Abstract
The Rhizobium etli rpoN1 gene, encoding the alternative sigma factor ς54 (RpoN), was recently characterized and shown to be involved in the assimilation of several nitrogen and carbon sources during free-living aerobic growth (J. Michiels, T. Van Soom, I. D’hooghe, B. Dombrecht, T. Benhassine, P. de Wilde, and J. Vanderleyden, J. Bacteriol. 180:1729–1740, 1998). We identified a second rpoN gene copy in R. etli, rpoN2, encoding a 54.0-kDa protein which displays 59% amino acid identity with the R. etli RpoN1 protein. The rpoN2 gene is cotranscribed with a short open reading frame, orf180, which codes for a protein with a size of 20.1 kDa that is homologous to several prokaryotic and eukaryotic proteins of similar size. In contrast to the R. etli rpoN1 mutant strain, inactivation of the rpoN2 gene did not produce any phenotypic defects during free-living growth. However, symbiotic nitrogen fixation was reduced by approximately 90% in the rpoN2 mutant, whereas wild-type levels of nitrogen fixation were observed in the rpoN1 mutant strain. Nitrogen fixation was completely abolished in the rpoN1 rpoN2 double mutant. Expression of rpoN1 was negatively autoregulated during aerobic growth and was reduced during microaerobiosis and symbiosis. In contrast, rpoN2-gusA and orf180-gusA fusions were not expressed aerobically but were strongly induced at low oxygen tensions or in bacteroids. Expression of rpoN2 and orf180 was abolished in R. etli rpoN1 rpoN2 and nifA mutants under all conditions tested. Under free-living microaerobic conditions, transcription of rpoN2 and orf180 required the RpoN1 protein. In symbiosis, expression of rpoN2 and orf180 occurred independently of the rpoN1 gene, suggesting the existence of an alternative symbiosis-specific mechanism of transcription activation.
Bacteria are able to activate or switch off specific sets of genes as they face changing environmental conditions. This can be achieved through the activities of RNA polymerases containing alternate ς factors and their cognate regulatory proteins. RNA polymerases containing the alternative sigma factor ς54 (ςN [RpoN]) recognize a conserved sequence motif centered at −24 and −12 nucleotides from the transcriptional start site (5′-TGGCAC-N5-TTGCA/T-3′ [see reference 9]). Transcription initiation from these promoters depends on the presence of specific activators which typically bind to DNA sequences located over 100 nucleotides upstream from the transcriptional start site (4). The activity of these activator proteins has been shown in several cases to be regulated by phosphorylation in response to physiological signals. In addition, evidence is accumulating that gene transcription by the ς54 RNA polymerase holoenzyme not only is regulated by the activator protein but also is regulated under certain conditions by ς54 abundance (3). In Caulobacter crescentus, transcription of rpoN is temporally regulated during the cell cycle and is 10-fold induced at the transition from swarmer to stalked cell (3). Upregulation of rpoN coincides with the onset of stalk formation and occurs before the ς54-dependent increase in flagellar gene expression. The rpoN gene from Rhodobacter capsulatus is organized in a nifU2-rpoN superoperon, and transcription of rpoN is under the control of two promoters. One promoter upstream of nifU2 is constitutively expressed with respect to nitrogen; a secondary nitrogen-dependent promoter is autoactivated by RpoN and NifA (5). Expression from the R. capsulatus rpoN secondary promoter is required for growth under certain stress conditions (5). Bradyrhizobium japonicum is presently the only bacterium in which two differentially regulated rpoN genes have been identified (16). Expression of the first copy, rpoN1, is activated in microaerobiosis by the FixLJ-FixK2 regulatory cascade, while the rpoN2 gene is negatively autoregulated (9, 16). In Azorhizobium caulinodans, one of the two genes coding for ς54, rpoF, is required for high-level nifA expression (19, 33). Indirect genetic evidence suggests that Rhodobacter sphaeroides (20) also contains a second rpoN gene.
Rhizobium etli is the nodulating symbiont of the common bean plant, Phaseolus vulgaris. It possesses at least two different regulatory cascades of nitrogen fixation genes. The first cascade is dependent on the transcriptional regulator nifA and controls the expression of both nifHDK operons, the third nifH gene, which is not linked to other nif genes, and the production of the black pigment melanin (23, 24, 34). Although the R. etli nifA gene is transcribed under aerobic and microaerobic conditions, NifA-dependent gene activation is only operative at low oxygen tensions (24). The second cascade is controlled by the R. etli fixLJ genes (6). Unlike the FixL protein from Sinorhizobium meliloti (14), R. etli FixL is not a hemoprotein (7). The physiological signal sensed by R. etli FixL is presently unknown. We have recently identified the R. etli rpoN gene (hereafter called rpoN1 [26]). Under free-living conditions, rpoN1 controls growth on C4-dicarboxylic acids and on several nitrogen sources. In addition, inactivation of rpoN1 abolishes microaerobic expression of nifH and the production of the black pigment melanin under free-living conditions (26).
Here, we describe the isolation of a second rpoN gene of R. etli CNPAF512, named rpoN2. This copy is located near a cluster of nitrogen fixation genes. Mutant analysis reveals that the rpoN1 and rpoN2 genes are both active, but under different physiological conditions. The rpoN1 gene is essential during free-living growth, while the second copy, rpoN2, is required for symbiosis. Transcription of the R. etli rpoN2 gene is highly induced under free-living microaerobic conditions by the RpoN1 and NifA proteins. In contrast, expression of rpoN2 in bacteroids occurs independently of the rpoN1 gene. We therefore propose that the activity of rpoN2 is regulated, in addition to oxygen, by a symbiosis-specific signal.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this work are listed in Table 1 or schematically presented in Fig. 1. Escherichia coli strains were grown at 37°C in Luria-Bertani medium. R. etli strains were cultured in liquid TY (0.5% tryptone, 0.3% yeast extract, 7 mM CaCl2) medium at 30°C or maintained on yeast-mannitol (YM) agar plates. Antibiotics were supplied to the medium to maintain selection for plasmids or to select recombinant strains at the following concentrations: 60 μg of nalidixic acid, spectinomycin, and neomycin per ml; 30 μg of kanamycin and gentamicin per ml; 100 μg of ampicillin per ml; and 1 μg (R. etli) or 10 μg (E. coli) of tetracycline per ml. Growth tests of R. etli on plates were carried out with acid minimal salts (AMS) medium (28) containing 1 mM CaCl2 and the carbon and nitrogen sources indicated in the text. Plates were incubated at 30°C under aerobic or microaerobic conditions (0.5% oxygen), and colony size was monitored over a period of 3 to 7 days. Triparental conjugations and site-directed mutagenesis of R. etli were done as described previously (6).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Gibco BRL | |
| HB101 | Smr | 30 |
| R. etli | ||
| CNPAF512 | Nalr, wild type | 26 |
| FAJ1154 | NmrrpoN1::Ω-Km | 26 |
| RpFAJ1002 | NmrfixL::Ω-Km | 6 |
| Rp1000 | NmrnifA::aphII | 24 |
| FAJ1169 | NmrrpoN2::Ω-Km | This work |
| FAJ1170 | Nmr SprrpoN1::Ω-Sp rpoN2::Ω-Km | This work |
| FAJ1172 | Nmr SprrpoN1::Ω-Sp rpoN2::pJQ200-UC1-Km | This work |
| FAJ1173 | Nmrorf180::Ω-Km | This work |
| FAJ1175 | SprrpoN1::Ω-Sp | This work |
| FAJ1176 | Nmr SprfixL::Ω-Km rpoN1::Ω-Sp | This work |
| CNPAF512:: pFAJ1187 | NmrrpoN2::pFAJ1187 | This work |
| CNPAF512:: pFAJ1188 | Nmrorf180::pFAJ1188 | This work |
| Plasmids | ||
| pUC18 | Apr, cloning vector | 27 |
| pUC18Not | Apr, cloning vector | 15 |
| pCR2.1 | Apr, PCR TA cloning vector | Invitrogen |
| pJQ200-UC1 | GmrsacB | 29 |
| pHP45Ω-Km | Apr Kmr | 8 |
| pHP45Ω-Sp | Apr Spr | 8 |
| pWM6 | Apr NmruidA2 | 22 |
| pLAFR1 | Tcr, broad-host-range vector | 12 |
| pLAFR3 | Tcr, broad-host-range vector | 32 |
| pFAJ21 | Tcr, A. brasilense pnifH-gusA | 35 |
| pFAJ1172 | Tcr, rpoN2 gene in pLAFR1 | This work |
| pFAJ1173 | Tcr, rpoN2 gene in pLAFR1 | This work |
| pFAJ1174 | Apr, 4.2-kb NotI fragment containing rpoN2 and orf180 cloned in pUC18Not | This work |
| pFAJ1175 | Tcr, rpoN2-gusA fusion in pLAFR3 | This work |
| pFAJ1176 | Tcr, orf180-gusA fusion in pLAFR3 | This work |
| pFAJ1180 | Tcr, rpoN2-gusA SmaI promoter deletion in pLAFR3 | This work |
| pFAJ1181 | Tcr, orf180-gusA SmaI promoter deletion in pLAFR3 | This work |
| pFAJ1183 | Gmr Kmr, pJQ200-UC1 containing rpoN2::Ω-Km | This work |
| pFAJ1184 | Gmr Kmr, pJQ200-UC1 containing orf180::Ω-Km | This work |
| pFAJ1185 | Apr, pCR2.1 containing a 0.6-kb internal PCR fragment from rpoN2 | This work |
| pFAJ1186 | Gmr Kmr, pJQ200-UC1 containing rpoN1::Ω-Sp | This work |
| pFAJ1187 | Gmr Kmr, pJQ200-UC1 containing rpoN2::gusA | This work |
| pFAJ1188 | Gmr Kmr, pJQ200-UC1 containing orf180::gusA | This work |
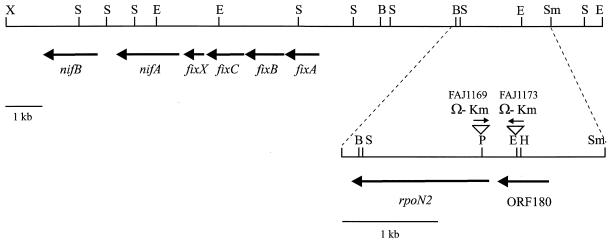
FIG. 1.
