Abstract
Two copper-binding compounds/cofactors (CBCs) were isolated from the spent media of both the wild type and a constitutive soluble methane monooxygenase (sMMOC) mutant, PP319 (P. A. Phelps et al., Appl. Environ. Microbiol. 58:3701–3708, 1992), of Methylosinus trichosporium OB3b. Both CBCs are small polypeptides with molecular masses of 1,218 and 779 Da for CBC-L1 and CBC-L2, respectively. The amino acid sequence of CBC-L1 is S?MYPGS?M, and that of CBC-L2 is SPMP?S. Copper-free CBCs showed absorption maxima at 204, 275, 333, and 356 with shoulders at 222 and 400 nm. Copper-containing CBCs showed a broad absorption maximum at 245 nm. The low-temperature electron paramagnetic resonance (EPR) spectra of copper-containing CBC-L1 showed the presence of a copper center with an EPR splitting constant between those of type 1 and type 2 copper centers (g⊥ = 2.087, g∥ = 2.42 G, |A∥| = 128 G). The EPR spectrum of CBC-L2 was more complex and showed two spectrally distinct copper centers. One signal can be attributed to a type 2 Cu2+ center (g⊥ = 2.073, g∥ = 2.324 G, |A∥| = 144 G) which could be saturated at higher powers, while the second shows a broad, nearly isotropic signal near g⊥ = 2.063. In wild-type strains, the concentrations of CBCs in the spent media were highest in cells expressing the pMMO and stressed for copper. In contrast to wild-type strains, high concentrations of CBCs were observed in the extracellular fraction of the sMMOC mutants PP319 and PP359 regardless of the copper concentration in the culture medium.
In methanotrophs, the relationship between the concentration of copper and expression of the two different methane monooxygenases (MMOs) is well characterized (8, 11, 45, 49, 50). Under low copper-to-biomass ratios, methane oxidation activity is observed in the soluble fraction, and the enzyme is referred to as the soluble methane monooxygenase (sMMO). At higher copper-to-biomass ratios, methane oxidation activity is observed in the membrane fraction, and the enzyme is referred to as the membrane-associated or particulate methane monooxygenase (pMMO). The polypeptides and structural genes for both enzymes have been characterized (4, 18–22, 24, 25, 32, 34–40, 43–45, 47–49, 51, 62, 63). In addition to expression of the two MMOs, four other physiological traits have been identified in cells expressing the pMMO that are affected by the copper concentration in the culture medium. First, the concentration of copper in the culture media is directly related to pMMO activity in cell-free fractions, although the levels of expression of pMMO polypeptides vary in different methanotrophs (1, 8, 30, 36, 50, 63). For example, the expression levels of the three pMMO polypeptides in Methylococcus capsulatus Bath remained constant with varying copper concentrations (8, 36), whereas in Methylomicrobium albus BG8, the expression level of the putative pMMO polypeptides increased with increased copper in the culture medium (8). Second, the concentrations of membrane-associated copper and iron show a proportional increase as the copper concentration in the culture medium is increased (36, 63). Third, the formation and level of intracytoplasmic membranes in cells cultured in copper-supplemented media are dependent on the copper concentration in the culture media (8, 11, 40, 48). Lastly, the Ks for methane oxidation by pMMO is altered by the copper concentration in the culture media (33a).
Berson and Lidstrom (1) have recently noted that in spite of the central role of copper in the physiology of methanotrophs, the mechanism(s) of copper acquisition remains vague. Although true, a few studies have suggested the existence of a specific copper acquisition system in M. capsulatus Bath and M. trichosporium OB3b. The first indication of a specific copper uptake system was provided from phenotypic characterization of the constitutive sMMO mutants (sMMOC) isolated by Phelps et al. (42). Fitch et al. (17) found that in M. trichosporium OB3b, these sMMOC mutants were defective in copper uptake and showed preliminary evidence for an extracellular copper-complexing agent. Working with the same mutants, Téllez et al. partially purified this copper-complexing agent and determined that it was a small molecule with a molecular mass of approximately 500 Da with an association constant with copper of 1.4 × 1016 M−1 (55). Other evidence for a specific copper uptake system was provided by the copper-binding cofactor (CBC) from M. capsulatus Bath (63). During the isolation of the pMMO from M. capsulatus Bath, CBC was identified in association with the purified enzyme, in the washed membrane fraction, and in the extracellular fraction. The CBC was determined to be a small polypeptide with a molecular mass of 1,232 Da. In M. capsulatus Bath, the cellular location of the CBC varied depending on the copper concentration in the culture medium and on the expression of the pMMO.
This paper ties together and extends these observations on specific copper acquisition systems in M. trichosporium OB3b and M. capsulatus Bath. Here we describe the initial isolation and characterization of two copper-complexing agents, called CBC-L1 and CBC-L2, from the M. trichosporium OB3b wild type and sMMOC mutant PP319. CBC-L1 from M. trichosporium OB3b was identical to the CBC previously identified during the isolation of the pMMO from M. capsulatus Bath. This paper is also the first report of a second CBC, CBC-L2, which may have been present as a contaminant in previous CBC preparations from M. capsulatus Bath. One or both of the CBCs appear to be the same copper-complexing agent partially purified by Téllez et al. (55). Lastly, this report describes the effect of the copper concentration in the culture medium on copper uptake, the expression of both MMOs, and extracellular concentration of the CBC in wild-type and sMMOC mutant strains of M. trichosporium OB3b.
MATERIALS AND METHODS
Organisms and cultivation.
Wild-type M. trichosporium OB3b and sMMOC mutants PP319 and PP358 were grown at 30°C in nitrate mineral salts (NMS) medium (9, 10) plus 0.0 or 5.0 μM CuSO4 under an atmosphere of 25% (vol/vol) methane and 75% (vol/vol) air. All cells were cultured by semicontinuous methods using a 3-liter fermentor (BIOFLO 3000; New Brunswick) sparged with methane and air in excess to maintain ambient oxygen concentrations at 75% saturation in air. A typical cell-culturing sequence involved culturing the cells in copper-free medium (no copper added) to an optical density at 600 nm (OD600) of between 0.6 and 0.7. The culture was then diluted with fresh medium to an OD600 of approximately 0.15 and cultured again to an OD of 0.7 to 0.75 before harvesting of 90% of the culture. At ODs of 0.6 and above, sMMO activities were measured prior to harvesting.
After harvesting, the remaining 10% of the culture was diluted with copper-free medium to an OD600 of about 0.15, and the concentration of copper in the culture medium was raised to 5.0 μM for the subsequent copper-amended experiments. The culture was then grown to an OD600 of 0.8 and diluted at least twice with medium containing 5 μM copper as described above prior to harvesting. The sMMO activity was monitored intermittently over this period. Wild-type cultures were harvested as before when no sMMO activity was detected for one dilution/growth cycle. Alternately, mutant cultures, which continually expressed sMMO activity in the presence of copper, were typically harvested after two dilution/growth cycles.
