ABSTRACT
Fusaric acid (FA) is a mycotoxin produced by several Fusarium species. Burkholderia ambifaria T16 is a rhizosphere bacterium, able to use FA as sole nitrogen, carbon, and energy source. By screening a transposon insertional library, combined with proteomic analysis, genes and enzymes involved in the microbial degradation of FA were identified for the first time. A functional 2-methylcitrate cycle, an anaplerotic pathway where propionyl-coenzyme A (CoA) is converted to pyruvate and succinate, was shown to be essential for growth in the presence of FA. The proteomic profile of B. ambifaria T16 showed that more than 50 enzymes (including those belonging to the 2-methylcitrate cycle, fatty acid metabolism, valine catabolism, and flavin biosynthesis) were significantly more abundant when growing on FA than on citrate. Flavin mononucleotide (FMN)-dependent luciferases like monooxygenase (LLM) are shown to catalyze the pyridine-ring cleavage reaction of several N-heterocyclic compounds. Deletion of a gene encoding a predicted LLM enzyme that was highly upregulated during growth on FA, completely abolished the capability of B. ambifaria T16 to grow with this mycotoxin as sole nitrogen, carbon, and energy source. Re-introduction of the wild type gene was able to restore growth. The mentioned gene is part of a gene cluster of unknown function that we termed fua, due to its probable role in fusaric acid catabolism. Our results suggest that the LLM encoded in the fua cluster catalyzes the pyridine-ring opening reaction during FA degradation, and that propionyl-CoA is one of the intermediates of FA catabolism in B. ambifaria T16.
IMPORTANCE
Fusaric acid (FA) is an important virulence factor produced by several Fusarium species. These fungi are responsible for wilt and rot diseases in a diverse range of crops. FA is toxic for animals, humans and soil-borne microorganisms. This mycotoxin reduces the survival and competition abilities of bacterial species able to antagonize Fusarium spp., due to its negative effects on viability and the production of antibiotics effective against these fungi. FA biodegradation is not a common characteristic among bacteria, and the determinants of FA catabolism have not been identified so far in any microorganism. In this study, we identified genes, enzymes, and metabolic pathways involved in the degradation of FA in the soil bacterium Burkholderia ambifaria T16. Our results provide insights into the catabolism of a pyridine-derivative involved in plant pathogenesis by a rhizosphere bacterium.
KEYWORDS: Burkholderia ambifaria T16, fusaric acid, two-component flavin-dependent monooxygenase, 2-methylcitrate cycle, detoxification, catabolism
INTRODUCTION
Fusaric acid (FA, 5-butylpyridine-2-carboxylic acid) is a secondary metabolite synthesized by several Fusarium species (e.g., F. oxysporum, F. solani, F. verticillioides, F. proliferatum, and F. subglutinans) (1). These fungi are responsible for important wilt and rot diseases in a diverse range of crops, including maize, cucurbits, legumes, tomato, onion, potato, and banana (2). Production of FA by different phytopathogenic Fusarium spp. has been evaluated in vitro and in planta. In most cases, a positive correlation between FA production and virulence was observed (3–8), which has led to the consideration of FA as one of the main virulence factors produced by Fusarium spp. FA has been described to possess several negative effects on plant cells, including reduction in the availability of metals (9), oxidative damage (5, 10, 11), loss of mitochondrial membrane potential (5, 12, 13), and a reduction in the content of photosynthetic pigments (11, 14).
Management of Fusarium wilt is mainly performed by crop rotation, cover cropping, soil disinfection, applying chemical fungicides, and cultivating-resistant varieties (15–18). However, several important crops do not possess Fusarium wilt-resistant varieties. Besides, the use of fungicides contributes to the development of new pathogen races that overcome host resistance, negatively impacting the environment.
One interesting and sustainable alternative to control Fusarium plant diseases is the use of biocontrol agents. However, FA is also harmful for soil-borne microorganisms (19). Gram-positive microorganisms (19–21) and mycobacteria (22) were shown to be highly sensitive to FA. Fluorescent pseudomonads, conversely, possess high tolerance to this mycotoxin (19, 21, 23). However, it was demonstrated that even low FA concentrations negatively affect growth rates (23), and the production of some antifungal metabolites implicated in the suppression of soil-borne fungal pathogens (24–27). These results highlight the important role of FA in the colonization of competitive environments by mycotoxigenic fungi (28), as this compound reduces the survival and competition abilities of soil microbes. In this context, the search of plant growth-promoting microorganisms able to detoxify FA, and the study of the mechanisms involved in mycotoxin detoxification, would greatly contribute to the design of novel biocontrol strategies. While fungi that are able to convert FA to less phytotoxic intermediates have been reported (29–31), the capability to catabolize FA seems to be much rare among soil-borne microorganisms. Burkholderia ambifaria T16 is a bacterial strain isolated from the rhizosphere of barley, able to use FA as the sole carbon, nitrogen, and energy source (32). When inoculated into barley seedlings exposed to FA, strain T16 was able to suppress the toxic effect of FA on the growth of roots and stems (32). In this study, we employed two different experimental approaches to explore FA catabolism in B. ambifaria T16. The construction of a mini-Tn5 insertional library, combined with genome-wide proteome analyses, allowed us to identify genes, enzymes, and metabolic pathways that support bacterial growth on FA.
MATERIALS AND METHODS
Bacterial strains and culture conditions
Bacterial strains used in this work are listed in Table 1. Escherichia coli strains were routinely grown in LB medium under aeration at 37°C. B. ambifaria strains were cultivated at 32°C in LB medium or M9 minimal medium (33) supplemented with 1 mM MgSO4, 0.1% (vol/vol) metal trace solution (34), and 0.2% (wt/vol) sodium citrate (M9 + CA), 0.2% (wt/vol) sodium propionate (M9 + PA) or 450 µg/mL FA (M9 +FA) as carbon sources. Solid media were prepared by adding 1.5% (wt/vol) agar. When needed, the following antibiotics were added to the culture medium: ampicillin (Amp) 100 µg/mL (E. coli), chloramphenicol (Cm) 34 µg/mL (E. coli), trimethoprim (Tmp) 10 µg/mL (E. coli) or 70 µg/mL (B. ambifaria), kanamycin (Km) 50 µg/mL (E. coli) or 400 µg/mL (B. ambifaria), gentamycin (Gm) 10 µg/mL (E. coli), tetracycline (Tc) 20 µg/mL (E. coli) or 200 µg/mL (B. ambifaria), and potassium tellurite (Tel) 20 µg/mL (E. coli and B. ambifaria).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Relevant genotype or description | Reference or source |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli | ||
| CC118 λpir | Cloning host; ∆(ara-leu) araD ∆lacX174 galE galK phoA thiE1 rpsE rpoB (Rifr) argE (Am) recA1; λpir lysogen | (35) |
| DH5α λpir | Cloning host; F-endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA-argF) U169; hsdR17(rK− mK+)/λpir | (36) |
| HB101 | Helper strain; F−λ−hsdS20 (rB−mB−) recA13 leuB6 (Am) araC14 ∆(gpt-proA)62 lacY1 galK2 (Oc) xyl-5 mtl-1 thiE1 | (35) |
| Burkholderia ambifaria | ||
| T16 | Wild type; isolated from the rhizosphere of barley; Tcr | (32) |
| FA2 | Derived of T16; prpB::mini-Tn5Tel;Tcr Telr | This work |
| FA3 | Derived of T16; mlaD::mini-Tn5Tel; Tcr Telr | This work |
| FA4 | Derived of T16; prpR::mini-Tn5Tel; Tcr Telr | This work |
| FA21 | Derived of T16; EIB72_16280::mini-Tn5Tel;Tcr Telr | This work |
| FA84 | Derived of T16, EIB72_11905::mini-Tn5Tel; Tcr Telr | This work |
| FA90 | Derived of T16; EIB72_05180–85::mini-Tn5Tel; Tcr Telr | This work |
| FA91 | Derived of T16; prpF::mini-Tn5Tel; Tcr Telr | This work |
| FA92 | Derived of T16; prpR::mini-Tn5Tel; Tcr Telr | This work |
| T42 | Derived of T16; ΔprpB; Tcr | This work |
| T800 | Derived of T16; ΔfuaC; Tcr | This work |
|
Mycobacterium smegmatis mc2 155 |
High-frequency transformation mutant of M. smegmatis ATCC 607 | (37) |
| Plasmids | ||
| pRK600 | Helper plasmid used for conjugation; oriRK2 (mob+tra+ ); Cmr | (38) |
| pJMT6 | mini-Tn5 delivery plasmid; Ampr Telr | (39) |
| pSEVA612S | oriR6K; mob+; lacZα-pUC19/I-SceI; Gmr | (40) |
| pSEVA712S | oriR6K; mob+; lacZα-pUC19/I-SceI; Tmpr | This work |
| pSEVAdprpB | pSEVA712S carrying a 1.2 kb in-frame deletion fragment of prpB | This work |
| pSEVA228S | oriRK2;mob+; xylS; PmI-sceI; Kmr | (40) |
| pSEVA234 | oripBBR1; mob+; lacIq-Ptrc; Kmr | (40) |
| pSEVAicl | pSEVA234 carrying the icl1gene of M. smegmatis mc2 155 | This work |
| pSEVAprpB | pSEVA234 carrying the prpB gene from B. ambifaria T16 |
This work |
| pSEVA731 | oripBBR1; mob+; Tmpr | (40) |
| pSEVAfuaC | pSEVA731 carrying the fuaC gene from B. ambifaria T16 |
This work |
| pGPI-SceI | oriR6K; Tmpr; mob+; carries I-SceI cut site | (41) |
| pGPIdfuaC | pGPI-SceI carrying a 1.1 kb in frame deletion fragment of fuaC | This work |
| pDAI-SceI | oripBBR1; Tcr, mob+, carries the gene encoding I-SceI | (41) |
| Oligonucleotides (5′-> 3′) | ||
| ARB2 | GGCCACGCGTCGACTAGTAC | (42) |
| ARB6 | GGCCACGCGTCGACTAGTACNNNNNNNNNNCGCC | (42) |
| TelBint | GTTGGGCTGGCAGTGTCGATCCGCAA | (32) |
| TelBext | TTGCGAAGCAGTACCAGCAGGAAT | (32) |
| KilAint | CCCATGTTCACGCTTTGTTCTTCCAT | (32) |
| KilAext | CAGATTCGACAGCTCGTTGAGGGA | (32) |
| prpB3Bamfor | ACGTGGATCCGTGCGAGAAGTGATACCAGTACA | This work |
| prpB3rev | GGCTATCACGAATACGAGC | This work |
| prpB5for2 | GCTCGTATTCGTGATAGCCCGACAGGTAGACGGCCTTGA | This work |
| prpB5Nherev | CTAGCTAGCCCTGCATCACGTCGAAGCC | This work |
| SEVA_marker_rv_U | ATTTAAAUACGTAAATCGTAATTATT | This work |
| SEVA_marker_fw_U | ACCGTTGUCCTTTTCCGCGCATAACT | This work |
| TmpR_rv_U | ACAACGGUCCAATTAATTATTAACCCTT | This work |
| TmpR_fw_U | ATTTAAAUTTGTGTCTCAAAATCTCT | This work |
| PS3 | GAACGCTCGGTTGCCGC | (40) |
| PS4 | AATGACCCCGAAGCAGGG | (40) |
| P224 | GGACAAATCCGCCGCCCT | (40) |
| P225 | AGGGCGGCGGATTTGTCC | (40) |
| P226 | GCGGCAACCGAGCGTTC | (40) |
| P274 | CCTGCTCTGCGAGGCTGG | (40) |
| P291 | CCCTGCTTCGGGGTCATT | (40) |
| P292 | CCAGCCTCGCAGAGCAGG | (40) |
| prpBcheckfor | TGCAGCAGGTGCAGGAAGTG | This work |
| prpBNco2 | CATGCCATGGATGAGCAACACGCATCTT | This work |
| prpBdelcheckrv3 | CGACGCGTGCGAATACAGGAAC | This work |
| prpBrevXbaI2 | TTACTTCTTGCCCTGCGCGAAC | This work |
| luc2EcoRIfw5 | GGAATTCGCAGACATCGCCGAAACCTC | This work |
| luc2rv5 | CGAGTAGCTATGCCCGACCT | This work |
| luc2fw3 | AGGTCGGGCATAGCTACTCGCGCAACAATATCGTCCGCCC | This work |
| luc2BamHIrv3 | CGGGATCCACACCGGCATTGACGTGCGT | This work |
| luccheckfw1 | AAGCGGGATGCTCGAAGCA | This work |
| luccheckrv1 | CAGCAGCTTGTCGCTTTCGA | This work |
| iclfwXbaI | GCTCTAGAAGGAGGAAAAACATATGGCAACAATCGAAGCCAAC | This work |
| iclrvHindIII | CCCAAGCTTTTTCAGTCCTCCTTGCGGAT | This work |
| prpBfwXbaI | GCTCTAGAAGGAGGCCGTTCATGAGCAACA | This work |
| prpBrvHindIII | CCCAAGCTTAAAGCGTTCCGGGAACCGT | This work |
| fuaCfwEcoRI | GGAATTCCGGCAAGGTCGCGGCATGAA | This work |
| fuaCrvBamHI | CGGGATCCACACCGGAGGACCGCTGTTCGA | This work |
To evaluate the growth of B. ambifaria strains with sodium citrate (CA) or sodium propionate (PA), an overnight culture in LB broth was inoculated at a 1/100 ratio into M9 + CA to prepare the inoculum. The inoculum for M9 + FA was prepared in M9 + CA supplemented with 90 µg/mL FA. After overnight growth, cells were centrifuged, washed two times with M9 without carbon source, and resuspended in 1 mL of the same medium. These concentrated cell suspensions were used to inoculate Erlenmeyer flasks containing M9 with the addition of different carbon sources (CA, PA, or FA) at an initial OD at 600 nm (OD600) of 0.05 for M9 + CA and M9 + FA, or 0.1 for M9 + PA. Antibiotics and 1 mM IPTG were added to the medium where necessary.
