Abstract
Although mRNA SARS-CoV-2 vaccines are generally safe and effective, in certain immunocompromised individuals they can elicit poor immunogenic responses. Among these individuals, people living with HIV (PLWH) have poor immunogenicity to several oral and parenteral vaccines. As the gut microbiome is known to affect vaccine immunogenicity, we investigated whether baseline gut microbiota predicts immune responses to the BNT162b2 mRNA SARS-CoV-2 vaccine in healthy controls and PLWH after two doses of BNT162b2. Individuals with high spike IgG titers and high spike-specific CD4+ T-cell responses against SARS-CoV-2 showed low α-diversity in the gut. Here, we investigated and presented initial evidence that the gut microbial composition influences the response to BNT162b2 in PLWH. From our predictive models, Bifidobacterium and Faecalibacterium appeared to be microbial markers of individuals with higher spike IgG titers, while Cloacibacillus was associated with low spike IgG titers. We therefore propose that microbiome modulation could optimize immunogenicity of SARS-CoV-2 mRNA vaccines.
Subject terms: Microbiome, Bacteriology
Introduction
The COVID-19 pandemic has caused more than 6 million deaths1. However, vaccines against SARS-CoV-2 have changed the course of the pandemic by reducing the lethality of the disease2,3 and the incidence of the post-acute COVID-19 syndrome4. Two messenger RNA (mRNA) vaccines (BNT162b2 from Pfizer-BioNTech and mRNA-1273 from Moderna) have reached an efficacy of upto 95% with minimal side effects in initial randomized clinical trials2,5. However, the vaccines do not fully protect against reinfection, especially with new variants6, with boosters being essential for enhancing host immunity. Therefore, it is important to understand the factors influencing the immunogenicity of SARS-CoV-2 vaccines, including their capacity for long-term protection against disease severity and death. Several known factors are essential for the vaccine response, such as age, medications, disease-associated comorbidities, disorders of the immune system, and inflammatory conditions7,8. An additional factor reported to regulate immunogenicity for several other oral and parenteral vaccines is the gut microbiota9. Without any bacterial stimulation, in fact, the mucosal immune system remains poorly developed, both anatomically and functionally10. The crosstalk between the microbial communities and immune system is crucial in maintaining homeostasis and their mutualistic relationship. In fact, gut dysbiosis has also been known to be associated with immunological disequilibrium11, which is often described as Th2 cell activation or T-regulatory cell deficiency11,12. Recent animal studies have also shed new light on the association between the gut microbiome and the immune system. In vitro studies have in fact reported the role of Bifidobacterium adolescentis in reducing the adhesion of Norovirus to Caco-2 and HT-29 cells13, two colon epithelial cell lines. Bifidobacterium has been shown to have broad immunomodulatory effects in humans. For example, the cellular immunity in the elderly has been reported to be augmented by Bifidobacterium14,15. In addition, Bifidobacterium animalis has been shown to increase NK cell activity and polymorphonuclear cell functionality in old-aged individuals. In addition, the Clostridium cluster XIVa and IV were shown to stimulate TGF-β for Treg cell activation16. In fact, butyrate, a fatty acid of which Clostridia are known key producers, is pivotal in activating Treg cells and their anti-inflammatory functionalities17,18. Interestingly, many animal and clinical studies have shown the role of the gut microbiome in vaccine immunogenicity19,20, such as the case of Bifidobacterium longum, which has been associated with antigen-specific T-cell immunity against tuberculosis and polio vaccines21. Actinobacteria, the bacterial phylum to which Bifidobacterium belongs, in fact, has been shown to have a positive correlation with adaptive immunity to certain oral (i.e., polio) and systemic vaccines (i.e., BCG, tetanus toxoid, hepatitis B virus) in infants from Bangladesh21. Another study has correlated the gut microbiome in infants with immune responses to the rotavirus vaccine22,23. Overall, the general mechanism with which the gut microbiome influences vaccine efficacy primarily involves its ability to use bacterial-derived natural adjuvants to activate specific immune pathways responsible for both innate and adaptive immune responses24. One such case is the expression of flagellin or LPS by the bacterial phylum Proteobacteria, leading to the activation of pattern-recognition receptors (PRRs) found on the antigen-presenting cells19. The concept of “endogenous adjuvant potential” was further highlighted in a study where an inactivated influenza vaccine elicited lower antibody responses in germ-free or antibiotic-treated mice25,26. Treating these mice with flagellated Escherichia coli restored the normal antibody titers. Similar results were also obtained from analogously adjuvanted vaccines against poliovirus26. In addition, activation of Nucleotide-binding oligomerization domain-containing protein 2 (Nod2) by gut commensals was observed to stimulate cholera toxin-mediated antigen-specific immune responses through high levels of IL-1β during oral vaccination27,28. Other studies have shown the immunomodulatory property of bacterial flagellin, which further activated TLR5 for improved antibody production24,26.
Notably, differences in gut microbiome profiles have also been associated with health status, metabolic abnormalities, and aging, which are key elements in influencing vaccine immunogenicity29. Studies have clearly shown a reduced efficacy of the Pfizer/BioNTech vaccine due to aging, obesity, and other comorbidities30–32. Elderly individuals with underlying comorbidities are at utmost risk of COVID-19 infections and display low immunogenicity against COVID-19 vaccinations, since aging leads to reduced immunity and chronic inflammation. A study compared BNT162b2 vaccine immunogenicity in two groups of patients aged <60 and >80 years, where the latter showed significantly lower spike antibody titers33.
Aside from the elderly, another COVID-19 risk group comprises individuals with underlying chronic diseases or disorders, which also affect the immunogenic responses to vaccines. Among them, immunocompromised individuals with primary immunodeficiencies or secondary immunodeficiencies such as HIV, are particularly prone to SARS-CoV-2 infections and display less immunogenicity to COVID-19 vaccines. Considering the significantly lower level of COVID-19 vaccine immunogenicity in immunocompromised individuals, such as people living with HIV (PLWH)34–36, we hypothesized that the gut microbiome might influence the efficacy of COVID-19 vaccines in these individuals. Elucidating the potential role that the gut microbiome plays in COVID-19 vaccine immunogenicity might thus offer the possibility to identify poor vaccine responders, as well as to develop potential microbiome-targeting therapies that may enhance vaccine responses. Several clinical trials, in fact, are aiming to increase the efficacy of COVID-19 vaccines by manipulating the gut microbiome. One involves the use of 5-ALA-phosphate, which is known to maintain gut homeostasis, to increase the immunogenic responses to COVID-19 vaccines37. Another trial involves the use of three Bifidobacterium strains to increase the immunogenicity of SARS-CoV-2 vaccines and reduce the side effects in elderly diabetic patients38. In the present work, we addressed the association between the gut microbiome and immune responses to the BNT162b2 mRNA SARS-CoV-2 vaccine. The current study presents an association between the gut microbiome and humoral/cellular COVID-19 vaccine responses in PLWH and healthy controls (HC).
Results
PLWH and elderly individuals displayed less immunogenicity to the BNT162b2 vaccine
All participants included were ≥18 years of age and without any prior history of SARS-CoV-2 infection, as determined by serological testing. PLWH were on antiretroviral therapy for an average of 10.8 years (33–66 years). Their median CD4+ T-cell count was 615 cells/mL and 86% of PLWH had less than 50 copies/mL of HIV RNA. There were no significant differences in age, BMI, or number of comorbidities between PLWH and HC (Table 1).
Table 1.