M. capsulatus Bath was grown in nitrate mineral salts medium plus 0 or 5 μM CuSO4 as previously described (63).
Induction of CBC in wild-type strains.
Wild-type M. trichosporium OB3b and M. capsulatus Bath were cultured in 12-liter fermentors sparged at flow rates of 100 to 150 ml of methane per min and 2,000 to 2,500 ml of air per min in NSM plus 5 μM CuSO4 at 30°C for M. trichosporium OB3b or NMS plus 5 μM CuSO4 at 42°C for M. capsulatus Bath. When the culture reached an OD600 of 0.8 to 1.1, 8 liters of the culture medium was removed and replaced with 8 liters of low-copper medium. If the culture developed a visual yellow color when cells reached an OD600 of 0.8 to 1.2, the cells were harvested; if it did not, 8 liters of the culture was harvested and replaced with 8 liters of low-copper medium. These procedures were repeated until the medium developed a yellow color indicating the presence of CBC in the extracellular fraction.
Harvesting media and cells.
Cells were harvested by centrifugation for 30 min at 9,000 × g. The supernatant was decanted, collected, and filtered through a 0.22-μm-pore-size filter. The filtrate was either lyophilized or loaded on Sep-Pak cartridges (Millipore Corp., Bedford, Mass.) which had been pretreated with 10 ml each of ethanol, dichloromethane, ethanol, and H2O. Both CBCs bound to the Sep-Pak cartridges and were washed with 30 ml of H2O and 30 ml of 25 mM Tris-HCl–1 M urea and eluted with 0.2% (vol/vol) trifluoroacetic acid–99.8% (vol/vol) acetonitrile; the sample was then lyophilized.
Isolation of the CBC from spent medium. (i) Method I.
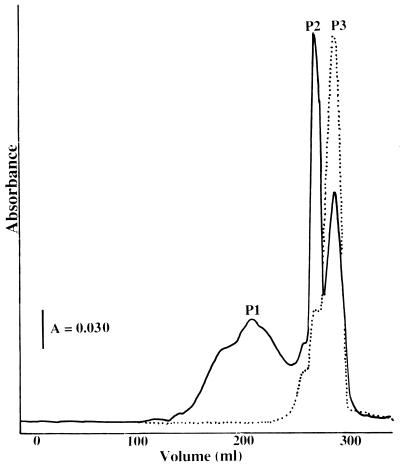
Lyophilized spent medium or lyophilized material extracted from Sep-Pak cartridges was resuspended in a minimal volume of 20 mM Tris-HCl (pH 8.0) plus 3 M urea and loaded on a 2.6- by 60-cm Superdex 30 (Pharmacia, Uppsala, Sweden) column equilibrated with 20 mM Tris-HCl (pH 8.0)–3 M urea. The yellow-colored sample migrated in three fractions with molecular masses of approximately 2,000 (P1), 1,000 (P2), and 500 (P3) Da (Fig. 1). Each colored fraction was concentrated in a stirred cell (YC05 filter) and individually loaded on a 2.6- by 60-cm Superdex 30 column equilibrated with 20 mM Tris-HCl (pH 8.0)–3 M urea. The CBCs were then loaded separately on a 1- by 10-cm 15RPC (Pharmacia) column equilibrated with 2 mM ammonium phosphate (pH 7.0) buffer. The sample was washed with four column volumes of 2 mM ammonium phosphate (pH 7.0) buffer, and the concentration of 2% (vol/vol) trifluoroacetic acid in acetonitrile increased to 75% over a 200-ml linear gradient. The CBC eluted at approximately 15% trifluoroacetic acid–2% acetonitrile mixture. The sample was lyophilized, resuspended in a minimal volume of H2O, and run a second time on the 1- by 10-cm 15RPC column. The sample was then lyophilized and resuspended in H2O.
FIG. 1.
Elution profile of the YM10 filtrate fraction separated on a Superdex 30 column. Fractions were monitored at 280 nm (——) and for conductivity (······).
(ii) Method II.
Lyophilized spent medium or lyophilized material extracted from Sep-Pak cartridges was resuspended in a minimal volume of 20 mM Tris-HCl (pH 7.5)–50 mM KCl (buffer A) and loaded on a 2.6- by 20-cm Chelating Sepharose Fast Flow (FF) (Pharmacia Biotech) column equilibrated with buffer A. Prior to sample addition, the first 1.0 cm of the column was charged with CuCl2, followed by a washing step with 2 column volumes of buffer A as described by Donat et al. (13). CBC-L2 bound to the column, and the sample was washed with 2 column volumes of buffer A. The concentration of buffer A plus 500 mM imidazole buffer (buffer B) was increased to 100% buffer B over a 500-ml linear gradient; CBC-L2 eluted at approximately 150 mM imidazole. The CBC column fractions were dialyzed and concentrated on a stirred cell (YC05 filter) and isolated individually following the column steps described for method I.
Isolation of the CBC from the washed membrane fraction.
Both CBCs were also extracted from the washed membrane by using a 0.1% (vol/vol) HCl-N,N′-dimethyl formamide solution as previously described (63). The extraction was repeated until the extraction solution was clear. Following 0.1% HCl–N,N′-dimethyl formamide extraction, the N,N′-dimethyl formamide was evaporated under vacuum and the sample was resuspended in a minimal volume of 20 mM Tris-HCl (pH 8.0)–3 M urea and purified as described for method II.
Reconstitution of CBCs with copper.
As isolated by method II, CBC-L2 contained two copper ions. CBCs isolated from the spent medium by method I were copper free. To these samples, copper was added as CuCl2 solutions (1 to 200 μM) by titration, monitoring saturation by UV-visible absorption or electron paramagnetic resonance (EPR) spectroscopy.
Enzyme activity.
sMMO activity was determined by using a modified version of the naphthalene assay described by Brusseau et al. (3), as follows. Between 3.0 and 20.0 ml of cells was centrifuged for 15 min at 9,000 × g and resuspended in 10 mM phosphate buffer (pH 7.0) plus 10 mM formate to an OD600 of between 0.6 and 0.7. Reaction mixtures contained the cell suspension (2.5 ml) plus 50 μl of aqueous o-dianisidine (5 mg/ml), and the reaction was initiated by the addition of 50 mg of crushed naphthalene crystals to the sample cuvettes. After naphthalene addition, the amended cuvette was hand shaken aggressively for 10 s. Following stabilization of the cell-naphthalene suspension (approximately 15 s), color formation was monitored for 20 min at 525 nm. Activities were normalized to the cell dry weight of the assay suspension.
Copper analysis.
Samples were collected for copper analysis in conjunction with each sample-harvesting event. Sample preparation for copper analysis was determined as described by Fitch et al. (17) except that the incubation time of cells in the EDTA extraction step was increased from 10 min to 3 h. Copper concentrations were measured in triplicate, and coefficients of variation were less than 0.1 in all experiments reported.
Mass spectroscopy.
