Abstract
Cyclin-dependent kinases (CDKs) regulate cell cycle progression and the transcription of a number of genes, including lipid metabolism-related genes, and aberrant lipid metabolism is involved in prostate carcinogenesis. Previous studies have shown that CDK13 expression is upregulated and fatty acid synthesis is increased in prostate cancer (PCa). However, the molecular mechanisms linking CDK13 upregulation and aberrant lipid metabolism in PCa cells remain largely unknown. Here, we showed that upregulation of CDK13 in PCa cells increases the fatty acyl chains and lipid classes, leading to lipid deposition in the cells, which is positively correlated with the expression of acetyl-CoA carboxylase (ACC1), the first rate-limiting enzyme in fatty acid synthesis. Gain- and loss-of-function studies showed that ACC1 mediates CDK13-induced lipid accumulation and PCa progression by enhancing lipid synthesis. Mechanistically, CDK13 interacts with RNA-methyltransferase NSUN5 to promote its phosphorylation at Ser327. In turn, phosphorylated NSUN5 catalyzes the m5C modification of ACC1 mRNA, and then the m5C-modified ACC1 mRNA binds to ALYREF to enhance its stability and nuclear export, thereby contributing to an increase in ACC1 expression and lipid deposition in PCa cells. Overall, our results disclose a novel function of CDK13 in regulating the ACC1 expression and identify a previously unrecognized CDK13/NSUN5/ACC1 pathway that mediates fatty acid synthesis and lipid accumulation in PCa cells, and targeting this newly identified pathway may be a novel therapeutic option for the treatment of PCa.
Subject terms: Cancer metabolism, Epigenetics, Oncogenes, Lipidomics
Introduction
Prostate cancer (PCa) is one of the most common cancers in middle-aged and older men [1]. Unlike other cancer types, PCa depends on the androgen receptor (AR) signaling for growth [2]. Therefore, androgen deprivation therapy (ADT) and anti-androgen therapy have become the main methods of clinical intervention in advanced PCa [3]. Although most patients initially respond well to ADT and anti-androgen therapy, after a period of treatment (18 months), a subset of patients become castration-resistant and no curative treatments exist [4]. Thus, there is an urgent need to discover new approaches to prevent, inhibit or even reverse the progression of castration-resistant PCa.
Tumor cell proliferation and survival depend upon their metabolic changes [5]. The Warburg effect is a common feature of many cancer cells, which facilitates cell growth and cancer progression by altering glucose, glutamine and lipid metabolism. However, PCa is an exception to this general process because prostate cells have truncated TCA cycles [6], which produces high level of citrate by inhibiting the Krebs cycle [7]. Accumulating evidence suggests that PCa cells highly express several enzymes associated with de novo lipogenesis, and the high expression of these enzymes and aberrant lipid metabolism are critical for PCa growth, hormone-refractory progression and treatment resistance [8]. However, little is known about how lipid metabolism is dysregulated in PCa pathogenesis.
Acetyl-CoA carboxylase 1 (ACC1) catalyzes the conversion of acetyl-CoA to malonyl-CoA, the first rate-limiting enzyme in the synthesis pathway of nascent fatty acids [9]. Growing evidence indicates that ACC1 is involved in the progression of various tumors. A preclinical study reported that inhibition of ACC1 can repress fatty acid synthesis and tumor progress in non-small cell lung cancer [10]. In prostate and breast cancer, siRNA-mediated silencing of the ACC1 expression reduces fatty acid synthesis, inhibits cell cycle progression and induces cell apoptosis [11, 12]. These observations suggest that ACC1 may play a crucial role in promoting fatty acid synthesis and PCa progression. Recent studies have found that ACC1 expression is regulated at the transcriptional and post-transcriptional levels. For instance, the level of m6A and its Reader HNRNPA2B1 is significantly increased in esophageal cancer, and HNRNPA2B1 promotes esophageal cancer progression by upregulating ACC1 [13]. The lncRNA CTD-2245E15.3 interacts with ACC1 protein and suppresses its phosphorylation at an inhibitory site for enzymatic activity, supporting non-small cell lung cancer growth [14]. In addition, 5-methylcytosine (m5C), as an important post-transcriptional modification, has been identified to be involved in the regulation of subcellular localization and stability of some mRNAs [15]. Despite increasing knowledge of post-transcriptional regulation of ACC1 expression, the precise mechanisms underlying the regulation of the ACC1 expression in PCa cells remain largely unknown.
Cyclin-dependent kinases (CDKs) promote cellular proliferation by driving cell cycle phase, and dysregulated CDKs often lead to abnormal proliferation of tumor cells [16]. Emerging evidence suggests that beyond the cell cycle, multiple CDKs participate in the regulation of cell metabolism and are involved in lipid metabolism [17]. For example, CDK8 is involved in inhibiting adipogenesis, and the absence of CDK8 leads to abnormal fat accumulation in Drosophila and mammals [18]. CDK9 acts as a key mediator of fatty acid metabolism in PCa cells, and inhibition of both CDK9 and CPT1/2 induces PCa cell death [19]. The fatty acid synthase inhibitor orlistat induces hepatoma Hep3B cell death by inhibiting the expression of cyclin A, cyclin B, CDK1, CDK2, and CDK4 [20]. CDK4/6 participate in mitochondrial fatty acid oxidation in melanoma cells [21]. CDK12 and CDK13 are evolutionarily related and structurally similar kinases, besides phosphorylating the carboxy-terminal domain (CTD) of RNA polymerase II (RNA Pol II), they also phosphorylate other substrates, which may be critical for their functions in metabolic regulation [17]. However, other targets of CDK12 and CDK13 have yet to be fully characterized. Our previous study found that CDK13 stimulates the formation of a positive feedback loop between circCDK13, miR-212-5p/miR-449a and E2F5, which contributes to the progression of PCa [22]. However, whether and how CDK13 is implicated in the regulation of ACC1 expression and fatty acid synthesis in PCa cells has not been well investigated. In the present study, we hypothesized that CDK13 may promote PCa progression via facilitating ACC1 expression and lipid accumulation in PCa cells.
Results
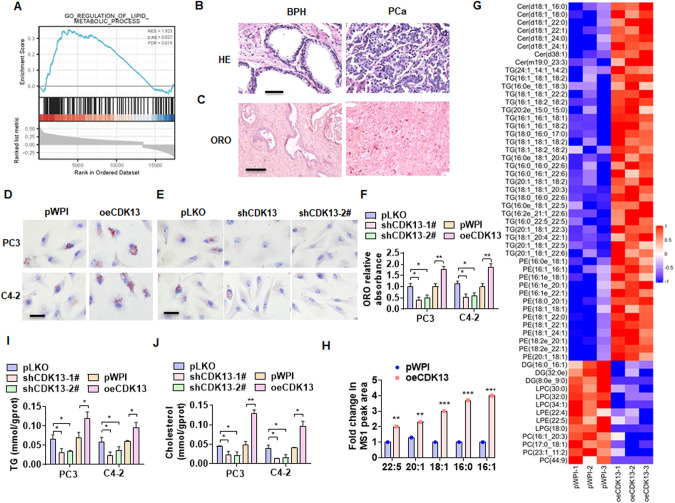
CDK13 promotes fatty acid synthesis and lipid deposition in PCa cells
The increased CDK13 expression and lipid accumulation have been associated with PCa development and progression [22]. Although high expression of CDK13 mRNA has not been found in our data to be associated with poor outcomes in patients, it is unclear whether CDK13 regulates PCa progression by affecting lipid metabolism. To clarify the causal relationship between CDK13 upregulation and lipid accumulation, we performed a gene set enrichment analysis (GSEA) using TCGA dataset and found that CDK13 has a close relationship with lipid metabolism process (Fig. 1A). Oil Red O (ORO) staining of the clinical specimens revealed the higher lipid deposition in PCa tissues than that in BPH tissues (Fig. 1B, C). Next, the above observations were further experimentally investigated by overexpressing or silencing CDK13 in PC3 and C4-2 cells. The results showed that CDK13 overexpression significantly increased and knockdown of CDK13 decreased lipid accumulation in both PC3 and C4-2 cells, as shown by ORO staining (Fig. 1D–F). To further validate these results, we performed mass spectrometry–based global lipidomic analysis to comparatively evaluate the lipid composition of CDK13-overexpressed PC3 cells and empty vector-infected cells. Heatmap analysis showed that among the top 58 significantly different lipid ions, and the fatty acyl chains and lipid classes significantly increased in CDK13-overexpressed PC3 cells (Fig. 1G, H, Supplementary table I). Subsequently, triglyceride (TG) and cholesterol contents were measured to examine the regulatory role of CDK13 on lipid metabolism in PCa cells. The results showed that PC3 and C4-2 cells stably overexpressed CDK13 had much higher levels of TG and cholesterol than the cells stably knocked down CDK13 (Fig. 1I, J). Together, these data support the idea that CDK13 plays a role in the regulation of lipid metabolism in PCa cells.
Fig. 1. CDK13 promotes fatty acid synthesis and lipid deposition in PCa cells.
A HE staining was used to detect the change of prostate histopathology in the BPH and PCa tissues. Scale bar = 50 μm. B Oil red O (ORO) staining detected lipid accumulation in the BPH and PCa tissues (n = 31). Scale bar = 100 μm. C GSEA was used to analyze lipid metabolic signaling pathways regulated by CDK13 in PCa. FDR < 0.019 and P = 0.027. D, E PC3 and C4-2 cells were transfected with CDK13-expressing vector (oeCDk13), 2 individual shRNAs targeting different regions of the CDK13 gene (shCDK13-1#, shCDK13-2#) or their corresponding control vectors, and then ORO staining was performed to detect lipid deposition in the cells. Scale bar = 20 μm. F Quantitative analysis of Oil Red O staining in (D) and (E). G Lipidomics analysis by LC/MS was performed to examine lipid components in PC3 cells with and without stable overexpression of CDK3 (n = 3). Heatmap showing lipid molecules with significant change, represented by red (upregulated) and green (downregulated). H A statistically significant increase in abundance of fatty acyl chains in CDK13-overexpressing cells. I, J TG and cholesterol contents were measured as quantitative indicators of lipid deposition in PC3 and C4-2 cells. All data are expressed as the mean ± SEM of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 vs. their corresponding controls.
