Abstract
Recepteur d’origine nantais (RON, MST1R) is a single-span transmembrane receptor tyrosine kinase (RTK) aberrantly expressed in numerous cancers, including various solid tumors. How naturally occurring splicing isoforms of RON, especially those which are constitutively activated, affect tumorigenesis and therapeutic response, is largely unknown. Here, we identified that presence of activated RON could be a possible factor for the development of resistance against anti-EGFR (cetuximab) therapy in colorectal cancer patient tissues. Also, we elucidated the roles of three splicing variants of RON, RON Δ155, Δ160, and Δ165 as tumor drivers in cancer cell lines. Subsequently, we designed an inhibitor of RON, WM-S1-030, to suppress phosphorylation thereby inhibiting the activation of the three RON variants as well as the wild type. Specifically, WM-S1-030 treatment led to potent regression of tumor growth in solid tumors expressing the RON variants Δ155, Δ160, and Δ165. Two mechanisms for the RON oncogenic activity depending on KRAS genotype was evaluated in our study which include activation of EGFR and Src, in a trimeric complex, and stabilization of the beta-catenin. In terms of the immunotherapy, WM-S1-030 elicited notable antitumor immunity in anti-PD-1 resistant cell derived mouse model, likely via repression of M1/M2 polarization of macrophages. These findings suggest that WM-S1-030 could be developed as a new treatment option for cancer patients expressing these three RON variants.
Subject terms: Oncogenes, Predictive markers
Introduction
In 2020, 1.93 million new colorectal cancer (CRC) cases were diagnosed worldwide, representing 10% of total global cancer incidence [1]. Although the National Cancer Comparison Network® (NCCN) guidelines (version 3.2021) offer insightful treatment options for CRC, it is crucial to acknowledge that these therapies do not cover all patients’ needs. One such treatment is cetuximab (Erbitux®), a monoclonal antibody targeting the epidermal growth factor receptor (EGFR), which has shown efficacy in metastatic CRC patients with wild-type KRAS (KRAS wt) tumors [2]. Nevertheless, a considerable number of patients (50–60%) fail to initially respond to cetuximab [3]. Moreover, a troubling 80% of patients who initially respond to cetuximab treatments develop drug resistance within 3 to 12 months [4, 5]. Cetuximab resistance is attributed to various factors, including gene mutations in KRAS, BRAF, and EGFR and gene amplification of KRAS, HER2, and MET [2]. These findings underscore the urgent need to identify novel druggable targets for colon cancer treatment, particularly those that can overcome the current limitations of conventional anti-EGFR therapies.
Recepteur d’origine nantais (RON, MST1R) is a member of the MET receptor tyrosine kinase family that is highly expressed in numerous cancers. However, despite its prevalence in cancer, the exact role and function of RON remains uncertain [6–8]. Previous research conducted on various primary cancer tissues and established cell lines revealed that aberrant activation of RON signaling can arise either through overexpression of RON wild-type (RON WT) or the direct generation of its constitutively active isoforms generated by alternative splicing of RON(RON variants), such as RONΔ160 [9]. Aberrant expression and activation of RON has also been found to be a significant prognostic factor impacting patient survival in several cancer types [10–14]. Interestingly, different variants of RON, including RONΔ160 (exon 5,6 deletion), RONΔ165 (exon 11 deletion), and RONΔ155 (exon 5,6,11 deletion), are thought to be naturally occurring tumorigenic RON variants [15]. RON variants are predominantly found in cancer tissues, and numerous reports have demonstrated their association with increased tumor growth [14–17].
In particular, altered expression of RON variants has been implicated in progression of colorectal cancer [18, 19]. The pattern of RON expression has also been found predictive of colorectal cancer patient outcomes [14]. RON expression has also been associated with various clinicopathological factors, including tumor size, lymphovascular invasion, depth of invasion, lymph node metastasis, distant metastasis, tumor state, and poor survival, in colon cancer patients [20]. Recent research has shown that activation of MET and RON are linked to colon cancer resistance to cetuximab [21, 22].
These significant findings have sparked numerous investigations into the potential of RON as a therapeutic target for cancer treatment, resulting in the development of several small molecule RON inhibitors (primarily targeting RON wild type rather than its variants), with some now in clinical trials. Notably, these compounds have demonstrated potent (low nanomolar range) inhibition of RON enzymatic activity, and high antitumor activity, in various tumor xenograft models. These RON inhibitors include AMG-458 (Amgen) [23], LCRF-0004 (MethylGene) [24], and BMS777607 (Bristol-Myers Squibb) [25]. There have been attempts to suppress RON activity by blocking interaction with its ligand, MSP, using antibody-based approaches, early clinical studies of these strategies were discontinued due to a lack of efficacy [26]. As of now, no drugs specifically targeting the three constitutively active splicing variants of RON (RONΔ155, RONΔ160, RONΔ165) have been developed [15, 27].
In the present study, we reveal the tumorigenic potential of the three RON variants and also demonstrate that our novel compound, WM-S1-030, induces cell death and effectively suppresses tumor growth in RON variant-positive solid tumors. These compelling results suggest that RON variants represent promising therapeutic targets for solid tumors, and WM-S1-030 could be an attractive therapeutic option for potential treatment strategies in cancer therapy.
Results
Development of WM-S1-030, a specific inhibitor of RON variants, as an anticancer agent
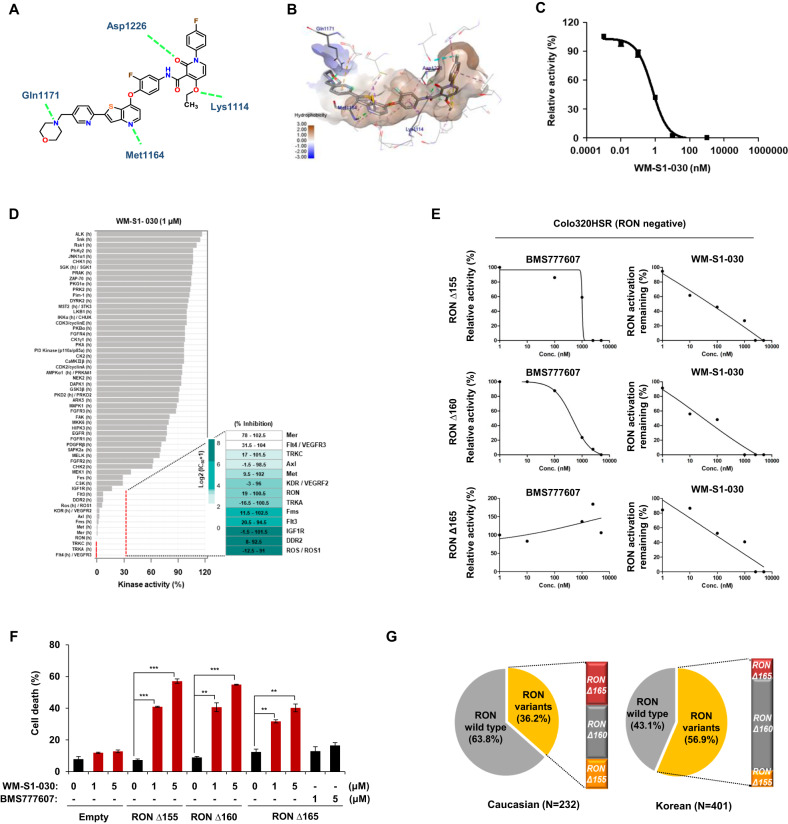
To identify cetuximab resistance factors in colorectal cancer, we obtained six tumor specimens from colorectal cancer (CRC) patients treated with cetuximab plus chemotherapy, and divided them into cetuximab responders and non-responders according to the primary response criteria for cetuximab treatment [4]. Next, we performed a receptor tyrosine kinase (RTK) array using tumor tissues and found out that phosphorylated RON(p-RON) was upregulated in the cetuximab non-responder group (Supplementary Fig. S1). According to a previous report, RON activity induces resistance to cetuximab, which can be overcome with RON-targeting drugs [21]. Consequently, we investigated whether activated RON could contribute to the responsiveness to cetuximab responsiveness using cetuximab-resistant (KM12C) and -sensitive (LIM1215) cell lines. We first generated a RON knock-out cell line in RONΔ160-positive, cetuximab resistant KM12C cell line and conducted (Supplementary Fig. S2A, B, Supplementary Table S1) xenograft mouse model experiment. As a result, RON knockout facilitated tumor suppression by cetuximab in these cell lines (Supplementary Fig. S2C). On the other hand, cetuximab resistance was induced when RONΔ155 or RONΔ160 was ectopically expressed in wild type RON expressing, and cetuximab-sensitive LIM1215 cells (Supplementary Fig. S2D, E). Therefore, these results raised the possibility of RON variants being an important factor in determining the response to cetuximab, and their therapeutic targeting could be a possible means of overcoming such resistance. As the first step, we explored the structure of RON variants in public repositories, for the purpose of developing small molecule inhibitors against RON variants. Unfortunately, there were no reports of a DFG-out (i.e., Asp oriented away from the ATP binding pocket) conformation structure, or the crystal structure of RON variants, in the Protein Data Bank (PDB). Therefore, we used a structure-based drug design approach via chemical modification and limited scaffold hopping to generate compounds that could inhibit the enzymatic activity of RON. We then performed in vitro cell-based screening of these compounds to screen selective compounds against RON variants showing significant anticancer effect against RON variant-positive cancer cells. Based on these findings, and through our structure-based studies, we finally developed a novel RON inhibitor specifically designed to target phosphorylated forms of RON, including the three RON variants as well as the activated RON by its ligand, macrophage -stimulating protein (MSP). The structure of this inhibitor, WM-S1-030, was based on a conformationally constrained 2-pyridone structure (Fig. 1A). A docking study was performed using a homology model established by ORCHESTRAR of Sybyl X-2.1.1 (Tripos, Inc.), using the crystal structure of c-MET (PDB code: 3F82), with 62% sequence homology with RON, as a template. The docking simulation of WM-S1-030 complexed with the RON kinase domain showed WM-S1-030 to occupy an ATP-binding site within the activation loop in an inactive conformation, with the Asp-Phe-Gly (DFG) motif (a well-known kinase domain) pointed outward from the complex. Here, WM-S1-030 was stabilized through four key hydrogen bonding interactions with Lys1114, Met1164, Gln1171 and Asp1226. The oxygen of ethoxy group and carbonyl group in 2-pyridone core interact with side chain NH of Lys1114 and backbone NH of Asp1226. The nitrogen on thieno[3,2-b]pyridine ring located in hinge region forms hydrogen bonding interaction with the backbone NH of Met1164. In particular, WM-S1-030 is further stabilized by the formation of additional hydrogen bond between the nitrogen of terminal morpholine ring, which occupied the solvent-exposed region and the side chain NH of Gln1171 (Fig. 1B) [28]. WM-S1-030 strongly suppressed (IC50 = 0.39 nM) the enzymatic activity of RON in in vitro enzyme activity (Fig. 1C) [28]. In addition, kinase profiling assay to measure the IC50 value of WM-S1-030 against a panel of kinases was performed, and kinase selectivity of WM-S1-030 was analyzed. WM-S1-030 inhibited the activity of Met family members and a few tyrosine kinases (TKs) (Axl, Flt4, Mer, and TRKC), with IC50 values < 10 nM. On the other hands, WM-S1-030 was > 100-fold selective for the TKs DDR2, Flt3, IGF1R, and ROS, and > 1000-fold selective for all other kinases in the panel, except Fms, KDR and TrkA (Fig. 1D and Supplementary Fig. S3A,B) [28]. To assess the efficacy of WM-S1-030 against RON variants, we analyzed the relative activation of RON in RONΔ160, RONΔ155, and RONΔ165 overexpressing Colo320HSR cells(RON negative cell line), following treatment with WM-S1-030 or BMS777607, a RON inhibitor (IC50 (wt RON) = 2.55 nM) [25] as the reference drug. WM-S1-030 showed strong potency against colo320HSR cells, with IC50 values of 27.41 nM, 98.7 nM, and 137.2 nM, in cell lines expressing RONΔ160, RONΔ155, and RONΔ165, respectively. BMS777607 showed relatively weak inhibition against RONΔ160 and RONΔ155 and RON Δ165, with IC50 values of 458.3 nM, 1406 nM, and 1366 nM, respectively (Fig. 1E). To investigate the cell death effect of WM-S1-030 on RON variants, we treated WM-S1-030 in Colo320HSR cells transfected with RONΔ155, RONΔ160 and RONΔ165. In these cell lines, dose-dependent cell death was caused by WM-S1-030, but not by BMS777607 (Fig. 1F and Supplementary Fig. S3C).
Fig. 1. Discovery of RON variants target inhibitor, WM-S1-030, as anticancer drug.
A 2D chemical structure of WM-S1-030 and key interactions with RON kinase domain. The green dashed lines represent four key hydrogen bonds with Lys1114, Met1164, Gln1171, and Asp1226, which stabilize WM-S1-030. B Docking study of WM-S1-030 located to the ATP-binding pocket in RON homology model based on X-ray crystal structure of c-Met. C In vitro RON kinase enzyme assay of WM-S1-030. D Representative kinase selectivity profiles and IC50 values of WM-S1-030. #Data collected from Eurofins Scientific and Thermo Scientific. E Graph of WM-S1-030/BMS-777607 concentration versus relative phosphorylation (%) of RON variants. RONΔ155, RONΔ160, and RONΔ165 were overexpressed in Colo320HSR cells treated with 0 ~ 1,000 nM BMS-777607 (left panel) and WM-S1-030 (right panel). Cell lysates were subjected to IP and Western blot analysis used to detect the bands representing phospho-RON (RON activity). Band intensities were quantified using ImageJ, and IC50 values calculated using Graphpad Prism 9 software. F Cell death rates of Colo320HSR cells transfected with RONΔ155, RONΔ160, and RONΔ165 and treated with 1 µM or 5 µM WM-S1-030 or BMS777607 for 48 hr, followed by trypan blue exclusion assay. The data are presented as means ± SEMs. (n = 3) *P < 0.01; **P < 0.001; ***P < 0.0001. G Frequencies of RON variants in Caucasian and Korean colorectal cancer patients, using RT-PCR and Sanger sequencing.