To analyze the growth with FA as the sole nitrogen source, a modified M9 medium without NH4Cl, and supplemented with 450 µg/mL FA, 0.2% (wt/vol) citrate, 1 mM MgSO4, and 0.1% (vol/vol) metal trace solution, was used.
The MIC of FA was determined after 24 h incubation in M9 + CA, supplemented with antibiotics and IPTG where necessary. All cultures were inoculated at an initial OD600 = 0.05 from inocula prepared as described above. The MIC was determined as the minimum concentration of FA that completely inhibited growth. Growth curves and MIC determinations were performed at least in biological triplicates.
Construction of a mini-Tn5 mutant library; screening and mapping of mini-transposon insertions
To construct a mini-Tn5 insertional library in B. ambifaria T16, plasmid pJMT6 was introduced into B. ambifaria by triparental mating as described by Martínez-García et al. (43), with the modifications detailed in Simonetti et al. (44). The selection of transconjugants was performed in LB agar plates containing 20 µg/mL of the antibiotics Tel and Tc. A total of 8,000 transconjugants were screened for the inability to grow with FA as the sole carbon source. Each colony was simultaneously streaked in M9 + CA and M9 + FA agar plates, supplemented with the antibiotics Tel and Tc. After incubation at 32°C for 48 h, colonies unable to grow in M9 + FA were selected. Mapping of mini-Tn5 insertions was performed by arbitrary primed PCR (42) as previously described (44).
DNA extraction
Genomic DNA from B. ambifaria T16 was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilton, Germany), according to the protocol provided by the manufacturer. To extract genomic DNA of Mycobacterium smegmatis mc2 155, cells from a fresh plate were resuspended in a buffer containing 100 mM NaCl, 10 mM Tris·HCl pH = 8.0, 1 mM EDTA, and 1% (vol/vol) Triton X-100. For cell lysis, the suspension was incubated at 95°C for 20 min and subjected to three thawing-freezing cycles (5 min at –70°C, followed by 5 min at 100°C). After centrifugation at 13,000 rpm for 5 min, the supernatant was transferred to a new tube, and the DNA was precipitated according to Ausubel et al. (45).
Construction of plasmids and markerless B. ambifaria deletion mutants
Primers and vectors used in this work are listed in Table 1. Restriction enzymes were purchased from New England Biolabs Inc. (Ipswich, USA) or Takara Bio Inc. (Shiga, Japan). To construct markerless gene deletions in the genome of B. ambifaria T16 two different systems based on the I-SceI endonuclease were used. The deletion of the prpB gene was performed by using a suicide vector belonging to the pSEVA (Standard European Vector Architecture) database harboring the Tmp selection marker (46). To construct this plasmid, named pSEVA712S, a DNA fragment carrying the dihydrofolate reductase gene (dhfrI) from E. coli (GenBank accession number X00926.1) conferring Tmp resistance, was cloned into the pSEVA backbone using the uracil excision methodology (47) as follows. The pSEVA backbone and the dhfrI gene, were amplified from pSEVA612S and from a gBlock Gene Fragment carrying dhfrI (IDT, Cambridge, USA), respectively, using Phusion U Hot Start DNA polymerase (Thermo Scientific, Waltham, USA) and the primers pairs SEVA_marker_rv_U/SEVA_marker_fw_U or TmpR_rv_U/TmpR_fw_U. Fragments were purified with the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel GmbH & Co, Düren, Germany). DNA concentrations were measured using NanoDrop 2000 (Thermo Scientific, Massachusetts, USA). The USER reaction was performed as described by Cavaleiro et al. (47), and incubated at 30°C for 30 min, followed by a touchdown step of 27 cycles at 37°C (−1°C/cycle) for 2 min. The total reaction was used to transform E. coli DH5α chemically competent cells. Transformants were selected in LB agar plates containing Tmp and screened for the presence of the Tmpr marker by colony PCR using primer pairs PS3 and PS4, One Taq DNA polymerase (New England Biolab Inc.), and the protocol provided by the manufacturer. Finally, the pSEVA plasmid carrying the Tmpr marker was extracted with the NucleoSpin Plasmid Easy Pure (Macherey-Nagel GmbH & Co) and its complete sequence was checked with primers P224, P225, P226, PS3, PS4, P274, P291, and P292 in Eurofins Genomics LLC (Ebersberg, Germany). The new vector, pSEVA712S, was added to the SEVA database (46).
To delete the prpB gene of B. ambifaria T16, two DNA fragments ≈600 bp flanking the prpB gene were amplified with the enzyme Phusion High-Fidelity DNA polymerase (New England Biolabs Inc.), using the primers pairsprpB5Nherev/prpB5for2 and prpB3Bamfor/prpB3rev. The amplified fragments were purified with the Accuprep purification kit (Bioneer Corporation, Korea) and 50 ng of each fragment was used in an overlap-PCR (48) to obtain the 1.2 kb fragment for in-frame deletion. After purification, the fused fragment was cloned in pSEVA712S with the restriction enzymes NheI and BamHI to obtain plasmid pSEVAdprpB. This plasmid was introduced into B. ambifaria T16 by triparental mating as described by Simonetti et al. (44), with minor modifications. Liquid cultures were grown overnight in 2 mL of LB broth supplemented with the corresponding antibiotics. Cells were washed with 10 mM MgSO4 and mixed in a 1:1:1 ratio into 5 mL of MgSO4 to obtain a final OD600 = 0.05 of each strain. Using a syringe, the mixture was concentrated onto a Millipore filter disk, which was placed onto the surface of an LB agar plate. After 16 h of incubation at 30°C, the filter was transferred into 1 mL of 10 mM MgSO4 and suspended by vortexing. The resulting suspension was serially diluted and plated onto LB agar plates containing Tmp and Tc. Plates were incubated for 48 h at 30°C. The obtained transconjugants colonies were transferred to a fresh plate containing antibiotics and the co-integration of pSEVAdprpB into the genome of B. ambifaria T16 was evaluated by colony PCR using the primers pairs prpBcheckfor/prpBNco2 and prpBdelcheckrv3/prpBrevXbaI2. Co-integrate resolution was achieved by the introduction of the plasmid pSEVA228S, using the protocol described by Martínez-García and de Lorenzo (49). The deletion of the prpB gene was checked by colony PCR using primers prpBcheckfor and prpBdelcheckrv3. One of these mutants was selected and named B. ambifaria T42 (ΔprpB). Finally, plasmid pSEVA228S was curated from B. ambifaria by performing three serial transfers in LB broth without antibiotics, plating serial dilutions in LB agar plates and selecting KmS colonies.
To delete the fuaC gene, DNA regions located upstream and downstream of the gene were amplified with the primers pairs luc2Ecofw5/luc2rv5 and luc2fw3/luc2BamHIrv3, respectively. Both fragments were fused using primers luc2Ecofw5 and luc2BamHIrv3. After digestion with EcoRI and BamHI, the obtained 1,127 bp fragment for in-frame deletion was ligated into pSEVA712S. It was not possible to obtain co-integrate clones using this construct, and plasmid pGPI-SceI was used for generating the fuaC deletion instead. Therefore, the 1,127 bp fragment was sub-cloned into pGPI-SceI using EcoRI and XbaI, and the resulting plasmid, pGPIdfuaC, was introduced into B. ambifaria T16 by triparental mating. The integration of pGPIdfuaC into the genome of B. ambifaria T16 was checked with primers pairs luccheckfw1/luc2BamHIrv3 and luc2Ecofw5/luccheckrv1. The co-integrate was resolved by the introduction of plasmid pDAI-SceI. Transconjugants were screened for the loss of Tmp resistance, and TmpS clones were subjected to colony-PCR with primers luccheckfw1 and luccheckrv1 in order to verify the deletion of the fuaC gene in the genome of B. ambifaria. One of these double-crossover mutants was selected and named B. ambifaria T800. Finally, curation of pDAI-SceI was performed as described by Flannagan et al. (41).
To construct plasmid pSEVAicl bearing the icl1 gene (encoding the enzyme isocitrate lyase from M. smegmatis mc2 155) under the lacIq-Ptrc inducible promoter, the gene was amplified using primers iclfwXba and iclrvHindIII, digested and ligated into the expression plasmid pSEVA234. To obtain the plasmid pSEVAprpB, the prpB gene of B. ambifaria T16 was amplified with primers prpBfwXbaI and prpBrvHindIII, digested and ligated into pSEVA234. Finally, complementation of the fuaC deletion was performed with plasmid pSEVAfuaC. In this case, a 1,475 bp fragment, which includes the fuaC coding sequence of B. ambifaria T16 and its promoter region, was amplified with the primer pair fuaCfwEcoRI/fuaCrvBamHI, digested and ligated into pSEVA731.
FA toxicity assay using barley seedlings
Barley seeds were surface-disinfected with 0.2% (vol/vol) NaClO, submerged in sterile deionized water for 1.5 h and placed inside a humid chamber for 24 h to allow germination. The effect of FA on the growth of barley seedlings was evaluated as described by Simonetti et al. (32) using M9, M9 + FA, and cell-free supernatants from stationary phase cultures of B. ambifaria strains cultivated in M9 + FA.
Identification of differentially induced proteins during growth with FA as the sole carbon source
Three independent cultures of B. ambifaria T16 were cultivated in M9 + CA and M9 + FA to an OD600 ≈ 0.7. Cells were harvested at 4°C, washed with ice-cold 10 mM Tris·HCl pH = 7.5, resuspended in Lysis Buffer (10 mM Tris·HCl pH = 7.5, 1 mM PMSF, 100 µg/mL lysozyme, 1 mM DMSO, and 1 mM EDTA), and mechanically disrupted using glass beads. The obtained protein extracts were lyophilized, and subjected to trypsin digestion and clean-up using the iST 8 × kit (PreOmics GmbH, Martinsried, Germany), and the protocol provided by the manufacturer. Protein samples were analyzed by LC-MS as described elsewhere (50).