Physical map of the R. etli nif region containing the rpoN2 gene. The physical map of the 2.8-kb fragment that was sequenced is given below. The positions and orientations of the different ORFs are presented below the restriction maps. Triangles represent genomic insertions of the Ω-Km interposon or the gusA gene. B, BamHI; E, EcoRI; H, HindIII; P, PstI; S, SalI; Sm, SmaI; X, XbaI.
DNA methods.
DNA preparation and recombinant DNA techniques were performed according to standard procedures (2, 30). DNA fragments were recovered from agarose gels with the Nucleotrap kit (Macherey-Nagel). Hybridizations were carried out with digoxigenin-labeled probes as described by the manufacturer (Boehringer Mannheim). To generate blunt ends to incompatible DNA fragments, DNA was incubated with Klenow or T4 DNA polymerases in the presence of the four deoxynucleoside triphosphates.
DNA sequencing.
Automated DNA sequencing was performed with an A.L.F. sequencer (Pharmacia Biotech) with fluorescein-labeled oligonucleotide primers. Sequence data analysis was performed with the PC/Gene software package (IntelliGenetics, Inc.). Homology searches were carried out with the BLAST server at the National Center for Biotechnology Information.
PCR-mediated isolation of cosmid clones carrying the R. etli rpoN2 gene.
A 0.6-kb fragment from the R. etli rpoN2 gene was amplified with the primers OJM081 (5′-CTGAGAATTCGACGCGGCTGACGGTGGATTCGTGC-3′) and OJM082 (5′-CTGAGAATTCGATCCGCCGGGTGTTTTCGC-3′) and genomic DNA from the R. etli rpoN1 mutant strain FAJ1154 as a template (26). Amplification of DNA fragments by PCR was performed as described previously (26), except that the annealing temperature was 50°C. The primers were chosen in conserved regions of rpoN genes and annealed at each site of the PstI restriction site of the rpoN1 gene used to insert the Ω-Km interposon, thereby avoiding preferential amplification of the rpoN1 gene. The 0.6-kb PCR product was subsequently cloned into the pCR2.1 TA cloning vector (Invitrogen). The nucleotide sequence of this fragment was different from that of rpoN1, but was clearly homologous to the rpoN1 and rpoN genes from other bacterial species.
Because we were unable to isolate a cosmid clone containing rpoN2 by colony PCR with OJM081 and OJM082, new primers perfectly matching the rpoN2 sequence were designed (OJM093, 5′-GAATCCATCATACCCGACGTC-3′; and OJM094, 5′-TGGTGGCGCACGATTTCGGTC-3′) to amplify a 240-bp fragment. These primers were used to screen a pLAFR1 cosmid library of R. etli CNPAF512, maintained in E. coli HB101 and containing approximately 2,000 clones (6), for plasmids carrying the R. etli rpoN2 gene. PCRs were first carried out with cell mixtures of 16 different clones. In a second step, pools of 16 clones yielding a fragment of the expected size were each analyzed separately. Two positive overlapping cosmid clones, pFAJ1172 and pFAJ1173, were selected for further analysis.
Expression analysis of gusA fusions and melanin production.
Precultures were grown overnight in TY medium and diluted 20-fold in AMS medium containing 10 mM NH4Cl and 10 mM succinate. Microaerobic induction of the different strains was carried out overnight at 0.3% oxygen as described previously (24). β-Glucuronidase activity and melanin production were quantified as described previously (26).
Plant culture and bacteroid isolation.
Seeds of Phaseolus vulgaris cv. Limburgse vroege were sterilized, and seedlings were inoculated as previously described (25). Bean plants were grown in 250-ml cylindrical flasks in a Sanyo Gallenkamp Fitotron plant growth room with a 12-h photoperiod (day/night temperature, 22°C/18°C; day/night relative humidity, 65%/75%). Acetylene reduction activities (ARAs) were determined 4 weeks after inoculation on a Hewlett-Packard 5890A gas chromatograph equipped with a “PLOT fused silica” column. Ethylene production was quantified with propane as an internal standard.
To determine symbiotic expression of the different gusA fusion plasmids, bacteroids were isolated from the nodules of 4-week-old plants. For this, all of the nodules of one plant were collected and treated essentially as described by Leyva et al. (18). The nodules were crushed on ice in 15-ml plastic tubes in the presence of 0.5 g of polyvinylpolypyrrolidone and 2 ml of Mg-phosphate buffer (2.5 mM MgCl2, 50 mM potassium phosphate [pH 6.8]). Next, the paste was diluted with 10 ml of ice-cold Mg-phosphate buffer. The tubes were centrifuged at 121 × g for 1 min to remove plant cell material. This step was repeated with the supernatant. Bacteroids were finally precipitated at 4°C by centrifugation at 4,500 × g for 5 min and resuspended in 100 μl to 1 ml of Mg-phosphate buffer. Dilutions of the bacteroids were immediately assayed for glucuronidase activity.
Construction of mutants.
To construct rpoN2 and orf180 mutants, a 4.2-kb fragment containing the entire rpoN2 gene, except the amino terminus, and the complete orf180 gene was amplified by PCR with primers OJM104 (5′-CTGAGGATCCGCGGCCGCGCCTGTTTCCTTGAGCTTG-3′) and OJM106 (5′-CT GAGGATCCGCGGCCGCTTTGTCCTGAATGTCAGTTC-3′) annealing near the 3′ end of rpoN2 and approximately 2 kb upstream from orf180, respectively. Both primers contain NotI recognition sites at their 5′ ends. The 4.2-kb DNA fragment was cloned into the NotI site of pUC18Not (15). The resulting plasmid, pFAJ1174, was digested with PstI (rpoN2) or EcoRI (orf180) and blunt end ligated to the 1.8-kb BamHI fragment from pHP45Ω-Km, thereby inactivating the rpoN2 gene or orf180, respectively. In both plasmids, the orientations of Ω-Km and rpoN2 or orf180 are opposed and the same, respectively (Fig. 1). Finally, the 6-kb NotI fragments from the resulting plasmids were cloned into the NotI site of the Rhizobium suicide plasmid pJQ200-UC1, yielding pFAJ1183 (rpoN2::ΩKm) and pFAJ1184 (orf180::ΩKm). These two mutations were finally recombined into the genome of R. etli CNPAF512. Insertion of the Ω-Km cassette at the correct site was confirmed by Southern hybridization with the appropriate DNA probes. The R. etli rpoN2 mutant was named FAJ1169, and the R. etli orf180 mutant was named FAJ1173.
