Abstract
The germinating conidia of many phytopathogenic fungi on hosts must differentiate into an infection structure called the appressorium in order to penetrate their hosts. Chemical signals, such as the host’s surface wax or fruit ripening hormone, ethylene, trigger germination and appressorium formation of the avocado pathogen Colletotrichum gloeosporioides only after the conidia are in contact with a hard surface. What role this contact plays is unknown. Here, we describe isolation of genes expressed during the early stage of hard-surface treatment by a differential-display method and report characterization of one of these cloned genes, chip1 (Colletotrichum hard-surface induced protein 1 gene), which encodes a ubiquitin-conjugating enzyme. RNA blots clearly showed that it is induced by hard-surface contact and that ethylene treatment enhanced this induction. The predicted open reading frame (ubc1Cg) would encode a 16.2-kDa ubiquitin-conjugating enzyme, which shows 82% identity to the Saccharomyces cerevisiae UBC4-UBC5 E2 enzyme, comprising a major part of total ubiquitin-conjugating activity in stressed yeast cells. UBC1Cg can complement the proteolysis deficiency of the S. cerevisiae ubc4 ubc5 mutant, indicating that ubiquitin-dependent protein degradation is involved in conidial germination and appressorial differentiation.
Many phytopathogenic fungi must differentiate from the germ tube into an infection structure called the appressorium in order to penetrate hosts (10, 33, 34). Chemical and/or physical signals are known to trigger germination of and appressorium formation by fungal conidia (7, 9, 16–18). Some of the molecular events triggered by the physical signal in the bean rust fungus Uromyces appendiculatus (4, 37, 38) and the rice rust fungus Magnaporthe grisea have been studied (24). It has been known for a long time that contact with a hard surface is necessary for many fungi to induce appressorium formation (10). Conidia of Colletotrichum gloeosporioides are induced to germinate and differentiate to form appressoria by chemical signals, including the host surface wax (30) and a fruit ripening hormone, ethylene (11). However, contact with a hard surface is necessary for the chemical signals to induce appressorium formation. Conidia resting on either a hydrophilic hard surface (glass) or a hydrophobic hard surface responded to the chemical signals only between 2 and 4 h after the initiation of contact with the hard surface (11, 12, 20). Recently, four genes expressed uniquely during appressorium formation induced by the host surface wax were cloned by differential screening of a library produced by a subtractive hybridization approach (19, 20). Disruption of one of these genes drastically decreased its virulence for the host (19). However, the nature of the genes expressed during the 2 h of contact with the hard surface that primes the conidia to respond to the chemical signals is unknown.
To study molecular events triggered by hard-surface contact, genes expressed in C. gloeosporioides conidia during hard-surface treatment were examined by an mRNA differential-display method (25, 26). Here, we report that one of the genes expressed during hard-surface treatment encodes a ubiquitin-conjugating enzyme, which shows very high homology to the Saccharomyces cerevisiae UBC4-UBC5 enzyme pair, comprising a major part of total ubiquitin-conjugating activity in stressed yeast cells. We show that the C. gloeosporioides gene expressed in S. cerevisiae can complement the proteolysis deficiency of an S. cerevisiae ubc4 ubc5 mutant. These results suggest that expression of this ubc gene triggered by hard-surface contact mediates ubiquitin-dependent protein degradation associated with germination and appressorium formation.
MATERIALS AND METHODS
Fungal and bacterial strains and materials.
C. gloeosporioides, an isolate from avocado, was kindly provided by Dov Prusky (Volcani Center, Bet-Dagan, Israel). Cultures were maintained at 25°C on potato dextrose agar. Conidia were obtained by gently scraping 5- to 7-day-old cultures in petri dishes flooded with sterilized distilled water, as described previously (19, 20). Escherichia coli DH5α was used for propagating all plasmids. Restriction and modification enzymes and Taq DNA polymerase were from Life Technologies, Inc. (Bethesda Research Laboratories [BRL]).
RNA preparation.
Conidia of C. gloeosporioides (∼5 × 106 conidia/dish) were spread into petri dishes (150 by 15 mm) containing 30 ml of water and were incubated for various periods of time. The conidia were harvested by scraping them off the petri dishes with a rubber policeman (Fisher Scientific, Cincinnati, Ohio) and were subjected to centrifugation at 12,000 × g for 15 min as described previously (19, 20). For large-scale total-RNA isolation, the conidia from at least 50 petri dishes were resuspended in a solution containing 4.5 M guanidinium thiocyanate, 50 mM EDTA (pH 8.0), 100 mM β-mercaptoethanol, 25 mM sodium citrate (pH 7.0), and 2% sodium N-lauroylsarcosine (3 to 5 ml) and disrupted for 5 min with 425- to 600-μm-diameter glass beads in a mini-bead beater (Biospec Products, Bartlesville, Okla.). The total RNA was isolated by density gradient centrifugation through CsCl (3). For small-scale total-RNA isolation, the conidia from ∼10 petri dishes were suspended in 500 μl of homogenization buffer (50 mM LiCl, 25 mM Tris-HCl [pH 8.0], 35 mM EDTA, 35 mM EGTA, 0.5% sodium dodecyl sulfate [SDS]) and 500 μl of phenol-chloroform (1:1) and disrupted for 5 min with 425- to 600-μm-diameter glass beads in a mini-bead beater. The aqueous phase was then extracted with 500 μl of chloroform, and RNA was precipitated with an equal volume of 4 M LiCl. The RNA pellet was washed with 500 μl of 2 M LiCl and then with 70% ethanol.
