Abstract
Temperature is a major factor that regulates plant growth and phenotypic diversity. To ensure reproductive success at a range of temperatures, plants must maintain developmental stability of their sexual organs when exposed to temperature fluctuations. However, the mechanisms integrating plant floral organ development and temperature responses are largely unknown. Here, we generated barley and rice loss-of-function mutants in the SEPALLATA-like MADS-box gene MADS8. The mutants in both species form multiple carpels that lack ovules at high ambient temperatures. Tissue-specific markers revealed that HvMADS8 is required to maintain floral meristem determinacy and ovule initiation at high temperatures, and transcriptome analyses confirmed that temperature-dependent differentially expressed genes in Hvmads8 mutants predominantly associate with floral organ and meristem regulation. HvMADS8 temperature-responsive activity relies on increased binding to promoters of downstream targets, as revealed by a cleavage under targets and tagmentation (CUT&Tag) analysis. We also demonstrate that HvMADS8 directly binds to 2 orthologs of D-class floral homeotic genes to activate their expression. Overall, our findings revealed a new, conserved role for MADS8 in maintaining pistil number and ovule initiation in cereal crops, extending the known function of plant MADS-box proteins in floral organ regulation.
MADS8 maintains pistil number and ovule initiation at high ambient temperature in barley and rice via a temperature-dependent increase in binding to its downstream targets.
IN A NUTSHELL.
Background: Temperature is a major factor regulating plant growth and phenotypic diversity. When exposed to temperature fluctuations, plants must maintain the developmental stability of their sexual organs in order to survive and reproduce, especially in the face of current and expected global warming trends. In the grass family, the development of the 4 concentric flower whorls including the pistil, the innermost female reproductive organ, contributes directly to reproductive success. However, the genes and mechanisms maintaining floral organ identity and development in response to high temperature remain underexplored.
Question: We wanted to know how female reproductive development is maintained in cereal crops at high ambient temperatures, and whether members of the SEPALLATA subfamily of floral organ regulators contribute.
Findings: We discovered that barley (Hordeum vulgare) and rice (Oryza sativa) MADS8 genes are indispensable for female reproductive development at high temperatures. Knockout mutants lacking functional MADS8 exhibited multiple pistils with no ovule at high ambient temperatures. We also showed that HvMADS8 increases its binding to the promoters of its downstream target genes in a temperature-responsive manner, which maintains floral meristem determinacy and ovule initiation at high temperatures. Our results shed light on the conserved molecular mechanism that stabilizes female reproductive development in grass species exposed to high temperatures, extending the known functions of floral organ identity regulators.
Next steps: We aim to stabilize and improve crop yields under high-temperature conditions. Further study will focus on understanding pathways regulating reproductive organ development in greater detail and engineering plants with improved developmental stability in the context of global warming by manipulating the expression of these key regulators.
Introduction
To survive and reproduce, plants must adjust their physiology and development to the changing environment, especially in the face of current and expected global warming (Zhao et al. 2017; Vu et al. 2019). High, but not stressful, ambient temperatures induce a series of morphological adjustments in plants, collectively termed thermomorphogenesis, such as reduced leaf area and elongated hypocotyls (Franklin 2009; Casal and Balasubramanian 2019). Several thermosensors and thermally responsive elements, such as phytochrome B (phyB), PHYTOCHROME INTERACTING FACTOR 4 (PIF4), and the histone variant H2A.Z, have been identified in the model plant Arabidopsis (Arabidopsis thaliana) (Kumar and Wigge 2010; Quint et al. 2016; Ding et al. 2020; Xue et al. 2021).
In the past 2 decades, molecular genetic studies have uncovered genes essential for reproductive development at high temperatures in grasses (Poaceae). These studies have focused on rice (Oryza sativa), where thermosensitive genic male sterile (TGMS) lines that are male sterile at high temperatures but have normal fertility at low temperatures are widely used in breeding (Chen et al. 2007; Zhou et al. 2014; Yu et al. 2017; Zhu et al. 2020; Wu et al. 2022). Recently, a maize (Zea mays) thermosensitive mutant, thermosensitive vanishing tassel1-R (tvt1-R), was characterized (Xie et al. 2020). However, compared to the extensively studied regulators of vegetative growth, the genes and mechanisms maintaining floral organ identity and development in response to high temperature are underexplored.
Another economically important Poaceae crop, barley (Hordeum vulgare), has flowers with a structure similar to those of rice. Within each barley floret are 4 concentric whorls; the outermost whorl produces 2 bract-like organs (lemma and palea) that surround the inner 3 whorls, which produce the 2 lodicules, 3 stamens, and a pistil born from the 3 fused carpel primordia, respectively (Kellogg 2001; Dreni et al. 2013). The mature pistil, also referred to as the gynoecium, is the female reproductive organ and consists predominantly of a carpel that bears a single ovule. Carpels develop on the basis of timed floral meristem (FM) termination (Shen et al. 2021). Unlike indeterminate barley inflorescence meristems that can continuously produce lateral primordia, the FM is determinate, which means its stem cells will be consumed by the final floral organ initiated from it (Chu et al. 2006; Dreni et al. 2007; Yamaki et al. 2011). Several mutants with loss of FM determinacy displaying extra pistils have been reported in cereal plants (Yang and Tucker 2021). FLORAL ORGAN NUMBER (FON) genes in rice control FM size, and loss-of-function fon mutants produce larger meristems with supernumerary anthers and pistils that can produce grain (Jiang et al. 2005; Chu et al. 2006; Moon et al. 2006; Suzaki et al. 2006). Florets from tri-pistil mutants in wheat (Triticum aestivum) produce 3 pistils that occasionally contain multiple ovules and generate 1 to 3 grains in a single floret (Peng et al. 2008; Li et al. 2020; Liu et al. 2022). The 2 extra pistils appear to be initiated late in floret development, and anthers are retained, suggesting that the tri-pistil phenotype may relate to meristem determinacy (Nagasawa et al. 2003; Li et al. 2020; Selva et al. 2021; Liu et al. 2022).
According to the ABCDE combinatorial model proposed to explain flower development in Arabidopsis, snapdragon (Antirrhinum majus), and petunia (Petunia hybrida), C-class and D-class homeotic genes are required for carpel and ovule development, while E-class genes are essential for the function of the other 4 classes (Colombo et al. 1995; Pelaz et al. 2000, 2001; Favaro et al. 2003; Pinyopich et al. 2003). Except for the A-class APETALA 2 (AP2) gene, all ABCDE genes encode MIKC-type MADS-box transcription factors (Jofuku et al. 1994). These proteins bind as dimers to DNA sequences named “CArG” boxes and interact to form heterotetrameric complexes that can recognize different CArG targets (Muiño et al. 2014; Gusewski et al. 2017). In rice, there are 2 duplicated D-lineage genes, OsMADS13 and OsMADS21; Osmads13 mutants result in the homeotic conversion of ovules into carpels (Dreni et al. 2007). Furthermore, reiterative carpel development in Osmads13 lines indicates a role for OsMADS13 in FM determinacy, confirmed by the prolonged expression of the meristematic cell marker ORYZA SATIVA HOMEOBOX 1 (OSH1), in Osmads13 florets (Yamaki et al. 2011). Recent transcriptome analysis revealed the upregulation of 3 rice carpel identity genes in Osmads13 FM and ovule primordia, namely DROOPING LEAF (DL) and the 2 C-class genes OsMADS3 and OsMADS58, indicating an important repressor role for OsMADS13 in carpel development (Osnato et al. 2021). In contrast, OsMADS21 appears to play no crucial role in the formation of reproductive organs (Dreni et al. 2007). Nevertheless, the Osmads13 phenotype can still be partially rescued by OsMADS21 expressed from the OsMADS13 promoter, suggesting it has retained some ovule identity determination activity (Dreni et al. 2011).
E-class proteins are encoded by SEPALLATA (SEP) subfamily MADS-box genes, which can be further divided into 2 subclades in all angiosperms: in rice, the LOFSEP clade contains OsMADS1, OsMADS5, and OsMADS34, while the SEP3 clade comprises OsMADS7 and OsMADS8 (also known as OsMADS45 and OsMADS24, respectively) (Malcomber and Kellogg 2005; Zahn et al. 2005; Arora et al. 2007). All rice SEP genes have a conserved and redundant role in pistil development. Pistils are either increased or completely missing in several combinations of mutants defective in LOFSEP clade members (Wu et al. 2018), and the simultaneous knockdown of OsMADS7 and OsMADS8 causes different degrees of defects in the inner 3 whorls, affecting carpel fusion and FM determinacy with additional reproductive organ-like structures initiated inside the mutant carpels (Cui et al. 2010).
HvMADS1 was recently shown to be responsible for maintaining high temperature-responsive inflorescence meristem determinacy in barley (Li et al. 2021). At high temperatures, HvMADS1 exhibits increased binding to A-tract CArG-box promoter elements in downstream target genes to regulate inflorescence differentiation and phytohormone signaling, thereby integrating temperature responses and cytokinin homeostasis to repress meristem activity. This research extended the role of SEP proteins in inflorescence thermomorphogenesis while raising more questions as to whether and how other SEP proteins are involved in thermally responsive development.
In this study, we demonstrate the conserved role of MADS8 in regulating the formation of a single pistil at high ambient temperature in 2 grass species, barley and rice. In their respective mads8-null mutants, floral organs develop normally under control temperatures, but extra carpels repeatedly form without ovules at high temperatures. Our results demonstrate that HvMADS8 is crucial in maintaining FM determinacy and ovule initiation under high ambient temperature by modulating the expression of downstream genes in flower development pathways in a temperature-dependent manner. Furthermore, our study offers new understanding and genetic materials that might be used in the context of managing plant responses to global warming.
Results
HvMADS8 regulates pistil number in response to high temperature
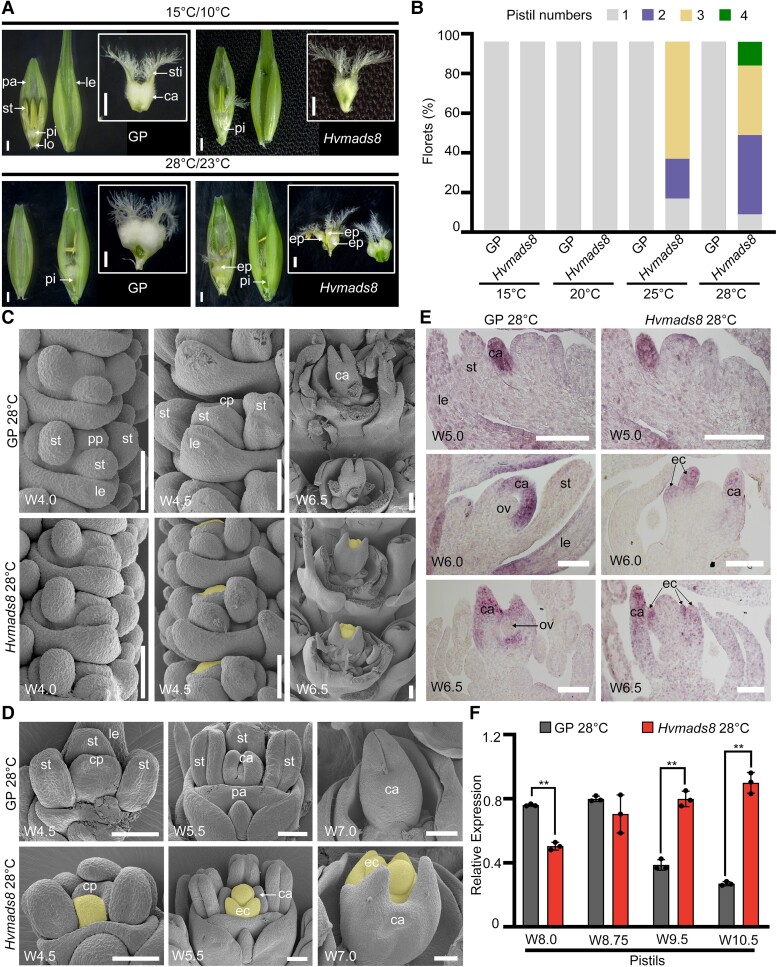
Having previously shown that spikes of Hvmads1 mutants (LOFSEP clade) produce ectopic spike/spikelet-like organs at high temperatures (Li et al. 2021), we examined the inflorescence phenotype of a SEP3 clade mutant at different temperature conditions. Using clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated gene editing (Ma et al. 2015), we generated loss-of-function Hvmads8 mutants in the commonly used UK barley variety Golden Promise (GP) and the Australian barley variety WI4330 (WI), which is adapted to a warmer climate (Supplemental Fig. S1A). Under control temperatures (15 °C day/10 °C night), the Hvmads8 mutant exhibited no visible changes in flower organ morphology in either background (Fig. 1A; Supplemental Fig. S1B). However, at high temperatures (28 °C day/23 °C night), the mutant lines produced extra pistil-like structures in the fourth (innermost) whorl in the GP and WI backgrounds, while other floral organs maintained a wild-type (WT) structure (Fig. 1A; Supplemental Fig. S1B).
