Abstract
Acetylation is an important posttranslational modification (PTM) that regulates almost all core processes of autophagy in yeast and mammals. However, the role of protein acetylation in plant autophagy and the underlying regulatory mechanisms remain unclear. Here, we show the essential role of the putative acetyltransferase HOOKLESS1 (HLS1) in acetylation of the autophagy-related protein ATG18a, a key autophagy component that regulates autophagosome formation in Arabidopsis (Arabidopsis thaliana). Loss of HLS1 function suppressed starvation-induced autophagy and increased plant susceptibility to nutrient deprivation. We discovered that HLS1 physically interacts with and directly acetylates ATG18a both in vitro and in vivo. In contrast, mutating putative active sites in HLS1 inhibited ATG18a acetylation and suppressed autophagy upon nutrient deprivation. Accordingly, overexpression of ATG18a mutant variants with lower acetylation levels inhibited the binding activity of ATG18a to PtdIns(3)P and autophagosome formation under starvation conditions. Moreover, HLS1-modulated autophagy was uncoupled from its function in hook development. Taken together, these findings shed light on a key regulator of autophagy and further elucidate the importance of PTMs in modulating autophagy in plants.
HOOKLESS1 is a bona fide lysine acetyltransferase in Arabidopsis that regulates activation of nutrient starvation-induced autophagy by interacting with and acetylating AUTOPHAGY-RELATED PROTEIN18a.
IN A NUTSHELL.
Background: Autophagy is a highly conserved process that delivers cytoplasmic components to the vacuole or lysosome for breakdown and recycling during stresses such as nutrient starvation. Almost every pivotal process of autophagy in yeast and mammals is regulated by an important posttranslational modification (PTM) called protein acetylation. Nevertheless, whether and how core autophagy proteins in plants are regulated by acetylation remains elusive. HOOKLESS1 (HLS1) is a putative N-acetyltransferase, but its biochemical function has remained largely unclear. The loss-of-function mutant hls1-1 displays similar phenotypes to autophagy-defective (atg) mutants in senescence and immune responses, suggesting that there may be a relationship between HLS1 and plant autophagy.
Question: Is HLS1 involved in plant autophagy? Does HLS1 regulate autophagy? How does HLS1 function in autophagy?
Findings: We mainly uncovered the following 4 findings: (i) HLS1 is crucial for triggering autophagy during nutrient starvation in Arabidopsis (Arabidopsis thaliana); (ii) HLS1 is a bona fide lysine acetyltransferase that can directly physically interact with and acetylate a key autophagy-related protein (ATG18a) in response to nutrient starvation; (iii) HLS1-mediated ATG18a acetylation affects the ATG2–ATG18a interaction and the binding of ATG18a to phosphatidylinositol 3-phosphate to promote autophagy activation and plant responses to nutrient deprivation; and (iv) the normal enzymatic activity of HLS1 is also important for apical hook development of etiolated seedlings, but HLS1-regulated autophagy by acetylating ATG18a is uncoupled from HLS1-mediated hook formation.
Next steps: We aim to explore how nutrient starvation modulates HLS1-mediated autophagy via ATG18a acetylation. Moreover, additional HLS1 substrates in multiple biological processes such as hook development are worthy of further investigation.
Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved degradative mechanism in eukaryotic cells that employs double-membrane vesicles called autophagosomes to encapsulate and deliver unnecessary or dysfunctional cytoplasmic components to vacuoles (in yeasts and plants) or lysosomes (in animals) for breakdown and recycling (Han et al. 2011; Li and Vierstra 2012; Liu and Bassham 2012; Michaeli et al. 2016; Marshall and Vierstra 2018; Qi et al. 2021). Autophagy occurs constitutively at a basal level for maintenance of cellular homeostasis and is induced by various stresses, including nutrient starvation, oxidative stress, heat, cold, drought, high salt, hypoxia, endoplasmic reticulum (ER) stress, and pathogen infections, leading to the degradation of intracellular materials into metabolites to support cell survival (Doelling et al. 2002; Xiong et al. 2005; Bassham et al. 2006; Xiong et al. 2007; Liu et al. 2009; Lai et al. 2011; Liu et al. 2012; Zhou et al. 2014; Chen et al. 2015; Luo et al. 2017; Islam et al. 2019; Bao et al. 2020; Chi et al. 2020; Zhang et al. 2021; Tang and Bassham 2022). As an important protein degradation pathway in plants, autophagy is necessary for seedling establishment, root meristem maintenance, senescence, response to stresses, metabolism, and reproduction (Michaeli et al. 2016; Marshall and Vierstra 2018; Signorelli et al. 2019; Huang et al. 2019a).
The occurrence of autophagy requires the coordination of multiple autophagy-related (ATG) proteins that function at different stages. To date, over 40 ATG proteins have been discovered in yeast (Saccharomyces cerevisiae), and their orthologs have subsequently been identified in animals and plants (Feng et al. 2014; Wen and Klionsky 2016; Marshall and Vierstra 2018; Qi et al. 2021). These proteins assemble into 4 complexes: (i) the ATG1–ATG13 protein kinase complex initiates autophagy (Suttangkakul et al. 2011; Li et al. 2014); (ii) the ATG9–ATG2–ATG18 complex mediates membrane enlargement and promotes phagophore nucleation and expansion (Xiong et al. 2005; Zhuang et al. 2017; Kang et al. 2018); (iii) the class III phosphatidylinositol 3-kinase (PI3K) complex mediates vesicle nucleation (Welters et al. 1994; Fujiki et al. 2007; Wang et al. 2012; Qi et al. 2017); and (iv) 2 ubiquitin-like conjugation complexes, ATG12–ATG5 (Doelling et al. 2002; Thompson et al. 2005; Phillips et al. 2008) and ATG8–phosphatidylethanolamine (PE) (Yoshimoto et al. 2004; Chung et al. 2010; Zhuang et al. 2013), regulate autophagosome formation. Ultimately, lipidated ATG8 located in the outer membrane of autophagosomes is released for recycling and to complete the formation of the autophagosome via delipidation by ATG4, whereas ATG8–PE embedded in the inner membrane is degraded in the vacuole (Woo et al. 2014).
In yeast, Atg18 is essential for autophagy as a component of the core autophagic machinery and functions as an effector of phosphatidylinositol 3,5-bisphosphate (PtdIns(3,5)P2) and phosphatidylinositol 3-phosphate (PtdIns(3)P) (Dove et al. 2004; Obara et al. 2008). In Arabidopsis (Arabidopsis thaliana), 8 proteins share sequence similarity with yeast Atg18, namely, ATG18a to ATG18h, although only ATG18a has been chosen for study because ATG18a is a unique gene whose expression can be induced by nutrition starvation and senescence (Xiong et al. 2005). Knockdown mutants of ATG18a are unable to accumulate autophagosomes and exhibit increased susceptibility to nutrient starvation, oxidative stress, and infection by necrotrophic pathogens (Xiong et al. 2005, 2007; Lai et al. 2011; Zhang et al. 2021). However, the underlying mechanisms by which ATG18a functions in nutrient starvation remain unclear.
Over the past decades, multiple posttranslational modifications (PTMs) have been shown to regulate autophagy (Füllgrabe et al. 2013; Wani et al. 2015; Qi et al. 2021). In yeast and mammals, autophagy is mainly regulated by 3 classes of PTMs, namely, phosphorylation, ubiquitination, and acetylation (McEwan and Dikic 2011; Bánréti et al. 2013). Among these PTMs, acetylation is an important and highly conserved modification that transfers the acetyl group of acetyl-CoA to the N-terminus or lysine residues in target proteins (Choudhary et al. 2009, 2014; A et al. 2020). Reversible lysine acetylation can take place on both histones and nonhistones and is tightly controlled by lysine acetyltransferases (KATs) and deacetylases (KDACs) within cells (Choudhary et al. 2009, 2014; A et al. 2020). Thus far, KAT families have been mainly divided into 3 classes: the GNAT (Gcn5-related N-acetyltransferase) family, the p300/CBP (CREB-binding protein) family, and the MYST (MOZ, Ybf2/Sas3, Sas2, and TIP60) family (Sterner and Berger 2000; Bánréti et al. 2013; Choudhary et al. 2014).
Increasing evidence has revealed that acetylation is involved in the regulation of autophagy, including its initiation, elongation, and fusion steps. In human cells, the acetyltransferase p300 interacts with ATG7 and inhibits autophagy by acetylating the key autophagy proteins ATG5, ATG7, ATG8, and ATG12 under nutrient-rich conditions (Lee and Finkel 2009). In contrast, deacetylation of ATG5, ATG7, and ATG8 by the NAD-dependent deacetylase Sirtuin 1 (Sirt1) stimulates autophagosome formation in mammals during starvation (Lee et al. 2008). Moreover, deacetylation of the microtubule-associated protein 1 light chain 3 (LC3), a mammalian homolog of yeast and plant ATG8, at lysine 49 (K49) and K51 by Sirt1 is required for LC3–ATG7 interaction, which then drives autophagy initiation upon starvation (Huang et al. 2015). Upon deprivation of growth factors, the autophagy initiation kinase ULK1 (unc-51-like kinase1), a mammalian homolog of yeast and plant ATG1, is acetylated by acetyltransferase TIP60 (HIV-1 Tat-interactive protein, 60 kD, from the MYST family) and induces autophagy (Lin et al. 2012). In yeast, the acetyltransferase Esa1 (essential SAS2-related acetyltransferase 1), which also belongs to the MYST family, acetylates Atg3 at K19 and K48, and higher K19–K48 acetylation levels enhance autophagy (Yi et al. 2012).
Arabidopsis HOOKLESS1 (HLS1) contains putative acetyltransferase domains conserved among members of the GNAT superfamily (Lehman et al. 1996; Neuwald and Landsman 1997), but its putative biochemical function has not been well characterized. Generally, HLS1 is a key regulator of apical hook formation in etiolated seedlings (Guzmán and Ecker 1990; Lehman et al. 1996). Loss-of-function hls1 mutants exhibit a typical hookless phenotype and are insensitive to exogenous application of ethylene for hook formation (Guzmán and Ecker 1990). HLS1 was also recently shown to play an important role in plant development and immunity. Indeed, hls1 mutants display early senescence and hypersensitivity to infection by the fungus Botrytis cinerea, and they are insensitive to thermomorphogenesis (Liao et al. 2016; Jin and Zhu 2019). Moreover, HLS1 modulates the expression of WRKY33 and ABA-INSENSITIVE5 (ABI5) by regulating histone H3 acetylation on the chromatin, but HLS1 did not directly acetylate histones in vitro (Liao et al. 2016). In another study, the transcription factor WRKY33 appeared to directly interact with ATG18a, the key protein of the autophagy pathway, to coordinately regulate plant resistance to necrotrophic fungal pathogens (Lai et al. 2011). Overall, the senescence and pathogen resistance phenotypes of hls1 mutants are very similar to those caused by autophagy deficiency, providing a hint that HLS1 may be involved in plant autophagy.
In this study, we show that HLS1 modulates autophagy via acetylation of ATG18a under nutrient starvation conditions. Our results uncover how HLS1 directly interacts with and acetylates ATG18a in vitro and in vivo. HLS1 genetic inactivation or mutations of the potential acetylation sites in ATG18a reduced ATG18a acetylation levels and increased plant susceptibility to carbon and nitrogen starvation. These data reveal the acetyltransferase activity of HLS1 and unravel an additional regulatory mechanism for autophagy in Arabidopsis. Moreover, we reveal that HLS1-regulated autophagy is uncoupled from its crucial function in hook formation.
Results
Loss of HLS1 function results in hypersensitivity to nutrient starvation
Recent studies have revealed that HLS1 participates in plant development and immune responses besides its importance in regulating hook formation (Liao et al. 2016; Jin and Zhu 2019). Indeed, loss-of-function hls1 mutants exhibit early senescence and increased susceptibility to B. cinerea, which is reminiscent of Arabidopsis autophagy-defective (atg) mutants (Hanaoka et al. 2002; Lai et al. 2011). Thus, we speculated that HLS1 may be involved in autophagy. Considering that the atg mutants commonly exhibit hypersensitivity to nutrient starvation (Doelling et al. 2002; Thompson et al. 2005; Phillips et al. 2008), we tested the sensitivity of the hls1-1 mutant to nutrient deprivation.
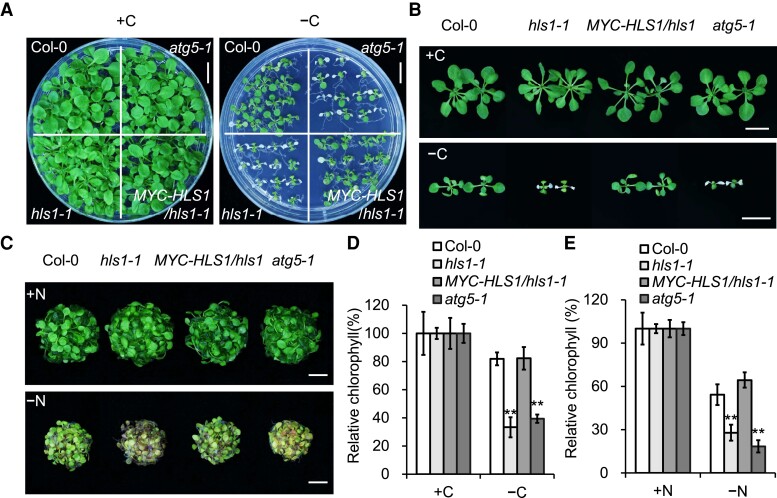
When grown under nutrient-rich conditions (MS growth medium supplemented with 1% [w/v] sucrose), the growth of the hls1-1 mutant appeared similar to that of the wild type (Col-0) (Fig. 1, A and B). In contrast, like the strong autophagy mutant atg5-1 (Thompson et al. 2005; Phillips et al. 2008), the hls1-1 mutant showed a hypersensitive phenotype to fixed-carbon starvation (growth on MS medium without sucrose and in the dark), with significantly lower relative chlorophyll contents, compared to Col-0 (Fig. 1, A, B, and D). Moreover, following nitrogen starvation for 6 d, the hls1 mutant exhibited increased yellowing of cotyledons and lower chlorophyll contents relative to Col-0 seedlings (Fig. 1, C and E). Together, the sensitivity of the hls1 mutant to the nutrient starvation conditions examined in this study was comparable to that of the well-characterized atg5-1, suggesting that HLS1 is crucial in regulating plant responses to nutrient starvation.
