Key Points
-
•
Parallel occurrence of resistance mechanisms, including preexisting epigenetic profiles and converging adaptation patterns across subclones.
-
•
Subclone-specific interactions of myeloma and bone marrow microenvironment cells.
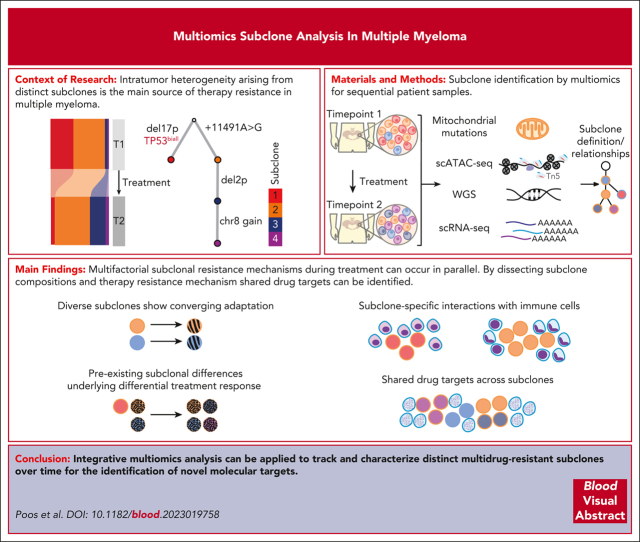
Visual Abstract
Abstract
Intratumor heterogeneity as a clinical challenge becomes most evident after several treatment lines, when multidrug-resistant subclones accumulate. To address this challenge, the characterization of resistance mechanisms at the subclonal level is key to identify common vulnerabilities. In this study, we integrate whole-genome sequencing, single-cell (sc) transcriptomics (scRNA sequencing), and chromatin accessibility (scATAC sequencing) together with mitochondrial DNA mutations to define subclonal architecture and evolution for longitudinal samples from 15 patients with relapsed or refractory multiple myeloma. We assess transcriptomic and epigenomic changes to resolve the multifactorial nature of therapy resistance and relate it to the parallel occurrence of different mechanisms: (1) preexisting epigenetic profiles of subclones associated with survival advantages, (2) converging phenotypic adaptation of genetically distinct subclones, and (3) subclone-specific interactions of myeloma and bone marrow microenvironment cells. Our study showcases how an integrative multiomics analysis can be applied to track and characterize distinct multidrug-resistant subclones over time for the identification of molecular targets against them.
Poos et al used a multiomics approach to define multiple myeloma subclones leading to chemotherapy resistance. The authors performed whole genome sequencing, single cell transcriptomics, chromosomal accessibility assays and utilized mitochondrial DNA mutations to demonstrate the complex heterogeneity of myeloma relapse. They identified 3 parallel mechanisms for relapse, including preexisting subclones with survival advantage, evolution of new subclones, and changing subclone interactions with the marrow microenvironment.
Introduction
Intratumor heterogeneity arising from genetically distinct subclones as well as their adaptation and evolution during treatment is the main source of therapy resistance.1 A paradigm disease for this process is multiple myeloma (MM), which is characterized by the clonal expansion of malignant plasma cells (PCs) in the bone marrow (BM).2, 3, 4, 5 MM intratumor heterogeneity as a clinical challenge becomes most evident during the course of several lines of treatment, when various resistance mechanisms accumulate.6 With the recent introduction of novel targeted immunotherapies7 for heavily pretreated relapsed or refractory MM (RRMM), the identification of common vulnerabilities of distinct multidrug-resistant subclones emerges as an urgent unmet need in the field. Toward this goal, MM subclones have been defined using single-cell RNA-sequencing (scRNA-seq) through the analysis of copy number aberrations (CNAs)8 and transcriptional clusters.9 Furthermore, mitochondrial DNA (mtDNA) mutations called from sc chromatin accessibility (scATAC-seq) data have been exploited to dissect and trace subclones in other hematologic malignancies.10, 11, 12 At the same time, genomic,13, 14, 15, 16, 17 transcriptomic,9,18, 19, 20 or epigenetic19 sequencing readouts revealed molecular mechanisms that govern the treatment response in MM. Moreover, scRNA-seq methods provided information on the interplay of MM cells with their BM microenvironment (BME), which have been shown to be an essential part of the disease phenotype.6,8,21 However, despite these advancements, it remains challenging to address the multifactorial nature of therapy resistance. Recent studies defined subclones based on a single readout and, therefore, focused only on individual aspects of molecular resistance. As such, it is not clear to what extend different resistance mechanisms can occur in combination in the same subclone and change during treatment.
In this study, we profiled serially collected BM samples from patients with RRMM using a multiomics approach that combines scRNA-seq, scATAC-seq, and bulk whole-genome sequencing (WGS) as well as bulk RNA-seq data. We show how copy number information from WGS and both sc modalities together with mtDNA mutations can be integrated to define subclones and their ancestral relationships at high resolution. Based on our matched transcriptome and epigenome analysis of individual subclones, we simultaneously address various layers of resistance, and specifically, we relate resistance to (1) subclones with preexisting epigenetic profiles that are associated with survival advantages, (2) converging phenotypic adaptation of genetically distinct subclones, and (3) subclone-specific interactions of MM and BME cells. From our analysis, it is apparent that multifactorial subclonal resistance mechanisms can occur in parallel, which has a number of implications for the development of strategies that target shared vulnerabilities across individual subclones.
Methods
Study cohort
Patient samples and related clinical information from both male (n = 7) and female (n = 8) patients with RRMM were obtained after written informed consent and in accordance with the Declaration of Helsinki. The study was approved by the ethics committee of the Medical Faculty at the University of Heidelberg (#S206/2011v13).
scRNA or ATAC-seq
Viably frozen CD138+-enriched or CD138+-depleted BM mononuclear cells were thawed at 37°C, resuspended in RPMI with 10% fetal bovine serum, and washed in cold phosphate-buffered saline, with cells being collected via centrifugation at 500g for 5 minutes. scRNA-seq was performed according to the Chromium Single Cell 3ʹ Reagent Kit version 2 user guide (10x Genomics). Generated gene expression libraries were pooled and paired-end sequenced (26 bp and 74 bp) on an Illumina NovaSeq 6000 S2. For scATAC-seq, cell pellets were carefully resuspended in an ice-cold NP-40 lysis buffer and spun down immediately. Nuclei were resuspended in the nuclei buffer provided by 10x Genomics, counted and subjected to Tn5 tagmentation. The subsequent steps were performed according to the manufacturer's instructions for the 10x Genomics Single Cell ATAC version 1.0. Generated libraries were pooled and paired-end sequenced (26 bp and 74 bp) on an Illumina NovaSeq 6000 S2.
Bulk WGS data analysis
Preprocessing was performed using the DKFZ OTP WGS pipeline.22 Calling of CNAs and single nucleotide variants (SNVs) was performed as previously described.23 For the supervised mutational signature analysis, we applied mmsig.24 In order to call SNVs in the mitochondrial genome, we established a bulk sequencing analysis workflow.
scRNA-seq data analysis
Preprocessing, data normalization, and CNA prediction were performed as previously described.8 Differential and pseudobulk gene expression analysis was performed with Find(All)Markers and AverageExpression in Seurat.25 Intercellular communication networks between tumor and BME cells were quantitatively inferred and analyzed with CellChat.26
scATAC-seq data analysis
Preprocessing was performed with CellRanger ATAC and downstream analysis with ArchR.27 All cells with a total number of <3000 fragments, a transcription start site enrichment score <7, a doublet enrichment score >6, and predicted doublet score >100 were excluded (supplemental Figure 1, available on the Blood website). PCs were distinguished from BME cells based on inferred gene-activity scores of specific marker genes, including TNFRSF17/CD138/SLAMF7/CD38, using MAGIC.28 Nonmalignant PCs from different patients clustered together (supplemental Figure 1F). Myeloma cells from all patients were normalized, dimensionality reduced using iterative latent semantic indexing,29,30 and clustered. Peak calling was performed with MACS2.31 Motif deviations were calculated based on the JASPAR database.32,33 For each transcription factor (TF) motif, a mean value was calculated over all cells per sample. Coaccessibility among genomic regions was separately calculated for time points and subclones, adjusting the ArchR-framework to sc resolution without the aggregation of cells, as described previously.34 We predicted CNAs by adopting an approach suggested by Lareau et al.10 Mitochondrial variants were called with mgatk.10
Subclone assignment
Subclones were identified separately in scRNA-seq and scATAC-seq data via supervised iterative hierarchical WGS-guided clustering of specific chromosomes or chromosomal regions using custom R-scripts (supplemental Figure 1K; supplemental Information; https://github.com/a-poos/MM_subclones).