Next, we assessed the frequency of RON variants in Caucasian and Korean colon cancer patient and normal tissues. For the analysis of specific RON variants in colon cancer patient samples and the cancer cell lines, we performed real-time PCR (Supplementary Fig. 4A, left lower panel) or RT-PCR & Sanger sequencing (Supplementary Fig. S4, right lower panel). Among 232 Caucasian patient samples, 84 (36.2%) expressed any of the three RON variants, with 20 expressing RONΔ155, 35 expressing RONΔ160, and 29 expressing RONΔ165. Similarly, out of 401 Korean patient samples, 228 (56.9%) had at least one of the three RON variants, with 28 expressing RONΔ155, 164 expressing RONΔ160, and 36 expressing RONΔ165 (Fig. 1G) [28]. In addition, we investigated the presence of RON variant transcripts in multiple cancers, by analyzing TCGA data. Only data for RONΔ165 (ENST00000344206) expression levels was available, showing significant elevation in various cancers including the colon adenocarcinoma (COAD), rectum adenocarcinoma (READ), lung adenocarcinoma (LUAD), cholangiocarcinoma (CHOL) and pancreatic adenocarcinoma (PAAD) tissues, when compared to normal tissues (Supplementary Fig. S4B). Tumorigenic RON variant expression appears to be indifferent of genotypic factors related to geography and ethnicity (e.g., Korean and Caucasian) suggesting that RON variants could be a potentially serve as predictive biomarkers for WM-S1-030 efficacy/response in various cancer patients.
Oncogenic function of RON variants and characterization of WM-S1-030 as its selective inhibitor
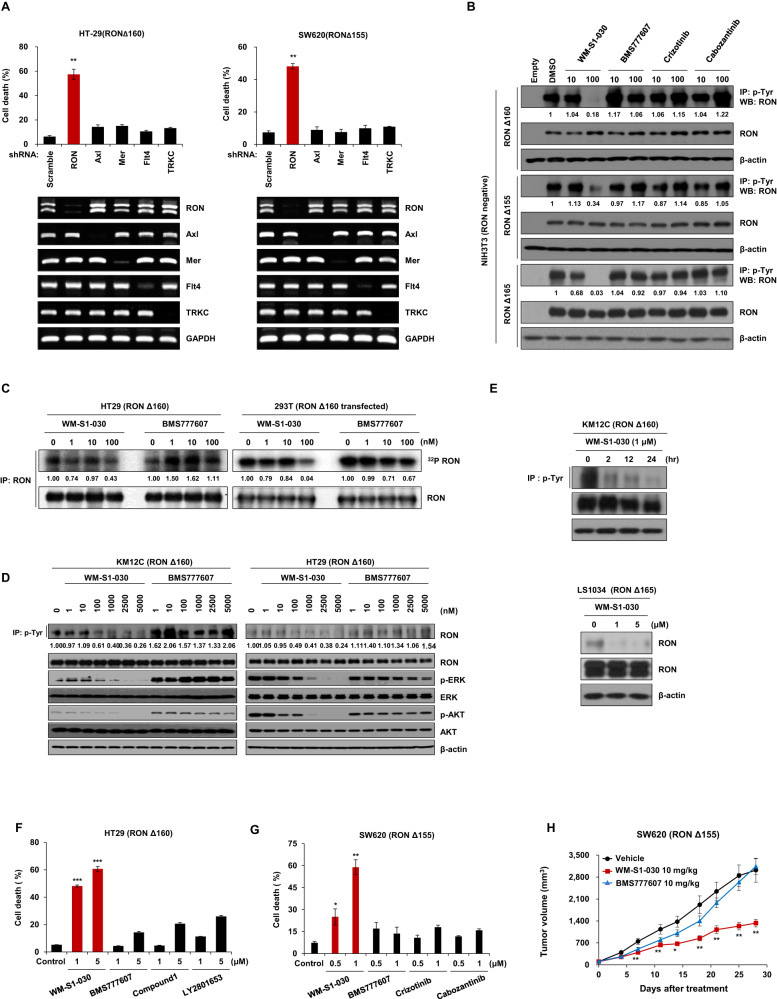
To examine the oncogenic function of RON variants, we transfected shRNA targeting RON variants in RONΔ160-, RONΔ155-, and RONΔ165-positive colon cancer cell lines to knockdown each RON variants. In LS513 (RONΔ165), SNU1047 (RONΔ155), and HT29 (RONΔ160) cell lines, knockdown of RON dramatically increased the cell death in these cell lines, with cell death rates of approximately 62%, 60%, and 75%, respectively(Supplementary Fig. S5A, B). Colony forming assays showed that knockdown of RON variants significantly reduced the colony-forming abilities of HCT15 (RONΔ160) and SW620 (RONΔ155) cell lines (Supplementary Fig. S5C). Also, knockdown of RON variants in KM12C and SNU1047 cell lines significantly decreased the numbers of colonies in soft agar assay (Supplementary Fig. S5D).
In order to find out whether these effects were specific to the knockdown of RON variants, we selected several off-target genes of WM-S1-030 (Axl, Mer, Flt4, TRKC), and knocked down each gene in HT-29 (RONΔ160) and SW620 (RONΔ155) cell lines, confirming that in both cell lines, cell death was induced only by knockdown of RON variants (Fig. 2A). We next evaluated whether WM-S1-030 could selectively inhibit the RON variants, and found out that WM-S1-030 strongly inhibited RONΔ160, RONΔ155, and RONΔ165 when compared to other kinase targeting drugs in RON variant expressing cell lines (Fig. 2B). Similar results were observed in the 32P-kinase assay with WM-S1-030 and BMS777607 (Fig. 2C). These results demonstrates that WM-S1-030 has potent activity against RON variants when compared to other reference drugs. Furthermore, we evaluated the downstream signaling pathway in KM12C and HT29 colon cancer cell lines, showing that WM-S1-030 suppressed phosphorylation of ERK and AKT, which are downstream effectors of RON (Fig. 2D). Moreover, phosphorylation of RON was decreased by WM-S1-030 treatment in a dose- and time-dependent manner (Fig. 2E). The in vitro cell death effect of WM-S1-030 and other kinase-targeting drugs was compared, showing significant induction of cell death by WM-S1-030 compared to other drugs (Fig. 2F, G). These effects were not shown in LIM1215, a wild type RON-expressing colon cancer cell line, demonstrating that expression of RON splicing variants determines the responsiveness to WM-S1-030 in in vitro (Supplementary Fig. S6A). Furthermore, the in vivo efficacy of WM-S1-030 against SW620 xenograft models was assessed. In the SW620 xenograft group treated with WM-S1-030, we observed significant inhibition of tumor growth compared to BMS777607 (Fig. 2H and Supplementary Fig. S7A). Aside from the RON variants, we also evaluated the effect of WM-S1-030 against wild type RON. WM-S1-030 effectively inhibited the phosphorylation of RON in wild type RON-transfected NIH3T3 cells (Supplementary Fig. S8A), and also in Colo201 and HCT8 cell line, which express wild type RON and RON is activated by the secreted MSP (Supplementary Fig. S8B, C, E) [19]. WM-S1-030 also showed sufficient tumor growth inhibition in the Colo201 and HCT8 xenografts (Supplementary Fig. S8D, F). These results demonstrate that WM-S1-030 is a novel potent inhibitor targeting RON variants (RONΔ155, RONΔ160 and RONΔ165) as well as the wild type RON activated by MSP.
Fig. 2. Characterization of WM-S1-030, a specific inhibitor of RON variants, and analysis of its antitumor effects.
A Cell death rates determined by trypan blue exclusion assay of HT29 and SW620 cells transfected with shRNA against RON, AXL, MERTK, FLT4, and NTRK3, after 48–72 hr. Knockdown of each gene was confirmed by RT-PCR. GAPDH, loading control. Data are presented as means ± SEMs. (n = 3) *P < 0.01 B Western blot results following immunoprecipitation showing inhibited phosphorylation of RON variants by treatment with 0, 10, or 100 nM of WM-S1-030, BMS777607, crizotinib, and cabozantinib for 2 hr in RON ∆155-, ∆160-, and ∆165-transfected NIH3T3 cells. β-Actin, loading control. C In vitro radioactive kinase assay results using 32P-γ-phosphate ATP to phosphorylate immunoprecipitated RON variants from HT29 and RON∆160-transfected 293 T cells treated with 1, 10, 100 nM of WM-S1-030 or BMS777607. D Western blot results of the downstream signaling molecules of RON in colon cancer cell lines (KM12C and HT29) expressing endogenous RON variants. β-actin, loading control. Colon cancer cell lines were treated with 0, 1, 10, 100, 1000, 2500, 5000 nM of WM-S1-030 or BMS777607 for 2 hr. E Phosphorylated RON in KM12C and LS1034 cell lines treated with WM-S1-030 in a time- and dose-dependent manner using immunoprecipitation and western blot analysis. β-actin, loading control. F, G Cell death rates of HT29 (left panel) and SW620 (right panel) cells were measured by trypan blue exclusion assay, following treatment with 1 µM or 5 µM of each compound, for 48 hr. (n = 3) *P < 0.05 ; **P < 0.01; *** P < 0.001. H Tumor volumes of SW620 xenografts (n = 6) treated with 10 mg/kg WM-S1-030 or BMS-777607. The data are presented as means ± SEMs. *P < 0.05; **P < 0.01.
WM-S1-030’s potential for overcoming primary resistance to cetuximab in vitro and in vivo
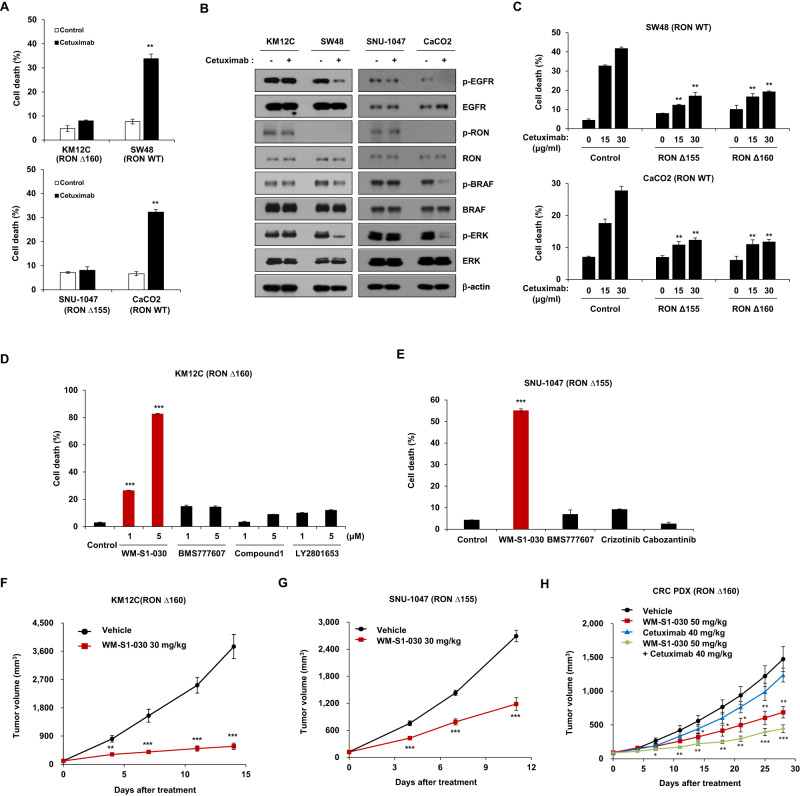
Since the discovery of RON variants in cetuximab-resistant patient samples, we investigated whether WM-S1-030 could potentially overcome this resistance. Based on the previous reports [29, 30] and our findings, KM12C and SNU1047 cell lines were found to be resistant to cetuximab treatment while SW48 and CaCO2 cell lines, are both found to be sensitive to cetuximab in the cell death assay (Fig. 3A). Interestingly, RON variants and phosphorylated RON were exclusively expressed in the resistant cell lines, with no cetuximab-induced changes in downstream signaling pathways (Fig. 3B). Moreover, expression of RONΔ155, RONΔ160 in cetuximab sensitive cell lines induced resistance to cetuximab treatment (Fig. 3C).Treatment of WM-S1-030 in KM12C and SNU1047 cell lines induced cell death, and these results were shown only in the WM-S1-030 treated cell lines when compared to other drugs (Fig. 3D, E). In addition, we confirmed that tumor growth was inhibited when WM-S1-030 was administered to xenograft models of these two cell lines (Fig. 3F, G). Also, additive effects by combination of WM-S1-030 and cetuximab in a RONΔ160-positive patient-derived xenograft (PDX) model were shown (Fig. 3H), demonstrating the efficacy of WM-S1-030 in overcoming the primary resistance to cetuximab treatment.
