Abstract
Purpose:
To demonstrate improvements in the precision of inverse probability-weighted estimators by use of auxiliary variables, i.e., determinants of the outcome that are independent of treatment, missingness or selection.
Methods:
First with simulated data, and then with public data from the National Health and Nutrition Examination Survey (NHANES), we estimated the mean of a continuous outcome using inverse probability weights to account for informative missingness. We assessed gains in precision resulting from the inclusion of auxiliary variables in the model for the weights. We compared the performance of robust and nonparametric bootstrap variance estimators in this setting.
Results:
We found that the inclusion of auxiliary variables reduced the empirical variance of inverse probability-weighted estimators. However, that reduction was not captured in standard errors computed using the robust variance estimator, which is widely used in weighted analyses due to the non-independence of weighted observations. In contrast, a nonparametric bootstrap estimator properly captured the precision gain.
Conclusions:
Epidemiologists can leverage auxiliary data to improve the precision of weighted estimators by using bootstrap variance estimation, or a closed-form variance estimator that properly accounts for the estimation of the weights, in place of the standard robust variance estimator.
Keywords: auxiliary variable, inverse probability weighting, missing data, precision, variance estimation, robust estimation, bootstrap, simulation
Estimators that incorporate inverse probability weights (IPW) are a multi-tool for the epidemiologist’s toolbox, offering a unified approach to handling confounding, selection bias, and explicit missing data, as well as generalizing study results. First appearing in the survey sampling literature, [1] and building on propensity score methods introduced by Rosenbaum and Rubin (1983), [2] IPW methods have received a great deal of attention in the epidemiologic literature since the development of marginal structural models by Robins, Hernán and Brumback (2000) [3-8]. IPW are typically constructed as the inverse probability of inclusion in a given group (e.g., the treated group, observed group, or uncensored group) conditional on the set of variables needed to block all backdoor paths between group membership and the outcome of interest [2,9,10]. However, Brookhart et al. (2006) suggest that including additional variables that are unrelated to group membership, but related to the outcome, when constructing IPW “will increase the precision of the estimated exposure effect without increasing bias” [11]. Such “auxiliary” variables could be useful for improving the statistical efficiency of parameter estimates, and have indeed long been used to improve precision in analyses that use multiple imputation to handle missing data [12,13].
However, the use of auxiliary variables is rarely reported in analyses involving IPW. One reason for the underutilization of auxiliary variables may be widespread use of the standard “robust” variance estimator to account for the non-independence of weighted observations [4,14]. The robust variance is a conservative approximation of the true variance of a weighted estimator, and is easier to estimate because it does not require knowledge of how the weights were generated. For this same reason, however – because it ignores the errors in the model for the weights – it hides any efficiency gains that can be achieved by using auxiliary variables to construct IPW.
In a survey of 411 original investigations published in the American Journal of Epidemiology and Epidemiology between January 2019 and December 2020, we found that 8% (n = 33) accounted for some type of bias using IPW. Of those, 11 reported using the standard robust variance estimator, 8 used a bootstrap estimator, 2 used generalized estimating equations with an unstructured covariance matrix, 1 used the expectation-maximization algorithm, and 11 used an unspecified variance estimator. None reported including auxiliary variables in the models used to estimate IPW.
Here, we demonstrate that precision can be gained by including auxiliary variables in IPW, but that such gains are lost by use of the standard robust variance estimator. This result has been demonstrated in the statistics literature in both observational and randomized settings, [15,16] but may be unfamiliar to epidemiologists. By drawing awareness to it, we aim to illustrate how epidemiologists can reduce uncertainty around estimates from epidemiologic studies through the use of auxiliary data, ultimately leading to better-informed clinical and public health decisions.
We demonstrate the inclusion of auxiliary variables in weighted analyses first using simulated data, and then using public data from the National Health and Nutrition Examination Survey (NHANES). In both cases, we consider a simple scenario in which we estimate the mean of a continuous outcome in a setting where a portion of outcomes are missing. We use IPW, constructed both with and without an auxiliary variable, to account for missing data. To assess gains in precision resulting from inclusion of the auxiliary variable, we apply the standard robust variance estimator and a nonparametric bootstrap variance estimator, and compare the estimated standard errors (SEs).
Methods
Simulation study
The aim of our simulation study was to compare the efficiency and confidence interval coverage of IPW estimators constructed with and without an auxiliary variable.
Data generation
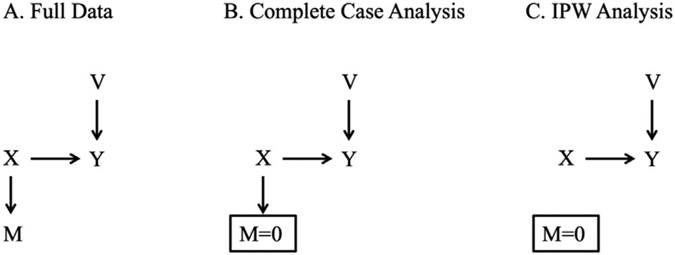
We simulated data according to the causal diagram depicted in Fig. 1 (Panel A). We generated continuous variable as normal conditional on covariate and auxiliary variable . and were generated as independent standard normal variables, associated with by the equation , where was a standard normal random error term. We generated an indicator that was missing, , as a Bernoulli random variable with mean . Thus, in the simulated data, about half of records were missing , and was missing at random conditional on . was strongly associated with (explaining 82% of the variance of ), but was not associated with except through .