For further details, see supplemental Information.
Results
A WGS-guided sc multiomics analysis integrates transcriptome and epigenome profiles of subclones
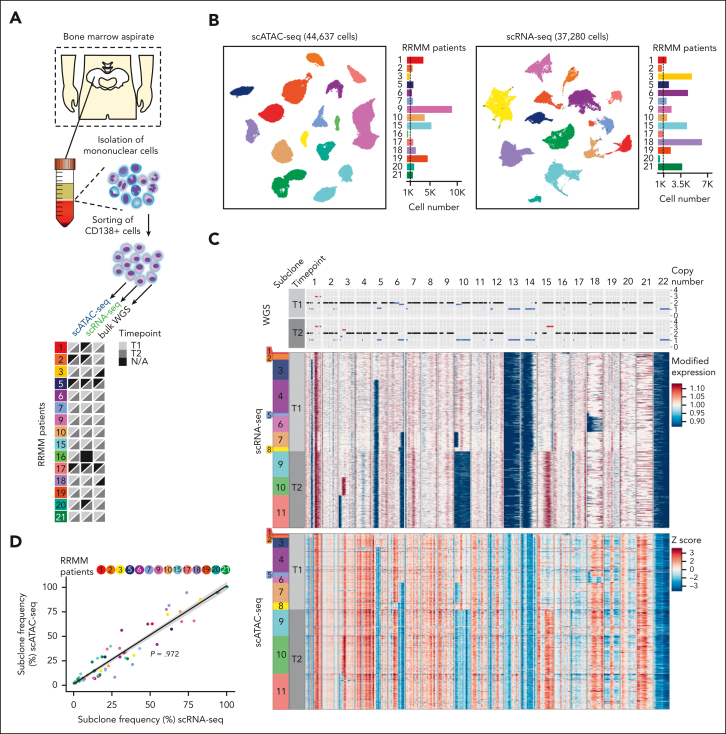
Previous studies have highlighted treatment-related subclonal dynamics in MM.13 However, how individual subclones respond to treatment and interact with the BME remains largely unknown, although this could provide crucial information of therapy resistance mechanisms. Accordingly, we performed droplet-based scATAC-seq and scRNA-seq in combination with WGS for CD138+-sorted BM aspirates of 15 patients with RRMM. Serial samples were collected for 12 of 15 patients before salvage therapy (T1) and at the time of subsequent relapse (T2; Figure 1A; supplemental Table 1), yielding 44 637 and 37 280 tumor cells after quality control, for scATAC-seq and scRNA-seq, respectively (Figure 1B; supplemental Figure 1; supplemental Tables 1 and 2). With clonal and subclonal CNAs occurring in virtually all patients with MM,36 we implemented a WGS-guided clustering strategy to resolve the subclonal structure in scRNA-seq and scATAC-seq data (Figure 1C; supplemental Figures 1M and 2; supplemental Table 3). The results obtained for both sc-seq modalities were highly correlated (ρ = 0.972, linear model; average deviation, ∼5), enabling us to combine transcriptional and epigenetic profiles of individual subclones (Figure 1D; supplemental Figure 2Q).
Figure 1.
A WGS-guided sc multiomic analysis integrates transcriptome and epigenome profiles of subclones. (A) Experimental overview of the patient cohort and available sequencing data. N/A indicates sample not available or sequencing yield <200 cells after quality control (QC). (B) UMAP embedding showing (left) scATAC- and (right) scRNA-seq data after QC, based on latent sematic indexing. Individual malignant PCs are colored according to patient identity. The bar plot on the right shows the number of cells per patient after QC. (C) Representative copy number profiles for the 22 autosomal chromosomes from (top) WGS, (middle) scRNA-, and (bottom) scATAC-seq of patient RRMM15. The color bar on the left indicates the respective subclonal population and sampling time point before relapse treatment (T1) and at subsequent relapse (T2). (Top) Copy number profile highlighting copy number gains (red), loss of heterozygosity (gray), and copy number losses (blue). Black represents a diploid copy number status. (Middle) Heatmap showing modified gene expression values generated using inferCNV.35 Gains (red) and losses (blue) are highlighted. The scales were limited to 0.9 and 1.1. (Bottom) Heatmap for z-scores from scATAC-seq, using the identical color code as shown for WGS and scRNA-seq. Z-scores were limited to 3 and –3. (D) Pearson correlation between the subclone frequency identified via scRNA- (x-axis) and scATAC-seq (y-axis) for all analyzed patients with both modalities. Each time point was plotted separately, with each subclone being colored based on the patient. Linear regression line is depicted in black, in which the gray area marks the 95% confidence interval. The deviation between the subclone frequency between scRNA-seq and scATAC-seq was from 0% to 23.9% (mean deviation = 5.08%).
mtDNA mutations further delineate the subclonal structure
Our initial approach assumed that MM cells with the same CNA profile belong to the same subclone. Next, we investigated whether mtDNA mutations further dissected CNA-based subclone definition in MM, because they have recently been proposed as an alternative approach for studying clonal dynamics in patients with chronic lymphocytic leukemia10,11 and those with acute myeloid leukemia.12 We called mtDNA mutations in the scATAC-seq data and confirmed the abundance of the respective variants in matched WGS (ρ = 0.98, linear model; supplemental Figure 3C-F). Overall, each patient with RRMM displayed a largely distinct mtDNA mutational profile (mean = 6 mutations; range 1-14 mutations; Figure 2A; supplemental Table 4). In line with previous reports, mutations followed a pattern equivalent to replication asymmetry and selective neutrality with C>T and T>C mutations11,37,38 (supplemental Figure 3D).
Figure 2.
Mitochondrial mutations further delineate the subclonal structure. (A) Mitochondrial DNA (mtDNA) mutations detected in scATAC-seq data across patients. Only mtDNA mutations with a mean heteroplasmy >1% in at least one of the paired samples were considered. The color bar at the top indicates the respective patient and sampling time point before treatment (T1) and at relapse (T2). (B) Inferred phylogenetic clonal tree for patient RRMM15. Top node denotes the common ancestor, from which subclones are derived according to their respective CNA and mtDNA mutation changes detected via WGS, scRNA-seq, and scATAC-seq. (Bottom) scATAC-seq heatmap of 1912 differential accessibility peaks and 3034 differential gene scores across subclones for patient RRMM15. Color indicates the column Z score of normalized peak accessibility and gene scores, respectively. (C) Heatmap of mtDNA mutations inferred from scATAC-seq data across subclones for patient RRMM15. Only mtDNA mutations with a mean heteroplasmy >1% in at least one of the 2 samples are shown. The proportion of cells per mtDNA mutation, subclone, and time point is shown. Clonal relationships are indicated by framing branches. Color legend indicates the heteroplasmy of mtDNA mutations. (D-E) UMAP plots of single RRMM cells clustered according to chromatin accessibility profiles for patient RRMM15. Examples for (D) subclone enriched and (E) branch enriched mtDNA mutations are shown. Each individual cell is colored according to (top) the respective subclone or (bottom) the heteroplasmy of the indicated mtDNA mutations, with red representing >10% and gray, 0%. (F) Heatmap of the most highly variable 72 TF motifs across all subclones of T1 and T2. Color indicates pseudobulk TF motif deviation score per subclone. (G) UMAP plots of single RRMM cells. Each cell is colored according to the TF motif deviation score of POU5F1B, TCF3, and IRF4. (H) Stability of mtDNA mutations from diagnosis to relapsed/refractory disease. Line plots showing the variant allele frequency of each mtDNA mutation in longitudinal samples of the indicated patients. Only mtDNA mutations with a mean heteroplasmy >1% in at least 1 of the paired samples (T1/T2) were considered. mtDNA mutations enriched in relapsed/refractory samples are colored in red.
