ABSTRACT
Acinetobacter baumannii is a common cause of healthcare-associated infections and hospital outbreaks, particularly in intensive care units. Much of the success of A. baumannii relies on its genomic plasticity, which allows rapid adaptation to adversity and stress. The capacity to acquire novel antibiotic resistance determinants and the tolerance to stresses encountered in the hospital environment promote A. baumannii spread among patients and long-term contamination of the healthcare setting. This review explores virulence factors and physiological traits contributing to A. baumannii infection and adaptation to the hospital environment. Several cell-associated and secreted virulence factors involved in A. baumannii biofilm formation, cell adhesion, invasion, and persistence in the host, as well as resistance to xeric stress imposed by the healthcare settings, are illustrated to give reasons for the success of A. baumannii as a hospital pathogen.
KEYWORDS: Acinetobacter baumannii, desiccation, metals uptake, secretion systems, tolerance, virulence factors
Introduction
Bacteria belonging to the genus Acinetobacter are Gram-negative coccobacilli that emerged in the last three decades as an increasingly frequent cause of healthcare-associated infections [1–3]. To date, genomic methods have identified 72 species within the Acinetobacter genus (https://pubmlst.org/bigsdb?db=pubmlst_abaumannii_isolates&page=query&genomes=1; accessed on 2 February 2023). Of these, A. baumannii, A. nosocomialis, A. pittii, A. seifertii, and A. lactucae, formerly A. dijkshoorniae, are the most clinically relevant species [1,2,4] They are evolutionarily related and phenotypically almost indistinguishable from the environmental species A. calcoaceticus and are therefore grouped into a species complex called A. calcoaceticus-A. baumannii (ACB) complex [1,2,4]. A. baumannii is the most important pathogen of the ACB complex and is responsible for various healthcare-associated infections, including pneumonia, bloodstream infection, urinary tract infections, and wound infections, and less frequently for community-acquired infections among predisposed patients [1,5]. Although A. baumannii is frequently isolated from the nosocomial environment, which is widely recognized as its primary reservoir [1], extra-hospital reservoirs have also been identified: Indeed, A. baumannii has been isolated from soil, freshwater [6–9], animals, foods, poultry livestock, wild birds, and human lice [10–12].
A. baumannii represents a showcase example of an antibiotic-resistant pathogen, as demonstrated by the frequent isolation of multidrug-resistant (MDR) or extensively drug-resistant (XDR) strains. The majority of these isolates are resistant to carbapenems and a fraction of them are also resistant to tigecycline and/or colistin [5]. Therefore, many A. baumannii infections show limited treatment options and high mortality, especially in intensive care unit patients [1,5], and this has led the World Health Organization to classify carbapenem-resistant A. baumannii as a “critical” pathogen among the antibiotic-resistant bacteria of global priority [13]. Accordingly, a recent comprehensive assessment of the global burden caused by drug-resistant bacteria in 2019 identified A. baumannii as one of the six leading pathogens for antibiotic resistance-associated deaths [14].
A. baumannii genomic epidemiology
Several studies investigated the phenotypic and genotypic characteristics of A. baumannii isolates and demonstrated the spread of MDR isolates in healthcare facilities at the national and international levels [1,13,15–18]. The availability of two multilocus sequence typing (MLST) schemes allowed to discriminate genetically distinct clonal lineages [18]. The global epidemiology of A. baumannii highlighted a clonal population structure dominated by three major international clonal lineages (ICLs), namely ICL I, II, and III, and 5 to 8 additional epidemic clonal lineages [15,16,18]. The epidemic A. baumannii lineages are frequently resistant to a broad range of antimicrobials, including carbapenems [15–17]. Moreover, they possess virulence traits which stimulate biofilm growth on abiotic surfaces, tolerance to desiccation, and adherence to human epithelial cells, and contribute to their persistence in the hospital environment and spread among infected patients [19]. However, an elevated phenotypic heterogeneity has recently been documented among recent MDR clinical isolates and reference A. baumannii strains, which show differences in colony morphology, capsule production, natural transformability, and virulence [20].
The high number of A. baumannii genomes currently available in the NCBI database highlighted the elevated genome plasticity of the species, witnessing the propensity to acquire exogenous genetic information through horizontal gene transfer and genomic recombination, and indicating that A. baumannii has an open pan-genome [17,21]. For example, Mangas et al. showed that the A. baumannii pan-genome consists of approximately 20,000 different genes, the majority of which (ca. 16000) are present in less than 20 % of the strains [22].
Acinetobacter baumannii virulence
The aim of this review is to provide an overview of recent insights into the pathogenicity and virulence of A. baumannii. Model systems designed to investigate A. baumannii-host interactions are discussed in relation to the impact of cell-associated and secreted extracellular virulence factors on pathogenicity. Additionally, this review explores the molecular responses implicated in desiccation and oxidative stress tolerance, since they are expected to contribute to the adaptation of A. baumannii to the hospital environment.
Cell-associated virulence factors
Lipooligosaccharide
A. baumannii surface appendages, adhesins, and glycoconjugates, like capsular polysaccharides, glycosylated proteins, lipooligosaccharide (LOS), and peptidoglycan are important virulence determinants [23] (Figure 1). The most abundant constituent of the Gram-negative bacterial outer membrane is lipopolysaccharide (LPS) or, alternatively, LOS. Like Campylobacter and Neisseria species, A. baumannii possesses a LOS consisting of a hepta-acylated disaccharide (lipid A) linked to an oligosaccharide core without the addition of repeat-unit polysaccharides known as O-antigen [23]. The hepta-acylated lipid A moiety of A. baumannii LOS has been demonstrated to stimulate the host innate immune response through activation of the pattern recognition receptor Toll-like receptor 4 (TLR4) and to promote resistance to cationic antimicrobial peptide and desiccation [24]. The structure of the LOS can undergo alterations to enable A. baumannii to withstand stressful conditions. Indeed, A. baumannii adapts to cold temperatures through the increased expression of the LpxS acyltransferase, which incorporates a short C 8:0 chain fatty acid instead of a C12:0 fatty acid at position 2 of the lipid A in order to maintain outer membrane fluidity and permeability during cold stress [25].
Figure 1.
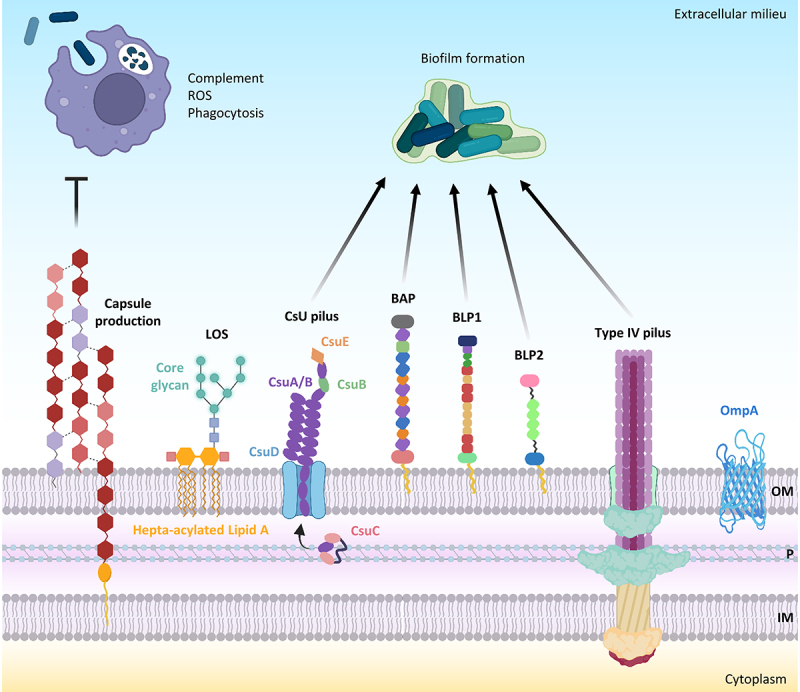
A. baumannii cell-associated virulence factors. negatively charged capsular polysaccharides hinder interactions with negatively charged surfaces of neutrophils and macrophages and protect against complement-mediated killing, peptidoglycan degradation by lysozyme, and ROS. Hepta-acylation of lipid A in LOS strengthens the outer membrane (OM) and protects A. baumannii from cationic antimicrobial peptides (CAMPs) and lysozyme. Csu pilus, BAP, BLP1, BLP2, and type IV pilus are the main proteins involved in biofilm formation. The OM protein A (OmpA) inhibits complement-mediated killing. Other abbreviations: P, periplasm; IM, inner membrane. Figure created with Biorender.
Capsule
A polysaccharide capsule consisting of high-molecular-weight carbohydrate repeats with a variable composition surrounds A. baumannii cells [23,26] and plays a major role in bacterial virulence [23,27,28] (Figure 1). The capsular polysaccharide is critical for growth in human ascites and resistance to the complement-mediated killing of A. baumannii AB307–0294 [27]. The transition from virulent opaque (VIR-O) highly capsulated to avirulent translucent (AV-T) low capsulated colony variants in A. baumannii AB5075 demonstrated that the capsule is essential for causing pulmonary infection of mice, while increasing A. baumannii resistance to antimicrobials, hospital disinfectants, and desiccation. A Tet-R type “master” transcriptional regulator is responsible for the switch from the VIR-O to the AV-T phenotype; its overexpression abrogates virulence and resistance to lysozyme, a cathelicidin-related antimicrobial peptide, and reactive oxygen species (ROS) [28]. Deletion of the wzc gene required for capsule production decreased the tolerance of A. baumannii AB5075 to disinfectants, lysozyme, and desiccation [29]. Further studies demonstrated that the switch from the VIR-O to AV-T phenotype in A. baumannii AB5075 and other clinical isolates is mediated by the stochastic activation of different TetR-type transcriptional regulators [30,31] and that the switching frequency from VIR-O to AV-T is positively influenced by the small RNA SrvS [31]. The activation of different combinations of TetR-type transcriptional regulators in the VIR-O state may generate distinct translucent subpopulations, whose unique phenotypic properties should confer survival advantages in various environments [31]. Overall, these studies point to the coordinated regulation of capsule production during invasive infection and the environmental persistence of A. baumannii [28–31].
Increasing evidence indicates that the capsule carbohydrate structure is responsible for the difference in virulence among A. baumannii clinical isolates. Loss of an N-acetyl-β-D-glucosamine group in the capsule carbohydrate due to a spontaneous transposon insertion in the gtr6 glycosyltransferase gene protects from CR3 receptor-mediated phagocytosis and increases the virulence in a bacteraemia mouse model of the clinical isolate HUMC1 assigned to ICL II [32]. Similarly, the clonal shift observed among A. baumannii clinical isolates assigned to epidemic ICL II in Portugal has been ascribed to the occurrence of ST218Oxf isolates displaying a peculiar KL7 capsular locus, containing legionaminic acid [33].
OmpA
The outer membrane protein A (OmpA) of A. baumannii is an abundant surface protein that plays a multifaceted role during bacterial infection [34–39] (Figure 1). OmpA has been demonstrated to bind host epithelial cells and primary monocyte-derived macrophages, translocate to the nucleus by a novel monopartite nuclear localization signal, and induce cell death of host cells in vitro [34]. OmpA has also been shown to stimulate ROS production originating from mitochondria and induces early-onset apoptosis and delayed-onset necrosis in dendritic cells [37]. Further studies demonstrated that A. baumannii ATCC 19606T OmpA induces biofilm growth on abiotic surfaces, adhesion, and invasion of host epithelial cells by a zipper-like mechanism, which involves microfilament and microtubule-rearrangements of the cytoskeleton [35,38].
The important role of OmpA during infection has been documented in different animal models [35,38,39]. In a murine pneumonia model, wild-type A. baumannii ATCC 19606T caused severe lung pathology and induced bacteraemia, whereas an OmpA-deficient mutant showed a significantly lower bacterial burden in blood [35]. Likewise, a knockout mutant in the ompA gene of ATCC 17978 showed greater lethal doses and reduced invasion, with a lower bacterial concentration in the spleen and lungs in a murine peritoneal sepsis model, relative to the wild-type counterpart [38]. Moreover, the ompA gene is expressed at higher levels in A. baumannii isolates from patients with pneumonia as compared to those from colonized patients, and isolates from bacteraemic patients showed higher ompA expression than those from nonbacteremic patients, suggesting that ompA overexpression positively contributes to pneumonia, bacteraemia, and A. baumannii-associated mortality [39].
Virulence determinants involved in biofilm formation
Biofilm formation has been associated with A. baumannii virulence [1,2,40]. The ability to form biofilm favours the colonization of host mucosa and growth of bacteria on abiotic surfaces, thus contributing to medical device-associated infections and survival in the environment [1,2,40]. A. baumannii epidemic strains show high biofilm-forming capacity on abiotic surfaces, which may contribute to their persistence in the hospital environment [19].
Biofilm formation is a multifactorial process, and several cell-associated factors contribute to A. baumannii sessile growth and adherence to host epithelial cells (Figure 1). An archaic chaperone-usher pili is the surface appendage that mediates the adhesion and sessile growth of A. baumannii [41,42]. The Csu pilus comprises the CsuA/B pilus rod, the CsuA and CsuB adaptor subunits, and the two-domain tip adhesin CsuE complexed with its CsuC chaperon, which binds to various abiotic surfaces through hydrophobic finger-like loops, thereby initiating biofilm formation [42,43]. The type IV pili is an additional surface appendage implicated in twitching motility, natural transformation [44], and adhesion to biotic or abiotic surfaces, or between bacterial cells [45]. The surface electrostatics of the major protein subunit PilA variants of type IV pilus modulate the equilibrium between motility and biofilm formation. Increasing the electronegativity of the alpha-beta group of PilA variant in A. baumannii AB5075, promotes repulsion between cells and twitching motility while decreasing the electronegativity of the alpha-beta group of PilA variant in A. baumannii ACICU promotes biofilm growth [45]. The above data are concordant with a previous study demonstrating that A. baumannii isolates assigned to ICL II, including the strain ACICU, show higher biofilm growth compared with A. baumannii isolates assigned to ICL I [19]. On the other hand, differences in surface electrostatics of PilA variants do not affect bacterial adhesion to host epithelial cells [45].
Most A. baumannii strains express at the cell surface a large multidomain protein, which is composed of distinct and repetitive immunoglobulin-like domains, named BAP (biofilm-associated protein) for its homology to the staphylococcal biofilm-associated protein BAP [46] (Figure 1). BAP induces biofilm growth and adherence to host epithelial cells [47]. In addition to BAP, A. baumannii strains express two other proteins, BLP1 and BLP2 (BAP-like proteins 1 and 2), which share with BAP immunoglobulin-like repeats and a sequence motif at the N-terminus, and positively regulate biofilm formation and adherence to epithelial cells [48] (Figure 1). The expression of BAP, BLP1, and BLP2 is very low during the exponential phase, while it is induced during the stationary growth phase [48].
Mounting evidence indicates that the expression of virulence factors involved in biofilm growth and adherence is finely tuned at the transcriptional level. A transcriptomic analysis comparing planktonic and biofilm growth in vitro of A. baumannii ATCC 17978 showed that the genes overexpressed in biofilm-forming cells are implicated in quorum-sensing regulation, and in the synthesis of Csu pilus proteins and BAP [49]. On the other hand, the expression of types I and IV pili, quorum sensing components, and other genes regulating biofilm formation was reduced in A. baumannii ATCC 17978 during systemic infection of mice [50]. This suggests that differential expression of genes relevant to biofilm formation likely occur in vivo, depending on the site and type of infection.