Fig. 1.
Causal diagram depicting the relationships between variables in the simulation study.
For the sake of illustration, we simulated datasets of size 200 to obtain relatively large SEs. We simulated 2,000 datasets to obtain estimates of confidence interval coverage that are reliably within plus or minus 1 percentage point of 95% [17].
Statistical analysis
In each simulated dataset, we fit an intercept-only model to the full data, regardless of the value of . We expect this correct linear model, fit to the full data, to yield unbiased estimates with maximum precision, given the data and model. We use the estimates from this “full data analysis” as a benchmark for our simulation study. However, because such an analysis would be impossible to conduct in a real dataset where a portion of outcomes are missing, we also conducted a “complete case” analysis in each simulated dataset, fitting an intercept-only linear model to the observed data (i.e., where ). In the complete case analysis, we expect biased estimates of the mean of because is not missing completely at random (MCAR) [18]. That is, because is a cause of both and , the mean of in the subset of observations with will not equal the mean of in the full data with . As depicted in Fig. 1 (Panel B), conditioning on opens the backdoor path between and through .
Next, to obtain unbiased estimates of in the presence of missing data that are not MCAR, we used IPW [7,19]. IPW account for the bias induced by informative missing data by upweighting observations with non-missing to represent observations with missing that share similar values of . This procedure removes the association between and in the weighted pseudo-population, thereby blocking the open backdoor path between and through , as depicted in Fig. 1 (Panel C). First, we ignored the auxiliary variable and fit a standard IPW model in which each observation was weighted by the inverse probability of being a complete case conditional only on . Second, we included the auxiliary variable in the IPW, fitting a model in which each observation was weighted by the inverse probability of being a complete case conditional on and .
We constructed 95% confidence intervals around our estimates of , using standard closed-form estimators of the variance to obtain SEs. Specifically, for the full data and complete case analyses, we used the “naïve” model-based variance estimator; for IPW analyses, we used the robust (i.e., Huber-White) variance estimator [14]. In addition to the standard closed-form estimators, we also estimated the variance using a nonparametric bootstrap procedure [20]. From each of the 2,000 simulated datasets, we resampled the 200 observations with replacement 1,000 times, and fit all models in each resample to obtain estimates of . For each model, we used the standard deviation of the 1,000 estimates as an estimate of the SE of to calculate a Wald-type 95% CI. Finally, for each estimation approach, we calculated the empirical coverage of 95% CIs for .
Applied example
The aim of our applied example was to illustrate how auxiliary data can be used to improve the precision of epidemiologic estimates. We chose a widely used, publicly available dataset: the National Health and Nutrition Examination Survey (NHANES). We sought to estimate average exposure to acrylamide, a carcinogenic chemical found in heat-processed foods, cigarette smoke, and industrial products, among adult study participants from 2003-2016 [21]. This time frame was selected to include all NHANES survey waves that measured acrylamide concentration: 2003-2004, 2005-2006, 2013-2014, and 2015-2016.
We considered three auxiliary variables in our analysis: current smoking status, sex, and age. Acrylamide concentration is 3 to 4 times higher in smokers than in non-smokers, [22] and is higher among males and adults aged 20-59 compared with adults older than 59, possibly due to occupational exposure [23]. In the 2013-2016 waves of NHANES, acrylamide was only measured in a randomly selected subset of participants, so data on acrylamide concentration were missing at random by design, and our selected auxiliary variables were not associated with missingness.
Participants and measures
Acrylamide concentration (pmoL/G Hb) was measured in all participants aged 3 and older from 2003-2006, and in a one-third random sample of participants aged 6 and older from 2013-2016. Smoking data were collected from all participants aged 20 and older from 2003-2004 and from all participants aged 12 and older from 2005-2006 and 2013-2016. To simplify our example, we restricted our target population to NHANES participants aged 20 or older, and excluded participants with missing data on current smoking status (n=26), for a final analytic sample size of 21,482.
Statistical analysis
We repeated the complete case, standard IPW, and auxiliary-variable IPW analyses described above using the data from NHANES. Because acrylamide concentration was missing completely at random conditional on survey wave, we included survey wave as an indicator variable in the model for the standard IPW. We additionally included current smoking status, male sex, and age >59 as indicator variables in the model for the auxiliary-variable IPW. For simplicity, we ignored the complex survey design and sampling weights; we return to this issue in the Discussion.
All analyses were performed using SAS 9.4 (SAS Institute, Cary, NC). Code to generate the simulated data and conduct the analyses is provided in the Appendix.
Results
In our simulated data, the mean of was 1.0 (Table 1). Accordingly, in the “full data” analysis, our estimates of had a mean of 1.0. The estimated SEs were distributed around mean 0.2, matching the standard deviation of the across simulated datasets (i.e., the empirical SE). As expected, the “complete case” analysis yielded biased estimates of the mean of , with an average estimate of 0.68. We recovered unbiased estimates when we estimated using standard IPW conditional on . These estimates were, however, slightly less precise than those from the complete case analysis, with an empirical SE of 0.36 in the standard IPW analysis compared to 0.34 in the complete case analysis. When using IPW conditional on alone, the average robust and bootstrap estimates of the SE were similar to the empirical SE. Adding the auxiliary variable to the IPW resulted in similarly unbiased estimates of . Moreover, upon including in the IPW, the empirical SE and the average bootstrap SE were each reduced by more than 20%. However, the robust SE did not shrink with the addition of to the IPW. When was included in the IPW, the coverage of 95% CIs was conservative when constructed using the robust SE (0.99), and nominal when constructed using the bootstrap SE (0.95).
