Abstract
Mycoplasma equirhinis is the predominant equine Mycoplasma sp. isolated from clinically normal horses and is suspected to be associated with inflammatory airway disease in which cough is the primary sign. Quantitative evaluation of bacterial counts is useful in assessing the association between the bacteria in samples and observed clinical signs, but this evaluation has been difficult with conventional culture methods of M. equirhinis given the need for pre-enrichment using liquid cultures. We established a quantitative real-time PCR (qPCR) assay for the quantification of M. equirhinis, targeting the hypothetical protein FJM08_00025. We confirmed its high species-specificity for M. equirhinis and a limit of detection of 2.9 copies/reaction. We quantified M. equirhinis in tracheal wash samples from 20 clinically normal horses and 22 coughing horses. The copy numbers detected by qPCR in 18 of the 22 samples from clinically affected horses were within the range detected in the 20 clinically normal horses (0–84 copies/reaction). The remaining 4 samples had considerably higher copy numbers (734–1,620,000 copies/reaction), suggesting the likely involvement of M. equirhinis infection. Quantitative evaluation of M. equirhinis over time using our qPCR assay may allow a more accurate assessment of M. equirhinis infection in coughing horses compared to culture methods.
Keywords: horses, Mycoplasma equirhinis, quantitative PCR assay
Mycoplasma spp., known for their small cellular and genomic size, are widespread among mammalian hosts and have been identified as pathogens in a variety of animal species. 12 Mycoplasma spp. are commonly isolated from horses with respiratory disease and, among Mycoplasma spp., M. equirhinis is the most frequently isolated mycoplasma from the respiratory tract of coughing horses.8,19 However, M. equirhinis can also be isolated from clinically normal horses, 17 and its pathogenic potential remains controversial. Mycoplasma infections have been suggested to be associated with the development of inflammatory airway disease in horses 19 ; however, another study found no association between Mycoplasma spp. isolation and clinical signs. 9
Serologic tests for Mycoplasma spp. can be performed in humans and animals to assess mycoplasma infection,7,10,13 but a combination of serologic testing with other tests would provide more accurate identification of mycoplasma infection. 15 Serologic tests for M. equirhinis, including the complement fixation test, indirect hemagglutination test, and ELISA, were reported in the 1970s and 1980s,5,6 but no reliable test methods are available, to date. Bacterial isolation, combined with pre-enrichment using a selective liquid medium, or conventional PCR assay are often used to detect M. equirhinis.8,17 However, because it is difficult to quantitatively assess the number of bacteria in a clinical sample by using these methods, even if M. equirhinis is detected by bacterial isolation or PCR assay, the association with clinical signs cannot be determined. 8 Quantitative real-time PCR (qPCR) assays have been used to detect various Mycoplasma spp., and their capacity for rapid quantitative evaluation is useful for directly assessing mycoplasma infection in clinical samples.1,14 Our aim was to develop and validate a qPCR assay for M. equirhinis, the Mycoplasma sp. most commonly isolated from the equine respiratory tract.
Identification of orthologous gene relationships among Mycoplasma spp. was performed to select specific genes for the development of a M. equirhinis–specific qPCR assay. Mycoplasma spp. files in FASTA format of 102 species were downloaded from the NCBI database, and their annotations were performed by using DFAST v.1.2.14. 16 SonicParanoid v.1.3.5 was used to identify orthologous genes 3 (Suppl. Table 1). We thereby identified 27 single-copy orthologous genes present only in M. equirhinis. To further select a target gene for the qPCR from among the 27 candidate genes, we checked intraspecific genetic diversity by using 28 M. equirhinis clinical strains stored in our laboratory. A gene encoding a hypothetical protein, FJM08_00025 (GenBank TPD99552.1), was selected as a target that is conserved in all M. equirhinis strains and has few single nucleotide variations. A set of primers and probe (Table 1) was designed in PCR Primer 3 v.0.4.0 software. 18 Bacterial DNA from a variety of species, as described below, was extracted with a commercial DNA extraction kit (InstaGene Matrix; Bio-Rad) in accordance with the manufacturer’s instructions. All qPCR tests were performed in triplicate (Probe qPCR mix with UNG; Takara Bio). Reaction mixtures contained 2 μL of DNA template, 10 μL of 2× Probe qPCR mix with UNG, oligonucleotide primers (0.8 µL each from 10 µM stock solution), and probe (0.2 µL from 10 µM stock solution), in a final volume of 20 μL. Amplification was performed by using an initial denaturation step of 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, and 56°C for 30 s (StepOnePlus; Thermo Fisher).
Table 1.
