ABSTRACT
Several studies indicate that CRISPR-Cas gene-editing systems can be used to inactivate the HIV-1 proviral DNA in infected cells. Such gene editing introduces mutations, mostly small insertions and deletions (indels), at the targeted sites in the HIV genome or causes excision or inversion of the proviral DNA fragment between two target sites. To investigate whether CRISPR-Cas treatment of latently infected T cell lines can also cause large unintended deletions, we designed a PCR-based sequencing strategy with primer binding sites at various positions in the chromosomal DNA surrounding the integrated proviral DNA genome. We, here, demonstrate that both continuous and transient CRISPR-Cas attack on the integrated HIV DNA does not only result in the expected small indels, but also frequently causes much larger deletions that can include flanking cellular DNA sequences. Analysis of the breakpoint junction sites indicates that the deletions were triggered by an initial on-target attack by CRISPR-Cas. Upon continuous CRISPR-Cas treatment, small microhomologies were frequently observed at the junction sites, which indicates that microhomology-mediated end-joining DNA repair is involved in the generation of the large deletions. As the loss of chromosomal sequences may cause oncogenic cell transformation, unintended large deletions form a potential safety risk in clinical application of this antiviral application.
IMPORTANCE
Although HIV replication can be effectively inhibited by antiretroviral therapy, this does not result in a cure as the available drugs do not inactivate the integrated HIV-1 DNA in infected cells. Consequently, HIV-infected individuals need lifelong therapy to prevent viral rebound. Several preclinical studies indicate that CRISPR-Cas gene-editing systems can be used to achieve permanent inactivation of the viral DNA. It was previously shown that this inactivation was due to small inactivating mutations at the targeted sites in the HIV genome and to excision or inversion of the viral DNA fragment between two target sites. We, here, demonstrate that CRISPR-Cas treatment also causes large unintended deletions, which can include surrounding chromosomal sequences. As the loss of chromosomal sequences may cause oncogenic transformation of the cell, such unintended large deletions form a potential safety risk in clinical application of this antiviral application and possibly all CRISPR-Cas gene-editing approaches.
KEYWORDS: HIV-1, latent reservoir, CRISPR-Cas, gene therapy, gene editing, deletion
INTRODUCTION
Current antiretroviral therapy with multiple antiviral drugs can effectively inhibit HIV replication, such that the viral load becomes undetectable, but does not lead to a cure as HIV proviral DNA remains present in cells that constitute the viral reservoir (1–3). As a consequence, HIV-infected individuals need lifelong therapy to prevent viral rebound (4, 5). To inactivate this viral reservoir, novel therapeutic strategies have been proposed, including targeting of the integrated HIV DNA with CRISPR-Cas gene-editing systems [reviewed in references (6, 7)]. Cas cleavage results in a double-stranded break in the DNA that can be repaired by cellular DNA repair mechanisms, in particular non-homologous end-joining and microhomology-mediated end-joining (MMEJ) (8, 9). Such repair usually introduces mutations at the Cas cleavage site, in particular small insertions and deletions (indels), which can inactivate the virus, especially when critical viral sequences are targeted.
We and others demonstrated that HIV replication in T cell cultures can be strongly inhibited when the cells continuously produce Cas9 or Cas12a and a single guide RNA (gRNA for Cas9 and crRNA for Cas12a) targeting the HIV DNA genome (10, 11). However, the DNA repair following Cas cleavage did not only result in virus-inactivating mutations, but also accelerated virus escape by generating mutations that prevented binding of the gRNA/crRNA, yet did not block virus replication (10, 12–15). More durable inhibition was achieved through a combined approach employing dual gRNAs or crRNAs that target distinct highly conserved and essential viral sequences (16–18). Some gRNA/crRNA pairs completely prevented virus escape and resulted in complete and permanent virus inactivation. More recently, we demonstrated that repeated transient treatment with Cas9 or Cas12a ribonucleoprotein (RNP) complexes can also achieve inactivation of all proviral DNA in latently infected T cells (19). Both continuous and transient dual-gRNA/crRNA attack on the proviral DNA resulted in mutations at the target sites, mostly small indels, but also in excision and, to a lesser extent, inversion of the fragment between the target sites (18–20). Notably, Cas12a treatment resulted less frequently in excision or inversion compared to Cas9 treatment, which is possibly related to the distinct cleavage pattern or differential kinetics of the Cas proteins. For instance, Cas9 cleaved DNA is blunt-ended, whereas Cas12a cleaved DNA has sticky ends with a 5´ overhang (21).
CRISPR-Cas-induced mutations are commonly detected by PCR amplification of the targeted DNA, followed by sequencing of the PCR products (16). Hereby, PCR primers annealing upstream and downstream of the target sites in the viral DNA are used to amplify the targeted DNA region, which will allow detection of the anticipated mutations. However, larger deletions extending beyond the position of the primers will not be detected. When such large deletions would extend beyond the proviral HIV DNA into flanking chromosomal DNA, expression of tumor suppressor genes or proto-oncogenes could be affected, potentially triggering oncogenic transformation of the cell and malignancy (22–24). We therefore designed assays to investigate whether CRISPR-Cas attack on the HIV provirus in latently infected T cell lines can cause unintended large deletions involving chromosomal sequences flanking the proviral DNA.
RESULTS
Detection of large deletions upon continuous CRISPR-Cas activity
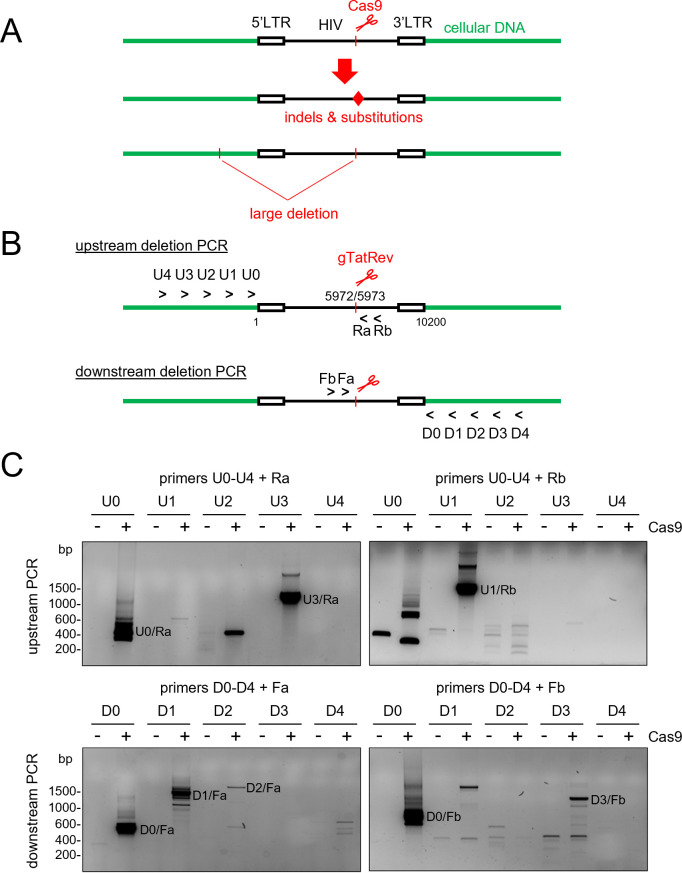
We previously demonstrated that continuous expression of Cas9 and a single antiviral gRNA in HIV-infected cells can result in the acquisition of small indels and nucleotide substitutions at the target site in the proviral DNA [(6, 10, 16, 20); Fig. 1A]. To investigate whether such treatment can also cause larger deletions, we targeted the HIV provirus in the latently infected Jurkat-derived cell lines J-Lat 8.4 and J-Lat 9.2 with Cas9 and a gRNA that directs Cas9 to the overlapping Tat and Rev coding regions (gTatRev; Fig. 1B). A single proviral DNA copy of an Env and Nef-inactivated HIV-1 variant, HIV-R7/E-/GFP, is present on chromosome 1 in J-Lat 8.4 cells and on chromosome 19 in J-Lat 9.2 cells (25). Cells were transduced with lentiviral vectors expressing Cas9 and gTatRev. Two weeks after transduction, intracellular DNA of the total cell culture was isolated and analyzed by PCR. To identify large deletions upstream of the gTatRev target site, forward primers that bind to the chromosomal DNA upstream of the integrated proviral DNA (U0–U4, Fig. 1B) were combined with reverse primers that bind to the HIV DNA downstream of the gTatRev target site (Ra and Rb). The original distance between the forward and reverse primers ranges from approximately 6 kb to 10 kb for the different primer combinations. Because a relatively short PCR extension time of 30 s is used, such long DNA fragments will, however, not be efficiently amplified and only truncated DNA fragments with large deletions upstream of the gTatRev target site will be detected. Similarly, to identify large downstream deletions, reverse primers binding to the chromosomal DNA downstream of the integration site (D0–D4) were combined with forward primers that bind to the HIV DNA upstream of the gTatRev site (Fa and Fb). The original distance between the forward and reverse primers now ranges from approximately 4 kb to 8 kb for the different primer combinations and only truncated proviral fragments with large deletions downstream of the Cas9 cleavage site will be efficiently amplified. PCR products were analyzed by agarose gel electrophoresis, followed by cloning and sequencing.
