ABSTRACT
Cancer associated fibroblasts (CAF) are known to orchestrate multiple components of the tumor microenvironment, whereas the influence of the whole stromal-fibroblast compartment is less understood. Here, an extended stromal fibroblast signature was investigated to define its impact on immune cell infiltration. The lung cancer adenocarcinoma (LUAD) data set of the cancer genome atlas (TCGA) was used to test whole stroma signatures and cancer-associated fibroblast signatures for their impact on prognosis. 3D cell cultures of the NSCLC cancer cell line A549 together with the fibroblast cell line SV80 were used in combination with infiltrating peripheral blood mononuclear cells (PBMC) for in-vitro investigations. Immune cell infiltration was assessed via flow cytometry, chemokines were analyzed by immunoassays and RNA microarrays. Results were confirmed in specimens from NSCLC patients by flow cytometry or immunohistochemistry as well as in the TCGA data set. The TCGA analyses correlated the whole stromal-fibroblast signature with an improved outcome, whereas no effect was found for the CAF signatures. In 3D microtumors, the presence of fibroblasts induced infiltration of B cells and CD69+CD4+ T cells, which was linked to an increased expression of CCL13 and CXCL16. The stroma/lymphocyte interaction was confirmed in NSCLC patients, as stroma-rich tumors displayed an elevated B cell count and survival in the local cohort and the TCGA data set. A whole stromal fibroblast signature was associated with an improved clinical outcome in lung adenocarcinoma and in vitro and in vivo experiments suggest that this signature increases B and T cell recruitment via induction of chemokines.
KEYWORDS: CAF, cancer, co-culture, immune cells, infiltration, microenvironment
1. Introduction
Fibroblast diversity has gained interest due to its varying functions on pathophysiological processes that are tightly inter-connected within the local cancer microenvironment.1,2 Tumor microenvironment-derived fibroblasts (also termed cancer-associated fibroblasts = CAF) mediate cancer-immune cell interactions by direct contact and by paracrine effects via secretion of cytokines and chemokines.2 Whereas CAFs are an emerging research field, the influence of the whole stromal fibroblast compartment using an extended stromal signature exceeding the standard known cancer associated fibroblast patterns and their chemokine patterns appears to be less clear, even though it might be of particular importance in early stage cancer, where transition of fibroblasts into CAFs remains less prominent.3 For example, the human fibroblast cell line SV80 induced cytotoxic T cell infiltration in microtumors in vitro.4 Recent work reveals B lymphocytes and recently activated tissue resident CD4+CD69+ T helper lymphocytes5,6 as essential players of antitumor immunity, partially by forming tertiary lymphoid structures (TIL) and performing local T cell priming.7 Their infiltration and activity highly depend on various cytokines and chemokines.7 Amongst others, CCL13 and CXCL16 are considered to induce immune cell recruitment and differentiation.8,9
We suggest that an extended stromal fibroblast signature is important for the lung adenocarcinoma outcome, as it is linked to a specific chemokine pattern and the intra-tumor abundance of B cells and CD4+ T helper cells in ex vivo microtumors.
2. Material & methods
2.1. Analysis of data from the cancer genome atlas (TCGA)
Gene expression profiles (RNA sequencing V2 data) and corresponding clinical information of 515 patients with primary lung adenocarcinoma and 58 adjacent normal lung tissue of The Cancer Genome Atlas cohort (LUAD-TCGA)10 were retrieved via firebrowse.org (courtesy Broad Institute of MIT & Harvard). Consistent survival outcome data were retrieved from a recent TCGA pan-cancer approach.11 Normalized expression data (TPM) from the RSEM pipeline were log2 transformed adding a pseudo count of 1. Single sample gene set enrichment analyses (ssGSEA) were performed using a cancer associated fibroblast (CAFs) marker gene set and a tumor stroma gene set using the R package GSVA (Table S1).12,13 The ESTIMATE algorithm14 was used as a whole stromal score. A deconvolution method (EPIC) based on RNA sequencing data were used to calculate the amount of CAF and tumor stroma and to estimate the infiltration of specific immune cell types using the R package immunedeconv.15,16 Patients were dichotomized based on median expression or median of estimated scores and visualized as Kaplan Meier curves. Differences between groups were tested with log-rank test and univariate Cox regression analyses. Spearman rank based correlation coefficient was used to identify association between different gene expression and infiltration scores. All analyses were performed using R version 4.1.0 (https://www.r-project.org/, The R project for Statistical Computing, Vienna, Austria) and R package survival.
2.2. Single cell analysis of stromal signatures in NSCLC
Expression of gene signatures and individual marker genes were mapped onto UMAP plots from our single-cell lung cancer atlas17 comprising more than 1.3 million cells from 318 patients with annotated cell types and stromal subpopulations (https://luca.icbi.at).
2.3. Cell lines
The human NSCLC cell lines A549 (Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), Germany: ACC107) and the human fibroblasts cell line SV80 (Cell line services, Germany: 300345) were cultured in DMEM (PAA, Pasching, Austria) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, Munich, Germany, Lot 010M3396) and 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM L-Glutamine (PAA). Cells were incubated at 37°C and 5% CO2.