Molecular masses of the CBCs were determined by time-of-flight mass spectrometry on a Finnigan (matrix-assisted laser desorption ionization [MALDI]) mass spectrometer, using sinapinic acid as the matrix, and on a Finnigan 972947 electrospray mass spectrometer as previously described (63). The molecular mass of CBC-L2 was also determined by fast atom bombardment (FAB)-mass spectrometry in the positive ion mode on a Finnigan TSQ-700, using a matrix of 3-nitrobenzyl alcohol in acetonitrile.
Amino acid and sequence analysis.
Amino acid analysis was carried out with an Applied Biosystems 420A derivatizer coupled to an Applied Biosystems 130A separation system. Samples were hydrolyzed in 6 M HCl for 1 h in a vacuum at 150°C. After hydrolysis, norleucine was added as an internal standard.
Amino acid sequence analyses were performed on samples bound to either Prosorb or Sequelon AA (Millipore) membranes by Edman degradation with an Applied Biosystems 492 protein sequencer coupled to a 140C analyzer.
Other methods.
Optical absorption spectroscopy, EPR spectra, labeling with [U-14C]acetylene, protein determinations, cytochrome c oxidase activity, electrophoresis, and immunoblot analysis were determined as previously described (12, 16, 26, 27, 30, 33, 63).
RESULTS
Isolation of CBC.
Culture conditions for optimal production and purification of CBC-L1 and CBC-L2 from the spent media or from the washed membrane fractions of M. trichosporium OB3b are described in Materials and Methods. In wild-type M. capsulatus Bath and M. trichosporium OB3b, the concentration of CBC in the spent media was highest in cells expressing the pMMO and stressed for copper. Under these conditions, the spent media have a light yellow color due to the high concentration of CBC. Once the cells switch from expression of pMMO to expression of sMMO, the spent media become clear and the concentrations of CBCs in the spent media decrease by over 75% (Table 1).
TABLE 1.
Copper localization, extracellular CBC concentrations, and sMMO activities in wild-type and copper uptake mutants
| Strain | Initial copper concn in culture medium (μM) | CBCs in spent culture medium (mg/liter) | sMMO activity (ng of naphthol · min−1 · mg of protein−1) |
|---|---|---|---|
| Wild type | 0.5 | 82 | 0 |
| 5.0 | 8 | 0 | |
| 0.09 | 34 | 105 ± 18 | |
| PP319 | 4.99 | 52 | 155 ± 23 |
| 0.12 | 58 | 122 ± 25 | |
| PP358 | 4.98 | 67 | 123 ± 28 |
| 0.10 | 54 | 118 ± 18 |
The two M. trichosporium OB3b sMMOC mutants, PP319 and PP358, have proven to be useful for the isolation of both CBCs. In contrast to the wild-type strains, the concentrations of both CBCs in the culture media of the two sMMOC mutants did not significantly change with the concentration of copper in the culture media (Table 1). The concentrations of CBCs in the mutant strains remained similar (65 to 80%) to the levels observed in wild-type M. trichosporium OB3b cultured in low-copper medium just before induction of the sMMO. Interestingly, also in contrast to wild-type strains, high concentrations of copper-containing CBCs could be isolated from the spent copper-containing media of the two sMMOC mutants PP319 and PP358. In cells cultured in high-copper media, the high concentration of copper containing CBC in the medium of the two sMMOC mutants was evident by a bright yellow color. The color of both CBCs became more intense with the binding of copper.
Separation of the concentrated spent media by gel filtration on a Superdex 30 column resulted in three distinct yellow-colored fractions with approximate molecular masses of 2,000 to 2,500, 1,000 to 1,500, and 500 to 750 Da (Fig. 1; Table 2). The concentration of each fraction varied with the preparation, but the fraction migrating in the 1,000- to 1,500-Da fraction (P2) was consistently the largest. The low-molecular-weight fraction (P3) showed high conductivity (Fig. 1). Each fraction was collected and purified by a second gel filtration and by reverse-phase chromatography as described in Materials and Methods.
TABLE 2.
Properties of the three copper-binding peptides isolated from the spent media of M. trichosporium OB3b
| Peptide | Property
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subunit molecular mass (Da) determined by:
|
N-terminal amino acid sequence | Absorption maxima (nm) | Fluores- cence | EPR signal
|
|||||
| MALDI mass spectroscopy | Electrospray mass spectroscopy | FAB mass spectroscopy | g⊥ | g∥ (G) | |A∥| (G) | ||||
| P1 | NDa | 1,093 | ND | S?MYPGS?M | 204, 222, 275, 333, 356, 400 | − | 2.087 | 2.42 | 128 |
| CBC-L1 | 1,238 | 1,216 | ND | S?MYPGS?M | 204, 222, 275, 333, 356, 400 | − | 2.087 | 2.42 | 128 |
| P3 | ND | 382 | ND | S?M | 194, 217, 260, 354, 390 | + | 2.087 | 2.42 | 128 |
| CBC-L2 | ND | ND | 779 | SPMP?S | 194, 217, 260, 354, 390 | − | 2.073 | 2.324 | 144 |
ND, not determined.
All three yellow-colored fractions isolated from the spent medium from M. trichosporium OB3b bound iron as well as copper. Based on the molecular weights, N-terminal amino acid sequences, and spectral properties, the P1 and P3 fractions from the Superdex 30 column were determined to be breakdown products of the CBC isolated from the P2 fraction, CBC-L1 (Table 2). This statement is based on the following observations. First, the results of N-terminal amino acid analysis for the P1 fraction were identical to those for CBC-L1; however, the subunit molecular mass of P1 was 122 Da less than that of CBC-L1. P1 migrated as a dimer on Superdex 30 gel filtration columns. Second, the lower molecular mass of P3 and the similarity in the first three amino acids from P3 to the first three amino acids of CBC-L1 suggest that P3 was the N-terminal fragment of CBC-L1 (Table 2).
Solution properties of the CBCs varied with metal binding and pH. Under acidic conditions (pH values below 6.5), both CBCs were very soluble and did not stick to reverse-phase columns such as 15RPC or C18. However, at neutral or alkaline pH values, or with the addition of copper, the CBCs showed a tendency to bind to most low-pressure column resins tested in the absence of high (2 to 3 M) concentrations of urea.
CBC-L2 was not readily identified during purification by method I. CBC-L2 comigrates on Superdex 30 columns with the P3 fraction. However, CBC-L2 could be separated from CBC-L1, as well as from the P1 and P3 fractions, on Chelating Sepharose FF columns due to the higher affinity of CBC-L2 for this resin. In contrast to CBC-L1, CBC-L2 eluted from the Chelating Sepharose column along with two copper atoms per CBC-L2 atom. Both copper ions remained bound to CBC-L2 during the subsequent purification steps. Removal of the copper ions from the iminodiacetic acid group on the Chelating Sepharose FF resin suggests the formation constant for CBC-L2 is greater than 11.1 (28).