Figure 1.
HvMADS8 regulates pistil number in response to high temperature. A) Pistil phenotypes of the WT GP and Hvmads8 mutants grown under control (15 °C day/10 °C night) or high (28 °C day/23 °C night)-temperature conditions. ca, carpel; ep, extra pistil; le, lemma; lo, lodicule; pa, palea; pi, pistil; st, stamen; sti, stigma. Scale bars, 1 mm. B) Pistil number in GP and Hvmads8 florets grown at different temperatures. C, D) SEM of GP and Hvmads8 spike C) and floret D) at 28 °C at different developmental stages. cp, carpel primordia; ec, extra carpel; pp, pistil primordia. Extra carpel primordia bulges and extra carpels are shaded in yellow. Scale bars, 100 μm. E) In situ hybridization showing the expression of the carpel marker gene, HvDL, in GP and Hvmads8 florets at 28 °C. ov, ovule. Scale bars, 100 μm. F) Relative expression of HvDL as determined by RT-qPCR at different stages in GP and Hvmads8 pistils at 28 °C, relative to HvActin7. Values are means ± Sd; n = 3 biological replicates. Asterisks indicate significant differences (2-way ANOVA test; **P < 0.01).
To confirm this temperature-associated phenotype, we grew WT GP and mutant plants at various temperatures, with control (15 °C) and higher ambient temperatures (20, 25, and 28 °C). Accordingly, we grew seedlings to Waddington stage 1 (W1, inflorescence meristem initiation; Waddington et al. 1983) at 15 °C before subjecting them to high-temperature treatment. After heat treatment, we dissected W9.5 (prior to pollination) florets from WT and Hvmads8 lines and analyzed pistil number. The Hvmads8 lines developed normal pistils with no detectable difference in seed setting rates or grain morphologies compared to WT at 15 or 20 °C (Fig. 1B; Supplemental Fig. S1, C and D). At 25 °C, only 18% (17, n = 96) of the flowers produced 1 pistil, while other flowers had 2 or 3 pistil-like structures. When temperatures increased to 28 °C, the maximum number of pistil-like structures per floret increased to 4, and the proportion of flowers with 1 pistil decreased to 9% (9, n = 96; Fig. 1B). Detailed phenotypic analysis revealed partial extra carpel structures such as stigmas within the single pistil with no distinguishable ovule structure within the ovary (Supplemental Fig. S1E). Consequently, Hvmads8 formed no mature grains at either 25 or 28 °C (Supplemental Fig. S1F). Overall, the number of extra pistils increased with temperature, indicating a dosage, but not a threshold, effect of temperature on the Hvmads8 phenotype, similar to Hvmads1.
We turned to scanning electron microscopy (SEM) to trace pistil development, particularly in the context of carpel growth and differentiation during early stages of inflorescence and floret development. In WT florets, pistil development begins with the initiation of pistil primordia at W4.0 (Fig. 1C). At W4.5, a bulge corresponding to the compound carpel primordium arises first on the lemma side of the FM (Fig. 1, C and D). At stages W5 to W5.5, the carpel primordia elongate to enclose the developing ovule primordium (Fig. 1D). In early-stage Hvmads8 inflorescences, the first pistil and carpel primordia initiated as in WT at W4.0 and W4.5; however, the FM in the center of the gynecium started to display abnormal expansion toward the palea side at W4.5 (Fig. 1, C and D). After W5.0, when the WT ovule primordium is enclosed by the fused compound carpel, extra carpels repeatedly initiated inside the Hvmads8 gynecium, with the innermost carpel maintaining an abnormal FM-like bulge even at W7.0 (Fig. 1D).
Using the barley ortholog of the rice carpel marker gene DL as a probe, we performed in situ hybridization to confirm the identities of extra Hvmads8 carpels. As in rice, the barley HvDL gene is specifically expressed in the lemma and carpel primordia in WT (Fig. 1E) (Yamaguchi et al. 2004). In the first initiated Hvmads8 carpel, we observed no significant changes in HvDL expression relative to WT (W5.0; Fig. 1E). However, at later stages, when the WT ovule primordium has already been enveloped by the carpel, we detected ectopic HvDL transcripts in extra Hvmads8 carpel primordia growing inside the first initiated carpels (Fig. 1E). We detected an overall increased level of HvDL transcripts in late-stage Hvmads8 mutant pistils (W10.5, stage after pollination) grown at high temperatures, in contrast the decreasing HvDL transcript levels seen in WT after completion of pistil development (Fig. 1F). Collectively, these results demonstrate that repeated carpel initiation occurs inside the previously formed carpels throughout gynecium development in Hvmads8 lines, indicating a deficiency in FM determinacy and ovule development.
HvMADS8 induces ovule initiation and establishes FM determinacy
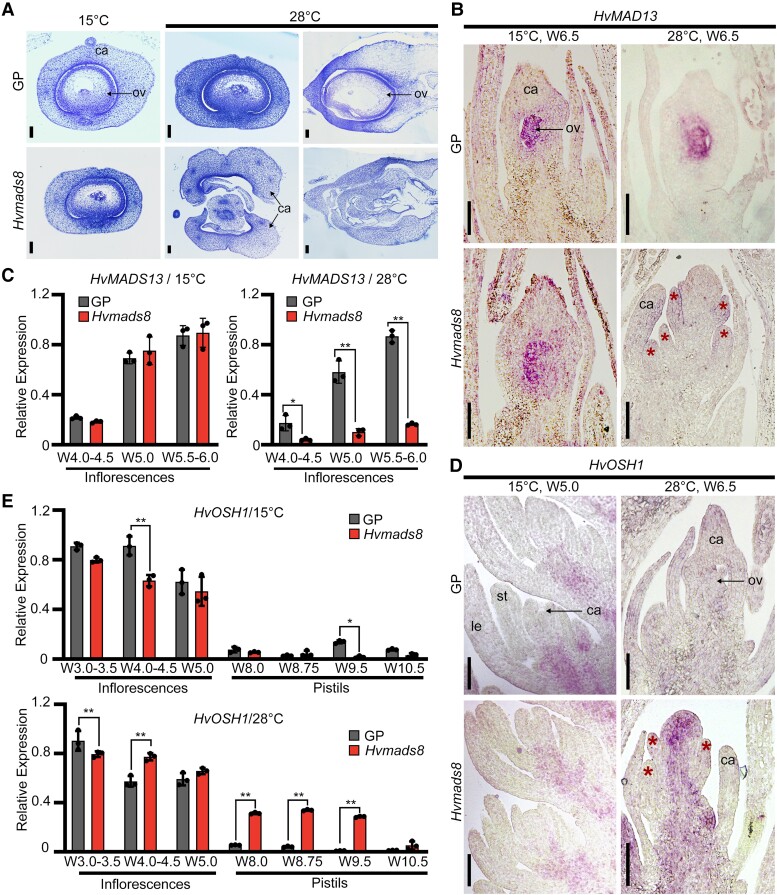
To further investigate Hvmads8 ovule development, we performed a detailed histological analysis using W9.5 florets. In WT W9.5 pistils, the fully mature ovule is composed of 3 tissues: the outer and inner integuments, nucellus, and embryo sac (Fig. 2A). At control temperatures, we observed the complete ovule structure in Hvmads8 pistils, whereas Hvmads8 pistils grown at high temperatures lacked distinguishable ovule tissues inside them (Fig. 2A). We used HvMADS13, the barley homolog of the rice ovule identity gene OsMADS13, to further elucidate ovule initiation status. In WT, HvMADS13 expression mirrored that of OsMADS13 in rice, which is highly specific to the developing ovule and inner layer of the carpel that develops into the ovary wall (Fig. 2B) (Dreni et al. 2007, 2011). HvMADS13 expression in Hvmads8 pistils at 15 °C resembled WT but at 28 °C, HvMADS13 expression was missing from the center of the pistil, with only weak expression detected in carpels (Fig. 2B). Reverse transcription quantitative PCR (RT-qPCR) analysis confirmed significantly decreased HvMADS13 expression levels in early-stage W4.0 to 6.0 inflorescences at high temperatures (Fig. 2C). Taken together, these data show that ovule initiation is severely disrupted in Hvmads8 lines at high temperatures, resulting in the disappearance of all ovule structures.
Figure 2.
HvMADS8 induces ovule initiation and establishes FM determinacy. A) Semi-thin sections of W9.5 GP and Hvmads8 pistils at 15 °C (horizontal sections) or 28 °C (left, horizontal sections; right, longitudinal sections). ca, carpel; ov, ovule. Scale bars, 100 μm. B) In situ hybridization showing expression of HvMADS13 (ovule marker gene) in GP and Hvmads8 florets at 15 and 28 °C. Asterisks indicate extra carpels in Hvmads8 lines at 28 °C. Scale bars, 100 μm. C) Relative expression of HvMADS13 at early stages of inflorescence development at 15 and 28 °C. Values are means ± Sd; n = 3 biological replicates. Asterisks indicate significant differences (2-way ANOVA test; *P < 0.05, **P < 0.01). D) In situ hybridization showing expression of HvOSH1 (meristematic cell marker gene) in GP and Hvmads8 inflorescences and florets at 15 and 28 °C. le, lemma; st, stamen. Asterisks indicate extra carpels in Hvmads8 lines at 28 °C. Scale bars, 100 μm. E) Relative expression of HvOSH1 at different stages of inflorescence and pistil development at 15 and 28 °C. Values are means ± Sd; n = 3 biological replicates. Asterisks indicate significant differences (2-way ANOVA test; *P < 0.05, **P < 0.01).
In the center of Hvmads8 carpels, ovules are repeatedly replaced by a new, indeterminate carpel-like tissue; this phenotype suggests that floral stem cells are not terminated correctly. The barley homolog of the rice FM marker gene OSH1 exhibited ectopic and prolonged expression in Hvmads8 carpels (Fig. 2, D and E). In situ hybridization analysis revealed that HvOSH1 continues to be expressed in the central region after several extra Hvmads8 carpels formed at high temperatures, whereas its expression disappeared from the flower center after carpel formation in WT and Hvmads8 florets at control temperatures (Fig. 2D). We confirmed this prolonged expression of HvOSH1 by RT-qPCR analysis, which revealed transcript levels up to 10-fold higher in late-stage (W8.0 to 10.5) mutant pistils collected at 28 °C (Fig. 2E). Notably, HvMADS13 expression (Fig. 2B) was completely missing from the expanded expression zone of HvOSH1 (Fig. 2D), suggesting a close link between ovule initiation and FM determinacy. Based on these results, we conclude that HvMADS8 functions to induce ovule initiation and establish meristem determinacy in the center of the FM, as loss of HvMADS8 activity leads to missing ovule structures and FM indeterminacy.
Temporal and spatial expression patterns of HvMADS8 at different temperatures
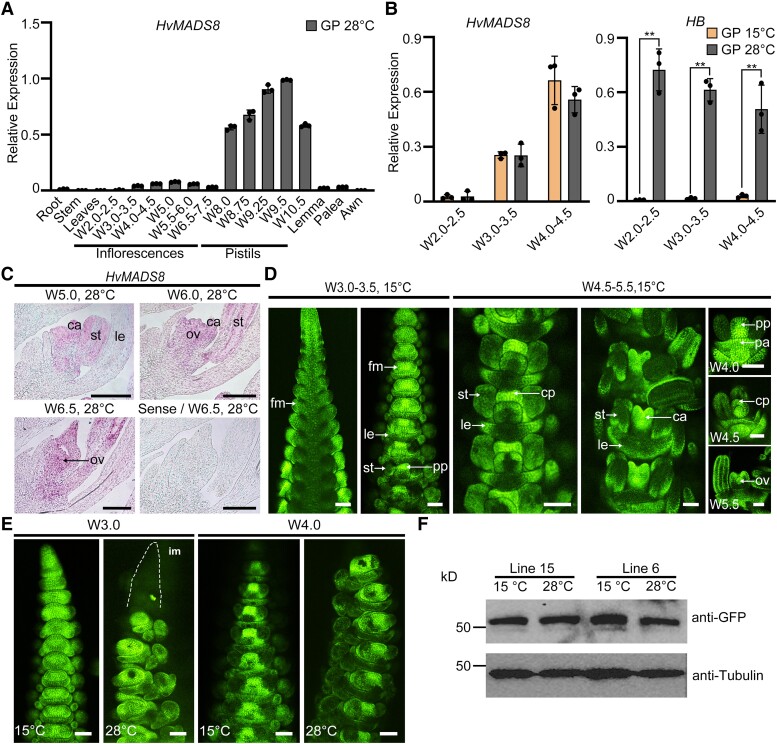
To elucidate how HvMADS8 regulates temperature-responsive pistil development, we analyzed its temporal and spatial expression pattern throughout flower development. We first detected HvMADS8 transcripts in inflorescence meristems from W3.0 to 3.5 (lemma and stamen primordia formation), accumulating later in carpels and ovules (Fig. 3, A and C). Strong transcript accumulation was subsequently detected in maturing pistils, which were dissected from florets from W8.0 onwards (Fig. 3A). Using the temperature-responsive gene Homeobox (HB) as a positive control for high temperature responses (Li et al. 2021), HvMADS8 expression at different temperatures was not significantly altered in early inflorescence meristems (W2.0 to 4.5; Fig. 3B). Similarly, HvMADS8 protein in proHvMADS8:HvMADS8-enhanced GFP (eGFP) transgenic lines showed a broad distribution in early-stage florets and enriched accumulation in pistils after the formation of pistil primordia at control temperatures (Fig. 3D), which did not change in distribution or amount at high temperatures (Fig. 3, E and F). Thus, neither HvMADS8 mRNA nor protein levels are affected by temperature during early development (W2.0 to W6.5).
