Abstract
Thiol hypersensitivity in a mutant of Escherichia coli (IS16) was reversed by complementation with a plasmid that carried the ubiX gene. The mutant had low ubiquinone content. Complementation elevated the ubiquinone level and eliminated thiol hypersensitivity. Analysis of chromosomal ubiX genes indicated that both parent and mutant strains were ubiX mutants. The low ubiquinone content of IS16 was possibly caused by a ubiD ubiX genotype. A ubiA mutant also exhibited thiol hypersensitivity. Neither IS16 nor the ubiA mutant strain could produce alkaline phosphatase (in contrast to their parent strains) after 2 h of induction, thus showing Dsb− phenotypes. The phenomena of thiol hypersensitivity and low ubiquinone content may be linked by their connections to the periplasmic disulfide bond redox machinery.
The neutral water-soluble thiols 2-mercaptoethanol, 1-thioglycerol, and dithiothreitol inhibit gram-positive and gram-negative bacteria at millimolar concentrations (19, 21, 24). Although the precise mechanism of growth inhibition is not understood, it is known that several processes are affected. These include the lowering of intracellular concentrations of S-adenosylmethionine (14), inhibition of aerobic respiration (15), and possibly interference with the formation of disulfide bonds of periplasmic and outer membrane proteins (4, 27).
Exposure of aerobically growing Escherichia coli to exogenous thiols also results in a pleiotropic reductive stress response, which includes elevation of riboflavin and porphyrin syntheses (16), blockage of septum formation (18), and changes in the expression of hundreds of genes (17, 20).
In an attempt to look for genes which may be regulating this complex response, we searched for thiol-hypersensitive mutants. The search was based on the rationale that should such a gene(s) exist, its (their) inactivation would likely yield a thiol-hypersensitive phenotype.
MATERIALS AND METHODS
Reagents.
Amino acids, nucleotides, monosaccharides, coenzyme Q10, and dithiothreitol were purchased from Sigma Chemical Co., St. Louis, Mo. Hexane, petroleum ether, and 1-thioglycerol were bought from Aldrich Chemical Co., Milwaukee, Wis. The Wizard genomic purification kit and the Wizard Minipreps and Wizard PCR Preps DNA purification systems were from Promega Corp., Madison, Wis. Cloned Pfu DNA polymerase was obtained from Stratagene, La Jolla, Calif.
Bacterial strains and plasmids.
Strains and plasmids are listed in Table 1.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant description or genotype | Source |
|---|---|---|
| E. coli strains | ||
| THU | thy-43 his-67 pyr-37; colicin resistant | S. S. Cohen (7) |
| IS16 | Mutagenized (1) derivative of THU | This study |
| IS16B1 | IS16 with pPZ2 | This study |
| AN387 | F−; wild-type E. coli | Catherine Clarke |
| AN385 | F−; ubiA420 | Catherine Clarke |
| JCB 570 | MC1000 [araD139 Δ(araABC-leu)7679 galU galK Δ(lac)X74 rpsL thi] phoR Tet zih-12::Tn10 | James C. A. Bardwell |
| JCB 571 | JCB 570 dsbA::kan1 | James C. A. Bardwell |
| JCB 758 | JCB 570 dsbA::kan1 dsbB::kan5 | James C. A. Bardwell |
| JCB 789 | JCB 570 dsbB::kan5 | James C. A. Bardwell |
| Plasmids | ||
| pPZ2 | pBR322 with a 1,282-bp E. coli chromosomal insert into the BamHI site | This study |
| pHZ1 | pBR322 with a 699-bp E. coli THU chromosomal insert into the EcoRI-HindIII site | This study |
| pHZ2 | pBR322 with a 699-bp E. coli IS16 chromosomal insert into the EcoRI-HindIII site | This study |
Growth media.
All experiments were performed on aerobically growing cells, at 37°C, in Davis minimal salts medium (12). Glucose (0.3% [wt/vol]) was the carbon source. The media were supplemented for strains THU and IS16 and its derivatives with uracil plus thymine at 20 μg/ml each and with histidine at 40 μg/ml. The growth media for strain IS16B1 and all plasmid-bearing strains were supplemented with 100 μg of ampicillin per ml. dsb mutant strains were supplemented with 25 μg each of leucine, isoleucine, and valine per ml and 2.5 μg of cystine per ml.
Growth inhibition studies.
Growth inhibition studies were essentially carried out as described before (15, 21), except that the absorbance was monitored at 540 nm (A540).
