Bugaut et al. report a comparative study of MAIT cells in six species spanning 110 million years of evolution. They identify a deeply conserved transcriptional program characterized by coexpression of type-1 and type-17 effector genes. These ancestral features are further conserved in human iNKT cells.
Abstract
Mucosal-associated invariant T (MAIT) cells harbor evolutionarily conserved TCRs, suggesting important functions. As human and mouse MAIT functional programs appear distinct, the evolutionarily conserved MAIT functional features remain unidentified. Using species-specific tetramers coupled to single-cell RNA sequencing, we characterized MAIT cell development in six species spanning 110 million years of evolution. Cross-species analyses revealed conserved transcriptional events underlying MAIT cell maturation, marked by ZBTB16 induction in all species. MAIT cells in human, sheep, cattle, and opossum acquired a shared type-1/17 transcriptional program, reflecting ancestral features. This program was also acquired by human iNKT cells, indicating common differentiation for innate-like T cells. Distinct type-1 and type-17 MAIT subsets developed in rodents, including pet mice and genetically diverse mouse strains. However, MAIT cells further matured in mouse intestines to acquire a remarkably conserved program characterized by concomitant expression of type-1, type-17, cytotoxicity, and tissue-repair genes. Altogether, the study provides a unifying view of the transcriptional features of innate-like T cells across evolution.
Introduction
Mucosal-associated invariant T (MAIT) cells are very abundant in humans (1–8% of T cells in blood and intestine, 20–40% in the liver) and are potentially involved in many pathologies (Godfrey et al., 2019; Legoux et al., 2020). MAIT cells are restricted by the major histocompatibility (MHC)-I–like protein, MHC-I–related protein, MR1, which is absent in birds and reptiles and appeared about 170 million years ago in the earliest ancestor of mammals. While classical MHC genes underwent diversifying selection during evolution, MR1 displays very limited polymorphism in humans and shows signs of purifying selection in mammals (Riegert et al., 1998; Huang et al., 2009; Boudinot et al., 2016), suggesting binding and presentation of a limited set of ligands. In particular, the lysine residue in position 43 of MR1, which forms a Schiff base with the canonical MAIT ligand 5-(2-oxoprop[or ethyn] ylideneamino)-6-d-ribitylaminouracil (5-OP/E-RU; Corbett et al., 2014), is maintained across mammalian species, suggesting a conserved ability to present 5-OP/E-RU. Other, much less potent agonist ligands as well as inhibitory compounds binding to MR1 have been described (Awad et al., 2023). Uncharacterized endogenous ligand(s) probably select MAIT cells in the thymus as a small number of mature MAIT cells develop in germ-free mice while no MAIT cells are present in MR1-deficient animals (Legoux, et al., 2019a).
In mice, cattle (Edmans et al., 2021), humans (Tilloy et al., 1999), non-human primates (Greene et al., 2017), and pigs (Xiao et al., 2019), MAIT cells are characterized by the expression of a semi-invariant T cell receptor (TCR) composed of a single TRAV1-TRAJ33 TCRα chain paired with β chains of limited diversity. TRAV1 and TRAJ33 TCR gene segments are highly conserved across species, suggesting important function(s) (Boudinot et al., 2016). Strikingly, the few species that have lost TRAV1, such as Lagomorphs, Carnivora, and Armadillo, have also lost functional MR1, suggesting that the main function of MR1 is to present antigens to TRAV1-expressing T cells (Boudinot et al., 2016). Thus, the presence of functional MR1 and TRAV1 genes in any given species suggests the existence of MR1-restricted T cells with specificity for 5-OP-RU.
MAIT cells develop in the murine thymus by interacting with MR1 expressed by CD4+CD8+ (DP) thymocytes (Seach et al., 2013). DP thymocytes also select another innate-like T cell subset, the invariant natural killer T (iNKT) cells, that recognize glycolipids presented by the MHC-I–like molecule, CD1d, through a semi-invariant (TRAV10-TRJA18) TCRα chain. For both iNKT and MAIT cells, selection by DP thymocytes leads to homotypic signaling lymphocyte activation molecule (SLAM) interactions transduced by the adaptor SLAM-associated protein (SAP; Griewank et al., 2007; Koay et al., 2019; Legoux et al., 2019b). Through this process, in humans and mice, thymocytes expressing an MR1- or CD1d-restricted TCR undergo intrathymic differentiation into effector cells marked by the expression of the master transcription factor Zinc finger and BTB domain containing 16 (ZBTB16, also referred to as PLZF; Savage et al., 2008; Koay et al., 2016). PLZF directly controls the expression of effector genes and inhibits genes of the naïve T cell program (Mao et al., 2016), and as such represents a lineage-defining transcription factor for innate-like T cells. PLZF expression induces a tissue residency program (Thomas et al., 2011). In species other than mice and humans, it is unclear whether MR1 restriction also instructs intrathymic expression of PLZF and acquisition of an innate-like T cell phenotype.
In addition to PLZF, MAIT cells express the transcription factors Tbet and RORγt that drive expression of type-1 and type-17 effector genes, respectively. Intriguingly, thymic maturation of MAIT cells results in distinct outcomes in humans and mice. Specifically, MAIT cells differentiate into a single population coexpressing Tbet and RORγt in humans (Leeansyah et al., 2015), while mouse MAIT cells undergo functional branching into either Tbet+ (MAIT1) or RORγt+ (MAIT17) cells with distinct transcriptional programs (Salou et al., 2018) and cytokine production abilities (Rahimpour et al., 2015). The degree of conservation, across species, of MAIT cell differentiation processes in the thymus is unclear. Since conserved genes are more likely to contribute important functions, defining a core transcriptional program for MR1-restricted T cells, i.e., conserved across species, would help decipher the mechanisms controlling MAIT cell maturation in the thymus.
Herein, we used single-cell RNA sequencing (scRNAseq) to characterize 5-OP-RU–specific thymocytes in six species spanning the mammalian phylogenetic tree from marsupials to humans. Thymocytes specific for peptide antigens and differentiating into naïve T cells were analyzed for comparison in mice and humans. The study identifies a deeply conserved, multifunctional transcriptional program shared by MAIT cells from all species. The evolutionarily conserved MAIT cell program is acquired in the thymus in all species except in rodents, in which it is acquired upon further differentiation in the mesenteric lymph nodes (LNs) and intestines.
Results
scRNAseq identifies immature and mature 5-OP-RU:MR1–specific T cell subsets in the thymus of six mammalian species
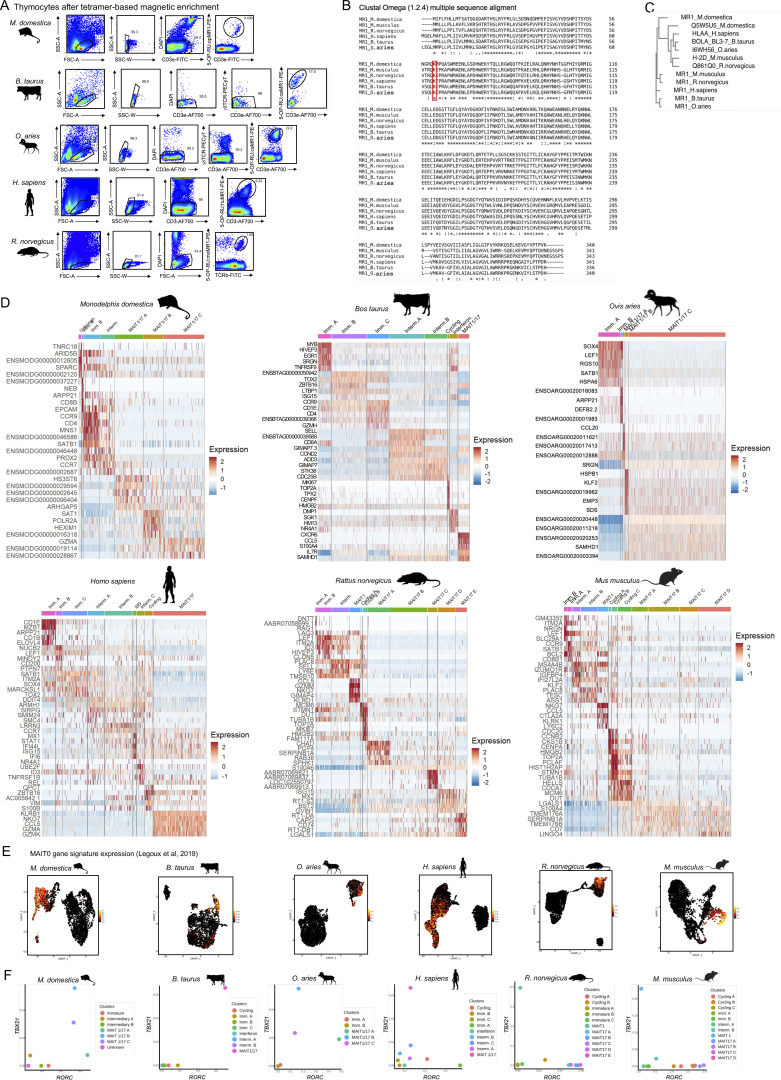
The coevolution of TRAV1 and MR1 in mammals strongly suggests a conserved presentation of 5-OP-RU to T cells. To study 5-OP-RU–specific T cells across evolution, we generated or obtained 5-OP-RU–loaded MR1 tetramers from various species (see Materials and methods). MR1 tetramers were used to identify 5-OP-RU–specific cells in the thymus of six mammalian species: Monodelphis domestica (opossum), Bos taurus (cattle), Ovis aries (sheep), Homo sapiens (human), Rattus norvegicus (rat), and Mus musculus (mouse; Fig. S1 A). Sheep 5-OP-RU–specific cells were identified using the cattle tetramer, owing to the high sequence identity between cattle and sheep MR1 (Fig. S1, B and C). For the same reason, the mouse tetramer was used to label rat 5-OP-RU–specific cells (Fig. S1, A–C). In all species tested, MR1 carries a conserved lysine in position 43 enabling formation of a Schiff base with 5-OP-RU (Corbett et al., 2014; Fig. S1 B).
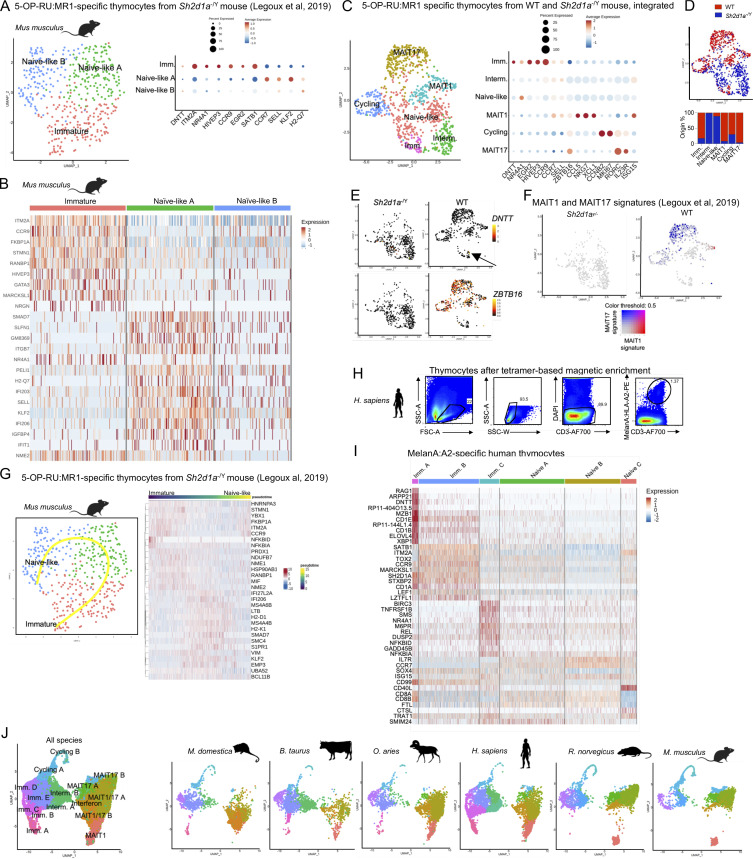
Figure S1.
scRNAseq identifies immature and mature MAIT cell subsets in the thymus of six mammalian species. (A) Flow cytometry identification of 5-OP-RU:MR1–specific thymocytes in the indicated species. Data representative of at least two independent experiments. (B) Clustal Omega alignment of MR1 protein sequences from the six species of the study. The lysine residue in position 43 is highlighted in the red box. (C) Clustal Omega phylogenetic tree showing homology in MR1 and MHC-I proteins from the indicated species. (D) Heatmap showing the top five most differentially expressed genes in scRNAseq data from 5-OP-RU–specific thymocytes from the indicated species. (E) Expression of the mouse MAIT0 gene signature defined in Legoux et al. (2019b), in 5-OP-RU:MR1–specific thymocytes from the indicated species. (F) Mean TBX21 and RORC expression in each cell cluster from the indicated species.
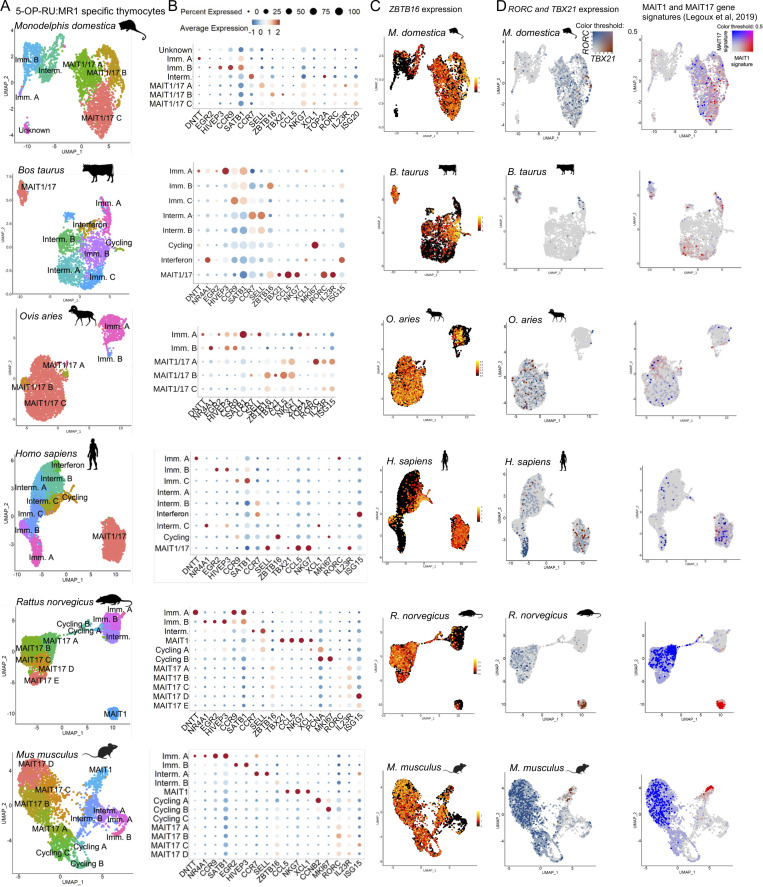
Only few antibodies are available against marsupial and bovid antigens. To characterize 5-OP-RU–specific thymocytes in the absence of antibodies and in a non-supervised fashion, 5-OP-RU:MR1 tetramer+ TCRβ+ (or CD3+) thymic cells were sorted by flow cytometry and analyzed by droplet-based scRNAseq. After quality controls and filtering steps (see Materials and methods), 1,814–6,023 cells with a median gene count of 1,602 genes per cell were retained for downstream analyses (Table S1). Cells from each dataset were displayed on a uniform manifold approximation and projection (UMAP; Hao et al., 2021; Fig. 1 A) at a resolution providing stable clusters, as determined using Clustree (Zappia and Oshlack, 2018). Analysis of the differentially expressed genes in each cluster (Fig. S1 D and Table S2, A–F) and expression of known marker genes (Fig. 1 B) were used to identify each cell population. Cell clusters with an expression of the immature thymocyte markers DNTT (coding for the DNA nucleotidyl exotransferase involved in TCR gene rearrangements), EGR2 (expressed early upon positive selection), or CCR9 (controlling thymocyte retention in the thymic cortex [Uehara et al., 2002; Kwan and Killeen, 2004]) were labeled as “immature.” RAG1 or RAG2 were detected in immature cells from opossum, human, and rat MAIT cells (Table S2). Immature cells expressed a gene signature of MAIT cell precursors (MAIT0) previously identified in mice (Legoux et al., 2019b; Table S3 A and Fig. S1 E), validating the assignment of cell clusters. Cell clusters with expression of CCR7 (controlling thymocyte migration to the thymic medulla [Kwan and Killeen, 2004; Ueno et al., 2004]) or SELL (Weinreich and Hogquist, 2008) were identified as “intermediary” (Fig. 1, A and B; Fig. S1 D; and Table S2, A–F), while cells expressing the proliferation markers MKI67, PCNA (proliferating cell nuclear antigen), or CCNB2 (Cyclin B2) were identified as “cycling.”