Biofilm formation in A. baumannii is regulated by several factors, including transcriptional regulators and efflux systems. The LysR-type transcriptional regulator LeuO has been demonstrated to downregulate the genes within the csuA/BABCDE chaperon-usher pili system and the genes involved in iron acquisition, and act as a repressor of biofilm synthesis. In fact, disruption of the leuO gene in A. baumannii ATCC 17987 increased biofilm formation and pathogenicity during systemic infection in neutropenic mice. Also, mutations within the leuO gene were observed in clinical A. baumannii isolates showing hyper-biofilm phenotype [51]. Similarly, the expression of Csu pili that are assembled through the archaic chaperone-usher pathway and biofilm growth are positively regulated by the phenylacetic acid produced in response to folate stress induced by subinhibitory concentrations of trimethoprim and sulfamethoxazole [52]. Coherently, inhibition of the phenylacetic acid degradation pathway resulted in attenuated virulence of A. baumannii in a murine catheter-associated urinary tract infection model [52]. Several A. baumannii strains were shown to form a tenacious biofilm, called surface pellicle, at the solid-liquid interface [53]. The over-expression of AdeABC and AdeIJK resistance-nodulation-cell division (RND)-type efflux systems have been demonstrated to negatively regulate biofilm and surface pellicle formation in A. baumannii BM4587 [54]. Along the same lines, a recent study demonstrated that the MacAB-TolC efflux pump is involved in the envelope stress response occurring in mature biofilm to maintain bacterial membrane rigidity and provide tolerance to high osmolarity conditions [55].
Impact of secretion systems on A. baumannii virulence
To date, four protein secretion systems have been identified in A. baumannii, and a contribution to infectivity has been proposed or experimentally validated for all of them. The genetics, biochemistry, and structural properties of these systems have previously been reviewed [56–58]. In this paragraph, we discuss the advances in the knowledge of the substrates secreted by these systems, along with the direct or indirect evidence of the role they play in A. baumannii pathogenicity. The type IV secretion system (T4SS) is not discussed as it has been only identified in conjugative plasmids [59,60], and there are no reports showing or suggesting its involvement in functions other than horizontal gene transfer in A. baumannii.
Type I secretion systems (T1SSs)
T1SSs are tripartite secretion systems consisting of an integral ATP-binding cassette (ABC) transporter in the inner membrane, which energizes the system, an inner membrane-anchored protein that protrudes into the periplasm (membrane fusion protein or MFP), and an outer membrane β-barrel protein [61]. These components form a transmembrane tunnel that spans the entire cell envelope of Gram-negative bacteria and transports unfolded protein substrates from the cytosol outside the cell (Figure 2). T1SSs are widespread in Gram-negative bacterial pathogens and, overall, can secrete different effectors, such as adhesins, lipolytic or proteolytic enzymes, iron-scavenger proteins, or pore-forming toxins [61]. Some bacteria possess multiple T1SSs, each dedicated to the secretion of specific protein substrates [62]. T1SS substrates are generally recognized via a C-terminal secretion domain that is not cleaved after transport [61].
Figure 2.
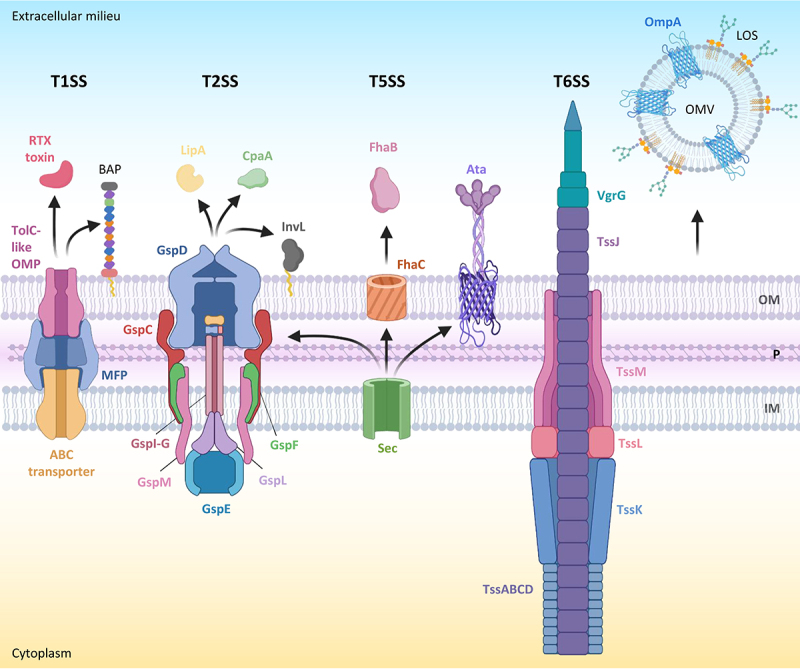
Secretion systems and released effector proteins associated with A. baumannii virulence. for each secretion system, the name of the main protein components is indicated. Outer membrane vesicles (OMVs) are also shown. LOS in the outer membrane (OM) is omitted. Other abbreviations: P, periplasm; IM, inner membrane. Proteins and protein subunits are not in scale. Figure created with Biorender.
Thus far, two papers have investigated the presence and role of the T1SS in Acinetobacter [63,64]. Harding and collaborators identified in A. nosocomialis M2 (formerly named A. baumannii M2) a homolog of the HlyBD-TolC T1SS of Escherichia coli and attempted to verify its functionality through the construction of T1SS-defective mutants. The comparison of the secretome of wild-type and mutant strains highlighted several proteins whose secretion was directly or indirectly affected by the lack of the T1SS. Two components of the secretome, namely a serralysin-like protein with a repeats-in-toxin (RTX) domain and the biofilm-associated protein BAP, carry a C-terminal secretion domain characteristic of T1SS substrates, leading to the proposal that the T1SS of A. nosocomialis can secrete two different effectors [63]. T1SS-deficient mutants were slightly attenuated in virulence in the Galleria mellonella insect model of infection and showed significant defects in biofilm formation [63], in line with the important role of BAP in promoting biofilm maturation and maintenance in A. baumannii [46,47,65]. In contrast, the function of the RTX toxin has not yet been evaluated in any Acinetobacter species.
A few years later, the contribution of the T1SS to virulence was further addressed using a recent urinary isolate of A. baumannii (UPAB1). This clinical isolate has the capability to persist and replicate inside macrophages, and genomic analysis revealed that it carries two operons predicted to encode for T1SSs [64]. The generation of single and double mutants in these operons led to the demonstration that both are required for replication inside macrophages. Notably, proteomic analysis of culture supernatants showed that the two T1SSs influence the secretion of different sets of proteins. One system (defined T1SSa) was found to be required for the secretion of RTX toxin and BAP, suggesting that it is the ortholog of the T1SS previously identified in A. nosocomialis. Many other proteins were less abundant in the supernatants of T1SS-defective mutants, including proteases, phosphatases, glycosidases, and a putative invasin [64]. Since the inactivation of the T1SS in A. nosocomialis resulted in the downregulation of other secretion systems [63], further experiments are needed to verify whether these proteins are true T1SS effectors and their actual role in cell invasion and intracellular replication. Notably, our search for T1SS genes in a large collection of completely sequenced genomes revealed that the presence of two T1SSs appears to be an exception in A. baumannii, as most isolates (>95%) have a single system (Table S1), as observed in A. nosocomialis M2 [63].
Type II secretion system (T2SS)
The T2SS is a complex macromolecular protein machinery for the release in the extracellular milieu of folded protein effectors which are translocated into the periplasm by the general secretory (Sec) or the twin-arginine translocation (TAT) pathway (Figure 2). T2SS substrates include toxins, lipases, metalloproteases, and digestive enzymes, and have been demonstrated to contribute to virulence and host adaptation in different bacterial pathogens [66–68].
The presence of a T2SS in A. baumannii was first proposed in 2014, based on bioinformatics and comparative genomic analyses [69]. Its role in virulence was experimentally proven through the demonstration that a T2SS-defective mutant of the reference strain ATCC 17978 was outcompeted by the wild-type strain when co-inoculated in a leucopenic mouse model of septicaemia [70]. Similar colonization and/or persistence defects in vivo were observed for an isogenic mutant unable to produce the LipA lipase, which was demonstrated to be a T2SS substrate [70], suggesting that this lipase may represent one of the main virulence factors secreted by the T2SS in A. baumannii. While LipA was required in vitro for A. baumannii ATCC 17978 growth with long-chain fatty acids as the sole carbon source [70], the mechanism by which LipA influences the in vivo fitness of A. baumannii remains unknown. The contribution of the T2SS in the pathogenicity of the A. baumannii strain ATCC 17978 was further confirmed in a murine model of pneumonia, as mice infected with a T2SS-defective mutant showed a significant reduction in the mean CFUs in the lung as compared to mice infected with the wild-type strain [71]. Concurrently, it has been demonstrated that the T2SS is also present and functional in other Acinetobacter species, including A. nosocomialis, A. pittii, A. calcoaceticus, and A. junii, and that is required in A. nosocomialis M2 for full lethality in G. mellonella larvae and for optimal lung colonization in a murine pulmonary infection model [72]. In contrast to most Gram-negative species, the structural and functional T2SS genes of Acinetobacter spp. are dispersed throughout the chromosome rather than being clustered in a single genetic locus [70,72]. More recently, the T2SS has also been reported to affect the virulence of the urinary isolate A. baumannii UPAB1 in a mouse model of catheter-associated urinary tract infection, as the T2SS-defective mutant exhibited a 10-fold decreased ability to bind to the catheter and colonize the bladder [73].
Several proteomics studies revealed that the T2SS has a major impact on the secretome of A. nosocomialis M2 and different A. baumannii strains, with dozens of proteins showing reduced levels in the supernatants of T2SS-defective mutants as compared to wild-type strains [71–74]. However, the direct role of the T2SS in secretion has only been validated for a few substrates. Besides LipA, the metallopeptidase CpaA, the lipases LipH and LipAN, and the adhesin InvL were confirmed as T2SS substrates in Acinetobacter spp [71–73]. Artificially induced overexpression of LipAN in A. baumannii ATCC 17978 was shown to slightly promote colonization of the murine lung upon intranasal infection [71], suggesting that this enzyme represents a T2SS-dependent virulence factor of A. baumannii. However, LipAN is encoded by the plasmid pAB2 which appears to be restricted to the ATCC 17978 strain [71], thus limiting its clinical relevance. While the role of LipH in virulence-related traits has not yet been investigated, the possible functions of CpaA and InvL during pathogenesis have been proposed based on in vitro and in vivo studies. InvL is predicted to be a surface-exposed lipoprotein that binds to several extracellular matrix components, facilitates A. baumannii UPAB1 adhesion to urinary tract cell lines, and promotes uropathogenesis in the murine model of catheter-associated urinary tract infection [73]. InvL homologs were observed in about 70% of isolates belonging to different Acinetobacter spp., with a very high prevalence (almost 100%) in the A. baumannii ICL II [73], a lineage of great clinical success [1,15,18,19]. CpaA was thoroughly characterized at the molecular and biochemical levels. It is a zinc-dependent metalloendopeptidase originally identified in some A. baumannii clinical isolates [75], and later demonstrated to be secreted by different medically relevant Acinetobacter species [76]. CpaA cleaves several O-linked human glycoproteins, some of which are involved in complement activation, thus affecting the intrinsic coagulation pathway [75,77,78]. The contribution of CpaA to virulence has been confirmed for A. nosocomialis M2, as a CpaA-deficient mutant showed a significantly reduced lethality in the G. mellonella model of infection, similar to that of a T2SS-defective mutant, and a slightly impaired ability to disseminate to the spleen after intranasal infection in mice [76].
Overall, evidence has been gained that A. baumannii and other medically relevant Acinetobacter species have a functional T2SS that significantly contributes to virulence by secreting a number of protein effectors. Some of them have been proven to play a role during experimental infections, but many others have not yet been investigated. Moreover, the specific contribution of each T2SS effector to virulence varies among different strains/species and/or models of infection.
Type V secretion system (T5SS)
The T5SS, also known as the autotransporter system, is a simple secretion mechanism based on a protein (the autotransporter) which consists of three major domains, namely the N-terminal signal peptide for transport across the cytoplasmic membrane through the Sec pathway, the extracellular “passenger” domain, and the “translocator” β barrel domain into the outer membrane. T5SSs have been classified into different subtypes based on differences in topology, domain order, and oligomeric states. The passenger is either anchored to the translocator domain and protrudes from the bacterial surface or it is released into the environment through an autoproteolytic mechanism or by a separate protease. Passenger domains can have a plethora of functions, including adhesion, cell-cell aggregation, biofilm formation, intracellular motility, and haemolysis, or be endowed with enzymatic activities, such as protease and lipase [79–81].
The first T5SS identified in A. baumannii belongs to the trimeric autotransporter family (type 5c) and was therefore named Ata (Acinetobacter trimeric autotransporter) (Figure 2). The Ata passenger domain is an adhesin that can bind to multiple proteins of the host extracellular matrix in vitro [82]. More recently, it has been proposed that the protein-binding activity of Ata is due to the ability to bind glycans, which are present on many of the proteins bound by Ata [83]. Genetic inactivation of the ata gene in A. baumannii ATCC 17978 drastically reduced biofilm formation on a plastic surface and abrogated the lethality of systemic infection in immunocompetent mice [82]. Similar results were obtained in A. baumannii ATCC 19606T, in which the loss of Ata reduced the adhesion to epithelial and endothelial cells and the pathogenicity in the G. mellonella [84]. Different studies have provided in vitro and in vivo evidence that Ata – or its C-terminal extracellular region – might represent a promising antigen for the development of anti-A. baumannii vaccines [85–87]. However, phylogenomic analyses revealed that Ata is present in most but not all (about 80%) A. baumannii isolates [84].
Genomic analysis of the A. baumannii AbH12O-A2 outbreak isolate led to the identification of a genomic island, which is present (complete or partial) in some A. baumannii strains, harbouring genes encoding a two-partner secretion (TPS) system, named AbFhaB/AbFhaC, belonging to the T5bSS family [88]. AbFhaB is an extracellular filamentous haemagglutinin-adhesin that requires the outer membrane β-barrel protein AbFhaC for transport across the outer membrane. Genetic inactivation of FhaC strongly impaired the ability of A. baumannii AbH12O-A2 to adhere to lung epithelial cells and slightly reduced the mortality associated with AbH12O-A2 intraperitoneal infection in mice [88]. A similar TPS system (named FhaB1/FhaC1) was previously identified in A. baumannii ATCC 19606T. FhaB1/FhaC1 was reported to promote cell adhesion and biofilm formation when ectopically expressed in E. coli and to confer protection from A. baumannii infection when used to immunize mice [89]. Sequence analysis confirms that AbFhaB/AbFhaC and FhaB1/FhaC1 have a low sequence identity (30–40%; Table S2), suggesting that different A. baumannii strains may have different TPS systems.
Finally, the majority of A. baumannii strains express functional contact-dependent inhibition (CDI) systems. These are TPS systems in which the outer membrane pore-forming CdiB protein mediates the translocation of a CdiA toxin to the cell surface. CdiA binds to specific receptors on susceptible bacteria and then delivers its toxin domain located at the C-terminus (CdiA-CT) into target cells. CDI systems are widely distributed among Acinetobacter species of the ACB complex and are used for intraspecies competition to eliminate bacteria that do not express the cognate immunity protein [63,90,91]. Two different CDI system groups were identified in A. baumannii strains, which differ in CdiA protein structure. Type II CdiA are large proteins (ranging from 3,700 to 5,700 residues) with a long array of 20-mer repeats, while type I CdiA are smaller (1,900–2,400 residues) with a central heterogeneity region [91]. At present, there is no evidence that CDI systems play a role in A. baumannii pathogenesis.
Type VI secretion system (T6SS)
The T6SS is a complex, bacteriophage-like protein machinery that spans the entire cell envelope of Gram-negative bacteria and injects proteins into neighbouring cells [92,93] (Figure 2). T6SS effectors are generally involved in killing or inhibiting the growth of prokaryotic cells, but some of them can also affect eukaryotic cell functions. Although the T6SS is mainly involved in interspecies competition among bacteria, it can also directly contribute to infectivity in many pathogenic bacteria by allowing them to colonize host tissues, disseminate, enter host cells, and evade the host immune response [94–96].
