ABSTRACT
Infection of the central nervous system by the Japanese encephalitis virus (JEV) is characterized by extensive neuronal cell death and neuroinflammation. Several protein-coding genes and microRNAs are implicated in JEV-induced neuronal cell death. However, the global expression patterns and functional contributions of long non-coding RNAs (lncRNAs) during JEV-induced neuronal cell death have not been explored. Here, we profiled the transcriptome of the JEV-infected neuronal cell line and identified several lncRNAs whose expression is altered during JEV infection. We functionally characterized a lncRNA named JINR1 (JEV-induced non-coding RNA 1), which is evolutionarily conserved in primates. JINR1 induction during JEV infection is regulated by nuclear factor-kappa B (NF-κB). Depletion of JINR1 during infection reduces flavivirus replication, neuronal cell death, and the expression of genes involved in ER stress and neuroinflammation. Interestingly, GRP78 overexpression prevents the decrease in flavivirus replication due to JINR1 knockdown. JINR1 interacts with RBM10 and NF-κB to regulate the transcription of virus-induced genes. In addition, RBM10 and JINR1 form a feed-forward loop to reciprocally promote each other’s expression by regulating NF-κB activity. Our results suggest the role of JINR1 in promoting flavivirus replication and flavivirus-induced neuronal cell death.
IMPORTANCE
Central nervous system infection by flaviviruses such as Japanese encephalitis virus, Dengue virus, and West Nile virus results in neuroinflammation and neuronal damage. However, little is known about the role of long non-coding RNAs (lncRNAs) in flavivirus-induced neuroinflammation and neuronal cell death. Here, we characterized the role of a flavivirus-induced lncRNA named JINR1 during the infection of neuronal cells. Depletion of JINR1 during virus infection reduces viral replication and cell death. An increase in GRP78 expression by JINR1 is responsible for promoting virus replication. Flavivirus infection induces the expression of a cellular protein RBM10, which interacts with JINR1. RBM10 and JINR1 promote the proinflammatory transcription factor NF-κB activity, which is detrimental to cell survival.
KEYWORDS: JEV, DENV, WNV, flavivirus, lncRNAs, ER stress, NF-κB, GRP78, RBM10, LINC01518
INTRODUCTION
Japanese encephalitis virus (JEV) is a mosquito-borne, positive single-stranded RNA-containing, neurotropic virus belonging to the genus Flavivirus (1, 2). This genus includes Dengue virus (DENV), West Nile virus (WNV), Yellow fever virus, Zika virus, and Tick-borne encephalitis virus. JEV is endemic in Asia-Pacific and causes viral encephalitis, known as Japanese encephalitis (JE), for which no cure is available (2). According to WHO estimates, JEV is responsible for ~50,000 cases annually, one-third of which are fatal, and around half result in permanent neurological damage in survivors (3, 4). Neurons are the most supportive cells for the infection and replication of JEV in the central nervous system (5, 6). Although all cases of JEV infection are not symptomatic, its infection of the central nervous system (CNS) is marked by an uncontrolled inflammatory response and neuronal cell death (3, 7–9). Other flaviviruses, such as DENV and WNV, also infect neurons, resulting in neuroinflammation and apoptosis (10–19). Despite the availability of vaccines, the disease burden of JE is high in India due to low vaccine usage and efficacy, suggesting the need for novel therapeutic agents to combat symptomatic JEV infection and prevent disease progression (7, 8).
Long non-coding RNAs (lncRNAs) are regulatory transcripts that play functional roles in development and disease, including viral pathogenesis, by regulating gene expression in cis and trans at both transcriptional and post-transcriptional levels (20, 21). LncRNAs are poorly conserved across species, have expanded widely in the primate brain, and likely contribute to neural diversification (22). LncRNAs show differential expression (DE) in response to viral replication or viral protein expression induced by infection (22). During viral life cycles, lncRNAs modulate viral gene expression, viral replication, genome packaging, virion release, and viral-induced host gene expression (22–24). LncRNAs also regulate innate immune responses by promoting or inhibiting viral replication, highlighting them as novel targets for developing anti-viral therapies (25–27). Moreover, lncRNAs play a significant role in inducing or suppressing cell death using diverse mechanisms (28–30).
JEV infection triggers endoplasmic reticulum (ER) stress and neuronal apoptosis through several pathways, including protein kinase R-like endoplasmic reticulum kinase (PERK), activating transcription factor 6, Inositol-requiring enzyme 1 (IRE1), and Forkhead box O signaling (31–33). Interestingly, JEV infection in the CNS of mice results in the upregulation of a nuclear virus-inducible ncRNA (VINC)/NEAT1, but its function in JEV pathogenesis is unknown (34). JEV-induced PERK activation during unfolded protein response (UPR) results in the upregulation of lncRNA MALAT1 in mouse neuroblastoma cells, but its exact role in JEV pathogenesis is unclear (35). Li et al., using a microarray screen, identified 618 DE lncRNAs upon JEV infection in mice’s brains; co-expression network analysis of DE lncRNAs suggested their involvement in host immune and inflammatory response signaling pathways (36). Furthermore, they found that silencing lncRNAs E52329 and N54010 in mouse microglial cells reduces the phosphorylation of JNK and MKK4 to regulate inflammatory responses (36). JEV-infected swine PK-15 cells (pigs are the amplifying host of JEV) initiate cell defense systems by upregulating lncRNA-SUSAJ1 expression, which reduces JEV replication (37). However, C–C chemokine receptor type 1 protein expression rapidly reduces lncRNA-SUSAJ1 transcript levels by decreasing the recruitment of transcription factor (TF) SP1 at the promoter of SUSAJ1 (37). SUSAJ1 activates the UPR pathway, ER stress, and apoptosis in PK-15 cells (38). However, the function and underlying mechanism of the action of lncRNAs participating in flavivirus-induced human neuronal cell death are unknown.
We examined lncRNA expression profiles in response to JEV infection in human neuronal cell line SH-SY5Y and characterized the role of LINC01518, named JINR1 (JEV-induced non-coding RNA 1) by us, in JEV, DENV, and WNV infection of the SH-SY5Y cells. JINR1 is among the lncRNA transcripts expressed during early cortical neuron differentiation, but it has no homolog in mice; however, its sequence is conserved across primates (39). JINR1 expression is upregulated in human glaucoma tissues, TGF-β1-treated human tenon capsule fibroblast (HTF) cells, and esophageal squamous cell carcinoma (ESCC) (40, 41). JINR1 acts as a competing endogenous RNA for miR-1-3p, and its silencing inhibits tumorigenicity in ESCC cells by suppressing PIK3CA/Akt pathway (40). JINR1 promotes proliferation, migration, and autophagy in TGF-β1-induced HTF cells by interacting with miR-216b-5p (41). However, the function of JINR1 in viral infection or neuronal cell death is unknown.
We show that NF-κB induces JINR1 expression during JEV infection. JINR1 promotes flavivirus replication, flavivirus-induced neuronal apoptosis, and the expression of NF-κB target genes involved in ER stress and neuroinflammation. JINR1 interacts with RNA-binding protein RBM10, promoting flavivirus replication, apoptosis, and NF-κB target gene expression. JINR1 and RBM10-mediated increase in GRP78 expression promotes flavivirus replication. Moreover, JINR1 also interacts with the p65 subunit of NF-κB. JINR1 and RBM10 regulate the expression of flavivirus-induced NF-κB target genes by modulating the binding of the p65 subunit of NF-κB to their promoters. Interestingly, JEV induces RBM10 expression via NF-κB. In line with their role in regulating NF-κB target gene expression, JINR1 and RBM10 promote self and each other’s expression by enhancing p65 binding to their promoters. Our results identify JINR1 as an integral player regulating flavivirus replication and neuronal cell death and suggest new targets for therapeutic intervention during flavivirus infection.
RESULTS
Identification of JEV-regulated lncRNAs in human neuronal cell line SH-SY5Y
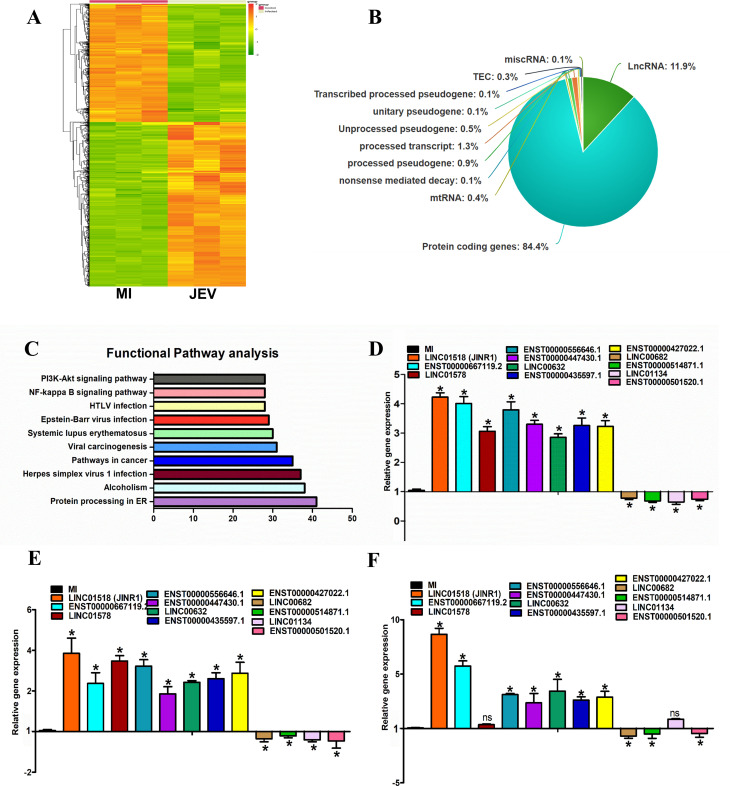
We sought to identify lncRNAs whose expression is responsive to JEV infection in human neuronal cells at a genome-wide scale. To this end, we performed paired-end, high-throughput RNA-sequencing of SH-SY5Y cells infected with or without the GP78 strain of JEV for 48 h in triplicates. JEV-infected SH-SY5Y cells showed significant upregulation of JEV RNA, whereas mock-infected cells (MI) showed no viral RNA expression (Fig. S1A). As expected, JEV infection reduced cell viability and increased caspase-3/7 activity in SH-SY5Y cells (Fig. S1B and C). After RNA sequencing, the QC passed reads were mapped onto the human reference genome (GRCh38.p7) using the HISAT2 aligner. On average, 82.40% of the reads aligned onto the reference genome. We identified differentially expressed genes (DEGs) by comparing mock or JEV-infected SH-SY5Y cells. Transcripts with absolute log base 2 (fold ≥ 0.6) and log base 2 (fold ≤ −0.6) and adjusted P-value ≤ 0.05 were considered significant. The expression profile of DE transcripts upon JEV infection is represented as a heatmap (Fig. 1A). We identified 2,290 (83.72%) DE protein-coding RNAs, 330 (11.9%) non-coding RNAs, and 91 (3.32%) DE pseudogenes during JEV infection of SH-SY5Y cells (Fig. 1B; Table S1).
Fig 1.
Identification of JEV-regulated lncRNAs in human neuronal cell line. (A) Heat map of DEG in SH-SY5Y cells 48 h after mock or JEV Infection (n = 3; DEGs ≥ 1.5-fold, P < 0.05). (B) Pie chart representing the class of DEGs (P < 0.05) identified from the whole transcriptome sequencing in SH-SY5Y cells at 48 hpi. (C) Pathway analysis of DEGs using Kyoto Encyclopedia of Genes and Genomes. The vertical axis shows the pathways in descending order by the deregulated genes, and the horizontal axis represents the fold enrichment. (D) Validation of JEV-regulated significant DE lncRNAs identified from whole transcriptome sequencing using quantitative real-time PCR (qRT-PCR) in SH-SY5Y cells. RNA samples were analyzed by qRT-PCR, and error bars represent the mean ± SEM from three independent experiments. *Significant change compared to MI. Statistical comparisons were made using Student’s t-test. (E) Expression of DE lncRNAs during DENV infection using qRT-PCR in SH-SY5Y cells. RNA samples were analyzed by qRT-PCR, and error bars represent the mean ± SEM from three independent experiments. *Significant change compared to MI. Statistical comparisons were made using Student’s t-test. (F) Expression of DE lncRNAs during WNV infection using qRT-PCR in SH-SY5Y cells. RNA samples were analyzed by qRT-PCR, and error bars represent the mean ± SEM from three independent experiments. *Significant change compared to MI. Statistical comparisons were made using Student’s t-test.
We found that the most significant GOs associated with upregulated transcripts were related to the regulation of cytokine production, response to ER stress, and type I interferon signaling pathway. Similarly, the most enriched upregulated pathways included protein processing in ER, NF-κB signaling pathway, and inflammatory response (Fig. 1C). We next verified the JEV-induced changes in levels of selected lncRNAs in SH-SY5Y cells using quantitative real-time PCR (qRT-PCR). In agreement with our sequencing results, LINC01518/JINR1, ENST00000580528.2, ENST00000447430.1, ENST00000556646.1, ENST00000427022.1, LINC00632, and ENST00000435597.1 get approximately three- to fourfold upregulated upon 48 h post-JEV infection (hpi) in SH-SY5Y cells (Fig. 1D). We also confirmed downregulation of lncRNAs LINC00682, ENST00000514871.1, LINC01134, and ENST00000501520.1 upon JEV infection by qRT-PCR (Fig. 1D). Next, we evaluated the expression of these JEV-regulated lncRNAs during DENV or WNV infection in SH-SY5Y cells using qRT-PCR (Fig. 1E and F). As with JEV infection, DENV infection also resulted in significant differential expression of all candidate lncRNAs in SH-SY5Y cells (Fig. 1E). WNV infection of SH-SY5Y cells also results in substantial changes in the expression of all JEV-regulated lncRNAs except LINC01578 and LINC01134 (Fig. 1F). We also confirmed the changes in the expression of DE mRNAs during JEV infection identified from the RNA sequencing using qRT-PCR (Fig. S1D). Among the JEV-induced lncRNAs, we further set out to characterize the role of lncRNA JINR1 in flavivirus pathogenesis.
NF-κB induces JINR1 expression during JEV infection
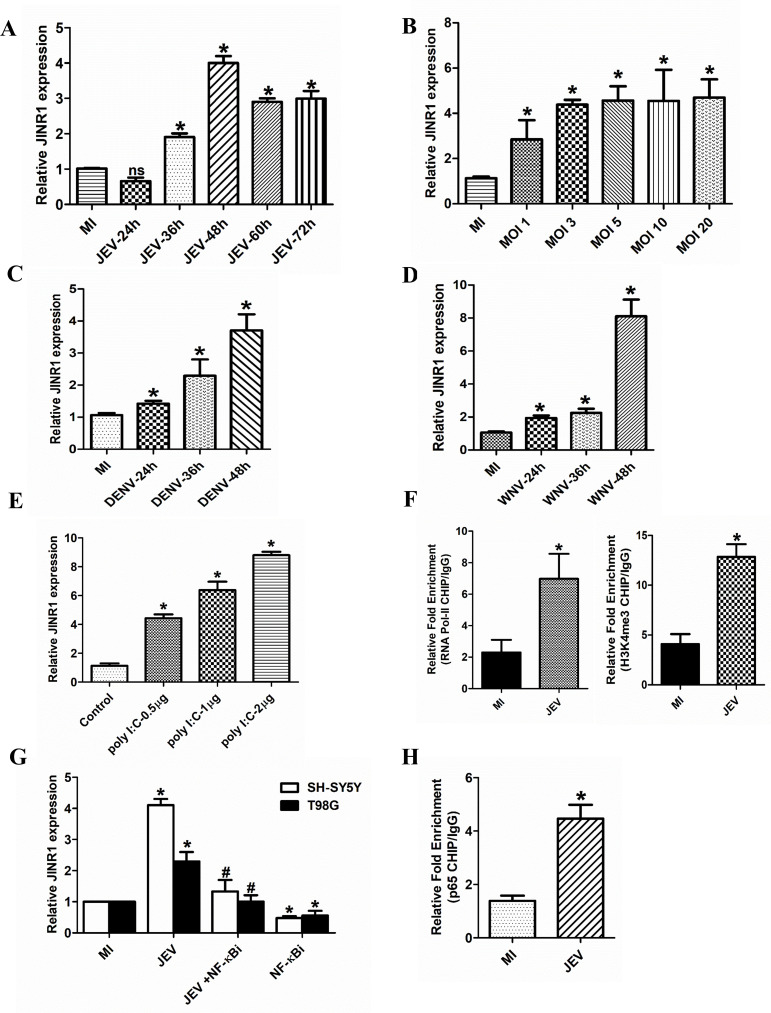
JINR1 is an intergenic intronless transcript of 1,856 bases located on human chromosome 10q11.21(-) between ZNF33b and HNRNPF genes. The time-course analysis identified JINR1 as a delayed transcript with induction of approximately twofold at 36 hpi, approximately fourfold at 48 hpi, and approximately threefold at 60 hpi, reaching a plateau after that (Fig. 2A). JINR1 induction was MOI dependent in response to JEV infection (Fig. 2B). JEV infection also results in a time-dependent increase of JINR1 expression in human astrocytoma cell line T98G (Fig. S1E). Interestingly, we failed to detect measurable expression or induction of JINR1 upon JEV infection using qRT-PCR in the human microglia HMC3 cell line. Time-course analysis of JINR1 expression upon DENV or WNV infection in the SH-SY5Y cell line also showed a time-dependent increase of JINR1 expression (Fig. 2C and D). Next, we evaluated JINR1 expression post-polyinosinic:polycytidylic acid [poly (I:C)] exposure which is a synthetic analog of double-stranded RNA that mimics viral infection and activates toll-like receptors (TLR3), retinoic acid-inducible gene I (RIG-I)-like receptors pathway, NF-κB signaling, and type I IFN secretion (42–44). JINR1 gets upregulated upon poly (I:C) treatment in a dose-dependent manner (Fig. 2E). These results suggest that apart from JEV, DENV, and WNV, JINR1 may get induced as a general host response to other RNA viruses in neuronal cells.
Fig 2.