Figure 1.
scRNAseq identifies immature and mature MAIT cell subsets in the thymus of six mammalian species. (A) UMAP of 5-OP-RU:MR1–specific thymocytes isolated from the indicated species and analyzed by scRNAseq. The mouse data were generated previously (Legoux et al., 2019b). (B) Dotplot showing expression of selected genes by MAIT cells from the indicated UMAP clusters. (C) Expression of ZBTB16 in 5-OP-RU:MR1–specific thymocytes from the indicated species. (D) Left: TBX21 (in red) and RORC (in blue) expression in 5-OP-RU:MR1–specific thymocytes from the indicated species. Right: Expression of the mouse MAIT1 (in red) and MAIT17 (in blue) gene signatures defined in Legoux et al. (2019b) in 5-OP-RU:MR1–specific thymocytes from the indicated species.
Interestingly, ZBTB16 was expressed in 5-OP-RU–specific thymocytes from all species (Fig. 1 C). ZBTB16 expression was detected in immature and intermediary cell clusters, but not in the most immature, DNTT-expressing cells, indicating ZBTB16 induction during maturation. In mice, PLZF expression is followed by the acquisition of the lineage-defining transcription factors TBX21 (coding Tbet) or RORC (coding RORγt; Koay et al., 2016; Legoux et al., 2019b), marking MAIT1 and MAIT17 cell subsets, respectively. To identify these subsets, we used previously defined gene signatures for mature MAIT1 and MAIT17 mouse thymocytes (Legoux et al., 2019b; Table S3, B and C). The MAIT1 signature included key type-1 genes such as TBX21, IFNG, and NK receptor genes. The MAIT17 signature included RORC, IL23R, and CCR6, among other type-17–associated genes. Importantly, TBX21 and RORC and the associated MAIT1 and MAIT17 gene signatures were expressed in 5-OP-RU–specific thymocytes from all species (Fig. 1 D), suggesting that the MAIT transcriptional program is highly conserved in mammals.
Cells expressing TBX21 and the MAIT1 gene signature were labeled MAIT1, while cells expressing RORC and the MAIT17 gene signature were labeled MAIT17 (Fig. 1 D). Interestingly, cells coexpressing TBX21 and RORC, together with both MAIT1 and MAIT17 signatures, were identified in M. domestica, B. taurus, O. aries, and H. sapiens (Fig. 1 D and Fig. S1 F). Although human MAIT1/17 thymocytes clearly expressed RORC (Fig. 1 D) as expected (Koay et al., 2016), RORC expression was highest in the most immature precursors (Fig. S1 F). By contrast, TBX21 and RORC and the associated MAIT1 and MAIT17 signature genes were expressed in distinct, non-overlapping clusters in R. norvegicus and M. musculus (Fig. 1 D and Fig. S1 F). Thus, rodents appear as an exception among the studied mammals, harboring two distinct MAIT1 and MAIT17 subsets, whereas the four other therians hold a single RORC+TBX21+ population, hereafter referred to as MAIT1/17 cells.
Conserved transcriptional regulation during thymic differentiation of 5-OP-RU:MR1–specific T cells
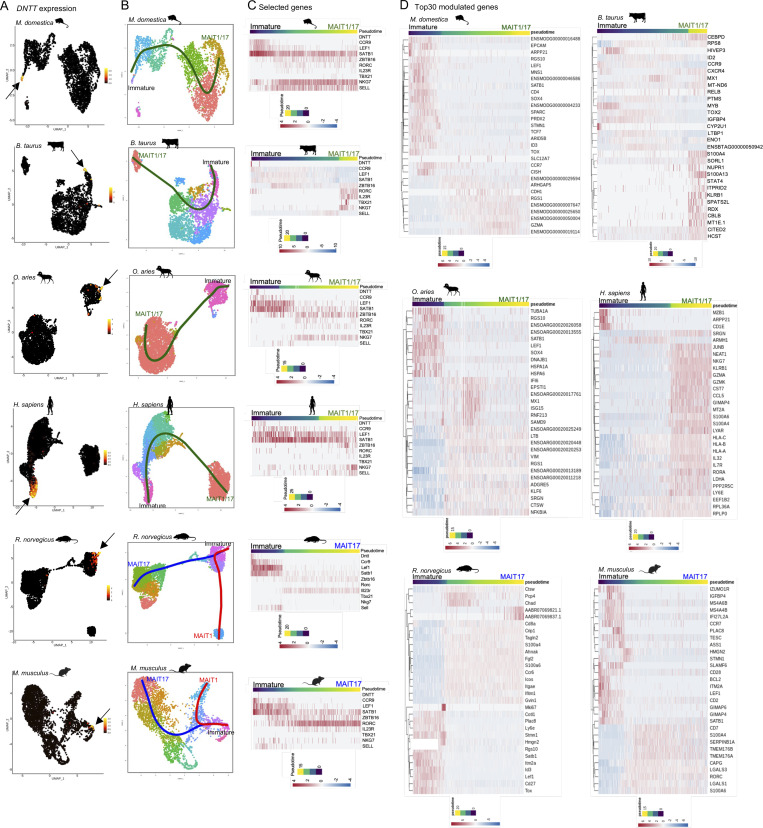
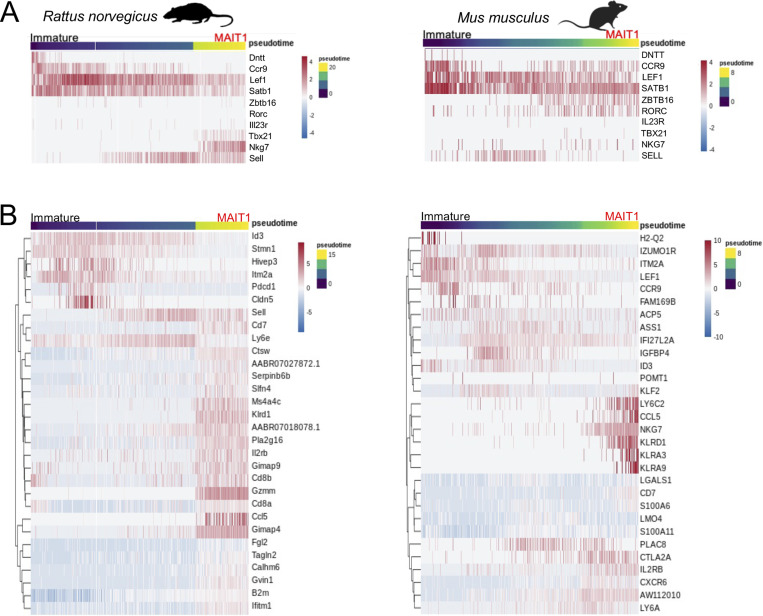
We next asked whether 5-OP-RU:MR1–specific thymocytes undergo similar transcriptional remodeling during thymic differentiation across species. Since each dataset contains both immature (DNTT+) and mature (TBX21/RORC+) cells, a succession of intermediate stages should also be present. To identify transcriptionally modulated genes along differentiation, pseudotemporal developmental trajectories were constructed with Slingshot (Street et al., 2018) using DNTT-expressing cells as a starting point as these cells represent the most immature precursors captured using our approach (Fig. 2, A and B). Trajectories were identified leading to the MAIT1/17 program (in opossum, cattle, sheep, and human) and to the MAIT1 and MAIT17 programs (in rat and mouse; Fig. 2 B). TradeSeq (Van den Berge et al., 2020) was then used downstream of Slingshot to identify all genes whose expression varies at any point along pseudotime. 958–2,258 genes were modulated along 5-OP-RU:MR1–specific T cell maturation depending on species (Table S4, A–H). To facilitate comparisons across species, expression of a selection of 10 evolutionarily conserved genes is displayed as a function of pseudotime (Fig. 2 C and Fig. S2 A). These genes were chosen based on a documented role in the development of conventional T cells or MAIT cells in human or mice. Key genes followed a conserved pattern of expression across species, with downregulation of DNTT, CCR9, and LEF1 and upregulation of ZBTB16 and RORC in MAIT1/17 and MAIT17 cells. Other genes followed various patterns of expression across species, suggesting variations in MAIT cell developmental processes from species to species. For instance, SELL was induced in mature MAIT cells in human and opossum, but not in cattle and sheep (Fig. 2 C). The most differentially expressed genes, for each species, are displayed in Fig. 2 D and Fig. S2 B, providing a global view of MAIT cell transcriptional maturation across mammals.
Figure 2.
Transcriptional maturation of 5-OP-RU:MR1–specific thymocytes across species. (A) Expression of the terminal deoxynucleotidyl transferase gene (DNTT) in 5-OP-RU:MR1–specific thymocytes from the indicated species. DNTT-expressing cells are indicated by an arrow. (B) Developmental trajectories of 5-OP-RU:MR1–specific thymocytes determined using Slingshot. In all species, DNTT-expressing cells were used as the starting point. (C) Heatmap showing expression of selected genes during pseudotimed MAIT cell differentiation in the indicated species. (D) Heatmap showing the 30 most differentially expressed genes during 5-OP-RU:MR1–specific T cell maturation in the indicated species. For rodents, only genes modulated during MAIT17 differentiation are displayed. Results for trajectories leading to MAIT1 differentiation are shown in Fig. S2.
Figure S2.
Developmental trajectory of MAIT1 thymocytes. (A) Heatmap showing expression of selected genes during pseudotimed MAIT1 cell differentiation in the indicated species. (B) Heatmap showing the 30 most differentially expressed genes during MAIT1 cell maturation in the indicated species.
Genes that are conserved across species are more likely to play important roles in a given biological process. To identify conserved genes that are regulated during MAIT cell development, we selected genes with one ortholog in each species using the orthology matrix (OMA) (Altenhoff et al., 2021) and generated a list of 54 genes that are both conserved and modulated along development of 5-OP-RU–specific thymocytes in all six species tested (Table S5). Of note, the selection also eliminated poorly annotated genes, which may be involved in MAIT cell development but lack orthologous annotation. This unsupervised analysis retrieved ZBTB16, as expected, but also HIVEP3, which was recently uncovered as a regulator of innate-like T cell maturation (Harsha Krovi et al., 2020). Genes associated with proliferation (TOP2A), type-1 (XCL1), and type-17 differentiation (IL23R) programs were also found modulated in all species, consistent with a conserved differentiation process for MAIT cells across species.
Transcriptional regulation during thymic development of mainstream antigen-specific T cells
Some of the genes modulated during thymic development of MAIT thymocytes, such as CCR9, may also be modulated during the thymic maturation of conventional T cells. To identify such genes, unsupervised analyses of transcriptional changes occurring during thymic development of mainstream antigen-specific thymocytes are needed. We previously reported that 5-OP-RU–specific thymocytes differentiate into naïve-like, mainstream T cells in the absence of functional SLAM-SAP signaling during positive selection (Legoux et al., 2019b). To identify genes modulated during mainstream T cell development in mice, we reanalyzed our scRNAseq data from 5-OP-RU–specific thymocytes isolated from the thymus of Sh2d1a (SAP)−/Y mice. As previously, cells were clustered on a UMAP (Seurat4) with a clustering resolution determined using Clustree (Zappia and Oshlack, 2018; Table S1). UMAP clustering identified subsets defined by the differential expression of ITM2A, CCR9, and EGR2 (immature cells) and CCR7, SELL, and KLF2 (naïve-like cells; Fig. S3, A and B). Next, 5-OP-RU:MR1–specific thymocytes from WT and Sh2d1a−/Y mice were integrated for direct subset comparisons. Integration revealed distinct populations according to mouse genotype (Fig. S3, C and D). In particular, Sh2d1a−/Y cells failed to express ZBTB16 (Fig. S3 E) and MAIT1 or MAIT17 gene signatures (Fig. S3 F), consistent with a mainstream T cell development. To define transcriptional changes occurring during thymic development of 5-OP-RU:MR1–specific mainstream-like T cells, a developmental trajectory starting from immature cells and leading to mature, naïve-like cells was inferred using Slingshot (Fig. S3 G). The 412 genes identified as modulated along development characterize the maturation of mainstream T cells in mice (Table S4 I).
Figure S3.
scRNAseq-based reconstruction of the development of conventional antigen-specific T cells in the mouse and human thymus. (A) Left: UMAP of an scRNAseq analysis of 5-OP-RU:MR1–specific thymocytes isolated from the thymus of Sh2d1a−/Y mice. The raw data were generated previously (Legoux et al., 2019b). Right: Dotplot showing expression of selected genes in the indicated UMAP clusters. (B) Heatmap showing the most differentially expressed genes in the indicated clusters by scRNAseq analysis of 5-OP-RU–specific thymocytes from Sh2d1a−/Y mice. (C) Left: Integrated UMAP of 5-OP-RU:MR1–specific thymocytes from WT and Sh2d1a−/Y mice. The WT dataset was down-sampled to obtain comparable numbers of cells as in the Sh2d1a−/Y sample (488 WT and 435 Sh2d1a−/Y cells). Right: Dotplot showing expression of selected genes in the indicated UMAP clusters. (D) UMAP showing integrated data as in C, with cells colored according to WT (blue) or Sh2d1a−/Y (red) origin. (E) Expression of DNTT (top) and ZBTB16 (bottom) in 5-OP-RU:MR1–specific thymocytes from the indicated mice. (F) Expression of MAIT1 and MAIT17 gene signatures in 5-OP-RU:MR1–specific thymocytes from the indicated mice. (G) Left: Developmental trajectory of 5-OP-RU:MR1–specific thymocytes in Sh2d1a−/Y mice determined using Slingshot. The raw data were generated previously (Legoux et al., 2019b). DNTT-expressing cells were used as the starting point. Right: Heatmap showing the 30 most differentially expressed genes during 5-OP-RU:MR1–specific T cell maturation in Sh2d1a−/Y mice. (H) MelanA:A2 tetramer staining of human HLA-A2+ thymocytes after tetramer-based enrichment. Representative of three independent experiments. (I) Heatmap of differentially expressed genes in the indicated clusters by scRNAseq analysis of MelanA:A2-specific thymocytes. (J) UMAP showing the distribution of 5-OP-RU:MR1–specific thymocytes by species of origin, after integration. Integration was performed in Seurat using expression of genes with a documented orthology in all six species.