MaxQuant 2.1.2.0 was used to identify and quantify the proteins by LFQ with the following parameters: Database NCBI_Burkholderia_ambifaria_RQYA01.1_20220630; MS tol, 10 ppm; MS/MS tol, 20 ppm Da; Peptide FDR, 0.1; Protein FDR, 0.01 Min. peptide Length, 7; Variable modifications, Oxidation (M); Fixed modifications, Carbamidomethyl (C); Peptides for protein quantitation, razor and unique; Min. peptides, 1; Min. ratio count, 2. The quantitative protein abundance data (MaxQuant LFQ values) were analyzed by performing an adjusted two-sample t test as described in Perseus (51). P values lower than 0.05 were considered significant. Finally, the proteomic data were uploaded in the pathway genome database (PGDB) of B. ambifaria T16 previously constructed (52), by using the Omics module of the Pathway Tool software v. 25 (53).
RESULTS
Transposon mutagenesis and mapping of mini-Tn5 insertion sites in mutants unable to grow with FA as the sole carbon and energy source
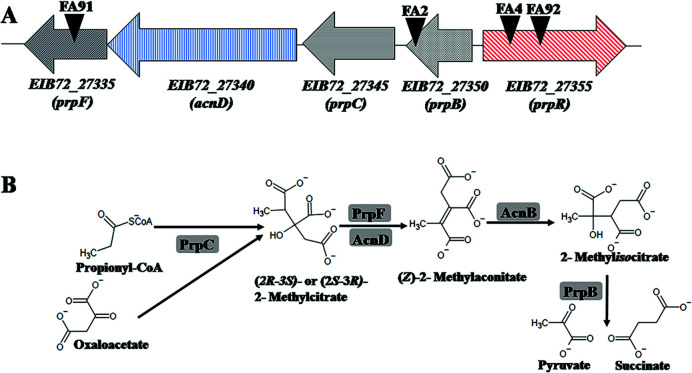
With the aim to find genes involved in FA catabolism and FA tolerance in B. ambifaria T16, two different experimental approaches were used. First, we constructed a mini-Tn5 insertional library to select mutants unable to grow in M9 + FA. After plating and incubation, eight clones from the insertional library were unable to grow on FA as carbon source. The coordinates of the mini-Tn5 insertions and the genes interrupted by the transposon in these mutants are shown in Table 2. In four of these insertional mutants (B. ambifaria FA2, B. ambifaria FA4, B. ambifaria FA91, and B. ambifaria FA92), the transposon insertion mapped in different genes encoding components of the 2-methylcitrate cycle (2-MCC; Table 2 and Fig. 1A). The 2-MCC is an anaplerotic pathway widely distributed among bacteria and fungi, where propionyl-coenzyme A (CoA) is converted to pyruvate and succinate. In B. ambifaria T16, the prp locus comprises the prpR gene, encoding the σ54-dependent transcriptional activator of the prp genes (54), and four structural genes, prpB, prpC, acnD, and prpF, encoding the enzymes involved in propionyl-CoA metabolism (Fig. 1A and B).
TABLE 2.
Location of mini-Tn5 Tel insertions in B. ambifaria mutants unable to grow with FA as the sole carbon and energy source
| Mutant | Interrupted gene (GenBank Locus Tag) (strand)a | Gene | Putative functionb | Coordinates of insertion (scaffold)c | |
|---|---|---|---|---|---|
| B. ambifaria FA2 | EIB72_27350 (−) | prpB | 2-methylisocitrate lyase | 236678 (scaffold 9) |
|
| B. ambifaria FA3 | EIB72_01775 (+) | mlaD | Outer membrane lipid asymmetry maintenance protein | 88315 (scaffold 2) |
|
| B. ambifaria FA4 | EIB72_27355 (+) | prpR | Propionate catabolism operon regulatory protein | 238068 (scaffold 9) |
|
| B. ambifaria FA21 | EIB72_16280 (+) | Acyl-CoA dehydrogenase | 115004 (scaffold 6) |
||
| B. ambifaria FA84 | EIB72_11905 (+) | Phosphoenolpyruvate carboxylase | 214877 (scaffold 4) |
||
|
B. ambifaria FA90 |
Intergenic region EIB72_05180–85 | - | 592108 (scaffold 3) |
||
| B. ambifaria FA91 | EIB72_27335 (−) | prpF | 2-methylaconitate isomerase | 232072 (scaffold 9) |
|
| B. ambifaria FA92 | EIB72_27355 (+) | prpR | Propionate catabolism operon regulatory protein | 238495 (scaffold 9) |
|
The sign + or − refers to the DNA strand encoding the gene.
Predicted functions are based on the annotation of B. ambifaria T16 genome using the RAST server (55).
Genomes coordinates in B. ambifaria T16 genome (GenBank accession number RQYA00000000).
Fig 1.
(A) Genomic organization of the prp locus in B. ambifaria T16 and localization of mini-Tn5 insertions in the mutants B. ambifaria FA2, FA4, FA92, and FA91. Genes are represented by arrows and black inverted triangles represent the localization of the mini-Tn5 insertions in each mutant. (B) 2-Methylcitrate cycle in B. ambifaria T16. Enzymes are shaded in gray rectangles. PrpC: methylcitrate synthase, AcnD: methylcitrate dehydratase, PrpF: methylaconitate cis-trans isomerase, AcnB: aconitase, PrpB: methylisocitrate lyase. The AcnB enzyme is not encoded in the prp locus.
In B. ambifaria FA3, the mini-Tn5 cassette interrupted the gene EIB72_01775, encoding the outer membrane lipid asymmetry maintenance protein MlaD. MlaD is part of the highly conserved Mla (maintenance of lipid asymmetry) system (56), involved in the maintenance of outer membrane lipid asymmetry in Gram-negative bacteria.
In B. ambifaria FA21, the mini-Tn5 mapped in locus EIB72_16280. This gene is predicted to encode an acyl-CoA dehydrogenase. In B. ambifaria FA84, the transposon interrupted gene EIB72_11905, which encodes the enzyme phosphoenolpyruvate carboxylase (PEPC). In close proximity to EIB72_11905, genes predicted to be involved in the transport and catabolism of aromatic compounds were found. Finally, the mutant B. ambifaria FA90 bears a mini-Tn5 insertion in an intergenic region between two genes encoding enzymes involved in RNA processing, degradation, and modification (RNase E/G family endoribonuclease and RluA family pseudouridine synthase) (57, 58).
Tolerance to FA and ability to grow on FA as the sole nitrogen source in B. ambifaria insertional mutants
As mentioned above, FA is toxic to different types of cells and negatively affects diverse cellular functions. Therefore, the growth impairment observed with FA as the sole carbon and energy source in the insertional mutants could be attributed to the inactivation of a gene involved in FA catabolism or a gene required to tolerate the exposure to this mycotoxin. For that reason, the MIC of FA was determined in cultures of the wild-type strain (B. ambifaria T16) and the insertional mutants. Table 3 shows that two insertional mutants, B. ambifaria FA2 (prpB::mini-Tn5Tel) and B. ambifaria FA3 (mlaD::mini-Tn5Tel), were more sensitive to the presence of FA in the growth medium than the wild-type strain. While B. ambifaria T16 showed a MIC of 1,600 µg/mL, the MIC obtained for B. ambifaria FA2 and FA3 was 200 µg/mL In the remaining insertional mutants, the tolerance to FA was similar to that obtained for the wild-type strain (Table 3). Next, we decided to analyze the growth of the insertional mutants with FA as the sole nitrogen source. With this purpose in mind, the strains were cultivated in a modified M9 minimal medium without the addition of NH4Cl, supplemented with 0.2% (wt/vol) citrate (carbon source) and 450 µg/mL FA (carbon source and sole nitrogen source) (32). Only B. ambifaria FA84, which possesses a mini-Tn5 insertion in a gene predicted to encode PEPC, was able to reach an OD600 similar to the wild-type strain (Table 3). As expected, B. ambifaria FA2 and FA3 were unable to grow with 450 µg/mL FA as sole nitrogen source. This result is in agreement with the FA MIC values obtained for these mutants (Table 3). Another strain unable to grow with FA as nitrogen source was B. ambifaria FA21. This strain possesses a mini-Tn5 insertion in EIB72_16280, which encodes a predicted acyl-CoA dehydrogenase. The cell densities of the remaining mutants were lower than those of the wild-type, indicating that the transposon insertions, not only completely prevent growth with FA as sole carbon source, but also negatively affect the capability to grow with FA as the sole nitrogen source.
TABLE 3.
MICs of FA and capability to grow with FA as carbon and nitrogen source for B. ambifaria T16 (wt) and mini-Tn5 insertional mutants
| B. ambifaria strain | MIC FA (µg/mL) | Growth with FA as carbon source | Growth with FA as nitrogen source Final OD600a |
|---|---|---|---|
| T16 (wild type) | 1,600 | + | 1.26 ± 0.27 |
| FA2 (prpB::mini-Tn5Tel) | 200 | - | 0.11 ± 0.05 |
| FA3 (mlaD::mini-Tn5Tel) | 200 | - | 0.05 ± 0.01 |
| FA4 (prpR::mini-Tn5Tel) | 1,600 | - | 0.65 ± 0.06 |
| FA21 (EIB72_16280::mini-Tn5Tel) | 1,600 | - | 0.07 ± 0.01 |
| FA84 (EIB72_11905::mini-Tn5Tel) | 1,600 | - | 1.22 ± 0.22 |
| FA90 (EIB72_05180–85::mini-Tn5Tel) | 1,600 | - | 0.81 ± 0.04 |
| FA91 (prpF::mini-Tn5Tel) | 1,600 | - | 0.81 ± 0.04 |
| FA92 (prpR::mini-Tn5Tel) | 1,600 | - | 0.62 ± 0.07 |
Mean ± SD of three independent cultures is shown.
Construction of a B. ambifaria prpB deletion mutant and evaluation of its ability to grow on FA
The results obtained with the insertional mutants pointed to an important role of the 2-MCC in the capability to grow with FA as the sole carbon source. To provide genetic evidence supporting this phenotype, the gene prpB, encoding methylisocitrate lyase (the last enzyme of the 2-MCC; Fig. 1), was deleted. The B. ambifaria ΔprpB strain obtained was named B. ambifaria T42. In agreement with the results obtained with all the mutants with insertions in the prp locus, B. ambifaria T42 was also unable to grow in M9 + FA. Furthermore, the prpB deletion also negatively affected the tolerance to FA (Table 4), as the MIC obtained for this strain was 400 µg/mL, much lower than the value obtained for the wild-type strain (1,600 µg/mL) (Table 3).
TABLE 4.
MICs of FA, capability to grow with FA as carbon source and specific growth rates (µ) calculated for B. ambifaria strains
| B. ambifaria strain/plasmid | MIC FA (µg/mL) |
µCA C. sourcea (h−1) |
µPA C. sourceb (h−1) |
µFA C. sourcec (h−1) |
µFA N. sourced (h−1) |
|---|---|---|---|---|---|
| T42 (ΔprpB) | 400 | NDe | ND | ND | ND |
| T16/pSEVA234 | 1,600 | 0.40 ± 0.020 | 0.04 ± 0.004 | 0.19 ± 0.013 | ND |
| T42/pSEVA234 | 400 | 0.42 ± 0.022 | 0 | 0 | ND |
| T42/pSEVAicl | 1,600 | 0.37 ± 0.020 | 0.03 ± 0.004 | 0.09 ± 0.010 | ND |
| T42/pSEVAprpB | 3,200 | 0.42 ± 0.024 | 0.19 ± 0.011 | 0.14 ± 0.047 | ND |
| T16/pSEVA731 | 1,600 | ND | ND | 0.20 ± 0.018 | 0.17 ± 0.006 |
| T800/pSEVA731 | 1,600 | ND | ND | 0 | 0 |
| T800/pSEVAfuaC | 1,600 | ND | ND | 0.24 ± 0.046 | 0.13 ± 0.005 |
µ Sodium citrate as carbon source.
