Abstract
Meaningful advances in targeted therapy for head and neck squamous cell carcinoma (HNSCC) have been hampered by limited availability of robust preclinical models for translational research. Using an impressive array of in vitro and in vivo preclinical HNSCC models, Smith and colleagues demonstrate the efficacy of alpelisib and tipifarnib combination therapy through sustained mTOR inhibition in PIK3CA/HRAS-dysregulated HNSCC, including preliminary evidence of robust antitumor activity in a patient enrolled on a precision medicine trial. These data illustrate the value of preclinical avatars for informing biomarker-driven clinical trials to advance precision medicine in HNSCC and other cancers.
Comment
Precision medicine for cancer involves personalizing therapeutic selection based on a patient’s individual tumor characteristics. While highly attractive in theory, biomarker-guided targeted therapy options remain limited in many malignancies, including head and neck squamous cell carcinoma (HNSCC)1. To date, the only molecular targeted agents approved for recurrent/metastatic (R/M) HNSCC are cetuximab, a monoclonal antibody against the epidermal growth factor receptor (EGFR), and the immune checkpoint inhibitors (ICIs) pembrolizumab and nivolumab, which target programmed cell death protein 1 (PD-1). Unfortunately, these agents benefit a minority of patients with R/M HNSCC, and with the exception of the combined positive score for ICIs, lack validated biomarkers for thoughtful selection of patients most likely to achieve a clinically meaningful therapeutic response.
A critical step in advancing precision medicine for HNSCC patients is the development and characterization of robust preclinical models of HNSCC. Historically, the majority of laboratory cancer research (including in HNSCC) has relied on immortalized cancer cell lines, which are widely utilized for in vitro and in vivo (via cell line-derived xenografts) studies. Despite their ubiquity, it is well established that immortalized cancer cell lines develop unique biologic changes when propagated in tissue culture which are not seen in primary human tumors. This genetic divergence and clonal evolution is problematic for translating preclinical findings to the clinic, as foundational laboratory discoveries in cell lines may be predicated on models which no longer reflect human biology2. For this reason, there has been an impetus in recent decades to develop more representative preclinical models for facilitating the bench-to-bedside pipeline.
Patient-derived xenografts (PDXs) are preclinical cancer models which far more accurately reflect primary human tumors than cell line-derived xenografts. In the PDX model, patient tumor tissue obtained from a biopsy or resection specimen is implanted into highly immunodeficient mice. Tumor fragments are then serially passaged from mouse to mouse, maintaining the intratumoral heterogeneity characteristic of human cancers. Importantly, HNSCC PDXs display a remarkable resemblance to human HNSCC at the genetic, transcriptomic, and proteomic levels3–5. Thus, the rationale for using PDX models as preclinical avatars is strong. However, there are also limitations that must be acknowledged. The generation of PDXs can be logistically challenging, requiring access to high quality, often fresh, patient samples. In addition, both PDXs and human cell line-derived xenografts require propagation in immunocompromised hosts, which limits investigation of the role of the immune system on tumor growth and response to therapies.
The disappointing results of targeted therapy in HNSCC to date is likely due, at least in part, to a lack of biomarker-guided recommendations for optimal treatment selection. Therefore, studies using well characterized preclinical HNSCC avatars, such as PDXs, provide an optimal opportunity to translate laboratory discoveries into real-world clinical benefit. For example, while there are currently no reliable biomarkers for cetuximab response in HNSCC (including EGFR expression, mutation, or amplification), our group employed an unsupervised classifier approach using proteomics in HNSCC PDXs to identify a minimalist protein signature for cetuximab response prediction, which was validated in tumor samples from a HNSCC patient cohort6. In this way, robust preclinical insights can potentially inform the design of future biomarker-guided clinical trials. In this issue of Cancer Research, Smith and colleagues illuminate the promise of a novel combination therapy across a spectrum of preclinical HNSCC models, combining in vitro cell line data with in vivo experiments using both cell line-derived xenografts and PDXs, in addition to describing encouraging results in a patient case from an ongoing clinical trial7.