Differential display of mRNA.
Total RNA was treated with amplification-grade RNase-free DNase I (BRL) at 37°C for 30 min to remove possible DNA contamination. The RNA concentration was calculated from the absorbance at 260 nm. The differential-display procedure recommended by the manufacturer (GenHunter Corporation, Brookline, Mass.) was followed. For first-strand cDNA synthesis, a 19-μl mixture containing 0.5 μg of total RNA, 4 pmol of oligo(dT) primer 5′-HT11M-3′ (where M may be G, A, or C), 400 pmol of deoxynucleoside triphosphate (dNTP), 25 mM Tris-HCl (pH 8.3), 37.6 mM KCl, 1.5 mM MgCl2, and 5 mM dithiothreitol was heated at 65°C for 5 min. The temperature was then reduced to 37°C, and after 10 min, 1 μl of Moloney murine leukemia virus reverse transcriptase (200 U) was added and incubation was continued at 37°C for another 50 min. Finally, the 20-μl reaction mixture was heated at 75°C for 5 min and then chilled to 4°C. For PCR, 2 μl of first-strand cDNA solution was added to a mixture (18 μl) containing 1.5 U of Taq DNA polymerase (BRL), 2.2 μM dNTP, 0.22 μM oligo(dT) primer 5′-HT11M-3′, 0.22 μM arbitrary decanucleotide primer, 11.1 μM Tris-Cl (pH 8.4), 55.6 mM KCl, 1.67 mM MgCl2, and 0.0011% gelatin. The reaction was carried out in a programmable thermal controller (MJ Research, Watertown, Mass.) as follows: 94°C (30 s), 40°C (2 min), and 72°C (30 s) for 40 cycles. The additional final extension step was performed at 72°C for 5 min. Each PCR product (3.5 μl) was mixed with 2 μl of loading dye (95% formamide, 10 mM EDTA [pH 8.0], 0.09% xylene cyanole FF, and 0.09% bromophenol blue) and incubated at 80°C for 2 min immediately before being loaded onto a 6% DNA sequencing gel. The gel was run at 60 W for ∼3 h, placed on a piece of 3M paper, vacuum dried at 80°C for 1 h, and exposed to X-ray film. For reamplification of the cDNA probe, gel segments representing DNA bands of interest were cut out with razors, each gel slice along with the 3M paper was soaked in 100 μl of water for 10 min, and DNA was eluted by boiling for 15 min and precipitated with ethanol in the presence of 50 μg of glycogen as a carrier. To reamplify the DNA fragments, 4 μl of the total 10 μl of eluted DNA was mixed with 36 μl of a reaction mixture containing 3 U of Taq DNA polymerase (BRL), 2.2 μM dNTP, 0.22 μM oligo(dT) primer 5′-T11M-3′, 0.22 μM arbitrary decanucleotide primer, 11.1 μM Tris-Cl (pH 8.4), 55.6 mM KCl, 1.67 mM MgCl2, and 0.0011% gelatin. The PCR conditions were the same as those described above. Finally, the amplified DNA fragments were cloned into a pCRII vector (Invitrogen, Carlsbad, Calif.). Double-stranded plasmid DNAs were prepared by the alkaline lysis-polyethylene glycol precipitation method (31) and used directly for automated sequencing with a model 373A sequencer from Applied Biosystems (Foster City, Calif.).
Isolation of C. gloeosporioides full-length cDNA by 5′ rapid amplification of cDNA ends (RACE) and sequence analysis.
To obtain the upstream nucleotide sequence, an internal specific primer (5′-GTG CTC CTA ACT CTG ATC GGT C-3′) and Lambda ZAP vector primers (T7 and T3) were used for PCR, with a Lambda ZAP cDNA library from hard-surface-treated conidia as a template. A Lambda ZAP cDNA library was prepared according to the manufacturer’s instructions (Stratagene). The PCR was initiated by denaturation at 94°C for 2.5 min and then carried out for 40 cycles as follows: 94°C (25 s), 54°C (35 s), and 72°C (1.5 min). The additional final extension step was performed at 72°C for 8 min. The ∼1-kb PCR product was purified from the 1% agarose gel with a Geneclean kit (Bio 101, Vista, Calif.), cloned into a pCRII vector (TA cloning kit; Invitrogen), and sequenced as indicated above. The DNA sequence from both strands was analyzed with DNA Stride 1.2. Amino acid homology searches were conducted with the BLAST program from the National Center for Biotechnology Information (1). Homology comparison was performed with the SeqApp program.