µ Sodium propionate as carbon source.
µ Fusaric acid as carbon source.
µ Fusaric acid as nitrogen source.
ND: not determined.
Then, we decided to perform two different complementation tests, one by introducing the prpB gene of B. ambifaria T16, and the other by introducing a gene from a different bacterial species encoding an enzyme with methylisocitrate lyase activity into the ΔprpB mutant. This functional complementation was performed with the icl1 gene of M. smegmatis mc2 155. This gene was chosen because it encodes an enzyme with reported isocitrate lyase and methylisocitrate lyase activities (59). The prpB and icl1 genes were separately cloned in an inducible expression plasmid and introduced into B. ambifaria T42. Table 4 indicates that the introduction of plasmids pSEVAicl and pSEVAprpB in the ΔprpB mutant increased the tolerance to FA. The MIC value obtained for B. ambifaria T42/pSEVAicl (1,600 µg/mL) was similar to that of the wild-type strain. Interestingly, B. ambifaria T42/pSEVAprpB showed a higher MIC value than the wild type (3,200 µg/mL).
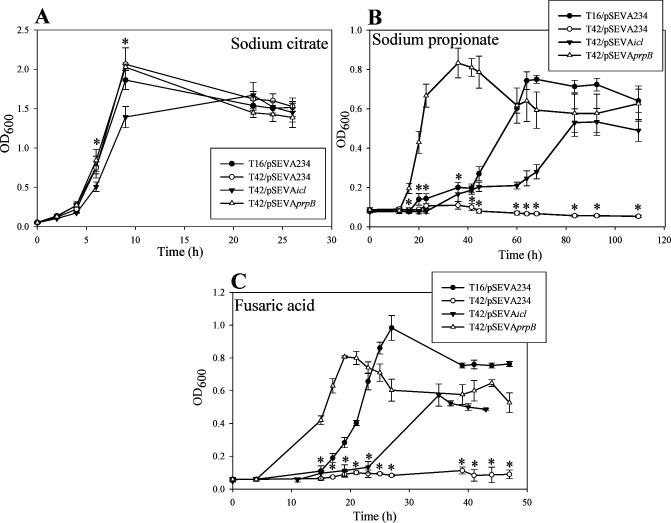
Next, we compared the growth of B. ambifaria T16/pSEVA234, B. ambifaria T42/pSEVA234, B. ambifaria T42/pSEVAicl, and B. ambifaria T42/pSEVAprpB in minimal medium with CA, PA, or FA as carbon and energy sources (Fig. 2 and Table 4). As shown in Fig. 2A and Table 4, the wild-type strain, the ΔprpB mutant carrying the vector plasmid and the mutant bearing pSEVAprpB showed very similar growth profiles and almost identical specific growth rates when they were cultivated with CA as the sole carbon source. Meanwhile, the growth rate of the ΔprpB strain carrying pSEVAicl was slightly lower than the obtained for the other strains. Different results were obtained when these strains were grown with PA as the sole carbon source (Fig. 2B and Table 4). Severe growth impairment was observed for B. ambifaria T42/pSEVA234. The introduction of prpB or the icl1 gene in the T42 strain restored growth in PA. Besides, as it was observed with CA, the growth rate of strain T42/pSEVAicl was lower than the growth rates obtained for strains T16/pSEVA234 and T42/pSEVAprpB. Moreover, B. ambifaria T42 carrying pSEVAprpB showed a much higher growth rate and a shortened lag phase compared to the wild type strain. Finally, similar results to the ones obtained with PA were observed when FA was used as the sole carbon and energy source (Fig. 2C and Table 4). The prpB deletion mutant carrying pSEVA234 was unable to grow using FA as carbon source. The introduction of pSEVAicl or pSEVAprpB in the ΔprpB strain was able to restore the growth with FA. Over-expression of prpB shortened the lag phase. These results indicate that a functional 2-MCC is essential for the growth of B. ambifaria T16 with FA as the sole carbon and energy source, and suggest that propionyl-CoA is generated during FA catabolism.
Fig 2.
Growth of B. ambifaria strains in M9 minimal medium with 0.2% (wt/vol) sodium citrate (A), 0.2% (wt/vol) sodium propionate (B) or 450 µg/mL fusaric acid (C) as sole carbon and energy source. The graphs show the means ± SD obtained from three biological replicates. The OD600 values obtained were subjected to Kruskal-Wallis analysis of variance followed by Dunnett’s test. The ΔprpB strain bearing the vector plasmid (T42/pSEVA234) was used as control in the test. P values were considered significant when they were lower than 0.05. Asterisks indicate that there is a statistically significant difference between OD600 values of strains T16/pSEVA234, T42/pSEVAicl or T42/pSEVAprpB and OD600 values of T42/pSEVA234.
Proteins differentially induced during growth with FA as the sole carbon source
A proteomic analysis of B. ambifaria T16 grown with CA or FA as the sole carbon and energy source was performed to identify proteins involved in FA catabolism. A detailed list of the identified proteins (1,998 proteins), and the difference in protein abundance between the two cultivation conditions are shown in Table S1 in the supplemental material. Several enzymes involved in fatty acid biosynthesis and cofactor biosynthesis (including coenzyme A and flavin biosynthesis) were upregulated during growth of B. ambifaria T16 with FA. Furthermore, a higher abundance of enzymes involved in degradation of amino acids, carboxylates, and lipids was observed when the bacteria used the mycotoxin as the sole carbon and energy source (Fig. S1; supplemental material).
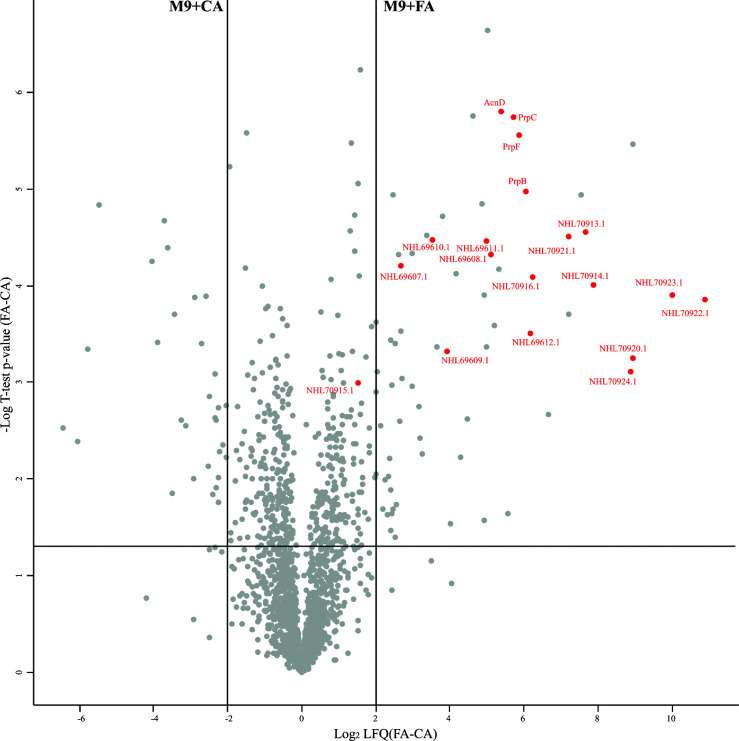
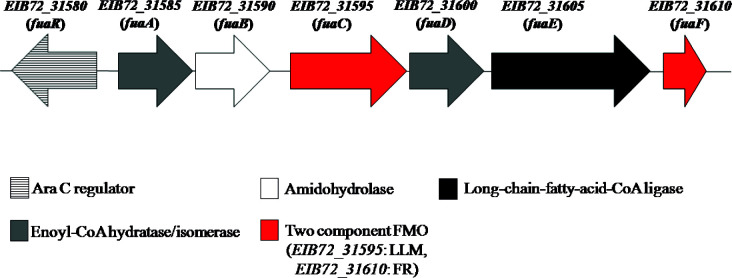
Fig. 3 shows the proteomic results as a volcano plot. Enzymes belonging to the 2-MCC showed a log2LFQ(FA-CA)=5–6, indicating a much higher intracellular level of these proteins during growth with FA compared to CA. Some proteins encoded in a gene cluster predicted to be involved in valine degradation (NHL69607.1 to NHL69612.1) were also found to be significantly upregulated during growth with FA (Fig. 3 ; Table S1). Other proteins that showed very high levels of abundance during the growth of B. ambifaria T16 with FA, with log2LFQ(FA-CA) values from 6.2 to 10.8, include NHL70913.1, NHL70914.1, NHL70916.1, and NHL70920.1 to NHL70924.1 (Fig. 3 ; Table S1). Protein NHL70916.1 is predicted to be a BdkR transcriptional factor involved in the regulation of isoleucine and valine catabolism, and proteins NHL70913.1 and NHL70914.1 belong to a 2-oxoacid dehydrogenase complex. These complexes convert 2-oxoacids to the corresponding acyl-CoA derivatives. Proteins NHL70920.1 to 70924.1, encoded in the gene cluster EIB72_31580 to EIB72_31610, have no known function. The genetic organization and the predicted functions of the proteins encoded in this cluster are shown in Fig. 4 and Table 5. The gene EIB72_31580 encodes a predicted transcriptional regulator of the AraC family. This regulator possesses 38% identity to the NimR regulator of E. coli involved in the tolerance to 2-nitroimidazole (60). Downstream of this regulatory gene, six genes encoding enzymes with different activities are found (Fig. 4). Among them, EIB72_31585 and EIB72_31600 are predicted to encode two enoyl-CoA hydratases. These proteins identified as NHL70920.1 and NHL70923.1, possess sequence identities values of 36 and 34%, respectively, to the 2,3-dehydroadipyl-CoA hydratase and 1,2-epoxyphenylacetyl-CoA isomerase of E. coli, which are involved in the catabolism of phenylacetate (61). The protein encoded by EIB72_31590 belongs to the creatininase family, which includes proteins catalyzing the hydrolysis of an amide bond (62, 63). The gene EIB72_31595 is predicted to encode a flavin-dependent oxidoreductase (NHL70922.1) from the luciferase-like monooxygenase (LLM) family. According to a classification based on their structural and functional characteristics, members of the LLM belong to group C Flavin-dependent monooxygenases (FMOs-C) (64). These proteins use reduced FMN for the activation of molecular oxygen, which is generated by a NAD(P)H-dependent flavin reductase (64). A gene predicted to encode a flavin-reductase (EIB72_31610) with an FMN binding site is also located in this gene cluster (Fig. 4). The flavin reductase encoded by EIB72_31610 has 45.6% identity to the NADH-FMN reductase RutF of E.coli, involved in the catabolism of uracil (65). Finally, the gen EIB72_31605 encodes a predicted AMP binding protein with high sequence identity to long-chain fatty acid-CoA ligases.
Fig 3.
Differential protein abundance in B. ambifaria T16 grown in M9 minimal medium with citrate (M9 + CA) or fusaric acid (M9 + FA) as sole carbon source. Red dots represent some upregulated proteins during growth with FA. Proteins IDs NHL69607.1 to NHL69612.1 correspond to proteins involved in valine degradation. Proteins NHL70913.1 to 70915.1: 2-oxo acid dehydrogenase complex. NHL70916.1: predicted BdkR transcriptional regulator involved in the regulation of isoleucine and valine catabolism. Proteins NHL70920.1 to 70924.1 are encoded in a gene cluster of unknown function.
Fig 4.
Organization of the gene cluster EIB72_31580-EIB72_31610 (fua cluster) encoding proteins NHL70920.1 to 70924. 1. Genes are represented by arrows. Locus IDs are shown above the genes. FR, flavin reductase; LLM, luciferase-like monooxygenase.
TABLE 5.
