ABSTRACT
Shotgun metagenomic sequencing has the potential to provide bacterial strain-level resolution which is of key importance to tackle a host of clinical questions. While bioinformatic tools that achieve strain-level resolution are available, thorough benchmarking is needed to validate their use for less investigated and low biomass microbiomes like those from the upper respiratory tract. We analyzed a previously published data set of longitudinally collected nasopharyngeal samples from Bangladeshi infants (Microbiota and Health study) and a novel data set of oropharyngeal samples from Swiss children with cystic fibrosis. Data from bacterial cultures were used for benchmarking the parameters of StrainPhlAn 3, a bioinformatic tool designed for strain-level resolution. In addition, StrainPhlAn 3 results were compared with metagenomic assemblies derived from StrainGE and newly derived whole-genome sequencing data. After optimizing the analytical parameters, we compared StrainPhlAn 3 results to culture gold standard methods and achieved sensitivity values of 87% (Streptococcus pneumoniae), 80% (Moraxella catarrhalis), 75% (Haemophilus influenzae), and 57% (Staphylococcus aureus) for 420 nasopharyngeal and 75% (H. influenzae) and 46% (S. aureus) for 260 oropharyngeal samples. Comparing the phylogenetic tree of the core genome of 50 S. aureus isolates with a corresponding marker gene tree generated by StrainPhlAn 3 revealed a striking similarity in tree topology for all but three samples indicating adequate strain resolution. In conclusion, a comparison of StrainPhlAn 3 results to data from bacterial cultures revealed that strain-level tracking of the respiratory microbiome is feasible despite the high content of host DNA when parameters are carefully optimized to fit low biomass microbiomes.
IMPORTANCE
The usage of 16S rRNA gene sequencing has become the state-of-the-art method for the characterization of the microbiota in health and respiratory disease. The method is reliable for low biomass samples due to prior amplification of the 16S rRNA gene but has limitations as species and certainly strain identification is not possible. However, the usage of metagenomic tools for the analyses of microbiome data from low biomass samples is not straight forward, and careful optimization is needed. In this work, we show that by validating StrainPhlAn 3 results with the data from bacterial cultures, the strain-level tracking of the respiratory microbiome is feasible despite the high content of host DNA being present when parameters are carefully optimized to fit low biomass microbiomes. This work further proposes that strain retention analyses are feasible, at least for more abundant species. This will help to better understand the longitudinal dynamics of the upper respiratory microbiome during health and disease.
KEYWORDS: respiratory tract, metagenomics, bacterial culture, strain resolution, genome analysis
INTRODUCTION
The development of high-throughput sequencing technologies has greatly facilitated the characterization of the human microbiome in health and disease. 16S rRNA gene amplicon sequencing has been used to characterize the human microbiota, including the one from the respiratory tract (1). Disordered microbiota patterns have been found in the respiratory tract of patients suffering from diseases with obvious involvement of microbes such as cystic fibrosis (CF) (2–4). Disordered microbiota patterns have also been observed in other conditions like asthma (5), chronic obstructive pulmonary disease (6–9), and interstitial lung disease (10), where bacterial respiratory infections are not known to be the main drivers of disease. However, 16S rRNA gene sequencing does not allow for the investigation of strain retention due to limited strain resolution within bacterial taxa.
Strain retention is usually investigated using traditional culturing methods combined with whole genome sequencing (WGS) from longitudinally collected isolates. As an example, convergent evolution and adaption of Pseudomonas aeruginosa and Staphylococcus aureus strains have been studied in patients with CF (11, 12). However, for most species, including mainly commensal or difficult to culture bacteria of the respiratory microbiome, strain retention remains understudied. Using shotgun metagenomic sequencing and appropriate bioinformatic tools could allow investigating strain retention of microbial species beyond selected culturable pathobionts. For example, StrainPhlAn has been developed to achieve microbial strain-level resolution using shotgun metagenomic data for more than 1,500 gut metagenomes (13). StrainPhlAn is based on reconstructing consensus sequence variants within species-specific marker genes and using them to estimate strain-level phylogenies. In contrast to other tools like StrainGE (14), short-read sequences do not have to be assembled into contigs. It is less clear if strain resolution can also be achieved on less investigated microbiomes, such as respiratory samples, which contain a high proportion of host-derived DNA (15). Therefore, using such strain-level resolution tools requires careful benchmarking to address low bacterial biomass (16).
Lower airway samples are considered to be clinically relevant for a number of respiratory diseases, but the sampling is invasive. However, it has been hypothesized that the lower airway microbiome is seeded by the oropharyngeal (OP) and nasopharyngeal (NP) microbiome (17).
Within this study, we aimed to optimize StrainPhlAn 3 to achieve strain resolution from low-biomass samples from the respiratory tract. To achieve this aim, we used culture and shotgun sequencing data from upper airway samples from a previously published NP data set from Bangladeshi infants (18) and a new data set from Swiss children with CF. To test the applicability of the approach on low biomass, we analyzed the retention over time of multiple strains of clinically relevant microbial species.
RESULTS
Data characteristics and bacterial species assignment
An NP and OP data set comprising both bacterial culture and shotgun metagenomic data were investigated for this study (Table 1). The read numbers of the NP data ranged from 40.1M reads to 186.5M reads, with a mean of 76.1M reads. After removing the human reads, the range of bacterial reads was between 0.3M and 74.8M, with a mean value of 11.7M. For the OP data, the number of reads varied between 14M and 199M with an average of 65.6M and between 0.07M and 158.7M with an average of 19.0M after removing human reads.
TABLE 1.
Characteristics of data setsa
| Type of samples | Age |
N (individuals) |
N (samples) |
Sequencing information | Culture results | Disease | Origin | Reference |
|---|---|---|---|---|---|---|---|---|
| NP | 2 mo | 223 | 223 | 2 × 150 bp paired-end sequencing; Illumina HiSeq; 11.4 Gb (mean) | 119 of 221 (54%; SP), 69 of 222 (31%; HI), 75 of 222 (33%; MC) 92 of 222 (41; SA) |
H | BD | (18) |
| NP | 4 mo | 199 | 199 | 2 × 150 bp paired-end sequencing; Illumina HiSeq; 11.4 Gb (mean) | 129 of 198 (65%; SP), 93 of 198 (47%; HI), 75 of 198 (38%; MC) 30 of 198 (15%; SA) |
H | BD | (18) |
| OP | <4 yr | 15 | 96 | NovaSeq 6000 PE150; 10 G of raw data per sample | 26 (27%; HI), 38 (40%; SA), 5 (5%; PA) |
CF | CH | This study |
| OP | 4–6 yr | 15 | 87 | NovaSeq 6000 PE150; 10 G of raw data per sample | 31 (36%; HI), 36 (41%; SA), 5 (6 %; PA) |
CF | CH | This study |
| OP | 6–8 yr | 14 | 60 | NovaSeq 6000 PE150; 10 G of raw data per sample | 19 (32%; HI), 34 (57%; SA), 1 (2%; PA) |
CF | CH | This study |
| OP | >8 yr | 8 | 16 | NovaSeq 6000 PE150; 10 G of raw data per sample | 8 (50%; HI), 15 (94%; SA) |
CF | CH | This study |
The total number of samples with available cultures from the NP samples are: M. catarrhalis (MC) (N = 378), H. influenzae (HI) (N = 380), S. pneumoniae (SP) (N = 377), and S. aureus (SA) (N = 375). Pseudomonas aeruginosa (PA) was only cultured for OP samples. For 1 OP sample, age was not available. Healthy individuals (H) and individuals with cystic fibrosis (CF) were included from Bangladesh (BD) and Switzerland (CH), respectively. Individuals were aged from 2 mo up to 10 yr.