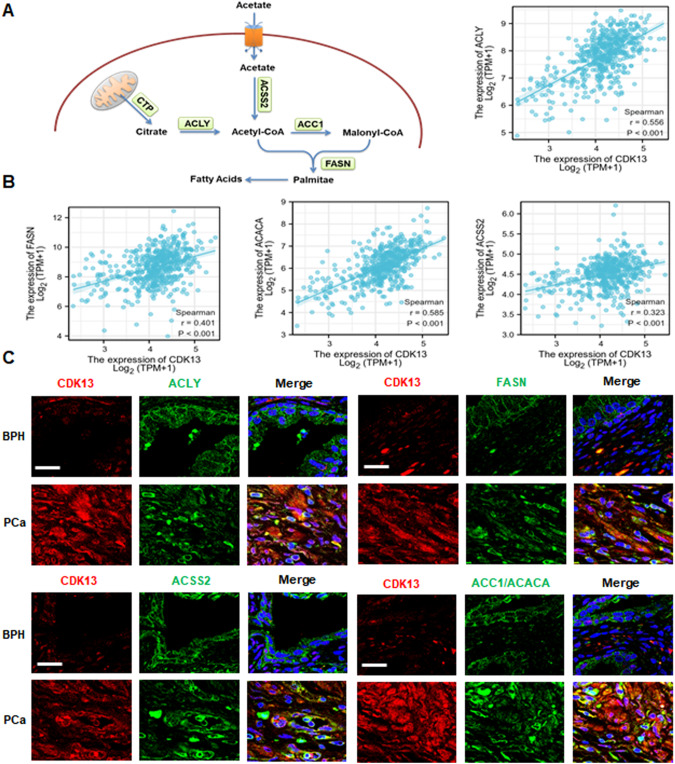
CDK13 is positively correlated with the expression of key genes involved in fatty acid synthesis
To investigate how CDK13 participates in lipid metabolism, we first analyzed the expression of several key enzymes associated with fatty acid synthesis (Fig. 2A). In the TCGA dataset of PCa, the expression of CDK13 was positively associated with ATP-citrate synthase (ACLY), acetyl-coenzyme A synthetase (ACSS2), acetyl-CoA carboxylase 1 (ACC1) (also name ACACA) and fatty acid synthase (FASN) (Fig. 2B). Subsequently, we preformed the double immunofluorescence staining to detect the co-expression of these genes and CDK13 in clinical samples. As shown in Fig. 2C, CDK13 was highly expressed in PCa tissues as compared with the BPH tissues. Double immunofluorescence staining showed that highly expressed CDK13 was accompanied by increased expression of ACLY, ACSS2, ACC1 and FASN genes in PCa tissues. These results suggest that CDK13 exerts its regulatory role on lipid metabolism possibly through regulating the expression of key genes involved in fatty acid synthesis in PCa cells.
Fig. 2. CDK13 is positively correlated with the expression of key fatty acid synthesis genes.
A Schematic showing the intermediates in fatty acid synthesis and enzymes involved in this metabolic pathway. B Correlation analysis between CDK13 expression and key fatty acid synthesis genes, such as ACLY, ACSS2, ACC1, and FASN in PCa from TCGA. C Immunofluorescence staining detected the co-expression of CDK13 and fatty acid synthases ACLY, ACSS2, ACC1, and FASN in PCa and BPH tissues. Scale bar = 20 μm.
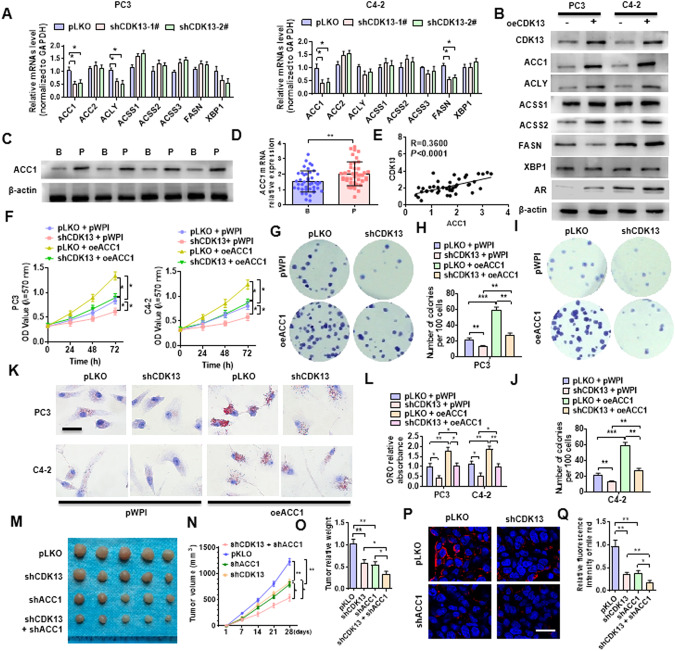
ACC1 mediates CDK13-induced lipid deposition and PCa progression by regulating lipid synthesis
To explore which enzyme in fatty acid synthesis pathway is responsible for CDK13 regulation of fatty acid synthesis, we introduced shRNA targeting CDK13 into PC3 and C4-2 cells by adenovirus and detected the expression of the above-mentioned four enzymes associated with fatty acid synthesis. As shown in Fig. 3A, depletion of CDK13 significantly decreased ACC1 and ACLY mRNA level in PC3 cells, and reduced ACC1 and FASN mRNA expression in C4-2 cells. Conversely, overexpression of CDK13 in both types of cells increased levels of ACC1, ACLY, ACSS2, and FASN proteins, as well as AR, an important regulator of lipid metabolism (Fig. 3B). Further, we examined the ACC1 expression in clinical specimens and found that the expressions of ACC1 protein and mRNA were significantly upregulated in PCa relative to the BPH tissues (Fig. 3C, D), and the expression level of ACC1 was positive relationship with CDK13 in PCa tissues (Fig. 3E). Because shCDK13-1# is more efficient than shCDK13-2# [22], and in subsequent experiments, we chose the shCDK13-1# to silence CDK13 expression, and for convenience, we named shCDK13-1# as shCDK13.
Fig. 3. ACC1 mediates CDK13-induced lipid deposition and PCa progression.
A PC3 and C4-2 cells were transfected with shCDK13-1# or shCDK13-2# or control vector, and then RT-qPCR was used to detect the expression of the indicated fatty acid synthase genes. B PC3 and C4-2 cells were transfected with oeCDK13 or control vector, and then Western blotting examined the indicated fatty acid synthases. C, D Western blot and RT-qPCR were used to detect the ACC1 expression in BPH (B) and PCa (P) tissues. E Correlation analysis between CDK13 and ACC1 expression in PCa tissues. F PC3 and C4-2 cells were transfected with shCDK13 or oeACC1 or both, and MTS assay was used to examine cell viability. G–J PC3 and C4-2 cells were transfected as in (F), and clone formation assay was used to detect cellular proliferation. K, L Cells were transfected as in (F), and ORO staining detected the lipid accumulation in cells. Scale bar = 20 μm. M PC3 cells were engineered to stably silence CDK13 or ACC1, alone or together, and xenograft tumor models were established by implanting these engineered cells into nude mice. Tumor sizes of each group were displayed after 21 days. N Tumor volumes were measured with calipers and calculated by the formula: (length × width2)/2. O, Wet weight of xenograft tumors was determined after tumor resection. P Nile red staining was used to detect the lipid deposition in xenograft tumors. Red: lipids; Blue: nuclei. Scale bar = 25 μm. Q Quantitative analysis of Nile red staining in (P). All data are expressed as the mean ± SEM of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 vs. their corresponding controls.
To define whether ACC1 mediates CDK13-induced PCa cell proliferation, we examined the effect of ACC1 on PC3 and C4-2 cell viability and growth. As shown in Fig. 3F, deletion of CDK13 significantly reduced, while ACC1 overexpression enhanced the proliferation of these two cells. Moreover, overexpression of ACC1 abrogated the inhibitory effect of CDK13 knockdown on cellular proliferation. In parallel, the clone formation assay revealed that overexpression of ACC1 markedly facilitated the growth of PC3 and C4-2 cells and reversed the suppressive effect of CDK13 depletion on cellular proliferation (Fig. 3G–J). Simultaneously, we observed a markedly increase in the level of lipid accumulation in ACC1-overexpressed PC3 and C4-2 cells. Also, overexpressing ACC1 weakened the inhibitory effect of CDK13 knockdown on lipid accumulation (Fig. 3K, L).
To determine whether ACC1 mediates CDK13-induced PCa carcinogenesis in vivo by the regulation of fat synthesis, we established a xenograft tumor model by implanting PC3 cells with stable knockdown of CDK13 or ACC1 or both together into nude mice. As shown in Fig. 3M, depletion of CDK13 or ACC1 alone obviously reduced the tumor growth, and this inhibitory role was further enhanced when both of them were knocked down simultaneously in PC3 cells. The tumor volumes and wet weight showed the same results (Fig. 3N, O). Moreover, Nile red staining revealed that depletion of CDK13 or ACC1 in PC3 cells significantly decreased lipid deposition, and silencing both of them further strengthened suppressive effect on lipid deposition (Fig. 3P, Q). The same results were obtained by flow cytometry in Nile red stained cells (Supplementary Fig. 1). Together, these data support the idea that ACC1 mediates CDK13-induced lipid accumulation and PCa progression by regulating lipid synthesis.
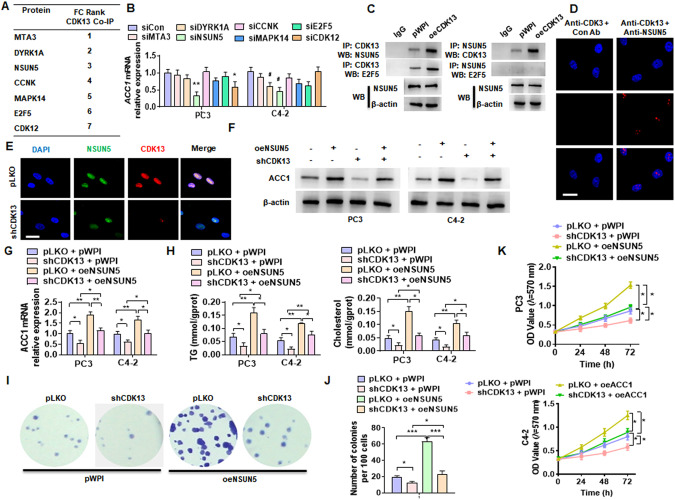
CDK13 interacts with NSUN5 to promote ACC1 expression
In the previous immunoprecipitation–mass spectrometry analysis, we found that CDK13 interacts with a range of proteins in PCa cells, including MTA3, DYRK1A, NSUN5, CCKN, MAPK14, E2F5, and CDK12 (Fig. 4A) [22]. To investigate whether CDK13 regulates ACC1 expression by interacting with one of these proteins, we first knocked down the expression of these seven genes (Supplementary Fig. 2) and examined ACC1 expression. Among these seven genes, only knockdown of NSUN5 significantly reduced ACC1 mRNA expression in PC3 and C4-2 cells (Fig. 4B). In addition, the expression of NSUN5 was significantly increased in PCa tissues compared with BPH tissues (Supplementary Fig. 3A). The analysis from TCGA database yielded the same results (Supplementary Fig. 3B). Next, we examined whether CDK13 interacts with NSUN5 by co-immunoprecipitation analysis using the specific antibodies for CDK13 and NSUN5. The results showed that there exists a strong association between CDK13 and NSUN5, and overexpression of CDK13 markedly increased their interaction (Fig. 4C). Further, a proximity ligation assay (PLA) confirmed the interaction between CDK13 and NSUN5 in the nucleus of PC3 cells (Fig. 4D). Double immunofluorescence staining also revealed the co-localization of CDK13 and NSUN5 in the nucleus (Fig. 4E).