For patient RRMM19, in line with WGS-based cancer clonal fractions for somatic nuclear mutations, 2 mtDNA mutations (15777G>A and 3749T>C) were distinctly enriched in single cells at T1 and T2, respectively. Thus, 2 subclones with the same CNA profile can be distinguished based on their mtDNA mutations (supplemental Figure 3A-B). We next compared subclones defined based on mtDNA mutations alone with those from our initial CNA-based assessment. Unique mtDNA mutations were detectable in 33% of the CNA-defined subclones (18 of 55; supplemental Figure 3G-L; supplemental Table 5), demonstrating that mtDNA mutations alone are not enough to resolve the subclonal architecture in RRMM.
By combining CNAs and mtDNA mutations, we were able to reveal further details of the subclonal architecture. Patient RRMM15 is the prime example for this finding (Figure 2B-G). In this patient, we initially identified 9 subclones using only the WGS-guided CNA approach (supplemental Figure 2P). By incorporating mtDNA mutation information, we identified 2 additional subclones in the scATAC-seq data (Figure 2B-D). These 2 subclones could be distinguished by the 2 lineage-defining mtDNA mutations, 10114T>C and 14769A>G (branch A) in scATAC-seq, and depicted unique chromosomal aberrations (supplemental Figure 3M). Importantly, these initially undetected CNAs could also be assigned to corresponding subclones in the scRNA-seq data (supplemental Figure 3N). In addition, we detected mtDNA mutations jointly enriched in multiple subclones that as such marked subclonal branches (eg, 15928G>A in subclones 1 and 2, 15356G>A and 12962G>A in subclones 7 and 8, and 13958G>A in subclones 9, 10, and 11; Figure 2B-C,E), which allowed us to delineate clonal relationships (Figure 2B-C,E; supplemental Figure 3G-L; supplemental Table 5). Cells with these jointly enriched mtDNA mutations segregated on the UMAP plot and marked distinct populations, including the branch-specific activity of POU5F1B, TCF3, and IRF4 TF motifs (Figure 2F-G), supporting the close relationship of those subclones.
Next, we assessed whether emerging relapse disease can be traced via mtDNA mutations. For this, sequencing data of tumor samples from 6 patients consecutively sampled over a time span of up to 3899 days were analyzed (Figure 2H; supplemental Figure 4A-C). Dynamic mtDNA mutations, including RRMM-enriched mutations, were detected in 4 of 6 patients (RRMM2/9/10/15; Figure 2H; supplemental Figure 4C). Remarkably, in these patients, RRMM-enriched mutations were seen up to 2464 days before T2 (Figure 2H). However, none of them were detectable at baseline (Figure 2H), suggesting clonal expansion of single tumor cells upon exposure to melphalan during first-line or relapse therapy (supplemental Table 1). Indeed, in our study, all patients with RRMM-enriched mtDNA mutations carried the single-base substitution MM1 mutational signature (supplemental Figure 4D), which is strongly linked with exposure to melphalan.39, 40, 41
Together, our results describe how mtDNA mutations can be best applied as lineage markers when combined with a CNA-based subclone assignment. This integrated approach yielded a comprehensive picture of the subclonal architecture with an average of 4 (range 1-11) subclones per patient (supplemental Figure 2Q) and matched transcriptome and epigenome profiles. In patients with paired samples, stable evolution, clonal selection, and branching evolution were seen in 5, 4, and 3 patients, respectively (supplemental Figure 2Q; supplemental Figure 5). It provides the foundation to investigate subclonal dynamics before and after treatment, as described in subsequent sections.
Parallel occurrence of resistance mechanisms across subclones
Having determined subclones with matching transcriptional and epigenetic profiles, we investigated the impact of treatment on individual subclones in all patients with multiple identical subclones that were present at both time points and defined based on our combined CNA- and mtDNA-based assignment (supplemental Figure 2Q). In patients with branching evolution, even at the sc level, we were unable to detect newly emerging subclones already at T1. Therefore, we focused on patients with stable evolution or clonal selection patterns to compare subclonal transcriptional and epigenetic changes over time (supplemental Figure 2Q). Although heterogeneous treatment regimens were applied (supplemental Table 1), a common pattern emerged in these patients: we found that the majority of therapy-induced changes in gene expression (median, 86%; range 64%-96%) and TF motif activity (median, 93%; range, 71%-95%) were shared across individual subclones for each patient (Figure 3A-G; supplemental Figure 6; supplemental Table 6). These converging adaptation mechanisms can be illustrated by known resistance mechanisms to 2 treatment approaches: proteasome inhibition via increased expression of heat-shock proteins (HSPs)42, 43, 44 (Figure 3B-D) and MEK/BRAF inhibition via NF-κB pathway45 upregulation (Figure 3E-G). In patient RRMM7, several HSPs known as resistance drivers, such as HSP90AA1,42, 43, 44 were significantly upregulated upon carfilzomib treatment in both subclones (5 of 25 genes; Padj ≤ .05; 1.5-fold enrichment both ways; Wilcoxon rank sum test; Figure 3C). In line with this finding, the motifs of TFs, including ZNF384, IRF1, and MEF2 family members, which can bind to the promoter of the upregulated HSPs,46 displayed a significantly higher activity after therapy in both subclones (Padj ≤ .05, Wilcoxon rank sum test; Figure 3D). In RRMM3, who was treated with MEK/BRAF inhibitors, both subclones demonstrated a significant upregulation of genes of the NF-κB pathway (18 of 37 upregulated genes; Padj ≤ .05; 1.5-fold enrichment; Figure 3F).47, 48, 49, 50 This was corroborated by a significantly higher activity of NF-κB motifs upon MEK/BRAF inhibition in both subclones (Padj ≤ .05; Figure 3G). Combined BRAF/MEK inhibition has previously been shown to induce high rates of clinical response.23,51 Accordingly, we analyzed an additional patient belonging to this treatment group, who exhibited major changes in its clonal composition (RRMM9; Figure 3H). Similar to RRMM3, 11 of 37 (30%) genes upregulated upon MEK/BRAF inhibition are part of the NF-κB pathway47 (Padj ≤ .05; log2 fold–change > 0.25; Wilcoxon rank sum test; Figure 3I). These genes included BCMA (TNFRSF17),51 CD79A,48,49 and BTG2.50 For RRMM9, only 1 of 4 subclones was still detectable after therapy (Figure 3H). This subclone depicted the highest NF-κB–family member motif activity of all subclones at T1 (Figure 3J). We conclude that, independent of subclonal composition, treatment resistance can be mediated either by converging phenotypic adaptation of all subclones or preexisting epigenetic profiles that do not necessarily have to be therapy-induced. This emphasizes the importance to assess MM on the level of individual subclones as well as the use of sc multiomics analysis for our understanding of the role of transcriptional and epigenetic features in treatment-induced resistance.
Figure 3.