Using the remaining bacterial reads of both data sets and comparing them with culture data, we first assessed species assignment (Table 2; Fig. S1). Compared to bacterial culture data of four species, MetaPhlAn 3 and Metagenome-Atlas showed comparable sensitivity, specificity, and F1 values (Table 2).
TABLE 2.
Performance of bioinformatics tools at the species levela
| Species (data set) | Analysis tool | TPb | TN | Specificity | Sensitivity | F1 score |
|---|---|---|---|---|---|---|
| S. pneumoniae (NP) | MetaPhlAn 3 | 240 | 121 | 71% | 97% | 0.89 |
| Metagenome-Atlas | 224 | 102 | 66% | 95% | 0.88 | |
| StrainGE | 73 | 151 | 97% | 31% | 0.47 | |
| StrainPhlan 3 df | 168 | 154 | 90% | 68% | 0.78 | |
| StrainPhlan 3 new | 215 | 126 | 74% | 87% | 0.85 | |
| S. aureus (NP) | MetaPhlAn 3.0 | 87 | 257 | 86% | 71% | 0.70 |
| Metagenome-Atlas | 74 | 253 | 90% | 67% | 0.70 | |
| StrainGE | 10 | 275 | 98% | 9% | 0.16 | |
| StrainPhlan 3 df | na | na | na | na | Na | |
| StrainPhlan 3 new | 70 | 278 | 93% | 57% | 0.66 | |
| S. aureus (OP) | MetaPhlAn 3 | 74 | 131 | 98% | 60% | 0.74 |
| StainPhlAn df | 40 | 134 | 100% | 33% | 0.49 | |
| StrainPhlan 3 new | 56 | 132 | 99% | 46% | 0.62 | |
| M. catarrhalis (NP) | MetaPhlAn 3 | 132 | 228 | 84% | 89% | 0.81 |
| Metagenome-Atlas | 119 | 218 | 87% | 85% | 0.82 | |
| StrainGE | 53 | 241 | 96% | 38% | 0.52 | |
| StrainPhlan 3 df | 89 | 256 | 94% | 60% | 0.70 | |
| StrainPhlan 3 new | 119 | 243 | 89% | 80% | 0.80 | |
| H. influenzae (NP) | MetaPhlAn 3 | 152 | 205 | 79% | 94% | 0.83 |
| Metagenome-Atlas | 150 | 182 | 77% | 96% | 0.84 | |
| StrainGE | 60 | 226 | 96% | 38% | 0.53 | |
| StrainPhlan 3 df | na | na | na | na | na | |
| StrainPhlan 3 new | 122 | 233 | 90% | 75% | 0.79 | |
| H. influenzae (OP) | MetaPhlAn 3 | 66 | 96 | 55% | 80% | 0.58 |
| StrainPhlan 3 df | na | na | na | na | na | |
| StrainPhlan 3 new | 62 | 121 | 70% | 75% | 0.63 |
Data for selected bacterial species from NP and OP are indicated for default (df) and new settings (new). Culture results were considered as the “golden standard.” For certain settings, no values were achievable for the available data (na). Strain resolution tools are indicated in italic.
TP = True Positive; TN = True negative. See text for details.
Adjusting StrainPhlAn 3 parameters for optimal species detection of low biomass samples
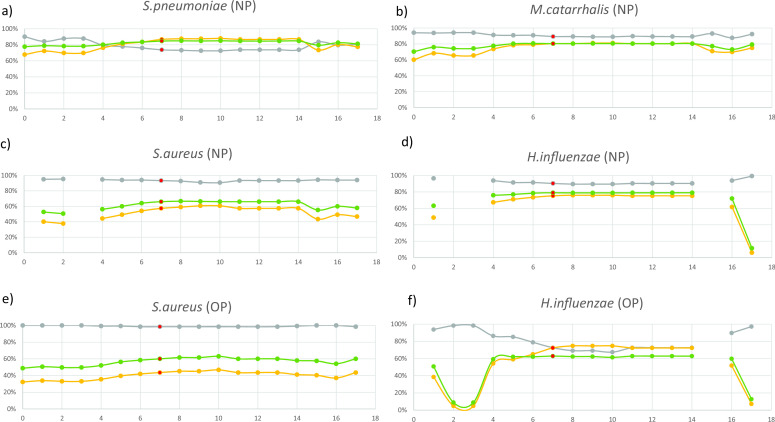
We next assessed how species detection performed when using higher coverage thresholds required by strain resolution tools StrainPhlAn 3 and StrainGE, comparing again the results to the culture data. Using default parameters, both tools showed low sensitivity (being less than 0.7) and F1 values for Streptococcus pneumoniae, Moraxella catarrhalis, S. aureus, and Haemophilus influenzae from NP samples (Table 2). However, StrainPhlAn performed better than StrainGE, presumably because, in contrast to StrainGE, it does not rely on an assembly expected to perform poor for low biomass samples. That is why, we focused our efforts on optimizing StrainPhlAn, hypothesizing that using less stringent parameters to further adapt for low coverage rates (and hence improving species detection), would not affect the performance of the tool for strain resolution. Benchmarking results of different parameter sets were received for the four (S. pneumoniae, M. catarrhalis, S. aureus, and H. influenzae) and the two species (S. aureus and H. influenzae) from the NP and OP data set, respectively (Fig. 1). The overall plot then suggested that the set of parameters No. 7 with a F1 value of 0.72 were the most optimal ones for further analysis (Fig. S2; Table S1). Table 2 shows the performance values for species detection using the newly set of parameters No. 7. Using these optimized parameters, we noticed that StrainPhlAn 3 analyses would also be feasible for frequently found commensal bacterial species in the NP data set, i.e., Streptococcus mitis (n = 281), Dolosigranulum pigrum (n = 240), and Corynebacterium pseudodiphtheriticum (n = 165) for which, however, no culture data were available to confirm the taxonomic assignment (Table S2).
Fig 1.
Benchmarking the different set of parameters. The default set of parameters is illustrated in run No. 0 and is BREADTH_THRESHOLD 80, TRIM_SEQUENCES 50, MARKER_IN_N_SAMPLES 80, and SAMPLE_WITH_N_MARKERS 20. Panels a, b, c, and d represent benchmarking for NP data; e and f for OP data. The default (No. 0) and 17 different sets of parameters (No. 1–17) are illustrated on the x-axis, and their characteristics are described in Table S1. Gray = specificity, yellow = sensitivity, and green = F1-score. The optimized sets of parameters are shown at No. 7 (indicated in red).