Bioinformatic analysis suggested that the T6SS is present in many but not all Acinetobacter species and that it is overall conserved in A. baumannii isolates [97]. However, the functionality of the A. baumannii T6SS in interbacterial competition varies depending on the strain. Indeed, while some strains did not show any T6SS-dependent killing in vitro, others employ the T6SS to efficiently out-compete different bacterial strains or species [97–99]. Expression of the T6SS varies significantly among different A. baumannii isolates [97,100] and is also affected by the presence of negative regulators of the T6SS in some conjugative resistance plasmids [101,102]. However, it has been shown that the activity of the T6SS in bacterial competition assays does not always correlate with the expression of T6SS genes [99]. Comparative genomic studies highlighted significant diversity in the range of putative T6SS effectors among A. baumannii isolates, many of which have unknown functions [103–105], leading to speculation that the different functionality of the T6SS might be related to the different sets of T6SS effectors carried by each strain. Such strain-dependent variability also makes it difficult to evaluate the impact of the T6SS on pathogenicity. An early report showed that the T6SS does not affect the virulence of the reference strain A. baumannii ATCC 17978 towards the amoeba Dictyostelium discoideum, the insect G. mellonella, and in a mouse model of pneumonia [97]. In contrast, delayed killing of G. mellonella larvae or decreased mortality in a mouse bacteraemia model were observed for T6SS-deficient mutants of A. baumannii DSM30011 and ATCC 19606T, respectively [99,106]. Moreover, the analysis of a large panel of A. baumannii isolates from patients with bacteraemia showed that strains with higher expression/activity of the T6SS better survive in human serum and were isolated more often from patients with catheter-related bloodstream infection and transplant recipients, arguing for the involvement of T6SS in A. baumannii pathogenicity [100]. Very recently, an A. baumannii T6SS effector with DNase activity (TafE) was demonstrated to be responsible for the T6SS-mediated killing of fungi [107]. However, TafE appears to enter the nucleus of fungal but not mammalian cells, consistent with its inability to affect human cell viability [108]. Thus, to what extent and by which mechanism(s) the T6SS contributes to A. baumannii fitness during infection remains an open question.
Outer membrane vesicles (OMVs)
Like other Gram-negative bacteria, Acinetobacter species release OMVs. Several studies have analysed the proteome of OMVs from different A. baumannii isolates, collectively identifying hundreds of OMV proteins [108–111]. As expected, many cell-associated virulence factors have been detected in OMVs, in line with the capability of A. baumannii OMVs to induce cytotoxicity and elicit pro-inflammatory responses in human cell lines [109,112]. The OmpA porin is the most abundant protein in A. baumannii OMVs. OmpA is important for OMV biogenesis and can be delivered by OMVs into the cytoplasm of host cells causing cytotoxicity [112–114], providing further evidence of the important role of this protein in A. baumannii pathogenicity. Notably, OMVs were also detected in lung tissues of mice with A. baumannii pulmonary infection [109], implying that OMVs could contribute to A. baumannii virulence in vivo. Finally, A. baumannii OMVs have also been shown to carry genetic material and efficiently mediate horizontal gene transfer, including the transmission of resistance plasmids [115,116], thus potentially contributing to the spread of antibiotic resistance.
Role of quorum sensing in A. baumannii pathogenicity
The population density and coordinated control of gene expression in A. baumannii are regulated by an intercellular communication quorum sensing (QS) system, that is homologous to the LuxI/LuxR QS system found in other Gram-negative bacteria. A. baumannii can release distinct N-acyl-homoserine lactones (AHLs) acting as QS signal molecules, that vary in length and oxidation state of the acyl chain and are likely produced by a single synthase, named AbaI. These signal molecules activate the intracellular cognate receptor AbaR, which controls at the transcriptional level the expression of QS-regulated genes, including the synthase gene abaI [117–119].
Several independent works have demonstrated that A. baumannii mutants defective in abaI or abaR are impaired in surface motility and in biofilm and pellicle formation [117–122], in line with the evidence that QS genes are overexpressed in biofilm cells [49]. Also, the phase-variable mechanism identified in A. baumannii AB5075, which results in the interconversion between the VIR-O and AV-T opaque and translucent colony phenotypes, was found to be affected by cell density and a diffusible extracellular signal, suggesting the involvement of QS system [123]. Recently, a genus-wide comparative genomic analysis found that the abaIR genes belong to an evolutionarily stable gene cluster (ESGC) that is conserved within the ACB complex, suggesting that the QS regulatory system might have played a role in the evolution of Acinetobacter towards pathogenicity [124].
On the other hand, the role of QS in A. baumannii virulence gene regulation is less clear. Several studies demonstrated that A. baumannii strains defective in QS signal production (abaI mutants) are less virulent in different animal models, including G. mellonella larvae, zebrafish embryos, and a mouse model of bacteraemia [122,125,126]. However, recent transcriptomic studies of the QS regulon in different A. baumannii strains (ATCC 17978, AB5075, and ATCC 19606T), showed that QS primarily controls a plethora of metabolic genes and the csu operon rather than well-known virulence determinants [126–128]. The only exception is the T6SS, which however was found to be downregulated only in the abaI mutant of ATCC 19606T [128]. Based on available evidence, the importance of QS for A. baumannii pathogenicity may be mainly due to its role in modulating bacterial physiology, biofilm formation, and in vivo fitness, rather than controlling the expression of secreted virulence factors as it is in other Gamma-proteobacteria.
Importance of metal metabolism for A. baumannii survival in vivo and pathogenicity
Transition metals, such as iron and zinc, are essential for the survival of all living organisms including bacterial pathogens. Metal ions, in addition to their structural and catalytic roles in proteins involved in various biological processes such as DNA replication, transcription, respiration, and oxidative stress responses, also exert a role in virulence gene regulation [129]. During infection, vertebrates undergo drastic alterations in metal metabolism, aiming to sequester and deprive pathogens of essential trace metals in a process recognized as nutritional immunity. To counteract the metal starvation imposed by the host, bacteria have evolved sophisticated strategies to withhold essential ions from the host metal-binding proteins [130].
Iron uptake systems
The complexity of A. baumannii iron uptake mechanisms reflects the exceptional adaptability of this organism in coping with iron limitation in various environments, including the host’s tissues and biological fluids. Up to six iron-acquisition gene clusters have been identified in A. baumannii, namely the Feo system, responsible for the ferrous iron uptake, three siderophore synthesis/transport systems, and two haem uptake systems [131–133].
The analysis of the distribution of A. baumannii iron uptake gene clusters on a large and representative genome dataset, composed of the most prevalent lineages, highlights that more than 98% of A. baumannii genomes have the Feo system, the clusters responsible for the synthesis and acquisition of the siderophores acinetobactin and baumannoferrin, and the HemT system for haem acquisition [133]
The Feo system is widely conserved among Gram-negative bacteria [134]. It consists of FeoA, a cytoplasmic protein with unknown function, FeoB, an integral membrane protein responsible for actively transporting of Fe2+ across the inner membrane, and FeoC, a putative transcriptional repressor (Figure 3). The contribution of the Feo system to A. baumannii virulence is still controversial, as independent studies provided apparently contrasting results. In one study, the deletion of feoB in A. baumannii ATCC 19606T resulted in a growth defect in human serum but did not reduce the lethality in G. mellonella larvae or intraperitoneally infected mice [135]. In contrast, other studies have demonstrated that the deletion of feoA in the ATCC 17978 strain significantly affects the capability of A. baumannii to adhere to human alveolar epithelial cells A549 and to kill intratracheally infected mice [136,137]. A transcriptomic analysis conducted on ATCC 17978 and the highly invasive MDR clinical isolate AbH12O-A2 also showed that the expression of iron uptake genes, including feoA, is upregulated during pneumonia in mice, supporting its role in sustaining A. baumannii growth in vivo [137]. The utilization of A. baumannii strains carrying different repertoires of iron uptake systems [133], and/or infection models can explain the apparently contrasting results obtained with feo mutants.
Figure 3.
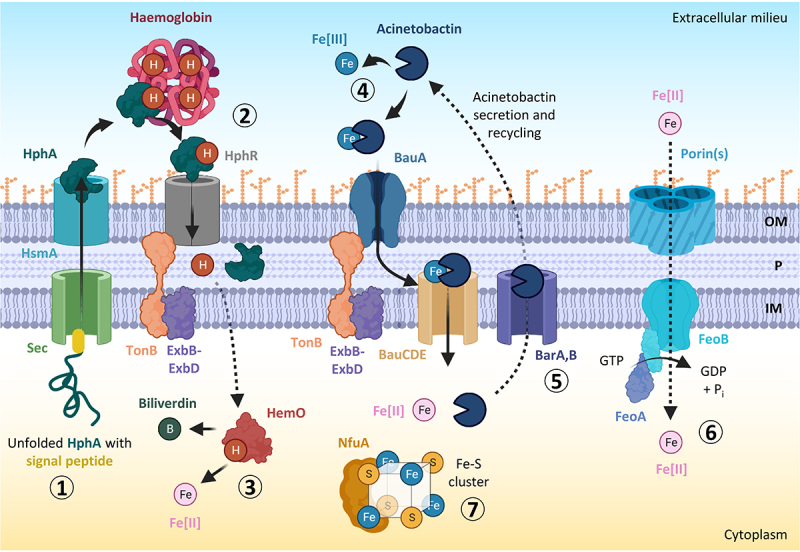
Iron acquisition, utilization, and storage mechanisms in A. baumannii. the haem scavenger HphA is translocated via the sec-dependent pathway and secreted outside the cell by the haemophilin secretion modulator HsmA (step 1). The haemophilin HphA delivers haem to its receptor, HphR, facilitating haem transport across the outer membrane by the concerted action of an inner membrane TonB complex (step 2). The enzyme HemO causes iron release from haem for use by the cell (step 3). The acinetobactin siderophore binds extracellular ferric iron (Fe[III]) with high affinity, and uses the BauA TonB-dependent outer membrane receptor and an inner membrane ATP-binding cassette (ABC) transporter composed of BauC, BauD, and BauE to deliver iron inside the cell (step 4). Acinetobactin can be recycled by the BarA/BarB siderophore secretion system (step 5). The GTP-dependent FeoAB system located in the cytoplasmic membrane is involved in the import of ferrous iron (Fe[II]) (step 6). NfuA is a cytoplasmic Fe-S cluster protein needed for intracellular iron storage and utilization (step 7). Abbreviations: OM, outer membrane; P, periplasm; IM, inner membrane. Figure created with Biorender.
Siderophores are high-affinity iron-chelators capable of scavenging iron from a variety of environmental sources. Besides acinetobactin and baumannoferrin, a gene cluster involved in the synthesis and acquisition of a third siderophore called fimsbactin has been identified in a few A. baumannii strains, including ATCC 17978 [131]. Although gene expression analysis in a mouse model of bacteriaemia demonstrated that A. baumannii ATCC 17978 expresses all siderophore gene clusters during infection, only the acinetobactin was found to be essential for the survival and proliferation in the host [51,138,139]. The crucial role of acinetobactin in A. baumannii virulence has also been demonstrated by using an acinetobactin-deficient ATCC 19606T mutant in G. mellonella larvae and mouse sepsis models [140]. The acinetobactin cluster consists of the genes basABCDEFGHIJ for the synthesis, bauABCDEF for the uptake, and barAB for the export of the siderophore [141] (Figure 3). Another essential gene for acinetobactin biosynthesis, entA, maps outside of the chromosomal region encompassing the acinetobactin gene cluster [141,142]. Acinetobactin is an extremely versatile chelator since it binds a range of metals in addition to iron, including Zn(II), Mn(II), Cu(II), Ni(II), and Co(II) [143]. The siderophore is composed of dihydroxybenzoic acid, threonine, and N-hydroxy histamine and exists as two pH-dependent isomers, one featuring oxazoline (Oxa), commonly called pre-acinetobactin, which is prevalent under acidic conditions, and the other with an isoxazolidinone (Isox), defined as acinetobactin, which is favoured under neutral and basic conditions [144]. Both isomers are capable of scavenging iron from transferrin and lactoferrin, thereby alleviating the nutritional challenge imposed by these proteins during infection [143]. The ability to prey iron from host proteins is not the only property of acinetobactin, since this siderophore has recently been shown to provide A. baumannii with a competitive advantage over some respiratory tract and skin commensal bacteria, such as Staphylococcus hominis and Corynebacterium striatum [145]. Interestingly, A. baumannii AB5075, a highly virulent isolate commonly used in pathogenicity studies, was able to inhibit the growth of the S. hominis and C. striatum, whereas acinetobactin-deficient mutants were not. In addition, purified acinetobactin exhibited growth inhibitory activity on both S. hominis and C. striatum [146]. Outcompeting commensal bacteria through iron competition may help to sustain A. baumannii colonization of the human host [145].
Most of the iron in the human body is retained by haem-containing proteins, primarily haemoglobin and other host proteins, such as serum albumin, haptoglobin, and haemopexin [147]. Consequently, the ability of pathogens to uptake haem from host proteins confers a significant advantage during infections. Two different haem uptake systems have been detected in A. baumannii, namely HemT and HemO [131,133]. All A. baumannii strains appear to possess the hemT genetic cluster, whereas the hemO cluster has only been found in some strains [131,133]. Although widely conserved in A. baumannii, the molecular properties of the HemT system and its role in A. baumannii pathogenesis remain elusive. On the other hand, several pieces of evidence indicate that the hemO cluster contributes to A. baumannii virulence. The hemO cluster is composed of two operons and two monocistronic genes. The monocistronic genes encode the TonB-dependent receptor, HphR, and the haemophore, a haem-scavenging secreted protein also named haemophilin or HphA. One of the operons encodes an extracytoplasmic function (ECF) sigma factor and its anti-sigma factor, while the other one encodes the haemophilin secretion modulator (HsmA), essential for HphA secretion, a TonB-related protein, the haem-degrading enzyme haem oxygenase (HemO), and a hypothetical protein with unknown function [148,149] (Figure 3). The role of the hemO cluster in haem uptake has been proven in the hypervirulent and MDR A. baumannii strain LAC-4, which grows in an iron-poor medium using haemin as the sole iron source, as opposed to A. baumannii ATCC 17978 which does not grow because lacking the hemO gene cluster [150]. The strain LAC-4 was isolated during a nosocomial outbreak in the Los Angeles country hospital in 2008 along with other 19 A. baumannii strains, all carrying the hemO cluster albeit belonging to different clonal lineages [151]. The pivotal role of the hemO cluster during infection has recently been demonstrated in A. baumannii AB5075, as it has been shown that an AB5075 knock-out mutant in the TonB-dependent receptor gene hphR was less virulent than the parental strain in both mouse sepsis and pulmonary infection models [149].
Unlike the Feo system, siderophore and haem uptake systems require the presence of a tripartite protein complex (TonB, ExbB, and ExbD) localized in the inner membrane and spanning the periplasmic space, called the TonB complex. The TonB complex converts the transmembrane proton gradient into the energy required to internalize the iron carrier (siderophore or haem) into the periplasm [152] (Figure 3). The TonB system is an attractive target for antimicrobials since its inhibition would impair growth under low iron conditions, hence during infection.
Intriguingly, three paralogues of the tonB gene (tonB1, tonB2, and tonB3) have been identified in A. baumannii genomes. Two of them (tonB1 and tonB3) are components of exbB-exbD-tonB operons, while tonB2 is a monocistronic gene. The analysis of individual tonB mutants in A. baumannii ATCC 19606T revealed that only tonB3 is essential for sustaining growth in conditions of limited iron availability [135]. Accordingly, tonB3 inactivation completely abrogated A. baumannii virulence both in G. mellonella and in a mice model of systemic infection, indicating that the TonB3 system, and consequently the ability to uptake iron from siderophores and/or haem, is essential for A. baumannii pathogenicity. The role of the other TonB systems is still elusive; it has been shown that TonB2 contributes by an unknown mechanism to the adhesion of A. baumannii to the human alveolar epithelial cells [153], while no information is available on TonB1. Intriguingly, while tonB1 and tonB2 individually are not required for virulence in the G. mellonella infection model, the disruption of both genes causes a significant reduction in the A. baumannii ability to kill the larvae [153].
Finally, the screening of a transposon library aimed at identifying genes essential for the growth of A. baumannii ATCC 19606T under iron-depleted conditions showed that the NfuA protein is also essential to ensure bacterial growth when iron is scarce [154]. NfuA is involved in the biogenesis of Fe-S clusters and is important for the utilization of intracellular iron and defence against oxidative stress [154]. Accordingly, the nfuA knock-out mutant showed reduced infectivity in G. mellonella, confirming the importance of intracellular iron metabolism for A. baumannii virulence [154].