NF-κB activates JINR1 expression during JEV infection. (A) JINR1 induction upon JEV infection plateaus. SH-SY5Y cells were infected with JEV (MOI 5), and JINR1 levels were measured at the indicated time points using qRT-PCR. (B) JINR1 induction upon JEV infection is MOI dependent. SH-SY5Y cells were infected with different MOI as indicated, and the JINR1 levels were measured by qRT-PCR at 48 hpi. (C) JINR1 induction upon DENV infection is time dependent. SH-SY5Y cells were infected with DENV (MOI 5), and the JINR1 levels were measured at the indicated time points using qRT-PCR. (D) JINR1 induction upon WNV infection is time dependent. SH-SY5Y cells were infected with WNV (MOI 5), and the JINR1 levels were measured at the indicated time points using qRT-PCR. (E) Poly (I:C) induces JINR1 expression. SH-SY5Y cells were transfected with different amounts of poly(I:C), as indicated for 24 h. JINR1 transcript levels were determined by qRT-PCR. (F) JEV promotes RNA Pol II and H3K4me3 recruitment at the promoter of JINR1. Relative enrichment of RNA Pol II and H3K4me3 at the JINR1 promoter region in MI or JEV-infected SH-SY5Y cells determined by ChIP-qRT-PCR 36 hpi. Enrichment values are relative to MI-IgG. (G) NF-κB inhibition abrogates JINR1 induction upon JEV infection. SH-SY5Y and T98G cells were pre-treated with 1 µM of BAY11-7085 for 2 h, followed by JEV infection (MOI 5). JINR1 transcript levels were determined by qRT-PCR at 48 hpi. (H) JEV promotes the p65 subunit of NF-κB recruitment at the promoter of JINR1. Relative enrichment of p65 at the JINR1 promoter in MI or JEV-infected SH-SY5Y cells determined by ChIP-qRT-PCR 36 hpi. Enrichment values are relative to MI-IgG. Error bars represent the mean ± SEM from three independent experiments. Statistical comparisons were made using Student’s t-test. (A–E and G) RNA samples were analyzed by qRT-PCR. (A–D) *Significant change compared to the MI. (E) *Significant change compared to the control. (F and H) ChIP purified DNA was analyzed by qRT-PCR, and error bars represent the mean ± SEM from three independent experiments. *Significant change compared to the MI. Statistical comparisons were made using Student’s t-test. (G) *Significant change compared to the respective MI sample. #Significant change from JEV-infected sample.
To confirm if the JINR1 promoter is transcriptionally active upon JEV infection. We performed ChIP qRT-PCR to assess the recruitment of RNA Pol II and H3K4me3 (a marker for active transcription) at the JINR1 promoter upon JEV infection. The recruitment of RNA Pol II and H3K4me3 to the promoter of the JINR1 was significantly enhanced in cells infected with JEV, suggesting that an increase in its expression upon JEV infection is due to transcription activation (Fig. 2F). To identify the TFs involved in JINR1 expression upon JEV infection, we analyzed the 2,000-bp putative promoter sequence upstream of the JINR1 transcription start site (TSS) for TF binding sites. In silico analysis using Alibaba2 revealed NF-κB binding sites in the JINR1 promoter region ~1.1 kb upstream of the TSS (45). Since poly (I:C) and JEV both activate the NF-κB pathway, we evaluated JINR1 expression upon NF-κB inhibition during JEV infection (46, 47). Treatment of SH-SY5Y and T98G cells with the NF-κB inhibitor BAY11-7085 before JEV infection strongly suppressed JEV-induced JINR1 expression (Fig. 2G). ChIP qRT-PCR assay confirmed increased occupancy of the p65 subunit of NF-κB on the JINR1 promoter upon JEV infection (Fig. 2H). These results suggest that the NF-κB induces the transcription of JINR1 upon JEV infection.
LncRNA-JINR1 inhibition attenuates JEV replication and neuronal apoptosis in SH-SY5Y cells
To test the role of JINR1 in JEV replication, we established JINR1 depletion in SH-SY5Y cells using three different antisense oligonucleotides (ASOs) (Fig. S2A). Depletion of JINR1 with ASO-JINR1-1, ASO-JINR1-2, and ASO-JINR1-3 resulted in ~83%, ~77%, and ~43% reduction of JINR1 expression, respectively, compared to the cells transfected with non-specific ASO (ASO-NS). Hence, we used ASO-JINR1-1 and ASO-JINR1-2 for the functional analysis of JINR1. We also cloned the full-length JINR1 in pcDNA3.1, and transfection of JINR1 in SH-SY5Y resulted in ~8.5-fold increase in JINR1 expression compared to cells transfected with empty vector (Fig. S2B).
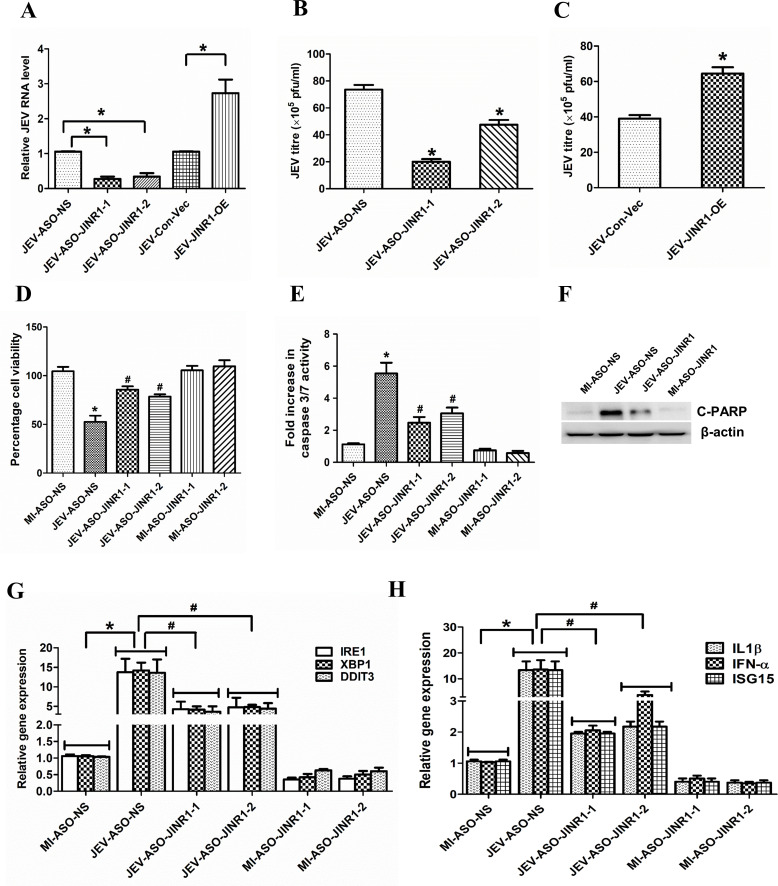
Assessment of JEV replication by measuring intracellular JEV RNA levels upon JINR1 depletion in JEV-infected SH-SY5Y cells indicated ~73% and ~66% reduction in JEV RNA levels in ASO-JINR1-1 and ASO-JINR1-2 transfected cells, respectively, in comparison to JEV-infected ASO-NS-transfected cells (Fig. 3A). Moreover, the overexpression of JINR1 during JEV infection in SH-SY5Y cells also increases intracellular JEV viral RNA levels by approximately twofold compared to the cells transfected with an empty vector (Fig. 3A). We also measured infectious viral particle release in the culture supernatant of JEV-infected SH-SY5Y cells upon JINR1 knockdown using the JEV plaque assay in PS cells. Supernatant from SH-SY5Y cells transfected with ASOs against JINR1 and infected with JEV for 48 h had lower viral titers than ASO-NS-transfected cells infected with JEV (Fig. 3B; Fig. S2C). JINR1 overexpression also increased viral particle release from JEV-infected cells by ~2.15-fold compared to JEV-infected cells transfected with empty vector (Fig. 3C; Fig. S2C). To further confirm the role of JINR1 in inhibiting viral replication, we also checked the viral RNA levels in cell culture supernatants from the JEV-infected SH-SY5Y cells transfected with ASOs against JINR1. In concordance with the above results, significant depletion in mature virion RNA release was observed in cells transfected with ASO-JINR1 compared to ASO-NS as evidenced by respective CT values (viral band detected at 180 bp, 1 kb ladder was used) (Fig. S2D). In addition, JINR1 overexpression significantly increased mature virion RNA levels (Fig. S2D).
Fig 3.
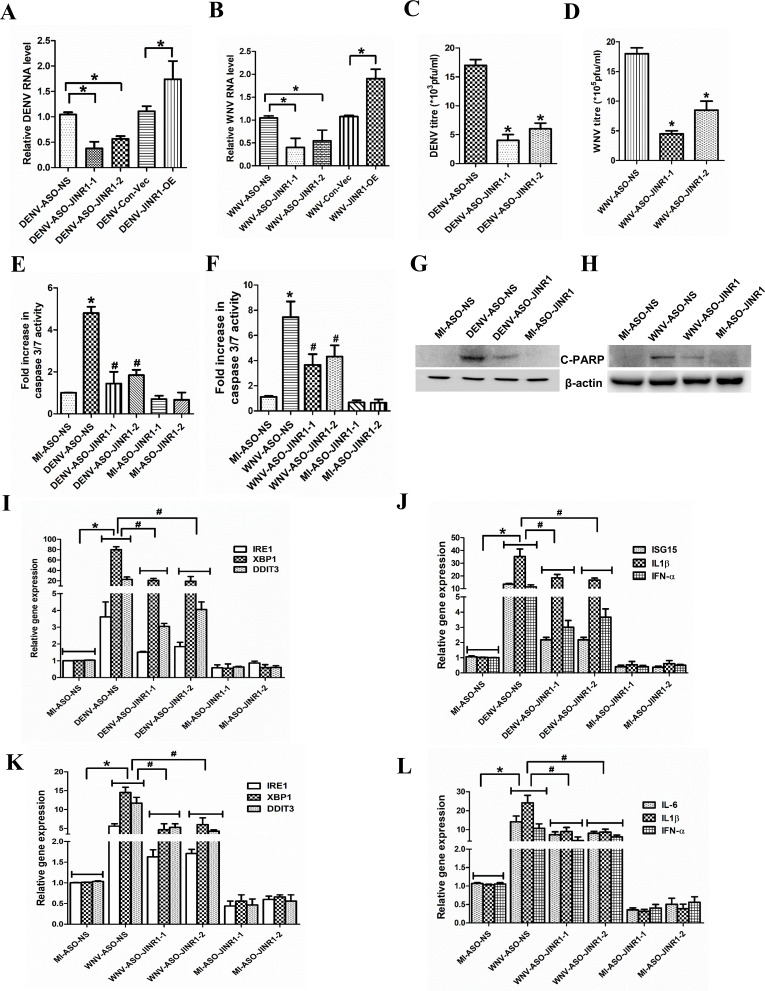
JINR1 inhibition attenuates JEV replication, neuronal apoptosis, and the expression of genes involved in ER stress and neuroinflammation. (A) JINR1 promotes JEV replication in SH-SY5Y cells. Cells were transfected with either ASO-NS, ASO-JINR1-1, ASO-JINR1-2, empty vector (Con-Vec), or pcDNA3.1 with full-length JINR1 (JINR1-OE), and viral replication was determined by quantifying the intracellular levels of JEV RNA using qRT-PCR at 48 hpi. (B) JINR1 silencing reduces JEV titer. Quantification of viral titer upon JINR1 depletion during JEV infection is shown in Fig. S2C (upper panel). (C) JINR1 overexpression increases JEV titer. Quantification of viral titer upon JINR1 overexpression during JEV infection is shown in Fig. S2C (lower panel). (D) JINR1 knockdown attenuates JEV-mediated reduction in cell proliferation. The proliferation of SH-SY5Y cells was assessed by WST1 assay upon JINR1 depletion at 60 hpi. The graph represents the percentage of viable cells. (E) JINR1 promotes apoptosis in SH-SY5Y cells. SH-SY5Y cells were transfected with either ASO-NS, ASO-JINR1-1, or ASO-JINR1-2, and the caspase-3/7 activity was determined at 60 hpi. (F) JINR1 depletion reduces JEV-induced cleaved PARP protein expression. SH-SY5Y cells were transfected with ASO-NS or ASO-JINR1-1, and protein lysates were collected at 60 hpi. Cleaved PARP protein levels were analyzed by western blotting. A representative blot is shown from three independent experiments with similar results. Blots were reprobed for β-actin to establish equal loading. (G) JINR1 depletion attenuates JEV-induced ER stress genes. qRT-PCR analysis of indicated ER stress genes upon JINR1 depletion in SH-SY5Y cells at 48 hpi. (H) JINR1 depletion reduces JEV-induced neuroinflammatory genes. qRT-PCR analysis of indicated neuroinflammatory genes upon JINR1 depletion in SH-SY5Y cells at 48 hpi. Error bars represent the mean ± SEM from three independent experiments. (A, G, and H) RNA samples were analyzed by qRT-PCR. (A and B) *Significant change compared to JEV-ASO-NS/JEV-Con-Vec. (C) *Significant change compared to JEV-Con-Vec. (D, E, G, and H) *Significant change compared to MI-ASO-NS. #Significant change compared to JEV-ASO-NS. Statistical comparison was made using the Student’s t-test.
Since JINR1 inhibition reduced JEV replication, we evaluated the impact of JINR1 depletion on JEV-induced changes in cell proliferation and death in SH-SY5Y cells. JINR1 knockdown using ASOs prevented JEV-mediated reduction in cell proliferation of SH-SY5Y cells (Fig. 3D). Moreover, JINR1 knockdown in JEV-infected SH-SY5Y significantly reduced neuronal apoptosis, as indicated by ~50% reduction in caspase-3/7 activity in JEV-infected cells transfected with ASO against JINR1 compared to JEV-infected cells transfected with ASO-NS (Fig. 3E). Consistent with this, JEV-infected SH-SY5Y cells, upon JINR1 knockdown, had significantly reduced cleaved PARP protein levels compared to JEV-infected ASO-NS-transfected cells (Fig. 3F; Fig. S3A). These results suggest that JINR1 enhances JEV-induced neuronal cell death.
JINR1 regulates JEV-induced increase in the expression of genes involved in ER stress and neuroinflammation
JEV-induced ER stress and neuroinflammation are crucial in neuronal cell death (3, 32, 48–50). Since JINR1 silencing reduced JEV replication and neuronal apoptosis, we evaluated the role of JINR1 in regulating the expression of JEV-regulated genes involved in ER stress (GRP78, XBP1, IRE1, DDIT3, and c-JUN) and neuroinflammation (IFNα, IL-1β, RIG-I, IL-8, IL6, CCL5, and ISG15). Compared with the MI, the ER stress-related transcripts level significantly increased in the JEV-infected group, and JINR1 silencing with both ASOs significantly attenuated JEV-induced ER stress gene expression (Fig. 3G; Fig. S3B). JINR1 overexpression significantly enhanced the basal and JEV-induced ER stress gene expression in SH-SY5Y cells (Fig. S3D and E). Furthermore, ASO-mediated JINR1 depletion also resulted in a marked reduction in the expression of JEV-induced inflammatory genes (Fig. 3H; Fig. S3C). As with ER genes, overexpression of JINR1 enhanced basal and JEV-induced expression of neuroinflammatory transcripts in SH-SY5Y cells compared to their respective controls (Fig. S3F and G). These findings suggest that JINR1 promotes the expression of genes associated with ER stress and neuroinflammation during JEV infection.