2.4. PBMCs
Anonymized leftover specimens of blood donations were provided by the blood bank of the Medical University of Innsbruck. Utilization of these specimens had been approved by the ethics committee of the Medical University of Innsbruck (AN2014–0353 345/4.1 - 3673a) and was conducted in accordance with the principles of the Declaration of Helsinki. PBMC were isolated via Ficoll density gradient centrifugation according as described previously.18
2.5. 3D cell culture and multicellular cultivation
3D cell culture was performed as described previously4,18 with the “GravityPLUSTM“microtissue culture system (InSphero AG, Zürich, Switzerland). A549 cells (2500 cells/well) with or without SV80 fibroblasts (5000 cells/well) were seeded in DMEM as described above, and cultivated for 10 days with medium exchange every second day. On day 10, 25000 PBMCs were added to each well (total media volume was kept constant). The tumor cell/PBMC co-cultures were incubated for 24 h to obtain immune cell infiltration.
2.6. Gene expression profiling of 3D cancer cell co-cultures
We used a dataset created previously19 for the analysis of gene (chemokine) expression profiles. In brief, Affymetrix HuGene 1.0 ST microarrays were used for 3D cancer cell cultures (A549 alone or in combination with SV80) to quantify chemokine expression. Differentially expressed genes were identified using the R package limma. In order to determine whether CAF or tumor stroma gene signatures were enriched in the tumor-fibroblast co-culture gene set enrichment analyses (GSEA) were performed on preranked log2 fold changes.20
2.7. Chemokine profiling of supernatants
To determine the concentration of cytokines and chemokines secreted by the microtissues, we harvested and pooled 24 single co-cultivated microtissues which were stored at −80°C until use. Concentrations were determined using ProcartaPlex™ Essential Human Th1/Th2 immunoassay (Affymetrix, eBioscience) and Bio-Plex ProTM Human Chemokine Assay (Bio-Rad, USA), following manufacturers’ instructions. Samples were measured using a Magpix (Luminex) and data were analyzed by the “xPONENT 4.2” software (Luminex).
2.8. Analysis of immune markers
Subpopulations of infiltrating PBMC in co-culture 3D microtissues were analyzed with flow cytometry (FACS Canto™ II). Single cell suspensions were generated from pooled microtissues by enzymatic disaggregation with 100 µg collagenase from clostridium histolyticum at a concentration of 1 mg/ml (Sigma-Aldrich) for 15 min at 37°C and 5% CO2. Reaction was stopped with medium and cells washed twice with PBS containing 10% FCS. Detailed information about antibodies used for staining can be found in the supplementary file (Table S2).
2.9. In-vivo NSCLC specimens
Immune infiltration of CD4+ T cell subpopulations and of CD20+ B cells was assessed using immunohistochemistry (IHC) in 45 patients who underwent surgical tumor eradication in curative intent (stage I-III). Ethical approval and written informed consent were obtained for all participating patients (AN2014–0293).
Beneath a conventional hematoxylin/eosin staining, the Ventana Benchmark Ultra and Ultra View DAB Kit automated staining system (Ventana Medical Systems, Tucson, AZ, USA) was performed on paraffinized sections (3 µm) according to the manufacturers´ instructions with slight optimization (sSCC31 and mSCC31 programs). Monoclonal antibodies were CD4 (Ventana, #790–4423), CD8 (Dako, #M7103), CD20 (Roche, clone: L26, #5267099001, mouse) and cytokeratin (Ventana #760–2595/#760–2135). Positive and negative tissues were used as quality controls. Staining was evaluated by a pathologist using a bright field microscope (Olympus BX 50) with a 10-fold magnification for whole slides and 40-fold for scoring of expression and localization.
Quantification of the stromal amount was performed by two trained specialists in pathology using a visual verification of cytokeratin stained slides. The stromal amount was defined as the entirety of non-cytokeratin expressing areas within the tumor mass. Quantification was accomplished in semi-quantitative steps: 0%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90% and 100% of the total tumor mass.
Concerning localization of B cells (CD 20+) and T cells (CD4+ and CD8+), we analyzed the tumor center (epithelial center) and the invasive margin (epithelial frontline), as well as the infiltration in the stroma (stromal center) and at the stromal margin (stromal frontline). Quantification was performed by classifying four groups of infiltrating cells: <1%, 1–5%, 5–50% and >50%. The cutoff for the stroma stratification was 40%, defining a stroma proportion <40% as low stromal compartment and ≥40% as high stromal compartment. This value was defined as the ideal cutoff point by ROC analysis.
Infiltrating lymphocyte subpopulations were quantified with flow cytometry. Flow cytometry data were available in 69 NSCLC specimens in our local cohort. Infiltrating subpopulations were expressed as percentage of the parental population. Patients were stratified by the percentage of investigated populations (high vs low percentage) with the following cutoff values: 9% of leukocytes for B cells, 45% of CD3+ lymphocytes for CD4+ T cells and 25% of CD4+ T cells for CD4+CD69+ T cells defined as optimal cutoff values by ROC analyses.
2.10. Statistical analysis
For statistical analysis of the experiments, mean value and the standard error of the mean (SEM) are specified. Statistical significance analyzed by paired Student’s-t-test (two-tailed) for the in-vitro data and by the Mann-Whitney-U-test for the in-vivo data. Significant differences were defined as P-values <.05. For survival analysis, logrank-test was used to determine statistical significance (p < .05) and Kaplan–Meier survival curves were used to express overall survival. Correlation of the stromal score and tumor purity was performed with a linear regression analysis. Regarding the data of the local patient cohort, optimal cutoff values for survival curves of the stromal amount in the IHC and immune cell infiltration in the flow cytometry experiments were defined by ROC-analyses. Heatmap visualization was generated with Genesis (v1.8.0, Graz University of Technology, Graz, Austria) based on z-score transformed data.