Smith et al. investigate a rational combination approach of alpelisib, a selective inhibitor of the p110α catalytic subunit of PI3Kα, and tipifarnib, a farnesyltransferase inhibitor targeting multiple oncogenic effector proteins, including HRAS and RHEB, an activator of mTOR signaling. Developing effective strategies for PI3K inhibition in HNSCC is particularly attractive, as the PI3K-AKT-mTOR signaling pathway is frequently dysregulated in HNSCC, most commonly due to activating mutations or amplifications of the PIK3CA gene8. Thus far, the efficacy of alpelisib monotherapy in HNSCC has been modest, in part due to reactivation of proteins downstream of PI3K (i.e., mTOR), and the activity of parallel signaling pathways (particularly RAS-RAF-MEK-MAPK) which also regulate PI3K9,10. Smith et al. first investigated the combination of alpelisib and tipifarnib in HNSCC cell lines with PIK3CA or HRAS dysregulation, revealing marked additive or synergistic inhibitory effects with the combination in cell lines harboring PIK3CA activating mutations/amplification or elevated HRAS activity. The synergy between these agents was also evident in vivo in a PIK3CA-mutant cell line-derived xenograft and nine PDXs with either PIK3CA mutation/amplification or elevated HRAS expression. The authors probe the putative molecular mechanism underlying this synergy, showing that tipifarnib treatment of PIK3CA/HRAS-dysregulated cell lines prior to alpelisib exposure dampened the rebound activation of AKT, mTOR, and ERK that is posited to beget resistance to alpelisib monotherapy. These in vitro observations were strengthened by in vivo cell line-derived xenograft findings that post-alpelisib mTOR rebound (assessed via 4EBP1 and S6K phosphorylation) was blunted by tipifarnib, leading to cell-cycle arrest and apoptosis induction. By demonstrating that small interfering RNA-mediated downregulation of HRAS and RHEB expression phenocopied the effects of tipifarnib on post-alpelisib mTOR rebound, the authors conclude that the defarnesylation of HRAS and RHEB are primarily responsible for tipifarnib’s anti-tumor effects and synergism with alpelisib (Figure).
Figure. Proposed mechanism of tipifarnib/alpelisib synergism in HNSCC.
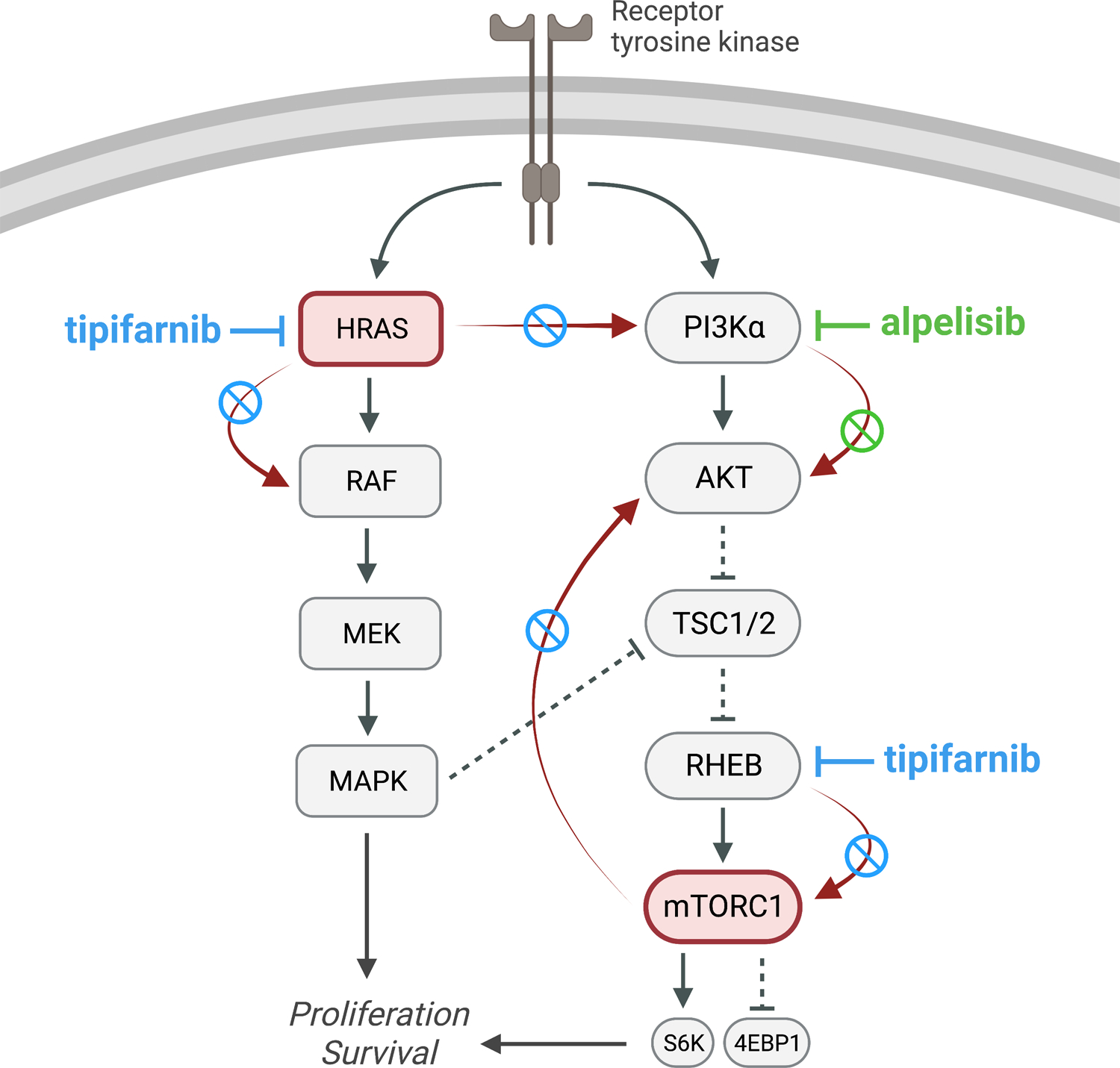
Alpelisib selectively targets PI3Kα, inhibiting downstream signaling of the AKT-mTOR pathway (green inhibitor line and green circle/slash overlying red arrow). Resistance to alpelisib monotherapy is multifactorial, including rebound activation of mTOR and parallel activation of the RAS-RAF-MEK-MAPK pathway, which also modulates PI3K (red arrows). Tipifarnib inhibits the activity of HRAS and RHEB (an mTORC1 activator) via defarnesylation, thereby blunting the rebound activation of mTOR and HRAS which are thought to principally underlie alpelisib resistance (blue inhibitor lines and blue circle/slashes overlying red arrows). Figure created with BioRender.