JINR1 associates with RBM10 and also regulates its expression to promote viral replication and neuronal apoptosis during JEV infection
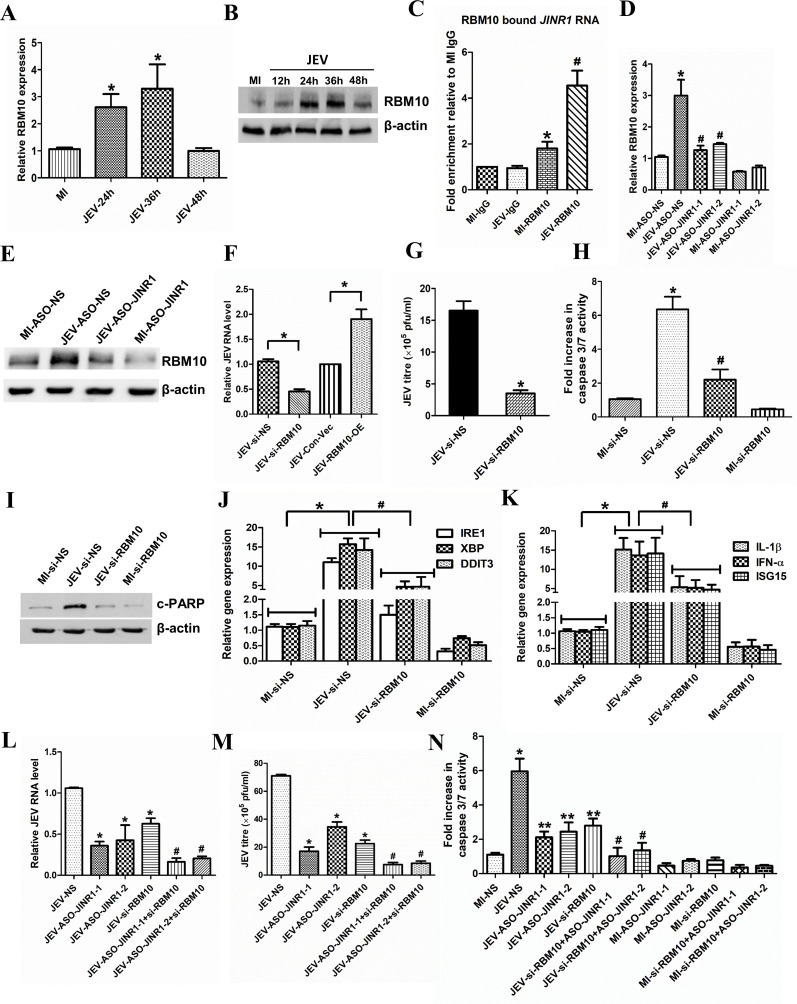
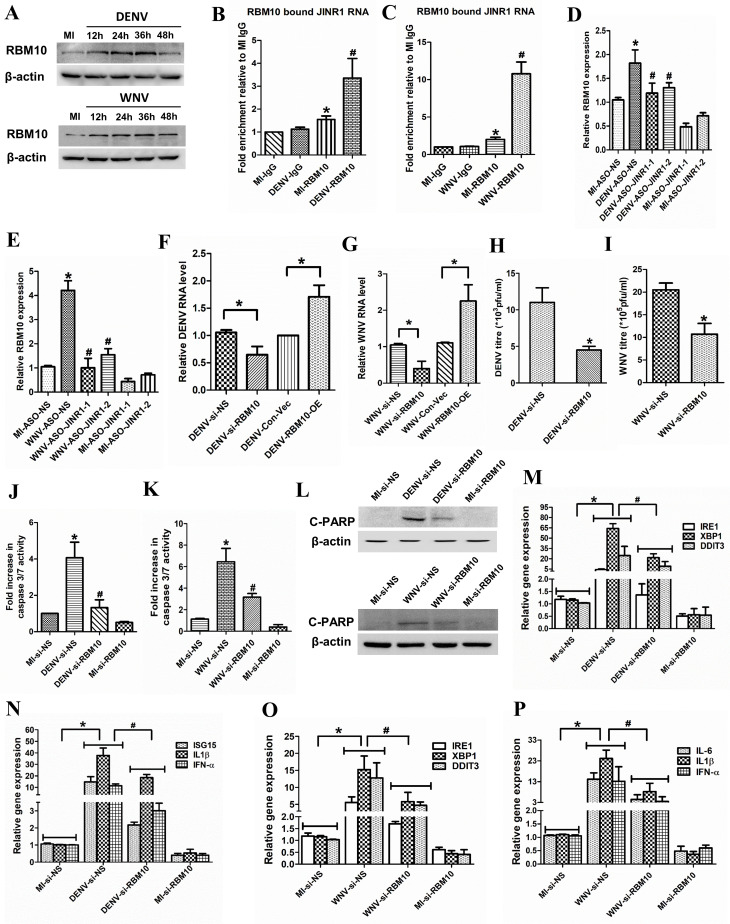
LncRNAs often exert their functions via interaction with protein complexes (51, 52). To identify proteins interacting with JINR1, we used the POSTAR database (53). Using it, we identified several proteins that interact with JINR1, such as CAPRIN1, CSTF2, FIP1L1, LIN28B, MOV10, YTHDF3, RBM10, and YTHDC1 (Table S2). Since RBM10 regulates inflammatory gene expression (54) and is also involved in regulating immune responses during dengue infection (55), we tested whether RBM10 plays any role in JEV pathogenesis (54, 55). First, we checked the mRNA and protein levels of the RBM10 upon JEV infection in SH-SY5Y cells. RBM10 transcript and protein levels increase in a time-dependent manner during JEV infection in SH-SY5Y (Fig. 4A and B; Fig. S4A). RBM10 transcripts levels were increased approximately twofold at 24 and 36 hpi and then returned to basal level at 48 hpi (Fig. 4A). JEV-infected SH-SY5Y cells had ~1.3-, ~2.1-, ~2.56-, and ~1.76-fold higher RBM10 protein levels at 12, 24, 36, and 48 hpi, respectively, in comparison to MI cells (Fig. 4B; Fig. S4A). JINR1 was among the RBM10 interacting transcripts identified using PAR-CLIP in HEK-293T cells (56). Hence, we confirmed the association between RBM10 and lncRNA JINR1 during JEV infection in SH-SY5Y cells using formaldehyde crosslinked RNA immunoprecipitation (RIP) with the RBM10 antibody. We detected RBM10 binding to JINR1 in MI SH-SY5Y cells (1.5-fold enrichment compared to MI IgG IP), and this association was further enhanced by ~4.5-fold in JEV-infected SH-SY5Y cells (Fig. 4C). Next, we evaluated the impact of JINR1 knockdown on RBM10 expression during JEV infection. JINR1 depletion significantly attenuated JEV-induced RBM10 transcript expression (Fig. 4D). JINR1 inhibition also reduces JEV-induced RBM10 protein expression (Fig. 4E; Fig. S4B). To study the functional relevance of RBM10 during JEV infection, we first established the RBM10 knockdown using the previously validated siRNA (56). siRNA mediated knock-down of RBM10 resulted in ~80% decrease in RNA and protein levels of RBM10 in SH-SY5Y cells (Fig. S4C and D). Interestingly, RBM10 depletion results in ~20% increases in cell proliferation of SH-SY5Y cells and T98G cells (Fig. S4E and F).
Fig 4.
JINR1 associates with RBM10 and also regulates its expression to promote viral replication and neuronal apoptosis during JEV infection. (A) JEV infection increases RBM10 mRNA expression. Time course analysis of RBM10 mRNA expression at indicated time points during JEV infection was measured using qRT-PCR. (B) JEV infection increases RBM10 protein levels. SH-SY5Y cells were infected with JEV (MOI 5) for the indicated time, and cell lysates were subjected to Western blot analysis. (C) JINR1 interacts with RBM10. JINR1 RNA levels in RBM10 immunoprecipitated from lysates of formaldehyde-crosslinked JEV or MI SH-SY5Y cells were measured by qRT-PCR analysis, normalized to input, and represented as fold enrichment relative to MI-IgG IP. Values represent mean ± SEM from three independent experiments. *Significant change compared to MI-IgG IP (P < 0.05). #Significant change compared to MI-RBM10 IP (P < 0.05). (D) JINR1 depletion attenuates JEV-induced RBM10. qRT-PCR analysis of RBM10 upon JINR1 depletion in SH-SY5Y cells at 36 hpi. (E) JINR1 promotes RBM10 protein expression during JEV infection. SH-SY5Y cells were transfected with either ASO-NS or ASO-JINR1, cell lysates were collected at 36 hpi, and subjected to western blot analysis. (F) RBM10 promotes JEV replication. SH-SY5Y cells were transfected with si-NS, si-RBM10, empty vector (Con-Vec), or pcDNA3.1 with full-length RBM10 (RBM10-OE), and viral replication was determined by quantifying the intracellular levels of JEV RNA using qRT-PCR at 36 hpi. (G) RBM10 depletion reduces JEV titer. Quantification of viral titer upon RBM10 silencing during JEV infection is shown in Fig. S5A. (H) RBM10 promotes apoptosis in SH-SY5Y cells. SH-SY5Y cells were transfected with si-NS or si-RBM10, and the caspase-3/7 activity was determined at 60 hpi. (I) RBM10 promotes cleaved PARP protein expression in JEV-infected SH-SY5Y cells. SH-SY5Y cells were transfected with si-NS or si-RBM10, and protein lysates were collected at 60 hpi. Cleaved PARP protein levels were analyzed by western blotting. (J) RBM10 depletion attenuates JEV-induced ER stress genes. qRT-PCR analysis of indicated ER stress genes upon RBM10 depletion in SH-SY5Y cells at 36 hpi. (K) RBM10 depletion reduces JEV-induced neuroinflammatory genes. qRT-PCR analysis of indicated neuroinflammatory genes upon RBM10 depletion in SH-SY5Y cells at 36 hpi. (L) Co-inhibition of JINR1 and RBM10 reduces JEV replication. SH-SY5Y cells were transfected either with ASO-JINR1 or si-RBM10 or both, and viral replication was determined by quantifying the intracellular levels of JEV RNA using qRT-PCR at 36 hpi. (M) Co-inhibition of JINR1 and RBM10 significantly reduces JEV titer. Quantification of viral titer upon JINR1 and RBM10 co-inhibition during JEV infection is shown in Fig. S7. (N) Co-inhibition of JINR1 and RBM10 promotes apoptosis in SH-SY5Y cells. SH-SY5Y cells were transfected with either ASO-JINR1 or si-RBM10 or both ASO-JINR1 and si-RBM10. Caspase-3/7 activity was determined at 60 hpi. Error bars represent the mean ± SEM from three independent experiments. Statistical comparison was made using the Student’s t-test. (A, D, F, and J–L) RNA samples were analyzed by qRT-PCR. (A) *Significant change compared to MI. (C) *Significant change compared to MI-IgG. #Significant change compared to MI-RBM10. (D) *Significant change compared to MI-ASO-NS, and #significant change compared to JEV-ASO-NS. (F) *Significant change compared to JEV-si-NS/JEV-con-Vec. (G) *Significant change compared to JEV-si-NS. (H) *Significant change compared to MI-si-NS. #Significant change compared to JEV-si-NS. (B, E, and I) A representative blot is shown from three independent experiments with similar results. Blots were reprobed for β-actin to establish equal loading. (J and K) *Significant change compared to MI-si-NS, and #significant change compared to JEV-si-NS. (L and M) *Significant change compared to JEV-NS, and #significant change compared to JEV-ASO-JINR1/2 or JEV-si-RBM10. (N) *Significant change compared to MI-NS, **Significant change compared to JEV-NS, and #significant change compared to JEV-ASO-JINR1/2 or JEV-si-RBM10.
Next, we evaluated the impact of RBM10 depletion on JEV replication in SH-SY5Y cells. RBM10 knockdown during JEV infection resulted in a ~40% reduction in JEV RNA levels compared to JEV-infected SH-SY5Y cells transfected with si-NS (Fig. 4F). Analysis of JEV titer and RNA in culture supernatant from RBM10-depleted cells also confirmed that RBM10 silencing inhibits JEV replication (Fig. 4G; Fig. S5A and B). In line with these results, we observed that overexpression of RBM10 promotes JEV replication (Fig. 4F; Fig. S5C and D).
Since RBM10 depletion attenuated JEV replication in SH-SY5Y cells, we evaluated the effect of RBM10 knockdown on JEV-mediated neuronal cell death. JEV-infected SH-SY5Y cells transfected with si-RBM10 had ~50% less caspase-3/7 activity than JEV-infected SH-SY5Y cells transfected with si-NS (Fig. 4H). Moreover, RBM10 depletion significantly attenuated the JEV-mediated increase in cleaved PARP levels (Fig. 4I; Fig. S6A). These results indicate that similar to JINR1, RBM10 also promotes JEV replication and JEV-mediated neuronal apoptosis in SH-SY5Y cells.
RBM10 is involved in regulating the expression of genes involved in ER stress and neuroinflammation during JEV infection
We next checked if RBM10 is involved in regulating JINR1-induced ER stress and inflammatory gene expression upon JEV infection in SH-SY5Y cells. RBM10 depletion in JEV-infected SH-SY5Y cells significantly reduces ER stress and inflammatory gene expression compared to JEV-infected SH-SY5Y cells transfected with si-NS (Fig. 4J and K; Fig. S6B and C). Overexpression of RBM10 induces not only the basal level of ER stress and neuroinflammatory transcripts but also significantly enhances JEV-triggered induction of these transcripts (Fig. S6D through G). These results indicate that, like JINR1, RBM10 promotes JEV replication, cell death, and transcription of genes involved in ER stress and neuroinflammation.
Next, we evaluated the impact of the double knockdown of JINR1 and RBM10 on JEV replication. Simultaneous inhibition of JINR1 and RBM10 during JEV infection in SH-SY5Y cells resulted in a significantly higher reduction in JEV RNA and viral titer levels compared to cells transfected only with ASO-JINR1 or si-RBM10 (Fig. 4L and M; Fig. S7). Co-inhibition of JINR1 and RBM10 significantly reduces JEV-induced caspase-3/7 activity compared to individual inhibition of either JINR1 or RBM10 (Fig. 4N). In line with the above results, co-inhibition of JINR1 and RBM10 significantly reduced ER stress and neuroinflammatory gene expression more than their individual inhibition (Fig. S8 and S9).
JINR1 depletion attenuates DENV and WNV replication in SH-SY5Y cells
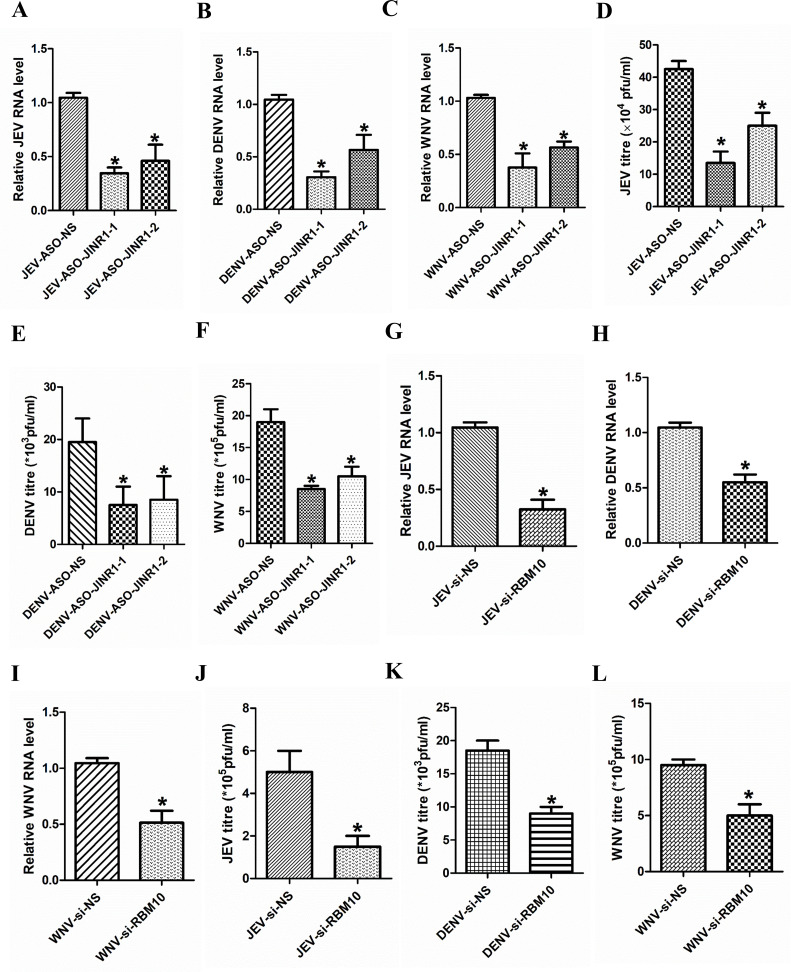
Next, we investigated the consequences of JINR1 depletion on DENV and WNV replication in SH-SY5Y cells. JINR1 knockdown during DENV or WNV infection significantly reduced viral RNA and titer levels (Fig. 5A and B). Moreover, overexpression of JINR1 increases intracellular DENV and WNV viral RNA levels by ~1.7- and ~1.9-fold, respectively, compared to infected cells transfected with an empty vector (Fig. 5A and B). Plaque assay from the supernatant of SH-SY5Y cells transfected with ASOs against JINR1 and infected with DENV or WNV-infected for 48 h had lower viral titers than DENV- or WNV-infected SH-SY5Y cells transfected with ASO-NS (Fig. 5C and D; Fig. S10A and D). JINR1 overexpressing cells also released significantly more DENV or WNV viral particles than cells transfected with an empty vector (Fig. S10B, C, E, and F).
Fig 5.
JINR1 inhibition attenuates DENV and WNV replication, neuronal apoptosis, and the expression of genes involved in ER stress and neuroinflammation. (A) JINR1 promotes DENV replication in SH-SY5Y cells transfected with either ASO-NS, ASO-JINR1-1, ASO-JINR1-2, empty vector, or JINR1-OE, and viral replication was determined by quantifying the intracellular levels of DENV RNA using qRT-PCR at 48 hpi. (B) JINR1 promotes WNV replication in SH-SY5Y cells transfected with either ASO-NS, ASO-JINR1-1, ASO-JINR1-2, empty vector, or JINR1-OE, and viral replication was determined by quantifying the intracellular levels of WNV RNA using qRT-PCR at 48 hpi. (C) JINR1 silencing reduces DENV titer. Quantification of viral titer upon JINR1 depletion during DENV infection is shown in Fig. S10A. (D) JINR1 silencing reduces WNV titer. Quantification of viral titer upon JINR1 depletion during WNV infection is shown in Fig. S10D. (E) JINR1 promotes apoptosis in DENV-infected SH-SY5Y cells. SH-SY5Y cells were transfected with either ASO-NS, ASO-JINR1-1, or ASO-JINR1-2, and the caspase-3/7 activity was determined at 60 hpi. (F) JINR1 promotes apoptosis in WNV-infected SH-SY5Y cells. SH-SY5Y cells were transfected with either ASO-NS, ASO-JINR1-1, or ASO-JINR1-2, and the caspase-3/7 activity was determined at 60 hpi. (G) JINR1 depletion reduces DENV-induced cleaved PARP protein expression. SH-SY5Y cells were transfected with ASO-NS or ASO-JINR1-1, and protein lysates were collected at 60 hpi. Cleaved PARP protein levels were analyzed by western blotting. A representative blot is shown from three independent experiments with similar results. Blots were reprobed for β-actin to establish equal loading. (H) JINR1 depletion reduces WNV-induced cleaved PARP protein expression. SH-SY5Y cells were transfected with ASO-NS or ASO-JINR1-1, and protein lysates were collected at 60 hpi. Cleaved PARP protein levels were analyzed by western blotting. A representative blot is shown from three independent experiments with similar results. Blots were reprobed for β-actin to establish equal loading. (I) JINR1 depletion attenuates DENV-induced ER stress genes. qRT-PCR analysis of indicated ER stress genes upon JINR1 depletion in SH-SY5Y cells at 48 hpi. (J) JINR1 depletion reduces DENV-induced neuroinflammatory genes. qRT-PCR analysis of indicated neuroinflammatory genes upon JINR1 depletion in SH-SY5Y cells at 48 hpi. (K) JINR1 depletion attenuates WNV-induced ER stress genes. qRT-PCR analysis of indicated ER stress genes upon JINR1 depletion in SH-SY5Y cells at 48 hpi. (L) JINR1 depletion reduces WNV-induced neuroinflammatory genes. qRT-PCR analysis of indicated neuroinflammatory genes upon JINR1 depletion in SH-SY5Y cells at 48 hpi. Error bars represent the mean ± SEM from three independent experiments. (A, B, I-L) RNA samples were analyzed by qRT-PCR. (A and B) *Significant change compared to DENV-ASO-NS/WNV-ASO-NS/ DENV-Con-Vec/WNV-Con-Vec. (C and D) *Significant change compared to DENV-ASO-NS/WNV-ASO-NS. (E, F, and I–L) *Significant change compared to MI-ASO-NS. #Significant change compared to DENV-ASO-NS/WNV-ASO-NS. Statistical comparison was made using the Student’s t-test.
