Abstract
Protein and mRNA levels of heat-labile enterotoxin (LT) of Escherichia coli are highest at 37°C, and they decrease gradually as temperature is decreased. This temperature effect is eliminated in an Hns− mutant. Deletion of portions of DNA coding for the LT A subunit also results in an increase in LT expression at low temperatures, suggesting that the H-NS protein causes inhibition of transcription at low temperatures by interacting with the LT A-subunit DNA. The region that interacts with H-NS is referred to as the downstream regulatory element (DRE). Plasmids in an hns strain from which the DRE has been deleted still produce elevated levels of LT at 18°C, suggesting that intact DRE is not required for transcription from the LT promoter.
A major proportion of infectious diarrhea in humans in third-world countries and in domestic animals worldwide is caused by enterotoxigenic strains of Escherichia coli (ETEC strains). ETEC strains carry transmissible plasmids, called Ent plasmids, that encode heat-stable enterotoxin (ST) or heat-labile enterotoxin (LT) or both.
LT is very similar antigenically and pharmacologically to cholera toxin (CT) produced by Vibrio cholerae. However, based on the considerable amount of information known about the regulation of CT (23) and from what has been learned about LT regulation, there is no evidence that LT, like CT, is regulated by a two-component regulatory system (26).
LT mRNA and protein levels are significantly affected by temperature but are only slightly affected by some of the other environmental conditions known to strongly influence CT expression (23). When LT is in a native plasmid (8) or when LT is subcloned into vectors, it is optimally expressed at 37°C and its expression decreases as temperature is decreased to 18°C (26).
Recently, much has been reported about the global regulator H-NS with respect to its ability to mediate the response of many operons to environmental changes (2, 28). HN-S mediates the temperature regulation of several virulence factor operons, including the CFA/I fimbriae, CS1 pili, and pap pili of E. coli and the virF and virB genes of enteroinvasive E. coli and Shigella species (10, 12, 18–20, 29). H-NS also mediates osmoregulation of the proU operon in E. coli and Salmonella species (9). Interestingly, it has been shown through extensive genetic analysis that H-NS influences proU expression by binding to a downstream regulatory element (the DRE) in the proU structural gene (11, 14, 15, 27).
We shall present results obtained with an H-NS mutant that produces a truncated H-NS protein with altered DNA binding capacities (5). Hns+ (GM37) and Hns− mutant (GM230) strains (9) carrying LT plasmids with various deletions located in the structural gene coding for the LT A subunit or upstream of the −35 and −10 promoter elements were grown at either 37 or 18°C to analyze the role that H-NS plays in mediating temperature control of LT expression.
Plasmid construction.
Standard DNA manipulations were carried out as described by Sambrook et al. (21). The plasmid pLT was derived from plasmid pJT2, which contains the entire LT operon on an HpaI-BamHI fragment from pEWD030 (24) subcloned into the EcoRV and BamHI sites of pBR322. Previously, we sequenced approximately 725 bp upstream of the LT promoter subcloned from pEWD030 (GenBank accession no. M61015). Bal31 mutagenesis and primer extension were performed to characterize the region upstream of the promoter elements and to confirm the precise locations of the promoter elements (26).
The 2,800 bp upstream of the LT mRNA start site from pJT2 was substituted with a fragment containing only 723 bp upstream of the LT mRNA start site. This fragment was isolated from one of the plasmids obtained from Bal31 deletion analysis of an LT–β-galactosidase translational fusion construct. Likewise, a second fragment containing only 34 bp upstream of the LT mRNA start site was used to construct pLTΔUCR (26).
pLTΔNC was constructed by excision of a 686-bp XbaI-EcoRI fragment from pLT, pLTΔN was constructed by excision of a 422-bp XbaI-AgeI fragment from pLT, and pLTΔC was constructed by excision of a 264-bp AgeI-EcoRI fragment from pLT. pLTΔUCRΔNC was constructed by excision of a 686-bp XbaI-EcoRI fragment from pLTΔUCR. The XbaI, AgeI, and EcoRI sites are located 184, 550, and 814 bp downstream of the LT mRNA start site, respectively.
Effect of temperature and H-NS on LT mRNA levels.
The promoter activity of the LT gene was measured in primer extension experiments. The segment of LT mRNA chosen for the primer extension experiments is shown in Fig. 1A. It is located between the promoter and the 5′ end of the LT A gene. This location avoids complications that we have previously found to arise in assessing promoter activity by using reporter genes in translational fusion plasmids that encode β-galactosidase or alkaline phosphatase (26).