Functional annotations of the proteins encoded by the EIB72_31580 to EIB72_31610 (fua) gene cluster
| Gene locus |
Protein ID | Protein length (aa) | Putative funcion | Conserved domains/intervala |
Nearest Homolog/ accession Nos°b |
Identity (%) |
|---|---|---|---|---|---|---|
| EIB72_31580 (fuaR) |
NHL70919.1 | 273 | AraC family transcriptional regulator | Cupin_NimR (40–121) AraC family (147–272) |
HTH-type transcriptional regulator NimR (E. coli K-12)/P76241.1 | 38.49 |
| EIB72_31585 (fuaA) |
NHL70920.1 | 254 | Enoyl-CoA hydratase/ Isomerase family protein |
Crotonase (7–194) |
2,3-dihydroadipyl-CoA hydratase (E. coli K-12)/P76082.1 | 36.62 |
| EIB72_31590 (fuaB) |
NHL70921.1 | 259 | Creatininase family protein (amido- hydrolase) |
Creatininase (22–250) |
Mycofactocin precursor peptide peptidase (Mycobacterium ulcerans Agy99)/A0PM51.1 |
33.33 |
| EIB72_31595 (fuaC) |
NHL70922.1 | 384 | LLM class flavin-dependent oxidoreductase |
Bac-luciferase (3–324) |
Uncharacterized protein y4vj (Synorhizobium fredii NGR234)/Q53218.2 | 25.22% |
| EIB72_31600 (fuaD) |
NHL70923.1 | 262 | Enoyl-CoA hydratase/isomerase family protein |
Crotonase (6–254) |
Probably enoyl-CoA hydratase (Caenorhabditis elegans)/P34559.1 |
35.25 |
| EIB72_31605 (fuaE) |
NHL70924.1 | 635 | AMP binding protein | Adenylate forming domain- ClassI superfamily (16–624) |
Putative fatty-acid-CoA ligase (Mycobacterium tuberculosis)/P9WQ52.1 |
33.79 |
| EIB72_31610 (fuaF) |
- | 154 | Flavin reductase (pseudogen) | Pyridoxine 5′-phosphate oxidase and flavin reductase (1–149) |
FMN reductase (NADH) RutF (Yersina enterocolitica 8081)/A1JMW4.1 | 49.04 |
The proteins sequences were analyzed against the Conserved Domain Database at the NCBI website. Intervals correspond to aminoacid sequence numbers.
Top BLASTP hit obtained using the UniProtKB/Swiss-Prot Database at the NCBI website.
Construction of a B. ambifaria deletion mutant in the gene encoding the LLM NHL70922.1 and evaluation of its capability to grow on FA
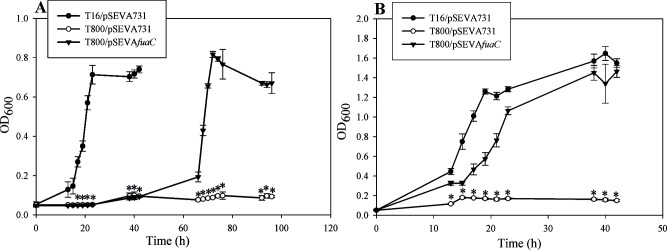
According to in silico predictions, the genes EIB72_31595 and EIB72_31610 encode a two-component FMO from the LLM family. Two-component FMOs catalyze the oxidation of several substrates, including N-heterocyclic compounds (65–68). The LLM NHL70922.1 possesses 25% identity to the pyrimidine monooxygenase RutA of E. coli (65), 34% identity to the tetramethyl pyrazine oxygenase TpdA of Rhodococcus sp. THP1 (67), 29% to the pyridine monooxygenase PyrA of Arthrobacter sp. 68b (66), and 23% to the trigonelline monooxygenase TgnB of Acitenobacter baylyi ADP1 (68). We hypothesized that the products of the gene cluster EIB72_31580-EIB72_31610 are involved in the first steps of FA catabolism in B. ambifaria T16. To find out if NHL70922.1 is involved in the catabolism of FA, we constructed a clean deletion mutant in the gene encoding this protein (EIB72_31595). This new strain was designated B. ambifaria T800. Next, a DNA fragment encompassing the gene locus EIB72_31595 and its promoter region was cloned in pSEVA731 and introduced into B. ambifaria T800. The MICs of FA and the capability to grow with the mycotoxin were analyzed in B. ambifaria T16 and T800 carrying pSEVA731 and in B. ambifaria T800 with the pSEVA plasmid carrying the EIB72_31595 gene fragment. B. ambifariaT800/pSEVA731 showed the same MIC for FA as T16/pSEVA731 (Table 4), indicating that deletion of EIB72_31595 does not affect the tolerance of B. ambifaria to this mycotoxin. However, elimination of EIB72_31595 completely abolished the capability of B. ambifaria to grow with FA as the sole carbon and energy source (Fig. 5A). Introduction of pSEVA731 carrying EIB72_31595 into B. ambifaria T800 was able to restore growth (Fig. 5A), demonstrating that EIB72_31595 is essential for FA catabolism in B. ambifaria T16. For that reason, we decided to designate the gene cluster encompassing EIB72_31580-EIB72_31610 as fua (fusaric acid) cluster and the gene EIB72_31595 as fuaC. As it can be observed in Fig. 5A and Table 4, the wild-type strain with the vector plasmid (B. ambifaria T16/pSEVA731) and the complemented strain (B. ambifaria T800/pSEVAfuaC) grew at very similar specific growth rates. A very long lag phase, which lasted more than 40 h, was observed when B. ambifariaT800/pSEVAfuaC was cultivated with FA as the sole carbon source (Fig. 5A).
Fig 5.
Growth of B. ambifaria strains with fusaric acid as the sole carbon and energy source (A) or as the sole nitrogen source (B). The graphs show the means ± SD obtained from three biological replicates. OD600 values were subjected to Kruskal-Wallis analysis of variance, followed by Dunnett’s test. B. ambifaria T800/pSEVA731 was used as a control in the test. P values were considered significant when they were lower than 0.05. Asterisks indicate that there is a statistically significant difference with B. ambifaria T800/pSEVA731.
As expected, the deletion of the fuaC gene also eliminated the capability of B. ambifaria to grow with FA as the sole nitrogen source (Fig. 5B). Growth was restored by the introduction of pSEVAfuaC. B. ambifaria T800 bearing pSEVAfuaC showed a slightly lower growth rate than the wild-type strain carrying the vector plasmid (Table 4).
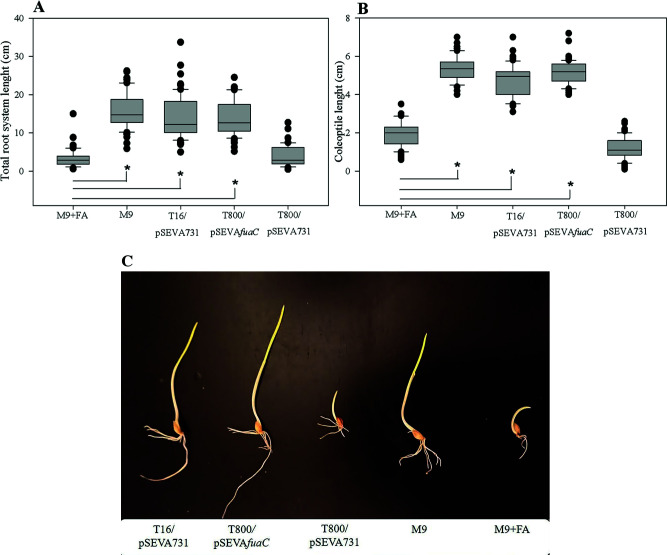
Due to its reported phytotoxic effects, the presence of FA can be detected by employing simple phytotoxicity assays (8, 10, 11, 13, 32). Taking this into account, we evaluated the presence of FA in the culture supernatants of B. ambifaria T16/pSEVA731, B. ambifaria T800/pSEVA731, and B. ambifaria T800/pSEVAfuaC cultivated with FA as the sole carbon and energy source, by applying the cell-free supernatants on germinated barley seedlings (32)(Fig. 6A and B). The growth medium with the addition of 450 µg/mL FA (M9 + FA) was used as a control of the experiment. As expected, the growth of the roots and the coleoptile of barley seedlings was negatively affected by the treatment with M9 + FA (Fig. 6A and B). Barley seedlings treated with cell-free-supernatants from stationary phase-cultures of B. ambifaria T800/pSEVA731 showed similar coleoptile’s and root’s lengths to the seeds treated with M9 + FA, indicating that FA is present in the supernatant and therefore has not been catabolized by this strain. Meanwhile, the barley seedlings exposed to the cell-free-supernatants from stationary phase-cultures of B. ambifaria T16/pSEVA731 or B. ambifaria T800/pSEVAfuaC cultivated with FA, showed coleoptiles and roots significantly longer than the barley seedlings exposed to M9 + FA, showing that FA has been degraded in cultures of these strains.
Fig 6.
FA toxicity assay using barley seedlings. Box plots of median, 1st and 3rd quartiles (boxes), and 1.5x quartiles (whiskers) corresponding to total root lenght (A) and coleoptile length (B) of seedlings (n=60) treated with cell-free supernatants from stationary phase-cultures of B. ambifaria strains, M9 minimal medium (M9), or M9 supplemented with 450 μg/mL FA (M9+FA). Three biological replicates were used. Dots denote observations outside the range of adjacent values. Data were subjected to Kruskal-Wallis analysis of variance and compared by Dunnet’s test. Seeds treated with M9 + FA were used as control. Asterisks indicate significant differences at P < 0.05 between values from the barley seedlings treated with bacterial supernatants or M9, and the barley seedlings treated with M9 + FA (control). (C) Photographic image showing barley seedlings treated with culture supernatants of B. ambifaria strains, M9 minimal medium (M9), or M9 supplemented with 450 μg/mL FA (M9 + FA).
DISCUSSION
This work describes the first example of bacterial genes essential for growth on FA as the sole carbon and energy source. By screening of a transposon insertional library, several mini-Tn5 mutants of B. ambifaria were found to be unable to grow in minimal medium with 450 µg/mL FA as the sole carbon source. Among these mutants, B. ambifaria FA2 and FA3 showed an enhanced sensitivity to FA. Moreover, none of these mutants were able to grow with 450 µg/mL FA as the sole nitrogen source.
In B. ambifaria FA3, the mini-Tn5 insertion mapped in mlaD, a gene encoding one of the periplasmic components of the Mla system. This system is composed of six proteins distributed across the cell envelope and functions as an intermembrane phospholipid transport system that maintains lipid asymmetry in the outer membrane (OM) of Gram-negative bacteria (56). This asymmetry, established by having phospholipids confined to the inner leaflet of the membrane and lipopolysaccharides to the outer leaflet, is crucial to the barrier function of the OM. Mutants defective in the Mla pathway are hypersensitive to detergents (56), hydrophobic compounds (69), antibiotics and human serum (70). Besides enhanced sensitivity to FA, the growth of B. ambifaria FA3 was also affected by the presence of toluene and SDS/EDTA (Supplemental material; Table S2 and Table S3). Although the effects of FA on the cell envelope of Gram-negative bacteria have not been evaluated so far, the structure of this mycotoxin, its ability to bind metal cations with high affinity (23), and its oxidative power (11), point to a probable OM disruptive effect. In this scenario, the Mla system would be very important to preserve the integrity of the OM (Fig. 7) . The importance of the OM in the tolerance to FA is illustrated by the fact that Gram-positive species exhibit lower MIC values for this mycotoxin compared to Gram-negative species (19–21, 23).