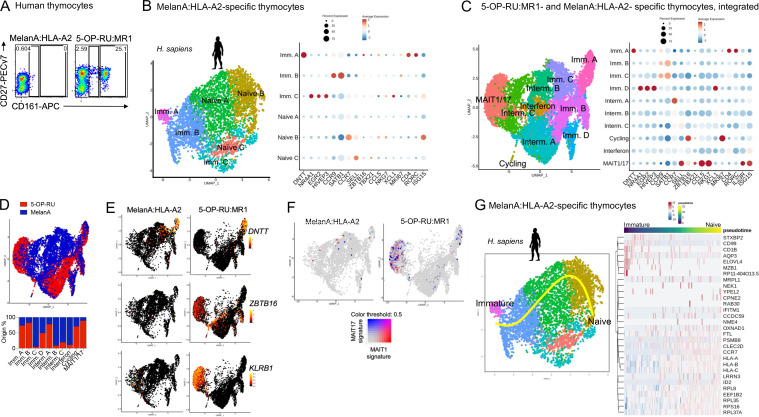
To assess the transcriptional maturation along development of human conventional T cells, we next studied thymocytes specific for MelanA, a melanocyte differentiation peptidic antigen presented by the classical MHC-I molecule HLA-A2. Consistent with a mainstream T cell lineage, MelanA:A2-specific thymocytes identified in human thymus using a MelanA:A2 tetramer (Fig. S3 H) lacked expression of the innate-like T cell marker CD161 (Fig. 3 A). To define transcriptional modulation during human mainstream T cell development, MelanA:A2-specific thymocytes were FACS-sorted and analyzed by scRNAseq. 4,685 MelanA:A2-specific cells with a median gene count of 1,195 genes per cell passed quality controls and were clustered using Seurat4, as performed previously (Fig. 3 B and Fig. S3 I; and Table S1). Cells expressing the immature thymocyte markers DNTT, EGR2, or CCR9 were labeled as immature, while cells expressing CCR7, SELL, or HLA-A were considered mature naïve T cells (Fig. 3 B and Table S2 G).
Figure 3.
Lack of ZBTB16 induction during thymic maturation of conventional antigen-specific thymocytes. (A) CD27 and CD161 expression by MelanA:HLA-A2– and 5-OP-RU:MR1–specific human thymocytes identified by flow cytometry using tetramers. (B) Left: UMAP of an scRNAseq analysis of MelanA:HLA-A2–specific cells isolated from the thymus of a HLA-A2+ donor. Right: Dotplot showing expression of selected genes in the indicated UMAP clusters. (C) Left: Integrated UMAP of 5-OP-RU:MR1– and MelanA:HLA-A2–specific human thymocytes. Right: Dotplot showing expression of selected genes in the indicated UMAP clusters. (D) UMAP showing integrated data as in C, with cells colored according to 5-OP-RU (red) or MelanA (blue) specificity. (E) Expression of DNTT, ZBTB16, and KLRB1 (coding CD161) in thymocytes from the indicated datasets. (F) Expression of MAIT1 (in red) and MAIT17 (in blue) gene signatures in thymocytes with the indicated specificity. (G) Left: Developmental trajectory of human MelanA:HLA-A2–specific thymocytes determined using Slingshot. DNTT-expressing cells were used as the starting point. Right: Heatmap showing the 30 most differentially expressed genes during MelanA:HLA-A2–specific T cell maturation.
Data from MelanA:A2-specific thymocytes were integrated together with human 5-OP-RU:MR1–specific thymocytes for side-by-side comparison (Fig. 3 C). Immature, intermediary, and mature cell subsets were identified using the same markers as for individual datasets (Fig. 3 C and Table S6 A). Interestingly, the most immature cell cluster (Imm. A) was composed of mixed MelanA- and 5-OP-RU–specific thymocytes, suggesting limited transcriptional differences in the earliest stages of MAIT cell development as compared to conventional T cell development (Fig. 3 D). The intermediary B-C clusters were mainly composed of MelanA-specific cells, while the MAIT1/17 cluster was composed of 5-OP-RU–specific cells, as expected (Fig. 3 D). MelanA-specific thymocytes lacked expression of ZBTB16, KLRB1 (coding CD161; Fig. 3 E), or MAIT1/MAIT17 gene signatures (Fig. 3 F), consistent with a mainstream lineage. To define genes modulated along conventional T cell development, pseudotemporal ordering of MelanA-specific thymocytes was performed using DNTT-expressing cells as the starting point and the naïve B cluster as the end point (Fig. 3 G). The 672 identified genes define the transcriptional maturation of conventional CD8+ T cells in the human thymus (Table S4 J). Genes modulated during development of mainstream antigen-specific T cells in both mice (based on the SAP−/Y dataset) and human (based on the MelanA-specific dataset) are listed in Table S4 K. Genes known to be involved in conventional T cell development were retrieved (such as CCR9, KLF2, or ID2) together with other genes whose functions in T cell development remain to be investigated.
Conserved transcriptional changes unique to MAIT cell differentiation
To identify genes involved in the differentiation of 5-OP-RU:MR1–specific T cells, and not conventional T cells, we selected genes modulated along the development of MAIT cells in all six species (Table S5) and excluded from this list all the genes modulated during MelanA-specific T cell development or during maturation of 5-OP-RU–specific T cells in Sh2d1a−/Y mice (Table S4, I and J). The resulting list contains 31 genes that define a conserved transcriptional program for developing MAIT cells (Table 1).
Table 1.
Genes modulated during thymic maturation of MAIT cells, but not conventional T cells, across the six studied species
| Downregulated ↘ |
Upregulated ↗ |
Up- then downregulated ↗ ↘ |
|---|---|---|
| EZH2 | ANXA2 | CASP8AP2 |
| MYB | CD40LG | CDC45 |
| CITED2 | CLSPN | |
| CXCR4 | DBF4 | |
| DUSP1 | DCTPP1 | |
| FOSB | EIF5A | |
| GPR183 | HMMR | |
| IL23R | MCM4 | |
| MYC | MELK | |
| RPLP0 | MTHFD1 | |
| S100A10 | NOLC1 | |
| XCL1 | PRDX4 | |
| ZBTB16 | RFC3 | |
| TOP2A | ||
| UHRF1 | ||
| WDHD1 |
Transcription factors are indicated in bold.
In all species, MAIT cell development involved downregulation of EZH2, the negative regulator of ZBTB16 (Vasanthakumar et al., 2017). Genes consistently induced during MAIT cell development included ZBTB16, but also genes associated with TCR signaling (ANXA2 [Dubois et al., 1995; Bharadwaj et al., 2021], FOSB [Jain et al., 1993], CD40LG [Stark et al., 2013], and DUSP1 [Zhang et al., 2009; Stanford et al., 2012]) or with type-1 (XCL1) and type-17 (IL23R) effector programs. Interestingly, MAIT cell maturation was also accompanied by expression of GPR183, whose product modulates homing to the gut (Emgård et al., 2018). Finally, a number of genes associated with cell cycle and DNA replication (such as CASP8A2, CDC45, CLSPN, DBF4, DCTPP1, HMMR, MCM4, MELK, MTHFD1, RFC3, TOP2A, UHRF1, and WDHD1) were consistently up- and then downmodulated during MAIT cell development, suggesting that intrathymic proliferation is a hallmark of MAIT cell maturation.
MAIT cell branched development is maintained across mouse genetic backgrounds and health status
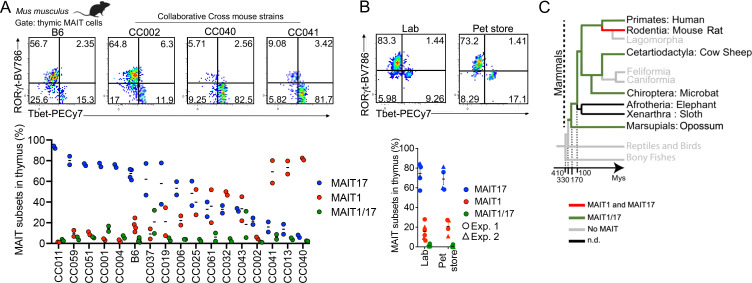
Having identified transcriptional features consistently modulated during MAIT cell development across species, we further explored the functional branching of MAIT cells in distinct mature subsets, a characteristic that we uniquely observed in MAIT cells from rat and mouse. The B6-MAITCAST mice used in the study harbor only a fraction of the genetic diversity of the M. musculus genome and were raised in specific pathogen–free (SPF) conditions and therefore cannot reflect the potential variability of MAIT cell development. To determine whether MAIT cells develop into distinct MAIT1 and MAIT17 subsets in genetically diverse mouse strains, we took advantage of the collaborative cross (CC; Srivastava et al., 2017), which represents a collection of inbred strains with high genetic diversity. CC strains harbor recombinant genetic backgrounds from eight founder strains, including three M. musculus subspecies (M. m. castaneus CAST/EiJ, M. m. musculus PWK/PhJ, and M. m. domesticus WSB/EiJ), thus capturing an estimated 90% of the total genetic diversity of the mouse species. The CC strains analyzed in this study were housed in the same animal facility, thus excluding potential housing-dependent factors. Expression of Tbet and RORγt was assessed by flow cytometry following MR1 tetramer-based magnetic enrichment of MAIT thymocytes from 16 CC strains. MAIT cells differentiated preferentially into RORγt-expressing MAIT17 cells in the thymus of B6 mice, as expected (Fig. 4 A). However, MAIT cell development in CC strains showed strong strain-to-strain variability, with preferential differentiation into Tbet-expressing MAIT1 cells in the thymus of several strains (Fig. 4 A). Thus, MAIT cell differentiation patterns can be very different from previously appreciated in B6 mice. Importantly, across the tested strains, MAIT cells always differentiated into either Tbet+ or RORγt+ subsets, with few to no detectable MAIT1/17 cells in the thymus (Fig. 4 A). Thus, MAIT cell maturation into mutually exclusive MAIT1 and MAIT17 subsets is independent of the mouse genetic background.
Figure 4.
Branched MAIT1 or MAIT17 differentiation is maintained across mouse genetics and health status. (A) Top: Representative flow cytometry showing RORγt and Tbet expression in MAIT cells (defined as TCRβ+ 5-OP-RU:MR1 tetramer+ CD24− CD44+) enriched from the thymus of the indicated mouse strains. Bottom: Percentages of mature (CD44+) MAIT cells expressing Tbet (MAIT1), RORγt (MAIT17), or both (MAIT1/17) in the thymus of the indicated strains. (B) Top: Representative flow cytometry showing RORγt and Tbet expression in MAIT cells enriched from the thymus of SPF (lab) or dirty (pet store) mice. Bottom: Summary representation of the results from two independent experiments. (C) Schematic representation of thymic MAIT cell differentiation pattern along mammalian evolution. Branches in which no functional MR1 gene was found are shown in gray (Boudinot et al., 2016). MAIT1/17 differentiation in bat MAIT cells was reported elsewhere (Leeansyah et al., 2020). n.d.: not determined.
To explore the possible role of SPF rearing in MAIT cell maturation, MAIT cells were phenotyped by flow cytometry in the thymus of pet store mice, which have a history of exposure to pathogens (Beura et al., 2016). Mice from the local pet store tested positive for the mouse hepatitis virus, for pinworm (Aspiculuris tetraptera), and for the protozoan pathogens Spironucleus muris and Giardia muris. Nevertheless, MAIT cells matured into either MAIT1 or MAIT17 cells in the thymus of these mice in proportions similar to those found in SPF B6 mice (Fig. 4 B). Thus, thymic development into distinct MAIT subsets in mice occurs independently of previous pathogen exposure.
Altogether, the generation of distinct MAIT1 and MAIT17 subsets in the mouse thymus appears independent of genetic backgrounds and rearing conditions and contrasts with the development of a unique MAIT subset coexpressing TBX21 and RORC in the opossum, cattle, sheep, and human (Fig. 1 D). Coexpression of Tbet and RORγt was reported in MAIT cells from Pteropus alecto (the black fruit bat; Leeansyah et al., 2020). Thus, the MAIT1/17 differentiation program probably appeared in the common ancestor of mammals, while branched development into either MAIT1 or MAIT17 represents a recent innovation in rodents (Fig. 4 C).
Defining a conserved transcriptional program for MAIT cells
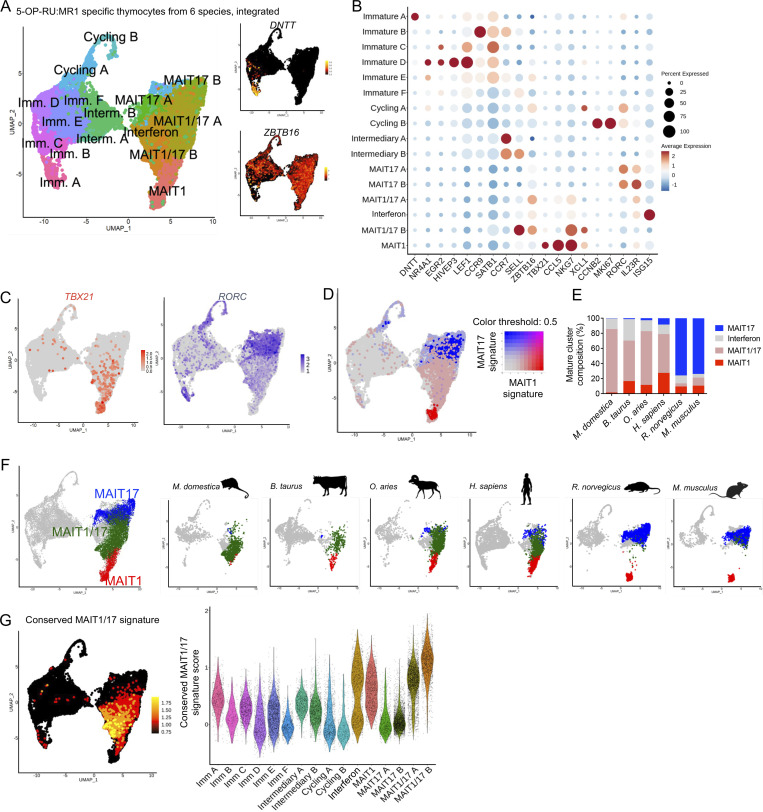
Since coexpression of TBX21 and RORC likely represents an ancestral feature of 5-OP-RU–specific T cells, we next sought to better characterize the associated transcriptional program along evolution. To directly compare gene expression in developing MAIT cells across species, we focused on the orthologous genes present in all six species and identified them using the OMA orthology inference algorithm (Roth et al., 2008; Table S7). scRNAseq data from MAIT thymocytes from the six mammalian species were filtered (Table S8) and integrated on a single UMAP based on orthologous gene expression. Integration was performed pairwise in Seurat, starting with M. musculus and R. norvegicus, and merging additional samples in the order of the phylogenetic tree (Materials and methods). Upon integration, MAIT cells clustered according to gene expression rather than species of origin (Fig. S3 J). Clusters corresponding to immature (with expression of DNTT, EGR2, or CCR9), intermediary (CCR7, SELL), or cycling (MKI67) MAIT cells were identified (Fig 5, A and B; and Table S9). Interestingly, an immature subset (Imm. C) was mostly composed of cells of human origin (Fig. S3 J). Mature MAIT cells clustered apart from immature and intermediary cells and followed a gradient according to the expression of TBX21 and RORC, which also matched the expression of the MAIT1 and MAIT17 gene signatures (Fig. 5, C and D). MAIT1/17 cells expressed genes associated with the MAIT1 program (such as NKG7) together with genes associated with the MAIT17 program (such as IL18R1 and IKZF3 coding for IKAROS) and clustered in between MAIT1 and MAIT17 cells from rodents (Fig. 5 D and Table S9). Mature MAIT cells from rat and mouse were mostly identified as MAIT17 cells, while mature MAIT cells from opossum, cattle, sheep, and human also expressed TBX21 and were mostly identified as MAIT1/17 cells (Fig. 5, E and F).
Figure 5.