Amino acid analysis, N-terminal sequence analysis, mass spectrometry, and nuclear magnetic resonance (NMR) spectroscopy.
The molecular masses of the CBC-L1 isolated fractions of wild-type M. trichosporium OB3b or sMMOC mutant PP319 were 1,218 ± 36 Da by electrospray mass spectroscopy and 1,236 ± 43 Da by MALDI mass spectroscopy. The reason for the variability has not been determined, but it may be due to the binding of ions in solution. The molecular mass of CBC-L2 was determined to be 779 Da by FAB mass spectroscopy. The molecular masses obtained from CBCs isolated from either the extracellular fraction or membrane fractions were within the range of intertrial error, indicating that the same molecule was isolated from both sources. In addition, the amino acid sequences of CBC-L1 and CBC-L2 isolated from the washed membrane fraction and from spent media were identical.
Amino acid analysis of CBC-L1 detected only the presence of S, G, P, Y, and M in a molar ratio of 2S:1G:1P:1Y:2M. Assuming a molecular mass of approximately 1,220 Da, the amino acid composition of CBC was 2 mol of S, 1 mol of G, 1 mol of P, 1 mol of Y, and 2 mol of M per mol of CBC-L1, which was consistent with the N-terminal amino acid sequence of S?1MYPGS?2M. Based on the molecular mass of CBC-L1 and on the ratio of amino acids obtained from amino acid analysis, the sequence of CBC-L1 appears to be complete. The molecular mass determined from the identified amino acid composition or N-terminal sequence was approximately 40% less than that determined by mass spectroscopy. The difference may reflect the presence of unidentified amino acids indicated as ?1 and ?2 (Table 2), modified amino acids, or the presence of additional side groups which are commonly observed in small metal-binding polypeptides (20, 45, 53, 57, 58). The N-terminal amino acid sequence of CBC-L2, SPMP?S, differed from that of CBC-L1. The amino acid compositions of both CBCs show some similarity, especially in the multiple S, to the iron-chelating pseudobactins, ferribactins, and pyoverdins produced by some species of Pseudomonas and Azotobacter (20, 53, 55) but showed no sequence similarities. Unlike many iron-binding polypeptides, the identified amino acids of both CBCs are all l-amino acids (20, 53, 58). The amino acid sequences of CBC-L1 and CBC-L2 also showed no sequence similarities to MMO polypeptides.
Preliminary NMR results for CBC and for the P3 fragment in aqueous solution at pH 3 indicate that the unknown functional group or amino acid designated ?1 gives rise to three singlet resonances of equivalent intensity in the aromatic region of the proton spectrum (30). One of these singlet resonances disappears from the spectrum of P3 in D2O solution, indicating that this resonance results from an exchangeable nitrogen-bound proton. These NMR results are consistent with a nitrogen-containing aromatic moiety and confirm that the ?1 functional group of CBC-L1 is retained in the P3 fragment. The presence of an aromatic amino acid is consistent with the UV-visible absorption spectra of CBC-L1 and CBC-L2 as well as of the P1 and P3 fractions (see below).
Spectral properties.
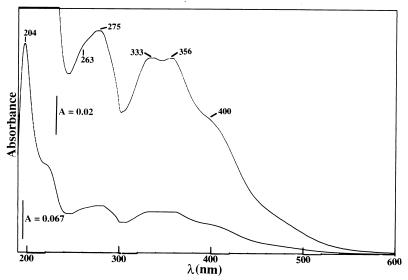
As observed with M. capsulatus Bath (63), the CBC-L1 isolated from the spent media of wild-type M. trichosporium OB3b was copper free. In the absence of copper, the CBC-L1 was EPR silent. The UV-visible absorption spectrum of copper-free CBC-L1 showed absorption maxima at 204, 275, 333, and 356 nm, with shoulders at 222 and 400 nm (Fig. 2). Addition of Cu2+ to CBC-L1 resulted in the increased absorption in the 200- to 290-nm range (Fig. 3). Copper titration experiments also demonstrated that CBC-L1 bound one copper ion. Previous reports of the CBC from M. capsulatus Bath binding two or three copper ions was an overestimate (63). As demonstrated below, previous purification procedures of CBC from M. capsulatus Bath contained contaminating levels of CBC-L2, P1, and P3. These lower-molecular-mass copper-binding polypeptides resulted in the overestimation in the ratio of copper to CBC in the earlier report (63).
FIG. 2.
Absorption spectra of the CBC-L1 isolated from spent media of M. trichosporium OB3b (15-mmol sample in 10 mM PIPES buffer [pH 7.0]).
FIG. 3.
Absorption spectra of CBC-L1 (a) with 15 μmol of CuCl2, plus 2 (b), 4 (c), 6 (d), 8 (e), 10 (f), and 12 (g) μmol of CuCl2.
The UV-visible spectral properties of CBC-L2 were identical to those of CBC-L1. The major difference between the absorption spectra of CBC from M. trichosporium OB3b and M. capsulatus Bath (59) and the spectra from the iron-binding polypeptides (53, 58, 59) was the lack of the 400-nm absorption maximum. In addition, with the exception of the CBC-L1 fragment isolated from the P3 fraction (Fig. 1), the CBC was not fluorescent, which is a common property of iron-binding polypeptides (20, 53, 58, 59).
The low-temperature X-band EPR spectrum of copper-containing CBC-L1 showed the presence of a copper center with a splitting constant between those for type 1 and type 2 centers (g⊥ = 2.087, g∥ = 2.42 G, |A∥| = 128 G [Fig. 4]) (24, 52, 57). The hyperfine signals of copper-containing samples isolated from both P1 and P3 were similar to those for CBC-L1, but less defined and complex. As isolated, CBC-L2 contains two spectrally distinct copper centers (Fig. 5). One signal could be attributed to a type 2 Cu2+ center (g⊥ = 2.073, g∥ = 2.324 G |A∥| = 144 G) and can be saturated at higher powers at low temperatures (8 K), while the second (g∥ = 2.063) is not. The individual EPR spectra of CBC-L1 and CBC-L2 from M. trichosporium OB3b were less complex than originally reported for CBC from M. capsulatus Bath (63). The difference can be accounted for by the improved purification procedure which separates the sample into four distinct fractions, CBC-L1, CBC-L2, P1, and P3. Previous purification methods used Bio-Gel P-2 (Bio-Rad Laboratories, Hercules, Calif.) gel filtration columns which enriched for CBC-L1 but failed in the complete separation of CBC-L1 for the three other peptides (63). A mixture of the four CBC fractions provides some of the spectral complexity previously noted in the CBC isolated from the M. capsulatus Bath membrane fraction (63).
FIG. 4.
EPR spectrum of CBC-L1 from wild-type M. trichosporium OB3b isolated from spent medium. Operating parameters were as follows: temperature, 8 K, modulation frequency, 100 kHz; modulation amplitude, 12.5 G; time constant, 100 ms. The microwave settings were a frequency of 9.42 GHz, a gain of 3.3, and a microwave power of 0.2 mW.