Primer and probe set used for Mycoplasma equirhinis detection.
| DNA oligos name | Sequence (5ʹ→3ʹ) | Location* |
|---|---|---|
| Mequi1719_for | GATTTTTGAATCCTTGCGAAT | 2716–2736 |
| Mequi1719_rev | TCCAATTTCGATTGCTTGTG | 2874–2893 |
| Mequi1719_probe | FAM-AAAGCCATTTTTAAGTCATTAA-BHQ1 | 2822–2843 |
Positions of each primer and probe from the start codon of Mycoplasma equirhinis NCTC 10148 (GenBank VFIX01000001.1).
We used 64 strains to confirm the analytical specificity of our assay: 29 strains of M. equirhinis, consisting of 1 strain of the type strain NCTC 10148 and 28 clinical strains isolated from horses in Japan; 3 strains of M. felis; 1 strain each of M. pulmonis, M. equigenitalium, M. bovis, M. pneumoniae, and Acholeplasma laidlawii; and 27 non-Mollicutes strains (Suppl. Table 2). Positive amplification of the 29 M. equirhinis strains was confirmed by qPCR. In contrast, no positive amplification occurred with the other Mycoplasma strains, the Acholeplasma strain, and the other non-Mollicutes strains. To assess nonspecific amplification by our qPCR assay in clinical samples, the nucleic acid sequences of the amplified DNA products were verified as follows. For DNA extraction, 1 mL of well-pipetted tracheal wash was centrifuged at 13,000 × g for 3 min. The supernatant was removed, and 100 µL of InstaGene Matrix was added. The procedure was then performed following the manufacturer’s instructions.
Four amplified samples from horses without M. equirhinis isolation (determined by conventional culture method), but with M. equirhinis DNA detected at 3.2, 6.1, 13.1, and 54.7 copies/reaction, were purified (ExoSAP-IT PCR product cleanup reagent; Thermo Fisher) and directly sequenced by a commercial service (Fasmac). These 4 specimens, low-load qPCR-positive and with no bacterial isolation, were selected because these specimens were less likely to have nonspecific reactions. All 4 amplicons had the target gene sequence (data not shown), indicating the high specificity of our qPCR assay, with no false positives.
To confirm the limit of detection (LOD), linearity, and efficiency of our qPCR assay, 10-fold serial dilutions of a suspension of a M. equirhinis clinical strain, Myco-24, were prepared. Bacterial DNA in the suspensions was then extracted (InstaGene matrix; Bio-Rad) in accordance with the manufacturer’s instructions. DNA concentration was quantified (Qubit 3.0 fluorometer; Thermo Fisher), and copy number was calculated. Analytical sensitivity tests were performed with triplicate samples; the lowest bacterial concentration (copies/reaction) that yielded positive results in ≥ 2 samples was considered as the LOD. As a result, the LOD of our qPCR assay was 2.9 copies/reaction (Suppl. Fig. 1A). The results also showed high linearity (R2 = 0.9998) and high efficiency (96%) with a standard curve slope of −3.42, indicating high amplification and a linear equation (Suppl. Fig. 1B).
When our qPCR assay is applied to clinical samples, it must be considered that M. equirhinis can also be detected from clinically normal horses. Because nucleic acid amplification tests, such as PCR-based assays, can detect M. equirhinis DNA even in clinically normal horse samples,8,9 quantitative assessment must be based on the DNA copy number, rather than a qualitative result. Therefore, we performed qPCR assays on tracheal wash samples from 20 clinically normal horses and 22 clinically affected horses, and compared the copy numbers. Clinically healthy or affected horses were examined by veterinarians: healthy horses were those with normal body temperatures and no clinical respiratory signs; affected horses were those with coughing as the main clinical respiratory sign, but lower respiratory tract disease such as pneumonia or pleuropneumonia had been ruled out. Tracheal wash samples were collected from 2019–2022 without duplication of subjects, and samples were stored at −20°C until DNA extraction, which was performed as described above.
Prior to DNA extraction, we thoroughly pipetted tracheal washes. Sputum has been reported to be suitable for the detection of Mycoplasma spp. in humans,2,11 and the tracheal wash samples from the clinically affected horses used in our study often contained large amounts of sticky mucus. We found that the copy number of M. equirhinis can vary widely, even within the same sample, depending on whether or not sticky mucus is included in the DNA extraction steps (data not shown). Therefore, we recommend that the sticky mucus in the equine tracheal wash sample be well homogenized, as we did, by thorough pipetting just prior to DNA extraction.