Assessment of StrainPhlAn 3 for species resolution
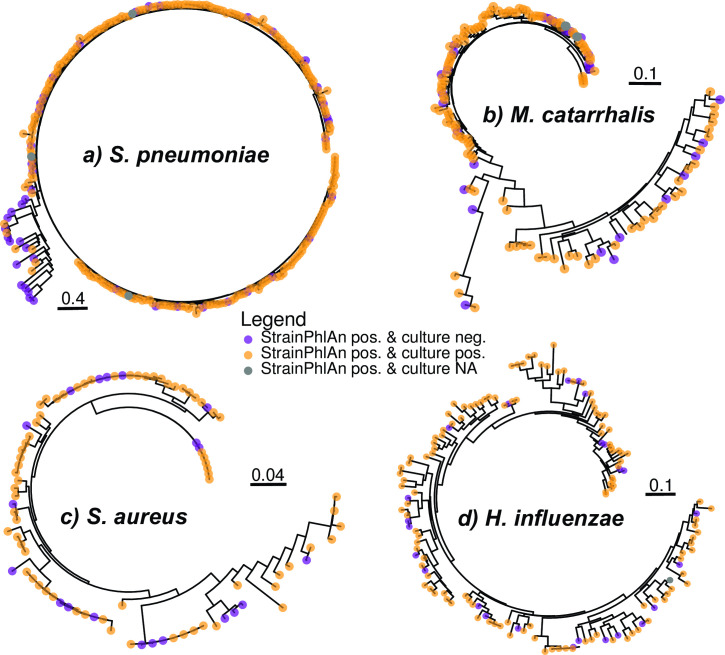
To further investigate the limitations of the specificity and sensitivity of bacterial species captured by StrainPhlAn 3, we subsequently inspected trees generated by StrainPhlAn 3 for S. pneumoniae, M. catarrhalis, S. aureus, and H. influenzae (Fig. 2A through D). For S. pneumoniae, we identified a small, separate cluster of 22 samples for which a positive culture of S. pneumoniae was only found in six samples (Fig. 2A). To investigate the latter, we additionally added the streptococcal metagenome-assembled genomes (MAGs) from the Metagenome-Atlas analyses run on the same samples as reference genomes into the StrainPhlAn 3 tree. We found that the tree indeed split into S. pneumoniae and non-S. pneumoniae strains (Fig. S3). S. pneumoniae was also frequently detected in the OP samples by StrainPhlAn 3 (n = 246; 94.6%; Table S3) despite only a single S. pneumoniae culture was observed (n = 1; 0.4%). Taken together, these analyses suggest that StrainPhlAn occasionally misidentifies S. pneumoniae, although we cannot completely rule out, that few S. pneumoniae isolates were missed with the applied culture conditions. The StrainPhlAn 3 tree for M. catarrhalis also splits into two clusters, but in contrast to S. pneumoniae, both were readily detected through culture (Fig. 2B). No obvious clustering has been observed for S. aureus and H. influenzae and samples which were culture negative but species positive according to StrainlPhlAn 3 were found randomly all along the trees (Fig. 2C and D).
Fig 2.
StrainPhlAn-derived trees from NP data sets. Trees are StrainPhAn 3 generated for (a) S. pneumoniae, (b) M. catarrhalis, (c) S. aureus. and (d) H. influenzae from NP data and were using the optimized parameters of StrainPhlAn 3. Samples are indicated in different colors depending on the presence/absence of culture data [i.e. purple = found in StrainPhlAn but not culture, yellow = found with StrainPhlAn and culture, and gray = found with StrainPhlAn but no culture data available (NA)].
The low detection rates of S. aureus using StrainPhlAn 3 (Table 2) were also further investigated in the OP samples. We investigated if false positives/negatives were caused by a lower coverage, but we did not observe any difference in the mean bacterial reads (Fig. S4A). We also compared quantitative culture information (from 0 to 4 according to number of colonies during culturing) with StrainPhlAn 3 output (Fig. S4B). We found that the higher the quantity of S. aureus detected by culture, the more likely S. aureus was also detected by StrainPhlAn 3 (P < 0.01). This correlation was independent of the total number of (bacterial) reads, i.e., read coverage.
Evaluating newly defined parameters of StrainPhlAn 3 for strain resolution
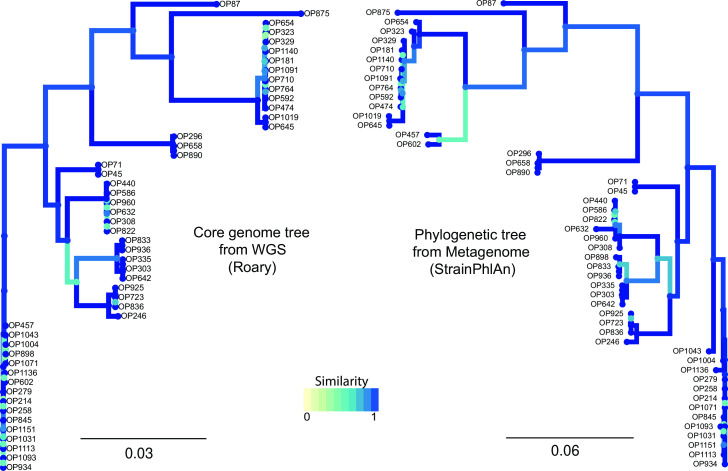
We next compared strain diversity of S. aureus captured by StrainPhlAn 3 to strain diversity represented by WGS of S. aureus isolated from OP samples by visualizing side by side the phylogenetic tree generated by each approach (Fig. 3). Despite using different data sources and analyses, both trees showed a strikingly congruent topology, with only three samples (OP457, OP602, and OP898) showing a difference in placement. This could be because the isolated strains differed from the most dominant one detected through StrainPhlAn 3.
Fig 3.
Phylogenetic trees of S. aureus from OP data sets. On the left, core genome tree from WGS data of S. aureus isolates. On the right, the phylogenetic tree was created from metagenomic data of OP samples using the adjusted parameters of StraiPhlAn. Sample IDs are indicated in black.
We also investigated whether the adjusted parameters of StrainPhlAn 3 (which improved species detection) would diminish the performance of strain resolution as compared to the default parameters. Comparing StrainPhlAn 3 trees of S. aureus between default and adjusted parameters showed comparable results, indicating that our change of parameters did not impact strain resolution (Fig. S5).
To further investigate if our chosen parameters are too relaxed and, therefore, can produce low-quality phylogenetic results, we have also created the StrainPhlAn 3 trees of M. catarrhalis between default and adjusted parameters from the NP samples (Fig. S6). Again, the results were congruent indicating that our parameters are not too relaxed and still produce similar quality phylogenetic results as compared to the standard settings and, at the same time, improve species detection. However, careful benchmarking is generally recommended for other bacterial species and data from other studies if non-default parameters are to be used.