FIG. 5.
EPR spectra for CBC-L2 fraction from wild-type M. trichosporium OB3b isolated from spent media. Operating parameters were as follows: temperature, 8 K; modulation frequency, 100 kHz; modulation amplitude, 12.5 G; time constant, 100 ms. The microwave settings were a frequency of 9.42 GHz, a gain of 3.3, and power increases from 0.063 (top trace) to 0.2 (second from top trace), 2.02 (third from top trace), and 20 (bottom trace) mW.
Properties of the CBC in sMMOC mutants PP319 and PP358 and wild-type M. trichosporium OB3b.
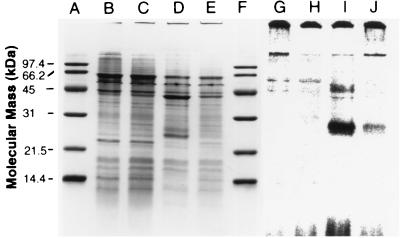
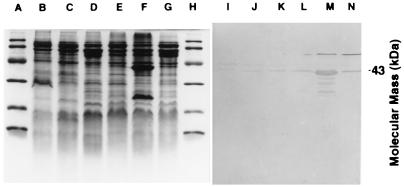
Although the phenotype was not well defined, several sMMOC mutants isolated by Phelps et al. (42) may provide keys to the mechanism of copper acquisition in M. trichosporium OB3b. Fitch et al. (17) demonstrated the presence of sMMO polypeptides in the sMMOC mutants in cells cultured under high- and low-copper conditions but failed to verify the absence of pMMO polypeptides. To address this question, we examined the expression of both MMOs in sMMOC mutants PP319 and PP358. Figure 6 shows the sodium dodecyl sulfate (SDS)-denaturing gels of the membrane fractions isolated from the wild type and sMMOC mutant PP319 from M. trichosporium OB3b cultured in high- and low-copper media and treated with [U-14C]acetylene. The results demonstrate the absence of the pMMO 27,000-Da acetylene-binding polypeptide in PP319 cells even when cultured in high-copper medium (Fig. 6, lane G). Similar results were obtained with sMMOC mutant PP358. Immunoblot analysis of sMMOC mutants PP319 and PP358 and wild-type M. trichosporium OB3b cultured in low- and high-copper media with antibodies against either pMMO (Fig. 7) or sMMO (results not shown) confirmed the constitutive expression of the sMMO polypeptide and little to no expression of the pMMO polypeptide in both sMMOC mutants.
FIG. 6.
SDS-polyacrylamide gel electrophoresis of the membrane fractions from copper uptake mutant PP319 (lanes B, C, G, and H) and wild-type (lanes D, E, I, and J) M. trichosporium OB3b cells treated with [U-14C]acetylene, using formate as the reductant. The cells were cultured under high (5 μM)-copper (lanes B, D, G, and I) or low-copper (lanes C, E, H, and J) conditions. Lanes A through F were stained for total protein with Coomassie brilliant blue R-250, and lanes G through J are phosphorescence images of [U-14C]acetylene-labeled polypeptides exposed for 3.5 days on a storage phosphorescence imaging screen. Bio-Rad low-range molecular mass standards are shown in lanes A and F.
FIG. 7.
Immunoblot analysis of copper uptake mutants 319 (lanes B, C, I, and J) and 358 (lanes D, E, K, and L) and wild-type (lanes F, G, M, and N) M. trichosporium OB3b cultured under high (lanes B, D, F, I, K, and M)- and low (lanes C, E, G, J, L, and N)-copper conditions. Lanes A through H are stained with Coomassie brilliant blue R-250. Lanes I through N are probed with antibodies to the 47,000-Da polypeptide of the pMMO.
Copper acquisition.
As previously observed by Fitch et al. (17), sMMOC mutant strains PP319 and PP358 bound less than 14% of the copper observed in the wild-type strains (Fig. 8). The copper uptake mutants also showed an increase in the nonprecipitable copper in the culture media (Fig. 8). The copper-complexing agent described here can explain the observations by Fitch et al. (17) and Téllez et al. (55) of a potential copper-binding ligand. In this study, as in these earlier studies, high concentrations of the copper-complexing agent or copper-free CBCs were observed in the spent media of cells expressing the pMMO cultured in low-copper media, i.e., in copper-stressed conditions. Cells cultured in high-copper media and expressing the pMMO showed lower concentrations of CBC in the extracellular media. In copper-containing solid media, the sMMOC mutants PP319 and PP358 produced a yellow halo around each colony as a result of high concentrations of extracellular copper-containing CBC.
FIG. 8.
Localization of copper in wild-type and sMMOC mutant PP319 and PP358 cultures of M. trichosporium OB3b grown in culture medium containing 5 μM copper. Culture densities ranged from 300 to 330 mg (dry weight)/liter. Nonprecipitable (□) copper is copper which is not cell associated; extractable copper (▧) is cell-associated copper which can be removed by NaEDTA washes; perceptible (▪) copper is cell-associated copper which cannot be removed by an NaEDTA wash.
DISCUSSION
In addition to regulating the expression of the two MMOs (11, 47, 52), the concentration of copper in the culture medium of methanotrophs affects a number of physiological traits in cells expressing the pMMO, such as membrane development (8, 11, 41, 49), metal acquisition (1, 8, 37, 63), methane oxidation activity (11, 31, 37, 49, 63), expression of membrane-associated polypeptides (2), and substrate affinity (33a). The results presented here add the extracellular concentration of the CBCs in M. trichosporium OB3b to this list. With respect to the extracellular concentration of CBCs, the response of wild-type M. trichosporium OB3b was similar to that observed in M. capsulatus Bath (63). The highest concentrations of copper-free CBCs were observed in the spent media of cells cultured under copper stress conditions just before the cells switched from expressing the pMMO to expressing the sMMO. Cells cultured in high-copper media and expressing the pMMO showed lower concentrations of CBCs in the spent media. Under higher-copper growth conditions, the majority of the CBCs were associated with the membrane fraction and contained bound copper.
