Abstract
Reactive oxygen species (ROS) play an essential role in plant growth and responses to environmental stresses. Plant cells sense and transduce ROS signaling directly via hydrogen peroxide (H2O2)–mediated posttranslational modifications (PTMs) on protein cysteine residues. Here, we show that the H2O2-mediated cysteine oxidation of NAC WITH TRANS-MEMBRANE MOTIF1-LIKE 1 (GmNTL1) in soybean (Glycine max) during salt stress promotes its release from the endoplasmic reticulum (ER) membrane and translocation to the nucleus. We further show that an oxidative posttranslational modification on GmNTL1 residue Cys-247 steers downstream amplification of ROS production by binding to and activating the promoters of RESPIRATORY BURST OXIDASE HOMOLOG B (GmRbohB) genes, thereby creating a feed-forward loop to fine-tune GmNTL1 activity. In addition, oxidation of GmNTL1 Cys-247 directly promotes the expression of CATION H+ EXCHANGER 1 (GmCHX1)/SALT TOLERANCE-ASSOCIATED GENE ON CHROMOSOME 3 (GmSALT3) and Na+/H+ Antiporter 1 (GmNHX1). Accordingly, transgenic overexpression of GmNTL1 in soybean increases the H2O2 levels and K+/Na+ ratio in the cell, promotes salt tolerance, and increases yield under salt stress, while an RNA interference–mediated knockdown of GmNTL1 elicits the opposite effects. Our results reveal that the salt-induced oxidation of GmNTL1 promotes its relocation and transcriptional activity through an H2O2-mediated posttranslational modification on cysteine that improves resilience of soybean against salt stress.
Oxidation of transcription factor GmNTL1 triggers its translocation to the nucleus, where it activates the transcription of several target genes to regulate soybean (Glycine max) salt tolerance.
IN A NUTSHELL.
Background: Worldwide, soybean (Glycine max) production is often adversely affected by salinity. Reactive oxygen species (ROS) are key signaling molecules that enable cells to rapidly respond to different stimuli, regulating plant growth and development by mediating protein oxidative post-translational modifications (OxiPTMs). Thus, identifying the underlying mechanisms of redox regulation and its contribution to various physiological processes is a current research hotspot.
Question: The membrane-bound NAC WITH TRANS-MEMBRANE MOTIF1-LIKE (NTL) transcription factors play critical roles in plant responses to various environmental stimuli. However, how GmNTLs translocate to the nucleus and regulate salt tolerance remains unclear.
Findings: Our study reports that GmNTL1 is released from the endoplasmic reticulum (ER) and translocates to the nucleus upon NaCl or hydrogen peroxide (H2O2) treatment. Salt-induced H2O2 production increases GmNTL1 nuclear import and DNA-binding activity by oxidizing cysteine 247. Furthermore, GmNTL1 oxidation directly activates the expression of RESPIRATORY BURST OXIDASE HOMOLOG B (GmRbohB) genes, affecting the production of H2O2, thereby forming a feed-forward loop to fine-tune GmNTL1 activity. In addition, the oxidation of GmNTL1 further activates the expression of CATION H+ EXCHANGER 1 (GmCHX1)/SALT TOLERANCE-ASSOCIATED GENE ON CHROMOSOME 3 (GmSALT3) and Na+/H+ EXCHANGER 1 (GmNHX1), reducing root Na+ accumulation and improving soybean salt tolerance.
Next steps: Given that OxiPTMs are highly dynamic and interconvertible, we will next focus on analyzing the reduction mechanisms of GmNTL1. Understanding how GmNTL1 regulates cellular redox dynamics will generate new ideas to elucidate soybean salt stress responses.
Introduction
Saline soils cover 3.1% (397 million hectares) of the total land area of the world (Setia et al. 2013). A large portion of cultivated land is damaged by salinity, and the situation worsens yearly (Rasheed et al. 2022). High salinity is an adverse environmental factor that limits the growth and yield of diverse crop plants (van Zelm et al. 2020). Soybean (Glycine max) is greatly affected by salinity at all growth stages (Cho et al. 2021). Salt stress causes ion toxicity, osmotic stress, and reactive oxygen species (ROS)–derived damage, with ion toxicity being the primary and fundamental issue (van Zelm et al. 2020; Zhao et al. 2021).
To protect themselves from salt and other stresses, plants deploy a variety of complex mechanisms to rapidly sense and adapt to their changing surroundings (Yang and Guo 2018; Zhao et al. 2021). The activities of many stress-related proteins are enhanced under abiotic stress, including ion channel proteins in soybean, such as Na+/H+ Antiporter 1 (GmNHX1) (Yang et al. 2017), CATION/PROTON EXCHANGER1 (GmCAX1) (Luo et al. 2005), cation/H+ antiporter (GmCHX1/GmSALT3) (Guan et al. 2014; Qi et al. 2014; Liu et al. 2016), CHLORIDE CHANNEL1 (GmCLC1) (Li et al. 2006), and ARABIDOPSIS K+ TRANSPORTER1(GmAKT1) (Wang et al. 2021). GmNHX1 sequesters Na+ from the cytoplasm into the vacuole, while GmCHX1 confers leaf Na+ exclusion to reduce the toxic effects of excess salt (Qu et al. 2022).
ROS are enzymatically produced in various subcellular organelles by oxidases such as RESPIRATORY BURST OXIDASE HOMOLOGS (Rbohs), or as byproducts of metabolic pathways (Waszczak et al. 2018). ROS function as a double-edged sword; excessive ROS affect redox signaling and disrupt cellular redox homeostasis to damage plant development (Sachdev et al. 2021; Zhang et al. 2021), but lower levels ROS promote plant growth and development in response to stresses (Niu and Liao 2016; Nazir et al. 2020). ROS can oxidize many biomolecules, including lipids, proteins, RNA, and DNA (Mittler 2017; De Smet et al. 2019; Huang et al. 2019; Smirnoff and Arnaud 2019; Nietzel et al. 2020; Sies and Jones 2020).
Hydrogen peroxide (H2O2) is the most stable ROS due to its relatively long half-life in living cells (Bi et al. 2022). H2O2 can directly oxidize proteins through oxidative posttranslational modifications (Oxi-PTMs), mainly on methionine and cysteine residues, that can induce changes in the protein conformation, subcellular localization, and/or activity of proteins including transcription factors to initiate specific H2O2 signaling pathways (Waszczak et al. 2015; Zhou et al. 2023). Protein cysteine residue (Cys) contains the sulfhydryl (-SH) group, also named thiol, on its side chain, which is highly redox reactive and thus can undergo different Oxi-PTMs including S-sulfenylation, S-gutathiolnylation, disulfide bond formation, and persulfidation.
Increasing evidence shows that Cys Oxi-PTMs are involved in multiple critical signaling pathways in plant stress responses and development and affect proteins including ACYL-PROTEIN THIOESTERASE 1 (APT1) (Ji et al. 2023), BRASSINOSTEROID-INSENSITIVE2 (BIN2) (Lu et al. 2022), PLASTID TRIOSE PHOSPHATE ISOMERASE (pdTPI) (Fu et al. 2023), cold-responsive C-REPEAT BINDING TRANSCRIPTION FACTORs (CBFs) (Lee et al. 2021), the cytosolic ENOLASE2 (ENO2) (Liu et al. 2022), BASIC LEUCINE ZIPPER 68 (bZIP68) (Li et al. 2019b), HEAT SHOCK FACTOR A8 (HSFA8) (Giesguth et al. 2015), MULTIPROTEIN BRIDGING ACTOR 1C (MBF1C) (Suzuki et al. 2013), and BRASSINAZOLE-RESISTANT1 (BZR1) (Tian et al. 2018).
The membrane-bound NAC WITH TRANS-MEMBRANE MOTIF1-LIKE (NTL) transcription factors harbor a C-terminal transmembrane motif and play critical roles in responses to various environmental stimuli (Nakashima et al. 2012; De Clercq et al. 2013; Shao et al. 2015; Duan et al. 2017; Meng et al. 2019; Lin et al. 2021). In a dormant state, NTLs are anchored at the plasma membrane (Tang et al. 2012; Sun et al. 2022) or the endoplasmic reticulum (ER) membrane (Li et al. 2016). When the plant is exposed to biotic or abiotic stress, NTLs are released from the membrane and enter the nucleus to activate their downstream target genes (Seo et al. 2010). In soybean, 15 ER-localized GmNTLs have been identified. Of them, several GmNTLs were shown to translocate to the nucleus in Arabidopsis (Arabidopsis thaliana) protoplasts treated with H2O2 or cold (Li et al. 2016); however, the molecular mechanisms of how GmNTLs perceive the oxidative stress or cold triggered signal and relocate to the nucleus remain unclear.
In this study, we show that overexpression of the salt-induced gene GmNTL1 leads to a significant improvement in soybean field–based salinity tolerance. We demonstrate that salt stress–induced H2O2 production results in the Oxi-PTM on residue Cys-247 of GmNTL1, leading to its translocation from the ER to the nucleus, where it activates the transcription of its target genes, including the GmNHX1 and GmCHX1. In addition, the oxidation of GmNTL1 promotes the expression of GmRbohBs, thereby increasing ROS levels. Our data reveal a feed-forward regulatory loop for the oxidation-induced translocation of GmNTL1 to initiate and amplify salt-induced H2O2 signaling via the transcriptional activation of GmRbohBs.
Results
The NaCl stress–induced expression of GmNTL1 is H2O2 dependent
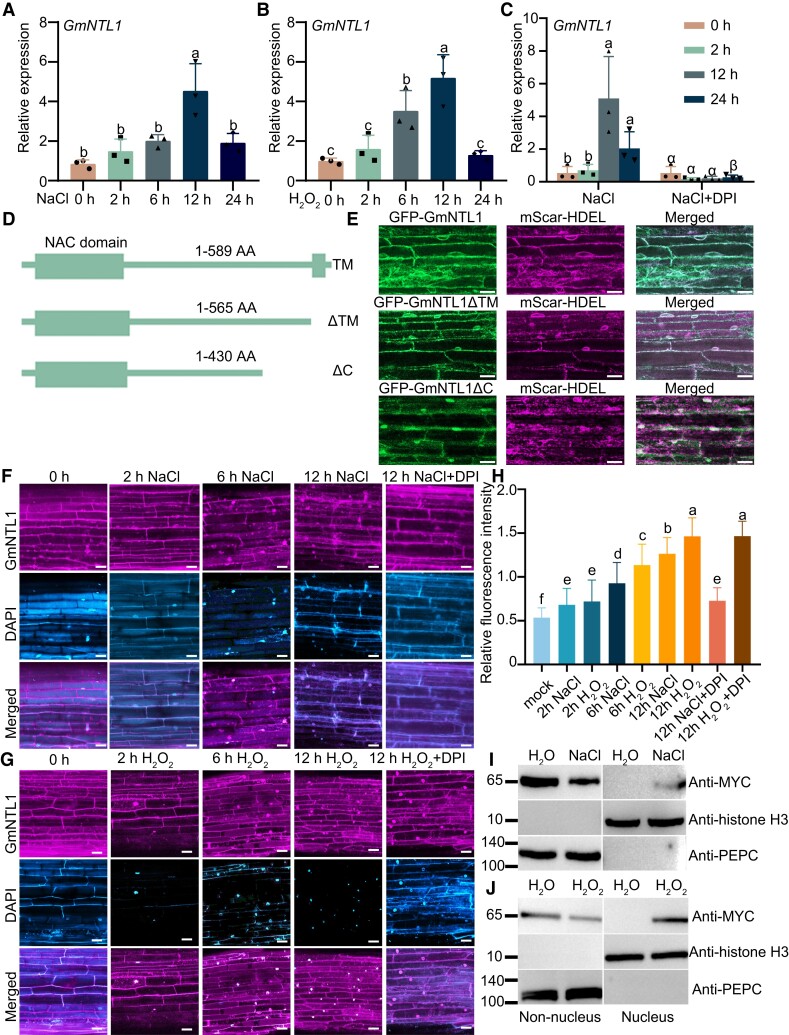
Our previous study reported that both NaCl and H2O2 induced the expression of GmNTL1 (Li et al. 2016). As salt stress can induce the production of ROS (Xing et al. 2022), we explored whether the expression of GmNTL1 induced by NaCl is H2O2 dependent. We analyzed the transcript level of this gene under NaCl, H2O2, and NaCl cotreatment with the NADPH oxidase inhibitor diphenyleneiodonium chloride (DPI) using reverse transcription quantitative PCR (RT-qPCR). The relative GmNTL1 transcript level was 4.5-fold higher upon 12 h of 150 mm NaCl treatment and 5.3 times higher after 12 h of 10 mm H2O2 treatment compared to that of nontreated samples (0 h) (Fig. 1A and B). When treated with DPI and NaCl simultaneously (Zhang et al. 2021), GmNTL1 transcript levels were significantly lower compared to NaCl treatment alone, suggesting that salt-induced GmNTL1 expression is largely H2O2 dependent (Fig. 1C).