Figure 1.
Mutation of HLS1 confers hypersensitivity to nutrient starvation. A, B) Phenotypes of wild type (Col-0), hls1-1, MYC–HLS1/hls1-1, and atg5-1 mutants in response to carbon starvation. One-week-old seedlings were transferred to MS agar with sucrose (+C) or MS agar plates without sucrose under constant dark treatment (−C) for 9 d. The photographs were taken after a 9-d recovery. Bars = 1 cm. C) Phenotypes of Col-0, hls1-1, MYC–HLS1/hls1-1, and atg5-1 mutants in response to nitrogen starvation. One-week-old seedlings were transferred to N-rich (+N) or N-deficient (−N) liquid medium and photographed at 6 d after treatment. Bars = 1 cm. D, E) Relative chlorophyll content in A) and C), respectively. The relative chlorophyll contents are expressed relative to those of the genotypes on +C A) or +N C) medium. Three independent experiments were performed with similar results. Values are means ± Sd (n = 4 replicates) from one experiment. For each experiment, leaves from 8 seedlings were used per one replicate. Asterisks indicate significant differences from the Col-0 (**P < 0.01, Student's t test).
To verify that the hypersensitivity of the hls1-1 mutant to nutrient limitation was due to the dysfunction of HLS1 protein, we introduced the cauliflower mosaic virus (CaMV) 35S promoter–derived MYC-tagged HLS1 into the hls1-1 mutant to obtain Pro35S:MYC–HLS1/hls1-1 (MYC–HLS1/hls1-1) transgenic plants and then examined the tolerance of MYC–HLS1/hls1-1 seedlings to nutrient deprivation. Overexpression of HLS1 restored the hypersensitive phenotype of the hls1-1 mutant under carbon or nitrogen starvation conditions to that seen for wild-type seedlings, as supported by the higher relative chlorophyll contents of these transgenic seedlings compared to hls1-1 (Fig. 1). Collectively, these findings indicate that HLS1 is required for plant response to nutrient limitation.
HLS1 is crucial for nutrient starvation–induced autophagic flux
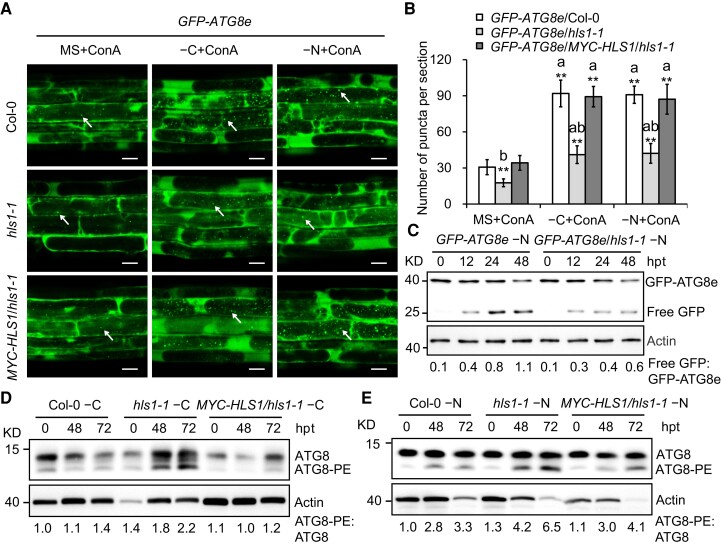
To investigate the potential role of HLS1 in autophagy, we crossed the GFP–ATG8e transgenic line, a well-characterized autophagosome marker line (Contento et al. 2005; Xiao et al. 2010), to the hls1-1 mutant and transgenic MYC–HLS1/hls1-1 to generate GFP–ATG8e/hls1-1 and GFP–ATG8e/MYC–HLS1/hls1-1 plants, respectively. Nutrient starvation induces the accumulation of many GFP–ATG8e-labeled punctate structures (autophagosomes or their intermediates) upon autophagy activation (Qi et al. 2017; Huang et al. 2019b). Accordingly, the Col-0, hls1-1, and MYC–HLS1/hls1-1 seedlings expressing GFP–ATG8e were grown on MS medium with sucrose for 5 d, after which we transferred the seedlings to carbon-limited (−C) or nitrogen-limited (−N) MS medium containing the autophagy inhibitor concanamycin A (ConA; Svenning et al. 2011) for 6 h. We then examined seedlings by confocal laser scanning microscopy to observe autophagosomes. After carbon or nitrogen starvation for the indicated times, we determined that the accumulation of GFP–ATG8e-labeled punctate autophagosomes markedly increases (∼300% after both carbon and nitrogen starvation) in Col-0 root cells, whereas this accumulation was more moderate in the hls1-1 background under either starvation condition tested. Consistent with the phenotypic analysis, overexpression of HLS1 restored the autophagosome accumulation in the root cells of hls1-1 mutant (Fig. 2, A and B).
Figure 2.
HLS1 is crucial for autophagic flux during nutrient starvation. A) Confocal analysis of GFP–ATG8e/Col-0, GFP–ATG8e/hls1-1, and GFP–ATG8e/MYC–HLS1/hls1-1 lines. Five-day-old seedlings were exposed to carbon (C)- and nitrogen (N)-sufficient (MS) or C- and N-deficient (−C and −N) liquid medium with the addition of 0.5 μm ConA for 6 h and then visualized by confocal laser scanning microscopy. The arrows indicate autophagic bodies. ConA, concanamycin A. Bars = 20 μm. B) Numbers of puncta per section in the root cells of the GFP–ATG8e/Col-0, GFP–ATG8e/hls1-1, and GFP–ATG8e/MYC–HLS1/hls1-1 in A). Three independent experiments were done with similar results. Values are means ± Sd (n = 15 sections) from one representative experiment. Asterisks with “a” indicate significant differences when compared to that of MS + ConA; asterisks with “b” indicates significant differences from that of the GFP–ATG8e/Col-0, respectively. Asterisks indicate significant differences (**P < 0.01, Student's t test). C) Immunoblotting showing the free GFP released during the cleavage of GFP–ATG8e reporter in response to nitrogen starvation (−N). One-week-old seedlings expressing GFP–ATG8e were exposed to −N liquid medium for the indicated times. Anti-GFP antibody was to examine the GFP–ATG8e fusion and free GFP levels. The Actin protein bands are shown below as the loading control. The ratio of free GFP to GFP–ATG8e is shown below. hpt, hours posttreatment. The experiment was repeated 3 times with similar results. D, E) ATG8 lipidation level in the Col-0, hls1-1, and MYC–HLS1/hls1-1 lines after carbon starvation (−C, D) or nitrogen starvation (−N, E) treatment for the indicated times. Anti-ATG8a antibody was used to detect the ATG8 and ATG8–PE protein levels. Relative intensity of ATG8–PE band normalized to the ATG8 is shown below. The relative intensities of each band on immunoblots were quantified using ImageJ. PE, phosphatidylethanolamine; hpt, hours posttreatment. The experiment was repeated 3 times with similar results.
We also evaluated autophagic flux by the examination of the release of free GFP from the GFP–ATG8e fusion protein. Upon induction of autophagy, GFP–ATG8e is rapidly digested to release free and relatively stable GFP once inside vacuoles; thus, the ratio between free GFP and GFP–ATG8e can reflect the rate of autophagy (Contento et al. 2005; Chung et al. 2010; Huang et al. 2019a). In agreement with the microscopy results, we detected substantial free GFP derived from GFP–ATG8e in the Col-0 background under nitrogen starvation conditions but obtained a lower GFP/GFP–ATG8e ratio in the hls1-1 mutant background under the same conditions (Fig. 2C). These findings suggest that HLS1 is crucial for autophagic flux.
Since conjugation of ATG8–PE occurs during autophagy activation and the ATG8–PE level has been widely regarded as another reliable indicator to assess the strength of autophagic activity in yeast and animals (Rubinsztein et al. 2009; Chung et al. 2010), we examined ATG8 and ATG8–PE abundance in wild-type and hls1-1 mutant transgenic lines carrying the MYC–HLS1 transgene using anti-ATG8a antibody. Interestingly, the ratio of ATG8–PE to ATG8 in hls1-1 mutant was higher than that in wild type upon carbon or nitrogen starvation for 48 and 72 h (Fig. 2, D and E), which is reminiscent of the hyperaccumulation of ATG8–PE in autophagy-defective mutant previously described (Zhuang et al. 2017; Kang et al. 2018). Consistent with the observed phenotypic rescue by the MYC–HLS1 transgene, ATG8–PE abundance was not evidently different between the MYC–HLS1/hls1-1 line and Col-0 (Fig. 2, D and E).
We next tested the expression of several ATG genes in Col-0 and the hls1-1 mutant when seedlings suffered from carbon starvation. Reverse transcription quantitative PCR (RT-qPCR) analysis revealed that relative ATG transcript levels induced by carbon starvation are comparable in the hls1-1 mutant and Col-0 (Supplemental Fig. S1A), suggesting that HLS1 is not involved in the transcriptional regulation of ATG genes. Taken together, these results suggest that HLS1 is crucial for nutrient starvation–induced autophagy in Arabidopsis.
HLS1 physically interacts with ATG18a
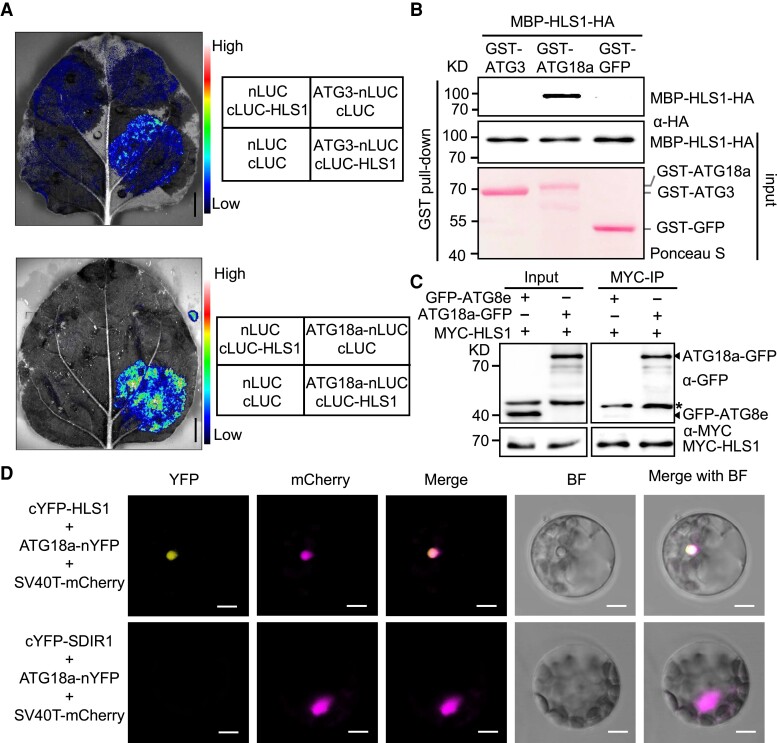
As HLS1 appeared to play an important role in autophagy, we hypothesized that HLS1 might associate or even directly interact with autophagy components. To test this possibility, we conducted split-luciferase (LUC) complementation assays to test the potential for interaction between known ATG proteins and HLS1. For this purpose, we generated constructs encoding full-length HLS1, ATG1a, ATG1b, ATG1c, ATG3, ATG5, ATG6, ATG7, ATG12, ATG18a, and ATG18b individually fused to the N-terminus of LUC (HLS1–nLUC and ATGs–nLUC), together with constructs encoding the LUC C-terminus fused to HLS1 (cLUC–HLS1) or ATG8e (cLUC–ATG8e). We then coinfiltrated appropriate nLUC and cLUC pairs into Nicotiana benthamiana leaves via Agrobacterium (Agrobacterium tumefaciens)–mediated infiltration and looked for reconstitution of LUC, as evidenced by luminescence.
Compared to the negative controls nLUC + cLUC and nLUC + cLUC–HLS1, we only detected luminescence when cLUC–HLS1 was coexpressed with ATG3–nLUC and observed strong luminescence for the ATG18a–nLUC + cLUC–HLS1 pair (Fig. 3A; Supplemental Fig. S2A). ATG18a belongs to an 8-protein family in Arabidopsis, prompting us to also examine the potential interaction between HLS1 and the other ATG18 members. Importantly, HLS1 specifically interacted only with ATG18a but not with the other ATG18 members (Supplemental Fig. S2B). To verify the physical interaction between HLS1 and ATG3 or ATG18a, we performed an in vitro pull-down assay. We produced and purified recombinant glutathione S-transferase (GST)–tagged GFP (GST–GFP), GST–ATG3, GST–ATG18a, and maltose-binding protein (MBP)– tagged HLS1 (MBP–HLS1) proteins in Escherichia coli. We found that GST–ATG18a, but neither GST–ATG3 nor GST–GFP, was able to pull down recombinant MBP–HLS1 (Fig. 3B), suggesting that HLS1 directly interacts with ATG18a in vitro. We further confirmed the interaction between HLS1 and ATG18a by coimmunoprecipitation (Co-IP) assay in transgenic Arabidopsis lines stably coexpressing MYC–HLS1 and ATG18a–GFP (Fig. 3C).
Figure 3.
HLS1 interacts with ATG18a in vitro and in vivo. A) Split-LUC complementation imaging assay to analyze the potential interaction between HLS1 and ATG proteins (ATG3 and ATG18a) in N. benthamiana leaves. Full-length ATG3 or ATG18a was fused with the LUC N-terminus (nLUC), and full-length HLS1 was fused with the LUC C-terminus (cLUC). Agrobacterium strain GV3101 harboring different construct combinations was injected into different N. benthamiana leaf regions. After 2 d of injection, LUC activities were examined in these regions, as indicated by the color-coded bar next to the images. Three biological replicates were done with similar results. Bars = 1 cm. B) Pull-down assay showing the in vitro interaction between HLS1 and ATG proteins. GST-tagged ATG3 (GST–ATG3), GS–ATG18a, and GST–GFP (negative control) were expressed in E. coli BL21 (DE3) cells and immobilized with GST beads then combined with MBP–HLS1–HA protein. Anti-HA antibody was used for protein detection. Ponceau staining indicates the loading of GST-tagged proteins. C) In vivo Co-IP analysis of the interaction between ATG proteins (ATG8e and ATG18a) and HLS1. MYC-tagged HLS1 (MYC–HLS1) was coexpressed in transgenic plants expressing GFP–ATG8e or ATG18a–GFP and immunoprecipitated by MYC nanobody coated agarose beads. GFP–ATG8e as the negative control. Anti-GFP and anti-MYC antibodies were used for protein detection. Asterisk indicates nonspecific bands. D) HLS1 interacts with ATG18a in the nucleus. cYFP–HLS1 or cYFP–SDIR1 was coexpressed with ATG18a–nYFP in Col-0 protoplasts and colocalized with the nuclear localization marker SV40T–mCherry. The fluorescence was observed after culturing under low light for 16 h. The vector pairs cYFP–SDIR1 + ATG18a–nYFP + SV40T–mCherry were cotransfected as negative control. BF, bright field. Bars = 20 μm.