Evaluating pairwise SNVs and strain retention in NP samples
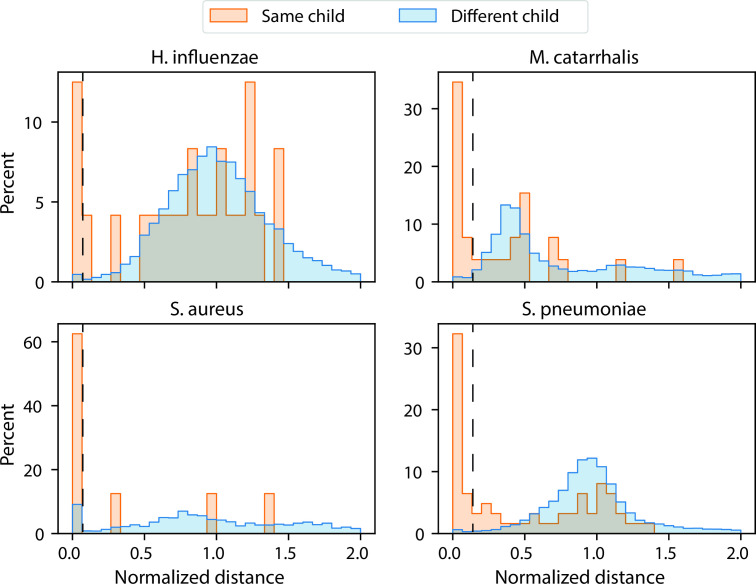
Having defined the optimized parameters for StrainPhlAn 3 for our data sets, we next investigated strain retention in the NP samples which were collected at two different time points for each subject. We first generated pairwise single nucleotide variation (SNV) rates for S. pneumoniae, M. catarrhalis, H. influenzae, and S. aureus of the NP data set (Fig. S7). We then binned the normalized distance values into intervals of 0.1 and visualized the results in a linear and log-transformed manner (Fig. 4; Fig. S8 and S9). For different infants (inter-comparisons), we found that values from different timepoints were strikingly comparable to values from the same timepoints (Fig. S8).
Fig 4.
Strain retention analyses based on normalized genetic distances of four different species of the NP data set. The all-versus-all normalized genetic distances have been separately calculated for S. pneumoniae, M. catarrhalis, H. influenzae, and S. aureus. Values were binned in intervals of 0.1. Bins with values 0.0–0.1 (for S. aureus and H. influenzae) and 0.0–0.2 (for S. pneumoniae and M. catarrhalis) were defined for strain retention (dotted lines; see text for details).
However, as expected, the distance values were lower in samples from the same infant as compared to different infant (Fig. 4; Fig. S9). Therefore, frequent longitudinal carriage of identical isolates, i.e., strain retention can be assumed. Based on inter- vs intra-comparisons, we suggest defining bins with values 0.0–0.1 (for S. aureus and H. influenzae) and 0.0–0.2 (for S. pneumoniae and M. catarrhalis) for strain retention for the four species. Using these definitions, the values for strain retention were 40.8% (S. pneumoniae), 41.9% (M. catarrhalis) 16.7% (H. influenzae), and 55.6% (S. aureus; Fig. 4).
Investigating strain retention of S. aureus over time in children with CF
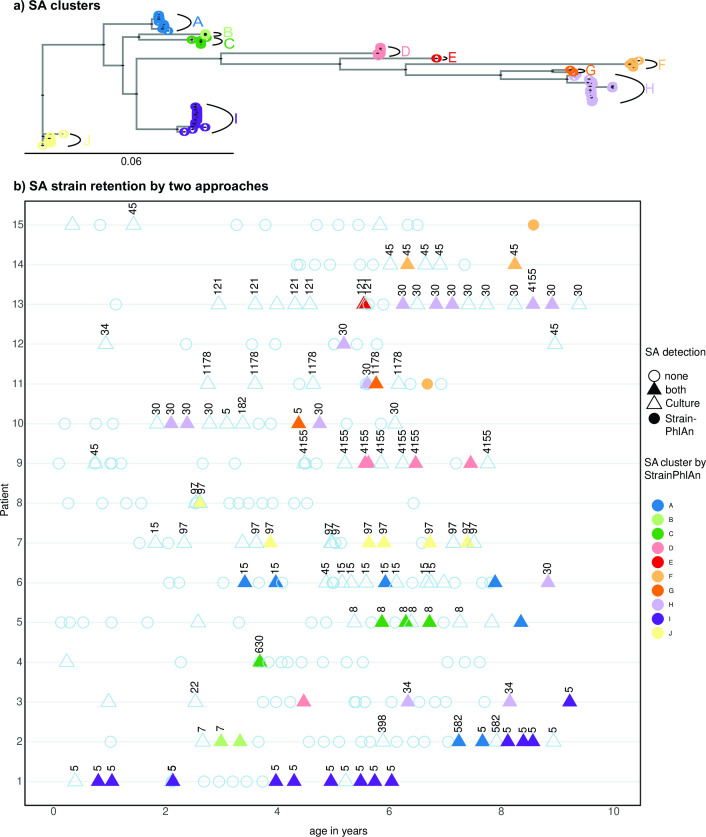
Retaining of S. aureus strains over time was also investigated in the OP samples. Based on the StrainPhlAn tree of S. aureus, we found 10 clusters of samples which were subsequently used as information for the definition of strain retention in the OP samples (Fig. 5A). Figure 5B shows this cluster assignment within samples over time and, in addition, Multi-Locus Sequence Typing (MLST) data (extracted from WGS data from isolates genomes). Generally, we found that up to three strains were retained for the majority of patients as seen for both metagenomic clusters and MLST. In some cases, we observed that the same strain was retained for many years (ST97 and cluster J for subject ID7; ST5 and cluster I for subject ID1). This analysis shows that strain resolution derived from metagenomic data is at least comparable to the traditionally used MLST scheme (at least for S. aureus).
Fig 5.
Clustering and longitudinal visualization of S. aureus strains of OP samples. Phylogenetic tree was created with StrainPhlAn using the new parameters from metagenomic data, highlighting 10 clusters (1-J) (Fig. 5A). Strain retention of S. aureus was investigated in a total of fifteen individuals with CF, sampled for up to the first 10 years of life (Fig. 5B). Strain retainment was investigated by (i) extracting the MLST from the WGS from S. aureus (SA) and (ii) using the clustering information from metagenomic data (shown as SA_clusters from A-J with different colors). For the detection of SA we used four categories. No S. aureus found by culture and metagenomic sequencing (indicated by an empty circle), S. aureus found by culture and metagenomic sequencing (filled triangle), S. aureus found by culture but not metagenomic sequencing (empty triangle), and S. aureus found by metagenomic sequencing but not by culture (filled circle). StrainPhlAn clusters are indicated with different colors. Numbers reflect the sequence types from MLST found in the respective S. aureus isolates. As for triangles without numbers, S. aureus by culture has been reported, but the isolate has not been kept for WGS.
DISCUSSION
Bacterial strain characterization is indispensable for, in particular, clinically relevant bacteria also called pathobionts (19). It is classically achieved by bacterial culture and subsequent genomic characterization. This is cumbersome if many members of the microbiome are to be characterized, and/or large sample sets are included. Metagenomic sequencing has the potential to provide bacterial strain resolution of multiple organisms at once. However, airway microbiome is low biomass, making metagenomic analysis a challenge and careful benchmarking necessary. In this study, using shotgun metagenomics, culture, and genomic data from OP and NP samples, we optimized species and strain resolution, enabling strain retention analyses in our longitudinal samples.
Despite low microbial biomass, metagenomics allows reliable species detection
Using NP and OP samples from infants and children, we first confirmed that the samples were highly dominated by host DNA despite high overall read counts. We therefore expected limitations in reaching high sensitivity for the detection of bacterial species and strain resolutions in metagenomic data. This problem is well recognized, and alternative strategies like different DNA extraction techniques (20) or the Plate coverage algorithm have been suggested (21). Despite this potential issue, sensitivity, and specificity in detecting S. pneumoniae, M. catarrhalis and H. influenzae at the species level were quite good for both MetaPhlAn 3 and MAGs-based Metagenome-Atlas, compared to culture data.