Fig. 3. Evaluation of WM-S1-030’s potential for overcoming cetuximab primary resistance, in vitro and in vivo.
A Cell death rates evaluated by trypan blue exclusion assay of cetuximab-sensitive or -resistant colorectal cancer cell lines following treatment with 30 µg/ml of cetuximab for 48 hr. Data are presented as means ± SEMs. (n = 3) *P < 0.01; **P < 0.001; ***P < 0.0001. B Expression of phosphorylated RON, EGFR, and downstream signaling in cetuximab- sensitive or -resistant colorectal cancer cell lines following treatment with 30 µg/ml cetuximab. β-actin, loading control. C Cell death rates evaluated by trypan blue exclusion assays of RONΔ155- and RONΔ160-transfected CaCO2 or SW48 cells, after cetuximab treatment. CaCO2 cell lines treated with 0, 15, and 30 μg/ml cetuximab for 48 h and SW48 cell lines treated with 0, 30, and 50 μg/ml cetuximab for 48 hr. Data are presented as means ± SEMs. (n = 3) **P < 0.01. D, E Trypan blue exclusion assay results of KM12C (left panel) and SNU1047 (right panel) cells, following treatment with each compound. KM12C cell lines treated with 1 and 5 μM of WM-S1-030, BMS-777607, Compound 1, or LY2801653, for 48 hr. SNU1047 cell lines treated with 1 μM WM-S1-030, BMS-777607, crizotinib, or cabozantinib, for 48 hr. (n = 3) **P < 0.01; ***P < 0.001. Effect of WM-S1-030 (30 mg/kg) on KM12C xenograft tumors (n = 5) (F) and SNU1047 xenograft model (n = 9-10) (G). H Average tumor growth curve of colorectal patient-derived xenograft (PDX) mouse model (RONΔ160) (n = 5). WM-S1-030 (50 mg/kg) and vehicle were given by oral gavage (p.o) once daily. Cetuximab (40 mg/kg) and vehicle were given intraperitoneally (i.p) twice a week. F–H Tumor volumes were measured twice a week, until mice were sacrificed 14 or 28 days after treatment. Data are presented as means ± SEMs. *P < 0.05; **P < 0.01; ***P < 0.001.
In vitro and in vivo effects of WM-S1-030 on RON variant-positive colon cancer
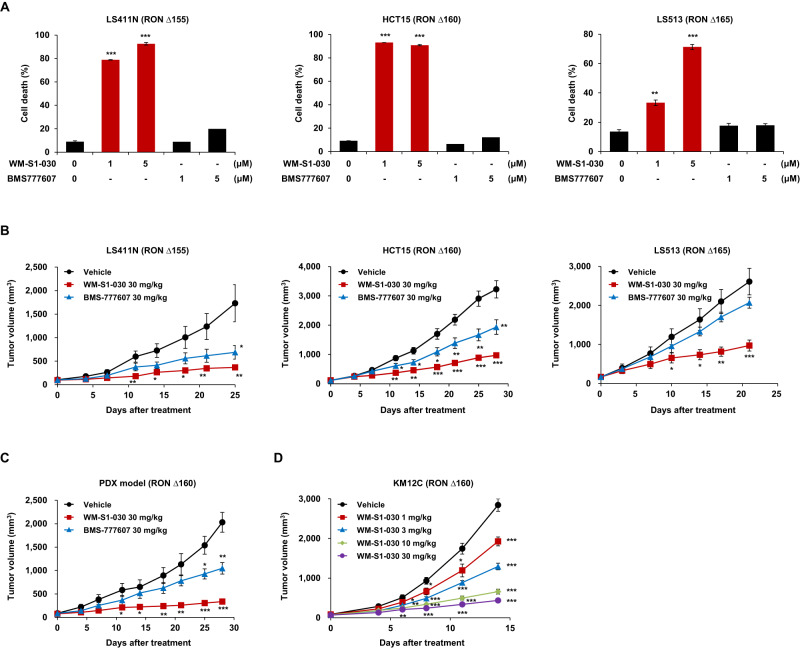
In vitro anticancer activity of WM-S1-030 with BMS777607 against various RON variant-positive colon cancer cells was explored by in vitro cell death assays following WM-S1-030 treatment in LS411N, HCT15, and LS513 cells expressing RON ∆155, ∆160, and ∆165, respectively. We confirmed that BMS777607 showed no effect, while WM-S1-030 significantly increased the cell death rate in these cell lines (Fig. 4A). In addition, we confirmed that tumor growth was significantly inhibited only by 30 mg/kg WM-S1-030, but not 30 mg/kg BMS-777607, in xenograft mouse models using the same cell lines (Fig. 4B and Supplementary Fig. S9A). Moreover, we confirmed the anticancer activity of WM-S1-030 in a RON ∆160-positive colon cancer PDX model, demonstrating that WM-S1-030, but not BMS-777607, significantly suppressed tumor growth (Fig. 4C). We further confirmed the dose dependent response of WM-S1-030 in a xenograft mouse model. Dose-dependent suppression of tumor growth was confirmed through treatment with WM-S1-030 ranging from 1 mg/kg to 30 mg/kg, exhibiting a correlation between the administered dosage and the inhibition of tumor growth (Fig. 4D and Supplementary Fig. S9B).
Fig. 4. In vitro and In vivo effect of WM-S1-030 on RON variant-positive colon cancer.
A Cell death rates of LS411N (trypan blue exclusion assay, left panel), HCT15 (PI staining, middle panel) and LS513 (trypan blue exclusion assay, right panel) cells following treatment with 1 and 5 μM WM-S1-030 or BMS-777607 for 48 hr. (n = 3) **P < 0.01; ***P < 0.001. B Tumor volumes of LS411N (left panel), HCT15 (middle panel) and LS513 (right panel) xenograft models (n = 6–11) treated with 30 mg/kg WM-S1-030 or BMS-777607. *P < 0.05; **P < 0.01; ***P < 0.001. C Tumor growth curves for vehicle, WM-S1-030 (30 mg/kg) or BMS-777607 (30 mg/kg) treated CRC PDX (RON Δ160) mouse models (n = 5). D KM12C xenograft tumor volumes following 0 to 30 mg/kg WM-S1-030 treatment (n = 7). Data are presented as means ± SEMs. *P < 0.05, **P < 0.01, ***P < 0.001.
Furthermore, to assess the potential toxicity associated with high dose or long term administration of WM-S1-030, we conducted an in vivo efficacy test using the SW620 xenograft model for 35 days at a 100 mg/kg dose, and in the A549 xenograft model for 49 days at a 30 mg/kg dose (Supplementary Fig. S9C, D). In these studies, the mice were well tolerated and their body weights were well maintained during the study period demonstrating that WM-S1-030 might have a manageable toxicity at high doses or during long term treatment.
Taken together, these results shows that WM-S1-030 effectively inhibits the RON variants and exerts potent anticancer efficacy in RON variant-positive colon cancer when compared to the reference drug, with manageable toxicity.
Mode of action of WM-S1-030 in colon cancer
In the previous results, we have discovered that WM-S1-030 exerts anticancer effects against RON variant-positive colon cancer cells, regardless of the KRAS genotype. Therefore, we tried to analyze the mode of action of WM-S1-030 in KRAS wt and mutant(mt) colon cancer cell lines.
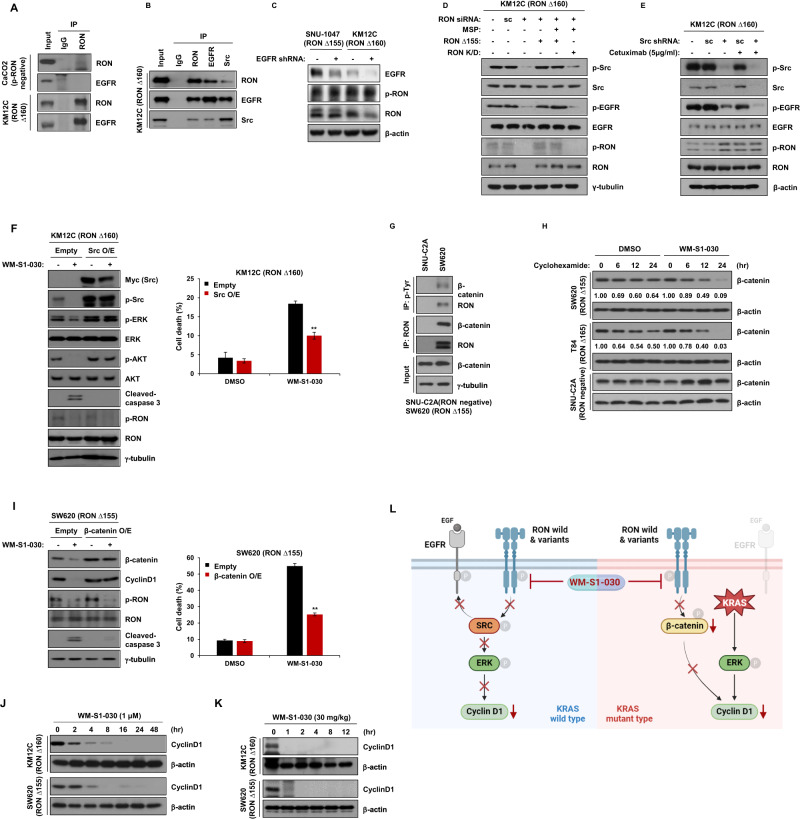
It is known that crosstalk between different receptor tyrosine kinases (RTKs) is thought to lead to oncogenic signal transduction and the acquisition of therapeutic resistance [31–33]. We hypothesized that the resistance to cetuximab treatment in KRAS wt cell lines might be due to the crosstalk between RON and EGFR. Therefore, we immunoprecipitated RON in KM12C(RONΔ160) and CaCO2(RON WT, p-RON negative) cell line. Results showed that RON binds to EGFR only in the KM12C cell line where RON is constitutively activated (Fig. 5A). The binding of EGFR with RON was mediated by EGFR domain III (Supplementary Fig. S10A). Based on the previous report that activation and binding of Src mediates the downstream signaling of RON and EGFR [34], we immunoprecipitated RON in KM12C cell line to see whether Src binds with RON and EGFR, and western blot results revealed that RON,EGFR and Src binds to each other (Fig. 5B) and this interaction occurred only in KRAS wt cell line not the KRAS mt cell line (Supplementary Fig. S10B). To define the detailed mechanism of the crosstalk between RON and EGFR, we knocked down EGFR in KM12C or SNU1047 cell lines, and it did not affect the phosphorylation of RON (Fig. 5C). However, in KM12C cell line, phosphorylation of EGFR was decreased by knockdown of RON (Fig. 5D) and Src (Fig. 5E), and was recovered by overexpression of RON Δ155 but not by the overexpression of RON Kinase Dead protein (Fig. 5D). Also, when Src was knocked down by shRNA, the phosphorylation of EGFR was decreased by cetuximab treatment in cetuximab resistant KM12C cell line (Fig. 5E) demonstrating that the activation of RON regulates the phosphorylation of Src to affect the phosphorylation of EGFR.
Fig. 5. Mode of action of WM-S1-030 against colon cancer.
A Possible interactions of RON with EGFR in CaCo-2 and KM12C cell lines were investigated by immunoprecipitation and western blot analysis. B A possible EGFR-RON-Src trimeric complex, in RON∆160-positive KM12C cells, was investigated by immunoprecipitation and western blot analysis. C Expression of phosphorylated RON after knockdown of EGFR in SNU1047 and KM12C cancer cell lines. Cells were harvested 48 hr after transfection. β-actin, loading control. D RON-dependent phosphorylation of EGFR and Src was evaluated by immunoprecipitation and western blot. Scrambled or anti-RON shRNA was transfected for 48 hr to knock down RON expression, as indicated. RON shRNA was transfected for 48 hr w/wo RON ∆155 or RON kinase-dead (KD) RON ∆155 and RON kinase-dead (KD), as indicated, and then treated with 50 ng/ml MSP for 30 min, before cell harvest. E KM12C cell line was transfected with scrambled or Src shRNA and phosphorylated EGFR and Src levels analyzed by western blot. Cells were treated with cetuximab (5 µg/ml) for 24 hr, after 48-hr shRNA transfection, and levels of phosphorylated EGFR and Src analyzed by western blot. F pcDNA3.1 myc/his(A)-Src was overexpressed in KM12C cells, and the effects of WM-S1-030 (1 μM) on activation of RON, Src, and various downstream signaling molecules, were assessed by western blot. γ-tubulin, loading control. (left panel) 24-hr and 48-hr drug treatments, after transfection, and the resulting cell death rates, as measured by trypan blue exclusion (right panel). (n = 3) **P < 0.01. G Assessment of interaction of RON variants (RON∆155) and β-catenin in SNUC2A and SW620 cell lines by immunoprecipitation and western blot analysis. γ-tubulin, loading control. H The half-life of β-catenin was determined in SW620, T84, and SNUC2A cell lines following treatment with WM-S1-030 (1 µM) and cycloheximide (50 µg/ml) for 0 to 24 hours, as evaluated by western blot. β-actin, loading control. I pcDNA3.1 myc/his(A)-β-catenin plasmid was transfected into SW620 cells and the effects of WM-S1-030 (1 μM) on the activation of RON and downstream signaling molecules were analyzed by western blot. γ-tubulin, loading control (left panel). Drugs treatments were for 24 hr after transfection, and WM-S1-030 cell death rates on β-catenin-overexpressing SW620 cells were measured by trypan blue exclusion 48 hr after drug treatment (right panel). (n = 3) **P < 0.01. J Western blot results of cyclinD1 in KM12C and SW620 cell lines after treatment with WM-S1-030 (1 μM) for 0 to 48 h. β-actin, loading control. K Western blot results of cyclinD1 after WM-S1-030 (30 mg/kg) treatment for 0 to 12 hr of KM12C and SW620 xenograft tumors. β-actin, loading control. Data are presented as means ± SEMs. ***P < 0.001. L Summary of the regulatory mechanism of WM-S1-030 in colon cancer.