Figure 3.
Temporal and spatial expression of HvMADS8 at different temperatures. A) Relative expression of HvMADS8 in different GP plant tissues and at different stages of development at 28 °C. Values are means ± Sd; n = 3 biological replicates. B) Relative expression of HvMADS8 at early stages of inflorescence development at 15 and 28 °C. Expression of HB (temperature-responsive gene) was used as positive control. Values are means ± Sd; n = 3 biological replicates. Asterisks indicate significant differences (2-way ANOVA test; **P < 0.01). C) In situ hybridization showing expression of HvMADS8 at stages W5.0 to 6.5 at 28 °C in longitudinal sections of GP florets. The sense probe served as a negative control. ca, carpel; le, lemma; ov, ovule; st, stamen. Scale bars, 100 μm. D) Accumulation of HvMADS8 in spikes and florets from W3.0 to 5.5 in proHvMADS8:HvMADS8-eGFP transgenic lines at 15 °C. cp, carpel primordia; fm, floral meristem; pa, palea; pp, pistil primordia. Scale bars, 100 μm. E) Accumulation of HvMADS8 in W3.0 and W4.0 proHvMADS8:HvMADS8-eGFP transgenic spikes grown at 15 and 28 °C. im, inflorescence meristem. Scale bars, 100 μm. F) Immunoblot analysis of HvMADS8-eGFP abundance in W5.5 to 6.5 spikes from 2 independent proHvMADS8:HvMADS8-eGFP lines grown at 15 and 28 °C. Tubulin served as loading control. All experiments were independently performed at least 3 times with similar results.
HvMADS8 coordinates thermal transcriptome programming of floral development
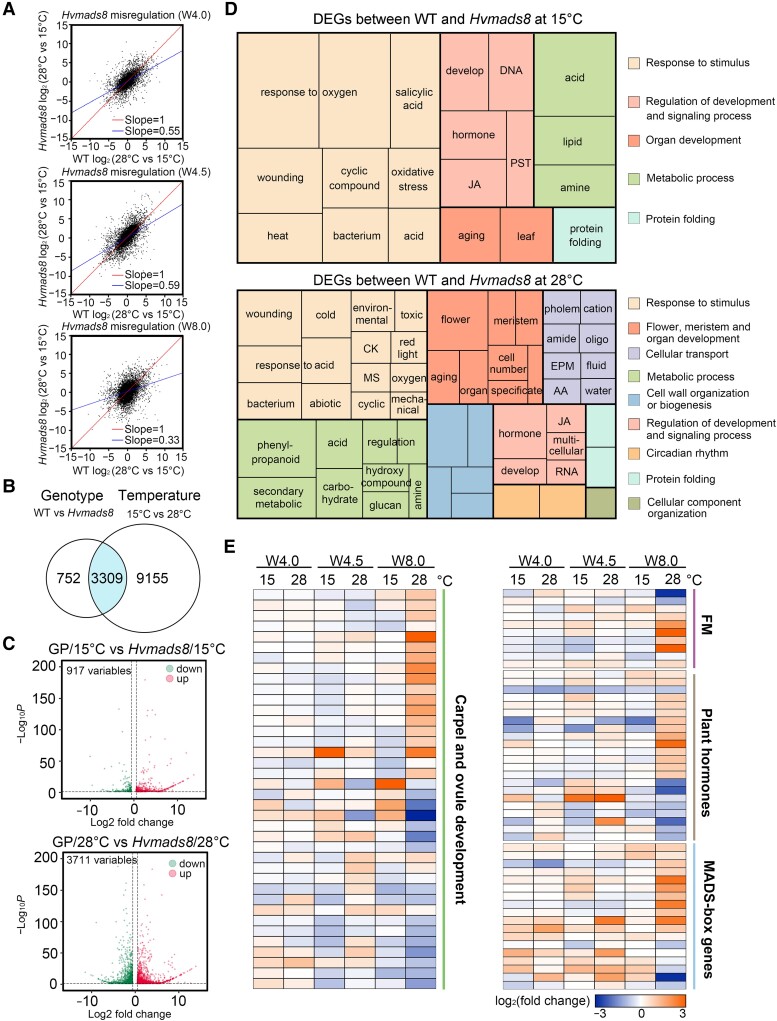
We compared the transcriptome patterns (transcriptome deep sequencing [RNA-seq]) of WT and Hvmads8 lines grown at 28 and 15 °C to examine downstream biological pathways or gene regulation programs influenced by the Hvmads8 mutation. The 3 stages analyzed covered early inflorescence meristems before and after the first extra carpel primordia bulge appears (stages W4.0 and W4.5), and developing pistils at W8.0 dissected (1 mm in length) from the young inflorescence. Correlation analyses using global transcriptional patterns at these stages revealed significant dysregulation of the expression of thermal response genes between WT and Hvmads8 lines (Fig. 4A). We identified 13,216 differentially expressed genes (DEGs) in total, with genes affected by genotype (WT compared to Hvmads8) or temperature (15 °C compared to 28 °C; Fig. 4B; Supplemental Data Set 1). Of the 4,061 DEGs responding to genotype, 3,309 (81.5%) were also affected by temperature (Fig. 4B). Changes in the expression of DEGs were much greater at 28 °C (Fig. 4C), highlighting the essential role of HvMADS8 in regulating gene expression at higher temperatures.
Figure 4.
HvMADS8 coordinates the thermal transcriptome programming of floral development. A) Correlation analyses of transcript levels at control and high temperatures at 3 stages of Hvmads8 and GP carpel development (W4.0, W4.5, and W8.0). B) Overlap of 13,216 DEGs responding to genotype and temperature. The blue-shaded region contains genes coregulated by genotype and temperature. C) Volcano plots showing differential gene expression in GP relative to Hvmads8 at 15 and 28 °C at the W4.0, W4.5, and W8.0 stages of inflorescence and carpel development. D) GO term enrichment analysis of DEGs between WT and Hvmads8 at 15 and 28 °C. CK, response to cytokinin; JA, regulation of jasmonic acid–mediated signaling; MS, response to monosaccharides; PST, phosphorelay signal transduction; EPM, export across the plasma membrane; AA, amino acid transport. E) Heatmap representation of expression levels for DEGs involved in carpel and ovule development, or FM regulation; plant hormone regulation; or that encode MADS-box proteins.
Gene ontology (GO) analysis revealed that compared to 15 °C, DEGs at high temperatures are largely associated with flower development and meristem fate, and also enriched for cellular transport and cell wall biogenesis, possibly due to the extra carpel growth in Hvmads8 florets (Fig. 4D; Supplemental Data Set 2). Consistent with the clear FM and ovule defects in high temperature-grown Hvmads8 lines, genes encoding MADS-box proteins and barley homologs of rice and Arabidopsis proteins involved in carpel and ovule development, FM regulation, and plant hormone pathways were overrepresented among the DEGs at 28 °C (Fig. 4E; Supplemental Data Set 3). For example, homologs of YABBY, AINTEGUMENTA (ANT), and SHI-RELATED SEQUENCE (SRS1) genes, which are all involved in the development of Arabidopsis carpel and ovules, displayed expression changes in Hvmads8 tissues at elevated temperature (Fig. 4E; Supplemental Fig. S2, A to C). Moreover, genes encoding meristem regulators such as CLAVATA-like, KNOX-like, and HvLFY (LEAFY) generally exhibited higher expression in Hvmads8 tissues, especially in pistils at 28 °C (Fig. 4E; Supplemental Fig. S2, D to F), supporting the observed changes in FM termination (Fig. 2, B and D).
Prominent expression changes in Hvmads8 mutants occurred in the genes encoding MADS-box proteins. C-class (HvMADS3 and HvMADS58) and E-class (HvMADS1) genes were upregulated at 28 °C (Supplemental Fig. S2, G to I). As the E-class proteins have been reported to upregulate C-class genes (Kaufmann et al. 2009; Wu et al. 2018), these expression changes of the 2 C-class members in Hvmads8 may be caused by an indirect effect due to the increased expression of HvMADS1 at W8.0. D-class (HvMADS13 and HvMADS21) and B-sister class (HvMADS31) genes were downregulated (Figs. 4E and 2C; Supplemental Fig. S2, J to L), suggesting that other MADS-box genes in the ABCDE model may contribute to extra pistil formation in the Hvmads8 mutant. In addition, the barley homologs of many thermal response genes, including those encoding heat shock proteins, showed significant changes (increases or decreases) in their expression based on genotype and/or temperature (Fig. 4A; Supplemental Fig. S2M). We conclude that HvMADS8 has a critical regulatory role in thermal transcriptome programming at high temperatures, especially in terms of flower and meristem regulation.
Increased HvMADS8 binding activity contributes to floral development transcriptome reprogramming
Stable HvMADS8 transcript and HvMADS8 protein levels at high temperatures (Fig. 3) were reminiscent of HvMADS1 under similar conditions; hence, we speculated that HvMADS8 may function in a similar manner as HvMADS1 (Li et al. 2021), through temperature-dependent changes in binding affinity to regulate downstream gene expression. We therefore used W4.5 inflorescence meristems from a proHvMADS8:HvMADS8-eGFP transgenic line to perform cleavage under targets and tagmentation (CUT&Tag) analyses to identify binding targets of HvMADS8 at low and high temperatures.
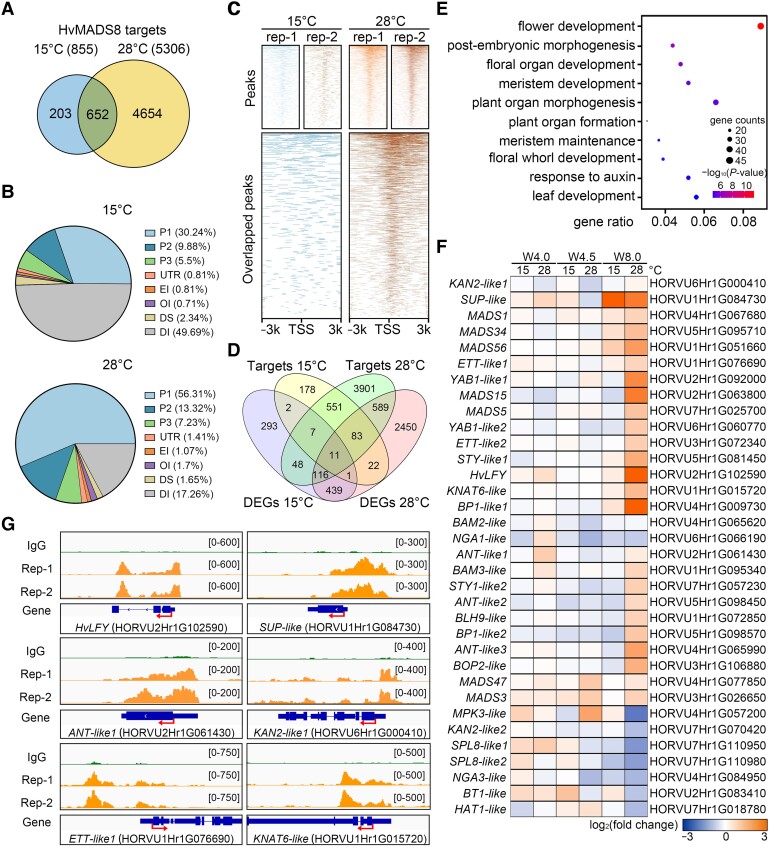
We identified far more HvMADS8 binding targets at high compared to low temperatures (5,306 and 855, respectively; Fig. 5A; Supplemental Data Set 5). Approximately 76% (652/855) of the targets identified at 15 °C were in the 28 °C target set (Fig. 5A), suggesting that an increase, rather than a switch, in HvMADS8 targets occurred at higher temperatures. Consistent with HvMADS8 functioning as a transcription factor, 46% of the total binding peaks were located <3 kb upstream of gene transcriptional start sites at 15 °C, a percentage that increased to 77% at 28 °C; 56% of binding sites were within 1 kb of the transcriptional start sites (Fig. 5B). Furthermore, the HvMADS8 binding region sites from 2 biological replicates were highly enriched around the transcription start sites at 28 °C, while the binding sites at 15 °C did not show this pattern (Fig. 5C). These data collectively indicate that HvMADS8 regulates temperature-responsive gene expression through differential binding to promoters of downstream targets.