Figure 1.
Salt and H2O2 induce the translocation of GmNTL1 from the ER to the nucleus. A–C) Relative GmNTL1 expression levels, as determined by RT-qPCR in response to treatment with 150 mm NaCl (A), 10 mm H2O2 (B), or 150 mm NaCl, and 100 µM DPI (C) for 0 to 24 h in the roots of 6-d-old seedlings of the soybean cultivar Ludou 11. Error bars denote SD (n = 3 from 3 independent experiments). D) Schematic diagram of GmNTL1, GmNTL1ΔTM (TM), and GmNTL1ΔC (ΔC, lacking the C-terminal region). Rectangle, N-terminal region with the highly conserved NAC domain. E) GFP-GmNTL1 localizes to the ER in transiently transfected soybean hairy roots. The truncated forms of GmNTL1 (GmNTL1ΔTM) proteins localize to the ER, and the truncated forms of GmNTL1 (GmNTL1ΔC) proteins localize to the ER and nucleus. Left, GFP signal (green); middle, mScarlet signal (magenta; an ER marker); right, merged images. Scale bar, 20 μm. F, G) Representative fluorescence images of soybean hairy roots expressing RFP-GmNTL1 treated with 150 mm NaCl (F) or 1 mm H2O2 (G) for 0 to 12 h in the absence or presence of 100 µM DPI. Scale bars, 20 µm. DAPI staining used to show the nucleus. H) Quantification of the ratio between nuclear and cytoplasmic RFP fluorescent intensities in (F, G). At least 10 seedling roots were examined for each biological repeat. Data shown are the results of 3 biological experiments. Error bars denote SD (n = 100 cells from 10 images). I, J) Immunoblot analysis of MYC-GmNTL1 in different subcellular fractions of transgenic soybean hairy roots treated with 150 mm NaCl or 1 mm H2O2 or water as control for 12 h. Phosphoenolpyruvate carboxylase (PEPC) and Histone H3 serve as cytosolic and nuclear markers, respectively. Each biological repeat contains 1 technical repeat. Data shown are the results of 2 biological experiments. Lowercase letters and Greek letters indicate significant differences between samples, as determined by 1-way ANOVA, P < 0.05 in (A), (B), (C), and (H). Please see Supplemental Data Set 4 for detailed statistical analyses.
Salt-induced GmNTL1 relocation is H2O2 dependent
GmNTL1 was shown to release from the ER to the nucleus in Arabidopsis protoplasts when treated with H2O2 (Li et al. 2016). We next investigated whether NaCl had the same effect by examining the subcellular localization of GmNTL1 in Arabidopsis protoplasts and soybean hairy roots. Under control conditions, GFP-GmNTL1 mainly localized to the ER; treatment with 150 mm NaCl for 1 h or 10 mm H2O2 for 10 min led to its nuclear accumulation in Arabidopsis protoplasts (Supplemental Fig. S1B). We expressed GFP-GmNTL1, the truncated NTL1 forms GmNTL1-ΔTM (lacking the transmembrane [TM], 1-565 amino acid), GmNTL1-ΔC (lacking the C-terminal region, 1-430 amino acid), and an ER marker (pCambia-mScar-HDEL) in soybean hairy roots. While both GFP-GmNTL1 and GmNTL1-ΔTM were localized to the ER membrane under untreated conditions (Fig. 1D and E), GmNTL1-ΔC was partially localized to the nucleus (Fig. 1D and E; Supplemental Fig. S1A).
To investigate whether NaCl and H2O2 affect the relocation of GmNTL1 in soybean hairy roots. We expressed RFP-GmNTL1 (encoding red fluorescent protein [RFP] fused to GmNTL1) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter in soybean hairy roots. RFP-GmNTL1 relocated to the nucleus when treated with 150 mm NaCl for 2 h, 6 h, or 12 h, as quantified by the rise of the nucleocytoplasmic fluorescence intensity ratio from 1.27 to 1.73 and 2.36, respectively, when compared to the mock controls (Fig. 1F and H). Similarly, under 1 mm H2O2 treatments for 2 h, 6 h, or 12 h, the nucleocytoplasmic fluorescence intensity ratios gradually increased from 1.35 to 2.12 and 2.73, respectively, when compared to the mock (Fig. 1G and H).
We further tested whether the inhibition of H2O2 production might suppress the salt-induced relocation of GmNTL1 using 100 µM DPI in addition to either 150 mm NaCl or 1 mm H2O2 for 12 h. We observed that the salt-induced nuclear relocation of GmNTL1 is repressed by DPI, but not when applying DPI together with H2O2 (Fig. 1F, G, and H). While DPI could not inhibit the nuclear relocation of GmNTL1 upon H2O2 treatment, the addition of glutathione (GSH, an antioxidant) resulted an exclusively ER-localized RFP signal (Supplemental Fig. S2A and D), suggesting that GmNTL1 relocation is redox regulated. Moreover, we further confirmed our observations by using the subcellular fractionation and immunoblotting of proteins extracted from soybean hairy roots expressing MYC-GmNTL1. The GmNTL1 was found to be present only in the cytoplasmic fraction under normal conditions but predominantly relocated in nuclear after a 12-h treatment with NaCl or H2O2 (Fig. 1I and J). Taken together, these results suggest that the NaCl-induced relocation of GmNTL1 to the nucleus is H2O2 dependent.
GmNTL1-mediated salt tolerance is H2O2 dependent
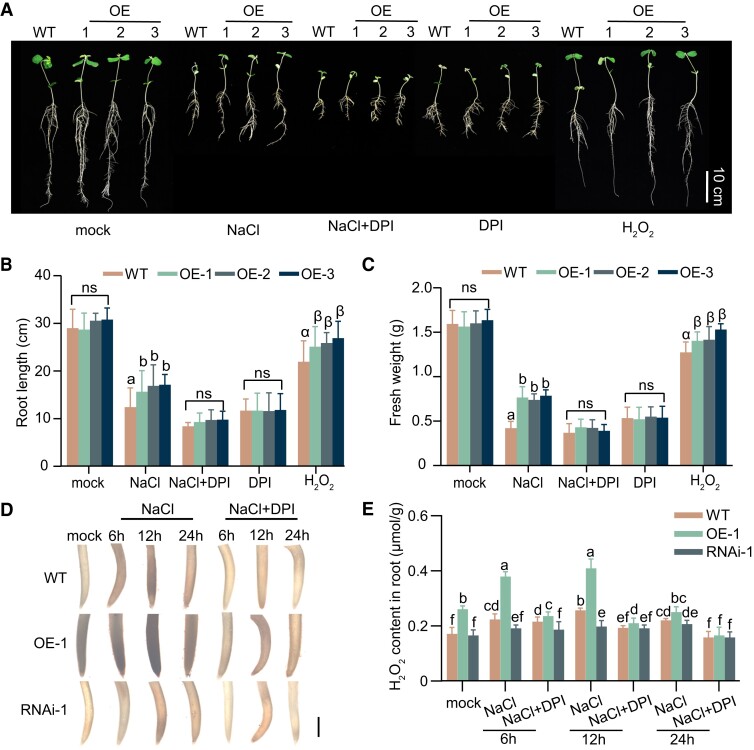
To further determine whether H2O2 is required for GmNTL1-mediated salt tolerance, we generated transgenic soybean plant overexpressing an active form of GmNTL1 (Pro35S:GmNTL1-ΔC; OE-1, OE-2, and OE-3), of which the similar protein levels of GmNTL1 were confirmed by immunoblotting using anti-GmNTL1 antibody (Supplemental Fig. S3A, C, and D). We also overexpressed a GmNTL1 RNA interference construct (Pro35S:GmNTL1-RNAi; lines RNAi-1, RNAi-2, and RNAi-3) in transgenic soybeans (Supplemental Fig. S3B, C, and D). We determined the transgenic or residual protein levels of GmNTL1 in the T2 RNAi lines. Immunoblotting showed that GmNTL1 decreased in RNAi lines (RNAi-1, RNAi-2, and RNAi-3) (Supplemental Fig. S3B, C, and D). Then, we performed a root growth assay under treatments consisting of NaCl only, NaCl coupled with DPI, DPI only, and H2O2 only. To this end, we transferred 4-d-old seedlings to Hoagland nutrient solution containing 175 mm salt or without salt (mock) for 7 d, after which we measured primary root length and fresh weight. Under mock treatment, no visible differences among 11-d-old wild-type (WT, Ludou11) and GmNTL1-ΔC OE seedlings are present (Fig. 2A), whereas OE seedlings displayed longer primary roots and higher fresh weights than the WT under 175 mm NaCl treatment (Fig. 2A–C).
Figure 2.
GmNTL1 promoted root growth and H2O2 was required GmNTL1-mediated salt tolerance. A) Root length of 14-d-old soybean seedlings grown with no NaCl (mock) or challenged with 175 mm NaCl, 175 mm NaCl, and 100 μM DPI, 100 μM DPI, or 200 μM H2O2. The treatments began 4 d after germination and lasted for 7 d. Scale bar, 10 cm. B, C) Primary root length (B) and fresh weight (C) of the roots from the seedlings shown in (A). Data are means ± SD (n = 20 from 3 biological experiments). D) DAB staining of roots from GmNTL1 verexpressor-1 (OE-1), GmNTL1 RNA interference-1 (RNAi-1), and WT plants exposed to either no NaCl (mock) or 150 mm NaCl for 6, 12, or 24 h. The staining intensity reflects the concentration of H2O2. Five seedling roots were examined for each biological repeat. Error bars denote SD (n = 15 from 3 biological experiments). Scale bar, 1 mm. E) H2O2 contents of the roots of 6-d-old GmNTL1 OE-1, GmNTL1 RNAi-1, and WT seedlings. Four seedling roots were examined for each biological repeat. Error bars denote SD (n = 12 from 3 biological experiments). Lowercase letters and Greek letters indicate significant differences between samples, as determined by 1-way ANOVA, P < 0.05 in (B) and (C). Lowercase letters indicate significant differences between samples, as determined by 2-way ANOVA, P < 0.05 in (E). Please see Supplemental Data Set 4 for detailed statistical analyses.
Next, we examined whether the salt tolerance of GmNTL1-ΔC OE seedlings was dependent on H2O2. When co-treated with 175 mm NaCl and 100 μM DPI or 100 μM DPI alone, GmNTL1-ΔC OE seedlings showed similar root lengths and fresh weights as the WT (Fig. 2B and C). Compared to the WT, the GmNTL1-ΔC OE lines produced longer primary roots and higher fresh weights under the H2O2 (200 μM) treatment condition (Fig. 2B and C). These data suggest that H2O2 is essential for GmNTL1-mediated salt tolerance.