Fig 1.
Detection of large deletions induced by CRISPR-Cas attack of HIV provirus. (A) CRISPR-Cas cleavage of HIV proviral DNA may not only result in mutations at the cleavage site (indels and nucleotide substitutions; indicated with the red diamond) but also in unintended large deletions that may include surrounding chromosomal sequences. (B) Detection of large upstream and downstream deletions by PCR analysis with primers annealing at several positions in the chromosomal DNA (U0–U4, forward primers annealing at ~0 kb to 4 kb upstream of the HIV integration site; D1–D4, reverse primers annealing at ~0 kb to 4 kb downstream of the HIV integration site) and proviral DNA (Ra and Rb, reverse primer a and b; Fa and Fb, forward primer a and b). (C) PCR analysis of the integrated HIV DNA in untreated and Cas9 + gTatRev-treated J-Lat 9.2 cells. The PCR products resulting from the different primer combinations were analyzed by agarose gel electrophoresis. Example gel images are shown (−, untreated samples; +, Cas9/gTatRev-treated samples; DNA molecular weight markers are shown on the left). PCR products specifically detected upon Cas treatment were isolated from gel and sequenced. PCR products resulting from large deletions, as confirmed by sequencing analysis, are labeled with the primer combination that was used.
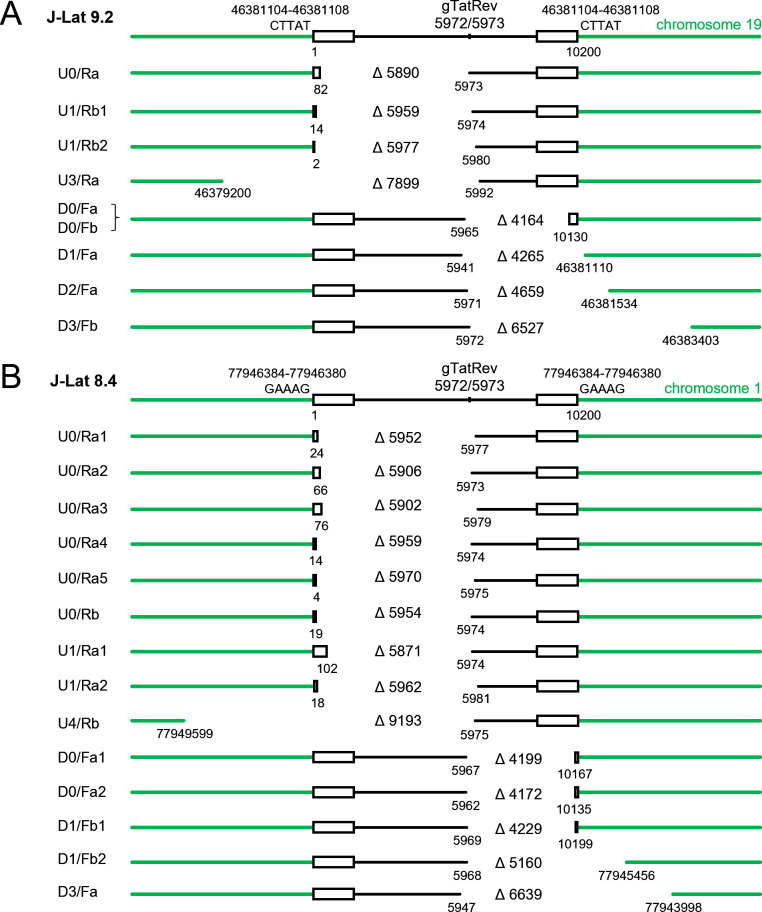
When cellular DNA isolated from Cas9/gTatRev-treated and control (untreated) J-Lat 9.2 cells was analyzed with the different primer combinations, PCR products varying in size from few hundred to several thousand base pair (bp) were detected for the treated cells but not for the control cells (Fig. 1C). Cloning and sequencing of these PCR products revealed that most of them corresponded to truncated DNA fragments in which relatively large DNA fragments upstream or downstream of the gTatRev target site had been deleted (Fig. 2A). The upstream deletions that were detected varied in size from 5,890 bp to 7,899 bp and all ended at or near the Cas9/gTatRev cleavage site, which suggests that Cas9 cleavage at the intended target had triggered the subsequent deletion. Similarly, downstream deletions varied from 4,164 bp to 6,527 bp and all started at or near the intended Cas9/gTatRev target site. One out of four upstream deletions and three out of four downstream deletions did not only remove proviral sequences but also the adjacent chromosomal sequences. Notably, not every primer combination successfully detected a deletion-containing DNA fragment and some PCR products corresponded to chromosomal sequences only. For example, the U2/Ra combination did not detect the deletion-containing fragment that was detected with the U3/Ra combination, while the U2 binding site was not removed by the deletion, but resulted in a ~400 bp chromosomal DNA fragment. The latter observations are likely due to specific characteristics of every primer, like its binding affinity and capacity to anneal to alternative positions in the chromosomal DNA, and the large variety of mutations (large deletions, small indels, and nucleotide substitutions) that can result from the CRISPR-Cas treatment.
Fig 2.
Large deletions detected in J-Lat cells upon continuous Cas9 attack of the HIV proviral DNA. Large deletions were detected in J-Lat 9.2 (A) and J-Lat 8.4 (B) cells by PCR analysis and sequencing of PCR products (as described in Fig. 1). The labels at the left side indicate the primer combination that was used to detect the DNA fragment (as described in Fig. 1). When multiple fragments were detected with the same primer combination, a number was added to the label. The position and size (indicated with ∆) of the deletions are shown. The position of the 5 bp repeat at the integration site (CTTAT in J-Lat 9.2 and GAAAG in J-Lat 8.4) and the Cas9/gTatRev cleavage site are indicated on top.
Similar analysis of Cas9/gTatRev-treated J-Lat 8.4 cells resulted in the identification of large upstream deletions varying in size from 5,871 bp to 9,193 bp and downstream deletions varying from 4,199 bp to 6,639 bp (Fig. 2B). Some of these deletions do not only remove HIV proviral DNA sequences but also flanking chromosomal sequences. All large deletions, either in the upstream or downstream direction, started at or near the original target site. Taken together, these results indicate that Cas9 cleavage at the HIV target site can result in both upstream and downstream large DNA deletions that do not only include proviral DNA sequences, but also the surrounding cellular DNA.