Table 1.
Estimated mean of across 5,000 simulated datasets under various estimation approaches
| Estimation Approach | Average | SD of (Empirical SE) |
Average (Estimated SE) |
95% Confidence Interval Coverage |
||
|---|---|---|---|---|---|---|
| Standard Closed-Form Estimatora |
Bootstrap Estimator | Standard Closed-Form Estimatora |
Bootstrap Estimator | |||
| Full Data | 1.00 | 0.23 | 0.23 | 0.23 | 0.95 | 0.95 |
| Complete Cases | 0.68 | 0.33 | 0.33 | 0.33 | 0.82 | 0.82 |
| Standard IPW | 1.00 | 0.36 | 0.35 | 0.35 | 0.95 | 0.94 |
| IPW with Auxiliary Variable | 1.00 | 0.27 | 0.36 | 0.27 | 0.99 | 0.95 |
, mean of ; SD, standard deviation; SE, standard error.
For the full data and complete case analyses, the standard closed-form estimator is the “naïve” model-based estimator. For both IPW models, the standard closed-form estimator is the robust (i.e., Huber-White) variance estimator [14].
In the data from NHANES, 44% of eligible participants were missing data on acrylamide concentration. Missingness was associated with survey wave: 19% of eligible participants were missing data on acrylamide concentration from 2003-2004 compared with 10% from 2005-2006, 71% from 2013-2014, and 70% from 2015-2016. Among complete cases, estimated average acrylamide concentration was upwardly biased due to a decrease in average acrylamide concentration over time and the higher proportion of participants missing data on acrylamide concentration in later waves (Table 2). Using IPW to correct for informative missingness, we estimated that average acrylamide concentration was 68.9 pmoL/G Hb (95% CI: 67.6, 70.2). When we included smoking status, sex, and age as auxiliary variables in the weights model, the bootstrap estimate of the SE decreased by 7%, from 0.68 to 0.63. This resulted in a slightly narrower estimated confidence interval around the estimate obtained using auxiliary-variable IPW compared with standard IPW. However, the robust standard error did not reflect the efficiency gain from including auxiliary variables in the IPW.
Table 2.
Estimated mean acrylamide concentration (pmoL/G Hb) among participants aged 20 and older in NHANES waves 2003-2016
| Estimation Approach | and 95% CI | ||||
|---|---|---|---|---|---|
| Standard Closed-Form Estimatora |
Bootstrap Estimator |
||||
| 95% CI | 95% CI | ||||
| Complete Cases | 72.33 | 0.62 | 71.12, 73.54 | 0.65 | 71.06, 73.60 |
| Standard IPW | 68.88 | 0.67 | 67.57, 70.19 | 0.68 | 67.54, 70.22 |
| IPW with Auxiliary Variable | 68.83 | 0.67 | 67.52, 70.14 | 0.63 | 67.59, 70.07 |
, mean of ; SD, standard deviation; SE, standard error
For the complete case analysis, the standard closed-form estimator is the “naïve” model-based estimator. For both IPW models, the standard closed-form estimator is the robust (i.e., Huber-White) variance estimator [14].
Discussion
In both our simulation study and our applied example, the standard robust variance estimator did not capture the improvement in precision achieved by adding auxiliary variables to an inverse probability-weighted model for the mean of a continuous outcome. In contrast, bootstrap variance estimation did capture the efficiency gain from including auxiliary variables in the IPW. This finding suggests that investigators wishing to improve the precision of their estimates can include auxiliary variables when estimating IPW. However, they should use alternative variance estimation methods, such as the nonparametric bootstrap, or a closed-form variance estimator that properly accounts for the estimation of the weights, [24] in place of the widely used robust variance estimator.
Our results are consistent with a prior simulation study by Williamson et al. (2012) [15]. Yet, a decade after the publication of their study in Statistics in Medicine, their findings remain under-appreciated by epidemiologists. We hope that our simple simulation study and applied example will help to amplify and translate their findings into practice. Below, we provide some intuition as to why auxiliary variables can improve the precision of weighted analyses, why the robust variance estimator hides those precision gains, and why the bootstrap variance estimator properly reflects them.
Because auxiliary variables are not causally associated with the exposure, selection, or missingness mechanism, including them in the model for the weights does not reduce bias due to confounding, selection, or missing data. However, including them in the weights model can improve the precision of estimates generated by the analysis model. Why? Even if auxiliary variables are not theoretically associated with the exposure, selection, or missingness mechanism in the target population, they are likely empirically associated in any given sample from that population [11]. That is, chance imbalances among variables in any given draw from the population create associations between the auxiliary variable and exposure, selection, or missingness, and accounting for those associations increases efficiency. The magnitude of the precision gains that can be achieved in practice depends on the strength of these chance associations, the relationship between the auxiliary variable and the outcome of interest, and the amount of confounding, selection, or missing data.
The standard robust SE is a simple sandwich SE, a conservative simplification of the proper (as in properly accounting for the estimation of the weights) sandwich SE [4,14]. Because the robust SE treats the weights as known rather than estimated, it ignores the random errors in the model for the weights, and thereby obviates any reduction in those errors that is achieved through the inclusion of auxiliary variables. Theory suggests that the bootstrap SE would be approximately equivalent to the proper sandwich SE [24]. However, perhaps because it is easier to implement, the robust SE appears to be more popular in the epidemiological literature than the bootstrap SE. Reticence to include auxiliary variables in IPW, when they are available, may be a consequence of widespread use of the robust SE, which, due to its conservative nature, hides any variance reduction that can be achieved by including auxiliary variables in IPW.