Subclone-specific adaptation mechanisms to treatment reflect the clonal evolution pattern. (A) (Left) Percentage of differentially expressed genes between T1 and T2 that are shared across subclones of the same patient (Padj ≤ .05; 1.5-fold enrichment both ways; Wilcoxon rank sum test). (Right) Percentage of TF motif activity changes between T1 and T2 (Padj ≤ .05) that are shared across subclones of the same patient. Motif deviation scores were calculated based on all differential peaks, with Padj ≤ .05 and 1.5-fold enrichment (Wilcoxon rank sum test). (B,E,H) Bar plot depicting the subclone frequency of patients (B) RRMM7, (E) RRMM3, and (H) RRMM9 and time points (T1/T2) based on scATAC-seq data. Individual stacks are colored according to the respective subclone identity. (C,F,I) Gene expression changes for patients (C) RRMM7, (F) RRMM3, and (I) RRMM9 across subclones from T1 to T2. Color indicates pseudobulk gene expression of the differentially expressed genes between T1 and T2 (Padj ≤ .05; RRMM3+7: 1.5-fold enrichment both ways, RRMM9: log2-fold change >0.25 both ways; Wilcoxon rank sum test). (Left) Heatmap of scRNA-seq pseudobulk expression across subclones and timepoints. Significant (C) HSPs or (F,I) genes of the NF-κB pathway commonly upregulated in all subclones that have previously been described in the context of drug resistance are labelled. (Right) Violin plots showing normalized gene expression per time point and subclone for HSPs or genes belonging to the NF-κB pathway. (D,G,J) Epigenetic changes for patients (D) RRMM7, (G) RRMM3, and (J) RRMM9 across subclones from T1 to T2. Color indicates pseudobulk TF motif activity of the top 50 most highly variable TFs (plus NF-κB TFs motifs for RRMM9). (Left) Heatmap of the top 50 of 54 TF motifs, which show a significant higher or lower motif deviation score at relapse (T2) compared with that at T1. (D) TFs putatively binding to the HSPs promoter or (G,J) TFs from the NF-κB family commonly upregulated in both subclones are labelled. (Right) Violin plots showing scATAC-seq TF motif activity per time point and subclone for 2 or 3 exemplary TFs.
Differential treatment response is associated with subclone-specific BME interaction
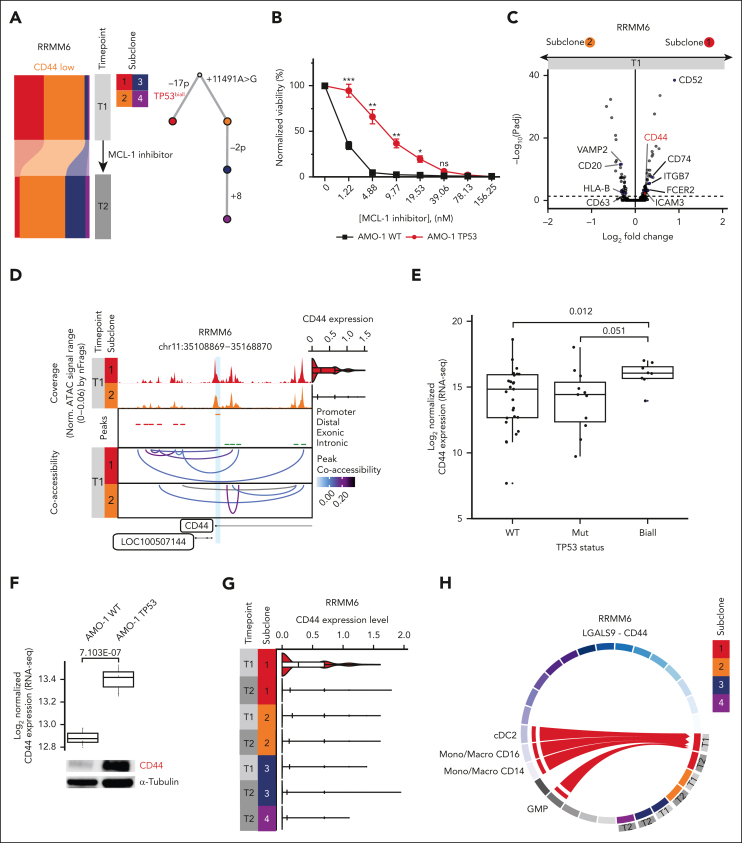
Based on the observation that subclonal properties can underlie differential treatment response, we next considered other patients in whom treatment-induced changes in the subclonal architecture were seen (supplemental Figure 2Q). We found that MCL-1 inhibition in patient RRMM6 resulted in the depletion of a subclone with a biallelic TP53 aberration (Figure 4A; supplemental Figures 4A and 5C). To address the molecular mechanism underlying this depletion, we first explored potential subclonal alterations of the target. We detected differences in the coaccessibility at the MCL-1 locus between the 2 main subclones (subclone 1 vs subclone 2) before initiation of MCL1 therapy. However, these differences did not result in significant MCL-1 expression changes (supplemental Figure 7). Next, we assessed whether TP53 aberrations directly affect the vulnerability of cells to MCL-1 induced cytotoxicity. Therefore, genetically engineered biallelic TP53 deleted + R175H mutated (del/mut)52,53 and wild-type (wt/wt) AMO-1 cell lines were exposed to different doses of an MCL-1 inhibitor in vitro (Figure 4B). In contrast to what is suggested by the findings for RRMM6, the biallelic TP53 AMO-1 cell line was less sensitive compared with the wt cell line (P < .05, Welch two-sample t test, Figure 4B; supplemental Figure 7B). Thus, inactivation of TP53 does not appear to underlie the depletion of subclone 1 in RRMM6. We, next, turned toward transcriptional differences between the 2 main subclones. The top differentially expressed genes between the 2 main subclones at T1 covered several surface markers (Figure 4C; supplemental Table 6). Among those, CD44 has been reported as a key cell adhesion–mediated drug resistance (CAM-DR)54 molecule and is a known mediator of tumor-BME interactions.55,56 In line with the differential gene expression analysis, a coaccessibility analysis of the scATAC-seq data showed a strong correlation between the CD44 promoter and a distal putative enhancer element for subclone 1 that was absent for subclone 2 at time point T1 (Figure 4D). Thus, activating an enhancer in one but not the other subclone could lead to the observed differences in CD44 expression. To test whether a higher CD44 expression is related to biallelic TP53 inactivation, we performed bulk RNA-seq in a set of 46 patients with RRMM.23,57 We found CD44 expression significantly increased in patients with TP53 biallelic mutation compared with that in patients with wt TP53 (P = .012, two-sided Wilcoxon rank sum test) as well as mutated TP53 (P = .051; Figure 4E). As a functional validation, we turned toward publicly available bulk RNA-seq data52,53 from genetically engineered biallelic TP53 del/mut (R175H) and wt/wt AMO-1 cell lines. We found a significantly higher CD44 gene expression in the cell line with inactivated TP53 than in wt TP53 (Figure 4F; adjusted P = 7.103e−07; log2 fold–change = 0.524, Wald test after Benjamini-Hochberg correction for multiple testing). This finding could also be confirmed using western blot analysis (Figure 4F). The high CD44 expression at T1 subsequently decreased upon MCL1-inhibition, suggesting a potential mechanism involving altered subclone-BM microenvironment interactions (Figure 4G). Indeed, CellChat26 predicted a specific interaction between subclone 1 and cDC2/GMPs/CD14+ as well as CD16+ monocytes via CD44 at T1 (Figure 4H; supplemental Table 7). Importantly, this respective interaction was lost upon MCL1-inhibition (Figure 4H). Together these results suggest differences in tumor-BME interactions among subclones with differential treatment response.
Figure 4.