Figure 5.
Changes in HvMADS8 binding activity at high temperatures contribute to transcriptome reprogramming for floral development. A) Venn diagram showing the numbers and overlap of target genes bound by HvMADS8 at 15 and 28 °C. B) Distribution of HvMADS8-bound regions in different barley genomic regions. P1, promoters (<1 kb upstream of ATG); P2, promoters (1 to 2 kb upstream of ATG); P3, promoters (2 to 3 kb upstream of ATG); UTR, 3′ untranslated region; EI, exons and first intron; OI, other introns; DS, downstream; DI, distal intergenic. C) Density heatmaps showing CUT&Tag peaks upstream and downstream of the TSS at 15 and 28 °C. D) Venn diagram showing the overlap of HvMADS8-regulated DEGs and binding targets at 15 and 28 °C. E) GO term enrichment analysis of 799 genes common to DEGs and CUT&Tag targets at 28 °C. F) Heatmap representation of the expression of HvMADS8-bound DEGs involved in pistil and FM development. G) Chromatin-binding profiles of HvMADS8 in the promoters of 6 selected pistil regulation genes. CUT&Tag reads of 2 GFP biological repeats (Rep-1 and Rep-2; orange) and the IgG negative control (green) were visualized using the Integrative Genomics Viewer. Arrows in the gene model indicate transcription start sites, the larger boxes indicate exons, the smaller boxes illustrate untranslated regions combined from all alternative transcripts, and the lines indicate introns.
Integrating DEG (RNA-seq) and binding target (CUT&Tag) data revealed 21 and 799 DEGs at 15 and 28 °C, respectively, that are bound by HvMADS8, which represent 2% and 15% of all HvMADS8 binding targets (Fig. 5D). GO analysis of the 799 gene targets at 28 °C revealed that 8 of the top 10 categories affect flower and meristem development or plant organ morphogenesis (Fig. 5E). Among these genes, we noticed a large number of the carpel, ovule, and FM regulators (Fig. 5F). Interestingly, the recently reported barley HvLFY gene showed a significant expression increase in Hvmads8 W8.0 pistils, with binding peaks in its promoter and second intron, as visualized by the Integrative Genomics Viewer (Fig. 5, F and G) (Robinson et al. 2011). Given that HvLFY controls barley floral organ specification in early stages, upregulation of HvLFY may further indicate the overactivation of FMs in Hvmads8 at higher temperatures (Selva et al. 2021). We also identified genes homologous to Arabidopsis SUPERMAN (SUP, which acts in the boundary specification between stamens in whorl 3 and carpels in whorl 4), ANT, and KANADI 2 (KAN2, which functions in Arabidopsis gynecium patterning) as DEGs with obvious binding peaks in their promoter regions at 28 °C (Fig. 5, F and G) (Cucinotta et al. 2014; Prunet et al. 2017; Wils and Kaufmann 2017). Moreover, homologs to Arabidopsis meristem regulators ETTIN (ETT) and KNOTTED1-LIKE HOMEOBOX GENE 6 (KNAT6), which function in ovule and gynecium development, both displayed direct binding by HvMADS8 (Fig. 5, F and G) (Ferrándiz et al. 2010; Kelley et al. 2012). Taken together, these data suggest that HvMADS8 affects transcriptome reprogramming through binding affinity changes that coordinate floral developmental pathways to inhibit the overactivation of FMs and stabilize female reproductive organ development at high temperatures.
HvMADS8 directs D-class genes at high temperature
OsMADS13 affects ovule initiation and FM termination (Dreni et al. 2007), and expression of the D-class genes HvMADS13 and HvMADS21 is affected by the Hvmads8 mutation (Fig. 2C; Supplemental Fig. S2, J and K), prompting us to examine how these genes might interact to control ovule establishment in response to temperature.
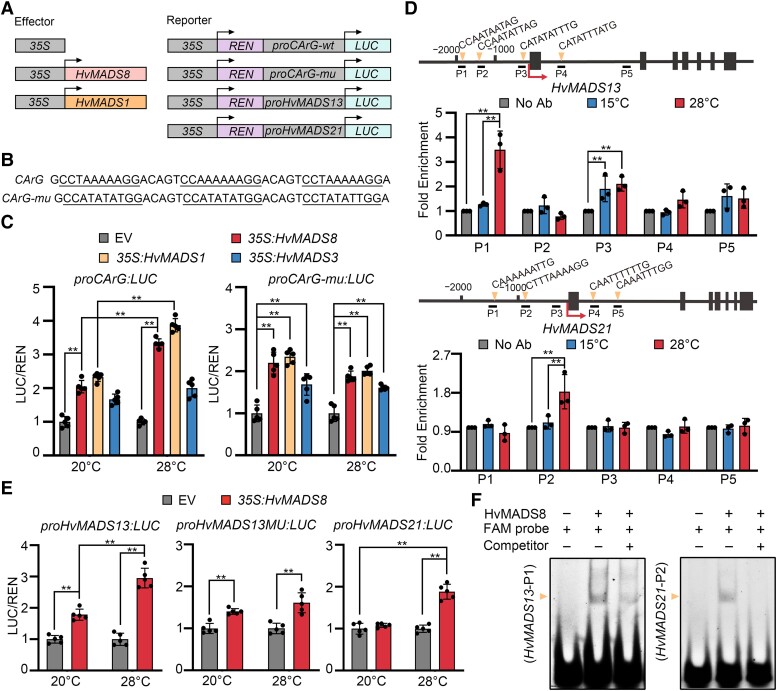
Using a dual-luciferase transient expression assay in Nicotiana benthamiana leaves (Fig. 6A), we tested the ability of HvMADS8 to activate gene expression at 2 temperatures, 20 °C as a control, which is a normal growth temperature for N. benthamiana and also a relatively low temperature for Hvmads8 with no obvious phenotypes shown (Fig. 1B), and 28 °C as the high temperature. Compared to the control, HvMADS3, which was previously shown to be temperature independent in regulating gene expression (Li et al. 2021), HvMADS8 exhibited temperature-dependent transcriptional activation only on artificial promoters carrying A-tract CArG-box motifs similar to HvMADS1 (Fig. 6, B and C). Using the inflorescence meristems of proHvMADS8:HvMADS8-eGFP plants grown at 15 and 28 °C, we investigated in vivo HvMADS8 binding targets by chromatin immunoprecipitation (ChIP) assays. HvMADS8 bound to promoter fragments harboring CArG-box sequences (primer set P1, P3 for HvMADS13; P2 for HvMADS21) of these 2 D-class genes, with significantly increased binding at high temperature (Fig. 6D). Transient luciferase expression assays also revealed that HvMADS8 activates the expression of HvMADS13 and HvMADS21 directly in a temperature-dependent manner, and the P1 motif of HvMADS13 promoter contributed directly to this temperature-responsive activation (Fig. 6E). We confirmed the binding to P1 in the HvMADS13 promoters and P2 in the HvMADS21 promoter by electrophoretic mobility shift assays (EMSAs). We observed clearly shifted bands when HvMADS8 was incubated with CArG-box motif-containing probes labeled by fluorescein amidite (FAM); moreover, the band intensity decreased when unlabeled competitor probes were added (Fig. 6F). Collectively, these results demonstrate that HvMADS8 can bind to the promoters of the 2 barley D-class genes directly to regulate their expression, especially at high temperatures, which may prevent FM indeterminacy and overgrowth of carpels in WT plants.
Figure 6.
HvMADS8 regulates D-class genes at high temperatures. A) Diagrams of effector and reporter constructs for dual-LUC assays. The HvMADS8 and HvMADS1 effector genes were under the control of the 35S promoter. An empty 35S effector construct was used as a negative control. LUC, firefly luciferase; REN, Renilla luciferase. B) Artificial CArG-boxes with A-tracts (underlined, CArG, WT) or non-A-tracts (underlined, CArG-mu, mutant) used to drive LUC expression. C) Normalized luciferase (LUC/REN) activity derived from the artificial CArG-box promoters in the presence of HvMADS8, HvMADS1, HvMADS3, or empty vector (EV, negative control) at 20 and 28 °C. Values are means ± Sd; n = 5 biological replicates. Asterisks indicate significant differences (2-way ANOVA test; **P < 0.01). D) In vivo binding of HvMADS13 and HvMADS21 CArG-boxes by HvMADS8 at 15 and 28 °C, as determined by ChIP-qPCR. Upper, HvMADS13 or HvMADS21 genomic regions, showing locations of 5 DNA fragments (P1 to P5) for ChIP-qPCR. Boxes indicate exons; lines indicate promoter or intron regions; arrows indicate transcription start sites. Lower, fold enrichment of 5 DNA fragments at 15 or 28 °C relative to no-antibody (Ab) control. Values are means ± Sd; n = 3 biological replicates. Asterisks indicate significant differences (2-way ANOVA test; **P < 0.01). E) Normalized luciferase activity driven by the HvMADS13 and HvMADS21 promoters in the presence of HvMADS8 or EV (negative control) at 20 and 28 °C. The CArG-box in P1 binding site of the HvMADS13 promoter is mutated to ATGTGAATAG in the proHvMADS13MU:LUC construct. Values are means ± Sd; n = 5 biological replicates. Asterisks indicate significant differences (2-way ANOVA test; **P < 0.01). F) EMSAs to detect interactions between HvMADS8 and the P1 (CArG-box motif of the HvMADS13 promoter) and P2 (CArG-box motif of the HvMADS21 promoter) probes.
Physical interactions of HvMADS8 with other floral homeotic proteins
As the identity of different floral organs is determined by combinations of floral homeotic MADS-box proteins in Arabidopsis and rice (Cui et al. 2010; Theissen et al. 2016; Wu et al. 2018), we tested the interactions of HvMADS8 with other barley homologs of homeotic proteins under different temperatures using a yeast 2-hybrid assay (Supplemental Fig. S3). We used 15 °C as the lowest temperature, and 34 °C as the highest, since yeast cotransformants will not grow normally on synthetic-defined (SD) medium lacking Trp and Leu (SD/–Leu/–Trp [SD–2]) at 36 °C (Xie et al. 2020). HvMADS8 formed homodimers and heterodimers with all C-class, D-class, and E-class proteins except with HvMADS34 and the AGAMOUS-LIKE 6 (AGL6)-like protein HvMADS6 on selective medium (SD/–Ade/–His/–Leu/–Trp [SD–4]). Most of these interactions were insensitive to temperature, except for HvMADS21, HvMADS1, HvMADS5, and HvMADS6, which showed stronger interactions at 20 to 30 °C (Supplemental Fig. S3). These results suggest that HvMADS8 can form protein complexes with other floral homeotic proteins, most of which were not affected by temperature.
Conserved OsMADS8 function in temperature-dependent pistil regulation
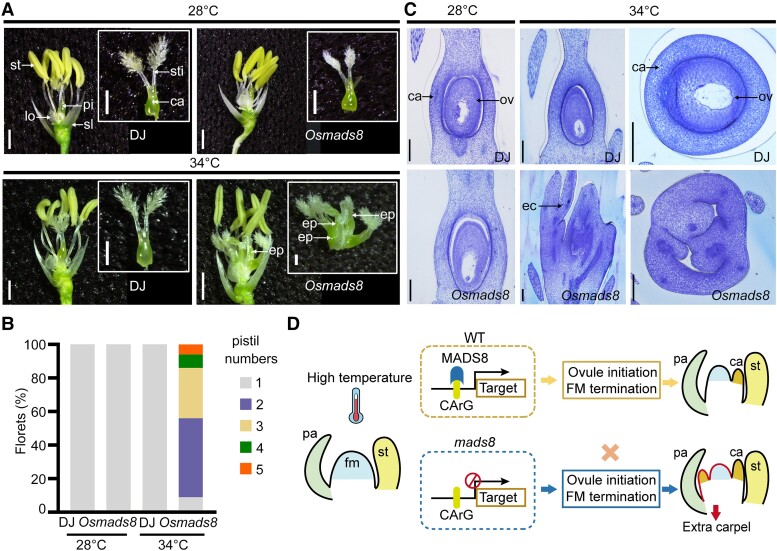
Previous studies have shown no obvious alterations in either vegetative or reproductive organ phenotypes in OsMADS8 single knockdown lines (Cui et al. 2010). To assess the function of OsMADS8 at different temperatures, we created a rice Osmads8 mutant that contains a 2-bp insertion within the first exon in the Dongjin (DJ) background (Supplemental Fig. S1A). Consistent with previous reports from RNA interference (RNAi) lines, the loss-of-function Osmads8 mutant obtained here displayed no significant floral organ defects at control temperatures (28 °C; Fig. 7A). However, we determined that Osmads8 plants display a lower seed setting rate and shrunken grain morphologies compared to WT at 28 °C (Supplemental Fig. S4, A to D), indicating a key role for OsMADS8 during postfertilization development. At higher temperatures (34 °C), the Osmads8 mutant also produced multiple pistils with no noticeable changes in other flower organs, as in barley (Fig. 7, A and B). Histological analysis of preanthesis rice florets also revealed an ovule initiation defect in Osmads8 mutant pistils at 34 °C, resulting in the failure of grain production at high temperatures (Fig. 7C; Supplemental Fig. S4E). Collectively, our data suggest a conserved function for MADS8 between rice and barley in maintaining the developmental stability of FM and pistil structures in a temperature-specific manner. As shown in our proposed model (Fig. 7D), downstream genes in flower development pathways are regulated by MADS8 to drive ovule initiation and FM determinacy in WT pistils at high temperatures. In mads8 mutants, the lack of MADS8 leads to the iterative formation of carpels and failure to initiate ovule primordia at high temperatures (Fig. 7D).