An R. etli rpoN1 rpoN2 double mutant was constructed as follows. A pJQ200-UC1 derivative containing the 1.8-kb EcoRI fragment from pFAJ1150 carrying part of the R. etli rpoN1 gene (26) was digested with PstI and blunt end ligated with the 1.7-kb Ω-Sp cassette from pHP45Ω-Sp, thereby inactivating the gene. The construct carrying rpoN1 with the Ω-Sp cassette in the opposite orientation (pFAJ1186) was used to mutagenize FAJ1169. The resulting mutants were finally tested for growth on AMS plates containing 10 g of mannitol and 2 g of alanine per liter. rpoN1 mutants are unable to grow on alanine (26). Appropriate insertion of the resistance cassette was verified by Southern hybridization. The R. etli rpoN1 rpoN2 double mutant was named FAJ1170.
To confirm the results obtained with FAJ1170, we also constructed a second, independent R. etli rpoN1 rpoN2 double mutant. R. etli CNPAF512 was mutagenized with plasmid pFAJ1186, yielding the R. etli rpoN1 mutant FAJ1175. Next, the rpoN2 internal 0.6-kb PCR fragment, amplified with primers OJM081 and OJM082 (described above), was cloned into pCR2.1. The resulting vector was digested with BamHI and ligated to the 3.8-kb BamHI fragment from pWM6, thereby inserting a Nmr gene in a site flanking the 0.6-kb insert. The 4.4-kb KpnI-NotI insert from this construct was finally ligated into SmaI-digested pJQ200-UCI, after the ends of the insert fragment had been blunted. This plasmid was used to mutagenize R. etli FAJ1175. In the resulting R. etli rpoN1 rpoN2 strain, FAJ1172, the rpoN2 gene is disrupted by insertion of the entire plasmid into the gene.
Finally, to determine whether the R. etli rpoN2 and fixLJ genes belong to the same regulatory cascade, a R. etli fixL rpoN1 double mutant was generated. For this, the R. etli fixL mutant RpFAJ1002 (6) was mutagenized with pFAJ1186. The resulting strain was named FAJ1176.
Construction of rpoN2-gusA and orf180-gusA plasmid and chromosomally integrated fusions.
To construct transcriptional rpoN2-gusA and orf180-gusA fusions, pFAJ1174 was digested with PstI (rpoN2) or EcoRI (orf180) and blunt end ligated to the 3.8-kb BamHI fragment from pWM6. The two 8-kb NotI fragments carrying the gusA gene insertion in the right orientation were cloned, after being blunted, into BamHI-digested pLAFR3 or into the NotI site of pJQ200-UC1. The resulting plasmids were named pFAJ1175 (pLAFR3-rpoN2-gusA), pFAJ1176 (pLAFR3-orf180-gusA), pFAJ1187 (pJQ200-UC1-rpoN2-gusA), and pFAJ1188 (pJQ200-UC1-orf180-gusA). To construct chromosomally integrated rpoN2-gusA and orf180-gusA fusions, pFAJ1187 and pFAJ1188 were integrated at the homologous site into the R. etli CNPAF512 genome, yielding CNPAF512::pFAJ1187 and CNPAF512::pFAJ1188. Promoter deletions of pFAJ1175 and pFAJ1176 were constructed as follows (also see Fig. 1). In pFAJ1180, the 6.3-kb SmaI fragment from pFAJ1175 was blunt end ligated into the BamHI site of pLAFR3. Similarly, to generate pFAJ1181, the 6.3-kb SmaI fragment from pFAJ1176 was blunt end ligated into the BamHI site of pLAFR3.
Nucleotide sequence accession number.
The nucleotide sequence of the R. etli rpoN2 gene locus has been deposited in the DDBJ-EMBL-GenBank nucleotide sequence databases under accession no. AJ005696.
RESULTS
Isolation and localization of the R. etli rpoN2 gene.
We have recently characterized the R. etli rpoN1 gene (26). Mutant analysis indicated that although the R. etli rpoN1 mutant had phenotypic defects under free-living conditions (26), symbiotic nitrogen fixation was not affected, suggesting the existence of a second rpoN copy. Therefore, based on the DNA sequence of R. etli rpoN1, several PCR primers were designed such that they annealed to conserved regions of rpoN genes belonging to different organisms. The primers were chosen to anneal at both sites of the rpoN1 Ω-Km insertion, making it possible to distinguish between PCR fragments originating from the rpoN1 and rpoN2 genes, because PCR was performed with genomic DNA isolated from the R. etli rpoN1 mutant, FAJ1154. A 0.6-kb internal fragment of the rpoN2 gene was amplified with one of these primer combinations (see also Materials and Methods). Based on the DNA sequence of this fragment, new primers, specific for the rpoN2 gene, were designed and used to screen a pLAFR1 genomic library of R. etli CNPAF512 (6), as detailed in Materials and Methods. Two overlapping cosmid clones, pFAJ1172 and pFAJ1173, were isolated. Partial DNA sequence analysis and restriction mapping of the DNA fragments common to both clones confirmed the identity of rpoN2 and allowed us to localize the rpoN2 gene approximately 3.5 kb from the previously identified nifB-nifA-fixABCX cluster of nitrogen fixation genes (23, 24). A physical map of this DNA region is presented in Fig. 1.
Sequence analysis of R. etli rpoN2.
To further characterize the rpoN homologous DNA region, the DNA sequence of a 2,739-bp fragment was established (Fig. 2). Two putative open reading frames (ORFs) were identified (Fig. 1 and 2). The first ORF codes for a protein that displays 59% amino acid identity with the previously characterized R. etli RpoN1 protein and was therefore named rpoN2. RpoN2 contains 483 amino acids and has a calculated molecular mass of 54,013 Da and a pI of 5.71, which is higher than that of RpoN1 (pI, 4.32). A putative ribosome-binding site (AGGAG) is located 13 bp upstream from the rpoN2 initiation codon. An alignment was made between R. etli RpoN1 and RpoN2 and the RpoN proteins from S. meliloti (1), B. japonicum (16), A. caulinodans (33), and Rhizobium sp. strain NGR234 (36). The three domains (as defined by van Slooten et al. [36]) of RpoN2 are amino-terminal glutamine-rich region I (M1 to E50), acidic region II (E50 to E118), and carboxy-terminal region III (E118 to G483). The latter domain contains a conserved helix-turn-helix motif involved in DNA binding and the RpoN box (36). All rhizobial RpoN proteins have a domain II of 98 to 110 amino acids, with two notable exceptions in which the acidic domain is considerably shorter, B. japonicum RpoN1 (62 amino acids [E50 to T112]) and R. etli RpoN2 (68 amino acids). Two hydrophobic heptad repeats have been proposed to form an intramolecular leucine zipper in the E. coli RpoN protein (31). Similar structures were found in rhizobial RpoN proteins and have the consensus L21-L28-L35-E42 (amino acid numbering refers to R. etli RpoN1) and (L/I)198-L205-V212-(L/I)219. An alignment of RpoN proteins indicated that while B. japonicum RpoN1 and RpoN2 are highly similar (87% amino acid identity), the RpoN proteins from R. etli are more divergent. The R. etli RpoN1 protein displays higher amino acid identity with the RpoN proteins of S. meliloti (68%) and Rhizobium sp. strain NGR234 (65%) compared to the R. etli RpoN2 protein. Also at the level of DNA sequence, the G+C contents of both R. etli rpoN genes differ (rpoN1, 64%; rpoN2, 56%). It is therefore likely that the rpoN2 gene did not arise by gene duplication but by lateral transfer.