RNA blot analysis.
Total RNA isolated from conidia or germinating conidia of C. gloeosporioides was dissolved in a solution containing 50% formamide, 16% formaldehyde, 20 mM MOPS [3-(N-morpholino)propanesulfonic acid], 5 mM sodium acetate, and 1 mM EDTA (pH 7.0), incubated for 15 min at 65°C, and chilled on ice. Denatured samples were subjected to electrophoresis on 1% agarose gels containing 2.2 M formaldehyde and were blotted onto Nytran membranes. The blots were prehybridized for ∼4 h at 65°C in a solution containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 7.6]), 2× Denhardt’s solution, 0.1% SDS, and 100 μg of sheared salmon DNA/μl and hybridized for ∼16 h in the same solution with 106 cpm of a 32P-labeled cDNA probe/ml prepared by randomly primed labeling. The membranes were washed twice for 10 min at room temperature in 2× SSC plus 0.1% SDS, briefly washed at 65°C with 0.2× SSC plus 0.1% SDS, and exposed to X-ray film at −80°C in the presence of an intensifying screen. 32P was quantitated with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Southern blot analysis.
Genomic DNA was isolated from mycelium grown in mineral medium (15) containing 1% yeast extract and 1% glucose with shaking (200 rpm) for 36 h. The genomic DNA was digested to completion with restriction enzymes, subjected to electrophoresis on 1% agarose gels, and transferred to Nytran membranes. The conditions for prehybridization, hybridization, and washing were the same as those described above for RNA blots. The ubcCg genomic DNA was amplified by PCR with a 5′ noncoding region primer (5′-GAC TCT CAC AAT CCA AAT CAA AAG-3′) and the internal specific primer (5′-GTG CTC CTA ACT CTG ATC GGT C-3′). The PCR was initiated by denaturation at 94°C for 2.5 min and was then carried out for 38 cycles as follows: 94°C (25 s), 54°C (35 s), and 72°C (1.5 min). The additional final extension step was performed at 72°C for 8 min.
Yeast complementation.
The S. cerevisiae ubc4 ubc5 double mutant [Y0096; his3-Δ200 leu2-3,2-112 lys2-801 trp1-1(Am) ura3-52 ubc4::HIS3 ubc5::LEU2] was kindly provided by Stefan Jentsch, Friedrich Miescher Laboratory, Heidelberg, Germany. The ubc1Cg cDNA was cloned into the EcoRI site in both orientations of a low-copy-number yeast expression vector, pBM272 (kindly provided by Douglas Johnson, University of Vermont, Burlington), under the control of a GAL10 promoter with a URA3 selectable marker. Plasmids with inserts in both orientations with regard to the GAL10 promoter, as well as the plasmid without any insert, were used to transform the Y0096 strain. Yeast transformation was carried out according to standard protocols (3). Ura+ transformants were obtained in synthetic complete medium lacking uracil with 2% glucose (SC−U) at 30°C. They were streaked onto SC−U plates and SC(Gal)−U plates (plates with synthetic complete medium lacking uracil with 2% galactose) and incubated at either 30 or 37°C.
Nucleotide sequence accession number.
The nucleotide sequence for the ubc1Cg cDNA is in the GenBank database under accession no. AF030296.
RESULTS
Differential display of RNA from C. gloeosporioides during hard-surface contact.
Total RNAs from hard-surface-treated (2 h) or control (untreated) conidia were reverse transcribed with primers as indicated in Materials and Methods. Products were amplified by using combinations of eight arbitrary 5′ decamers and three oligo(dT) HT11M primers. Figure 1 shows the area of a differential-display gel including the amplified products obtained with primer combination HT11A and H-AP2 or HT11A and H-AP3. A band representing an enhanced level of expression of a gene during the hard-surface treatment is present at ∼190 bp. The same pattern was observed when PCR and electrophoresis were repeated. When the ∼190-bp DNA band recovered from the gel was amplified by PCR and used as a probe for Northern blot analysis, two transcripts were found: a strongly hybridizing band at ∼1 kb and a much less strongly hybridizing band at ∼2.4 kb. Both transcripts were induced by 2 h of hard-surface treatment (data not shown). The reamplified PCR product was used directly as the substrate for automated sequencing with the 5′ decamer as the primer. The sequence is shown in Fig. 2. When the PCR product was cloned and independent clones were sequenced, four different sequences were found; one of them was identical to that underlined in Fig. 2. Since the direct sequencing of the PCR product gave this sequence, further studies were focused on this clone, which we designated chip1 (for Colletotrichum hard-surface-induced protein 1).