Besides, we performed bimolecular fluorescence complementation (BiFC) assay in Col-0 protoplasts to determine the subcellular localization of the interaction between HLS1 and ATG18a. As shown in Fig. 3D, coexpressing cYFP–HLS1 and ATG18a–nYFP, but not cYFP–SDIR1 and nYFP–ATG18a, reconstituted YFP signals that colocalizing with the nuclear marker SV40T–mCherry (Jin et al. 2016). Together, these findings demonstrate that HLS1 physically interacts with ATG18a in planta and in vitro, raising the possibility that ATG18a function might be directly modulated by HLS1 during autophagy.
HLS1 acetylates ATG18a in vitro and in vivo
To unravel the regulatory relationship between HLS1 and ATG18a, we investigated the effect of HLS1 on ATG18a protein abundance. To this end, we introduced a Pro35S:ATG18a–GFP transgene into the wild type to obtain 35Spro:ATG18a–GFP/Col-0 (ATG18a–GFP/Col-0) line and generated the ATG18a–GFP/hls1-1 line by crossing the hls1-1 mutant to the ATG18a–GFP transgenic line. In line with the relative ATG18a transcript levels in Col-0 and the hls1-1 mutant, the accumulation of ATG18a–GFP induced by carbon starvation was not affected by the loss of HLS1 function (Supplemental Fig. S1B). This result suggests that HLS1 likely affects ATG18a activity through PTMs, rather than from simple transcriptional or translational regulation.
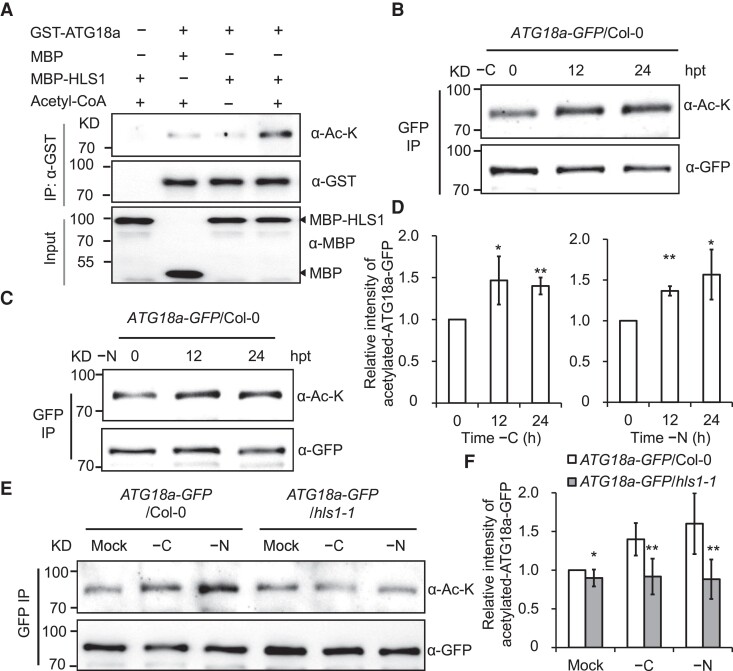
HLS1 has previously been proposed to be a putative N-acetyltransferase because of the high similarity displayed by its N-terminal 158 amino acids to N-acetyltransferases from yeast, bacteria, and mammals (Guzmán and Ecker 1990; Lehman et al. 1996). However, the acetyltransferase activity of HLS1 is not well characterized. We therefore speculated that HLS1 might transfer acetyl groups to lysine residues of ATG18a. To test this idea, we performed an in vitro acetylation assay with recombinant proteins incubated in the presence of the acetyl donor acetyl-CoA. Indeed, we detected the acetylation of recombinant GST–ATG18a by MBP–HLS1, but not by MBP alone, in the presence of acetyl-CoA (Fig. 4A), indicating that HLS1 functions as a bona fide protein KAT, with ATG18a being one of its substrates.
Figure 4.
HLS1 acetylates ATG18a in vitro and in vivo. A) HLS1 can acetylate ATG18a in vitro. The recombinant proteins GST–ATG18a and MBP–HLS1 were immunoprecipitated by GST and MBP beads, respectively. Anti-acetyl-lysine (α-Ac-K) and anti-GFP (α-GFP) antibodies were used to detect the acetylation level and the loading of ATG18a, respectively. Anti-MBP antibody was used to detect the loading of MBP and MBP–HLS1 proteins. IP, immunoprecipitation. B, C) Acetylation of ATG18a–GFP in ATG18a–GFP/Col-0 transgenic seedlings after fixed-C starvation (−C, B) or nitrogen starvation (−N, C) treatment for the indicated times. hpt, hours posttreatment. D) Quantification of the relative intensity of acetylated ATG18a–GFP during carbon starvation (−C, left) in B) or nitrogen starvation (−N, right) in C). Data are means ± Sd calculated from 3 independent experiments. Asterisks indicate significant differences from the 0 h (*P < 0.05; **P < 0.01, Student's t test). E) Acetylation of ATG18a–GFP in ATG18a–GFP/Col-0 and ATG18a–GFP/hls1-1 transgenic seedlings after −C or −N treatment for 24 h. ATG18a–GFP was immunoprecipitated by GFP beads. α-Ac-K and α-GFP antibodies were used to detect the acetylation level and the loading of ATG18a–GFP, respectively. F) Relative intensity of acetylated ATG18a–GFP in E). Data are means ± Sd calculated from 3 independent experiments. Asterisks indicate significant differences from ATG18a–GFP/Col-0 (*P < 0.05; **P < 0.01, Student's t test).
To determine whether ATG18a protein was acetylated by HLS1 in planta, we grew stable transgenic seedlings expressing ATG18a–GFP in the Col-0 and hls1-1 backgrounds and subjected them to nutrient starvation for up to 24 h. At each of the indicated time points, we immunoprecipitated ATG18a–GFP with anti-GFP nanobody agarose beads and determined its acetylation level with a specific anti-acetyl-K antibody. Compared with no substantial changes in acetylation of GFP (Supplemental Fig. S3), we observed an increase in acetylation levels of ATG18a–GFP in Col-0 seedlings over time following carbon starvation (Fig. 4, B and D) and nitrogen starvation (Fig. 4, C and D). In contrast, the acetylation level of ATG18a–GFP remained relatively constant in the hls1-1 mutant background under both carbon and nitrogen starvation conditions (Fig. 4, E and F). Collectively, these results confirm that ATG18a is acetylated by HLS1, and this acetylation is promoted when seedlings are subjected to nutritional starvation.
ATG18a acetylation enhances ATG2–ATG18a interaction and the binding affinity of ATG18 with PtdIns(3)P
To explore the mechanism by which HLS1 acetylates ATG18a in more detail, we wanted to identify the acetylation sites in ATG18a. To this end, we immunoprecipitated ATG18a–GFP from ATG18a–GFP transgenic seedlings in the Col-0 background exposed to nutrient starvation for MS analysis. However, this initial approach failed, possibly due to technical limitations associated with the identification of acetylation sites in plants (Xia et al. 2022).
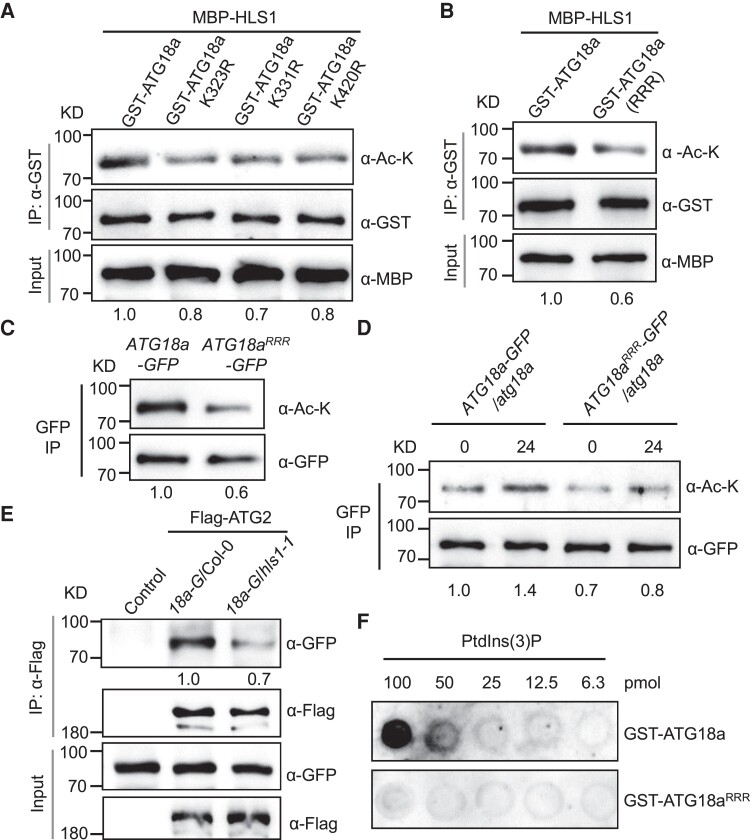
Next, we used the acetylation site prediction software GPS-PAIL to predict potential lysine acetylation sites in ATG18a (Deng et al. 2016). This analysis highlighted lysine 323 (K323), K331, and K420 from ATG18a as candidate acetylated residues (Supplemental Fig. S4). As K-to-arginine (R) substitutions are reported to prevent acetylation without affecting side chain charges of the residues (Barlev et al. 2001), we mutated 3 K residues (K323, K331, and K420) to R, respectively, and generated 3 recombinant ATG18a variant proteins GST–ATG18aK323R, GST–ATG18aK331R, and GST–ATG18aK420R. Subsequently, in vitro acetylation assay showed that each K-to-R mutation substantially reduced the acetylation level of ATG18a (Fig. 5A), suggesting that K323, K331, and K420 are all the acetylation sites in ATG18a.
Figure 5.
HLS1-mediated ATG18a acetylation enhances ATG2–ATG18a interaction and the binding of ATG18a to PtdIns(3)P. A) Amino acid substitutions (K323R, K331R, and K420R) reduce ATG18a acetylation in vitro. GST-tagged and MBP-tagged target proteins were enriched by GST and MBP beads, respectively, and then coincubation for acetylation analysis. Relative intensity of acetylated GST–ATG18a band normalized to the loading control GST–ATG18a is shown below. K, lysine; R, arginine; IP, immunoprecipitation. The experiment was repeated 3 times with similar results. B) In vitro acetylation analysis of GST–ATG18a and GST–ATG18aK323R–K331R–K420R (GST–ATG18aRRR) in the presence of MBP–HLS1. GST-tagged and MBP-tagged target proteins were enriched by GST and MBP beads, respectively, and then coincubation for acetylation analysis. Relative intensity of acetylated GST–ATG18a band normalized to the loading control GST–ATG18a is shown below. The experiment was repeated 3 times with similar results. C) Acetylation analysis of ATG18a–GFP and ATG18aRRR–GFP after transient expressions in N. benthamiana. GFP fusion proteins were immunoprecipitated by GFP beads. α-Ac-K and α-GFP antibodies were used to detect the acetylation level and the loading of corresponding proteins, respectively. The numbers below the lanes represent the ratio of acetylated ATG18a–GFP relative to the loading control ATG18a–GFP. The experiment was repeated 3 times with similar results. D) Acetylation analysis of ATG18a–GFP in the ATG18a–GFP/atg18a and ATG18aRRR–GFP/atg18a transgenic seedlings upon carbon starvation for 24 h. The acetylated and loading ATG18a–GFP proteins were detected using specific antibodies as indicated. Relative intensity of acetylated ATG18a–GFP or ATG18aRRR–GFP band normalized to the loading control ATG18a–GFP or ATG18aRRR–GFP is shown below. The experiment was repeated 3 times with similar results. E) Mutation of HLS1 weakens ATG2–ATG18a interaction. One-week-old ATG18a–GFP/Col-0 (18a-G/Col-0) and ATG18a–GFP/hls1-1 (18a-G/hls1-1) were exposed to carbon-deficient liquid medium for 24 h. The total proteins were extracted from whole seedlings and incubated with Flag beads that had not (control) or had been prebound to the Flag-tagged ATG2 (Flag–ATG2) protein. Anti-GFP and anti-Flag antibodies were used to detect the proteins. The numbers below the lanes represent the ratio of immunoprecipitated ATG18a–GFP relative to Flag–ATG2. F) Lipid binding of recombinant GST–ATG18a and GST–ATG18aRRR proteins on membranes. Purified GST–ATG18a or GST–ATG18aRRR proteins were incubated with a membrane containing serial diluted amount of PtdIns(3)P. PtdIns(3)P, phosphatidylinositol 3-phosphate. Anti-GST antibody was used to detect the bound protein. The experiment was repeated twice with similar results.
We then mutated all 3 K residues to R simultaneously and purified recombinant GST–ATG18a, GST–ATG18aK323R–K331R–K420R (GST–ATG18aRRR), and MBP–HLS1 from E. coli. As expected, the acetylation levels of ATG18a were markedly lower ∼0.4 when K323, K331, and K420 were mutated to R simultaneously (Fig. 5B), and this was not due to the disruption of the interaction between HLS1 and ATG18a (Supplemental Fig. S5A). Consistent with this result, in vivo acetylation assay by transient expression also showed that the acetylation in ATG18aRRR–GFP samples was much weaker than that of samples expressing intact ATG18a–GFP (Fig. 5C), indicating the importance of the K323, K331, and K420 sites for acetylation of ATG18a protein.