Alkaline phosphatase assay.
Cultures were grown in Davis minimal salts medium until the A540 was 0.4 to 0.5. The bacteria were centrifuged and washed twice with low-phosphate-containing Davis medium (Davis medium containing only 10−4 M K2HPO4) at room temperature. The cells were resuspended in low-phosphate Davis medium containing all growth supplements at their initial volumes and were incubated at 37°C with shaking. At 20- or 30-min intervals, 0.1-ml aliquots were collected into 1.9 ml of ice-cold 1 M Tris-HCl buffer (pH 8.0). The enzyme assays were done as described by Brickman and Beckwith (6) except that A540 was used instead of A600.
Measurement of coenzyme Q8.
Coenzyme Q8 was routinely extracted by the method of Krivankova and Dadak (23). For its use as a high-performance liquid chromatography (HPLC) marker and for the purpose of quantitation, an extracted sample was further purified by thin-layer chromatography. On a Whatman linear-K silica gel thin-layer plate, with a 70:30 chloroform-light petroleum ether solvent mixture, coenzyme Q8 had an Rf value of 0.71. This substance was eluted from the plate with methanol, and its concentration was determined by its ΔA (oxidized-reduced)275, with 12.7 as the millimolar extinction coefficient (10).
Quantitation of coenzyme Q8 was performed by the HPLC method of Andersson (3). A Spherisorb octyldecyl silane 2 column (manufactured by Phenomenex Co., Torrance, Calif.) was used. The mobile phase was 10% (vol/vol) n-hexane in methanol, at a flow rate of 0.6 ml/min. Ubiquinone was detected at 275 nm, with a V4 variable UV-visible detector (manufactured by ISCO, Inc., Lincoln, Nebr.) under the control of Dynamax software from Rainin Instrument, Inc., Woburn, Mass.
PCR.
Genomic DNA was isolated with the Promega Wizard Genomic DNA kit. Cloned Pfu DNA polymerase purchased from Stratagene Cloning Systems was used. The ubiX genes of strains THU and IS16 were isolated by PCR, with the following primers: forward, 5′-ttcgaagcagtgcaacgtcagagcg-3′, and reverse, 5′-gaattcaaacagggcaacagcggag-3′. (The 5′ end of the forward primer was given a HindIII cutting site, and that of the reverse primer was given an EcoRI cutting site). The primers were synthesized on an Applied Biosystems 394 DNA synthesizer. The products of PCR were purified with the Promega Wizard PCR Preps DNA purification kit. They were sequenced on an Applied Biosystems Model 373 sequencer at the Center for Molecular Biology and Gene Therapy of Loma Linda University.
The PCR fragments were ligated into pBR322 plasmids by a standard protocol (30).
RESULTS
Isolation of a thiol-hypersensitive derivative of E. coli THU.
A culture of strain THU was mutagenized by nitrosoguanidine (1), and the cells were grown on minimal glucose plates containing 25 mM 1-thioglycerol. The smallest colonies were replica plated onto minimal medium- and minimal medium-plus-25 mM-1-thioglycerol-containing dishes. For controls, nonmutagenized THU colonies were replica plated as well. Colonies which were normal size on minimal plates but small on thioglycerol plates were isolated for further study. The isolate exhibiting the greatest hypersensitivity to 1-thioglycerol, named IS16, was used for further studies.
Strain IS16 had no additional growth requirements. Its mean growth rate (0.53 doubling/h) was lower than that of THU (0.87 doubling/h) in minimal glucose medium. It had a longer adaptation lag period when shifting down from glucose to lactate or malate and could not utilize succinate for growth.
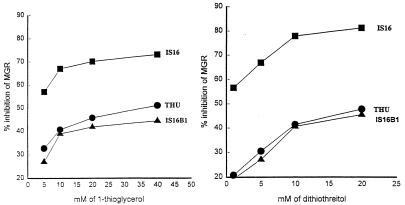
Inhibitory activities by exogenous 1-thioglycerol and dithiothreitol for strains THU and IS16 were compared. Percents inhibition of the mean growth rates were determined in a series of growth experiments, as described earlier (14), and the results are shown in Fig. 1. The experiments were performed on cells growing aerobically in minimal salts medium, with glucose as the carbon energy source. It can be seen that strain IS16 was substantially more inhibited than its parent strain THU by both thiols. This hypersensitivity was seen only under aerobic conditions. Thiols are less inhibiting to E. coli under anaerobic conditions (15), and both strains were inhibited to a similar extent (results not shown).