Baseline demographic and clinical characteristics of the study participants.
| HC (n = 75) | PLWH (n = 68) | Overall (n = 143) | p value | |
|---|---|---|---|---|
| Sex, n (%) | ||||
| Man | 32 (43) | 40 (59) | 72 (50) | 0.066 |
| Woman | 43 (57) | 28 (41) | 71 (50) | |
| Age (years) | 52 (43.5–63) | 54 (33–65.75) | 54 (38–64) | 0.36 |
| BMI (kg/m2) | 25.1 (22.6–29.2) | 25 (23–27) | 25 (23–28.3) | 0.49 |
| Total IgG | 10.9 (9.66–12.8) | 12.45 (10.77–14.5) | 11.7 (9.895–13.575) | 0.003 |
| Lymphocytes (mL) | 1.75 (1.5–2.2) | 1.8 (1.4–2.3) | 1.8 (1.5–2.3) | 0.59 |
| Creatinine (µmol/L) | 72 (63–80) | 84 (68–99.3) | 75 (65–86) | 0.0004 |
| Ethnicity | ||||
| Caucasian | 65 (87) | 40 (59) | 105 (73) | |
| Latin | 1 (1) | 2 (3) | 3 (2) | |
| Asian | 3 (4) | 13 (19) | 16 (11) | |
| Black | 1 (1) | 11 (16) | 12 (9) | |
| Other/unknown | 5 (7) | 2 (3) | 7 (5) | |
| Diet | ||||
| Omnivorous | 66 (88) | 63 (92) | 129 (90) | |
| Vegetarian | 6 (8) | 4 (6) | 10 (7) | |
| Others/unknown | 3 (4) | 1 (2) | 4 (3) | |
| Individuals with comorbiditiesa | 22 (29%) | 24 (35%) | 46 (32%) | 0.48 |
| Duration of ART (years) | NA | 9 (4–16) | ||
| CD4+ T cell count | NA | 615 (290–730) | ||
| CD4/CD8 ratio | NA | 0.95 (0.47–1.35) | ||
| CD4 nadir | NA | 270 (117–437.5) | ||
Mann–Whitney U test was applied to compare the continuous variables and Fisher’s exact test to analyze the categorical variables. P value < 0.05 was considered significant. All baseline characteristics are illustrated as median (interquartile range) and demographic characters are illustrated as n (%).
BMI body mass index, ART antiretroviral treatment.
aComorbidities included diabetes, hypertension, cardiovascular diseases and hypercholesterolemia.
Two weeks after the second mRNA vaccine dose (day 35) (Fig. 1a), the HC showed a significantly higher spike IgG titers as compared to the PLWH (p = 0.0001) (Supplementary Fig. 1a). Gender, BMI, and baseline total IgG levels did not affect the spike antibody titers in the whole cohort (Supplementary Fig. 1b–d). Furthermore, we observed significantly higher antibody titers in younger individuals (18–39 years) compared with middle-aged (40–59 years; p = 0.003) and older participants (>60 years; p < 0.0001) (Supplementary Fig. 1e). Age was therefore negatively correlated with spike IgG (p = 0.04). In addition, in PLWH, spike IgG levels were not affected by the CD4+ T-cell counts or CD4/CD8 ratio (Supplementary Fig. 1f, g).
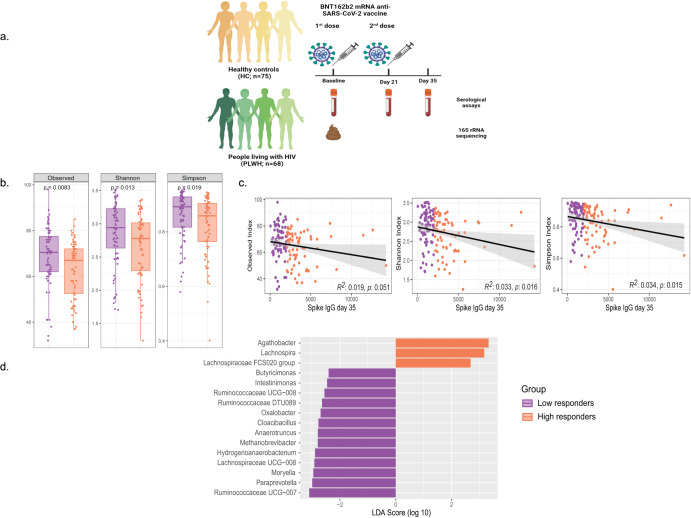
Fig. 1. Changes in alpha diversity and bacterial genus from fecal samples collected at the baseline, to predict spike IgG titers in individuals.
a Study design. b Alpha diversity decreased significantly in individuals with high spike IgG titers in the whole cohort (n = 143). P values were given by paired Mann–Whitney U test. c Linear regression relationship between alpha diversity values and spike IgG levels in all the individuals. d Differentially abundant bacterial genus between individuals with high and low antibodies as detected by LEfSe. LEfSe Linear Discriminant Analysis Effect Size.
Bacterial diversity is lower in individuals with higher spike IgG titers
To better discriminate the different levels of vaccine immunogenicity among the whole cohort, individuals from both PLWH and HC group were divided into high and low responders, based on the median spike IgG titer (1972 U/mL). Notably, high responders displayed significantly lower bacterial α-diversity (richness and evenness) than low responders (Fig. 1b). All the α-diversity indices negatively correlated with spike IgG titers for the whole cohort (Fig. 1c, Observed p = 0.05, Shannon p = 0.016, Simpson p = 0.01). In addition, the phylogenetic diversity in high responders was reduced compared with low responders (Faith’s PD, p = 0.02). However, there were no major changes in β-diversity shifts in the whole cohort (Supplementary Fig. 2a), while NMDS2 scores differed significantly between high and low responders in the HC group (Supplementary Fig. 2b, p = 0.0002). However, no significant cluster differences were observed in the PLWH (Supplementary Fig. 2c).
In addition to bacterial diversity, we also observed significant changes in the microbiota composition between individuals with high and low spike IgG titers. A total of 258 bacterial genera were detected in the whole cohort. At the genus level, Agathobacter (p = 0.02), Lachnopsira (p = 0.03), and Lachnospiraceae FCS020 group (p = 0.03) were markers for high responders, according to linear discriminant analysis (LDA) scores (LDA > 2; p < 0.05) (Fig. 1d). Butyricimonas (p = 0.02), Cloacibacillus (p = 0.009), Intestinimonas (p = 0.02), Ruminococcaceae DTU089 (p = 0.006), and Paraprevotella (p = 0.02) were significantly enriched in low responders (LDA > 2; p < 0.05) (Fig. 1d).
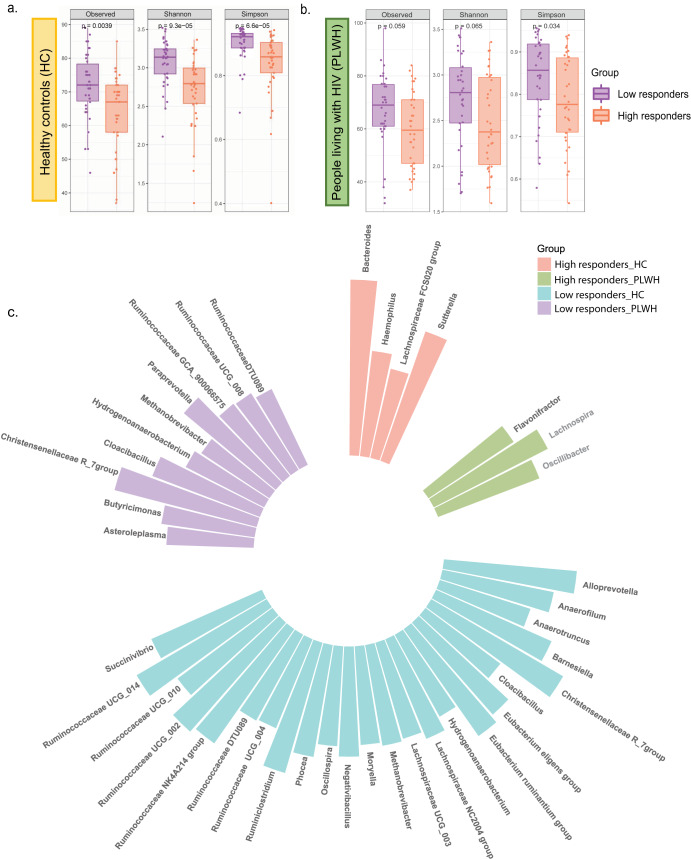
Likewise, negative associations between α-diversity and antibody titers were observed within the HC (Supplementary Fig. 3a) and the PLWH (Supplementary Fig. 3b) when individuals from these groups were divided into high and low responders. High responders with HC (Observed p = 0.004, Shannon and Simpson p < 0.0001) and PLWH (Simpson p = 0.034) displayed significantly reduced bacterial diversity than low responders within the respective groups (Fig. 2a, b). Enrichment of Bacteroidetes and depletion of Firmicutes were observed in the higher responders of the HC (p < 0.01). In addition, higher populations of Bacteroides (p < 0.05), Sutterella (p < 0.05), Lachnospiraceae FCS020 group (p < 0.05), and reduced numbers of Alloprevotella (p < 0.05), Anaerofilum (p < 0.05), Succinivibrio (p < 0.05), Moryella (p < 0.01), Negativibacillus (p < 0.05), and certain members of the Ruminococcaceae family at the genus level (p < 0.05) were observed in the high responders within the HC (LDA > 2.3) (Fig. 2c). Within the PLWH group, on the other hand, Flavonifractor, Lachnospira, and Oscillibacter were increased in high responders (p < 0.05, LDA > 2.6), whereas Butyricimonas and Paraprevotella were depleted in low responders (p < 0.05, LDA > 2) (Fig. 2c). Both the HC and the PLWH groups displayed an increased abundance of Hydrogenoanaerobacterium, Methanobrevibacter, Cloacibacillus, and Ruminococcaceae DTU089 in individuals with low spike IgG titers.