Defining an evolutionarily conserved gene signature for the thymic MAIT1/17 program. (A) Left: UMAP showing integrated scRNAseq analyses of 5-OP-RU:MR1–specific thymocytes from six species. Individual datasets from opossum, cattle, sheep, human, rat, and mouse are described in Fig. 1. Right: Expression of DNTT and ZBTB16 in 5-OP-RU:MR1–specific thymocytes after cross-species integration. (B) Dotplot showing expression of selected genes in the indicated UMAP clusters. (C) Expression of TBX21 and RORC in 5-OP-RU:MR1–specific thymocytes after cross-species integration. (D) Expression of MAIT1 (in red) and MAIT17 (in blue) gene signatures in 5-OP-RU:MR1–specific thymocytes after cross-species integration. (E) Distribution of mature cells from the indicated datasets in the UMAP clusters identified as MAIT1, MAIT17, MAIT1/17, or interferon. (F) UMAP showing MAIT1 (red), MAIT17 (blue), and MAIT1/17 (green) clusters in the indicated individual datasets, after cross-species integration. (G) Left: Expression of the conserved MAIT1/17 gene signature in 5-OP-RU:MR1–specific thymocytes after cross-species integration. The signature is defined as genes overexpressed in the MAIT1/17 cluster as compared with MAIT1 or MAIT17 clusters. Right: Violin plot showing expression of the conserved MAIT1/17 gene signature in the indicated 5-OP-RU:MR1–specific cells after cross-species integration.
To define a conserved signature for MAIT1/17 cells, we calculated the overexpressed genes in MAIT1/17 clusters as compared with MAIT1 or MAIT17 clusters (Fig. 5 G). The 16 identified genes (Table 2) are conserved across species and thus represent an evolutionarily conserved signature for thymic MAIT1/17 cells. The signature contains genes associated with T cell activation (BATF3 [Ataide et al., 2020], DUSP1 [Stanford et al., 2012]) and TCR signaling (genes coding for the AP-1 complex FOS and JUN [Jain et al., 1993; Yukawa et al., 2020], but also PDE4D [Peter et al., 2007]), indicating strong TCR stimulation in thymic MAIT1/17 cells across species. In addition, overexpression of GZMM (coding granzyme M) and NKG7 (coding the NK cell granule protein 7) in MAIT1/17 cells suggests a cytotoxicity potential.
Table 2.
Genes conserved in the six species and upregulated (Pval_adj <0.05) in MAIT1/17 cells as compared to MAIT1 and MAIT17 cells
| Gene | Pval_adj |
|---|---|
| ATF3 | 1.20E-05 |
| B2M | 2.69E-07 |
| BATF3 | 2.99E-13 |
| DUSP1 | 6.98E-07 |
| FOS | 5.64E-22 |
| GZMM | 1.63E-29 |
| JUN | 1.72E-05 |
| KLF6 | 0.0001407 |
| LTB | 0.00399221 |
| MACF1 | 4.34E-05 |
| NKG7 | 6.40E-14 |
| NUCB2 | 2.19E-24 |
| PDE4D | 0.00068253 |
| RPS25 | 0.00104954 |
| SH2D1A | 1.80E-08 |
| TNFAIP3 | 0.00088657 |
Transcription factors are indicated in bold.
Human MAIT and iNKT cells acquire an identical 1/17 transcriptional program in the thymus
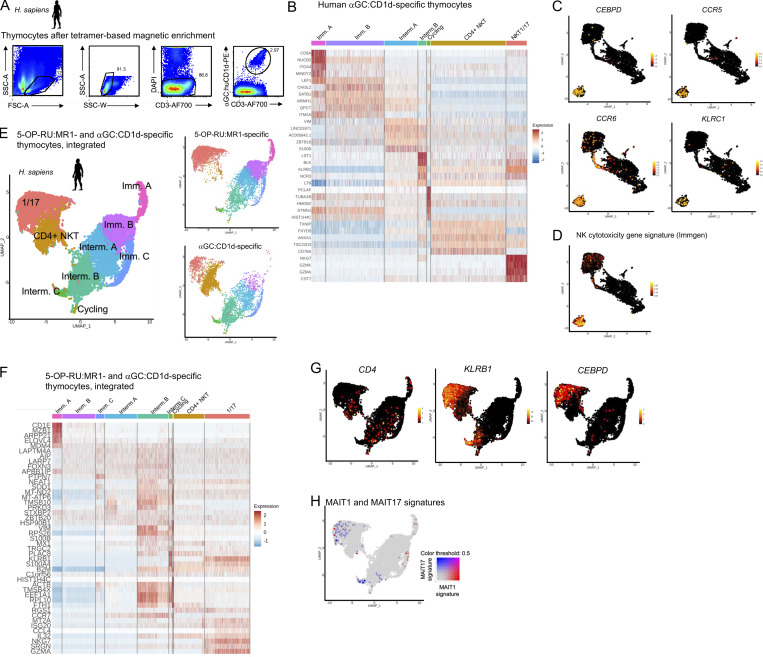
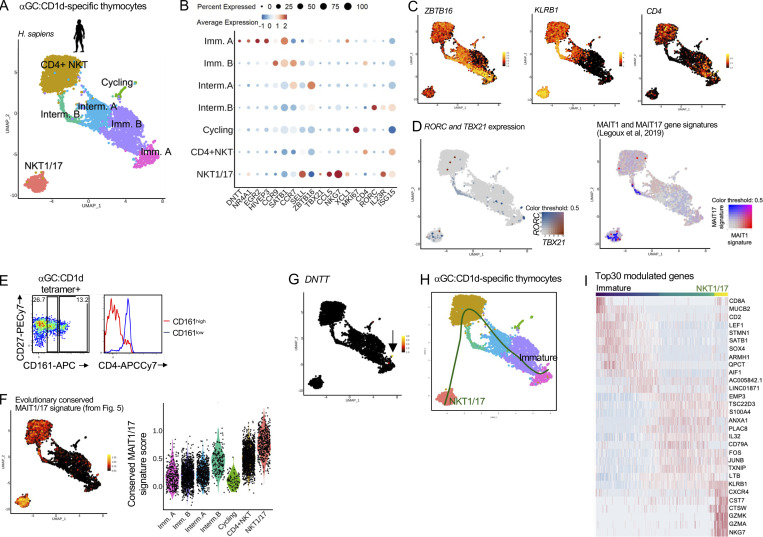
To determine whether the MAIT cell transcriptional program is shared across innate-like T cell populations, we extended our analyses to iNKT cells, which recognize α-galactosylceramide (αGC) presented by CD1d. Human αGC:CD1d-specific thymocytes were sorted by flow cytometry (Fig. S4 A) and analyzed by scRNAseq. 4,359 cells were retained after quality control and filtering steps and were clustered on a UMAP (Fig. 6 A). Previously described markers and differentially expressed genes (Table S2 H and Fig. S4 B) were used to label each cluster (Fig. 6, A and B). Immature cells (expressing DNTT or CCR9) were identified, as well as a few cycling (MKI67+) cells and mature cells (expressing ZBTB16 but not CCR9). Two distinct subsets of iNKT cells were described in human peripheral blood: a CD4neg, type-1-polarized subset and a CD4pos subset with IL-4 and IL-13 production capacity (Gumperz et al., 2002; Lee et al., 2002). Similarly, in the thymus, two populations could be identified as mature: the first one lacked CD4 but expressed high levels of KLRB1 (CD161) together with TBX21, RORC, and the MAIT1 and MAIT17 gene signatures (Fig. 6, C and D) and thus was labeled “NKT1/17.” The second one expressed low levels of KLRB1, TBX21, and RORC but high levels of CD4 and thus was labeled “CD4+ NKT.” Flow cytometry confirmed the presence of mature (CD27+) CD161low and CD161high αGC:CD1d-specific thymocytes (Fig. 6 E). CD161low cells expressed CD4, while CD161high did not (Fig. 6 E), confirming the scRNAseq results. A description of iNKT subset-specific genes is provided in Table S2 H. By contrast with CD4+ iNKT cells, iNKT1/17 cells expressed CEBPD, suggesting the ability to cross the inflamed endothelium (Lee et al., 2018), as well as CCR5 and CCR6, suggesting the ability to migrate to inflamed tissues (Fig. S4 C). iNKT1/17 cells also expressed genes encoding NK receptors such as KLRC1, together with a strong cytotoxicity gene signature (Immgen signature defined in Table S3 F; Fig. S4 D). Interestingly, iNKT1/17 thymocytes also overexpressed the evolutionarily conserved MAIT1/17 gene signature defined previously (Fig. 6 F and Table 2), suggesting a shared transcriptional program between MAIT and a subset of iNKT cells in humans.
Figure S4.
Human MAIT and iNKT cells acquire an identical 1/17 transcriptional program upon thymic development. (A) αGC:CD1d tetramer staining of human thymocytes after tetramer-based enrichment. Representative of two independent experiments. (B) Heatmap showing the top five most differentially expressed genes in the indicated clusters by scRNAseq analysis of human αGC:CD1d-specific thymocytes. (C) Expression of CEBPD, CCR5, CCR6, and KLRC1 in human αGC:CD1d-specific thymocytes. (D) Expression of an NK cytotoxicity gene signature (Immgen; defined in Table S3) in αGC:CD1d-specific thymocytes. (E) Left: UMAP showing integrated scRNAseq analyses of 5-OP-RU:MR1– and αGC:CD1d-specific human thymocytes. Right: Data split by cell origin. (F) Heatmap showing the top five most differentially expressed genes in the indicated clusters by scRNAseq analysis of integrated 5-OP-RU:MR1– and αGC:CD1d-specific thymocytes. (G) Expression of CD4, KLRB1, and CEBPD in 5-OP-RU:MR1– and αGC:CD1d-specific thymocytes after integration. (H) Expression of MAIT1 (in red) and MAIT17 (in blue) gene signatures in 5-OP-RU:MR1– and αGC:CD1d-specific thymocytes after integration.
Figure 6.
Human MAIT and iNKT cells acquire an identical 1/17 transcriptional program upon thymic development. (A) UMAP of human αGC:CD1d-specific thymocytes analyzed by scRNAseq. (B) Dotplot showing expression of selected genes by cells from the indicated UMAP clusters. (C) Expression of ZBTB16, KLRB1 (coding CD161), and CD4 in human αGC:CD1d-specific thymocytes. (D) Left: TBX21 (in red) and RORC (in blue) expression in αGC:CD1d-specific thymocytes. Right: Expression of the mouse MAIT1 (in red) and MAIT17 (in blue) gene signatures in αGC:CD1d-specific thymocytes. (E) CD27 and CD161 expression by αGC:CD1d-specific human thymocytes by flow cytometry. CD4 expression is shown in CD161high (in red) and CD161low (in blue) αGC:CD1d tetramer+ thymocytes. Data are representative of two independent experiments with different donors. (F) Expression of the conserved MAIT1/17 gene signature (defined in Fig. 5) in αGC:CD1d-specific thymocytes. (G) Expression of DNTT in αGC:CD1d-specific thymocytes. (H) Developmental trajectory of αGC:CD1d-specific thymocytes determined using Slingshot. DNTT-expressing cells were used as a starting point. (I) Heatmap showing the 30 most differentially expressed genes during iNKT cell intra-thymic maturation.
To directly compare the transcriptional programs acquired in the thymus by human iNKT and MAIT cells, the two scRNAseq datasets were integrated and analyzed jointly on a UMAP (Fig. S4 E). Cell clusters were labeled based on the expression of marker genes as described previously (Fig. S5 F and Table S6 B). iNKT1/17 thymocytes clustered together with mature MAIT thymocytes in a cluster marked by the coexpression of KLRB1, CEBPD, and MAIT1 and MAIT17 gene signatures (Fig. S4, G and H). Thus, thymic maturation leads to the acquisition of a shared 1/17 program in 5-OP-RU:MR1– and in a subset of αGC:CD1d-specific thymocytes.
Figure S5.
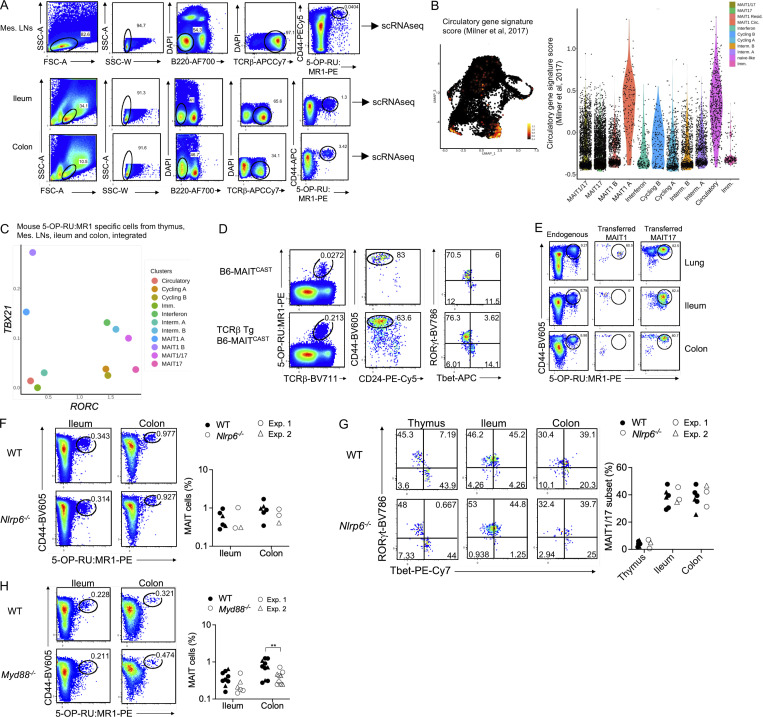
Requirements for the development of MAIT1/17 cells in the mouse intestine. (A) Gating strategy used for flow cytometry sorting of MAIT cells from the mesenteric LNs (Mes. LNs), ileum, and colon of B6-MAITCAST mice prior to scRNAseq analysis. MAIT cells were identified as live B220− TCRβ+ CD44+ MR1 tetramer+ cells. (B) Left: UMAP representation of the expression of a gene signature defined by Milner et al. (2017) for circulatory T lymphocytes. Right: Violin plot showing expression of the circulatory gene signature (described in Table S3) in the indicated clusters. (C) Mean TBX21 and RORC expression in each cell cluster from the integrated mouse datasets (Fig. 7 B). (D) Normal MAIT cell development in the B6-MAITCAST × TCRβ transgenic mouse. Left: Flow cytometry showing 5-OP-RU:MR1-tetramer staining in the thymus of the indicated mice. Center: CD24 and CD44 expression in tetramer+ thymocytes from the indicated strain. Right: Tbet and RORγt expression in CD44+ MAIT thymocytes from the indicated mice. Dotplots are representative of at least four independent experiments. (E) Flow cytometry showing 5-OP-RU:MR1-tetramer staining in the indicated tissues of B6-MAITCAST × TCRβ transgenic mouse (endogenous) or 8 wk after MAIT adoptive transfers into RAG2−/− recipients. The data are representative of two independent experiments. (F) Left: 5-OP-RU:MR1-tetramer staining in the ileum and colon of WT and Nlrp6−/− mice. Right: MAIT cell frequencies in the indicated tissues. Data pooled from two independent experiments. (G) Left: Representative Tbet and RORγt staining in MAIT (CD44+ tetramer+) cells from the indicated tissues. Right: Tbet and RORγt expression in 5-OP-RU:MR1 tetramer+ cells from the indicated tissues of B6 and Nlrp6−/− mice. Results pooled from two independent experiments. (H) Left: 5-OP-RU:MR1-tetramer staining in the ileum and colon of WT and Myd88−/− mice. Right: MAIT cell frequencies in the indicated tissues. Data were pooled from two independent experiments.
