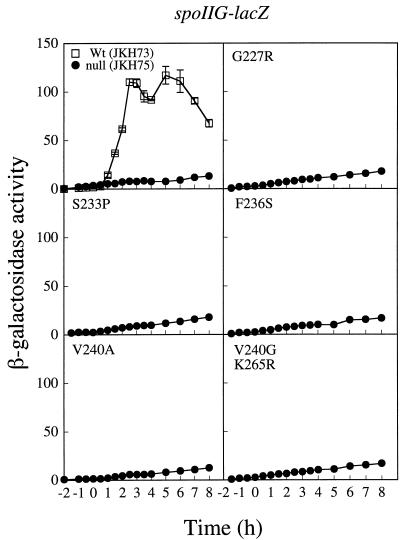
To establish whether the effect on spoIIE transcription was due to a general defect in utilization of ςA-dependent promoters under Spo0A control, we transformed each of the mutant spo0A alleles into a spoIIG-lacZ fusion strain. The spoIIG promoter is a second ςA-dependent promoter that is also dependent on Spo0A for activation (20). Transcriptional activation of this promoter was severely decreased (Fig. 5). At 2 h after the onset of sporulation, levels of activity at the spoIIG promoter in our mutant stains were indistinguishable from that of the null strain, JKH75 (approximately 11% of the wild-type level).
FIG. 5.
Verification of a defect in transcriptional activation by spo0A mutants of ςA-dependent promoters. The effect of the spo0A mutants on the expression of a spoIIG-lacZ transcriptional fusion is shown. Samples were collected and assayed as described in the legend to Fig. 2. In each panel, the filled circles represent the mutant indicated. Open squares represent the wild type (JKH73). Data were averaged from three independent trials. Error bars, SEM. One unit of activity is defined as described in the legend to Fig. 2.
Spo0A protein levels in wild-type and mutant cell extracts.
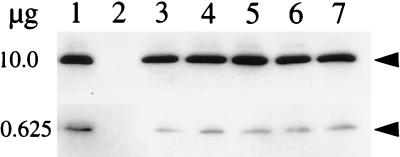
Although some of the spo0A mutants retained nearly wild-type levels of spoIIA expression, some reduction was detectable in all mutant strains, and the F236S mutant retained only 25% of the wild-type level of expression. To determine whether any portion of the reduction in spoIIA transcription in the mutants might be attributable to a decrease in Spo0A protein levels, we examined Western blots prepared with mutant and wild-type extracts, probed with polyclonal antibody to Spo0A. The results indicated slight reductions in the Spo0A protein levels of the mutants, ranging from two- to fourfold at most (Fig. 6).
FIG. 6.
Immunoblot analyses of wild-type and mutant Spo0A proteins. Culture samples were collected and harvested at 1 h after the end of exponential growth (T1). Samples containing 10 and 0.625 μg of total cellular protein were subjected to electrophoresis through an SDS–12% polyacrylamide gel. Samples were electroblotted to a PVDF-Plus membrane and the Spo0A protein was probed with anti-Spo0A antibody as described in Materials and Methods. Lane 1 in each panel contains wild-type Spo0A (JKH73), and lanes 2 through 7 contain the following Spo0A mutants: null (JKH75) (lane 2), G227R (lane 3), S233P (lane 4), F236S (lane 5), V240A (lane 6), and V240G K265R (lane 7). Arrowheads indicate Spo0A.
DISCUSSION
The results reported here and in complementary work in the accompanying paper of Buckner et al. (8) strongly implicate a 14-aa segment of Spo0A, extending from G227 to V240 (the ςA activating region in Fig. 3), as a region of the protein critical for transcription at promoters positively regulated by Spo0A. Motivated initially by the existence of a single mutation at G227 that appeared to cause a sigma-specific defect (24), we have subjected the entire spo0A coding sequence to intensive, random mutagenesis by PCR and have characterized four additional mutants exhibiting a similar phenotype. In all cases, the phenotype was caused by mutational changes within the 14-aa segment. The work of Buckner et al. (8) converged on precisely the same small segment of the spo0A coding sequence from a completely independent direction. In that study, the investigators started with a mutation in sigA (ςA H359R) that specifically affected transcription from Spo0A-dependent promoters, which therefore caused a Spo− phenotype, and sought a phenotypic suppressor after random localized mutagenesis (8). The suppressor they recovered (Spo0A S231F) fell within the 14-aa segment identified in our study. This segment is immediately adjacent to the region of Spo0A (residues 194 to 224) proposed to comprise the helix-turn-helix motif responsible for binding to Spo0A recognition sites in DNA (7). Taken together, our results and those reported by Buckner et al. (8) strongly support a model in which Spo0A stimulates transcription through direct contact with the ς subunit of RNAP holoenzyme, involving a surface or domain of Spo0A that includes residues 227 to 240.