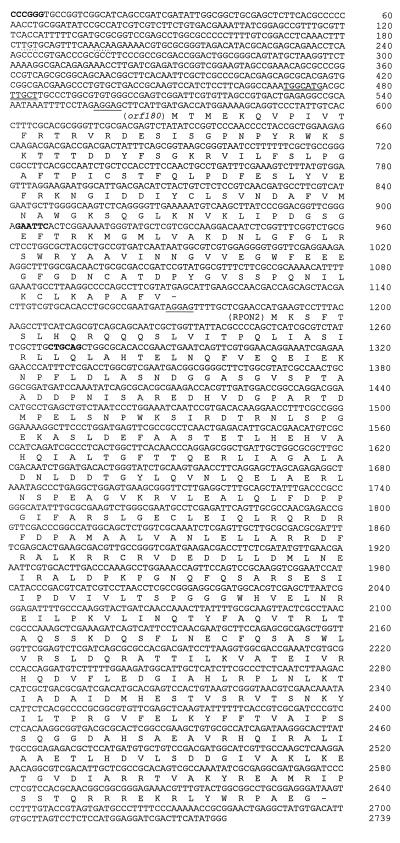
FIG. 2.
DNA sequence of the 2,739-bp DNA fragment containing R. etli rpoN2 and orf180. The deduced amino acid sequence of these genes is indicated below the DNA sequence. Putative ribosome-binding sites are underlined. Sequences resembling the consensus of a −24/−12 promoter and a NifA-binding site are marked with a double line and with dots, respectively. The SmaI, EcoRI, and PstI restriction sites are indicated in boldface.
Sequence analysis of R. etli orf180.
Upstream from the R. etli rpoN2 gene, a second ORF was identified (Fig. 1 and 2). This ORF codes for a putative protein of 180 amino acids with a predicted molecular mass of 20,180 Da and a pI of 7.82. orf180 is located 78 bp upstream from rpoN2 and might therefore be cotranscribed with the rpoN2 gene. A putative Shine-Dalgarno sequence (AGGAG) is found 8 bp upstream from orf180. In addition, putative regulatory sequences were identified in the 5′ region of orf180. A sequence (TGGCA-N6-TTGCT [underlined in Fig. 2]) resembling the consensus of a −24/−12 promoter (9) was located 84 nucleotides upstream from the putative ATG initiation codon. Approximately 270 bp upstream from this −24/−12 type of promoter, a possible NifA-binding (TGT-N10-ACA) site was found. Homology searches in databases revealed homology between the gene product of R. etli orf180 and a protein encoded by the y4vd ORF located on the symbiotic plasmid of Rhizobium sp. strain NGR234 (74% amino acid identity [Q53212]), a protein from Haemophilus influenzae (44% identity [P44758]), and a putative membrane protein from Synechocystis sp. strain PCC6803 (60% identity [D90909]). An alignment of these proteins is shown in Fig. 3. In addition, lower homology was detected with yeast peroxisomal proteins from Lipomyces kononenkoae (26% identity [U11244]), Candida boidinii (26% identity [P14292] and 25% identity [P14293]), Schizosaccharomyces pombe (35% identity [G2598045]), and an unknown protein from Arabidopsis thaliana (32% identity [AC002292]). Homology is found over the entire protein length, except for the amino terminus (the first 44 amino acids in the R. etli protein encoded by orf180). Amino acids conserved between the prokaryotic proteins and the yeast peroxisomal proteins are underlined in Fig. 3. The conserved amino acids are also found in the A. thaliana protein, with the exception of the strongly conserved PGAFTP(T/I/P)C motif, in which only the G and F residues are maintained. The positive and negative charges in the protein encoded by orf180 are evenly distributed throughout the protein, with the exception of a stretch of 17 amino acids (overlined in Fig. 3) predicted to form a putative transmembrane segment (17). Interestingly, this hydrophobic segment is conserved among the prokaryotic and eukaryotic (except A. thaliana) proteins and contains the PGAFTP(T/I/P)C motif.
FIG. 3.
Alignment of the prokaryotic protein sequences homologous to the gene product of R. etli orf180. RE, protein encoded by R. etli orf180; NGR234, Rhizobium sp. strain NGR234 21.4-kDa protein translated from the ORF y4vd (Q53212); SYN, Synechocystis protein encoded by ORF sII1621 (D90909); HI, H. influenzae 26.7-kDa protein (P44758). Identical and similar amino acids (S-T-A, L-V-I-M, K-R, Q-N, and F-Y-W) are indicated below the protein sequences by stars and dots, respectively. Residues that are also conserved in the yeast proteins from Lipomyces kononenkoae (U11244), C. boidinii (P14292 and P14293), and S. pombe (G2598045) are underlined. The putative transmembrane hydrophobic domain in the protein encoded by R. etli orf180 is overlined.
Analysis of R. etli rpoN mutants.
To further characterize the rpoN genes of R. etli, a phenotypical analysis of R. etli rpoN1 (FAJ1154), rpoN2 (FAJ1169), rpoN1 rpoN2 (FAJ1170 and FAJ1172), and rpoN1 fixL (FAJ1176) mutants was carried out under free-living and symbiotic conditions. First, growth of these mutants was tested under aerobic and microaerobic conditions on defined media containing ammonium, nitrate, and alanine as nitrogen sources (Table 2). These compounds were previously shown to reduce growth of an R. etli rpoN1 mutant during aerobic growth (26). In contrast to the wild type, growth of FAJ1154 was clearly reduced on alanine and mannitol both aerobically and microaerobically, while growth was not affected on the complex TY medium and on AMS minimal medium containing mannitol (carbon source) and glutamine (nitrogen source) (Table 2). Similar phenotypes were observed with the double mutants rpoN1 rpoN2 (FAJ1170 and FAJ1172) and rpoN1 fixL (FAJ1176). However, the R. etli rpoN2 mutant displayed wild-type growth on each of the different media tested under aerobic as well as microaerobic conditions. Growth phenotypes were different when nitrate or ammonium was used as an N source and mannitol was used as a C source (data not shown). Aerobic growth and microaerobic growth of the wild type and the rpoN2 mutant were indistinguishable when nitrate or ammonium was used as an N source. Also, the growth rates of the rpoN1, rpoN1 fixL, and rpoN double mutants were identical both aerobically and microaerobically on these media. Growth of these mutants was clearly reduced compared to that of the wild type during aerobic growth. Under microaerobic conditions, these mutants produced increased amounts of exopolysaccharides compared to the rpoN2 mutant and the wild-type strain. Excessive production of exopolysaccharides by the rpoN1 mutants may indicate a low intracellular concentration of fixed nitrogen, as can be expected from nitrate assimilatory mutants. These results indicate that under free-living conditions, the phenotypes of the R. etli wild type and the rpoN2 mutant are similar, and also the growth rates of the rpoN1 and rpoN double mutant strains are indistinguishable from each other on the different media tested.
TABLE 2.
Growth of R. etli rpoN mutants under free-living aerobic and microaerobic conditions
| Straina | Relevant genotype | Result for growth onb:
|
|||||
|---|---|---|---|---|---|---|---|
| Glutamine + mannitol
|
TY medium
|
Alanine + mannitol
|
|||||
| Aerobic | Microaerobic | Aerobic | Microaerobic | Aerobic | Microaerobic | ||
| CNPAF512 | Wild type | +++ | ++ | ++ | + | +++ | + |
| FAJ1154 | rpoN1 | +++ | ++ | ++ | + | +/− | +/− |
| FAJ1169 | rpoN2 | +++ | ++ | ++ | + | +++ | + |
| FAJ1170 | rpoN1 rpoN2 | +++ | ++ | ++ | + | +/− | +/− |
| FAJ1172 | rpoN1 rpoN2 | +++ | ++ | ++ | + | +/− | +/− |
| FAJ1173 | orf180 | +++ | ++ | ++ | + | +++ | + |
| FAJ1176 | fixL rpoN1 | +++ | ++ | ++ | + | +/− | +/− |
R. etli strains CNPAF512 (wild type), FAJ1154 (rpoN1), FAJ1169 (rpoN2), FAJ1170 and FAJ1172 (rpoN1 rpoN2), FAJ1173 (orf180 mutant), and FAJ1176 (rpoN1 fixL).