The relative proportions of the CD4+ and CD161+ iNKT subsets vary with age: CD4+ iNKT cells predominate in neonatal thymus and blood, while CD161+ iNKT cells increase over time and make up the majority of the iNKT cell population in adults (Berzins et al., 2005), raising the possibility that CD161+ iNKT cells arise from the CD4+ subset. To explore this possibility, pseudo-temporal ordering of αGC:CD1d-specific thymocytes was performed using Slingshot, with DNTT-expressing cells as starting point (Fig. 6, G and H). The proposed developmental trajectory leads to CD4+ iNKT thymocytes prior to iNKT1/17, indeed suggesting a possible precursor-product relationship between these two populations. The genes identified as differentially expressed during human iNKT cell development are shown in Fig. 6 I and Table S4 L. Together, CD4+ iNKT cells may give rise to mature CD161+ iNKT cells expressing a 1/17 program shared with MAIT cells.
MAIT cells acquire an evolutionarily conserved transcriptional program in the mouse intestine
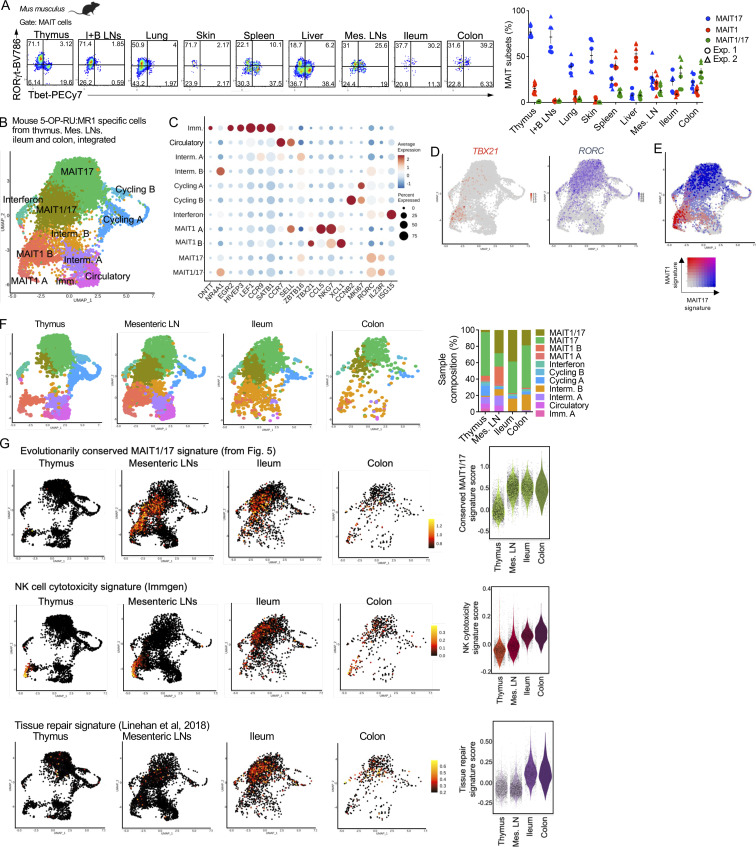
In mice, MAIT cells co-expressing Tbet and RORγt were described in lungs upon Salmonella typhimurium or Legionella longbeachae infections (Chen et al., 2017; Wang et al., 2018), indicating that the functional program of MAIT cells is malleable and varies in response to pathogen challenge and tissue cues. To define MAIT phenotypes in peripheral tissues at steady-state, we measured Tbet and RORγt expression in mouse MAIT cells from lungs, skin, spleen, LNs, ileum, and colon by flow cytometry. While MAIT cells expressed either RORγt or Tbet in inguinal and brachial LNs, lung, and skin (like in thymus), a population of MAIT cells co-expressed Tbet and RORγt in the mesenteric LNs, ileum, and colon (Fig. 7 A). A small subset of Tbet+RORγt+ MAIT cells was also detectable in the spleen and liver.
Figure 7.
MAIT cells acquire an evolutionarily conserved MAIT1/17 program in the mouse intestine at steady-state. (A) Left: Representative flow cytometry showing RORγt and Tbet expression in MAIT cells (defined as TCRβ+ 5-OP-RU:MR1 tetramer+ CD24− CD44+) from the indicated tissues of B6-MAITCAST mice. Right: Percentages of MAIT cells expressing Tbet (MAIT1), RORγt (MAIT17), or both (MAIT1/17) in the indicated tissues. Data pooled from two independent experiments. (B) Integrated scRNAseq data from MAIT cells isolated from mouse thymus, mesenteric LNs, ileum, and colon. (C) Dotplot showing expression of selected genes in the indicated UMAP clusters. (D) Expression of TBX21 and RORC in MAIT cells after cross-tissue integration. (E) Expression of MAIT1 (in red) and MAIT17 (in blue) gene signatures in MAIT cells after cross-tissue integration. (F) Left: UMAP showing the individual scRNAseq datasets of MAIT cells after cross-tissue integration. Right: Distribution of MAIT cells from the indicated tissues in the individual UMAP clusters after cross-tissue integration. (G) Expression of the indicated gene signatures in MAIT cells from the indicated mouse tissues. Gene signatures are listed in Table S3. The evolutionarily conserved signature is defined in Fig. 5.
To characterize the transcriptome of mouse intestinal MAIT cells, MR1:5-OP-RU tetramer+ TCRβ+ CD44+ cells were isolated from mesenteric LNs, ileum, and colon by flow cytometry (Fig. S5 A) and profiled by scRNAseq. After quality controls and filtering steps, 4,000 cells from the mesenteric LNs, 2,299 cells from the ileum, and 446 cells from the colon were retained for downstream analyses. Data from peripheral MAIT cells were integrated with scRNAseq data from mouse thymic MAIT cells (Legoux et al., 2019b) to directly assess MAIT cell peripheral maturation (Fig. 7 B). UMAP identified cell populations, which were labeled according to expression of known marker genes, as performed previously. Cells expressing DNTT or EGR2 (Fig. 7 C and Table S10) were only found in the thymus (Fig. 7 F) and therefore were identified as immature. A subset of cells expressed SELL and CCR7 but lacked ZBTB16 expression, resembling central memory T cells. These cells were present in the thymus and the mesenteric LNs, but not in the ileum or colon. They expressed a gene signature associated with T cell recirculation (Milner et al., 2017; Table S3 G; Fig. S5 B), and therefore were labeled as “circulatory.” Cells expressing CCNB2 or MKI67 were identified as cycling. Cells expressing ISG15 (among other interferon stimulated genes) were labeled “interferon” and could represent MAIT cells recently stimulated with interferon. Finally, cells expressing TBX21 but not RORC were identified as MAIT1 cells, while cells expressing RORC but not TBX21 were labeled MAIT17 (Fig. 7 D and Fig. S5 C). In agreement with flow cytometry data, a subset of cells co-expressed TBX21 and RORC and were therefore labeled as MAIT1/17 cells (Fig. 7, B–D; and Fig. S5 C). On the UMAP, MAIT1/17 cells localized in-between MAIT1 and MAIT17 cells and expressed both MAIT1 and MAIT17 gene signatures (Fig. 7 E). MAIT1/17 cells also expressed high levels of NR4A1, indicative of TCR signaling (Fig. 7 C). Additional genes differentially expressed between cell clusters are presented in Table S10. Partition of the cells by tissue of origin revealed that immature cells originated from the thymic sample, while naïve-like cells originated from the thymic and mesenteric LN sample (Fig. 7 F). MAIT1/17 cells were absent from the thymus and were found in the mesenteric LNs, the ileum, and the colon (Fig. 7 F).
Since MAIT cells from mesenteric LNs and intestine co-expressed Tbet and RORγt, we then asked whether these cells also express the evolutionarily conserved MAIT transcriptional gene signature defined previously (Fig. 5 and Table 2). The signature was not expressed in thymic MAIT cells but was strongly expressed in MAIT cells from mesenteric LNs and intestine (Fig. 7 G). Thus, MAIT1/17 cells with an evolutionarily conserved transcriptional program are lacking in the mouse thymus but exist at steady-state in mouse mesenteric LNs and intestine. To further explore the post-thymic maturation of MAIT cells in mice, we assessed expression of gene signatures associated with cytotoxicity (Immgen) and tissue repair (Linehan et al., 2018) in MAIT cells (Table S3, E and F). While cytotoxicity-associated genes were only expressed by MAIT1 cells in the thymus, MAIT cells from the intestine expressed cytotoxicity-associated genes and upregulated a tissue repair gene signature (Fig. 7 G), indicating maturation outside the thymus to acquire additional functionalities.
Mouse thymic MAIT17 cells give rise to intestinal MAIT1/17 cells in a partially Myd88- and Il23-dependent process
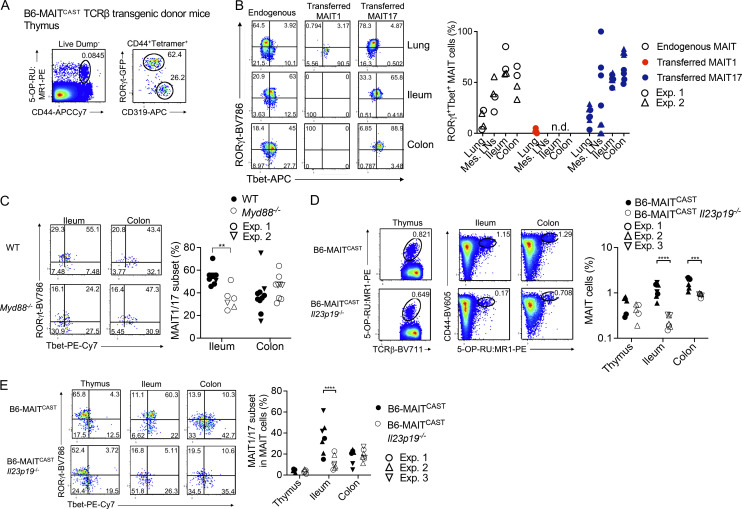
To decipher the ontogeny of intestinal MAIT cells in mice, we next asked which of the thymic MAIT cell subsets give rise to intestinal MAIT1/17 cells. To this end, we adoptively transferred MAIT1 (identified as CD44+ CD319+) and MAIT17 (CD44+ RORγt−GFP+) thymocytes into RAG2−/− recipients (Fig. 8 A). To obtain enough donor MAIT cells, we used B6-MAITCAST mice crossed to a MAIT TCRβ transgenic mouse strain (Martin et al., 2009), which present higher frequencies of MAIT thymocytes with a differentiation pattern identical to that of B6 mice (Fig. S5 D). MAIT cells were tracked and phenotyped in peripheral organs of recipient mice 8 wk after adoptive transfer (Fig. S5 E). Adoptively transferred MAIT1 cells were recovered in small numbers from the lungs and remained Tbet+RORγt−. No donor cell could be recovered from mesenteric LNs or intestines. By contrast, adoptively transferred MAIT17 cells were recovered from lungs, mesenteric LNs, ileum, and colon (Fig. 8 B). Transferred MAIT17 cells remained RORγt+Tbet− in the lungs but acquired a RORγt+Tbet+ phenotype in mesenteric LNs, ileum, and colon (Fig. 8 B). Thus, thymic MAIT17, but not MAIT1 cells, can give rise to intestinal MAIT1/17 cells.
Figure 8.
Mouse intestinal MAIT1/17 cells arise from thymic MAIT17 cells and are maintained through a Myd88 and Il23-dependent mechanism. (A) Representative gating strategy used to FACS-sort MAIT1 (RORγt-GFP− CD319+) and MAIT17 (RORγt-GFP+ CD319−) cells from the thymus of TCRβ transgenic B6-MAITCAST mice. (B) Left: Representative Tbet and RORγt expression in MAIT1 and MAIT17 cells recovered from the indicated tissues 8 wk after adoptive transfer into RAG2−/− recipients. Endogenous MAIT cells from B6-MAITCAST TCRβ transgenic mice are shown as controls. Right: Percentages of MAIT cells co-expressing Tbet and RORγt in the indicated conditions. Data are pooled from two independent experiments. (C) Left: Representative Tbet and RORγt staining in MAIT cells from tissues of WT and Myd88−/− mice. Right: Percentages of MAIT cells coexpressing Tbet and RORγt in the indicated tissues. Data pooled from two independent experiments. (D) Left: Representative 5-OP-RU:MR1 tetramer staining in tissues from WT and Il23p19−/− B6-MAITCAST mice. Right: MAIT cell frequencies in the indicated tissues. Data pooled from three independent experiments. (E) Left: Representative Tbet and RORγt staining in MAIT cells from tissues of WT and Il23p19−/− B6-MAITCAST mice. Right: Percentages of MAIT cells co-expressing Tbet and RORγt in the indicated tissues. Data pooled from three independent experiments. n.d.: not detected. **P < 0.01, ***P < 0.001, ****P < 0.0001 by multiple unpaired t tests with false discovery rate = 1%.
We next looked for the tissue-specific cues that would sustain the RORγt+Tbet+ phenotype of MAIT cells in the intestine. Given the proximity to commensal microbes living in the gut, MAIT cell phenotype could be driven by receptors for microbe-associated molecular patterns such as Toll-like receptors (TLR) or Nod-like receptors (NLR). MAIT cell frequencies (Fig. S5 F) and phenotype (Fig. S5 G) were unaffected in the ileum and colon of mice deficient for the inflammasome-forming NLR Nlrp6. By contrast, the percentage of Tbet+ RORγt+ MAIT cells was reduced in the ileum of Myd88−/− mice (Fig. 8 C), which lack the TLR adaptor MyD88. MAIT cell frequencies were only slightly reduced in the colon of Myd88−/− mice (Fig. S5 H). Of note, WT and Myd88−/− mice used in these experiments were housed in the same room but were not littermates. MyD88 controls signal transduction downstream of most TLRs (except for TLR3), but also downstream of cytokine receptors from the IL1 and IL18 family. In addition, MyD88 expression drives IL23 production in the gut (Hoshi et al., 2012; Friedrich et al., 2017). Because IL23 is critical for Tbet/RORγt co-expression in lung MAIT cells upon pulmonary infections (Wang et al., 2019), and since IL23 is constitutively expressed by intestinal dendritic cells in response to the microbiota (Becker et al., 2003), we further characterized MAIT cells in Il23p19−/− mice, which lack IL23. MAIT cells developed normally in the thymus of B6-MAITCAST Il23p19−/− mice as compared to B6-MAITCAST controls but their frequencies were reduced in the ileum and colon (Fig. 8 D). In addition, MAIT cells failed to co-express Tbet and RORγt in the ileum in Il23p19−/− mice (Fig 8 E). Thus, IL23 is dispensable for thymic MAIT cell development but required for maintaining Tbet+RORγt+ MAIT cells in the small intestine.
Discussion
Here, we used scRNAseq to characterize the sequential transcriptional changes occurring after positive selection of 5-OP-RU:MR1–specific thymocytes in six mammalian species. The panel of species included five eutherian species (human, cattle, sheep, rat, and mouse) and the marsupial opossum, whose oldest common ancestor with eutherians lived during the early Cretaceous (110 million years ago; Bi et al., 2018). The approach identified 5-OP-RU:MR1–specific thymocytes at various stages of development, ranging from recently selected RAG-expressing cells to mature lymphocytes expressing effector genes such as TBX21 or RORC. In each species, pseudo-time modeling identified a sequence of genes induced and repressed at individual stages of 5-OP-RU:MR1–specific thymocyte maturation. An evolutionary conserved intra-thymic development sequence was identified leading to 5-OP-RU:MR1–specific T cells expressing both TBX21 and RORC, which was, however, only acquired in the periphery, in the gut, in the case of rodents.