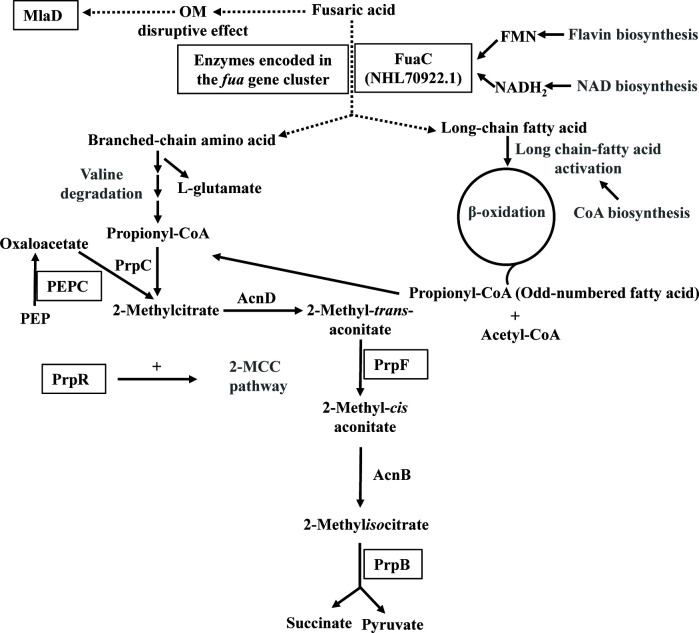
Fig 7.
Schematic representation of key proteins and metabolic pathways likely required for fusaric acid catabolism in B. ambifaria T16. Proteins belonging to metabolic pathways written in gray were significantly more abundant during growth on fusaric acid compared to sodium citrate. Proteins essential for growth in the presence of fusaric acid are black-framed. OM, outer membrane; PEP, phosphoenolpyruvate.
The transposon insertion in the gene EIB72_16280 (B. ambifaria FA21), encoding a predicted acyl-CoA dehydrogenase (NHL67938.1), completely abolished the capability of B. ambifaria T16 to use FA as carbon and nitrogen source. However, the sensitivity to FA was not affected by this insertion, suggesting that this acyl-CoA dehydrogenase could be implicated in FA catabolism. The gene EIB72_16280 clusters with a gene encoding a LysR-type transcriptional regulator (NHL67939.1), two genes for short-chain dehydrogenases, a gene encoding an aminoglycoside phosphotransferase and a gene predicted to encode a protein belonging to the histidine phosphatase superfamily (NHL67935.1). It is noteworthy that the LysR regulator NHL6739.1 has a C-terminal substrate binding domain involved in the catabolism of nitroaromatic/naphthalene compounds (71).
The interruption of the 2-MCC genes prpR, prpF, and prpB (mutants B. ambifaria FA4, FA92, FA91, and FA2) completely abolished the capability of B. ambifaria T16 to grow with FA as the sole carbon and energy source. Moreover, these insertional mutants were also affected in the capability to use FA as sole nitrogen source, but to different extents. According to the proteomic analysis, all 2-MCC enzymes were significantly more abundant during growth with FA compared to CA. Altogether, these results indicated that the 2-methylcitrate pathway plays a very important function during the growth of B. ambifaria T16 on FA as carbon source (Fig. 7) . The role of the 2-MCC in the detoxification of propionyl-CoA and its derived catabolites (e.g., 2-methylcitrate, 2-methylisocitrate) has been extensively demonstrated in bacteria and fungi (59, 72–80). A non-functional 2-MCC causes severe growth inhibition when propionate is added to the growth medium. Propionyl-CoA can be generated by the activation of propionate or from the catabolism of different molecules, such as odd-chain fatty acids, branched-chain amino acids, or some aromatic compounds (81–84). The inability of the 2-MCC insertional mutants to grow with FA, led us to think that propionyl-CoA was generated during FA catabolism (Fig. 7). When the tolerance to FA was evaluated in these mutants, mutant B. ambifaria FA2 which possesses the mini-Tn5 insertion in the gene encoding 2-methylisocitrate lyase (2-MCL)-showed a very high sensitivity to FA. For the remaining 2-MCC mutants, the MIC values obtained were similar to that of the wild-type strain. In agreement with this observation, the loss of 2-MCL activity in M. smegmatis was more detrimental to growth on propionate than the absence of all 2-MCC enzymes (59). Moreover, Eoh and Rhee (75) have demonstrated that the extreme vulnerability of 2-MCL deficient strains during growth on propionate is due to several metabolic defects resulting from a truncated 2-MCC pathway. These defects include depletion of oxaloacetate from the TCA cycle and gluconeogenic pathways, as well as accumulation of 2-methylisocitrate, a noncompetitive inhibitor of the enzyme fructose 1,6-bisphosphatase (80). Our results showed that the absence of the gene encoding 2-MCL in B. ambifaria T16 caused a high sensitivity to FA and the incapability to grow on propionate and FA as carbon sources. The higher sensitivity to FA of the insertional mutant FA2 compared to the ΔprpB strain could be attributed to polar effects on the expression of the genes located downstream prpB (prpC, prpF, or acnD) due to the mini-transposon insertion. The introduction of prpB from B. ambifaria T16 or a gene (icl1) encoding an enzyme of M. smegmatis with dual 2-MCL and isocitrate lyase activities into the ΔprpB mutant of B. ambifaria, increased the tolerance to FA and the ability to grow on FA as sole carbon source, confirming the essentiality of the 2-MCL activity for FA catabolism. Moreover, overexpression of prpB shortened the long lag phase observed for the wild type strain during growth on PA and FA, which reinforces the importance of 2-MCL activity for adaptation under these growth conditions. The lower growth rates obtained for the ΔprpB strain expressing the icl1 gene in comparison with the wild-type strain could be attributed to the augmented isocitrate lyase levels, which competes with the enzyme isocitrate dehydrogenase (85).
Insertion of the mini-Tn5 in the locus EIB72_11905, encoding PEPC, eliminated the capability to grow with FA as carbon source but did not alter the capability to use FA as nitrogen source. The essentiality of PEPC on FA catabolism could be explained considering our hypothesis that propionyl-CoA is generated during FA degradation, and taking into account that the role of PEPC is to replenish oxaloacetate. This compound is one of the substrates of the 2-methylcitrate synthase PrpC, the enzyme that catalyzes the condensation of propionyl-CoA to form 2-methylcitrate (Fig. 7).
The analysis of the proteomic results using the Pathway Tools Omics module (86) showed that several enzymes predicted to be involved in the generation of propionyl-CoA, including acyl-CoA synthetases and proteins involved in valine catabolism were upregulated during the growth on FA as carbon source. Besides, enzymes involved in fatty acid catabolism were also highly overexpressed in M9 + FA. Taking this into account, it is reasonable to assume that a long-chain fatty acid and a branched-chain amino acid would be generated as intermediates during FA catabolism (Fig. 7).
As it was mentioned in the Introduction, FA is toxic for most soil inhabiting microbes (19). According to a screening performed on soil-inhabiting bacteria, the ability to grow on FA as carbon and energy source was not a common characteristic in this environment (32). The enzymes that showed the greatest differences in their intracellular levels during cultivation of B. ambifaria T16 with FA compared with CA (NHL70920.1–70924. 1) are encoded in a gene cluster that we designate fua, due to its likely role in FA catabolism. Interestingly, the fua cluster is located in a genomic region that is highly variable in Burkholderia species, and it was predicted to be part of a genomic island (52).
As the cleavage of the pyridine ring would require the action of an oxygenase and as FMOs were found to be involved in the initial step in the biodegradation of different N-heterocyclic aromatic compounds (65–68), we hypothesize that the flavin-monooxygenase enzyme encoded by fuaC, is involved in the catabolism of FA in B. ambifaria T16. The strain B. ambifaria ΔfuaC (T800) was unable to grow with FA as the sole carbon and nitrogen source. Introduction of fuaC into T800 was able to restore growth, confirming that this gene is essential for FA catabolism. The sensitivity to FA was not affected by the absence of fuaC, suggesting that B. ambifaria T16 possesses mechanisms to cope with the toxic effects of FA, such as an OM disruptive effect and iron sequestration (23). Moreover, the proteomic analysis showed a marked upregulation of some enzymes involved in flavin biosynthesis during cultivation of B. ambifaria T16 with FA, which reinforces the hypothesis of the implication of flavin-dependent enzymes in the degradation of the mycotoxin (Fig. 7).
FA is a pyridine derivative, and although several environmental bacteria are able to degrade different pyridine-containing molecules, there is not much information about the enzymes that catalyze the first step in the catabolism of these compounds. The hydroxylation of the pyridine ring was proposed to be the key initial step involved in the metabolism of this N-heterocyclic compound (87–90). Multi-component molybdenum-containing monooxygenases and mono-component FAD-dependent monooxygenases were reported to catalyze this reaction (87–92). In 2018, Perchant et al. (68) found that the catabolism of N-methylnicotinate started by the pyridine ring cleavage by a two-component FMO from the LLM family. These enzymes were also shown to catalyze the initial ring-opening reaction of other N-heterocyclic compounds, such as uracil (65) and tetramethylpyrazine (67). More recently, Časaité et al. (66) demonstrated that the two-component LLM PyrA catalyzes the first step in the pyridine biodegradation pathway of Arthrobacter sp. 68b. As described in the Results section, the FuaC LLM from B. ambifaria T16 shares between 23% and 34% identity to others LLMs involved in the catabolism of different N-heterocyclic compounds. It is worth mentioning that members of the LLM family usually share low amino acid sequences identity (93).
In conclusion, we report the identification of genes involved in the biodegradation of the mycotoxin FA by the soil bacterium B. ambifaria T16. We were able to demonstrate that a functional 2-MCC is essential for growth on FA as carbon and energy source, suggesting that propionyl-CoA is an intermediate in FA catabolism. Moreover, a two-component FMO of unknown function, encoded in a genomic region predicted to be located in a genomic island, was shown to be fundamental to support the growth of B. ambifaria T16 with FA as sole nitrogen, carbon and energy source (Fig. 7). This enzyme is a good candidate to catalyze the pyridine ring-opening reaction during the biodegradation of FA.
ACKNOWLEDGMENTS
The authors thank Dr. Miguel Valvano for providing plasmids pGPI-SceI and pDAI-SceI, and Victor de Lorenzo for the gift of plasmid pJMT6. The authors are grateful to Dr. Mariana Piuri for Mycobacterium smegmatis mc2 155.
This work was supported by the National Agency for Scientific Promotion, Technological Development and Innovation (AGENCIA I + D + i. Grants PICT-2017-1500 and PICT-2020-SERIEA-02598), the National Research Council (CONICET, PUE 0136 and PIP 0307), and the Alexander von Humboldt Foundation. P.I.N. received financial support from The Novo Nordisk Foundation through grants NNF20CC0035580, LiFe (NNF18OC0034818), and TARGET (NNF21OC0067996), the Danish Council for Independent Research (SWEET, DFF-Research Project 8021-00039B), and the European Union’s Horizon 2020 Research and Innovation Program under grant agreement no. 814418 (SinFonia).
Contributor Information
Jimena A. Ruiz, Email: jruiz@agro.uba.ar.
Arpita Bose, Washington University in St. Louis, Missouri, USA.
DATA AVAILABILITY
The data sets generated and analyzed during the current study are available from the corresponding author on reasonable request.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.00630-23.