We then ordered an atg18a T-DNA insertion mutant (GK-651D08) from the ABRC and did not detect ATG18a transcript in the atg18a mutant, as determined by RT-qPCR (Supplemental Fig. S6, A to D). Then, we tested the sensitivity of the atg18a mutant to nitrogen and carbon starvation. Consistent with the ATG18a–RNAi lines (Xiong et al. 2005), the atg18a mutant was more susceptible to nutrient starvation with lower chlorophyll contents compared to Col-0 (Supplemental Fig. S6, E to G). To further confirm the role of the K323, K331, and K420 sites in ATG18a acetylation, we generated stable transgenic lines expressing ATG18a–GFP or ATG18aRRR–GFP in the atg18a mutant background. Immunoblotting assays revealed that the acetylation level of ATG18aRRR was reduced under both normal and carbon starvation conditions relative to intact ATG18a (Fig. 5D), suggesting that K323, K331, and K420 are the regulatory acetylation sites of ATG18a.
To understand how HLS1-mediated acetylation of ATG18a regulates its function, we first tested whether acetylation affects the subcellular localization of ATG18a upon nutrient starvation due to the finding that HLS1 interacts with ATG18a in nucleus. Confocal microscopy analyses showed that ATG18a–GFP in all the ATG18a–GFP/Col-0, ATG18a–GFP/hls1-1, ATG18a–GFP/atg18a, and ATG18aRRR–GFP/atg18a transgenic seedlings localized to the nucleus and the cytoplasm in both leaf epidermal cells and root cells under either normal or carbon starvation conditions (Supplemental Fig. S7), suggesting that acetylation modification is unlikely to change subcellular localization of ATG18a.
Given that Atg2–Atg18 interaction plays a vital role in autophagosome formation during autophagy in yeast (Obara et al. 2008; Kotani et al. 2018), we investigated the effect of the mutation of HLS1 on ATG2–ATG18a interaction. To this end, we purified recombinant Flag–ATG2 protein and GST–ATG18a proteins and found that GST–ATG18a, not GST–GFP, interacted Flag–ATG2 in vitro (Supplemental Fig. S5B). We then examined the ATG2–ATG18a interaction in the ATG18a–GFP/Col-0 and ATG18a–GFP/hls1-1 plants under carbon starvation, due to the finding that ATG18a acetylation is significantly reduced in the hls1-1 background (Fig. 4, E and F). As shown in Fig. 5E, the ability of recombinant Flag–ATG2 to pull down ATG18a–GFP in plant was reduced in hls1-1 background, suggesting that HLS1-mediated ATG18a acetylation enhances ATG2–ATG18a interaction.
Given that Atg2–Atg18 interaction facilitates the binding of Atg18 to PtdIns(3)P, which is required for Atg2–Atg18 to localize at the preautophagosomal structure (PAS) in yeast (Watanabe et al. 2012; Kotani et al. 2018), we speculated that ATG18a acetylation may affect its binding to PtdIns(3)P. To test this, we examined the binding ability of GST–ATG18a and GST–ATG18aRRR to PtdIns(3)P in vitro. As expected, the protein–lipid binding assay showed that the binding of ATG18aRRR to PtdIns(3)P was markedly inhibited compared to ATG18a (Fig. 5F; Supplemental Fig. S8). Taken together, these finding suggest that ATG18a acetylation modulated by HLS1 enhances ATG2–ATG18a interaction and the binding affinity of ATG18a with PtdIns(3)P in Arabidopsis.
Reduced ATG18a acetylation impairs autophagy
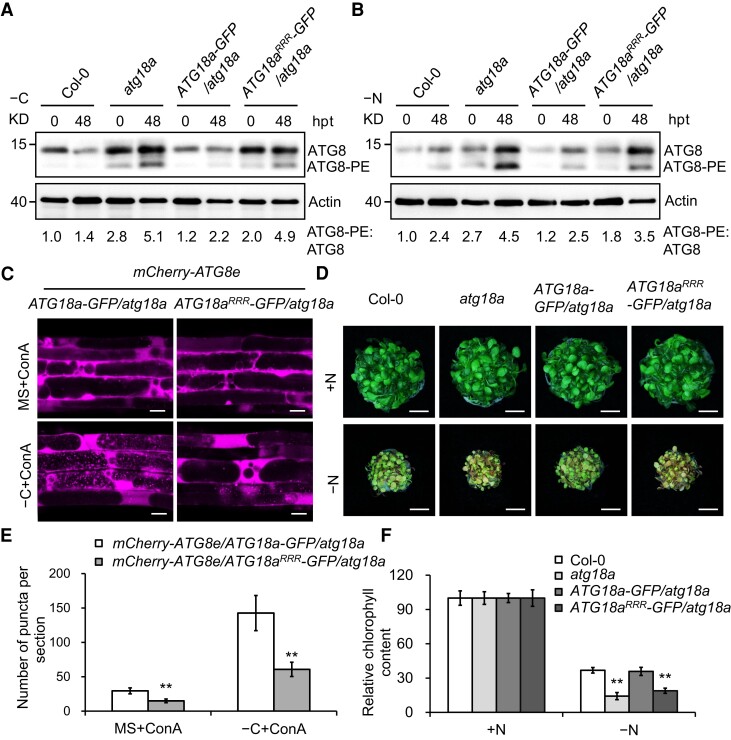
To elucidate whether preventing ATG18a acetylation might impair autophagy, we examined ATG8 lipidation in the Col-0, atg18a, ATG18a–GFP/atg18a, and ATG18aRRR–GFP/atg18a seedlings using an anti-ATG8a antibody to immunoprecipitate the protein. As reported for atg18a mutant (Kang et al. 2018), ATG8–PE was more abundant in the atg18a mutant compared with Col-0 (Fig. 6, A and B). Overexpression of ATG18a–GFP restored the higher ratio of ATG8–PE to ATG8 in atg18a mutant under either nutrient-rich or starvation conditions to that of Col-0. In contrast, the ratio of ATG8–PE to ATG8 in ATG18aRRR–GFP/atg18a line was comparable to that of atg18a mutant (Fig. 6, A and B). These results suggest that reduced acetylation of ATG18a at the 3 K sites blocks the turnover of ATG8 protein.
Figure 6.
ATG18a acetylation positively regulates autophagy. A, B) ATG8 lipidation level in the Col-0, atg18a, and transgenic lines after carbon starvation (−C, A) or nitrogen starvation (−N, B) treatment for the indicated times. Anti-ATG8a antibody was used to detect the ATG8 and ATG8–PE protein levels. Relative intensity of ATG8–PE band normalized to the ATG8 is shown below. The relative intensities of each band on immunoblots were quantified using ImageJ. PE, phosphatidylethanolamine; hpt, hours posttreatment. The experiment was repeated 3 times with similar results. C) Confocal analysis of mCherry–ATG8e/ATG18a-GFP/atg18a and mCherry–ATG8e/ATG18aK323R–K331R–K420R–GFP/atg18a (mCherry–ATG8e/ATG18aRRR–GFP/atg18a) transgenic plants. Five-day-old seedlings were exposed to carbon (C)-sufficient (MS) or C-deficient (−C) liquid medium with the addition of 1 μm ConA for 6 h and then visualized by confocal laser scanning microscopy. K, lysine; R, arginine; ConA, concanamycin A. Bars = 20 μm. D) Phenotypes of Col-0, atg18a, and transgenic seedlings in response to nitrogen starvation. Seven-day-old seedlings were transferred to N-rich (+N) or N-deficient (−N) liquid medium and photographed at 6 d after treatment. Bars = 1 cm. E) Numbers of puncta per section in the root cells of the transgenic plants in C). Three independent experiments were done with similar results. Values are means ± Sd (n = 15) from one representative experiment. Asterisks indicate significant differences from that of the mCherry–ATG8e/ATG18a–GFP/atg18a (**P < 0.01, Student's t test). F) Relative chlorophyll content in D). The relative chlorophyll contents are expressed relative to those of the genotypes on +N medium. Three independent experiments were performed with similar results. Values are means ± Sd (n = 4 replicates) from one experiment. For each experiment, leaves from eight seedlings were used per one replicate. Asterisks indicate significant differences from the wild type (**P < 0.01, Student's t test).
To further assess the effect of ATG18a acetylation on autophagosome formation, we crossed the autophagosome marker line mCherry–ATG8e (Zhuang et al. 2017) to transgenic ATG18a–GFP/atg18a and ATG18aRRR–GFP/atg18a lines to generate mCherry–ATG8e/ATG18a–GFP/atg18a and mCherry–ATG8e/ATG18aRRR–GFP/atg18a plants, respectively. After carbon starvation for 6 h, the accumulation pattern of ATG18a–GFP and ATG18aRRR–GFP was essentially the same (Supplemental Fig. S9), but fewer GFP–ATG8e-labeled punctate autophagosomes accumulated in root cells of ATG18aRRR–GFP/atg18a than that in ATG18a–GFP/atg18a plants (Fig. 6, C and E).
Furthermore, we tested the responses of Col-0, atg18a, ATG18a–GFP/atg18a, and ATG18aRRR–GFP/atg18a seedlings to nitrogen deficiency. After a 6-d treatment, the hypersensitive phenotype of the atg18a mutant to nitrogen starvation was fully rescued by overexpression of intact ATG18a (Fig. 6, D and F). However, atg18a seedlings expressing ATG18aRRR still exhibited increased susceptibility to starvation conditions compared to Col-0 (Fig. 6, D and F), indicating that ATG18a acetylation at K323–K331–K420 is crucial for autophagy in Arabidopsis.
Mutations of V108 and L151 residues in HLS1 severely impair its activity
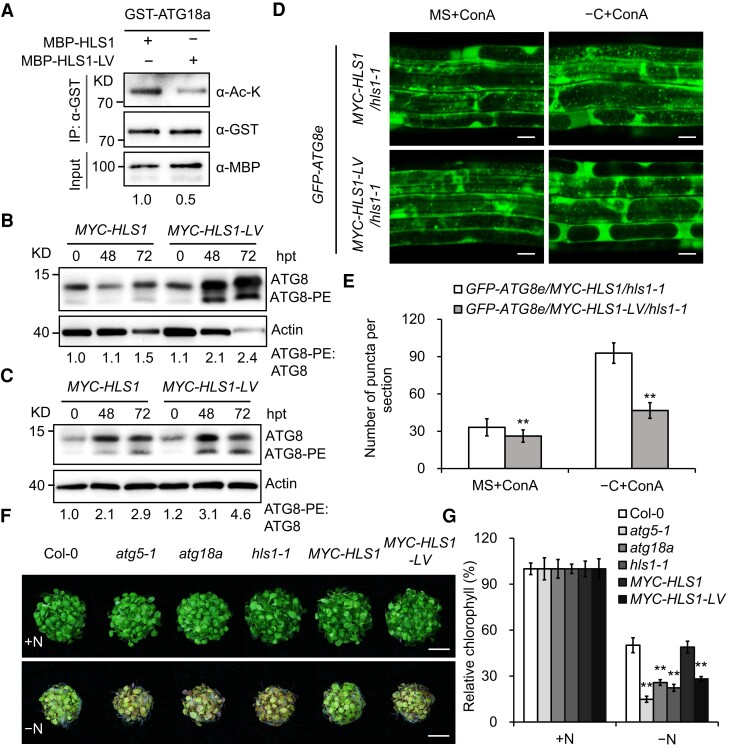
Given the finding that HLS1 acetylates ATG18a in planta, we then aimed to identify the domain or residues required for HLS1 activity. To this end, we performed a sequence alignment between HLS1 and several acetyltransferases from Arabidopsis (Supplemental Fig. S10) and mutated the 2 most conserved amino acids (V108 and L151) of HLS1 (Supplemental Fig. S10), located in the active center regions of previously characterized N-acetyltransferases (Tercero et al. 1992; Coon et al. 1995), to alanine (A). We then constructed vectors for protein purification of recombinant MBP–HLS1V108A–L151A (MBP–HLS1–LV, harboring the V108A and L151A mutations). To assess the effect of the V108–L151 mutations on HLS1 enzymatic activity, we performed an in vitro acetylation assay using recombinant GST–ATG18a and MBP–HLS1 or MBP–HLS1–LV in the presence of acetyl-CoA. We again detected the acetylation of ATG18a by HLS1, but HLS1 acetylation was largely abolished in the presence of HLS1–LV (Fig. 7A), indicating that mutating the 2 conserved sites leads to lower HLS1 enzymatic activity.
Figure 7.
The V108 and L151 sites in HLS1 are required for its enzymatic activity and induction of autophagy. A) In vitro acetylation analysis showing mutations of V108 and L151 residues in HLS1 inhibited the acetylation of GST–ATG18a. GST-tagged and MBP-tagged target proteins were enriched by GST and MBP beads, respectively, and then coincubation for acetylation analysis. IP, immunoprecipitation; L, leucine; V, valine; A, alanine; MBP–HLS1–LV, MBP–HLS1V108A–L151A. Ac-K, GST, and MBP antibodies were used for immunoblotting. Relative intensity of acetylated GST–ATG18a band normalized to the loading control GST-–ATG18a is shown below. The experiment was repeated 3 times with similar results. B, C) ATG8 lipidation level in the MYC–HLS1/hls1-1 (MYC–HLS1) and MYC–HLS1V108A–L151A/hls1-1 (MYC–HLS1–LV) lines after carbon starvation (−C, B) or nitrogen starvation (−N, C) treatment for the indicated times. Anti-ATG8a antibody was used to detect the ATG8 and ATG8–PE protein levels. Relative intensity of ATG8–PE band normalized to the ATG8 is shown below. The relative intensities of each band on immunoblots were quantified using ImageJ. PE, phosphatidylethanolamine; hpt, hours posttreatment. The experiment was repeated 3 times with similar results. D) Confocal analysis of GFP–ATG8e/MYC–HLS1/hls-1 and GFP–ATG8e/MYC–HLS1–LV/hls-1 transgenic plants. Five-day-old seedlings were exposed to carbon (C)-sufficient (MS) or C-deficient (−C) liquid medium with the addition of 0.5 μm ConA for 6 h and then visualized by confocal laser scanning microscopy. ConA, concanamycin A. Bars = 20 μm. E) Numbers of puncta per section in the root cells of the transgenic plants in D). Three independent experiments were done with similar results. Values are means ± Sd (n = 15) from one representative experiment. Asterisks indicate significant differences from that of the GFP–ATG8e/MYC–HLS1/hls-1 (**P < 0.01, Student's t test). F) Phenotypes of wild type (Col-0), atg5-1, atg18a, hls1-1, MYC–HLS1/hls1-1 (MYC–HLS1), and MYC–HLS1V108A–L151A/hls1-1 (MYC–HLS1–LV) lines in response to nitrogen starvation. Seven-day-old seedlings were transferred to N-rich (+N) or N-deficient (−N) liquid medium, and the photographs were taken at 6 d after treatment. Bars = 1 cm. G) Relative chlorophyll content in F). The relative chlorophyll contents are expressed relative to those of the genotypes on +N medium. Three independent experiments were conducted with similar results. Values are means ± Sd (n = 4 replicates) from one experiment. For each experiment, leaves from 8 seedlings were used per one replicate. Asterisks indicate significant differences from the wild type (**P < 0.01, Student's t test).