In addition to 5-OP-RU:MR1–specific thymocytes, we report here the scRNAseq analysis of human thymocytes specific for the melanocyte differentiation antigen peptide MelanA presented by HLA-A2, which identifies cells at various stages of maturation. Integrated analysis with 5-OP-RU:MR1–specific thymocytes revealed immature cells with quasi-identical transcriptomes (marked by RAG1, RAG2, DNTT, and CD1 expression) irrespective of their specificity, confirming that the earliest steps of innate-like T cell differentiation are shared with conventional T cells. Following RAG downregulation, MAIT precursors from all species expressed ZBTB16 and subsequently MAIT1 and MAIT17-associated genes, while MelanA:A2-specific thymocytes acquired CCR7 and LRRN3 consistent with a naïve T cell program. The genes uniquely induced or repressed during development of MAIT cells across species, but not altered during maturation of conventional T cells, were associated with TCR signaling and cell cycle, which suggests that strong TCR signals and intra-thymic expansion are hallmarks of MAIT cell maturation. Strong TCR signals may result from the engagement of the SLAM-SAP pathway during positive selection, which amplifies TCR signaling in mouse thymocytes (Dutta et al., 2013) and is required for MAIT cell development (Koay et al., 2019; Legoux et al., 2019b).
The negative regulator of ZBTB16 (Vasanthakumar et al., 2017), EZH2, was consistently downregulated in MAIT precursors from all species, suggesting that the control of PLZF induction may be conserved. In mice, PLZF can be induced in vitro in DP thymocytes by concomitant CD3 and Slamf6 stimulations (Dutta et al., 2013; Tuttle et al., 2018; Legoux et al., 2019b), pointing to the SLAM-SAP signaling pathway as a determinant of PLZF induction upon positive selection. However, although MAIT cells fail to induce PLZF in SAP−/Y mice (Koay et al., 2019; Legoux et al., 2019b), MAIT cells expressing PLZF are found in the blood of SAP-deficient patients (Martin et al., 2009), suggesting that additional, species-specific mechanisms may control the induction of PLZF. Since PLZF drives intra-thymic expansion in mice (Savage et al., 2008), it is plausible that MAIT cell proliferation is controlled by PLZF across species.
Although this is not as clear in humans, mice harbor discrete subsets of effector helper T cells that are distinguished by the exclusive expression of master transcription factors (such as Tbet or RORγt, among others) associated to distinctive functional features (Abbas et al., 1996). Although MAIT cells isolated from the mouse thymus present two completely distinct subsets in line with this paradigm, MAIT cells in the mouse intestine formed a more homogeneous population displaying a continuum of transcriptomic features ranging from type-1 to type-17. This observation fits with the description of a similar continuum in conventional CD4+ T cells from the mouse intestine (Kiner et al., 2021). Given that MAIT17 cells also upregulate Tbet in the mouse lungs upon bacterial infection (Chen et al., 2017), the blended MAIT1/17 phenotype of mouse MAIT cells may represent a general response to bacterial exposure in both mainstream and innate-like T cells.
A conserved MAIT1/17 program is acquired in the thymus in the marsupial, human, cattle, and sheep, and in the periphery in rodents. This program is characterized by co-expression of genes associated with distinct and seemingly opposite functions, notably cytotoxicity and tissue repair. The acquisition of a common transcriptional program by MAIT cells in all species suggests that a blended functional potential could be an early evolutionarily conserved feature. In agreement with this hypothesis, 5-OP-RU:MR1–specific T cells co-express PLZF, RORγt, and Tbet in the bat P. alecto (Leeansyah et al., 2020), suggesting a similar MAIT1/17 program. Analysis of human αGC:CD1-specific thymocytes revealed acquisition of an identical, polyfunctional program in a subset of human iNKT cells, indicating that the mixed 1/17 differentiation program is a preserved and common feature of innate-like T cells during evolution.
The origin of the differences in MAIT cell development between rodents and other species is unclear. In mice, microbial 5-OP-RU is presented in the thymus, leading to increased numbers of immature and mature MAIT cells (Legoux et al., 2019a). In human cord blood, MAIT cells lack expression of the human memory marker CD45RO, contrasting with the acquisition of CD44 in mouse MAIT cells directly in the thymus. Human MAIT cells become memory-like a few months after birth and reach adult frequencies at around 6 years of age (Ben Youssef et al., 2018), suggesting a lack of antigenic activation directly in the human fetal thymus. We previously suggested (Legoux et al., 2020) that the size of the animal could be a determining factor for 5-OP-RU concentration in the body: the concentration of 5-OP-RU reaching the thymus would be inversely correlated to the size of the animal. In this model, MAIT cells from large animals would not encounter 5-OP-RU in the thymus, even more so during fetal life, and selection would exclusively rely on an unknown endogenous MR1 ligand. Since MAIT cells acquire a MAIT1/17 program in the opossum, whose size is similar to rats, a role for 5-OP-RU availability in determining thymic MAIT cell programing appears unlikely. The reason for the two-step maturation of MAIT cells in rodents is unclear. It could be beneficial to trigger a type-17 immunity in the thymus in some circumstances, as proposed for IL4-secreting iNKT2 cells acting on conventional naïve CD8 T cells to generate natural memory T cells (Lee et al., 2013).
In summary, we highlight a conserved transcriptional program in 5-OP-RU–specific T cells across mammals. MAIT cells are characterized by co-expression of functional modules associated with tissue repair, cytotoxicity, type-1 and type-17 effector genes, pointing to versatile and context-dependent responses that are shared across innate-like T cell populations.
Materials and methods
Thymic samples
Human thymic samples were obtained as surgical tissue discards with informed consent from the parents and with approval of the ethical review board of Necker Enfants Malades Hospital at Paris Descartes. The patients were 1–15-mo-old children undergoing corrective cardiac surgery at the Necker Hospital (Paris). 5-OP-RU:MR1–specific and αGC:CD1d-specific thymocytes were obtained from the thymus of a 4-mo-old female patient. MelanA:A2-specific thymocytes were obtained from a 13-mo-old HLA-A2+ male patient. Opossums were bred and maintained under UK Home Office Regulations, UK Animals (Scientific Procedures) Act 1986, and according to ethical guidelines at the Francis Crick Institute. Permission for animal experiments was granted by The Crick Biological Research Facility Strategic Oversight Committee incorporating the Animal Welfare and Ethical Review Body (Project Licence P8ECF28D9). Thymic samples from three males were used for the experimental setup. The thymus from a 12-wk-old male was used for scRNAseq analyses of 5-OP-RU:MR1–specific thymocytes. Cattle and sheep thymic samples were obtained from 2–6-mo-old Charolais calves and pre-Alpes sheep euthanized at the Biosurgical Research lab of the Georges Pompidou European Hospital in Paris, France (protocol registration: AC.108.12 and 01425.04) in accordance with guidelines for the care and use of laboratory animals. Thymi from 11-wk-old Sprague Dawley rat females were purchased from Janvier labs.
Mice and crosses
Transgenic mice were maintained on a C57BL/6J background in SPF conditions at the Institut Curie central facility. B6-MAITCAST mice were generated as described previously (Cui et al., 2015). Tg(Rorc-EGFP)1Ebe (RORγt-GFP, MGI:3829387) mice (Lochner et al., 2008; obtained from G. Eberl, Institut Pasteur, Paris, France) and Il23atm1Ngh (Il23p19−/−, MGI:3036163) mice (Ghilardi et al., 2004) were crossed to B6-MAITCAST mice. Myd88tm1Aki (Myd88−/−, MGI:2385681; Adachi et al., 1998) and Nlrp6tm1Macha (Nlrp6−/−, MGI:2141990; Normand et al., 2011) mice were obtained from the University of Orléans, France. Previously described MAIT TCRβ Tg mice (Martin et al., 2009) were crossed to the B6-MAITCAST strain for use as a source of MAIT thymocytes for adoptive transfer experiments (see below). CC mice were bred and maintained in the animal facility of the Institut Pasteur under SPF conditions. Tissues were collected from 6–10-wk-old males of 16 CC strains. Two age- and sex-matched C57BL/6N mice raised in the same animal facility were included as controls in each experiment. Pet store mice were purchased from Animalis Bercy. Cutaneous swabs, mouth swabs, and feces were sampled from pet mice and from control SPF mice for the detection of common pathogens by PCR (Envigo). All mouse and rat experiments were performed according to national and international guidelines, approved by the Institut Curie Ethics Committee and authorized by the French Ministry of Research (project #251802020042118535702).
Cell isolation from thymi
Freshly isolated thymi from all species were maintained in CO2-independent medium (Gibco) at 4°C until processing. For all samples, processing started <6 h after euthanasia. Mouse thymi were mashed over 40-μm cell strainers to create single-cell suspensions. Thymi from all other species were minced with a sterile scalpel in a petri dish containing 5 ml of CO2-independent medium. Tissue pieces were then washed with 40 ml of FACS buffer (PBS supplemented with 2% fetal calf serum [FCS] and 2.5 mM EDTA) to recover single cell suspensions of thymocytes.
Cell isolation from mouse peripheral tissues
Mouse spleen, thymus, and LNs were mashed over 40-μm nylon mesh to create single cell suspensions. Lungs were incubated with 100 μg/ml Liberase TL (Roche) and 100 μg/ml DNAse I (Sigma-Aldrich) for 30 min at 37°C, and dissociated with a gentleMACS Dissociator (Miltenyi). Livers were mashed over 100-μm cell strainer. Ileum and colon samples were flushed with PBS, opened longitudinally, and cut into ∼0.5 cm pieces. Dissociation of epithelial cells was performed by incubation with constant stirring in HBSS without Ca/Mg (Life Technologies) containing 5 mM EDTA (Thermo Fisher Scientific), 1 mM DTT (Euromedex), and 5% FCS twice for 20 min at 37°C. After each step, samples were vortexed and the epithelial fraction discarded. Tissue fragments were then washed in HBSS (Thermo Fisher Scientific) and enzymatic digestion was performed in CO2-independent medium (Life Technologies) containing collagenase D (1 mg/ml; Roche), Liberase TM (0.17 U/ml; Roche), and DNase I (100 µg/ml; Roche) on a shaker for 30 min at 37°C. Colon fragments were then dissociated with a gentleMACS dissociator according to the manufacturer’s instructions (Miltenyi). Skin samples were processed as described (du Halgouet et al., 2023). Lymphocytes from lung, liver, ileum, and colon samples were collected at the interface of a 40%/80% Percoll gradient (GE Healthcare).
Tetramers
PE-labeled human and mouse MR1 tetramers loaded with 5-OP-RU and PE-labeled human CD1d tetramer loaded with PBS-57 were provided by the National Institutes of Health (NIH) tetramer core facility (Emory University, Atlanta, GA, USA). B. taurus tetramers were generated in-house. M. domestica tetramers were generated by the Nantes University recombinant protein production core facility (Nantes, France) as previously described for a mouse MR1 tetramer (Legoux et al., 2019b). Briefly, DNA coding for the soluble M. domestica heavy chain of MR1 fused with a biotinylation tag and the M. domestica β2 microglobuline sequence were synthetized and cloned in pET24 vector by Geneart (Thermo Fisher Scientific). The two chains were produced as inclusion bodies in Escherichia coli and refolded in the presence of 5-OP-RU, as described for mouse MR1 tetramer production (Legoux et al., 2019b). Biotinylated MelanA:A2 monomers were purchased from the Nantes recombinant protein production facility and tetramerized on a PE-conjugated streptavidin (PJRS25; Agilent).
Cell sorting and scRNAseq
Mouse thymic scRNAseq data were generated in a previous study (Legoux et al., 2019b). To generate scRNAseq data from the other species, up to 109 thymocytes were stained with the appropriate PE-labeled MR1, CD1d, or HLA-A2 tetramer for 30 min at room temperature (RT; for opossum, cattle, and mouse tetramers) or on ice (for human tetramers), then washed and stained with 70 μl anti-PE microbeads (Miltenyi). The opossum MR1 tetramer was used to label opossum thymocytes, the cattle MR1 tetramer was used to label cattle and sheep thymocytes, the human MR1 tetramer was used on human thymocytes, and the mouse MR1 tetramer was used on rat thymocytes. Magnetic enrichment of tetramer-labeled thymocytes was then performed as described (Legoux and Moon, 2012). Following magnetic enrichment, thymocytes were stained for 30 min at 4°C with the following primary antibodies: Opossum cells were stained with polyclonal rabbit anti-human CD3e (A0452; Dako) and with mouse anti-CD79A (clone HM57, GTX74022; GeneTex). Cattle and sheep cells were stained with mouse anti-bovine CD3 (clone MM1A, MCA6080; Bio-rad) and with mouse anti-bovine γδ TCR (clone GB21A, BOV2058; Monoclonal Antibody Center). Human cells were stained with anti-CD3 AF700 (clone HIT3a, 300324; Biolegend), anti-CD161 APC (clone HP-3G10, 339912; Biolegend), and anti-CD27 PECy7 (clone O323, 302838; Biolegend). Rat cells were stained with anti-rat TCRαβ FITC (clone R73, 201105; Biolegend) and anti-rat CD161 APC (clone 3.2.3, 205606; Biolegend). After washing with FACS buffer, opossum cells were further stained for 30 min on ice with the following secondary antibodies: donkey anti-rabbit IgG AF488 (406416; Biolegend) and anti-mouse IgG1 AF700 (clone RMG1-1, 406631; Biolegend). Cattle and sheep cells were stained with secondary anti-mouse IgG1 AF700 (clone RMG1-1, 406631; Biolegend) and anti-mouse IgG2b PE-Cy7 (clone RMG2b-1, 406713; Biolegend). Cells were then washed with FACS buffer. All samples were stained with DAPI (Sigma-Aldrich). 5-OP-RU:MR1–specific, αGC:CD1d-specific, and MelanA:A2-specific thymocytes were defined as DAPI− Tetramer-PE+ CD3 (or TCRβ)+. A population of γδ thymocytes from cattle and sheep were found to bind to the cattle MR1 tetramer and were gated out and excluded from the FACS-sorted population. Cells were flow-sorted (Aria III; BD) into PBS supplemented with 0.35% BSA (Sigma-Aldrich). Sorted cells were centrifuged and counted, and 5,000–10,000 cells were loaded onto the Chromium 3′ chip. Reverse transcription, library preparation, and sequencing were performed according to manufacturer’s recommendations (10x Genomics).
Flow cytometry analyses
For flow cytometry analysis of mouse MAIT cells, single-cell suspensions from the thymus, LNs, spleen, liver, lung, skin, ileum, and colon were stained in the presence of Fc block (clone 2.4G2 produced in-house) for 30 min on ice with a fixable live-dead marker (Aqua, L34965; Thermo Fisher Scientific) and the following fluorescently labeled antibodies: anti-TCRβ PECy5 (clone H57-597, 553173; BD), anti-CD319 APC (clone 4G2, 152004; Biolegend), anti-CD11c AF700 (clone N418, 117320; Biolegend), anti-CD19 AF700 (clone 6D5, 115528; Biolegend), anti-B220 AF700 (clone RA3-6B2, 56-0452-82; Invitrogen), anti-CD44 APC-Cy7 (clone IM7, 103028; Biolegend). The cells were then washed with FACS buffer. For transcription factor staining, cells were fixed and permeabilized using the Foxp3-staining buffer set (eBioscience) following the manufacturer’s instructions. The cells were first washed with PBS and then incubated on ice for 20 min with 100 μl Fix/Perm solution. Cells were then washed twice with 200 μl wash buffer prior to staining for intranuclear proteins at 4°C overnight in wash buffer. The following intranuclear antibodies were used: anti-RORγt BV786 (clone Q31-378, 564723; BD) and anti-Tbet PECy7 (clone eBio4B10, 25-5825-82; Invitrogen). Cells were then washed with FACS buffer and analyzed on an LSR Fortessa (BD) or Ze5 (Bio-Rad). Data were analyzed using FlowJo V10.2 software (Treestar).