Figure 7.
MADS8 function in temperature-dependent pistil regulation is conserved between barley and rice. A) Pistil phenotypes of DJ (WT) and Osmads8 mutants grown at control (28 °C average) and high (34 °C average) temperatures. ca, carpel; ep, extra pistil; lo, lodicule; pi, pistil; sl, sterile lemma; st, stamen; sti, stigma. Scale bars, 900 μm. B) Pistil number in Osmads8 florets grown at different temperatures. C) Semi-thin sections of stage 12 DJ and Osmads8 pistils at 28 or 34 °C. ca, carpel; ec, extra carpel; ov, ovule. Scale bars, 100 μm. D) Proposed model of HvMADS8-mediated pistil development regulation at high temperatures. In WT flowers, MADS8 binds target genes to induce ovule initiation and FM termination at the appropriate developmental stage under a specific high-temperature signaling pathway. In the absence of MADS8, continued FM development triggers the repeated formation of extra carpels in the innermost whorl. fm, floral meristem; pa, palea.
Discussion
Barley SEP members stabilize reproductive development through changes in their DNA binding affinity at high ambient temperature
Several studies have identified MADS-box transcription factors as key components of a plant regulatory network that integrates environmental cues to affect responses at the cellular level but most are involved in flowering time control (Trevaskis et al. 2003; Sureshkumar et al. 2016; Whittaker and Dean 2017). Here, we report that Hvmads8 exhibits temperature-dependent defects in female organs and FM during gynecium development (Figs. 1 and 2). Moreover, using the CUT&Tag approach, we revealed the genome-wide downstream binding targets of a MADS-box protein at high temperatures (Fig. 5, A to C). By integrating these results with RNA-seq data, we propose that at high temperatures, increased binding of HvMADS8 to the promoters of genes in flower and meristem development pathways contributes to pistil developmental stability (Figs. 5 and 7D).
Compared to our previous discovery that the LOFSEP member HvMADS1 regulates barley inflorescence shape stability (Li et al. 2021), here we show that the SEP3 member MADS8 modulates temperature-responsive female organ development. Unlike the branch-like inflorescence of Hvmads1 under high temperatures, the defects of Hvmads8 mutants are restricted to the innermost floral whorl, i.e. meristem indeterminacy and extra pistil-like structures at high temperatures (Figs. 1 and 2). Similarly, HvMADS8 and HvMADS1 have stable transcript and protein levels at different temperatures (Fig. 3, B and F); they fulfill their thermal response functions through temperature-sensitive binding to the promoters of their downstream target genes (Fig. 5). These findings on the barley SEP family extend the known function of floral homeotic proteins in linking them to thermal responsiveness during reproductive development. Further investigations on higher-order mutants of SEP family members, and other floral homeotic genes at different temperatures, may provide more information about the mechanistic framework underlying reproductive organ thermal response. The single mutant Hvmads7 displays no noticeable differences in pistil development at either control or high temperatures (Supplemental Fig. S5, A and B). However, double knockout mutants for the 2 SEP3-like genes HvMADS7 and OsMADS8 display extra lemma and palea in florets, and lack inner floral organs at the control temperature (15 °C), suggesting a conserved redundant function among E-class genes of barley, rice, and Arabidopsis in controlling the inner floral organ identities at normal growth temperature conditions (Supplemental Fig. S5, A and C) (Pelaz et al. 2000, 2001; Wu et al. 2018). The contribution of HvMADS7 and other floral homeotic proteins in temperature-responsive floral development remains to be explored.
MADS8 directs ovule initiation and FM determinacy at high temperatures
The pattern of FM termination is diverse among angiosperm species. In Arabidopsis, stem cell maintenance ceases during carpel primordium initiation, and then ovules differentiate from the placenta inside carpels (Cucinotta et al. 2014). The carpel identity gene AG can, directly and indirectly, repress the key FM regulator WUSCHEL to arrest floral stem cell activity for proper carpel development (Liu et al. 2011; Sun et al. 2019). In rice, after stamen and carpel differentiation, the FM is exhausted and ovules are finally produced from the terminating FM (Yamaki et al. 2011). Loss of function of the ovule identity gene, OsMADS13, leads to FM termination defects (Dreni et al. 2007). Thus, the central region of the FM is consumed by the carpel or ovule primordium in Arabidopsis and in rice, respectively, and carpel identity genes in the innermost whorl play indispensable roles in FM determinacy control. Barley may share a similar FM termination mechanism as rice. Unlike the extra carpels produced in rice fon mutants and wheat tri-pistil mutants that can produce multiple grains within a single floret (Jiang et al. 2005; Chu et al. 2006; Moon et al. 2006; Suzaki et al. 2006; Liu et al. 2022), extra carpels in Hvmads8 florets form at the expense of ovules at high temperatures (Fig. 2A), indicating a close link between ovule initiation and FM determinacy during the floral developmental program. Based on similar defects observed in Osmads13 mutant lines, we speculate that barley D-class genes also function in the ovule and FM regulatory pathway and contribute to the Hvmads8 phenotype, where their expression is downregulated at high temperatures (Fig. 2, B and C; Supplemental Fig. S2, J and K). We confirmed the direct regulation of HvMADS13 and HvMADS21 by HvMADS8 (Fig. 6), although we failed to identify binding sites in the promoters of these 2 genes by CUT&Tag analyses at a relatively strict threshold (P ≤ 0.01). One possible explanation is that the strong direct binding of HvMADS8 to these 2 D-lineage genes occurs at later stages, as there is a stage difference between the samples used for CUT&Tag (W4.5 inflorescence) and ChIP-qPCR (W5.5 to W6.5 inflorescence). Nevertheless, single and double mutants of barley D-class genes are needed to uncover their precise genetic functions in ovule initiation and FM development.
The potential roles of protein interaction networks in HvMADS8 function
MADS-box proteins interact with each other to determine flower formation and floral organ specification (Theissen et al. 2016; Abraham-Juárez et al. 2020). In Arabidopsis, SEP3 functions as a central hub to drive protein–protein interaction with other MADS-box transcription factors (Immink et al. 2009). Furthermore, the heterotetramer formed by SEP3 and the C-class protein AG is necessary for meristem determinacy through the activation of KNUCKLES and CRABSCLAW (Hugouvieux et al. 2019). Additionally, the variant SEP3Δtet produced from a splice variant in Arabidopsis, lacking heterotetramerization capacity, was able to restore the first 3 whorls in the sep1 sep2 sep3 triple mutant background but failed to rescue the unfused carpel and indeterminate flower-within-a-flower phenotype of this triple mutant. Thus, tetramerization of SEP3 with other floral homoeotic proteins, probably AG, may be required to activate a subset of target genes to terminate the FM (Hugouvieux et al. 2019).
Our work further reveals that HvMADS8 can interact with other C-, D-, E-class, and AGL6 subfamily homeotic proteins, so it is possible that protein–protein interactions also contribute to HvMADS8-regulated gene expression at high temperatures (Supplemental Fig. S3). Although we observed limited temperature-sensitive differences in protein–protein interactions in yeast, other assays, such as bimolecular fluorescence complementation (BiFC), or truncated HvMADS8 variants that affect protein–protein interactions, may be needed to provide clarity regarding interaction networks between HvMADS8 and floral homeotic proteins at various temperatures. Besides, sequence analyses comparing the barley E-class proteins to the B- and the C-class proteins, which have no temperature-responsive regulation to CArG-box elements (Muiño et al. 2014; Li et al. 2021; Selva et al. 2023), revealed a very conserved DNA-binding MADS domain from all families, while the C-terminal domain, involved in transcriptional activity (Muiño et al. 2014; Callens et al. 2018), showed a huge diversity among these temperature-dependent or temperature-independent members (Supplemental Fig. S6).
Conserved MADS8 function in barley and rice during carpel development
Studies in key model plants have shown that the formation of various floral organs is coordinated by the actions and interactions among MADS-box family proteins (Gramzow and Theissen 2010; Theissen et al. 2016). Interestingly, many of these factors show conserved function in flower development among species (Ciaffi et al. 2011; Murai 2013; Wu et al. 2018; Kuijer et al. 2021). In this study, we analyzed loss-of-function mads8 mutants in barley and rice, with both mutants displaying increased carpels and loss of ovules specifically at high temperatures (Figs. 1 and 7). Although the normal growth temperature differs between barley and rice (15 to 20 and 28 °C, respectively) due to their distinct domestication trajectories (Fuller et al. 2009; Pankin and von Korff 2017), MADS8 has a conserved temperature-dependent function (Figs. 1 and 7). The unaffected levels of HvMADS1 and HvMADS8 mRNA and HvMADS1 and HvMADS8 protein (Fig. 3, B and F) (Li et al. 2021) under high temperature led to our speculation that these MADS-proteins may undergo conformational changes to enhance their DNA binding affinity and facilitate global transcriptional adjustments to high temperatures during reproduction, which remains to be investigated.
Analyses of single nucleotide polymorphisms (SNPs) using the rice SNP-Seek Database derived from 3,000 rice genomes revealed no SNP within the coding regions of OsMADS8 (Wang et al. 2018); analyses of 267 barley genomes also revealed strong conservation of the MADS8 sequence among different accessions (Russell et al. 2016) (Supplemental Table S1). Overall, our study indicates that MADS8 appears to have a conserved function in determining pistil development and terminating FM in a temperature-dependent manner in at least 2 grass species. In Arabidopsis, SEP3 also plays a similar role as MADS8 in grasses in terms of FM determination and carpel regulation. Floral organs are converted to sepaloid-like structures in Arabidopsis sep1 sep2 sep3 triple mutants with a lack of FM determinacy, resulting in carpels being replaced by continuous generation of sepals (Pelaz et al. 2000).
It will be interesting to examine whether more conserved regulators in flower developmental pathways are shared between crop species. Knowledge of the mechanisms governing floral organ development in restrictive temperatures, and whether they are conserved across cereal crops, might offer crucial insights that can be used in the context of global warming to engineer plants with improved developmental stability.
Materials and methods
Plant materials, CRISPR design, and plant transformation
The barley (H. vulgare) cultivars used in this study were ‘GP’ (United Kingdom) and ‘WI’ (Australia). The monocot CRISPR/Cas9 system (Ma et al. 2015) was used to obtain barley edited lines. Two single guide RNAs (sgRNAs) were designed to target the first exon of HvMADS8 and HvMADS7 within the sequence encoding the MADS domain (http://skl.scau.edu.cn/targetdesign/) (Supplemental Figs. S1A and S5A). Target specificity was confirmed by BLAST of the barley genome (http://plants.ensembl.org/Hordeum_vulgare/Tools/Blast). Target sites sequenced in GP and WI all showed 100% identity to the ‘Morex’ reference genome. The sgRNA expression cassettes driven by the promoters OsU6a and OsU6b were cloned into the PYLCRISPR-Cas9Pubi-H binary vector (Li et al. 2021). Agrobacterium (Agrobacterium tumefaciens)-mediated transformation (strain AGL1) of immature barley embryos was performed for both cultivars (Harwood 2014). Independent T0 plants were genotyped using a Phire Plant Direct PCR Kit (Thermo Fisher Scientific), and the amplified target sites were Sanger sequenced (AGRF, Australia).
The rice (O. sativa) cultivar ‘DJ’ (ssp. japonica) was used to generate the Osmads8 CRISPR/Cas9 knockout mutant as previously described (Xie et al. 2015). Genotyping of Osmads8 lines was performed by PCR amplification from genomic DNA extracted from the T1 plants, followed by Sanger sequencing (Personalbio, Shanghai).
To investigate HvMADS8 protein accumulation and function, proHvMADS8:HvMADS8-eGFP transgenic lines were created by Agrobacterium-mediated transformation as described above. The 2,870 bp HvMADS8 promoter and full-length cDNA were cloned in-frame and upstream of the eGFP sequence into pCAMBIA1301, using the In-Fusion (Takara) cloning technology with KpnI and NcoI sites. The resulting construct was transformed into the GP WT barley background. The primers are listed in Supplemental Data Set 5.