Differential treatment response is associated with subclone-specific BME interaction. (A) (Left) Bar plot depicting the proportion of subclones per time point for patient RRMM6, detected via scATAC-seq. The individual stacks are colored according to the respective subclone identity. (Right) Phylogenetic tree for patient RRMM6. Top node at baseline denotes the most recent common ancestor with subsequent decedents being shown according to the combined CNA and mtDNA mutation subclone assignment. (B) Viability of the MM cell line AMO-1 based on normalized CellTiter-Glo luminescence reads on exposure to increasing concentrations of MCL-1 inhibitor MIK665 (Novartis) for 24 hours in cells with wt TP53 or biallelic TP53 (TP53 del/mut [R175H]). Data are represented as means ± standard deviation; n = 3. Welch two-sample t test: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. (C) Volcano plot summarizing the global changes in gene expression levels between subclones 1 and 2 for patient RRMM6. An absolute Padj value of .05 is indicated by a dashed line (Wilcoxon rank sum test). Surface markers are labeled. (D) Chromatin accessibility at the CD44 promoter plus 50 kb upstream and downstream in patient RRMM6. The CD44 promoter peak is highlighted in light blue. (Top) Aggregated pseudobulk scATAC-seq tracks at both timepoints. (Right) Violin plots showing normalized CD44 expression from scRNA-seq data per timepoint with individual cells plotted as dots. (Middle) Peaks are colored based on the location of the peak in either promoter (orange), distal (red), exonic (blue), or intronic (green) regions; (bottom) peak coaccessibility in the CD44 region at both timepoints colored based on the Pearson correlation coefficient. (E) CD44 expression based on bulk RNA-seq data in a set of 46 patients with RRMM with biallelic-inactivated (n = 8), monoallelic-depleted (n = 11) or wt TP53 (n = 27). (F) CD44 expression in genetically engineered biallelic TP53 del/mut (R175H) and wt/wt AMO-1 cell lines. (Top) Publicly available bulk RNA-seq data (GSE13234052,53) were analyzed for CD44 expression. Box plot of normalized expression values for n = 3 replicates. (Bottom) Western blot analysis of CD44 protein levels in the indicated MM cell lines. Tubulin was used as a loading control. This blot is representative of 3 independent experiments. (G) Violin and box-whisker plots showing normalized CD44 expression per timepoint and subclone for patient RRMM6. (H) Circos plot and chord interaction diagram of LGALS9-CD44–mediated interactions between subclones of patient RRMM6 and BME cells. The links start at a sender and end at a receiver. Sender-receiver interactions seen in all subclones as well as between BME cells are not indicated.
Shared BME interactions across subclones indicate potentially actionable therapeutic targets
The results described above prompted us to broaden our interaction predictions to all patients with at least 2 coexisting subclones per time point and matched scRNA-seq of the BME (n = 7). Across these patients, CellChat predicted, on average, 32 ligand-receptor interactions (range 23-45) between tumor and BME cells, of which ∼20% (median = 7; range 3-12) were not predicted in all subclones per patient (unshared interaction, Figure 5A-B; supplemental Table 7). The intercellular adhesion molecule (ICAM) signaling network was found to mediate most of the unshared interactions with the BM microenvironment across our patient cohort (Figure 5C-D; supplemental Figure 8A-B). In particular, the interaction between ICAM2 on the myeloma cells and several integrins (ITGB2/ITGAL/ITGAM/ITGAX) on the BME cells was predicted to be subclone-specific in 5 patients.
Figure 5.
Shared BME interactions across subclones indicate potentially actionable therapeutic targets. (A) Total number of cellular interactions between myeloma and BME cells in patients with RRMM. Predicted unshared (subclone-specific) and shared (interactions predicted in all subclones per patient) interactions are shown. (B) Total number of cellular interactions between specific subclones and BME cells in RRMM patients. Unshared interactions are colored based on the interaction type: (1) subclonal, interactions predicted to be specific for a subclone; (2) temporal, time point–specific interactions that are shared between ≥ 2 subclones; or (3) subclonal + temporal, interactions of a subclone that differ between time points T1 and T2. Sender-receiver interactions seen in all subclones as well as between BME cells are not indicated. (C) Circos plot and chord interaction diagram of ICAM1-(ITGAX + ITGB2) mediated interactions between subclones of patient RRMM15 and BME cells. The links start at a sender and end at a receiver. (D) Pathways based on CellChatDB26 ranked based on the number of subclone-specific interactions across our patient cohort. (E) Bulk RNA-seq differential expression (log2-fold change) of ICAM1 between time point T1 (before treatment initiation) and T2 (at relapse) of CD138+-enriched PCs isolated from serial samples of 16 patients with RRMM. (F) Chromatin accessibility at the ICAM1 promoter plus 50 kb upstream and downstream in patient RRMM15. The ICAM1 promoter peak is highlighted in light blue. (Top) Aggregated pseudobulk scATAC-seq tracks at both time points. (Right) Violin plots showing normalized ICAM1 expression per time point, with individual cells plotted as dots. (Middle) Peaks are colored based on the location of the peak in either promoter (orange), distal (red), exonic (blue), or intronic (green) regions. IRF4 ChIP-seq peaks from the MM cell line KMS12BM are shown in purple. (Bottom) Peak coaccessibility in the ICAM1 region at both time points colored based on the Pearson correlation coefficient. (G) The top TF motifs for patient RRMM15, which show a significantly higher motif deviation score at relapse (T2) compared with that at T1. TFs putatively binding to the ICAM1 promoter are labeled in blue.
All patients with longitudinal samples showed interactions that changed between time points (temporal changes; Figure 5B-C; supplemental Table 7). For example, in patient RRMM15, all subclones at T2 showed a temporal interaction mediated by the ICAM1 pathway, which was not detected at T1 (Figure 5C; supplemental Figure 8B). ICAM1-targeted immunotherapies in the form of anti-ICAM1 monoclonal antibody bersanlimab (BI-505, BioInvent)58 or CD38 × ICAM1 bispecific antibody59 are already being tested in clinical trials (NCT01838369) or have been suggested for MM and B-cell non-Hodgkin lymphoma.60, 61, 62 Accordingly, we further investigated ICAM1 temporal expression changes in an extended bulk RNA-seq data set of 16 patients with RRMM. We found that ICAM1 expression was significantly upregulated at T2 (P = .02, paired Wilcoxon signed rank test), particularly in patients RRMM6 and RRMM15 (Figure 5E). To further delineate the mechanism underlying increased ICAM1 expression, we assessed the chromatin accessibility at the ICAM1 locus through our scATAC-seq data. We observed a higher coaccessibility at the ICAM1 promoter region for cells at T2 than those at T1, suggesting a therapy-induced higher activity of ICAM1 (Figure 5F; supplemental Figure 8C). Moreover, RRMM cells also showed higher accessibility of TF motifs previously described in MM pathogenesis, including IRF,63, 64, 65, 66, 67, 68, 69, 70, 71, 72 NF-κB,73, 74, 75 and STAT76, 77, 78 family members at T2 (Figure 5G; supplemental Figure 8D). Within these families, IRF4, IRF1, RELA, STAT1, and STAT2 are known TFs that drive ICAM1 expression.46 This upregulation mechanisms derived from our scATAC-seq data was in agreement with chromatin immunoprecipitation assays followed by sequencing (ChIP-seq) analysis of the MM cell line KMS12BM.72 We found an overlap of the ICAM1 promoter peak with IRF4 ChIP-seq peaks (Figure 5F; supplemental Figure 8C). These orthogonal data sets suggest a differential ICAM1 expression driven by TFs that include IRF4.
In summary, we observed subclone-specific interactions with BME cells that change upon treatment in all patients. Some interactions, such as those mediated by ICAM1, were shared among subclones at a given time point and, thus, represent potentially actionable immunotherapy targets across subclones.
Discussion
The assessment of MM subclones and their molecular characterization during treatment are key for advancing patient stratification and therapy response prediction. In this study, we implemented an integrative multiomics approach that combines information from different bulk and sc readouts in a longitudinal analysis of RRMM to determine subclones at unprecedented resolution. Firstly, using WGS as a reference largely improved the accuracy of the CNA-based subclone definition from sc-seq data. Secondly, integrating 2 sc-seq approaches enabled us to further cross-validate our WGS-guided subclone definition and to analyze subclone-specific transcriptome and epigenome plasticity. Thirdly, including mtDNA mutations called from scATAC-seq resolved subclones with identical CNA profile. Our framework increased the ability to capture the subclonal diversity and their ancestry. For example, we identified 11 subclones for patient RRMM15 when using all readouts, although only 4 subclones were detected by clustering based on CNAs of all chromosomal regions. This was because CNA subclones from the same ancestral branch clustered together, suggesting that defining subclones based on transcriptional and epigenetic clusters alone may lead to an underestimation of the subclonal diversity. In addition, we conclude that mtDNA mutations on their own are insufficient for MM subclone definition, with only one-third of CNA-defined subclones in this study displaying unique mtDNA mutations. This observation could also be relevant for other hematologic malignancies, including chronic lymphocytic leukemia 10,11 and acute myeloid leukemia,12 in which mtDNA mutations have been used to define subclones.