3. Results
3.1. Whole stromal fibroblast signature correlates with survival
NSCLC data of “The Cancer Genome Atlas” (TCGA) were used to investigate the influence of a whole stromal fibroblast signature within the tumor on the probability of overall survival (OS). The ”ESTIMATE stromal score” showed a significant correlation regarding the OS of LUAD patients (Figure 1). A higher ESTIMATE stromal score (which suggests higher extent of stromal tissue within a tumor) associates with a prolonged OS (hazard ratio (HR) ESTIMATE stromal scorehigh vs low 0.74; p = 0.04). In contrast, we could not identify OS difference between high vs. low CAF marker signatures (Figure 1). The gene sets used for the ESTIMATE score and the CAF markers are shown in the supplementary file (Table S1).
Figure 1.
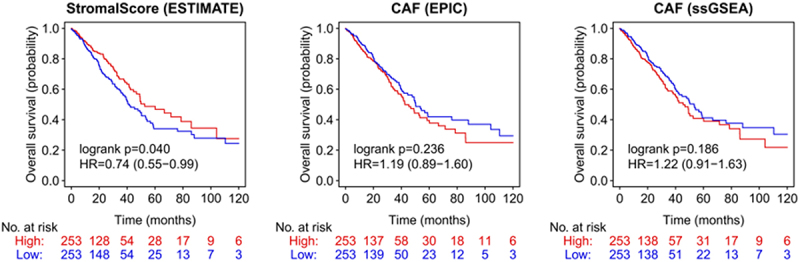
Stromal score and CAF in NSCLC patients (TCGA dataset).
Non-small-cell lung cancer (NSCLC) data of The Cancer Genome Atlas (TCGA) were used to determine the influence of the tumor stroma and cancer associated fibroblasts (CAF) on the probability of overall survival (OS). CAF = cancer associated fibroblasts.
To further evaluate how the CAF signature and the whole stromal signature (ESTIMATE score) are distributed in single cell analyses, we used our single-cell lung cancer atlas.15 In this NSCLC cohort, the CAF signature is mainly upregulated in stromal cells and bronchus fibroblasts (Figure S1). The gene signature of the ESTIMATE score was also upregulated in the same cell compartments, but was additionally expressed in lung fibroblasts and macrophages. Thereby, we are able to show that the ESTIMATE stromal score not only covers the CAF signature, but exceeds it by comprising also other parts of the stromal compartment, especially non cancer associated lung fibroblasts (Figure S1).
3.2. Three-dimensional (3D) co-cultures mimic fibroblast gene signatures and impact immune cell infiltration
To define the influence of fibroblasts in a reproducible in vitro system, 3D microtissues solely consisting of A594 cells were compared with microtissues additionally containing fibroblasts (SV80 cell line). To confirm the fibroblast signature of SV80 fibroblasts, a gene set enrichment analyses of SV80 microtissues was performed, showing an explicit fibroblast/stromal signature (Figure S2). In order to determine whether genes associated with an extended stromal fibroblast signature or the CAF signature are enriched in the co-cultures, we performed gene set enrichment analysis (Table S1). In microtissues comprising stromal cells, the in vivo defined extended whole stromal fibroblast signature (ESTIMATE score) was more prominent than CAF signatures, even though both were enriched (Figure S3). We conclude that the fibroblast-containing microtumors at least in part mimic the situation in patients and thus represents an appropriate tool to perform functional analyses with respect to the impact of the stromal cells on immune cell recruitment. For this purpose, we co-cultured preformed cancer cell/fibroblast microtissues with PBMCs and analyzed the infiltration of immune cells. In addition to the already known induction of CD8+ T cell infiltration4; we now also observed an increased abundance of CD19+ B cells and activated CD69+CD4+ T-helper cells (gating strategy see Figure S4) in the co-culture of A549/SV80/PBMCs (Figure 2a).
Figure 2.
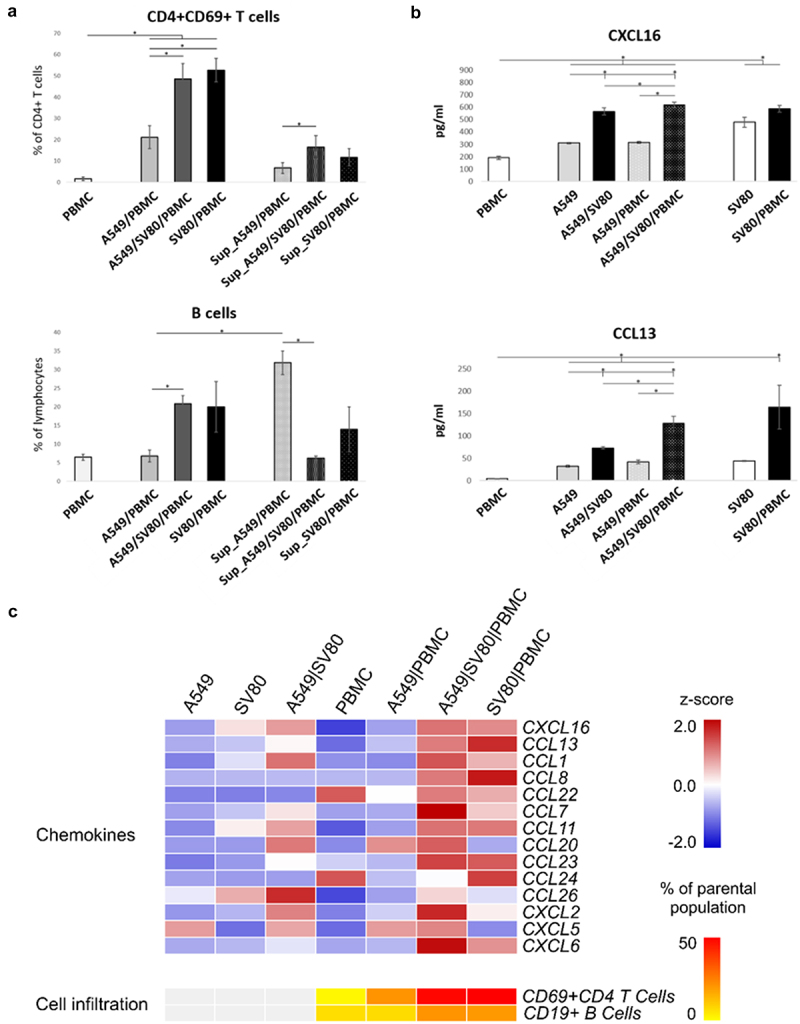
In-vitro chemokine expression and CD4+CD69+ T cell and B cell infiltration.