Next, we stained roots with 3,3-diaminobenzidine (DAB), which forms a brown precipitate in the presence of H2O2. Under untreated conditions, we observed stronger DAB staining in the GmNTL1-ΔC OE plants and weaker DAB staining in the RNAi plants than in the WT (Fig. 2D). After NaCl treatment for 6 to 12 h, the OE plants also showed more staining than the WT, while the RNAi plants showed less staining than the WT. By contrast, in the OE plants, RNAi plants, and WT plants, there is no obvious DAB staining, in the presence of NaCl and DPI (Fig. 2D). These observations consolidated that the induction of DAB staining by NaCl treatment is due to the elevated levels of H2O2.
The levels of H2O2 in the roots of OE, RNAi, and WT plants under NaCl and the combined NaCl and DPI treatments were measured by using the H2O2 Assay Kit. Under normal growth conditions, H2O2 levels in the roots were approximately 1.52-fold higher in OE plants, but 0.96-fold lower in the RNAi plants compared to WT plants (Fig. 2E). Upon 12 h NaCl treatment, H2O2 levels in the roots rose by approximately 1.59-fold in the OE plants but diminished approximately 0.77-fold in the RNAi plants compared to WT plants (Fig. 2E). However, H2O2 levels were significantly lower in the OE, RNAi, and WT roots after 24 h of NaCl treatment than 12 h of NaCl treatment (Fig. 2E). By contrast, the extent of H2O2 increase was noticeably diminished in the OE, RNAi, and WT roots under NaCl treatment in the presence of DPI (Fig. 2E). Taken together, our results demonstrate GmNTL1-mediated salt tolerance is H2O2 dependent.
Overexpressing of GmNTL1 promotes soybean yield under salt stress
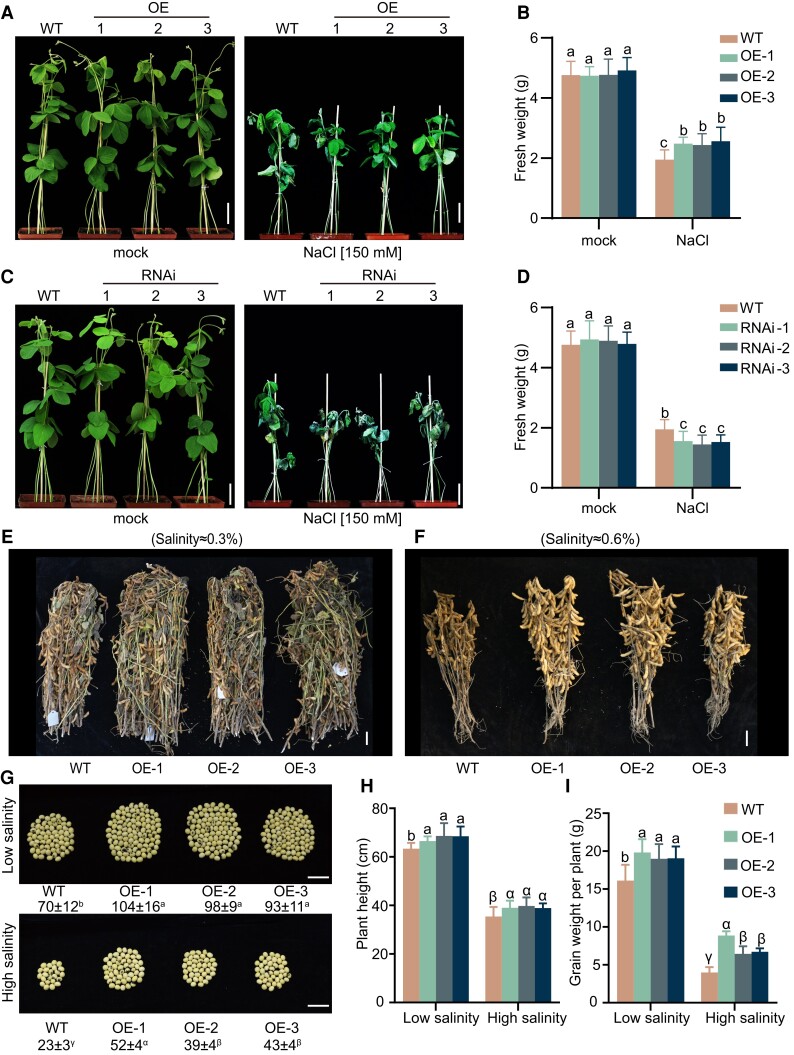
To confirm the role of GmNTL1 in salt tolerance in soybean, 14-d-old OE (lines OE-1, OE-2, and OE-3) and RNAi lines soybean seedlings were treated with 150 mm NaCl daily. To test the effect of overexpression and silencing of GmNTL1, we examined the fresh weight of plants. The fresh weight levels were approximately 1.28-fold, 1.25-fold, and 1.32-fold higher in the OE-1, OE-2, and OE-3 plants than in WT upon NaCl treatment, while the fresh weights of RNAi-1, RNAi-2, and RNAi-3 lines were 0.78-fold, 0.73-fold, and 0.78-fold of that WT plants upon NaCl treatment, demonstrating that the OE plants are more salt tolerant and the RNAi lines are more salt sensitive than the WT plants (Fig. 3, A–D).
Figure 3.
GmNTL1-enhanced salt tolerance. A–D) Phenotype of GmNTL1ΔC transgenic plants challenged with either 0 mm (mock) or 150 mm NaCl. Representative photographs of soil-grown WT, GmNTL1 OE (A), and GmNTL1 RNAi (C) plants under mock and NaCl treatment, taken 10 d after the onset of salt treatment. Scale bars, 5 cm. B, D) Quantification of fresh weight from the plants shown in (A, B). Five seedlings were examined for each biological repeat. Error bars denote SD (n = 15 from 3 biological experiments). Lowercase letters indicate significant differences between samples labeled as determined by 2-way ANOVA, P < 0.05. E, F) Representative photographs of main stems with pods from 15 plants each for GmNTL1-OE and WT grown in fields with 0.3% (E) or 0.6% (F) salinity. Scale bars, 5 cm. G) Number of seeds produced per plant for the indicated genotypes grown on low or high salinity. Scale bars, 2 cm. H, I) Plant height and grain weight per plant. Data are means ± SD (n = 18 to 22). Lowercase letters or Greek letters indicate significant differences between samples labeled as determined by 1-way ANOVA, P < 0.05. Please see Supplemental Data Set 4 for detailed statistical analyses.
According to reports, salt stress can affect physiological indicators such as the concentration of malonaldehyde (MDA), a marker of lipid peroxidation, and the activities of the antioxidant enzymes including superoxide dismutase (SOD) and peroxidase (POD) (Li et al. 2019a; Li et al. 2020; Fal et al. 2022). Here, we examined the concentration of MDA and the activities of the antioxidant enzymes including SOD and POD among the OE lines under salt stress. After a 10-d NaCl treatment, in the OE plants, MDA content dropped to 67.7%, 71.1%, and 73.2%, respectively. The activities of SOD and POD increased significantly in the OE plants compared with the WT plants (Supplemental Fig. S4A–C). The results suggest that GmNTL1 likely promotes salt tolerance through redox regulation.
Next, we performed a field trial experiment to assess salt tolerance of the transgenic lines within a natural environment context. Three GmNTL1-ΔC OE lines and WT plants were grown under low-salinity (salinity concentration 0.2% to 0.3%) or high-salinity (salinity concentration 0.4% to 0.6%) field conditions. After harvest, the grain number and weight per plant, together with plant height, were measured. GmNTL1-ΔC OE plants were taller than WT (Fig. 3E, F, and H); the grain number per plant and the grain weight were higher in the GmNTL1-ΔC OE plants than WT under both low- and high-salinity conditions (Fig. 3G and I). Together, these data show that GmNTL1 participates in salt stress tolerance in soybean seedlings, with the overexpression of this gene increasing yield of plants growing under both low-salinity and high-salinity soils.
H2O2 induces the Oxi-PTM of GmNTL1 at Cys-247
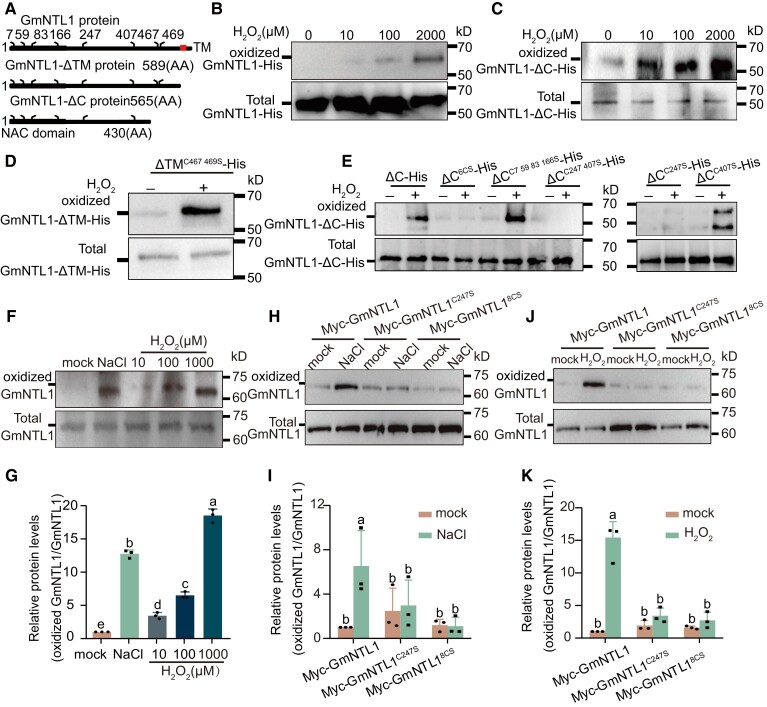
Since H2O2 is required for the relocation of GmNTL1 and its accumulation is essential for GmNTL1-mediated salt stress tolerance, we next examined whether H2O2 could directly modify GmNTL1 via Oxi-PTMs on its 8 Cys residues (Cys-7, Cys-59, Cys-83, Cys-166, Cys-247, Cys-407, Cys-467, and Cys-469) (Fig. 4A). To test this hypothesis, we purified recombinant GmNTL1-His and GmNTL1-ΔC-His proteins and performed an adapted biotin switch assay (Tian et al. 2018). Here, the free -SHs were primarily alkylated with N-Ethylmaleimide (NEM); then, the reversible Cys Oxi-PTMs were reduced by dithiothreitol (DTT) to -SHs and the newly generated -SHs were sequentially labeled by a biotin-conjugated iodoacetamide (BIAM) for enrichment. Enriched proteins were finally detected by anti-His immunoblotting.
Figure 4.