Plant growth conditions and temperature treatments
All barley plants were germinated on cocopeat soil at control temperatures (15 °C day/10 °C night), with 50% humidity and a 16-h light/8-h dark photoperiod in growth chambers with a light intensity of ∼190 μmol m−2 s−1 (The Plant Accelerator, University of Adelaide; Conviron GR48, Shanghai Jiao Tong University). For high-temperature treatments, plants were grown at the control temperature until Waddington stage W1 and then were moved to higher temperatures (20, 25, or 28 °C) for phenotyping or sample harvesting. Night (dark) temperatures were 5 °C below the day (light) temperature for all temperature treatments.
Rice plants were grown at an average temperature of 28 °C with 14-h light/10-h dark photoperiod and 75% relative humidity in a growth chamber (Conviron GR48) as control conditions. For high-temperature treatments, plants were first grown in control conditions; at the onset of reproductive development, plants were moved to a growth chamber with an average temperature of 34 °C.
Morphological analysis and microscopy observation
Fresh pistils from WT and mutant florets were photographed using a stereomicroscope equipped with a digital camera (Leica, MZ FLIII and S8AP0). Immature inflorescence and pistil tissues from plants grown at different temperatures were collected at various stages and prepared for SEM (Philips, XL30 FEG) as previously described (Wang et al. 2021). For cytological analysis, pistils at W9.5 were collected and fixed in formalin–aceto–alcohol (FAA) solution (50% [v/v] ethanol, 10% [v/v] formalin, and 5% [v/v] acetic acid) overnight and then dehydrated in a graded ethanol series. Samples were embedded in Technovit 7100 resin (Hereaus Kulzer). Transverse 2-μm sections were cut with an Ultratome III ultramicrotome (LKB) and then stained with 1% (w/v) toluidine blue and photographed using an optical microscope (Ni-E, Nikon and E600, Nikon). All confocal fluorescent images for proHvMADS8:HvMADS8-eGFP lines were captured using an Olympus IXplore SpinSR microscope from inflorescences and florets at various stages of development (excitation 488 nm; emission 500 to 530 nm).
RNA extraction and RT-qPCR analysis
Total RNA was extracted from 3 biological replicates using TRIzol reagent (Invitrogen) from barley and rice samples. Samples included different stages of barley inflorescence (W2.0 to 7.5) and pistil (W8.0 to 10.5) development; the lemma, palea, awn, stem, and leaves from W9.5 florets and plants; and root samples from young seedlings. The young inflorescences in Hvmads8 mutants were collected based on the awn and stamen primordia stages as in WT plants, and the various stages of pistils dissected from Hvmads8 were determined according to the lemma and anther morphological changes in WT florets.
Two micrograms of total RNA was used for first-strand cDNA synthesis using a FastKing RT kit with gDNase (Tiangen Biotech). qPCR of diluted cDNA was performed using a 384-well QuantStudio Flex 6 (Thermo Fisher Scientific) instrument with a QuantiNova SYBR Green PCR Kit (Qiagen). The HvActin7 gene was used as internal control as described previously (Li et al. 2021). All primers are listed in Supplemental Data Set 5.
Immunoblotting of HvMADS8
Nuclear protein was extracted from W5.5 to 6.5 inflorescences collected from 2 independent proHvMADS8:HvMADS8-eGFP lines using nuclear protein extraction buffers as described previously for ChIP assays (Bowler et al. 2004). The concentrations of isolated protein samples were measured using a Bradford Protein Assay Kit (Sangon Biotech). Equal amounts of protein from each sample were separated on 12.5% (w/v) SDS-PAGE using a PAGE Gel Fast Preparation Kit (EpiZyme Biotechnology) and transferred onto PVDF membrane (Bio-Rad). The membrane was incubated with primary antibodies against GFP (1:1,000 dilution; Proteintech 66002-1-Ig) or tubulin (1:2,000 dilution; Beyotime AT819) at 4 °C overnight and with secondary antibodies conjugated to horseradish peroxidase (1:5,000 dilution; Abmart M21001) for 1 h at room temperature. Immunoreactive bands were visualized with a Chemidoc MP Imaging System (Bio-Rad) using SuperSignal West Pico chemiluminescent substrate (Pierce).
In situ hybridization
Barley inflorescences were fixed immediately and embedded for in situ hybridization as previously described (Wu et al. 2018). Fragments of HvDL (314 bp), HvMADS13 (350 bp), HvOSH1 (327 bp), and HvMADS8 (302 bp) were amplified from WT cDNA as probe templates with specific primers fused to the T7 promoter (Supplemental Data Set 5). Digoxigenin-labeled antisense and sense probes were transcribed using a DIG RNA Labeling Kit (SP6/T7; Roche) according to the manufacturer's instructions. An InsituPro VSi robot (Intavis) was used to perform the in situ hybridizations automatically following a standard protocol, as described before (Javelle et al. 2011; Li et al. 2021). Images were photographed by an optical microscope (Ni-E, Nikon).
RNA sequencing and data analysis
Inflorescence (W4.0 and W4.5) and pistil (W8.0) samples were collected from WT (GP) and Hvmads8 plants grown at control and high temperatures. Total RNA from 3 biological replicates was extracted using TRIzol reagent (Invitrogen). RNA-seq library preparation and sequencing were performed on an Illumina HiSeq4000 platform by Novogene (China). The quality of raw reads was examined and reads were trimmed; clean reads were mapped to the barley reference genome (Barley cv. Morex IBSC 2 V1 2016, https://galaxy-web.ipk-gatersleben.de/libraries/folders/Ff2db41e1fa331b3e) as described previously (Mascher et al. 2017; Li et al. 2021). The count data were calculated using HTSeq. Transcripts per million (TPM) values were normalized using a Python script. Further analysis was based on 39,734 high-confidence genes. The scatterplots for correlation analyses were generated using log2 fold change of gene expression at 28 °C compared to 15 °C at each stage, and a custom R script was used to calculate linear relationships (Jung et al. 2016). The R package DESeq2 (version 1.32.0) was used to identify DEGs that correlated with genotype (WT relative to Hvmads8) or temperature (28 °C compared to 15 °C). The false discovery rate (FDR)-adjusted P-values were computed by the Benjamini–Hochberg model. Genes were considered to be differentially expressed if both the |log2 fold change| > 0.585 and the P-adjust < 0.05 (Supplemental Data Set 1). DEGs were annotated using the BioMart database of Ensembl Plants. Information on homologs in Arabidopsis (TAIR10_peptide; http://www.arabidopsis.org/) and rice (MSU7_peptide; http://rice.plantbiology.Msu.edu/) was retrieved from the listed websites. Volcano plots were created using log2 fold change of DEGs at 15 and 28 °C with the R package EnhancedVolcano (Blighe et al. 2019).
GO analysis was performed with the R package (topGO) for DEGs according to Arabidopsis annotations (Supplemental Data Set 2). GO terms were considered to be significantly enriched with a corrected FDR < 0.05. Treemaps were used to visualize GO term results (Li et al. 2021). Genes related to meristem regulation, flower development, and temperature response were selected for heatmap analysis using MeV software (mev.tm4.org; Supplemental Data Set 3).
ChIP-qPCR
Approximately 1 g of W5.5 to 6.5 inflorescence was collected from one line of proHvMADS8:HvMADS8-eGFP plants grown at control or high temperatures. Samples were crosslinked with 1% (v/v) formaldehyde in extraction buffer (0.4 m sucrose, 10 mm Tris-HCl, pH 8.0, 5 mm β-mercaptoethanol, and 0.1 mm PMSF), and the ChIP assay was performed as described previously (Zhu et al. 2022). GFP-Trap and Binding Control Magnetic Agarose (ChromoTek) were used for chromatin precipitation at the same time. Crosslinks were reversed with 0.2 m sodium chloride and overnight incubation at 65 °C. The extracted DNA was diluted 1:4, and the abundance of the promoter fragments from HvMADS13 and HvMADS21 was quantified by qPCR using a LightCycler 96 Real-Time PCR system (Roche) with a SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biology). The HvActin7 gene was used as a negative control. All primers are listed in Supplemental Data Set 5.
Dual-luciferase assays
The dual-luciferase (LUC) assay was performed in N. benthamiana leaves, from plants grown and treated as described before (Li et al. 2021). Full-length HvMADS8 and HvMADS3 cDNA was cloned into the pGreenII-0000 vector containing the cauliflower mosaic virus (CaMV) 35S promoter for effector plasmid construction. For reporter constructs, the promoters of HvMADS13 and HvMADS21 from GP were cloned upstream of the firefly luciferase (LUC) reporter gene into the pGreenII-0800-LUC vector, which also harbors the Renilla luciferase (REN) gene driven by the CaMV 35S promoter. The proHvMADS13MU:LUC construct contains a mutation in the CArG-box sequence in the P1 binding motif, changing the original CArG-box sequence from CCAATAATAG to ATGTGAATAG. The 35S:HvMADS1, proCArG, and proCArG-mu effector and reporter constructs were used as described previously (Li et al. 2021). Primers are listed in Supplemental Data Set 5.
Effectors and reporters were individually transformed into Agrobacterium strain GV3101 containing the helper plasmid pSoup-P19, which encodes a repressor of cosuppression. Agrobacterium colonies harboring each effector construct were then coinfiltrated into N. benthamiana leaves with Agrobacterium colonies carrying the reporter construct. The empty effector vector pGreenII-0000 was used as negative control. N. benthamiana plants were grown in the dark for around 48 h at relatively low temperature (20 °C, no phenotypes shown for Hvmads8) or high (28 °C) temperatures before sample collection. LUC and REN activities were measured with a Dual-Luciferase reporter kit (Promega) according to the manufacturer's instructions using a GloMax-96 Microplate Luminometer (Promega). Five biological replicates were used for each effector and reporter combination.
EMSAs
The full-length HvMADS8 cDNA was amplified from inflorescence cDNA and cloned into the pGADT7 vector (Clontech) for in vitro translation using a TNT T7/SP6 Coupled Wheat Germ Extract System (Promega). The CArG-box and its flanking sequence in the promoter region of HvMADS13 and HvMADS21 were synthesized as FAM-labeled primers at Generay Biotech (China). 10× unlabeled probes were used as competitor. The probe preparation and assays were performed as described previously (Zhu et al. 2022). Primers are listed in Supplemental Data Set 5.
CUT&Tag assay and data processing
The CUT&Tag assay was performed as previously described (Li et al. 2022), with minor modifications. Briefly, antibodies specific to GFP (Abcam, ab290), H3K4me3 (Abcam, ab8580; positive control), and IgG (Sigma, 12-370; negative control) were used as primary antibodies at 1:50 dilutions. Guinea pig antirabbit IgG (Abnova, PAB9407) was used as the secondary antibody at 1:100 dilution. Sequencing was performed on a NovaSeq instrument as 150-bp paired-end reads by Personalbio (China). All sequencing reads were cleaned with NGmerge to move adapters before mapping the clean reads to the barley reference genome (Barley cv. Morex IBSC 2 V1 2016, https://galaxy-web.ipk-gatersleben.de/libraries/folders/Ff2db41e1fa331b3e) using Bowtie2 with --end-to-end --very-sensitive --no-mixed -I 10 -X 700 as parameters (Mascher et al. 2017). The aligned BAM files were sorted for further analysis using Samtools; the peaks were called using Genrich with the threshold P-value set to 0.01. Peaks were merged using Bedtools between groups (Quinlan and Hall 2010). The ChIPseeker package was used to analyze the HvMADS8 binding positions (Yu et al. 2015). Genes identified in both biological replicates with peaks located within ±3 kb of the gene translation start site (TSS) were deemed potential targets (Supplemental Data Set 4). Bound peaks were visualized using Integrative Genomics Viewer (version 2.11.2) (Robinson et al. 2011).
Yeast 2-hybrid assays
The full-length cDNAs of HvMADS3, HvMADS58, HvMADS13, HvMADS21, HvMADS1, HvMADS5, HvMADS34, HvMADS7, HvMADS8, and HvMADS6 were individually amplified with gene-specific primers from inflorescence cDNA (Supplemental Data Set 5). The amplified HvMADS8 sequence was then cloned into the prey activation domain (Ad) vector pGADT7; the other sequences were cloned into the bait DNA-binding domain (BD) vector pGBKT7 (Clontech), using the In-Fusion (Takara) cloning technology at the EcoRI and BamHI sites. These constructs were transformed using one-step buffer (0.1 m dithiothreitol; 0.3 m lithium acetate and 40% [w/v] polyethylene glycol 4000) followed by a 30-min incubation at 45 °C into yeast (Saccharomyces cerevisiae) strain AH109 (BD Biosciences, USA), and the yeast 2-hybrid assay was performed according to the MATCHMAKER GAL4 Two-Hybrid System 3 & Libraries User Manual (Clontech). Yeast transformants were selected on SD medium lacking Leu and Trp (SD/–Leu/–Trp, SD–2). Protein interactions were tested, with no detectable self-activation for each of the single constructs on SD medium with high stringency (SD–Ade/–His/–Leu/–Trp, SD–4). To compare interactions at different temperatures, transformants grown on SD–2 medium were used as a reference. Transformants containing empty plasmids pGADT7 (prey) and pGBKT7 (bait) were used as negative controls.