Although iron is essential for bacterial growth and proliferation, excessive iron catalyzes the Fenton reaction, leading to the production of ROS that damage cellular macromolecules [155]. For this reason, the expression of genes involved in iron metabolism and uptake is tightly regulated by the ferric uptake regulator (Fur) repressor protein. Under iron-replete conditions, the iron-loaded Fur repressor binds the promoter of its target genes by recognizing a specific sequence called Fur box and blocks their transcription [156]. In silico analysis predicts the presence of Fur box in the promoter of ca. 80 genes of the A. baumannii strain ATCC 17978, including the genes involved in siderophore synthesis, tonB3, and in 15 genes encoding putative siderophore receptors [157]. Gene expression analysis in ATCC 17978 confirmed that the genes having the Fur box are overexpressed when A. baumannii grows under iron-depleted conditions [157].
Interestingly, two genes encoding phospholipases (plc1 and plc2) have been identified among the Fur-regulated genes. These genes were detected in different A. baumannii strains, including ATCC 19606T, ATCC 17978, and AYE. The study conducted on ATCC 19606T showed that mutation in plc1 or plc2 does not affect the cytolytic activity against erythrocytes or A549 alveolar epithelial cells, while the double mutant lacking both phospholipases has strongly reduced cytolytic activity [158]. The in vivo experiments demonstrated that only plc1 significantly contributes to A. baumannii ATCC 19606T virulence in G. mellonella caterpillars [158]. Although phospholipases are not directly involved in iron acquisition, their activity could result in the lysis of erythrocytes and the subsequent release of haemoglobin, which can be used to acquire haem as an iron source. Of note, PLC1 and PLC2 are not the only phospholipases identified in A. baumannii ATCC 19606T, since other non-Fur regulated genes encoding type D phospholipases were found (namely pld1, pld2, and pld3). The contribution of these phospholipases to host colonization has been investigated, demonstrating that the combined activity of all three phospholipases is required for the complete virulence of the A. baumannii type strain. Indeed, only the pld1, pld2, and pld3 triple knockout mutant exhibited reduced invasion of A549 alveolar basal epithelial cells and decreased virulence in G. mellonella [159].
Zinc metabolism
Zinc is the metal cofactor of numerous proteins involved in many essential processes, including transcription, amino acid biosynthesis, carbon metabolism, and DNA repair [129]. As part of nutritional immunity, zinc is sequestered by zinc-binding proteins in mammals, such as calprotectin. In a murine model of pneumonia using the A. baumannii strain ATCC 17978, it has been observed that calprotectin is present in the lungs within 6 hours following infection and disappears after bacterial clearance, consistent with the role of calprotectin in nutritional immunity [160].
A. baumannii overcomes zinc limitations by producing a high-affinity zinc-acquisition system. The genetic locus involved in zinc uptake consists of an operon, which encodes the zinc uptake regulator Zur, the ATPase ZnuC and the inner membrane transporter ZnuB, and two monocistronic genes for the periplasmic protein ZnuA and the TonB-dependent receptor ZnuD [161] (Figure 4). All these genes are required for A. baumannii growth under low zinc conditions or in the presence of calprotectin, except for znuD2 [161,162]. In vivo studies using sepsis and pneumonia mouse models demonstrated that deletion of the TonB-dependent receptor znuD in A. baumannii ATCC 17978 determines a decrease of bacterial burdens in the spleen, liver, and kidney, demonstrating the importance of zinc uptake for A. baumannii dissemination within the host [161].
Figure 4.
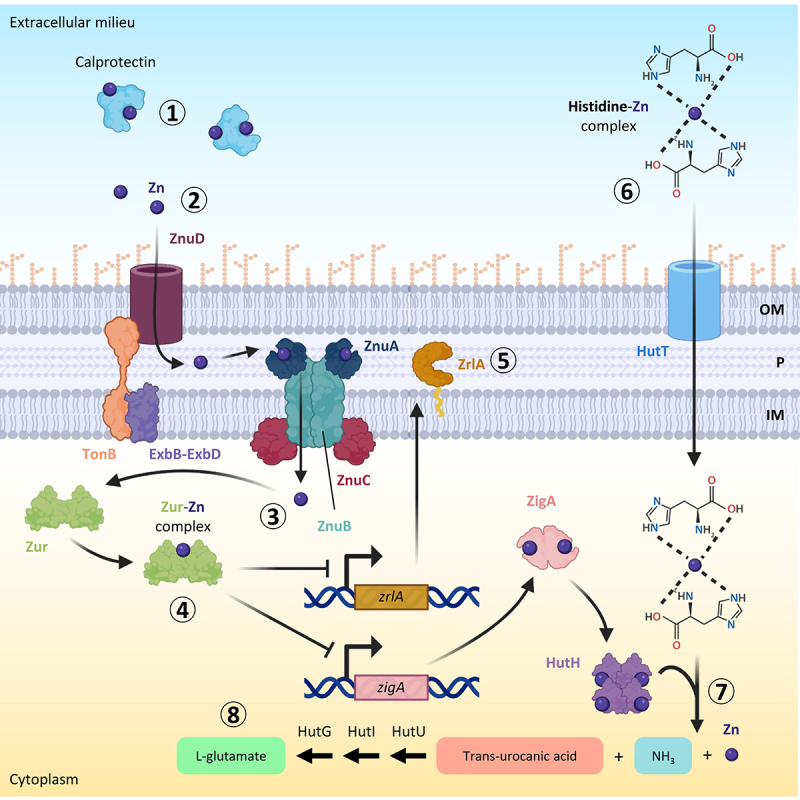
A. baumannii zinc uptake system and mechanism of in vivo zinc uptake. during infections, zinc is sequestered mainly by calprotectin (step 1). The A. baumannii TonB-dependent receptor ZnuD transports free zinc through the outer membrane (OM) (step 2). Once in the periplasm (P), zinc crosses the inner membrane (IM) and enters the cytoplasm through the ZnuABC system (step 3). Zur binds intracellular zinc and acts as a transcriptional repressor, modulating the expression of zinc-regulated genes, including zigA and zrlA (step 4). ZrlA is a putative peptidase involved in peptidoglycan remodelling and cell envelope integrity (step 5). The zinc-His complex can enter the cell through the HutT transporter (step 6). Once in the cytoplasm, this complex is cleaved into trans-urocanic acid, ammonia, and zinc by the Zn-dependent His ammonia-lyase HutH, activated by the metallochaperone ZigA with a still unknown mechanism (step 7). Trans-urocanic acid is eventually converted into L-glutamate by sequential reactions catalysed by HutU, HutI, and HutG (step 8). Figure created with Biorender.
The expression of zinc uptake genes is tightly regulated by Zur to balance the nutritional need for zinc with its toxicity. Similar to the role of Fur in iron uptake regulation, Zur acts as a transcriptional repressor of the zinc-regulated genes when the intracellular concentration of the metal is high, while under zinc-depleted conditions Zur repression is relieved and zinc uptake genes are expressed [163]. Zur was found to be essential for A. baumannii ATCC 17978 dissemination in a mouse model of pneumonia, indicating that both zinc-sensing and regulation of zinc acquisition are critical processes during infection [163]. Transcriptomic analysis of a Zur-defective A. baumannii ATCC 17978 mutant revealed that this protein not only regulates the Znu system but also other genes involved in zinc homoeostasis [163]. Among them, the most upregulated gene in the zur mutant or in the presence of calprotectin was zigA (Zur-induced GTPase A). ZigA binds zinc and has a zinc-stimulated GTPase activity. The zigA gene is located near the genes encoding the histidine utilization (Hut) system. This system consists of a putative histidine importer (HutT), and enzymes converting L-histidine to L-glutamate (HutH, HutU, HutI, HutG). Histidine coordinates free zinc in the cells and ZigA stimulates histidine catabolism via HutH during zinc starvation, making zinc free from histidine chelation [164]. ZigA was shown to contribute to A. baumannii growth under zinc-limiting conditions and the dissemination of bacterial cells in a mouse model of pneumonia [164].
The transcriptomic analysis of the A. baumannii zur mutant showed that Zur also regulates the expression of the gene zrlA [163]. ZrlA (Zur-regulated lipoprotein A) is a zinc-binding peptidase critical for cell envelope maintenance and cell morphology [165,166] (Figure 4). The role of ZrlA in virulence has been demonstrated in a mouse model of pneumonia, in which a zrlA mutant showed significantly reduced lung colonization [165]. Moreover, in line with the crucial role of ZrlA in the maintenance of membrane integrity, the mutant defective in this gene showed susceptibility to carbenicillin and vancomycin, whereas the wild-type strain was completely resistant [165].
Metabolic adaptation to the human host
The high A. baumannii genomic plasticity parallels its ability to utilize a broad spectrum of carbon and nitrogen sources, showcasing its adaptation to different environments, including the human host [167]. Although the complete list of potential carbon sources for Acinetobacter spp. is currently unavailable, more than 20 different metabolic pathways have been identified in the ACB complex [124], many of these shuttling metabolites that are prevalent in the human body into the carbohydrate metabolism of the bacterium [124]. Notably, despite the undisputed importance of glucose in human physiology, glucose serves as a carbon and energy source for a few environmental species in the genus Acinetobacter (e.g., Acinetobacter soli, Acinetobacter apis, and Acinetobacter baylyi), while no pathways for glucose degradation were identified in the ACB pathogenic species [124]. However, several A. baumannii strains that cannot utilize glucose are still able to degrade glucose to gluconolactone/gluconate [124], as demonstrated by the acidification of the medium in the presence of glucose [168,169].
Another example of human host adaptation is the capability of A. baumannii to use carnitine as sole carbon source [170,171], an amino acid essential for the oxidative catabolism of fatty acids in humans [172]. However, the importance of carnitine metabolism in A. baumannii virulence is only speculative, and no direct evidence has been currently provided [171].
The use of aromatic carbon sources represents a metabolic benefit for bacteria colonizing the human host, due to the high abundance of aromatics, such as hormones and amino acids [173]. Recent findings demonstrated that A. baumannii strains AYE and ATCC 19606T were able to grow on quinate, benzoate, kynurenic acid, and p-hydroxybenzoate as sole carbon and energy sources [174]. Interestingly, A. baumannii AYE mutants in pcaHG and catA, genes involved in p-hydroxybenzoate and benzoate catabolism, were impaired in complement resistance, underlying the deep intertwining between utilization of aromatic compounds and survival in the host [174]. Concerning the degradation of other aromatic compounds, it is noteworthy to mention the capability of A. baumannii to catabolize the phenylacetic acid, an intermediate in phenylalanine degradation. The paa operon involved in the phenylacetic acid degradation was found to be critical for antibiotic resistance and virulence in mice of the uropathogenic strain UPAB1 [52]. Similarly, previous findings have linked phenylacetic acid catabolism to virulence in the reference strains ATCC 19606T and ATCC 17978 [175,176]. Indeed, it was demonstrated that phenylacetic acid produced by A. baumannii is sensed by zebrafish neutrophils, which are attracted to the site of infection, leading to increased bacterial clearance and host survival [175].
Many genes putatively involved in the degradation of taurine, xanthine, 2-aminoethylphosphonate, vanillin/valinate, acyclic terpenes, and anthranilate were identified in A. baumannii [124], although experimental evidence is currently needed to confirm the capability to use these new potential carbon sources [124].
Overall, these results indicate that A. baumannii uses various strategies to acquire nutrients, even in the face of nutrient restriction by the host, and the intricate metabolism of this species may reveal still unexplored host-pathogen interaction mechanisms [167].
Resistance to xeric stress
Xeric stress, which is caused either by water evaporation (desiccation or matric stress) or excessive solute concentrations (hypertonicity or osmotic upshock) in the surrounding environment, subtracts water from cells [176–178], thereby triggering multiple mechanical and structural alterations of the cell. The complex nature of the consequences of this stress complicated the understanding of the main cause of death in bacteria undergoing desiccation. Different studies showed that the removal of water affects a wide array of macromolecules [176–178]. Bacterial membranes, proteins, and DNA endure modifications reflecting cellular stress in many ways. In strictly aerobic bacteria, like A. baumannii, mechanical shrinkage of membranes due to water loss causes respiratory complexes’ misfunction, leading to ROS formation. ROS can induce the formation of Maillard reactions, disulphide bridges, and oxidation of proteins. Moreover, due to crowding and hydration shell removal, damaged proteins can misfold or undergo denaturation, generating protein aggregates [177,178].
Xeric stress induces several DNA damages such as covalent modifications (e.g., the formation of hydantoin rings), cross-linking, and double-strand breaks, impairing DNA transcription and/or replication. At the same time, several membrane modifications occur, including i) transition from the liquid-crystalline to the gel phase, which generates membrane discontinuities that cause leakage of the cytoplasmic content, ii) aberrant interaction between inner and outer phospholipid leaflets, and iii) ROS-mediated chemical modifications of lipids (peroxidation and de-esterification) [176–179].
A. baumannii has evolved multiple strategies to survive when only scarce amounts of water molecules are available, enough to be considered a Gram-negative model organism to study osmostress responses [40,180,181] (Figure 5). Interestingly, xeric stress resistance has profound clinical implications, allowing A. baumannii to persist in hospital settings, and facilitating its transmission from contaminated surfaces to susceptible patients [182]. The intertwining between resistance to water loss and colonization of the nosocomial environment is testified by the fact that clinical isolates usually present higher desiccation resistance compared to laboratory-adapted strains [19,183,184]. However, the high variability in resistance to xeric stress [19,183] suggests that some of the main resistance mechanisms are not shared by all A. baumannii strains, likely due to differences in their genetic background.
Figure 5.
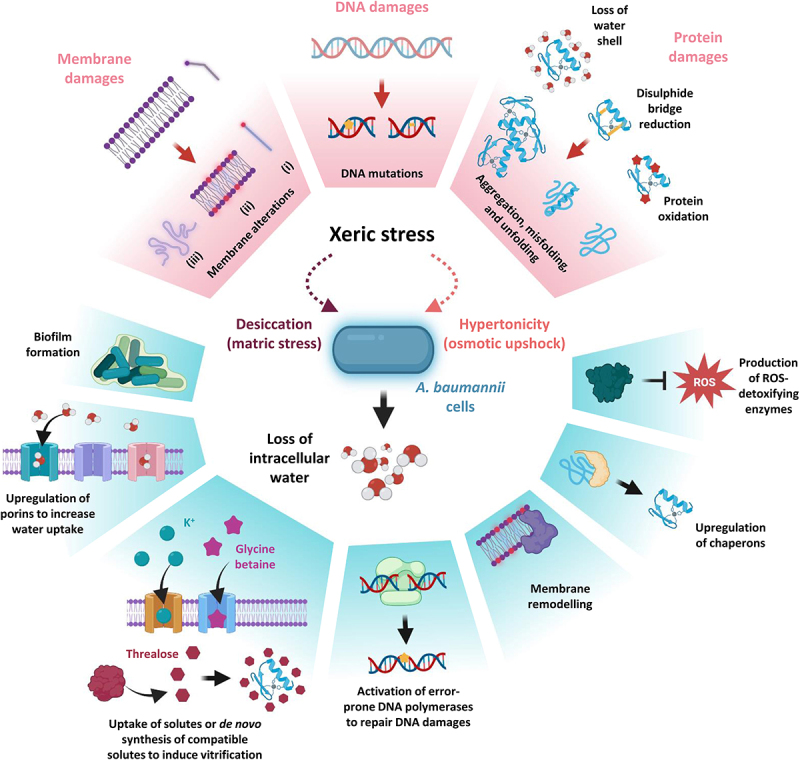
The effects of xeric stress on A. baumannii cells. The effects of xeric stress at the molecular level and A. baumannii responses are boxed in pink and cyan, respectively. During xeric stress, water loss causes lesions to membranes, DNA, and proteins (upper part of the figure). Among membrane damages, the saturation of aliphatic chains of phospholipids (i) results in the transition from a liquid crystalline to a gel phase (ii) causing membrane leakage and ruffling (iii). ROS increasing during dehydration causes DNA damage occurring through covalent modifications, single- and double-strand breaks, and cross-linking. ROS can induce the formation of disulphide bridges, Maillard reactions, and oxidation of proteins. Together with crowding and the removal of the hydration shell, damaged proteins can misfold, denature or lead to the formation of protein aggregates. The lower part of the figure shows the A. baumannii strategies to counteract the molecular damages caused by xeric stress (see text for details). Figure created with Biorender.