To rule out the possibility that HLS1–LV no longer acetylates ATG18a due to a loss of interaction, we conducted an in vitro pull-down assay. As shown in Supplemental Fig. S11A, both MBP–HLS1 and MBP–HLS1–LV were pulled down by GST–ATG18a. In addition, we performed a split-LUC complementation assay to verify the interaction between each protein pair. We consistently observed an interaction between HLS1–LV and ATG18a in N. benthamiana leaves, based on luminescence (Supplemental Fig. S11B). We further confirmed the interaction between HLS1–LV and ATG18a by Co-IP analysis (Supplemental Fig. S11C). Collectively, these results suggest that mutations in the conserved sites (V108 and L151) of the putative acetyltransferase domain of HLS1 do not affect its interaction with ATG18a. Together, these results indicate that V108 and L151 are required for HLS1 enzymatic activity.
The enzymatic activity of HLS1 is necessary for regulation of autophagy under nutrient starvation
To determine whether the enzymatic activity of HLS1 is required for the induction of autophagy under starvation conditions, we introduced a MYC–HLS1V108A–L151A transgene into the hls1-1 mutant to generate transgenic lines MYC–HLS1V108A–L151A/hls1-1 (MYC–HLS1–LV/hls1-1) for ATG8 lipidation analysis. Compared with MYC–HLS1/hls1-1 plants, the ratio of ATG8–PE to ATG8 was higher in MYC–HLS1–LV/hls1-1 plants after nutrient starvations (Fig. 7, B and C), which is comparable to the pattern between hls1-1 and Col-0.
To further evaluate the role of HLS1–LV in autophagy, we crossed the autophagosome marker line GFP–ATG8e to MYC–HLS1–LV/hls1-1 to generate GFP–ATG8e/MYC–HLS1–LV/hls1-1 line. Confocal microscopy analysis showed that carbon starvation–induced autophagosome accumulation was significantly reduced in MYC–HLS1–LV/hls1-1 roots than that in MYC–HLS1/hls1-1 roots (Fig. 7, D and E). Together, these results indicate that impaired enzymatic activity of HLS1 via V108–L151 mutations suppresses the autophagy activity under nutrient starvation.
We next tested the response of MYC–HLS1–LV/hls1-1 to nutrient starvation for comparative phenotypic analyses with Col-0 and MYC–HLS1/hls1-1 plants. We had already demonstrated that MYC–HLS1/hls1-1 exhibits a similar tolerance to nutrient starvations as Col-0. Conversely, MYC–HLS1–LV/hls1-1 plants displayed an increased susceptibility to individual carbon or nitrogen starvation, similar to the hls1-1 mutant (Fig. 7, F and G; Supplemental Fig. S12), indicating that MYC–HLS1–LV cannot rescue the hls1-1 mutant. Thus, mutations in the putative acetyltransferase domains of HLS1 affect its functions in response to nutrient limitation. Moreover, we compared the responses of MYC–HLS1–LV/hls1-1 seedlings to atg5-1 and atg18a mutants under nutrient starvation conditions. We observed similar hypersensitive phenotypes in MYC–HLS1–LV/hls1-1, atg5-1, and atg18a seedlings following starvation (Fig. 7, F and G; Supplemental Fig. S12). These results indicate that the V108 and L151 residues of HLS1 are required for plants to cope with nutrient starvation, and this function of HLS1 is likely mediated through autophagy.
Since atg mutants are characterized by premature leaf senescence in addition to hypersensitivity to nutrient deprivation (Doelling et al. 2002; Hanaoka et al. 2002), we subsequently investigated leaf senescence at different time points in the genetic materials generated here. After 4 wk of growth, MYC–HLS1–LV/hls1-1, like hls1-1 and atg mutants, showed premature senescence compared with Col-0 or MYC–HLS1/hls1-1 plants (Supplemental Fig. S13). These phenotypes became more visible in 5-wk-old plants (Supplemental Fig. S13).
To verify the connections between HLS1–LV and autophagy, we also tested the susceptibility of hls1-1 and MYC–HLS1–LV/hls1-1 lines to the reactive oxygen species (ROS)–inducing reagent methyl viologen (MV), which can strongly induce autophagy (Xiong et al. 2007). As reported previously (Xiong et al. 2007), the atg18a mutant displayed a strong hypersensitivity to MV treatment, manifesting as bleached cotyledons, compared to Col-0 (Supplemental Fig. S14A). Importantly, we determined that atg5-1, hls1-1, and MYC–HLS1–LV/hls1-1 seedlings display the same susceptibility to MV as the atg18a mutant, as evidenced by their lower chlorophyll contents relative to Col-0 (Supplemental Fig. S14B). Collectively, the enzymatic activity of HLS1 appears to be necessary for its regulation of autophagy in response to stresses, including but not limited to nutrient starvation.
HLS1-regulated autophagy is uncoupled from HLS1-mediated hook formation
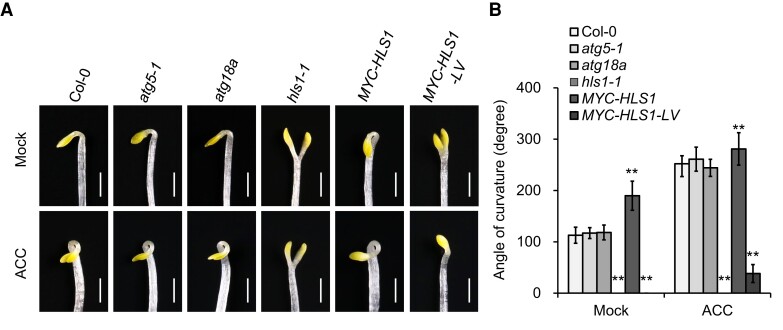
HLS1 was previously described as a key regulator of apical hook development (Guzmán and Ecker 1990; Lehman et al. 1996). Our observations that HLS1 also plays an important role in the regulation of autophagy prompted us to ask whether autophagy might be involved in hook formation of etiolated seedlings. To this end, we investigated the hook phenotypes of hls1-1, atg5-1, and atg18a mutant seedlings grown under air (mock) and 1-aminocyclopropane-1-carboxylic acid (ACC; ethylene biosynthesis precursor) treatment in darkness for 3.5 d. Consistent with previous studies (Guzmán and Ecker 1990; Lehman et al. 1996), the hls1-1 mutant was completely insensitive to exogenous ACC application, with its typical hookless phenotype compared to the dramatic exaggerated hook curvature of Col-0 seedlings (Fig. 8). In contrast, atg5-1 and atg18a mutant seedlings showed similar hook phenotypes as Col-0 under both air and exogenous ACC conditions (Fig. 8), indicating that autophagy is not involved in HLS1-regulated apical hook formation.
Figure 8.
HLS1-regulated autophagy is uncoupled from HLS1-mediated hook formation. A) Phenotypes of 3.5-d-old etiolated wild-type (Col-0), atg5-1, atg18a, hls1-1, MYC–HLS1/hls1-1 (MYC–HLS1), and MYC–HLS1V108A–L151A/hls1-1 (MYC–HLS1–LV) seedlings grown on MS medium supplemented without (mock) or with 10 μm ACC. L, leucine; V, valine; A, alanine; MS, Murashige and Skoog; ACC, 1-aminocyclopropane-1-carboxylic acid. Bars = 1 mm. B) The angles of curvature in A). The angles of curvature were measured using ImageJ software. The experiments were conducted 3 times with similar results. Values are means ± Sd (n ≥ 15) from one experiment. Asterisks indicate significant differences from the Col-0 (**P < 0.01, Student's t test).
To further explore whether the enzymatic activity of HLS1 has any effect on hook formation, we analyzed hook formation in MYC–HLS1/hls1-1 and MYC–HLS1–LV/hls1-1 etiolated seedlings. The overexpression of MYC–HLS1 restored the hookless phenotype of the hls1-1 mutant after exogenous ACC treatment and produced seedlings with an enhanced hook curvature even under normal air conditions (Fig. 8). In contrast, MYC–HLS1–LV/hls1-1 seedlings showed no obvious sign of rescue for the hook formation under either air or ACC treatment (Fig. 8), suggesting that the normal enzymatic activity of HLS1 is also necessary for apical hook development. Taken together, although the V108 and L151 residues of HLS1 play key roles in both autophagy and hook formation, HLS1-regulated autophagy is uncoupled from HLS1-mediated apical hook development.
Discussion
HLS1 is a well-known key regulator of apical hook formation and is also involved in thermomorphogenesis, pathogen defense, sugar signaling, and abscisic acid (ABA) responses (Guzmán and Ecker 1990; Lehman et al. 1996; Ohto et al. 2006; Liao et al. 2016; Jin and Zhu 2019; Guo et al. 2023). Although HLS1 was previously considered as a putative N-acetyltransferase due to the sequence similarity it shares with this class of enzymes (Lehman et al. 1996), its biochemical function has remained largely unclear. Here, we demonstrate that HLS1 is required for autophagy activation during nutrient starvation and reveal the biological and biochemical roles of HLS1 in plant autophagy through acetylation of ATG18a.
In this study, we aimed to study the enzymatic function of HLS1 and verified that HLS1 exerts its functions as a bona fide acetyltransferase based on the following evidence. First, in vitro acetylation assays revealed that HLS1 acetylated ATG18a (Fig. 4A). Second, the acetylation levels of ATG18a decreased significantly in the hls1-1 mutant background (Fig. 4, E and F). Third, mutations at the conserved sites (V108 and L151) in the putative acetyltransferase active domains of HLS1 did not affect its interaction with ATG18a but markedly blocked the acetylation of ATG18a and autophagy activation (Fig. 7; Supplemental Fig. S11). Prior to this study, HLS1 had been reported to function as a putative histone acetyltransferase (HAT) required for histone H3 acetylation at ABI5 and WRKY33 chromatin, although HLS1 protein did not exhibit HAT activity in vitro (Liao et al. 2016). Considering that a growing number of HATs appear to have a wide range of substrates in addition to histones (Narita et al. 2019), our work does not exclude the possibility that HLS1 might also function as a HAT to acetylate histones as well as nonhistones. Thus, the acetyltransferase activity of HLS1 on histones should be revisited.
The acetyl group at the core of lysine acetylation is provided by acetyl-CoA, which is a central integrator of the nutritional status at the metabolic crossroads of sugar, fat, and proteins (Mariño et al. 2014; Xia et al. 2022). In human cells, nutrient starvation causes the rapid depletion of acetyl-CoA and induces protein deacetylation (Mariño et al. 2014). In contrast, acetylation of Atg3 increases after nutrient starvation in yeast, while the acetylation levels of other Atg proteins are lower or unchanged (Yi et al. 2012). Here, we found that the acetylation of ATG18a, similar to Atg3 in yeast, was strongly promoted under carbon or nitrogen starvation (Fig. 4, B to F), suggesting that ATG18a acetylation could be tightly regulated in response to nutrient deprivation, possibly due to the alleviation of substrate inhibition (Reed et al. 2010). Given that HLS1 plays an important role in hook development of dark-grown seedlings that themselves would undergo nutrient starvation, we speculate that HLS1 might also be affected by prolonged darkness and nutrient starvation through the regulation of its activity. Therefore, whether the activity of HLS1 is modulated by changes in acetyl-CoA levels caused by nutrient starvation should be carefully examined.
Protein acetylation is one of most important PTMs that regulates autophagy initiation and autophagosome formation by targeting core components in yeast and mammals (McEwan and Dikic 2011; Bánréti et al. 2013; Füllgrabe et al. 2013). A recent study demonstrated that deacetylation of histone H3 at the K9 and K27 residues by HISTONE DEACETYLASE9 (HDA9) represses the expression of ATG5 and ATG8e in Arabidopsis (Yang et al. 2020), providing a clue for the modulation of plant autophagy at the transcriptional level by acetylation. However, whether core autophagy proteins are directly regulated by acetylation and what the underlying mechanisms in plants are is still poorly understood.
In this study, we showed that HLS1-mediated acetylation regulated autophagy activation by targeting ATG18a (Figs. 4, A, E, and F, and 7A), a core autophagy component that forms a conserved ATG9–ATG2–ATG18 complex across eukaryotes (Reggiori et al. 2004; Marshall and Vierstra 2018). Our findings also show how ATG18a acetylation affects its function in autophagy activation. Disruption of ATG18a acetylation suppressed ATG2–ATG18a interaction under carbon starvation and inhibited the binding of ATG18a to PtdIns(3)P (Fig. 5, E and F), which is necessary for PAS localization of Atg2–Atg18 complex in yeast (Kotani et al. 2018). In Arabidopsis, ATG9 is essential for the trafficking of ATG18a on the autophagosomal membrane in a PtdIns(3)P-dependent manner (Zhuang et al. 2017). Therefore, further studies addressing whether and how ATG18a acetylation affects the trafficking of ATG9 vesicles are crucial for an in-depth understanding of autophagy modulated by HLS1 in planta.
Different from the acetylation of ATG18a, phosphorylation and persulfidation of Arabidopsis ATG18a were shown to negatively regulate autophagy upon infection by necrotrophic pathogens (Zhang et al. 2021) and under ER stress (Aroca et al. 2021), respectively. In contrast, we observed that reduced acetylation of ATG18a blocked autophagy activity and increased plant susceptibility to nitrogen starvation (Fig. 6), suggesting that acetylation of ATG18a positively regulates its function in autophagy induction during nutrient starvation. This result indicates that ATG18a is modified by different PTMs when plants are subjected to changes in external conditions, and these modifications must be finely controlled.