JINR1 regulates DENV/WNV-mediated neuronal apoptosis and expression of genes involved in ER stress and neuroinflammation
DENV/WNV infection of the CNS also results in neuronal cell death (11, 14, 57–61). Hence, we evaluated the impact of JINR1 depletion on DENV/WNV-induced neuronal cell death. JINR1 knockdown reduces DENV/WNV-mediated neuronal apoptosis, as indicated by a reduction in caspase-3/7 activity (Fig. 5E and F) and cleaved PARP protein levels in virus-infected cells transfected with ASO against JINR1 compared with virus-infected cells transfected with ASO-NS (Fig. 5G and H; Fig. S11A and B).
Next, we evaluated the role of JINR1 in regulating infection-mediated changes in gene expression upon JINR1 knockdown during DENV or WNV infection. JINR1 knockdown reduces the virus infection-mediated expression of genes involved in ER stress and neuroinflammation (Fig. 5I through L; Fig. S12A through D). Moreover, JINR1 overexpression during DENV or WNV infection markedly enhanced ER stress and neuroinflammatory gene expression (Fig. S13 and 14). These findings suggest that JINR1 promotes ER stress and neuroinflammation during flavivirus infection.
RBM10 promotes virus replication, apoptosis, and gene expression during DENV and WNV infection
Next, we tested the role of RBM10 in DENV and WNV infection in SH-SY5Y cells. First, we performed a time course of the RBM10 transcript and protein levels during DENV or WNV infection in SH-SY5Y cells (Fig. S15A and B; Fig. 6A). DENV-induced RBM10 transcripts peak at 24 hpi and then return to basal level at 48 hpi (Fig. S15A). WNV infection also results in peak expression of RBM10 mRNA at 24 hpi and then taper to basal levels at 48 hpi (Fig. S15B). DENV-infected SH-SY5Y cells have ~1.5-, ~2-, ~2.5-, and ~1.5-fold higher RBM10 protein levels at 12, 24, 36, and 48 hpi, respectively, compared to MI cells (Fig. 6A; Fig. S15C). WNV-infected SH-SY5Y cells have ~2.4-fold at 12 hpi, ~2.9-fold at 24 hpi, ~3.56-fold at 36 hpi, and ~2.06-fold at 48 hpi higher RBM10 protein levels compared to MI cells (Fig. 6A; Fig. S15D). Moreover, RIP analysis confirmed RBM10 binding to JINR1 during DENV or WNV infection in SH-SY5Y cells (Fig. 6B and C). Since JINR1 depletion negatively regulates JEV-induced RBM10 expression (Fig. 4D and E), we evaluated the impact of JINR1 knockdown on RBM10 expression during DENV/WNV infection. JINR1 depletion attenuated DENV/WNV infection-mediated increase in RBM10 mRNA levels in SH-SY5Y cells (Fig. 6D and E).
Fig 6.
JINR1 associates with RBM10 and also regulates its expression to regulate DENV and WNV viral replication, cell death, and gene expression. (A) DENV or WNV infection increases RBM10 protein levels. SH-SY5Y cells were infected with DENV or WNV (MOI 5) for the indicated time, and cell lysates were subjected to Western blot analysis. (B) JINR1 interacts with RBM10 during DENV infection. JINR1 RNA levels in RBM10 immunoprecipitated from lysates of formaldehyde-crosslinked DENV or MI SH-SY5Y cells were measured by qRT-PCR analysis, normalized to input, and represented as fold enrichment relative to MI-IgG IP. Values represent mean ± SEM from three independent experiments. *Significant change compared to MI-IgG IP (P < 0.05). #Significant change compared to MI-RBM10 IP (P < 0.05). (C) JINR1 interacts with RBM10 during WNV infection. JINR1 RNA levels in RBM10 immunoprecipitated from the lysates of formaldehyde-crosslinked WNV or MI SH-SY5Y cells were measured by qRT-PCR analysis, normalized to input, and represented as fold enrichment relative to MI-IgG IP. Values represent mean ± SEM from three independent experiments. *Significant change compared to MI-IgG IP (P < 0.05). #Significant change compared to MI-RBM10 IP (P < 0.05). (D) JINR1 depletion attenuates DENV-induced RBM10. qRT-PCR analysis of RBM10 upon JINR1 depletion in SH-SY5Y cells at 36 hpi. (E) JINR1 depletion attenuates WNV-induced RBM10. qRT-PCR analysis of RBM10 upon JINR1 depletion in SH-SY5Y cells at 36 hpi. (F) RBM10 promotes DENV replication. SH-SY5Y cells were transfected with si-NS, si-RBM10, empty vector, or RBM10-OE, and viral replication was determined by quantifying the intracellular levels of DENV RNA using qRT-PCR at 36 hpi. (G) RBM10 promotes WNV replication. SH-SY5Y cells were transfected with si-NS, si-RBM10, empty vector, or RBM10-OE, and viral replication was determined by quantifying the intracellular levels of WNV RNA using qRT-PCR at 36 hpi. (H) RBM10 silencing reduces DENV titer. Quantification of viral titer upon RBM10 depletion during DENV infection is shown in Fig. S16A. (I) RBM10 silencing reduces WNV titer. Quantification of viral titer upon RBM10 depletion during WNV infection is shown in Fig. S16C. (J) RBM10 promotes apoptosis in DENV-infected SH-SY5Y cells. SH-SY5Y cells were transfected with si-NS or si-RBM10, and the caspase-3/7 activity was determined at 60 hpi. (K) RBM10 promotes apoptosis in WNV-infected SH-SY5Y cells. SH-SY5Y cells were transfected with si-NS or si-RBM10, and the caspase-3/7 activity was determined at 60 hpi. (L) RBM10 promotes cleaved PARP protein expression in DENV or WNV-infected SH-SY5Y cells. SH-SY5Y cells were transfected with si-NS or si-RBM10, and protein lysates were collected at 60 hpi. Cleaved PARP protein levels were analyzed by western blotting. (M) RBM10 depletion attenuates DENV-induced ER stress genes. qRT-PCR analysis of indicated ER stress genes upon RBM10 depletion in SH-SY5Y cells at 36 hpi. (N) RBM10 depletion reduces DENV-induced neuroinflammatory genes. qRT-PCR analysis of indicated neuroinflammatory genes upon RBM10 depletion in SH-SY5Y cells at 36 hpi. (O) RBM10 depletion attenuates WNV-induced ER stress genes. qRT-PCR analysis of indicated ER stress genes upon RBM10 depletion in SH-SY5Y cells at 36 hpi. (P) RBM10 depletion reduces WNV-induced neuroinflammatory genes. qRT-PCR analysis of indicated neuroinflammatory genes upon RBM10 depletion in SH-SY5Y cells at 36 hpi. Error bars represent the mean ± SEM from three independent experiments. Statistical comparison was made using the Student’s t-test. (B, D–G, and M–P) RNA samples were analyzed by qRT-PCR. (B and C) *Significant change compared to MI-IgG. #Significant change compared to MI-RBM10. (D and E) *Significant change compared to MI-ASO-NS. #Significant change compared to DENV-ASO-NS/WNV-ASO-NS. (F and G) *Significant change compared to DENV-si-NS/WNV-si-NS/DENV-Con-Vec/WNV-Con-Vec. (H and I) *Significant change compared to DENV-si-NS/WNV-si-NS. (J and K) *Significant change compared to MI-si-NS. #Significant change compared to DENV-si-NS/WNV-si-NS. (A and L) A representative blot is shown from three independent experiments with similar results. Blots were reprobed for β-actin to establish equal loading. (M–P) *Significant change compared to MI-si-NS. #Significant change compared to DENV-si-NS/WNV-si-NS.
RBM10 depletion in SH-SY5Y cells during DENV infection results in ~62% reduction in DENV RNA levels in si-RBM10 transfected cells compared to si-NS transfected cells (Fig. 6F). RBM10 inhibition during WNV infection results in ~67% reduction in WNV RNA levels compared to si-NS transfected SH-SY5Y cells (Fig. 6G). Plaque assays also confirmed that RBM10 silencing inhibits DENV and WNV replication (Fig. 6H and I; Fig. S16A and B). RBM10 overexpression analysis in SH-SY5Y cells during DENV or WNV infection also indicated the positive role of RBM10 in promoting DENV and WNV replication (Fig. 6F and G; Fig. S16C through F). There was a significant decrease in caspase-3/7 activity and cleaved PARP protein levels during DENV or WNV infection in si-RBM10 transfected cells compared to si-NS transfected cells (Fig. 6J through L; Fig. S17). As in the case of JEV infection, RBM10 silencing during DENV or WNV infection significantly reduced the expression of ER stress and neuroinflammatory genes (Fig. 6M through P; Fig. S18), and the overexpression of RBM10 enhanced the expression of these genes during DENV (Fig. S19) or WNV (Fig. S20) infection in SH-SY5Y cells. Our results suggest that RBM10 and JINR1 promote flavivirus replication, cell death, and expression of genes involved in ER stress and neuroinflammation.
JINR1 and RBM10 promote flavivirus replication in human astrocytoma cells
Apart from SH-SY5Y cells, JINR1 expression is also induced in human astrocytoma cells (T98G) upon JEV infection (Fig. S1E). Hence, we evaluated the impact of JINR1 and RBM10 depletion on flavivirus replication in T98G cells. Assessment of flavivirus replication by measuring intracellular flavivirus RNA levels during JINR1 depletion in flavivirus-infected T98G cells indicated a significant reduction in flavivirus RNA levels in ASO-JINR1-1/ASO-JINR1-2 transfected cells in comparison to flavivirus-infected ASO-NS transfected T98G cells (Fig. 7A through C). Plaque assays also confirmed that JINR1 silencing inhibits JEV, DENV, and WNV replication (Fig. 7D through F; Fig. S21). RBM10 depletion during flavivirus infection of T98G cells results in significantly lower flavivirus RNA levels in comparison to flavivirus infected si-NS-transfected cells (Fig. 7G through I). Plaque assay from the supernatant of T98G cells transfected with siRNA against RBM10 and infected with flavivirus for 36 h had lower viral titers than flavivirus-infected T98G cells transfected with si-NS (Fig. 7J through L; Fig. S22). These results suggest that JINR1 and RBM10 promote flavivirus replication in cells of astrocytic lineage.
Fig 7.
JINR1 and RBM10 promote flavivirus replication in human astrocytoma cells. (A) JINR1 promotes JEV replication in T98G cells. Cells were transfected with either ASO-NS, ASO-JINR1-1, or ASO-JINR1-2, and viral replication was determined by quantifying the intracellular levels of JEV RNA at 48 hpi. (B) JINR1 promotes DENV replication in T98G cells. Cells were transfected with either ASO-NS, ASO-JINR1-1, or ASO-JINR1-2, and viral replication was determined by quantifying the intracellular levels of DENV RNA at 48 hpi. (C) JINR1 promotes WNV replication in T98G cells. Cells were transfected with either ASO-NS, ASO-JINR1-1, or ASO-JINR1-2, and viral replication was determined by quantifying the intracellular levels of WNV RNA at 48 hpi. (D) JINR1 silencing reduces JEV titer. Quantification of viral titer upon JINR1 depletion during JEV infection in T98G cells is shown in Fig. S21A. (E) JINR1 silencing reduces DENV titer. Quantification of viral titer upon JINR1 depletion during DENV infection in T98G cells is shown in Fig. S21B. (F) JINR1 silencing reduces WNV titer. Quantification of viral titer upon JINR1 depletion during WNV infection in T98G cells is shown in Fig. S21C. (G) RBM10 promotes JEV replication in T98G cells. Cells were transfected with either si-NS or si-RBM10, and viral replication was determined by quantifying the intracellular levels of JEV RNA at 36 hpi. (H) RBM10 promotes DENV replication. T98G cells were transfected with si-NS or si-RBM10, and viral replication was determined by quantifying the intracellular levels of DENV RNA at 36 hpi. (I) RBM10 promotes WNV replication. T98G cells were transfected with either si-NS or si-RBM10, and viral replication was determined by quantifying the intracellular levels of WNV RNA at 36 hpi. (J) RBM10 silencing reduces JEV titer. Quantification of viral titer upon RBM10 depletion during JEV infection in T98G cells is shown in Fig. S22A. (K) RBM10 silencing reduces DENV titer. Quantification of viral titer upon RBM10 depletion during DENV infection in T98G cells is shown in Fig. S22B. (L) RBM10 silencing reduces WNV titer. Quantification of viral titer upon RBM10 depletion during WNV infection in T98G cells is shown in Fig. S22C. Error bars represent the mean ± SEM from three independent experiments. Statistical comparison was made using the Student’s t-test. (A–C and G–I) RNA samples were analyzed by qRT-PCR. (A and D) *Significant change compared to JEV-ASO-NS. (B and E) *Significant change compared to DENV-ASO-NS. (C and F) *Significant change compared to WNV-ASO-NS. (G and J) *Significant change compared to JEV-si-NS. (H and K) *Significant change compared to DENV-si-NS. (I and L) *Significant change compared to WNV-si-NS.
Increase in flavivirus replication mediated by JINR1 and RBM10 is regulated by GRP78 in SH-SY5Y cells
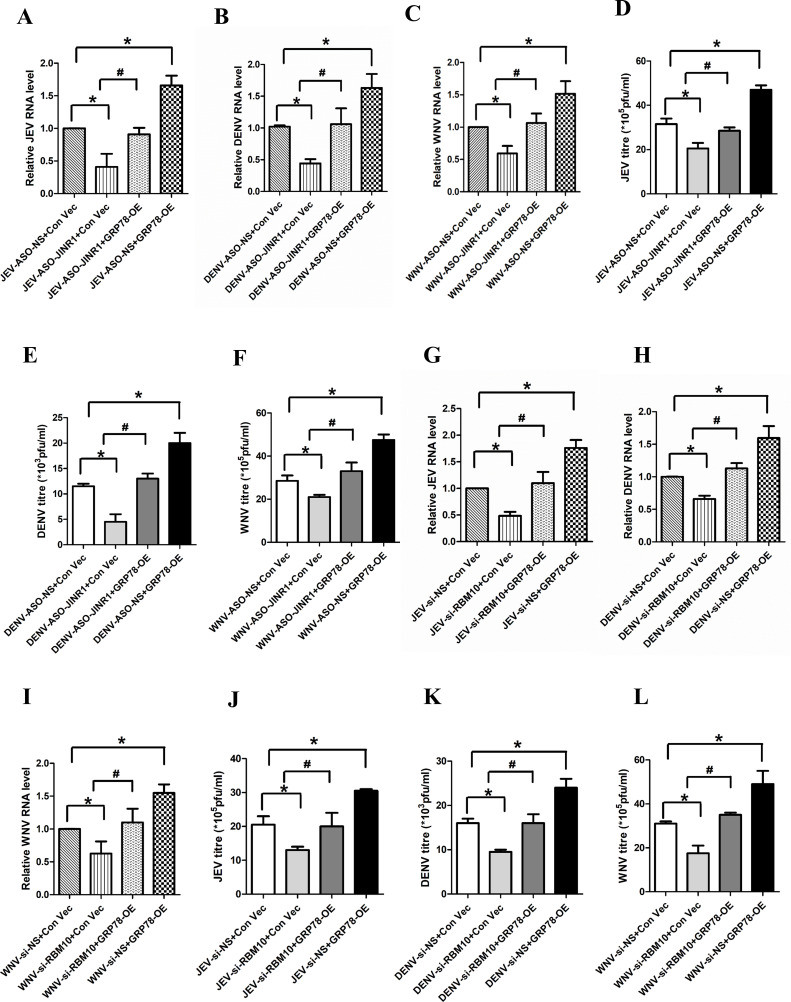
Next, we wanted to explore the mechanism behind JINR1/RBM10-mediated increase in flavivirus replication. Since JINR1 and RBM10 promote GRP78 mRNA expression during flavivirus infection (Fig. S3B, S6B, S12A, C, S18A, and C) and because GRP78 is involved in promoting flavivirus replication and entry in host cells (62–64), we hypothesized that JINR1/RBM10 mediated increase in GRP78 expression during flavivirus infection promotes viral replication in SH-SY5Y cells. To test this, we evaluated the impact of GRP78 overexpression on flavivirus replication in SH-SY5Y cells during JINR1/RBM10 depletion. Interestingly, GRP78 overexpression completely prevents the decrease in intracellular flavivirus RNA levels due to JINR1 depletion (Fig. 8A through C). Plaque assay also confirmed that GRP78 overexpression significantly rescues the decrease in infectious viral particle release in the culture supernatant of flavivirus-infected SH-SY5Y cells due to JINR1 knockdown (Fig. 8D through F; Fig. S23). GRP78 overexpression also prevents the decrease in flavivirus replication due to RBM10 knockdown (Fig. 8G through L; Fig. S24). These results suggest that JINR1/RBM10-mediated increase in GRP78 expression promotes flavivirus replication.
Fig 8.