Strain resolution tools require optimization for species detection first
Ultimately, the goal of the study was to investigate bacterial strain retention. To this end, we assessed strain-level resolution tools such as StrainGE and StrainPhlAn 3 (13, 14). The first step was to assess whether the species assignation of these tools was performed correctly. We showed that performance using default parameters was insufficient since they require a higher sequencing coverage of the target genomes, this being especially limiting for assembly-based tools.
Despite these improvements, challenges still remained in the correct species detection. Reference-based tools such as StrainPhlAn reflect the currently accepted taxonomic structures and their inherent discrepancies. For example, despite being classified into several distinct species, S. mitis-like bacteria (including S. pneumoniae) are highly related (22) with homologous housekeeping genes (23), which constitutes a challenge for proper classification. Therefore, it comes as no surprise that we detected a number of samples as positive for S. pneumoniae using the StrainPhlAn 3 pipeline, while most likely, these samples contained other S. mitis-like strains. This problem is even more accentuated in our OP samples, and we therefore recommend to critically assess metagenomics-derived results for the identification of S. pneumoniae in OP samples. Of note, unlike the NP, the challenges of using OP samples to measure pneumococcal carriage are known (24, 25).
Another example, it has been described that there are seroresistant and serosensitive M. catarrhalis with separate evolutionary histories (26, 27). In our NP data set, this is reflected as two separate clusters indicating that M. catarrhalis should probably better be considered as two different subspecies. In addition, we also obtained somewhat low sensitivity values for S. aureus. This is not explained by the difficulties in the taxonomic species assignment using StrainPhlAn but rather by the rigidity of the cell wall of S. aureus. To achieve higher sensitivity values of S. aureus, a more stringent DNA extraction procedure would be needed (28) which may, however, lead to DNA loss for other species.
Strain resolution is challenging with low genomic coverage but achievable with adequate tools optimization
Systematically capturing whole microbiome diversity, especially at the strain level is challenging for such low biomass samples, whether through metagenomics or classical approach, i.e., isolation and genotyping of the isolates (13). Nonetheless, analyses targeting specific pathobionts are highly relevant in a clinical context as information from pathobionts in combination with basic information from commensal bacteria could prove sufficient to address a number of clinical research questions (19). Being able to reconstruct full genomes from metagenomic data sets would allow efficient tracking of co-occurring strains with dedicated tools such as StrainGE. However, our study showed the limitations of such an approach in a low biomass setting context, where low genomic coverage does not allow the reconstruction of metagenomic assemblies with high enough quality for strain resolution. Detecting only the dominant strain of a sample is often considered a limitation of StrainPhlAn, but despite this, StrainPhlAn permitted strain retention analyses once the parameters were carefully optimized.
Strain resolution allows longitudinal strain tracking
Strain identification and retention analyses are very important, especially in studies with longitudinal design (29). It allows the calculation of carriage duration of distinct strains and to study their association with health and disease. Though there are only two timepoints in the NP data set, strain retention, e.g., S. pneumoniae, was found to be 40.8% which is in a similar range as found in other studies using the same age (2 and 4 months of age) and time distance between samples (2 months; 29, 30).
In chronic diseases like CF, the ability to perform strain-resolved analyses in longitudinally collected samples may help identifying strains that are associated with disease exacerbation. In our study, we detected considerable long-term colonization of distinct S. aureus strains in children with CF. Though the sensitivity of the detection of S. aureus with the metagenomic data is somewhat reduced in some samples, the fact that the CF patients were sampled every 2–3 months for up to 10 years allowed to thoroughly assess strain retention even if one or the other sample was found to be negative (based on culture and/or metagenomic sequencing). In the future, this allows more in-depth investigation with clinical relevance such as on the consequences of antibiotic treatment on the strain retention, e.g., comparing recolonization vs strain retention.
Strengths and limitations
The strength of this study is the parallel investigation of culture, whole genome, and metagenomic data of two different sets of samples. While many benchmarking studies are focusing on simulations and metanalyses of already existing data, comparisons of culture and metagenomic data are rather rare. Bacterial cultures are not only considered as the golden standard for species assignation; successful culture demonstrates that metagenomic reads originate from viable bacterial cells. However, this study has some limitations. We only cultured four and two species from the NP and OP samples, respectively. Also, we only sequenced S. aureus isolates from the OP samples as isolates from the NP samples were not available for sequencing. Finally, we have not analyzed data from the lower airway microbiome which has been shown to be clinically relevant but not easy to retrieve due to the invasive nature of sampling (5). However, it has also been shown that the lower airways are seeded by both the OP and NP microbiome (17) which were included in our study. Finally, the StrainPhlAn pipeline enables strain retention analyses but does not allow the analyses for structural genome variations including, e.g., the genomic insertion and loss of phages typically associated with shifts of virulence and antimicrobial susceptibility in the airways of CF patients
Conclusions
We have performed benchmarking analyses for StrainPhlAn and provided strain retention results for two different metagenomic data sets from the upper respiratory tract. However, while shotgun metagenomic data analysis remains challenging for low biomass samples, it allows for strain-level resolution in contrast to 16S rRNA gene sequencing (1). Strain level information is indispensable to associate the presence of microbial strains or subclades of microbial species with disease phenotypes, but a careful evaluation of tools and parameters is a prerequisite, especially for low biomass samples (16).
MATERIALS AND METHODS
Characteristics of the data sets
We reanalyzed previously published shotgun metagenomic data from NP samples of the Microbiota and Health study from infants conducted in Dhaka, Bangladesh (18). The study design has been described and included the collection of NP samples from 267 infants at bi-monthly intervals during scheduled visits (18). Therefore, data were available for infants being asymptomatic for respiratory infections at 2 and 4 months of age. NP samples were initially stored at −20°C but then transferred to −80°C for shipment until further analysis (31). In total, an average of 38.1 million (M) read pairs per sample was re-processed for 422 samples. Culture data for H. influenzae, S. aureus, S. pneumoniae, and M. catarrhalis were available for all samples (31).
OP samples were obtained from a prospective follow-up study of infants diagnosed with CF by newborn screening at the university hospital (Inselspital) in Bern, Switzerland (32, 33). Ethical approval has been obtained (KEK-ethics no. 114/11). For this project, OP samples from 15 children who were followed up until 10 years of age were included. OP swabs were collected approximately every 3 months during clinical visits and, upon arrival, kept at −80°C until DNA extraction was done (see below). In parallel, routine culturing of each OP sample was performed during which a broad range of “clinical relevant” bacterial species were received. In brief, OP swabs from CF patients were streaked out onto Columbia sheep blood agar, which was used for the cultivation of non-fastidious and fastidious microbes as a screen for overall growth (34). In addition, MacConkey Agar was used for the selection of Gram-negative bacteria (i.e., P. aeruginosa). Most importantly for this study, Chocolate agar with the addition of bacitracin and Mannitol salt agar plates were used for the investigation of H. influenzae and S. aureus, respectively. Species identification was done using MALDI-TOF mass spectrometry analysis and Antibiogram and semi-quantitative information of bacterial growth from all isolates were also reported. In this study, we focused on culture results from H. influenzae and S. aureus due to high numbers in the OP swabs. Also, isolates identified as S. aureus were picked and kept at −80°C until further (WGS) analysis (see below for details).