GmNTL1 is oxidized under H2O2 treatment. A) Schematic diagram of GmNTL1, GmNTL1ΔTM (TM), and GmNTL1ΔC (ΔC, lacking the C-terminal region). The highly conserved NAC domain is in the N-terminal region (black boxes). Red box, α-helical TM. The numbers indicate the position of cysteine residues. B) In vitro analysis of the oxidative modification of GmNTL1 by the biotin switch assay. Recombinant GmNTL1-His was pretreated with different H2O2 concentrations. Biotin-conjugated iodoacetamide (BIAM)-tagged proteins in the samples were captured with streptavidin beads and detected by immunoblot using an anti-His antibody. Data shown are the results of 3 biological experiments. C) In vitro analysis of the oxidative modification of GmNTL1ΔC by the biotin switch assay. Data shown are the results of 2 biological experiments. D, E) In vitro analysis of the oxidation of GmNTL1ΔTM and GmNTL1ΔC variants after the recombinant proteins were treated with water (0 µM) or 2 mm H2O2 for 30 min. Data shown are the results of 3 biological experiments. F, G) Immunodetection of oxidized GmNTL1 in roots from 5-d-old wild-type seedlings treated with 150 mm NaCl for 6 h, 0, 10, 100, or 1000 µM H2O2 for 12 h using the anti-GmNTL1 antibody. Error bars denote SD (n = 3 from 3 biological experiments). The intensity of each band was measured using ImageJ, and relative protein levels were normalized against those in untreated controls, which were set to 1. Lowercase letters indicate significant differences between labeled samples as determined by 1-way ANOVA, P < 0.05. H–K) In vivo analysis of the oxidative modification of GmNTL1, GmNTL18CS (with all 8 Cys residues mutated to Ser), and GmNTL1C247S (with the Cys-247 residue mutated to Ser) proteins in plants by the biotin-switch assay. Total proteins from MYC-GmNTL1, MYC-GmNTL18CS, and MYC-GmNTL1C247S transgenic soybean hairy roots treated with or without 150 mm NaCl (H) for 6 and 1 mm H2O2 (J) for 12 h were sequentially treated with N-Ethylmaleimide (NEM), dithiothreitol (DTT), and BIAM and then analyzed for biotin label. The intensity of each band was measured using ImageJ, and relative protein levels were normalized against those in untreated controls, which were set to 1. Error bars denote SD (n = 3 from 3 biological experiments). Lowercase letters indicate significant differences between labeled samples determined by 2-way ANOVA, P < 0.05 in (I) and (K). Please see Supplemental Data Set 4 for detailed statistical analyses.
With this method, we were able to examine the level of reversible Cys Oxi-PTMs that can be considered as the Cys oxidation level. We observed a dose-dependent increase of Cys oxidation when GmNTL1-His was treated for 30 min with H2O2 (10, 100 µM, and 2 mm; Fig. 4B). Moreover, a similar effect of H2O2-induced Cys oxidation was observed for GmNTL1-ΔC-His not only by using the modified biotin-switch assay (Fig. 4C) but also via the standard BIAM labeling assay, which labels the total free -SHs, showing a decreased level of reduced Cys when treated with increasing concentration of H2O2 (Supplemental Fig. S5). These results indicated that H2O2 induces Cys Oxi-PTMs in GmNTL1.
To confirm the occurrence of oxidation-sensitive Cys sites, we examined Cys oxidation by biotin switch assay on different Cys mutants of GmNTL1. We first mutated Cys-467 and Cys-469 of GmNTL1-ΔTM, which are located near the TM domain for analysis, and found that replacing Cys-467 and Cys-469 with serine (Ser) did not affect the H2O2-induced Cys oxidation of GmNTL1 (Fig. 4D). We then examined the Cys oxidation level for the other 6 Cys residues (Cys-7, Cys-59, Cys-83, Cys-166 Cys-247, and Cys 407). We found that simultaneously mutating the 6 Cys to Ser of GmNTL1-ΔC-His (designated GmNTL1-ΔC6CS) abolished the H2O2-induced Cys oxidation, as well as the mutant of both Cys-247 and Cys-407 (GmNTL1-ΔCC247 407S), while the simultaneous mutation of the first 4 Cys residues (GmNTL1-ΔCC7, 59, 83,166 S) did not (Fig. 4E). We further determined the Cys oxidation level of the single variants GmNTL1-ΔCC247S and GmNTL1-ΔCC407S, revealing that only GmNTL1-ΔCC407S was able to be oxidized by H2O2 but not GmNTL1-ΔCC247S (Fig. 4E), indicating that the Cys-247 residue is essential for H2O2-induced oxidation on GmNTL1.
To determine whether GmNTL1 is modified via Oxi-PTMs in vivo, we isolated total proteins from NaCl- and H2O2-treated and untreated Williams 82 WT seedlings, respectively, followed by the detection of oxidized GmNTL1 with the anti-GmNTL1 antibody via the biotin-switch method. Seedlings treated with 150 mm NaCl or 10, 100, or 1,000 μM H2O2 accumulated more oxidized GmNTL1 than the untreated control (Fig. 4F and G; Supplemental Fig. S3D). Total protein was also extracted from the MYC-GmNTL1 transgenic soybean hairy roots pretreated with or without 150 mm NaCl and 1000 μM H2O2. The biotin switch assay showed that NaCl and H2O2 induced the oxidation of GmNTL1 (Fig. 4H–K).
To further investigate whether the Cys-247 site of GmNTL1 was oxidized by H2O2 in vivo, MYC-GmNTL1 (GmNTL1 fused to MYC driven by the CaMV 35S), MYC-GmNTL18CS (with all 8 Cys residues mutated to Ser and fused to MYC driven by the CaMV 35S), and MYC-GmNTL1C247S (with Cys-247 residue mutated to Ser and fused to MYC driven by the CaMV 35S) transgenic soybean hairy roots were obtained. Further experiments showed that MYC-GmNTL18CS and MYC-GmNTL1C247S decreased oxidative modification, indicating that the H2O2-induced oxidation occurs mainly on Cys-247 of GmNTL1 (Fig. 4H–K).
Oxidation of GmNTL1 at Cys-247 is required for its salt-induced relocation to the nucleus and important for salt tolerance
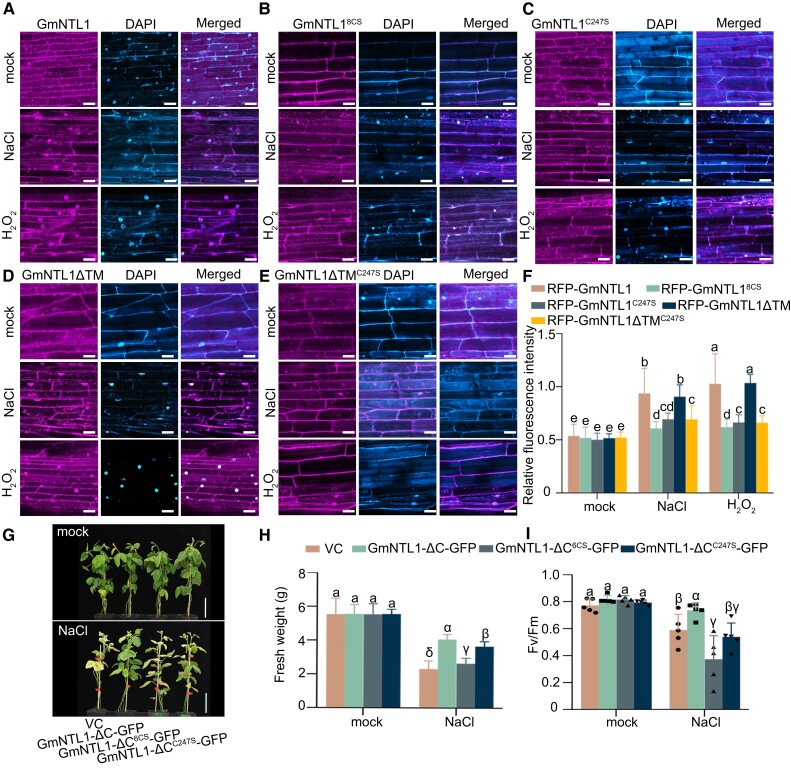
To investigate whether NaCl- and H2O2-triggered relocation of GmNTL1 was mediated via Oxi-PTMs of Cys residues, we expressed a construct encoding RFP-GmNTL18CS (with all 8 Cys residues mutated to Ser and fused to RFP) and RFP-GmNTL1C247S (with Cys-247 residue mutated to Ser and fused to RFP) in soybean hairy roots. Under untreated conditions, both RFP-GmNTL18CS and RFP-GmNTL1C247S were localized to the ER membrane (Fig. 5B and C). Under 150 mm NaCl treatments or 1 mm H2O2 for 6 h, the nucleocytoplasmic fluorescence intensity ratios of RFP-GmNTL18CS decreased to 0.65-fold and 0.60-fold, respectively, compared with RFP-GmNTL1 (Fig. 5A, B, and F). The nucleocytoplasmic fluorescence intensity ratios of RFP-GmNTL1C247S decreased 0.74-fold and 0.65-fold, respectively, compared with RFP-GmNTL1 (Fig. 5A, C, and F; Supplemental Fig. S2B, C, E, and F), indicating that the oxidation of Cys-247 in GmNTL1 is essential for its nuclear import during plant responses to salt stress.
Figure 5.
Cys-247 oxidation of GmNTL1 by H2O2 is critical for plant salt response. A–E) Subcellular localization of GmNTL1 (A), GmNTL18CS (B), GmNTL1C247S (C), GmNTL1ΔTM (TM, transmembrane) (D), and GmNTL1ΔTM C247S (E) in soybean hairy roots treated with 150 mm NaCl and 1 mm H2O2 for 6 h. Scale bars, 20 μm. DAPI staining used to show the nucleus. F) Quantification of the ratio between nuclear and cytoplasmic RFP fluorescent intensities in (A–E). At least 10 seedling roots were examined for each biological repeat. Data shown are the results of 3 biological experiments. Error bars denote SD (n = 100 cells from 10 images). Lowercase letters indicate significant differences between samples, as determined by 2-way ANOVA with P < 0.05. Please see Supplemental Data Set 4 for detailed statistical analyses. G–I) Phenotypes (G), fresh weights (H), and photosystem II photochemical potential (Fv/Fm), n = 5. I) of vector control (VC), GmNTL1-ΔC-GFP (ΔC, lacking the C-terminal region), GmNTL1-ΔC6CS-GFP, and GmNTL1-ΔCC247S-GFP transgenic hairy root soybean plants under normal and NaCl conditions. In (G), scale bars, 10 cm. In (H), 5 seedlings roots were examined for each biological repeat. Error bars denote SD (n = 15 from 3 biological experiments). In (I), error bars denote SD (n = 5 from 1 biological experiment). Lowercase letters and Greek letters indicate significant differences between samples, as determined by 1-way ANOVA with P < 0.05. Please see Supplemental Data Set 4 for detailed statistical analyses.
To further investigate whether the GmNTL1-TM domain determines GmNTL1 translocation, we expressed a construct encoding RFP-GmNTL1ΔTM (lacking only the TM and fused to RFP) and RFP-GmNTL1ΔTMC247S (lacking the TM and Cys-247 residue mutated to Ser and fused to RFP) in soybean hairy roots. Under 150 mm NaCl treatments or 1 mm H2O2 for 6 h, RFP-GmNTL1ΔTM exhibited similar nucleocytoplasmic fluorescence intensity ratio levels with RFP-GmNTL1, but RFP-GmNTL1ΔTMC247S showed lower levels compared with RFP-GmNTL1ΔTM (Fig. 5D, E, and F). The results indicated that Cys247 oxidation rather than the GmNTL1-TM domain determines GmNTL1 translocation.
In addition, we obtained GmNTL1-ΔC-GFP, GmNTL1-ΔC6CS-GFP, GmNTL1-ΔCC247S-GFP, and empty vector control (VC) transgenic soybean hairy root plants. Once the emerged hairy roots can support the plants, the main roots are removed, and we performed a growth assay with these plants subjected to a 100 mm NaCl treatment for 14 d. We examined the fresh weight and photosynthetic parameters affected by salt stress. The fresh weights of VC, GmNTL1-ΔC-GFP, GmNTL1-ΔC6CS-GFP, and GmNTL1-ΔCC247S-GFP plants showed no obvious differences under control conditions (Fig. 5G, H, and I).
Under salt stress, the fresh weights were approximately 0.57-fold, 0.65-fold, and 0.89-fold lower in VC, GmNTL1-ΔC6CS-GFP, and GmNTL1-ΔCC247S-GFP than in GmNTL1-ΔC-GFP plants (Fig. 5G, H, and I). For photosynthesis, the photosystem II photochemical potential (Fv/Fm) was 0.90-fold, 0.76-fold, and 0.87-fold lower in VC, GmNTL1-ΔC6CS-GFP, and GmNTL1-ΔCC247S-GFP than in GmNTL1-ΔC-GFP (Fig. 5I), demonstrating that the salt tolerance of GmNTL1-ΔC6CS-GFP and GmNTL1-ΔCC247S-GFP is lower than that of GmNTL1-ΔC-GFP. Taken together, these results demonstrate that oxidation of GmNTL1 at Cys-247 is required for its salt-induced relocation to the nucleus and important for improving salt stress tolerance.