By taking advantage of bioinformatic tools, we identified 3 predicted acetylation sites (K323, K331, and K420) of ATG18a protein and demonstrated that they are essential for ATG18a acetylation by HLS1. Mutations of K323, K331, and K420 sites of ATG18a simultaneously exhibited distinctly reduced acetylation levels of ATG18a and attenuated autophagy induced by nutrient starvation (Figs. 5 and 6). Of note, the combined mutations of the 3 lysine residues in ATG18a did not eliminate all acetylation of ATG18a, suggesting other acetylation sites also exist in ATG18a. Indeed, our initial attempt to identify total acetylation sites using immunopurified ATG18a from Arabidopsis was unsuccessful. This failure may be due to technological limitations in the identification of acetylation sites by MS, which is still far from being routine in plant research (Xia et al. 2022). Moreover, acetylation usually occurs at a low stoichiometry amount, which may therefore fall below the detection threshold of the method (Choudhary et al. 2014; O'Leary et al. 2020). Nevertheless, our findings contribute to elucidating the mechanism by which ATG18a acetylation regulates autophagy.
Our previous studies have validated the important role of HLS1 in hook development (An et al. 2012; Zhang et al. 2014, 2018; Huang et al. 2020; Peng et al. 2022). Here, we extended our knowledge to assess the importance of acetylation in apical hook development and determined that the enzymatic activity of HLS1 was also necessary for this process (Fig. 8). However, we noticed that the atg18a mutant exhibited a similar phenotype as the wild type for hook formation (Fig. 8), suggesting that ATG18a-involved autophagy is not required for HLS1-regulated apical hook development, and HLS1 may target other substrates.
Furthermore, hls1-1 plants also displayed similar phenotypes as atg mutants in plant senescence (Supplemental Fig. S13) and oxidative stress responses (Supplemental Fig. S14). Therefore, it is reasonable to speculate that HLS1 may regulate aging and plant responses to oxidative stress by modulating autophagy via acetylation of ATG18a or other ATG proteins. In addition, whether HLS1 regulates thermomorphogenesis, pathogen defense, and sugar and ABA responses through acetylation of distinct substrates also needs to be addressed. Hence, efforts are required to focus on identifying more substrates of HLS1 in multiple biological processes.
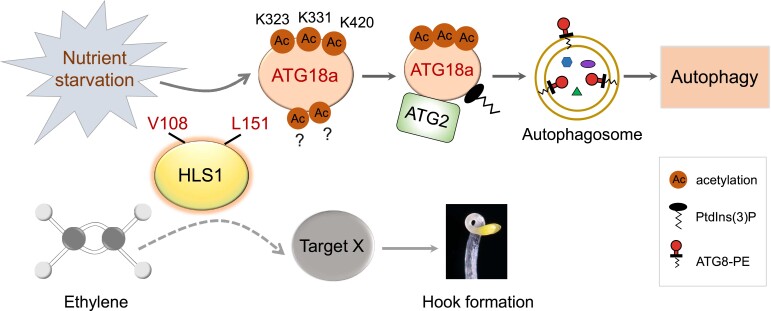
In summary, our study unveils the crucial role of the acetyltransferase HLS1 in autophagy regulation through affecting the acetylation status of ATG18a during the plant response to nutrient starvation (Fig. 9). When plants are exposed to nutrient starvation, the acetylation levels of ATG18a at potential sites increase, likely through the interaction between ATG18a and HLS1. Then, increased ATG18a acetylation promotes the ATG2–ATG18a interaction and the binding of ATG18a to PtdIns(3)P and ultimately activates autophagy. During this process, the enzymatic activity of HLS1 is necessary for ATG18a acetylation (Fig. 9), implying that HLS1 is an important regulator of plant autophagy during nutrient deprivation. Furthermore, HLS1-modulated autophagy is uncoupled from the well-known HLS1-mediated hook formation in Arabidopsis. Taken together, these results unravel a key PTM of a core autophagy protein and further elucidate the importance of PTMs on autophagy regulation in plant.
Figure 9.
A working model of HLS1-modulated autophagy activation through acetylation of ATG18a protein. Under nutrient (carbon or nitrogen) starvations, the acetylation level of ATG18a at potential sites (K323, K331, and K420) increases, and this increase promotes the ATG2–ATG18a interaction and the binding of ATG18a to PtdIns(3)P and ultimately activates autophagy to enable plants to survive. Among this process, V108 and L151 sites in HLS1 are necessary for its function for ATG18a acetylation, as well as the hook formation. Whereas the autophagy route modulated by HLS1 is uncoupled from its regulated apical hook formation. L, leucine; K, lysine; V, valine; PtdIns(3)P, phosphatidylinositol 3-phosphate; PE, phosphatidylethanolamine. The question marks indicate unknown acetylation sites in ATG18a; solid arrows indicate well-defined processes; and dashed arrow indicates unknown process.
Materials and methods
Plant materials, growth conditions, and treatments
The A. thaliana accession Col-0 was used as the wild type line in this study. The T-DNA insertion knockout mutant atg18a (GK-651D08) was obtained from the Arabidopsis Biological Resource Center (ABRC; http://www.arabidopsis.org). hls1-1 (Lehman et al. 1996) and atg5-1 (Chen et al. 2015) mutants used in this study were described previously. The mutants were identified by genomic PCR using gene-specific primers paired with a T-DNA border-specific primer (Supplemental Data Set 1). The mutants in this study are listed in Supplemental Table S1. Transgenic plant GFP–ATG8e (Xiao et al. 2010) and mCherry–ATG8e (Zhuang et al. 2017) have been described previously.
All Arabidopsis seeds were surface sterilized with 75% (v/v) ethanol containing 0.05% Triton X-100 for 15 min, washed with distilled water at least 5 times, then plated on MS (Sigma-Aldrich, M5519) agar (0.7%, w/v) medium containing 1% (w/v) sucrose. After stratification at 4 °C for 3 d in darkness, the plates were cultured in a plant growth room under a 16-h light/8-h dark photoperiod at 22 °C, and the light intensity was 120 to 150 μmol/m2/s using fluorescent bulbs (PHILIPS, TL5 21W/865). For N. benthamiana growth, the seeds were scattered directly on the soil and grown in controlled greenhouse conditions with 16-h light (120 to 150 μmol/m2/s)/8-h dark photoperiod at 25 °C using fluorescent bulbs (PHILIPS, TL5 21W/865). For carbon starvation, 1-wk-old seedlings grown on MS medium supplemented with 1% (w/v) sucrose were transferred to fresh MS liquid or solid medium under continuous darkness for the indicated time points. For nitrogen starvation, 1-wk-old seedlings grown on MS medium containing 1% sucrose (w/v) were transferred to nitrogen-free MS (Caisson, MSP07-50LT) liquid medium and grown under normal conditions for indicated duration. For chemical treatment, 1-wk-old seedlings grown on solid MS medium containing 1% sucrose (w/v) were transferred to liquid MS medium containing 1 μm MV (Sigma-Aldrich, 856177) for indicated times. The effect of ACC (Sigma-Aldrich, A3903) on apical hook formation was determined according to Huang et al. (2020).
Confocal microscopy
Monitoring the autophagosomes in transgenic plants expressing GFP–ATG8e was performed using a Zeiss LSM880 confocal laser microscope (Carl Zeiss, Germany). Five-day-old seedlings were transferred to carbon- or nitrogen-deficient MS medium containing 0.5 or 1.0 μm ConA (APExBIO, A8633) for indicated times. Then, the primary root cells were observed using the microscope. For GFP fluorescence, the excitation wavelength was 488 nm with 10% to 20% output signal intensity, and the emission was detected at 500 to 530 nm with gain values of 650 to 680. For mCherry fluorescence, the excitation wavelength was 516 nm with 10% to 20% output signal intensity, and the emission was detected at 560 to 610 nm with gain values of 650 to 680. For DAPI fluorescence, the excitation wavelength was 405 nm with 5% to 10% output signal intensity, and the emission was detected at 415 to 515 nm with gain values of 620 to 650.
Vector construction
Primers for all vector constructs are listed in Supplemental Data Set 1. For split-LUC complementation assay, the coding sequences of ATG1a, ATG1b, ATG1c, ATG3, ATG5, ATG6, ATG7, ATG12, ATG18a, ATG18b, ATG18c, ATG18d, ATG18e, ATG18f, ATG18g, ATG18h, and HLS1 were inserted into the pCAMBIA1300–nLUC (Chen et al. 2008) using the Mlu and Pac restriction sites. The full-length coding sequences of ATG8e, HLS1, and HLS1V108A–L151A were inserted into the pCAMBIA1300–cLUC at Kpn–Pac site. To generate vectors for the BiFC assay, the full coding sequences of HLS1 or ATG18a were inserted into the multiple cloning sites of pSAT6 cEYFP-C1 and pSAT6 nEYFP-N1 (Citovsky et al. 2006) at Kpn–Smal and EcoRI–Smal sites, respectively. cYFP–SDIR1 was constructed as described previously (Hao et al. 2021). To generate vectors for recombinant protein expression, the coding sequences of HLS1–HA and HLS1V108A–L151A–HA were inserted into BamHI- and EcoRI-digested pMAL-p2X vector (Huang et al. 2020), and the full-length coding sequences of 6×His–ATG3 and 6×His–ATG18a were inserted into the pGEX5x-1 vector (Huang et al. 2020) digested by BamHI and EcoRI to generate GST–ATG3 and GST–ATG18a, respectively. Similarly, the full-length coding sequence of Flag–ATG2 was inserted into the pET28a at BamHI–EcoRI site. For stable expression, the full-length coding sequences of HLS1 and HLS1V108A–L151A were inserted into the pCAMBIA1307 (Huang et al. 2020) vector using BamH and Sal sites to generate MYC–HLS1 and MYC–HLS1V108A–L151A, respectively. Similarly, the GFP coding sequence was inserted into the binary vector pQG110 (Hao et al. 2021) digested by Sal and Sac under 35S promoter to generate Pro35S:GFP. The complete coding sequences of ATG18a and ATG18aK323R–K331R–K420R were inserted into the Pro35S:GFP vector at BamH–Kpn site to obtain ATG18–GFP and ATG18aK323R–K331R–K420R–GFP, respectively.
Mutations derived from HLS1 and ATG18a were generated by site-directed mutagenesis of aforementioned corresponding constructs using a KOD-Plus-Mutagenesis kit (TOYOBO, SMK-101) with the specific primers listed in Supplemental Data Set 1.
Generation of transgenic lines
For plant transformation, the vectors of ATG18a–GFP, ATG18aK323R–K331R–K420R–GFP, MYC–HLS1, and MYC–HLS1V108A–L151A were introduced into A. tumefaciens strain GV3101. Then, the floral dip method (Clough and Bent 1998) was used for generating transgenic plants. T2 transgenic plants of ATG18a–GFP/Col-0, ATG18a–GFP/atg18a, and ATG18aK323R–K331R–K420R–GFP/atg18a with single insertion sites were selected on MS medium containing 50 mg/L kanamycin. T2 transgenic plants expressing MYC–HLS1 and MYC–HLS1V108A–L151A in hls1-1 background with single insertion sites were selected on MS medium containing 50 μg/mL hygromycin B.
To generate GFP–ATG8e/hls1-1, GFP–ATG8e/MYC–HLS1/hls1-1, and GFP–ATG8e/MYC–HLS1V108A–L151A/hls1-1 transgenic lines, GFP–ATG8e was crossed with hls1-1, MYC–HLS1/hls1-1, and MYC–HLS1V108A–L151A/hls1-1. The transgenic F2 seedlings were selected on MS medium containing kanamycin and hygromycin B as described above.
To generate mCherry–ATG8e/ATG18a–GFP/atg18a and mCherry–ATG8e/ATG18aK323R–K331R–K420R–GFP/atg18a transgenic lines, mCherry–ATG8e was crossed with ATG18a–GFP/atg18a and ATG18aK323R–K331R–K420R–GFP/atg18a. The transgenic F2 seedlings were selected on MS medium containing kanamycin and hygromycin B as described above, and atg18a locus was confirmed using PCR-based genotyping using the gene-specific primers listed in Supplemental Data Set 1.
To generate ATG18a–GFP/hls1-1 transgenic line, homozygous ATG18a–GFP/Col-0 was crossed with the hls1-1 mutant. The transgenic F2 seedlings were selected on MS containing kanamycin as described above and genotyped for the hls1-1 mutation by sequencing using the gene-specific primers listed in Supplemental Data Set 1.
To generate ATG18a–GFP/MYC–HLS1 and ATG18a–GFP/MYC–HLS1V108A–L151A, MYC–HLS1/hls1-1 and MYC–HLS1V108A–L151A/hls1-1 were crossed with ATG18a–GFP/hls1-1. The F2 seedlings were selected on MS containing kanamycin and hygromycin B as described above. The transgenic lines generated in this study are listed in Supplemental Table S2.
Measurement of chlorophyll content
To measure the chlorophyll contents from the samples, Arabidopsis leaves were harvested after nutrient starvation or MV treatment. Chlorophyll was extracted by immersing the samples in 95% (v/v) ethanol for 48 h at 4 °C in the dark. Absorbances of the supernatants were measured at 649 and 664 nm, and the total chlorophyll content was calculated as described previously (Lichtenthaler 1987).
RNA extraction and RT-qPCR
Total RNA of whole Arabidopsis seedlings was extracted using the Eastep Super Total RNA Extraction Kit (Promega), and the isolated RNA was reverse transcribed using M-MLV Reverse Transcriptase (Promega). RT-qPCR was performed on the Light Cycler 480 system (Roche) using SYBR Premix ExTaq reagents (Takara). Four technical replicates were performed for each sample. ACTIN2 was used as the reference gene. The primers for qPCR analysis are listed in Supplemental Data Set 1.