GRP78 overexpression prevents the decrease in flavivirus infection due to JINR1/RBM10 knockdown. (A) GRP78 overexpression prevents the reduction in JEV RNA levels due to JINR1 depletion in SH-SY5Y cells. JEV-infected SH-SY5Y cells were co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of JEV RNA at 48 hpi. (B) GRP78 overexpression prevents the reduction in DENV RNA levels due to JINR1 depletion in SH-SY5Y cells. DENV-infected SH-SY5Y cells were co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of DENV RNA at 48 hpi. (C) GRP78 overexpression prevents the reduction in WNV RNA levels due to JINR1 depletion in SH-SY5Y cells. WNV-infected SH-SY5Y cells were co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of WNV RNA at 48 hpi. (D) GRP78 overexpression prevents the reduction in JEV titer due to JINR1 depletion in SH-SY5Y cells. Quantification of viral titer from JEV-infected SH-SY5Y cells co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE is shown in Fig. S23A. (E) GRP78 overexpression prevents the reduction in DENV titer due to JINR1 depletion in SH-SY5Y cells. Quantification of viral titer from DENV-infected SH-SY5Y cells co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE is shown in Fig. S23B. (F) GRP78 overexpression prevents the reduction in WNV titer due to JINR1 depletion in SH-SY5Y cells. Quantification of viral titer from WNV-infected SH-SY5Y cells co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE is shown in Fig. S23C. (G) GRP78 overexpression prevents the reduction in JEV RNA levels due to RBM10 depletion in SH-SY5Y cells. JEV-infected SH-SY5Y cells were co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of JEV RNA at 36 hpi. (H) GRP78 overexpression prevents the reduction in DENV RNA levels due to RBM10 depletion in SH-SY5Y cells. DENV-infected SH-SY5Y cells were co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of DENV RNA at 36 hpi. (I) GRP78 overexpression prevents reduction in WNV RNA levels due to RBM10 depletion in SH-SY5Y cells. WNV-infected SH-SY5Y cells were co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of WNV RNA at 36 hpi. (J) GRP78 overexpression prevents the reduction in JEV titer due to RBM10 depletion in SH-SY5Y cells. Quantification of viral titer from JEV-infected SH-SY5Y cells co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE is shown in Fig. S24A. (K) GRP78 overexpression prevents the reduction in DENV titer due to RBM10 depletion in SH-SY5Y cells. Quantification of viral titer from DENV-infected SH-SY5Y cells co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE is shown in Fig. S24B. (L) GRP78 overexpression prevents the reduction in WNV titer due to RBM10 depletion in SH-SY5Y cells. Quantification of viral titer from WNV-infected SH-SY5Y cells co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE is shown in Fig. S24C. Error bars represent the mean ± SEM from three independent experiments. Statistical comparison was made using the Student’s t-test. (A–C and G–I) RNA samples were analyzed by qRT-PCR. (A and D) *Significant change compared to JEV-ASO-NS + Con-Vec. #Significant change compared to JEV-ASO-JINR1 + Con-Vec. (B and E) *Significant change compared to DENV-ASO-NS + Con-Vec. #Significant change compared to DENV-ASO-JINR1 + Con-Vec. (C and F) *Significant change compared to WNV-ASO-NS + Con-Vec. #Significant change compared to WNV-ASO-JINR1 + Con-Vec. (G and J) *Significant change compared to JEV-si-NS + Con-Vec. #Significant change compared to JEV-si-RBM10 + Con-Vec. (H and K) *Significant change compared to DENV-si-NS + Con-Vec. #Significant change compared to DENV-si-RBM10 + Con-Vec. (I and L) *Significant change compared to WNV-si-NS + Con-Vec. #Significant change compared to WNV-si-RBM10 + Con-Vec.
JINR1 and RBM10 regulate the transcription of ER stress and inflammatory genes during JEV infection
Since lncRNA and protein complexes are involved in the regulation of gene transcription (65, 66), we checked if JINR1 and RBM10 regulate ER stress and inflammatory gene expression at the transcription level. For this, we carried out ChIP-qRT-PCR analysis to assess the recruitment of RNA Pol II and H3K4me3 at target gene promoter upon RBM10 and JINR1 depletion during JEV infection in SH-SY5Y cells. JEV infection significantly enhanced the recruitment of RNA Pol II and H3K4me3 at the promoters of the ER stress and inflammatory genes in SH-SY5Y cells (Fig. S25 to S27). RBM10 knockdown in JEV-infected cells reduced RNA Pol II and H3K4me3 binding at the promoters of ER stress (Fig. S25) and neuroinflammatory genes (Fig. S26) compared to JEV-infected cells transfected with si-NS. As with RBM10 depletion, JINR1 silencing also reduced the recruitment of RNA Pol II and H3K4me3 at target gene promoters during JEV infection compared to JEV-ASO-NS transfected cells (Fig. S27). Our results suggest that JINR1 and RBM10 regulate the transcription of ER stress and inflammatory genes during JEV infection.
JINR1 interacts with the p65 subunit of NF-κB and regulates its recruitment to the promoter of genes involved in ER stress and neuroinflammation
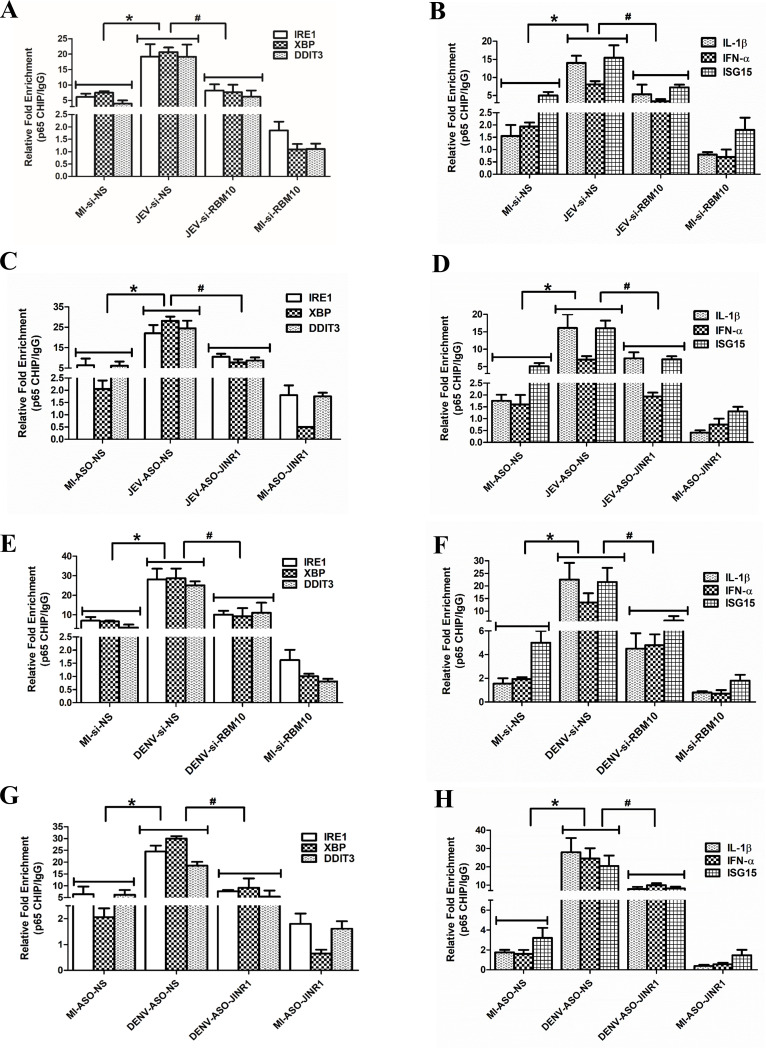
We further explored the mechanism of gene transcription by RBM10 and JINR1 during flavivirus infection. Since RBM10 regulates gene expression by regulating NF-κB transcriptional activity (54), we hypothesized that NF-κB might regulate JINR1 and RBM10-mediated changes in gene expression during flavivirus infection. Consistent with this, in silico analysis using Alibaba2 (45) revealed multiple NF-κB binding sites in the promoters of genes regulated by JINR1 and RBM10 during JEV infection. Hence, we evaluated the role of RBM10 and JINR1 in regulating the recruitment of the p65 subunit of NF-κB to the promoter of genes involved in ER stress and neuroinflammation during JEV infection. ChIP qRT-PCR analysis revealed enhanced binding of the p65 subunit of NF-κB on the promoter of JEV-induced ER and inflammatory genes in SH-SY5Y cells (Fig. 9A through 9D; Fig. S28 and S29). RBM10 depletion during JEV infection significantly reduced the binding of p65 on the promoter of these genes (Fig. 9A and B; Fig. S28A and B).
Fig 9.
RBM10 and JINR1 regulate the recruitment of the p65 subunit of NF-κB to the promoters of ER stress and neuroinflammation genes. (A) RBM10 promotes the p65 recruitment at the promoter of ER stress genes. Relative enrichment of p65 at the promoter of indicated genes in MI or JEV-infected SH-SY5Y cells transfected with si-NS or si-RBM10 was determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-si-NS IgG IP. (B) RBM10 promotes the p65 recruitment at the promoter of inflammatory genes. Relative enrichment of p65 at the promoter of indicated genes in MI or JEV-infected SH-SY5Y cells transfected with si-NS or si-RBM10 was determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-si-NS IgG IP. (C) JINR1 promotes the p65 recruitment at the promoter of ER stress genes during JEV infection. Relative enrichment of p65 at the promoter of indicated ER stress genes in MI or JEV-infected SH-SY5Y cells transfected with ASO-NS or ASO-JINR1 was determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-ASO-NS-IgG IP. (D) JINR1 promotes the p65 recruitment at the promoter of inflammatory genes during JEV infection. Relative enrichment of p65 at the promoter of indicated neuroinflammatory genes in MI or JEV-infected SH-SY5Y cells transfected with ASO-NS or ASO-JINR1 was determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-ASO-NS-IgG IP. (E) RBM10 promotes the p65 recruitment at the promoter of ER stress genes during DENV infection. Relative enrichment of p65 at the promoter of indicated genes in MI or DENV-infected SH-SY5Y cells transfected with si-NS or si-RBM10 was determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-si-NS IgG IP. (F) RBM10 promotes the p65 recruitment at the promoter of inflammatory genes during DENV infection. Relative enrichment of p65 at the promoter of indicated genes in MI or DENV-infected SH-SY5Y cells transfected with si-NS or si-RBM10 was determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-si-NS IgG IP. (G) JINR1 promotes the p65 recruitment at the promoter of ER stress genes during DENV infection. Relative enrichment of p65 at the promoter of indicated genes in MI or DENV-infected SH-SY5Y cells transfected with ASO-NS or ASO-JINR1 was determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-ASO-NS-IgG IP. (H) JINR1 promotes the p65 recruitment at the promoter of inflammatory genes during DENV infection. Relative enrichment of p65 at the promoter of indicated genes in MI or DENV-infected SH-SY5Y cells transfected with ASO-NS or ASO-JINR1 was determined by ChIP-qRT-PCR 36 hpi. Enrichment values are relative to MI-ASO-NS-IgG IP. Error bars represent the mean ± SEM from three independent experiments. Statistical comparison was made using the Student’s t-test. (A and B) *Significant change compared to MI-si-NS, and #significant change compared to JEV-si-NS. (C and D) *Significant change compared to MI-ASO-NS, and #significant change compared to JEV-ASO-NS. (E and F) *Significant change compared to MI-si-NS, and #significant change compared to DENV-si-NS. (G and H) *Significant change compared to MI-ASO-NS, and #significant change compared to DENV-ASO-NS.
RBM10 overexpression enhanced the recruitment of p65 to the promoters of NF-kB genes in both mock-infected and JEV-infected SH-SY5Y cells (Fig. S29). These results suggest that RBM10 promotes ER stress and neuroinflammatory gene expression by modulating the recruitment of p65 to the promoters of ER stress and neuroinflammatory genes (Fig. 9). We next evaluated the impact of JINR1 depletion on the binding of p65 at the promoters of JEV-induced genes. JINR1 silencing also reduced p65 binding at the promoters of NF-κB-responsive genes involved in ER stress and neuroinflammation during JEV infection (Fig. 9C and D; Fig. S30). Next, we checked if this reduction in binding of p65 to the promoters of ER stress and neuroinflammatory genes upon JINR1/RBM10 depletion during JEV infection is due to a reduction in the p65 expression. However, we did not see any significant change in p65 protein expression upon JINR1 or RBM10 depletion during JEV infection (Fig. S31).
RBM10/JINR1 silencing during DENV or WNV infection also negatively impacts p65 recruitment to the promoters of NF-κB target genes (Fig. 9E through H; Fig. S32). Our results suggest that the JINR1 and RBM10 complex regulates the transcription of ER stress and neuroinflammatory genes during flavivirus infection by controlling p65 recruitment to their promoters.
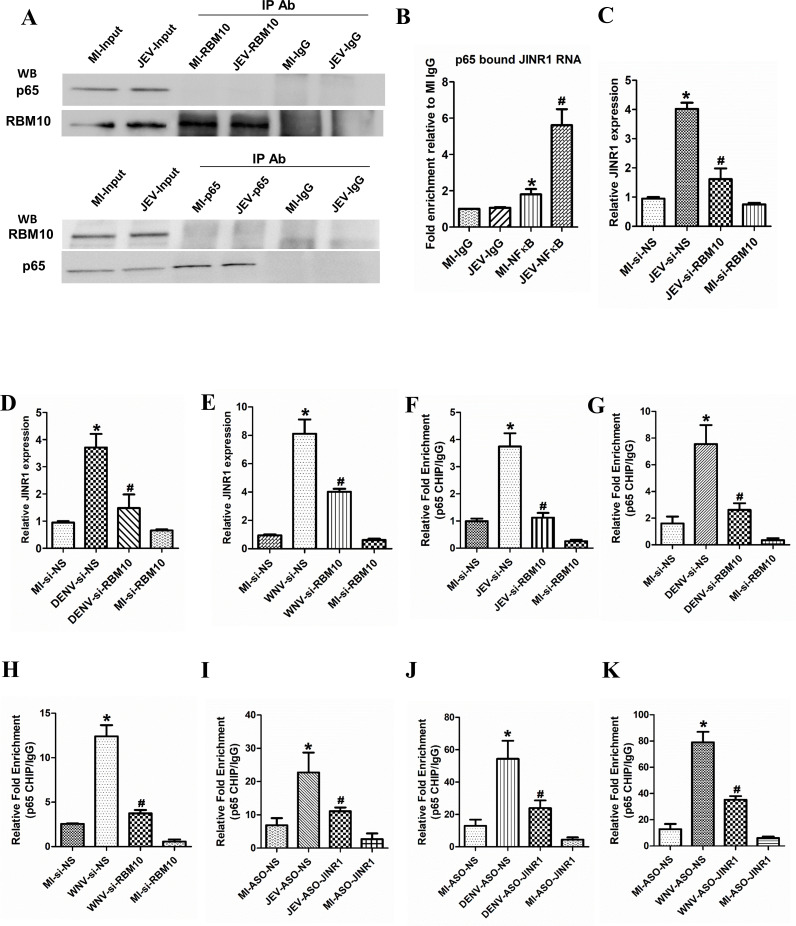
Since RBM10 regulates p65 binding to the promoter of target genes, we next examined whether RBM10 interacts with p65. Hence, we performed RBM10/p65 co-immunoprecipitation (Co-IP) assays with or without JEV infection in SH-SY5Y cells. Surprisingly, p65 was not pulled down together with RBM10 using anti-RBM10 antibodies in SH-SY5Y cells (Fig. 10A). Reciprocal Co-IP assays with anti-65 antibodies also confirmed that RBM10 does not directly interact with p65 (Fig. 10A). Consistent with our Co-IP assays, we did not identify any direct interaction between p65 and RBM10 using the Search Tool for the Retrieval of Interacting Genes (STRING) database (67) (Fig. S33).
Fig 10.
JINR1 interacts with the p65 subunit of NF-κB, JINR1 and RBM10 reciprocally regulate each other’s expression through NF-κB. (A) Upper panel: cell lysates from SH-SY5Y cells were immunoprecipitated with anti-RBM10 or IgG antibody. The immune complexes and the input were analyzed by immunoblotting with antibodies specific to p65, and the same membrane was stripped and reprobed to detect RBM10. Lower panel: protein prepared from SH-SY5Y cells was also immunoprecipitated with anti-p65 or IgG antibodies. The immune complexes and the input were analyzed by immunoblotting with an antibody specific to RBM10; the same membrane was stripped and reprobed to detect p65. A representative blot is shown from two independent experiments with similar results. (B) JINR1 interacts with the p65 subunit of NF-κB. JINR1 RNA levels in p65 immunoprecipitated from the lysates of formaldehyde-crosslinked MI or JEV-infected SH-SY5Y cells were measured by qRT-PCR analysis, normalized to input, and represented as fold enrichment relative to MI-IgG IP. Values represent mean ± SD from three independent experiments. *Significant change compared to MI-IgG IP (P < 0.05). #Significant change compared to MI-p65 IP (P < 0.05). (C) RBM10 depletion attenuates JEV-induced JINR1. qRT-PCR analysis of JINR1 upon RBM10 depletion in SH-SY5Y cells at 36 hpi. (D) RBM10 depletion attenuates DENV-induced JINR1. qRT-PCR analysis of JINR1 upon RBM10 depletion in SH-SY5Y cells at 36 hpi. (E) RBM10 depletion attenuates WNV-induced JINR1. qRT-PCR analysis of JINR1 upon RBM10 depletion in SH-SY5Y cells at 36 hpi. (F) RBM10 promotes the p65 recruitment at the promoter of JINR1 during JEV infection. Relative enrichment of p65 at the JINR1 promoter in MI or JEV-infected SH-SY5Y cells transfected with si-NS or si-RBM10 determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-si-NS-IgG. (G) RBM10 promotes the p65 recruitment at the promoter of JINR1 during DENV infection. Relative enrichment of p65 at the JINR1 promoter in MI or DENV-infected SH-SY5Y cells transfected with si-NS or si-RBM10 determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-si-NS-IgG. (H) RBM10 promotes the p65 recruitment at the promoter of JINR1 during WNV infection. Relative enrichment of p65 at the JINR1 promoter in MI or WNV-infected SH-SY5Y cells transfected with si-NS or si-RBM10 determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-si-NS-IgG. (I) JINR1 promotes the p65 recruitment at the promoter of RBM10 during JEV infection. Relative enrichment of p65 at the RBM10 promoter in MI or JEV-infected SH-SY5Y cells transfected with ASO-NS or ASO-JINR1 determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-ASO-NS-IgG. (J) JINR1 promotes the p65 recruitment at the promoter of RBM10 during DENV infection. Relative enrichment of p65 at the RBM10 promoter in MI or DENV-infected SH-SY5Y cells transfected with ASO-NS or ASO-JINR1 determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-ASO-NS-IgG. (K) JINR1 promotes the p65 recruitment at the promoter of RBM10 during WNV infection. Relative enrichment of p65 at the RBM10 promoter in MI or WNV-infected SH-SY5Y cells transfected with ASO-NS or ASO-JINR1 determined by ChIP-qRT-PCR at 36 hpi. Enrichment values are relative to MI-ASO-NS-IgG. Error bars represent the mean ± SEM from three independent experiments. Statistical comparison was made using the Student’s t-test. (C and F) *Significant change compared to MI-si-NS, and #significant change compared to JEV-si-NS. (D and G) *Significant change compared to MI-si-NS, and #significant change compared to DENV-si-NS. (E and H) *Significant change compared to MI-si-NS, and #significant change compared to WNV-si-NS. (I) *Significant change compared to MI-ASO-NS, and #significant change compared to JEV-ASO-NS. (J) *Significant change compared to MI-ASO-NS, and #significant change compared to DENV-ASO-NS. (K) *Significant change compared to MI-ASO-NS, and #significant change compared to WNV-ASO-NS.