FIG. 1.
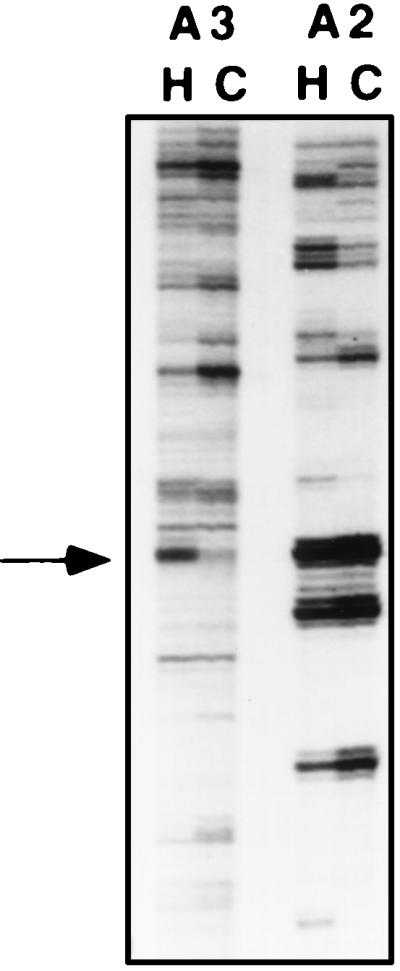
Area of a differential-display gel showing the amplified products obtained with primer combinations of oligo(dT) primer HT11A and arbitrary 5′ decamer H-AP2 (A2) or H-AP3 (A3) by using as templates cDNAs derived from conidia treated on a hard surface for 2 h (H) and an untreated control (C). The amplified product of interest is indicated by an arrow at ∼190 bp. Experimental details are provided in the text.
FIG. 2.
Nucleotide sequence and deduced amino acid sequence of the cloned cDNA fragment showing the ORF coding for UBC1Cg. The sequence of the differential-display product is underlined, and the specific internal primer used for 5′ RACE is in boldface and is underlined, as is the active site, Cys, required for ubiquitin-thioester formation.
Isolation of full-length cDNA for CHIP1 by 5′ RACE and sequence analysis.
To obtain the upstream nucleotide sequence, an internal specific primer (5′-GTG CTC CTA ACT CTG ATC GGT C-3′) and Lambda ZAP vector primers (T7 and T3) were used for PCR, with a Lambda ZAP cDNA library of hard-surface-treated conidia as a template. An ∼1-kb PCR product was obtained with the internal specific primer and T7 vector primer. Cloning and sequencing of this product revealed an open reading frame (ORF) that would encode a 147-amino-acid protein with a deduced molecular mass of 16.2 kDa (Fig. 2). The DNA sequence surrounding the ATG translation start site (underlined) (GCCAAAATGGC) has a conserved Kozak sequence found in filamentous fungi (CA[C/A][A/C]ATGNC) and closely resembles the Kozak sequence from mammals (GCC[A/G]CCATGG) (14, 23).
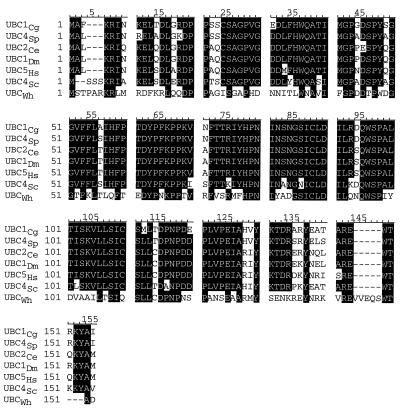
The protein that is predicted to be encoded by this ORF shows very high homology to ubiquitin-conjugating enzymes from various organisms (Fig. 3): 91.2% to UBC4Sp of Saccharomyces pombe (5), 85.7% to UBC1Dm of Drosophila (32), 8.4% to UBC2Ce of Caenorhabditis elegans (38), 83.0% to human UBC5Hs (21), 82% to UBC4Sc of S. cerevisiae (32), and 42.2% to UBCWh of wheat (13). Therefore, we designate CHIP1 C. gloeosporioides ubiquitin-conjugating enzyme 1, or UBC1Cg.
FIG. 3.
Homology comparison of UBC1Cg with UBC4Sp of S. pombe, UBC2Ce of C. elegans, UBC1Dm of Drosophila, human UBC5Hs, and UBCWh of wheat. The homologous residues are shaded.
ubc1Cg transcript levels induced by hard-surface and ethylene treatment.