MAIT cell adoptive transfers
MAIT cells from the thymus of 6–8-wk-old B6-MAITCAST RORγt-GFP+ TCRβ Tg mice were identified as live (Aqua−) CD11c-AF700− CD19-AF700− B220-AF700− TCRβ-PECy5+ 5-OP-RU:MR1 tetramer+ CD44-APC-Cy7+. MAIT1 cells were further defined as RORγt-GFP− CD319-APC+ while MAIT17 cells were identified as RORγt-GFP+ CD319-APC−. MAIT cell subsets were FACS-sorted (Aria) into sterile PBS supplemented with 10% FCS. Sorted cells were counted, washed, resuspended in sterile PBS, and injected intravenously into sublethally (4 Gy) irradiated 10–12-wk-old RAG2−/− recipients. Each recipient received either 5,000 MAIT1 or 60,000 MAIT17 cells. All recipient mice were euthanized 8 wk later for analysis of MAIT cells by flow cytometry in lungs, mesenteric LNs, ileum, and colon.
scRNAseq analyses
Alignments to reference genomes and feature-barcode matrices were performed with Cell Ranger (Table S1). Downstream analyses were performed using R version 4.2.2 Patched (2022-11-10 r83330) and the following packages: stringr 1.5.0; Seurat 4.3.0; dplyr 1.1.2; Matrix 1.5-4; slingshot 2.4.0; tradeSeq 1.10.0; scales 1.2.1; ggplot2 3.4.2; clustree 0.5.0; biomaRt 2.52.0; tidyr 1.3.0; and ComplexHeatMap 2.12.1. The human MAIT and NKT thymic datasets were sequenced twice to improve sequencing coverage. For each dataset except SAP−/Y thymocytes, which contain few cells, only genes present in at least three cells were retained for analysis. Dataset-specific filters were applied (listed in Table S1) to remove empty droplets, dying cells, and cells expressing B, myeloid, or γδ T cell markers. Data were then internally normalized using the NormalizeData() function, followed by identification of the most variable features with the FindVariableFeatures() function with the vst selection method. The number of variable features retained for each sample is listed in Table S1. Data were then scaled with the ScaleData() function followed by principal component analysis with the RunPCA() function on variable features. The ElbowPlot() function was used to determine the optimal number of dimensions for each sample (listed in Table S1). Clustree (Zappia and Oshlack, 2018) was used to identify a clustering resolution yielding stable clusters (Table S1). The FindCluster() function was used with the chosen resolution for each dataset to identify cell clusters, which were displayed on UMAP using the RunUMAP() function. Differentially expressed genes between clusters were identified with the FindAllMarker() function with the following parameters: only.pos = FALSE, min.pct = 0.1, logfc.threshold = 0.25, base = 10. Heatmaps were built using the DoHeatmap() function to display top overexpressed genes ordered by avg_log10FC within each cluster. Gene signatures were displayed using the AddModuleScore() function. Gene signatures used in the study are listed in Table S3. Pseudotemporal ordering of cells was performed in Slingshot (Street et al., 2018) using the Seurat clusters and the following parameters: reduction = “UMAP,” stretch = 0, extend = “n.” DNTT-expressing cells were used as a starting point for all trajectories. No final destination cluster was selected. For relevant trajectories leading to MAIT1/17, MAIT1, or MAIT17 subsets, a new Seurat object was created with only the cells present on the trajectory. ComplexHeatmap() was then used to build pseudotime-dependent heatmaps, in which cells were ordered according to their pseudotime score. TradeSeq (Van den Berge et al., 2020) was used downstream of Slingshot to identify genes whose expression varied along each pseudotime trajectory.
For integration of datasets from the same species (human or mouse), filtered datasets were normalized individually and the most variable features were selected prior to merging using the merge() function with merge.data = TRUE. Mouse thymic and Mes. LN samples were integrated together, prior to integration to an object made of integrated colon and ileum samples. The integrated object was further processed as an individual dataset: data were scaled with the ScaleData() function followed by principal component analysis with the RunPCA() function on variable features. The ElbowPlot() function was used to determine the optimal number of dimensions. Clustree was used to identify a clustering resolution yielding stable clusters. The FindCluster() function was used with the chosen resolution to identify cell clusters, which were displayed on UMAP using the RunUMAP() function. Differentially expressed genes between clusters were identified with the FindAllMarker() function with the following parameters: only.pos = FALSE, min.pct = 0.1, logfc.threshold = 0.25, base = 10.
For cross-species integration, mouse genes with orthologs present in all six species were selected based on OMA groups (Altenhoff et al., 2021) and are listed in Table S7. All the other genes were deleted from the datasets. Genes in all datasets were renamed according to the mouse nomenclature. Integration was then performed in Seurat using the merge() function with merge.data = TRUE. The integration was guided and intermediary Seurat objects were created following the phylogenetic tree: the mouse and rat datasets were integrated to generate a “Rodent” object, which was integrated with the human dataset to obtain the “Euarchontoglires” object. Cattle and sheep datasets were integrated, generating the “Bovine” object, which was integrated with the “Euarchontoglires” object to obtain the “Boreoeutheria” object. Integration of the opossum dataset with the “Boreoeutheria” object created the “Mammals” object described in Fig. 5. The integrated object was then analyzed as described for individual datasets.
Statistical analyses
Statistical analyses were performed with Prism software (GraphPad). Two-tailed P values were determined using Wilcoxon’s and Mann–Whitney’s tests for paired and non-paired samples, as appropriate. A false discovery rate of 1% was calculated for multiple t tests and multiple Mann–Whitney’s tests using the two-stage step-up method of Benjamini, Krieger, and Yekutieli.
Online supplemental material
Fig. S1 shows analyses of individual scRNAseq datasets from each species. Fig. S2 shows developmental trajectories of MAIT1 thymocytes. Fig. S3 shows supplemental data regarding the analysis of conventional T cells and merged MAIT thymocytes analyses. Fig. S4 shows iNKT cell supplemental data. Fig. S5 shows mouse MAIT supplemental data. Table S1 shows basic descriptions of each scRNAseq dataset. Table S2 shows the differentially expressed genes in each UMAP cluster from each individual scRNAseq dataset. Table S3 shows the gene signatures used or defined in the study. Table S4 shows TradeSeq results from all developmental trajectories. Table S5 shows the conserved genes found modulated along MAIT cell development in all species. Table S6 shows the differentially expressed genes in UMAP clusters after integration of MAIT and conventional datasets. Table S7 shows the ortholog genes present in the studied species. Table S8 shows the preprocessing parameters used for integration of MAIT cells from the six individual datasets. Table S9 shows the differentially expressed genes in UMAP clusters after integration of MAIT from the six species. Table S10 shows the differentially expressed genes in UMAP clusters after integration of mouse MAIT datasets.
Supplementary Material
shows basic descriptions of each scRNAseq dataset.
shows the differentially expressed genes in each UMAP cluster from each individual scRNAseq dataset.
shows the gene signatures used or defined in the study.
shows TradeSeq results from all developmental trajectories.
shows the conserved genes found modulated along MAIT cell development in all species.
shows the differentially expressed genes in UMAP clusters after integration of MAIT and conventional datasets.
shows the ortholog genes present in the studied species.
shows the preprocessing parameters used for integration of MAIT cells from the six individual datasets.
shows the differentially expressed genes in UMAP clusters after integration of MAIT from the six species.
shows the differentially expressed genes in UMAP clusters after integration of mouse MAIT datasets.
Acknowledgments
We thank M. Garcia, V. Dangles-Marie, the mouse facility technicians, and the flow cytometry core at Institut Curie. We thank the ICGex NGS platform of the Institut Curie for technical help with scRNAseq experiments. ICGex is supported by the grants ANR10EQPX03 (Equipex) and ANR10INBS0908 (France Génomique Consortium) from the Agence Nationale de la Recherche (“Investissements d’Avenir” program), by the ITMO-Cancer Aviesann (Plan Cancer III), and by the SiRIC-Curie program (INCa-DGOS-465 and INCa-DGOS-Inserm_12554). We thank T. Penel and the Institut Pasteur facility staff for the breeding of the Collaborative Cross mice. We thank the National Institutes of Health (NIH) tetramer core facility (Emory University, Atlanta, GA, USA) for providing mouse and human MR1 tetramers. The MR1:5-OP-RU tetramer technology was developed jointly by J. McCluskey, J. Rossjohn, and D. Fairlie, and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne.
This work was supported by the Institut National de la Santé et de la Recherche Médicale (O. Lantz, F. Legoux, H. Bugaut), Institut Curie (O. Lantz), Institut Pasteur (X. Montagutelli), Agence Nationale de la Recherche Grant JCJC ANR-19-CE15-0002-01 MAIT (F. Legoux), Agence Nationale de la Recherche Grants MAIT (ANR-16-CE15-0020-01), diabMAIT (ANR-17-CE14-0002-02), MAITrepair (ANR-20-CE15-0028-01), and ANR-10-IDEX-0001-02 PSL (O. Lantz), European Research Council (ERC-2019-AdG-885435) (O. Lantz), Chaire de recherche from Rennes Métropole—22C0451 (F. Legoux), Société Française de Dermatologie (H. Bugaut), Novartis (H. Bugaut), and Fondation pour la Recherche Médicale (SPF202209015773) (R.A. Paiva). Work in the Turner lab is supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (CC2052), UK Medical Research Council (CC2052), and Wellcome Trust (CC2052); and European Research Council (CoG 647971).
Author contributions: Conceptualization: F. Legoux; Methodology: F. Legoux, H. Bugaut, M. Mestdagh; Investigation: H. Bugaut, Y. El Morr, M. Mestdagh, A. Darbois, R.A. Paiva, M. Salou, L. Perrin, M. Fürstenheim, L. Bilonda-Mutala, A.-L. Le Gac, M. Arnaud, F. Legoux; Formal analysis: M. Mestdagh, H. Bugaut, Y. El Morr, F. Legoux; Funding acquisition: O. Lantz, F. Legoux; Critical material provision: A. El Marjou, C. Guerin, A. Chaiyasitdhi, J. Piquet, D.M. Smadja, A. Cieslak, B. Ryffel, V. Maciulyte, J.M.A. Turner, K. Bernardeau, X. Montagutelli; Supervision: O. Lantz, F. Legoux; Writing—original draft: H. Bugaut, F. Legoux; Writing—review & editing: O. Lantz, F. Legoux.
Data availability
The sequencing data generated in this study are available at GEO NCBI under the accession number GSE239558. All the other data are available in the main text or the supplementary materials.
References
- Abbas, A.K., Murphy K.M., and Sher A.. 1996. Functional diversity of helper T lymphocytes. Nature. 383:787–793. 10.1038/383787a0 [DOI] [PubMed] [Google Scholar]
- Adachi, O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., and Akira S.. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 9:143–150. 10.1016/S1074-7613(00)80596-8 [DOI] [PubMed] [Google Scholar]
- Altenhoff, A.M., Train C.M., Gilbert K.J., Mediratta I., Mendes de Farias T., Moi D., Nevers Y., Radoykova H.S., Rossier V., Warwick Vesztrocy A., et al. 2021. OMA orthology in 2021: Website overhaul, conserved isoforms, ancestral gene order and more. Nucleic Acids Res. 49:D373–D379. 10.1093/nar/gkaa1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataide, M.A., Komander K., Knöpper K., Peters A.E., Wu H., Eickhoff S., Gogishvili T., Weber J., Grafen A., Kallies A., et al. 2020. BATF3 programs CD8+ T cell memory. Nat. Immunol. 21:1397–1407. 10.1038/s41590-020-0786-2 [DOI] [PubMed] [Google Scholar]
- Awad, W., Ciacchi L., McCluskey J., Fairlie D.P., and Rossjohn J.. 2023. Molecular insights into metabolite antigen recognition by mucosal-associated invariant T cells. Curr. Opin. Immunol. 83:102351. 10.1016/j.coi.2023.102351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, C., Wirtz S., Blessing M., Pirhonen J., Strand D., Bechthold O., Frick J., Galle P.R., Autenrieth I., and Neurath M.F.. 2003. Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells. J. Clin. Invest. 112:693–706. 10.1172/JCI17464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Youssef, G., Tourret M., Salou M., Ghazarian L., Houdouin V., Mondot S., Mburu Y., Lambert M., Azarnoush S., Diana J.S., et al. 2018. Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. J. Exp. Med. 215:459–479. 10.1084/jem.20171739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berzins, S.P., Cochrane A.D., Pellicci D.G., Smyth M.J., and Godfrey D.I.. 2005. Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur. J. Immunol. 35:1399–1407. 10.1002/eji.200425958 [DOI] [PubMed] [Google Scholar]
- Beura, L.K., Hamilton S.E., Bi K., Schenkel J.M., Odumade O.A., Casey K.A., Thompson E.A., Fraser K.A., Rosato P.C., Filali-Mouhim A., et al. 2016. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 532:512–516. 10.1038/nature17655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj, A., Kempster E., and Waisman D.M.. 2021. The annexin A2/S100A10 complex: The mutualistic symbiosis of two distinct proteins. Biomolecules. 11:1849. 10.3390/biom11121849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, S., Zheng X., Wang X., Cignetti N.E., Yang S., and Wible J.R.. 2018. An Early Cretaceous eutherian and the placental-marsupial dichotomy. Nature. 558:390–395. 10.1038/s41586-018-0210-3 [DOI] [PubMed] [Google Scholar]
- Boudinot, P., Mondot S., Jouneau L., Teyton L., Lefranc M.P., and Lantz O.. 2016. Restricting nonclassical MHC genes coevolve with TRAV genes used by innate-like T cells in mammals. Proc. Natl. Acad. Sci. USA. 113:E2983–E2992. 10.1073/pnas.1600674113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Wang H., D’Souza C., Sun S., Kostenko L., Eckle S.B., Meehan B.S., Jackson D.C., Strugnell R.A., Cao H., et al. 2017. Mucosal-associated invariant T-cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol. 10:58–68. 10.1038/mi.2016.39 [DOI] [PubMed] [Google Scholar]
- Corbett, A.J., Eckle S.B., Birkinshaw R.W., Liu L., Patel O., Mahony J., Chen Z., Reantragoon R., Meehan B., Cao H., et al. 2014. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 509:361–365. 10.1038/nature13160 [DOI] [PubMed] [Google Scholar]
- Cui, Y., Franciszkiewicz K., Mburu Y.K., Mondot S., Le Bourhis L., Premel V., Martin E., Kachaner A., Duban L., Ingersoll M.A., et al. 2015. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J. Clin. Invest. 125:4171–4185. 10.1172/JCI82424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Halgouet, A., Darbois A., Alkobtawi M., Mestdagh M., Alphonse A., Premel V., Yvorra T., Colombeau L., Rodriguez R., Zaiss D., et al. 2023. Role of MR1-driven signals and amphiregulin on the recruitment and repair function of MAIT cells during skin wound healing. Immunity. 56:78–92.e6. 10.1016/j.immuni.2022.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois, T., Oudinet J.P., Russo-Marie F., and Rothhut B.. 1995. In vivo and in vitro phosphorylation of annexin II in T cells: Potential regulation by annexin V. Biochem. J. 310. 243–248. 10.1042/bj3100243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, M., Kraus Z.J., Gomez-Rodriguez J., Hwang S.-H., Cannons J.L., Cheng J., Lee S.-Y., Wiest D.L., Wakeland E.K., and Schwartzberg P.L.. 2013. A role for Ly108 in the induction of promyelocytic zinc finger transcription factor in developing thymocytes. J. Immunol. 190. 2121–2128. 10.4049/jimmunol.1202145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmans, M.D., Connelley T.K., Jayaraman S., Vrettou C., Vordermeier M., Mak J.Y.W., Liu L., Fairlie D.P., Maze E.A., Chrun T., et al. 2021. Identification and phenotype of MAIT cells in cattle and their response to bacterial infections. Front. Immunol. 12:627173. 10.3389/fimmu.2021.627173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emgård, J., Kammoun H., García-Cassani B., Chesné J., Parigi S.M., Jacob J.M., Cheng H.W., Evren E., Das S., Czarnewski P., et al. 2018. Oxysterol sensing through the receptor GPR183 promotes the lymphoid-tissue-inducing function of innate lymphoid cells and colonic inflammation. Immunity. 48:120–132.e8. 10.1016/j.immuni.2017.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, C., Mamareli P., Thiemann S., Kruse F., Wang Z., Holzmann B., Strowig T., Sparwasser T., and Lochner M.. 2017. MyD88 signaling in dendritic cells and the intestinal epithelium controls immunity against intestinal infection with C. rodentium. PLoS Pathog. 13:e1006357. 10.1371/journal.ppat.1006357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghilardi, N., Kljavin N., Chen Q., Lucas S., Gurney A.L., and De Sauvage F.J.. 2004. Compromised humoral and delayed-type hypersensitivity responses in IL-23-deficient mice. J. Immunol. 172. 2827–2833. 10.4049/jimmunol.172.5.2827 [DOI] [PubMed] [Google Scholar]