To further assess whether activation of Src is involved in mediating the anti-cancer effect of WM-S1-030, we overexpressed Src in KM12C cell line and the cell death was analyzed following treatment with WM-S1-030. As a result, cell death by WM-S1-030 was decreased by the overexpression of Src (Fig. 5F right panel) and in concordance with the cell death results, cleaved caspase-3 was not detected in the same group. Western blot results showed that the downstream signaling molecule of Src, phosphorylation of p-ERK and p-Akt, was not decreased by WM-S1-030 in Src overexpressed KM12C cell line (Fig. 5F left panel). Treatment of WM-S1-030 in KM12C and Colo201, a KRAS wt and RON WT cell line reduced the phosphorylation of Src demonstrating that Src mediates the effect of WM-S1-030 in KRAS wt, RON variant-positive cell lines (Fig. 5F and Supplementary Fig. S8E).
Mode of action of WM-S1-030 in KRAS mt colon cancer cell lines was also evaluated. In colon cancer, mutation of KRAS contributes to tumorigenesis by the interaction with β-catenin [35]. Also in breast cancer, RON binds to β-catenin and stabilizes β-catenin by the phosphorylation of Tyr654 and Tyr670 residue of β-catenin [36]. To seek whether β-catenin is also involved in KRAS mt colon cancer, we immunoprecipitated RON in the two RON variant-positive KRAS mt cell line, SW620 and LS513 cell line, and it was revealed that β-catenin binds with RON in these KRAS mt cell lines (Supplementary Fig. S10B). Moreover, it was shown that β-catenin was phosphorylated and stabilized only when RON was activated either by MSP dependent phosphorylation or autophosphorylation (Fig. 5G and Supplementary Fig. S10D). Knockdown of RON in the SW620 and T84(RON Δ165), another RON variant-positive KRAS mt cell line, lead to the decrease in the phosphorylation of β-catenin (Supplementary Fig. S10E). WM-S1-030 decreased the stability of β-catenin in these cell lines (Fig. 5H) and when β-catenin was overexpressed in SW620 cells, the cell death rate and cleaved caspase 3 was significantly decreased upon the treatment of WM-S1-030 (Fig. 5I), indicating that β-catenin mediates the downstream signaling pathway of RON, and WM-S1-030 exerts its effect by regulating this pathway in KRAS mt colon cancer cell lines. Similar results were shown in KRAS wt and RON wt-expressing colon cancer cell line (Supplementary Fig. S8C). Pharmacodynamic(PD) marker of WM-S1-030 was confirmed to be cyclin D1, by in vitro and in vivo assays. After the treatment of WM-S1-030 in KM12C and SW620 colon cancer cell lines, suppression of cyclin D1 levels up to 48 hours after drug treatment was shown in in vitro (Fig. 5J). Also, in xenograft mouse models of KM12C and SW620 cell lines cyclin D1 expression levels were inhibited up to 12 hours after WM-S1-030 administration in each tumor (Fig. 5K and Supplementary Fig. S10F).
Mode of action of WM-S1-030 could be summarized as illustrated in Fig. 5L. In the KRAS wt, activated RON binds with Src and induce the phosphorylation of EGFR. The both RON WT and splice variants binds with Src, but the RON variants tend to bind with Src stronger than the RON WT. WM-S1-030 effectively suppress this resistance mechanism, thereby providing an opportunity to overcome the resistance to cetuximab in KRAS wt colon cancer. In the case of KRAS mt colon cancer, the stability of β-catenin was reduced by when WM-S1-030 was treated. And in the β-catenin-overexpressed KRAS mt cell line, expression of cyclin D1, the PD marker of WM-S1-030 was maintained although the phosphorylation of RON was decreased upon the treatment of WM-S1-030.
Anti-cancer efficacy of WM-S1-030 in various RON variant-positive solid tumors
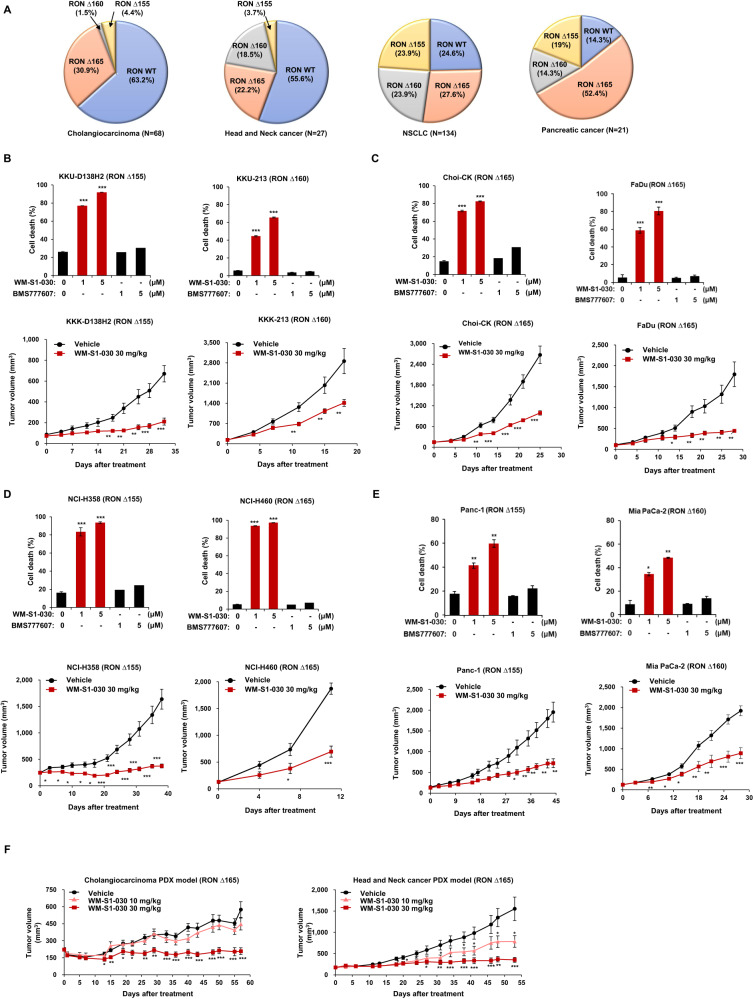
To investigate the anticancer efficacy of WM-S1-030 in RON variant-positive solid tumors other than colon cancer, we evaluated the prevalence of RON variants in cholangiocarcinoma, head and neck cancer, non-small cell lung cancer, and pancreatic cancer (Fig. 6A) [28] and we treated these various cancer cell lines with WM-S1-030 to determine the potential effect of WM-S1-030 against these cell lines. In the case of cholangiocarcinoma, in vitro cell death following WM-S1-030 treatment was measured in KKU-D138H2, KKU-213, and Choi-CK cells expressing RON variants ∆155, ∆160, and ∆165, respectively (Fig. 6B upper panels); tumor growth was suppressed by WM-S1-030 in each cholangiocarcinoma cell line-derived xenograft model (Fig. 6B lower panels). In head and neck cancer, in vitro cell death following WM-S1-030 treatment was measured in FaDu cells expressing RON variant ∆165 (Fig. 6C upper panel); tumor growth was likewise suppressed by WM-S1-030 in FaDu cell line-derived xenografts (Fig. 6C lower panel). In non-small cell lung cancer (NSCLC), in vitro cell death occurred following WM-S1-030 treatment in NCI-H358 and NCI-H460 cells expressing RON variants ∆155 and ∆165, respectively (Fig. 6D upper panels); tumor growth was likewise suppressed by WM-S1-030 in each non-small cell lung cancer cell line-derived xenograft model (Fig. 6D lower panels). Similar results were seen for pancreatic cancer, with in vitro cell death occurring following WM-S1-030 treatment in Panc1 and MiaPaca-2 cells expressing RON variants ∆155 and ∆160, respectively (Fig. 6E upper panels), and tumor growth suppressed by WM-S1-030 treatment of each pancreatic cancer cell line-derived xenograft model (Fig. 6E lower panels). Moreover, we measured the anticancer activity of WM-S1-030 in cholangiocarcinoma and head and neck cancer PDX models, showing significant suppression of tumor growth by WM-S1-030 (Fig. 6F). Based on these results, it could be concluded that WM-S1-030 could potentially be effective in numerous solid tumors beyond colorectal cancer.
Fig. 6. Anticancer efficacy of WM-S1-030 in various RON variants-positive solid tumors.
A The frequencies of RON variants in cholangiocarcinoma, head and neck cancer, NSCLC, and pancreatic cancer patient tissues were analyzed using RT-PCR and Sanger sequencing. B Trypan blue exclusion assay result of KKK-D138-H2 (left upper panel), KKU-213 (middle upper panel), and Choi-CK (right upper panel) cholangiocarcinoma cells were measured following treatment with 1 and 5 μM of WM-S1-030 or BMS-777607 for 72–96 hr. (n = 3) *P < 0.05; **P < 0.01, ***P < 0.001. Antitumor activity of WM-S1-030 (30 mg/kg) in mice harboring xenografts derived from KKK-D138-H2 (left bottom panel), KKU-213 (middle bottom panel), and Choi-CK (right bottom panel) cells (n = 5–9). *P < 0.05; **P < 0.01; ***P < 0.001. C Cell death rates of FaDu (upper panel) cells as measured by trypan blue exclusion assay, following 48-hr treatment with 1 and 5 μM WM-S1-030 or BMS-777607. (n = 3) **P < 0.01; ***P < 0.001. Antitumor activity of WM-S1-030 (30 mg/kg) in mice with xenografts derived from FaDu (bottom panel, n = 5–9). *P < 0.05; **P < 0.01; ***P < 0.001. D Cell death rates of NCI-H358 (left upper panel) and NCI-H460 (right upper panel) cells were measured following treatment with 1 and 5 μM WM-S1-030 or BMS-777607 for 48–72 hr. (n = 3) ***P < 0.001. Antitumor activity of WM-S1-030 (30 mg/kg) in mice with xenografts derived from NCI-H358 (left lower panel) and NCI-H460 (right lower panel) non-small cell lung cancer cells (n = 5–9). *P < 0.05; **P < 0.01; ***P < 0.001. E Cell death rates of Panc-1 (left upper panel) and MiaPaca-2 (right upper panel) pancreatic cells were measured following treatment with 1 and 5 μM of WM-S1-030 or BMS-777607 for 72–96 hr. (n = 3) ***P < 0.001. Antitumor activity of WM-S1-030 (30 mg/kg) in mice with xenografts derived from Panc-1 (left lower panel) and MiaPaca-2 (right lower panel) pancreatic cancer cells (n = 5–9). *P < 0.05; **P < 0.01; ***P < 0.001. F Antitumor activity of WM-S1-030 (10 and 30 mg/kg) in cholangiocarcinoma (left panel) and head and neck cancer (right panel) and PDX models (n = 8–10). Data are presented as means ± SEMs. *P < 0.05, **P < 0.01, ***P < 0.001.
Immunomodulatory activity of WM-S1-030 and its combination effect with anti-PD-1
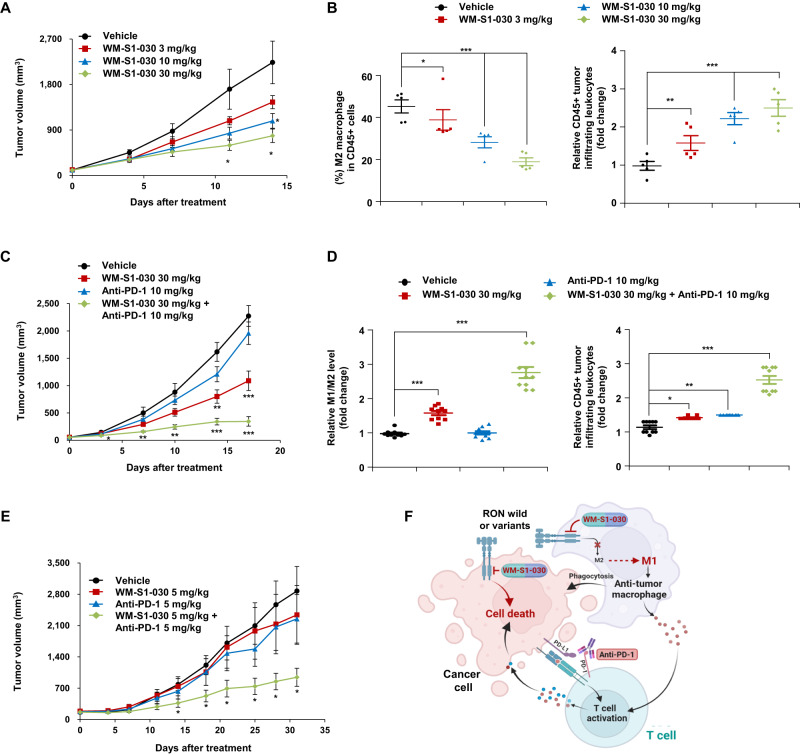
The receptor tyrosine kinase RON (also known as macrophage-stimulating 1 receptor, MST1R) is expressed in macrophages [8] and promotes M2 macrophage polarization, which makes the tumor microenvironment immunosuppressive [37–40]. We hypothesized that inhibition of RON signaling in macrophages by WM-S1-030 might reverse the immunosuppressive state of the tumor micro environment (TME) induced by M2 macrophages. To test this hypothesis, we constructed a CT26 mouse colon cancer syngeneic model in immunocompetent mice, and evaluated the effect of WM-S1-030 on the properties of macrophages. The results showed that compared to vehicle alone, WM-S1-030 significantly decreased tumor growth in a dose-dependent manner (Fig. 7A). WM-S1-030 significantly and dose-dependently increased CD45+ tumor-infiltrating leukocytes, while significantly decreasing M2 (CD45 + F4/80+Arg1 + ) macrophages (Fig. 7B) in these tumors. We also investigated whether WM-S1-030 combined with the immune checkpoint inhibitor anti-PD-1 could exert synergistic antitumor effects in the CT26 syngeneic mouse model. As a result, the combination of WM-S1-030 and anti-PD-1 and synergistically inhibited tumor growth (Fig. 7C).