Fig. 4. CDK13 interacts with NSUN5 to promote ACC1 expression.
A The proteins interacting with CDK13 in PC3 cells identified by co-immunoprecipitation coupled with mass spectrometry (CoIP-MS). B The indicated seven genes were knocked down in PC3 and C4-2 cells by their corresponding siRNAs, and the RT-qPCR detected the ACC1 mRNA expression. C CoIP coupled with Western blot was performed to verify the interaction between CDK13 and NSUN5. D Proximity ligation assay (PLA) detected CDK13 and NSUN5 interaction by using the indicated antibodies. E PC3 cells were transfected with shCDK13 or control vector, and then double immunofluorescence staining was used to detect CDK13 and NSUN5 co-localization. F, G PC3 and C4-2 cells were transfected with oeNSUN5 or shCDK13 or both together, and then Western blot and RT-qPCR detected ACC1 and ACLY expression. H TG and cholesterol contents were measured as quantitative indicators of lipid accumulation in PC3 and C4-2 cells. K PC3 and C4-2 cells were transfected as in (F), and MTS assay detected cell viability. I, J Cells were transfected as in (F), and clone formation assay detected cell growth. All data are expressed as the mean ± SEM of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 vs. their corresponding controls.
To explore whether NSUN5 participates in CDK13 regulation of ACC1 expression, PC3 and C4-2 cells were transfected with shCDK13 or oeNSUN5 or both and then the expression of ACC1 was detected. As shown in Fig. 4F, G, CDK13 knockdown significantly attenuated, while overexpression of NSUN5 increased ACC1 protein and mRNA expression levels and partially reversed the suppressive effect of CDK13 knockdown on ACC1 expression. Remarkably, alterations of TG and cholesterol contents caused by CDK13 knockdown and NSUN5 overexpression were consistent with the ACC1 expression changes caused by CDK13 knockdown and NSUN5 overexpression (Fig. 4H). Moreover, the clone formation and cell viability assays revealed that CDK13 knockdown obviously attenuated, while NSUN5 overexpression increased the PC3 and C4-2 cell clone formation and cell viability. However, NSUN5 overexpression-promoted PCa cell proliferation and clone formation were greatly counteracted by CDK13 knockdown (Fig. 4I–K). A large number of studies have shown that AR is up-regulated in PCa (Supplementary Fig. 4) and plays a crucial role in lipid metabolism [23–25]. Moreover, overexpression of AR partially reversed the lipid droplet formation inhibited by knockdown of CDK13 (Supplementary Fig. 5). These results indicate that AR is involved in CDK13 regulation of lipid metabolism. However, AR is involved in CDK13-regulated ACLY expression rather than ACC1 (Supplementary Fig. 6). Collectively, these findings suggested that CDK13 promotes ACC1 expression and PCa cell proliferation by co-operating with NSUN5.
NSUN5 promotes ACC1 expression and lipid deposition by regulating the m5C modification of ACC1 mRNA
Considering the fact that NSUN5 regulates the synthesis of multiple proteins by promoting RNA m5C modification [26], we investigated whether NSUN5 regulates ACC1 expression by regulating the m5C modification of mRNA. Therefore, we first detected the m5C modification level of ACC1 mRNA in PCa specimens. As shown in Fig. 5A, the m5C-modified ACC1 mRNA level was markedly elevated in PCa tissues relative to BPH tissues. Next, we knocked down NSUN5 expression in PC3 and C4-2 cells and measured the m5C modification of ACC1 mRNA. The results revealed that depletion of NSUN5 in PCa cells not only reduced the level of m5C-modified ACC1 mRNA, but also decreased the rate of m5C-modified mRNA in total mRNA (Fig. 5B, C). To verify whether NSUN5 directly modifies ACC1 mRNA, we constructed a Flag-NSUN5 vector and confirmed that ACC1 mRNA was significantly enriched in the immunoprecipitates pulled down by anti-Flag (Fig. 5D, E). Consistently, NSUN5, but not CDK13, was pulled down by biotinylated probe specifically recognizing ACC1 mRNA (Fig. 5F). These results suggested that NSUN5 can directly bind to and modify the ACC1 mRNA.
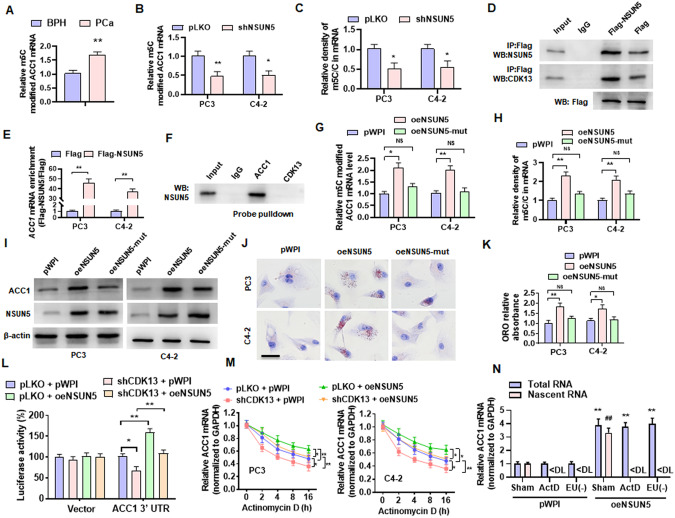
Fig. 5. NSUN5 promotes ACC1 expression and lipid deposition by regulating the m5C modification of ACC1 mRNA.
A Methylated RNA was immunoprecipitated by an m5C specific antibody (MeRIP), and then qPCR detected the level of m5C-modified ACC1 mRNA in the BPH and PCa tissues. B PC3 and C4-2 cells were transfected with shNSUN5 or control vector, and then MeRIP-qPCR examined m5C-modified ACC1 mRNA. C PC3 and C4-2 cells were ransfected as in (B) and then the UHPLC-MRM-MS/MS analysis detected the m5C methylation levels in the total mRNA. D PC3 cells were transfected with Flag-NSUN5 or a control vector (Flag), followed by CoIP to examine the interaction between CDK13 and NSUN5 and the efficacy pulled down by anti-Flag antibody. E PC3 and C4-2 cells were transfected as in (D), and RNA immunoprecipitation (RIP) with anti-Flag antibody detected the NSUN5 binding to ACC1 mRNA. F Lysates of PC3 cells were pulled down with biotinylated probe recognizing CDK13 or ACC1 mRNA, and then NSUN5 in the precipitates was detected by western blotting. G PC3 and C4-2 cells were transfected with a vector encoding wild-type or mutant forms (oeNSUN5-mut, K65A/C404, L405 del) of NSUN5, and then MeRIP-qPCR examined m5C-modified ACC1 mRNA. H PC3 and C4-2 cells were treated as in (G), and the UHPLC-MRM-MS/MS measured the m5C methylation levels in the total mRNA. I PC3 and C4-2 cells were treated as in (G), Western blot detected ACC1 and NSUN5 protein level. J, K ORO staining examined lipid accumulation in PC3 and C4-2 cells transfected with oeNSUN5 or mutant oeNSUN5. Scale bar = 20 μm. L ACC1 3’UTR was cloned into a psiCHECK™-2 Vector and then co-transfected into HEK-293T cells with shCDK13 or oeNSUN5, alone or together, and then a luciferase reporter assay detected the luciferase activity of ACC1 3ʹ-UTR luciferase reporter. M PC3 and C4-2 cells were transfected with the indicated constructs, followed by stimulation with actinomycin D for 0, 2, 4, 8 and 16 h. The ACC1 mRNA was determined by RT-qPCR. N RT-qPCR detected total and nascent ACC1 mRNA in oeNSUN5-overexpressed PC3 cells treated with or without actinomycin D or EU. ActD: actinomycin D; EU(-): without EU treatment. All data are expressed as the mean ± SEM of 3 independent experiments. *P < 0.05, #P < 0.05, ##P < 0.01 and **P < 0.01 vs. their corresponding controls.
Subsequently, PC3 and C4-2 cells were transfected with a vector encoding wild-type (wt) or mutant forms (oeNSUN5-mut, K65A/C404, L405 del) of NSUN5, and the m5C-modified ACC1 mRNA was detected by MeRIP. Overexpression of NSUN5 (wt), but not its mutant, significantly elevated the m5C-modified ACC1 mRNA level and the rate of m5C-modified mRNA in total mRNA (Fig. 5G, H). Simultaneously, overexpression of NSUN5, but not its mutant, markedly increased ACC1 protein level (Fig. 5I). As expected, the lipid deposition was significantly increased in the NSUN5-overexpressing cells, as indicated by ORO staining (Fig. 5J, K). We then measured the activity of a luciferase reporter fused to ACC1-3’ UTR, and the results showed that depletion of CDK13 inhibited, while NSUN5 overexpression enhanced luciferase activity and reversed the inhibitory effect of CDK13 knockdown on luciferase activity (Fig. 5L). Further, to investigate whether the m5C modification of ACC1 mRNA by NSUN5 facilitates mRNA stability, we blocked new RNA synthesis with actinomycin D and then examined the stability of CDK13 and NSUN5 mRNA. The results showed that CDK13 knockdown significantly reduced, while NSUN5 overexpression increased ACC1 mRNA stability, abrogating the decrease in the stability of ACC1 mRNA caused by CDK13 knockdown (Fig. 5M). Also, the similar results were obtained by using a nascent RNA capture assay (Fig. 5N). Taken together, the above results indicated that the m5C modification of ACC1 mRNA by NSUN5 promotes ACC1 expression and lipid deposition in PCa cells.