Fig. 2. Differences in diversity and composition between high and low responders in healthy controls (HC) and people living with HIV (PLWH).
a Alpha diversity indices between the high and low responders in the healthy controls (HC). b α-diversity and richness between the high and low responders in the people living with HIV (PLWH). c Circular plot depicting the significant microbial biomarkers abundant in either the high responders or low responders in both the HC and PLWH group via LEfSe analysis with pairwise comparison.
Spike-specific CD4+ T-cell response shows negative association with α-diversity
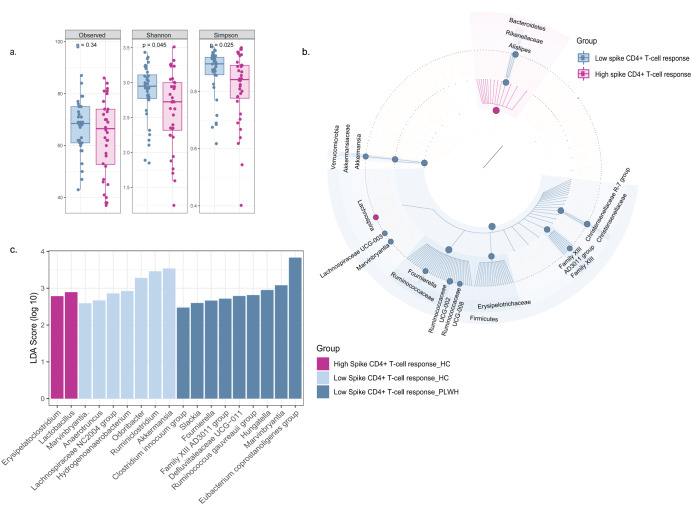
CD4+ T-cell response to the spike protein of SARS-CoV-2 was evaluated in the HC (n = 45) and PLWH (n = 45) at day 35. We stratified all individuals (n = 90) into two groups based on the magnitude of their spike CD4+ T-cell response (median 0.36%). A significant decline in the α-diversity was found in individuals with higher levels of spike-specific CD4+ T-cells (Fig. 3a) as compared to individuals with low levels of spike-specific CD4+ T-cells (Shannon p = 0.045, Simpson p = 0.025). Similar to spike IgG titers, we also found a negative association between α-diversity and spike-specific CD4+ T-cell response (observed p = 0.003, Shannon p = 0.001, Simpson p = 0.005) from linear regression analysis (Supplementary Fig. 4). There were shifts in the β-diversity (p = 0.01) with unique clustering patterns specific to each group (Supplementary Fig. 5). In addition, individuals with low spike-specific CD4+ T cell response had more Firmicutes (p = 0.005, LDA > 4.5) and less Bacteroidetes (p = 0.005, LDA > 4) compared with high responders (Fig. 3b). Moreover, Ruminococcaceae (p = 0.02, LDA > 4.5), Erysipelotrichaceae (p = 0.04, LDA > 3.5), and Akkermansiaceae (p = 0.03, LDA > 3.5) were enriched in individuals with low CD4+ T cell responses (Fig. 3b). Individuals eliciting a higher magnitude of CD4+ T cell response had an increased abundance of Lachnospira (p = 0.035, LDA > 3). In contrast, the low responders showed an increased abundance of Akkermansia (p = 0.035, LDA > 3), Fournierella (p = 0.014, LDA > 3), and Alistipes (p = 0.029, LDA > 3.5) (Fig. 3b).
Fig. 3. Changes in microbial diversity and composition affected by spike-specific CD4+ T-cell response.
a Alpha diversity changes between individuals with high and low spike-specific CD4+ T-cell response (n = 90). b GraPhlAn plot depicting the significant changes in bacterial populations in the individuals with high or low levels of CD4+ T-cell response. c Differences in the richness of bacterial genus between the people with high and low CD4+ T-cell responses to spike protein within the HC and PLWH group via LEfSe analysis.
Similarly to the whole cohort, a consistent negative association between spike-specific CD4+ T cell response and α-diversity was also observed within the HC (Supplementary Fig. 6a, b) and PLWH (Supplementary Fig. 6c, d). The low responders in the PLWH were enriched with Firmicutes (p = 0.01, LDA > 4.5) and depleted of Bacteroidetes (p = 0.046, LDA > 4.5). Erysipelotrichaceae (p = 0.01), Eggerthellaceae (p = 0.006), and Succinivibrionaceae (p = 0.036) were detected as signatures of low responders. Both the HC and PLWH had a high abundance of Marvinbryantia in communities with low CD4+ T cell response (p < 0.05). Higher numbers of Fournierella (p = 0.03) were identified in low responders within the PLWH (Fig. 3c). On the other hand, the high responders in the HC showed a higher abundance of Lactobacillaceae (p = 0.01, LDA > 3) and a decline of Akkermansiaceae (p = 0.02, LDA > 3.5). At the genus level, the high responders in the HC displayed an enrichment of Lactobacillus (p = 0.014). Instead, Akkermansia (p = 0.017), Ruminiclostridium (p = 0.037), and Hydrogenoanaerobacterium (p = 0.047) were more abundant in low spike-specific CD4+ T cell responders (Fig. 3c).
Furthermore, we explored the correlation of bacterial taxa associated with spike CD4+ T-cell response and spike IgG titers (Supplementary Table 1). In the whole cohort, Sutterella (p = 0.01), Bifidobacterium (p = 0.015), Bacteroides, Lachnospira, and Lactobacillus showed possible positive correlation, while Escherichia-Shigella (p = 0.015), Marvinbryantia (p = 0.002), Ruminococcaceae DTU089 (p = 0.018), Methanobrevibacter (p = 0.028), and Cloacibacillus (p = 0.015) showed negative correlation with antibody levels (Supplementary Fig. 7).
Gut microbiome diversity is affected by age
A relative decline in diversity and richness was observed in young adults (18–39 years, n = 37) as compared to older individuals (>60 years, n = 50) (Observed, p < 0.001 and Shannon, Simpson, p < 0.0001, Supplementary Fig. 8a). Moreover, there was a significant positive association between age and α-diversity (Supplementary Fig. 8b, observed p = 0.0002, Shannon p = 0.00001, Simpson p = 0.0001). Similarly, a significant shift in β-diversity was observed among the different age groups (p = 0.03) (data not shown). At the genus level, certain members of the Ruminococcaceae family (p < 0.05), Butyricimonas (p = 0.01), Ruminiclostridium (p = 0.005), Hydrogenoanaerobacterium (p = 0.005), Fournierella (p = 0.009), Christensenellaceae R_7 group (p = 0.007) and Methanobrevibacter (p = 0.007) showed increased abundance in older individuals (>60 years) (LDA > 2.5, Supplementary Fig. 8c). In young adults we observed an enrichment of Lachnospira (p = 0.02), Bacteroides (p = 0.02), and Agathobacter (p = 0.009) which were also enriched in individuals with high spike IgG titers (LDA > 3). Within the HC and PLWH groups, we observed a similar pattern of relative decrease in the α-diversity indices of young adults compared to the elderly and middle-aged (Supplementary Fig. 8d, e). Predominantly, the Bacteroidetes population was significantly enriched in younger adults as compared to the older individuals (p = 0.001), who harbored a higher abundance of Firmicutes (p = 0.006; LDA > 4.5) (Supplementary Fig. 8f). The Firmicutes/Bacteroidetes (F/B) ratio was highest in the elderly and lowest in the youths.