Unlike the standard robust SE, the nonparametric bootstrap SE appropriately captures the reduction in random error achieved by adding auxiliary variables to the model for the IPW [15,16]. A related benefit of the bootstrap SE is that it tends to yield confidence intervals with correct coverage, whereas the robust SE tends to yield conservative confidence intervals (i.e., intervals with greater than 95% coverage), as seen in Table 1 [25]. While the robust SE is easily estimated in statistical software and may be appropriate for many weighted analyses, investigators should be aware that it does not incorporate variance reductions due to the inclusion of auxiliary variables in IPW.
Epidemiologists who rely on data sources like surveys and medical records often have access to auxiliary data that are not fundamental to their research question. Rather than simply discarding those data, investigators may wish to leverage characteristics that are associated with the outcome of interest but not with the exposure, selection, or missingness mechanism to improve the precision of their estimates, using the appropriate variance estimator. Maximizing the precision of epidemiologic estimates is an important goal because reducing uncertainty around answers to questions of public health importance leads to better-informed decisions. Methods to improve precision using data that are already available should be welcomed and widely adopted by epidemiologists as cost-effective alternatives to increasing a study’s sample size. An incremental increase in precision achieved through the use of auxiliary data may represent thousands of dollars saved when compared to the recruitment and testing of additional participants to achieve the same degree of precision.
While bootstrap variance estimation may be too computationally intensive for some practical applications, procedures that output the proper sandwich variance estimate are available in some software, such as PROC CAUSALTRT in SAS (demonstrated in the Appendix). It is worth noting that the inclusion of auxiliary variables in IPW may not be beneficial when the analysis must account for a complex survey design. This is because estimation of the weights occurs outside of the built-in variance estimators that are available in survey analysis packages. We demonstrate this result in the Appendix using the NHANES data and the bootstrap variance estimator available in SAS’s PROC SURVEYREG.
As the use of auxiliary variables becomes more widespread, their capacity to improve precision should be further explored in a broader range of settings, including in studies of rare outcomes, studies with small sample sizes, and other settings where it is difficult to obtain precise estimates. The estimation of exposure-outcome associations is another important setting in epidemiology, and one which may entail unique considerations for the selection of auxiliary variables, such as the inclusion of variables that are effect modifiers on the scale of interest, rather than simply predictors of the outcome, in the model for the weights.
Acknowledgements
The authors are grateful to Ashley Naimi for helpful feedback on a draft.
Funding
This work was supported by the National Institutes of Health [R01-AI157758 to S.C. and J.E.; K01-AI125087 to J.E.].
Appendix
SAS Code to Simulate and Analyze Data
*IPW for Missing Data with an Auxiliary
Variable;xs
*Simulation Study;
*Simulate data: M <- X -> Y <- V;
data a;
call streaminit(7);
do j=1 to 2000;
do i=1 to 200;
v=rand("normal",0,1); *auxiliary variable;
x=rand("normal",0,1); *covariate;
y=1+1*x+3*v+rand("normal",0,1); *outcome;
m=rand("bernoulli",1/(1+exp(−(0+log(2)*x))));
*indicator that outcome is missing;
if m then y2=.; else y2=y;
output;
end;
end;
*Examine simulated data;
proc means data=a maxdec=3 fw=8;
title1 "IPW for Missing Data with an
Auxiliary Variable";
title2 "Data"; run;
*Draw bootstrap sample;
proc surveyselect data=a out=boot
(drop=expectedhits numberhits samplingweight
rename=(replicate=k))
seed=7 method=urs samprate=1 outhits
rep=200 noprint; strata j; run;
*Append original dataset, setting k=0;
data a; set a; k=0; run;
data data.boot; set a boot; run;
proc delete data=work.boot work.a; run;
*For each of k bootstrap samples of n=200,
estimate E[Y];
proc sort data=data.boot; by j k; run;
ods exclude all; options nonotes; run;
*Estimate IPMW;
proc logistic data=data.boot; by j k; model
m=x; output out=data.ipw p=pi; run;
data data.ipw (keep=j k i wt1); set data.ipw;
if m=0 then wt1=1/pi; else wt1=0; run;
proc logistic data=data.boot noprint; by j k;