ALYREF facilitates the nuclear export of ACC1 mRNA mediated by NSUN5 in an m5C-dependent manner
Because it is known that ALYREF, the main nuclear m5C reader, promotes mRNA nuclear export [27], we hypothesized that the m5C modification of mRNA mediated by NSUN5 facilitates the nuclear export of ACC1 mRNA. To test this, we knocked down the m6A “reader” YTHDF2 or m5C “readers” YBX1 and ALYREF (Supplementary Fig. 7) and detected ACC1 mRNA expression. As shown in Fig. 6A, only knockdown of ALYREF reduced ACC1 mRNA expression level in these two cells. Western blot analysis also revealed that depletion of ALYREF, but not YTHDF2 or YBX1, down-regulated ACC1 protein level, without affecting the expression of ACLY (Fig. 6B). Next, a RNA immunoprecipitation (RIP)-PCR was performed to examine the binding between ALYREF and ACC1 mRNA, and the results showed that ACC1 mRNA was significantly enriched in the anti-ALYREF immunoprecipitates (Fig. 6C). As expected, deletion of NSUN5 in PC3 and C4-2 cells markedly reduced the m5C-modified ACC1 mRNA level but did not affect the total ACC1 mRNA (Fig. 6D). Consistently, a biotinylated probe recognizing ACC1 mRNA, but not CDK13 mRNA, effectively pulled down the ALYREF and NSUN5 protein (Fig. 6E). These data suggested that ALYREF directly binds to m5C-modified ACC1 mRNA.
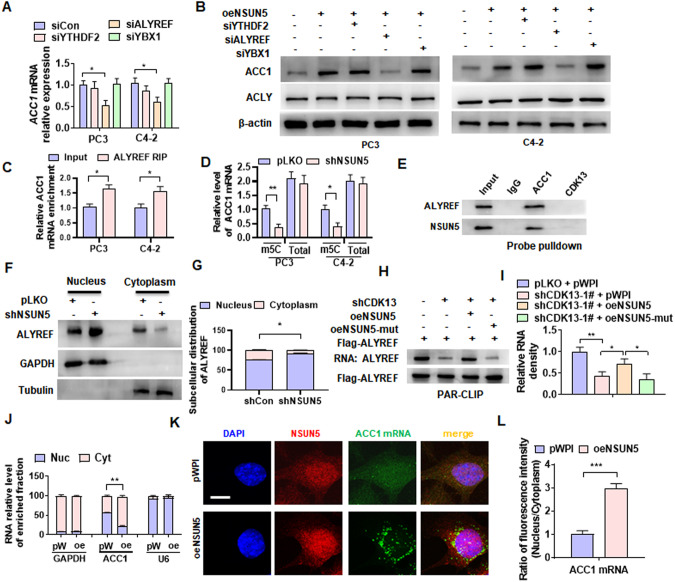
Fig. 6. ALYREF facilitates the nuclear export of ACC1 mRNA mediated by NSUN5.
A PC3 and C4-2 cells were transfected with specific siRNA against YTHDF2 (siYTHDF2), YBX1 (siYBX1) or ALYREF (siALYREF), and then RT-qPCR detected ACC1 mRNA expression. B PC3 and C4-2 cells were transfected with oeNSUN5 alone or in combination with the indicated siRNAs, and then Western blotting examined ACC1 expression. C RIP-PCR detected the ACC1 mRNA interaction with ALYREF in PC3 and C4-2 cells. D PC3 and C4-2 cells were transfected with shNSUN5 or control vector (pLKO), m5C-modified and total RNA of ACC1 were detected by RT-qPCR. E Lysates of PC3 cells were pulled down with biotinylated probe recognizing CDK13 or ACC1 mRNA. ALYREF and NSUN5 in the precipitates were detected by western blotting. F, G PC3 cells were transfected as in (D), and the nuclear and cytoplasmic fractions were isolated using NE-PER nuclear and cytoplasmic extraction reagents. Western blotting detected the subcellular distribution of ALYREF. H, I PAR-CLIP assay detected the ALYREF RNA-binding affinity. PC3 cells were transfected with the indicated constructs, and RNA was labeled with biotin at its 3’ end. RNA pulled down by Flag-ALYREF was detected and visualized by a chemiluminescent nucleic acid module. J PC3 cells were transfected with oeNSUN5 (oe) or pWPI (pW), and then RNA was isolated from the nucleus (Nuc) and cytoplasm (Cyt). mRNA of GAPDH, ACC1 and U6 was detected by RT-qPCR. K PC3 cells were treated as in (J), and the expression of NSUN5 protein and ACC1 mRNA as well as their distribution in the nucleus and cytoplasm were observed by immunofluorescence combined with FISH. Anti-NSUN5 antibody was used to detect NSUN5 protein (red), while the FITC-labeled probe was used to detect ACC1 mRNA (green). Scale bar = 5 μm. L Quantitative analysis of ACC1 mRNA fluorescence intensity in (K). All data are expressed as the mean ± SEM of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 vs. their corresponding controls.
Because ALYREF regulates gene expression by promoting nuclear-cytoplasmic shuttling of mRNA and the nuclear-cytoplasmic shuttling of ALYREF is regulated by NSUN5 [27, 28], we wanted to test whether ALYREF stimulates the nuclear export of ACC1 mRNA through its nuclear-cytoplasmic shuttling. Therefore, we isolated the nuclear and cytoplasmic fractions from the NSUN5-depleted PC3 cells and detected the subcellular distribution of ALYREF. As shown in Fig. 6F, G, knockdown of NSUN5 blocked the nuclear export of ALYREF and thus decreased its cytoplasmic distribution. Furthermore, we examined whether the RNA-binding affinity of ALYREF is regulated by CDK13 and NSUN5. The PAR-CLIP experiment revealed that knockdown of CDK13 in PC3 cells decreased ALYREF RNA-binding affinity, which could be restored by overexpressing the wild-type, but not mutant forms (mut) of NSUN5 (Fig. 6H, I).
In the further experiments, we isolated the cytoplasmic and nuclear RNA from PC3 cells transfected with oeNSUN5 or empty vector and examined the subcellular distribution of ACC1 mRNA. We found that ACC1 mRNA was equally distributed within both the cytoplasm and nucleus in empty vector-transfected cells, but upon overexpression of NSUN5, the distribution of ACC1 mRNA in the cytoplasm was significantly increased, while the distribution in the nucleus decreased correspondingly, without significant change in the distribution of GAPDH and U6 mRNA between the cytoplasm and nucleus (Fig. 6J). Also, we performed an immunofluorescence staining combined with FISH by using the NSUN5 antibody and FITC-labeled probe of ACC1 mRNA and confirmed that overexpression of NSUN5 significantly increased the distribution of ACC1 mRNA in the cytoplasm (Fig. 6K, L). These findings demonstrated that ALYREF facilitates the nuclear export of ACC1 mRNA mediated by NSUN5 in an m5C-dependent manner.
Phosphorylation of NSUN5 by CDK13 enhances lipid deposition
Given the important role of CDK13 in the phosphorylation of downstream target molecules [29], we sought to determine whether CDK13 promotes the lipid deposition in PCa cells by phosphorylating NSUN5. To do this, CDK13-overexpressing PC3 and C4-2 cells were treated with or without 1NM-PP1, a potent inhibitor of Src family kinases, and NSUN5 phosphorylation and ACC1 expression were detected. The results showed that CDK13 overexpression increased, while 1NM-PP1 treatment decreased the level of the phosphorylated NSUN5 and ACC1 protein expression, and partially abolished the promoting effect of CDK13 overexpression on NSUN5 phosphorylation and ACC1 expression (Fig. 7A). Moreover, we found that the expression level of total and phosphorylated NSUN5 obviously increased in PCa specimens compared with BPH tissues (Fig. 7B). Because several potential phosphorylation sites in the NSUN5, including Ser167, Ser290, Ser315, Thr325 and Ser327, and Ser408, were predicted by GPS 5.0 program [30], we sought to determine which site is phosphorylated by CDK13 to mediate the expression of ACC1. Therefore, we constructed a series of the phosphorylation-deficient NSUN5 mutants. As shown in Fig. 7C, the S290A, T325A and S327A mutants of NSUN5 remarkably attenuated the levels of NSUN5 phosphorylation. Intriguingly, among these three mutants, only the NSUN5-S327A mutant inhibited the promoting effect of NSUN5 on ACC1 expression. This suggested that NSUN5 phosphorylation at Ser327 is responsible for the up-regulation of ACC1 expression.
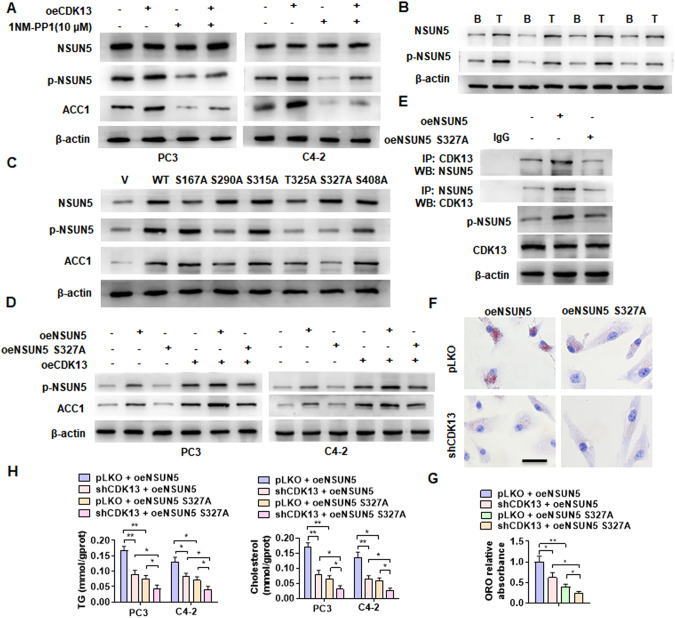
Fig. 7. Phosphorylation of NSUN5 by CDK13 enhances lipid deposition.
A PC3 and C4-2 cells were transfected with oeCDK13 and then treated with 1NM-PP1 (10 μM). Western blot examined NSUN5, p-NSUN5 and ACC1 protein expression. B Western blot analysis detected NSUN5 and p-NSUN5 expression in BPH and PCa tissues. C C4-2 cells were transfected with the vector encoding NSUN5 (WT) or different phosphorylation-deficient NSUN5 mutants, and then Western blotting examined NSUN5, p-NSUN5 and ACC1 protein level. D PC3 and C4-2 cells were co-transfected with or without oeNSUN5, oeNSUN5-S327A mutant or oeCDK13, and then Western blotting detected p-NSUN5 and ACC1 expression. E C4-2 cells were transfected with oeNSUN5 or oeNSUN5-S327A, and then CoIP-Western blotting detected the interaction between CDK13 and NSUN5. F C4-2 cells were co-transfected with the indicated constructs and then lipid accumulation was detected by ORO straining. Scale bar = 20 μm. G Quantitative analysis of ORO staining in (F). H PC3 and C4-2 cells were treated as in (F), and then TG and cholesterol contents were measured as quantitative indicators of lipid deposition. All data are expressed as the mean ± SEM of 3 independent experiments. *P < 0.05, **P < 0.01 vs. their corresponding controls.