Gut microbiome profiles are affected by total IgG levels at baseline
In addition, we studied whether total IgG levels at baseline were associated with specific microbiome profiles, since we observed a borderline significant association with spike IgG levels in our cohort (p = 0.09). Stratifying the individuals into those with high and low total IgG levels at baseline, we found a decline in the bacterial diversity of the individuals with high total IgG levels (data not shown). The most differential genera among all samples from our analysis (individuals with high and low total IgG levels) were Anaerostipes (p < 0.01), Fournierella (p < 0.01), Mitsuokella (p < 0.05), and Lactobacillus (p < 0.05) (Supplementary Fig. 9a). Interestingly, an increased abundance of Anaerostipes (p < 0.01, LDA > 3), Bacteroides, and Bifidobacterium were detected in the individuals with high levels of total IgG at baseline, and these genera were also likely to be positively correlated with spike IgG titers (Supplementary Fig. 9b, c).
Gut microbiota α-diversity and composition are associated with spike IgG levels irrespective of age and disease status
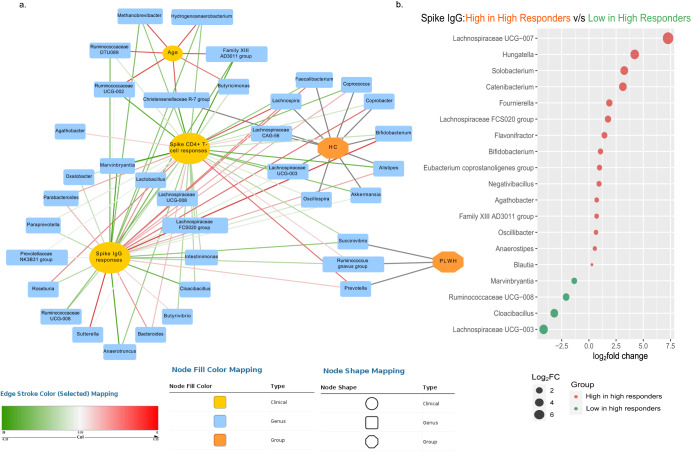
We further integrated the cellular and humoral immune responses on day 35, with selected baseline factors, such as age and disease group, into a network and correlated the microbial taxa, which were significantly associated with said clinical characteristics and immunological responses. Overall, from our predictive models, we observed a positive association of spike IgG levels with Lachnospira, Faecalibacterium, and Bifidobacterium, which were also markers of the HC group (Fig. 4a). Agathobacter, Lactobacillus, Bacteroides, and Lachnospira positively correlated with both spike IgG levels and spike-specific CD4+ T-cell responses (Fig. 4a). The microbes positively linked with age (Hydrogenoanaerobacterium, Methanobrevibacter, Ruminococcaceae DTU089, Butyricimonas) showed a negative correlation with both spike IgG levels and CD4+ T-cell responses. Spike IgG levels and CD4+ T-cell responses were negatively associated with Methanobrevibacter, Ruminococcaceae DTU089, Paraprevotella, Marvinbryantia, Cloacibacillus, and Succinivibrio (Fig. 4a).
Fig. 4. Association of gut microbiota with spike IgG levels.
a Network analysis of linking different bacterial genus to clinical factors such as spike IgG levels on day 35, age, and spike-specific CD4+ T-cell responses. b DESeq2 analysis to observe the differential abundance through estimation of fold change of each microbial taxon associated with antibody levels, irrespective of different clinical parameters, such as age, gender, and disease group.
Furthermore, we carried out a DESeq2 analysis to model the absolute abundance of each microbial taxon associated with antibody levels irrespective of other clinical parameters (such as age, gender, disease group) to determine the log2fold change (effect size estimate) for differential changes in microbial abundance as true absolute counts. We observed significant changes in the abundance of several gut microbes directly associated with spike IgG. Thus, individuals with high spike IgG titers revealed enrichment of Bifidobacterium (p = 0.03), Faecalibacterium (p = 0.03), Blautia (p = 0.03), Catenibacterium (p = 0.03), and Hungatella (p = 0.005) from this analysis (Fig. 4b). Bifidobacterium was increased in subjects with high spike IgG titers and high baseline total IgG. Conversely, individuals with low antibody titers showed a higher abundance of Cloacibacillus (p = 0.0001) (Fig. 4b).
In univariate analysis, the number of observed species, Shannon diversity, Simpson index, and age were all significantly associated with spike IgG levels (Table 2). To reveal the independent association between α-diversity and humoral response to COVID-19 vaccination, we conducted multiple regression analysis. We found a significant association of bacterial abundance (Shannon p = 0.02, Simpson p = 0.019) with spike IgG levels, independent of age (Table 2 and Supplementary Table 2). However, age showed a significant impact when the number of observed species and age were correlated to spike IgG response (p = 0.04). The results suggest that microbial abundance is independent of age and influences spike IgG levels. Nonetheless, the observed richness shows age dependency to affect spike IgG levels, while richness estimates are unaffected by species abundance.
Table 2.
Univariate and multivariate regression analysis for analyzing the linear relationship between baseline characteristics and Spike IgG response among individuals.
| p value | p valuea (FDR-corrected) | Beta coefficient | |
|---|---|---|---|
| Observed | 0.05 | 0.09 | –0.16 |
| Shannon | 0.01 | 0.06 | –0.20 |
| Simpson | 0.01 | 0.06 | –0.20 |
| Age | 0.03 | 0.09 | –0.17 |
| Gender | 0.73 | 0.78 | 0.02 |
| Total IgG | 0.09 | 0.13 | –0.14 |
| BMI | 0.78 | 0.78 | –0.02 |
| Observed, age | 0.04 | 0.04 | –0.76 |
| Shannon, age | 0.02 | 0.03 | –0.04 |
| Simpson, age | 0.019 | 0.03 | 0.22 |
aSignificance of difference (p < 0.05) expressed as false discovery rate-corrected p value.
Using multivariate analysis, we further analyzed the effect of other baseline clinical factors, such as gender and total IgG, and observed that the α-diversity significantly affected the spike IgG response (Supplementary Table 3, observed p = 0.02, Shannon p = 0.006, Simpson, p = 0.005). Gender showed no effect on antibody titers, followed by age and IgG. In addition, the α-diversity significantly impacted the spike-specific CD4+ T-cell responses in the cohort irrespective of age, gender, and total IgG (Supplementary Table 3, observed p = 0.006, Shannon p = 0.0009, Simpson, p = 0.004).
Discussion
SARS-CoV-2 mRNA vaccines are highly immunogenic and effective in protecting from severe COVID-192. However, several factors, such as increasing age, underlying medical conditions, and concomitant medications, are associated with lower vaccine efficacy7,8,39. Increasing evidence, from clinical cohorts, interventional studies, and animal models, show that gut microbiota plays a significant role in modulating responses to vaccination9,40.
In our study, we investigated whether the baseline gut microbial diversity and composition would affect the immunogenicity of mRNA BNT162b2 SARS-CoV-2 vaccine in PLWH, as immunocompromised individuals possess lower immunogenic responses to these vaccines. Interestingly, α-diversity metrics were negatively associated with spike IgG titers post-vaccination, both in the whole cohort and within the subgroups of HC and PLWH. In addition, we found that the elevated levels of spike-specific CD4+ T-cell responses were associated with reduced microbiome diversity, thus further validating the association of the microbiome with the immunogenicity of the BNT162b2 mRNA vaccine. These results are in line with data presented by previous studies using vaccines against other viral diseases. A collection of studies reported that similar vaccines (both oral and parenteral) elicit impaired immunogenicity in individuals living in low-income countries compared with people from high-income countries, due to their differences in gut microbiome profiles19. Moreover, two studies on oral polio vaccine, in India and China, showed a low bacterial diversity in individuals with high vaccine efficacy41,42. Furthermore, the role of microbiota in modulating T-cell responses has been shown in influenza infection43. Lastly, antibiotic treatment prior to H1N1 vaccination altered the microbial diversity, resulting in decreased influenza-specific IgG1 and IgA antibody titers44.