Fig. 7. Immunomodulatory activity of WM-S1-030 and its potential combinatorial effect with anti-PD-1.
A Antitumor activity of WM-S1-030 (0, 3, 10, and 30 mg/kg) in the CT26 syngeneic mouse model (n = 5). Data are presented as means ± SEMs. *P < 0.05. B The proportions of M2 macrophages (CD45 + /CD11b + /Arginase + ) out of total CD45+ immune cells (CD45 + ) (left panel) and CD45+ tumor-infiltrating leukocytes (right panel) were analyzed in mouse CT26 tumor tissues treated with WM-S1-030 (0, 3, 10, and 30 mg/kg), followed by flow cytometry. Data are presented as means ± SEMs. *P < 0.05, **P < 0.01, ***P < 0.001. C Antitumor activity of WM-S1-030 (30 mg/kg), anti-PD-1 (10 mg/kg), singly or in combination, in the CT26 syngeneic mouse model (n = 5). Data are presented as means ± SEMs. *P < 0.05, **P < 0.01, ***P < 0.001. D Relative M1 (CD45 + /CD11b + /iNOS + )/M2 (CD45 + /CD11b + /Arginase + ) macrophages (left panel) out of total CD45+ immune cells (CD45 + ), and CD45+ tumor-infiltrating leukocytes (right panel), were analyzed by flow cytometry in mouse CT26 tumor tissues treated with WM-S1-030 (30 mg/kg), anti-PD-1(10 mg/kg), singly or in combination. Data are presented as means ± SEMs. *P < 0.05, ** P < 0.01, ***P < 0.001. E Antitumor activity of WM-S1-030 (5 mg/kg), anti-PD-1 (5 mg/kg), singly or in combination, in the NCI-H358 CD34+ humanized mouse model (n = 4). Data are presented as means ± SEMs. *P < 0.05. F Schematic representation of a potential mechanism underlying the combination effect of WM-S1-030 and anti-PD-1.
We also investigated the combined effect of WM-S1-030 and anti-PD-1 on the tumor microenvironment by analyzing changes in the intra-tumoral immune cell populations. We found that CD45+ tumor-infiltrating leukocytes and M1/M2 macrophage ratios were increased by the combination of WM-S1-030 and anti-PD-1 (Fig. 7D). A similar effect was observed in CD34+ humanized mouse model with NCI-H358 non-small cell lung cancer cells, where the combination of WM-S1-030 and anti-PD-1 inhibited tumor growth more strongly than either WM-S1-030 or anti-PD-1 alone (Fig. 7E). We further demonstrated that WM-S1-030 and anti-PD-1 exerted synergistic effects in SW620 CRC cell-derived CD34+ humanized mice, including the upregulation of numerous immunocytokines (Supplementary Fig. S11).
Next, we induced the differentiation of macrophages from PBMCs of 8 healthy donors and 17 cancer patients and analyzed the RON expression by RT-PCR and Sanger sequencing. Our findings revealed that RON variants are expressed only in the PBMC-derived macrophages of cancer patients (Supplementary Fig. S12A). Based on those findings, we next tried to find out the impacts of WM-S1-030 on the differentiation of macrophages by analyzing the expression level of ARG1 and MRC1(marker of M2 macrophages), and CXCL10 (marker of M1 macrophages). In RON WT macrophages, WM-S1-030 increased the M1 macrophage marker while decreasing the M2 macrophage marker in a MSP dependent manner. However, in RON Δ155, Δ160, and Δ165)-positive macrophages, WM-S1-030 increased the M1 macrophage marker while decreasing the M2 macrophage marker in a MSP independent manner. (Supplementary Fig. S12B). Thus, we concluded that WM-S1-030 suppresses both MSP-dependent and MSP-independent RON signaling in macrophages, preventing their polarization toward the M2 phenotype, and likely alleviating the immunosuppressive tumor microenvironment. Therefore, both the direct killing effect of WM-S1-030 against tumor cells and reprogramming of macrophages from M2 to M1 by WM-S1-030 might contribute to the tumor growth inhibition shown in the syngeneic and humanized mouse model (Fig. 7F).
Discussion
The receptor tyrosine kinase RON is expressed and activated in numerous cancer types, and many studies have reported its oncogenic potential, carcinogenic effects, and association with poor prognosis [10–14]. However, the oncogenic functions of RON and alternative splicing forms and which of them act as oncogenic drivers are unclear. Currently, there are many means of stratifying CRC patients, including sensitivity to cetuximab, a representative CRC tyrosine kinase inhibitor [39, 40] with a response rate < 50%. Over half of all CRC patients, particularly those who lack KRAS wt (a biomarker for cetuximab efficacy), or who acquire resistance, do not respond to cetuximab, greatly limiting further treatment options [41–44]. In this study, we identified that RON variants, such as RONΔ155, RONΔ160, and RONΔ165, are constitutively active (independent of the RON ligand MSP) [18] and phosphorylated, with oncogenic activity in colorectal cancer (CRC) patients and numerous solid tumor cell lines [18, 38]. Also identified three RON variants (Δ155, Δ160, and Δ165) in cetuximab-resistant patients, and evaluated their roles in cell proliferation and tumor growth. Based on these results, these variants can be considered new potential targets for the treatment of CRC. We also found that RON variant expression is independent of KRAS genotype.
Why did previous approaches targeting RON fail? This could be explained by couple reasons. First of all, most RON-targeted antibodies prevent MSP binding to RON wild type. One of these antibodies, narnatumab, showed little efficacy in a phase I trial, and its use has since been discontinued [26]. Likewise, an anti-RON monoclonal antibody, Zt/f2, failed to bind the Δ155 and Δ165 variants [45]. Secondly, we found that BMS777607 inhibits wild type RON but was ineffective in inducing cell death in RON variant-positive cells of various cancer types in in vitro and in vivo experiments. In conclusion, previous drugs had limitations in that they only inhibited RON wild type and did not inhibit RON variants with oncogenic driver functions. WM-S1-030 showed potent efficacy against various RON variant-positive colon cancer cell lines and in xenograft models compared to BMS777607 (Fig. 4, Supplementary Table S1), demonstrating that WM-S1-030 is more potent against the RON variants than the reference drug, BMS777607. According to our docking study, it seems that compounds that bind to Gln1171 including WM-S1-030, tend to show better efficacy in inhibiting RON variants. However, despite our efforts, we were unable to obtain the crystal structures of wild-type RON and RON variants, so identifying the differences between WM-S1-030 and the reference compound based on the structure of RON variants remains a task.
Our molecular studies of mode of action indicate thatWM-S1-030 has two different mode of action depending on the KRAS genotype, and Src and β-catenin are the likely downstream factors affected by WM-S1-030 (Fig. 5L). The reason for the difference in the mode of action in KRAS wt and mt colon cancer is yet unclear which needs further study. But we assume that this difference would be resulting from the strong downstream signal transduction to MAPK and PI3K pathway in KRAS mt cells. Further studies in the downstream signaling mechanism of RON in RON variant-positive KRAS wt and mt cells would be needed to define the relations of Src and β-catenin with the effect of WM-S1-030.
The RON (MST1R) receptor tyrosine kinase is also well known to be expressed in macrophages, and RON signaling induces polarization of macrophages toward the M2 phenotype, suppresses the activity of CD8 + T cells, leading to an immunosuppressive tumor microenvironment(TME) that promotes tumor progression [37–40, 46]. RON tyrosine kinase deleted (Ron TK − /−) mouse model shows morphology of M1 macrophages and shows increased CD8 + T cell activity and tumor-infiltrating lymphocytes [47, 48]. This indicates that the expression and activation of RON in macrophages play an important role in the intersection of tumor growth, macrophage polarization, and T cell activity [49]. Nevertheless, the application of existing RON inhibitors as the immunotherapy agents is limited due to the insufficient efficacy. We induced the differentiation macrophages in patient-derived PBMC and analyzed the expression of RON variants. As a result, it was confirmed that RON variants were expressed in about 30% of cancer patients. Also, herein, we showed that WM-S1-030 significantly decreased tumor volume in a syngeneic mouse model, by increasing the number of tumor-infiltrating lymphocytes and decreasing the number of M2 macrophages. Although WM-S1-030 (RON inhibitor) does not directly affect T cell activation, but it could influence the TME by reprograming the M2 macrophages to M1 macrophages. WM-S1-030-increased M1/M2 macrophage ratios and this might indirectly affect the T cell activation by increasing the cytokines such as CXCL10 produced by M1 macrophages (Supplementary Fig. S12B) [50, 51].
In summary, as far as we know, WM-S1-030 is the first small molecule inhibitor of both RON WT and RON variants, RON RONΔ155, RONΔ160, and RONΔ165. Our findings provide experimental evidence that RON variants have potential to be an attractive target for anticancer agents and WM-S1-030 could be applicated as a novel therapeutic agent for RON variant-positive tumors. Based on these preclinical studies, a phase I clinical trial assessing the efficacy of our drug is currently ongoing (ClinicalTrials.gov Identifier: NCT04801095) to elucidate the safety of WM-S1-030 in humans. Furthermore, our preclinical studies, support the possibility of WM-S1-030 being effective in various RON variant (RONΔ155, RONΔ160, and RONΔ165)-positive cancers of humans.
Materials and methods
Ethics statement
All animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals and protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Wellmarker Bio. All operations were performed under anesthesia with isoflurane to minimize animal suffering, in compliance with all relevant ethical regulations. Colon tumor specimens were obtained from patients at Asan Medical Center with Institutional Review Board approval (IRB: S2018-0033-0001 and S2017-0440-0016). We also obtained PBMCs (peripheral blood mononuclear cells) from NSCLC patients from Inje University Busan Paik Hospital, with Institutional Review Board approval (IRB: 2022-03-009-007).
Cell lines
The LIM1215 colorectal cancer (CRC) cell line was purchased from Sigma-Aldrich and cultured in RPMI 1640 medium (Welgene) supplemented with 10% FBS (Gibco), 1% penicillin/streptomycin (Welgene), and 0.6 μg/ml bovine insulin (Sigma-Aldrich) at 37 °C under 5% CO2. The KKU-213, KKKD138, and Choi-CK cholangiocarcinoma cell lines were purchased from the Japanese Collection of Research Bioresources Cell Bank (JCRB) and cultured in DMEM (Welgene) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin (Welgene) at 37 °C under 5% CO2. The CT-26 (mouse colon adenocarcinoma), KM12C (colon carcinoma), MiaPaCa-2 (pancreatic cancer), FaDu (head and neck squamous cell carcinoma), and SNU1047 (colon adenocarcinoma) cell lines were obtained from the Korean Cell Line Bank (KCLB) and cultured in MEM (Welgene), DMEM, or RPMI-1640, supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C under 5% CO2. 293 T, Col201, Colo320HSR, HCT8, HCT15, NCI-H358, NCI-H417, NCI-H460, HT-29, LS513, LS411N, LS1034, SW48, CaCO2, NIH3T3, SW620 and T84 cell lines were purchased from American Type Cell Collection (ATCC) and cultured in DMEM or RPMI 1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C under 5% CO2. Cell lines were periodically tested for mycoplasma contamination and discarded if positive. All the cancer cell lines we used endogenously express RON variants except Colo320HSR and SNUC2A cells, which are negative for RON expression (RON WT and variants) (Supplementary table 1).
shRNA, plasmids and transfection reagents
The pcDNA3.1(+) myc/his(A) vector was purchased from Invitrogen, into which the RON isoforms Δ155 (exons 5, 6, and 11 deleted), RONΔ160 (exons 5 and 6 deleted) and RONΔ165 (exon 11 deleted) were subcloned by WMBIO. RON kinase-dead (KD), pcDNA3.1(+) myc/his(A)-RON(K1114M) was generated by WMBIO. pCMV6-entry-EGFR (Origene, RC214877) vector was purchased from Origene, and EGFR wild type and deletion mutants were subcloned into the vector pEGFP-N1. pcDNA3.1 myc/his(A)-Src and pcDNA3.1 myc/his(A)- β-catenin plasmids were generated by WMBIO. RON, restriction enzymes (XhoI, AgeI), and DNA ligase were obtained from Enzynomics, and all other materials used for DNA amplification were purchased from Bioneer. Colonies were cultured in LB growth medium, and plasmids purified using DNA midi prep kits (Qiagen). The following shRNAs were used in this study:
| Gene | shRNA Sequence |
|---|---|
| scramble | 5′-GGUAGAACGGAGAGGGGUA-3′ |
| RON | 5′-CAGCUAGUUCUUCCUCCCAACCUGA-3′ |
| 5′-UCUC UCAGGUUGGGAGGAAGAACUAGCUG-3′ | |
| EGFR | 5′-GCUGAGAAUGUGGAAUACCUA -3′ |
| Src | 5′-GGGCGAACCACCUGAACAAUU-3′ |
| Axl | 5′-CACUGAAGCUACCUUGAACAGCCTGUU-3′ |
| MERTK | 5′-GUAUCUGAGCAACAGGAAUUUUCTTUU-3′ |
| FLT | 5′-GCACCGAGGUCAUUGUGCAUGAAAAUU-3′ |
| NTRK3 | 5′-GGGCGAAUCUGCUAGUGAAGAUTGUU-3′ |
DNA constructs or shRNAs were transfected using Lipofectamine 2000 (Thermo Fisher) or jetPRIME® (Polyplus), following the manufacturer’s instructions. MSP was from R&D Systems (352MS050/CF) and cycloheximide was from Sigma Aldrich (C4859).