Detection of large deletions upon transient CRISPR-Cas activity
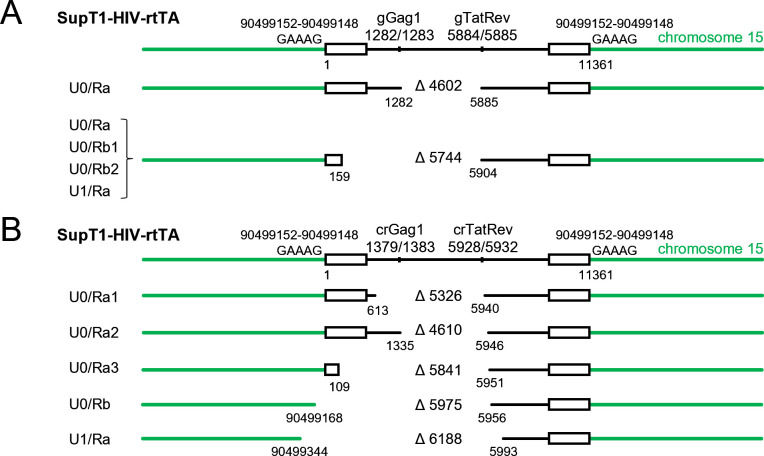
The same PCR strategy was used to detect large CRISPR-Cas-induced deletions in latently infected SupT1 T cells after transient treatment with the Cas9 or Cas12a endonuclease. These cells were previously generated by infection of SupT1 T cells with a doxycycline (dox)-inducible HIV-1 variant (HIV-rtTA-GFP) and a clonal cell line was selected in which the integrated provirus is latent in the absence of dox and transcriptionally active upon dox administration, leading to high virus production and replication (19). We recently demonstrated that repeated transfection of these cells with Cas9 or Cas12a protein combined with gRNAs/crRNAs targeting the Gag and TatRev sites resulted in inactivation of the proviral genome, which prevented dox-induced virus production. Previous DNA analysis demonstrated that this dual-gRNA treatment frequently resulted in indels at the Cas cleavage sites, but also excision and, to a lesser extent, inversion of the viral DNA fragment between the two target sites was observed (19). To detect large Cas-induced upstream deletions, we first performed an Alu PCR analysis (26) to identify provirus integration sites in these cells, which revealed an integration site in the IQ motif containing GTPase activating protein 1 gene locus on chromosome 15 (Fig. S1). We subsequently designed PCR primers that bind to chromosome 15 at different positions upstream of the integration site and to the proviral DNA downstream of the gTatRev and crTatRev target sites. The distance between these reverse and forward primers ranges from 6 kb to 10 kb for the different primer combinations and, because a short PCR extension time of 30 s was used, only truncated proviral fragments with large deletions upstream of the Cas cleavage site will be efficiently amplified by PCR. PCR products were analyzed by agarose gel electrophoresis, followed by cloning and sequencing, to identify large deletions. Analysis of the Cas9-treated cells with different primer combinations identified a 5,744 bp deletion in the proviral DNA that likely resulted from initial Cas9 cleavage at the gTatRev site (Fig. 3A). In addition, the excision product resulting from simultaneous cleavage at both the gGag1 and gTatRev sites and subsequent ligation of the DNA ends was detected (Δ4602). Similarly, analysis of the Cas12a-treated cells (Fig. 3B) identified several large proviral DNA deletions (Δ5326, Δ4610, Δ5841), but also larger deletions that included surrounding chromosomal sequences (Δ5975, Δ6188). These data demonstrate that also transient CRISPR-Cas9 or Cas12a treatment can result in large chromosomal deletions beyond the integrated HIV-rtTA-GFP genome.
Fig 3.
Large deletions detected upon transient Cas9 or Cas12a attack of HIV proviral DNA. Upon transient treatment of SupT1-HIV-rtTA cells with Cas9 + gGag1 + gTatRev (A) or Cas12a + crGag1 + crTatRev (B), large deletions were detected by PCR analysis and sequencing of PCR products. The large deletions are labeled, and their position and size are indicated as described in Fig. 2. The position of the 5 bp repeat at the integration site (GAAAG) and the gRNA/crRNA target sites are indicated on top.
Origin of Cas-induced large deletions
For all observed large deletions, one of the breakpoint junctions was located at or near the original HIV target site (Fig. 2 and 3), which suggests that on-target Cas cleavage induced the large deletion. The other breakpoint junction was located in a more distal proviral DNA region or in the surrounding chromosomal DNA. Comparison of the joined sequences to the original sequences (upstream and downstream breakpoint junction sites; Table 1) revealed the acquisition of short indels in 14 of the 29 joined sequences. Such indels are typical for the error-prone DNA repair following Cas cleavage (11, 14, 15, 27).
TABLE 1.
Analysis of breakpoint junction sites
| Experiment | Large deletion | Joined sequenceb | 5' Breakpoint junction site | 3' Breakpoint junction site | Microhomologya | |||
|---|---|---|---|---|---|---|---|---|
| Not included in deletion | Included in deletion | Included in deletion | Not included in deletion | nt number | Sequence | |||
| J-Lat 9.2 + Cas9 + gTatRev |
U0/Ra | TCCCTGATΔGGCAGGAAGAA | TCCCTGATTG | GCAGAACTAC | ATCTCCTATG | GCAGGAAGAA | 4 | GCAG |
| U1/Rb1 | GTCCTTATTGAATTACTGAAGCGGAGA | GTCCTTATTG | GAAGGGCTAA | ATGGCAGGAA | GAAGCGGAGA | 4 | GAAG | |
| U1/Rb2 | AGGGCTAATTΔCAGGAAGAAG | AGGGCTAATT | CACTCCCAAC | TCTCCTATGG | CAGGAAGAAG | 2 | CA | |
| U3/Ra | CTACAGGTGTΔGCGACGAAGA | CTACAGGTGT | GCACCACCAT | AGCGGAGACA | GCGACGAAGA | 2 | GC | |
| D0/Fa | CTTAGGCATCΔCCGTCTGTTG | CTTAGGCATC | TCCTATGGCA | TAGTGTGTGC | CCGTCTGTTG | 1 | C | |
| D1/Fa | CCAAGTTTGTATTCACTGATT | CCAAGTTTGT | TTCATGACAA | AGCACTTATG | TTCACTGATT | 4 | TTCA | |