Since lncRNAs are known to interact with NF-κB to regulate its transcriptional activity (68, 69), we hypothesized that JINR1 directly interacts with p65 and regulate its recruitment to the promoters of the ER stress and neuroinflammatory genes. To investigate whether JINR1 interacts with p65, we used bioinformatic analysis using RPISeq to predict the binding affinity between p65 and JINR1. The RPI Seq suggested a strong (>80%) interaction probability between JINR1 and NF-κB. We next used RIP assay to check the direct interaction between JINR1 and p65. RIP analysis indicated p65 binding to JINR1 in MI SH-SY5Y cells (~1.8-fold enrichment compared to MI IgG IP), and this association was further enhanced by approximately fivefold in JEV-infected SH-SY5Y cells (Fig. 10B). Collectively, our findings indicate that JINR1 and RBM10 regulate p65 recruitment to the promoter of genes involved in ER stress and neuroinflammation during JEV infection (Fig. 11).
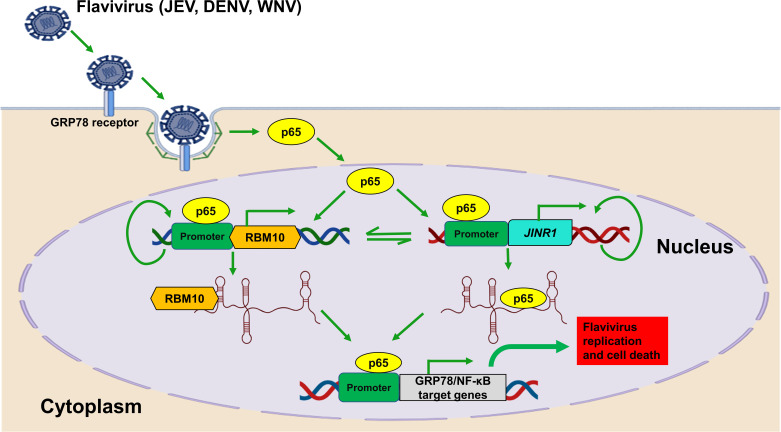
Fig 11.
Flavivirus infection results in NF-κB mediated increase in JINR1 expression. It promotes JEV/DENV/WNV replication and neuronal cell death by promoting the expression of GRP78 and other NF-κB target genes. JINR1 interacts with RBM10 and p65 to promote p65 binding to its target genes.
JINR1 and RBM10 reciprocally regulate each other’s expression through NF-κB
NF-κB activates JINR1 expression (Fig. 2F), and since RBM10 regulates p65 binding to the promoters of its target genes, we tested the role of RBM10 in regulating JINR1 expression. As anticipated, RBM10 silencing attenuates JEV-induced JINR1 expression (Fig. 10C). RBM10 depletion during DENV or WNV infection also reduces JINR1 expression (Fig. 8D and E). Since JINR1 depletion attenuates the flavivirus-mediated increase in RBM10 expression (Fig. 4D and E, Fig. 6D and E) and because JINR1 regulates target gene expression by regulating p65 binding, we tested if it regulates RBM10 expression via NF-κB. We first analyzed the promoter of RBM10 for the presence of NF-κB binding sites. Interestingly, in silico analysis using Alibaba2 revealed multiple NF-κB binding sites in the RBM10 promoter region ~1.1 kb upstream of the TSS. Hence, we evaluated RBM10 expression upon NF-κB inhibition during JEV infection. Treatment of SH-SY5Y cells with the NF-κB inhibitor BAY11-7085 before JEV infection strongly suppressed JEV-induced RBM10 expression (Fig. S34A). Next, we assessed the recruitment of p65 at the promoters of JINR1 and RBM10 during flavivirus infection upon RBM10/JINR1 depletion. Cells transfected with siRNA against RBM10 during flavivirus infection had significantly less p65 binding at the JINR1 promoter than flavivirus-infected cells transfected with si-NS (Fig. 10F through H). JINR1 silencing also reduced the flavivirus-mediated recruitment of p65 at the RBM10 promoter (Fig. 10I through K). These results indicate that JINR1 and RBM10 reciprocally regulate each other’s induction during flavivirus infection via NF-κB. Moreover, depletion of either JINR1 or RBM10 during infection reduces p65 binding at their own promoters (Fig. S34B through G). These findings suggest that apart from co-regulating each other, JINR1 and RBM10 also promote self-expression during JEV infection via NF-κB (Fig. 11).
DISCUSSION
Flavivirus infection results in a complex signaling cascade leading to ER stress and neuronal apoptosis (14, 48, 58, 61, 70). It also leads to a robust immune response and expression of inflammatory cytokines and chemokines (12, 19, 49). However, an exacerbated inflammatory response is often detrimental as it induces neuronal cell death (49, 50, 71, 72). The role of miRNAs in gene regulation during JEV-induced neuroinflammation, apoptosis, and viral replication is well-studied (8, 73, 74). However, knowledge about the functions and mechanisms of action of lncRNAs in JEV pathogenesis is in its infancy (8). Moreover, there are no studies about the role of lncRNAs in regulating JEV-induced human neuronal cell death. Our study provides the first evidence of the role of primate-specific host lncRNA JINR1 in flavivirus replication and neuronal cell death.
JEV infection in SH-SY5Y cells elicits NF-κB-mediated expression of JINR1. NF-κB also induces the expression of RBM10 during JEV infection. RBM10 is a nuclear RNA-binding protein that regulates pre‐mRNA splicing, mRNA stabilization, and transcription (75, 76). RBM10 inhibits cell proliferation and promotes apoptosis (54, 76, 77). PAR-CLIP studies have shown that RBM10 interacts with JINR1 in HEK-293T cells (56). We observed the interaction of JINR1 with RBM10 in SH-SY5Y cells during flavivirus infection. JINR1 or RBM10 depletion results in (i) inhibition of flavivirus replication, (ii) reduction in flavivirus-induced neuronal cell death, and (iii) attenuated expression of NF-κB target genes. Interestingly, RBM10 overexpression in A549 cells inhibits DENV replication, and DENV infection decreases RBM10 protein levels (55). The anti-viral activity of RBM10 observed by Pozzi et al. in the case of DENV and the pro-viral action of RBM10 observed by us in the case of JEV, WNV, and DENV infection are likely due to the contrasting impact of RBM10 depletion on cell proliferation on the different cell types used to study viral infection. Pozzi et al. used A549 cells for studying DENV replication, in which RBM10 depletion reduces cell proliferation of A549 cells (78), and we used SH-SY5Y and T98G cells in which RBM10 depletion promotes cell proliferation (Fig. S4E and F). The differing impact of RBM10 depletion on DENV replication in different cell types may also depend on the differential ability of cells (neuronal-SHSY5Y) versus (lung-A549) to clear virus infection. We also show that JINR1 and RBM10-mediated increase in GRP78 expression promotes flavivirus replication in SH-SY5Y cells. Host factors also regulate viral replication by interaction with viral proteins. Interestingly, Zika virus-NS5 and DENV-NS5 proteins interact with RBM10 (55, 79). Moreover, Pozzi et al. have shown that DENV NS5 binds to RBM10 and mediates its proteasomal degradation in A549 cells. Considering the pro-viral activity of RBM10 and JINR1 complex in neuronal cells, further studies are required to prove if RBM10 and JINR1 interact with flaviviral proteins in neuronal cells and decipher the consequence of this interaction, if any, on the stability of viral proteins and vice versa.
JINR1 and RBM10 promote the expression of NF-κB target genes involved in ER stress and neuroinflammation during flavivirus infection. RBM10 is known to regulate NF-κB transcriptional activity (54, 77). RBM10 depletion studies during flavivirus infection suggest that it promotes NF-κB target gene expression by enhancing p65 binding to its target gene promoters (Fig. 11). JINR1 knockdown also reduces p65 binding to its target genes during flavivirus infection. Interestingly, RBM10-mediated changes in NF-κB transcription are indirect via the regulation of alternative splicing of DNA (cytosine-5)-methyltransferase 3b (DNMT3B) (54). RBM10 depletion promotes the splicing of active DNMT3B2 isoform, which increases DNA methylation at the promoters of NF-κB-target genes, inhibiting NF-κB-mediated transcriptional activity (54). Further investigations are required to understand whether RBM10 and JINR1 play any role in regulating DNMT3B splicing during flavivirus infection. Interestingly, we observed that apart from interacting with RBM10, JINR1 interacts with the p65 subunit of NF-κB and regulates its recruitment to its target genes.
JINR1 and RBM10 are NF-κB target genes that enhance their own expression and also reciprocally regulate each other’s expression by promoting the recruitment of p65 at their promoters. These findings indicate the existence of an NF-κB-mediated autocrine loop promoting JINR1 and RBM10 expression during JEV infection. Even though JINR1 and RBM10 regulate each other’s expression, considering that they interact with each other, we speculate that JINR1 and RBM10 are part of an RNA-protein complex that regulates the recruitment of the NF-κB to the promoters of JEV-regulated genes. However, further studies are required to decipher if DNA methylation plays any role in JINR1-mediated recruitment of NF-κB to its target genes during flavivirus infection.
We also observed that JINR1 is induced upon poly (I:C) treatment (RNA virus mimic) in SH-SY5Y cells. This suggests that other neurotropic RNA viruses might also induce JINR1, which may be part of a general host response. Hence, further studies are needed to understand the role of JINR1 in the pathogenesis of other RNA viruses. As such, JINR1 represents a novel component of the host response that regulates flavivirus replication, ER stress, inflammation, and neuronal cell death. Further characterization of its regulatory networks and studies in primary human neurons and the JEV primate model will likely reveal better opportunities for therapeutic intervention during flavivirus infection.
MATERIALS AND METHODS
Cell culture
SH-SY5Y, Vero cells, PS, and C6/36 cells were purchased from the National Centre for Cell Science, Pune, India. HMC3 and T98G were purchased from the American Type Culture Collection (Manassas, VA, USA). The SH-SY5Y cells were maintained in DMEM-F12, while other cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM of glutamine, and penicillin/streptomycin (Gibco) at 37°C and 5% CO2. C6/36 cells were grown in complete Eagle’s minimum essential medium (EMEM) with 10% heat-inactivated FBS, 2 mM of glutamine, and penicillin/streptomycin (Gibco) at 28°C and 5% CO2. All cell lines used in this study have been tested for potential mycoplasma contamination. Cells post-infection with JEV multiplicity of infection (MOI 5) were cultured in media with 5% FBS.
Virus propagation
GP78 strain of JEV was propagated in Vero cells. Briefly, Vero cell monolayers were inoculated with JEV at 0.1 MOI and incubated at 37°C and 5% CO2 for 72 hpi until the infected monolayers showed the cytopathic effect. Subsequently, the culture supernatant was harvested by centrifugation at 2,000 rpm for 20 min at 4°C followed by filtration using syringe filters with 0.8/0.2 µm (Cat. No. 4658, Acrodisc Syringe Filters) and aliquoted to generate a virus stock. JEV titer was determined by plaque assay in PS cells. Dengue virus serotype 2 (DENV2) strain IND/P23085/1960 was a kind gift from Dr. Arup Banerjee, Regional Centre for Biotechnology, Faridabad, India. DENV2 was propagated by infecting Aedes albopictus C6/36 cell monolayers at 0.2 MOI, and after that the cells were incubated with 2% FBS-containing media at 28°C and 5% CO2 for 5 days until the infected monolayers showed the cytopathic effect. Subsequently, the supernatant was harvested and concentrated by passing through a 100 kDa Amicon filter (Millipore; UFC9100) by centrifugation at 5,000 rpm for 5 min at 4°C and aliquoted to generate a virus stock (80). DENV2 titer was determined by performing the plaque assay on PS cells (81). Eg101 strain of West Nile Virus was propagated by infecting C6/36 cell monolayers at 0.2 MOI, and after that the cells were incubated with 2% FBS-containing media at 28°C and 5% CO2 for 5 days until the infected monolayers showed the cytopathic effect. Subsequently, the supernatant was harvested by centrifugation at 5,000 rpm for 5 min at 4°C to generate a virus stock (82). All virus stocks were aliquoted and stored at −80°C.
Plaque assay analysis of virion production
A total of 2 × 105 PS or Vero cells were seeded in 6-well tissue culture plates, and the next day the cells were infected for 2 h with serially diluted cell culture-grown JEV (GP78), DENV-2, and WNV viruses. PS cells were rinsed twice with PBS to remove the excess virus and were topped with a 2% low-melting agarose/growth media mixture (1:1). After growing for 4 days at 37°C with 5% CO2, cells were fixed with 4% formaldehyde at 37°C for 1 h. Cells were stained for 5 min at 37°C by adding 0.5% crystal violet solution and washed thrice with RO water, and then plaques were counted to calculate the concentration of virus in plaque-forming units (PFU) per mL (83). Similarly, to determine WNV, the protocol mentioned above was followed in Vero cells. [PFU = N × DF/V (N, number of plaques; DF, virus dilution factor; and V, the volume of the inoculum)].
RNA sequencing
Total RNA from GP78 strain JEV (MOI 5) infected or MI SH-SY5Y cells was isolated at 48 hpi using an MN Nucleo Spin RNA Plus isolation kit (Cat. No. 740984.5). RNA samples were treated with recombinant DNAseI (Invitrogen Thermo Scientific—Cat. No. EN0521) as per the manufacturer’s instructions. The RNA samples were then column purified using the MN Nucleo Spin column purification kit. Library preparation and RNA sequencing analyses were done at Clevergene Biocorp Private Limited, Bangalore, as described below. The DNAseI-treated RNA was treated with Invitrogen’s RiboMinusTM Eukaryote kit according to the manufacturer’s instructions to remove ribosomal RNAs. RNA-seq libraries were made using NEBNext UltraTM II Directional RNA Library prep kit for Illumina according to the manufacturer’s instructions. One hundred base pair-long strand-specific sequencing was performed on an Illumina HiSeq 2500 instrument at a depth of ∼100 million paired-end reads per sample. The sequence data quality was checked using FastQC and MultiQC software, and raw sequence reads were processed to remove adapter sequences and low-quality bases using fastp. The QC passed reads were mapped onto the indexed Human reference genome (GRCh38.p7) using the HISAT2 aligner. Transcript-level expression values were obtained as read counts using feature counts software. Differential expression analysis was done using the edgeR package after normalizing the data based on trimmed mean of M values. DEGs were selected based on log base 2 (fold ≥ 0.6) and log base 2 (fold ≤ −0.6) with a statistical significance of P-value < 0.05.
Gene ontology and pathway analysis
Gene ontology (GO) enrichment analysis for biological process, molecular function, cellular component, and Kyoto Encyclopedia of Genes and Genomes Pathway was performed using the ClusterProfiler R package. GO and pathway terms with multiple tests adjusted P-value ≤ 0.05 are considered significant. To visualize the GO enrichment results and calculate the z-score, GOplot R package was used. The pathways were visualized using the Pathview package to check the differential expression level of the genes.
RNA isolation and real-time PCR
RNA isolation and real-time PCR were performed as described previously (84, 85). Briefly, total RNA was extracted from cells using the MN NucleoSpin RNA plus isolation kit (Cat. No. 740984.5). One microgram of RNA was converted into cDNA using the PrimeScript first-strand cDNA kit from Takara (#6110A). qRT-PCR was performed with the SYBR Green PCR Kit (#RR820A, Takara) in the LightCycler 480 Real-Time PCR system. All reactions were performed in triplicates and normalized with GAPDH as an internal control. The relative gene expression of each sample was calculated using the 2-ddct formula. The gene primer sequences are listed in (Table S3).
Virus and inhibitor treatment
One-day post-seeding, SH-SY5Y and T98G cells were incubated with a virus at an MOI of 5 in an incomplete DMEM-F12 at 37°C for 2 h. After virus adsorption, the inoculum was removed, and the cells were washed with PBS. The time of virus removal was taken as 0 hpi for all experiments. After removing the virus-containing medium, the cells were grown in growth media with reduced serum (5%) for the duration indicated in the figures and legends. For poly (I:C) (Cat. No. P9582, Sigma-Aldrich) transfection, 5 × 105 SH-SY5Y cells were seeded in six wells and transfected with poly (I:C) using RNAimax (Invitrogen) according to the manufacturer’s protocol for 24 h. Cells were pre-treated with Bay 11-7085 (Tocris #1743), an inhibitor of NF-κB, before JEV infection at a concentration of 1 µM.
Viral RNA isolation from culture supernatant
Takara MiniBEST Viral RNA/DNA Extraction Kit (Cat. No. # 9766) was used to isolate JEV viral RNA from cell culture supernatants as per the manufacturer’s protocol. An equal amount of RNA from each transfected sample was used for cDNA synthesis, followed by qRT-PCR of viral RNA to get Ct values. qRT-PCR products were also analyzed on the agarose gel.