As previously observed by Phelps et al. (42), the two sMMOC mutants PP319 and PP358 constitutively expressed the sMMO regardless of the copper concentration in the culture medium. The results presented here confirm the constitutive expression of the sMMO and little to no expression of the three pMMO polypeptides. With respect to the CBCs, the two sMMOC mutants PP319 and PP358 differed from wild-type M. trichosporium in a number of basic properties. In contrast to wild-type M. trichosporium OB3b, the CBC isolated from the spent media of sMMOC mutants cultured in high-copper media contained copper. Also in contrast to the wild-type strain, no CBCs were detected in the membrane fraction of sMMOC mutants regardless of the culture conditions. The mutations in sMMOC mutants PP319 and PP358 appear to be in the regulation of either the pMMO, the pMMO structural genes, or the copper acquisition system. Mutations in the pMMO structural genes seem unlikely since all three structural genes of the pMMO have been found in duplicate copies (51). Thus, the mutation in PP319 and PP358 appears to be in the regulation of either the two MMOs or a component of the copper acquisition system. The data presented here indicate the mutations in PP319 and PP358 are in the cellular mechanism utilized to bind copper-containing CBCs from the extracellular fraction. Whether the pMMO itself is the membrane-associated binding protein for copper-containing CBCs or whether a different uptake protein is involved has not been determined. Both mutants still acquire copper, but at only 15 to 20% of the copper uptake level observed in wild-type M. trichosporium OB3b. With the exception of pMMO expression, the mutants appear to show no negative effects of the lower concentrations of cell-associated copper. For example, the terminal oxidase activity in acetylene-treated PP319 or PP358, as measured by ascorbate-N,N,N′,N′-tetramethyl-p-phenylenediamine oxidase assay (results not shown), did not significantly differ from that of acetylene-treated wild-type M. trichosporium OB3b expressing either MMO. Thus, CBCs do not appear to be a general mechanism of copper acquisition but appear to be associated with expression of the pMMO or a secondary copper acquisition system.
Although the molecular structures of both CBCs are still under investigation, the results of N-terminal amino acid sequencing have provided some structural information about CBC-L1, CBC-L2, and the P1 and P3 fragment polypeptides. The fact that N-terminal sequencing proceeded normally suggests that the unknown residues, denoted by ?’s in Table 2, contain amino acid-like structural features. The known portion of the primary structure consists of amino acids that are not usually associated with the formation of strong copper complexes, although the thioether group of Met, the phenol and phenolate groups of Tyr, and the hydroxyl group of Ser are all metal-coordinating ligands (24, 52, 57). This observation suggests that the unknown functional groups may be responsible for siderophore properties of both CBCs while the native amino acids probably contribute to the formation of a conformation that facilitates copper complexation. In particular, the amino acid sequence YPG is known to promote β-turn formation in short linear peptides (14). A stable folded β-turn conformation would explain the pH-dependent changes in the hydrophobicity observed during the chromatographic purification of CBC as well as the peptide’s resistance to carboxypeptidase.
The physiological role of the CBCs in M. trichosporium OB3b or M. capsulatus Bath is still unknown. Both CBCs showed siderophore-like properties, but this activity was observed only in association with expression of the pMMO and may prove to be a major component of a pMMO-specific copper acquisition system. On the other hand, one or both CBCs may be more directly involved in methane oxidation by the pMMO. On SDS-denaturing gels, the purified pMMO consists of three polypeptides with molecular masses of 47,000, 27,000, and 25,000 Da (38, 63). Depending on the report, metal analysis of the purified enzyme from M. capsulatus Bath indicates the purified enzyme contains 0 to 2 nonheme iron and 12 to 15 copper atoms (38, 63). Depending on the current pMMO models, the catalytic site involves either both iron and copper or just copper (39, 63, 64). Solubilization of pMMO polypeptides in Triton X-100 or 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate followed by separation on a sucrose gradient revealed that two to four of the copper atoms are tightly associated with the two larger pMMO polypeptides, which are believed to be the site of methane oxidation by the pMMO (12, 46, 63). The X-band EPR spectra of the coppers associated with the two larger polypeptides is consistent with Cu2+ in a square planar configuration in which the copper is bound to three or four nitrogen atoms (24, 52, 57, 63). The 11 histidines residues in the two larger pMMO polypeptides can account for the ligation of the 2 to 4 tightly associated copper ions but not for the remaining 10 to 12 copper ions. The remaining copper ions are loosely associated with the three larger polypeptides (39, 63). Nguyen et al. (39) propose that the remaining copper ions are “ligated to the nitrogen atoms in the peptide backbone or other side chain ligands, such as carboxylates of glutamates and aspartates.” In contrast, results from this laboratory have suggested the loosely bound copper ions are associated with the CBCs. Copurification with the pMMO and the loss of methane oxidation activity with the removal of the CBCs from the three larger pMMO polypeptides indicate the CBCs may be a cofactor of the pMMO (63). The CBC may also provide a secondary function such as enzyme stabilization, protection from oxygen radicals as observed with other Cu complexes (15), maintaining a particular redox state, or sequestering copper.
Currently studies are focusing on the properties and structures of the two CBCs and their roles in copper acquisition in methanotrophs and in the environmental biochelation of metals. With respect to biochelation of metals, the properties of the two CBCs are similar to those of the copper-complexing ligands described by Gordon (21) and may represent the first isolated examples of organic chelators believed to be responsible for copper complexation in marine environments (5, 6, 13, 21).
ACKNOWLEDGMENTS
We thank A. B. Hooper (University of Minnesota), J. D. Semrau (University of Michigan), and D. J. Thiele (University of Michigan) for useful discussions, R. Arnold (University of Arizona) for providing unpublished data, R. S. Hanson (University of Minnesota) for antibodies to the reductase and B subunits of M. trichosporium OB3b sMMO, and J. Nott (ISU Protein Facility) for technical assistance.
This work was supported by Department of Energy 02-96ER20237 (A.A.D.), the Iowa State University Office of Biotechnology (A.A.D.), an Iowa State University Professional Advancement Grant (J.A.Z.), and National Science Foundation BES 9504383 (D.W.G. and A.T.).