| D2/Ra | CATCTCCTATAGCCCCACCCAGTAGTT | CATCTCCTAT | GGCAGGAAGA | ATCGCTTGAG | CCCAGTAGTT | − | − | |
| D3/Fb | ATCTCCTATGTACCTTCCAGAT | ATCTCCTATG | GCAGGAAGAA | GTCTGCCCTG | CCTTCCAGAT | 2 | TG | |
| J-Lat 8.4 + Cas9 + gTatRev |
U0/Ra1 | CACTCCCAACΔGAAGAAGCGG | CACTCCCAAC | GAAGACAAGA | CCTATGGCAG | GAAGAAGCGG | 5 | GAAGA |
| U0/Ra2 | CACACACAAGΔGCAGGAAGAA | CACACACAAG | GCTACTTCCC | ATCTCCTATG | GCAGGAAGAA | 2 | GC | |
| U0/Ra3 | GCTACTTCCCΔAGAAGCGGAG | GCTACTTCCC | TGATTGGCAG | TATGGCAGGA | AGAAGCGGAG | − | − | |
| U0/Ra4 | AGGGCTAATTCTCCGCTTCAGGAAGAAGCGGAG | AGGGCTAATT | CACTCCCAAC | TCTCCTATGG | CAGGAAGAAG | 2 | CA | |
| U0/Ra5 | GGAAAGTGGAΔAGGAAGAAGC | GGAAAGTGGA | AGGGCTAATT | CTCCTATGGC | AGGAAGAAGC | 3 | AGG | |
| U0/Rb | TAATTCACTCTCAGGAAGAAG | TAATTCACTC | CCAACGAAGA | TCTCCTATGG | CAGGAAGAAG | 1 | C | |
| U1/Ra1 | ACACCAGGGCΔCAGGAAGAAG | ACACCAGGGC | CAGGGATCAG | TCTCCTATGG | CAGGAAGAAG | 4 | CAGG | |
| U1/Ra2 | CTAATTCACTACCAGCGAATTCAAAGCGGAGAC | CTAATTCACT | CCCAACGAAG | TGGCAGGAAG | AAGCGGAGAC | − | − | |
| U4/Rb | CAAAAAGAAAΔAGGAAGAAGC | CAAAAAGAAA | AGAGCGTTCT | CTCCTATGGC | AGGAAGAAGC | 2 | AG | |
| D0/Fa1 | TAGGCATCTCΔAGACCCTTTT | TAGGCATCTC | CTATGGCAGG | GAGATCCCTC | AGACCCTTTT | 3 | CTC | |
| D0/Fa2 | AGCCTTAGGCΔTGTTGTGTGA | AGCCTTAGGC | ATCTCCTATG | TGTGCCCGTC | TGTTGTGTGA | 1 | C | |
| D1/Fb1 | GGCATCTCCTTACAGAAAGTCT | GGCATCTCCT | ATGGCAGGAA | AAATCTCTAG | CAGAAAGTCT | − | − | |
| D1/Fb2 | CCTTAGGCATCTCCTTAGGCATCTCCCTAATATGGTA | AGGCATCTCC | TATGGCAGGA | TTCCCATACA | TAATATGGTA | 2 | TA | |
| D3/Fa | TTGTTTCATGGTAAAATGGTTCCTCACTGGGTTTT | TTGTTTCATG | ACAAAAGCCT | TTTCTACTTG | ACTGGGTTTT | 2 | AC | |
| SupT1-HIV-rtTA + Cas9 + gGag1/gTatRev |
U0/Ra1c | GAGACCATCAΔGCAGGAAGAA | GAGACCATCA | ATGAGGAAGC | ATCTCCTATG | GCAGGAAGAA | − | − |
| U0/Ra2 | AAGGGACTTTGGCGACGAAGA | AAGGGACTTT | CCGCTGGGGA | AGCGGAGACA | GCGACGAAGA | − | − | |
| SupT1-HIV-rtTA + Cas12a + crGag1/crTatRev |
U0/Ra1 | GCGAGGGGAGTGGAGTTTCTCTA | GCGAGGGGAG | GCGACTGGTG | AGACTCATCA | AGTTTCTCTA | − | − |
| U0/Ra2 | AGGGCCTATTATCTATCAAAG | AGGGCCTATT | GCACCAGGCC | ATCAAGTTTC | TCTATCAAAG | − | − | |
| U0/Ra3 | GAGAGCTGCAACAAAGCAGTA | GAGAGCTGCA | TCCGGAGTAC | GTTTCTCTAT | CAAAGCAGTA | − | − | |
| U0/Rb | CTGAACTTTAΔCAGTAAGTAG | CTGAACTTTA | TAAGAGTGGT | TCTATCAAAG | CAGTAAGTAG | − | − | |
| U1/Ra | AGGTGCGATTΔCAATAGCAGC | AGGTGCGATT | ATTCTAACTT | ATACAAATAG | CAATAGCAGC | − | − | |
Microhomology at junction sites as determined with mhScanR; –, no microhomology.
nt deletions indicated with Δ; nt insertions underlined; repeated and inverted repeated sequences in bold.
Excision product.
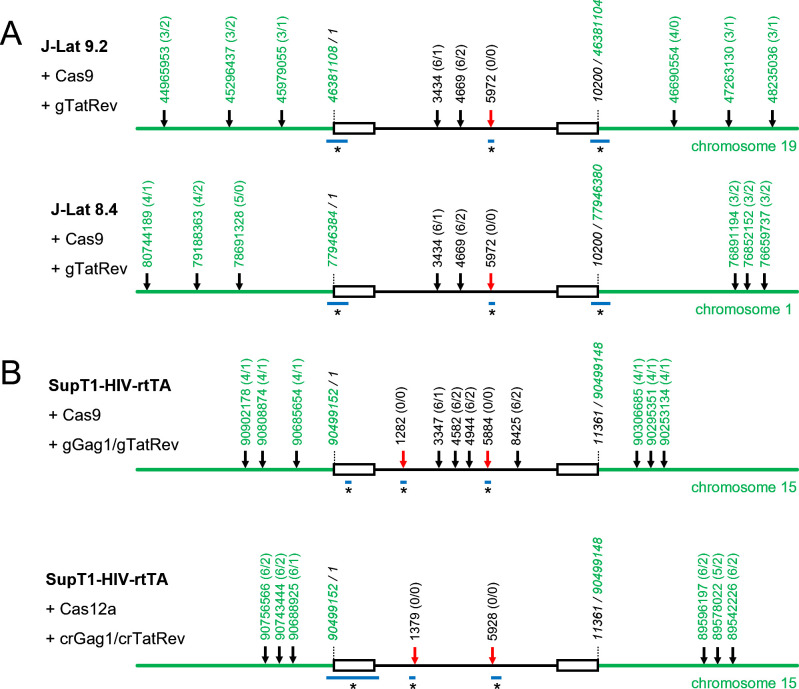
A possible scenario for the creation of such large deletions would be a dual-cleavage event, with simultaneous cleavage at the intended target in HIV and at an off-target site in the flanking chromosomal DNA, followed by excision of the fragment between these two sites. We analyzed the proviral and surrounding chromosomal DNA with the Cas-OFFinder algorithm (28) to identify potential off-target sites with incomplete sequence complementarity to the gRNA/crRNA (29, 30). When allowing up to 6 nt mismatches between the DNA and gRNA/crRNA and up to 2 nt insertions or deletions in the target DNA sequence, this analysis resulted in the identification of several potential off-target sites (Fig. 4). However, none of these sites were located at or near the observed breakpoint junction sites (Fig. 2 and 3), which indicates that the large deletions are not due to off-target Cas cleavage.
Fig 4.
Breakpoint junction sites of large deletions do not coincide with potential CRISPR-Cas off-target sites. For every latently infected cell and Cas-gRNA/crRNA combination, the chromosomal and proviral sequences were analyzed with the Cas-OFFinder algorithm (28) to identify potential off-target sites, allowing up to 6 nt mismatches between the DNA and gRNA/crRNA and up to 2 nt insertions or deletions in the DNA. The position of potential off-target sites in the chromosomal DNA (only the three upstream and three downstream sites closest to the proviral integration site are shown; positions in green) and the position of the on-target and potential off-target sites in the proviral DNA (positions in black) are indicated (line figure is not on scale), with the number of mismatches (varying from 0 to 6) and the number of nt insertions or deletions (varying from 0 to 2) in the target DNA indicated between parentheses (x/y; x, number of nt mismatches; y, number of nt insertions or deletions). Red arrow, on-target cleavage site. Black arrow, potential off-target cleavage site. The horizontal blue lines (labeled with an asterisk) indicate the region where the breakpoint junction sites of the large deletions (as described in Fig. 2 and 3) are located.