Antisense oligos/siRNA transfection
SH-SY5Y cells were reverse transfected with 40 nM antisense oligos (ASOs) targeting JINR1 using Lipofectamine RNAiMAX (Life Technologies-Invitrogen, Cat. No.: 13778-075) as per the manufacturer’s instructions. The following ASOs from Qiagen were used in this study: ASO-JINR1-1: 5′-TACAGCGCCTTTGGTA-3′ (Cat. No: LG00781145-DDA), ASO-JINR1-2: 5′-GAGGGAGAACAACGTT-3′ (Cat. No: LG00781146-DDA), ASO-JINR1-3: 5′-GTAGAGGGCTTTGAAT-3′ (Cat. No: LG00781147-DDA), and non-specific ASO-NS: 5′-AACACGTCTATACGC-3′ (Cat. No: LG00000002-DDA). The sequence of siRNA duplexes against RBM10 (Applied biosystem) is sense 5′-CCGCUGUGCUCAAAUCUGATT-3′ and antisense 5′-UCAGAUUUGAGCACAGCGGGT-3′. Non-specific siRNA that does not target any known mammalian gene was purchased from (Dharmacon, Cat. No.: D-001810-10-20. siRNAs were reverse transfected at a final concentration of 40 nM with lipofectamine RNAiMAX (Invitrogen). ASOs/siRNA co-transfection with plasmids was done using lipofectamine 3000 (Cat. No: L3000008) following the manufacturer’s instructions.
Plasmid construction
The full-length human JINR1 (1,856 bp) cDNA was amplified by RT-PCR from total RNA isolated from SH-SY5Y cells using the primers listed in Table S3. Amplified JINR1 was cloned into the pcDNA3.1 plasmid at the NheI and BamHI sites. Empty plasmid (pcDNA3.1) was used as a control. The full-length RBM10 construct (pDest26-RBM10) was a kind gift from Prof. Juan Valcárcel, Center for Genomic Regulation, Pompeu Fabra University, Spain (86). The pcDNA3.1(+)-GRP78/BiP construct was purchased from addgene (Cat. No. 32701). All constructs were verified by sequencing. Plasmids were transfected in SH-SY5Y cells using jetOPTIMUS Polyplus transfection reagent (Cat. No. 101000051) according to the manufacturer’s instructions.
Western blot analysis
Protein isolation and western blotting were done as described previously (84). The following primary antibodies were used: Cleaved PARP (1: 2,000, CST #5625), RBM10 (1:1,500, Abcam #ab72423), NF-κB p65 (1:2,000) (CST #8242), secondary antibodies (1:20,000)—HRP conjugated anti-rabbit (Vector Laboratories Cat. No.: PI 2000) or anti-mouse IgG (Invitrogen, Cat. No.: A16072). The blot signals were quantified using ImageJ software for Microsoft Windows (National Institutes of Health, Bethesda, MD, USA).
Cell proliferation assay
Cell proliferation assay was done using the WST-1 reagent (Cat. No. 05015944001, Roche) at the indicated times according to the manufacturer’s instructions. Briefly, SH-SY5Y or T98G cells were seeded in 96-well plates and reverse transfected with ASO-NS, ASO-JINR1-1, ASO-JINR1-2, si-NS and si-RBM10 at 40 nM, and cell proliferation was estimated by taking an OD at 450 nm.
Caspase-3/7 Assay
A luminometric assay kit for caspase-3/7 (Promega, Cat. No. G8090) was used to determine the enzymatic activity of caspase-3/7 in SH-SY5Y cells transfected with ASO-JINR1-1, ASO-JINR1-2, ASO-NS, si-NS and si-RBM10, as per the manufacturer’s instructions.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described previously (84, 85). Briefly, MI or JEV-infected cells were fixed with 1% formaldehyde for 8 min at room temperature, followed by quenching with 0.125 M glycine for 5 min. Cells were washed twice with ice-cold phosphate-buffered saline, harvested by scraping, pelleted, and resuspended in 250 µL of ChIP lysis buffer [50 mM Tris–HCl (pH 8.1), 0.9% SDS, 10 mM EDTA, protease inhibitor], and samples were incubated on ice for 1 h. Samples were sonicated using Bioruptor Plus (Diagenode) with sonication conditions: 30 s on and 30 s off for 25 cycles. After sonication, samples were centrifuged at 14,000 rpm at 4°C for 20 min. Supernatants were diluted fivefold in ChIP dilution buffer (167 mM NaCl, 16.7 mM Tris–HCl pH 8.1, 1.2 mM EDTA, 1.1% Triton X-100, and protease inhibitors), 25 µg chromatin samples were used for IP, and 5% of the sample was taken as input. Samples were incubated at 4°C overnight with NF-κB p65 (1:200) (CST #8242), RNA Polymerase II (1:100) (RNA Pol II; CST #2629), Histone H3 trimethylated at Lysine 4 (H3K4me3; CST #9751), or anti-rabbit IgG (CST #2729S) antibody and anti-mouse IgG (CST #5415). The following day, Dynabeads Protein G (Invitrogen #10004D) 50 µL per sample was added to the IP tubes and incubated for 2 h at 4°C. The beads were then washed once each with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1, and 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris HCl pH 8.1, and 500 mM NaCl), LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris–HCl pH 8.1), and TE buffer (pH 8.0). Following this, samples were reverse crosslinked using decrosslinking buffer (222 mM NaCl, 50 mM Tris, 10 mM EDTA, and 0.025% SDS), containing 5 µL proteinase K (NEB #P8107S, 800 U/mL) per sample with an overnight incubation at 65°C. Genomic DNA was then extracted with a DNA purification kit (Zymo Cat. No.: D3020), and the DNA in immunoprecipitated samples was measured using qRT-PCR. Primers for ChIP are listed in Table S3.
Formaldehyde-cross-linked RNA immunoprecipitation
RIP was performed as described (45), with minor modifications. Briefly, 15 × 106 SH-SY5Y cells infected with or without JEV (MOI 5) for 24 h were fixed with 1% formaldehyde to crosslink protein-bound RNA/DNA complexes, and formaldehyde was quenched by the addition of glycine to a concentration of 0.125 M. Cells were then washed with PBS and incubated in 1 mL collection buffer [100 mM Tris–HCl (pH 9.4) and 100 mM DTT, protease inhibitor, RNaseOUT] on ice for 15 min. Nuclei were isolated by resuspension in 1 mL buffer A [10 mM EDTA, 0.5 mM ethyleneglycol- bis (β-aminoethyl)-N,N,Nʹ,Nʹ-tetraacetic acid (EGTA), 10 mM HEPES, and 0.25% Triton X-100, protease inhibitor, RNaseOUT] and centrifugation at 3,000 g at 4°C. Nuclear pellets were then sonicated using bioruptor (Diagenode) with sonication conditions: 30 s on and 30 s off for 25 cycles in 900 µL Shearing buffer AM2 (Active motif 53024) to produce DNA/RNA fragments of about 500 nucleotides. The sonicated extract was split into equal aliquots, made up to a total of 1.5 mL with IP buffer (1% Triton X-100, 0.1% DOC, 1× TE, RNaseOUT), 50 µL of the Dynabeads Protein G, RBM10 (1:40) (ab72423; Abcam), NF-κB p65 (1:200) (CST #8242), or rabbit IgG (CST #2729S) antibodies, and incubated overnight at 4°C. The beads-bound RNA/DNA complexes were washed six times with RIPA buffer (50 mM HEPES pH 8, 1 mM EDTA, 1% NP-40, 0.7% DOC, 0.5 M LiCl, and protease inhibitor mix, RNaseOUT) and once with TE buffer (protease inhibitor mix, RNaseOUT). The DNA/RNA was extracted from the beads by adding 100 µL of elution buffer (10 mM Tris pH 8, 1 mM EDTA, 1% SDS, RNaseOUT) and incubated at 65°C with gentle shaking for 15 min. For reversing the crosslinking, elution buffer containing RNA/DNA was supplemented with 2 µL of 5 M NaCl and 2 µL proteinase K (NEB) and made up to a total volume of 170 µL and incubated for 5 h at 70°C. After reverse crosslinking, 750 µL of TRIzol was added per sample. RNA was extracted using the Direct zol RNA kit (Zymo R2050) per the manufacturer’s instructions. cDNA synthesis was carried out using the PrimeScript first-strand cDNA kit from Takara (#6110A). qRT-PCR was performed in triplicates with 0.5 µL cDNA using SYBR Green PCR Kit (#RR820A, Takara) on a LightCycler 480 Real-Time PCR System. The primers used for JINR1 are provided in Table S3. The results are expressed as fold enrichment over input and normalized to MI IgG.
Construction of protein-protein interaction network
The Search Tool for the Retrieval of Interacting Genes (STRING) is an online tool that assesses protein-protein interaction (PPI) network information (67). STRING (version 11.5) was used to evaluate the potential PPI relationships for the p65 (RelA) and RBM10. Then, the primary PPI network was constructed using predicted and experimentally validated interacting proteins with a confidence score ≥ 0.7, and the network was grouped into four clusters using k-means.
Co-immunoprecipitation
5 × 106 SH-SY5Y cells were infected with or without JEV (MOI 5). Thirty-six-hour post-infection cells were washed with PBS and scraped using 250 µl of ice-cold IP lysis buffer [25 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, 0.5% sodium deoxycholate, 0.05% SDS, and protease and phosphatase inhibitors] on ice, transferred to 1.5 mL microcentrifuge tubes and rotated for 30 min at 4°C. Lysates were then passed through the 26-gauge needle and incubated on ice for 5 min. The supernatant was collected by centrifuging the lysates 14,000 g for 15 min at 4°C, and the protein concentration was determined by BCA assay. Cell lysates were pre-cleared by mixing with 30 µL Dynabeads Protein G (Invitrogen #10004D) and incubating at 4°C for 2 h with gentle agitation. One hundred fifty micrograms of pre-cleared protein was incubated with RBM10 (1:100) (ab72423; Abcam), NF-κB p65 (1:100) (CST #8242), or rabbit IgG (CST #2729S) antibodies overnight at 4°C. The following day lysates with antibodies were incubated with Dynabeads Protein G (Invitrogen #10004D) for 2 h at 4°C with gentle shaking. The beads-bound protein complexes were washed three times with IP wash buffer [25 mM Tris–HCl (pH 7.5), 0.1% (vol/vol) NP-40, 1 mM EDTA, 125 mM NaCl] and resuspended in 2× Laemmli buffer and boiled for 5 min to release the bound protein. The samples were then analyzed by Western blot with a specific antibody. A fifteen µg aliquot of cell lysate was used as input control. The same membrane was stripped by incubating at 50°C for half an hour in stripping buffer and reprobed with the corresponding IP antibody.
Statistical analysis
The results are presented as mean ± SEM unless otherwise stated. We used paired Student’s t-test for comparisons between two experimental groups. Additional statistical test information is described in the Figure legends. P < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Dr. Arup Banerjee and Dr. Prasenjit Guchhait (Regional Centre for Biotechnology, Faridabad) for providing DENV strains and sharing protocols for DENV propagation. We thank Prof. Juan Valcárcel (Centre for Genomic Regulation, Pompeu Fabra University, Spain) for providing a full-length RBM10 construct. We thank Dr. Kanhaiya Lal Kumawat (NBRC) and Swati (BITS, Hyderabad) for their technical help and support.
This work was supported by extramural grants from the Department of Biotechnology ,Government of India, DBT-BT/PR27371/MED/122/122/2017 to V.S. and A.B.; DBT-RLS 102/IFD/SAN/3499/2016–17 to V.S.; J. C. Bose Fellowship (JCB/2020/000037) from Science and Engineering Research Board, DST to A.B.; OPERA award from BITS Pilani to V.S. V.S. would like to thank Prof. G. Sundar, Director of BITS Pilani, Hyderabad, for funding support. S.T. received a Ph.D. fellowship from BITS Pilani.
S.T. was involved in conceptualization, methodology, validation, formal analysis, investigation, visualization, and writing of the original draft. S.S., B.S., and S.M. devised the methodology, validated the study, performed the investigation, and visualized the study. A.B. conceptualized the study, acquired funding, and reviewed the manuscript. V.S. was involved in conceptualization, methodology, investigation, visualization, formal analysis, resources, supervision, project administration, funding acquisition, and writing of original draft, review, and editing.
Contributor Information
Anirban Basu, Email: anirban@nbrc.ac.in.
Vivek Sharma, Email: viveksharma@hyderabad.bits-pilani.ac.in.
Mark T. Heise, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA
DATA AVAILABILITY
The NCBI Gene Expression Omnibus accession number for the RNA seq data is GSE221680.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.01183-23.
Figures S1 to S34.
List of differentially expressed genes upon JEV infection in SH-SY5Y cells identified by RNA sequencing.
List of JINR1 interacting proteins identified from POSTAR database.
List of primers for qRT-PCR.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Pierson TC, Diamond MS. 2020. The continued threat of emerging flaviviruses. Nat Microbiol 5:796–812. doi: 10.1038/s41564-020-0714-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mulvey P, Duong V, Boyer S, Burgess G, Williams DT, Dussart P, Horwood PF. 2021. The ecology and evolution of Japanese encephalitis virus. Pathogens 10:1534. doi: 10.3390/pathogens10121534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sharma KB, Vrati S, Kalia M. 2021. Pathobiology of Japanese encephalitis virus infection. Mol Aspects Med 81:100994. doi: 10.1016/j.mam.2021.100994 [DOI] [PubMed] [Google Scholar]
- 4. Quan TM, Thao TTN, Duy NM, Nhat TM, Clapham H. 2020. Estimates of the global burden of Japanese encephalitis and the impact of vaccination from 2000-2015. Elife 9:e51027. doi: 10.7554/eLife.51027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kimura-Kuroda J, Ichikawa M, Ogata A, Nagashima K, Yasui K. 1993. Specific tropism of Japanese encephalitis virus for developing neurons in primary rat brain culture. Arch Virol 130:477–484. doi: 10.1007/BF01309676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang J, Li M, Yuan M, Bian P, Dong Y, Zhang H, Luo C, Xue Z, Wang Y, Zhang F, Shen L, Lei Y. 2022. Axl-/- neurons promote JEV infection by dampening the innate immunity. Virus Res 307:198605. doi: 10.1016/j.virusres.2021.198605 [DOI] [PubMed] [Google Scholar]
- 7. Wongchitrat P, Samutpong A, Lerdsamran H, Prasertsopon J, Yasawong M, Govitrapong P, Puthavathana P, Kitidee K. 2019. Elevation of cleaved p18 BAX levels associated with the kinetics of neuronal cell death during Japanese encephalitis virus infection. Int J Mol Sci 20:5016. doi: 10.3390/ijms20205016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ashraf U, Ding Z, Deng S, Ye J, Cao S, Chen Z. 2021. Pathogenicity and virulence of Japanese encephalitis virus: neuroinflammation and neuronal cell damage. Virulence 12:968–980. doi: 10.1080/21505594.2021.1899674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Turtle L, Solomon T. 2018. Japanese encephalitis - the prospects for new treatments. Nat Rev Neurol 14:298–313. doi: 10.1038/nrneurol.2018.30 [DOI] [PubMed] [Google Scholar]
- 10. Desprès P, Frenkiel MP, Ceccaldi PE, Duarte Dos Santos C, Deubel V. 1998. Apoptosis in the mouse central nervous system in response to infection with mouse-neurovirulent dengue viruses. J Virol 72:823–829. doi: 10.1128/JVI.72.1.823-829.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jan JT, Chen BH, Ma SH, Liu CI, Tsai HP, Wu HC, Jiang SY, Yang KD, Shaio MF. 2000. Potential dengue virus-triggered apoptotic pathway in human neuroblastoma cells: arachidonic acid, superoxide anion, and NF-kappaB are sequentially involved. J Virol 74:8680–8691. doi: 10.1128/jvi.74.18.8680-8691.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niranjan R, Muthukumaravel S, Jambulingam P. 2019. The involvement of neuroinflammation in dengue viral disease: importance of innate and adaptive immunity. Neuroimmunomodulation 26:111–118. doi: 10.1159/000501209 [DOI] [PubMed] [Google Scholar]
- 13. Diniz JAP, Da Rosa A, Guzman H, Xu F, Xiao S-Y, Popov VL, Vasconcelos PFC, Tesh RB. 2006. West Nile virus infection of primary mouse neuronal and neuroglial cells: the role of astrocytes in chronic infection. Am J Trop Med Hyg 75:691–696. doi: 10.4269/ajtmh.2006.75.691 [DOI] [PubMed] [Google Scholar]
- 14. Peng B-H, Wang T. 2019. West Nile virus induced cell death in the central nervous system. Pathogens 8:215. doi: 10.3390/pathogens8040215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Debiasi RL, Tyler KL. 2006. West Nile virus meningoencephalitis. Nat Clin Pract Neurol 2:264–275. doi: 10.1038/ncpneuro0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kobayashi S, Orba Y, Yamaguchi H, Kimura T, Sawa H. 2012. Accumulation of ubiquitinated proteins is related to West Nile virus-induced neuronal apoptosis. Neuropathology 32:398–405. doi: 10.1111/j.1440-1789.2011.01275.x [DOI] [PubMed] [Google Scholar]
- 17. Fulton CDM, Beasley DWC, Bente DA, Dineley KT. 2020. Long-term, West Nile virus-induced neurological changes: a comparison of patients and rodent models. Brain Behav Immun Health 7:100105. doi: 10.1016/j.bbih.2020.100105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Medigeshi GR, Lancaster AM, Hirsch AJ, Briese T, Lipkin WI, Defilippis V, Früh K, Mason PW, Nikolich-Zugich J, Nelson JA. 2007. West Nile virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J Virol 81:10849–10860. doi: 10.1128/JVI.01151-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Constant O, Barthelemy J, Nagy A, Salinas S, Simonin Y. 2022. West Nile virus neuroinfection in humans: peripheral biomarkers of neuroinflammation and neuronal damage. Viruses 14:756. doi: 10.3390/v14040756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yi K, Zhang Y, Wang Y, Wang Z, Xie M, Jin Z, Zhao T. 2019. Long noncoding RNA and its role in virus infection and pathogenesis. Front Biosci (Landmark Ed) 24:777–789. doi: 10.2741/4750 [DOI] [PubMed] [Google Scholar]