Growth tests were carried out on TY or AMS plates containing glutamine or alanine at 2 g/liter and mannitol at 10 g/liter. Plates were incubated aerobically or microaerobically (0.5% oxygen), and colony size was determined after 5 days of incubation at 30°C. +/−, colony size <0.5 mm; +, size 0.5 to 1 mm; ++, size 1 to 2 mm; +++, size >2 mm.
The different mutant strains were also inoculated on P. vulgaris, the common bean plant, and tested for nodulation and symbiotic nitrogen fixation activity (Table 3). The shape and size of the nodules induced by the wild type and the different mutant strains were similar. Also, nodule dry weight and nodule number of plants inoculated with the rpoN2 and rpoN1 rpoN2 mutants were not different from those of plants nodulated by the wild type. In contrast, the nodule number was significantly higher on plants inoculated with the nifA mutant, although the nodule dry weight was not statistically different. A marked difference in symbiotic nitrogen fixation activity of both rpoN mutants was observed. While the rpoN1 mutant shows wild-type ARA, nitrogen fixation is reduced by more than 90% in the rpoN2 mutant strain FAJ1169 and completely abolished in the rpoN1 rpoN2 mutants FAJ1170 and FAJ1172. From these results, it can be concluded that the rpoN2 gene is essential for nitrogen fixation while rpoN1 is dispensable. In addition, ARA of an rpoN1 fixL double mutant is not abolished, indicating that the R. etli fixL gene is not required for rpoN2 transcription during symbiosis. Taken together, the free-living and symbiotic data indicate that the activity of both R. etli rpoN genes is required under distinct physiological conditions. While R. etli rpoN1 controls the assimilation of several nitrogen and carbon sources during free-living growth, rpoN2 function is essential for symbiotic nitrogen fixation.
TABLE 3.
Nitrogenase activity and nodulation of R. etli wild type and rpoN mutants
| Strain | Relevant genotype | ARA (μmol of ethylene/ plant/h)a | No. of nodules/ plantb | Nodule dry wt/plant (mg)b |
|---|---|---|---|---|
| CNPAF512 | Wild type | 3.2 (1.3) | 170 (35) | 48 (19) |
| Rp1000 | nifA | 0 | 212 (45) | 60 (24) |
| FAJ1154 | rpoN1 | 2.8 (0.7) | 143 (53) | 41 (23) |
| FAJ1169 | rpoN2 | 0.3 (0.3) | 149 (41) | 38 (19) |
| FAJ1170 | rpoN1 rpoN2 | 0 | 188 (47) | 63 (14) |
| FAJ1176 | rpoN1 fixL | 3.5 (1.1) | NDc | ND |
| FAJ1173 | orf180 | 0.6 (0.5) | ND | ND |
| FAJ1173/ | 0.9 (0.3) | ND | ND | |
| pFAJ1175 |
Data are the means of at least five plants. Standard deviations are given in parentheses.
Data are the means of 11 (FAJ1169), 10 (CNPAF512 and Rp1000), and 8 (FAJ1170) plants. Standard deviations are given in parentheses.
ND, not determined.
Analysis of an R. etli orf180 mutant.
Immediately upstream from R. etli rpoN2, an ORF coding for a 180-amino-acid protein was identified. To determine its function, orf180 was inactivated by insertion of the Ω-Km cassette into the EcoRI restriction site of the gene, yielding strain FAJ1173 (Fig. 1). This mutant was tested for growth on several nitrogen sources under aerobic and microaerobic conditions (Table 2). No difference between the wild type and mutant FAJ1173 was observed on these media. In addition, the symbiotic phenotype of this mutant strain was determined (Table 3). The nodulation phenotypes of the wild type and mutant were indistinguishable. However, nitrogen fixation activity was strongly reduced compared with the wild-type level and was not different from that of the R. etli rpoN2 mutant. The phenotypes of the R. etli orf180 mutant and the rpoN2 mutant, as shown in Tables 2 and 3, are identical. Since the Ω-Km interposon is known to create polar mutations, it is possible that the observed phenotypes of FAJ1173 are due to absence of transcription of the rpoN2 gene, which is located downstream from orf180. This hypothesis was tested by complementation of the R. etli orf180 mutant strain, FAJ1173, with plasmid pFAJ1175 containing R. etli orf180 and an inactivated rpoN2 gene (Table 3). No complementation of the symbiotic phenotype of FAJ1173 was observed, indicating that orf180 and rpoN2 are probably transcribed from a promoter located upstream from orf180.
Expression analysis of nifH-gusA and production of melanin.
The effect of mutations introduced into the rpoN1 and rpoN2 genes and orf180 on the RpoN/NifA-dependent expression of nifH and production of melanin was tested under free-living aerobic and microaerobic (0.3% oxygen) conditions and in bacteroids (Table 4). Expression of nifH was monitored by using the Azospirillum brasilense translational nifH-gusA fusion plasmid, pFAJ21 (35). The production of the black pigment melanin in R. etli was previously shown to be dependent on the presence of NifA and RpoN1 (24, 26). From Table 4, it can be seen that as expected, expression of the nifH gene and the synthesis of melanin are abolished under aerobic conditions in all strains tested but are highly induced under microaerobic conditions in the wild type. No expression of pFAJ21 under microaerobic conditions or production of melanin during microaerobiosis or symbiosis is observed in the R. etli nifA mutant and in both rpoN1 rpoN2 double mutants. However, levels of β-glucuronidase activity of pFAJ21 and melanin production during microaerobic growth were identical in the rpoN2 mutant, the orf180 mutant, and the wild type. These results indicate that rpoN1, but not rpoN2, is required for free-living expression of the A. brasilense nifH-gusA fusion and for the production of melanin. During symbiosis, melanin production was abolished only in the nifA mutant and in the rpoN double mutant strain, suggesting that both rpoN genes can substitute for each other with respect to synthesis of melanin. Induction of pFAJ21 was also assayed in complex TY medium and in AMS medium containing mannitol as a carbon source and NH4Cl or nitrate as a nitrogen source (data not shown). In contrast to the wild type, no activity of the fusion could be observed in the rpoN1 mutant strain under any of the growth conditions tested.
TABLE 4.
Expression of the nifH-gusA fusion plasmid pFAJ21 and production of melanin in R. etli wild type and mutants
| Strain | Relevant genotype | β-Glucuronidase activity of pFAJ21 (Miller units)a
|
Melanin productionb
|
|||
|---|---|---|---|---|---|---|
| Aerobic | Microaerobic | Aerobic | Microaerobic | Symbiotic | ||
| CNPAF512 | Wild type | 19 (6) | 1,841 (327) | 0 (2) | 93 (4) | 114 (56) |
| Rp1000 | nifA | 0 (1) | 0 (2) | 0 (5) | 0 (2) | 4 (7) |
| RpFAJ1002 | fixL | 9 (7) | 1,778 (125) | NDc | ND | 104 (14) |
| FAJ1154 | rpoN1 | 0 (10) | 0 (14) | 0 (5) | 10 (11) | 118 (66) |
| FAJ1169 | rpoN2 | 54 (14) | 2,149 (243) | 0 (2) | 82 (21) | 121 (69) |
| FAJ1170 | rpoN1 rpoN2 | 0 (17) | 0 (21) | 0 (3) | 15 (21) | 11 (11) |
| FAJ1172 | rpoN1 rpoN2 | 0 (6) | 0 (11) | 0 (5) | 14 (19) | 8 (9) |
| FAJ1173 | orf180 | 23 (6) | 2,107 (167) | 0 (1) | 89 (12) | 122 (28) |
β-Glucuronidase activities are the means of at least four replicates. Standard deviations are given within parentheses.
Melanin production was quantified essentially as described by Michiels et al. (26). Units are expressed as the ratio of the change in optical density at 340 nm (ΔOD340)/OD595 (cell density)/time (minutes). Standard deviations are given in parentheses.
ND, not determined.
Expression analysis of R. etli rpoN1, rpoN2, and orf180.