In addition to bacterial diversity, this study showed that the baseline microbiota composition could potentially influence the immunological effects of the SARS-CoV-2 vaccines. We observed that certain bacterial genera at the baseline were associated with vaccine immunogenicity, measured by IgG spike and spike-specific CD4+ T-cell responses. Interestingly, in the network analysis, Agathobacter, Lactobacillus, Bacteroides, and Lachnospira were positively correlated with both spike IgG levels and spike-specific CD4+ T-cell responses. These bacteria with known immunomodulatory properties could contribute to higher immune responses to vaccinations45–47. Bacteroides, in fact, have been shown to have a key role in vaccine responses due to the induction of homeostatic immune priming. For instance, the LPS of Bacteroides thetaiotaomicron acts as an adjuvant, enhancing the production of hepatitis B virus antigen-specific antibodies48. In addition, another study revealed the association of different Bacteroides strains with diverse response levels to rotavirus vaccine49. Overall, Bacteroides are resident commensals, acting as major polysaccharide degraders in the gut for butyrate production50. Conversely, Methanobrevibacter, Ruminococcaceae DTU089, Marvinbryantia, Cloacibacillus, and Succinivibrio were enriched in the baseline microbiome of individuals with low IgG titers and low spike-specific CD4+ T-cell responses. This is in line with previous reports, as Cloacibacillus has been previously associated with bacteremia51, Succinivibrio was found in inflammatory conditions52 and previous studies have reported the enrichment of Marvinbryantia spp., which are SCFA-producers, in the elderly53, who experience a lower response to vaccinations, as observed in our study. Similarly, in our study, Marvinbryantia was observed to be less abundant in the younger group in the whole cohort and within the HC and PLWH groups. Moreover, our findings are also consistent with results from Borgognone et al.54, in which lower α-diversity and gene richness were observed in PLWH, who showed better immunogenic response to an HIV vaccine. These individuals, dubbed viremic controllers, were observed to have significantly higher Bacteroidales/Clostridiales ratio and lower Methanobacteriales than the non-controllers54. Our study also shows similar findings in individuals with high spike IgG levels, with less abundance of bacterial groups belonging to Clostridiales and Methanobacteriales, such as Ruminococcaceae DTU089 and Methanobrevibacter, respectively. The abundance of certain bacterial groups belonging to Clostridiales in low responders might be potentially associated with metabolic pathways related to methanogenesis and carbohydrate synthesis.
Notably, in the present work, Bifidobacterium and Faecalibacterium showed significant association with high spike IgG titers, irrespective of other factors, from our predictive models. This is in line with earlier studies, where enrichment of Bifidobacterium was correlated with CD4+ T-cell responses and higher antibody titers to parenteral and oral vaccinations41,55. In addition, B. adolescentis was also associated with a higher rate of neutralizing antibodies after CoronaVac immunization56. Current clinical trials are exploring the effect of supplementation with different Bifidobacterium species on immune responses to SARS-CoV-2 vaccine38. Interestingly, our findings on microbiome associations with mRNA vaccine immunogenicity in the whole cohort were also consistent within the PLWH group. This is of note, since the gut microbial composition of PLWH is altered due to gut dysbiosis occurring during the course of HIV-1 infection and further changed during antiretroviral therapy, as shown by our previous studies57,58. Interestingly, in our study, we observed different bacterial groups from Lachnospiraceae to be associated with either high or low spike IgG levels. Generally, the bacterial members belonging to the family Lachnospiraceae are well-known producers of SCFAs in colon mucosa-associated microbiota. Lachnospiracea FSC020, which shows a positive association with spike IgG levels in our study, has been reported to have a potential positive association with the production of acetate and propionate59 and to be linked to circulating lipid metabolites in blood60. In addition, some members of Lachnospiraceae family, such as Lachnospiraceae UCG-003 can potentially protect against colon cancer by butyrate production61. While existing literature shows Lachnospiraceae UCG-008 to be negatively associated with age62 and saturated fatty acid intake63 and has decreased abundance in conditions like liver cirrhosis and hepatocellular carcinoma, our findings reveal the abundance of this taxon in low responders to COVID-19 vaccine64.
The key findings of this study link α-diversity with spike IgG responses, which in turn is also influenced by age. Although α-diversity has been known to play a crucial role in association studies, we believe that the gut bacterial composition and its functional aspects play a much more important role in influencing the landscape of gut balance. In this study, individuals with low α-diversity showed better immunogenic responses and an abundance of Bifidobacterium, which is known to have a protective function in the gut. In this study, we also observed younger adults with low α-diversity to have better vaccine immunogenicity compared to elderly individuals with higher α-diversity. In addition, individuals with low immunogenic responses were observed to have high α-diversity. This overall demonstrates a negative association between diversity and spike IgG levels and a positive association between age and α-diversity. Higher α-diversity in elderly individuals has also been observed in a few other studies65–67. Moreover, when investigating the gut composition, we observed bacterial genera specific to younger participants, such as Lachnospira, Bacteroides, and Agathobacter to be positively associated with spike IgG levels. Similarly to the findings from another study, we also detected enrichment of potential pathobionts in elderly individuals, which have higher α-diversity, such as an increase of Enterobacteriaceae which has been correlated with frailty in elder individuals65. At the genus level, we also observed some microbial signatures of pathobionts such as Escherichia-Shigella, and Enterococcus in the elderly group, as corroborated by the literature66,67. Similarly to other studies65–68, our analyses also detected in this group an abundance of certain members of the Ruminococcaceae family, Butyricimonas, Christensenellaceae R_7 group, Akkermansia, members of Erysipelotrichaceae and a reduction of Faecalibacterium, a pattern which has been linked to inflammatory disorders69. Ruminococcaceae are well-known symbionts present in the human gut generating SCFA70 to enhance the protective functions of the intestinal epithelium and prevent infections from opportunistic pathogens71. An enrichment of Ruminococcaceae diversity (Supplementary Fig. 10) might be associated with better metabolic plasticity and versatility of the gut in elderly individuals65. Notably, while the elderly population has been reported to have a lower F/B ratio72, our study shows it to display a higher F/B ratio. This has also been observed in another study cohort from Ukraine73. In fact, the LefSe data shows the abundance of Firmicutes in elderly and Bacteroidetes in younger individuals, both of which are the predominant bacterial phyla colonizing the gut. As a matter of fact, certain key species within the immunomodulatory Gram-negative Bacteroidetes phylum have been reported to exert anti-inflammatory effects through T-cell modulation74. Lastly, the elderly population showed an abundance of gut microbes, such as Hydrogenoanaerobacterium, Methanobrevibacter, Negativibacillus, which were also observed in individuals with low spike IgG levels, which is in line with the observed negative association between age and spike IgG levels in our study.
The mechanisms by which the gut microbiota modulates immunological responses to vaccines are not yet fully understood. Several potential mechanisms, however, have been anticipated. Microbiota-derived flagellin, peptidoglycan, and lipopolysaccharide can act as natural adjuvants to vaccination, recognized by PRRs19,75. Another important mechanism concerns gut integrity, which is crucial in regulating immune responses, and is disrupted in a state of inflammation, malnutrition, or by antibiotic treatment76. The gut–barrier integrity is modulated by certain bacterial metabolites, which induce the expression of tight junction proteins, thereby maintaining the epithelial integrity77. Microbiota-derived metabolites, such as SCFAs (acetate, butyrate, and propionate), tryptophan, and secondary bile acids can, therefore, directly and indirectly alter immune responses19. They provide energy sources for enterocytes, strengthen the epithelial barrier, and can act as signaling molecules in regulatory pathways of intestinal and systemic immunity. SCFAs also regulate T-cell metabolism and can augment B-cell metabolism, enhancing pathogen-specific antibody responses78,79. In the present study, certain butyrate producers, such as Lachnospira, Catenibacterium, and Faecalibacterium, were enriched in individuals with higher immunogenic responses to the BNT162b2 vaccine, and metabolites derived from these bacteria might likely have an association with higher vaccine immunogenic responses. As a critical butyrate producer, Faecalibacterium prausnitzii has been previously added to probiotic formulations for restoring gut health80,81. Overall, we hypothesize that microbiota-derived metabolites, such as butyrate, indole, and bile acids, might possibly have a strong potential to improve vaccine responses, and quantifying such metabolites could be an interesting topic for future research.
The primary limitation of the present study lies in its cross-sectional design, preventing definitive conclusions from being drawn due to the sample being collected at a single time point. Another constraint pertains to the specific sequencing approach used on the samples. Employing 16S RNA sequencing, indeed, did not yield a high resolution of the microbiome, lacking species- and strain-level information along with not being able to evaluate and understand their functional and metabolic potential. However, one of the strengths of the study is the substantial number of individuals for each of the two groups that were enrolled in the clinical study. The participants were followed using a strict protocol, where we carefully selected the individuals who did not undergo antibiotic treatment nor showed signs of prior or ongoing SARS-CoV-2 infection.
Conclusively, we observed that the baseline gut microbiota diversity and abundance significantly affected immunologic responses to SARS-CoV-2 vaccines, irrespective of age, gender, and total IgG. Although age had a significant association with both the microbial diversity and spike IgG levels, results from multiple regression analyses showed that baseline microbial abundance was independent of age and had a dominant impact on modulating antibody response to the COVID-19 vaccine.