model m=x v; output out=data.ipwa p=pi; run;
data data.ipwa (drop=_LEVEL_ pi); set
data.ipwa; if m=0 then wt2=1/pi; else wt2=0;
run;
proc delete data=data.boot; run;
proc sort data=data.ipw; by j k i; run; proc
sort data=data.ipwa; by j k i; run;
data data.boot; merge data.ipw data.ipwa; by
j k i; run;
proc delete data=data.ipw data.ipwa; run;
*Full Data;
proc genmod data=data.boot; model y=; by j
k;
ods output parameterestimates=m1(keep=j k
parameter estimate stderr); run;
data m1; length model $32.; set m1; if
parameter="Intercept" then do; model="Full
Data";
b=estimate; se=stderr;
lcl=b−1.96*se;
ucl=b+1.96*se;
if lcl<=1<=ucl then cover=1; else cover=0;
output; end;
keep model j k b se lcl ucl cover;
run;
*Complete Case;
proc genmod data=data.boot; model y2=; by j
k;
ods output parameterestimates=m2(keep=j k
parameter estimate stderr); run;
data m2; length model $32.; set m2; if
parameter="Intercept" then do; model="Complete
Case";
b=estimate; se=stderr;
lcl=b−1.96*se;
ucl=b+1.96*se;
if lcl<=1<=ucl then cover=1; else cover=0;
output; end;
keep model j k b se lcl ucl cover;
run;
*Standard IPW, Robust SE;
proc genmod data=data.boot; class i; model
y2=; weight wt1; by j k;
repeated subject=i/type=ind;
ods output geeemppest=m3(keep=j k parm
estimate stderr); run;
data m3; length model $32.; set m3; if
parm="Intercept" then do; model="Standard IPW";
b=estimate; se=stderr;
lcl=b−1.96*se;
ucl=b+1.96*se;
if lcl<=1<=ucl then cover=1; else cover=0;
output; end;
keep model j k b se lcl ucl cover;
run;
*IPW with Auxiliary Variable, Robust SE;
proc genmod data=data.boot; class i; model
y2=; weight wt2; by j k;
repeated subject=i/type=ind;
ods output geeemppest=m4(keep=j k parm
estimate stderr); run;
data m4; length model $32.; set m4; if
parm="Intercept" then do; model="IPW with
Auxiliary Variable";
b=estimate; se=stderr;
lcl=b−1.96*se;
ucl=b+1.96*se;
if lcl<=1<=ucl then cover=1; else cover=0;
output; end;
keep model j k b se lcl ucl cover;
run;
*IPW with Auxiliary Variable, Proper Sandwich
SE;
proc causaltrt data=data.boot method=ipw;
psmodel m=x v;
model y= / dist=normal;
by j k;
ods output CausalEffects=m5;
run;
data m5; length model $32.; set m5;
if Parameter="POM" and Level=0 then do;
model="PROC CAUSALTRT"; b=Estimate; se=stderr;
lcl=LowerWaldCL;
ucl=UpperWaldCL;
if lcl<1<ucl then cover=1; else cover=0;
output; end;
keep model j k b se lcl ucl cover;
run;
*Stack estimates from all models;
data m; set m1 m2 m3 m4 m5; run;
proc delete data=work.m1 work.m2 work.m3
work.m4 work.m5; run;
*Within jth simulated dataset, obtain BSE
(i.e. SD of E[Y]) across all k>0 bootstrap
samples;
proc sort data=m; by model j; run;
proc means data=m fw=8; by model j; where
k>0; var b; output out=bse; run;
data bse; set bse; if _STAT_="STD" then
output; rename b=bse; drop _TYPE_ _FREQ_
_STAT_; run;
*Merge BSEs with estimates of E[Y];
data estimates; set m; where k=0; run;
proc sort data=estimates; by model j; run;
proc sort data=bse; by model j; run;
data boot_out; merge estimates bse; by model
j; run;
*Calculate 95% CI and coverage using BSE;
data boot_out; set boot_out;
lcl_b=b−1.96*bse;
ucl_b=b+1.96*bse;
if lcl_b<=1<=ucl_b then cover_b=1; else
cover_b=0;
run;
ods exclude none;
*Print summary of estimates;
proc means data=boot_out n mean std stderr
maxdec=3 fw=8; where model="Full Data"; var b
se lcl ucl cover bse lcl_b ucl_b cover_b;
title2 "Full Data (Bootstrap SE) Summary";
run;
proc means data=boot_out n mean std stderr
maxdec=3 fw=8; where model="Complete Case";
var b se lcl ucl cover bse lcl_b ucl_b cover_b;
title2 "Complete Case (Bootstrap SE)
Summary"; run;
proc means data=boot_out n mean std stderr
maxdec=3 fw=8; where model="Standard IPW"; var
b se lcl ucl cover bse lcl_b ucl_b cover_b;
title2 "Standard IPW (Bootstrap SE) Summary";
run;
proc means data=boot_out n mean std stderr
maxdec=3 fw=8; where model="IPW with Auxiliary
Variable"; var b se lcl ucl cover bse lcl_b
ucl_b cover_b;
title2 "IPW with Auxiliary Variable
(Bootstrap SE) Summary"; run;
proc means data=boot_out n mean std stderr
maxdec=3 fw=8; where model="PROC CAUSALTRT";
var b se lcl ucl cover bse lcl_b ucl_b cover_b;
title2 "PROC CAUSALTRT (Bootstrap SE)
Summary"; run;
*IPW for Missing Data with an Auxiliary
Variable;
*Applied Example using Data from NHANES;
*Estimate mean concentration of acrylamide,
a probable carcinogen found in industrial
products, cigarette smoke, and foods cooked at
high temperatures;
*Acrylamide concentration was measured in all
participants ages 3+ in NHANES waves 2003-2004
and 2005-2006 and in a one-third random sample
ages 6+ in waves 2013-2014 and 2015-2016;