Of the tracheal wash samples collected from the 20 clinically normal horses, 10 were negative (0 copies/reaction) by qPCR assay, whereas 10 were positive (1–84 copies/reaction; Fig. 1). In 18 of the 22 tracheal wash samples from clinically affected horses, the copy numbers detected by our qPCR assay were within the copy number range of clinically normal horses (0–84 copies/reaction). In contrast, 4 samples had markedly higher copy numbers (734–1,620,000 copies/reaction). Our results using clinical samples indicated that 1) M. equirhinis DNA could be detected in clinically normal horses at relatively low copy numbers (< 100 copies/reaction); 2) some coughing horses had as few copies of M. equirhinis DNA as clinically normal horses, suggesting that their clinical signs may have been caused by pathogens other than M. equirhinis or by non-infectious factors; and 3) some samples from clinically affected horses contained markedly higher levels of M. equirhinis DNA, and there was a high level of suspicion that M. equirhinis infection was associated with these horses’ clinical signs.
Figure 1.
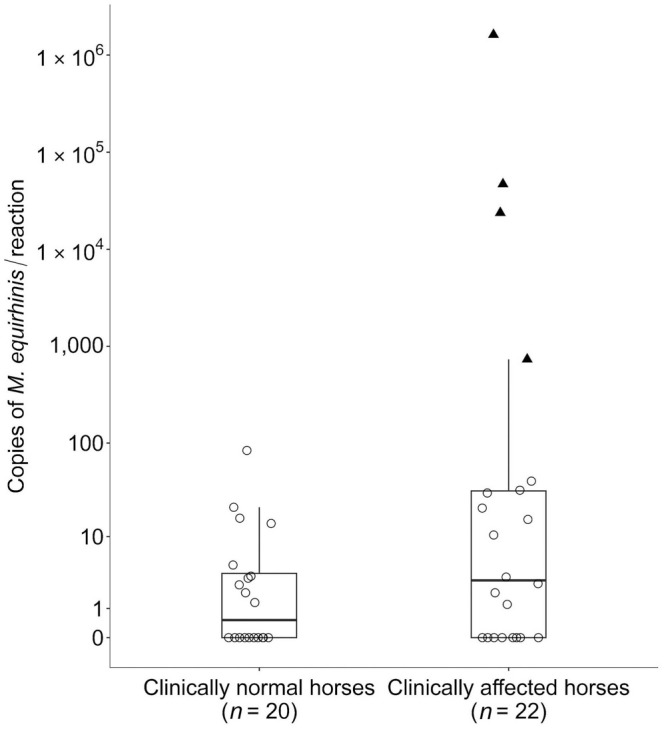
Boxplot and scatterplot comparison of copy numbers of Mycoplasma equirhinis detected in tracheal washes from clinically normal and affected horses. Box plots show the interquartile range (IQR; i.e., the bottom of the box indicates the 25th percentile, a black line inside the box indicates the median, and the top of the box indicates the 75th percentile). The whisker extending above the box indicates a range of the 3rd quartile + 1.5 × IQR. Each scatterplot represents a single sample. Samples that fall within the range of clinically normal horses (≤ 84 copies/reaction) are shown as open circles, and those above are shown as black triangles.
The results of our qPCR assay of samples from the clinically normal and affected horse are consistent with the fact that M. equirhinis can be isolated from clinically normal horses 17 and that there are both infectious and non-infectious causes of coughing.4,19 As well, a high copy number does not mean that M. equirhinis is definitively the cause of the clinical signs. Therefore, judgments about whether M. equirhinis infection is the cause of the disease should be combined with the detection of, or failure to detect, other pathogens, with or without consistency between the clinical signs and copy number changes over time.
Following is a clinical case that we experienced in which we tested samples with our M. equirhinis–specific qPCR assay over time. The 22 clinically affected horses described above did not include this horse. A horse had been coughing for ~10 d, but other clinical signs, such as nasal discharge, body temperature, and appetite, were absent or very mildly changed. Veterinary management was initiated with 3 tracheal wash samplings and 3 endoscopic examinations. The frequency of coughing and the amount of yellow tracheal mucus were both linked to an increase or decrease in M. equirhinis copy number, which increased from 150 copies/reaction at initial examination to 16,200 copies/reaction at 12 d, when the clinical signs were most severe, to 2 copies/reaction at 20 d as the clinical signs abated. In this case, no notable bacteria other than the genus Mycoplasma were isolated from the tracheal washes, and no Getah virus, equine influenza A viruses, or equine rhinitis viruses A or B were detected, strongly suggesting that the clinical signs were caused by M. equirhinis infection. Note that, in this case, the clinical signs improved after the start of nebulized 100 mg of minocycline hydrochloride, although its efficacy must be verified in the future.