Primers
The sequence of primers used for expression analysis by RT-PCR were as follows:
| Gene | Forward | Reverse |
|---|---|---|
| RON | 5′-ATCTGTGGCCAGCATCTAAC-3′ | 5′-CTTGGTATCCTGCTGCCTTT-3′ |
| AXL | 5′-GCTAATGGACATAGGGCTAAGG-3′ | 5′-CAAGGAAGAGAGCCAAGATGAG-3′ |
| MERTK | 5′-GACTGCCTGGATGAACTGTATG-3′ | 5′-GGAGTGCAGGAGGCAATTATAG-3′ |
| FLT4 | 5′-GAAAGCGACGTGGTGAAGAT-3′ | 5′-GGCAGAACTCCTCATTGATCTG-3′ |
| NTRK3 | 5′-GCATGTCCAGAGATGTCTACAG-3′ | 5′-ACCTCCGTGTTTGAGAGTTG-3′ |
| GAPDH | 5′-GGACTGAGGCTCCCACCTTT-3′ | 5′-CCTGCAGCGTACTCCCCACA-3′ |
The sequence of primers used for the expression analysis by real time PCR are as follows:
| Gene | Forward | Reverse |
|---|---|---|
| ARG-1 | 5′- GTGAAGAACCCACGGTCTGT-3′ | 5′- GCCAGAGATGCTTCCAACTG-3′ |
| MRC-1 | 5′- AGCCAACACCAGCTCCTCAAGA -3′ | 5′- CAAAACGCTCGCGCATTGTCCA -3′ |
| CXCL10 | 5′- GAAAGCAGTTAGCAAGGAAAGG-3′ | 5′- ATGTAGGGAAGTGATGGGAGAG-3′ |
| GAPDH | 5′- GGACTGAGGCTCCCACCTTT-3′ | 5′- CCTGCAGCGTACTCCCCACA -3′ |
RNA isolation and reverse transcription PCR for the analysis of RON variants
Total RNA was extracted from human colon cancer patient tissues using TRIzol (Thermo Scientific) and reverse transcribed using a PrimeScript™ 1st Strand cDNA Synthesis Kit (6110 A, Takara). The reaction conditions were as follows: denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, and elongation at 72 °C for 45 s, for 40 cycles. PCR products were electrophoresed on a 1.5% agarose gel containing RedSafe™ (Intron) stain. After cleanup, the PCR products were sequenced by Macrogen, and variants analyzed using Mutation Surveyor software (SoftGenetics). Primers were as follows:
| Gene | Forward | Reverse |
|---|---|---|
| RON 5/6 deletion | 5′-GAGCTGGTCAGGTCACTAAAC-3′ | 5′-CAGACACTCAGTCCCATTGAC-3′ |
| RON 11 deletion | 5′-ATCTGTGGCCAGCATCTAAC-3′ | 5′-AAAGGCAGCAGGATACCAAG-3′ |
| Axl | 5′-GCTAATGGACATAGGGCTAAGG-3′ | 5′-CAAGGAAGAGAGCCAAGATGAG-3′ |
| MERTK | 5′-GACTGCCTGGATGAACTGTATG-3′ | 5′-GGAGTGCAGGAGGCAATTATAG-3′ |
| FLT4 | 5′-GAAAGCGACGTGGTGAAGAT-3′ | 5′-GGCAGAACTCCTCATTGATCTG-3′ |
| NTRK3 | 5′-GCATGTCCAGAGATGTCTACAG-3′ | 5′-ACCTCCGTGTTTGAGAGTTG-3′ |
| GAPDH | 5′-GGAGTCAACGGATTTGGT-3′ | 5′-GTGATGGGATTTCCATTGAT-3′ |
Real-time PCR to detect RON variants
Total RNA was isolated from tissues using a Qiashredder and RNeasy Mini Kits (QIAGEN, MD, USA), and converted to cDNA with PrimeScript™ 1st strand cDNA Synthesis Kit (6110 A, Takara). Quantitative real-time PCR was performed using a LightCycler 480 Probes Master Kit (Roche Applied Science, Mannheim, Germany), according to the manufacturer’s protocol. The primers were as follows:
| Gene | Forward | Reverse |
|---|---|---|
| RON 5/6 deletion | 5′-CTGGCTCCTGGCAACAG-3′ | 5′-TGGTGCCTACAGACAGACT-3′ |
| RON 11 deletion | 5′-GCCAACCTAGTTCCACTGAAG-3′ | 5′-AGCCACAAGCAGCAGCAAAG-3′ |
| GAPDH | 5′-GGACTGAGGCTCCCACCTTT-3′ | 5′-CCTGCAGCGTACTCCCCACA-3′ |
Gene expression in various cancer
The gene expression profiling interactive analysis (GEPIA) tool was used to explore RNA sequence expression between normal and cancer samples from the TCGA projects. Differentially expressed genes (DEGs) were identified using the Limma R package, which analyzed the expression differences between normal and cancer samples. The analysis employed criteria of an adjusted p-value of less than 0.05 and a log2FC greater than 1. Additionally RNA-sequence data was log2 (TPM + 1) transformed for differential analysis.
Kinase profiling
All data for the kinase profiling assay were obtained by the commercially available services KinaseProfilerTM and IC50ProfilerTM (Eurofins Scientific). The inhibition percentage of 1 µM of the experimental drug against each kinase was determined in the presence of an ATP concentration equal to the Km value of the kinase (n = 2). For kinases whose activity was inhibited by more than 70%, and those that are known anticancer targets, 50% inhibitory concentrations (IC50s) were estimated from the percentage of remaining activity in the presence of nine concentrations (10, 3, 1, 0.3, 0.1, 0.03, 0.01, 0.003, and 0.001 µM) of the experimental drug, in duplicate, using GraphPad Prism™ software.
In vitro RON kinase assay
RON (Carna Biosciences, 08–152, GST-tagged human RON cytoplasmic domain [979–1400 (end) amino acids of accession number NP_002438.1]) was diluted in kinase assay buffer (50 mM HEPES (pH 7.5), 1 mM EGTA, 10 mM MgCl2, 2 mM DTT, and 0.01% Tween-20) to a final concentration of 0.2 nM, and added to white 384-well Optiplates™ (PerkinElmer). Serial dilutions of WM-S1-030 and BMS777607, ranging from 0.001–10 µM in 1% DMSO, were incubated with RON for 15 min at room temperature (RT). The kinase reaction was started by adding a mixture of poly-GT (PerkinElmer) and ATP to a 384-well Optiplate, which was then covered with TopSeal-A film and incubated at RT for 1 hr. After 1 hr, the kinase reaction was stopped by adding EDTA diluted to a final concentration of 10 mM in 1X LANCE Detection Buffer (PerkinElmer), and the plate was incubated for 5 min at RT. Then, a LANCE® Eu-W1024 anti-phosphotyrosine antibody (PT66, PerkinElmer), prepared to a final concentration of 2 nM in 1X LANCE Detection Buffer, was added to the wells, and the plates incubated for 1 hr at RT and covered with TopSeal-A film. After 1 hr, the film was removed, and the signal was read using a Victor X5 multiplate reader (PerkinElmer) in TR-FRET mode (excitation wavelength of 320 or 340 nm and emission wavelength of 665 nm). IC50 values of WM-S1-030 and BMS777607 against RON were calculated using GraphPad Prism software. In addition, in vitro kinase assays using 32P-γ-phosphate ATP were performed as follows. First, to conjugate RON protein to protein A agarose beads, immunoprecipitation was performed using an anti-RON antibody. Then, the RON-conjugated agarose bead was mixed with kinase buffer, 32P-γ-phosphate ATP, and WM-S1-030 or BMS777607, and reacted for 30 min in a water bath at 30 °C. After the reaction, the agarose beads were washed 4 times, subjected to SDS-PAGE, and the gel stained with Coomassie blue and dried. After exposing film to the dried gel, it was developed to check the signal.
In vitro radioactive kinase assay
Cells were lysed in immunoprecipitation lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 0.5% NP-40) containing a protease and phosphatase inhibitor cocktail (GenDEPOT, TX, USA). Precleared cell lysates were then incubated with anti-RON antibody (Santa Cruz), for 24 hr at 4 °C, with gentle rotation followed by incubation with protein A/G-agarose beads (Santa Cruz) for 2 hr at 4 °C, with gentle rotation. The antibody-protein-bead complexes were then washed 4 times with lysis buffer and incubated in kinase assay buffer (20 mM Tris pH 7.4, 5 mM EGTA, and 20 mM β-glycerol phosphate in 1X PBS) containing 32P-γ-phosphate ATP, and WM-S1-030 or BMS777607. Reactions were initiated by addition of activation buffer to final concentrations of 0.1 mM 32P-γ-phosphate ATP and 20 mM MgCl2, and incubated at 30 °C with shaking for 30 min. Reactions were terminated by adding 2X sample buffer and boiled at 95 °C for 5 min. Proteins were resolved by SDS-PAGE and the gels stained with Coomassie blue and dried. The dried gel was then exposed to X-ray films to detect 32P-labeled bands.
In vitro MTS assay
Cells (3 × 103 cells/well) were plated in 96-well plates and incubated overnight. The next day, 10 µl of test compound at doses ranging from 0.01 nM–10 µM in 0.1% DMSO was added to each well, and the cells incubated at 37 °C in a 5% CO2 incubator for 72 hr. 20 µl of CellTiter 96® AQueous One Solution (Promega) was then added to the wells, to determine the number of viable cells, and the cells incubated at 37 °C in a 5% CO2 incubator for 1–4 hr, depending on the experimental conditions. After incubation, absorbance at 490 nm was measured by a Victor X5 multiplate reader (PerkinElmer), and the IC50 value of each drug was calculated with GraphPad Prism software.
In vitro cell death assay
To evaluate cell death rates, in vitro trypan blue and propidium iodide (Invitrogen) staining was performed. 1 × 106 cells were seeded in 10 cm cell culture dishes and cultured overnight in growth media. The next day, cells were exposed to the appropriate compounds for the experiment. 48 hr after the initial seeding, cells were harvested by trypsinization and centrifugation at 1,500 rpm for 5 min at 4 °C. Pelleted cells were resuspended in cold PBS and mixed 1:1 with trypan blue solution (Gibco). Live cells and dead cells were counted using a hemocytometer, and the percentage of dead cells (relative to total cells) was calculated. For PI staining, cells were harvested by trypsinization and centrifuged at 1,500 rpm for 5 min. Pelleted cells were then resuspended in PI staining solution containing 2% FBS, 0.05% sodium azide, and 1 µg/ml PI in PBS, incubated for 10 min at RT, and analyzed on a BD LSRFortessa™ cell analyzer.
In vitro colony-forming assays
Cells were transfected with scrambled or RON shRNA for 48 hr, seeded in a 6-well plate at 100 cells/well, and cultured for 14 days. Cell culture medium (containing drugs) was changed every 2-3 days. The colonized cells were fixed with 10% formalin, stained with 0.01% crystal violet, and photographed. The number of colonies larger than 1 mm diameter was counted.
Soft agar assay
To assess anchorage independence, transfected cells (5 × 104) were resuspended in complete medium containing 0.3% agarose and layered on 0.6% base agarose-containing complete medium. The cells were then cultured to allow colony formation, and media added once a week to prevent the upper layer from drying out. After two weeks, colonies were observed by a microscope.
Immunoprecipitation
Cells were lysed in immunoprecipitation lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 0.5% NP-40) containing a protease and phosphatase inhibitor cocktail (GenDEPOT). Precleared cell lysates were then incubated with the appropriate primary antibodies, and incubated for 24 hr at 4 °C, with gentle rotation. The next day, protein A/G-agarose beads (Santa Cruz) were added to the mixtures and incubated for 2 hr at 4 °C, with gentle rotation. The antibody-protein complexes were then washed five times, and bound proteins eluted by boiling in 30 µl SDS sample buffer, followed by Western blot analysis.
Western blot analysis
Cell or tissue lysates were electrophoresed, transferred, and immunoblotted with specific antibodies. Membranes were incubated with specific primary antibodies against phospho-Tyr (4G10, Millipore), RON β (sc-374626, Santa Cruz), Erk (4695 S, Cell Signaling), phospho-Erk (4370 S, Cell Signaling), Akt (9272 S, Cell Signaling), phospho-Akt (4060 S, Cell Signaling), cyclin D1 (sc-8396, Santa Cruz), c-Myc (sc-40, Santa Cruz), Myc-tag (Ab9108, Abcam), β-catenin (8480 S, Cell Signaling), p-EGFR (2234 S, Cell Signaling), EGFR (2232 S, Cell Signaling), cleaved caspase-3 (9664 S, Cell Signaling), Src (2108 S, Cell Signaling), p-Src (2101 S, Cell Siganling), γ-tubulin (SC-17787, Santa Cruz), and β-actin (sc-47778, Santa Cruz). Anti-rabbit and anti-mouse secondary antibodies were obtained from Cell Signaling Technology. Detailed information of antibodies used is also described in Supplementary Table S2. Uncropped western blots are provided in Supplementary Material.