Protein isolation and immunoblot analysis
Protein extraction and immunoblotting were carried out as described previously (Huang et al. 2020). Samples were frozen and ground in liquid nitrogen and homogenized in the same volume of protein extraction buffer (100 mm Tris-HCl [pH 6.8], 4% [w/v] SDS, 10% [v/v] glycerol, 50 mm DTT, and 0.02% [w/v] bromophenol blue). Samples were incubated on ice for 15 min and then heated at 65 °C for 10 min. After centrifugation at 13,000 × g for 15 min, the supernatants were separated using SDS–PAGE. To separate ATG8 and ATG8–PE, a Urea–Tricine SDS–PAGE system was utilized as described previously (Schägger and von Jagow 1987). Anti-GFP (ABclonal, AE012; 1:5,000 dilution), anti-ATG8a (Abcam, ab77003; 1:2,000 dilution), anti-MYC–HRP (ABclonal, AE026; 1:10,000 dilution), anti-HA–HRP (Roche, 3F10; 1:10,000 dilution), anti-Actin (ABclonal, AC009; 1:10,000 dilution), anti-GST (Tiangen, AB101; 1:10,000 dilution), anti-MBP (ABclonal, AE016; 1:10,000 dilution), anti-acetyl-lysine (anti-Ac-K) (PTM BioLab, PTM-101; 1:5,000 dilution), and anti-Flag–HRP (Sigma, A8592; 1:10,000 dilution) antibodies were used for immunoblotting.
Split-LUC complementation assay
The split-LUC complementation assay was performed in N. benthamiana leaves as previously described (Huang et al. 2020). Briefly, the cLUC and nLUC plasmids were transformed into A. tumefaciens GV3101 cells. The bacteria containing individual constructs were suspended in IFB buffer (0.5% [w/v] glucose, 10 mm MES, 10 mm MgCl2, and 150 μm acetosyringone, pH 5.7) at OD600 = 0.5 and then mixed equally in each pair as indicated. The mixed culture was then infiltrated into N. benthamiana leaves. After infiltration for 2 d, the LUC activity was detected using the LB 985 NightSHADE system (Berthold Technologies).
Co-IP assay
For Co-IP assay in vivo, the seedlings were ground in liquid nitrogen and homogenized in IP buffer (50 mm Tris-HCl [pH 7.5], 150 mm NaCl, 1 mm DTT, 1× protease inhibitor cocktail Complete Minitablets [Roche] (0.1% (v/v) NP40)). Anti-MYC nanobody coated agarose beads (AlpaLife by KangTi, KTSM1306) were added to the extracts for precipitation. Then mixtures of extracts and beads were incubated for 2 h at 4 °C, washed at least 5 times with TBST buffer (50 mm Tris-HCl [pH7.5], 150 mm NaCl, and 0.1% [v/v] Tween-20), and followed by elution in protein extraction buffer before immunoblotting analysis.
BiFC assay
For BiFC assay, pairs of cYFP and nYFP fusion constructs and the nuclear marker SV40T–mCherry were cotransformed into Col-0 protoplast as previously described (Yoo et al. 2007). After culturing the protoplasts for 16 h, the fluorescence was detected by confocal microscopy (Zeiss LSM880). For YFP fluorescence, the excitation wavelength was 514 nm with 5% to 10% output signal intensity, and the emission spectra were collected at 520 to 567 nm with a gain value of 650. For mCherry fluorescence, the excitation wavelength was 516 nm with 5% to 10% output signal intensity, and the emission was detected at 560 to 610 nm with gain values of 600 to 630.
Pull-down assay
All relevant constructs were transformed into E. coli BL21 (DE3) competent cells. The expression of target proteins was induced by 0.3 mm isopropyl-β-d-thiogalactopyranoside, and cells were cultured at 22 °C for 3 h before collection. GST-tagged, MBP-tagged, and Flag-tagged target proteins were enriched by Glutathione Resin (GenScript, L00206-50), Amylose Resin (New England Biolabs, 10087424), and DYKDDDDK G1 Affinity Resin (GenScript, L00432-10), respectively, following the manufacturer's instructions. For pull-down assay, MBP-tagged or Flag-tagged proteins were incubated with beads loaded with corresponding purified GST-tagged proteins, or GST-tagged proteins were incubated with beads loaded with corresponding purified MBP-tagged proteins in pull-down buffer (50 mm Tris-HCl [pH 7.5], 150 mm NaCl, and 0.1% [v/v] NP40) for 2 h at 4 °C with gentle rotation. After washing at least 5 times with pull-down buffer, the protein-bound beads were collected by centrifugation at 1,200 × g for 2 min and then eluted in protein extraction buffer. Proteins were detected by immunoblotting using anti-HA–HRP antibody, anti-GST antibody, anti-Flag–HRP antibody, or by Ponceau staining.
In vitro acetylation analysis
The constructs expressing GST–ATG18a, MBP–GFP, MBP–HLS1, and MBP–HLS1V108A–L151A were transformed into E. coli BL21 (DE3) competent cells. Expression of target proteins was induced by adding 0.3 mm isopropyl-β-d-thiogalactopyranoside and incubating at 16 °C overnight, after which the cells were harvested. GST-tagged and MBP-tagged target proteins were enriched by Glutathione Resin (GenScript, L00206-50) and Amylose Resin (New England Biolabs, 10087424), respectively. For acetylation assay in vitro, MBP–GFP, MBP–HLS1, and MBP–HLS1V108A–L151A were eluted with 10 mm maltose and then incubated with beads loaded with GST–ATG18a proteins in acetylation buffer (50 mm HEPES [pH 6.8], 50 mm NaCl, 1 mm acetyl-CoA, 1 mm DTT, and 5 mm nicotinamide) for 3 h at 30 °C. Samples were subjected to SDS–PAGE for immunoblotting. Anti-Ac-K and anti-GFP antibodies were used to detect the acetylation level and the loading of ATG18a, respectively.
In vivo acetylation assay
For acetylation assay in vivo, 1-wk-old transgenic seedlings expressing ATG18a–GFP (WT or K323R–K331R–K420R) were exposed to carbon or nitrogen starvation for indicated times. The samples were ground in liquid nitrogen and homogenized in IP buffer (detailed in the Co-IP section) containing 5 mm nicotinamide. Different types of ATG18a–GFP proteins were precipitated by anti-GFP nanobody agarose beads (AlpaLife by KangTi, KTSM1301). The beads were washed at least 5 times with TBST buffer (50 mm Tris-HCl [pH7.5], 150 mm NaCl, and 0.1% [v/v] Tween-20) and then subjected to SDS–PAGE for immunoblotting analysis using anti-Ac-K and anti-GFP antibodies.
Protein–lipid binding assay
The protein–lipid binding assay was performed as described previously (Han et al. 2020; Tan et al. 2020) with modification. Briefly, membranes containing PIP arrays (Echelon Biosciences, P-6100) or PVDF membranes overlaid with 1 mm solution of PtdIns(3)P (Echelon Biosciences) were blocked in 3% (w/v) fatty acid–free BSA in TBST (50 mm Tris-HCl [pH 7.5], 150 mm NaCl, and 0.1% [v/v] Tween-20) for 1 h. The membranes were then incubated in the same solution with purified protein for 1 h at 4 °C with gentle agitation. The membranes were washed 3 times with TBST buffer (50 mm Tris-HCl [pH 7.5], 150 mm NaCl, and 0.1% [v/v] Tween-20) and incubated with SDS sample buffer for 10 min. The eluates were separated by SDS–PAGE, and membrane-bound GST–ATG18a was analyzed by using anti-GST antibody.
Hook curvature measurement
Hook curvature was measured following Huang et al. (2020). The etiolated seedlings were photographed using a Canon camera (EOS 760D), and the hook curvature angles between the cotyledons and hypocotyls were measured from digital images by ImageJ software (http://rsbweb.nih.gov/ij/).
Statistical analysis
In this study, the significance of the difference between 2 noted samples was determined using a 2-tailed Student's t test. Data are presented as mean ± Sd. The level of statistical significance is indicated by asterisks (*P < 0.05 and **P < 0.01). The relative intensities of each band on immunoblots were quantified using ImageJ (http://rsbweb.nih.gov/ij/). The detailed statistical results are listed in Supplemental Data Set 2.
Accession numbers
Sequence information from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ATG1a (AT3g61960), ATG1b (AT3G53930), ATG1c (AT2G37840), ATG2 (AT3G19190), ATG3 (AT5G61500), ATG5 (AT5G17290), ATG6 (AT3G61710), ATG7 (AT5G45900), ATG8e (AT2G45170), ATG12 (AT1G54210), ATG18a (AT3G62770), ATG18b (AT4G30510), ATG18c (AT2G40810), ATG18d (AT3G56440), ATG18e (AT5G05150), ATG18f (AT5G54730), ATG18g (AT1G03380), ATG18h (AT1G54710), HLS1 (AT4G37580), MAK3 (AT2G38130), NAA10 (AT5G13780), NAA20 (AT1G03150), NAA60 (AT5G16800), F21P24.12 (AT2G23060), GNA1 (AT5G15770), MCC1 (AT3G02980), SAT1 (AT1G55920), SAT3 (AT3G13110), and SDIR1(AT3G55530).
Supplementary Material
Acknowledgments
We thank the ABRC (www.arabidopsis.org) for providing atg18a mutant seed pools. We thank Prof. Shi Xiao (Sun Yat-sen University, China) for kindly providing the GFP–ATG8e line and atg5-1 mutant. We thank Prof. Liwen Jiang (The Chinese University of Hong Kong, China) for kindly providing the mCherry–ATG8e line. We thank Prof. Ziqiang Zhu (Nanjing Normal University, China) for sharing plasmids. We thank Dr. Wei Yan (Southern University of Science and Technology, China) for analyzing mass spectrometry data.
Contributor Information
Li Huang, New Cornerstone Science Laboratory, Institute of Plant and Food Science, Department of Biology, School of Life Sciences, Southern University of Science and Technology (SUSTech), Shenzhen, Guangdong 518055, China.
Xing Wen, New Cornerstone Science Laboratory, Institute of Plant and Food Science, Department of Biology, School of Life Sciences, Southern University of Science and Technology (SUSTech), Shenzhen, Guangdong 518055, China.
Lian Jin, New Cornerstone Science Laboratory, Institute of Plant and Food Science, Department of Biology, School of Life Sciences, Southern University of Science and Technology (SUSTech), Shenzhen, Guangdong 518055, China.
Huihui Han, New Cornerstone Science Laboratory, Institute of Plant and Food Science, Department of Biology, School of Life Sciences, Southern University of Science and Technology (SUSTech), Shenzhen, Guangdong 518055, China.
Hongwei Guo, New Cornerstone Science Laboratory, Institute of Plant and Food Science, Department of Biology, School of Life Sciences, Southern University of Science and Technology (SUSTech), Shenzhen, Guangdong 518055, China.
Author contributions
H.G., L.H., and X.W. conceived the program and designed the research plan. L.H. carried out most experiments and data analysis. X.W. participated in acetylation assays and protein–lipid binding assays. L.J. participated in acetylation assays. H.H. participated in genotyping, vector construction, and protein expression. L.H., H.G., and X.W. wrote the article.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Loss of HLS1 function does not affect carbon starvation–induced ATG18a accumulation.
Supplemental Figure S2. HLS1 interacts with ATG3 and ATG18a in N. benthamiana leaves.
Supplemental Figure S3. Effect of nutrient starvation on the acetylation of GFP.
Supplemental Figure S4. Predictive analysis of potential acetylation sites in ATG18a protein.
Supplemental Figure S5. ATG18a interacts with both HLS1 and ATG2 in vitro.
Supplemental Figure S6. Molecular identification and phenotypic analysis of atg18a mutants.
Supplemental Figure S7. Subcellular localization of ATG18a or ATG18a acetylation variant in leaves and roots in response to carbon starvation.
Supplemental Figure S8. Binding of recombinant GST–ATG18a and GST–ATG18aRRR proteins to PtdIns(3)P.
Supplemental Figure S9. Protein abundance of ATG18a–GFP and ATG18aRRR–GFP upon carbon starvation.
Supplemental Figure S10. Amino acid sequence alignment of HLS1 with acetyltransferases in Arabidopsis.
Supplemental Figure S11. Mutations in the putative acetyltransferase domain of HLS1 do not affect its interaction with ATG18a.
Supplemental Figure S12. Phenotypes of Col-0, atg5-1, atg18a, hls1-1, and transgenic seedlings in response to carbon starvation.
Supplemental Figure S13. Phenotypes of leaf senescence in Col-0, atg5-1, atg18a, hls1-1, and transgenic lines.
Supplemental Figure S14. Mutations of both V108 and L151 sites in HLS1 affect plant response to oxidative stress.
Supplemental Table S1. Mutants generated in this study.
Supplemental Table S2. Transgenic A. thaliana plants generated in this study.
Supplemental Data Set 1. Primers used in this study.
Supplemental Data Set 2. Details of the t test analyses in this study.
Funding
This work was funded by the National Natural Science Foundation of China (32230008 to H.G. and 31970308 to X.W.), Shenzhen Science and Technology Innovation Program (20200925161843002 to H.G.), the New Cornerstone Science Foundation (NCI202235 to H.G.), and the China Postdoctoral Science Foundation (2019M662939 to L.H.).
Data availability
The data underlying this article are available in the article and in its online supplementary material.