Our approach provided a high-resolution map of the dynamic subclonal architecture with matched RNA and ATAC profiles for individual subclones before and after therapeutic interventions. We find that relapse can occur via different resistance mechanisms that include the expansion of subclones with preexisting epigenetic profiles associated with resistance, plasticity, and adaptation of gene expression programs in individual subclones as well as changes in MM-BME interactions. Plasticity in gene expression is in line with previously described resistance mechanisms to proteasome inhibitors via the upregulation of HSPs42, 43, 44 and activation of the NF-κB pathway45 upon MEK/BRAF inhibition. Thus, it will be important to monitor these pathways in relation to subclone composition for assessing treatment response.
In several patients, we found a branching evolution pattern as identified previously as the major evolution pattern in deep responding patients with MM.13 This pattern is characterized by depletion of previously dominant subclones and emergence of new subclones, which makes it challenging to study subclones over time in these patients. In this study, we were unable to detect the new subclones present at T2 at sc level at T1 in patients with branching evolution. To account for this issue, we envision our approach to be applied to tumor cells purified from patients receiving therapy, for example, as previously performed 48 hours after therapy start,79 or from patients tested positive for minimal residual disease at various time points during the course of disease.80 In patients with stable evolution and clonal selection, we were able to longitudinally track individual subclones over time. Future studies focusing on larger cohorts with uniform treatment will be crucial to understand common transcriptomic or epigenetic features among subclones that have disappeared or emerged.
Previous work has characterized the BME of different MM disease stages,6,8,81 providing evidence for deregulation of cell surface marker expression as resistance mechanisms.8,19 Through the combined analysis of tumor subclones and their BME, we show that individual subclones can have distinct interaction profiles with the BME, which further increases the complexity of MM heterogeneity. This includes subclone-specific expression and transcriptional plasticity for CD44, a key cell adhesion–mediated drug resistance54 molecule and promising immunotherapy target.55,56 In future studies, subclone-specific cellular interactions, such as CD44 on MM cells and LGALS9 on BME cells, could be validated by emerging spatially resolved techniques.82,83 Thus, at least in some patients, subclones might display individual resistance mechanisms and respond differently to immunotherapy.7,84,85 Consequently, to improve the outcome of available therapies, the identification of shared targets across subclones is essential and remains one of the biggest challenges to achieve long-term remissions. In this context, we show highly similar transcriptional and epigenetic changes upon treatment in patients with multiple distinct subclones, indicating converging adaptation mechanisms. The repertoire of shared treatment-induced changes included upregulation of ICAM1, another key mediator of cell-cell interactions.86 As BME cells nurture and facilitate the proliferation of MM subclones,87 such shared mediators could constitute actionable candidates for immunotherapeutic targeting of a heterogenous disease.2 In summary, we anticipate that the integrative multiomics analysis, introduced in this article for RRMM, will prove useful for therapy response prediction based on the subclone composition as well as the identification of novel molecular targets against multidrug-resistant subclones.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Acknowledgments
The authors are grateful to the patients for participation in this study and thank the Sample Processing Lab, the High Throughput Sequencing unit of the Genomics & Proteomics Core Facility and the Omics IT and Data Management Core Facility of the German Cancer Research Center (DKFZ), the DKFZ–Heidelberg Center for Personalized Oncology (DKFZ-HIPO) office, the Biobank Multiple Myeloma UKHD and the Myeloma Registry for excellent services.
Support and funding of the project via the NCT Heidelberg Molecular Precision Oncology Program through projects HIPO H067, K08K and K43R (H.G., K.R., M.S.R.) is gratefully acknowledged. N.P. was supported by a European Union Horizon Europe Marie Skłodowska-Curie Postdoctoral Fellowship (101068158); S.H., H.G., M.S.R., the Biobank Multiple Myeloma UKHD, and the Myeloma Registry are supported by the Dietmar-Hopp Foundation; K.R. is supported by the German Research Foundation (DFG) grant for subproject Z1 within SFB1074. Data storage service via SDS@hd is supported by the Ministry of Science, Research and the Arts Baden-Württemberg and the DFG through grants INST 35/1314-1 FUGG and INST 35/1503-1 FUGG.
Authorship
Contribution: A.M.P., N.P., M.S.R., K.R., and N.W. conceptualized the study; N.P., J.-P.M., S.M.T., K.B., A.B., and J.R. performed experiments; A.M.P., M.J.P., S.S., I.S., and N.W. analyzed data; A.M.P., N.P., M.J.P., K.R., and N.W. interpreted the data; N.P. drafted the figures; A.M.P. and N.P. wrote the original draft; I.S., M.J.P., M.S.R., K.R., and N.W. edited the manuscript; C.M.-T., H.G., M.S.R., and K.R. acquired funding; L.J., U.M., L.R., K.M.K., P.R., S.H., N.G., and M.S.R. acquired resources; O.S., M.S.R., K.R., and N.W. supervised the study; and all authors gave final approval.
Footnotes
∗A.M.P., N.P., and M.J.P. contributed equally to this work.
†M.S.R., K.R., and N.W. codirected this work.
The data produced in this study are deposited in the European Genome-phenome Archive (EGA) (accession numbers EGAS00001006538, EGAS00001005973, EGAS00001004805, and EGAS00001004363). The processed scATAC-seq and scRNA-seq data used in this study are publicly available at Gene Expression Omnibus (accession numbers GSE235783 and GSE161801) (see Resources Table in supplemental Information).
Data are available on request from the corresponding authors, Karsten Rippe (karsten.rippe@dkfz-heidelberg.de) and Niels Weinhold (niels.weinhold@med.uni-heidelberg.de).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Contributor Information
Karsten Rippe, Email: karsten.rippe@dkfz-heidelberg.de.
Niels Weinhold, Email: niels.weinhold@med.uni-heidelberg.de.