Amino acid sequence alignment
The full-length amino acid sequence of HvMADS2, HvMADS4, HvMADS16, HvMADS3, HvMADS58, HvMADS1, HvMADS5, HvMADS34, HvMADS7, and HvMADS8 were imported into SnapGene (Version 4.3.7), and the amino acid alignment was conducted using MUSCLE alignment. The HvMADS8 protein sequence was used as the reference.
Variation of HvMADS8
The exome-sequencing data derived from 267 barley accessions were analyzed (https://www.ebi.ac.uk/ena/data/view/PRJEB8044) (Russell et al. 2016). Morex_contig_350004, Morex_contig_6154, and Morex_contig_2548971, which contain the HvMADS8 gene sequence, were used for SNP identification. SNP calling was performed by visual inspection of the sequences (Supplemental Table S1).
Quantification and statistical analysis
Statistical analysis was performed with Excel (Microsoft) or GraphPad Prism 8.0.2. For the comparison of 2 data groups, 2-way ANOVA was used to evaluate significance, with values given as means ± Sd. At least 3 biological repeats are displayed. All analyses are summarized in Supplemental Data Set 6.
Accession numbers
The sequences for all barley genes used in this study are available in the Phytozome database (https://phytozome-next.jgi.doe.gov/). Rice gene sequences are available in the Rice Genome Annotation Project database (http://rice.uga.edu/analyses_search_locus.shtml). The accession numbers of the key genes mentioned are as follows: HvMADS8 (HORVU5Hr1G076400), HvDL (HORVU4Hr1G067780), HvMADS13 (HORVU1Hr1G023620), HvOSH1 (HORVU4Hr1G009730), HvMADS21 (HORVU1Hr1G064150), HvMADS1 (HORVU4Hr1G067680), and OsMADS8 (LOC_Os09g32948). The accession numbers of genes used in RT-qPCR and yeast 2-hybrid assay are listed in Supplemental Data Set 5. The raw data files for the CUT&Tag analysis reported in this paper have been deposited in the SRA database (accession no. PRJNA1005699).
Supplementary Material
Acknowledgments
The authors wish to dedicate this paper to Prof. Dabing Zhang who tragically passed away in June 2023. Dabing's exceptional vision, energy, and friendship made this work possible, and he will be sorely missed. We thank Mr. C. Ma (University of Adelaide) for assistance with in situ hybridization; Prof. Y. Liu (South China Agricultural University) for providing vectors for CRISPR/Cas9 editing; Dr. H. Kuijer for providing some of the RT-qPCR primers; Dr. G. Mayo (Adelaide Microscopy) for assisting with the microscopy work; and Miss A. Chieng and Miss H. Zhou (University of Adelaide) for managing the plant materials and assistance with barley transformation.
Contributor Information
Chaoqun Shen, Joint International Research Laboratory of Metabolic and Developmental Sciences, State Key Laboratory of Hybrid Rice, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 20040, China; Waite Research Institute, School of Agriculture, Food and Wine, The University of Adelaide, Waite campus, Adelaide, South Australia 5064, Australia.
Yueya Zhang, Joint International Research Laboratory of Metabolic and Developmental Sciences, State Key Laboratory of Hybrid Rice, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 20040, China.
Gang Li, Waite Research Institute, School of Agriculture, Food and Wine, The University of Adelaide, Waite campus, Adelaide, South Australia 5064, Australia.
Jin Shi, Joint International Research Laboratory of Metabolic and Developmental Sciences, State Key Laboratory of Hybrid Rice, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 20040, China.
Duoxiang Wang, Joint International Research Laboratory of Metabolic and Developmental Sciences, State Key Laboratory of Hybrid Rice, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 20040, China.
Wanwan Zhu, Joint International Research Laboratory of Metabolic and Developmental Sciences, State Key Laboratory of Hybrid Rice, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 20040, China.
Xiujuan Yang, Waite Research Institute, School of Agriculture, Food and Wine, The University of Adelaide, Waite campus, Adelaide, South Australia 5064, Australia.
Ludovico Dreni, Joint International Research Laboratory of Metabolic and Developmental Sciences, State Key Laboratory of Hybrid Rice, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 20040, China.
Matthew R Tucker, Waite Research Institute, School of Agriculture, Food and Wine, The University of Adelaide, Waite campus, Adelaide, South Australia 5064, Australia.
Dabing Zhang, Joint International Research Laboratory of Metabolic and Developmental Sciences, State Key Laboratory of Hybrid Rice, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 20040, China; Waite Research Institute, School of Agriculture, Food and Wine, The University of Adelaide, Waite campus, Adelaide, South Australia 5064, Australia.
Author contributions
D.Z. and G.L. initiated the project. C.S., G.L., and D.Z. conceived the projects and designed the experiments. C.S. and Y.Z. carried out most of the experiments. G.L. created the Hvmads8 mutant lines in the GP background. L.D. created the Osmads8 mutant line in the DJ background. J.S. and D.W. analyzed the RNA-seq and CUT&Tag data. G.L. and C.S. performed the dual-LUC assay. C.S., Y.Z., and W.Z. conducted the ChIP-qPCR experiment. C.S. and X.Y. conducted the in situ hybridization. C.S., G.L., J.S., Y.Z., D.W., X.Y., L.D., M.R.T., and D.Z. analyzed and discussed the results. C.S., D.Z., and M.R.T. wrote the manuscript with input from all authors.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Genotypes and phenotypes of Hvmads8 mutants in the GP and WI WT backgrounds.
Supplemental Figure S2 . Mutation of HvMADS8 alters expression of floral- and meristem-related genes at high temperature.
Supplemental Figure S3 . Interaction of HvMADS8 with barley homologs of homeotic regulatory proteins at different temperatures.
Supplemental Figure S4 . Seed setting of Osmads8 at different temperatures.
Supplemental Figure S5 . Genotypes and phenotypes of Hvmads7 single and Hvmads7 Hvmads8 double mutants in the GP background.
Supplemental Figure S6 . Amino acid sequence alignment of barley E-class family proteins with B- and C-class members.
Supplemental Table S1 . Variation of HvMADS8 coding sequences.
Supplemental Data Set 1 . DEGs of Hvmads8 in terms of different temperatures or genotypes.
Supplemental Data Set 2 . GO analysis of DEGs in WT and Hvmads8 at 15 and 28 °C.
Supplemental Data Set 3 . Curated list of genes for heatmaps in RNA-seq data.
Supplemental Data Set 4 . Target genes of HvMADS8 at 15 and 28 °C from CUT&Tag.
Supplemental Data Set 5 . Primers used in this study.
Supplemental Data Set 6 . Summary of statistical analyses.
Funding
This work was financially supported by the National Natural Science Foundation of China (32130006 and 31970803), a Yazhou Bay Seed Laboratory Project (B21HJ8104), the Jiangsu Provincial Key R&D Programme (BE2021323), the Australian Research Council Discovery Project grants (DP210100956 and DP230102476), the Australian Research Council Training Center for Future Crops Development (IC210100047), the Australian Research Council Linkage Project (LP210301062), the China Innovative Research Team (Ministry of Education), and the Programme of Introducing Talents of Discipline to Universities (111 Project, B14016) to D.Z.
Data availability
The authors declare that all data supporting the finding of this study are available within the manuscript and its supplementary information files or are available from the corresponding author upon request.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Abraham-Juárez MJ, Schrager-Lavelle A, Man J, Whipple C, Handakumbura P, Babbitt C, Bartlett M. Evolutionary variation in MADS box dimerization affects floral development and protein abundance in maize. Plant Cell 2020:32(11):3408–3424. 10.1105/tpc.20.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 2007:8(1):242. 10.1186/1471-2164-8-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blighe K, Rana S, Lewis M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R Package version. 2019:1:0.
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. Chromatin techniques for plant cells. Plant J. 2004:39(5):776–789. 10.1111/j.1365-313X.2004.02169.x [DOI] [PubMed] [Google Scholar]
- Callens C, Tucker MR, Zhang D, Wilson ZA. Dissecting the role of MADS-box genes in monocot floral development and diversity. J Exp Bot. 2018:69(10):2435–2459. 10.1093/jxb/ery086 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Balasubramanian S. Thermomorphogenesis. Annu Rev Plant Biol. 2019:70:321–346. 10.1146/annurev-arplant-050718-095919 [DOI] [PubMed] [Google Scholar]
- Chen R, Zhao X, Shao Z, Wei Z, Wang Y, Zhu L, Zhao J, Sun M, He R, He G. Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 2007:19(3):847–861. 10.1105/tpc.106.044123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Qian Q, Liang W, Yin C, Tan H, Yao X, Yuan Z, Yang J, Huang H, Luo D, et al. The FLORAL ORGAN NUMBER4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 2006:142(3):1039–1052. 10.1104/pp.106.086736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaffi M, Paolacci AR, Tanzarella OA, Porceddu E. Molecular aspects of flower development in grasses. Sex Plant Reprod. 2011:24(4):247–282. 10.1007/s00497-011-0175-y [DOI] [PubMed] [Google Scholar]
- Colombo L, Franken J, Koetje E, Van Went J, Dons HJ, Angenent GC, Van Tunen AJ. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 1995:7(11):1859–1868. 10.1105/tpc.7.11.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta M, Colombo L, Roig-Villanova I. Ovule development, a new model for lateral organ formation. Front Plant Sci. 2014:5:117. 10.3389/fpls.2014.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z. Functional conservation and diversification of class e floral homeotic genes in rice (Oryza sativa). Plant J. 2010:61(5):767–781. 10.1111/j.1365-313X.2009.04101.x [DOI] [PubMed] [Google Scholar]
- Ding Y, Shi Y, Yang S. Molecular regulation of plant responses to environmental temperatures. Mol Plant. 2020:13(4):544–564. 10.1016/j.molp.2020.02.004 [DOI] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PB, An G, Colombo L, Kater MM. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 2007:52(4):690–699. 10.1111/j.1365-313X.2007.03272.x [DOI] [PubMed] [Google Scholar]
- Dreni L, Osnato M, Kater MM. The ins and outs of the rice AGAMOUS subfamily. Mol Plant. 2013:6(3):650–664. 10.1093/mp/sst019 [DOI] [PubMed] [Google Scholar]
- Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D, Kater MM. Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 2011:23(8):2850–2863. 10.1105/tpc.111.087007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colombo L. MADS-box protein complexes control carpel and ovule development in arabidopsis. Plant Cell 2003:15:(11):2603–2611. 10.1105/tpc.015123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C, Fourquin C, Prunet N, Scutt CP, Sundberg E, Trehin C, Vialette-Guiraud CM. Carpel development. Adv Bot Res. 2010:55:1–74. 10.1016/B978-0-12-380868-4.00001-6 [DOI] [Google Scholar]
- Franklin KA. Light and temperature signal crosstalk in plant development. Curr Opin Plant Biol. 2009:12(1):63–68. 10.1016/j.pbi.2008.09.007 [DOI] [PubMed] [Google Scholar]
- Fuller DQ, Qin L, Zheng Y, Zhao Z, Chen X, Hosoya LA, Sun GP. The domestication process and domestication rate in rice: spikelet bases from the Lower Yangtze. Science 2009:323(5921):1607–1610. 10.1126/science.1166605 [DOI] [PubMed] [Google Scholar]
- Gramzow L, Theissen G. A hitchhiker's guide to the MADS world of plants. Genome Biol. 2010:11(6):214 . 10.1186/gb-2010-11-6-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusewski S, Melzer R, Rüempler F, Gafert C, Theissen G. The floral homeotic protein SEPALLATA3 recognizes target DNA sequences by shape readout involving a conserved arginine residue in the MADS-domain. bioRxiv 133678. 10.1101/133678. 2017. [DOI]
- Harwood WA. A protocol for high-throughput Agrobacterium-mediated barley transformation. In: Henry RJ, Furtado A, editors. Cereal genomics: methods and protocols. Totowa: (NJ: ): Humana Press; 2014. p. 251–260 [DOI] [PubMed] [Google Scholar]