Proteomic data set and tolerance to outer membrane disruptive agents
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Bacon CW, Porter JK, Norred WP, Leslie JF. 1996. Production of fusaric acid by Fusarium species. Appl Environ Microbiol 62:4039–4043. doi: 10.1128/aem.62.11.4039-4043.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ma L-J, Geiser DM, Proctor RH, Rooney AP, O’Donnell K, Trail F, Gardiner DM, Manners JM, Kazan K. 2013. Fusarium pathogenomics. Annu Rev Microbiol 67:399–416. doi: 10.1146/annurev-micro-092412-155650 [DOI] [PubMed] [Google Scholar]
- 3. Pegg GF. 1981. Biochemistry and physiology of pathogenesis, p 193–246. In Mace ME, Bell AA, Beckman CH (ed), Fungal wilt diseases of plants. Academic Press Inc, New York. [Google Scholar]
- 4. El-Hassan KI, El-Saman MG, Mosa AA, Mostafa M 2. 2007. Variation among Fusarium spp., the causal of potato tuber dry rot in their pathogenicity and mycotoxins production. Egypt J Phytopathol 35:53–68. [Google Scholar]
- 5. Liu S, Li J, Zhang Y, Liu N, Viljoen A, Mostert D, Zuo C, Hu C, Bi F, Gao H, Sheng O, Deng G, Yang Q, Dong T, Dou T, Yi G, Ma LJ, Li C 2. 2020. Fusaric acid Instigates the invasion of banana by Fusarium oxysporum f. sp. cubense TR4. New Phytol 225:913–929. doi: 10.1111/nph.16193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ding Z, Yang L, Wang G, Guo L, Liu L, Wang J, Huang J. 2018. Fusaric acid is a virulence factor of Fusarium oxysporum f.sp. cubense on banana plantlets. Trop plant pathol 43:297–305. doi: 10.1007/s40858-018-0230-4 [DOI] [Google Scholar]
- 7. López-Díaz C, Rahjoo V, Sulyok M, Ghionna V, Martín-Vicente A, Capilla J, Di Pietro A, López-Berges MS. 2018. Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol Plant Pathol 19:440–453. doi: 10.1111/mpp.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oubraim S, Sedra MH, Lazrek1 HB. 2018. Fusaric acid and phytotoxic metabolites produced by Fusarium oxysporum f.sp. albedinis: effects on date palm and their use for resistance screening trial. J Mater Environ Sci 9:1622–1629. doi: 10.26872/jmes.2018.9.5.180 [DOI] [Google Scholar]
- 9. Chakrabarti DK, Ghosal S 1. 1989. The disease cycle of mango malformation induced by Fusarium moniliforme var. subglutinans and the curative effects of mangiferinmetal chelates. Journal of Phytopathology 125:238–246. doi: 10.1111/j.1439-0434.1989.tb01065.x [DOI] [Google Scholar]
- 10. Singh VK, Upadhyay RS. 2014. Fusaric acid induced cell death and changes in oxidative metabolism of Solanum lycopersicum L. Bot Stud 55:66. doi: 10.1186/s40529-014-0066-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh VK, Singh HB, Upadhyay RS. 2017. Role of fusaric acid in the development of ‘Fusarium wilt symptoms in tomato: physiological, biochemical and proteomic perspectives. Plant Physiol Biochem 118:320–332. doi: 10.1016/j.plaphy.2017.06.028 [DOI] [PubMed] [Google Scholar]
- 12. Pavlovkin J, Mistrík I, Prokop M. 2004. Some aspects of the phytotoxic action of fusaric acid on primary Ricinus roots. Plant Soil Environ 50:397–401. doi: 10.17221/4050-PSE [DOI] [Google Scholar]
- 13. Wang M, Ling N, Dong X, Liu X, Shen Q, Guo S 2. 2014. Effect of fusaric acid on the leaf physiology of cucumber seedlings. Eur J Plant Pathol 138:103–112. doi: 10.1007/s10658-013-0306-4 [DOI] [Google Scholar]
- 14. Nurcahyani E, Sholekhah S, Qudus HI. 2021. Analysis of total carbohydrate and chlorophyll content of the orchid plantlet [Phalaenopsis amabilis (L.) BI.] resistant Fusarium wilt disease. J Phys: Conf Ser 1751. doi: 10.1088/1742-6596/1751/1/012061 [DOI] [Google Scholar]
- 15. Arie T. 2019. Fusarium diseases of cultivated plants, control, diagnosis, and molecular and genetic studies. J Pestic Sci 44:275–281. doi: 10.1584/jpestics.J19-03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fravel D, Olivain C, Alabouvette C. 2003. Fusarium oxysporum and its biocontrol. New Phytol 157:493–502. doi: 10.1046/j.1469-8137.2003.00700.x [DOI] [PubMed] [Google Scholar]
- 17. Yates IE, Arnold JW, Hinton DM, Basinger W, Walcott RR 2. 2003. Fusarium verticillioides induction of maize seed rot and its control. Can J Microbiol 81:422–428. doi: 10.1139/b03-034 [DOI] [Google Scholar]
- 18. Dita M, Barquero M, Heck D, Mizubuti ESG, Staver CP. 2018. Fusarium wilt of banana: current knowledge on epidemiology and research needs toward sustainable disease management. Front Plant Sci 9:1468. doi: 10.3389/fpls.2018.01468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crutcher FK, Puckhaber LS, Stipanovic RD, Bell AA, Nichols RL, Lawrence KS, Liu J. 2017. Microbial resistance mechanisms to the antibiotic and phytotoxin fusaric acid. J Chem Ecol 43:996–1006. doi: 10.1007/s10886-017-0889-x [DOI] [PubMed] [Google Scholar]
- 20. Bacon CW, Hinton DM, Hinton A. 2006. Growth-inhibiting effects of concentrations of fusaric acid on the growth of Bacillus mojavensis and other biocontrol Bacillus species. J Appl Microbiol 100:185–194. doi: 10.1111/j.1365-2672.2005.02770.x [DOI] [PubMed] [Google Scholar]
- 21. Landa BB, Cachinero-Díaz JM, Lemanceau P, Jiménez-Díaz RM, Alabouvette C. 2002. Effect of fusaric acid and phytoanticipins on growth of rhizobacteria and Fusarium oxysporum. Can J Microbiol 48:971–985. doi: 10.1139/w02-094 [DOI] [PubMed] [Google Scholar]
- 22. Sao Emani C, Williams MJ, Wiid IJ, Baker B, Carolis C. 2018. Compounds with potential activity against Mycobacterium tuberculosis. Antimicrob Agents Chemother 62:e02236-17. doi: 10.1128/AAC.02236-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruiz JA, Bernar EM, Jung K. 2015. Production of siderophores increases resistance to fusaric acid in Pseudomonas protegens Pf-5. PLoS One 10:e0117040. doi: 10.1371/journal.pone.0117040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duffy BK, Keel C, Défago G 2. 2004. Potential role of pathogen signaling in multitrophic plant-microbe interactions involved in disease protection. Appl Environ Microbiol 70:1836–1842. doi: 10.1128/AEM.70.3.1836-1842.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Notz R, Maurhofer M, Dubach H, Haas D, Défago G. 2002. Fusaric acid-producing strains of Fusarium oxysporum alter 2,4-diacetylphloroglucinol biosynthetic gene expression in Pseudomonas fluorescens CHA0 in vitro and in the rhizosphere of wheat. Appl Environ Microbiol 68:2229–2235. doi: 10.1128/AEM.68.5.2229-2235.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quecine MC, Kidarsa TA, Goebel NC, Shaffer BT, Henkels MD, Zabriskie TM, Loper JE. 2015. An interspecies signaling system mediated by fusaric acid has parallel effects on antifungal metabolite production by Pseudomonas protegens strain Pf-5 and antibiosis of Fusarium spp. Appl Environ Microbiol 82:1372–1382. doi: 10.1128/AEM.02574-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Rij ET, Girard G, Lugtenberg BJJ, Bloemberg GV. 2005. Influence of fusaric acid on phenazine-1-carboxamide synthesis and gene expression of Pseudomonas chlororaphis strain PCL1391. Microbiology 151:2805–2814. doi: 10.1099/mic.0.28063-0 [DOI] [PubMed] [Google Scholar]
- 28. Venkatesh N, Keller NP. 2019. Mycotoxins in conversation with bacteria and fungi. Front Microbiol 10:403. doi: 10.3389/fmicb.2019.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Crutcher FK, Liu J, Puckhaber LS, Stipanovic RD, Duke SE, Bell AA, Williams HJ, Nichols RL. 2014. Conversion of fusaric acid to fusarinol by Aspergillus tubingensis: a detoxification reaction. J Chem Ecol 40:84–89. doi: 10.1007/s10886-013-0370-4 [DOI] [PubMed] [Google Scholar]
- 30. Crutcher FK, Puckhaber LS, Bell AA, Liu J, Duke SE, Stipanovic RD, Nichols RL. 2017. Detoxification of fusaric acid by the soil microbe Mucor rouxii. J Agric Food Chem 65:4989–4992. doi: 10.1021/acs.jafc.7b01655 [DOI] [PubMed] [Google Scholar]
- 31. Fakhouri W, Walker F, Armbruster W, Buchenauer H. 2003. Detoxification of fusaric acid by a nonpathogenic Colletotrichum sp. Physiological and Molecular Plant Pathology 63:263–269. doi: 10.1016/j.pmpp.2004.03.004 [DOI] [Google Scholar]
- 32. Simonetti E, Roberts IN, Montecchia MS, Gutierrez-Boem FH, Gomez FM, Ruiz JA. 2018. A novel Burkholderia ambifaria strain able to degrade the mycotoxin fusaric acid and to inhibit Fusarium spp. growth. Microbiol Res 206:50–59. doi: 10.1016/j.micres.2017.09.008 [DOI] [PubMed] [Google Scholar]
- 33. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor. NY. [Google Scholar]
- 34. Lageveen RG, Huisman GW, Preusting H, Ketelaar P, Eggink G, Witholt B. 1988. Formation of polyesters by Pseudomonas oleovorans: effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl Environ Microbiol 54:2924–2932. doi: 10.1128/aem.54.12.2924-2932.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Herrero M, de Lorenzo V, Timmis KN. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol 172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller VL, Mekalanos J 1. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x [DOI] [PubMed] [Google Scholar]
- 38. Fernández-Tresguerres ME, Martín M, García de Viedma D, Giraldo R, Díaz-Orejas R. 1995. Host growth temperature and a conservative amino acid substitution in the replication protein of pPS10 influence plasmid host range. J Bacteriol 177:4377–4384. doi: 10.1128/jb.177.15.4377-4384.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanchez-Romero JM, Diaz-Orejas R, De Lorenzo V. 1998. Resistance to tellurite as a selection marker for genetic manipulations of Pseudomonas strains. Appl Environ Microbiol 64:4040–4046. doi: 10.1128/AEM.64.10.4040-4046.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Silva-Rocha R, Martínez-García E, Calles B, Chavarría M, Arce-Rodríguez A, de Las Heras A, Páez-Espino AD, Durante-Rodríguez G, Kim J, Nikel PI, Platero R, de Lorenzo V. 2013. The standard European vector architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res 41:D666–75. doi: 10.1093/nar/gks1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Flannagan RS, Linn T, Valvano MA. 2008. A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ Microbiol 10:1652–1660. doi: 10.1111/j.1462-2920.2008.01576.x [DOI] [PubMed] [Google Scholar]
- 42. Das S, Noe JC, Paik S, Kitten T. 2005. An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J Microbiol Methods 63:89–94. doi: 10.1016/j.mimet.2005.02.011 [DOI] [PubMed] [Google Scholar]
- 43. Martínez-García E, Calles B, Arévalo-Rodríguez M, de Lorenzo V. 2011. PBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol 11:38. doi: 10.1186/1471-2180-11-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simonetti E, Alvarez F, Feldman N, Vinacour M, Roberts IN, Ruiz JA. 2021. Genomic insights into the potent antifungal activity of B. ambifaria T16. Biological Control 155:104530. doi: 10.1016/j.biocontrol.2020.104530 [DOI] [Google Scholar]
- 45. Ausubel FM, Brent R, Kingston R, Moore DD, Seiman J, Smith JA, Struhl K. 1987. Current protocols in molecular biology. John Wiley & Sons, New York. [Google Scholar]
- 46. Martínez-García E, Fraile S, Algar E, Aparicio T, Velázquez E, Calles B, Tas H, Blázquez B, Martín B, Prieto C, Sánchez-Sampedro L, Nørholm MHH, Volke DC, Wirth NT, Dvořák P, Alejaldre L, Grozinger L, Crowther M, Goñi-Moreno A, Nikel PI, Nogales J, de Lorenzo V. 2023. SEVA 4.0: an update of the standard European vector architecture database for advanced analysis and programming of bacterial phenotypes. Nucleic Acids Res 51:D1558–D1567. doi: 10.1093/nar/gkac1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cavaleiro AM, Kim SH, Seppälä S, Nielsen MT, Nørholm MHH. 2015. Accurate DNA assembly and genome engineering with optimized uracil excision cloning. ACS Synth Biol 4:1042–1046. doi: 10.1021/acssynbio.5b00113 [DOI] [PubMed] [Google Scholar]