During desiccation, A. baumannii AbH12O-A2 increases its membrane permeability, as demonstrated by the overproduction of the outer membrane proteins Omp25, DcaP-like, and CarO [185], leading to hypothesize that the increased water uptake represents an attempt to recruit water molecules from atmospheric moisture.
To counteract the water loss due to hypertonicity, A. baumannii ATCC 19606T accumulates or synthesizes de novo compatible solutes, i.e., osmotically active compounds that do not interfere with physiological processes [186,187]. During hyperosmotic shock, A. baumannii cells firstly internalize K+ cations [188], a strategy also known as “salt-in-cytoplasm” that allows counterbalancing the extracellular osmolarity [186,189]. On the other hand, facing prolonged salt stress, A. baumannii accumulates, in the order, glutamate, mannitol, and trehalose or, if present, takes up glycine betaine from the environment [186,190,191]. The prominent role of compatible solutes in A. baumannii ATCC 19606T resistance to osmostress is corroborated by the fact that the growth of a ΔmtlD ΔotsB double mutant, a deletion mutant defective in the biosynthesis of both mannitol and trehalose, is significantly impaired at high NaCl concentrations [186]. However, unlike salt stress where solutes play a role in A. baumannii survival, not all compatible solutes positively affect resistance to desiccation. In fact, A. baumannii ATCC 19606T cells suspended in trehalose-containing media before desiccation displayed a significant advantage over those suspended in water or water supplemented with other compatible solutes [186]. This indicates that the responses to desiccation and hypertonicity overlap only in part, since in the former condition cells are in contact with air and not immersed in a solution, thus substance uptake is probably ineffective, and compatible solutes must be synthesized de novo. Moreover, while water stress can be mimicked by introducing solutes into a liquid environment, this method does not accurately replicate the conditions microorganisms face when exposed to dehydration. Dehydration entails the transition of water from the liquid phase to the gaseous phase, which this approach inadequately simulates [176,186,192].
The intracellular accumulation of trehalose observed in the A. baumannii type strain represents a common phenomenon in microorganisms adapted to xeric stress [177,178,193]. It was hypothesized that, when accumulated intracellularly in large quantities, trehalose can replace water molecules, determining a dramatic decrease in diffusion rates inside the cell, a phenomenon known as vitrification. Vitrification is considered a critical mechanism for conferring resistance to dehydration, with trehalose playing multiple roles as a chemical chaperone, osmolyte, and metabolite. Trehalose can directly or indirectly stabilize membranes and preserve protein folding. Additionally, it helps to slow down metabolic activity and prevent the accumulation of ROS [177,178,193].
Resistance to ROS-induced damage represents one of the main challenges that bacteria must overcome to cope with low water availability. For this reason, A. baumannii cells increase the expression of ROS-detoxifying enzymes during xeric conditions. In line with this observation, the two catalases KatG and KatE, able to convert the ROS-producing agent hydrogen peroxide into water and oxygen, were demonstrated to offer protection against desiccation [184,185,194] and oxidative burst of macrophages in different A. baumannii clinical isolates [195], highlighting the interconnection between osmostress resistance and host-pathogen interaction. In addition, A. baumannii ATCC 17978 has a superoxide dismutase (Sod2343), which can convert the superoxide anion O2− into the less toxic hydrogen peroxide, that was found to play a key role in oxidative stress resistance, virulence, and antibiotic susceptibility [196]. However, the role of Sod2343 in xeric stress resistance is still undetermined.
Many intracellular proteins, especially those that are denaturation-prone, lose their functional conformation because of cell dehydration. To face protein misfolding, different chaperones are upregulated in many A. baumannii clinical isolates experiencing both oxidative (GrpE, DnaK, GroES, GroEL) and desiccation (TF, GrpE, GroES, DnaK, DnaJ, ClpB, ClpX,) stresses (reviewed by Monem et al., 2020 [193]). Among the upregulated chaperones, the two predicted intrinsically disordered proteins (IDPs) DtpA and DtpB were found to promote desiccation tolerance in the A. baumannii strains ABUW 5075 and ATCC 17978, preventing protein denaturation or aggregation during dehydration conditions [197]. These hydrophilins, which are hydrophilic and highly charged, act as molecular shields protecting proteins and other cellular components from damage caused by desiccation and have been identified to be associated with xerotolerance in diverse organisms such as yeast, nematodes, tardigrades, and plant seeds [197]. The investigation of dtpA and dtpB orthologues in other members of the Acinetobacter genus led to the discovery of a desiccation resistance cassette [197]. This cassette contains genes encoding DtpA, DtpB, KatE, and umuCD, the latter coding for the error-prone DNA polymerase V (Pol V). Desiccation and rehydration cause various DNA lesions, including oxidation, alkylation, base removal, cross-linking, and strand breaks [176,177]. These DNA lesions would result in elevated mutagenesis rates when cells are rehydrated. To help prevent some of the DNA damages induced by desiccation and rehydration, A. baumannii relies on the protective role of the RecA protein, required for homologous recombination and recombination repair, and other proteins involved in the repairing of DNA lesions, such as the error-prone DNA polymerases Pol IV (encoded by dinB) and Pol V [198,199]. Interestingly, A. baumannii ATCC 17978 subjected to desiccation generated a higher frequency of rifampin-resistant mutants [198], leading to the provocative hypothesis that desiccation resistance impacts antibiotic resistance. The association between antibiotic resistance and desiccation tolerance is also supported by the fact that MDR clinical isolates are commonly more resistant to desiccation [19,183,184]. Interestingly, matric stress endurance is significantly reduced when desiccation-resistant strains are subjected to serial culturing in liquid media [184], corroborating the notion that environmental and clinical isolates are more resistant to dehydration compared to laboratory-adapted strains [19,183,184]. Furthermore, the A. baumannii biofilm-forming capacity, which is usually more pronounced in MDR clinical isolates [19,200], strongly contributes not only to boosting antibiotic resistance but also to desiccation survival [201,202]. Likewise, matric stress induces the biofilm-associated protein BAP [194], confirming the link between biofilm formation and defence from water loss.
Matric stress resistance strictly depends on environmental conditions, including relative humidity and temperature. Moreover, the growth phase at which the cells are harvested before dehydration can dramatically change bacterial viability: when bacteria are taken from the logarithmic phase, a dramatic reduction in their ability to survive desiccation is observed compared to those harvested from the stationary phase [180,181,184]. These results suggest that factors important for desiccation tolerance in A. baumannii are regulated in a growth phase-dependent manner. Among them, the two-component system BfmS/BfmR controls the expression of stress-related proteins during the stationary phase of growth [184]. Phosphorylation, which leads to the activation of BfmR, is controlled by the sensor histidine kinase BfmS and determines the upregulation of different stress-related proteins such as KatE, and the A. baumannii stress-related protein A (AbsA), whose role is not yet defined [184]. Accordingly, isogenic mutants in bfmR are more sensitive to nutrient starvation, NaCl-induced hyperosmotic shock, and desiccation compared to the wild-type strain [184]. Intriguingly, otsB (coding for the trehalose‐phosphatase involved in trehalose biosynthesis) and bfmS mutants showed impaired virulence in G. mellonella caterpillars [203], suggesting that A. baumannii in vivo survival may depend on the ability to cope with environmental stresses.
The importance of the BfmR/BfmS two-component system in bolstering desiccation resistance highlights the critical role of gene expression reshaping, enabling A. baumannii to mitigate the impacts of water loss [204]. During desiccation, it was observed that several genes responsible for transcription and translation processes showed altered expression levels in A. baumannii AbH12O-A2 [185]. Specifically, the RNA polymerase subunits RpoA and RpoC, proteins associated with ribosomes, and the elongation factor Tu were found to be downregulated [185]. Conversely, the ribosomal recycling factor (RRF), the integration host factor (IHF), and the histone-like protein HU exhibited upregulation [185]. RRF plays a role in facilitating ribosome disassembly upon completion of translation, while IHF and HU are involved in transcriptional regulation, crucial for maintaining DNA supercoiling and compaction [185]. Taken together these observations indicate a profound transcriptomic and proteomic readaptation during xeric stress. However, other regulatory mechanisms have recently been identified in different A. baumannii strains. In particular, the RNA-binding protein CsrA (carbon storage regulator A, also known as RsmA) is essential in desiccation resistance [194,205] by modulating the mRNA half-life of genes associated with xeric stress resistance such as katE, uspA (encoding the universal stress protein A UspA that plays a significant role in protecting A. baumannii from hydrogen peroxide and low pH [206]), bap, dtpA, and dtpB [194,199] (Figure 6).
Figure 6.
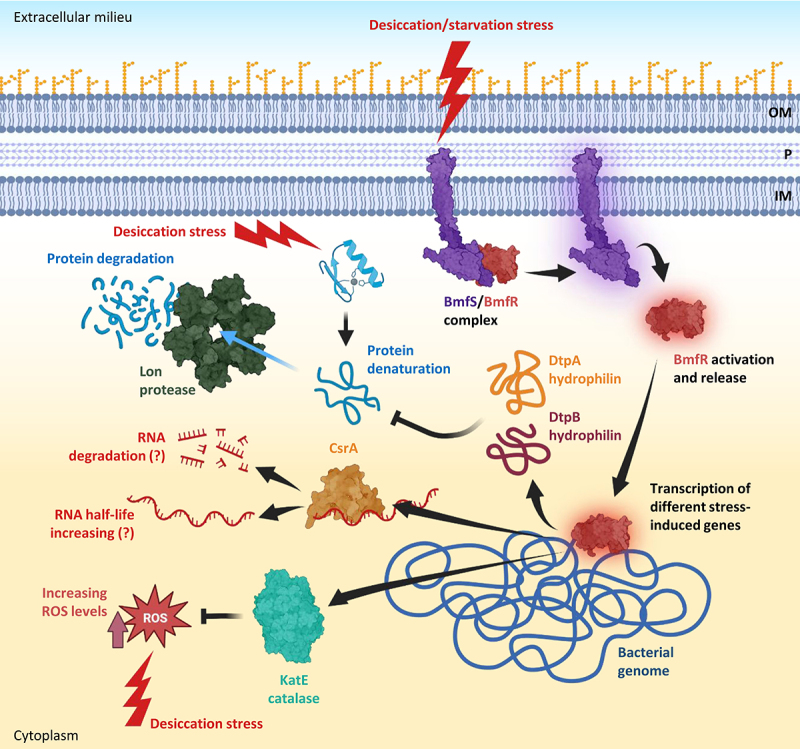
Molecular pathways induced by desiccation and starvation stresses in A. baumannii. desiccation and starvation stresses activate the two-component system BmfS/BmfR leading to the release of BmfR that acts as a transcriptional regulator. BmfR activates the expression of different genes, including those coding for the catalase KatE implicated in ROS detoxification, CsrA, an RNA-binding protein acting as a post-transcriptional regulator through modulation of the half-life of different mRnas, and the two hydrophilins DtpA and DtpB, acting as molecular chaperones that prevent protein denaturation. Denatured proteins that cannot be re-folded are degraded by the lon protease. Abbreviations: OM, outer membrane; P, periplasm; IM, inner membrane. Figure created with Biorender.
Dehydration stops cell growth, as inferred from the repression of genes involved in cell division in desiccated A. baumannii AbH12O-A2 cells [185]. For instance, the levels of FtsZ and MinD, proteins responsible for the formation of cell division septum, rapidly decrease during desiccation [185]. Moreover, matric stress induces the upregulation of HipA/B, a bacterial toxin−antitoxin system, responsible for the cell cycle arrest by translation inhibition: the HipA toxin acts as a kinase phosphorylating EF-TU, the translation elongation factor, whose levels decrease during desiccation, determining growth arrest [185,207]. These observations suggest that A. baumannii cells exposed to desiccation stress enter a dormant state [208]. Interestingly, the entrance in the dormancy phenotype is usually correlated with antibiotic persistence. Bacterial persister cells constitute a small fraction of bacterial populations able to survive to a bactericidal drug concentration without being resistant and are responsible for host clearance failure of the infection upon antibiotic treatment [209]. Although no direct comparison between antibiotic- and desiccation-induced metabolic quiescence mechanisms is available, the similarity between these two conditions raises the intriguing hypothesis that desiccation may induce the transition to persistence.
Concluding remarks and outlook
The ever-growing impact of A. baumannii infection on healthcare systems and the impressively rapid expansion of antimicrobial resistance in this organism have been paralleled by significant advances in the knowledge of its virulence potential, unveiling new features of its pathogenicity and environmental fitness.
A crucial feature of investigations of A. baumannii pathogenicity is the strain-dependent variability of virulence factors content. Many studies have been conducted on the reference strains ATCC 19606T and ATCC 17978, which were isolated from clinical specimens almost 70 years ago, and still represent valuable models to understand the biology of this pathogen. However, it is now clear that these reference strains do not possess the prototypical characteristics of currently circulating clinical A. baumannii strains. The use of the hypervirulent strain AB5075 was proposed a few years ago as a model to study A. baumannii pathogenesis and antibiotic resistance [146], but even this strain, belonging to ICL I, cannot be considered representative of the most successful clonal lineages circulating worldwide [15,16,18]. The difficulty in identifying a prototype strain for studying A. baumannii pathobiology lies in the great genetic diversity found within the A. baumannii species, as evidenced by the presence of approximately 20,000 genes constituting its pangenome [22], meaning that no single strain can be representative of the entire species. Such diversity poses the need for comparative studies, at both genome and phenotype levels, to assess the actual distribution and expression of individual factors in the global A. baumannii population [124]. Another critical issue in the assessment of A. baumannii pathogenicity is the animal models used to compare wild-type and attenuated isogenic mutants. While G. mellonella is a universally recognized and widely used animal model to verify the role of individual virulence factors in vivo, different mouse models of infection have also been proposed, and it is now clear that the contribution of virulence factors to A. baumannii pathogenicity varies depending on the animal model and the type of infection.
The success of A. baumannii in hospital settings is not only based on its ability to infect and spread among predisposed patients but also on its distinctive capacity to persist in the environment. As for other opportunistic pathogens, A. baumannii pathogenicity is multifactorial and the clinical success of this species relies on a combination of characteristics acting in concert to ensure bacterial survival inside and outside the human host. While several cell-associated and secreted virulence factors have been shown to influence pathogenicity, it is still difficult to define which A. baumannii virulence factors contribute most to the severity of A. baumannii infection.
Protein secretion systems are important for A. baumannii pathogenicity. Although proteomic studies demonstrated that these systems directly or indirectly affect the secretion of many extracellular and cell-associated virulence factors, the specific mechanism of secretion has been experimentally demonstrated for only a few of them. Moreover, experimental evidence suggests that there is some variability among strains at the level of secreted effectors, suggesting that each system may differently contribute to virulence, depending on the isolate and/or the site of infection. Since secretion systems have been proposed as targets for the development of drugs against A. baumannii drugs [59], their actual role in pathogenicity should be confirmed for a larger number of isolates and in different infection models.
Transition metals, particularly iron and zinc, have an essential nutritional function also in vivo, and the ability to acquire these metals during infection makes A. baumannii able to grow and proliferate in the host. Experimental evidence indicates that iron acquisition systems such as the acinetobactin siderophore, the hemO gene cluster implicated in haem uptake, and the TonB3 energy transducing system are crucial for A. baumannii pathogenicity in both insect and mouse models. Likewise, zinc uptake via the Znu system has experimentally been demonstrated to be crucial for the proliferation of A. baumannii during infection. The expression of iron- and zinc-uptake genes are controlled at transcriptional levels by Fur and Zur, respectively. Interestingly both the transcriptional regulators also modulate the expression of genes not directly related to the metal uptake, but essential for A. baumannii pathogenicity, such as phospholipases (Plc1 and Plc2) and zinc-binding peptidase (ZrlA), highlighting the interconnection between metal homoeostasis and virulence gene expression. The key role of metals in the pathogenicity and in the regulation of virulence gene expression makes metal metabolism the Achilles’ heel of A. baumannii that has already been exploited to develop new antimicrobial compounds, like siderophore-antibiotic conjugates, iron chelators, and iron mimetics [210–215].