In summary, we describe novel findings on a strong association between the intestinal microbial diversity, composition, and immunogenicity of a mRNA SARS-CoV-2 vaccine. These findings may influence the development of microbiota-centered therapies to optimize immunogenicity and durability of vaccination. For example, the augmentation of vaccination with concomitant treatment with probiotics, prebiotics, or diet could be a scalable intervention for individuals with lower vaccine responses, like elderly or immunocompromised individuals, such as PLWH. The feasibility of such therapies is considerable, as a systematic review of results from 26 interventional studies in humans using probiotics to enhance the efficacy of 17 different vaccines revealed positive outcomes in half of the trials82. Furthermore, a recent preclinical study showed the effectiveness of synbiotics in enhancing immunogenicity of the cholera vaccine in mice that were colonized with a poorly immunogenic infant’s microbiota83. In addition, the presented microbiome association with the vaccination outcome should warrant more careful use of treatments that influence the gut microbiome, like antibiotics76. Overall, these results pave the way for future studies, which could lead to a deeper understanding of microbiota modulation of vaccine responses in different populations and disease conditions.
Methods
SARS-CoV-2 COVAXID vaccine cohort and sample collection
An open-label, non-randomized clinical trial was initiated during spring 2021 at the Karolinska University Hospital, Stockholm, Sweden, to investigate the safety and clinical efficacy of the mRNA BNT162b2 vaccine (Comirnaty®, Pfizer/BioNTech) in healthy controls and immunocompromised patients (EudraCT no. 2021-000175-37, clinicaltrials.gov no. 2021-000175-37). The ethical permit was granted by the Swedish Ethical Review Authority (ID 2021-00451), and all participants provided written informed consent (Table 1)84. The study cohort of that study included individuals belonging to six groups84; however, the current study focused on two of these groups, PLWH (n = 90) and healthy controls (HC) (n = 90). Figure 1a displays the study design, with the fecal samples from the PLWH and HC group being collected at baseline for DNA extraction before they had received two doses of the mRNA vaccine three weeks apart. The extracted DNA was further sent for 16S rRNA sequencing. Humoral and cellular responses to SARS-CoV-2 vaccination were evaluated on day 35 after the 1st vaccine dose. Subjects with detectable baseline spike antibodies against SARS-CoV-2, antibiotic treatment (3 months before vaccination), and those with missing spike IgG data at day 35 were excluded from further analysis (PLWH: n = 22; HC: n = 15). The fecal samples were collected in RNA/DNA shield (Stratec, Germany). DNA was extracted using ZymoBIOMICS™ DNA Kit (Zymo Research, USA), according to the manufacturer’s specifications, and sequencing analysis was performed on the MiSeq platform.
Detection of spike IgG and spike CD4+ T-cell responses
Blood samples were collected at baseline and on day 35. Elecsys® Anti-SARS-CoV-2 S RBD (Roche Diagnostics) was used to identify and quantify antibodies specific to SARS-CoV-2 spike protein in serum samples84. Spike-specific CD4+ T-cell responses were quantified using activation-induced marker assays via up-regulation of CD69 and CD40L (CD154), as previously described85. Total IgG levels were analyzed by routine diagnostic methods with the Roche Elecsys anti-SARS-CoV-2 S enzyme immunoassay, at the Clinical Immunology laboratory of Karolinska University Hospital84. For PLWH, CD4+ and CD8+ T-cell counts and HIV viral load (VL) were determined by flow cytometry and Cobas Amplicor (Roche Molecular Systems Inc., USA), respectively86.
Microbiome analysis
Paired-end Illumina reads were checked for quality using FastQC87 and trimmed using Cutadapt88. During the pre-processing step, primers, adapters, and low-quality (Q < 30) reads were removed. The taxonomic classification and analysis of the trimmed reads were performed using dada289 within Qiime290 in combination with SILVA database (SILVA v132)91. DADA2 was used for denoising, read pair merging and PCR chimera removal which reduces sequence errors and dereplicates sequences (Supplementary Table 4). α-diversity of the samples was estimated using the R function estimate_richness in R package phyloseq (v1.30.0)92 and visualized using R package ggplot2 (v3.3.5)93. The diversity indices such as Observed, Shannon, and Simpson were performed to calculate the richness and diversity of the samples. The distances between the samples were clustered based on the Bray-Curtis distance metrics and visualized using non-metric multidimensional scaling (NMDS) ordination plots, and the significance of the different factors on the beta-diversity was calculated based on PERMANOVA using the vegan package (v2.5.7) (Adonis function). The relative abundance of the samples was calculated using Qiime2. Linear discriminant analysis Effect Size (LEfSe) was employed to determine the significant microbial communities between the groups94. All the barplots were visualized using the R package ggplot2. LefSe plot of spike CD4+ T-cell response and specific microbial biomarkers were visualized using GraPhlAn95. Multivariate regression analysis was used to predict the factors related to spike IgG. α-diversity indices, age, gender, total baseline IgG, and BMI were used as covariates in the regression analysis (R package). The models were predicted using stepwise backward selection, and p values < 0.05 were considered significant. Furthermore, we performed multiple hypothesis testing using a false discovery rate (FDR). Multivariate analysis of variance (MANOVA) was used to access the different patterns between the multiple dependent variables simultaneously and it was performed using the R function manova(). The Kruskal–Wallis test was performed for the post hoc analysis for MANOVA. The OTUs belonging to Ruminococcaceae were segregated to assess the richness and diversity of Ruminococcaceae family between elderly and younger individuals (Supplementary Fig. 10).
Correlation and network analysis
Correlation analyses were based on the Spearman correlation method using R package psych (v2.2.3)96,97. Benjamini–Hochberg method was used to adjust the p values for the multiple testing. The results were visualized using the R package corrplot (v0.92)98. The input variables for the networks were bacterial genus, age, spike CD4+ T-cell response, and the antibody level at day 35, visualized using Cytoscape (v3.6.1)99. Differential abundance analysis between high responders and low responders was performed using R package DESeq2 (v1.26.0)100 and visualized using bubble plots. Bacterial taxa with p values less than 0.05 were considered significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank all the participants enrolled in this study, and all the research and clinical staff at Karolinska University Hospital. We would like to thank SciLifeLab National Genomics Infrastructure in Stockholm, and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with sequencing and access to the UPPMAX computational infrastructure. The authors also acknowledge the assistance and support from the Centre for Bioinformatics and Biostatistics (CBB). The study was supported by grants from the Swedish Physicians Against AIDS research fund (FOa2021-0009; P.N., FOb2020-0019; S.R.), Stockholm County Council (SLL-KI for P.N.; ALF nr 20190451), Karolinska Institute Research Foundation Grants (2020-02198; S.R.), Swedish Research Council (Dnr 2016-01675; A.S.), EuCARE project “European cohorts of patients and schools to advance response to epidemics” funded by the EC under HORIZON-HLTH-2021-CORONA-01 Grant No. 101046016.
Author contributions
Conception, planning, fund acquisition: P.N., S.A., M.B., H.-G.L., A.S. Coordinated the sample collection: P.N., J.V., O.B., P.C., S.A. Designed the experiments: P.N., M.B. and S.R. Performed experiments and analyzed the data: S.R., A.N., G.G., Y.G., M.S.C., and L.M. Wrote the paper: S.R., A.N., and P.N. Reviewed and/or edited the manuscript: all authors.
Funding
Open access funding provided by Karolinska Institute.
Data availability
The metadata and raw 16S rRNA gene sequence data generated and analyzed during this study are deposited at the NCBI SRA database (Project number: PRJNA902956).
Code availability
All the codes are available at GitHub: https://github.com/Asw614/Gut-microbiome-on-immunological-responses-to-COVID-vaccination.git.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The ethical permit was reviewed and approved by the Swedish Ethical Review Authority (ID 2021-00451), and all participants provided written informed consent.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-023-00461-w.
References
- 1.World Health Organization (accessed 14 July 2023). https://covid19.who.int/.