GST pulldown assay
500 ng of SRC-GST(Sino biological,10755-H20B) and RONΔ155, RONΔ160, RONΔ165 (manufactured by Peak Proteins Ltd, Macclesfield, Cheshire, UK) recombinant proteins were incubated according to each combination with reaction buffer (20 mM Tris-HCl pH8.0, 500 mM NaCl, 1% Triton X-100, 0.02% bovine serum albumin and 5 mM 2-Mercaptoethanol), 120 mM NaCl) in a 30 °C water bath for 30 minutes. Immunoprecipitation and western blot were performed by adding glutathione beads instead of antibody and agarose beads to the reaction sample.
Tumor-infiltrating lymphocyte (TIL) isolation
Primary tumors were dissected and cut into small pieces in ice-cold PBS, incubated in 50 mL conical tubes with 1 mL media supplemented with 5% FBS and 2.4 mg/mL collagenase B (11088815001, Roche), and incubated for 2–4 hr, at 37 °C, on a shaker. After incubation, samples were centrifuged at 1200 rpm for 3 min, and supernatants collected for cytokine analysis. The residual pellets were resuspended in 20 mL PBS, dissociated further by 4x pipetting, centrifuged for 7 min at 300 × g at room temperature, resuspended again in 6 mL media containing 0.3 mg/mL DNase I (11284932001, Roche), and then incubated for 15–30 min at 37 °C on a shaker. Tissues were then centrifuged for 7 min at 300 × g at 4 °C, resuspended in 10 mL Trypsin-EDTA solution, and cells dissociated by gentle pipetting. Single cells were prepared by filtering through a 70-μm cell strainer.
Analysis of single cells by flow cytometry
All flow cytometry data were collected on an LSRFortessa™ Flow Cytometer (BD Biosciences) equipped with blue, red, and violet lasers. For sample acquisition, human PBMCs were first incubated with Human Fc block (# 564219, BD) and stained with the following fluorochrome-conjugated antibodies in FACS buffer (2% FBS and 0.05% sodium azide in PBS): anti-human CD45, anti-human CD3, anti-human CD8, anti-human CD11b, anti-human CD86, and anti-human CD206, for 30 min at 4 °C, and analyzed by flow cytometry. Mouse cells were first blocked with mouse Fc block (#553141, BD) and stained with the following fluorochrome-conjugated antibodies in FACS buffer (2% FBS and 0.05% sodium azide in PBS): anti-mouse CD45, anti-mouse T lymphocyte subset cocktail (CD3, CD4, CD8), and anti-mouse CD11b. Next, the cells were fixed and permeabilized with Fixation/Permeabilization kit (# 554714, BD) for 20 min at 4 °C, stained with anti-mouse iNOS or anti-mouse arginase 1, and analyzed by flow cytometry. All flow cytometry data was analyzed with FlowJo software (BD Biosciences). All electronic gating was performed downstream of an FSC-H x FSC-A “singlet” gate. Detailed information of antibodies used is also described in Supplementary Table S2.
Cytometric bead array (CBA) by flow cytometry
Cytokine levels of tumor-derived supernatants were analyzed by flow cytometry. To measure secreted cytokine levels, Cytometric Bead Array (CBA) Flex Sets (BD Biosciences), to detect human TNF-α, IFN-γ and granzyme B, were used according to the manufacturer’s instructions. Briefly, 50 μL of each sample was transferred to a FACS tube, and 50 μL of the capture bead mixture added to each tube and incubated in the dark for 1 hr at room temperature (RT). After 1 hr, 50 μL of PE detection reagent mixture was added and incubated in the dark for 2 hrs at RT. After 2 h, 1 mL of wash buffer was added and centrifuged at 200 g for 5 min. The supernatant was then removed, 200 μL of wash buffer added, and the cells gently resuspended. Data were acquired using a LSRFortessa™ Flow Cytometer (BD Biosciences), and analyzed with FlowJo software (BD Biosciences).
Mice
Six-week-old female BALB/c nude mice were purchased from Charles Rivers Japan and GemBiosciences, while 6-week-old female wild-type BALB/c mice were purchased from Orient Bio. Mice were housed at a consistent temperature (21 ± 2 °C) and relative humidity (55 ± 10%) on a 12-hr light/dark cycle at a fixed brightness (150–300 lux). Radiation-sterilized food (laboratory diet) and reverse osmosis-purified water were provided ad libitum. hCD34+ HSC mice were purchased from GemBiosciences. Four-week-old female NSG mice were used to generate humanized mice following myeloablation and transplantation of hCD34+ HSCs. Bone marrow cell production was suppressed by 1.5 Gy radiation generated by an XRAD 320 biological irradiator (Precision X-ray, CT) for 12 hr, and 1 × 105 hCD34+ HSCs then administered via tail vein injection. The success of humanization was confirmed by FACS analysis, and the procedure was considered successful when the ratio of human CD45+ PBMCs was greater than 25%. To evaluate human cell engraftment, 50 µl of blood retro-orbital sinus was collected in heparin-coated capillary tubes. Cells were incubated with APC-conjugated mouse anti-human CD45, FITC-conjugated mouse antihuman CD3, and PE-conjugated mouse anti-human CD19 antibodies (BD Pharmingen), and FACS analysis performed on a BD FACS Canto II flow cytometer.
In vivo syngeneic mouse model
A CT26 syngeneic model was constructed by subcutaneously injecting 2.5 × 105 suspended cells diluted in 100 µl PBS into the flanks of 6-week-old female BALB/c mice. When tumor volumes reached 100 mm3, the mice were randomly divided into four groups. WM-S1-030 was given once daily by oral gavage and anti-PD-1 (RMP1-14, BioXcell) given by intraperitoneal injection twice a week. Tumor volumes were measured twice a week. Tumor volumes, weights, and body weight data were collected and analyzed statistically after the end of the experiment.
In vivo xenograft mouse model
Xenograft tumors were established by right flank subcutaneous injection of 1.5 × 106 KM12C cells suspended in 100 µl PBS, 2 × 106 SNU1047 cells suspended in 100 µl PBS, 5 × 106 SW620, LS411N, or HCT15 cells suspended in 100 µl PBS, 2 × 106 LS513 cells suspended in 200 µl PBS mixed 1:1 with Matrigel, 1 × 107 LIM1215 cells suspended in 200 µl PBS mixed 1:1 with Matrigel. The mice were randomly divided into groups when tumor volumes reached approximately 100 mm3 and treated daily with vehicle control, WM-S1-030, or BMS-777607 by gavage for several days. Cetuximab (Merck) was administered intraperitoneally (i.p.) twice a week, and tumor volumes measured two or three times a week by a digital caliper and quantified by the following formula: tumor volume = 0.5 × length × width2.
In vivo patient-derived xenograft mouse model
Colon tumor specimens were obtained from patients at Asan Medical Center with Institutional Review Board approval. Cholangiocarcinoma and head and neck cancer patient-derived xenograft (PDX) experiments were performed by Champions Oncology. To establish PDX tissues, fresh tumor fragments were transplanted subcutaneously into the right or left flanks of anesthetized BALB/c-nude mice and athymic nude mice. When tumor sizes reached 800–1500 mm3, mice were sacrificed, and tumors harvested. For the main study of PDX model mice, 1–2 mm3 tumor fragments were implanted subcutaneously into the lateral flanks of female BALB/c nude mice and athymic nude mice. The mice were allocated into different groups in a random manner once tumor volumes reached ~100 mm3, after which they received daily oral treatments of either vehicle control, WM-S1-030, or BMS-777607, for five consecutive days.
Generation of humanized CD34 + HSC mice
NCI-H358 and SW620 humanized mouse models were constructed by subcutaneously injecting 5 × 106 cells (suspended in 200 µl of a 1:1 mixture of PBS and Matrigel, or 100 µl of PBS) into each flank of 13-week-old female hCD34+ HSC mice. Tumor volumes were calculated twice weekly, and when tumor volumes reached ~100 mm3, mice were randomly divided into four groups. WM-S1-030 (3 mg/kg) or vehicle was given daily by oral gavage. Anti-PD-1 (pembrolizumab) (Merck Sharp & Dohme, MSD) was administered twice a week by intraperitoneal injection. Tumor volumes, weights, and body weight data were collected and analyzed statistically after the end of the experiments.
Assessing tumor development and changes in body weight
Tumor volume was measured twice a week by digital caliper and quantified by the following formula: tumor volume = 0.5 × length × width2.
Animals were weighed at the same time points as tumor volume measurements. Average mouse body weights were recorded as percentages of their initial weights.
Immunohistochemistry
5 μm sections were cut from a tissue block and immunohistochemically analyzed. The sections were incubated overnight with antibodies against RON, p-RON, cyclin D1, and cleaved caspase 3, diluted at 1:100 in Antibody Diluent Solution. Sections were then incubated at RT for 1 hr with biotinylated secondary horse anti-mouse IgG (VECTASTAIN Elite mouse ABC HRP Kit, # PK-6102, Vector Labs). Avidin-biotin complex-horseradish peroxidase (VECTASTAIN Elite mouse ABC HRP Kit) was applied to sections and incubated for 1 hr at RT and exposed to 3,3′-diaminobenzidine substrate solution (# SK-4100, Vector labs) for 5 min. Tissue sections were stained with Mayer’s hematoxylin for 10 s and rinsed in tap water and mounted for viewing. All tissues were scanned using a Precipoint M8 microscope and scanner.
RTK arrays
Receptor tyrosine kinase (RTK) phosphorylation statuses, between cetuximab responders and non-responders, were analyzed using a Phospho-RTK Array Kit (# ARY001B, R&D Systems), according to the manufacturer’s protocol. Tumor tissues from cetuximab responders and non-responders were obtained from Asan Medical Center, Seoul, Korea. For analysis of phosphorylation statuses of multiple RTKs in tissue samples and cell lines, membranes were developed on X-ray film and the densities of the developed spots for each RTK were measured using ImageJ software. The RTK spot density for RTKs of cetuximab-responsive patient tissue lysate was set as 100, and the relative RTK spot densities for proteins from cetuximab-nonresponsive patient tissue lysates then calculated.
MSP ELISA assay
Human MSP ELISA kit (# DY352, R&D Systems) was performed according to the instructions suggested by the manufacturer. 100 μL per well of capture antibody diluted in PBS was coated on a 96-well microplate and incubated overnight at room temperature. The coated wells were washed three times with wash buffer and blocked with 300 μL Reagent Diluent buffer for 1 hour at room temperature. The well was washed three times with wash buffer, 100 μL of the medium in which HCT8 cells were cultured and standards samples were added, and cultured at room temperature for 2 hours. The well was washed three times with wash buffer, 100 μL of diluted detection antibody was added, and incubated at room temperature for 2 hours. Each well was again washed three times with wash buffer, 100 μL of diluted Streptavidin-HRP was added, and incubated at room temperature for 20 minutes. Each well was washed with wash buffer, 100 μL of Substrate Solution was added, and the reaction was stopped with 50 μL Stop solution after incubation at room temperature for 20 minutes. Determine the optical density of each well immediately, using a microplate reader set to 450 nm.
Statistical analysis
Unless otherwise stated, data are expressed as means ± standard errors of the mean. To assess significance of differences between groups, we used two-tailed Student’s t tests and one-way analysis of variance (ANOVA). Dunnett’s multiple comparisons test was used to evaluate the significance of differences among multiple group means.
Supplementary information
Author contributions
Conceptualization: JK, DIK, and DHJ. Methodology, formal analysis and investigation: JK, SWH, DIK, JP, SCH, ML, YSP, JSS, HK, HR, SMK, MSL, MHK, JHL, JJ, SB, JKH, HRJ, YSR, MC, DYH, and DHJ. Writing-original draft: JK, SWH, DIK, and JP. Writing, review and editing: JK, DIK, JP, and DHJ. Supervision: DHJ.
Funding
This research was supported by the Korea Drug Development Fund funded by the Ministry of Science and ICT, Ministry of Trade, Industry, and Energy, and Ministry of Health and Welfare (KDDF201612-12 and KDDF201812-22, Republic of Korea). The biospecimens and data used in this study were provided by the Asan Bio-Resource Center, Korea Biobank Network (2021-14(231)).
Data availability
The RTK array raw data has been deposited at datadryad.org and are available from 10.5061/dryad.8sf7m0cvc. Additional analytical raw data are available upon reasonable request to the corresponding author.
Materials availability
All unique reagents generated in this study are available from the corresponding author with a completed Materials Transfer Agreement.
Code availability
All analysis algorithm codes follow official tutorials. Relevant information is available upon reasonable request to the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Joseph Kim, Dong-In Koh.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01235-9.