*Read in data;
%macro import(dataset=);
libname &dataset. xport "&data.\&dataset..xpt";
proc copy in=&dataset. out=data; run;
proc contents data=data.&dataset.; run;
proc sort data=data.&dataset.; by seqn; run;
%mend;
%import(dataset=DEMO_C);
%import(dataset=DEMO_D);
%import(dataset=DEMO_H);
%import(dataset=DEMO_I);
%import(dataset=L06AGE_C);
%import(dataset=AMDGYD_D);
%import(dataset=AMDGYD_H);
%import(dataset=AMDGYD_I);
%import(dataset=SMQ_C);
%import(dataset=SMQ_D);
%import(dataset=SMQ_H);
%import(dataset=SMQ_I);
data nhanes0304; merge data.DEMO_C
data.L06AGE_C data.SMQ_C; by SEQN; run;
data nhanes0506; merge data.DEMO_D
data.AMDGYD_D data.SMQ_D; by SEQN; run;
data nhanes1314; merge data.DEMO_H
data.AMDGYD_H data.SMQ_H; by SEQN; run;
data nhanes1516; merge data.DEMO_I
data.AMDGYD_I data.SMQ_I; by SEQN; run;
data nhanes (keep=SEQN SDDSRVYR RIDSTATR
RIAGENDR RIDAGEYR SMQ040 LBD: LBX: WT: SDM: smkr
male over59 eligible svywt m);
set nhanes0304 nhanes0506 nhanes1314
nhanes1516;
*Indicator of current smoking status;
if SMQ040 in(1,2) then smkr=1; else if
SMQ040=3 or SMQ020=2 then smkr=0;
*Indicator of male sex;
if RIAGENDR=1 then male=1; if RIAGENDR=2
then male=0;
*Categorize age as 0-59 or >=60;
if RIDAGEYR<60 then over59=0; if
RIDAGEYR>=60 then over59=1;
*Indicator of missing acrylamide
concentration;
if LBXACR=. then m=1; else m=0;
*Use 1/3 random sample weights for waves
2013-2014 and 2015-2016;
if WTSA2YR ne . then svywt=WTSA2YR; else
svywt=WTMEC2YR;
*Restrict target population to ages 20+ since
smoking data only collected from individuals
aged 20+ from 2003-2004;
*Exclude if missing smoking status;
if RIDAGEYR>=20 and smkr ne . then
eligible=1; else eligible=0;
run;
proc contents data=nhanes; run;
*Examine missing data on smoking status;
proc freq data=nhanes; tables smkr; where
RIDAGEYR>=20; run;
*Analytic sample size;
proc freq data=nhanes; tables eligible; run;
proc freq data=nhanes; tables
eligible*SDDSRVYR / nopercent norow; run;
***Examine associations among variables;
*Total n/% missing acrylamide concentration;
proc means data=nhanes n nmiss; var LBXACR;
where eligible; run;
*Missing acrylamide concentration is
associated with survey wave;
proc sort data=nhanes; by SDDSRVYR; run;
proc means data=nhanes n nmiss; var LBXACR;
by SDDSRVYR; where eligible; run;
*Missing acrylamide concentration is not
associated with current smoking status,
age>=60, or male sex;
proc freq data=nhanes; tables m*(smkr over59
male) / missing norow nopercent; where eligible;
run;
*Mean acrylamide concentration is associated
with survey wave (higher concentrations in
earlier waves);
proc sort data=nhanes; by SDDSRVYR; run;
proc means data=nhanes n nmiss mean; var
LBXACR; where eligible; by SDDSRVYR; run;
*Mean acrylamide concentration is strongly
associated with smoking status (higher
concentrations among smokers);
proc sort data=nhanes; by smkr; run;
proc means data=nhanes n nmiss mean; var
LBXACR; where eligible; by smkr; run;
*Mean acrylamide concentration is associated
with age (higher concentrations among adults
under 60);
proc sort data=nhanes; by over59; run;
proc means data=nhanes n nmiss mean; var
LBXACR; where eligible; by over59; run;
*Mean acrylamide concentration is associated
with sex (higher concentrations among males);
proc sort data=nhanes; by male; run;
proc means data=nhanes n nmiss mean; var
LBXACR; where eligible; by male; run;
*Draw bootstrap sample;
proc delete data=work.boot; run;
proc surveyselect data=nhanes out=boot
seed=7 method=urs samprate=1 outhits rep=1000
noprint; run;
data boot; set boot; rename replicate=k; drop
NumberHits; run;
*Append original dataset, setting k=0;
data nhanes; set nhanes; k=0; run;
data data.boot; set nhanes boot; run;
*Estimate IPMW;
proc logistic data=data.boot;
by k;
where eligible;
class SDDSRVYR;
model m=SDDSRVYR;
output out=ipw(keep=k seqn m pi) p=pi;
run;
data ipw; set ipw; if m=0 then wt1=1/pi;
else wt1=0; run;
proc means data=ipw n sum mean; var wt1; by
k; run;
*Estimate IPMW+Auxiliary Variable;
proc logistic data=data.boot;
by k;
where eligible;
class SDDSRVYR;
model m=SDDSRVYR smkr male over59;
output out=ipwa(keep=k seqn m pi) p=pi;
run;
data ipwa; set ipwa; if m=0 then wt2=1/pi;
else wt2=0; run;
proc means data=ipwa n sum mean; var wt2; by
k; run;
proc sort data=data.boot; by k seqn; run;
proc sort data=ipw; by k seqn; run; proc sort
data=ipwa; by k seqn; run;
data weighted;
merge data.boot ipw ipwa;
by k seqn;
ipmwt1=svywt*wt1;
ipmwt2=svywt*wt2;
run;
proc means data=weighted; var svywt ipmwt1
ipmwt2; where eligible; run;
*Estimate mean acrylamide concentration