Sample preparation and sequencing of the OP microbiome
OP swabs were swiped in a 1.5 mL tube containing 500 µL sterile phosphate-buffered saline (PBS). 200 µL of the bacterial suspension was used for DNA extraction using Qiacube running the DNA Mini extraction program. The shotgun metagenomic sequencing of the OP microbiome was done with a NovaSeq 6000 PE150 yielding approximately 10 G of raw data per sample. A negative template control sample was also included but yielded insufficient read quality.
Processing of metagenomic reads
For the present study, shotgun metagenomic reads from both data sets were analyzed. Human reads were removed in two runs of BBmap v38.84 (35): a first coarse run to remove a majority of human reads (parameters: mode fast = true, minratio = 0.9; maxindel = 3, minhits = 2, kmer length = 14). The second run used default parameters for more sensitivity in removing remaining human reads. Moreover, all reads mapping to Escherichia coli from NP data set were not considered for this study following recommendations of the original analysis publication (18). The quality of the reads was checked with FastQC (36) and MultiQC (37).
Taxonomic profiling through MetaPhlAn 3 and MAGs
Taxonomic profiling was done with MetaPhlAn 3 using the default parameters (38). In addition, the shotgun metagenomic data from the NP samples was re-analyzed using Metagenome-Atlas v 2.12 (38) to generate MAGs. In short, using tools from the BBMap suite v39 (35), reads were quality trimmed, and contaminations from the human genome were filtered out. Reads were error corrected and merged before assembly with metaSPAdes v3.15 (39). Contigs were binned using MetaBAT v2.15 (40) and MaxBin v2.2 (41), and their predictions were combined using DAS Tool v1.1 (42). The predicted MAGs, which had at least 50% completeness and <10% contamination based on the estimation by CheckM v1.1 (43), were clustered at 95% average nucleotide identity using dRep v3.2. The resulting 71 species representative MAGs were taxonomically annotated with GTDB-tk v2.1 to the Genome Taxonomy Data Base release 207 (44) and quantified using by the median of coverage in 1 kb windows along the genomes. A genome was called present if the median abundance was larger than 0. Reads were mapped using Minimap v2.24 (45).
Strain resolution analyses
Based on MAGs, strain inference was attempted using StrainGE v.1.3.3 (14) according to the documentation. In brief, the reads were mapped using bwa mem2 (46) to the detected strains starting from all complete RefSeq genomes from NCBI GenBank of the genera Streptococcus (n = 536; accessed on 24 October 2022), Staphylococcus (n = 317; accessed on 24 October 2022), Moraxella (n = 31; accessed on 20 December 2022), and Haemophilus (n = 87; accessed on 20 December 2022). The recommended threshold of 0.5 coverage and the detection using k-mers were used as the detection limit.
Based on MetaPhlAn 3 output, StrainPhlAn 3 was run with different sets of parameters for benchmarking (38). In total, four parameters can be changed: BREADTH_THRESHOLD, TRIM_SEQUENCES, MARKER_IN_N_SAMPLES, and SAMPLE_WITH_N_MARKERS. BREADTH_THRESHOLD describes the breadth of coverage threshold for the consensus markers and is used on the step of creating a marker file for each sample. The default value is 80%, and we used the range from 20% to 80%. TRIM_SEQUENCES sets the number of bases to be removed from both ends of the marker. The default value is 50, and we checked for the range from 30 to 50. This parameter was kept default, as the change in this parameter didn’t have an impact on the results. MARKER_IN_N_SAMPLES defines how many samples should have a marker so that it is kept for the analysis. The default value is 80%, and it should be lowered if species of interest are not expected to be found in less than 80% of the samples. SAMPLE_WITH_N_MARKERS defines the minimum number of markers to keep a sample in the analysis. The default value is 20, but some species have less markers available in the database (e.g., H. influenzae for which there were only 15 markers available in the database). Therefore, we investigated the performance of StrainPhlAn 3. Altogether, the default and 17 different sets of parameters were assessed, and the analyses were individually performed for four (S. pneumoniae, M. catarrhalis, H. influenzae, and S. aureus) and two (H. influenzae and S. aureus) species from the NP and OP data set, respectively. These species were chosen as the culture data were available for them. Also, an overall plot for all the bacterial species was created, and adjusted parameters were finally chosen based on the highest F1 value while being closest to default parameters (therefore, we would not expect a major impact on strain resolution). The full set of parameters can be seen in the Table S1. Using the adjusted parameters, phylogenetic trees were produced by StrainPhlAn3 based on a comparison of marker genes.
Whole genome shotgun analysis of from OP samples and phylogenetic comparison to StrainPhlAn 3 marker genes
To compare metagenomic-based phylogenetic resolution with genomic-based phylogenetic resolution, we used the WGS of S. aureus isolates from the OP samples. In brief, S. aureus cells were added into 200 µL PrepMan Ultra reagent in a microcentrifuge tube. Cells were then vortexed and boiled for 10 min at 100°C. Subsequently, we centrifuged at 8,500 × g for 3 min and transferred the supernatant to a clean 1.5 mL Eppendorf tube. The sequencing was done with a NovaSeq PE150. The microbial whole genome library preparation was performed for 350 bp. As for the Q30 of the PE150, this was indicated as Q30 >80%, and 1 G of raw data per S. aureus isolate was received. The quality of the reads was assessed with FastQC and MultiQC, and assembly was performed with SPAdes v3.15.2 (47) with default parameters. The core genome tree was created with Roary v3.11.0 (48) and visualized next to the StrainPhlAn3 tree using phylo.io (48). For this visualization, 50 OP samples were included for which both metagenomic and whole genome data were available for S. aureus.
Strain retention analyses in the NP data set
In order to trace strain retention, we first calculated the pairwise SNV rates using PhyloPhlAn integrated into StrainPhlAn. In brief, PhyloPhlAn computes the SNV rates as the number of bases that differ between each pair of samples (using the alignment of used marker genes) divided by the number of positions shared (i.e., containing no gaps) between the samples. Subsequently, the normalized distances were calculated from the SNV rate for each species by normalizing the values with the median of the SNV rate. The all-versus-all normalized genetic distances were generated for the four bacterial species (S. pneumoniae, M. catarrhalis, H. influenzae, and S. aureus) from the NP data set and grouped according to three different categories: comparing different timepoints for different infants (inter-comparison), same timepoint for different infants (inter-comparison), and different timepoints for same infants (intra-comparison). Values were binned into intervals of 0.1 in a histogram. Bins with values 0.0–0.1 (for S. aureus and H. influenzae) and 0.0–0.2 (for S. pneumoniae and M. catarrhalis) were defined for strain retention. These values were chosen based on the comparisons of within with between values with the latter being low as a condition.
Strain retention analyses in the OP data set
To analyze strain retention of S. aureus in the OP data set, a tree of shotgun metagenomic data were first created by StrainPhlAn and again visualized with phylo.io (48). Clusters of strains were then assigned based on the tree, using a threshold 0.01 branch length. The MLST profiles were extracted from the assembled S. aureus genomes using the center for genomic epidemiology website (https://cge.food.dtu.dk/services/MLST/). The resulting MLSTs were used for strain retention analyses of the S. aureus isolates. Strain clusters were longitudinally plotted together with the ST types of S. aureus to allow direct comparison of the two approaches for their potential to determine strain retention.
ACKNOWLEDGMENTS
We would like to thank the study nurses of the SCILD cohort for their involvement in the sample and data collection process as well as the participants. We also acknowledge the people working in the diagnostic unit of the Institute of Infectious Diseases for their work in receiving bacterial isolates.