REFERENCES
- 1.Berson O, Lidstrom M E. Study of copper accumulation by the type I methanotroph Methylomicrobium albus BG8. Environ Sci Technol. 1996;30:802–809. [Google Scholar]
- 2.Berson O, Lidstrom M E. Cloning and characterization of corA, a gene encoding a copper-repressible polypeptide in the type I methanotroph, Methylomicrobium albus BG8. FEMS Lett. 1997;148:169–174. doi: 10.1111/j.1574-6968.1997.tb10284.x. [DOI] [PubMed] [Google Scholar]
- 3.Brusseau G A, Tsien H-C, Hanson R S, Wackett L P. Optimization of trichloroethylene oxidation by methanotrophs and the use of colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation. 1990;1:19–29. doi: 10.1007/BF00117048. [DOI] [PubMed] [Google Scholar]
- 4.Cardy D J N, Laidler V, Salmond G P C, Murrell J C. Molecular analysis of the membrane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol Microbiol. 1991;5:335–342. doi: 10.1111/j.1365-2958.1991.tb02114.x. [DOI] [PubMed] [Google Scholar]
- 5.Coale K H, Bruland K W. Copper complexation in the northeast Pacific. Limnol Oceanogr. 1988;33:1084–1101. [Google Scholar]
- 6.Coale K H, Bruland K W. Spatial and temporal variability in copper complexation in the north Pacific. Deep-Sea Res. 1990;37:317–336. [Google Scholar]
- 7.Cody Y S, Gross D C. Characterization of pyoverdinpss, the fluorescent siderophore produced by Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1987;53:982–934. doi: 10.1128/aem.53.5.928-934.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins M L P, Buchholz L A, Remsen C C. Effect of copper on Methylomonas albus BG8. Appl Environ Microbiol. 1991;57:1261–1264. doi: 10.1128/aem.57.4.1261-1264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornish A, Nicholls K M, Scott P, Hunter B K, Aston W J, Higgins I J, Sanders J K M. In vivo13C NMR investigation of methanol oxidation by the obligate methanotroph Methylosinus trichosporium OB3b. J Gen Microbiol. 1984;130:2565–2575. [Google Scholar]
- 10.Cornish A, MacDonald J, Burrows K J, King T S, Scott D, Higgans I J. Succinate as an in vitro electron donor for the particulate methane monooxygenase of Methylosinus trichosporium OB3b. Biotechnol Lett. 1985;7:319–324. [Google Scholar]
- 11.Dalton H, Prior S D, Stanley S H. Regulation and control of methane monooxygenase. In: Crawford R L, Hanson R S, editors. Microbial growth on C1 compounds. Washington, D.C: American Society for Microbiology; 1984. pp. 75–82. [Google Scholar]
- 12.DiSpirito A A, Gulledge J, Shiemke A K, Murrell J C, Lidstrom M E, Krema C L. Trichloroethylene oxidation by the membrane-associated methane monooxygenase in type I, type II and type X methanotrophs. Biodegradation. 1992;2:151–164. [Google Scholar]
- 13.Donat J R, Kango R A, Gordon A S. Evaluation of immobilized metal affinity chromatography (IMAC) for isolation and recovery of strong copper-complexing ligands from marine waters. Mar Chem. 1997;57:1–10. [Google Scholar]
- 14.Dyson H, Rance J, Houghten M, Lerner R A, Wright P E. Folding of immunogenic peptide fragments of proteins in water solution. Sequence requirements for the formation of a reverse turn. J Mol Biol. 1998;201:161–200. doi: 10.1016/0022-2836(88)90446-9. [DOI] [PubMed] [Google Scholar]
- 15.El-Naggar M M. Protective action of some Cu(II) complexes against photohemolysis induced by m. chloroperbenzoic acid. J Inorg Biochem. 1997;65:263–266. doi: 10.1016/s0162-0134(96)00141-9. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson-Miller S, Brautigan D L, Margoliash E. Correlation of the kinetics of electron transfer activity of various eukaryotic cytochromes c with binding to mitochondrial cytochrome c oxidase. J Biol Chem. 1976;251:1104–1115. [PubMed] [Google Scholar]
- 17.Fitch M W, Graham D W, Arnold R G, Agarwal S K, Phelps P, Speitel G E, Jr, Georgiou G. Phenotypic characterization of copper-resistant mutants of Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1993;59:2771–2776. doi: 10.1128/aem.59.9.2771-2776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox B G, Surerus K K, Münck E, Lipscomb J D. Evidence for a μ-oxo-bridged binuclear iron cluster in the hydroxylase component of methane monooxygenase. J Biol Chem. 1988;263:10553–10556. [PubMed] [Google Scholar]
- 19.Fox B G, Froland W A, Dege J E, Lipscomb J D. Methane monooxygenase from Methylosinus trichosporium OB3b. J Biol Chem. 1989;264:10023–10033. [PubMed] [Google Scholar]
- 20.Fukasawa K, Goto M, Sasaki K, Hirata Y, Sato S. Structure of the yellow-green fluorescent peptide produced by iron-deficient Azotobacter vinelandii strain O. Tetrahedron. 1972;28:5359–5365. [Google Scholar]
- 21.Gordon A S. Microbial control of trace metals in the oceans. ASM News. 1998;64:79–83. [Google Scholar]
- 22.Green J, Dalton H. Protein B of soluble methane monooxygenase from Methylococcus capsulatus (Bath). A novel regulatory protein of enzyme activity. J Biol Chem. 1985;260:15795–15802. [PubMed] [Google Scholar]
- 23.Hendrich M P, Münck E, Fox B G, Lipscomb J D. Inter-spin EPR studies of the fully reduced methane monooxygenase hydroxylase component. J Am Chem Soc. 1990;112:5861–5865. [Google Scholar]
- 24.Holm R H, Kennopohl P, Solomon D I. Structural and functional aspects of metal sites in biology. Chem Rev. 1996;96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 25.Holmes A J, Costello A, Lidstrom M E, Murrell J C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 26.Hyman M R, Arp D J. The small-scale production of [U-14C] acetylene from Ba14CO3: application to labeling of ammonia monooxygenase in autotrophic nitrifying bacteria. Anal Biochem. 1990;190:348–353. doi: 10.1016/0003-2697(90)90206-o. [DOI] [PubMed] [Google Scholar]
- 27.Hyman M R, Arp D J. 14C2H2- and 14CO2-labeling studies of the de novo synthesis of polypeptides by Nitrosomonas europaea during recovery from acetylene and light inactivation of ammonia monooxygenase. J Biol Chem. 1992;267:1534–1545. [PubMed] [Google Scholar]
- 28.Janson J-C, Rydén C. Protein, principles, high resolution methods, and applications. Toronto, Ontario, Canada: VCH; 1997. [Google Scholar]
- 29.Koch K A, Peña M M O, Thiele D J. Copper-binding motifs in catalysis, transport, detoxification and signaling. Chem Biol. 1997;4:549–560. doi: 10.1016/s1074-5521(97)90241-6. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Larive, C. K. Unpublished results.