(a) Shows the infiltration of CD4+CD69+ T cells and B cells in microtissues with A549 cancer cells with and without SV80 fibroblasts. (b) Shows the secretion of CCL13 and CXCL16 in microtissues with A549 cancer cells, SV80 fibroblasts and peripheral blood mononuclear cells (PBMC). (c) The heatmap shows the expression of the measured chemokines. Values are expressed as z-scores between 2.0 and -2.0. Asterisks indicate significant results (p < .05).
3.3. CD19+ B lymphocytes
A significant increase of B cells was observed in the presence of fibroblasts with similar results for triple and co-cultures (A549/PBMC 7% vs. A549/SV80/PBMC 21%, p = 0.017; A549/SV80/PBMC 21% vs. SV80/PBMC 20%, p = 0.91). Non-infiltrating B cells are reduced in the supernatant of microtissues with fibroblasts (A549/SV80/PBMC 21% vs. Sup_A549/SV80/PBMC 6%, p = 0.032; SV80/PBMCs 20% vs Sup_SV80/PBMC 14%, p = 0.721), whereas the opposite was observed in supernatants of co-cultures of tumor cells with PBMC (A549/PBMC 7% vs Sup_A549/PBMCs 32%, p = 0.043). Concordantly, differences are significant for the B cell amount in the supernatant between microtissues without and with fibroblasts (Sup_A549/PBMC 32% vs. Sup_A549/SV80/PBMC 6%, p = 0.022). Thus, evidence is provided that the presence of fibroblasts within cancer microtissues triggered the recruitment of CD19+ B cells into the microtissues (Figure 2a).
3.4. Recently activated tissue resident T helper lymphocytes
An upregulation of recently activated tissue resident T helper lymphocytes (CD69+CD4+) was already observed in tumor cell microtissues cultured together with PBMCs (Figure 2a) compared to the PBMC only controls (A549/PBMC 21% vs PBMC 2%, p = 0.035). This effect was even more evident in cultures with fibroblasts (A549/PBMC 21% vs. A549/SV80/PBMC 49%, p = 0.0076; SV80/PBMC 53% vs A549/PBMC 21%, p = 0.0037). However, no significant difference between cancer cell/fibroblast/PBMC tri-cultures and fibroblast/PBMC co-cultures was observed regarding the amount of infiltrating CD69+CD4+ T cells. A similar, albeit reduced effect was observed in cells from the supernatant that did not migrate into the microtissues, suggesting that direct cellular interactions as well as soluble factors seem to induce activation of T helper lymphocytes (CD4+CD69+ T cells in Sup_A549 21% vs. Sup_A549/SV80 49%, p = 0.084)
3.5. In vitro secretion of chemokines
Chemokine expression in “PBMC-only” controls were compared to co- and triplet-cultures with equal cell counts (Figure 2b,c). Chemokine levels were very low in monocultures of cancer cells, fibroblasts or PBMC. Only CCL22 and CCL24 were detectable in PBMC monocultures (Figure 2c). Co-incubation of PBMC with cancer cells resulted in an increased secretion of CXCL5 and CCL20. In the triple culture, all B cell and CD4+ T cell-associated chemokines were elevated, with the exception of CCL24 (Figure 2c). Interestingly, a secretion of CCL24 and an even stronger increase in CCL13 and CCL8 was found, when PBMC were added to fibroblast microtissues. CCL20, CCL26, CXCL2 and CXCL5 were not induced in comparison to the triple cultures (Figure 2c). The most prominent differences were observed for CCL13 and CXCL16 (Figure 2b).
CCL13 was slightly expressed in PBMC-only controls and monocultures (A549 and SV80, Figure 2b). After the addition of fibroblasts to tumor cells, the levels of CCL13 increased about 2-fold (A549 vs. A549/SV80 p = 0.03), furthermore CCL13 concentrations increased another 2-fold in triple cultures, which, however, did not reach statistical significance (A549/SV80 vs. A549/SV80/PBMC p = 0.06). Actually, when PBMCs were cultivated together with tumor cells but without additional fibroblasts no effect was observed, suggesting a synergistic effect of fibroblasts and PBMCs on the production of CCL13. This is supported by the observation that SV80 cultures together with PBMCs have CCL13 amounts comparable to the triple culture (A549/SV80/PBMC).