- 48. Hilgarth RS, Lanigan TM. 2020. Optimization of overlap extension PCR for efficient transgene construction. MethodsX 7:100759. doi: 10.1016/j.mex.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martínez-García E, de Lorenzo V. 2011. Engineering multiple genomic deletions in gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ Microbiol 13:2702–2716. doi: 10.1111/j.1462-2920.2011.02538.x [DOI] [PubMed] [Google Scholar]
- 50. Gasperotti A, Göing S, Fajardo-Ruiz E, Forné I, Jung K. 2020. Function and regulation of the pyruvate transporter CstA in Escherichia coli. Int J Mol Sci 21:9068. doi: 10.3390/ijms21239068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. doi: 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 52. Alvarez F, Simonetti E, Draghi WO, Vinacour M, Palumbo MC, Do Porto DF, Montecchia MS, Roberts IN, Ruiz JA. 2022. Genome mining of Burkholderia ambifaria strain T16, a rhizobacterium able to produce antimicrobial compounds and degrade the mycotoxin fusaric acid. World J Microbiol Biotechnol 38:114. doi: 10.1007/s11274-022-03299-0 [DOI] [PubMed] [Google Scholar]
- 53. Paley S, Parker K, Spaulding A, Tomb J-F, O’Maille P, Karp PD. 2017. The omics dashboard for interactive exploration of gene-expression data. Nucleic Acids Res 45:12113–12124. doi: 10.1093/nar/gkx910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Suvorova IA, Ravcheev DA, Gelfand MS. 2012. Regulation and evolution of malonate and propionate catabolism in proteobacteria. J Bacteriol 194:3234–3240. doi: 10.1128/JB.00163-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. doi: 10.1073/pnas.0903229106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Deutscher MP. 2009. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci 85:369–391. doi: 10.1016/S0079-6603(08)00809-X [DOI] [PubMed] [Google Scholar]
- 58. Del Campo M, Kaya Y, Ofengand J. 2001. Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA 7:1603–1615. [PMC free article] [PubMed] [Google Scholar]
- 59. Upton AM, McKinney JD. 2007. Role of the methylcitrate cycle in propionate metabolism and detoxification in Mycobacterium smegmatis. Microbiology 153:3973–3982. doi: 10.1099/mic.0.2007/011726-0 [DOI] [PubMed] [Google Scholar]
- 60. Ogasawara H, Ohe S, Ishihama A. 2015. Role of transcription factor NimR (YeaM) in sensitivity control of Escherichia coli to 2-nitroimidazole. FEMS Microbiol Lett 362:1–8. doi: 10.1093/femsle/fnu013 [DOI] [PubMed] [Google Scholar]
- 61. Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G. 2010. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci U S A 107:14390–14395. doi: 10.1073/pnas.1005399107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bruender NA, Bandarian V. 2017. The creatininase homolog MftE from Mycobacterium smegmatis catalyzes a peptide cleavage reaction in the biosynthesis of a novel ribosomally synthesized post-translationally modified peptide (RiPP). J Biol Chem 292:4371–4381. doi: 10.1074/jbc.M116.762062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsuru D, Oka I, Yoshimoto T 1. 1976. Creatinine decomposing enzymes in Pseudomonas putida. Agric Biol Chem 40:1011–1018. doi: 10.1271/bbb1961.40.1011 [DOI] [Google Scholar]
- 64. Huijbers MME, Montersino S, Westphal AH, Tischler D, van Berkel WJH. 2014. Flavin dependent monooxygenases. Arch Biochem Biophys 544:2–17. doi: 10.1016/j.abb.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 65. Mukherjee T, Zhang Y, Abdelwahed S, Ealick SE, Begley TP. 2010. Catalysis of a flavoenzyme-mediated amide hydrolysis. J Am Chem Soc 132:5550–5551. doi: 10.1021/ja9107676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Časaitė V, Stanislauskienė R, Vaitekūnas J, Tauraitė D, Rutkienė R, Gasparavičiūtė R, Meškys R. 2020. Microbial degradation of pyridine: a complete pathway in Arthrobacter sp. strain 68b deciphered. Appl Environ Microbiol 86:e00902-20. doi: 10.1128/AEM.00902-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kutanovas S, Stankeviciute J, Urbelis G, Tauraite D, Rutkiene R, Meskys R. 2013. Identification and characterization of a tetramethylpyrazine catabolic pathway in Rhodococcus jostii TMP1. Appl Environ Microbiol 79:3649–3657. doi: 10.1128/AEM.00011-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Perchat N, Saaidi P-L, Darii E, Pellé C, Petit J-L, Besnard-Gonnet M, de Berardinis V, Dupont M, Gimbernat A, Salanoubat M, Fischer C, Perret A. 2018. Elucidation of the trigonelline degradation pathway reveals previously undescribed enzymes and metabolites. Proc Natl Acad Sci U S A 115:E4358–E4367. doi: 10.1073/pnas.1722368115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim K, Lee S, Lee K, Lim D. 1998. Isolation and characterization of toluene-sensitive mutants from the toluene-resistant bacterium Pseudomonas putida GM73. J Bacteriol 180:3692–3696. doi: 10.1128/JB.180.14.3692-3696.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bernier SP, Son S, Surette MG. 2018. The MLA pathway plays an essential role in the intrinsic resistance of Burkholderia cepacia complex species to antimicrobials and host innate components. J Bacteriol 200:e00156-18. doi: 10.1128/JB.00156-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A. 2020. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268. doi: 10.1093/nar/gkz991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Brock M. 2005. Generation and phenotypic characterization of Aspergillus nidulans methylisocitrate lyase deletion mutants: methylisocitrate inhibits growth and conidiation. Appl Environ Microbiol 71:5465–5475. doi: 10.1128/AEM.71.9.5465-5475.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Brock M, Buckel W 2. n.d. On the mechanism of action of the antifungal agent propionate propionyl-CoA inhibits glucose metabolism in Aspergillus nidulans. Eur J Biochem 271:3227–3241. doi: 10.1111/j.1432-1033.2004.04255.x [DOI] [PubMed] [Google Scholar]
- 74. Brock M, Fischer R, Linder D, Buckel W. 2000. Methylcitrate synthase from Aspergillus nidulans: implications for propionate as an antifungal agent. Mol Microbiol 35:961–973. doi: 10.1046/j.1365-2958.2000.01737.x [DOI] [PubMed] [Google Scholar]
- 75. Eoh H, Rhee KY. 2014. Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids. Proc Natl Acad Sci U S A 111:4976–4981. doi: 10.1073/pnas.1400390111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Horswill AR, Dudding AR, Escalante-Semerena JC. 2001. Studies of propionate toxicity in Salmonella enterica identify 2-methylcitrate as a potent inhibitor of cell growth. J Biol Chem 276:19094–19101. doi: 10.1074/jbc.M100244200 [DOI] [PubMed] [Google Scholar]
- 77. Lee JJ, Lim J, Gao S, Lawson CP, Odell M, Raheem S, Woo J, Kang SH, Kang SS, Jeon BY, Eoh H. 2018. Glutamate mediated metabolic neutralization mitigates propionate toxicity in intracellular Mycobacterium tuberculosis. Sci Rep 8:8506. doi: 10.1038/s41598-018-26950-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Maruyama K, Kitamura H. 1985. Mechanisms of growth inhibition by propionate and restoration of the growth by sodium bicarbonate or acetate in Rhodopseudomonas sphaeroides S. J Biochem 98:819–824. doi: 10.1093/oxfordjournals.jbchem.a135340 [DOI] [PubMed] [Google Scholar]
- 79. Plassmeier J, Barsch A, Persicke M, Niehaus K, Kalinowski J. 2007. Investigation of central carbon metabolism and the 2-methylcitrate cycle in Corynebacterium glutamicum by metabolic profiling using gas chromatography-mass spectrometry. J Biotechnol 130:354–363. doi: 10.1016/j.jbiotec.2007.04.026 [DOI] [PubMed] [Google Scholar]
- 80. Rocco CJ, Escalante-Semerena JC. 2010. In Salmonella enterica, 2-methylcitrate blocks gluconeogenesis. J Bacteriol 192:771–778. doi: 10.1128/JB.01301-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Johnson GR, Jain RK, Spain JC. 2000. Properties of the trihydroxytoluene oxygenase from Burkholderia cepacia R34: an extradiol dioxygenase from the 2,4-dinitrotoluene pathway. Arch Microbiol 173:86–90. doi: 10.1007/s002039900111 [DOI] [PubMed] [Google Scholar]
- 82. Nazarova EV, Montague CR, La T, Wilburn KM, Sukumar N, Lee W, Caldwell S, Russell DG, VanderVen BC. 2017. Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. Elife 6:e26969. doi: 10.7554/eLife.26969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Suen WC, Spain JC. 1993. Cloning and characterization of Pseudomonas sp. strain DNT genes for 2,4-dinitrotoluene degradation. J Bacteriol 175:1831–1837. doi: 10.1128/jb.175.6.1831-1837.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang Y-Q, Brock M, Keller NP. 2004. Connection of Propionyl-CoA metabolism to polyketide biosynthesis in Aspergillus nidulans. Genetics 168:785–794. doi: 10.1534/genetics.104.027540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cozzone AJ. 1998. Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Annu Rev Microbiol 52:127–164. doi: 10.1146/annurev.micro.52.1.127 [DOI] [PubMed] [Google Scholar]
- 86. Karp PD, Midford PE, Billington R, Kothari A, Krummenacker M, Latendresse M, Ong WK, Subhraveti P, Caspi R, Fulcher C, Keseler IM, Paley SM. 2021. Pathway tools version 23.0 update: software for pathway/genome informatics and systems biology. Brief Bioinform 22:109–126. doi: 10.1093/bib/bbz104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jiménez JI, Canales A, Jiménez-Barbero J, Ginalski K, Rychlewski L, García JL, Díaz E. 2008. Deciphering the genetic determinants for aerobic nicotinic acid degradation: the NIC cluster from Pseudomonas putida KT2440. Proc Natl Acad Sci U S A 105:11329–11334. doi: 10.1073/pnas.0802273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li H, Xie K, Yu W, Hu L, Huang H, Xie H, Wang S, Löffler FE. 2016. Nicotine dehydrogenase complexed with 6-hydroxypseudooxynicotine oxidase involved in the hybrid nicotine-degrading pathway in Agrobacterium tumefaciens S33. Appl Environ Microbiol 82:1745–1755. doi: 10.1128/AEM.03909-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Qiu J, Liu B, Zhao L, Zhang Y, Cheng D, Yan X, Jiang J, Hong Q, He J, Kivisaar M. 2018. A novel degradation mechanism for pyridine derivatives in Alcaligenes faecalis JQ135. Appl Environ Microbiol 84:e00910-18. doi: 10.1128/AEM.00910-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yu H, Tang H, Li Y, Xu P, Kivisaar M. 2015. Molybdenum-containing nicotine hydroxylase genes in a nicotine degradation pathway that is a variant of the pyridine and pyrrolidine pathways. Appl Environ Microbiol 81:8330–8338. doi: 10.1128/AEM.02253-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Qiu J, Zhao L, Xu S, Chen Q, Chen L, Liu B, Hong Q, Lu Z, He J. 2019. Identification and characterization of a novel pic gene cluster responsible for picolinic acid degradation in Alcaligenes faecalis JQ135. J Bacteriol 201:e00077-19. doi: 10.1128/JB.00077-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yu H, Hausinger RP, Tang H-Z, Xu P. 2014. Mechanism of the 6-hydroxy-3-succinoyl-pyridine 3-monooxygenase flavoprotein from Pseudomonas putida S16. J Biol Chem 289:29158–29170. doi: 10.1074/jbc.M114.558049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ellis HR. 2010. The FMN-dependent two-component monooxygenase systems. Arch Biochem Biophys 497:1–12. doi: 10.1016/j.abb.2010.02.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteomic data set and tolerance to outer membrane disruptive agents
Data Availability Statement
The data sets generated and analyzed during the current study are available from the corresponding author on reasonable request.