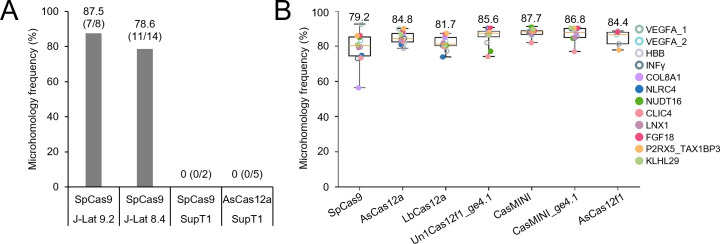
Alternatively, upon Cas cleavage at the intended target site, MMEJ DNA repair may have caused the large deletions (31–35). Hence, we checked for the presence of microhomology at the breakpoint junction sites using the mhScanR application. This analysis resulted in the identification of short microhomologies varying in size from 1 nt to 5 nt at the junction sites for seven out of eight (87.5%) of the large deletions in J-Lat 9.2 cells and for 11 out of 14 (78.6%) of the large deletions in J-Lat 8.4 cells (Table 1; Fig. 5A), which indicates involvement of MMEJ DNA repair in the generation of the deletions. Remarkably, this analysis did not reveal any microhomology at the junction sites for the large deletions detected upon transient Cas9 and Cas12a treatment of the latently infected SupT1 T cells.
Fig 5.
Microhomology at the breakpoint junction sites of Cas-induced large deletions. (A) The frequency of microhomologies at the breakpoint junction sites for the large deletions detected in J-Lat 9.2 and J-Lat 8.4 cells upon continuous Cas9 attack and in SupT1-HIV-rtTA cells upon transient Cas9 or Cas12a attack is shown. Between parentheses, the number of deletions with microhomology at the breakpoint junction sites is indicated as a fraction of the total number of identified deletions. (B) Microhomology frequency at breakpoint junction sites for large deletions induced by different Cas nucleases upon targeting of cellular genes in 293T cells. Xin et al. (36) recently reported large deletions in human embryonic kidney 293T cells upon targeting 12 cellular genes with seven different Cas variants. In their supplementary data (NCBI Gene Expression Omnibus database; accession code GSE213149), they indicated the presence or absence of microhomology at the breakpoint junction sites for every editing event. After filtering for deletions larger than 100 bp in size with both breakpoint junction sites located on the same chromosome and with the same strand orientation to exclude more complex chromosome rearrangement events, we calculated the microhomology frequency for every Cas and target combination. The numbers above the whiskers refer to the average value for all targets. Values from minimum to maximum are shown by the whiskers, and the bounds of the box indicate the first and third quartile.
Recently, Xin et al. (36) analyzed the formation of large deletions in human embryonic kidney 293T (HEK-293T) cells upon transient treatment with different Cas nucleases, including the Cas9 (SpCas9) and Cas12a (AsCas12a) variants that we used in our study. Large deletions were detected in all targeted cellular genes and the frequency varied only slightly for the different Cas systems, with the frequency observed for SpCas9 being 1.9% and for AsCas12a 1.4% of the total editing events. Xin et al. indicated the presence or absence of microhomology at the breakpoint junction sites for every editing event (36). We used these data (NCBI Gene Expression Omnibus database; accession code GSE213149) to calculate the microhomology frequency for every Cas and target combination (Fig. 5B). This analysis revealed a similar high microhomology frequency (79.2% to 87.7%) for large deletions induced by the different Cas variants. This result indicates that for all tested Cas variants, the MMEJ DNA repair pathway was likely involved in the generation of the large deletions in the transiently treated 293T cells.
DISCUSSION
Several previous studies in which the HIV proviral DNA was attacked with CRISPR-Cas9 or Cas12a systems demonstrated that Cas cleavage frequently results in the accumulation of small indels and nucleotide substitutions at the target sites. When the provirus was simultaneously attacked at two or more positions, excision or inversion of the DNA fragment between target sites was also observed (11, 16, 18). We, here, used a PCR-based sequencing strategy to demonstrate that CRISPR-Cas9 and Cas12a treatment can also result in much larger deletions that may include the surrounding chromosomal DNA sequences. As the observed deletions start at the intended gRNA target, they are likely induced by on-target Cas cleavage activity. We demonstrate the frequent presence of microhomology at the breakpoint junction sites of large deletions, which indicates the involvement of the MMEJ DNA repair mechanism in the formation of the large deletions.
Such large deletions were not detected in the earlier HIV studies , which can simply be explained by the fact that most studies used PCR-based methods with primers annealing at a relatively short distance from the intended DNA cleavage site, while the large deletions remove at least one of these primer binding sites. By using more distal primers annealing to the chromosomal DNA surrounding the proviral DNA, we were able to detect Cas-induced large deletions up to 9.2 kb. Larger deletions will likely also occur, but will also be missed in our analysis because one of the primer binding sites will be lost. Thus, extreme caution is warranted in the analysis of Cas editing products by means of PCR-based strategies. Complementary use of RNA analysis may be warranted to assess whether aberrant transcripts are formed, which could confirm deleterious effects of CRISPR-Cas treatment.
Our HIV editing findings agree with other studies in which the CRISPR-Cas9 system was used for other genome editing purposes (37–39). Also in these studies, large deletions could be detected upstream or downstream of the cleavage site. For example, Wen et al. reported that over 80% of the large deletions observed upon Cas9/gRNA treatment of human T cells varied in size between 100 and 1,000 bp, but deletions can extend over many kilobases (39). Cas cleavage can even result in chromosome truncations (40) and rearrangements (41) and trigger genome instability (42). We observed microhomologies at the breakpoint junction sites for most of the Cas9-induced large deletions in J-Lat cells (82%), which is in agreement with the high frequency of microhomology marks in other CRISPR-Cas studies (31, 35, 36). Recently, Xin et al. (36) reported the generation of large deletions in HEK-293T cells upon targeting cellular genes with different Cas nucleases. Analysis of their supplementary data revealed a similar high frequency of small microhomologies at the breakpoint junction sites of the large deletions induced by the different Cas variants, with a frequency of 79% and 85% for the SpCas9- and AsCas12a-induced deletions, respectively (Fig. 5B). Taken together, these results suggest that large deletions frequently result from the MMEJ DNA repair that follows Cas cleavage. Surprisingly, we did not identify any microhomology at the breakpoint junction sites for the large deletions detected in the infected SupT1 T cells upon transient Cas9 and Cas12a treatment. Our limited data may suggest that the cell type could affect the cellular DNA repair process and formation of large deletions, but further analysis will be required to understand this observation.
We here demonstrate that CRISPR-Cas attack of integrated HIV DNA can cause large unintended deletions that can include surrounding chromosomal DNA sequences. Such large deletions may lead to the loss of tumor suppressor genes or activation of proto-oncogenes and thereby result in oncogenic transformation (22–24). The frequency of such a detrimental event may be low, in particular when targeting central positions in the HIV genome. However, this frequency may increase when targeting positions close to the 5´ or 3´ end of the provirus, such as the long-terminal repeat (LTR) region. The LTR is in fact a popular target because it is present at both the 5´ and 3´ end of the proviral DNA and dual cleavage may allow excision of the nearly complete viral genome with a single guide RNA (43–45). Furthermore, HIV integrates in a random manner on any of the chromosomes, which increases the chance of hitting a nearby proto-oncogene or tumor suppressor gene (22–24, 46). Several strategies can be considered to reduce the large deletion frequency during CRISPR-Cas treatment, including the use of alternative Cas nucleases like CasMINI (a modified Un1Cas12f1) and AsCas12f1 (36), or drugs that inhibit the MMEJ pathway (47–49). However, these strategies have thus far demonstrated limited success and do not prevent the formation of large deletions (36, 47–49). Clinical trials in which CRISPR-Cas9 is used to inactivate HIV in infected persons have recently started (50, 51). It will be important to closely monitor the generation of unintended large deletions and oncogenic transformation of cells in the treated persons, as this would pose a serious safety risk.
MATERIALS AND METHODS
Cell lines and culturing
J-Lat 8.4 cells and J-Lat 9.2 cells with an integrated HIV-R7/E-/GFP genome were kindly provided by Dr. Eric Verdin through the National Institutes of Health (NIH) HIV Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH (52). J-Lat cells were maintained in advanced RPMI-1640 medium (Gibco, Life Technologies, Bleiswijk, The Netherlands) with 1% fetal bovine serum, 15 U/mL penicillin, 15 U/mL streptomycin mix, and 1% L-glutamine. Production and culturing of the SupT1-HIV-rtTA T cells with an integrated dox-controlled HIV variant (HIV-rtTA-GFP) were described previously (19). Human embryonic kidney 293T cells were cultured in Dulbecco's modified Eagle medium (DMEM; Gibco, Life Technologies, Bleiswijk, The Netherlands) supplemented with 10% fetal calf serum, penicillin (100 U/mL), and streptomycin (100 mg/mL), as described previously (53).