From the data concerning growth and symbiotic phenotypes, expression of nifH, and production of melanin in single and double R. etli rpoN mutant strains, it appears that both rpoN genes are active under different physiological conditions. The rpoN1 gene is essential during free-living growth, and the presence of rpoN2 is required for an effective symbiosis. We tested whether these observations could be explained by differential expression of both genes. First, the expression of genomic gusA fusions, integrated at their homologous site, was monitored (Table 5). Expression of the R. etli rpoN1 gene was previously shown to occur independently of the nitrogen concentration (26). In addition, activity of a chromosomally integrated rpoN1-gusA fusion was shown to be negatively autoregulated. Here, we observed that the level of expression of rpoN1-gusA under free-living microaerobic and symbiotic conditions in a wild-type background was reduced approximately fourfold compared to free-living aerobic expression (Table 5). On the other hand, microaerobic expression of rpoN1-gusA in the rpoN1 mutant strain FAJ1156 was only slightly reduced (approximately 40%) compared to the aerobic expression level. In contrast to the expression of rpoN1-gusA, wild-type genomic orf180-gusA and rpoN2-gusA fusions were not expressed aerobically but were strongly induced during microaerobic growth and symbiosis (Table 5). From this expression analysis, it can be concluded that in the R. etli wild type, rpoN1 but not rpoN2 is expressed during aerobic growth, while the rpoN2 gene, but not rpoN1, is strongly transcribed in bacteroids.
TABLE 5.
Expression of genomic rpoN1-gusA, rpoN2-gusA, and orf180-gusA fusions
| Strain | Relevant genotype | β-Glucuronidase activity (Miller units)a
|
||
|---|---|---|---|---|
| Aero- bic | Micro- aerobic | Sym- biotic | ||
| FAJ1156 | rpoN1, rpoN1-gusA | 185 (46) | 116 (63) | 8 (1) |
| FAJ1156/pFAJ1150 | Wild type, rpoN1-gusA | 27 (10) | 7 (10) | 7 (1) |
| CNPAF512::pFAJ1187 | Wild type, rpoN2-gusA | 0 (2) | 674 (42) | 1,487 (366) |
| CNPAF512::pFAJ1188 | Wild type, orf180-gusA | 0 (2) | 67 (14) | 503 (169) |
β-Glucuronidase activities are the means of at least eight replicates. Standard deviations are given within parentheses.
To investigate in more detail the regulation of rpoN2 and orf180, plasmid-borne gusA fusions were constructed with both genes as detailed in Materials and Methods. Plasmid pFAJ1175 carries a 2.6-kb rpoN2 promoter fragment, and pFAJ1176 carries a 1.9-kb DNA fragment from the orf180 promoter. The gusA gene was inserted into the PstI site of rpoN2 at 28 codons from the presumptive 5′ terminus of the gene. In pFAJ1176, the gusA cassette was cloned in the EcoRI site of orf180 at 111 codons from the proposed ATG initiation codon. Expression of rpoN2-gusA (pFAJ1175) and orf180-gusA (pFAJ1176) was monitored in the CNPAF512 wild type and in R. etli rpoN1 (FAJ1154), rpoN2 (FAJ1169), rpoN1 rpoN2 (FAJ1170 and FAJ1172), orf180 (FAJ1173), nifA (Rp1000), and fixL (RpFAJ1002) mutant strains under free-living aerobic and microaerobic conditions and in bacteroids. From Table 6, it can be seen that both fusion plasmids are not expressed aerobically and are strongly induced in the wild-type strain upon incubation at 0.3% oxygen and during symbiosis. These data are in accordance with the results obtained in strains CNPAF512::FAJ1187 and CNPAF512::FAJ1188 carrying the rpoN2 and orf180 gusA genomic fusions. Wild-type expression levels are also observed in the fixL mutant strain RpFAJ1002, confirming the observation that nitrogen fixation is not abolished in the R. etli rpoN1 fixL double mutant, FAJ1176. No expression under any condition is obtained in the R. etli nifA mutant and in the rpoN double mutants, indicating that transcription of rpoN2 and orf180 is controlled by RpoN and NifA during free-living growth and symbiosis. No gusA activity of pFAJ1175 and pFAJ1176 was found in the rpoN1 mutant under free-living microaerobic conditions, whereas wild-type activity was obtained in bacteroids. On the other hand, the inactivation of the rpoN2 gene or orf180 had no effect on free-living or symbiotic expression of both rpoN2-gusA and orf180-gusA fusions. To confirm these results, two additional gusA fusion plasmids derived from pFAJ1175 and pFAJ1176 were constructed. These plasmids carry a deletion of approximately 1.5 kb from the SmaI site (Fig. 1). The putative −24/−12 promoter and the NifA-binding site in the promoters of rpoN2 and orf180 were not deleted in these plasmids. These fusion plasmids were pFAJ1180 (rpoN2) and pFAJ1181 (orf180). Similar expression patterns to those obtained with pFAJ1175 and pFAJ1176 were observed in the different mutant strains during free-living growth (Table 6).
TABLE 6.
Expression of rpoN2-gusA and orf180-gusA fusions in R. etli wild-type and mutant strains
| Straina | β-Glucuronidase activity (Miller units)b
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
rpoN2-gusA
|
orf180-gusA
|
|||||||||
| pFAJ1175
|
pFAJ1179
|
pFAJ1176
|
pFAJ1181
|
|||||||
| Aerobic | Microaerobic | Symbiotic | Aerobic | Microaerobic | Aerobic | Microaerobic | Symbiotic | Aerobic | Microaerobic | |
| CNPAF512 | 16 (7) | 1,686 (507) | 1,638 (401) | 202 (27) | 897 (160) | 15 (2) | 217 (31) | 290 (92) | 0 (5) | 267 (166) |
| Rp1000 | 0 (19) | 32 (21) | 0 (0) | 177 (8) | 175 (17) | 9 (4) | 4 (13) | 0 (21) | 8 (7) | 0 (6) |
| RpFAJ1002 | 11 (1) | 1,320 (105) | 2,454 (878) | 141 (14) | 694 (35) | 4 (6) | 181 (21) | 395 (215) | 4 (4) | 188 (36) |
| FAJ1154 | 0 (14) | 0 (35) | 1,836 (655) | 205 (37) | 189 (30) | 11 (6) | 0 (18) | 409 (133) | 0 (20) | 0 (18) |
| FAJ1169 | 32 (4) | 1,195 (105) | 1,724 (667) | 213 (56) | 875 (86) | 1 (1) | 322 (166) | NDc | 0 (1) | 111 (14) |
| FAJ1170 | 20 (22) | 0 (23) | 51 (37) | 103 (49) | 125 (56) | 0 (17) | 0 (18) | ND | 0 (0) | 0 (33) |
| FAJ1172 | 0 (8) | 0 (6) | 0 (0) | 140 (63) | 190 (54) | 0 (4) | 0 (25) | ND | 2 (9) | 0 (14) |
| FAJ1173 | 32 (1) | 1,194 (288) | 1,483 (601) | 190 (44) | 731 (161) | 0 (1) | 343 (146) | ND | 2 (5) | 194 (23) |
R. etli strains CNPAF512 (wild type), Rp1000 (nifA), RpFAJ1002 (fixL), FAJ1154 (rpoN1), FAJ1169 (rpoN2), FAJ1170 and FAJ1172 (rpoN1 rpoN2), and FAJ1173 (orf180 mutant).
Expression was determined as defined in footnote a of Table 4. Standard deviations are given in parentheses.
ND, not determined.
Several conclusions can be drawn from these observations. First, the expression patterns of rpoN2 and orf180 are identical, suggesting coordinate expression of both genes. Second, transcription of rpoN2 can be highly induced under free-living microaerobic conditions and depends on the presence of a functional rpoN1 gene and on NifA. Under these conditions, no effect of rpoN2 inactivation on the expression level of rpoN2 is observed. Third, rpoN2 is highly expressed during symbiosis. Transcription is abolished in nifA and rpoN double mutant strains, but not in R. etli rpoN1 or rpoN2 single mutants, indicating that both rpoN genes can functionally substitute for each other to activate rpoN2 and orf180 expression during symbiosis. Finally, expression of rpoN1 is negatively autoregulated and reduced in a wild-type background during microaerobic growth and in bacteroids. It is possible that in addition to RpoN1, RpoN2 negatively regulates rpoN1 expression. First, compared with aerobic conditions, rpoN1 expression is strongly reduced microaerobically in the wild type, but not in an rpoN1 mutant strain. Second, the rpoN2 gene is expressed microaerobically in the wild type, but not in the R. etli rpoN1 mutant.