Resistance against xeric stress, including both desiccation and hypertonicity, is crucial for the persistence of A. baumannii in healthcare settings. The increasing number of A. baumannii factors contributing to both host colonization and survival to xeric stress should stimulate the research of common mechanisms between these two processes. Proteins implicated in persistence under xeric stress conditions can represent promising targets for new inhibitory agents. However, our current understanding of the pathways and regulatory circuitry underlying this process remains incomplete. Since the xeric stress is inductive of A. baumannii dormancy [208] future research should be directed to unveil the cellular and molecular mechanisms behind the transition to the dormant state. This research could pave the way to the discovery of novel mechanisms of resistance to disinfectants and antibiotics, which is of undisputed importance for the prevention and control of A. baumannii infections.
Supplementary Material
Acknowledgements
We apologize to those colleagues whose work could not be cited in this paper due to space limitations.
Funding Statement
The authors acknowledge funding support from the Italian Ministry of University and Research (MUR), grants PRIN 2017 (no. 2017SFBER to R.Z. and no. 20177J5Y3P to P.V. and F.I.), grant PRIN 2020 (no. 20208LLXEJ to F.I.) and Excellence Departments grant to the Department of Science of the Roma Tre University (art. 1, comma 314-337 Legge 232/2016), from Regione Lazio (grant no. A0375-2020-36558 GAVAP to P.V.) and from the Rome Technopole (PNRR grant M4-C2-Inv). The authors also acknowledge the support of NBFC to Department of Science of the Roma Tre University (MUR PNRR, Missione 4 Componente 2, “Dalla ricerca all’impresa”, Investimento 1.4, Project CN00000033).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability statement
The authors confirm that the data supporting the findings of this study are available within the article or from the corresponding author [R.Z.] upon request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2023.2289769
References
- [1].Wong D, Nielsen TB, Bonomo RA, et al. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev. 2017;30(1):409–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sarshar M, Behzadi P, Scribano D, et al. Acinetobacter baumannii: an ancient commensal with weapons of a pathogen. Pathogens. 2021;10(4):387. doi: 10.3390/pathogens10040387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Whiteway C, Breine A, Philippe C, et al. Acinetobacter baumannii. Trends Microbiol. 2022;30(2):199–200. [DOI] [PubMed] [Google Scholar]
- [4].Migliaccio A, Bray J, Intoccia M, et al. Phylogenomics of Acinetobacter species and analysis of antimicrobial resistance genes. Front Microbiol. 2023;14:1264030. doi: 10.3389/fmicb.2023.1264030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].AIDA Study Group, Dickstein Y, Lellouche J, Ben Dalak Amar M, et al. Treatment outcomes of colistin- and carbapenem-resistant Acinetobacter baumannii infections: an exploratory subgroup analysis of a randomized clinical trial. Clin Infect Dis. 2019;69(5):769–776. [DOI] [PubMed] [Google Scholar]
- [6].Yusuf I, Skiebe E, Wilharm G.. Evaluation of CHROMagar Acinetobacter and MacConkey media for the recovery of Acinetobacter baumannii from soil samples. Lett Appl Microbiol. 2023;76(2):ovac051. doi: 10.1093/lambio/ovac051 [DOI] [PubMed] [Google Scholar]
- [7].Suresh S, Aditya V, Deekshit VK, et al. A rare occurrence of multidrug-resistant environmental Acinetobacter baumannii strains from the soil of Mangaluru, India. Arch Microbiol. 2022;204(7):422. doi: 10.1007/s00203-022-03035-0 [DOI] [PubMed] [Google Scholar]
- [8].Hamidian M, Maharjan RP, Farrugia DN, et al. Genomic and phenotypic analyses of diverse non-clinical Acinetobacter baumannii strains reveals strain-specific virulence and resistance capacity. Microb Genom. 2022;8(2):000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Furlan JPR, Pitondo-Silva A, Stehling EG. New STs in multidrug-resistant Acinetobacter baumannii harbouring β-lactamases encoding genes isolated from Brazilian soils. J Appl Microbiol. 2018;125(2):506–512. doi: 10.1111/jam.13885 [DOI] [PubMed] [Google Scholar]
- [10].Eveillard M, Kempf M, Belmonte O, et al. Reservoirs of Acinetobacter baumannii outside the hospital and potential involvement in emerging human community-acquired infections. Int J Infect Dis. 2013;17(10):e802–5. [DOI] [PubMed] [Google Scholar]
- [11].Lupo A, Vogt D, Seiffert SN, et al. Antibiotic resistance and phylogenetic characterization of Acinetobacter baumannii strains isolated from commercial raw meat in Switzerland. J Food Prot. 2014;77(11):1976–1981. [DOI] [PubMed] [Google Scholar]
- [12].Wilharm G, Skiebe E, Higgins PG, et al. Relatedness of wildlife and livestock avian isolates of the nosocomial pathogen Acinetobacter baumannii to lineages spread in hospitals worldwide. Environ Microbiol. 2017;19(10):4349–4364. doi: 10.1111/1462-2920.13931 [DOI] [PubMed] [Google Scholar]
- [13].WHO . Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of New antibiotics. Geneva: World Health Organization; 2017. [Google Scholar]
- [14].Murray CJ, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zarrilli R, Pournaras S, Giannouli M, et al. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents. 2013;41(1):11–19. doi: 10.1016/j.ijantimicag.2012.09.008 [DOI] [PubMed] [Google Scholar]
- [16].Higgins PG, Dammhayn C, Hackel M, et al. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;65(2):233–238. [DOI] [PubMed] [Google Scholar]
- [17].Hamidian, Nigro, Hamidian M, et al. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom. 2019;5(10):e000306. doi: 10.1099/mgen.0.000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gaiarsa S, Batisti Biffignandi G, Esposito EP, et al. Comparative analysis of the two Acinetobacter baumannii Multilocus sequence typing (MLST) schemes. Front Microbiol. 2019;10:930. doi: 10.3389/fmicb.2019.00930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Giannouli M, Antunes LC, Marchetti V, et al. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis. 2013;13:282. doi: 10.1186/1471-2334-13-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Valcek A, Philippe C, Whiteway C, et al. Phenotypic characterization and heterogeneity among modern clinical isolates of Acinetobacter baumannii. Microbiol Spectr. 2023;11(1):e0306122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chan AP, Sutton G, DePew J, et al. A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii. Genome Biol. 2015;16(1):143. doi: 10.1186/s13059-015-0701-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mangas EL, Rubio A, Álvarez-Marín R, et al. Pangenome of Acinetobacter baumannii uncovers two groups of genomes, one of them with genes involved in CRISPR/Cas defence systems associated with the absence of plasmids and exclusive genes for biofilm formation. Microb Genom. 2019;5(11):e000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Geisinger E, Huo W, Hernandez-Bird J, et al. Acinetobacter baumannii: Envelope Determinants That Control Drug Resistance, Virulence, and Surface Variability. Annu Rev Microbiol. 2019;73:481–506. doi: 10.1146/annurev-micro-020518-115714 [DOI] [PubMed] [Google Scholar]
- [24].Boll JM, Tucker AT, Klein DR, et al. Reinforcing lipid A acylation on the cell surface Of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. MBio. 2015;6(3):e00478–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Herrera CM, Voss BJ, Trent MS. Homeoviscous adaptation of the Acinetobacter baumannii outer membrane: alteration of lipooligosaccharide structure during cold stress. MBio. 2021;12(4):e0129521. doi: 10.1128/mBio.01295-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wyres KL, Cahill SM, Holt KE, et al. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb Genom. 2020;6(3):e000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Russo TA, Luke NR, Beanan JM, et al. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307–0294 is a major virulence factor. Infect Immun. 2010;78(9):3993–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chin CY, Tipton KA, Farokhyfar M, et al. A high-frequency phenotypic switch links bacterial virulence and environmental survival in Acinetobacter baumannii. Nat Microbiol. 2018;3:563–569. doi: 10.1038/s41564-018-0151-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tipton KA, Chin C-Y, Farokhyfar M, et al. Role of capsule in resistance to disinfectants, host antimicrobials, and desiccation in Acinetobacter baumannii. Antimicrob Agents Chemother. 2018;62:e01188–18. doi: 10.1128/AAC.01188-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tierney ARP, Chin CY, Weiss DS, et al. A LysR-Type transcriptional regulator controls multiple phenotypes in Acinetobacter baumannii. Front Cell Infect Microbiol. 2021;11:778331. doi: 10.3389/fcimb.2021.778331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pérez-Varela M, Tierney AR, Dawson E, et al. Stochastic activation of a family of TetR type transcriptional regulators controls phenotypic heterogeneity in Acinetobacter baumannii. PNAS Nexus. 2022;1(5):pgac231. doi: 10.1093/pnasnexus/pgac231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Talyansky Y, Nielsen TB, Yan J, et al. Capsule carbohydrate structure determines virulence in Acinetobacter baumannii. PLOS Pathog. 2021;17(2):e1009291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Silva L, Grosso F, Rodrigues C, et al. The success of particular Acinetobacter baumannii clones: accumulating resistance and virulence inside a sugary shield. J Antimicrob Chemother. 2021;76(2):305–31. [DOI] [PubMed] [Google Scholar]
- [34].Choi CH, Hyun SH, Lee JY, et al. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell Microbiol. 2008;10(2):309–319. doi: 10.1111/j.1462-5822.2007.01041.x [DOI] [PubMed] [Google Scholar]
- [35].Choi CH, Lee JS, Lee YC, et al. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol. 2008;8:216. doi: 10.1186/1471-2180-8-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gaddy JA, Tomaras AP, Actis LA. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun. 2009;77(8):3150–3160. doi: 10.1128/IAI.00096-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee JS, Choi CH, Kim JW, et al. Acinetobacter baumannii outer membrane protein a induces dendritic cell death through mitochondrial targeting. J Microbiol. 2010;48(3):387–392. [DOI] [PubMed] [Google Scholar]
- [38].Smani Y, Dominguez-Herrera J, Pachón J. Association of the outer membrane protein Omp33 with fitness and virulence of Acinetobacter baumannii. J Infect Dis. 2013;208(10):1561–1570. doi: 10.1093/infdis/jit386 [DOI] [PubMed] [Google Scholar]
- [39].Sánchez-Encinales V, Álvarez-Marín R, Pachón-Ibáñez ME, et al. Overproduction of outer membrane protein a by Acinetobacter baumannii as a risk factor for nosocomial pneumonia, bacteremia, and mortality rate increase. J Infect Dis. 2017;215(6):966–974. [DOI] [PubMed] [Google Scholar]
- [40].Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91–102. doi: 10.1038/nrmicro.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tomaras AP, Dorsey CW, Edelmann RE, et al. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology. 2003;149:3473–3484. doi: 10.1099/mic.0.26541-0 [DOI] [PubMed] [Google Scholar]
- [42].Pakharukova N, Malmi H, Tuittila M, et al. Archaic chaperone-usher pili self-secrete into superelastic zigzag springs. Nature. 2022;609(7926):335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pakharukova N, Tuittila M, Paavilainen S, et al. Structural basis for Acinetobacter baumannii biofilm formation. Proc Natl Acad Sci, USA. 2018;115(21):5558–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Harding CM, Tracy EN, Carruthers MD, et al. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. MBio. 2013;4(4):e00360–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ronish LA, Lillehoj E, Fields JK, et al. The structure of PilA from Acinetobacter baumannii AB5075 suggests a mechanism for functional specialization in Acinetobacter type IV pili. J Biol Chem. 2019;294(1):218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Loehfelm TW, Luke NR, Campagnari AA. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J Bacteriol. 2008;190:1036–1044. doi: 10.1128/JB.01416-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Brossard KA, Campagnari AA, Blanke SR. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect Immun. 2012;80(1):228–233. doi: 10.1128/IAI.05913-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].De Gregorio E, Del Franco M, Martinucci M, et al. Biofilm-associated proteins: news from Acinetobacter. BMC Genomics. 2015;16:933. doi: 10.1186/s12864-015-2136-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rumbo-Feal S, Gómez MJ, Gayoso C, et al. Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS One. 2013;8(8):e72968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Murray GL, Tsyganov K, Kostoulias XP, et al. Global Gene Expression Profile of Acinetobacter baumannii During Bacteremia. J Infect Dis. 2017;215(suppl_1):S52–S57. [DOI] [PubMed] [Google Scholar]
- [51].Islam MM, Kim K, Lee JC, et al. LeuO, a LysR-Type Transcriptional Regulator, Is Involved in Biofilm Formation and Virulence of Acinetobacter baumannii. Front Cell Infect Microbiol. 2021;11:738706. doi: 10.3389/fcimb.2021.738706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hooppaw AJ, McGuffey JC, Di Venanzio G, et al. The phenylacetic acid catabolic pathway regulates antibiotic and oxidative stress responses in Acinetobacter. MBio. 2022;13(3):e0186321. doi: 10.1128/mbio.01863-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Martí S, Rodríguez-Baño J, Catel-Ferreira M, et al. Biofilm formation at the solid-liquid and air-liquid interfaces by Acinetobacter species. BMC Res Notes. 2011;4:5. doi: 10.1186/1756-0500-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yoon EJ, Chabane YN, Goussard S, et al. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. MBio. 2015;6(2):e00309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Robin B, Nicol M, Le H, et al. MacAB-TolC contributes to the development of Acinetobacter baumannii biofilm at the solid–liquid interface. Front Microbiol. 2022;12:785161. doi: 10.3389/fmicb.2021.785161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Weber BS, Kinsella RL, Harding CM, et al. The secrets of Acinetobacter secretion. Trends Microbiol. 2017;25(7):532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Elhosseiny NM, Attia AS. Acinetobacter: an emerging pathogen with a versatile secretome. Emerg Microbes Infect. 2018;7(1):33. doi: 10.1038/s41426-018-0030-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li P, Zhang S, Wang J, et al. Uncovering the secretion systems of Acinetobacter baumannii: structures and functions in pathogenicity and antibiotic resistance. Antibiotics. 2023;12(2):195. doi: 10.3390/antibiotics12020195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Iacono M, Villa L, Fortini D, et al. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother. 2008;52(7):2616–2625. doi: 10.1128/AAC.01643-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Liu CC, Kuo HY, Tang CY, et al. Prevalence and mapping of a plasmid encoding a type IV secretion system in Acinetobacter baumannii. Genomics. 2014;104(3):215–223. doi: 10.1016/j.ygeno.2014.07.011 [DOI] [PubMed] [Google Scholar]
- [61].Spitz O, Erenburg IN, Beer T, et al. Type I secretion systems-One mechanism for all? Microbiol Spectr. 2019;7(2). doi: 10.1128/microbiolspec.PSIB-0003-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Abby SS, Cury J, Guglielmini J, et al. Identification of protein secretion systems in bacterial genomes. Sci Rep. 2016;6:23080. doi: 10.1038/srep23080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Harding CM, Pulido MR, Di Venanzio G, et al. Pathogenic Acinetobacter species have a functional type I secretion system and contact-dependent inhibition systems. J Biol Chem. 2017;292(22):9075–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sycz G, Di Venanzio G, Distel JS, et al. Modern Acinetobacter baumannii clinical isolates replicate inside spacious vacuoles and egress from macrophages. PLOS Pathog. 2021;17(8):e1009802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Goh HM, Beatson SA, Totsika M, et al. Molecular analysis of the Acinetobacter baumannii biofilm-associated protein. Appl Environ Microbiol. 2013;79(21):6535–6543. doi: 10.1128/AEM.01402-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Cianciotto NP, White RC, Maurelli AT. Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect Immun. 2017;85(5):e00014–17. doi: 10.1128/IAI.00014-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Korotkov KV, Sandkvist M. Architecture, function, and substrates of the type II secretion system. EcoSal Plus. 2019;8(2):10.1128. doi: 10.1128/ecosalplus.ESP-0034-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Naskar S, Hohl M, Tassinari M, et al. The structure and mechanism of the bacterial type II secretion system. Mol Microbiol. 2021;115(3):412–424. [DOI] [PubMed] [Google Scholar]