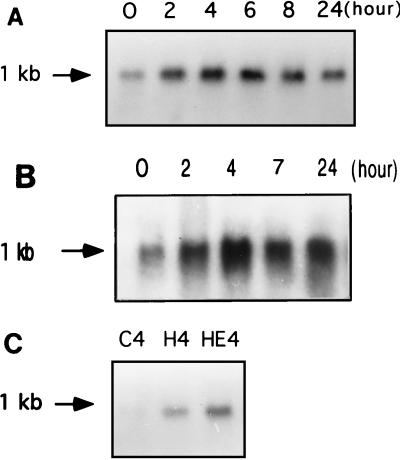
When the ∼1-kb PCR product was used as a probe for Northern blot analysis, a single transcript of ∼1 kb was found, indicating that the cloned cDNA represents a nearly full-length transcript. Analysis of the time course of induction by hard-surface treatment showed that induction of ubc1Cg was readily detectable in 2 h, increased until about 6 h of hard-surface treatment, and subsequently decreased (Fig. 4A).
FIG. 4.
(A) Northern blots showing time course of induction of ubc1Cg transcription by hard-surface contact in C. gloeosporioides conidia. The equal loading amounts of total RNAs (20 μg/lane) were reflected by ethidium bromide staining of 28S and 18S rRNA. (B) Northern blots showing time course of induction of ubc1Cg transcription by 10 μM ethephon on a hard surface in C. gloeosporioides conidia. The amount of total RNAs used was 20 μg/lane. (C) Northern blots showing induction of ubc1Cg by a hard surface with or without ethylene treatment. Total RNAs (10 μg/lane) isolated from C. gloeosporioides conidia that had been on a hard surface for 4 h (H4), on a hard surface with 10 μM ethephon (HE4) for 4 h, or left in the tube at room temperature for 4 h (C4) were subjected to electrophoresis and blotted onto Nytran membranes. 32P-labeled ∼1.0-kb cDNA containing the coding sequence of ubc1Cg was used as a probe. Estimation of RNA sizes was based on the 0.24- to 9.5-kb RNA ladder (BRL). Experimental details are provided in the text.
Since ethylene is known to induce germination and appressorium formation by C. gloeosporioides conidia on a hard surface (11), the effect of ethylene on induction of ubc1Cg in conidia resting on a hard surface was tested. The time course of induction was quite similar to that observed on a hard surface in the absence of ethylene, with maximal induction occurring at ∼4 h (Fig. 4B). The degree of induction on a hard surface with ethylene was higher than that observed on a hard surface without ethylene. A direct comparison of the RNA blots shown in Fig. 4C demonstrates that the ubc1Cg transcript level was higher on a hard surface with ethylene than that reached on a hard surface without ethylene. Quantitation showed that the hard-surface treatment alone caused a twofold increase in transcript level, whereas hard-surface and ethylene treatment caused a sixfold increase in transcript level.
Southern blot analysis of ubc1Cg.
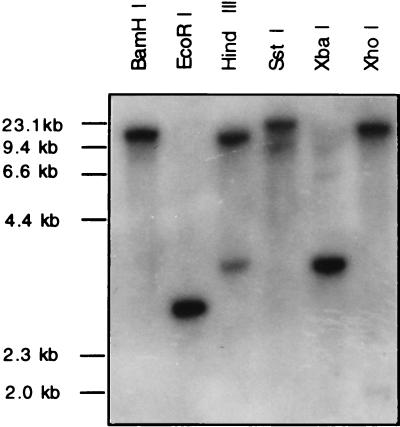
The genomic DNA isolated from C. gloeosporioides was digested with BamHI, EcoRI, HindIII, SstI, XbaI, or XhoI, and Southern blots of the digests were hybridized with the cDNA clone. The results showed only one band in the case of all digests except the HindIII digest, which showed two bands (Fig. 5). However, the restriction map of the cDNA clone showed that there is no HindIII site within the cDNA. To test whether there is a HindIII site in the genomic DNA, PCR-amplified ∼1.5-kb genomic DNA was digested with HindIII. This digestion yielded ∼0.9- and ∼0.6-kb fragments, indicating that there is a HindIII site in this genomic DNA (data not shown). Apparently, there is an intron containing a HindIII site in this genomic DNA. Thus, the Southern blot analysis indicates that the genome of C. gloeosporioides contains one copy of the ubc1Cg gene.
FIG. 5.
Southern hybridization of restriction enzyme-digested genomic DNA (10 μg/lane) from C. gloeosporioides with a 32P-labeled ∼1.0-kb cDNA containing the coding sequence of UBC1Cg as the probe. Molecular sizes (determined with a λ DNA HindIII size marker) are shown on the left. Experimental details are provided in the text.
Complementation of ubc yeast mutant with ubc1Cg.