DISCUSSION
We have isolated the second R. etli rpoN gene, rpoN2. This rpoN copy is clustered with nitrogen fixation genes and is located approximately 3.5 kb upstream from the R. etli fixA gene (23). Kullik et al. (16) have previously demonstrated that B. japonicum contains two differentially regulated rpoN genes. None of the genes maps closely to one of the previously identified nif gene regions. The B. japonicum rpoN2 gene, but not rpoN1, is flanked by ORFs that are also conserved in other species (21). In A. caulinodans, the rpoN gene is flanked by conserved ORFs (33). A second rpoN copy, named rpoF, was shown to control the expression of the regulatory nifA gene (19). On the basis of indirect evidence, a second rpoN gene has also been suggested to exist in the photosynthetic bacteria R. sphaeroides (20) and R. capsulatus (10). In R. sphaeroides, the identified rpoN gene is not linked to conserved ORFs but is adjacent to the 3′ end of the nifUSVW gene cluster and is probably cotranscribed with the latter genes (20). Because the inactivation of the rpoN gene in R. sphaeroides does not impair diazotrophic growth, the existence of a second copy was postulated.
The R. etli rpoN2 gene product displays 59% amino acid identity with the RpoN1 protein. This is considerably lower than the RpoN proteins from B. japonicum, which are 87% identical to each other. Also, R. etli RpoN1 is more closely related to the RpoN proteins from S. meliloti and Rhizobium sp. strain NGR234 than to the R. etli RpoN2 protein. In addition, the G+C contents of the R. etli rpoN1 and rpoN2 genes differ considerably. It is therefore likely that the R. etli rpoN2 gene did not arise by duplication of a common ancestral rpoN gene, but may be the result of lateral transfer.
The protein encoded by orf180 is homologous to several prokaryotic (Rhizobium sp. strain NGR234, H. influenzae, and Synechocystis sp. strain PCC6803) and eukaryotic (yeast and A. thaliana) proteins. The H. influenzae protein possesses an additional C-terminal glutaredoxin domain and is therefore assumed to participate in redox reactions. The highest similarity is found with the gene product of Rhizobium sp. strain NGR234 y4vd, which maps on the symbiotic plasmid pNGR234a approximately 1.4 kb upstream from fixA (11). The promoter region of y4vd contains a consensus −24/−12 promoter sequence and a NifA-binding site, as observed in the R. etli orf180 promoter region. In C. boidinii, two PMP20-like peroxisomal proteins are also homologous to the R. etli orf180-encoded protein. These proteins are abundantly synthesized when methanol is used as the sole carbon source and are believed to be associated with the membrane of peroxisomes (13). The function of orf180 in R. etli is unclear, but because it is coregulated with nitrogen fixation genes, it is tempting to speculate that it is involved in processes related to symbiotic nitrogen fixation.
R. etli orf180 is probably cotranscribed with the rpoN2 gene. First, no recognizable terminator structure is found in the 78-bp intergenic region between orf180 and rpoN2. Second, under free-living conditions, expression of both genes is controlled by NifA and RpoN1, in agreement with the presence of RpoN- and NifA-binding sites in the promoter region of orf180. Finally, the reduced nitrogen fixation activity of an R. etli orf180 insertion mutant cannot be restored to the wild-type level by a plasmid carrying only orf180.
During aerobic growth, the R. etli rpoN1 gene was shown to be expressed at a constant level independently of the nitrogen source and was negatively autoregulated. Under these conditions, the rpoN2 gene is not expressed. The R. etli rpoN1 mutant strain displays growth defects on C4-dicarboxylic acids and on the nitrogen sources ammonium, nitrate, alanine, and serine (26).
When cells were cultured under free-living microaerobic conditions, expression of the R. etli rpoN1 gene was strongly decreased and transcription of rpoN2 was activated. The latter process was strictly dependent on the presence of RpoN1 and NifA proteins. Therefore, also under microaerobic conditions, no growth of the rpoN1 mutant strain was observed when alanine was used as a nitrogen source. Similarly, microaerobic production of melanin or expression of a nifH-gusA fusion plasmid was abolished in the rpoN1 mutant strain. Therefore, no active RpoN2 protein is formed under free-living aerobic or microaerobic conditions in the rpoN1 mutant. In B. japonicum, both rpoN genes are functional during free-living growth. The rpoN2 gene is required aerobically for growth on nitrate, while both copies are active under microaerobic conditions (16).
We have previously proposed that the consensus of a −24/−12 promoter overlapping the −10 promoter region and the transcription start site of rpoN1 is responsible for the observed negative autoregulation (26). RpoN1 may bind to this sequence and thereby inhibit transcription initiation from the −35/−10 promoter of the rpoN1 gene. If active RpoN2 protein is formed during microaerobiosis in the wild-type strain, this protein could account for the strongly decreased expression level of rpoN1 by directly interacting with the rpoN1 promoter. This is not the case in B. japonicum, because repression of rpoN2 transcription has been specifically attributed to the presence of RpoN2 and not RpoN1 (16).
Microaerobic expression of rpoN genes has been reported in two other organisms. In B. japonicum, rpoN1 is activated by an Fnr-like protein, FixK2. The expression of the fixK2 gene is controlled by the oxygen sensor proteins FixLJ (9, 16). The R. capsulatus rpoN gene is organized in a nifU2-rpoN superoperon structure (5). A primary promoter upstream from rpoN is expressed constitutively. The secondary promoter, which is NifA and RpoN dependent, accounts for increased induction under low-nitrogen and low-oxygen conditions. R. etli rpoN2 constitutes the second example of a NifA-controlled rpoN gene.
Expression of the R. etli rpoN1 gene is shut off during symbiosis. The RpoN2 protein could account for this effect, because it is strongly expressed under these conditions. In addition, an R. etli rpoN2 mutant strain still produces wild-type levels of melanin and activates rpoN2-gusA and orf180-gusA fusions, indicating that in this mutant, the rpoN1 gene is no longer repressed. In contrast to rpoN1, transcription of rpoN2 is highly activated during symbiosis. Transcription activation of rpoN2 requires the NifA protein, but in contrast to free-living conditions, is not abolished in an R. etli rpoN1 mutant strain. Therefore, symbiotic expression of rpoN2 involves a symbiosis-specific mechanism, as suggested for other Rhizobium genes (37). It is possible that this mechanism accounts for a low basal level of expression of the rpoN2 gene. After initial transcription, autoactivation by NifA/RpoN2 may then amplify rpoN2 transcription. The mechanism involved is at present unclear, but it does not involve the R. etli fixLJ genes, because an rpoN1 fixL double mutant still fixes nitrogen.
Nitrogenase activity of nodules induced by the rpoN1 mutant strain is not different from that of the wild type. On the other hand, nitrogen fixation is reduced by approximately 90% in the rpoN2 mutant. At present, it is unclear whether the difference between the symbiotic phenotypes of rpoN1 and rpoN2 mutants is due to a difference in expression level of the rpoN genes. The rpoN2 gene is highly induced in bacteroids, while rpoN1 is negatively autoregulated and can therefore not reach the high expression levels that may be required during symbiosis to activate nitrogen fixation genes. Alternatively, the RpoN1 and RpoN2 proteins may have distinct affinities for different −24/−12 promoters, depending on their DNA sequence, as was previously reported for A. caulinodans (19).