Substitutions in four different positions spanning residues 227 to 240 were found to produce a sigma-specific effect. The S233 position was identified twice in our screen. The change in each case was from serine to proline. In the work of Buckner et al. (8), a mutant in which serine was replaced by an alanine rather than proline at this position was found not to exhibit any defect. This suggests that the S233 residue is not directly involved in transcriptional activation; most likely, the S233P substitution causes a local conformational change in the peptide backbone that repositions other residues that make critical contacts with ς. However, the effect is unlikely to be profoundly disruptive, as the level of spoIIA expression for this mutant was nearly the same as the wild-type level. The most likely candidate for direct involvement in transcription activation and perhaps an interaction with the transcription machinery is the valine at position 240. Conservative changes at this residue cause a drastic decrease in the expression of the ςA-dependent promoters while still maintaining expression at the ςH-dependent promoters. One point that we did not address directly in this study was the contribution of the K265 residue to the defect seen in the V240G K265R mutant. However, the isolation of a second change at position 240, a valine-to-alanine single substitution, implies that the residue at position 240 is important.
Despite our success in identifying a region that is implicated in activation of one class of Spo0A-dependent promoters, we cannot rule out the possibility that other residues may be involved in transcription activation at the ςA-dependent promoters. The screen, although remarkable in identifying a narrow region of the Spo0A protein as important for activation of ςA-dependent promoters, was not a saturating screen. Moreover, C-terminal mutations were more likely to be isolated by integration of a vector carrying promoterless copies of mutant spo0A genes into the B. subtilis chromosome. We are confident, however, that our screen did include representatives of mutations in the N-terminal end of the protein, because some of the mutants we isolated also contained additional silent mutations in the N-terminal ends of their coding sequences (data not shown). Subtle changes in Spo0A function were most likely overlooked by our screen, since we focused on those strains that exhibited the strongest phenotypes in our primary screen. In addition, our screen failed to identify mutants of Spo0A specifically defective at ςH-dependent promoters. This result might be due to the complex regulatory relationship between Spo0A and ςH because of the roles they play in each other’s expression (36). Therefore, it is possible that mutations that affect Spo0A function at ςH-dependent promoters affect levels of Spo0A itself and perhaps result in a null phenotype. However, the existence of the spo0A9V mutant indicates that it is possible to obtain Spo0A mutants defective in stimulating transcription at ςH-dependent promoters that still maintain the ability to bind DNA (28).
The existence of mutant forms of Spo0A that are specifically defective in their ability to activate transcription from promoters utilized by ςA-associated holoenzyme is consistent with two mechanistic models. One possibility is that Spo0A binds differently to promoters recognized by the two forms of RNAP holoenzyme. Although this possibility cannot be excluded, and 0A boxes most critical for transcription activation actually overlap polymerase-binding sites, affinity for 0A boxes depends upon a consensus sequence that is independent of promoter context. We therefore favor the alternative explanation that mutations producing a sigma-specific phenotype do so by disrupting a sigma-specific contact between Spo0A and RNAP. The results of Buckner et al. (8) are also most easily explained by such a model. Nevertheless, we note the suggestion by Bird et al. (6) that the target for phosphorylated Spo0A binding is not DNA per se but rather DNA plus RNAP, and we acknowledge the possibility that holoenzyme-specific defects might reflect a more complex combination of DNA and protein interactions.
In this study, we have identified an activating region of Spo0A specific for ςA-dependent transcription which may interact directly with the sigma subunit of RNAP to activate transcription. The isolation of a region specific for interaction with ςA and promotion of ςA-dependent transcription suggests that a separate surface of Spo0A may exist for interaction with the ςH factor at ςH-dependent promoters. The ability of transcriptional activators and components of RNAP to interact in many different ways and on many different surfaces could greatly increase the available combinations of potential regulatory interactions between these proteins. The elucidation of the mechanism of transcriptional activation by Spo0A still waits on the identification of the DNA binding domain and residues important for activation at ςH-dependent promoters.