To test whether the sequence similarity of UBC1Cg to yeast UBC4 is also reflected in its function, we tried to complement the S. cerevisiae ubc4 ubc5 mutant by expression of ubc1Cg. The yeast ubc4 ubc5 mutant is heat sensitive; it cannot grow at 37°C and can grow only very slowly at 30°C (32). The ubc1Cg cDNA was cloned into the EcoRI site in both orientations in a low-copy-number yeast expression vector, pBM272, under the control of the GAL10 promoter with a URA3 selectable marker. Plasmids with inserts in both orientations, as well as pBM272 without any insert, were used to transform the yeast ubc4 ubc5 mutant strain. Transformants were streaked onto SC−U plates or SC(Gal)−U plates and incubated at either 30 or 37°C. When yeast ubc4 ubc5 mutant cells were transformed with plasmids with ubc1Cg inserted in the proper orientation, they grew relatively quickly at 37°C on inducible medium (containing galactose) but not on noninducible medium (containing glucose) (data not shown). Plasmids alone or with a ubc1Cg insert in the opposite orientation did not grow at 37°C on either inducible medium (containing galactose) or noninducible medium (containing glucose). Therefore, UBC1Cg complemented the growth deficiency and heat sensitivity of the ubc4 ubc5 mutant on inducible medium but not on noninducible medium. Thus, UBC1Cg is not only structurally but also functionally similar to yeast UBC4.
DISCUSSION
The formation of appressoria is essential for penetration of the avocado pathogen C. gloeosporioides into its host. Contact with a hard surface is necessary for the chemical signals ethylene and avocado wax to induce appressorium formation in C. gloeosporioides. C. gloeosporioides conidia can form appressoria on both a hydrophilic cover glass and a hydrophobic polystyrene petri dish when exposed to the chemical signals. On the other hand, on soft hydrophilic or hydrophobic substrates, such as 2% agar or petrolatum, respectively, only germination occurs (27). The hydrophilicity or hydrophobicity of the surface does not play an important role in appressorium formation by C. gloeosporioides conidia. The molecular mechanism by which hard-surface treatment assists appressorium formation remains unknown. Elucidation of the nature of genes uniquely expressed during hard-surface treatment could help in understanding the molecular basis of the early events in plant-fungus interaction. Chemical signals, such as ethylene or avocado wax, showed no effect on appressorium formation in C. gloeosporioides conidia during the first 2 h. Treatment for the next 2 to 3 h with chemical signals induced appressorium formation, but subsequent treatment had no effect (11, 12, 20). These observations suggest that a chain of molecular events that ultimately leads to differentiation of the germ tubes into appressoria is initiated upon contact with a hard surface. Breaking the chain of events at any critical stage should interfere with appressorium formation. The early contact with a hard surface presumably initiates molecular changes that prime the conidia to respond to chemical signals, such as the host wax or ethylene. Although some of the genes induced by the chemical signals have been cloned, nothing is known about genes expressed in the early phase. Therefore, we chose to concentrate on transcripts induced during 2 h of hard-surface treatment.
By using a differential-display method, we found eight genes, designated chip genes, expressed during the hard-surface treatment of conidia of C. gloeosporioides. chip1 encodes a ubiquitin-conjugating enzyme, which shows very high homology to the yeast UBC4-UBC5 enzyme pair. To test whether this clone, obtained from RNA from conidia subjected to hard-surface treatment, represents the transcript induced during hard-surface treatment, Northern blot analyses were performed. The results showed that the transcript reached its maximum level after 4 to 6 h of treatment with a hard surface and then decreased. ubc1Cg was induced to a higher level by exposure to an ethylene-generating compound, ethephon, on a hard surface. The increase ceased by 6 h, just before appressorium formation began to be detectable, and the transcript level decreased quite rapidly during the next few hours. The genes discovered by the present approach are probably involved in the induction of appressorium formation, although there is no direct proof that the cloned transcripts induced by hard-surface treatment are actually involved in the chain of events that lead to appressorium formation.
Since C. gloeosporioides ubc complemented the ubc4 ubc5 yeast mutant, it is clear that ubc1Cg is functionally equivalent to yeast ubc4 ubc5. In eukaryotes, the ubiquitin-proteasome system is involved in degradation of various proteins. The ubiquitination of target proteins is catalyzed by a ubiquitin-activating enzyme (E1) and ubiquitin-conjugating enzymes (E2) and in some cases also requires auxiliary substrate recognition proteins (E3). The targets of this degradation pathway include calmodulin and subunits of trimeric G protein (28, 29). A calmodulin gene was cloned from Colletotrichum trifolii (8). When an antisense strategy was used to reduce the expression of this calmodulin gene, appressoria were formed at a reduced frequency (6). Calmodulin was recently found to be involved in appressorium formation in C. gloeosporioides (22), and G protein was found to be essential for appressorium formation in M. grisea (6).