- 32.Lidstrom M E, Semrau J D. Metals and microbiology: the influence of copper on methane oxidation. In: Huang C P, editor. Advances in chemistry: aquatic chemistry. Washington, D.C: American Chemical Society; 1995. pp. 195–202. [Google Scholar]
- 33.Lipscomb J D. Biochemistry of the soluble methane monooxygenase. Annu Rev Biochem. 1994;48:371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- 33a.Lontoh S, Semrau J D. Methane and trichloroethylene degradation by Methylosinus trichosporium OB3b expressing particulate methane monooxygenase. Appl Environ Microbiol. 1998;64:1106–1114. doi: 10.1128/aem.64.3.1106-1114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 35.Lund J, Dalton H. Further characterization of the FAD and Fe2S2 redox centers of component C, the NADH: acceptor reductase of the soluble methane monooxygenase of Methylococcus capsulatus (Bath) Eur J Biochem. 1985;147:291–296. doi: 10.1111/j.1432-1033.1985.tb08749.x. [DOI] [PubMed] [Google Scholar]
- 36.Murrell J C, Hikes A J. Molecular biology of particulate methane monooxygenase. In: Lidstrom M E, Tabita F R, editors. Proceedings of the 8th International Symposium on Microbial Growth on C1 Compounds. Boston, Mass: Kluwer Academic Publishers; 1996. pp. 133–140. [Google Scholar]
- 37.Nguyen H-N T, Shiemke A K, Jacobs S J, Hales B J, Lidstrom M E, Chan S I. The nature of the copper ions in the membranes containing the particulate methane monooxygenase from Methylococcus capsulatus (Bath) J Biol Chem. 1994;269:14995–15005. [PubMed] [Google Scholar]
- 38.Nguyen H-N T, Nakagawa K H, Hedman B, Elliott S, Lidstrom M E, Hodgson K O, Chan S I. X-ray absorption and EPR studies on the copper ions associated with the particulate methane monooxygenase from Methylococcus capsulatus (Bath). Cu (I) ions and their implications. J Am Chem Soc. 1996;118:12766–12776. [Google Scholar]
- 39.Nguyen H-N T, Elliott S J, Yip J H-K, Chan S I. The particular methane monooxygenase from Methylococcus capsulatus (Bath) is a novel copper-containing three-subunit enzyme. J Biol Chem. 1998;273:7957–7966. doi: 10.1074/jbc.273.14.7957. [DOI] [PubMed] [Google Scholar]
- 40.Patel R N, Savas J C. Purification and properties of the hydroxylase component of methane monooxygenase. J Bacteriol. 1987;169:2313–2317. doi: 10.1128/jb.169.5.2313-2317.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peltola P, Priha P, Laakso S. Effect of copper on membrane lipids and on methane monooxygenase activity of Methylococcus capsulatus (Bath) Arch Microbiol. 1993;159:521–525. [Google Scholar]
- 42.Phelps P A, Agarwal S K, Speitel G E, Jr, Georgiou G. Methylosinus trichosporium OB3b mutants having constitutive expression of soluble methane monooxygenase in the presence of high levels of copper. Appl Environ Microbiol. 1992;58:3701–3708. doi: 10.1128/aem.58.11.3701-3708.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Philson S B, Llinás M. Siderochromes from Pseudomonas fluorescens. II. Structural homology as revealed by NMR spectroscopy. J Biol Chem. 1982;257:8086–8090. [PubMed] [Google Scholar]
- 44.Pilkington S J, Dalton H. Purification and characterization of the soluble methane monooxygenase from Methylosinus sporium demonstrates the highly conserved nature of the enzyme in methanotrophs. FEMS Microbiol Lett. 1991;78:103–108. [Google Scholar]
- 45.Premuzic T E, Francis A J, Lin M, Schubert J. Induced formation of chelating agents by Pseudomonas aeruginosa grown in presence of thorium and uranium. Arch Environ Contam Toxicol. 1985;14:759–768. doi: 10.1007/BF01055783. [DOI] [PubMed] [Google Scholar]
- 46.Prior S D, Dalton H. Acetylene as a suicide substrate and active site probe for membrane monooxygenase from Methylococcus capsulatus (Bath) FEMS Microbiol Lett. 1985;29:105–109. [Google Scholar]
- 47.Prior S D, Dalton H. The effect of copper ions on membrane content and methane monooxygenase activity in methanol-grown cells of Methylococcus capsulatus (Bath) J Gen Microbiol. 1985;131:155–163. [Google Scholar]
- 48.Rosenzweig A C, Frederick C A, Lippard S J, Norlund P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature (London) 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 49.Scott D, Brannan J, Higgins I J. The effect of growth conditions on intracytoplasmic membranes and methane monooxygenase activities in Methylosinus trichosporium OB3b. J Gen Microbiol. 1981;125:63–72. [Google Scholar]
- 50.Semrau J D, Zolandz D, Lidstrom M E, Chan S I. The role of copper in the pMMO of Methylococcus capsulatus Bath: a structural vs. catalytic function. J Inorg Chem. 1995;58:235–244. doi: 10.1016/0162-0134(94)00056-g. [DOI] [PubMed] [Google Scholar]
- 51.Semrau J D, Chistoserdov A, Lebron J, Costello A, Davagnino J, Kenna E, Holmes A J, Finch R, Murrell J C, Lidstrom M E. Particulate methane monooxygenase genes in methanotrophs. J Bacteriol. 1995;177:3071–3079. doi: 10.1128/jb.177.11.3071-3079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solomon E I, Sundaram U M, Machonkin T E. Multicopper oxidases and oxygenases. Chem Rev. 1996;96:2563–2605. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 53.Stanley S H, Prior S D, Leak D J, Dalton H. Copper stress underlies the fundamental change in intracellular location of methane mono-oxygenase in methane utilizing organisms: studies in batch and continuous cultures. Biotechnol Lett. 1983;5:487–492. [Google Scholar]
- 54.Teintze M, Leong J. Structure of pseudobactin A, a second siderophore from plant growth promoting Pseudomonas B10. Biochemistry. 1981;20:6457–6462. doi: 10.1021/bi00525a026. [DOI] [PubMed] [Google Scholar]
- 55.Téllez C M, Gaus K P, Graham D W, Arnold R G, Guzman R Z. Isolation of copper biochelates from Methylosinus trichosporium OB3b and sMMOC mutants. Appl Environ Microbiol. 1998;64:1115–1122. doi: 10.1128/aem.64.3.1115-1122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torres L, Pérez-Ortí L E, Tordera V, Beltrán J. Isolation and characterization of an Fe(II)-chelating compound produced by Pseudomonas syringae. Appl Environ Microbiol. 1986;52:157–160. doi: 10.1128/aem.52.1.157-160.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vänngård, T. Copper proteins, p. 411–447. In H. M. Swartz, J. R. Bolton, and D. C. Borg (ed.), Biological applications of electron spin resonance. Wiley-Interscience, New York, N.Y.
- 58.Wasserman A E. Absorption and fluorescence of water-soluble pigments produced by four species of Pseudomonas. Appl Microbiol. 1965;13:175–180. doi: 10.1128/am.13.2.175-180.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wendenbaum S, Demange P, Dell A, Meyer J M, Abdallah M A. The structure of pyoverdine Pa, the siderophore of Pseudomonas aeruginosa. Tetrahedron Lett. 1983;24:4877–4880. [Google Scholar]
- 60.Woodland M P, Dalton H. Purification and characterization of component A of the methane monooxygenase from Methylococcus capsulatus (Bath) J Biol Chem. 1984;259:53–60. [PubMed] [Google Scholar]
- 61.Yang C-C, Leong J. Structure of pseudobactin 7SR1, a siderophore from a plant-deleterious Pseudomonas. Biochemistry. 1984;23:3534–3540. doi: 10.1021/bi00310a023. [DOI] [PubMed] [Google Scholar]
- 62.Yuan H, Collins M L P, Antholine W E. Low-frequency EPR of the copper in particulate methane monooxygenase from Methylomicrobium albus BG8. J Am Chem Soc. 1997;119:5073–5074. [Google Scholar]
- 63.Zahn J A, DiSpirito A A. The membrane-associated methane monooxygenase from Methylococcus capsulatus Bath. J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zahn J A, Arcier D M, Hooper A B, DiSpirito A A. Evidence for an iron center in the ammonia monooxygenase from Nitrosomonas europaea. FEBS Lett. 1996;397:35–38. doi: 10.1016/s0014-5793(96)01116-7. [DOI] [PubMed] [Google Scholar]