Furthermore, we examined the effect of CDK13-mediated phosphorylation of NSUN5 on ACC1 expression by overexpressing NSUN5 and the NSUN5-S327A mutant in PC3 and C4-2 cells. Overexpression of NSUN5 or CDK13 alone significantly enhanced the level of p-NSUN5 and ACC1 expression, and concurrent overexpression of both NSUN5 and CDK13 had an additive effect. As expected, overexpression of the phosphorylation-deficient NSUN5-S327A mutant not only reduced the level of p-NSUN5, but also largely lost the ability to upregulate ACC1 expression (Fig. 7D). Simultaneously, compared with wild-type NSUN5, the mutation of phosphorylation site Ser327 obviously interfered with its interaction with CDK13 (Fig. 7E). These results indicated that CDK13 promotes ACC1 expression by interacting with NSUN5 and thus phosphorylating NSUN5 at Ser327.
Subsequently, we examined the effect of the mutation of NSUN5 phosphorylation site Ser327 on the lipid deposition in PCa cells, and found that the lipid deposition was much lower in the NSUN5-S327A mutant-overexpressing C4-2 cells than in wild-type NSUN5-overexpressing cells, and knockdown of CDK13 partially counteracted the promotion of lipid deposition by overexpression of NSUN5 (Fig. 7F, G). Consistently, compared with NSUN5-overexpressing cells, the contents of TG and cholesterol were significantly decreased in the NSUN5-S327A mutant-overexpressing PC3 and C4-2 cells, with CDK13 knockdown further reducing TG and cholesterol contents (Fig. 7H). Taken together, the above results suggested that NSUN5 phosphorylation at Ser327 by CDK13 is required for lipid deposition in PCa cells.
Blockade of CDK13/NSUN5/ACC1 pathway-mediated fatty acid synthesis inhibits PCa progression
Next, we sought to evaluate the therapeutic relevance of CDK13/NSUN5/ACC1 regulatory pathway in PCa. Considering that 1NM-PP1 inhibits the phosphorylation of several proteins by Src family kinases, including CDK13 [29], and we found that the phosphorylation of NSUN5 by CDK13 facilitated lipid deposition in PCa cells, we explored whether 1NM-PP1 could block CDK13/NSUN5/ACC1 pathway-mediated lipid deposition and PCa cell proliferation. Therefore, we performed ORO staining and cell viability assay to examine the effect of 1NM-PP1 on lipid deposition and cellular proliferation in PCa cells. As shown in Fig. 8A, B, 1NM-PP1 treatment or NSUN5 knockdown significantly depressed lipid accumulation in PC3 cells, and this suppressive effect was further enhanced by 1NM-PP1 treatment combined with NSUN5 knockdown. Cell viability assay revealed that 1NM-PP1 treatment or NSUN5 depletion significantly repressed cellular proliferation in PC3 and C4-2 cells, and the combination of both of them further enhanced the inhibitory effect on cellular proliferation (Fig. 8C). To verify the in vivo relevance of these in vitro findings and tested whether manipulating NSUN5 phosphorylation with 1NM-PP1 could regulate lipid deposition and PCa progression, we established a xenograft tumor model by implanting PC3 cells and investigated the effect of 1NM-PP1 treatment on lipid deposition and PCa progression. As shown in Fig. 8D, 1NM-PP1 treatment or NSUN5 depletion in PC3 cells drastically reduced xenograft tumor growth to a similar extent, and 1NM-PP1 treatment combined with NSUN5 knockdown further strengthened the inhibitory effect on tumor growth. In parallel, the same changes in the volume and wet weight of xenograft tumor were observed (Fig. 8E, F). Further, we examined the effect of 1NM-PP1 on NSUN5 phosphorylation and ACC1 expression in xenograft tumor, and found that 1NM-PP1 treatment substantially reduced the expression level of p-NSUN5 and ACC1, and 1NM-PP1 treatment combined with NSUN5 depletion further enhanced the inhibitory effect on ACC1 expression, without influence on the ACLY expression (Fig. 8G). Immunofluorescence staining of p-NSUN5 and ACC1 in the xenograft tumor yielded the same results as Western blot analysis (Fig. 8H, I). Notably, 1NM-PP1 treatment or NSUN5 depletion in PC3 cells strikingly attenuated lipid deposition in xenograft tumor, and this inhibitory effect was further enhanced upon 1NM-PP1 treatment combined with NSUN5 knockdown (Fig. 8J, K).
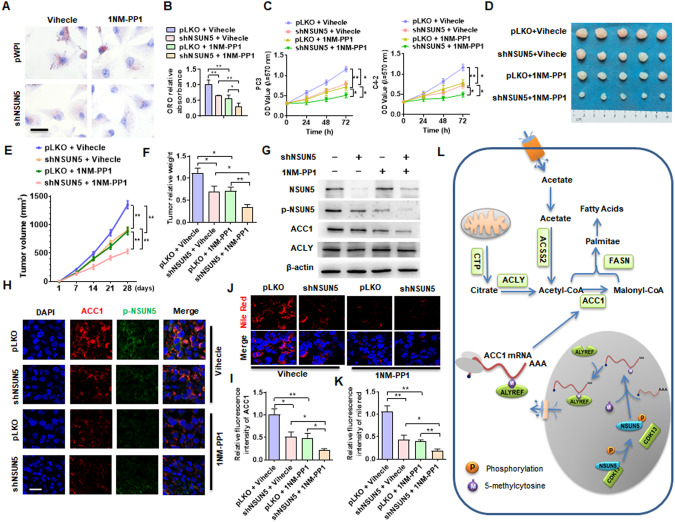
Fig. 8. Blockade of CDK13/NSUN5/ACC1 pathway-mediated fatty acid synthesis inhibits PCa progression.
A C4-2 cells were transfected with shNSUN5 and then treated with or without 1NM-PP1 (10 μM), lipid accumulation was measured by ORO straining. B Quantitative analysis of ORO staining in (A). C PC3 and C4-2 cells were treated as in (A), and cell viability was assessed by MTS assay. D Xenograft tumor models in nude mice were established by implanting PC3 cells with stable knockdown of NSUN5. From the first day, the mice were intraperitoneally injected with 20 mg/kg 1NM-PP1 every two days. Representative tumor sizes in each group were shown. E Tumor volumes were measured with calipers and calculated by the formula: (length × width2)/2. F Wet weight of the xenograft tumors was determined after tumor resection. G Western blotting detected the expression of NSUN5, p-NSUN5, ACC1 and ACLY proteins in xenograft tumors. H The expression of ACC1 and p-NSUN5 in xenograft tumors was detected by double immunofluorescence staining. Red: ACC1; green: p-NSUN5; blue: DAPI. Scale bar = 25 μm. I Quantitative analysis of fluorescence intensity of ACC1in (H). J Nile red staining detected the lipid deposition in xenograft tumors. Red: lipids; Blue: nuclei. Scale bar = 25 μm. K Quantitative analysis of Nile red staining in (J). L A proposed model illustrating the CDK13/NSUN5/ACC1 regulatory pathway-mediated fatty acid synthesis and lipid deposition. All data are expressed as the mean ± SEM of 3 independent experiments. *P < 0.05, **P < 0.01 vs. their corresponding controls.
Taken together, our results provide a novel insight into the functional roles of CDK13/NSUN5/ACC1 regulatory pathway in the development and progression of PCa, and targeting this pathway may be a novel therapeutic strategy for the treatment of PCa (Fig. 8L).
Discussion
CDKs can be broadly divided into two major subclasses: cell cycle–associated CDKs that directly regulate cell cycle progression and transcription-associated CDKs, and CDK13 belongs to the latter [17]. Transcription-related CDKs, such as CDK13, promote the transcription of genes by phosphorylating the carboxyl-terminal domain (CTD) of RNA Pol II (RNA Pol II). In addition to the CTD that phosphorylates RNA Pol II, these CDKs may also function through phosphorylation with factors critical to transcriptional or other processes. A variety of pre-mRNA processing and RNA splicing factors have been identified as potential substrate and protein binding partners by phosphorylated proteomics and affinity purification mass spectrometry [17, 31]. However, other substrates for these CDKs have yet to be identified. Here, we identified RNA-methyltransferase NSUN5 as a novel target of CDK13, and characterized that a previously unrecognized CDK13/NSUN5/ACC1 regulatory pathway mediates the fatty acid synthesis and lipid accumulation in PCa cells.
Acetyl-CoA is a precursor for the synthesis of fatty acids, and fatty acids are the main raw materials for lipid synthesis [32]. Since acetyl-CoA synthesized in mitochondria cannot be transported directly across the inner mitochondrial membrane to the cytosol, its carbon atoms first form citrate by combining with oxaloacetate. The citrate is transported from mitochondria to the cytoplasm via citrate transporter (CTP)-mediated citrate shuttle, where it is cleaved by ATP citrate lyase (ACLY) to generate acetyl-CoA [33]. In the cytoplasm, acetate can also be converted to acetyl-CoA by acetyl-CoA synthase (ACSS) [34]. Then, acetyl-CoA carboxylase (ACC) is the first rate-limiting enzyme in fatty acid synthesis, which carboxylates acetyl-CoA to malonyl-CoA [35]. Under the action of fatty acid synthase (FASN) and other enzymes, malonyl-CoA combines with acetyl-CoA to generate 16-carbon saturated fatty acid palmitate [36]. Palmitate is then elongated by elongases and/or desaturated by desaturases to produce various fatty acids. Despite increasing understanding of the fatty acid synthesis, the underlying mechanism of the dysregulation in fatty acid biosynthesis during PCa tumorigenesis and development is not fully understood. Our previous study found that increased CDK13 facilitates circCDK13 production and promotes PCa cell proliferation and tumor progression by interacting with E2F5 [22]. Surprisingly, this study showed that increased expression of CDK13 in PCa cells promotes lipid deposition. Lipidomics analysis revealed that CDK13 upregulation in PCa cells significantly increases fatty acyl chains and lipid classes. Additionally, the expression level of CDK13 is positively correlated with the expression of key genes involved in fatty acid synthesis, such as ACLY, ACSS2, ACC1 and FASN. But until now, it is unclear how CDK13 is involved in the dysregulation of lipid metabolism in PCa cells.