The samples used in this study are part from the SCILD cohort study, which is funded by the Swiss National Science Foundation. This work was supported by grants from the Research Fund of the Swiss Lung Association (M.H), the "Stiftung Lindenhof" (M.H.) and "Cystic Fibrosis Switzerland" (I.K. and M.H). A part of this work has also been funded by the Swiss National Science Foundation (grant No. 159791) to M.H.
N.M., L.S., O.S., and M.H. designed the study. I.K., R.S., and P.L. have been in charge of the sampling of the patients with cystic fibrosis. T.M.W. prepared the samples for sequencing. N.M. performed the bioinformatic analyses with the help of L.S., S.K., O.S., and M.H. The study was supervised by M.H. who also wrote the manuscript with input from the co-authors. All authors read and approved the final manuscript.
Contributor Information
Markus Hilty, Email: markus.hilty@unibe.ch.
Nicola Segata, Universita degli Studi di Trento, Trento, Italy.
DATA AVAILABILITY
All raw Illumina sequencing reads for the whole genome sequencing data were deposited in the European Nucleotide Archive (ENA) under study accession number PRJNA930445. All raw Illumina sequencing reads from the shotgun metagenomic sequencing runs (OP data) were deposited in the European Nucleotide Archive (ENA) under study accession number PRJNA931830.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00724-23.
Figures S1 to S9.
Tables S1 to S3.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Ritchie AI, Singanayagam A. 2020. Metagenomic characterization of the respiratory microbiome. a piece de resistance. Am J Respir Crit Care Med 202:321–322. doi: 10.1164/rccm.202005-1686ED [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mika M, Korten I, Qi W, Regamey N, Frey U, Casaulta C, Latzin P, Hilty M, SCILD study group . 2016. The nasal microbiota in infants with cystic fibrosis in the first year of life: a prospective cohort study. Lancet Respir Med 4:627–635. doi: 10.1016/S2213-2600(16)30081-9 [DOI] [PubMed] [Google Scholar]
- 3. de Koff EM, Groot KM de W, Bogaert D. 2016. Development of the respiratory tract microbiota in cystic fibrosis. Curr Opin Pulm Med 22:623–628. doi: 10.1097/MCP.0000000000000316 [DOI] [PubMed] [Google Scholar]
- 4. Prevaes SMPJ, de Winter-de Groot KM, Janssens HM, de Steenhuijsen Piters WAA, Tramper-Stranders GA, Wyllie AL, Hasrat R, Tiddens HA, van Westreenen M, van der Ent CK, Sanders EAM, Bogaert D. 2016. Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am J Respir Crit Care Med 193:504–515. doi: 10.1164/rccm.201509-1759OC [DOI] [PubMed] [Google Scholar]
- 5. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF, Cookson WOC. 2010. Disordered microbial communities in asthmatic airways. PLoS One 5:e8578. doi: 10.1371/journal.pone.0008578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mayhew D, Devos N, Lambert C, Brown JR, Clarke SC, Kim VL, Magid-Slav M, Miller BE, Ostridge KK, Patel R, Sathe G, Simola DF, Staples KJ, Sung R, Tal-Singer R, Tuck AC, Van Horn S, Weynants V, Williams NP, Devaster J-M, Wilkinson TMA. 2018. Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax 73:422–430. doi: 10.1136/thoraxjnl-2017-210408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garcia-Nuñez M, Millares L, Pomares X, Ferrari R, Pérez-Brocal V, Gallego M, Espasa M, Moya A, Monsó E. 2014. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J Clin Microbiol 52:4217–4223. doi: 10.1128/JCM.01967-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mika M, Nita I, Morf L, Qi W, Beyeler S, Bernasconi E, Marsland BJ, Ott SR, von Garnier C, Hilty M. 2018. Microbial and host immune factors as drivers of COPD. ERJ Open Res 4:00015–02018. doi: 10.1183/23120541.00015-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, Martinez FJ, Huffnagle GB, Bereswill S. 2011. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE 6:e16384. doi: 10.1371/journal.pone.0016384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garzoni C, Brugger SD, Qi W, Wasmer S, Cusini A, Dumont P, Gorgievski-Hrisoho M, Mühlemann K, von Garnier C, Hilty M. 2013. Microbial communities in the respiratory tract of patients with interstitial lung disease. Thorax 68:1150–1156. doi: 10.1136/thoraxjnl-2012-202917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marvig RL, Sommer LM, Molin S, Johansen HK. 2015. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet 47:57–64. doi: 10.1038/ng.3148 [DOI] [PubMed] [Google Scholar]
- 12. Long DR, Wolter DJ, Lee M, Precit M, McLean K, Holmes E, Penewit K, Waalkes A, Hoffman LR, Salipante SJ. 2021. Polyclonality, shared strains, and convergent evolution in chronic cystic fibrosis Staphylococcus aureus airway infection. Am J Respir Crit Care Med 203:1127–1137. doi: 10.1164/rccm.202003-0735OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. 2017. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27:626–638. doi: 10.1101/gr.216242.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olm MR, Crits-Christoph A, Bouma-Gregson K, Firek BA, Morowitz MJ, Banfield JF. 2021. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat Biotechnol 39:727–736. doi: 10.1038/s41587-020-00797-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whiteside SA, McGinniss JE, Collman RG. 2021. The lung microbiome: progress and promise. J Clin Invest 131:e150473. doi: 10.1172/JCI150473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kennedy KM, de Goffau MC, Perez-Muñoz ME, Arrieta M-C, Bäckhed F, Bork P, Braun T, Bushman FD, Dore J, de Vos WM, Earl AM, Eisen JA, Elovitz MA, Ganal-Vonarburg SC, Gänzle MG, Garrett WS, Hall LJ, Hornef MW, Huttenhower C, Konnikova L, Lebeer S, Macpherson AJ, Massey RC, McHardy AC, Koren O, Lawley TD, Ley RE, O’Mahony L, O’Toole PW, Pamer EG, Parkhill J, Raes J, Rattei T, Salonen A, Segal E, Segata N, Shanahan F, Sloboda DM, Smith GCS, Sokol H, Spector TD, Surette MG, Tannock GW, Walker AW, Yassour M, Walter J. 2023. Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies. Nature 613:639–649. doi: 10.1038/s41586-022-05546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prevaes SMPJ, de Steenhuijsen Piters WAA, de Winter-de Groot KM, Janssens HM, Tramper-Stranders GA, Chu MLJN, Tiddens HA, van Westreenen M, van der Ent CK, Sanders EAM, Bogaert D. 2017. Concordance between upper and lower airway microbiota in infants with cystic fibrosis. Eur Respir J 49:1602235. doi: 10.1183/13993003.02235-2016 [DOI] [PubMed] [Google Scholar]