Similar results were observed for CXCL16, even though the baseline levels were higher in “PBMC-only” controls. CXCL16 was highest secreted in triple cultures (A549 vs. A549/SV80/PBMC p = 0.006, Figure 2b), but comparably high in all cultures containing SV80 including their monoculture (A549 vs. A549/SV80 p = 0.013), whereas all cultures without fibroblast secreted lower amounts (A549, PBMCs, A549/PBMCs). Subsuming all approaches, CXCL16 showed a constitutive expression in fibroblast-containing cultures, assuming that the stromal tissues secreted the chemokine CXCL16 without interaction with cancer cells or immune cells (Figure 2b).
3.6. Infiltration of lymphocyte subpopulations in human lung cancer
To validate the in vitro data regarding the immune cell infiltration in association with the stromal compartment, we evaluated the infiltration of CD4+ T lymphocytes and B lymphocytes in tumor tissues from NSCLC patients, using immunohistochemistry (n = 45) and flow cytometry (n = 69).
An increased infiltration of CD20+ B cells was observed in both the central epithelial (p = 0.002) and central stromal region (p = .006, Figure 3a,b) in cancer tissues with high stromal content. Tumor tissues with a low stromal content showed a decreased infiltration of B cells within the stromal compartment and remarkably no infiltration in the epithelial fraction. Furthermore, an enhanced CD4/D8 ratio, reflecting an increased CD4+ T cell proportion, was associated with an increased infiltration of CD20+ B cells (Figure 3b).
Figure 3.
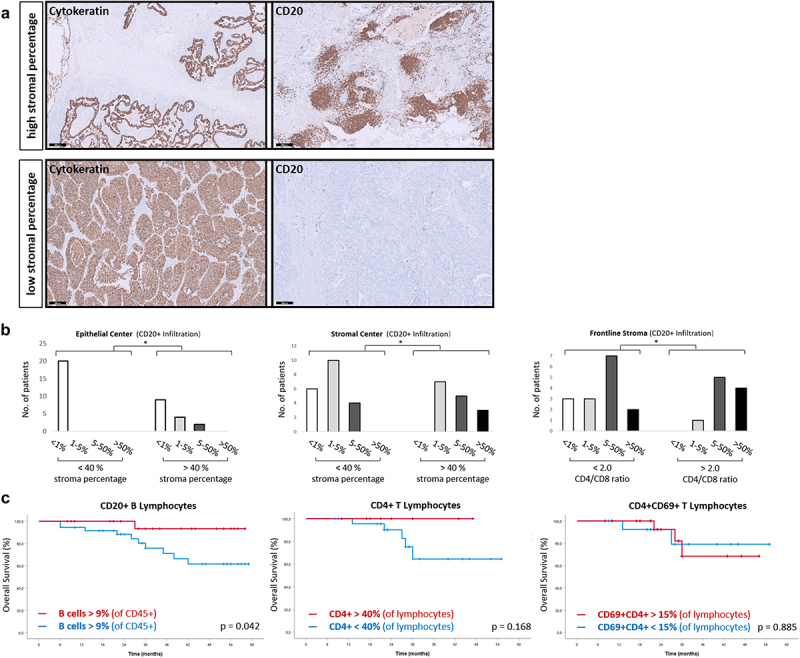
In-vivo infiltration of lymphocyte subpopulations.
(a) Cancer biopsies of patients with non-small cell lung cancer (NSCLC) of our local patient cohort were stained for cytokeratin and CD20 (brown), counterstaining was performed with hematoxylin (blue). (b) Patients were stratified by the stroma percentage and CD4/CD8 ratio. Infiltration of CD20+ lymphocytes is expressed as percentage of the parental population (CD5+ leukocytes). The CD4 ratio cutoff was defined at 2.0 (CD4:CD8 = 2:1). (c) NSCLC patients of the same cohort were investigated for the probability of survival regarding the infiltration of CD4+ T cells, CD4+CD69+ T cells and CD20+ B cells. Black bar = 200 µm.
To further validate these findings and test for the prognostic influence of CD20+ B cells and CD4+CD69+ T cells in this cohort, available flow cytometry results of 69 patients were investigated (Figure 3c). A CD20+ B cells percentage >9% of leukocytes within in the tumor specimen was associated with a significant increased OS (p = 0.042). Increased infiltration of B cells showed a 5-year survival of >90% of patients. In contrast, only 60% of the patients were alive after 5 years if the percentage of B cells was below 9%. Finally, the infiltration of CD4+ T cells (cutoff 40%, p = 0.566) and CD4+CD69+ T cells (cutoff 15%, p = 0.436) in flow cytometry was not linked to survival.
3.7. Combined effect of B lymphocytes and tumor stroma on survival using TCGA data
To investigate the influence of the stromal compartment on chemokine signaling and chemotaxis of B and CD4+ T cells, expression of CXCL16, CCL8, CCL13 and CCL22 was correlated with the stromal score and tumor purity using TCGA data (Figure 4a). A higher stromal score significantly correlated with an enhanced expression of these chemokines. Concordantly, tumor purity correlated negatively with the expression of the same chemokines. Among the investigated chemokines, the strongest correlation was observed for CCL13 (R2 = 0.312, p < .001, Figure 4a).
Figure 4.