- Hugouvieux V, Silva CS, Jourdain A, Stigliani A, Charras Q, Conn V, Conn SJ, Carles CC, Parcy F, Zubieta C. Tetramerization of MADS family transcription factors SEPALLATA3 and AGAMOUS is required for floral meristem determinacy in Arabidopsis. Nucleic Acids Res. 2019:46(10):4966–4977. 10.1093/nar/gky205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RG, Tonaco IA, de Folter S, Shchennikova A, van Dijk AD, Busscher-Lange J, Borst JW, Angenent GC. SEPALLATA3: the “glue” for MADS box transcription factor complex formation. Genome Biol. 2009:10(2):R24. 10.1186/gb-2009-10-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle M, Marco CF, Timmermans M. In situ hybridization for the precise localization of transcripts in plants. J Vis Exp. 2011:3(57):e3328. 10.3791/3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Qian Q, Mao L, Zhou QY, Zhai WX. Characterization of the rice floral organ number mutant fon3. J Integr Plant Biol. 2005:47(1):100–106. 10.1111/j.1744-7909.2005.00017.x [DOI] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994:6(9):1211–1225. 10.1105/tpc.6.9.1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016:354(6314):886–889. 10.1126/science.aaf6005 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. Target genes of the MADS transcription factor sepallata3: integration of developmental and hormonal pathways in the arabidopsis flower. PLoS Biol. 2009:7(4):e1000090. 10.1371/journal.pbio.1000090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 2012:139(6):1105–1109. 10.1242/dev.067918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA. Evolutionary history of the grasses. Plant Physiol. 2001:125(3):1198–1205. 10.1104/pp.125.3.1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijer HNJ, Shirley NJ, Khor SF, Shi J, Schwerdt J, Zhang D, Li G, Burton RA. Transcript profiling of MIKCc MADS-box genes reveals conserved and novel roles in barley inflorescence development. Front Plant Sci. 2021:12:705286. 10.3389/fpls.2021.705286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. H2a.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 2010:140(1):136–147. 10.1016/j.cell.2009.11.006 [DOI] [PubMed] [Google Scholar]
- Li G, Kuijer HNJ, Yang X, Liu H, Shen C, Shi J, Betts N, Tucker MR, Liang W, Waugh R, et al. MADS1 maintains barley spike morphology at high ambient temperatures. Nat Plants. 2021:7(8):1093–1107. 10.1038/s41477-021-00957-3 [DOI] [PubMed] [Google Scholar]
- Li S, Lin D, Zhang Y, Deng M, Chen Y, Lv B, Li B, Lei Y, Wang Y, Zhao L, et al. Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 2022:602(7897):455–460. 10.1038/s41586-022-04395-9 [DOI] [PubMed] [Google Scholar]
- Li Z, Ma S, Liu D, Zhang L, Du X, Xia Y, Song Q, Li Y, Zhang Y, Li Z, et al. Morphological and proteomic analysis of young spikes reveals new insights into the formation of multiple-pistil wheat. Plant Sci. 2020:296:110503. 10.1016/j.plantsci.2020.110503 [DOI] [PubMed] [Google Scholar]
- Liu T, Sun Y, Ma C, Jiang W, Wu H, Jiang Q, Xiao E, Wang Y, Kang Z, Shi X, et al. Morphological characterization and genetic analysis of tri-pistil trait, a precisely regulated pistil number mutation in bread wheat. Res Sq. 2022:1:1–25. 10.21203/rs.3.rs-1217258/v1 [DOI] [Google Scholar]
- Liu X, Kim YJ, Müller R, Yumul RE, Liu C, Pan Y, Cao X, Goodrich J, Chen X. AGAMOUS terminates floral stem cell maintenance in arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 2011:23(10):3654–3670. 10.1105/tpc.111.091538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015:8(8):1274–1284. 10.1016/j.molp.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. SEPALLATA gene diversification: brave new whorls. Trends Plant Sci. 2005:10(9):427–435. 10.1016/j.tplants.2005.07.008 [DOI] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017:544(7651):427–433. 10.1038/nature22043 [DOI] [PubMed] [Google Scholar]
- Moon S, Jung KH, Lee DE, Lee DY, Lee J, An K, Kang HG, An G. The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size. Mol Cells. 2006:21:147–152. https://www.molcells.org/journal/view.html?year=2006&volume=21&number=1&spage=147 [PubMed] [Google Scholar]
- Muiño JM, Smaczniak C, Angenent GC, Kaufmann K, van Dijk AD. Structural determinants of DNA recognition by plant MADS-domain transcription factors. Nucleic Acids Res. 2014:42(4):2138–2146. 10.1093/nar/gkt1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai K. Homeotic genes and the ABCDE model for floral organ formation in wheat. Plants (Basel) 2013:2(3):379–395. 10.3390/plants2030379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 2003:130(4):705–718. 10.1242/dev.00294 [DOI] [PubMed] [Google Scholar]
- Osnato M, Lacchini E, Pilatone A, Dreni L, Grioni A, Chiara M, Horner D, Pelaz S, Kater MM. Transcriptome analysis reveals rice MADS13 as an important repressor of the carpel development pathway in ovules. J Exp Bot. 2021:72(2):398–414. 10.1093/jxb/eraa460 [DOI] [PubMed] [Google Scholar]
- Pankin A, von Korff M. Co-evolution of methods and thoughts in cereal domestication studies: a tale of barley (Hordeum vulgare). Curr Opin Plant Biol. 2017:36:15–21. 10.1016/j.pbi.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000:405(6783):200–203. 10.1038/35012103 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Tapia-López R, Alvarez-Buylla ER, Yanofsky MF. Conversion of leaves into petals in Arabidopsis. Curr Biol. 2001:11(3):182–184. 10.1016/S0960-9822(01)00024-0 [DOI] [PubMed] [Google Scholar]
- Peng ZS, Martinek P, Kosuge K, Kuboyama T, Watanabe N. Genetic mapping of a mutant gene producing three pistils per floret in common wheat. J Appl Genet. 2008:49(2):135–139. 10.1007/BF03195606 [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003:424(6944):85–88. 10.1038/nature01741 [DOI] [PubMed] [Google Scholar]
- Prunet N, Yang W, Das P, Meyerowitz EM, Jack TP. SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proc Natl Acad Sci U S A. 2017:114(27):7166–7171. 10.1073/pnas.1705977114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010:26(6):841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, Van Zanten M. Molecular and genetic control of plant thermomorphogenesis. Nat Plants. 2016:2(1):1–9. 10.1038/nplants.2015.190 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011:29(1):24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J, Mascher M, Dawson IK, Kyriakidis S, Calixto C, Freund F, Bayer M, Milne I, Marshall-Griffiths T, Heinen S, et al. Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat Genet. 2016:48(9):1024–1030. 10.1038/ng.3612 [DOI] [PubMed] [Google Scholar]
- Selva C, Shirley NJ, Houston K, Whitford R, Baumann U, Li G, Tucker MR. HvLEAFY controls the early stages of floral organ specification and inhibits the formation of multiple ovaries in barley. Plant J. 2021:108(2):509–527. 10.1111/tpj.15457 [DOI] [PubMed] [Google Scholar]
- Selva C, Yang X, Shirley NJ, Whitford R, Baumann U, Tucker MR. HvSL1 and HvMADS16 promote stamen identity to restrict multiple ovary formation in barley. J Exp Bot. 2023:74(17):5039–5056. 10.1093/jxb/erad218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Li G, Dreni L, Zhang D. Molecular control of carpel development in the grass family. Front Plant Sci. 2021:12:635500. 10.3389/fpls.2021.635500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Zhou Y, Cai J, Shang E, Yamaguchi N, Xiao J, Looi LS, Wee WY, Gao X, Wagner D, et al. Integration of transcriptional repression and polycomb-mediated silencing of WUSCHEL in floral meristems. Plant Cell 2019:31(7):1488–1505. 10.1105/tpc.18.00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshkumar S, Dent C, Seleznev A, Tasset C, Balasubramanian S. Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat Plants. 2016:2(5):16055. 10.1038/nplants.2016.55 [DOI] [PubMed] [Google Scholar]
- Suzaki T, Toriba T, Fujimoto M, Tsutsumi N, Kitano H, Hirano HY. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 2006:47(12):1591–1602. 10.1093/pcp/pcl025 [DOI] [PubMed] [Google Scholar]
- Theissen G, Melzer R, Rümpler F. MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 2016:143(18):3259–3271. 10.1242/dev.134080 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci U S A. 2003:100(22):13099–13104. 10.1073/pnas.1635053100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu LD, Xu X, Gevaert K, De Smet I. Developmental plasticity at high temperature. Plant Physiol. 2019:181(2):399–411. 10.1104/pp.19.00652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington SR, Cartwright PM, Wall PC. A quantitative scale of spike initial and pistil development in barley and wheat. Ann Bot 1983:51(1):119–130. 10.1093/oxfordjournals.aob.a086434 [DOI] [Google Scholar]
- Wang D, Li J, Sun L, Hu Y, Yu J, Wang C, Zhang F, Hou H, Liang W, Zhang D. Two rice MYB transcription factors maintain male fertility in response to photoperiod by modulating sugar partitioning. New Phytol. 2021:231(4):1612–1629. 10.1111/nph.17512 [DOI] [PubMed] [Google Scholar]
- Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 2018:557(7703):43–49. 10.1038/s41586-018-0063-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker C, Dean C. The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu Rev Cell Dev Biol. 2017:33(1):555–575. 10.1146/annurev-cellbio-100616-060546 [DOI] [PubMed] [Google Scholar]
- Wils CR, Kaufmann K. Gene-regulatory networks controlling inflorescence and flower development in Arabidopsis thaliana. Biochim Biophys Acta Gene Regul Mech. 2017:1860(1):95–105. 10.1016/j.bbagrm.2016.07.014 [DOI] [PubMed] [Google Scholar]
- Wu D, Liang W, Zhu W, Chen M, Ferrándiz C, Burton RA, Dreni L, Zhang D. Loss of LOFSEP transcription factor function converts spikelet to leaf-like structures in rice. Plant Physiol. 2018:176(2):1646–1664. 10.1104/pp.17.00704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Jing X, Zhang B, Chen S, Xu R, Duan P, Zou D, Huang S, Zhou T, An C, et al. A natural allele of OsMS1 responds to temperature changes and confers thermosensitive genic male sterility. Nat Commun. 2022:13(1):2055. 10.1038/s41467-022-29648-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A. 2015:112(11):3570–3575. 10.1073/pnas.1420294112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Luo H, Huang Y, Wang Y, Ru W, Shi Y, Huang W, Wang H, Dong Z, Jin W. A missense mutation in a large subunit of ribonucleotide reductase confers temperature-gated tassel formation. Plant Physiol. 2020:184(4):1979–1997. 10.1104/pp.20.00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Zhang H, Zhao F, Zhao T, Li H, Jiang D. The INO80 chromatin remodeling complex promotes thermomorphogenesis by connecting H2A.Z eviction and active transcription in Arabidopsis. Mol Plant. 2021:14(11):1799–1813. 10.1016/j.molp.2021.07.001 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. The yabby gene drooping leaf regulates carpel specification and midrib development in Oryza sativa. Plant Cell 2004:16(2):500–509. 10.1105/tpc.018044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S, Nagato Y, Kurata N, Nonomura K. Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Dev Biol. 2011:351(1):208–216. 10.1016/j.ydbio.2010.12.006 [DOI] [PubMed] [Google Scholar]
- Yang X, Tucker MR. Establishing a regulatory blueprint for ovule number and function during plant development. Curr Opin Plant Biol. 2021:63:102095. 10.1016/j.pbi.2021.102095 [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015:31(14):2382–2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Yu J, Han J, Kim YJ, Song M, Yang Z, He Y, Fu R, Luo Z, Hu J, Liang W, et al. Two rice receptor-like kinases maintain male fertility under changing temperatures. Proc Natl Acad Sci U S A. 2017:114(46):12327–12332. 10.1073/pnas.1705189114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, DePamphilis CW, Ma H. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005:169(4):2209–2223. 10.1534/genetics.104.037770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Liu Bing, Piao S, Wang X, Lobell DB, Huang Y, Huang M, Yao Y, Bassu S, Ciais P, et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci U S A. 2017:114(35):9326–9331. 10.1073/pnas.1701762114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhou M, Yang Y, Li J, Zhu L, Jiang D, Dong J, Liu Q, Gu L, Zhou L, et al. RNase ZS1 processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nat Commun. 2014:5(1):4884. 10.1038/ncomms5884 [DOI] [PubMed] [Google Scholar]
- Zhu J, Lou Y, Shi Q-S, Zhang S, Zhou W-T, Yang J, Zhang C, Yao X-Z, Xu T, Liu J-L, et al. Slowing development restores the fertility of thermo-sensitive male-sterile plant lines. Nat Plants. 2020:6(4):360–367. 10.1038/s41477-020-0622-6 [DOI] [PubMed] [Google Scholar]
- Zhu W, Yang L, Wu D, Meng Q, Deng X, Huang G, Zhang J, Chen X, Ferrándiz C, Liang W, et al. Rice SEPALLATA genes OsMADS5 and OsMADS34 cooperate to limit inflorescence branching by repressing the TERMINAL FLOWER1-like gene RCN4. New Phytol. 2022:233(4):1682–1700. 10.1111/nph.17855 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the finding of this study are available within the manuscript and its supplementary information files or are available from the corresponding author upon request.