In total, all nine PIK3CA-altered or HRAS-overexpressing HNSCC PDXs evaluated in this study exhibited substantial inhibition, even regression, of tumor growth when treated with the combination of tipifarnib and alpelisib. Since scheduling of drug administration can play an important role in the effectiveness and practicality of combination treatment in humans, the authors also investigated different treatment schedules in the PDXs. Synchronous treatment was necessary for effective anti-tumor activity, and weekly on/off synchronous administration was supported. These findings suggest practical feasibility for delivery of this combination with flexible dosing protocols in the clinic. However, it should again be noted that the preclinical models used in this study lack intact immune systems, and findings obtained in syngeneic HNSCC models in immunocompetent hosts are likely to be informative.
The data obtained by Smith et al. in both mechanistic and therapeutic studies using well-characterized preclinical models have provided a strong rationale for evaluation of the tipifarnib/alpelisib combination in R/M HNSCC with HRAS overexpression and/or PIK3CA mutation/amplification, as is being assessed in the ongoing KURRENT-HN clinical trial (NCT049979012). The authors report remarkable results from one patient on this trial, a 35-year-old man with metastatic HPV+ HNSCC (harboring a R88Q PIK3CA mutation). This patient experienced a striking tumor reduction of two metastatic lung lesions following a 28-day cycle of tipifarnib and alpelisib, a response which was maintained as of the last trial follow-up prior to publication. While the case presented is certainly enticing, it is difficult to draw generalizable clinical conclusions based on a single patient, and testing in additional patients will be essential. However, if successful, this combination therapy has the potential to benefit up to approximately 45% of patients with R/M HNSCC, an estimation extrapolated by the authors from current mutational and gene expression patterns in The Cancer Genome Atlas (including 30% of R/M HNSCC with PIK3CA mutation/amplification and 15% with high HRAS mRNA expression).
Taken together, Smith and colleagues provide compelling preclinical evidence for the efficacy of tipifarnib and alpelisib combination therapy in PIK3CA/HRAS-dysregulated R/M HNSCC. More broadly, this study is a testament to the value of well-characterized preclinical cancer models (including PDXs) for both mechanistic investigation of therapeutic targeting strategies and clinical translation of plausible biomarkers from the bench to the bedside.
References
- 1.Wise-Draper TM, Bahig H, Tonneau M, Karivedu V & Burtness B Current Therapy for Metastatic Head and Neck Cancer: Evidence, Opportunities, and Challenges. Am Soc Clin Oncol Educ Book 42, 1–14 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Ben-David U et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 560, 325–330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuch LF et al. Head and neck cancer patient-derived xenograft models – A systematic review. Crit Rev Oncol Hematol 155, (2020). [DOI] [PubMed] [Google Scholar]
- 4.Facompre ND et al. Identifying predictors of HPV-related head and neck squamous cell carcinoma progression and survival through patient-derived models. Int J Cancer 147, 3236–3249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee RH et al. Therapeutic implications of transcriptomics in head and neck cancer patient-derived xenografts. PLoS One 18, e0282177 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouhaddou M et al. Caveolin-1 and Sox-2 are predictive biomarkers of cetuximab response in head and neck cancer. JCI Insight 6, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith AE et al. Tipifarnib Potentiates the Antitumor Effects of PI3Kα Inhibition in PIK3CA- and HRAS-Dysregulated HNSCC via Convergent Inhibition of mTOR Activity. Cancer Res 83, 3252–3263 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vander Broek R, Mohan S, Eytan DF, Chen Z & Van Waes C The PI3K/Akt/mTOR axis in head and neck cancer: functions, aberrations, cross-talk, and therapies. Oral Dis 21, 815–25 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Carracedo A et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest 118, 3065–74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright SCE, Vasilevski N, Serra V, Rodon J & Eichhorn PJA Mechanisms of Resistance to PI3K Inhibitors in Cancer: Adaptive Responses, Drug Tolerance and Cellular Plasticity. Cancers (Basel) 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