ACKNOWLEDGMENTS
We are grateful to Petra Levin and Richard Losick for their gift of the B. subtilis spo0A225 strain. We thank Charles Moran and Cindy Buckner for sharing data prior to publication. We also thank Sidney Kushner, Jaideep Behari, Paul Fawcett, Andrea Milenbachs, and Tad Seyler for critical reading of the manuscript and Dave Brown for technical assistance.
This work was supported by Public Health Services grant GM35495 from the National Institutes of Health.
REFERENCES
- 1.Albright L M, Huala E, Ausubel F M. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu Rev Genet. 1989;23:311–336. doi: 10.1146/annurev.ge.23.120189.001523. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K A. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1994. [Google Scholar]
- 3.Backman K, Ptashne M, Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci USA. 1976;73:4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldus J M, Buckner C M, Moran C P., Jr Evidence that the transcriptional activator Spo0A interacts with two sigma factors in Bacillus subtilis. Mol Microbiol. 1995;17:281–290. doi: 10.1111/j.1365-2958.1995.mmi_17020281.x. [DOI] [PubMed] [Google Scholar]
- 5.Baldus J M, Green B D, Youngman P, Moran C P., Jr Phosphorylation of Bacillus subtilis transcription factor Spo0A stimulates transcription from the spoIIG promoter by enhancing binding to weak 0A boxes. J Bacteriol. 1994;176:296–306. doi: 10.1128/jb.176.2.296-306.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird T H, Grimsley J K, Hoch J A, Spiegelman G B. The Bacillus subtilis response regulator Spo0A stimulates transcription of the spoIIG operon through modification of RNA polymerase promoter complexes. J Mol Biol. 1996;256:436–448. doi: 10.1006/jmbi.1996.0099. [DOI] [PubMed] [Google Scholar]
- 7.Brown D P, Ganova-Raeva L, Green B D, Wilkinson S R, Young M, Youngman P. Characterization of spo0A homologues in diverse Bacillus and Clostridium species identifies a probable DNA-binding domain. Mol Microbiol. 1994;14:411–426. doi: 10.1111/j.1365-2958.1994.tb02176.x. [DOI] [PubMed] [Google Scholar]
- 8.Buckner C M, Schyns G, Moran C P., Jr A region in the Bacillus subtilis transcription factor Spo0A that is important for spoIIG promoter activation. J Bacteriol. 1998;180:3578–3583. doi: 10.1128/jb.180.14.3578-3583.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burbulys D, Trach K A, Hoch J A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 10.de Crombrugghe B, Busby S, Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984;224:831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- 11.Dubnau E, Weir J, Nair G, Carter III L, Moran C, Jr, Smith I. Bacillus sporulation gene spo0H codes for ς30 (ςH) J Bacteriol. 1988;170:1054–1062. doi: 10.1128/jb.170.3.1054-1062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzmán P, Westpheling J, Youngman P. Characterization of the promoter region of the Bacillus subtilis spoIIE operon. J Bacteriol. 1988;170:1598–1609. doi: 10.1128/jb.170.4.1598-1609.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley and Sons Ltd.; 1990. [Google Scholar]
- 14.Hoch J A. The phosphorelay signal transduction pathway in the initiation of Bacillus subtilis sporulation. J Cell Biochem. 1993;51:55–61. doi: 10.1002/jcb.240510111. [DOI] [PubMed] [Google Scholar]
- 15.Igarashi K, Hanamura A, Makino K, Aiba H, Aiba H, Mizuno T, Nakata A, Ishihama A. Functional map of the alpha subunit of Escherichia coli RNA polymerase: two modes of transcription activation by positive factors. Proc Natl Acad Sci USA. 1991;88:8958–8962. doi: 10.1073/pnas.88.20.8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ireton K, Grossman A D. Coupling between gene-expression and DNA-synthesis early during development in Bacillus subtilis. Proc Natl Acad Sci USA. 1992;89:8808–8812. doi: 10.1073/pnas.89.18.8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ireton K, Rudner D Z, Siranosian K J, Grossman A D. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 18.Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karow M L, Piggot P J. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene. 1995;163:69–74. doi: 10.1016/0378-1119(95)00402-r. [DOI] [PubMed] [Google Scholar]
- 20.Kenney T J, Moran C P., Jr Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J Bacteriol. 1987;169:3329–3339. doi: 10.1128/jb.169.7.3329-3339.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar A, Grimes B, Fujita N, Makino K, Malloch R A, Hayward R S, Ishihama A. Role of the sigma 70 subunit of Escherichia coli RNA polymerase in transcription activation. J Mol Biol. 1994;235:405–413. doi: 10.1006/jmbi.1994.1001. [DOI] [PubMed] [Google Scholar]
- 22.LeBlanc D J, Lee L N, Inamine J M. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1804–1810. doi: 10.1128/aac.35.9.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung D W, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 24.Levin, P., and R. Losick. Personal communication.