Lentiviral vector transduction for continuous CRISPR-Cas editing
The lentiviral vectors LentiCas9-Blast (Addgene; 52962), containing a human codon-optimized Streptococcus pyogenes Cas9-expression cassette, and LentiGuide-Puro (Addgene; 52963), containing a gRNA expression cassette, were gifts from Feng Zhang (54). Construction of the gTatRev-expressing LentiGuide-Puro vector was previously described (10). LentiCas9-Blast and LentiGuide-Puro-gTatRev viral vector particles were produced as previously described (20, 55). Briefly, 293T cells were seeded in standard DMEM medium and transfected with the lentiviral vector plasmid and packaging plasmids pSYNGP, pRSV-rev, and pVSV-g using Lipofectamine 2000. After transfection for 6 h, the medium was replaced with Opti-MEM (Gibco) supplemented with penicillin (100 U/mL) and streptomycin (100 mg/mL) and the cells were cultured for 48 h at 37°C with 5% CO2. The lentiviral particles containing supernatant was briefly centrifuged at 250 × g for 5 min to remove cells, followed by filtration (0.45 µM) and concentration at 1,750 × g using a Vivaspin 20 ultrafiltration spin column (100 kDa molecular weight cutoff; Sartorius). Aliquots were stored at −80°C. For transduction of J-Lat 8.4 and J-Lat 9.2 cells, 2 × 105 cells in 1 mL culture medium were first transduced with LentiCas9-Blast virus particles (30 ng of CA-p24) and cultured with blasticidin (3 µg/mL) for 14 days to select LentiCas9-transduced cells. Cells were subsequently transduced with LentiGuide-Puro-gTatRev vector particles (30 ng of CA-p24) and cultured with puromycin (0.5 µg/mL) to select double-transduced cells. Intracellular DNA of the total cell culture was isolated at 14 days after the second transduction for analysis of the integrated proviral DNA and flanking chromosomal sequences.
Nucleofection of CRISPR-Cas reagents for transient editing
SupT1-HIV-rtTA T cells were transfected three times (at days 1, 6, and 29) with Cas9 and Cas12a ribonucleoprotein complexes, as previously described (19). In brief, for Cas9 RNP production, gRNA duplexes were prepared by mixing equal amounts of 200 µM crRNA (IDT) and 200 µM tracrRNA (IDT), heating at 95°C for 5 minutes, followed by slow cooling. One hundred picomoles gRNA duplex was subsequently combined with 60 pmol SpCas9 protein (TrueCut Cas9 Protein v2 NLS; Invitrogen A36499, Waltham, MA, USA). For Cas12a RNP production, 100 pmol crRNA (IDT) was combined with 60 pmol AsCas12a protein [Alt-R A.s. Cas12a (Cpf1) Ultra nuclease; IDT, Leuven, Belgium; 10001273]. Cells were transfected with the RNP complexes using the 4D-Nucleofector X Unit and SF cell line Kit S (Lonza) with the CA-137 program. Total cellular DNA was isolated at 6 days after the third nucleofection for analysis of the integrated proviral DNA and flanking chromosomal sequences.
Alu PCR for determination of HIV integration site
Alu PCR analysis (26) was used to identify HIV-rtTA-GFP integration sites in the SupT1-HIV-rtTA cells. Cellular DNA was isolated using the DNeasy Blood and Tissue Kit (QIAGEN) and a QIAshredder microcentrifuge spin column (QIAGEN), as previously described (20). In a first PCR step, 100 ng total DNA was mixed with DreamTaq Green PCR Master Mix (K1081, Thermo Scientific), 50 pmol primer Alu-R (5′-TGCTGGGATTACAGGCGTGAG-3′; binding the Alu repetitive element), and 50 pmol primer A0653 (5′-TTGCTACAAGGGACTTTCCGCTGG-3′; binding the U3 domain in the viral LTR) in a total volume of 50 µL. Samples were denatured at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s. After an additional extension step at 72°C for 10 min, 1 µL of the PCR product was used as template in a semi-nested PCR, using the DreamTaq Green PCR Master Mix, 50 pmol primer Alu-R, and 50 pmol primer A1995 (5′-TCAATAAAGCTTGCCTTGAGTGC-3′; binding the R domain in the LTR). After 25 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 10 min, the PCR product was analyzed by agarose gel electrophoresis. PCR products were isolated from gel, cloned in a pCRII-TOPO TA cloning vector (Thermo Fisher Scientific, USA), and sequenced using the BigDye Terminator v1.1 Cycle Sequencing Kit (Perkin Elmer Applied Biosystem). HIV sequences were identified using CodonCode Aligner software and the HIV-rtTA-GFP reference sequence, and chromosomal sequences were identified using the NCBI nucleotide BLAST tool (http://www.ncbi.nlm.nih.gov/blast) and the “human genomic plus transcript” databases. To confirm the HIV integration site in chromosome 15, the HIV-cellular DNA junctions were amplified by PCR using a primer binding HIV DNA downstream of the 5´LTR (A1311, 5′-CCTGTTCGGGCGCCACTGCTA-3′) or upstream of the 3´LTR (A1995) in combination with a primer binding the chromosomal DNA upstream (U0, 5′-ACACTTTCTATCCCGAAGG-3′) or downstream (D1, 5′-CAGCCAAGCCATAATAGG-3′) of the HIV DNA, respectively, followed by sequencing of the PCR product. The Alu repeat sequence was identified using the RepeatMasker web server (https://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) and the open database Dfam 3.0 (A.F.A. Smit, R. Hubley, and P. Green, unpublished data. Version: open-4.0.9).
PCR and sequencing analysis for detection of large deletions
Total cellular DNA was isolated from control and Cas-treated cells using the DNeasy Blood and Tissue Kit (QIAGEN) with a QIAshredder microcentrifuge spin column (QIAGEN). Large deletions were detected by nested PCR. Primers (Table S1) were designed using the OligoAnalyzer Tool (IDT) and the HIV-R7/E-/GFP sequence (56), HIV-rtTA-GFP sequence (19, 57, 58), and reference human genome sequence (GRCh38.p14, RefSeq assembly accession GCF_000001405.40). For the first PCR step, 100 ng DNA was mixed with 1 × DreamTaq Green PCR Master Mix, 50 pmol forward primer 1, and 50 pmol reverse primer 1 (primers listed in Table S1) in a total volume of 50 µL. After heating at 95°C for 5 min, 35 PCR cycles of 30 s at 95°C, 30 s at 55°C, 30 s at 72°C, and a final extension for 7 min at 72°C, 1 µL of the PCR mix was used as template in a second PCR containing 1 × DreamTaq Green PCR Master Mix, 50 pmol forward primer 2, and 50 pmol primer reverse primer 2. After 25 cycles of 30 s at 95°C, 30 s at 55°C, 30 s at 72°C, and a final extension for 7 min at 72°C, the PCR product was analyzed by agarose gel electrophoresis. PCR products uniquely observed for the Cas-treated cells were cloned in a pCRII-TOPO TA cloning vector and sequenced. Sequences were aligned to reference sequences using CodonCode Aligner (version 10.0.2). The NCBI BLAST tool was used to identify chromosomal sequences.