Methylation modification of mRNA plays an important regulatory role in a variety of tumor processes by regulating RNA stability, subcellular localization, and the efficiency of its translation into protein [37, 38]. Depending on the sites of methylation modification, RNA methylation mainly includes m6A, m1A, m5C, m7G, and 2-O-methylation modifications [37]. The level of RNA methylation is regulated by writers, readers and erasers, and various writers and readers play crucial roles in cancer progression [39]. Several recent studies have confirmed that RNA methylation participates in the regulation of the lipid metabolism, which provides new ideas for future drug development for the therapy of tumor and metabolic diseases [40]. Indeed, Guo et al. found that m6A Reader HNRNPA2B1 promotes esophageal cancer progression by upregulating ACLY and ACC1 [13]. The methyltransferase METTL3 inhibits hepatic insulin sensitivity and promotes fatty acid synthesis through N6-methyladenosine modification of FASN mRNA [41]. 5-Methylcytosine (m5C) is one of the most abundant RNA modifications in eukaryotic cells and is widely distributed in RNAs involved in various biological processes [39]. Studies have found that the targets of m5C modification include cytoplasmic and mitochondrial ribosomal RNA (rRNA), transfer RNA (tRNA), messenger RNA (mRNA), enhancer RNA (eRNA), and some non-coding RNAs [42]. In eukaryotic cells, the RNA m5C modification is generally catalyzed by the NOL1/NOP2/SUN domain (NSUN) family of enzymes and the DNA methyltransferase homolog DNMT2 [43]. NSUN5 is an important member of the NOL1/NOP2/SUN domain (NSUN) family. Studies have found that NSUN5 acts as a ribosomal RNA methyltransferase to regulate global protein synthesis and normal growth [26]. NSUN5 and ALYREF expression is upregulated in metastasis stage of head and neck squamous cell carcinoma [44]. However, the m5C modification of mRNA by NSUN5 is less reported. In this study, we found that CDK13 interacts with NSUN5 to promote its phosphorylation at Ser327. In turn, phosphorylated NSUN5 catalyzes the m5C modification of ACC1 mRNA, and then m5C-modified ACC1 mRNA binds to ALYREF and is transported to the cytoplasm, leading to an increase in ACC1 expression. Thus, NSUN5 mediates the promoting effect of CDK13 upregulation on ACC1 expression and lipid deposition in PCa cells. As a transcription-associated kinase [45], CDK13 promotes the m5C modification of ACC1 mRNA by NSUN5 via inducing NSUN5 phosphorylation at Ser327. Our findings reveal a novel function of CDK13 in regulating the ACC1 expression and identify a previously unrecognized CDK13/NSUN5/ACC1 pathway that mediates lipid accumulation and PCa progression.
Although 5-methylcytosine (m5C) is a widespread modification in RNAs, its regulatory and biological roles in cancer remain largely unknown [46]. The modification of m5C in RNAs is dynamically regulated by its related enzymes, mainly catalyzed by NSUNs, DNMTs, and TRDMTs family members (m5C writers), while demethylases (m5C erasers) TET and ALKBH1 promote demethylation, and m5C readers ALYREF, YTHDF2 and YBX1 bind to m5C [47]. As “Readers”, ALYREF, YTHDF3 and YBX1 play important roles in the regulation of mRNA stabilization and the nucleus export. Among them, the mRNA export factor ALYREF specifically recognizes m5C and binds to the m5C modification sites in the 5’ and 3’ regions of mRNA to promote its nuclear export [48]. It has been demonstrated that NSUN2 regulates ALYREF’s nuclear-cytoplasmic shuttling, RNA-binding affinity and associated mRNA export [27]. In the present study, we found that NSUN5 catalyzes the m5C modification of ACC1 mRNA, and as the “Reader” of m5C, ALYREF binds with the m5C-modified ACC1 mRNA to increase the mRNA stability and promote its nuclear export, thereby contributing to the ACC1 expression up-regulation and fatty acid synthesis increase in PCa cells.
In summary, our findings suggest that CDK13 promotes fatty acid synthesis and lipid deposition in PCa cells through up-regulating the expression of ACC1, a key gene for fatty acid biosynthesis. Mechanistically, increased CDK13 in PCa cells interacts with NSUN5 to promote its phosphorylation at Ser327. In turn, phosphorylated NSUN5 catalyzes the m5C modification of ACC1 mRNA, and then the m5C-modified ACC1 mRNA binds to ALYREF to enhance its stability and nuclear export, thus leading to ACC1 expression up-regulation and lipid deposition in PCa cells. Our findings reveal a novel function of CDK13 in regulating ACC1 expression and identify a previously unrecognized CDK13/NSUN5/ACC1 pathway that mediates fatty acid synthesis and lipid accumulation in PCa cells, and targeting this newly identified pathway may be a novel therapeutic option for the treatment of PCa.
Materials and methods
Tumor samples
All patients (median age, 63 years, range, 49 to 81 years) were treated in the Second Hospital of Hebei Medical University, among 2015.08 ~ 2021.12. For the collection of samples, 82 cases of primary prostate cancer tissue were obtained by radical prostatectomy. 43 cases of BPH tissue were obtained from transurethral resection of the prostate, as described previously [22, 49]. PCa tissue specimens were examined and confirmed by two pathologists. The patient characteristics are summarized in Supplementary Table II. The research protocol has been approved by the ethics committee of the Second Hospital of Hebei Medical University. Each patient enrolled in the study also signed written informed consent.
Cell lines and transfection
PC3 (CRL-1435; ATCC) and C4-2 (CRL-1740; ATCC) were grown in RPMI 1640 medium (Gibco, USA) containing 10% fetal bovine serum (Clark Bio, Claymont, DE, USA), penicillin (100 units/ml) and streptomycin (100 μg/ml). Cell transfection is performed using Lipofectamine 2000 (Invitrogen) according to the specific steps recommended by the manufacturer [22, 49]. All siRNAs, control siRNAs, mRNA probes, etc. for knockdown genes are purchased from GenePharma Co., Ltd. (Shanghai, China).
Lentiviral expression plasmid construction and luciferase assay
Vectors were constructed as previously described [22, 50]. pLKO.1-shCDK13, pWPI-oeCDK13, pLKO.1-shACC1, pWPI-oeACC1, pLKO.1-shNSUN5, pWPI-oeNSUN5, psPAX2 packaging plasmids and pMD2G envelope plasmids were co-transfected into HEK-293 cells. Lentiviral load was amplified and extracted using standard calcium chloride transfection 293 A method. Cell cultured lentiviral was collected and centrifuged at a density gradient for concentration, then stored at -80 °C for use. Point mutations of NSUN5 mutant plasmids were introduced into the wild-type plasmids by using the Quik-Change Lightning Site-Directed Mutagenesis Kit (Agilent). The ACC1 3’ UTR sequences containing m5C modification site were inserted into the NotI and XhoI digested-psiCHECKTM Luciferase Vector (Promega, USA). All plasmids were verified by DNA sequencing and isolated using the OMEGA Plasmid Mini Kit (OMEGA, D6942). PC3 cells were co-transfected with ACC1 3’UTR reporter construct or oeNSUN5 or shCDK13, or control vectors for 24 h and performed luciferase assay [49]. Collect and lyse cells according to the Dual-Glo Luciferase Assay System (Promega, Madison, WI) procedure. Luciferase activity was measured on cell supernatants using Flash and Glow (LB955, Berthold Technologies).
Cell viability assay
MTS and colony formation assay were performed to examine cell viability. For MTS assay, PC3 or C4-2 cells were transfected with indicated vectors in 96-well plates, and then cultured for 24, 48 or 72 h and counted as previously described [22]. MTS reagent (Promega, USA) was added to each well and the absorbance at 495 nm was measured using a microplate reader (Thermo Fisher, USA). We seed PC3 or C4-2 cells from 100 cells/well into a well plate for colony formation assay. PC3 or C4-2 were cultured for 7-10 days and fixed with glacial methanol solution. It is then stained with 0.5% crystalline violet court colonies. Finally, count the colonies with a microscope.
Hematoxylin and eosin, and immunofluorescence staining
Fresh PCa and BPH tissues were fixed in formalin and sliced 5-μm thick. Tissue sections were used to hematoxylin and eosin and immunofluorescence staining. For immunofluorescent staining [22, 49], cultured cells and tissue slides were permeabilized with Triton X-100, blocked with goat serum and then treated with indicated primary antibody. Fluorescein-conjugated secondary antibodies were reacted with slides, and nuclei were stained with DAPI. Acquire images using a Leica microscope (Leica DM6000B, Switzerland) and digitized using LAS V.4.4 (Leica).
Immunofluorescence combined with fluorescence in situ hybridization (FISH)
Cultured PC3 cell slides were fixed in 4% paraformaldehyde. In situ hybridization was performed by using the miRCURY LNATM microRNA ISH Optimization Kit (Exiqon), as previously described [51]. Fluorescently labeled ACC1 mRNA probes were hybridized with hybridization buffer (Exiqon). Rigorous gradient washes were performed with SSC buffer. Slides were blocked with goat serum, and reaction with anti-NSUN5 primary antibody (ab121633) for 1 h. Finally, the slides were treated with secondary antibody (KPL, USA) and images are obtained by using a Leica microscope (Leica DM6000B).
Western blot analysis
Total protein was extracted using RIPA buffer and protease inhibitor cocktail, as previously described [22, 49]. Quantify the lysate supernatant and load an equal amount of protein onto the gel. After separation by SDS-PAGE, proteins were electrotransferred onto polyvinylidene fluoride (PVDF) membranes (Millipore). After block with skim milk for 2 h, the membrane was incubated with the primary antibody as follows: ACC1 (1:1000, ab45174), ACLY (1:1000, ab40793), ACSS1 (1:500, 17138-1-AP), ACSS2 (1:500, ab66038), SLC25A1 (1:1000, 15235-1-AP), XBP1 (1:1000, ab37152), CDK13 (1:500, PA5-67681), NSUN5 (1:500, 15449-1-AP), Flag (1:1000, 20543-1-AP), ALYREF (1:1000, ab202894), GAPDH (1:1000, 60004-1-Ig), Tubulin (1:1000, 11224-1-AP), Phospho-(Ser) CDKs Substrate Antibody (1:500, #2324), CDK6 (1:1000, ab124821), and β-actin (1:1000, sc-47778). After reaction with HRP-conjugated secondary antibody (1:10000, Rockland), the membrane was incuated with Chemiluminescent HRP Substrate (Millipore) and detected by Fuazon Fx (Vilber Lourmat).
Quantitative real-time PCR
The Total RNA Kit II (Omega, #R6934) was used to isolate total RNA, and RNA Subcellular Isolation Kit (Active Motif, 25501) was used to extract the cytoplasmic and nuclear RNA. Nascent RNA transcripts were captured and isolated by using Click-iT Nascent RNA Capture Kit (Invitrogen) [52]. RNA were measured using a NanoDrop 2000 Spectrophotometer. First-strand cDNA was synthesized using the M-MLV First Strand Kit (Life Technologies). qRT-PCR analysis of mRNA using diluted cDNA was performed on an ABI 7500 FAST system using the Platinum SYBR Green qPCR Super Mix UDG Kit (Invitrogen). GAPDH was used for total RNA or cytoplasmic RNA, while U6 was used as the reference gene for nuclear RNA. Calculated using the 2-ddCt formula as previously described [53, 54]. All primers used in this study are summarized in Supplementary Table III.