- 25.Li M, Moyle H, Susskind M M. Target of the transcriptional activation function of phage lambda cI protein. Science. 1994;263:75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson W L, Setlow P. Sporulation, germination and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1990. pp. 391–450. [Google Scholar]
- 27.Olmedo G, Ninfa E G, Stock J, Youngman P. Novel mutations that alter the regulation of sporulation in Bacillus subtilis—evidence that phosphorylation of regulatory protein Spo0A controls the initiation of sporulation. J Mol Biol. 1990;215:359–372. doi: 10.1016/s0022-2836(05)80357-2. [DOI] [PubMed] [Google Scholar]
- 28.Perego M, Wu J J, Spiegelman G B, Hoch J A. Mutational dissociation of the positive and negative regulatory properties of the Spo0A sporulation transcription factor of Bacillus subtilis. Gene. 1991;100:207–212. doi: 10.1016/0378-1119(91)90368-l. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Casal J, Price J A, Maguin E, Scott J R. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol Microbiol. 1993;8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 30.Piggot P J, Curtis C A. Analysis of the regulation of gene expression during Bacillus subtilis sporulation by manipulation of the copy number of spo-lacZ fusions. J Bacteriol. 1987;169:1260–1266. doi: 10.1128/jb.169.3.1260-1266.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satola S, Kirchman P A, Moran C P., Jr Spo0A binds to a promoter used by ςA RNA polymerase during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1991;88:4533–4537. doi: 10.1073/pnas.88.10.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satola S W, Baldus J M, Moran C P., Jr Binding of Spo0A stimulates spoIIG promoter activity in Bacillus subtilis. J Bacteriol. 1992;174:1448–1453. doi: 10.1128/jb.174.5.1448-1453.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schyns G, Buckner C M, Moran C P., Jr Activation of the Bacillus subtilis spoIIG promoter requires interaction of Spo0A and the sigma subunit of RNA polymerase. J Bacteriol. 1997;179:5605–5608. doi: 10.1128/jb.179.17.5605-5608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strauch M, Webb V, Spiegelman G, Hoch J A. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci USA. 1990;87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strauch M A, Trach K A, Day J, Hoch J A. Spo0A activates and represses its own synthesis by binding at its dual promoters. Biochimie. 1992;74:619–626. doi: 10.1016/0300-9084(92)90133-y. [DOI] [PubMed] [Google Scholar]
- 37.Trach K, Burbulys D, Strauch M, Wu J J, Dhillon N, Jonas R, Hanstein C, Kallio P, Perego M, Bird T, Spiegelman G, Fogher C, Hoch J A. Control of the initiation of sporulation in Bacillus subtilis by a phosphorelay. Res Microbiol. 1991;142:815–823. doi: 10.1016/0923-2508(91)90060-n. [DOI] [PubMed] [Google Scholar]
- 38.Wu J J, Howard M G, Piggot P J. Regulation of transcription of the Bacillus subtilis spoIIA locus. J Bacteriol. 1989;171:692–698. doi: 10.1128/jb.171.2.692-698.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J J, Piggot P J, Tatti K M, Moran C P., Jr Transcription of the Bacillus subtilis spoIIA locus. Gene. 1991;101:113–116. doi: 10.1016/0378-1119(91)90231-y. [DOI] [PubMed] [Google Scholar]
- 40.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 41.York K, Kenney T J, Satola S, Moran C P, Jr, Poth H, Youngman P. Spo0A controls the ςA-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J Bacteriol. 1992;174:2648–2658. doi: 10.1128/jb.174.8.2648-2658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youngman P. Plasmid vectors for recovering and exploiting Tn917 transpositions in Bacillus and other gram-positive bacteria. In: Hardy K, editor. Plasmids: a practical approach. Oxford, United Kingdom: IRL Press; 1987. pp. 79–103. [Google Scholar]
- 43.Youngman P, Perkins J B, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]