Transcriptome analysis of Pro35S:GmNTL1 transgenic soybeans
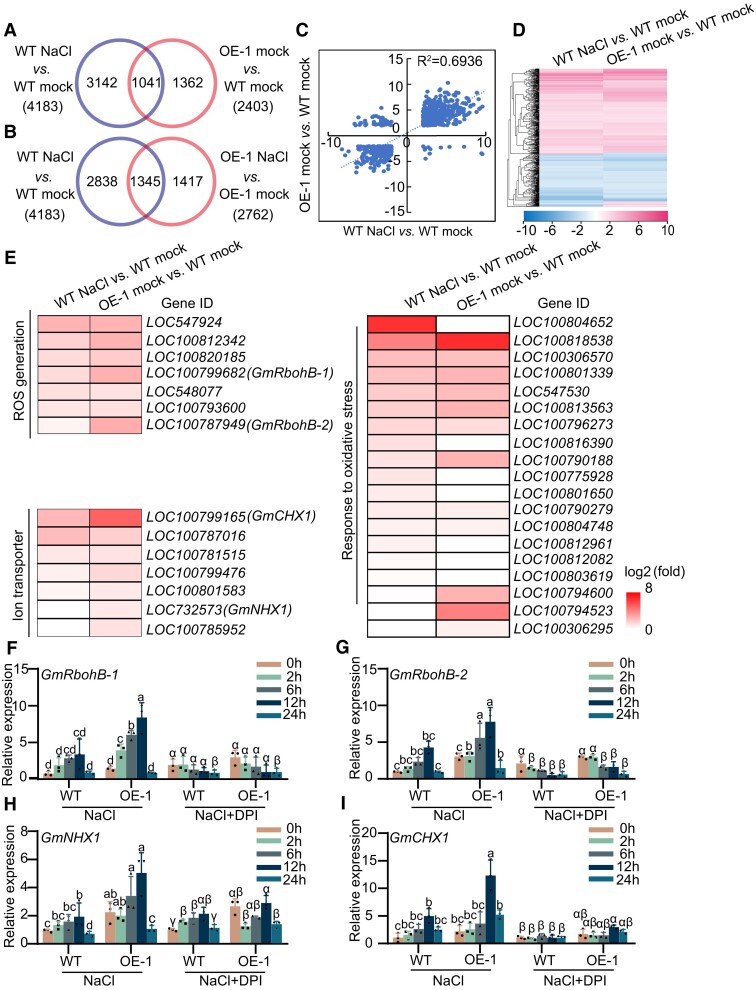
To understand the molecular consequences of salt stress–mediated nuclear translocation of GmNTL1 (Supplemental Fig. S3A–D), we performed a transcriptome sequencing analysis (RNA-seq) on 14-d-old transgenic GmNTL1-ΔC OE and WT roots subjected to 150 mm NaCl for 0 and 12 h. In the NaCl treatment, 4,183 genes were differentially regulated (fold change ≥ 2, and q-value ≤ 0.05) relative to mock treatment in WT plants; of these, 2,317 were upregulated and 1,866 were downregulated (Fig. 6A; Supplemental Fig. S6A). However, NaCl treatment did not cause a remarkable transcriptomic change in GmNTL1-ΔC OE plants relative to WT plants, with only 2,762 genes were differentially regulated (Fig. 6B; Supplemental Fig. S6A and Supplemental Data Set 1). This suggested that OE plants are less sensitive to NaCl treatment than WT plants.
Figure 6.
RNA-seq analysis of GmNTL1 OE-1 transgenic soybean. A, B) Venn diagrams showing the extent of overlap for DEGs between GmNTL1 OE-1 transgenic soybeans and WT under control (mock) and NaCl-treated seedlings. C) Correlation of the overlapping genes from (A) differentially expressed in NaCl-treated WT versus mock-treated WT and mock-treated GmNTL1 OE-1 versus mock-treated WT. D) Hierarchical cluster analysis of the overlapping genes from (A) differentially expressed in NaCl-treated WT versus mock-treated WT and mock-treated GmNTL1 OE-1 versus mock-treated WT. The numerical values in the gradient bar represent log2-fold change relative to the control sample. E) Heatmap representation of expression levels for ROS generation, ion transport-related genes and responding to oxidative stress genes in NaCl-treated WT versus mock-treated WT and mock-treated GmNTL1 OE-1 versus mock-treated WT. Values are log2 (fold change). F–I) Relative transcript levels of GmRbohB-1 (F), GmRbohB-2 (G), GmNHX1 (H), and GmCHX1 (I) in the roots of 10-d-old seedlings of GmNTL1-OE (OE-1) or WT exposed to either no NaCl (0 h), 150 mm NaCl, or 150 mm NaCl and 100 μM DPI for 2, 6, 12, or 24 h. Error bars denote SD (n = 3 from 3 biological experiments). Lowercase letters and Greek letters indicate significant differences between samples, as determined by 2-way ANOVA with P < 0.05. Please see Supplemental Data Set 4 for detailed statistical analyses.
Between mock-treated GmNTL1-ΔC OE plants and WT plants, we identified 2,403 differentially expressed genes (DEGs). Of these, we detected 1,041 DEGs in both the WT-mock versus WT-NaCl and OE-mock versus WT-mock comparisons (Fig. 6A). To further dissect the relationship of DEGs caused by GmNTL1 overexpression and by NaCl treatment, the expression pattern of the 1,041 DEGs was analyzed by using linear regression and hierarchical clustering methods. This showed that the correlation was high and the expression pattern was the same between WT-mock versus WT-NaCl and OE-mock versus WT-mock (Fig. 6C and D). These results suggest that some of the transcriptional changes caused by salt stress are mediated by GmNTL1.
A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed that the DEGs in the mock-treated GmNTL1-ΔC OE-1 and WT plants indicate the significance level in the categories “plant hormone signal transduction,” “phenylpropanoid biosynthesis,” “glutathione metabolism,” “autophagy,” and “isoflavonoid biosynthesis” (Supplemental Fig. S6B). In the NaCl-treated GmNTL1-ΔC OE-1 and WT plants, KEGG pathway enrichment analysis showed that DEGs were further enriched in the categories “plant hormone signal transduction,” “phenylpropanoid biosynthesis,” and “isoflavonoid biosynthesis” (Supplemental Fig. S6C). These results suggest that GmNTL1 mainly participates in the biological pathways of secondary metabolites. To investigate which biological pathways are involved in NaCl OE-1 versus NaCl WT cluster, we performed Gene Ontology (GO) analysis. The cluster was enriched in biological processes involved in response to chitin, regulation of defense response, and flavonoid biosynthetic process (Supplemental Data Set 2). These data suggest that GmNTL1 regulates salt tolerance by modulating the expression of a set of oxidative stress–related genes.
Notably, the heatmaps showed that 7 genes in ROS generation, 7 genes in the ion transport system, and 19 genes responding to oxidative stress were upregulated both in WT-mock versus WT-NaCl and OE-mock versus WT-mock (q-value ≤ 0.05) (Fig. 6E). Among these genes, we selected the GmRbohB-1 and GmRbohB-2 oxidative stress–related genes, GmNHX1 and GmCHX1 from ion transport system genes, which are known to positively regulate plant salt tolerance, for further analysis (Li et al. 2006; Guan et al. 2014; Yang et al. 2017; Li et al. 2019a).
First, to confirm the RNA-seq results, we performed RT-qPCR for some DEGs by using GmNTL1-ΔC OE and WT roots: GmRbohB-1, GmRbohB-2, GmNHX1, and GmCHX1. This analysis showed that these 4 genes are all upregulated by salt stress in the GmNTL1-ΔC OE plants relative to mock controls, which was consistent with the fold changes seen in the RNA-seq analysis (Fig. 6F–I). The RNAi plants displayed lower expression levels for these 4 genes by RT-qPCR (Supplemental Fig. S7). Additionally, these genes were also induced by the 12 h NaCl treatment, although this salinity-mediated induction was suppressed by the addition of DPI in both WT and GmNTL1-ΔC OE plants (Fig. 6F–I), suggesting that GmNTL1 transcriptionally regulates ion transport and ROS signaling pathways under NaCl treatment.
Oxidation of GmNTL1 at Cys-247 promotes GmRbohBs transcription to increase H2O2 content
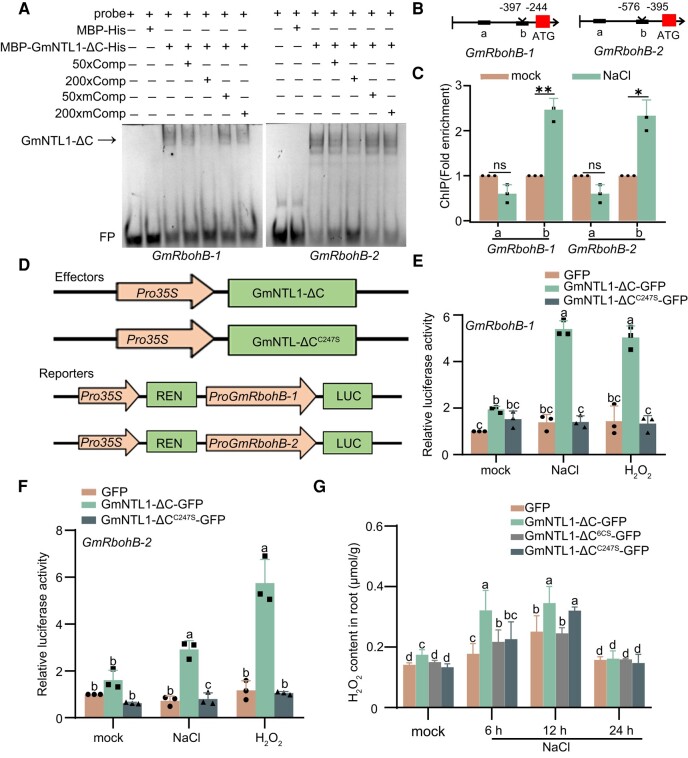
In order to understand how oxidation of GmNTL1 affects its downstream signaling, we assessed whether GmNTL1-ΔC can bind to the GmRbohB promoters to activate their transcription using electrophoretic mobility shift assay (EMSA). Recombinant MBP-GmNTL1-ΔC-His protein showed binding activity to the GmRbohB-1 and GmRbohB-2 DNA probes (Fig. 7A). The binding was efficiently competed off by unlabeled wild-type probes but not by unlabeled mutated probe (Fig. 7A). Next, in a chromatin immunoprecipitation (ChIP) assay using a specific anti-GmNTL1 antibody, we explored whether NaCl treatments affect the binding affinity of GmNTL1. Two-wk-old Ludou 11 seedlings were treated with 150 mm NaCl (or H2O as a mock-treated control) for 12 h. DNA fragments covering the possible binding motifs in GmRbohB promoters were specifically enriched (Fig. 7B and C).
Figure 7.