Off-target site prediction and microhomology analysis
The reference human genome (GRCh38/hg38) was analyzed with Cas-OFFinder (http://www.rgenome.net/cas-offinder) (28) to identify potential off-target sites for Cas9 and Cas12a cleavage. For SpCas9, the algorithm searched for 20 base pair gRNA target sequences followed by the 5′-NGG-3′ protospacer adjacent motif (PAM) sequence. For Cas12a, the algorithm searched for 23 base pair gRNA target sequences adjacent to the 5′-TTTV-3′ PAM sequence. Mismatches up to six base pairs and insertions (DNA bulge) or deletions (RNA bulge) up to two nucleotides were allowed. The reference HIV-1 sequence (NC_001802.1) was similarly analyzed with Cas-OFFinder, followed by alignment with the HIV-R7/E-/GFP and HIV-rtTA-GFP sequences. The R package mhscanR (https://github.com/d0minicO/mhscanR) (31) was used to identify microhomologies at the breakpoint junction sites at Cas9- and Cas12a-induced large deletions.
ACKNOWLEDGMENTS
Ye Liu is the recipient of a fellowship of the China Scholarship Council (CSC). This research is financially supported by the Aidsfonds, The Netherlands, grant P-22605, and the TKI-PPP grant Target2Cure from Health Holland/Aidsfonds (LSHM19101-SGF).
Contributor Information
Atze T. Das, Email: a.t.das@amsterdamumc.nl.
Frank Kirchhoff, Ulm University Medical Center, Ulm, Germany.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.01334-23.
Fig. S1. Identification of the HIV integration site on chromosome 15 in SupT1-HIV-rtTA cells. Table S1. DNA oligonucleotides used in PCR analyses.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 9:727–728. doi: 10.1038/nm880 [DOI] [PubMed] [Google Scholar]
- 2. Blankson JN, Persaud D, Siliciano RF. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu Rev Med 53:557–593. doi: 10.1146/annurev.med.53.082901.104024 [DOI] [PubMed] [Google Scholar]
- 3. Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 94:13193–13197. doi: 10.1073/pnas.94.24.13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davey RT, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, Kovacs JA, Polis MA, Walker RE, Falloon J, Masur H, Gee D, Baseler M, Dimitrov DS, Fauci AS, Lane HC. 1999. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A 96:15109–15114. doi: 10.1073/pnas.96.26.15109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colby DJ, Trautmann L, Pinyakorn S, Leyre L, Pagliuzza A, Kroon E, Rolland M, Takata H, Buranapraditkun S, Intasan J, et al. 2018. Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat Med 24:923–926. doi: 10.1038/s41591-018-0026-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang G, Zhao N, Berkhout B, Das AT. 2018. CRISPR-Cas based antiviral strategies against HIV-1. Virus Res 244:321–332. doi: 10.1016/j.virusres.2017.07.020 [DOI] [PubMed] [Google Scholar]
- 7. Atkins AJ, Allen AG, Dampier W, Haddad EK, Nonnemacher MR, Wigdahl B. 2021. HIV-1 cure strategies: why CRISPR? Expert Opin Biol Ther 21:781–793. doi: 10.1080/14712598.2021.1865302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O, Cassa CA, Liu DR, Gifford DK, Sherwood RI. 2018. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563:646–651. doi: 10.1038/s41586-018-0686-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Allen F, Crepaldi L, Alsinet C, Strong AJ, Kleshchevnikov V, De Angeli P, Páleníková P, Khodak A, Kiselev V, Kosicki M, Bassett AR, Harding H, Galanty Y, Muñoz-Martínez F, Metzakopian E, Jackson SP, Parts L. 2018. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat Biotechnol. doi: 10.1038/nbt.4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang G, Zhao N, Berkhout B, Das AT. 2016. CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mol Ther 24:522–526. doi: 10.1038/mt.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao Z, Fan M, Das AT, Herrera-Carrillo E, Berkhout B. 2020. Extinction of all infectious HIV in cell culture by the CRISPR-Cas12a system with only a single crRNA. Nucleic Acids Res 48:5527–5539. doi: 10.1093/nar/gkaa226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoder KE, Bundschuh R. 2016. Host double strand break repair generates HIV-1 strains resistant to CRISPR/Cas9. Sci Rep 6:29530. doi: 10.1038/srep29530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ueda S, Ebina H, Kanemura Y, Misawa N, Koyanagi Y. 2016. Anti-HIV-1 potency of the CRISPR/Cas9 system insufficient to fully inhibit viral replication. Microbiol Immunol 60:483–496. doi: 10.1111/1348-0421.12395 [DOI] [PubMed] [Google Scholar]
- 14. Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S, Wainberg MA, Liang C. 2016. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep 15:481–489. doi: 10.1016/j.celrep.2016.03.042 [DOI] [PubMed] [Google Scholar]
- 15. Liang C, Wainberg MA, Das AT, Berkhout B. 2016. CRISPR/Cas9: a double-edged sword when used to combat HIV infection. Retrovirology 13:37. doi: 10.1186/s12977-016-0270-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang G, Zhao N, Berkhout B, Das AT. 2016. A combinatorial CRISPR-Cas9 attack on HIV-1 DNA extinguishes all infectious provirus in infected T cell cultures. Cell Rep 17:2819–2826. doi: 10.1016/j.celrep.2016.11.057 [DOI] [PubMed] [Google Scholar]
- 17. Lebbink RJ, de Jong DCM, Wolters F, Kruse EM, van Ham PM, Wiertz E, Nijhuis M. 2017. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci Rep 7:41968. doi: 10.1038/srep41968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fan M, Berkhout B, Herrera-Carrillo E. 2022. A combinatorial CRISPR-Cas12a attack on HIV DNA. Mol Ther Methods Clin Dev 25:43–51. doi: 10.1016/j.omtm.2022.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Y, Jeeninga RE, Klaver B, Berkhout B, Das AT. 2021. Transient CRISPR-Cas treatment can prevent reactivation of HIV-1 replication in a latently infected T-cell line. Viruses 13:2461. doi: 10.3390/v13122461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Binda CS, Klaver B, Berkhout B, Das AT. 2020. CRISPR-Cas9 dual-gRNA attack causes mutation, excision and inversion of the HIV-1 proviral DNA. Viruses 12:330. doi: 10.3390/v12030330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771. doi: 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schröder ARW, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521–529. doi: 10.1016/s0092-8674(02)00864-4 [DOI] [PubMed] [Google Scholar]
- 23. Wagner TA, McLaughlin S, Garg K, Cheung CYK, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. 2014. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345:570–573. doi: 10.1126/science.1256304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. 2014. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345:179–183. doi: 10.1126/science.1254194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Symons J, Chopra A, Malatinkova E, De Spiegelaere W, Leary S, Cooper D, Abana CO, Rhodes A, Rezaei SD, Vandekerckhove L, Mallal S, Lewin SR, Cameron PU. 2017. HIV integration sites in latently infected cell lines: evidence of ongoing replication. Retrovirology 14:23. doi: 10.1186/s12977-017-0340-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Butler SL, Hansen MS, Bushman FD. 2001. A quantitative assay for HIV DNA integration in vivo. Nat Med 7:631–634. doi: 10.1038/87979 [DOI] [PubMed] [Google Scholar]