FIG. 1.
Inhibition by 1-thioglycerol and dithiothreitol of growth of strains THU, IS16, and IS16B1. The mean growth rates (MGR) of cultures in the presence of the indicated concentrations of thiols were compared with the MGR of untreated cultures.
Exogenous methionine provides significant protection against thiols (14) by increasing the S-adenosylmethionine pool of the cells. This protection remained for strain IS16, so that in the presence of 50 μg of methionine per ml thiol hypersensitivity could not be shown (results not shown).
Complementation of thiol hypersensitivity. Strain IS16 was transformed with a pBR322-based E. coli chromosomal library of BglII-cut fragments, and a transformant which lost its thiol hypersensitivity was isolated. This strain, IS16B1, contained a pBR322 plasmid with a 1,267-bp chromosomal fragment inserted in the BamHI site. This plasmid was named pPZ2.
The chromosomal insert in pPZ2 was sequenced, and with the help of the Blast program of the National Biomedical Library, its location on the E. coli chromosome was determined. It was at the 50-min segment of the E. coli chromosome and contained the 3′ end of the purF gene and the dedF gene (minus 8 codons from its 3′ end) (29) (Fig. 2). The nucleotide sequence of dedF shown in Fig. 2 is somewhat at variance with that in an earlier publication (29) but is in complete agreement with the latest sequence data (5).
FIG. 2.
Nucleotide sequence of dedF (ubiX) and its flanking regions. The BglII cutting sites are double underlined. Plasmid pPZ2’s chromosomal insert sequence begins at the 5′ end with G and ends at the 3′ end with A. The abbreviated 3′ end of purF shows only 69 nucleotides. The single-underlined sequences bracket the portions of the chromosome that were isolated by PCR from IS16 and THU and were inserted into pBR322 to form pHZ1 and pHZ2. Codon 98 in dedF (AGC), printed in boldface, is the site of the mutation in IS16 (AGA).
The dedF gene’s sequence is identical to that of the ubiX gene of Salmonella typhimurium, which codes for the enzyme polyprenyl p-hydroxybenzoate carboxylase (13, 25). Located at around 86 min of the E. coli chromosome is the ubiD gene, which codes for 3-octaprenyl-4-hydroxybenzoate decarboxylase, an enzyme functionally analogous to the product of Salmonella’s ubiX gene. It appears, therefore, that E. coli possesses two distinct genes, whose products catalyze the conversion of 3-octaprenyl-4-hydroxybenzoate to 2-octaprenyl phenol in the ubiquinone biosynthetic pathway (25). Studies with ubiD mutants suggest that in wild-type cells 80% of the enzyme activity is due to the ubiD gene product and 20% is due to the ubiX product (25). Although the ubiD mutation has been mapped to the 86-min segment of the E. coli chromosome (9), and the nucleotide sequence of that region is known (5, 11), the actual location of the gene is yet to be found (26).
Measurement of ubiquinone content of strain IS16.
Since the complementing plasmid contained only one functional gene, dedF (ubiX), a ubiquinone-biosynthetic gene, the ubiquinone contents of strains THU, IS16, and IS16B1 were measured. The results are shown in Fig. 3. Strain IS16 contained 85% less ubiquinone than did THU, and strain IS16B1, carrying multiple copies of ubiX, had 1.5 times as much ubiquinone.
FIG. 3.
Analysis of lipid extracts for ubiquinone by HPLC. Shown are the tracings of the absorption of the effluents at 275 nm. The arrows represent the peaks of ubiquinone Q8. From the areas under the peaks, the ubiquinone contents were calculated. The results are 0.21, 0.03, and 0.31 nmol/mg (dry weight) for strains THU, IS16, and IS16B1, respectively.
Search for the locus of mutation on ubiX in strain IS16.
Chromosomal ubiX genes of strain IS16 and strain THU were isolated by PCR, as described in Materials and Methods. Cloned Pfu polymerase was used, because of its considerably greater fidelity than Taq polymerase. The isolated fragments were sequenced from both directions. The ubiX sequence of IS16 differed from the published sequence only at codon 98 (shown in boldface in Fig. 2), where a serine residue (AGC) changed to arginine (AGA). Surprisingly, the ubiX sequence of the parent strain THU was identical to that of strain IS16.
To test the biological activity of the mutated ubiX gene, it was cloned into pBR322, and the new plasmid (pHZ1), when transformed into strain IS16, could not restore the ubiquinone levels of the parent strain. The thiol sensitivity of this strain was intermediate between that of IS16 and that of THU.