- [69].Eijkelkamp BA, Stroeher UH, Hassan KA, et al. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics. 2014;15(1):1020. doi: 10.1186/1471-2164-15-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Johnson TL, Waack U, Smith S, et al. Acinetobacter baumannii is dependent on the type II secretion system and its substrate LipA for lipid utilization and in vivo fitness. J Bacteriol. 2015;198(4):711–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Elhosseiny NM, El-Tayeb OM, Yassin AS, et al. The secretome of Acinetobacter baumannii ATCC 17978 type II secretion system reveals a novel plasmid encoded phospholipase that could be implicated in lung colonization. Int J Med Microbiol. 2016;306(8):633–641. [DOI] [PubMed] [Google Scholar]
- [72].Harding CM, Kinsella RL, Palmer LD, et al. Medically Relevant Acinetobacter Species Require a Type II Secretion System and Specific Membrane-Associated Chaperones for the Export of Multiple Substrates and Full Virulence. PLOS Pathog. 2016;12(1):e1005391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Jackson-Litteken CD, Di Venanzio G, Le NH, et al. InvL, an Invasin-Like Adhesin, Is a Type II Secretion System Substrate Required for Acinetobacter baumannii Uropathogenesis. MBio. 2022;13(3):e0025822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Elhosseiny NM, Elhezawy NB, Attia AS. Comparative proteomics analyses of Acinetobacter baumannii strains ATCC 17978 and AB5075 reveal the differential role of type II secretion system secretomes in lung colonization and ciprofloxacin resistance. Microb Pathog. 2019;128:20–27. doi: 10.1016/j.micpath.2018.12.039 [DOI] [PubMed] [Google Scholar]
- [75].Tilley D, Law R, Warren S, et al. CpaA a novel protease from Acinetobacter baumannii clinical isolates deregulates blood coagulation. FEMS Microbiol Lett. 2014;356(1):53–61. doi: 10.1111/1574-6968.12496 [DOI] [PubMed] [Google Scholar]
- [76].Kinsella RL, Lopez J, Palmer LD, et al. Defining the interaction of the protease CpaA with its type II secretion chaperone CpaB and its contribution to virulence in Acinetobacter species. J Biol Chem. 2017;292(48):19628–19638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Waack U, Warnock M, Yee A, et al. CpaA is a glycan-specific adamalysin-like protease secreted by Acinetobacter baumannii that inactivates coagulation factor XII. MBio. 2018;9(6):e01606–18. doi: 10.1128/mBio.01606-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Haurat MF, Scott NE, Di Venanzio G, et al. The Glycoprotease CpaA secreted by medically relevant Acinetobacter species targets multiple O-Linked host glycoproteins. MBio. 2020;11(5):e02033–20. doi: 10.1128/mBio.02033-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Bernstein HD. Type V secretion in Gram-negative bacteria. EcoSal Plus. 2019;8(2). doi: 10.1128/ecosalplus.ESP-0031-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Meuskens I, Saragliadis A, Leo JC, et al. Type V secretion systems: An overview of passenger domain functions. Front Microbiol. 2019;10:1163. doi: 10.3389/fmicb.2019.01163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Clarke KR, Hor L, Pilapitiya A, et al. Phylogenetic classification and functional review of autotransporters. Front Immunol. 2022;13:921272. doi: 10.3389/fimmu.2022.921272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bentancor LV, Camacho-Peiro A, Bozkurt-Guzel C, et al. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J Bacteriol. 2012;194(15):3950–3960. doi: 10.1128/JB.06769-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Tram G, Poole J, Adams FG, et al. The Acinetobacter baumannii autotransporter adhesin Ata recognizes host glycans as high-affinity receptors. ACS Infect Dis. 2021;7(8):2352–2361. [DOI] [PubMed] [Google Scholar]
- [84].Weidensdorfer M, Ishikawa M, Hori K, et al. The Acinetobacter trimeric autotransporter adhesin Ata controls key virulence traits of Acinetobacter baumannii. Virulence. 2019;10(1):68–81. doi: 10.1080/21505594.2018.1558693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bentancor LV, Routray A, Bozkurt-Guzel C, et al. Evaluation of the trimeric autotransporter Ata as a vaccine candidate against Acinetobacter baumannii infections. Infect Immun. 2012;80(10):3381–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hatefi Oskuei R, Alipour Astaneh S D, Rasooli I. A conserved region of Acinetobacter trimeric autotransporter adhesion, Ata, provokes suppression of Acinetobacter baumannii virulence. Arch Microbiol. 2021;203(6):3483–3493. doi: 10.1007/s00203-021-02343-1 [DOI] [PubMed] [Google Scholar]
- [87].Sun P, Li X, Pan C, et al. A Short Peptide of Autotransporter Ata Is a Promising Protective Antigen for Vaccination Against Acinetobacter baumannii. Front Immunol. 2022;13:884555. doi: 10.3389/fimmu.2022.884555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Pérez A, Merino M, Rumbo-Feal S, et al. The FhaB/FhaC two-partner secretion system is involved in adhesion of Acinetobacter baumannii AbH12O-A2 strain. Virulence. 2017;8(6):959–974. doi: 10.1080/21505594.2016.1262313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Darvish Alipour Astaneh S, Rasooli I, Mousavi Gargari SL. The role of filamentous hemagglutinin adhesin in adherence and biofilm formation in Acinetobacter baumannii ATCC19606(T). Microb Pathog. 2014;74:42–9. doi: 10.1016/j.micpath.2014.07.007 [DOI] [PubMed] [Google Scholar]
- [90].De Gregorio E, Esposito EP, Zarrilli R, et al. Contact-dependent growth inhibition proteins in Acinetobacter baylyi ADP1. Curr Microbiol. 2018;75(11):1434–1440. doi: 10.1007/s00284-018-1540-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].De Gregorio E, Zarrilli R, Di Nocera PP. Contact-dependent growth inhibition systems in Acinetobacter. Sci Rep. 2019;9(1):154. doi: 10.1038/s41598-018-36427-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Nguyen VS, Douzi B, Durand E, et al. Towards a complete structural deciphering of type VI secretion system. Curr Opin Struct Biol. 2018;49:77–84. doi: 10.1016/j.sbi.2018.01.007 [DOI] [PubMed] [Google Scholar]
- [93].Cherrak Y, Flaugnatti N, Durand E, et al. Structure and activity of the type VI secretion system. Microbiol Spectr. 2019;7(4). doi: 10.1128/microbiolspec.PSIB-0031-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Monjarás Feria J, Valvano MA. An overview of anti-eukaryotic T6SS effectors. Front Cell Infect Microbiol. 2020;10:584751. doi: 10.3389/fcimb.2020.584751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Jurėnas D, Journet L. Activity, delivery, and diversity of type VI secretion effectors. Mol Microbiol. 2021;115(3):383–394. doi: 10.1111/mmi.14648 [DOI] [PubMed] [Google Scholar]
- [96].Gallegos-Monterrosa R, Coulthurst SJ. The ecological impact of a bacterial weapon: microbial interactions and the type VI secretion system. FEMS Microbiol Rev. 2021;45(6):fuab033. doi: 10.1093/femsre/fuab033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Weber BS, Miyata ST, Iwashkiw JA, et al. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One. 2013;8(1):e55142. doi: 10.1371/journal.pone.0055142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Carruthers MD, Nicholson PA, Tracy EN, et al. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One. 2013;8(3):e59388. doi: 10.1371/journal.pone.0059388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Repizo GD, Gagné S, Foucault-Grunenwald ML, et al. Differential Role of the T6SS in Acinetobacter baumannii Virulence. PLoS One. 2015;10(9):e0138265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kim J, Lee JY, Lee H, et al. Microbiological features and clinical impact of the type VI secretion system (T6SS) in Acinetobacter baumannii isolates causing bacteremia. Virulence. 2017;8(7):1378–1389. doi: 10.1080/21505594.2017.1323164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Weber BS, Ly PM, Irwin JN, et al. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci U S A. 2015;112(30):9442–9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Di Venanzio G, Moon KH, Weber BS, et al. Multidrug-resistant plasmids repress chromosomally encoded T6SS to enable their dissemination. Proc Natl Acad Sci U S A. 2019;116(4):1378–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Fitzsimons TC, Lewis JM, Wright A, et al. Identification of novel Acinetobacter baumannii type VI secretion system Antibacterial effector and immunity pairs. Infect Immun. 2018;86(8):e00297–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lewis JM, Deveson Lucas D, Harper M, et al. Systematic Identification and analysis of Acinetobacter baumannii type VI secretion system effector and immunity components. Front Microbiol. 2019;10:2440. doi: 10.3389/fmicb.2019.02440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Repizo GD, Espariz M, Seravalle JL, et al. Bioinformatic analysis of the type VI secretion system and its potential toxins in the Acinetobacter genus. Front Microbiol. 2019;10:2519. doi: 10.3389/fmicb.2019.02519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Wang J, Zhou Z, He F, et al. The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS One. 2018;13(2):e0192288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Luo J, Chu X, Jie J, et al. Acinetobacter baumannii Kills Fungi via a Type VI DNase Effector. MBio. 2023;14(1):e0342022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Kwon SO, Gho YS, Lee JC, et al. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol Lett. 2009;297(2):150–156. [DOI] [PubMed] [Google Scholar]
- [109].Jin JS, Kwon SO, Moon DC, et al. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS One. 2011;6(2):e17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Mendez JA, Soares NC, Mateos J, et al. Extracellular proteome of a highly invasive multidrug-resistant clinical strain of Acinetobacter baumannii. J Proteome Res. 2012;11(12):5678–5694. [DOI] [PubMed] [Google Scholar]
- [111].Dhurve G, Madikonda AK, Jagannadham MV, et al. Outer Membrane Vesicles of Acinetobacter baumannii DS002 Are Selectively Enriched with TonB-Dependent Transporters and Play a Key Role in Iron Acquisition. Microbiol Spectr. 2022;10(2):e0029322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Jun SH, Lee JH, Kim BR, et al. Acinetobacter baumannii outer membrane vesicles elicit a potent innate immune response via membrane proteins. PLoS One. 2013;8(8):e71751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Moon DC, Choi CH, Lee JH, et al. Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J Microbiol. 2012;50(1):155–160. [DOI] [PubMed] [Google Scholar]
- [114].Tiku V, Kofoed EM, Yan D, et al. Outer membrane vesicles containing OmpA induce mitochondrial fragmentation to promote pathogenesis of Acinetobacter baumannii. Sci Rep. 2021;11(1):618. doi: 10.1038/s41598-020-79966-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Rumbo C, Fernández-Moreira E, Merino M, et al. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55(7):3084–3090. doi: 10.1128/AAC.00929-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Chatterjee S, Mondal A, Mitra S, et al. Acinetobacter baumannii transfers the blaNDM-1 gene via outer membrane vesicles. J Antimicrob Chemother. 2017;72(8):2201–2207. [DOI] [PubMed] [Google Scholar]
- [117].Niu C, Clemmer KM, Bonomo RA, et al. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J Bacteriol. 2008;190(9):3386–3392. doi: 10.1128/JB.01929-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Bhargava N, Sharma P, Capalash N. Quorum sensing in Acinetobacter: an emerging pathogen. Crit Rev Microbiol. 2010;36(4):349–360. doi: 10.3109/1040841x.2010.512269 [DOI] [PubMed] [Google Scholar]
- [119].Oh MH, Han K. AbaR is a LuxR type regulator essential for motility and the formation of biofilm and pellicle in Acinetobacter baumannii. Genes Genomics. 2020;42(11):1339–1346. doi: 10.1007/s13258-020-01005-8 [DOI] [PubMed] [Google Scholar]
- [120].Clemmer KM, Bonomo RA, Rather PN. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology (Reading). 2011;157(Pt 9):2534–2544. doi: 10.1099/mic.0.049791-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Mayer C, Muras A, Parga A, et al. Quorum sensing as a target for controlling surface associated motility and biofilm formation in Acinetobacter baumannii ATCC® 17978TM. Front Microbiol. 2020;11:565548. doi: 10.3389/fmicb.2020.565548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Sun X, Ni Z, Tang J, et al. The abaI/abar quorum sensing system effects on pathogenicity in Acinetobacter baumannii. Front Microbiol. 2021;12:679241. doi: 10.3389/fmicb.2021.679241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Tipton KA, Dimitrova D, Rather PN, et al. Phase-variable control of multiple phenotypes in Acinetobacter baumannii strain AB5075. J Bacteriol. 2015;197(15):2593–2599. doi: 10.1128/JB.00188-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Djahanschiri B, Di Venanzio G, S DJ, et al. Evolutionarily stable gene clusters shed light on the common grounds of pathogenicity in the Acinetobacter calcoaceticus-baumannii complex. PLoS Genet. 2022;18(6):e1010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Fernandez-Garcia L, Ambroa A, Blasco L, et al. Relationship between the quorum network (sensing/quenching) and clinical features of pneumonia and bacteraemia caused by A. baumannii. Front Microbiol. 2018;9:3105. doi: 10.3389/fmicb.2018.03105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Bhuiyan MS, Ellett F, Murray GL, et al. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc Natl Acad Sci U S A. 2016;113(34):9599–9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].López-Martín M, Dubern JF, Alexander MR, et al. AbaM Regulates Quorum Sensing, Biofilm Formation, and Virulence in Acinetobacter baumannii. J Bacteriol. 2021;203(8):e00635–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Xiong L, Yi F, Yu Q, et al. Transcriptomic analysis reveals the regulatory role of quorum sensing in the Acinetobacter baumannii ATCC 19606 via RNA-seq. BMC Microbiol. 2022;22(1):198. doi: 10.1186/s12866-022-02612-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Chandrangsu P, Rensing C, Helmann JD. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol. 2017;15(6):338–350. doi: 10.1038/nrmicro.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Murdoch CC, Skaar EP. Nutritional immunity: the battle for nutrient metals at the host-pathogen interface. Nat Rev Microbiol. 2022;20(11):657–670. doi: 10.1038/s41579-022-00745-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Antunes LC, Imperi F, Towner KJ, et al. Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res Microbiol. 2011;162(3):279–84. doi: 10.1016/j.resmic.2010.10.010 [DOI] [PubMed] [Google Scholar]
- [132].Cook-Libin S, Sykes EME, Kornelsen V, et al. Iron acquisition mechanisms and their role in the virulence of Acinetobacter baumannii. Infect Immun. 2022;90(10):e0022322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Artuso I, Poddar H, Evans BA, et al. Genomics of Acinetobacter baumannii iron uptake. Microb Genom. 2023;9(8):mgen001080. doi: 10.1099/mgen.0.001080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Lau CK, Krewulak KD, Vogel HJ. Bacterial ferrous iron transport: the Feo system. FEMS Microbiol Rev. 2016;40(2):273–98. doi: 10.1093/femsre/fuv049 [DOI] [PubMed] [Google Scholar]
- [135].Runci F, Gentile V, Frangipani E, et al. Contribution of active iron uptake to Acinetobacter baumannii pathogenicity. Infect Immun. 2019;87(4):e00755–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Álvarez-Fraga L, Vázquez-Ucha JC, Martínez-Guitián M, et al. Pneumonia infection in mice reveals the involvement of the feoA gene in the pathogenesis of Acinetobacter baumannii. Virulence. 2018;9(1):496–509. doi: 10.1080/21505594.2017.1420451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Martínez-Guitián M, Vázquez-Ucha JC, Álvarez-Fraga L, et al. Global Transcriptomic Analysis During Murine Pneumonia Infection Reveals New Virulence Factors in Acinetobacter baumannii. J Infect Dis. 2021;223(8):1356–1366. [DOI] [PubMed] [Google Scholar]
- [138].Sheldon JR, Skaar EP, Weiss DS. Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLOS Pathog. 2020;16(10):e1008995. doi: 10.1371/journal.ppat.1008995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Conde-Pérez K, Vázquez-Ucha JC, Álvarez-Fraga L, et al. In-depth analysis of the role of the acinetobactin cluster in the virulence of Acinetobacter baumannii. Front Microbiol. 2021;12:752070. doi: 10.3389/fmicb.2021.752070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Gaddy JA, Arivett BA, McConnell MJ, et al. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, galleria mellonella caterpillars, and mice. Infect Immun. 2012;80(3):1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Mihara K, Tanabe T, Yamakawa Y, et al. Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology. 2004;150(Pt 8):2587–2597. [DOI] [PubMed] [Google Scholar]