(pmol/g Hb);
*First, ignore the complex survey design;
*Complete Case;
proc genmod data=weighted;
by k;
where eligible;
model LBXACR=;
ods output parameterestimates=cc(keep=k
parameter estimate stderr);
run;
data cc; length model $32.; set cc; if
parameter="Intercept" then do;
model="Complete Case"; b=estimate;
se=stderr;
lcl=b−1.96*se;
ucl=b+1.96*se;
output; end;
keep model k b se lcl ucl;
run;
*Standard IPW, Robust SE;
proc genmod data=weighted;
by k;
where eligible;
class seqn;
model LBXACR=;
weight wt1;
repeated subject=seqn/type=ind;
ods output geeemppest=ipmw(keep=k parm
estimate stderr); run;
data ipmw; length model $32.; set ipmw; if
parm="Intercept" then do;
model="Standard IPW"; b=estimate; se=stderr;
lcl=b−1.96*se;
ucl=b+1.96*se;
output; end;
keep model k b se lcl ucl;
run;
*IPW + Auxiliary Variable, Robust SE;
proc genmod data=weighted;
by k;
where eligible;
class seqn;
model LBXACR=;
weight wt2;
repeated subject=seqn/type=ind;
ods output geeemppest=ipmwa(keep=k parm
estimate stderr); run;
data ipmwa; length model $32.; set ipmwa; if
parm="Intercept" then do;
model="IPW + AV"; b=estimate; se=stderr;
lcl=b−1.96*se;
ucl=b+1.96*se;
output; end;
keep model k b se lcl ucl;
run;
*Stack estimates from all models;
data m; set cc ipmw ipmwa; run;
*Obtain BSE (i.e. SD of E[Y]) across all k>0
bootstrap samples;
proc sort data=m; by model; run;
proc means data=m fw=8; by model; where k>0;
var b; output out=bse; run;
data bse; set bse; if _STAT_="STD" then
output; rename b=bse; drop _TYPE_ _FREQ_
_STAT_; run;
*Merge BSEs with estimates of E[Y];
data estimates; set m; where k=0; run;
proc sort data=estimates; by model; run; proc
sort data=bse; by model; run;
data boot_out; merge estimates bse; by model;
run;
*Calculate 95% CI using BSE;
data boot_out;
length proc $32.;
set boot_out;
lcl_b=b−1.96*bse;
ucl_b=b+1.96*bse;
proc="GENMOD";
run;
*Now take into account the complex survey
design;
proc sort data=data.boot; by k seqn; run;
proc sort data=ipw; by k seqn; run; proc sort
data=ipwa; by k seqn; run;
data weighted2; merge data.boot ipw ipwa; by
k seqn; run;
proc means data=weighted2; var wt1 wt2; where
eligible; run;
*Complete Case;
proc surveyreg data=weighted2;
by k;
domain eligible;
weight svywt;
strata SDMVSTRA;
cluster SDMVPSU;
model LBXACR=;
ods output parameterestimates=cc2; run;
data cc2; length model $32.; set cc2;
where eligible;
if parameter="Intercept" then do;
model="Complete Case"; b=estimate;
se=stderr;
lcl=b−1.96*se;
ucl=b+1.96*se;
output; end;
keep model k b se lcl ucl;
run;
*IPMW;
proc surveyreg data=weighted2;
by k;
domain eligible;
weight ipmwt1;
strata SDMVSTRA;
cluster SDMVPSU;
model LBXACR=;
ods output parameterestimates=ipmw2; run;
data ipmw2; length model $32.; set ipmw2;
where eligible;
if parameter="Intercept" then do;
model="Standard IPW"; b=estimate; se=stderr;
lcl=b−1.96*se;
ucl=b+1.96*se;
output; end;
keep model k b se lcl ucl;
run;
*IPMW + Auxiliary Variable;
proc surveyreg data=weighted2;
by k;
domain eligible;
weight ipmwt2;
strata SDMVSTRA;
cluster SDMVPSU;
model LBXACR=;
ods output parameterestimates=ipmwa2; run;
data ipmwa2; length model $32.; set ipmwa2;
where eligible;
if parameter="Intercept" then do;
model="IPW + AV"; b=estimate; se=stderr;
lcl=b−1.96*se;
ucl=b+1.96*se;
output; end;
keep model k b se lcl ucl;
run;
/*IPMW + Auxiliary Variable - Bootstrap
Variance Estimation;
*This bootstrap variance estimator does
not approximate the proper sandwich variance
estimator because estimation of the weights
occurs outside of the bootstrap;
proc surveyreg data=weighted2
varmethod=bootstrap;
by k;
domain eligible;
weight ipmwt2;
strata SDMVSTRA;
cluster SDMVPSU;
model LBXACR=;
ods output parameterestimates=ipmwa_bs; run;
data ipmwa_bs; length model $32.; set
ipmwa_bs; if parm="Intercept" then do;
model="IPW + AV, Bootstrap"; b=estimate;
se=stderr;
lcl=b−1.96*se;
ucl=b+1.96*se;
output; end;
keep model k b se lcl ucl;
run;*/
*Stack estimates from all models;
data m2; set cc2 ipmw2 ipmwa2 /*ipmwa_bs*/;
run;
*Obtain BSE (i.e. SD of E[Y]) across all k>0
bootstrap samples;
proc sort data=m2; by model; run;
proc means data=m2 fw=8; by model; where
k>0; var b; output out=bse2; run;
data bse2; set bse2; if _STAT_="STD" then
output; rename b=bse; drop _TYPE_ _FREQ_
_STAT_; run;
*Merge BSEs with estimates of E[Y];
data estimates2; set m2; where k=0; run;
proc sort data=estimates2; by model; run;
proc sort data=bse2; by model; run;
data boot_out2; merge estimates2 bse2; by
model; run;
*Calculate 95% CI using BSE;
data boot_out2;
length proc $32.;
set boot_out;
lcl_b=b−1.96*bse;
ucl_b=b+1.96*bse;