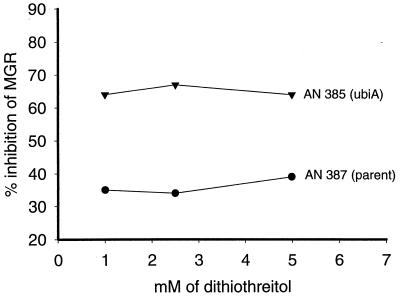
The correlation between low ubiquinone content and thiol sensitivity was extended to another E. coli strain, AN385, which carries a ubiA mutation. The ubiquinone content of this strain was 0.06 nmol/mg (dry weight), 26% of that of its parent strain AN387, which had 0.23 nmol/mg (dry weight). Their sensitivities to dithiothreitol were compared, and the results are shown in Fig. 4. It can be seen that the ubiA mutant strain was also hypersensitive to this thiol.
FIG. 4.
Inhibition by dithiothreitol of growth of strains AN387 and AN385 (ubiA). MGR, mean growth rate.
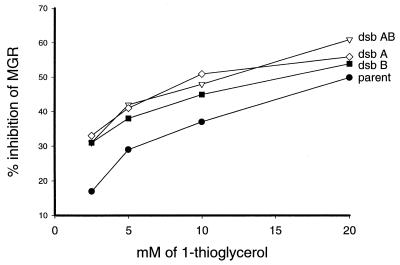
All previously reported thiol-hypersensitive E. coli strains belong to the dsb class of mutants (4, 27). These mutants are deficient in forming disulfide bonds in periplasmic proteins. The operational definition of hypersensitivity for these strains is their lack of ability to grow on Luria-Bertani agar plates containing 7 or 10 mM dithiothreitol (27). It was of interest to compare the thiol hypersensitivity of some dsb mutant strains with that of IS16, with liquid cultures and minimal growth medium. The results in Fig. 5 confirm the thiol hypersensitivities of the dsb mutant strains. A comparison with the results of Fig. 1 indicates that these cells were less sensitive to thiols than was strain IS16.
FIG. 5.
Inhibition of dsb mutant strains by 1-thioglycerol. Closed circles, parent JCB 570; open diamonds, dsbA mutant JCB 571; closed squares, dsbB mutant JCB 789; open inverted triangles, dsbAB mutant JCB 758. The cultures were grown in minimal medium as described in Materials and Methods. MGR, mean growth rate.
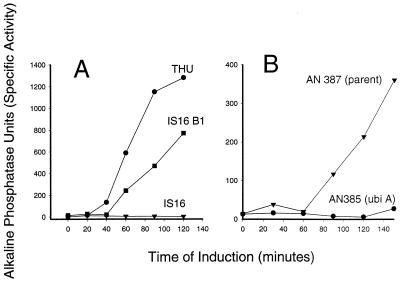
Strains IS16, IS16B1, and AN385 were tested for any evidence of the Dsb− phenotype. The test was the induction of the periplasmic enzyme, alkaline phosphatase, which needs two disulfide bonds for activity (2). The results are shown in Fig. 6. Strains IS16 and AN385 could not produce functional enzyme within 120 min of induction. In contrast, the parent strains and strain IS16B1 began making alkaline phosphatase within an hour. It was concluded that the low-ubiquinone-containing strains exhibited Dsb− phenotypes.
FIG. 6.
Induction of alkaline phosphatase in strains THU, IS16, IS16B1, AN385, and AN387. Cultures were grown in repressing, high-phosphate-containing medium until A540 was 0.4 to 0.5. Following washing and resuspension in low-phosphate, inducing medium, the enzyme levels were determined at the indicated times. (A) Strains THU (closed circles), IS16 (closed inverted triangles), and IS16B1 (closed squares). (B) Strains AN385 (closed circles) and AN387 (closed inverted triangles).
DISCUSSION
The low ubiquinone level of strain IS16 was puzzling, since E. coli has two genes, ubiD and ubiX, for the decarboxylation of octaprenyl p-hydroxybenzoate. It required multiple copies of the wild-type ubiX gene to increase this strain’s ubiquinone content. The ubiX gene of IS16 had a single deviation from the wild-type sequence, and increasing the number of copies of this variant gene was insufficient to elevate its ubiquinone content to near that of the wild type. It appears that the mutation at codon 98 of the ubiX gene practically inactivated the enzyme. Therefore, serine residue 98 is likely essential for the catalytic role of polyprenyl p-hydroxybenzoate lyase.