Selective protein degradation by the ubiquitin-proteasome system has been found to play a critical role in many situations, such as the cellular stress response and differentiation, that involve reprogramming of protein synthesis (36). In yeast, at least 12 different ubc genes encode ubiquitin-conjugating enzymes, which mediate strikingly diverse functions. One of the best-characterized yeast E2 enzymes is the UBC4-UBC5 pair. ubc4 ubc5 mutants are sensitive to heat shock, canavanine (an arginine analog), and cadmium, suggesting that the UBC4-UBC5 enzyme pair mediates selective degradation of short-lived and abnormal proteins (32). The UBC4-UBC5 enzyme pair comprises a major part of total ubiquitin-conjugating activity in stressed yeast cells (2). UBC4-UBC5 homologs have been found in several organisms. In C. elegans, UBC2 is developmentally regulated by becoming specific to the nervous system in L4 larvae and adults (40), and unlike the yeast UBC4-UBC5 enzyme pair, it is not induced by heat shock (39). UBC1Dm in Drosophila is also involved in selective protein degradation (35). Our finding that UBC1Cg can complement the proteolysis deficiency of the yeast ubc4 ubc5 mutant indicates that it may also mediate selective proteolysis pathways. In the present case, hard-surface contact probably signals a chain of molecular events that involve reprogramming of protein synthesis needed for conidial germination and differentiation into appressoria. The signal transduction processes involved in transmitting the hard-surface contact to the cellular machinery remain to be elucidated. It is possible that the physical signals and the chemical signals share some signal transduction pathways involved in the differentiation process that are essential for infection by many fungi. Such pathways could serve as targets of antifungal strategies to protect plants.
ACKNOWLEDGMENTS
We thank Daoxin Li and Yeon-ki Kim for many helpful discussions.
This work was supported by National Science Foundation grant IBN-9318554.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arnason T, Ellison M J. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol. 1994;14:7876–7883. doi: 10.1128/mcb.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1990. [Google Scholar]
- 4.Bhairi S M, Staples R C, Frene P, Yoder O C. Characterization of an infection structure-specific gene from the rust fungus. Gene. 1989;81:237–243. doi: 10.1016/0378-1119(89)90184-4. [DOI] [PubMed] [Google Scholar]
- 5.Damagnez V, Rolfe M, Cottarel G. Schizosaccharomyces pombe and Candida albicans cDNA homologues of the Saccharomyces cerevisiae UBC4 gene. Gene. 1995;155:137–138. doi: 10.1016/0378-1119(94)00926-j. [DOI] [PubMed] [Google Scholar]
- 6.Dean R A. Signal pathways and appressorium morphogenesis. Annu Rev Phytopathol. 1997;35:211–234. doi: 10.1146/annurev.phyto.35.1.211. [DOI] [PubMed] [Google Scholar]
- 7.Dickinson S. Studies in the physiology of obligate parasitism. X. Induction of response to a thigmotropic stimulus. Phytopathol Z. 1977;89:97–115. [Google Scholar]
- 8.Dickman M B, Buhr T L, Warwar V, Truesdell G M, Huang C X. Molecular signals during the early stages of alfalfa anthracnose. Can J Bot. 1995;73:1169–1177. [Google Scholar]
- 9.Dickson S. Growth of Erysiphe germinis on artificial membranes. Physiol Plant Pathol. 1979;15:219–221. [Google Scholar]
- 10.Emmett R W, Parbery D G. Appressoria. Annu Rev Phytopathol. 1975;13:147–167. [Google Scholar]
- 11.Flaishman M A, Kolattukudy P E. Timing of fungal invasion using host’s ripening hormone as a signal. Proc Natl Acad Sci USA. 1994;91:6579–6583. doi: 10.1073/pnas.91.14.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaishman M A, Hwang C-S, Kolattukudy P E. Involvement of protein phosphorylation in the induction of appressorium formation in Colletotrichum gloeosporioides by its host surface wax and ethylene. Physiol Mol Plant Pathol. 1995;47:103–117. [Google Scholar]
- 13.Girod P A, Vierstra R D. A major ubiquitin conjugation system in wheat germ extracts involves a 15-kDa ubiquitin-conjugating enzyme (E2) homologous to the yeast UBC4/UBC5 gene products. J Biol Chem. 1993;268:955–960. [PubMed] [Google Scholar]
- 14.Gurr S J, Unkles S E, Kinghorn J R. Structure and organization of nuclear genes of filamentous fungi. In: Kinghorn J R, editor. Gene structure in eukaryotic microbes. Oxford, England: IRL Press; 1987. pp. 93–139. [Google Scholar]