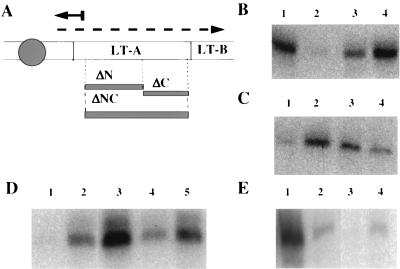
FIG. 1.
(A) Schematic representation of the LT operon and location of the DRE deletions with respect to the location of the primer extension product. LT A and LT B code for subunits of LT. Vertical bars, locations of the LT A and B structural genes; gray circle, the promoter region; dashed arrow, the LT mRNA; solid arrow, the LT primer extension product; solid bar, the LT primer; filled-in gray bars, approximate locations of the DRE deletions described in the text. (B) Effect of temperature and H-NS on LT mRNA production. Lanes: 1, hns+ pLT at 37°C; 2, hns+ pLT at 18°C; 3, hns pLT at 37°C; and 4, hns pLT at 18°C. (C) Effect of DRE deletions on LT mRNA production in an hns+ strain at 18°C. Lanes: 1, hns+ pLT; 2, hns+ pLTΔNC; 3, hns+ pLTΔN; and 4, hns+ pLTΔC. (D) Effect of DRE deletions on LT mRNA production in hns strains at 18°C. Lanes: 1, hns+ pLT; 2, hns pLT; 3, hns pLTΔNC; 4, hns pLTΔN; and 5, hns pLTΔC. (E) Effect of UCR deletion or UCR and DRE deletions on LT mRNA production in hns+ strains at 37 and at 18°C. Lanes: 1, pLT at 37°C; 2, pLTΔUCR at 37°C; 3, pLTΔUCR at 18°C; and 4, pLTΔUCRΔNC at 18°C. Exposure of the autoradiograph for panel E was five times longer than that for panels B to D.
A variation of the hot phenol procedure (1) was used to isolate RNA from cells grown to mid-log phase (optical density at 580 nm [OD580] of 0.3 as determined by a model 401A Lumetron colorimeter) in LB broth (17) containing ampicillin (30 μg/ml) at either 37 or 18°C. RNA purity, quantity, and quality were assessed by measuring the OD260/OD280 in a Beckman DU-40 spectrophotometer and electrophoresis on a 1.5% agarose gel before use for primer extension.
Primer extension on total RNA (10 μg per sample) was performed under the conditions described by Curtis (3), except that the oligonucleotide 5′-AGTCAGCACGGTATAATCTG-3′ (the binding site on RNA is at positions +119 through +138 relative to the mRNA start site) was end labeled with [γ-32P]dATP by using T4 polynucleotide kinase, and none of the added nucleotides was radioactive. Reaction products were analyzed on a 6% acrylamide-urea gel. The same primer was used for dideoxy chain termination sequencing reactions (with an AmpliCycle Sequencing kit from Perkin-Elmer Corp. and [α-33P]dATP) to serve as a size marker for the extended fragments (data not shown).
Promoter activities of the LT gene were determined at 37 and 18°C in an H-NS mutant and the corresponding wild-type strain. As shown in Fig. 1B, the wild type produces much less mRNA at the lower temperature (lane 2), whereas in the H-NS mutant, mRNA production is not inhibited at the lower temperature (lane 4). We also showed that the mRNA start sites were the same in the wild type and in the Hns− mutant (Fig. 1B).
To test if the difference seen between the wild type and the mutant was due to differences in plasmid levels, we measured the respective plasmid yields. Plasmid DNA was isolated from 4.5 ml of cultures grown to an OD580 of 0.3, linearized, and electrophoresed on a 1% agarose gel which was stained with ethidium bromide. We found plasmid levels to be essentially the same regardless of the growth conditions and host strains (data not shown). We have found this method to be sensitive enough to detect a twofold difference.
Effect of DRE deletions on mRNA levels.
To test for the presence of a site that interacts with H-NS in the structural part of the LT gene, we generated the three deletions shown in Fig. 1A. The effect of these deletions on promoter activity at 18°C was tested. As shown in Fig. 1C, the deletions alleviate the inhibition seen in the undeleted plasmid. Two of the plasmids, pLTΔN and pLTΔNC, eliminate the inhibition almost completely. pLTΔC has a weaker effect. We conclude from these experiments that H-NS protein interacts with the region of LT A DNA encoding the N terminus to cause the inhibition, presumably by binding to this region.