Since strain THU, the parent strain of IS16, also carried this mutation, both strains must have been ubiX negative. A reasonable guess is that the mutation in IS16, which resulted in very low ubiquinone levels, had to occur in the ubiD gene. This will have to be confirmed when the ubiD gene is identified.
Ubiquinone deficiency in E. coli is known to give rise to a pleiotropic phenotype of increased resistance to some antibiotics and heat inactivation; the inability to grow on succinate as the sole carbon source; and increased sensitivity to hydrogen peroxide, methyl methanesulfonate, and gamma radiation (8). The results reported here add another phenotypic characteristic to low ubiquinone content, thiol hypersensitivity.
Correlation between low ubiquinone content and thiol hypersensitivity was shown by (i) the loss of thiol hypersensitivity when the ubiquinone level was raised via complementation, (ii) the lessening of thiol hypersensitivity when ubiquinone levels were slightly elevated with multiple copies of the mutated ubiX gene, and (iii) the demonstration of thiol hypersensitivity in a ubiA mutant strain.
Previously published screens for thiol hypersensitivity, with dithiothreitol as the reducing agent, turned up dsbA dsbB, as well as trxA thioredoxin and trxB (thioredoxin reductase), mutants (27). The screen reported here employed 1-thioglycerol, a weaker reducing agent than dithiothreitol. This could be the explanation for finding a thiol-hypersensitive mutant which differed from the previously reported varieties.
A recent report suggests that there is a direct link between the respiratory chain and the DsbA-DsbB disulfide bond-forming system (22). Electrons removed from the periplasmic cysteine residues during disulfide bond formation pass first to the DsbA protein, then to the cytoplasmic membrane-associated DsbB protein, and finally to the respiratory chain. In support of their thesis, the authors showed that a ubiA menA double mutant, when deprived of para-hydroxybenzoate, slowed its growth (presumably because of reduction of ubiquinone content) and accumulated first reduced forms of DsbA and DsbB proteins and then the DsbA-DsbB complex.
This finding could explain how continually low ubiquinone levels such as those seen in strains IS16 and AN385 would reduce the ability of the respiratory chain to accept electrons from the DsbA-DsbB complex. Accumulation of reduced DsbA and DsbB proteins and the DsbA-DsbB complex in turn would result in a Dsb− phenotype.
The thiol hypersensitivity of dsb mutants could be the result of accumulation of reduced periplasmic and outer membrane proteins. Dysfunctional (reduced) DsbA proteins would prevent the appropriate folding and integration of outer membrane proteins (28). This scenario gains further credence from an earlier observation that exposure to 1-thioglycerol causes a rapid shift-up in the synthesis of outer membrane proteins OmpA and OmpF (17).
Thiol hypersensitivity of low-ubiquinone-containing cells would be the final outcome of the inability of the respiratory chain to absorb electrons from reduced Dsb proteins in the presence of excess exogenous thiols. Here millimolar concentrations of 1-thioglycerol or dithiothreitol outcompete the cysteine residues of periplasmic proteins for oxidation by DsbA. Growth inhibition would be for the same reasons as in the case of dsb mutants.
It was of interest that multiple copies of the ubiX gene in strain IS16B1 resulted in a 1.5-fold increase in ubiquinone content over that of the parent strain of THU. This implied that, in strain THU, the decarboxylation of octaprenyl p-hydroxybenzoate was rate limiting. However, since strain THU itself is a ubiX mutant, this observation may be applicable only to this strain. Further work will be required to determine whether this reaction is one of the flux-determining steps in ubiquinone biosynthesis in E. coli.
ACKNOWLEDGMENTS
We thank James Bardwell and Catherine Clarke for sending us bacterial strains and for helpful discussions. We also thank R. Meganathan for advising us and sharing unpublished results from his laboratory.