- 18. Binia A, Siegwald L, Sultana S, Shevlyakova M, Lefebvre G, Foata F, Combremont S, Charpagne A, Vidal K, Sprenger N, Rahman M, Palleja A, Eklund AC, Nielsen HB, Brüssow H, Sarker SA, Sakwinska O. 2021. The influence of Fut2 and Fut3 polymorphisms and nasopharyngeal microbiome on respiratory infections in breastfed Bangladeshi infants from the microbiota and health study. mSphere 6:e0068621. doi: 10.1128/mSphere.00686-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brugger SD, Bomar L, Lemon KP. 2016. Commensal-pathogen interactions along the human nasal passages. PLoS Pathog 12:e1005633. doi: 10.1371/journal.ppat.1005633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nelson MT, Pope CE, Marsh RL, Wolter DJ, Weiss EJ, Hager KR, Vo AT, Brittnacher MJ, Radey MC, Hayden HS, Eng A, Miller SI, Borenstein E, Hoffman LR. 2019. Human and extracellular DNA depletion for metagenomic analysis of complex clinical infection samples yields optimized viable microbiome profiles. Cell Rep 26:2227–2240. doi: 10.1016/j.celrep.2019.01.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Whelan FJ, Waddell B, Syed SA, Shekarriz S, Rabin HR, Parkins MD, Surette MG. 2020. Culture-enriched metagenomic sequencing enables in-depth profiling of the cystic fibrosis lung microbiota. Nat Microbiol 5:379–390. doi: 10.1038/s41564-019-0643-y [DOI] [PubMed] [Google Scholar]
- 22. Sadowy E, Bojarska A, Kuch A, Skoczyńska A, Jolley KA, Maiden MCJ, van Tonder AJ, Hammerschmidt S, Hryniewicz W. 2020. Relationships among streptococci from the mitis group, misidentified as Streptococcus pneumoniae. Eur J Clin Microbiol Infect Dis 39:1865–1878. doi: 10.1007/s10096-020-03916-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. 2009. The bacterial species challenge: making sense of genetic and ecological diversity. Science 323:741–746. doi: 10.1126/science.1159388 [DOI] [PubMed] [Google Scholar]
- 24. Boelsen LK, Dunne EM, Gould KA, Ratu FT, Vidal JE, Russell FM, Mulholland EK, Hinds J, Satzke C. 2020. The challenges of using oropharyngeal samples to measure pneumococcal carriage in adults. mSphere 5:e00478-20. doi: 10.1128/mSphere.00478-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Turner P, Sá-Leão R, Greenhill A, Leach A, Satzke C. 2022. World health organization (WHO) standard methods for pneumococcal carriage studies. Clin Infect Dis 75:924–925. doi: 10.1093/cid/ciac221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Earl JP, de Vries SPW, Ahmed A, Powell E, Schultz MP, Hermans PWM, Hill DJ, Zhou Z, Constantinidou CI, Hu FZ, Bootsma HJ, Ehrlich GD. 2016. Comparative genomic analyses of the Moraxella catarrhalis serosensitive and seroresistant lineages demonstrate their independent evolution. Genome Biol Evol 8:955–974. doi: 10.1093/gbe/evw039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wirth T, Morelli G, Kusecek B, van Belkum A, van der Schee C, Meyer A, Achtman M. 2007. The rise and spread of a new pathogen: seroresistant Moraxella catarrhalis. Genome Res 17:1647–1656. doi: 10.1101/gr.6122607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hassanzadeh S, Pourmand MR, Afshar D, Dehbashi S, Mashhadi R. 2016. TENT: a rapid DNA extraction method of Staphylococcus aureus. Iran J Public Health 45:1093–1095. [PMC free article] [PubMed] [Google Scholar]
- 29. Oyewole OR-A, Latzin P, Brugger SD, Hilty M. 2022. Strain-level resolution and pneumococcal carriage dynamics by single-molecule real-time (SMRT) sequencing of the plyNCR marker: a longitudinal study in swiss infants. Microbiome 10:152. doi: 10.1186/s40168-022-01344-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lees JA, Croucher NJ, Goldblatt D, Nosten F, Parkhill J, Turner C, Turner P, Bentley SD. 2017. Genome-wide identification of lineage and locus specific variation associated with pneumococcal carriage duration. Elife 6:e26255. doi: 10.7554/eLife.26255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vidal K, Sultana S, Patron AP, Binia A, Rahman M, Deeba IM, Brüssow H, Sakwinska O, Sarker SA. 2019. Microbiota and health study: a prospective cohort of respiratory and diarrheal infections and associated risk factors in Bangladeshi infants under two years. Pediatrics. doi: 10.1101/19000505 [DOI]
- 32. Neumann RP, Hilty M, Xu B, Usemann J, Korten I, Mika M, Müller L, Latzin P, Frey U. 2018. Nasal microbiota and symptom persistence in acute respiratory tract infections in infants. ERJ Open Res 4:00066-2018. doi: 10.1183/23120541.00066-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Korten I, Mika M, Klenja S, Kieninger E, Mack I, Barbani MT, Gorgievski M, Frey U, Hilty M, Latzin P, Ellis Dutch R. 2016. Interactions of respiratory viruses and the nasal microbiota during the first year of life in healthy infants. mSphere 1. doi: 10.1128/mSphere.00312-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frey PM, Marti GR, Droz S, de Roche von Arx M, Suter-Riniker F, Aujesky D, Brugger SD. 2019. Bacterial colonization of handheld devices in a tertiary care setting: a hygiene intervention study. Antimicrob Resist Infect Control 8:97. doi: 10.1186/s13756-019-0546-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bushnell B. 2014. BBMap: a fast, accurate, Splice-Aware Aligner. [Google Scholar]
- 36. Andrews S. 2010. Fastqc: a quality control tool for high throughput sequence data
- 37. Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beghini F, McIver LJ, Blanco-Míguez A, Dubois L, Asnicar F, Maharjan S, Mailyan A, Manghi P, Scholz M, Thomas AM, Valles-Colomer M, Weingart G, Zhang Y, Zolfo M, Huttenhower C, Franzosa EA, Segata N. 2021. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 10:e65088. doi: 10.7554/eLife.65088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu YW, Simmons BA, Singer SW. 2016. MaxBIN 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607. doi: 10.1093/bioinformatics/btv638 [DOI] [PubMed] [Google Scholar]
- 42. Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, Banfield JF. 2018. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol 3:836–843. doi: 10.1038/s41564-018-0171-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil PA, Hugenholtz P. 2022. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res 50:D785–D794. doi: 10.1093/nar/gkab776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vasimuddin M, Misra S, Li H, Aluru S. 2019. Efficient architecture-aware acceleration of BWA-MEM for Multicore systems. 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS); Rio de Janeiro, Brazil. doi: 10.1109/IPDPS.2019.00041 [DOI] [Google Scholar]
- 47. Demin KA, Lakstygal AM, Krotova NA, Masharsky A, Tagawa N, Chernysh MV, Ilyin NP, Taranov AS, Galstyan DS, Derzhavina KA, Levchenko NA, Kolesnikova TO, Mor MS, Vasyutina ML, Efimova EV, Katolikova N, Prjibelski AD, Gainetdinov RR, de Abreu MS, Amstislavskaya TG, Strekalova T, Kalueff AV. 2020. Understanding complex dynamics of behavioral, neurochemical and transcriptomic changes induced by prolonged chronic unpredictable stress in Zebrafish. Sci Rep 10:19981. doi: 10.1038/s41598-020-75855-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S9.
Tables S1 to S3.
Data Availability Statement
All raw Illumina sequencing reads for the whole genome sequencing data were deposited in the European Nucleotide Archive (ENA) under study accession number PRJNA930445. All raw Illumina sequencing reads from the shotgun metagenomic sequencing runs (OP data) were deposited in the European Nucleotide Archive (ENA) under study accession number PRJNA931830.