We also tested the effect of the three deletions at 18°C in the H-NS mutant. It can be seen (Fig. 1D) that all promoters in the mutant are active (lanes 2, 3, 4, and 5), in contrast to the undeleted plasmid in the H-NS+ strain (lane 1). There is some variability among the plasmids in the mutant. LTΔN has about the same activity as the undeleted plasmid (lanes 2 and 4), whereas activities are stronger in LTΔNC and LTΔC. So far, we have not found the cause for these differences. One possible explanation is that they are due to the H-NS homolog StpA. It has been shown that StpA production is increased in H-NS− strains (25, 30). Our results indicate that StpA binds to the C-terminal part of LT A and that this leads to inhibition of LT synthesis. However, in contrast to the inhibition by H-NS, this inhibition is not dependent on temperature. If anything, it is stronger at 37 than at 18°C (Fig. 1B).
A similar deletion analysis of the region upstream of the −35 element of the promoter (the upstream control region [UCR]) was performed. Deletion of 300 bp or more immediately upstream of the −35 element of the promoter and not including the promoter elements results in a considerable decrease in LT production at the protein and mRNA levels (only mRNA data shown [Fig. 1E, lane 2]). This effect is also H-NS dependent (data not shown) but is not affected by temperature. Hns+ E. coli cells carrying an LT plasmid with deletion of the UCR still manifest an observable decrease in LT protein and mRNA production at low temperatures (only mRNA data shown [Fig. 1E, lane 3]). Furthermore, Hns+ E. coli cells carrying a plasmid containing deletions in both the DRE and the UCR do not result in LT levels fully restored to wild-type levels at high temperatures (Fig. 1E, lane 4), suggesting that there is an additional H-NS-sensitive but temperature-independent control region in the UCR. Since the present paper is concerned with temperature control and since the UCR effect is independent of temperature and the interaction of H-NS with the DRE, it will not be considered further.
Effect of temperature and H-NS on LT protein synthesis.
In parallel with the above-described studies of mRNA synthesis, we measured LT protein synthesis. Cells were grown in modified K medium containing 171 mM NaCl, Casamino Acids (7), and ampicillin (30 μg/ml) or LB (17) and ampicillin (30 μg/ml) at 18°C and subcultured at either 37 or 18°C. After cultures had reached desired cell densities, polymyxin B (90 μg per ml of culture) (6) and MgSO4 (1.725 mg per ml of culture) were added directly to cell aliquots and were incubated at their respective temperatures until the cells appeared to be completely lysed. Determinations of amounts of protein in the extracts were made with the Bio-Rad system. Cell extracts were assayed by a GM1-enzyme-linked immunosorbent assay under conditions similar to those described by Scotland et al. (22). Microtiter plates were analyzed with a Dynatech MR5000 Microplate Reader at OD410.
The results of experiments with intact LT genes are shown in Table 1. At 37°C, both the H-NS mutant and its wild-type parent produce approximately the same amounts of LT (rows 1 and 3). At 18°C, LT formation is considerably less in the wild type but not in the mutant. These results confirm our findings on mRNA synthesis. It should be noted that the inhibition is greater at the end of growth than during the exponential phase (Table 1, row 2).
TABLE 1.
Effects of temperature and H-NS mutation on LT expression
| Genotype | Strain (plasmid) | Temp (°C) | OD410a
|
|
|---|---|---|---|---|
| Mid-log phaseb | Late-log phaseb | |||
| hns+ | GM37(pLT) | 37 | 0.32 | 0.45 |
| hns+ | GM37(pLT) | 18 | 0.13 | 0.001 |
| hns | GM230(pLT) | 37 | 0.50 | 0.60 |
| hns | GM230(pLT) | 18 | 0.60 | 0.40 |
OD410s measure the amounts of LT protein determined by GM1-ELISA. See text for details.
Sample extracts were derived from the same cultures at different points in the growth curve. Replicate OD410s for each sample extract were obtained. The reported values are the averages of these determinations.
In these experiments, the bacteria were grown in a minimal medium (modified K medium) rather than in the rich medium used for the mRNA experiments. This medium was used because it permits LT extraction directly from cells suspended in the culture medium with polymyxin B, thereby allowing us to measure all of the fully and partially assembled LT that had accumulated in the periplasmic space and in the supernatant (6). We also carried out these experiments with the bacteria grown in rich medium and obtained similar results (data not shown).
Effect of DRE deletions on the inhibition of LT protein synthesis.