REFERENCES
- 1.Adelberg E A, Mandel M, Chen G C C. Optimal conditions for mutagenesis by N-methyl-N′-nitro-N-nitrosoguanidine in Escherichia coli K12. Biochem Biophys Res Commun. 1965;18:788–795. [Google Scholar]
- 2.Akiyama Y, Kamitani S, Kusukawa N, Ito K. In vitro catalysis of oxidative folding of disulfide-bonded proteins by Escherichia coli dsbA (ppfA) gene product. J Biol Chem. 1992;267:22440–22445. [PubMed] [Google Scholar]
- 3.Andersson S. Determination of coenzyme Q by non-aqueous reversed-phase liquid chromatography. J Chromatogr. 1992;606:272–276. doi: 10.1016/0021-9673(92)87036-8. [DOI] [PubMed] [Google Scholar]
- 4.Bardwell J C A, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 5.Blattner R R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletion and φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 7.Cohen S S, Sekiguchi M, Stern J L, Barner H D. The synthesis of messenger RNA without protein synthesis in normal and phage-infected thymineless strains of Escherichia coli. Proc Natl Acad Sci USA. 1963;49:699–703. doi: 10.1073/pnas.49.5.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collis C M, Grigg G W. An Escherichia coli mutant resistant to phleomycin, bleomycin, and heat inactivation is defective in ubiquinone synthesis. J Bacteriol. 1989;171:4792–4798. doi: 10.1128/jb.171.9.4792-4798.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox G B, Young I G, McCann L M, Gibson F. Biosynthesis of ubiquinone in Escherichia coli K-12; location of genes affecting the metabolism of 3-octaprenyl-4-hydroxybenzoic acid and 2-octaprenyl phenol. J Bacteriol. 1969;99:450–458. doi: 10.1128/jb.99.2.450-458.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crane F L, Barr R. Determination of ubiquinones. Methods Enzymol. 1971;18C:137–165. [Google Scholar]
- 11.Daniels D L, Plunkett III G, Burland V, Blattner F R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992;257:771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- 12.Davis B D, Mingioli E S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950;60:17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howlett B J, Bar-Tana J. Polyprenyl p-hydroxybenzoate carboxylyase in flagellation of Salmonella typhimurium. J Bacteriol. 1980;143:644–651. doi: 10.1128/jb.143.2.644-651.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Javor G T. Depression of adenosylmethionine content of Escherichia coli by thioglycerol. Antimicrob Agents Chemother. 1983;24:860–867. doi: 10.1128/aac.24.6.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javor G T. Inhibition of respiration of Escherichia coli by thioglycerol. Antimicrob Agents Chemother. 1983;24:868–870. doi: 10.1128/aac.24.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Javor G T. Thiol-stimulated secretion of riboflavin and porphyrins by Escherichia coli. FEMS Microbiol Lett. 1985;27:243–245. [Google Scholar]
- 17.Javor G T. Thiol-sensitive genes of Escherichia coli. J Bacteriol. 1989;171:5607–5613. doi: 10.1128/jb.171.10.5607-5613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Javor G T. Morphological changes in thioglycerol treated Escherichia coli. Curr Microbiol. 1990;20:57–62. [Google Scholar]
- 19.Javor G T. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C: American Society for Microbiology; 1995. Growth inhibition of Escherichia coli by dithiothreitol, abstr. A-43; p. 151. [Google Scholar]
- 20.Javor G T, Stringer C D, Ryu J I. Thiol-sensitive promoters of Escherichia coli. J Bacteriol. 1988;170:3291–3293. doi: 10.1128/jb.170.7.3291-3293.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen K, Javor G T. Inhibition of Escherichia coli by thioglycerol. J Bacteriol. 1981;19:556–561. doi: 10.1128/aac.19.4.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi T, Kishigami S, Sone M, Inkuchi H, Mogi T, Ito K. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc Natl Acad Sci USA. 1997;94:11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krivankova L, Dadak V. Semimicro extraction of ubiquinone and menaquinone from bacteria. Methods Enzymol. 1980;67:111–114. doi: 10.1016/s0076-6879(80)67016-5. [DOI] [PubMed] [Google Scholar]
- 24.Limbosch-Rolin S. Les effets du β-mercaptoethanol et du dithioglycol sur la croissance de Escherichia coli et de Saccharomyces cerevisiae. Exp Cell Res. 1963;24:61–72. doi: 10.1016/0014-4827(63)90358-6. [DOI] [PubMed] [Google Scholar]
- 25.Meganathan R. Biosynthesis of the isoprenoid quinones menaquinone (vitamin K2) and ubiquinone (coenzyme Q) In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 642–656. [Google Scholar]
- 26.Meganathan, R. 1997. Personal communication.
- 27.Missiakas D, Georgopoulos C, Raina S. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci USA. 1993;90:7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Missiakas D, Raina S. Protein folding in the bacterial periplasm. J Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nonet M L, Marvel C C, Tolan D R. The hisT-purF region of the Escherichia coli K-12 chromosome. J Biol Chem. 1987;262:12209–12217. [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]