References
- A M, Latario CJ, Pickrell LE, Higgs HN. Lysine acetylation of cytoskeletal proteins: emergence of an actin code. J Cell Biol. 2020:219(12):e202006151. 10.1083/jcb.202006151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, Li M, Guo H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012:22(5):915–927. 10.1038/cr.2012.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A, Yruela I, Gotor C, Bassham DC. Persulfidation of ATG18a regulates autophagy under ER stress in Arabidopsis. Proc Natl Acad Sci U S A. 2021:118(20):e2023604118. 10.1073/pnas.2023604118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bánréti A, Sass M, Graba Y. The emerging role of acetylation in the regulation of autophagy. Autophagy 2013:9(6):819–829. 10.4161/auto.23908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Song W-M, Wang P, Yu X, Li B, Jiang C, Shiu S-H, Zhang H, Bassham DC. COST1 regulates autophagy to control plant drought tolerance. Proc Natl Acad Sci U S A. 2020:117(13):7482–7493. 10.1073/pnas.1918539117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001:8(6):1243–1254. 10.1016/S1097-2765(01)00414-2 [DOI] [PubMed] [Google Scholar]
- Bassham DC, Laporte M, Marty F, Moriyasu Y, Ohsumi Y, Olsen LJ, Yoshimoto K. Autophagy in development and stress responses of plants. Autophagy 2006:2(1):2–11. 10.4161/auto.2092 [DOI] [PubMed] [Google Scholar]
- Chen L, Liao B, Qi H, Xie L-J, Huang L, Tan W-J, Zhai N, Yuan L-B, Zhou Y, Yu L-J, et al. Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 2015:11(12):2233–2246. 10.1080/15548627.2015.1112483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou J-M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008:146(2):368–376. 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C, Li X, Fang P, Xia X, Shi K, Zhou Y, Zhou J, Yu J. Brassinosteroids act as a positive regulator of NBR1-dependent selective autophagy in response to chilling stress in tomato. J Exp Bot. 2020:71(3):1092–1106. 10.1093/jxb/erz466 [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009:325(5942):834–840. 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014:15(8):536–550. 10.1038/nrm3841 [DOI] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 2010:62(3):483–493. 10.1111/j.1365-313X.2010.04166.x [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. Journal of molecular biology. 2006:362(5):1120–1151. 10.1016/j.jmb.2006.08.017 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998:16(6):735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Contento AL, Xiong Y, Bassham DC. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005:42(4):598–608. 10.1111/j.1365-313X.2005.02396.x [DOI] [PubMed] [Google Scholar]
- Coon SL, Roseboom PH, Baler R, Weller JL, Namboodiri MAA, Koonin EV, Klein DC. Pineal serotonin N-acetyltransferase: expression cloning and molecular analysis. Science 1995:270(5242):1681–1683. 10.1126/science.270.5242.1681 [DOI] [PubMed] [Google Scholar]
- Deng W, Wang C, Zhang Y, Xu Y, Zhang S, Liu Z, Xue Y. GPS-PAIL: prediction of lysine acetyltransferase-specific modification sites from protein sequences. Sci Rep. 2016:6(1):39787. 10.1038/srep39787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002:277(36):33105–33114. 10.1074/jbc.M204630200 [DOI] [PubMed] [Google Scholar]
- Dove SK, Piper RC, McEwen RK, Yu JW, King MC, Hughes DC, Thuring J, Holmes AB, Cooke FT, Michell RH, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004:23(9):1922–1933. 10.1038/sj.emboj.7600203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014:24(1):24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007:143(3):1132–1139. 10.1104/pp.106.093864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllgrabe J, Klionsky DJ, Joseph B. Histone post-translational modifications regulate autophagy flux and outcome. Autophagy 2013:9(10):1621–1623. 10.4161/auto.25803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Wen X, Zhang W, Huang L, Peng Y, Jin L, Han H, Zhang L, Li W, Guo H. Arabidopsis EIN2 represses ABA responses during germination and early seedling growth by inactivating HLS1 protein independently of the canonical ethylene pathway. Plant J. 2023:115(6):1514–1527. 10.1111/tpj.16335 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 1990:2(6):513–523. 10.1105/tpc.2.6.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang Y, Zhao F, Zhang T, Yu X. An improved protein lipid overlay assay for studying lipid-protein interactions. Plant Methods 2020:16(1):33. 10.1186/s13007-020-00578-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Yu B, Wang Y, Liu Y. Role of plant autophagy in stress response. Protein Cell 2011:2(10):784–791. 10.1007/s13238-011-1104-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002:129(3):1181–1193. 10.1104/pp.011024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Jin L, Wen X, Yu F, Xie Q, Guo H. The RING E3 ligase SDIR1 destabilizes EBF1/EBF2 and modulates the ethylene response to ambient temperature fluctuations in Arabidopsis. Proc Natl Acad Sci U S A. 2021:118(6):e2024592118. 10.1073/pnas.2024592118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Dong Z, Guo P, Zhang X, Qiu Y, Li B, Wang Y, Guo H. Salicylic acid suppresses apical hook formation via NPR1-mediated repression of EIN3 and EIL1 in Arabidopsis. Plant Cell 2020:32(3):612–629. 10.1105/tpc.19.00658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015:57(3):456–466. 10.1016/j.molcel.2014.12.013 [DOI] [PubMed] [Google Scholar]
- Huang L, Yu L-J, Zhang X, Fan B, Wang F-Z, Dai Y-S, Qi H, Zhou Y, Xie L-J, Xiao S. Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy 2019a:15(3):407–422. 10.1080/15548627.2018.1520547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zheng C, Liu F, Yang C, Zheng P, Lu X, Tian J, Chung T, Otegui MS, Xiao S, et al. Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. Plant Cell 2019b:31(12):2973–2995. 10.1105/tpc.19.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS, Proshad R, Kormoker T, Tusher TR. Autophagy-mediated nutrient recycling and regulation in plants: a molecular view. J Plant Biol. 2019:62(5):307–319. 10.1007/s12374-019-0213-0 [DOI] [Google Scholar]
- Jin L, Qin Q, Wang Y, Pu Y, Liu L, Wen X, Ji S, Wu J, Wei C, Ding Bet al. Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathog. 2016:12(9):e1005847. 10.1371/journal.ppat.1005847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Zhu Z. HOOKLESS1 is a positive regulator in Arabidopsis thermomorphogenesis. Sci China Life Sci. 2019:62(3):423–425. 10.1007/s11427-018-9418-2 [DOI] [PubMed] [Google Scholar]
- Kang S, Shin KD, Kim JH, Chung T. Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep. 2018:37(4):653–664. 10.1007/s00299-018-2258-9 [DOI] [PubMed] [Google Scholar]
- Kotani T, Kirisako H, Koizumi M, Ohsumi Y, Nakatogawa H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc Natl Acad Sci U S A. 2018:115(41):10363–10368. 10.1073/pnas.1806727115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Wang F, Zheng Z, Fan B, Chen Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011:66(6):953–968. 10.1111/j.1365-313X.2011.04553.x [DOI] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008:105(9):3374–3379. 10.1073/pnas.0712145105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009:284(10):6322–6328. 10.1074/jbc.M807135200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 1996:85(2):183–194. 10.1016/S0092-8674(00)81095-8 [DOI] [PubMed] [Google Scholar]
- Li F, Chung T, Vierstra RD. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 2014:26(2):788–807. 10.1105/tpc.113.120014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012:17(9):526–537. 10.1016/j.tplants.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Liao C-J, Lai Z, Lee S, Yun DJ, Mengiste T. Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. Plant Cell 2016:28(7):1662–1681. 10.1105/tpc.16.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic membranes. Methods Enzymol. 1987:148:350–382. 10.1016/0076-6879(87)48036-1 [DOI] [Google Scholar]
- Lin S-Y, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang S-M, Lian G, Liu Q, Ruan K, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 2012:336(6080):477–481. 10.1126/science.1217032 [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012:63(1):215–237. 10.1146/annurev-arplant-042811-105441 [DOI] [PubMed] [Google Scholar]
- Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 2012:24(11):4635–4651. 10.1105/tpc.112.101535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009:5(7):954–963. 10.4161/auto.5.7.9290 [DOI] [PubMed] [Google Scholar]
- Luo L, Zhang P, Zhu R, Fu J, Su J, Zheng J, Wang Z, Wang D, Gong Q. Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front Plant Sci. 2017:8:1459. 10.3389/fpls.2017.01459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell. 2014:53(5):710–725. 10.1016/j.molcel.2014.01.016 [DOI] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD. Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol. 2018:69(1):173–208. 10.1146/annurev-arplant-042817-040606 [DOI] [PubMed] [Google Scholar]
- McEwan DG, Dikic I. The three musketeers of autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol. 2011:21(4):195–201. 10.1016/j.tcb.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S, Galili G, Genschik P, Fernie AR, Avin-Wittenberg T. Autophagy in plants—what’s new on the menu? Trends Plant Sci. 2016:21(2):134–144. 10.1016/j.tplants.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Narita T, Weinert BT, Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019:20(3):156–174. 10.1038/s41580-018-0081-3 [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997:22(5):154–155. 10.1016/S0968-0004(97)01034-7 [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008:283(35):23972–23980. 10.1074/jbc.M803180200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M-A, Hayashi S, Sawa S, Hashimoto-Ohta A, Nakamura K. Involvement of HLS1 in sugar and auxin signaling in Arabidopsis leaves. Plant Cell Physiol. 2006:47(12):1603–1611. 10.1093/pcp/pcl027 [DOI] [PubMed] [Google Scholar]
- O'Leary BM, Scafaro AP, Fenske R, Duncan O, Ströher E, Petereit J, Millar AH. Rubisco lysine acetylation occurs at very low stoichiometry in mature Arabidopsis leaves: implications for regulation of enzyme function. Biochem J. 2020:477(19):3885–3896. 10.1042/BCJ20200413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Zhang D, Qiu Y, Xiao Z, Ji Y, Li W, Xia Y, Wang Y, Guo H. Growth asymmetry precedes differential auxin response during apical hook initiation in Arabidopsis. J Integr Plant Biol. 2022:64(1):5–22. 10.1111/jipb.13190 [DOI] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 2008:178(3):1339–1353. 10.1534/genetics.107.086199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Xia F, Xiao S. Autophagy in plants: physiological roles and post-translational regulation. J Integr Plant Biol. 2021:63(1):161–179. 10.1111/jipb.12941 [DOI] [PubMed] [Google Scholar]
- Qi H, Xia F-N, Xie L-J, Yu L-J, Chen Q-F, Zhuang X-H, Wang Q, Li F, Jiang L, Xie Q, et al. TRAF family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell 2017:29(4):890–911. 10.1105/tpc.17.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MC, Lieb A, Nijhout HF. The biological significance of substrate inhibition: a mechanism with diverse functions. Bioessays 2010:32(5):422–429. 10.1002/bies.200900167 [DOI] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. Thef Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004:6(1):79–90. 10.1016/S1534-5807(03)00402-7 [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk VI, Kaushik S, Klionsky DJ. In search of an “autophagomometer”. Autophagy 2009:5(5):585–589. 10.4161/auto.5.5.8823 [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Analytical Biochemistry. 1987:166(2):368–379. 10.1016/0003-2697(87)90587-2 [DOI] [PubMed] [Google Scholar]
- Signorelli S, Tarkowski LP, Van den Ende W, Bassham DC. Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 2019:24(5):413–430. 10.1016/j.tplants.2019.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000:64(2):435–459. 10.1128/MMBR.64.2.435-459.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 2011:23(10):3761–3779. 10.1105/tpc.111.090993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning S, Lamark T, Krause K, Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011:7(9):993–1010. 10.4161/auto.7.9.16389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Zhang X, Kong W, Yang X-L, Molnár G, Vondráková Z, Filepová R, Petrášek J, Friml J, Xue H-W. The lipid code-dependent phosphoswitch PDK1-D6PK activates PIN-mediated auxin efflux in Arabidopsis. Nat Plants. 2020:6(5):556–569. 10.1038/s41477-020-0648-9 [DOI] [PubMed] [Google Scholar]
- Tang J, Bassham DC. Autophagy during drought: function, regulation, and potential application. Plant J. 2022:109(2):390–401. 10.1111/tpj.15481 [DOI] [PubMed] [Google Scholar]
- Tercero JC, Riles LE, Wickner RB. Localized mutagenesis and evidence for post-transcriptional regulation of MAK3. J Biol Chem. 1992:267(28):20270–20276. 10.1016/S0021-9258(19)88696-9 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005:138(4):2097–2110. 10.1104/pp.105.060673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W-Y, Zhang L, Xing S, Ma Z, Liu J, Gu H, Qin G, Qu L-J. Arabidopsis AtVPS15 plays essential roles in pollen germination possibly by interacting with AtVPS34. J Genet Genomics. 2012:39(2):81–92. 10.1016/j.jgg.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Wani WY, Boyer-Guittaut M, Dodson M, Chatham J, Darley-Usmar V, Zhang J. Regulation of autophagy by protein post-translational modification. Lab Invest. 2015:95(1):14–25. 10.1038/labinvest.2014.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Kobayashi T, Yamamoto H, Hoshida H, Akada R, Inagaki F, Ohsumi Y, Noda NN. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem. 2012:287(38):31681–31690. 10.1074/jbc.M112.397570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welters P, Takegawa K, Emr SD, Chrispeels MJ. AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc Natl Acad Sci U S A. 1994:91(24):11398–11402. 10.1073/pnas.91.24.11398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Klionsky DJ. An overview of macroautophagy in yeast. J Mol Biol. 2016:428(9):1681–1699. 10.1016/j.jmb.2016.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J, Park E, Dinesh-Kumar SP. Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc Natl Acad Sci U S A. 2014:111(2):863–868. 10.1073/pnas.1318207111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Kong X, Song H, Han Q, Zhang S. Advances in proteome-wide analysis of plant lysine acetylation. Plant Commun. 2022:3(1):100266. 10.1016/j.xplc.2021.100266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen Q-F, Chan S-W, Zheng S-X, Ma J, Wang M, Welti R, Chye M-L. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 2010:22(5):1463–1482. 10.1105/tpc.110.075333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005:42(4):535–546. 10.1111/j.1365-313X.2005.02397.x [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007:143(1):291–299. 10.1104/pp.106.092106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Shen W, Yang L, Sun Y, Li X, Lai M, Wei J, Wang C, Xu Y, Li F, et al. HY5-HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol Plant. 2020:13(3):515–531. 10.1016/j.molp.2020.02.011 [DOI] [PubMed] [Google Scholar]
- Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, Ma C, Sun Y, Zhang S, Feng W, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science 2012:336(6080):474–477. 10.1126/science.1216990 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature protocols. 2007:2(7):1565–1637. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004:16(11):2967–2983. 10.1105/tpc.104.025395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ji Y, Xue C, Ma H, Xi Y, Huang P, Wang H, An F, Li B, Wang Y, et al. Integrated regulation of apical hook development by transcriptional coupling of EIN3/EIL1 and PIFs in Arabidopsis. Plant Cell 2018:30(9):1971–1988. 10.1105/tpc.18.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Shao L, Wang J, Zhang Y, Guo X, Peng Y, Cao Y, Lai Z. Phosphorylation of ATG18a by BAK1 suppresses autophagy and attenuates plant resistance against necrotrophic pathogens. Autophagy 2021:17(9):2093–2110. 10.1080/15548627.2020.1810426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhu Z, An F, Hao D, Li P, Song J, Yi C, Guo H. Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 2014:26(3):1105–1117. 10.1105/tpc.113.122002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang J, Yu J-Q, Chen Z. Role and regulation of autophagy in heat stress responses of tomato plants. Front Plant Sci. 2014:5:174. 10.3389/fpls.2014.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Chung KP, Cui Y, Lin W, Gao C, Kang B-H, Jiang L. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci U S A. 2017:114(3):E426–E435. 10.1073/pnas.1616299114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Wang H, Lam SK, Gao C, Wang X, Cai Y, Jiang L. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 2013:25(11):4596–4615. 10.1105/tpc.113.118307 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.