- [142].Penwell WF, Arivett BA, Actis LA, et al. The Acinetobacter baumannii entA gene located outside the acinetobactin cluster is critical for siderophore production, iron acquisition and virulence. PLoS One. 2012;7(5):e36493. doi: 10.1371/journal.pone.0036493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Kim M, Kim DY, Song WY, et al. Distinctive roles of two acinetobactin isomers in challenging host nutritional immunity. MBio. 2021;12(5):e0224821. doi: 10.1128/mBio.02248-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Shapiro JA, Wencewicz TA. Acinetobactin Isomerization Enables Adaptive iron acquisition in Acinetobacter baumannii through pH-Triggered siderophore swapping. ACS Infect Dis. 2016;2(2):157–168. doi: 10.1021/acsinfecdis.5b00145 [DOI] [PubMed] [Google Scholar]
- [145].Knauf GA, Powers MJ, Herrera CM, et al. Acinetobactin-mediated inhibition of commensal bacteria by Acinetobacter baumannii. mSphere. 2022;7(1):e0001622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Jacobs AC, Thompson MG, Black CC, et al. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the Evaluation of pathogenesis and antimicrobial treatments. MBio. 2014;5(3):e01076–14. doi: 10.1128/mBio.01076-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Choby JE, Skaar EP. Heme synthesis and acquisition in bacterial pathogens. J Mol Biol. 2016;428(17):3408–3428. doi: 10.1016/j.jmb.2016.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Giardina BJ, Shahzad S, Huang W, et al. Heme uptake and utilization by hypervirulent Acinetobacter baumannii LAC-4 is dependent on a canonical heme oxygenase (abHemo). Arch Biochem Biophys. 2019;672:108066. doi: 10.1016/j.abb.2019.108066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Bateman TJ, Shah M, Ho TP, et al. A slam-dependent hemophore contributes to heme acquisition in the bacterial pathogen Acinetobacter baumannii. Nat Commun. 2021;12(1):6270. doi: 10.1038/s41467-021-26545-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].de Léséleuc L, Harris G, KuoLee R, et al. Serum resistance, gallium nitrate tolerance and extrapulmonary dissemination are linked to heme consumption in a bacteremic strain of Acinetobacter baumannii. Int J Med Microbiol. 2014;304(3–4):360–9. doi: 10.1016/j.ijmm.2013.12.002 [DOI] [PubMed] [Google Scholar]
- [151].Ou HY, Kuang SN, He X, et al. Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: epidemiology, resistance genetic determinants and potential virulence factors. Sci Rep. 2015;5:8643. doi: 10.1038/srep08643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Wandersman C, Stojiljkovic I. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr Opin Microbiol. 2000;3(2):215–220. doi: 10.1016/s1369-5274(00)00078-3 [DOI] [PubMed] [Google Scholar]
- [153].Zimbler DL, Arivett BA, Beckett AC, et al. Functional features of TonB energy transduction systems of Acinetobacter baumannii. Infect Immun. 2013;81(9):3382–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Zimbler DL, Park TM, Arivett BA, et al. Stress response and virulence functions of the Acinetobacter baumannii NfuA Fe-S scaffold protein. J Bacteriol. 2012;194(11):2884–2893. doi: 10.1128/JB.00213-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373(1):1–6. doi: 10.1006/abbi.1999.1518 [DOI] [PubMed] [Google Scholar]
- [156].Daniel C, Haentjens S, Bissinger MC, et al. Characterization of the Acinetobacter baumannii Fur regulator: cloning and sequencing of the fur homolog gene. FEMS Microbiol Lett. 1999;170(1):199–209. [DOI] [PubMed] [Google Scholar]
- [157].Eijkelkamp BA, Hassan KA, Paulsen IT, et al. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics. 2011;12:126. doi: 10.1186/1471-2164-12-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Fiester SE, Arivett BA, Schmidt RE, et al. Iron-regulated phospholipase C activity contributes to the cytolytic activity and virulence of Acinetobacter baumannii. PLoS One. 2016;11(11):e0167068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Stahl J, Bergmann H, Göttig S, et al. Acinetobacter baumannii virulence is mediated by the concerted action of three phospholipases D. PLoS One. 2015;10(9):e0138360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Moore JL, Becker KW, Nicklay JJ, et al. Imaging mass spectrometry for assessing temporal proteomics: analysis of calprotectin in Acinetobacter baumannii pulmonary infection. Proteomics. 2014;14(7–8):820–828. doi: 10.1002/pmic.201300046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Hesse LE, Lonergan ZR, Beavers WN, et al. The Acinetobacter baumannii Znu System Overcomes Host-Imposed Nutrient Zinc Limitation. Infect Immun. 2019;87(12):e00746–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Hood MI, Mortensen BL, Moore JL, et al. Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLOS Pathog. 2012;8(12):e1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Mortensen BL, Rathi S, Chazin WJ, et al. Acinetobacter baumannii response to host-mediated zinc limitation requires the transcriptional regulator Zur. J Bacteriol. 2014;196(14):2616–2626. doi: 10.1128/JB.01650-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Nairn BL, Lonergan ZR, Wang J, et al. The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe. 2016;19(6):826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Lonergan ZR, Nairn BL, Wang J, et al. An Acinetobacter baumannii, Zinc-Regulated Peptidase Maintains Cell Wall Integrity during Immune-Mediated Nutrient Sequestration. Cell Rep. 2019;26(8):2009–2018.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Lee EK, Choi CH, Oh MH. Zur-regulated lipoprotein a contributes to the fitness of Acinetobacter baumannii. J Microbiol. 2020;58(1):67–77. doi: 10.1007/s12275-020-9531-7 [DOI] [PubMed] [Google Scholar]
- [167].Ren X, Palmer LD, Ottemann KM. Acinetobacter Metabolism in Infection and Antimicrobial Resistance. Infect Immun. 2023;91(6):e0043322. doi: 10.1128/iai.00433-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Juni E. Genetics and physiology of Acinetobacter. Annu Rev Microbiol. 1978;32:349–371. doi: 10.1146/annurev.mi.32.100178.002025 [DOI] [PubMed] [Google Scholar]
- [169].Schie BJ, Dijken JP, Kuenen JG. Non-coordinated synthesis of glucose dehydrogenase and its prosthetic group PQQ in Acinetobacter and pseudomonas species. FEMS Microbiol Lett. 1984;24(1):133–138. doi: 10.1111/j.1574-6968.1984.tb01259.x [DOI] [Google Scholar]
- [170].Zhu Y, Jameson E, Crosatti M, et al. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc Natl Acad Sci U S A. 2014;111(11):4268–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Breisch J, Waclawska I, Averhoff B. Identification and characterization of a carnitine transporter in Acinetobacter baumannii. Microbiologyopen. 2019;8(6):e00752. doi: 10.1002/mbo3.752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Reuter SE, Evans AM. Carnitine and acylcarnitines: pharmacokinetic, pharmacological and clinical aspects. Clin Pharmacokinet. 2012;51(9):553–572. doi: 10.1007/BF03261931 [DOI] [PubMed] [Google Scholar]
- [173].Teufel R, Mascaraque V, Ismail W, et al. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci U S A. 2010;107(32):14390–14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Breisch J, Huber LS, Kraiczy P, et al. The ß-ketoadipate pathway of Acinetobacter baumannii is involved in complement resistance and affects resistance against aromatic antibiotics. Environ Microbiol Rep. 2022;14(1):170–178. doi: 10.1111/1758-2229.13042 [DOI] [PubMed] [Google Scholar]
- [175].Cerqueira GM, Kostoulias X, Khoo C, et al. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J Infect Dis. 2014;210(1):46–55. [DOI] [PubMed] [Google Scholar]
- [176].Potts M. Desiccation tolerance of prokaryotes. Microbiol Rev. 1994;58(4):755–805. doi: 10.1128/mr.58.4.755-805.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lebre PH, De Maayer P, Cowan DA. Xerotolerant bacteria: surviving through a dry spell. Nat Rev Microbiol. 2017;15:285–296. doi: 10.1038/nrmicro.2017.16 [DOI] [PubMed] [Google Scholar]
- [178].Greffe VRG, Michiels J. Desiccation-induced cell damage in bacteria and the relevance for inoculant production. Appl Microbiol Biotechnol. 2020;104:3757–3770. doi: 10.1007/s00253-020-10501-6 [DOI] [PubMed] [Google Scholar]
- [179].Fardelli E, Lucidi M, Di Gioacchino M, et al. Bio-physical mechanisms of dehydrating membranes of Acinetobacter baumannii linked to drought-resistance. Biochim Biophys Acta (Biomembranes). 2022;1864(12):184045. [DOI] [PubMed] [Google Scholar]
- [180].Zeidler S, Müller V. Coping with low water activities and osmotic stress in Acinetobacter baumannii: significance, current status and perspectives. Environ Microbiol. 2019;21(7):2212–2230. doi: 10.1111/1462-2920.14565 [DOI] [PubMed] [Google Scholar]
- [181].Bashiri S, Lucidi M, Visaggio D, et al. Growth phase- and desiccation-dependent Acinetobacter baumannii morphology: an atomic force microscopy investigation. Langmuir. 2021;37(3):1110–1119. doi: 10.1021/acs.langmuir.0c02980 [DOI] [PubMed] [Google Scholar]
- [182].Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5(12):939–951. doi: 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- [183].Jawad A, Heritage J, Snelling AM, et al. Influence of relative humidity and suspending menstrua on survival of Acinetobacter spp. On dry surfaces. J Clin Microbiol. 1996;34(12):2881–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Farrow JM, Wells G, Pesci EC, et al. Desiccation tolerance in Acinetobacter baumannii is mediated by the two-component response regulator BfmR. PLoS One. 2018;13(10):e0205638. doi: 10.1371/journal.pone.0205638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Gayoso CM, Mateos J, Méndez JA, et al. Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J Proteome Res. 2014;13(2):460–476. [DOI] [PubMed] [Google Scholar]
- [186].Zeidler S, Müller V. The role of compatible solutes in desiccation resistance of Acinetobacter baumannii. Microbiologyopen. 2019;8(5):e00740. doi: 10.1002/mbo3.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Zeidler S, Müller V. Unusual deprivation of compatible solutes in Acinetobacter baumannii. Environ Microbiol. 2020;22(4):1370–1380. doi: 10.1111/1462-2920.14951 [DOI] [PubMed] [Google Scholar]
- [188].König P, Averhoff B, Müller V. A first response to osmostress in Acinetobacter baumannii: transient accumulation of K+ and its replacement by compatible solutes. Environ Microbiol Rep. 2020;12(4):419–423. doi: 10.1111/1758-2229.12857 [DOI] [PubMed] [Google Scholar]
- [189].Gunde-Cimerman N, Plemenitaš A, Oren A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol Revi. 2018;42(3):353–375. doi: 10.1093/femsre/fuy009 [DOI] [PubMed] [Google Scholar]
- [190].Zeidler S, Hubloher J, Schabacker K, et al. Trehalose, a temperature- and salt-induced solute with implications in pathobiology of Acinetobacter baumannii. Environ Microbio. 2017;19(12):5088–5099. [DOI] [PubMed] [Google Scholar]
- [191].Zeidler S, Hubloher J, König P, et al. Salt induction and activation of MtlD, the key enzyme in the synthesis of the compatible solute mannitol in Acinetobacter baumannii. Microbiologyopen. 2018;7(6):e00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Billi D, Potts M. Life and death of dried prokaryotes. Res Microbiol. 2002;153(1):7–12. doi: 10.1016/s0923-2508(01)01279-7 [DOI] [PubMed] [Google Scholar]
- [193].Monem S, Furmanek-Blaszk B, Łupkowska A, et al. Mechanisms protecting Acinetobacter baumannii against multiple stresses triggered by the host immune response, antibiotics and outside-host environment. Int J Mol Sci. 2020;21(15):5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Oda Y, Shapiro MM, Lewis NM, et al. CsrA-controlled proteins reveal new dimensions of Acinetobacter baumannii desiccation tolerance. J Bacteriol. 2022;204(4):e0047921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Sato Y, Unno Y, Miyazaki C, et al. Multidrug-resistant Acinetobacter baumannii resists reactive oxygen species and survives in macrophages. Sci Rep. 2019;9(1):17462. doi: 10.1038/s41598-019-53846-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Heindorf M, Kadari M, Heider C, et al. Impact of Acinetobacter baumannii superoxide dismutase on motility, virulence, oxidative stress resistance and susceptibility to antibiotics. PLoS One. 2014;9(7):e101033. doi: 10.1371/journal.pone.0101033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Green ER, Fakhoury JN, Monteith AJ, et al. Bacterial hydrophilins promote pathogen desiccation tolerance. Cell Host Microbe. 2022;30(7):975–987.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Norton MD, Spilkia AJ, Godoy VG. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J Bacteriol. 2013;195(6):1335–1345. doi: 10.1128/JB.02176-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Aranda J, López M, Leiva E, et al. Role of Acinetobacter baumannii UmuD homologs in antibiotic resistance acquired through DNA damage-induced mutagenesis. Antimicrob Agents Chemother. 2014;58(3):1771–1773. doi: 10.1128/AAC.02346-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Gurung J, Khyriem AB, Banik A, et al. Association of biofilm production with multidrug resistance among clinical isolates of Acinetobacter baumannii and Pseudomonas aeruginosa from intensive care unit. Indian J Crit Care Med. 2013;17(4):214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Espinal P, Martí S, Vila J. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J Hosp Infect. 2012;80(1):56–60. doi: 10.1016/j.jhin.2011.08.013 [DOI] [PubMed] [Google Scholar]
- [202].Choudhary M, Shrivastava R, Vashistt J. Acinetobacter baumannii biofilm formation: association with antimicrobial resistance and prolonged survival under desiccation. Curr Microbiol. 2022;79(12):361. doi: 10.1007/s00284-022-03071-5 [DOI] [PubMed] [Google Scholar]
- [203].Gebhardt MJ, Gallagher LA, Jacobson RK, et al. Joint transcriptional control of virulence and resistance to antibiotic and environmental stress in Acinetobacter baumannii. MBio. 2015;6(6):e01660–15. doi: 10.1128/mBio.01660-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Palethorpe S, Farrow JM, Wells G, et al. Acinetobacter baumannii regulates its stress responses via the BfmRS two-component regulatory system. J Bacteriol. 2022;204(2):e0049421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Farrow JM, Wells G, Palethorpe S, et al. CsrA supports both environmental persistence and host-associated growth of Acinetobacter baumannii. Infect Immun. 2020;88(12):e00259–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Elhosseiny NM, Amin MA, Yassin AS, et al. Acinetobacter baumannii universal stress protein A plays a pivotal role in stress response and is essential for pneumonia and sepsis pathogenesis. Int J Med Microbiol. 2015;305(1):114–123. [DOI] [PubMed] [Google Scholar]
- [207].Scherrer R, Moyed HS. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J Bacteriol. 1988;170(8):3321–6. doi: 10.1128/jb.170.8.3321-3326.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].König P, Wilhelm A, Schaudinn C, et al. The VBNC state: a fundamental survival strategy of Acinetobacter baumannii. MBio. 2023;e0213923(5). doi: 10.1128/mbio.02139-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Balaban NQ, Helaine S, Lewis K, et al. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol. 2019;17(7):441–448. doi: 10.1038/s41579-019-0196-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Bohac TJ, Fang L, Banas VS, et al. Synthetic mimics of native siderophores disrupt iron trafficking in Acinetobacter baumannii. ACS Infect Dis. 2021;7(8):2138–2151. [DOI] [PubMed] [Google Scholar]
- [211].Thompson MG, Corey BW, Si Y, et al. Antibacterial activities of iron chelators against common nosocomial pathogens. Antimicrob Agents Chemother. 2012;56(10):5419–5421. doi: 10.1128/AAC.01197-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].de Léséleuc L, Harris G, KuoLee R, et al. In vitro and in vivo biological activities of iron chelators and gallium nitrate against Acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56(10):5397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Hijazi S, Visaggio D, Pirolo M, et al. Antimicrobial activity of gallium compounds on ESKAPE pathogens. Front Cell Infect Microbiol. 2018;8:316. doi: 10.3389/fcimb.2018.00316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Antunes LC, Imperi F, Minandri F, et al. In vitro and in vivo antimicrobial activities of gallium nitrate against multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56(11):5961–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Abdul-Mutakabbir JC, Nguyen L, Maassen PT, et al. In Vitro Antibacterial Activity of Cefiderocol against Multidrug-Resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2021;65(9):e0264620. doi: 10.1128/AAC.02646-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article or from the corresponding author [R.Z.] upon request.