References
- 1.Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14:101174. doi: 10.1016/j.tranon.2021.101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Misale S, Nicolantonio FD, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4:1269–80. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 3.Dahabreh IJ, Terasawa T, Castaldi PJ, Trikalinos TA. Systematic review: Anti-epidermal growth factor receptor treatment effect modification by KRAS mutations in advanced colorectal cancer. Ann Intern Med. 2011;154:37–19. doi: 10.7326/0003-4819-154-1-201101040-00006. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–45. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 5.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254–61. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 6.Maggiora P, Marchio S, Stella MC, Giai M, Belfiore A, Bortoli MD, et al. Overexpression of the RON gene in human breast carcinoma. Oncogene. 1998;16:2927–33. doi: 10.1038/sj.onc.1201812. [DOI] [PubMed] [Google Scholar]
- 7.Wang MH, Lee W, Luo YL, Weis MT, Yao HP. Altered expression of the RON receptor tyrosine kinase in various epithelial cancers and its contribution to tumourigenic phenotypes in thyroid cancer cells. J Pathol. 2007;213:402–11. doi: 10.1002/path.2245. [DOI] [PubMed] [Google Scholar]
- 8.Yao HP, Zhou YQ, Zhang R, Wang MH. MSP-RON signalling in cancer: pathogenesis and therapeutic potential. Nat Rev Cancer. 2013;13:466–81. doi: 10.1038/nrc3545. [DOI] [PubMed] [Google Scholar]
- 9.Yao HP, Zhuang CM, Zhou YQ, Zeng JY, Zhang RW, Wang MH. Oncogenic variant RON160 expression in breast cancer and its potential as a therapeutic target by small molecule tyrosine kinase inhibitor. Curr Cancer Drug Targets. 2013;13:686–97. doi: 10.2174/15680096113139990038. [DOI] [PubMed] [Google Scholar]
- 10.Comperat E, Roupret M, Chartier-Kastler E, Bitker MO, Richard F, Camparo P, et al. Prognostic value of MET, RON and histoprognostic factors for urothelial carcinoma in the upper urinary tract. J Urol. 2008;179:868–72. doi: 10.1016/j.juro.2007.10.079. [DOI] [PubMed] [Google Scholar]
- 11.Hunt BG, Wicker CA, Bourn JR, Lower EE, Takiar V, Waltz SE. MST1R (RON) expression is a novel prognostic biomarker for metastatic progression in breast cancer patients. Breast Cancer Res Treat. 2020;181:529–40. doi: 10.1007/s10549-020-05653-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molife LR, Dean EJ, Blanco-Codesido M, Krebs MG, Brunetto AT, Greystoke AP, et al. A phase I, dose-escalation study of the multitargeted receptor tyrosine kinase inhibitor, golvatinib, in patients with advanced solid tumors. Clin Cancer Res. 2014;20:6284–94. doi: 10.1158/1078-0432.CCR-14-0409. [DOI] [PubMed] [Google Scholar]
- 13.Wang MH, Padhye SS, Guin S, Ma Q, Zhou YQ. Potential therapeutics specific to c-MET/RON receptor tyrosine kinases for molecular targeting in cancer therapy. Acta Pharmacol Sin. 2010;31:1181–8. doi: 10.1038/aps.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CT, Chow NH, Su PF, Lin SC, Lin PC, Lee JC. The prognostic significance of RON and MET receptor coexpression in patients with colorectal cancer. Dis Colon Rectum. 2008;51:1268–74. doi: 10.1007/s10350-008-9297-1. [DOI] [PubMed] [Google Scholar]
- 15.Mayer S, Hirschfeld M, Jaeger M, Pies S, Iborra S, Erbes T, et al. RON alternative splicing regulation in primary ovarian cancer. Oncol Rep. 2015;34:423–30. doi: 10.3892/or.2015.3995. [DOI] [PubMed] [Google Scholar]
- 16.Chakedis J, French R, Babicky M, Jaquish D, Mose E, Cheng P, et al. Characterization of RON protein isoforms in pancreatic cancer: implications for biology and therapeutics. Oncotarget. 2016;7:45959–75. doi: 10.18632/oncotarget.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–90. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Zhou YQ, He C, Chen YQ, Wang D, Wang MH. Altered expression of the RON receptor tyrosine kinase in primary human colorectal adenocarcinomas: generation of different splicing RON variants and their oncogenic potential. Oncogene. 2003;22:186–97. doi: 10.1038/sj.onc.1206075. [DOI] [PubMed] [Google Scholar]
- 19.Chen YQ, Zhou YQ, Angeloni D, Kurtz AL, Qiang XZ, Wang MH. Overexpression and activation of the RON receptor tyrosine kinase in a panel of human colorectal carcinoma cell lines. Exp Cell Res. 2000;261:229–38. doi: 10.1006/excr.2000.5012. [DOI] [PubMed] [Google Scholar]
- 20.Park YL, Lee GH, Kim KY, Myung E, Kim JS, Myung DS, et al. Expression of RON in colorectal cancer and its relationships with tumor cell behavior and prognosis. Tumori. 2012;98:652–62. doi: 10.1177/030089161209800517. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Singh B, Graves-Deal R, Ma H, Starchenko A, Fry WH, et al. Three-dimensional culture system identifies a new mode of cetuximab resistance and disease-relevant genes in colorectal cancer. Proc Natl Acad Sci USA. 2017;114:E2852–E2861. doi: 10.1073/pnas.1618297114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graves-Deal R, Bogatcheva G, Rehman S, Lu Y, Higginbotham JN, Singh B. Broad-spectrum receptor tyrosine kinase inhibitors overcome de novo and acquired modes of resistance to EGFR-targeted therapies in colorectal cancer. Oncotarget. 2019;10:1320–33. doi: 10.18632/oncotarget.26663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Siegmund A, Xi N, Kaplan-Lefko P, Rex K, Chen A, et al. Discovery of a potent, selective, and orally bioavailable c-Met inhibitor: 1-(2-hydroxy-2-methylpropyl)-N-(5-(7-methoxyquinolin-4-yloxy)pyridin-2-yl)-5-meth yl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide (AMG 458) J Med Chem. 2008;51:3688–91. doi: 10.1021/jm800401t. [DOI] [PubMed] [Google Scholar]
- 24.Raeppel S, Gaudette F, Mannion M, Claridge S, Saavedra O, Isakovic L, et al. Identification of a novel series of potent RON receptor tyrosine kinase inhibitors. Bioorg Med Chem Lett. 2010;20:2745–9. doi: 10.1016/j.bmcl.2010.03.073. [DOI] [PubMed] [Google Scholar]
- 25.Schroeder GM, An Y, Cai ZW, Chen XT, Clark C, Cornelius LAM, et al. Discovery of N-(4-(2-amino-3-chloropyridin-4-yloxy)-3-fluorophenyl)-4-ethoxy-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide (BMS-777607), a selective and orally efficacious inhibitor of the Met kinase superfamily. J Med Chem. 2009;52:1251–4. doi: 10.1021/jm801586s. [DOI] [PubMed] [Google Scholar]
- 26.LoRusso PM, Gounder M, Jalal SI, André V, Kambhampati SRP, Loizos N, et al. Phase 1 study of narnatumab, an anti-RON receptor monoclonal antibody, in patients with advanced solid tumors. Investig New Drugs. 2017;35:442–50. doi: 10.1007/s10637-016-0413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang K, Karnad A, Zhao S, Freeman JW. Roles of c-Met and RON kinases in tumor progression and their potential as therapeutic targets. Oncotarget. 2015;6:3507–18. doi: 10.18632/oncotarget.3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu H, Kim H, Park I, Lee M, Park YS, Jin DH, et al. Discovery of Novel, Thienopyridine-Based Tyrosine Kinase Inhibitors Targeting Tumorigenic RON Splice Variants. ACS Med Chem Lett. 2023;14:1198–207. doi: 10.1021/acsmedchemlett.3c00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015;6:7002. doi: 10.1038/ncomms8002. [DOI] [PubMed] [Google Scholar]
- 30.Liang L, Liu M, Sun X, Yuan Y, Peng K, Rashid K, et al. Identification of key genes involved in tumor immune cell infiltration and cetuximab resistance in colorectal cancer. Cancer Cell Int. 2021;21:135. doi: 10.1186/s12935-021-01829-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun C, Bernards R. Feedback and redundancy in receptor tyrosine kinase signaling: relevance to cancer therapies. Trends Biochem Sci. 2014;39:465–74. doi: 10.1016/j.tibs.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Linklater ES, Tovar EA, Essenburg CJ, Turner L, Madaj Z, Winn ME, et al. Targeting MET and EGFR crosstalk signaling in triple-negative breast cancers. Oncotarget. 2016;7:69903–15. doi: 10.18632/oncotarget.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metibemu DS, Akinloye OA, Akamo AJ, Ojo DA, Okeowo OT, Omotuyi IO. Exploring receptor tyrosine kinases-inhibitors in Cancer treatments. Egypt J Med Hum Genet. 2019;20:35. [Google Scholar]
- 34.Danilkovitch-Miagkova A, Angeloni D, Skeel A, Donley S, Lerman M, Leonard EJ. Integrin-mediated RON growth factor receptor phosphorylation requires tyrosine kinase activity of both the receptor and c-Src. J Biol Chem. 2000;275:14783–6. doi: 10.1074/jbc.C000028200. [DOI] [PubMed] [Google Scholar]
- 35.Jeong WJ, Ro EJ, Choi KY. Interaction between Wnt/β-catenin and RAS-ERK pathways and an anti-cancer strategy via degradations of β-catenin and RAS by targeting the Wnt/β-catenin pathway. NPJ Precis Oncol. 2018;2:5. doi: 10.1038/s41698-018-0049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagh PK, Gray JK, Zinser GM, Vasiliauskas J, James L, Monga SP, et al. β-Catenin is required for Ron receptor-induced mammary tumorigenesis. Oncogene. 2011;30:3694–704. doi: 10.1038/onc.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan C, Brown NE, Vasiliauskas J, Pathrose P, Starnes SL, Waltz SE. Prostate Epithelial RON Signaling Promotes M2 Macrophage Activation to Drive Prostate Tumor Growth and Progression. Mol Cancer Res. 2020;18:1244–54. doi: 10.1158/1541-7786.MCR-20-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang MH, Ronsin C, Gesnel MC, Coupey L, Skeel A, Leonard EJ, et al. Identification of the ron Gene Product as the Receptor for the Human Macrophage Stimulating Protein. Science. 1994;266:117–9. doi: 10.1126/science.7939629. [DOI] [PubMed] [Google Scholar]
- 39.Gaudino G, Follenzi A, Naldini L, Collesi C, Santoro MM, Gallo K, et al. RON is a heterodimeric tyrosine kinase receptor activated by the HGF homologue MSP. EMBO J. 1994;13:3524–32. doi: 10.1002/j.1460-2075.1994.tb06659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshimura T, Yuhki N, Wang MH, Skeel A, Leonard EJ. Cloning, sequencing, and expression of human macrophage stimulating protein (MSP, MST1) confirms MSP as a member of the family of kringle proteins and locates the MSP gene on chromosome 3. J Biol Chem. 1993;268:15461–8. [PubMed] [Google Scholar]
- 41.Jonker DJ, O’Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–8. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 42.Xie YH, Chen YX, Fang JY. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther. 2020;5:22. doi: 10.1038/s41392-020-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liévre A, Bachet JB, Corre DL, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–5. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 44.Zhao B, Wang L, Qiu H, Zhang M, Sun L, Peng P, et al. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget. 2017;8:3980–4000. doi: 10.18632/oncotarget.14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao HP, Zhou YQ, Ma Q, Guin S, Padhye SS, Zhang RW, et al. The monoclonal antibody Zt/f2 targeting RON receptor tyrosine kinase as potential therapeutics against tumor growth-mediated by colon cancer cells. Mol Cancer. 2011;10:82. doi: 10.1186/1476-4598-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Song Y, Du W, Gong L, Chang H, Zou Z. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci. 2019;26:78. doi: 10.1186/s12929-019-0568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ekiz HA, Lai SCA, Gundlapalli H, Haroun F, Williams MA, Welm AL. Inhibition of RON kinase potentiates anti-CTLA-4 immunotherapy to shrink breast tumors and prevent metastatic outgrowth. Oncoimmunology. 2018;7:e1480286. doi: 10.1080/2162402X.2018.1480286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruiz-Torres SJ, Bourn JR, Benight NM, Hunt BG, Lester C, Waltz SE. Macrophage-mediated RON signaling supports breast cancer growth and progression through modulation of IL-35. Oncogene. 2022;41:321–33. doi: 10.1038/s41388-021-02091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cazes A, Childers BG, Esparza E, Lowy AM. The MST1R/RON Tyrosine Kinase in Cancer: Oncogenic Functions and Therapeutic Strategies. Cancers. 2022;14:2037. doi: 10.3390/cancers14082037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belgiovine C, Digifico E, Anfray C, Ummarino A, Andón FT. Targeting Tumor-Associated Macrophages in Anti-Cancer Therapies: Convincing the Traitors to Do the Right Thing. J Clin Med. 2020;9:3226. doi: 10.3390/jcm9103226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H, Liu L, Liu J, Dang P, Hu S, Yuan W, et al. Roles of tumor-associated macrophages in anti-PD-1/PD-L1 immunotherapy for solid cancers. Mol Cancer. 2023;22:58. doi: 10.1186/s12943-023-01725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RTK array raw data has been deposited at datadryad.org and are available from 10.5061/dryad.8sf7m0cvc. Additional analytical raw data are available upon reasonable request to the corresponding author.
All unique reagents generated in this study are available from the corresponding author with a completed Materials Transfer Agreement.
All analysis algorithm codes follow official tutorials. Relevant information is available upon reasonable request to the corresponding author.