Co‑immunoprecipitation (CoIP) assay
CoIP assay was performed by a Pierce™ Classic Magnetic IP/Co-IP Kit (Thermo, #88804). Briefly, cultured cells were treated by Lysis/Wash Buffer in ice and the lysates were immunoprecipitated with anti-CDK13 or anti-NSUN5 or Flag for 1 h at room temperature. The pre-washed pierce protein A/G magnetic beads were added to the lysates for incubating 1 h at room temperature. After wash, magnetic beads-antigen-antibody complexes were collected by magnetic stand. Wash the complex with elution buffer and finally use western blot for detection of bound proteins.
Proximity ligation assay (PLA)
PLA was performed to examine protein interactions [22]. Briefly, PC3 cells were cultured and fixed using 4% paraformaldehyde. Slides were then subjected to PLA assay using anti-CDK13 and anti-NSUN5. PLA kits (Roche, 4898117001) for In Situ were used to explore the interaction proteins. Fluorescence was detected using Leica microscope (Leica DM6000B, Switzerland).
Xenograft animal model
The animals involved in this study were approved by the ethics committee of the Second Hospital of Hebei Medical University (2021-R069). Every effort was made to minimize pain during the experiment. 5 × 106 PC3 cells stably expressing shCDK13 or shACC1 or shCDK13 and shACC1, or shNSUN5, were resuspended in 0.2 mL PBS and mixed with 50% Matrigel (BD, 356234); the suspension was then injected subcutaneously into the back of both sides of nude mice (Weitong Lihua, 4–6 weeks, 18–22 g, 12 mice in each group). After the experiment, mice were injected with 20 mg/kg of 1NM-PP1 every two days. The tumor volume was measured with calipers and calculated using (volume = [(length×width2)/2]) formula. At the end of the experiment, mice were euthanized by cervical dislocation. Finally, the tumor tissue was immediately fixed with 4% formaldehyde or frozen in liquid nitrogen for later use.
Mass spectrometry–based lipidomics
Cell lipidomics assays were performed as previously described [55]. Cultured cells were lysed and homogenized with ice-cold phosphate-buffered saline (PBS). A small amount of cell lysate was set aside to quantify DNA for normalization. Use a modified Bligh-Dyer protocol by adding equal volumes of CHCl3, 900 mL of 1 N HCl:CH3OH 1:8 (v/v), and 500 mg of the antioxidant 2,6-di-tert-butyl-4-methylphenol (Sigma). After thorough mixing and centrifugation until phase separation, the lower organic fraction was collected and the lipid particles were stored at 20 °C under argon. Reconstitute lipid particles based on adding 1 mL of diluent per 1 mg of DNA. The ratio of diluent was (CH3OH:CHCl3:NH4OH, 90:10:1.25). Lipids were analyzed by electrospray tandem mass spectrometry on an ion trap mass spectrometer (4000 QTRAP system, AB SCIEX).
Oil red O staining and Nile red staining
Cultured cell or tissue cryosections (8 μm) were fixed in 4% formalin. Stain with freshly prepared Oil Red O working solution for 15 min. Alum hematoxylin is used to stain the nucleus. Neutral resin mounts the tissue. For Nile Red staining, the fixed tissue sections were incubated with Nile Red solution. After wash, section was incubated with 4’,6-diamidino-2-phenylindole fluorescent mounting medium and image was acquired with a confocal laser scanning microscope (D6000, Leica, Germany).
TG and cholesterol measurement assay
Measurements of TG and cholesterol levels in cells or tissues were performed as previously described [23]. TG Assay Kit (Bioengineering, Catalog A110-1-1) was used to detect total TG and the Total Cholesterol Assay Kit (Bioengineering, Catalog A111-1-1) was used to examine Cholesterol. Cultured cells or tumor tissues were lysed in saline using a sonicator or abrader, and centrifuged at 4 °C. The supernatant and mix it with reagent in a 96-well plate in the dark. Finally, samples were detected on a microplate reader (Thermo Fisher, USA) with a wavelength of 510 nm. The protein concentration was detected as a control using a BSA kit.
PAR-CLIP
PAR-CLIP was performed as previously description [27]. PC3 cells treated with the different constructs were cultured in medium supplemented with 4-thiouridine (4-SU) (Sigma). CL-1000 Ultraviolet Crosslinker (UVP) was then added and crosslinked by irradiation at 365 nm with 400 mJ/cm2. Cells were harvested and lysed by spinning. The supernatant was collected by centrifugation at 4 °C. The supernatant was digested with RNase T1. Then, the supernatant was incubated with magnetic beads. The beads were digested again with RNase T1, then washed and resuspended in dephosphorylation buffer. Add calf intestinal alkaline phosphatase (CIP, NEB) and incubate at 37 °C with gentle rotation. Then wash the beads with wash buffer. Follow the manufacturer’s instructions to label the protein-RNA-bead complex with biotin using the RNA 3’ Terminal Biotinylation Kit (Thermo). About one-sixth of the samples detected protein-RNA interactions via SDS-PAGE. The relative density of RNA bound to a specific protein is analyzed by quantity one.
RNA immunoprecipitation (RIP) assay
RNA and protein interaction was examined by RIP according to Dynabeads™ Protein G Immunoprecipitation Kit (10007D, Thermo Fisher) instruction [56]. Briefly, cells were harvested and lysed using NETN buffer. Next, the Dynabeads bead-RNA complexes were washed 3 times with NETN buffer. RNA isolation and purification were then performed using an RNA purification kit (RNAeasy Mini Elute Kit, QIAGEN). Finally, the RNA fractions obtained were detected and quantified using NanoDrop 2000 (Thermo-Fisher) and subsequently used for RT-qPCR analysis.
Evaluation of m5C methylation
m5C methylation analysis in cells was performed as previously described [56]. Briefly, incubate 1 µg of anti-m5C antibody (ab10805), 20 µg of cellular RNA, and 20 µL of protein G agarose in IPP buffer containing 200 µL of 1 U/µL RNase inhibitor for 2 h at 4 °C. After that, the beads were washed five times with IPP buffer. RNA isolated from IP beads was reverse-transcribed (RT) followed by RT-qPCR analysis.
Statistical analysis
All data are expressed as the mean ± SEM of three independent experiments. Student’s t test was used to analyze differences between two groups. Values of P < 0.05 were considered statistically significant. GraphPad Prism 8.0 software was used for the statistical analysis (GraphPad Software).
Supplementary information
Author contributions
ZY, CQ and YZ carry out the design and conception. ZY, XC, HG ZH and BS carry out the methodology. YZ, ZH, JG, CZ, and YZ carry out the acquisition of the data. YZ, XC, ZY, JW and ZH carry out analysis and interpretation of the data. DW, HG, BS, and JG carry out animal models. ZY, XC, JW, and YZ carry out writing, review, and/or revision of the manuscript. All authors read and approved the final manuscript.
Funding
This study was partially supported by The National Natural Science Foundation of China (No. 82072842, 81972411, 81902573 and 81970216), and The Natural Science Foundation of Hebei Province (H2020206601, H2022206185).
Data availability
All data is included within the main text and supplementary files.
Competing interests
The authors declare no competing interests.
Ethics approval
Animal experimental procedures and operations approved by the ethics committee of the Second Hospital of Hebei Medical University, complied with the Guide for the Care and Use of Laboratory Animal, National Research Council.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yong Zhang, Xiao-Nan Chen.
Contributor Information
Yong Zhang, Email: zhangyong2022@cicams.ac.cn.
Chang-Bao Qu, Email: quchangbao@hb2h.com.
Zhan Yang, Email: yangzhan@hebmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01223-z.