GmNTL1 modulates hydrogen peroxide (H2O2) contents by directly binding to the GmRbohB-1 and GmRbohB-2 promoters. A) EMSA showing the binding of recombinant GmNTL1-ΔC (ΔC, lacking the C-terminal region) to its target gene promoters. Lanes from left to right: (i) free probe (digoxin [DIG]-labeled probe with no protein added), (ii) labeled probe with maltose binding protein-His (MBP-His) protein as negative control, (iii) labeled probe with MBP-GmNTL1ΔC-His protein, (iv) and (v) MBP-GmNTL1ΔC-His protein binding to the labeled probe was competed with 50× or 200× unlabeled WT probes, (vi) and (vii) binding was competed with unlabeled mutant probe sequences. Comp, the competitor probe; mComp, the mutant competitor probe; FP, free probe. Data shown are the results of 2 biological experiments. B) Schematic diagram of the GmRbohB-1 and GmRbohB-2 promoters. Black boxes indicate “a” and “b” fragments. Red blocks indicate the ATG translation initiation site. Roman numbers indicate the different potential binding fragments in the promoters. C) ChIP-qPCR analysis of GmNTL1 binding to the GmRbohB-1 and GmRbohB-2 promoters in vivo. One-wk-old Ludou11 seedlings after a 12 h NaCl treatment (150 mm) or mock treatment were used, and IP was performed with the specific anti-GmNTL1 antibody. Data are means ± SD of 3 independent experiments. Asterisks indicate statistically significant differences between samples (Student's t-test, *P < 0.05). ns, not significant. D) Schematic diagram of the vectors used for the transient expression assay. 1.5-kb fragment from the GmRbohB-1 or GmRbohB-2 promoter was cloned upstream of the LUC reporter gene. Arrows indicate promoters and boxes indicate coding sequences. E, F) Dual-luciferase reporter assays examining the effects of Pro35S:GmNTL1-ΔC or Pro35S:GmNTL1-ΔCC247S on GmRbohB-1 or GmRbohB-2 transcription under NaCl or H2O2 treatment. Error bars denote SD (n = 3 from 3 biological experiments). Lowercase letters indicate significant differences between samples, as determined by 2-way ANOVA with P < 0.05. Please see Supplemental Data Set 4 for detailed statistical analyses. G) H2O2 contents of the roots of GFP, GmNTL1-ΔC-GFP, GmNTL1-ΔC6CS-GFP, and GmNTL1-ΔCC247S-GFP overexpression transgenic hairy root soybean plants. Error bars denote SD (n = 9 from 3 biological experiments). Lowercase letters indicate significant differences between samples, as determined by 2-way ANOVA with P < 0.05. Please see Supplemental Data Set 4 for detailed statistical analyses.
We further evaluated the effect of GmNTL1 on the expression of GmRbohBs in vivo by dual-luciferase (LUC) reporter gene assays, in which the effector construct Pro35S:GmNTL1-ΔC and the reporter constructs ProGmRbohB-1:LUC and Pro GmRbohB-2:LUC were transiently transfected in Arabidopsis protoplasts (Fig. 7D). The expression of GmNTL1-ΔC, but not that of GFP from the Pro35S: GFP vector, increased LUC activity (Fig. 7E and F). Furthermore, a 150 mm NaCl treatment for 30 min or a 1 mm H2O2 treatment for 30 min increased LUC activity derived from the GmRbohB promoters (Fig. 7E and F). These results show that transcriptional activity of GmNTL1 for GmRbohBs was increased under NaCl and H2O2 treatments. To examine the role of oxidation on Cys-247 residue, we compared the effects of GmNTL1-ΔC and GmNTL1-ΔCC247S expression using the ProGmRbohB-1:LUC and ProGmRbohB-2:LUC reporters (Fig. 7E and F). The C247S mutation markedly inhibited the increase in LUC activity when GmNTL1 was expressed under both NaCl and H2O2 treatments, revealing that the H2O2-mediated GmNTL1 oxidation at Cys-247 plays a vital role in GmNTL1-mediated GmRbohBs transcriptional activation in response to NaCl and H2O2.
To investigate whether the mutation in the Cys-247 site affects H2O2 homeostasis, we measured the H2O2 content in GmNTL1-ΔC-GFP, GmNTL1-ΔC6CS-GFP, and GmNTL1-ΔCC247S-GFP overexpression transgenic soybean hairy root plants. Under 6 h NaCl treatment, the H2O2 content in WT, GmNTL1-ΔC6CS-GFP, and GmNTL1-ΔCC247S-GFP were 0.56-fold, 0.68-fold, and 0.70-fold lower than in GmNTL1-ΔC-GFP (Fig. 7G), revealing that oxidation on Cys-247 residue is required for H2O2 accumulation in response to salt stress.
Taken together, these data show that GmNTL1 binds to GmRbohBs promoters and that salt and H2O2 promote GmNTL1-mediated transcriptional activation, affecting the production of H2O2.
H2O2-mediated GmNTL1 oxidation promotes the expression of GmNHX1 and GmCHX1 to improve soybean salt tolerance
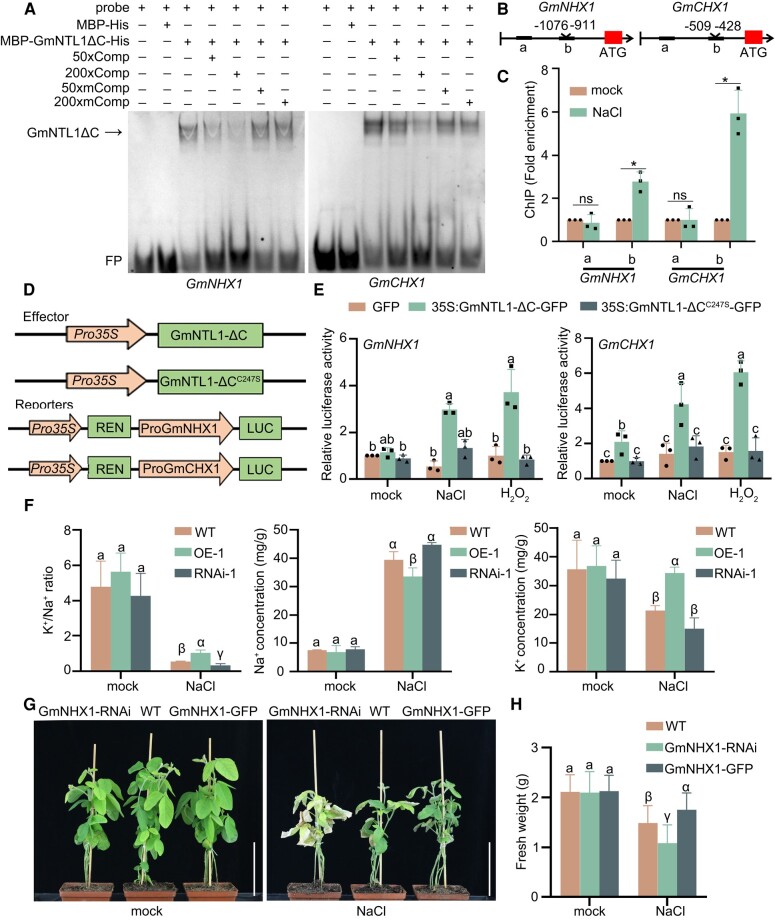
As GmNHX1 and GmCHX1 were upregulated in GmNTL1-ΔC OE seedlings (Fig. 6H and I), we speculated that these 2 genes might be direct targets of GmNTL1. We performed EMSA, which confirmed that GmNTL1 binds to the GmNHX1 and GmCHX1 promoters in vitro (Fig. 8A). Moreover, a ChIP-qPCR for GmNTL1 showed that 1 fragment each of the GmNHX1 and GmCHX1 promoters is enriched in the precipitated chromatin under salt treatment (Fig. 8B and C). To extend this result in vivo, we performed a LUC activity assay in Arabidopsis protoplasts, using Pro35S:GmNTL1-ΔC as the effector construct and ProGmNHX1:LUC or ProGmCHX1:LUC as the reporter. We established that GmNTL1-ΔC expression stimulates LUC activity derived from both reporters (Fig. 8D and E); however, NaCl- or H2O2-treated Arabidopsis protoplasts expressing Pro35S:GmNTL1C247S-ΔC displayed lower LUC activity levels than those expressing Pro35S:GmNTL1-ΔC, indicating that the H2O2-mediated GmNTL1 oxidation at Cys-247 enhances its binding to the GmNHX1 and GmCHX1 promoters (Fig. 8D and E).
Figure 8.
GmNTL1 induces GmNHX1 and GmCHX1 transcription by binding to their promoters. A) EMSA experiments showing the binding of recombinant GmNTL1ΔC (ΔC, lacking the C-terminal region) to probes for the GmNHX1 and GmCHX1 promoters. Lanes from left to right: (i) free probe (labeled probe with no protein added), (ii) labeled probe with maltose binding protein-His (MBP-His) protein as negative control, (iii) labeled probe with MBP-GmNTL1ΔC-His protein, (iv) and (v) MBP-GmNTL1ΔC-His protein binding to the labeled probe was competed with 50× or 200× unlabeled wild-type probes, (vi) and (vii) binding was competed with unlabeled mutant probe sequences. Comp, the competitor probe; mComp, the mutant competitor probe; FP, free probe. B) Schematic diagram of the GmNHX1 and GmCHX1 promoters. C) ChIP-qPCR assay showing the binding of GmNTL1 to the GmNHX1 and GmCHX1 promoters in vivo. One-wk-old Ludou11 seedlings treated with 150 mm NaCl for 12 h or subjected to mock treatment were used. Error bars denote SD (n = 3 from 3 biological experiments). Asterisks indicate statistically significant differences between samples (Student's t-test, *P < 0.05). ns, not significant. D) Schematic diagrams of constructs used for the transient expression assay. A 1.5-kb fragment of the GmNHX1 or GmCHX1 promoter was cloned upstream of the LUC reporter gene. Arrows indicate promoters; boxes indicate coding sequences. E) Dual-LUC activity assay showing the effects of GmNTL1ΔC or GmNTL1ΔCC247S on GmNHX1 and GmCHX1 transcription under NaCl or H2O2 treatment. Relative luciferase activity was calculated as firefly luciferase activity normalized to Renilla luciferase activity. Error bars denote SD (n = 3 from 3 biological experiments). Lowercase letters indicate significant differences between samples, as determined by 2-way ANOVA with P < 0.05. Please see Supplemental Data Set 4 for detailed statistical analyses. F) Determination of K+ and Na+ contents in plants. GmNTL1ΔC OE-1, GmNTL1 RNAi-1, and WT plants were treated with no NaCl or 150 mm NaCl for 7 d. Roots were collected to measure Na+ and K+ contents using Atomic Absorption Spectroscopy. Error bars denote SD (n = 9 from 3 biological experiments). G) Five-wk-old transgenic hairy root plants (GmNHX1-RNAi and GmNHX1-OE) and WT plants irrigated with 100 mm NaCl 3 times a week. The photographs were taken 1 wk after the onset of salt treatment. Scale bars, 10 cm. H) Fresh weight of the transgenic hairy root plants (GmNHX1-RNAi and GmNHX1-OE) and WT plants shown in (G). Error bars denote SD (n = 20 from 3 biological experiments). Lowercase letters and Greek letters indicate significant differences between samples, as determined by 1-way ANOVA with P < 0.05 in (F) and (H). Please see Supplemental Data Set 4 for detailed statistical analyses.
To clarify how GmNTL1 mediates the tolerance to salt stress through GmNHX1 and GmCHX1, we measured the K+ and Na+ contents in the roots of WT, GmNTL1-ΔC OE, and RNAi seedlings using Atomic Absorption Spectroscopy. Under control conditions, the K+ and Na+ contents of OE and WT or RNAi seedlings were not significantly different. When seedlings were subjected to a 150 mm NaCl treatment for 7 d, the Na+ contents in GmNTL1-ΔC OE roots and GmNTL1 RNAi roots were 0.85-fold and 1.13-fold than in WT roots. Under the same treatment, the K+ content in GmNTL1-ΔC OE roots and GmNTL1 RNAi roots were 1.61-fold and 0.70-fold than in WT roots, leading to lower Na+ concentration, higher K+ concentration, and a higher K+/Na+ ratio in the OE plants and higher Na+ concentration, lower K+ concentration, and a lower K+/Na+ ratio in the RNAi plants (Fig. 8F).