A limitation of our study is that, except for the one clinical case described, we were unable to evaluate the association of pathogens other than M. equirhinis. Whether there is a difference in the detection rate or copy number of M. equirhinis depending on the presence or absence of other pathogens, or whether there is a difference in pathogenicity between single and mixed infections, remains to be investigated in future studies.
Supplemental Material
Supplemental material, sj-pdf-1-vdi-10.1177_10406387231207631 for A real-time PCR assay for the quantification of Mycoplasma equirhinis in tracheal wash samples from Thoroughbred horses by Yuta Kinoshita, Hidekazu Niwa, Eri Uchida-Fujii and Takanori Ueno in Journal of Veterinary Diagnostic Investigation
Acknowledgments
We thank Michiyo Yamazaki (Equine Research Institute, Japan Racing Association) for her valuable technical help.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors declare that they received no financial support for their research and/or authorship of this article.
ORCID iDs: Yuta Kinoshita  https://orcid.org/0000-0002-4581-9322
https://orcid.org/0000-0002-4581-9322
Eri Uchida-Fujii  https://orcid.org/0000-0002-4127-1435
https://orcid.org/0000-0002-4127-1435
Supplemental material: Supplemental material for this article is available online.
References
- 1. Ade J, et al. Quantitative analysis of Mycoplasma wenyonii and ‘Candidatus Mycoplasma haemobos’ infections in cattle using novel gapN-based realtime PCR assays. Vet Microbiol 2018;220:1–6. [DOI] [PubMed] [Google Scholar]
- 2. Cho M-C, et al. Comparison of sputum and nasopharyngeal swab specimens for molecular diagnosis of Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Legionella pneumophila. Ann Lab Med 2012;32:133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cosentino S, Iwasaki W. SonicParanoid: fast, accurate and easy orthology inference. Bioinformatics 2019;35:149–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Couëtil LL, et al. Inflammatory airway disease of horses–revised consensus statement. J Vet Intern Med 2016;30:503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hooker JM, Butler M. The development of complement-fixing antibody in horses after infection with Mycoplasma equirhinis. J Comp Pathol 1977;87:281–286. [DOI] [PubMed] [Google Scholar]
- 6. Kirchhoff H, et al. Serological investigation of horse sera for antibodies against mycoplasmas and acholeplasmas. Vet Microbiol 1982;7:147–156. [DOI] [PubMed] [Google Scholar]
- 7. Lee W-J, et al. Role of serum Mycoplasma pneumoniae IgA, IgM, and IgG in the diagnosis of Mycoplasma pneumoniae-related pneumonia in school-age children and adolescents. Clin Vaccine Immunol 2017;24:e00471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martineau M, et al. Detection of Mycoplasma spp. in horses with respiratory disorders. Equine Vet J 2023;55:747–754. [DOI] [PubMed] [Google Scholar]
- 9. Mete A, Özgür NY. Investigation of the presence of Mycoplasma as an etiologic agent of inflammatory airway diseases in thoroughbred racehorses in İstanbul Province. Turk J Vet Anim Sci 2017;41:365–371. [Google Scholar]
- 10. Peyraud A, et al. An international collaborative study to determine the prevalence of contagious caprine pleuropneumonia by monoclonal antibody-based cELISA. BMC Vet Res 2014;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Räty R, et al. Sample type is crucial to the diagnosis of Mycoplasma pneumoniae pneumonia by PCR. J Med Microbiol 2005;54:287–291. [DOI] [PubMed] [Google Scholar]
- 12. Razin S, et al. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev 1998;62:1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Register KB, et al. Serological evidence for historical and present-day exposure of North American bison to Mycoplasma bovis. BMC Vet Res 2021;17:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sonalio K, et al. Genetic diversity of emerging hemotropic mycoplasmas in domestic pigs from Brazil. Transbound Emerg Dis 2021;68:1162–1174. [DOI] [PubMed] [Google Scholar]
- 15. Tang M, et al. Comparison of different detection methods for Mycoplasma pneumoniae infection in children with community-acquired pneumonia. BMC Pediatr 2021;21:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tanizawa Y, et al. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018;34:1037–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uchida-Fujii E, et al. High prevalence of Mycoplasma equirhinis in Thoroughbred horses with respiratory symptoms in autumn 2018. J Vet Med Sci 2021;83:1907–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Untergasser A, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res 2012;40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wood JLN, et al. Association between respiratory disease and bacterial and viral infections in British racehorses. J Clin Microbiol 2005;43:120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-vdi-10.1177_10406387231207631 for A real-time PCR assay for the quantification of Mycoplasma equirhinis in tracheal wash samples from Thoroughbred horses by Yuta Kinoshita, Hidekazu Niwa, Eri Uchida-Fujii and Takanori Ueno in Journal of Veterinary Diagnostic Investigation