- 2.Polack FP, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El Sahly HM, et al. Efficacy of the mRNA-1273 SARS-CoV-2 vaccine at completion of blinded phase. N. Engl. J. Med. 2021;385:1774–1785. doi: 10.1056/NEJMoa2113017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antonelli M, et al. Risk factors and disease profile of post-vaccination SARS-CoV-2 infection in UK users of the COVID Symptom Study app: a prospective, community-based, nested, case-control study. Lancet Infect. Dis. 2022;22:43–55. doi: 10.1016/S1473-3099(21)00460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baden LR, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carazo S, et al. Estimated protection of prior SARS-CoV-2 infection against reinfection with the Omicron variant among messenger RNA–vaccinated and nonvaccinated individuals in Quebec, Canada. JAMA Netw. Open. 2022;5:e2236670. doi: 10.1001/jamanetworkopen.2022.36670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connors J, Bell MR, Marcy J, Kutzler M, Haddad EK. The impact of immuno-aging on SARS-CoV-2 vaccine development. Geroscience. 2021;43:31–51. doi: 10.1007/s11357-021-00323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soiza RL, Scicluna C, Thomson EC. Efficacy and safety of COVID-19 vaccines in older people. Age Ageing. 2021;50:279–283. doi: 10.1093/ageing/afaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynn DJ, Pulendran B. The potential of the microbiota to influence vaccine responses. J. Leukoc. Biol. 2018;103:225–231. doi: 10.1189/jlb.5MR0617-216R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDermott AJ, Huffnagle GB. The microbiome and regulation of mucosal immunity. Immunology. 2014;142:24–31. doi: 10.1111/imm.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baradaran Ghavami S, et al. Cross-talk between immune system and microbiota in COVID-19. Expert Rev. Gastroenterol. Hepatol. 2021;15:1281–1294. doi: 10.1080/17474124.2021.1991311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mezouar S, et al. Microbiome and the immune system: from a healthy steady-state to allergy associated disruption. Hum. Microbiome J. 2018;10:11–20. doi: 10.1016/j.humic.2018.10.001. [DOI] [Google Scholar]
- 13.Li D, Breiman A, Le Pendu J, Uyttendaele M. Anti-viral effect of Bifidobacterium adolescentis against noroviruses. Front. Microbiol. 2016;7:864. doi: 10.3389/fmicb.2016.00864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang B-L, Sheih Y, Wang L, Liao C, Gill H. Enhancing immunity by dietary consumption of a probiotic lactic acid bacterium (Bifidobacterium lactis HN019): optimization and definition of cellular immune responses. Eur. J. Clin. Nutr. 2000;54:849–855. doi: 10.1038/sj.ejcn.1601093. [DOI] [PubMed] [Google Scholar]
- 15.Gill HS, Rutherfurd KJ, Cross ML, Gopal PK. Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am. J. Clin. Nutr. 2001;74:833–839. doi: 10.1093/ajcn/74.6.833. [DOI] [PubMed] [Google Scholar]
- 16.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Vitetta L. Inflammation-modulating effect of butyrate in the prevention of colon cancer by dietary fiber. Clin. Colorectal Cancer. 2018;17:e541–e544. doi: 10.1016/j.clcc.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 19.Lynn DJ, Benson SC, Lynn MA, Pulendran B. Modulation of immune responses to vaccination by the microbiota: implications and potential mechanisms. Nat. Rev. Immunol. 2022;22:33–46. doi: 10.1038/s41577-021-00554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vitetta L, Saltzman ET, Thomsen M, Nikov T, Hall S. Adjuvant probiotics and the intestinal microbiome: enhancing vaccines and immunotherapy outcomes. Vaccines. 2017;5:50. doi: 10.3390/vaccines5040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huda MN, et al. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134:e362–e372. doi: 10.1542/peds.2013-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris VC, et al. Effect of antibiotic-mediated microbiome modulation on rotavirus vaccine immunogenicity: a human, randomized-control proof-of-concept trial. Cell Host Microbe. 2018;24:197–207.e194. doi: 10.1016/j.chom.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris VC, et al. Significant correlation between the infant gut microbiome and rotavirus vaccine response in rural Ghana. J. Infect. Dis. 2017;215:34–41. doi: 10.1093/infdis/jiw518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pabst O, Hornef M. Gut microbiota: a natural adjuvant for vaccination. Immunity. 2014;41:349–351. doi: 10.1016/j.immuni.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh JZ, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, et al. Recognition of the microbiota by Nod2 contributes to the oral adjuvant activity of cholera toxin through the induction of interleukin‐1β. Immunology. 2019;158:219–229. doi: 10.1111/imm.13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, et al. Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin. Nat. Med. 2016;22:524–530. doi: 10.1038/nm.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turroni F, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 2009;75:1534–1545. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koff WC, et al. Development and deployment of COVID-19 vaccines for those most vulnerable. Sci. Transl. Med. 2021;13:eabd1525. doi: 10.1126/scitranslmed.abd1525. [DOI] [PubMed] [Google Scholar]
- 31.Pellini R, et al. Initial observations on age, gender, BMI and hypertension in antibody responses to SARS-CoV-2 BNT162b2 vaccine. EClinicalMedicine. 2021;36:100928. doi: 10.1016/j.eclinm.2021.100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefan N, Birkenfeld AL, Schulze MB. Global pandemics interconnected—obesity, impaired metabolic health and COVID-19. Nat. Rev. Endocrinol. 2021;17:135–149. doi: 10.1038/s41574-020-00462-1. [DOI] [PubMed] [Google Scholar]
- 33.Müller L, et al. Age-dependent immune response to the Biontech/Pfizer BNT162b2 coronavirus disease 2019 vaccination. Clin. Infect. Dis. 2021;73:2065–2072. doi: 10.1093/cid/ciab381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dandachi D, et al. Characteristics, comorbidities, and outcomes in a multicenter registry of patients with human immunodeficiency virus and coronavirus disease 2019. Clin. Infect. Dis. 2021;73:e1964–e1972. doi: 10.1093/cid/ciaa1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geretti AM, et al. Outcomes of coronavirus disease 2019 (COVID-19) related hospitalization among people with human immunodeficiency virus (HIV) in the ISARIC World Health Organization (WHO) clinical characterization protocol (UK): a prospective observational study. Clin. Infect. Dis. 2021;73:e2095–e2106. doi: 10.1093/cid/ciaa1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann C, et al. Immune deficiency is a risk factor for severe COVID‐19 in people living with HIV. HIV Med. 2021;22:372–378. doi: 10.1111/hiv.13037. [DOI] [PubMed] [Google Scholar]
- 37.Chang M, et al. Changes of gut microbiota in pregnant sows induced by 5-Aminolevulinic acid. Res. Vet. Sci. 2021;136:57–65. doi: 10.1016/j.rvsc.2021.01.021. [DOI] [PubMed] [Google Scholar]
- 38.Mak, J. W. Y. Modulation of gut microbiota to enhance health and immunity (accessed 2 July 2023). https://clinicaltrials.gov/ct2/show/NCT04884776 (2020).