To further explore the possibility that GmNHX1 is involved in the GmNTL1-mediated salt response, the positive hairy roots of GmNHX1-GFP and GmNHX1 RNAi transgenic soybean plants were screened, and then, the original roots were removed. Then, we performed a growth assay with GmNHX1-GFP and GmNHX1 RNAi transgenic soybean hairy root plants subjected to a 100 mm NaCl treatment. The plants of all genotypes showed no obvious differences under normal growth conditions (Fig. 8G). When irrigated with 100 mm NaCl solution 3 times over the course of a week, the fresh weight of GmNHX1-GFP and GmNHX1 RNAi whole plants was 1.24-fold and 0.86-fold than that of WT, respectively (Fig. 8H), indicating that GmNHX1 enhances the salt tolerance of soybean hairy roots. Taken together, these results demonstrate that the oxidation of GmNTL1 upon salt stress promotes the expression of ion stress responsive genes and ion homeostasis, which is associated with improved tolerance to salt stress.
Discussion
Many transcription factors have been shown to participate in salt tolerance (Dong et al. 2017; Baillo et al. 2019; Debbarma et al. 2019; Zhang et al. 2020; Djemal and Khoudi 2021), including the membrane-bound NAC transcription factor family members (Baillo et al. 2019; Li et al. 2021). For example, NAC WITH TRANSMEMBRANE MOTIF 2 (NTM2), NAC WITH TRANSMEMBRANE MOTIF-LIKE 4 (NTL4), NAC WITH TRANSMEMBRANE MOTIF-LIKE 6 (NTL6), and NAC WITH TRANSMEMBRANE MOTIF-LIKE 8 (NTL8) mediate salt response via different mechanisms (Kim et al. 2007; Seo and Park 2010; Park et al. 2011). Moreover, overexpression of several NAC transcription factors in soybean, such as GmNAC06, GmNAC11, GmNAC20, GmNAC085, and GmNAC109, increases salt tolerance (Hao et al. 2011; Yang et al. 2019; Hoang et al. 2021; Li et al. 2021; Yarra and Wei 2021). Our previous study also found that a soybean (Glycine max) salinity–induced NAM/ATAF1/2/CUC2 (NAC) transcription factor encoded by SALT INDUCED NAC1 (GmSIN1) directly upregulated (9-cis-epoxycarotenoid dioxygenase coding genes) GmNCED3s and GmRbohBs to increase ABA and ROS contents and thus enhance soybean salt tolerance (Li et al. 2019a). In this study, we found that H2O2-induced oxidation of GmNTL1 at Cys-247 was essential for its function. In addition, the oxidation of GmNTL1 upregulates the expression of GmNHX1 and GmCHX1, which promotes the efflux of Na+ ions, thereby enhancing soybean salt tolerance (Figs. 6H and I and 8; Supplemental Fig. S8).
H2O2 is a key signaling molecule within cell response to different stresses (Golldack et al. 2014; Mittler et al. 2022). Apoplastic H2O2 activates a cytoplasmic membrane-localized leucine-rich repeat receptor kinase HYDROGEN-PEROXIDE-INDUCED Ca2+ INCREASES1 (HPCA1), triggering an influx of Ca2+ ions in Arabidopsis (Wu et al. 2020). Unlike many other signal transduction molecules that have specific receptors, such as phytohormones or peptides, H2O2 can directly oxidize multiple proteins through posttranslational modifications (PTMs) and thereby alter their conformation, location, and function (Leferink et al. 2009; De Smet et al. 2019; Huang et al. 2019; Nietzel et al. 2020), as shown for PROTEIN PHOSPHATASE 2A (PP2A), NONEXPRESSER OF PR GENES1 (NPR1), and BZR1 (Miao et al. 2006; Lindermayr et al. 2010; Yin et al. 2018).
Here, we demonstrate that the Cys-247 residue of GmNTL1 was oxidized by H2O2, promoting its release from the ER membrane to the nucleus to initiate and amplify H2O2 signaling under salt stress (Figs. 4 and 5A–F). Notably, GmNTL1-ΔCC247S-GFP was slightly more resistant to salt compared with VC (Fig. 5G). Considering the importance of translocation of GmNTL1 from endoplasmic reticulum to nucleus for salt tolerance, we speculated that this phenomenon might be related to the fact that the translocation of RFP-GmNTL1C247S was not completely inhibited under salt stress (Fig. 5A and C). Moreover, we also found that GFP-GmNTL1-ΔC was localized in both ER and nucleus under normal condition (Fig. 1E), suggesting that the oxidative modification is not the only mechanism to control the relocalization of GmNTL1. In addition, the oxidation of GmNTL1 also promoted plant salt tolerance by enhancing its transcriptional activity to the promoters of GmRbohBs, GmNHX1, and GmCHX1, thereby inducing the accumulation of H2O2 and the efflux of Na+ ions (Figs. 7 and 8).
To act as transcription factors, membrane-bound transcription factors (MTFs) must relocate to the nucleus. This process involves releasing, intracellular movements, and nuclear import events. MTFs are released by a proteolytic cleavage event or ubiquitin/proteasome-dependent processing or alternative splicing to omit the transmembrane domain (TMD) from the transcript (Hoppe et al. 2001; Seo et al. 2008; Li et al. 2012; Lu et al. 2012; Takahashi et al. 2012). The current observation indicates that PTMs are indispensable for the nuclear import of MTFs. For example, phosphorylation of NTL6 at Thr142 by the SNF1-RELATED PROTEIN KINASE 2.8 (SnRK2.8) kinase is crucial for its nuclear import (Kim et al. 2012); the plant NTL11, phosphorylation by the type II phosphatidylinositol 4-kinase, PI4Kγ5 is indispensable for its release and relocalization (Tang et al. 2016). In addition to phosphorylation, other PTMs, such as depalmitoylation of the Medicago truncatula transcription factor MfNACsa, are crucial for its nuclear import (Duan et al. 2017). A K-acetylation (in RINGLET 2, RLT2) and an N-glycosylation (in AT5G63280) sites also were found between the transcription factor family domain (TFFD) and the TMD domain (De Backer et al. 2022). In our study, we demonstrate that the oxidative PTM of GmNTL1 at cys-247 under salt stress enhances its translocation to the nucleus (Fig. 9). Taken together, controlled relocalization adds a level of complexity to MTF regulation.
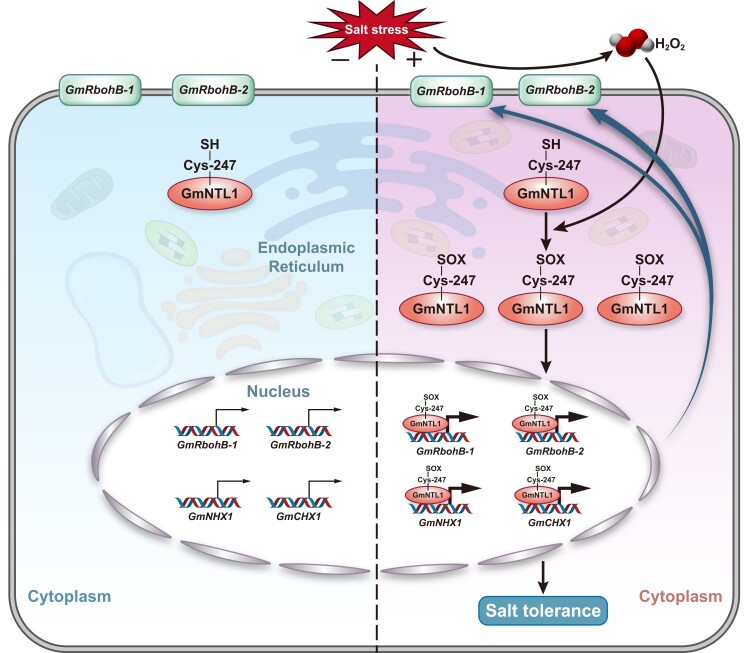
Figure 9.
A model for the role of GmNTL1 in H2O2-mediated induction of gene expression under salt stress. Under normal conditions, GmNTL1 localizes to the ER membrane. In response to salt stress, GmNTL1 translocates to the nucleus through its oxidation at the Cys-247 residue, where GmNTL1 directly activates the transcription of GmRbohB-1, GmRbohB-2, GmNHX1, and GmCHX1 in an H2O2-dependent manner and modulates H2O2 homeostasis and ion balance. H2O2, hydrogen peroxide; Cys, cysteine; -SH, sulfhydryl; -SOX, S-sulfenylation; S-sulfinylation, S-sulfonylation; ER, endoplasmic reticulum. The thin/thick arrows indicate the strength of target gene transcriptional activity. The blue arrow indicates a H2O2-GmNTL1-GmRbohBs-positive feed-forward regulatory loop. The black arrows indicate direct regulation.
Many studies have indicated that salt-tolerant plants can scavenge endogenous H2O2 more effectively using antioxidant enzymes to maintain H2O2 homeostasis in response to salt stress. For example, H2O2 is required for GmMYB84 to modulate primary root elongation in response to drought (Wang et al. 2017). A few proteins are known to participate in the H2O2 accumulation to amplify abiotic stress signals; for instance, Arabidopsis NTL4 promotes ROS production by binding directly to the promoters of AtrbohC and AtrbohE genes encoding ROS biosynthetic enzymes during drought-induced leaf senescence (Lee et al. 2012). Moreover, a positive feed-forward loop between transcription factor GmSIN1 and H2O2 was shown to initiate and amplify the salt stress signal (Li et al. 2019a). An appropriate amount of H2O2 therefore plays an important role in plant abiotic stress tolerance (Li et al. 2019a).
In this study, salt induces H2O2 accumulation, the oxidation, and the rapid nuclear import of GmNTL1 at the early stage of the stress response (Fig. 1F–J; Supplemental Fig. S2), in line with the increased expression of GmRbohBs (Fig. 6F and G). Moreover, the oxidation of GmNTL1 Cys-247 is required for the transcriptional activity of GmNTL1 (Figs. 7E and F and 8E). These findings reveal a H2O2-dependent feed-forward loop between GmNTL1 and GmRbohBs that enables the accumulation of H2O2, effectively improving transcriptional activity of GmNTL1, to amplify H2O2 signaling during the early stage of the salt stress response.
Excessive H2O2 accumulation may disturb intracellular ion homeostasis under saline conditions by activating a broad array of H2O2-sensitive ion channels (Liu et al. 2019). The inhibition of NADPH oxidase activity by DPI markedly decreased the PM H+-ATPase activities which enhances Na+ exclusion from the root (Niu et al. 2018). Here, we found that GmNTL1 directly promotes the expression of the ion transporter–related genes GmNHX1 and GmCHX1 in a salt induced H2O2-dependent manner (Fig. 6H and I). Furthermore, the oxidation of GmNTL1 activated its transcriptional factor activity and binding ability for GmNHX1 and GmCHX1 in salt and H2O2 treatments (Fig. 8C, D and E). Accordingly, the overexpression of GmNTL1 in soybean increased the K+/Na+ ratio in the cell and promoted salt tolerance and increased yield under salt stress (Fig. 3 and Fig. 8F). Our results reveal that the salt-induced oxidation of GmNTL1 enhances Na+ exclusion to improve salt tolerance.
Based on this study, we propose a working model for the function of GmNTL1 in the oxidation as a mechanism of salt tolerance (Fig. 9). GmNTL1 is anchored to the ER membrane in a dormant state under nonstressed conditions. Under salt stress, GmNTL1 is induced and translocated to the nucleus following the oxidative modification of its Cys-247 residue. The oxidation of GmNTL1 enhances its transcriptional binding to the GmRbohBs promoters and initiates a H2O2-GmNTL1-GmRbohBs-positive feed-forward regulatory loop. Moreover, the release of GmNTL1 promotes soybean salt tolerance by enhancing its transcriptional binding to the GmNHX1 and GmCHX1 promoters, which induces the efflux of Na+ ions (Fig. 9). Notably, overexpressing GmNTL1 could provide a viable strategy for improving salt tolerance in crops. Our study sheds some light on the mechanisms underlying salt stress response in plants and provides a target for molecular breeding in the future.