References
- 1.Coughlin SS, Vernon M, Klaassen Z, Tingen MS, Cortes JE. Knowledge of prostate cancer among African American men: a systematic review. Prostate. 2021;81:202–13. doi: 10.1002/pros.24097. [DOI] [PubMed] [Google Scholar]
- 2.Rebello RJ, Oing C, Knudsen KE, Loeb S, Johnson DC, Reiter RE, et al. Prostate cancer. Nat Rev Dis Primers. 2021;7:9. doi: 10.1038/s41572-020-00243-0. [DOI] [PubMed] [Google Scholar]
- 3.Nevedomskaya E, Baumgart SJ, Haendler B. Recent advances in prostate cancer treatment and drug discovery. Int J Mol Sci. 2018;19. 10.3390/ijms19051359. [DOI] [PMC free article] [PubMed]
- 4.Teo MY, Rathkopf DE, Kantoff P. Treatment of advanced prostate cancer. Annu Rev Med. 2019;70:479–99. doi: 10.1146/annurev-med-051517-011947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu JS, Cui W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016;143:3050–60. doi: 10.1242/dev.137075. [DOI] [PubMed] [Google Scholar]
- 6.Wang WL, Tenniswood M. Vitamin D, intermediary metabolism and prostate cancer tumor progression. Front Physiol. 2014;5:183. doi: 10.3389/fphys.2014.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quiros-Gonzalez I, Gonzalez-Menendez P, Mayo JC, Hevia D, Artime-Naveda F, Fernandez-Vega S et al. Androgen-dependent prostate cancer cells reprogram their metabolic signature upon GLUT1 upregulation by manganese superoxide dismutase. Antioxidants (Basel). 2022;11. 10.3390/antiox11020313. [DOI] [PMC free article] [PubMed]
- 8.Xu H, Chen Y, Gu M, Liu C, Chen Q, Zhan M et al. Fatty acid metabolism reprogramming in advanced prostate cancer. Metabolites. 2021;11. 10.3390/metabo11110765. [DOI] [PMC free article] [PubMed]
- 9.Li EQ, Zhao W, Zhang C, Qin LZ, Liu SJ, Feng ZQ, et al. Synthesis and anti-cancer activity of ND-646 and its derivatives as acetyl-CoA carboxylase 1 inhibitors. Eur J Pharm Sci. 2019;137:105010. doi: 10.1016/j.ejps.2019.105010. [DOI] [PubMed] [Google Scholar]
- 10.Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med. 2016;22:1108–19. doi: 10.1038/nm.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chajes V, Cambot M, Moreau K, Lenoir GM, Joulin V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res. 2006;66:5287–94. doi: 10.1158/0008-5472.CAN-05-1489. [DOI] [PubMed] [Google Scholar]
- 12.Brusselmans K, De Schrijver E, Verhoeven G, Swinnen JV. RNA interference-mediated silencing of the acetyl-CoA-carboxylase-alpha gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer Res. 2005;65:6719–25. doi: 10.1158/0008-5472.CAN-05-0571. [DOI] [PubMed] [Google Scholar]
- 13.Guo H, Wang B, Xu K, Nie L, Fu Y, Wang Z, et al. m(6)A Reader HNRNPA2B1 promotes esophageal cancer progression via up-regulation of ACLY and ACC1. Front Oncol. 2020;10:553045. doi: 10.3389/fonc.2020.553045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, Meng X, Zhou Y, Yu J, Li Q, Liao Z, et al. Long noncoding RNA CTD-2245E15.3 promotes anabolic enzymes ACC1 and PC to support non-small cell lung cancer growth. Cancer Res. 2021;81:3509–24. doi: 10.1158/0008-5472.CAN-19-3806. [DOI] [PubMed] [Google Scholar]
- 15.Song J, Zhai J, Bian E, Song Y, Yu J, Ma C. Transcriptome-wide annotation of m(5)C RNA modifications using machine learning. Front Plant Sci. 2018;9:519. doi: 10.3389/fpls.2018.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 17.Chou J, Quigley DA, Robinson TM, Feng FY, Ashworth A. Transcription-associated cyclin-dependent kinases as targets and biomarkers for cancer therapy. Cancer Discov. 2020;10:351–70. doi: 10.1158/2159-8290.CD-19-0528. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X, Feng D, Wang Q, Abdulla A, Xie XJ, Zhou J, et al. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J Clin Invest. 2012;122:2417–27. doi: 10.1172/JCI61462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itkonen HM, Poulose N, Walker S, Mills IG. CDK9 inhibition induces a metabolic switch that renders prostate cancer cells dependent on fatty acid oxidation. Neoplasia. 2019;21:713–20. doi: 10.1016/j.neo.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You BJ, Chen LY, Hsu PH, Sung PH, Hung YC, Lee HZ. Orlistat displays antitumor activity and enhances the efficacy of paclitaxel in human hepatoma Hep3B cells. Chem Res Toxicol. 2019;32:255–64. doi: 10.1021/acs.chemrestox.8b00269. [DOI] [PubMed] [Google Scholar]
- 21.Santiappillai NT, Abuhammad S, Slater A, Kirby L, McArthur GA, Sheppard KE et al. CDK4/6 inhibition reprograms mitochondrial metabolism in BRAF(V600) melanoma via a p53 dependent pathway. Cancers (Basel). 2021;13. 10.3390/cancers13030524. [DOI] [PMC free article] [PubMed]
- 22.Qi JC, Yang Z, Lin T, Ma L, Wang YX, Zhang Y, et al. CDK13 upregulation-induced formation of the positive feedback loop among circCDK13, miR-212-5p/miR-449a and E2F5 contributes to prostate carcinogenesis. J Exp Clin Cancer Res. 2021;40:2. doi: 10.1186/s13046-020-01814-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou L, Song Z, Hu J, Liu L, Hou Y, Zhang X, et al. ACSS3 represses prostate cancer progression through downregulating lipid droplet-associated protein PLIN3. Theranostics. 2021;11:841–60. doi: 10.7150/thno.49384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoykova GE, Schlaepfer IR. Lipid metabolism and endocrine resistance in prostate cancer, and new opportunities for therapy. Int J Mol Sci. 2019;20. 10.3390/ijms20112626. [DOI] [PMC free article] [PubMed]
- 25.Hamid A, Kusuma Putra HW, Sari NP, Diana P, Sesari SS, Novita E, et al. Early upregulation of AR and steroidogenesis enzyme expression after 3 months of androgen-deprivation therapy. BMC Urol. 2020;20:71. doi: 10.1186/s12894-020-00627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heissenberger C, Liendl L, Nagelreiter F, Gonskikh Y, Yang G, Stelzer EM, et al. Loss of the ribosomal RNA methyltransferase NSUN5 impairs global protein synthesis and normal growth. Nucleic Acids Res. 2019;47:11807–25. doi: 10.1093/nar/gkz1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606–25. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickramasinghe VO, Laskey RA. Control of mammalian gene expression by selective mRNA export. Nat Rev Mol Cell Biol. 2015;16:431–42. doi: 10.1038/nrm4010. [DOI] [PubMed] [Google Scholar]
- 29.Fan Z, Devlin JR, Hogg SJ, Doyle MA, Harrison PF, Todorovski I et al. CDK13 cooperates with CDK12 to control global RNA polymerase II processivity. Sci Adv. 2020;6. 10.1126/sciadv.aaz5041. [DOI] [PMC free article] [PubMed]
- 30.Wang C, Xu H, Lin S, Deng W, Zhou J, Zhang Y, et al. GPS 5.0: an update on the prediction of kinase-specific phosphorylation sites in proteins. Genomics Proteomics Bioinformatics. 2020;18:72–80. doi: 10.1016/j.gpb.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang K, Gao X, Gilmore JM, Florens L, Washburn MP, Smith E, et al. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol Cell Biol. 2015;35:928–38. doi: 10.1128/MCB.01426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng M, Yang D, Hou Y, Liu S, Zhao M, Qin Y, et al. Intracellular citrate accumulation by oxidized ATM-mediated metabolism reprogramming via PFKP and CS enhances hypoxic breast cancer cell invasion and metastasis. Cell Death Dis. 2019;10:228. doi: 10.1038/s41419-019-1475-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Sena LA, Denmeade SR. Fatty acid synthesis in prostate cancer: vulnerability or epiphenomenon? Cancer Res. 2021;81:4385–93. doi: 10.1158/0008-5472.CAN-21-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim CW, Addy C, Kusunoki J, Anderson NN, Deja S, Fu X, et al. Acetyl CoA carboxylase inhibition reduces hepatic steatosis but elevates plasma triglycerides in mice and humans: a bedside to bench investigation. Cell Metab. 2017;26:394–406 e396. doi: 10.1016/j.cmet.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fhu CW, Ali A. Fatty acid synthase: an emerging target in cancer. Molecules. 2020;25. 10.3390/molecules25173935. [DOI] [PMC free article] [PubMed]
- 37.Zhang M, Song J, Yuan W, Zhang W, Sun Z. Roles of RNA methylation on tumor immunity and clinical implications. Front Immunol. 2021;12:641507. doi: 10.3389/fimmu.2021.641507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q, Liu F, Chen W, Miao H, Liang H, Liao Z, et al. The role of RNA m(5)C modification in cancer metastasis. Int J Biol Sci. 2021;17:3369–80. doi: 10.7150/ijbs.61439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engel M, Chen A. The emerging role of mRNA methylation in normal and pathological behavior. Genes Brain Behav. 2018;17:e12428. doi: 10.1111/gbb.12428. [DOI] [PubMed] [Google Scholar]
- 41.Xie W, Ma LL, Xu YQ, Wang BH, Li SM. METTL3 inhibits hepatic insulin sensitivity via N6-methyladenosine modification of Fasn mRNA and promoting fatty acid metabolism. Biochem Biophys Res Commun. 2019;518:120–6. doi: 10.1016/j.bbrc.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 42.Bohnsack KE, Hobartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m(5)C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel). 10, 10.3390/genes10020102 (2019). [DOI] [PMC free article] [PubMed]
- 43.Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B, et al. NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. 2021;12:842. doi: 10.1038/s41419-021-04127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue M, Shi Q, Zheng L, Li Q, Yang L, Zhang Y. Gene signatures of m5C regulators may predict prognoses of patients with head and neck squamous cell carcinoma. Am J Transl Res. 2020;12:6841–52. [PMC free article] [PubMed] [Google Scholar]
- 45.Even Y, Escande ML, Fayet C, Geneviere AM. CDK13, a kinase involved in pre-mRNA splicing, is a component of the perinucleolar compartment. PLoS One. 2016;11:e0149184. doi: 10.1371/journal.pone.0149184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haruehanroengra P, Zheng YY, Zhou Y, Huang Y, Sheng J. RNA modifications and cancer. RNA Biol. 2020;17:1560–75. doi: 10.1080/15476286.2020.1722449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. 2021;12:e1639. doi: 10.1002/wrna.1639. [DOI] [PubMed] [Google Scholar]
- 48.Shi M, Zhang H, Wu X, He Z, Wang L, Yin S, et al. ALYREF mainly binds to the 5’ and the 3’ regions of the mRNA in vivo. Nucleic Acids Res. 2017;45:9640–53. doi: 10.1093/nar/gkx597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z, Qu CB, Zhang Y, Zhang WF, Wang DD, Gao CC, et al. Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene. 2019;38:2516–32. doi: 10.1038/s41388-018-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Yang Z, Gu J, Zhang Y, Wang X, Teng Z, et al. Estrogen receptor beta increases clear cell renal cell carcinoma stem cell phenotype via altering the circPHACTR4/miR-34b-5p/c-Myc signaling. FASEB J. 2022;36:e22163. doi: 10.1096/fj.202101645R. [DOI] [PubMed] [Google Scholar]
- 51.Yang Z, Zheng B, Zhang Y, He M, Zhang XH, Ma D, et al. miR-155-dependent regulation of mammalian sterile 20-like kinase 2 (MST2) coordinates inflammation, oxidative stress and proliferation in vascular smooth muscle cells. Biochim Biophys Acta. 2015;1852:1477–89. doi: 10.1016/j.bbadis.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 52.Hunziker M, Barandun J, Petfalski E, Tan D, Delan-Forino C, Molloy KR, et al. UtpA and UtpB chaperone nascent pre-ribosomal RNA and U3 snoRNA to initiate eukaryotic ribosome assembly. Nat Commun. 2016;7:12090. doi: 10.1038/ncomms12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong XY, Huang YX, Yang Z, Chu XY, Wu J, Wang S, et al. Downregulation of ROR2 promotes dental pulp stem cell senescence by inhibiting STK4-FOXO1/SMS1 axis in sphingomyelin biosynthesis. Aging Cell. 2021;20:e13430. doi: 10.1111/acel.13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren LX, Qi JC, Zhao AN, Shi B, Zhang H, Wang DD, et al. Myc-associated zinc-finger protein promotes clear cell renal cell carcinoma progression through transcriptional activation of the MAP2K2-dependent ERK pathway. Cancer Cell Int. 2021;21:323. doi: 10.1186/s12935-021-02020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Centenera MM, Scott JS, Machiels J, Nassar ZD, Miller DC, Zinonos I, et al. ELOVL5 is a critical and targetable fatty acid elongase in prostate cancer. Cancer Res. 2021;81:1704–18. doi: 10.1158/0008-5472.CAN-20-2511. [DOI] [PubMed] [Google Scholar]
- 56.He X, Yang Z, Chu XY, Li YX, Zhu B, Huang YX, et al. ROR2 downregulation activates the MSX2/NSUN2/p21 regulatory axis and promotes dental pulp stem cell senescence. Stem Cells. 2022;40:290–302. doi: 10.1093/stmcls/sxab024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is included within the main text and supplementary files.