- 27. Zhu W, Lei R, Le Duff Y, Li J, Guo F, Wainberg MA, Liang C. 2015. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology 12:22. doi: 10.1186/s12977-015-0150-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bae S, Park J, Kim J-S. 2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30:1473–1475. doi: 10.1093/bioinformatics/btu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang X-H, Tee LY, Wang X-G, Huang Q-S, Yang S-H. 2015. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids 4:e264. doi: 10.1038/mtna.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murugan K, Seetharam AS, Severin AJ, Sashital DG. 2020. CRISPR-Cas12a has widespread off-target and dsDNA-nicking effects. J Biol Chem 295:5538–5553. doi: 10.1074/jbc.RA120.012933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Owens DDG, Caulder A, Frontera V, Harman JR, Allan AJ, Bucakci A, Greder L, Codner GF, Hublitz P, McHugh PJ, Teboul L, de Bruijn M. 2019. Microhomologies are prevalent at Cas9-induced larger deletions. Nucleic Acids Res 47:7402–7417. doi: 10.1093/nar/gkz459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seol J-H, Shim EY, Lee SE. 2018. Microhomology-mediated end joining: good, bad and ugly. Mutat Res 809:81–87. doi: 10.1016/j.mrfmmm.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sfeir A, Symington LS. 2015. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem Sci 40:701–714. doi: 10.1016/j.tibs.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carvajal-Garcia J, Cho J-E, Carvajal-Garcia P, Feng W, Wood RD, Sekelsky J, Gupta GP, Roberts SA, Ramsden DA. 2020. Mechanistic basis for microhomology identification and genome scarring by polymerase theta. Proc Natl Acad Sci U S A 117:8476–8485. doi: 10.1073/pnas.1921791117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kosicki M, Allen F, Steward F, Tomberg K, Pan Y, Bradley A. 2022. Cas9-induced large deletions and small indels are controlled in a convergent fashion. Nat Commun 13:3422. doi: 10.1038/s41467-022-30480-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xin C, Yin J, Yuan S, Ou L, Liu M, Zhang W, Hu J. 2022. Comprehensive assessment of miniature CRISPR-Cas12f nucleases for gene disruption. Nat Commun 13:5623. doi: 10.1038/s41467-022-33346-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kosicki M, Tomberg K, Bradley A. 2018. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36:765–771. doi: 10.1038/nbt.4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, Hughes J, Pomerantz RT, Thomas PQ. 2018. Large deletions induced by Cas9 cleavage. Nature 560:E8–E9. doi: 10.1038/s41586-018-0380-z [DOI] [PubMed] [Google Scholar]
- 39. Wen W, Quan Z-J, Li S-A, Yang Z-X, Fu Y-W, Zhang F, Li G-H, Zhao M, Yin M-D, Xu J, Zhang J-P, Cheng T, Zhang X-B. 2021. Effective control of large deletions after double-strand breaks by homology-directed repair and dsODN insertion. Genome Biol 22:236. doi: 10.1186/s13059-021-02462-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cullot G, Boutin J, Toutain J, Prat F, Pennamen P, Rooryck C, Teichmann M, Rousseau E, Lamrissi-Garcia I, Guyonnet-Duperat V, Bibeyran A, Lalanne M, Prouzet-Mauléon V, Turcq B, Ged C, Blouin J-M, Richard E, Dabernat S, Moreau-Gaudry F, Bedel A. 2019. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun 10:1136. doi: 10.1038/s41467-019-09006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rayner E, Durin M-A, Thomas R, Moralli D, O’Cathail SM, Tomlinson I, Green CM, Lewis A. 2019. CRISPR-Cas9 causes chromosomal instability and rearrangements in cancer cell lines, detectable by cytogenetic methods. CRISPR J 2:406–416. doi: 10.1089/crispr.2019.0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alanis-Lobato G, Zohren J, McCarthy A, Fogarty NME, Kubikova N, Hardman E, Greco M, Wells D, Turner JMA, Niakan KK. 2021. Frequent loss of heterozygosity in CRISPR-Cas9-edited early human embryos. Proc Natl Acad Sci U S A 118:e2004832117. doi: 10.1073/pnas.2004832117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liao H-K, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, Suzuki K, Xu R, Hishida T, Chang C-J, Esteban CR, Young J, Izpisua Belmonte JC. 2015. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun 6:6413. doi: 10.1038/ncomms7413 [DOI] [PubMed] [Google Scholar]
- 44. Dash PK, Kaminski R, Bella R, Su H, Mathews S, Ahooyi TM, Chen C, Mancuso P, Sariyer R, Ferrante P, et al. 2019. Sequential LASER ART and CRISPR treatments eliminate HIV-1 in a subset of infected humanized mice. Nat Commun 10:2753. doi: 10.1038/s41467-019-10366-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lai M, Maori E, Quaranta P, Matteoli G, Maggi F, Sgarbanti M, Crucitta S, Pacini S, Turriziani O, Antonelli G, Heeney JL, Freer G, Pistello M. 2021. CRISPR/Cas9 ablation of integrated HIV-1 accumulates proviral DNA circles with reformed long terminal repeats. J Virol 95:e0135821. doi: 10.1128/JVI.01358-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Swaminathan S, Huang C, Geng H, Chen Z, Harvey R, Kang H, Ng C, Titz B, Hurtz C, Sadiyah MF, Nowak D, Thoennissen GB, Rand V, Graeber TG, Koeffler HP, Carroll WL, Willman CL, Hall AG, Igarashi K, Melnick A, Müschen M. 2013. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med 19:1014–1022. doi: 10.1038/nm.3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roidos P, Sungalee S, Benfatto S, Serçin Ö, Stütz AM, Abdollahi A, Mauer J, Zenke FT, Korbel JO, Mardin BR. 2020. A scalable CRISPR/Cas9-based fluorescent reporter assay to study DNA double-strand break repair choice. Nat Commun 11:4077. doi: 10.1038/s41467-020-17962-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schimmel J, Muñoz-Subirana N, Kool H, van Schendel R, van der Vlies S, Kamp JA, de Vrij FMS, Kushner SA, Smith GCM, Boulton SJ, Tijsterman M. 2023. Modulating mutational outcomes and improving precise gene editing at CRISPR-Cas9-induced breaks by chemical inhibition of end-joining pathways. Cell Rep 42:112019. doi: 10.1016/j.celrep.2023.112019 [DOI] [PubMed] [Google Scholar]
- 49. Yuan B, Bi C, Wang J, Jin Y, Alsayegh K, Tehseen M, Yi G, Hamdan S, Huang Y, Li M. 2022. Modulation of the microhomology-mediated end joining pathway suppresses large deletions and enhances homology-directed repair following CRISPR-Cas9-induced DNA breaks. bioRxiv. doi: 10.1101/2022.11.16.516713 [DOI] [PMC free article] [PubMed]
- 50. BioTherapeutics E . 2022. Study of EBT-101 in aviremic HIV-1 infected adults on stable ART. Available from: https://ClinicalTrials.gov/show/NCT05144386
- 51. BioTherapeutics E . 2023. Long-term follow-up study of HIV-1 infected adults who received EBT-101. Available from: https://ClinicalTrials.gov/show/NCT05143307
- 52. Jordan A, Bisgrove D, Verdin E. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22:1868–1877. doi: 10.1093/emboj/cdg188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Das AT, Harwig A, Berkhout B. 2011. The HIV-1 Tat protein has a versatile role in activating viral transcription. J Virol 85:9506–9516. doi: 10.1128/JVI.00650-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. ter Brake O, Konstantinova P, Ceylan M, Berkhout B. 2006. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther 14:883–892. doi: 10.1016/j.ymthe.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 56. Chung C-H, Mele AR, Allen AG, Costello R, Dampier W, Nonnemacher MR, Wigdahl B. 2020. Integrated human immunodeficiency virus type 1 sequence in J-Lat 10.6. Microbiol Resour Announc 9:e00179-20. doi: 10.1128/MRA.00179-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Das AT, Verhoef K, Berkhout B. 2004. A conditionally replicating virus as a novel approach toward an HIV vaccine. Meth Enzymol 388:359–379. doi: 10.1016/S0076-6879(04)88028-5 [DOI] [PubMed] [Google Scholar]
- 58. Zhou X, Vink M, Berkhout B, Das AT. 2006. Modification of the Tet-on regulatory system prevents the conditional-live HIV-1 variant from losing doxycycline-control. Retrovirology 3:82. doi: 10.1186/1742-4690-3-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Identification of the HIV integration site on chromosome 15 in SupT1-HIV-rtTA cells. Table S1. DNA oligonucleotides used in PCR analyses.