- 15.Hankin L, Kolattukudy P E. Metabolism of a plant wax paraffin (n-nonacosane) by a soil bacterium (Micrococcus certificans) J Gen Microbiol. 1968;51:457–463. doi: 10.1099/00221287-51-3-457. [DOI] [PubMed] [Google Scholar]
- 16.Hoch H C, Staples R C. Signaling for infection structure formation in fungi. In: Cole G T, Hoch H C, editors. The fungal spore and disease initiation in plants and animals. New York, N.Y: Plenum Press; 1991. pp. 25–46. [Google Scholar]
- 17.Hoch H C, Staples R C, Bourett T M. Chemically induced appressoria in Uromyces appendiculatus are formed aerially, apart from the substrate. Mycologia. 1987;79:418–424. [Google Scholar]
- 18.Hoch H C, Staples R C, Whitehead B, Comeau J, Wolfe E D. Signaling for growth orientation and cell differentiation by surface topography in Uromyces. Science. 1987;234:1659–1662. doi: 10.1126/science.235.4796.1659. [DOI] [PubMed] [Google Scholar]
- 19.Hwang C-S, Flaishman M A, Kolattukudy P E. Cloning of a gene expressed during appressorium formation by Colletotrichum gloeosporioides and a marked decrease in virulence by disruption of this gene. Plant Cell. 1995;7:183–193. doi: 10.1105/tpc.7.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang C-S, Kolattukudy P E. Isolation and characterization of genes expressed uniquely during appressorium formation by Colletotrichum gloeosporioides conidia induced by the host surface wax. Mol Gen Genet. 1995;247:282–294. doi: 10.1007/BF00293196. [DOI] [PubMed] [Google Scholar]
- 21.Jensen J P, Bates P W, Yang M, Vierstra R D, Weissman A M. Identification of a family of closely related human ubiquitin conjugating enzymes. J Biol Chem. 1995;270:30408–30414. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- 22.Kim, Y.-K., D. Li, and P. E. Kolattukudy. Induction of Ca2+-calmodulin signaling by hard surface contact primes Colletotrichum gloeosporioides to form appressoria. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 23.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y H, Dean R A. Stage-specific gene expression during appressorium formation of Magnaporthe grisea. Exp Mycol. 1993;17:215–222. [Google Scholar]
- 25.Liang P, Averboukh L, Pardee A B. Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 1993;21:3269–3275. doi: 10.1093/nar/21.14.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang P, Pardee A B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Z.-M., and P. E. Kolattukudy. Unpublished data.
- 28.Madura K, Varsharsky A. Degradation of G alpha by the N-end rule pathway. Science. 1994;265:1451–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 29.Parag H A, Dimitrovsky D, Raboy B, Kulka R G. Selective ubiquitination of calmodulin by UBC4 and a putative ubiquitin protein ligase (E3) from Saccharomyces cerevisiae. FEBS Lett. 1993;325:242–246. doi: 10.1016/0014-5793(93)81081-a. [DOI] [PubMed] [Google Scholar]
- 30.Polida G K, Rogers L M, Kolattukudy P E. Chemical signals from avocado surface wax trigger germination and appressorium formation in Colletotrichum gloeosporioides. Plant Physiol. 1993;103:267–272. doi: 10.1104/pp.103.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staples R C, Hoch H C. Infection structure, form and function. Exp Mycol. 1987;11:163–169. [Google Scholar]
- 34.Staples R C, Macko V. Formation of infection structures as a recognition response in fungi. Exp Mycol. 1980;4:2–16. [Google Scholar]
- 35.Treier M, Seufert W, Jentsch S. Drosophila UbcD1 encodes a highly conserved ubiquitin-conjugating enzyme involved in selective protein degradation. EMBO J. 1992;11:367–372. doi: 10.1002/j.1460-2075.1992.tb05059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 37.Xuei X, Bhairi S, Staples R C, Yoder O C. Characterization of INF56, a gene expressed during infection structure development of Uromyces appendiculatus. Gene. 1992;110:49–55. doi: 10.1016/0378-1119(92)90443-s. [DOI] [PubMed] [Google Scholar]
- 38.Xuei X, Bhairi S, Staples R C, Yoder O C. Differentiation-specific genes of rust fungi have limited distribution among fungi. Exp Mycol. 1992;16:320–323. [Google Scholar]
- 39.Zhen M, Heinlein R, Jones D, Jentsch S, Candido E P M. The ubc-2 gene of Caenorhabditis elegans encodes a ubiquitin-conjugating enzyme involved in selective protein degradation. Mol Cell Biol. 1993;13:1371–1377. doi: 10.1128/mcb.13.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhen M, Schein J E, Baillie D L, Candido E P M. An essential ubiquitin-conjugating enzyme with tissue and developmental specificity in the nematode Caenorhabditis elegans. EMBO J. 1996;15:3229–3237. [PMC free article] [PubMed] [Google Scholar]