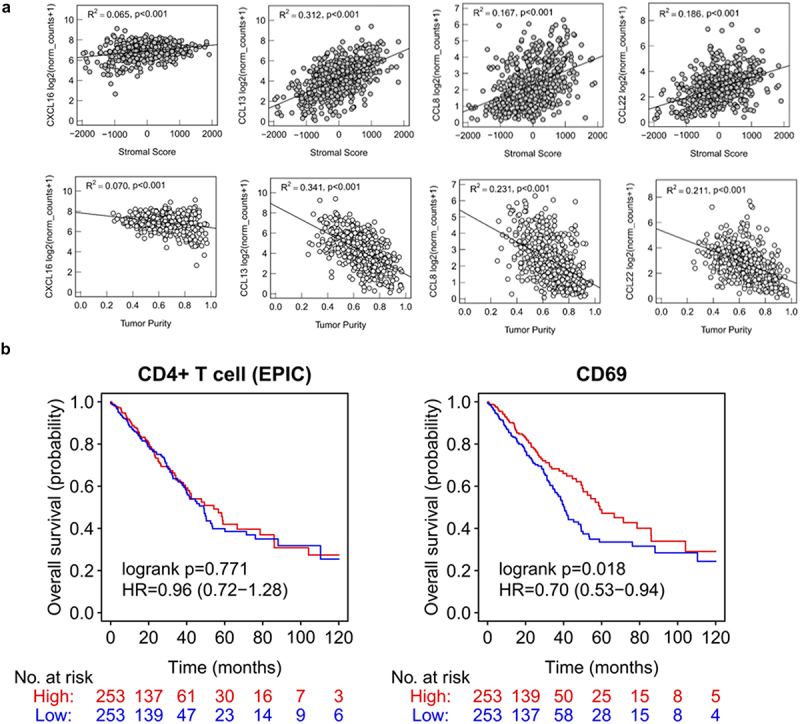
Gene expression of chemokines, CD4 and CD69 (TCGA dataset).
(a) Linear regression analyses showing chemokines CXCL16, CCL13, CCL8 and CCL22 and their association with tumor purity and stromal score (estimated by ESTIMATE). Significant results are defined by a p-value <.05. (b) Non-small-cell lung cancer (NSCLC) data of “The Cancer Genome Atlas” (TCGA) were used to determine the influence of CD4+ T cell infiltration and CD69 expression on the overall survival (OS).
We next estimated the infiltration of CD4+ T helper cells and CD19+ B cells and reassessed their prognostic relevance in NSCLC using bulk RNA sequencing data from the TCGA cohort (Figure 4b +5). Moreover, we correlated the expression of CD69 and prognosis of NSCLC patients (Figure 4b). An increased B cell infiltration was associated with a beneficial prognosis in NSCLC (B cell high vs. low: HR of 0.62, p = 0.001, EPIC). We further categorized the NSCLC TCGA dataset into four different groups with different stroma/B cell content (stroma high/B cell high, stroma high/B cell low, stroma low/B cell high and stroma low/B cell low; Figure 5a,b). Only a minor proportion of patients (6 out of 208) with a low stromal content showed a high B cell infiltration. OS was significantly increased in the stroma high/B cell high compared to those with low B cell infiltration independent of the stroma content. CD4+ T helper cells did not show any correlation with overall survival in the same patient cohort. However, we found a significant correlation between CD69 expression and an improved OS in NSCLC (HR = 0.70, p = .018, Figure 4b).
Figure 5.
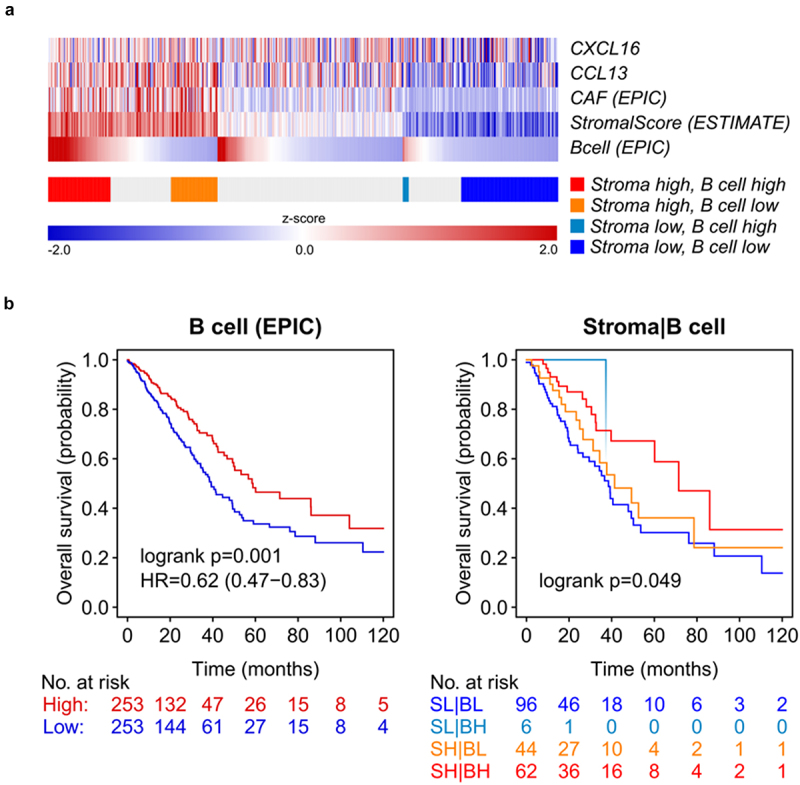
In-vivo B cell infiltration shows an improved overall survival in NSCLC patients.
(a) Using data of the non-small-cell lung cancer (NSCLC) data of “The Cancer Genome Atlas” (TCGA), the heatmap shows the expression of the chemokines CCL13 and CXCL16, B cell infiltration and the scores for the tumor stroma (ESTIMATE) and expression of markers related with cancer associated fibroblasts (CAF) using a deconvolution approach (EPIC). Values are expressed as z-scores between 2.0 and -2.0. (b) Estimated tumor stroma and infiltrating B lymphocytes were used to determine the influence on overall survival (OS). Patients were stratified into four different groups: Stroma Low/B cell Low, Stroma Low/B cell High, Stroma High/B cell Low, Stroma High/B cell High.