Supplementary Material
References
- 1.Marusyk A, Janiszewska M, Polyak K. Intratumor heterogeneity: the rosetta stone of therapy resistance. Cancer Cell. 2020;37(4):471–484. doi: 10.1016/j.ccell.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12(5):335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 3.Bolli N, Avet-Loiseau H, Wedge DC, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziccheddu B, Biancon G, Bagnoli F, et al. Integrative analysis of the genomic and transcriptomic landscape of double-refractory multiple myeloma. Blood Adv. 2020;4(5):830–844. doi: 10.1182/bloodadvances.2019000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manier S, Salem KZ, Park J, Landau DA, Getz G, Ghobrial IM. Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol. 2017;14(2):100–113. doi: 10.1038/nrclinonc.2016.122. [DOI] [PubMed] [Google Scholar]
- 6.Dutta AK, Alberge JB, Sklavenitis-Pistofidis R, Lightbody ED, Getz G, Ghobrial IM. Single-cell profiling of tumour evolution in multiple myeloma - opportunities for precision medicine. Nat Rev Clin Oncol. 2022;19(4):223–236. doi: 10.1038/s41571-021-00593-y. [DOI] [PubMed] [Google Scholar]
- 7.Da Vià MC, Dietrich O, Truger M, et al. Homozygous BCMA gene deletion in response to anti-BCMA CAR T cells in a patient with multiple myeloma. Nat Med. 2021;27(4):616–619. doi: 10.1038/s41591-021-01245-5. [DOI] [PubMed] [Google Scholar]
- 8.Tirier SM, Mallm JP, Steiger S, et al. Subclone-specific microenvironmental impact and drug response in refractory multiple myeloma revealed by single-cell transcriptomics. Nat Commun. 2021;12(1):6960. doi: 10.1038/s41467-021-26951-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen YC, Zada M, Wang SY, et al. Identification of resistance pathways and therapeutic targets in relapsed multiple myeloma patients through single-cell sequencing. Nat Med. 2021;27(3):491–503. doi: 10.1038/s41591-021-01232-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lareau CA, Ludwig LS, Muus C, et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat Biotechnol. 2021;39(4):451–461. doi: 10.1038/s41587-020-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penter L, Gohil SH, Lareau C, et al. Longitudinal single-cell dynamics of chromatin accessibility and mitochondrial mutations in chronic lymphocytic leukemia mirror disease history. Cancer Discov. 2021;11(12):30483063. doi: 10.1158/2159-8290.CD-21-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velten L, Story BA, Hernández-Malmierca P, et al. Identification of leukemic and pre-leukemic stem cells by clonal tracking from single-cell transcriptomics. Nat Commun. 2021;12(1):1366. doi: 10.1038/s41467-021-21650-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasche L, Schinke C, Maura F, et al. The spatio-temporal evolution of multiple myeloma from baseline to relapse-refractory states. Nat Commun. 2022;13(1):4517. doi: 10.1038/s41467-022-32145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keats JJ, Chesi M, Egan JB, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120(5):1067–1076. doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinhold N, Ashby C, Rasche L, et al. Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood. 2016;128(13):1735–1744. doi: 10.1182/blood-2016-06-723007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maura F, Bolli N, Angelopoulos N, et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat Commun. 2019;10(1):3835. doi: 10.1038/s41467-019-11680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corre J, Cleynen A, Robiou du Pont S, et al. Multiple myeloma clonal evolution in homogeneously treated patients. Leukemia. 2018;32(12):2636–2647. doi: 10.1038/s41375-018-0153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ledergor G, Weiner A, Zada M, et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat Med. 2018;24(12):1867–1876. doi: 10.1038/s41591-018-0269-2. [DOI] [PubMed] [Google Scholar]
- 19.Frede J, Anand P, Sotudeh N, et al. Dynamic transcriptional reprogramming leads to immunotherapeutic vulnerabilities in myeloma. Nat Cell Biol. 2021;23(11):1199–1211. doi: 10.1038/s41556-021-00766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldschmidt JM, Kloeber JA, Anand P, et al. Single-cell profiling reveals metabolic reprogramming as a resistance mechanism in BRAF-mutated multiple myeloma. Clin Cancer Res. 2021;27(23):6432–6444. doi: 10.1158/1078-0432.CCR-21-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sklavenitis-Pistofidis R, Aranha MP, Redd RA, et al. Immune biomarkers of response to immunotherapy in patients with high-risk smoldering myeloma. Cancer Cell. 2022;40(11):1358–1373.e8. doi: 10.1016/j.ccell.2022.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reisinger E, Genthner L, Kerssemakers J, et al. OTP: an automatized system for managing and processing NGS data. J Biotechnol. 2017;261:53–62. doi: 10.1016/j.jbiotec.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Giesen N, Chatterjee M, Scheid C, et al. A phase II clinical trial of combined BRAF/MEK inhibition for BRAF V600E-mutated multiple myeloma. Blood. 2023;141(14):1685–1690. doi: 10.1182/blood.2022017789. [DOI] [PubMed] [Google Scholar]
- 24.Rustad EH, Nadeu F, Angelopoulos N, et al. mmsig: a fitting approach to accurately identify somatic mutational signatures in hematological malignancies. Commun Biol. 2021;4(1):424. doi: 10.1038/s42003-021-01938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Granja JM, Corces MR, Pierce SE, et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat Genet. 2021;53(3):403–411. doi: 10.1038/s41588-021-00790-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dijk D, Sharma R, Nainys J, et al. Recovering gene interactions from single-cell data using data diffusion. Cell. 2018;174(3):716–729.e27. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satpathy AT, Granja JM, Yost KE, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol. 2019;37(8):925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granja JM, Klemm S, McGinnis LM, et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat Biotechnol. 2019;37(12):1458–1465. doi: 10.1038/s41587-019-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan A, Fornes O, Stigliani A, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2021;46(D1):D1284. doi: 10.1093/nar/gkx1188. D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50(D1):D165–D173. doi: 10.1093/nar/gkab1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallm J-P, Iskar M, Ishaque N, et al. Linking aberrant chromatin features in chronic lymphocytic leukemia to transcription factor networks. Mol Syst Biol. 2019;15(5):e8339. doi: 10.15252/msb.20188339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melchor L, Brioli A, Wardell CP, et al. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia. 2014;28(8):1705–1715. doi: 10.1038/leu.2014.13. [DOI] [PubMed] [Google Scholar]
- 37.Ju YS, Alexandrov LB, Gerstung M, et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife. 2014;3 doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan Y, Ju YS, Kim Y, et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat Genet. 2020;52(3):342–352. doi: 10.1038/s41588-019-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landau HJ, Yellapantula V, Diamond BT, et al. Accelerated single cell seeding in relapsed multiple myeloma. Nat Commun. 2020;11(1):3617. doi: 10.1038/s41467-020-17459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maura F, Weinhold N, Diamond B, et al. The mutagenic impact of melphalan in multiple myeloma. Leukemia. 2021;35(8):2145–2150. doi: 10.1038/s41375-021-01293-3. [DOI] [PubMed] [Google Scholar]
- 41.Rustad EH, Yellapantula V, Leongamornlert D, et al. Timing the initiation of multiple myeloma. Nat Commun. 2020;11(1):1917. doi: 10.1038/s41467-020-15740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jannuzzi AT, Arslan S, Yilmaz AM, et al. Higher proteotoxic stress rather than mitochondrial damage is involved in higher neurotoxicity of bortezomib compared to carfilzomib. Redox Biol. 2020;32 doi: 10.1016/j.redox.2020.101502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sha Z, Goldberg AL. Multiple myeloma cells are exceptionally sensitive to heat shock, which overwhelms their proteostasis network and induces apoptosis. Proc Natl Acad Sci U S A. 2020;117(35):21588–21597. doi: 10.1073/pnas.2001323117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah SP, Nooka AK, Jaye DL, Bahlis NJ, Lonial S, Boise LH. Bortezomib-induced heat shock response protects multiple myeloma cells and is activated by heat shock factor 1 serine 326 phosphorylation. Oncotarget. 2016;7(37):59727–59741. doi: 10.18632/oncotarget.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arozarena I, Wellbrock C. Overcoming resistance to BRAF inhibitors. Ann Transl Med. 2017;5(19):387. doi: 10.21037/atm.2017.06.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Q, Liu W, Zhang HM, et al. hTFtarget: a comprehensive database for regulations of human transcription factors and their targets. Genomics Proteomics Bioinformatics. 2020;18(2):120–128. doi: 10.1016/j.gpb.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.University, B NFKB target genes. https://www.bu.edu/nf-kb/gene-resources/target-genes/ Accessed 11 October 2022.