- 21. Yao R-W, Wang Y, Chen L-L. 2019. Cellular functions of long noncoding RNAs. Nat Cell Biol 21:542–551. doi: 10.1038/s41556-019-0311-8 [DOI] [PubMed] [Google Scholar]
- 22. Fortes P, Morris KV. 2016. Long noncoding RNAs in viral infections. Virus Res 212:1–11. doi: 10.1016/j.virusres.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ouyang J, Hu J, Chen J-L. 2016. lncRNAs regulate the innate immune response to viral infection. Wiley Interdiscip Rev RNA 7:129–143. doi: 10.1002/wrna.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meng X-Y, Luo Y, Anwar MN, Sun Y, Gao Y, Zhang H, Munir M, Qiu H-J. 2017. Long non-coding RNAs: emerging and versatile regulators in host–virus interactions. Front Immunol 8:1663. doi: 10.3389/fimmu.2017.01663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ding Y, Zhang Z, Liu Y, Shi C, Zhang J, Zhang Y. 2016. Relationship of long noncoding RNA and viruses. Genomics 107:150–154. doi: 10.1016/j.ygeno.2016.01.007 [DOI] [PubMed] [Google Scholar]
- 26. Vierbuchen T, Fitzgerald KA. 2021. Long non-coding RNAs in antiviral immunity. Semin Cell Dev Biol 111:126–134. doi: 10.1016/j.semcdb.2020.06.009 [DOI] [PubMed] [Google Scholar]
- 27. Basavappa MG, Ferretti M, Dittmar M, Stoute J, Sullivan MC, Whig K, Shen H, Liu KF, Schultz DC, Beiting DP, Lynch KW, Henao-Mejia J, Cherry S. 2022. The lncRNA ALPHA specifically targets chikungunya virus to control infection. Mol Cell 82:3729–3744. doi: 10.1016/j.molcel.2022.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rossi MN, Antonangeli F. 2014. LncRNAs: new players in apoptosis control. Int J Cell Biol 2014:473857. doi: 10.1155/2014/473857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang N, Zhang X, Gu X, Li X, Shang L. 2021. Progress in understanding the role of lncRNA in programmed cell death. Cell Death Discov 7:30. doi: 10.1038/s41420-021-00407-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tripathi S, Shree B, Mohapatra S, Basu A, Sharma V. 2021. The expanding regulatory mechanisms and cellular functions of long non-coding RNAs (lncRNAs) in neuroinflammation. Mol Neurobiol 58:2916–2939. doi: 10.1007/s12035-020-02268-8 [DOI] [PubMed] [Google Scholar]
- 31. Guo F, Yu X, Xu A, Xu J, Wang Q, Guo Y, Wu X, Tang Y, Ding Z, Zhang Y, Gong T, Pan Z, Li S, Kong L. 2018. Japanese encephalitis virus induces apoptosis by inhibiting Foxo signaling pathway. Vet Microbiol 220:73–82. doi: 10.1016/j.vetmic.2018.05.008 [DOI] [PubMed] [Google Scholar]
- 32. Wang Q, Xin X, Wang T, Wan J, Ou Y, Yang Z, Yu Q, Zhu L, Guo Y, Wu Y, Ding Z, Zhang Y, Pan Z, Tang Y, Li S, Kong L. 2019. Japanese encephalitis virus induces apoptosis and encephalitis by activating the PERK pathway. J Virol 93:e00887-19. doi: 10.1128/JVI.00887-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okamoto T, Suzuki T, Kusakabe S, Tokunaga M, Hirano J, Miyata Y, Matsuura Y. 2017. Regulation of apoptosis during flavivirus infection. Viruses 9:243. doi: 10.3390/v9090243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saha S, Murthy S, Rangarajan PN. 2006. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J Gen Virol 87:1991–1995. doi: 10.1099/vir.0.81768-0 [DOI] [PubMed] [Google Scholar]
- 35. Bhattacharyya S, Vrati S. 2015. The Malat1 long non-coding RNA is upregulated by signalling through the PERK axis of unfolded protein response during flavivirus infection. Sci Rep 5:17794. doi: 10.1038/srep17794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Y, Zhang H, Zhu B, Ashraf U, Chen Z, Xu Q, Zhou D, Zheng B, Song Y, Chen H, Ye J, Cao S. 2017. Microarray analysis identifies the potential role of long non-coding RNA in regulating neuroinflammation during Japanese encephalitis virus infection. Front Immunol 8:1237. doi: 10.3389/fimmu.2017.01237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou X, Yuan Q, Zhang C, Dai Z, Du C, Wang H, Li X, Yang S, Zhao A. 2021. Inhibition of Japanese encephalitis virus proliferation by long non-coding RNA SUSAJ1 in PK-15 cells. Virol J 18:29. doi: 10.1186/s12985-021-01492-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yuan Q, Fan J, Wang H, Li X, Yang S, Zhao A, Zhou X. 2022. LncRNA-SUSAJ1 activates the ER stress pathway inhibiting JEV proliferation by promoting PK15 cells apoptosis. Front Biosci (Landmark Ed) 27:260. doi: 10.31083/j.fbl2709260 [DOI] [PubMed] [Google Scholar]
- 39. Field AR, Jacobs FMJ, Fiddes IT, Phillips APR, Reyes-Ortiz AM, LaMontagne E, Whitehead L, Meng V, Rosenkrantz JL, Olsen M, Hauessler M, Katzman S, Salama SR, Haussler D. 2019. Structurally conserved primate LncRNAs are transiently expressed during human cortical differentiation and influence cell-type-specific genes. Stem Cell Reports 12:245–257. doi: 10.1016/j.stemcr.2018.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang D, Zhang H, Wang X, Hu B, Zhang F, Wei H, Li L. 2019. LINC01518 knockdown inhibits tumorigenicity by suppression of PIK3CA/Akt pathway in oesophageal squamous cell carcinoma. Artif Cells Nanomed Biotechnol 47:4284–4292. doi: 10.1080/21691401.2019.1699815 [DOI] [PubMed] [Google Scholar]
- 41. Kong N, Bao Y, Zhao H, Kang X, Tai X, Chen X, Guo W, Shen Y. 2022. Long noncoding RNA LINC01518 modulates proliferation and migration in TGF-β1-treated human tenon capsule fibroblast cells through the regulation of hsa-miR-216b-5p. Neuromolecular Med 24:88–96. doi: 10.1007/s12017-021-08662-2 [DOI] [PubMed] [Google Scholar]
- 42. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732–738. doi: 10.1038/35099560 [DOI] [PubMed] [Google Scholar]
- 43. Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. doi: 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 44. Vedagiri D, Gupta D, Mishra A, Krishna G, Bhaskar M, Sah V, Basu A, Nayak D, Kalia M, Valiya Veettil M, Harshan KH. 2021. Retinoic acid-inducible gene I-like receptors activate snail to limit RNA viral infections. J Virol 95:e0121621. doi: 10.1128/JVI.01216-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sharma V, Khurana S, Kubben N, Abdelmohsen K, Oberdoerffer P, Gorospe M, Misteli T. 2015. A BRCA 1‐interacting lnc RNA regulates homologous recombination. EMBO Rep 16:1520–1534. doi: 10.15252/embr.201540437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abraham S, Nagaraj AS, Basak S, Manjunath R. 2010. Japanese encephalitis virus utilizes the canonical pathway to activate NF-kappaB but it utilizes the type I interferon pathway to induce major histocompatibility complex class I expression in mouse embryonic fibroblasts. J Virol 84:5485–5493. doi: 10.1128/JVI.02250-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tung W-H, Tsai H-W, Lee I-T, Hsieh H-L, Chen W-J, Chen Y-L, Yang C-M. 2010. Japanese encephalitis virus induces matrix metalloproteinase-9 in rat brain astrocytes via NF-κB signalling dependent on MAPKs and reactive oxygen species. Br J Pharmacol 161:1566–1583. doi: 10.1111/j.1476-5381.2010.00982.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Su H-L, Liao C-L, Lin Y-L. 2002. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J Virol 76:4162–4171. doi: 10.1128/jvi.76.9.4162-4171.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lannes N, Summerfield A, Filgueira L. 2017. Regulation of inflammation in Japanese encephalitis. J Neuroinflammation 14:158. doi: 10.1186/s12974-017-0931-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Das S, Ghosh D, Basu A. 2009. Japanese encephalitis virus induce immuno-competency in neural stem/progenitor cells. PLoS One 4:e8134. doi: 10.1371/journal.pone.0008134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jonas K, Calin GA, Pichler M. 2020. RNA-binding proteins as important regulators of long non-coding RNAs in cancer. Int J Mol Sci 21:2969. doi: 10.3390/ijms21082969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ferrè F, Colantoni A, Helmer-Citterich M. 2016. Revealing protein-lncRNA interaction. Brief Bioinform 17:106–116. doi: 10.1093/bib/bbv031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao W, Zhang S, Zhu Y, Xi X, Bao P, Ma Z, Kapral TH, Chen S, Zagrovic B, Yang YT, Lu ZJ. 2022. POSTAR3: an updated platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res 50:D287–D294. doi: 10.1093/nar/gkab702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Atsumi T, Suzuki H, Jiang J-J, Okuyama Y, Nakagawa I, Ota M, Tanaka Y, Ohki T, Katsunuma K, Nakajima K, Hasegawa Y, Ohara O, Ogura H, Arima Y, Kamimura D, Murakami M. 2017. Rbm10 regulates inflammation development via alternative splicing of Dnmt3b. Int Immunol 29:581–591. doi: 10.1093/intimm/dxx067 [DOI] [PubMed] [Google Scholar]
- 55. Pozzi B, Bragado L, Mammi P, Torti MF, Gaioli N, Gebhard LG, García Solá ME, Vaz-Drago R, Iglesias NG, García CC, Gamarnik AV, Srebrow A. 2020. Dengue virus targets RBM10 deregulating host cell splicing and innate immune response. Nucleic Acids Res 48:6824–6838. doi: 10.1093/nar/gkaa340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang Y, Gogol-Döring A, Hu H, Fröhler S, Ma Y, Jens M, Maaskola J, Murakawa Y, Quedenau C, Landthaler M, Kalscheuer V, Wieczorek D, Wang Y, Hu Y, Chen W. 2013. Integrative analysis revealed the molecular mechanism underlying RBM 10‐mediated splicing regulation. EMBO Mol Med 5:1431–1442. doi: 10.1002/emmm.201302663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Samuel MA, Morrey JD, Diamond MS. 2007. Caspase 3-dependent cell death of neurons contributes to the pathogenesis of West Nile virus encephalitis. J Virol 81:2614–2623. doi: 10.1128/JVI.02311-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shrestha B, Gottlieb D, Diamond MS. 2003. Infection and injury of neurons by West Nileencephalitisvirus. J Virol 77:13203–13213. doi: 10.1128/jvi.77.24.13203-13213.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Parquet MC, Kumatori A, Hasebe F, Morita K, Igarashi A. 2001. West Nile virus-induced bax-dependent apoptosis. FEBS Lett 500:17–24. doi: 10.1016/s0014-5793(01)02573-x [DOI] [PubMed] [Google Scholar]
- 60. Amaral DCG, Rachid MA, Vilela MC, Campos RDL, Ferreira GP, Rodrigues DH, Lacerda-Queiroz N, Miranda AS, Costa VV, Campos MA, Kroon EG, Teixeira MM, Teixeira AL. 2011. Intracerebral infection with dengue-3 virus induces meningoencephalitis and behavioral changes that precede lethality in mice. J Neuroinflammation 8:23. doi: 10.1186/1742-2094-8-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee Y-R, Kuo S-H, Lin C-Y, Fu P-J, Lin Y-S, Yeh T-M, Liu H-S. 2018. Dengue virus-induced ER stress is required for autophagy activation, viral replication, and pathogenesis both in vitro and in vivo. Sci Rep 8:489. doi: 10.1038/s41598-017-18909-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ibrahim IM, Abdelmalek DH, Elfiky AA. 2019. GRP78: a cell’s response to stress. Life Sci 226:156–163. doi: 10.1016/j.lfs.2019.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nain M, Mukherjee S, Karmakar SP, Paton AW, Paton JC, Abdin MZ, Basu A, Kalia M, Vrati S. 2017. GRP78 is an important host factor for Japanese encephalitis virus entry and replication in mammalian cells. J Virol 91:e02274-16. doi: 10.1128/JVI.02274-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wati S, Soo M-L, Zilm P, Li P, Paton AW, Burrell CJ, Beard M, Carr JM. 2009. Dengue virus infection induces upregulation of GRP78, which acts to chaperone viral antigen production. J Virol 83:12871–12880. doi: 10.1128/JVI.01419-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Long Y, Wang X, Youmans DT, Cech TR. 2017. How do lncRNAs regulate transcription? Sci Adv 3:eaao2110. doi: 10.1126/sciadv.aao2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yao Z-T, Yang Y-M, Sun M-M, He Y, Liao L, Chen K-S, Li B. 2022. New insights into the interplay between long non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun (Lond) 42:117–140. doi: 10.1002/cac2.12254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. 2021. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49:D605–D612. doi: 10.1093/nar/gkab835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge R-M, Chang HY. 2013. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2:e00762. doi: 10.7554/eLife.00762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xue Z, Zhang Z, Liu H, Li W, Guo X, Zhang Z, Liu Y, Jia L, Li Y, Ren Y, Yang H, Zhang L, Zhang Q, Da Y, Hao J, Yao Z, Zhang R. 2019. lincRNA-Cox2 regulates NLRP3 inflammasome and autophagy mediated neuroinflammation. Cell Death Differ 26:130–145. doi: 10.1038/s41418-018-0105-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huang M, Xu A, Wu X, Zhang Y, Guo Y, Guo F, Pan Z, Kong L. 2016. Japanese encephalitis virus induces apoptosis by the IRE1/JNK pathway of ER stress response in BHK-21 cells. Arch Virol 161:699–703. doi: 10.1007/s00705-015-2715-5 [DOI] [PubMed] [Google Scholar]
- 71. Chen C-J, Ou Y-C, Lin S-Y, Raung S-L, Liao S-L, Lai C-Y, Chen S-Y, Chen J-H. 2010. Glial activation involvement in neuronal death by Japanese encephalitis virus infection. J Gen Virol 91:1028–1037. doi: 10.1099/vir.0.013565-0 [DOI] [PubMed] [Google Scholar]
- 72. Ye J, Zhang H, He W, Zhu B, Zhou D, Chen Z, Ashraf U, Wei Y, Liu Z, Fu ZF, Chen H, Cao S. 2016. Quantitative phosphoproteomic analysis identifies the critical role of JNK1 in neuroinflammation induced by Japanese encephalitis virus. Sci Signal 9:ra98. doi: 10.1126/scisignal.aaf5132 [DOI] [PubMed] [Google Scholar]
- 73. Cai W, Pan Y, Cheng A, Wang M, Yin Z, Jia R. 2022. Regulatory role of host microRNAs in flaviviruses infection. Front Microbiol 13:869441. doi: 10.3389/fmicb.2022.869441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Majumdar A, Basu A. 2022. Involvement of host microRNAs in flavivirus-induced neuropathology: an update. J Biosci 47:54. doi: 10.1007/s12038-022-00288-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Loiselle JJ, Sutherland LC. 2018. RBM10: harmful or helpful-many factors to consider. J Cell Biochem 119:3809–3818. doi: 10.1002/jcb.26644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jung JH, Lee H, Zeng SX, Lu H. 2020. RBM10, a new regulator of p53. Cells 9:2107. doi: 10.3390/cells9092107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Inoue A. 2021. RBM10: structure, functions, and associated diseases. Gene 783:145463. doi: 10.1016/j.gene.2021.145463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sun X, Jia M, Sun W, Feng L, Gu C, Wu T. 2019. Functional role of RBM10 in lung adenocarcinoma proliferation. Int J Oncol 54:467–478. doi: 10.3892/ijo.2018.4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, Mammi P, Mancini E, Yanovsky MJ, Andino R, Krogan N, Srebrow A, Gamarnik AV. 2016. The dengue virus NS5 protein intrudes in the cellular spliceosome and modulates splicing. PLoS Pathog 12:e1005841. doi: 10.1371/journal.ppat.1005841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Saini J, Thapa U, Bandyopadhyay B, Vrati S, Banerjee A. 2023. Knockdown of NEAT1 restricts dengue virus replication by augmenting interferon alpha-inducible protein 27 via the RIG-I pathway. J Gen Virol 104. doi: 10.1099/jgv.0.001823 [DOI] [PubMed] [Google Scholar]
- 81. Carneiro PH, Mendes-Monteiro L, Mohana-Borges R. 2022. Virus propagation and plaque assay for dengue virus. Methods Mol Biol 2409:3–9. doi: 10.1007/978-1-0716-1879-0_1 [DOI] [PubMed] [Google Scholar]
- 82. Brien JD, Lazear HM, Diamond MS. 2013. Propagation, quantification, detection, and storage of West Nile virus. Curr Protoc Microbiol 31:15D.3.1-15D.3.18. doi: 10.1002/9780471729259.mc15d03s31 [DOI] [PubMed] [Google Scholar]
- 83. Sharma H, Tripathi A, Kumari B, Vrati S, Banerjee A. 2018. Artificial microrna-mediated inhibition of Japanese encephalitis virus replication in neuronal cells. Nucleic Acid Ther 28:357–365. doi: 10.1089/nat.2018.0743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Shree B, Tripathi S, Sharma V. 2021. Transforming growth factor-beta-regulated LncRNA-MUF promotes invasion by modulating the miR-34a snail1 axis in glioblastoma multiforme. Front Oncol 11:788755. doi: 10.3389/fonc.2021.788755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shree B, Sengar S, Tripathi S, Sharma V. 2023. LINC01711 promotes transforming growth factor-beta (TGF-β) induced invasion in glioblastoma multiforme (GBM) by acting as a competing endogenous RNA for miR-34a and promoting ZEB1 expression. Neurosci Lett 792:136937. doi: 10.1016/j.neulet.2022.136937 [DOI] [PubMed] [Google Scholar]
- 86. Bechara EG, Sebestyén E, Bernardis I, Eyras E, Valcárcel J. 2013. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell 52:720–733. doi: 10.1016/j.molcel.2013.11.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S34.
List of differentially expressed genes upon JEV infection in SH-SY5Y cells identified by RNA sequencing.
List of JINR1 interacting proteins identified from POSTAR database.
List of primers for qRT-PCR.
Data Availability Statement
The NCBI Gene Expression Omnibus accession number for the RNA seq data is GSE221680.