4. Discussion
Fibroblasts within the tumor microenvironment (TME) are a highly heterogeneous cell population with diverse functions.21,22 Based on analyses of TCGA data, we here provide evidence that in NSCLC patients with an enriched ESTIMATE score prognosis is superior14, whereas known CAF signatures did not indicate a survival disadvantage, which is contrary to recent data.23 Using our single-cell lung cancer atlas, we are able to show that that the gene signature of the ESTIMATE score exceeds the CAF signature by comprising also non-cancer associated lung fibroblasts and represents rather a whole stromal fibroblast compartment. We speculated that the positive effect of a stromal compartment with a whole stromal,fibroblast signature outweighs the impact of CAF signatures, what might be induced by the modulation of the tumor immune-milieu.
The humoral axis with antigen-presenting abilities consisting of CD4+ T helper cells and B cells are of growing interest.24,25 Especially in combination with T helper cells, B cells were described to form tertiary lymphoid structures and to prime cytotoxic T cells.26,27
In our 3D in vitro model, immune cell infiltration is measurable and can be induced by chemokine-mediated attraction of PBMC. Moreover, we were able to show the extended stromal fibroblast signature to remain more prominent in the chosen setting. Notably, our 3D model has certain weaknesses, as the lack of HLA-matching of circulating immune cells and the use of the artificial SV80 fibroblast cell line as a substitute for the whole tumor stroma. With these limitations to consider, we were able to show that a whole stromal fibroblast signature is linked to an increased infiltration of B lymphocytes and recently activated tissue resident CD4+CD69+ T helper lymphocytes in fibroblast-containing microtumors.
The secretion of specific chemokines attracting T helper cell and B cells, proliferation and activation were enhanced in supernatants of fibroblast-enriched 3D tumors and to an even higher extent in the presence of PBMCs. Of note, CCL13 and CXCL16 are most up-regulated in our in vitro system. Analysis of the LUAD data set confirms the importance of chemokines on the prognosis in lung cancer. Moreover, also a higher expression of CD69 within the tumor specimens was associated with an improved survival. Our in vivo findings are supported by positive correlations of the in vitro identified chemokines with the stroma score in the NSCLC TCGA data set.
A higher number of infiltrating CD20+ B cells was found in NSCLC tissue specimens of a local cohort containing a prominent stromal compartment. Remarkably, survival probability was significantly increased in tumors with a prominent stromal compartment combined with an increased B cell infiltration. A higher B cell score calculated by ESTIMATE or EPIC algorithms in the LUAD data set of the TCGA confirmed beneficial prognosis for these patients. Based on our data, we speculate that chemokine expression patterns dictated by fibroblasts orchestrate the link between T helper cells, B cells and tertiary lymphoid structures formation.
In conclusion, our work shows an OS benefit for the B cell-rich tumor microenvironment with an extended stromal fibroblast signature and suggests an underlying chemokine pattern to translate into a consecutive inflammation phenotype with helper T and B cell infiltration. Therefore, the beneficial survival effect of a stroma-rich compartment might be just a surrogate for its potential to activate the chemotaxis of B cells leading to an improved OS by modulating immune response mechanisms.28
Supplementary Material
Acknowledgement
We thank PD Dr. Claudia Manzl, Ao.Univ.-Prof.Dr. Bettina Zelger and Dr. Georg Schaefer from the Department of Pathology, Neuropathology and Molecular Pathology of the Medical University of Innsbruck, Tyrol, Austria for their support with tissue preparation and immunohistochemical stainings.
Funding Statement
This work was funded by means of a PhD grant of the Austrian Society of Haematology and Oncology (OeGHO, www.oegho.at) for SK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
Conceived and designed experiments: GG SK JK HZ AA SSo. Performed experiments: SK MZ JK EL AA SSo SSp FM HH. Aquired and analyzed data: SK JK GG AA MS HH HM FA AP GP DW. Wrote the paper: SK GG.