- 39.Falahi S, Kenarkoohi A. Host factors and vaccine efficacy: implications for COVID‐19 vaccines. J. Med. Virol. 2022;94:1330–1335. doi: 10.1002/jmv.27485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Vitetta L, Henson JD, Hall S. The intestinal microbiota and improving the efficacy of COVID-19 vaccinations. J. Funct. Foods. 2021;87:104850. doi: 10.1016/j.jff.2021.104850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao T, et al. Influence of gut microbiota on mucosal IgA antibody response to the polio vaccine. npj Vaccines. 2020;5:47. doi: 10.1038/s41541-020-0194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Praharaj I, et al. Influence of nonpolio enteroviruses and the bacterial gut microbiota on oral poliovirus vaccine response: a study from South India. J. Infect. Dis. 2019;219:1178–1186. doi: 10.1093/infdis/jiy568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl Acad. Sci. USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagan T, et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178:1313–1328.e1313. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells, J. M. Immunomodulatory mechanisms of lactobacilli. Microbial Cell Factories 1–15 (BioMed Central, 2011). [DOI] [PMC free article] [PubMed]
- 46.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright EK, et al. Microbial factors associated with postoperative Crohn’s disease recurrence. J. Crohns Colitis. 2017;11:191–203. doi: 10.1093/ecco-jcc/jjw136. [DOI] [PubMed] [Google Scholar]
- 48.Chilton PM, Hadel DM, To TT, Mitchell TC, Darveau RP. Adjuvant activity of naturally occurring monophosphoryl lipopolysaccharide preparations from mucosa-associated bacteria. Infect. Immun. 2013;81:3317–3325. doi: 10.1128/IAI.01150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fix J, et al. Association between gut microbiome composition and rotavirus vaccine response among Nicaraguan infants. Am. J. Tropical Med. Hyg. 2020;102:213. doi: 10.4269/ajtmh.19-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zafar H, Saier MH., Jr Gut Bacteroides species in health and disease. Gut Microbes. 2021;13:1848158. doi: 10.1080/19490976.2020.1848158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Domingo M-C, et al. Cloacibacillus sp., a potential human pathogen associated with bacteremia in Quebec and New Brunswick. J. Clin. Microbiol. 2015;53:3380–3383. doi: 10.1128/JCM.01137-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong, T. S. et al. Gut microbiome profiles associated with steatosis severity in metabolic associated fatty liver disease. Hepatoma Res. 7, 10.20517/2394-5079.2021.55 (2021). [DOI] [PMC free article] [PubMed]
- 53.Bian G, et al. The gut microbiota of healthy aged Chinese is similar to that of the healthy young. Msphere. 2017;2:e00327–00317. doi: 10.1128/mSphere.00327-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borgognone A, et al. Gut microbiome signatures linked to HIV-1 reservoir size and viremia control. Microbiome. 2022;10:59. doi: 10.1186/s40168-022-01247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huda, M. N. et al. Bifidobacterium abundance in early infancy and vaccine response at 2 years of age. Pediatrics143, e20181489 (2019). [DOI] [PMC free article] [PubMed]
- 56.Ng SC, et al. Gut microbiota composition is associated with SARS-CoV-2 vaccine immunogenicity and adverse events. Gut. 2022;71:1106–1116. doi: 10.1136/gutjnl-2021-326563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ray S, et al. Altered gut microbiome under antiretroviral therapy: Impact of efavirenz and zidovudine. ACS Infect. Dis. 2020;7:1104–1115. doi: 10.1021/acsinfecdis.0c00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nowak P, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. Aids. 2015;29:2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 59.Ma T, et al. Altered mucosa-associated microbiota in the ileum and colon of neonatal calves in response to delayed first colostrum feeding. J. Dairy Sci. 2019;102:7073–7086. doi: 10.3168/jds.2018-16130. [DOI] [PubMed] [Google Scholar]
- 60.Vojinovic D, et al. Relationship between gut microbiota and circulating metabolites in population-based cohorts. Nat. Commun. 2019;10:5813. doi: 10.1038/s41467-019-13721-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gryaznova M, et al. Dynamics of changes in the gut microbiota of healthy mice fed with lactic acid bacteria and bifidobacteria. Microorganisms. 2022;10:1020. doi: 10.3390/microorganisms10051020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei Z-Y, et al. Characterization of Changes and Driver Microbes in Gut Microbiota During Healthy Aging Using A Captive Monkey Model. Genomics, Proteomics Bioinformatics. 2022;20:350–365. doi: 10.1016/j.gpb.2021.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsumoto N, et al. Relationship between nutrient intake and human gut microbiota in monozygotic twins. Medicina. 2021;57:275. doi: 10.3390/medicina57030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang H, et al. Identification reproducible microbiota biomarkers for the diagnosis of cirrhosis and hepatocellular carcinoma. AMB Express. 2023;13:1–14. doi: 10.1186/s13568-023-01539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tuikhar N, et al. Comparative analysis of the gut microbiota in centenarians and young adults shows a common signature across genotypically non-related populations. Mech. Ageing Dev. 2019;179:23–35. doi: 10.1016/j.mad.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 66.Kong F, et al. Gut microbiota signatures of longevity. Curr. Biol. 2016;26:R832–R833. doi: 10.1016/j.cub.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 67.Yu X, et al. Analysis of the intestinal microbial community structure of healthy and long-living elderly residents in Gaotian Village of Liuyang City. Appl. Microbiol. Biotechnol. 2015;99:9085–9095. doi: 10.1007/s00253-015-6888-3. [DOI] [PubMed] [Google Scholar]
- 68.Biagi E, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Tongeren SP, Slaets JP, Harmsen HJ, Welling GW. Fecal microbiota composition and frailty. Appl. Environ. Microbiol. 2005;71:6438–6442. doi: 10.1128/AEM.71.10.6438-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 71.Chassard C, Bernalier‐Donadille A. H2 and acetate transfers during xylan fermentation between a butyrate‐producing xylanolytic species and hydrogenotrophic microorganisms from the human gut. FEMS Microbiol. Lett. 2006;254:116–122. doi: 10.1111/j.1574-6968.2005.00016.x. [DOI] [PubMed] [Google Scholar]
- 72.Mariat D, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:1–6. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vaiserman A, et al. Differences in the gut Firmicutes to Bacteroidetes ratio across age groups in healthy Ukrainian population. BMC Microbiol. 2020;20:1–8. doi: 10.1186/s12866-020-01903-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neff CP, et al. Diverse intestinal bacteria contain putative zwitterionic capsular polysaccharides with anti-inflammatory properties. Cell Host Microbe. 2016;20:535–547. doi: 10.1016/j.chom.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ashkar AA, Mossman KL, Coombes BK, Gyles CL, Mackenzie R. FimH adhesin of type 1 fimbriae is a potent inducer of innate antimicrobial responses which requires TLR4 and type 1 interferon signalling. PLoS Pathog. 2008;4:e1000233. doi: 10.1371/journal.ppat.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheung K-S, et al. Association between recent usage of antibiotics and immunogenicity within six months after COVID-19 vaccination. Vaccines. 2022;10:1122. doi: 10.3390/vaccines10071122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scott SA, Fu J, Chang PV. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl Acad. Sci. USA. 2020;117:19376–19387. doi: 10.1073/pnas.2000047117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garrett WS. Immune recognition of microbial metabolites. Nat. Rev. Immunol. 2020;20:91–92. doi: 10.1038/s41577-019-0252-2. [DOI] [PubMed] [Google Scholar]
- 79.Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016;20:202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gautier T, et al. Next-generation probiotics and their metabolites in COVID-19. Microorganisms. 2021;9:941. doi: 10.3390/microorganisms9050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Toole PW, Marchesi JR, Hill C. Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017;2:1–6. doi: 10.1038/nmicrobiol.2017.57. [DOI] [PubMed] [Google Scholar]
- 82.Zimmermann P, Curtis N. The influence of probiotics on vaccine responses–a systematic review. Vaccine. 2018;36:207–213. doi: 10.1016/j.vaccine.2017.08.069. [DOI] [PubMed] [Google Scholar]
- 83.Di Luccia B, et al. Combined prebiotic and microbial intervention improves oral cholera vaccination responses in a mouse model of childhood undernutrition. Cell Host Microbe. 2020;27:899–908.e895. doi: 10.1016/j.chom.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bergman, P. et al. Safety and efficacy of the mRNA BNT162b2 vaccine against SARS-CoV-2 in five groups of immunocompromised patients and healthy controls in a prospective open-label clinical trial. EBioMedicine74, 103705 (2021). [DOI] [PMC free article] [PubMed]
- 85.Gao Y, et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022;28:472–476. doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu X, Vesterbacka J, Aleman S, Nowak P. High seroconversion rate after vaccination with mRNA BNT162b2 vaccine against SARS-CoV-2 among people with HIV–but HIV viremia matters? AIDS. 2022;36:479–481. doi: 10.1097/QAD.0000000000003135. [DOI] [PubMed] [Google Scholar]
- 87.Andrews S. Babraham Bioinformatics. Cambridge, United Kingdom: Babraham Institute; 2010. [Google Scholar]
- 88.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 89.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS one. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ginestet, C. ggplot2: Elegant Graphics for Data Analysis. Journal of the Royal Statistical SocietySeries A: Statistics in Society174, 245–246 (2011).
- 94.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:1–18. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ. 2015;3:e1029. doi: 10.7717/peerj.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Revelle, W. & Revelle, M. W. Package ‘psych’. The Comprehensive R Archive Network337, 338 (2015).
- 97.Revelle, W. psych: Procedures for Psychological, Psychometric, and Personality Research. Northwestern University, Evanston, Illinois. R package version 2.2.3 (2022). Retrieved from https://CRAN.R-project.org/package=psych.
- 98.Wei, T. et al. package “corrplot”: Visualization of a Correlation Matrix. 2017. Version 0.84 (2021).
- 99.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metadata and raw 16S rRNA gene sequence data generated and analyzed during this study are deposited at the NCBI SRA database (Project number: PRJNA902956).
All the codes are available at GitHub: https://github.com/Asw614/Gut-microbiome-on-immunological-responses-to-COVID-vaccination.git.