- Godfrey, D.I., Koay H.F., McCluskey J., and Gherardin N.A.. 2019. The biology and functional importance of MAIT cells. Nat. Immunol. 20:1110–1128. 10.1038/s41590-019-0444-8 [DOI] [PubMed] [Google Scholar]
- Greene, J.M., Dash P., Roy S., McMurtrey C., Awad W., Reed J.S., Hammond K.B., Abdulhaqq S., Wu H.L., Burwitz B.J., et al. 2017. MR1-restricted mucosal-associated invariant T (MAIT) cells respond to mycobacterial vaccination and infection in nonhuman primates. Mucosal Immunol. 10:802–813. 10.1038/mi.2016.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griewank, K., Borowski C., Rietdijk S., Wang N., Julien A., Wei D.G., Mamchak A.A., Terhorst C., and Bendelac A.. 2007. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 27:751–762. 10.1016/j.immuni.2007.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumperz, J.E., Miyake S., Yamamura T., and Brenner M.B.. 2002. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J. Exp. Med. 195:625–636. 10.1084/jem.20011786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y., Hao S., Andersen-Nissen E., Mauck W.M. III, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. 2021. Integrated analysis of multimodal single-cell data. Cell. 184:3573–3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsha Krovi, S., Zhang J., Michaels-Foster M.J., Brunetti T., Loh L., Scott-Browne J., and Gapin L.. 2020. Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells. Nat. Commun. 11:6238. 10.1038/s41467-020-20073-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi, N., Schenten D., Nish S.A., Walther Z., Gagliani N., Flavell R.A., Reizis B., Shen Z., Fox J.G., Iwasaki A., and Medzhitov R.. 2012. MyD88 signalling in colonic mononuclear phagocytes drives colitis in IL-10-deficient mice. Nat. Commun. 3:1120. 10.1038/ncomms2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., Martin E., Kim S., Yu L., Soudais C., Fremont D.H., Lantz O., and Hansen T.H.. 2009. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc. Natl. Acad. Sci. USA. 106:8290–8295. 10.1073/pnas.0903196106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, J., McCaffrey P.G., Miner Z., Kerppola T.K., Lambert J.N., Verdine G.L., Curran T., and Rao A.. 1993. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 365:352–355. 10.1038/365352a0 [DOI] [PubMed] [Google Scholar]
- Kiner, E., Willie E., Vijaykumar B., Chowdhary K., Schmutz H., Chandler J., Schnell A., Thakore P.I., LeGros G., Mostafavi S., et al. 2021. Gut CD4+ T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nat. Immunol. 22:216–228. 10.1038/s41590-020-00836-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koay, H.-F., Gherardin N.A., Enders A., Loh L., Mackay L.K., Almeida C.F., Russ B.E., Nold-Petry C.A., Nold M.F., Bedoui S., et al. 2016. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat. Immunol. 17:1300–1311. 10.1038/ni.3565 [DOI] [PubMed] [Google Scholar]
- Koay, H.-F., Su S., Amann-Zalcenstein D., Daley S.R., Comerford I., Miosge L., Whyte C.E., Konstantinov I.E., d’Udekem Y., Baldwin T., et al. 2019. A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci. Immunol. 4:eaay6039. 10.1126/sciimmunol.aay6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan, J. and Killeen N.. 2004. CCR7 directs the migration of thymocytes into the thymic medulla. J. Immunol. 172. 3999–4007. 10.4049/jimmunol.172.7.3999 [DOI] [PubMed] [Google Scholar]
- Lee, P.T., Benlagha K., Teyton L., and Bendelac A.. 2002. Distinct functional lineages of human V(alpha)24 natural killer T cells. J. Exp. Med. 195:637–641. 10.1084/jem.20011908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.J., Holzapfel K.L., Zhu J., Jameson S.C., and Hogquist K.A.. 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14:1146–1154. 10.1038/ni.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.H., Zhang H.H., Singh S.P., Koo L., Kabat J., Tsang H., Singh T.P., and Farber J.M.. 2018. C/EBPδ drives interactions between human MAIT cells and endothelial cells that are important for extravasation. Elife. 7:e32532. 10.7554/eLife.32532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeansyah, E., Svärd J., Dias J., Buggert M., Nyström J., Quigley M.F., Moll M., Sönnerborg A., Nowak P., and Sandberg J.K.. 2015. Arming of MAIT cell cytolytic antimicrobial activity is induced by IL-7 and defective in HIV-1 infection. PLoS Pathog. 11:e1005072. 10.1371/journal.ppat.1005072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeansyah, E., Hey Y.Y., Sia W.R., Ng J.H.J., Gulam M.Y., Boulouis C., Zhu F., Ahn M., Mak J.Y.W., Fairlie D.P., et al. 2020. MR1-Restricted T cells with MAIT-like characteristics are functionally conserved in the pteropid bat Pteropus alecto. iScience. 23:101876. 10.1016/j.isci.2020.101876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoux, F.P., and Moon J.J.. 2012. Peptide:MHC tetramer-based enrichment of epitope-specific T cells. J. Vis. Exp.:4420. 10.3791/4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoux, F., Bellet D., Daviaud C., El Morr Y., Darbois A., Niort K., Procopio E., Salou M., Gilet J., Ryffel B., et al. 2019a. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. 366:494–499. 10.1126/science.aaw2719 [DOI] [PubMed] [Google Scholar]
- Legoux, F., Gilet J., Procopio E., Echasserieau K., Bernardeau K., and Lantz O.. 2019b. Molecular mechanisms of lineage decisions in metabolite-specific T cells. Nat. Immunol. 20:1244–1255. 10.1038/s41590-019-0465-3 [DOI] [PubMed] [Google Scholar]
- Legoux, F., Salou M., and Lantz O.. 2020. MAIT cell development and functions: The microbial connection. Immunity. 53:710–723. 10.1016/j.immuni.2020.09.009 [DOI] [PubMed] [Google Scholar]
- Linehan, J.L., Harrison O.J., Han S.J., Byrd A.L., Vujkovic-Cvijin I., Villarino A.V., Sen S.K., Shaik J., Smelkinson M., Tamoutounour S., et al. 2018. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell. 172:784–796.e18. 10.1016/j.cell.2017.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochner, M., Peduto L., Cherrier M., Sawa S., Langa F., Varona R., Riethmacher D., Si-Tahar M., Di Santo J.P., and Eberl G.. 2008. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J. Exp. Med. 205:1381–1393. 10.1084/jem.20080034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, A.-P., Constantinides M.G., Mathew R., Zuo Z., Chen X., Weirauch M.T., and Bendelac A.. 2016. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc. Natl. Acad. Sci. USA. 113:7602–7607. 10.1073/pnas.1601504113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, E., Treiner E., Duban L., Guerri L., Laude H., Toly C., Premel V., Devys A., Moura I.C., Tilloy F., et al. 2009. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 7:e54. 10.1371/journal.pbio.1000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, J.J., Toma C., Yu B., Zhang K., Omilusik K., Phan A.T., Wang D., Getzler A.J., Nguyen T., Crotty S., et al. 2017. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. 552:253–257. 10.1038/nature24993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand, S., Delanoye-Crespin A., Bressenot A., Huot L., Grandjean T., Peyrin-Biroulet L., Lemoine Y., Hot D., and Chamaillard M.. 2011. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc. Natl. Acad. Sci. USA. 108:9601–9606. 10.1073/pnas.1100981108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, D., Catherine Jin S.L., Conti M., Hatzelmann A., and Zitt C.. 2007. Differential expression and function of phosphodiesterase 4 (PDE4) subtypes in human primary CD4+ T cells: Predominant role of PDE4D. J. Immunol. 178. 4820–4831. 10.4049/jimmunol.178.8.4820 [DOI] [PubMed] [Google Scholar]
- Rahimpour, A., Koay H.F., Enders A., Clanchy R., Eckle S.B., Meehan B., Chen Z., Whittle B., Liu L., Fairlie D.P., et al. 2015. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J. Exp. Med. 212:1095–1108. 10.1084/jem.20142110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegert, P., Wanner V., and Bahram S.. 1998. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J. Immunol. 161. 4066–4077. 10.4049/jimmunol.161.8.4066 [DOI] [PubMed] [Google Scholar]
- Roth, A.C., Gonnet G.H., and Dessimoz C.. 2008. Algorithm of OMA for large-scale orthology inference. BMC Bioinformatics. 9:518. 10.1186/1471-2105-9-518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salou, M., Legoux F., Gilet J., Darbois A., du Halgouet A., Alonso R., Richer W., Goubet A.-G., Daviaud C., Menger L., et al. 2018. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J. Exp. Med. 216:133–151. 10.1084/jem.20181483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., and Bendelac A.. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403. 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seach, N., Guerri L., Le Bourhis L., Mburu Y., Cui Y., Bessoles S., Soudais C., and Lantz O.. 2013. Double-positive thymocytes select mucosal-associated invariant T cells. J. Immunol. 191. 6002–6009. 10.4049/jimmunol.1301212 [DOI] [PubMed] [Google Scholar]
- Srivastava, A., Morgan A.P., Najarian M.L., Sarsani V.K., Sigmon J.S., Shorter J.R., Kashfeen A., McMullan R.C., Williams L.H., Giusti-Rodríguez P., et al. 2017. Genomes of the mouse collaborative cross. Genetics. 206:537–556. 10.1534/genetics.116.198838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford, S.M., Rapini N., and Bottini N.. 2012. Regulation of TCR signalling by tyrosine phosphatases: From immune homeostasis to autoimmunity. Immunology. 137:1–19. 10.1111/j.1365-2567.2012.03591.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, R., Hartung A., Zehn D., Frentsch M., and Thiel A.. 2013. IL-12-mediated STAT4 signaling and TCR signal strength cooperate in the induction of CD40L in human and mouse CD8+ T cells. Eur. J. Immunol. 43:1511–1517. 10.1002/eji.201243218 [DOI] [PubMed] [Google Scholar]
- Street, K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., and Dudoit S.. 2018. Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 19:477. 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S.Y., Scanlon S.T., Griewank K.G., Constantinides M.G., Savage A.K., Barr K.A., Meng F., Luster A.D., and Bendelac A.. 2011. PLZF induces an intravascular surveillance program mediated by long-lived LFA-1-ICAM-1 interactions. J. Exp. Med. 208:1179–1188. 10.1084/jem.20102630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilloy, F., Treiner E., Park S.H., Garcia C., Lemonnier F., de la Salle H., Bendelac A., Bonneville M., and Lantz O.. 1999. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J. Exp. Med. 189:1907–1921. 10.1084/jem.189.12.1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle, K.D., Krovi S.H., Zhang J., Bedel R., Harmacek L., Peterson L.K., Dragone L.L., Lefferts A., Halluszczak C., Riemondy K., et al. 2018. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat. Commun. 9:2650. 10.1038/s41467-018-05026-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara, S., Song K., Farber J.M., and Love P.E.. 2002. Characterization of CCR9 expression and CCL25/thymus-expressed chemokine responsiveness during T cell development: CD3(high)CD69+ thymocytes and gammadeltaTCR+ thymocytes preferentially respond to CCL25. J. Immunol. 168. 134–142. 10.4049/jimmunol.168.1.134 [DOI] [PubMed] [Google Scholar]
- Ueno, T., Saito F., Gray D.H., Kuse S., Hieshima K., Nakano H., Kakiuchi T., Lipp M., Boyd R.L., and Takahama Y.. 2004. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J. Exp. Med. 200:493–505. 10.1084/jem.20040643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berge, K., Roux de Bézieux H., Street K., Saelens W., Cannoodt R., Saeys Y., Dudoit S., and Clement L.. 2020. Trajectory-based differential expression analysis for single-cell sequencing data. Nat. Commun. 11:1201. 10.1038/s41467-020-14766-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasanthakumar, A., Xu D., Lun A.T., Kueh A.J., van Gisbergen K.P., Iannarella N., Li X., Yu L., Wang D., Williams B.R., et al. 2017. A non-canonical function of Ezh2 preserves immune homeostasis. EMBO Rep. 18:619–631. 10.15252/embr.201643237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., D’Souza C., Lim X.Y., Kostenko L., Pediongco T.J., Eckle S.B.G., Meehan B.S., Shi M., Wang N., Li S., et al. 2018. MAIT cells protect against pulmonary Legionella longbeachae infection. Nat. Commun. 9:3350. 10.1038/s41467-018-05202-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Kjer-Nielsen L., Shi M., D’Souza C., Pediongco T.J., Cao H., Kostenko L., Lim X.Y., Eckle S.B.G., Meehan B.S., et al. 2019. IL-23 costimulates antigen-specific MAIT cell activation and enables vaccination against bacterial infection. Sci. Immunol. 4. 10.1126/sciimmunol.aaw0402 [DOI] [PubMed] [Google Scholar]
- Weinreich, M.A. and Hogquist K.A.. 2008. Thymic emigration: When and how T cells leave home. J. Immunol. 181. 2265–2270. 10.4049/jimmunol.181.4.2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X., Li K., Ma X., Liu B., He X., Yang S., Wang W., Jiang B., and Cai J.. 2019. Mucosal-associated invariant T cells expressing the TRAV1-TRAJ33 chain are present in pigs. Front. Immunol. 10:2070. 10.3389/fimmu.2019.02070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa, M., Jagannathan S., Vallabh S., Kartashov A.V., Chen X., Weirauch M.T., and Barski A.. 2020. AP-1 activity induced by co-stimulation is required for chromatin opening during T cell activation. J. Exp. Med. 217:e20182009. 10.1084/jem.20182009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia, L., and Oshlack A.. 2018. Clustering trees: A visualization for evaluating clusterings at multiple resolutions. Gigascience. 7:giy083. 10.1093/gigascience/giy083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Reynolds J.M., Chang S.H., Martin-Orozco N., Chung Y., Nurieva R.I., and Dong C.. 2009. MKP-1 is necessary for T cell activation and function. J. Biol. Chem. 284:30815–30824. 10.1074/jbc.M109.052472 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows basic descriptions of each scRNAseq dataset.
shows the differentially expressed genes in each UMAP cluster from each individual scRNAseq dataset.
shows the gene signatures used or defined in the study.
shows TradeSeq results from all developmental trajectories.
shows the conserved genes found modulated along MAIT cell development in all species.
shows the differentially expressed genes in UMAP clusters after integration of MAIT and conventional datasets.
shows the ortholog genes present in the studied species.
shows the preprocessing parameters used for integration of MAIT cells from the six individual datasets.
shows the differentially expressed genes in UMAP clusters after integration of MAIT from the six species.
shows the differentially expressed genes in UMAP clusters after integration of mouse MAIT datasets.
Data Availability Statement
The sequencing data generated in this study are available at GEO NCBI under the accession number GSE239558. All the other data are available in the main text or the supplementary materials.