Ethical approval
Utilization of these specimens had been approved by the ethics committee of the Medical University of Innsbruck (approval number: AN2014–0353 345/4.1 - 3673a and AN 2014–0293 342/4.5) and was conducted in accordance with the principles of the Declaration of Helsinki.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2023.2274130
References
- 1.Labani-Motlagh A, Ashja-Mahdavi M, Loskog A.. The tumor microenvironment: a milieu hindering and obstructing antitumor immune responses. Front Immunol. 2020 May 15;11:940. doi: 10.3389/fimmu.2020.00940. PMID: 32499786; PMCID: PMC7243284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An Y, Liu F, Chen Y, Yang Q. Crosstalk between cancer-associated fibroblasts and immune cells in cancer. J Cell Mol Med. 2020;24(1):13–10. doi: 10.1111/jcmm.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galbo PM Jr., Zang X, Zheng D. Molecular features of cancer-associated fibroblast subtypes and their implication on cancer pathogenesis, prognosis, and immunotherapy resistance. Clinical Cancer Research. 2021;27(9):2636–2647. doi: 10.1158/1078-0432.CCR-20-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koeck S, Kern J, Zwierzina M, Gamerith G, Lorenz E, Sopper S, Zwierzina H, Amann A. The influence of stromal cells and tumor-microenvironment-derived cytokines and chemokines on CD3+CD8+ tumor infiltrating lymphocyte subpopulations. Oncoimmunology. 2017 May 8;6(6):e1323617. PMID: 28680763; PMCID: PMC5486171. 10.1080/2162402X.2017.1323617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller S, Mackay L. Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol. 2016;16(2):79–89. doi: 10.1038/nri.2015.3. [DOI] [PubMed] [Google Scholar]
- 6.Sato Y, Silina K, van den Broek M, Hirahara K, Yanagita M. The roles of tertiary lymphoid structures in chronic diseases. Nat Rev Nephrol 19, 525–537 (2023). 10.1038/s41581-023-00706-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo FF, Cui JW. The role of tumor-infiltrating B cells in tumor immunity. J Oncol. 2019;2592419. Published 2019 Sep 24. doi: 10.1155/2019/2592419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohli K, Pillarisetty VG, Kim TS. Key chemokines direct migration of immune cells in solid tumors. Cancer Gene Ther. 2022;29(1):10–21. doi: 10.1038/s41417-021-00303-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendez-Enriquez E, García-Zepeda EA. The multiple faces of CCL13 in immunity and inflammation. Inflammopharmacology. 2013. Dec;21(6):397–406. Epub 2013 Jul 12. PMID: 23846739. doi: 10.1007/s10787-013-0177-5. [DOI] [PubMed] [Google Scholar]
- 10.Hojo S, Koizumi K, Tsuneyama K, Arita Y, Cui Z, Shinohara K, Minami T, Hashimoto I, Nakayama T, Sakurai H, et al. High-level expression of chemokine CXCL16 by tumor cells correlates with a good prognosis and increased tumor-infiltrating lymphocytes in colorectal cancer. Cancer Res. 2007 May 15;67(10):4725–4731. doi: 10.1158/0008-5472.CAN-06-3424. PMID: 17510400. [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network . Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009 Nov 5;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013 Jan 16;14(1):7. doi: 10.1186/1471-2105-14-7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4(1):2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. 2017;6:e26476. doi: 10.7554/eLife.26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, List M, Aneichyk T. Comprehensive evaluation of transcriptome-based cell-type Quantification methods for immuno-oncology. Bioinformatics. 2019;35(14):i436–i445. doi: 10.1093/bioinformatics/btz363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salcher S, Sturm G, Horvath L, Untergasser G, Kuempers C, Fotakis G, Panizzolo E, Martowicz A, Trebo M, Pall G, et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell. 2022 Dec 12;40(12):1503–1520.e8. doi: 10.1016/j.ccell.2022.10.008. Epub 2022 Nov 10. PMID: 36368318; PMCID: PMC9767679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koeck S, Zwierzina M, Huber JM, Bitsche M, Lorenz E, Gamerith G, Dudas J, Kelm JM, Zwierzina H, Amann A, et al. Infiltration of lymphocyte subpopulations into cancer microtissues as a tool for the exploration of immunomodulatory agents and biomarkers. Immunobiology. 2016. May;221(5):604–617. Epub 2016 Feb 1. PMID: 26876590. doi: 10.1016/j.imbio.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Gamerith G, Rainer J, Huber JM, Hackl H, Trajanoski Z, Koeck S, Lorenz E, Kern J, Kofler R, Kelm JM, et al. 3D-cultivation of NSCLC cell lines induce gene expression alterations of key cancer-associated pathways and mimic in-vivo conditions. Oncotarget. 2017 Nov 6;8(68):112647–112661. doi: 10.18632/oncotarget.22636. PMID: 29348853; PMCID: PMC5762538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, Bassez A, Decaluwé H, Pircher A, Van den Eynde K, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018. Aug;24(8):1277–1289. Epub 2018 Jul 9. PMID: 29988129. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 23.Hu H, Piotrowska Z, Hare PJ, Chen H, Mulvey HE, Mayfield A, Noeen S, Kattermann K, Greenberg M, Williams A, et al. Three subtypes of lung cancer fibroblasts define distinct therapeutic paradigms. Cancer Cell. 2021;39(11):1531–47.e10. doi: 10.1016/j.ccell.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min KW, Kim DH, Noh YK, Son BK, Kwon MJ, Moon J-Y. Cancer-associated fibroblasts are associated with poor prognosis in solid type of lung adenocarcinoma in a machine learning analysis. Sci Rep. 2021;11(1):16779. doi: 10.1038/s41598-021-96344-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaud D, Steward CR, Mirlekar B, Pylayeva‐Gupta Y. Regulatory B cells in cancer. Immunol Rev. 2021. Jan;299(1):74–92. doi: 10.1111/imr.12939. Epub 2020 Dec 23. PMID: 33368346; PMCID: PMC7965344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosser EC, Mauri C. Regulatory B cells: origin, phenotype, and function. Immunity. 2015 Apr 21;42(4):607–612. PMID: 25902480. doi: 10.1016/j.immuni.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Garaud S, Dieu-Nosjean MC, Willard-Gallo K. T follicular helper and B cell crosstalk in tertiary lymphoid structures and cancer immunotherapy. Nat Commun. 2022;13(1):2259. doi: 10.1038/s41467-022-29753-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Germain C, Devi-Marulkar P, Knockaert S, Biton J, Kaplon H, Letaïef L, Goc J, Seguin-Givelet A, Gossot D, Girard N, et al. Tertiary lymphoid structure-B cells narrow Regulatory T cells impact in lung cancer patients. Front Immunol. 2021 Mar 8;12:626776. PMID: 33763071; PMCID: PMC7983944. 10.3389/fimmu.2021.626776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.