We measured LT protein production in strains carrying plasmids with deletions at 18°C and compared them with the same strain carrying a complete plasmid. We found a restoration of LT formation in the strains with deleted plasmids similar to restoration of mRNA synthesis (Fig. 1C). However, the restoration was only approximately twofold, which is not as great as the restoration of mRNA levels. We believe that this may be due to technical factors, because it is likely that with the deleted LT A subunits, the incomplete toxin molecules gave values in the ELISA, which measures the amount of LT B subunit, that were lower than the values obtained with complete LT molecules (4, 16).
Mechanism of H-NS temperature regulation.
It has been shown that H-NS is a member of the cold shock regulon and that its expression increases by three- to fourfold when the bacteria are shifted from 37 to 10°C (13). Here, we have shown that H-NS inhibits LT synthesis at the transcriptional level at low temperatures by interacting with a DNA sequence located at the part of the LT A gene encoding the N terminus, which is referred to as the DRE. The inhibition is exerted at the LT promoter upstream of the LT A gene.
Our results permit us to discriminate between two possible explanations for the inhibitory action of H-NS, with inhibition starting at the promoter or within the DRE. Since the inhibition of mRNA formation extends to the start site of transcription, the inhibition occurs at the promoter rather than in the DRE. Presumably, it prevents the action of RNA polymerase in initiating transcription.
We can also distinguish between an action of H-NS as an antagonist in preventing stimulation of promoter activity by the DRE and a cooperative action with DRE in inhibiting promoter activity. For the first explanation to be correct, DRE must by itself activate the promoter. We have shown that in the H-NS mutant, deletion of the DRE does not diminish mRNA synthesis (Fig. 1D). Therefore, the DRE is not required for activation of transcription but is required for inhibition by H-NS. Just how the interaction between DRE and H-NS brings about inhibition of promoter activity is not known at present. Presumably, H-NS has to bind to DRE in a specific manner to exert its effect. In the similar case of the proU operon (11, 14, 15, 27), it has been shown that H-NS also has to bind specifically to a DRE in the first structural gene in order to inhibit promoter activity and that other H-NS binding DNA segments could not replace DRE.
Acknowledgments
Much of the preliminary work for this publication was performed while J. D. Trachman was supported by NIH National Research Service award 5 T32 AI-07180 from the National Institute of Allergy and Infectious Diseases. This work was supported by Public Health Service grant GM-06048 from the National Institute of General Medical Sciences to W. K. Maas. The National Science Foundation is thanked for its support of computing grant BIR 9318128.
We acknowledge the valuable contributions of D. Lim, R. Maas, and H. Niersbach to our studies. We also thank R. Holmes for his generous supply of goat anti-LT serum and C. F. Higgins for sending us the congenic strains GM37 (hns+ strain) and GM230 (hns strain).
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact E. coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 3.Curtis S. Genes encoding the beta and epsilon subunits of the proton-translocating ATPase from Anabaena sp. strain PCC 7120. J Bacteriol. 1987;169:80–86. doi: 10.1128/jb.169.1.80-86.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Haan L, Verweij W R, Feil I K, Lijnema T H, Hol W G J, Agsteribbe E, Wilschut J. Mutants of the Escherichia coli heat-labile enterotoxin with reduced ADP-ribosylation activity or no activity retain the immunogenic properties of the native holotoxin. Infect Immun. 1996;64:5413–5416. doi: 10.1128/iai.64.12.5413-5416.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dersch P, Kneip S, Bremer K. The nucleoid associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol Gen Genet. 1994;245:255–259. doi: 10.1007/BF00283274. [DOI] [PubMed] [Google Scholar]
- 6.Evans D J, Jr, Evans D G, Gorbach S L. Polymyxin B-induced release of low-molecular weight, heat-labile enterotoxin from Escherichia coli. Infect Immun. 1974;10:1010–1017. doi: 10.1128/iai.10.5.1010-1017.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gowrishankar J. Identification of osmoresponsive genes in E. coli: evidence for participation of potassium and proline transport systems in osmoregulation. J Bacteriol. 1985;164:434–445. doi: 10.1128/jb.164.1.434-445.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gyles C L, Palchaudhuri S, Maas W K. Naturally occurring plasmid carrying genes for enterotoxin production and drug resistance. Science. 1977;198:198–199. doi: 10.1126/science.333581. [DOI] [PubMed] [Google Scholar]