Transcriptional regulation of sigma factors ensures that bacteria express specific sets of genes during specific physiological conditions (38). In recent years, it has become clear that expression levels of ς54 may vary according to the physiological need. Organisms seem to have evolved two different mechanisms. One unique rpoN gene may be subject to different control mechanisms. In C. crescentus, expression of the rpoN gene is increased at the swarmer-to-stalked-cell transition, where ς54 is required for stalk biosynthesis and transcription of flagellar genes (3). Expression of the rpoN gene from R. capsulatus is controlled by two promoters (5). The first promoter is expressed constitutively and allows cells to grow under normal nitrogen-limiting conditions. The secondary promoter is autoactivated and is required for growth on nitrogen-limiting high-salt or low-iron medium. A second mechanism that bacteria use to cope with an increased demand of ς54 is a differential regulation of two rpoN gene copies. In B. japonicum, rpoN2 is expressed under all conditions tested. In contrast, transcription of rpoN1 is activated during microaerobiosis and symbiosis, allowing rpoN2 to boost its expression during nitrogen fixation (16). A. caulinodans also possesses two genes coding for ς54 (19, 33). Their regulation has not yet been studied. Also in R. etli, two differentially regulated rpoN genes were identified. The first copy, rpoN1, is required for housekeeping functions and allows cells to grow on several nitrogen and carbon sources under free-living conditions. The second copy is essential for symbiotic nitrogen fixation.
ACKNOWLEDGMENTS
J.M. is a Postdoctoral Fellow of the Fund for Scientific Research—Flanders. We acknowledge financial support from the Fund for Scientific Research—Flanders (FWO G.0220.97).
REFERENCES
- 1.Albright L M, Ronson C W, Nixon B T, Ausubel F M. Identification of a gene linked to Rhizobium meliloti ntrA whose product is homologous to a family of ATP-binding proteins. J Bacteriol. 1989;171:1932–1941. doi: 10.1128/jb.171.4.1932-1941.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 3.Brun Y V, Shapiro L. A temporally controlled ς-factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev. 1992;6:2395–2408. doi: 10.1101/gad.6.12a.2395. [DOI] [PubMed] [Google Scholar]
- 4.Buck M, Miller S, Drummond M, Dixon R. Upstream activator sequences are present in the promoters of nitrogen fixation genes. Nature. 1986;320:374–378. [Google Scholar]
- 5.Cullen P J, Foster-Harnett D, Gabbert K K, Kranz R G. Structure and expression of the alternative sigma factor, RpoN, in Rhodobacter capsulatus: physiological relevance of an autoactivated nifU2-rpoN superoperon. Mol Microbiol. 1994;11:51–65. doi: 10.1111/j.1365-2958.1994.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 6.D’hooghe L, Michiels J, Vlassak K, Verreth C, Waelkens F, Vanderleyden J. Structural and functional analysis of the fixLJ genes of Rhizobium leguminosarum biovar phaseoli CNPAF512. Mol Gen Genet. 1995;249:117–126. doi: 10.1007/BF00290243. [DOI] [PubMed] [Google Scholar]
- 7.D’hooghe I, Michiels J, Vanderleyden J. The Rhizobium etli FixL protein differs in structure from other known FixL proteins. Mol Gen Genet. 1998;257:576–580. doi: 10.1007/s004380050684. [DOI] [PubMed] [Google Scholar]
- 8.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 9.Fischer H-M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster-Harnett D, Kranz R G. Analysis of the promoters and upstream sequences of nifA1 and nifA2 in Rhodobacter capsulatus: activation requires ntrC but not rpoN. Mol Microbiol. 1992;6:1049–1060. doi: 10.1111/j.1365-2958.1992.tb02170.x. [DOI] [PubMed] [Google Scholar]
- 11.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 12.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 13.Garrard L J, Goodman J M. Two genes encode the major membrane-associated protein of methanol-induced peroxisomes from Candida boidinii. J Biol Chem. 1989;264:13929–13937. [PubMed] [Google Scholar]
- 14.Gilles-Gonzales M A, Ditta G S, Helinski D R. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature. 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 15.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosome insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kullik I, Fritsche S, Knobel H, Sanjuan J, Hennecke H, Fischer H-M. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the ς54 gene (rpoN) J Bacteriol. 1991;173:1125–1138. doi: 10.1128/jb.173.3.1125-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein hydropathy scale. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 18.Leyva A, Palacios J M, Murillo J, Ruiz-Argüeso T. Genetic organization of the hydrogen uptake (hup) cluster from Rhizobium leguminosarum. J Bacteriol. 1990;172:1647–1655. doi: 10.1128/jb.172.3.1647-1655.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loroch A I, Nguyen B G, Ludwig R A. Interactive regulation of Azorhizobium nifA transcription via overlapping promoters. J Bacteriol. 1995;177:7210–7221. doi: 10.1128/jb.177.24.7210-7221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meijer W G, Tabita F R. Isolation and characterization of the nifUSVW-rpoN gene cluster from Rhodobacter sphaeroides. J Bacteriol. 1992;174:3855–3866. doi: 10.1128/jb.174.12.3855-3866.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 22.Metcalf W W, Wanner B L. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene. 1993;129:17–25. doi: 10.1016/0378-1119(93)90691-u. [DOI] [PubMed] [Google Scholar]
- 23.Michiels J, Vanderleyden J. Cloning and sequence of the Rhizobium leguminosarum biovar phaseoli fixA gene. Biochim Biophys Acta. 1993;1144:232–233. doi: 10.1016/0005-2728(93)90179-j. [DOI] [PubMed] [Google Scholar]
- 24.Michiels J, D’Hooghe I, Verreth C, Pelemans H, Vanderleyden J. Characterization of the Rhizobium leguminosarum biovar phaseoli nifA gene, a positive regulator of nif gene expression. Arch Microbiol. 1994;161:404–408. doi: 10.1007/BF00288950. [DOI] [PubMed] [Google Scholar]
- 25.Michiels J, Pelemans H, Vlassak K, Verreth C, Vanderleyden J. Identification and characterization of a Rhizobium leguminosarum bv. phaseoli gene that is important for nodulation competitiveness and shows structural homology to a Rhizobium fredii host-inducible gene. Mol Plant-Microbe Interact. 1995;8:468–472. doi: 10.1094/mpmi-8-0468. [DOI] [PubMed] [Google Scholar]
- 26.Michiels J, Van Soom T, D’hooghe I, Dombrecht B, Benhassine T, de Wilde P, Vanderleyden J. The Rhizobium etli rpoN locus: DNA sequence analysis and phenotypical characterization of rpoN, ptsN, and ptsA mutants. J Bacteriol. 1998;180:1729–1740. doi: 10.1128/jb.180.7.1729-1740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norrander K, Kempe T, Messing K. Construction of improved M13 vectors using oligonucleotide directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 28.Poole P S, Schofield N A, Reid C J, Drew E M, Walshaw D L. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology. 1994;140:2797–2809. doi: 10.1099/00221287-140-10-2797. [DOI] [PubMed] [Google Scholar]
- 29.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Sasse-Dwight S, Gralla J. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor ς54. Cell. 1990;62:945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- 32.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stigter J, Schneider M, de Bruijn F J. Azorhizobium caulinodans nitrogen fixation (nif/fix) gene regulation: mutagenesis of the nifA −24/−12 promoter element, characterization of a ntrA (rpoN) gene, and derivation of a model. Mol Plant-Microbe Interact. 1993;6:238–252. doi: 10.1094/mpmi-6-238. [DOI] [PubMed] [Google Scholar]
- 34.Valderama B, Dávalos A, Girard L, Morett E, Mora J. Regulatory proteins and cis-acting elements involved in the transcriptional control of Rhizobium etli reiterated nifH genes. J Bacteriol. 1996;178:3119–3126. doi: 10.1128/jb.178.11.3119-3126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vande Broek A, Michiels J, de Faria S M, Milcamps A, Vanderleyden J. Transcription of the Azospirillum brasilense nifH gene is positively regulated by NifA and NtrA and is negatively controlled by the cellular nitrogen status. Mol Gen Genet. 1992;232:279–283. doi: 10.1007/BF00280007. [DOI] [PubMed] [Google Scholar]
- 36.van Slooten J C, Cervantes E, Broughton W J, Wong C H, Stanley J. Sequence and analysis of the rpoN sigma factor gene of Rhizobium sp. strain NGR234, a primary coregulator of symbiosis. J Bacteriol. 1990;172:5563–5574. doi: 10.1128/jb.172.10.5563-5574.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Soom C, Vanderleyden J. A quest for symbiosis specific genes urges itself upon Rhizobium geneticists. Mol Microbiol. 1997;24:669–670. doi: 10.1046/j.1365-2958.1997.3581927.x. [DOI] [PubMed] [Google Scholar]
- 38.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]