proc="SURVEYREG";
run;
*Print all estimates;
data data.boot_1000; set boot_out boot_out2;
run;
proc print data=data.boot_1000; var proc
model b se lcl ucl bse lcl_b ucl_b; run;
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Horvitz DG, Thompson DJ. A Generalization of Sampling Without Replacement from a Finite Universe. J Am Stat Assoc 1952;47(260):663–85. [Google Scholar]
- [2].Rosenbaum PR, Rubin DB. The central role of the propensity score in observational studies for causal effects. Biometrika 1983;70(1):41–55. [Google Scholar]
- [3].Robins JM, Hernán MÁ, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology 2000;11(5):550–60. [DOI] [PubMed] [Google Scholar]
- [4].Hernán MA, Brumback B, Robins JM. Marginal structural models to estimate the causal effect of zidovudine on the survival of HIV-positive men. Epidemiology 2000;11(5):561–70. [DOI] [PubMed] [Google Scholar]
- [5].Cole SR, Hernan MA. Constructing Inverse Probability Weights for Marginal Structural Models. Am J Epidemiol 2008;168(6):656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Austin PC, Stuart EA. Moving towards best practice when using inverse probability of treatment weighting (IPTW) using the propensity score to estimate causal treatment effects in observational studies 2015;34:3661–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Seaman SR, White IR. Review of inverse probability weighting for dealing with missing data. Stat Methods Med Res 2011;22(3):278–95. [DOI] [PubMed] [Google Scholar]
- [8].Sato T, Matsuyama Y. Marginal Structural Models as a Tool for Standardization. Epidemiology 2003;14(6):680–6. [DOI] [PubMed] [Google Scholar]
- [9].Hernán MA, Robins JM. Causal Inference: What If. CRC Press; 2020. [Google Scholar]
- [10].Howe CJ, Cain LE, Hogan JW. Are All Biases Missing Data Problems? Curr Epidemiol Rep 2015;2(3):162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brookhart MA, Schneeweiss S, Rothman KJ, Glynn RJ, Avorn J, Stürmer T. Variable Selection for Propensity Score Models. Am J Epidemiol 2006;163(12):1149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Collins LM, Schafer JL, Kam C-M. A comparison of inclusive and restrictive strategies in modern missing data procedures. Psychol Methods 2001;6(4):330–51. [PubMed] [Google Scholar]
- [13].Lynch J. Efficiency Gains from Using Auxiliary Variables in Imputation. arXiv 2013;1311(5249):10. [Google Scholar]
- [14].Lin DY, Wei LJ. The Robust Inference for the Cox Proportional Hazards Model. J Am Stat Assoc 1989;84(408):1074–8. [Google Scholar]
- [15].Williamson EJ, Morley R, Lucas A, Carpenter JR. Variance estimation for stratified propensity score estimators. Stat Med 2012;31(15):1617–32. [DOI] [PubMed] [Google Scholar]
- [16].Williamson EJ, Forbes A, White IR. Variance reduction in randomised trials by inverse probability weighting using the propensity score. Stat Med 2014;33(5):721–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Morris TP, White IR, Crowther MJ. Using simulation studies to evaluate statistical methods. Stat Med 2019;38(11):2074–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Little RJA, Rubin. Statistical Analysis with Missing Data. 3rd Edition. John Wiley & Sons; 2019. [Google Scholar]
- [19].Greenland S, Finkle W. A Critical Look at Methods for Handling Missing Covariates in Epidemiologic Regression Analyses. Am J Epidemiol 1995;142(12):1255–64. [DOI] [PubMed] [Google Scholar]
- [20].Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Boca Raton, FL: Chapman & Hall, Inc; 1993. [Google Scholar]
- [21].National Toxicology Program Report on Carcinogens. Fifteenth Edition. Durham, NC: Department of Health and Human Services; 2021. [Internet]Available from: https://ntp.niehs.nih.gov/whatwestudy/assessments/cancer/roc/index.html. [Google Scholar]
- [22].National Biomonitoring Program Biomonitoring Summary: Acrylamide [Internet]. Centers for Disease Control and Prevention; 2017. Report No.: CAS No. 79-06-1Available from: https://www.cdc.gov/biomonitoring/Acrylamide_BiomonitoringSummary.html.
- [23].Centers for Disease Control and Prevention National Report on Human Exposure to Environmental Chemicals [Internet]. US Department of Health and Human Services; 2022. Available from: https://www.cdc.gov/exposurereport/. [Google Scholar]
- [24].Stefanski LA, Boos DD. The Calculus of M-Estimation. Am Stat 2002;56(1):29–38. [Google Scholar]
- [25].Austin PC. Variance estimation when using inverse probability of treatment weighting (IPTW) with survival analysis. Stat Med 2016;35(30):5642–55. [DOI] [PMC free article] [PubMed] [Google Scholar]