- 48.Luger D, Yang YA, Raviv A, et al. Expression of the B-cell receptor component CD79a on immature myeloid cells contributes to their tumor promoting effects. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0076115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monroe JG. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat Rev Immunol. 2006;6(4):283–294. doi: 10.1038/nri1808. [DOI] [PubMed] [Google Scholar]
- 50.Kawakubo H, Carey JL, Brachtel E, et al. Expression of the NF-kappaB-responsive gene BTG2 is aberrantly regulated in breast cancer. Oncogene. 2004;23(50):8310–8319. doi: 10.1038/sj.onc.1208008. [DOI] [PubMed] [Google Scholar]
- 51.Demchenko YN, Kuehl WM. A critical role for the NFkB pathway in multiple myeloma. Oncotarget. 2010;1:59–68. doi: 10.18632/oncotarget.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munawar U, Roth M, Barrio S, et al. Assessment of TP53 lesions for p53 system functionality and drug resistance in multiple myeloma using an isogenic cell line model. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-54407-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munawar U, Rasche L, Müller N, et al. Hierarchy of mono- and biallelic TP53 alterations in multiple myeloma cell fitness. Blood. 2019;134(10):836–840. doi: 10.1182/blood.2019000080. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki R, Ogiya D, Ogawa Y, Kawada H, Ando K. Targeting CAM-DR and mitochondrial transfer for the treatment of multiple myeloma. Curr Oncol. 2022;29(11):8529–8539. doi: 10.3390/curroncol29110672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bjorklund CC, Baladandayuthapani V, Lin HY, et al. Evidence of a role for CD44 and cell adhesion in mediating resistance to lenalidomide in multiple myeloma: therapeutic implications. Leukemia. 2014;28(2):373–383. doi: 10.1038/leu.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neri P, Bahlis NJ. Targeting of adhesion molecules as a therapeutic strategy in multiple myeloma. Curr Cancer Drug Targets. 2012;12(7):776–796. doi: 10.2174/156800912802429337. [DOI] [PubMed] [Google Scholar]
- 57.Giesen N, Paramasivam N, Toprak UH, et al. Comprehensive genomic analysis of refractory multiple myeloma reveals a complex mutational landscape associated with drug resistance and novel therapeutic vulnerabilities. Haematologica. 2022;107(8):1891–1901. doi: 10.3324/haematol.2021.279360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansson M, Gimsing P, Badros A, et al. A phase I dose-escalation study of antibody BI-505 in relapsed/refractory multiple myeloma. Clin Cancer Res. 2015;21(12):2730–2736. doi: 10.1158/1078-0432.CCR-14-3090. [DOI] [PubMed] [Google Scholar]
- 59.Chen X, Wong OK, Post L. CD38 x ICAM1 bispecific antibody is a novel approach for treating multiple myeloma and lymphoma [abstract] Blood. 2021;138(suppl 1):2413. doi: 10.1158/1535-7163.MCT-23-0052. [DOI] [PubMed] [Google Scholar]
- 60.Sherbenou DW, Su Y, Behrens CR, et al. Potent activity of an anti-ICAM1 antibody-drug conjugate against multiple myeloma. Clin Cancer Res. 2020;26(22):6028–6038. doi: 10.1158/1078-0432.CCR-20-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.ICAM1-targeted immunotherapy is effective in multiple myeloma. Cancer Discov. 2013;3:602. [Google Scholar]
- 62.Veitonmäki N, Hansson M, Zhan F, et al. A human ICAM-1 antibody isolated by a function-first approach has potent macrophage-dependent antimyeloma activity in vivo. Cancer Cell. 2013;23(4):502–515. doi: 10.1016/j.ccr.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 63.Agnarelli A, Chevassut T, Mancini EJ. IRF4 in multiple myeloma-Biology, disease and therapeutic target. Leuk Res. 2018;72:52–58. doi: 10.1016/j.leukres.2018.07.025. [DOI] [PubMed] [Google Scholar]
- 64.Shaffer AL, Emre NCT, Lamy L, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454(7201):226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu YX, Shi CX, Bruins LA, et al. Identification of lenalidomide resistance pathways in myeloma and targeted resensitization using cereblon replacement, inhibition of STAT3 or targeting of IRF4. Blood Cancer J. 2019;9(2):19. doi: 10.1038/s41408-019-0173-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mondala PK, Vora AA, Zhou T, et al. Selective antisense oligonucleotide inhibition of human IRF4 prevents malignant myeloma regeneration via cell cycle disruption. Cell Stem Cell. 2021;28(4):623–636.e9. doi: 10.1016/j.stem.2020.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ueno N, Nishimura N, Ueno S, et al. 1 acts as tumor suppressor for myeloma cells through direct transcriptional repression of IRF4. Oncogene. 2017;36(31):4481–4497. doi: 10.1038/onc.2017.79. [DOI] [PubMed] [Google Scholar]
- 68.Ohguchi H, Hideshima T, Bhasin MK, et al. The KDM3A-KLF2-IRF4 axis maintains myeloma cell survival. Nat Commun. 2016;7 doi: 10.1038/ncomms10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fedele PL, Liao Y, Gong JN, et al. The transcription factor IRF4 represses proapoptotic BMF and BIM to licence multiple myeloma survival. Leukemia. 2021;35(7):2114–2118. doi: 10.1038/s41375-020-01078-0. [DOI] [PubMed] [Google Scholar]
- 70.Morelli E, Leone E, Cantafio MEG, et al. Selective targeting of IRF4 by synthetic microRNA-125b-5p mimics induces anti-multiple myeloma activity in vitro and in vivo. Leukemia. 2015;29(11):2173–2183. doi: 10.1038/leu.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li N, Johnson DC, Weinhold N, et al. Multiple myeloma risk variant at 7p15.3 creates an IRF4-binding site and interferes with CDCA7L expression. Nat Commun. 2016;7 doi: 10.1038/ncomms13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin Y, Chen K, De Paepe A, et al. Active enhancer and chromatin accessibility landscapes chart the regulatory network of primary multiple myeloma. Blood. 2018;131(19):2138–2150. doi: 10.1182/blood-2017-09-808063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ordoñez R, Kulis M, Russiñol N, et al. Chromatin activation as a unifying principle underlying pathogenic mechanisms in multiple myeloma. Genome Res. 2020;30(9):1217–1227. doi: 10.1101/gr.265520.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12(2):115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hideshima T, Chauhan D, Richardson P, et al. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem. 2002;277(19):16639–16647. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- 76.Nelson EA, Walker SR, Kepich A, et al. Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3. Blood. 2008;112(13):5095–5102. doi: 10.1182/blood-2007-12-129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ogiya D, Liu J, Ohguchi H, et al. The JAK-STAT pathway regulates CD38 on myeloma cells in the bone marrow microenvironment: therapeutic implications. Blood. 2020;136(20):2334–2345. doi: 10.1182/blood.2019004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang Y-H, Molavi O, Alshareef A, et al. Constitutive activation of STAT3 in myeloma cells cultured in a three-dimensional, reconstructed bone marrow model. Cancers (Basel) 2018;10(6) doi: 10.3390/cancers10060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yaccoby S, Epstein J, Qu P, et al. Melphalan affects genes critical for myeloma survival, homing, and response to cytokines and chemokines. Blood. 2015;126(23):1808. [Google Scholar]
- 80.Goicoechea I, Puig N, Cedena MT, et al. Deep MRD profiling defines outcome and unveils different modes of treatment resistance in standard- and high-risk myeloma. Blood. 2021;137(1):49–60. doi: 10.1182/blood.2020006731. [DOI] [PubMed] [Google Scholar]
- 81.Manier S, Kawano Y, Bianchi G, Roccaro AM, Ghobrial IM. Cell autonomous and microenvironmental regulation of tumor progression in precursor states of multiple myeloma. Curr Opin Hematol. 2016;23(4):426–433. doi: 10.1097/MOH.0000000000000259. [DOI] [PubMed] [Google Scholar]
- 82.Zhao T, Chiang ZD, Morriss JW, et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature. 2022;601(7891):85–91. doi: 10.1038/s41586-021-04217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Black S, Phillips D, Hickey JW, et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat Protoc. 2021;16(8):3802–3835. doi: 10.1038/s41596-021-00556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Samur MK, Fulciniti M, Aktas Samur A, et al. Biallelic loss of BCMA as a resistance mechanism to CAR T cell therapy in a patient with multiple myeloma. Nat Commun. 2021;12(1):868. doi: 10.1038/s41467-021-21177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nijhof IS, Casneuf T, van Velzen J, et al. CD38 expression and complement inhibitors affect response and resistance to daratumumab therapy in myeloma. Blood. 2016;128(7):959–970. doi: 10.1182/blood-2016-03-703439. [DOI] [PubMed] [Google Scholar]
- 86.Frick C, Odermatt A, Zen K, et al. Interaction of ICAM-1 with beta 2-integrin CD11c/CD18: characterization of a peptide ligand that mimics a putative binding site on domain D4 of ICAM-1. Eur J Immunol. 2005;35(12):3610–3621. doi: 10.1002/eji.200425914. [DOI] [PubMed] [Google Scholar]
- 87.Moser-Katz T, Joseph NS, Dhodapkar MV, Lee KP, Boise LH. Game of bones: how myeloma manipulates its microenvironment. Front Oncol. 2020;10:625199. doi: 10.3389/fonc.2020.625199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.