- 9.Higgins C F, Dorman C J, Stirling D A, Waddell L, Booth I R, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 10.Jordi B J A M, Dagberg B, de Haan L A M, Hamers A M, van der Zeijst B A M, Gaastra W, Uhlin B E. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 1992;11:2627–2632. doi: 10.1002/j.1460-2075.1992.tb05328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordi B J A M, Fielder A E, Burns C M, Hinton J C D, Dover N, Ussery D W, Higgins C F. DNA binding is not sufficient for H-NS-mediated repression of proU expression. J Biol Chem. 1997;272:12083–12090. doi: 10.1074/jbc.272.18.12083. [DOI] [PubMed] [Google Scholar]
- 12.Jordi B J A M, van der Zeijst B, Gaastra W. Regions of the CFA/I promoter involved in the activation by the transcriptional activator CfaD and repression by the histone-like protein H-NS. Biochimie. 1994;76:1052–1054. doi: 10.1016/0300-9084(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 13.La Teana A, Brandi A, Falconi M, Spurio R, Pon C L, Gualerzi C O. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc Natl Acad Sci USA. 1991;88:10907–10911. doi: 10.1073/pnas.88.23.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucht J M, Bremer E. Characterization of mutations affecting the osmoregulated proU promoter of E. coli and identification of 5′ sequences required for high-level expression. J Bacteriol. 1991;173:801–809. doi: 10.1128/jb.173.2.801-809.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucht J M, Dersch P, Kempf B, Bremer E. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J Biol Chem. 1994;269:6578–6586. [PubMed] [Google Scholar]
- 16.Magagnoli C, Manetti R, Fontana M R, Giannelli V, Giuliani M M, Rappouli R, Pizza M. Mutations in the A subunit affect yield, stability, and protease sensitivity of nontoxic derivatives of heat-labile enterotoxin. Infect Immun. 1996;64:5434–5438. doi: 10.1128/iai.64.12.5434-5438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 18.Murphee D, Froehlich B, Scott J R. Transcriptional control of genes encoding CS1 pili: negative regulation by a silencer and positive regulation by Rns. J Bacteriol. 1997;179:5736–5743. doi: 10.1128/jb.179.18.5736-5743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porter M E, Dorman C J. Differential regulation of the plasmid-encoded genes in the Shigella flexneri virulence regulon. Mol Gen Genet. 1997;256:93–103. doi: 10.1007/s004380050550. [DOI] [PubMed] [Google Scholar]
- 20.Prosseda G, Fradiani P A, DiLorenzo M, Falconi M, Micheli G, Casalino M, Nicoletti M, Colonna B. A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. Res Microbiol. 1998;149:15–25. doi: 10.1016/s0923-2508(97)83619-4. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Scotland S M, Gross R J, Rowe B. Methods for enterovirulence factors. In: Sussman M, editor. The virulence of E. coli: reviews and methods for enterovirulence factors. Orlando, Fla: Academic Press, Inc.; 1985. pp. 395–405. [Google Scholar]
- 23.Skorupski K, Taylor R K. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 24.So M, Dallas W S, Falkow S. Characterization of an E. coli plasmid for synthesis of heat-labile enterotoxin: molecular cloning of the toxin determinant. Infect Immun. 1978;21:405–411. doi: 10.1128/iai.21.2.405-411.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonden B, Uhlin B E. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 1996;15:4970–4980. [PMC free article] [PubMed] [Google Scholar]
- 26.Trachman J D. Genetic and environmental factors involved in the regulation of expression of porcine heat-labile enterotoxin of Escherichia coli. Ph.D. thesis. New York, N.Y: Sackler Institute of Graduate Biomedical Sciences, New York University School of Medicine; 1990. [Google Scholar]
- 27.Tupper A E, Owen-Hughes T A, Ussery D W, Santos D S, Ferguson D J P, Sidebotham J M, Hinton J C D, Higgins C F. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 1994;13:258–268. doi: 10.1002/j.1460-2075.1994.tb06256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ussery D W, Hinton J C D, Jordi B J A M, Granum P E, Seirafi A, Stephen R J, Tupper A E, Berridge G, Sidebotham J M, Higgins C F. The chromatin-associated protein H-NS. Biochimie. 1994;76:968–980. doi: 10.1016/0300-9084(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 29.van der Woude M, Braaten B, Low D. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 1996;4:5–9. doi: 10.1016/0966-842x(96)81498-3. [DOI] [PubMed] [Google Scholar]
- 30.Zhang A, Rimsky S, Reaban M E, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]