Materials and methods
Plant materials and growth conditions
The soybean (Glycine max) cultivar Williams 82 was used for determining the oxidation of GmNTL1 in vivo and the gene cloning. Cultivar Ludou 11 was used for phenotypic assays. The soybean plants were grown in a culture room at 25 °C under long-day conditions (16-h light/8-h dark) with 800 µmol m−2 s−1 illumination provided by a fluorescent lamp (Philips, T5-28W) and a relative humidity of 60%. The soybean plants were grown in pots (10 cm × 10 cm × 9 cm; length × width × depth) containing vermiculite and Pindstrup soil mix (Ryomgaard, Denmark) (1:3, w/w) for phenotyping assays, grown hydroponically in half-strength Hoagland solution for the RNA preparation assay, or grown in plastic root growth bags soaked with water for root growth phenotype assays. Arabidopsis (Arabidopsis thaliana) plants were grown in a culture room at 22 °C under long-day conditions (16-h light/8-h dark) with 120 µmol m−2 s−1 illumination (Philips, T5-28W).
Abiotic stress was applied to soybean seedlings through the addition of 100, 150, or 175 mm NaCl or 1 mm H2O2. For the root growth phenotyping assays, salt stress was imposed on 4-d-old soybean seedlings grown in plastic root growth bags soaked with water. Salt stress was applied to soybean seedlings through the addition of 175 mm NaCl to the hydroponic solution, and the cover was removed 7 d later. Three independent measurements consisted of 20 soybean seedlings.
Plasmid construction and plant transformation
To generate the transgenic overexpression lines in soybean, the GmNTL1-ΔC coding sequence was amplified and cloned into the Gateway pDONR221 vector (Thermo Fisher Scientific, Waltham, MA, USA) using a BP reaction. The construct was recombined into the pB7FWG2 binary vector (under the control of the 35S promoter) (PSB, Ghent University, Belgium) in an LR reaction.
The RNAi construct was designed to target the center of the GmNTL1 coding sequence. A 500-bp fragment was amplified and cloned in reverse orientation into the binary vector pB7GWIWG2(II) under the control of the 35S promoter (VIB-UGent Center for Plant Systems Biology) to knock down GmNTL1. The binary plasmids were transferred into Agrobacterium (Agrobacterium tumefaciens) strain GV3101 using the freeze-thaw method. Soybean plants were transformed following the protocol described previously (Cui et al. 2013). A DNA fragment from the 35S promoter region present in pB7FWG2 and pB7GWIWG2(II) was amplified with the primers 35S-F and 35S-R and labeled as the probe. The primer sequences used for the above studies are listed in Supplemental Data Set 3.
Site-directed mutagenesis was used to construct gene mutants, including GmNTL1-ΔC6CS-GFP, and GmNTL1-ΔCC247S-GFP. The construction of the GmNTL1-ΔC6CS-GFP, GmNTL1-ΔCC247S-GFP, GmNHX1-GFP, and GmNHX1 RNAi vectors was consistent with that of the GmNTL1 vectors (PB7FWG2 and pB7GWIWG2(II)) described above. After sequence verification, the resulting plasmids, vector control (VC, with empty vector pB7FWG2), and pB7GWIWG2(II) were transformed into Agrobacterium rhizogenes K599 (WEIDI, China).
To generate prokaryotic expression vector (MBP-GmNTL1-ΔC-His), the GmNTL1-ΔC coding sequence was amplified and cloned into the Gateway pDONR221 vector recombined into the pMAL2CGW(N-MBP). To obtain the GmNTL1-His, GmNTL1-ΔC-His, GmNTL1-ΔC6CS-His, GmNTL1-ΔCC7,59,83,166S-His, GmNTL1-ΔTMC467,469S-His, GmNTL1-ΔCC247,407S-His, GmNTL1-ΔCC247S-His, and GmNTL1-ΔC407S-His plasmids, the coding sequences were cloned into the EcoRI and XhoI site of pET28a(+)/His. To obtain MYC-GmNTL1, MYC-GmNTL18CS, and MYC-GmNTL1C247S plasmids, GmNTL1 and mutated coding sequences were cloned in-frame and downstream of the 4xMYC Tags and then inserted into the PB2GW7 binary vector (under the control of the 35S promoter) (PSB, Ghent University, Belgium).
cDNA synthesis and RT-qPCR
TRIzol reagent (Thermo Fisher Scientific) was used to extract total RNA from the roots of 2-wk-old seedlings treated with 150 mm NaCl, 150 mm NaCl coupled with 100 μM DPI, or 10 mm H2O2 for 2, 6, 12, or 24 h. RNA quality was monitored based on its absorbance at 260 and 280 nm wavelengths, as measured using a spectrophotometer. The RT-qPCR was carried out in 96-well blocks using the MonScript RTIII All-in-One Mix with dsDNase (Monad, Shanghai, China) in a volume of 20 μL. The cDNA synthesis, RT-qPCR, and data analysis were conducted as previously described (Li et al. 2016). Gene-specific primers (sequences given in Supplemental Data Set 3) were designed using Beacon Designer v7.90 (http://www.premierbiosoft.com). Gm60S was used as the internal reference gene for profiling across the plant (Le et al. 2012). Three biological replicates from independent experiments were included for each treatment. For RT-qPCR, relative gene expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen 2001).
Subcellular localization assay
To obtain RFP-GmNTL1, RFP-GmNTL18CS, RFP-GmNTL1C247S, RFP-GmNTL1ΔTM, RFP-GmNTL1ΔC, and RFP-GmNTL1ΔTMC247 plasmids. The full-length GmNTL1, GmNTL1-ΔTM, and GmNTL1-ΔC coding sequences and the mutated sequences were cloned in-frame and downstream of the RFP sequence and then inserted into the pART27-GFP plant expression vector, which contained the GFP coding sequence driven by the CaMV 35S promoter. For subcellular localization assay, the GmNTL1, GmNTL1-ΔTM, and GmNTL1-ΔC coding sequences were amplified and cloned into the Gateway pDONR221 vector (Thermo Fisher Scientific, Waltham, MA, USA) using a BP reaction. The constructs were recombined into the pB7WGF2 binary vector (under the control of the 35S promoter) (PSB, Ghent University, Ghent, Belgium) in an LR reaction, yielding the GFP-GmNTL1, GFP-GmNTL1ΔTM, and GFP-GmNTL1ΔC construct.
The pMDC32-1A-BES1n-mCherry construct was used as a nucleus marker (Liang et al. 2015). To create mScarlet-HDEL, the ER signal peptide and the HDEL sequence were added to the N and C termini of mScarlet with PCR and then cloned into pCambia1300 (Brandizzi et al. 2003). The detection of GFP and RFP was carried out by laser scanning confocal microscopy (LSM 900; Carl Zeiss, Oberkochen, Germany) The GFP fluorescence was excited at 488 nm, and emission was recorded at 542 nm. The TaRFP or mScarlet fluorescence was excited at 555 nm or 587 nm, and emission was recorded at 584 to 610 nm. The 4′,6-diamidino-2-phenylindole (DAPI) staining fluorescence was excited at 350 to 360 nm, and emission was recorded at 450 to 460 nm.
Subcellular fractionation of nuclear and nonnuclear proteins
We fractionate nuclear and nonnuclear proteins from MYC-GmNTL1 soybean hairy root plant with or without 150 mm NaCl/1 mm H2O2 treatment for 12 h. Plant root tissues were frozen in liquid N2, ground to a fine powder, and mixed with 1× nuclei isolation buffer (Extraction Buffer I) (0.4 m sucrose, 10 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 15 mm β-mercaptoethanol, and 1 mm PMSF). Filter the solution into a new centrifuge tube by using a magical filter cloth. The samples were centrifuged at 5,500 rpm for 20 min, and the supernatant containing the nonnuclear protein fraction was separated from the pellet, which contained the nucleus and other subcellular organelles. The pellet was resuspended in 1× Extraction Buffer II (0.25 m sucrose, 10 mm Tris-HCl, pH 8.0, 5 mm β-mercaptoethanol, and 1 mm PMSF) containing 10% Triton X-10 (v/v). Organelle membranes were lysed by adding 10% Triton X-100 to a final concentration of 0.3%, and the lysates in 1× Extraction Buffer II were applied to a 1.5 m sucrose cushion. Centrifugation of the solution at 10,000 g for 10 min resulted in a semipure preparation of nuclei. The pellet was resuspended in Extraction Buffer I and used as the nuclear fraction. The purity of the nuclear and nonnuclear fractions was confirmed by western blotting using 1:5,000 diluted anti-histone H3 (Proteintech, 17168-1-AP) and 1:3,000 diluted anti-PEPC (Phyto, PHY2038S) antibodies, representing nuclear and nonnuclear standard markers, respectively.
Hairy root transformation
The soybean hairy root transformation system was conducted as described previously (Kereszt et al. 2007). First, keep the seeds in the chlorine gas atmosphere for 16 h. Then, place the sterilized seeds into wet vermiculite at a depth of 1 to 2 cm. After 5 d, collect Agrobacterium rhizogenes strain K599 from the plates and inoculate the seedlings by stabbing at the cotyledonary node and/or at the hypocotyl proximal to the cotyledon. When the hairy roots are approximately 5 to 10 cm in length, remove the primary root by cutting the hypocotyl 1 cm under the wounding site where the hairy roots are formed. Successful transformation was indicated by the presence of fluorescence from GFP, which were used as visual markers. The detection of GFP was carried out by laser scanning confocal microscopy (LSM 900; Carl Zeiss, Oberkochen, Germany). The GFP fluorescence was excited at 488 nm, and emission was recorded at 542 nm.
BIAM labeling assay
GmNTL1ΔC-His proteins were purified from Escherichia coli and then treated with different concentrations of H2O2 at room temperature for 30 min. The proteins were precipitated by adding 1 volume of acetone at −20°C for 20 min and centrifuged at 6,000 × g for 5 min. The pellets were washed 3 times with 50% (v/v) acetone and dissolved in 500 μL labeling buffer (50 mm MES-NaOH, pH 6.5, 100 mm NaCl, 1% TritonX-100 [v/v]) and then incubated with 100 µM BIAM at room temperature in the dark for 1 h. The labeling reactions were terminated by the addition of β-mercaptoethanol to a final concentration of 20 mm. The reaction mixtures were precipitated by adding 1 volume of acetone at −20°C for 20 min and centrifuged at 6,000 × g for 5 min. The pellets were dissolved in 50 μL SDS sample buffer and subjected to separate on SDS–PAGE. Proteins labeled with BIAM were detected with Anti-biotin, HRP-linked Antibody (Cell Signaling #7075, China, 1:5000 dilution). An antibody against His was used to show the total GmNTL1ΔC-His proteins (Proteintech, catalog # 6605-1, China, 1:5,000 dilution) (Tian et al. 2018).
In vitro biotin–switch assay
GmNTL1ΔC-His was treated with 10, 100 µM, and 2 mm H2O2 at room temperature for 30 min. Then, the GmNTL1ΔC was incubated with 100 μM NEM at room temperature for 30 min (Tian et al. 2018). The samples were precipitated with 1 volume of acetone and washed 3 times with 50% acetone. The pellets were dissolved in 500 µL REB buffer (20 mm HEPES, pH 8.0, 40 mm KCl, 5 mm EDTA, 0.5% TritonX-100 (v/v), 1% SDS (v/v), and 1 mm PMSF), plus 20 mm DTT, and then incubated at 37°C for 30 min to reduce the oxidized thiols. The reaction mixtures were precipitated by adding 1 volume of acetone at −20°C for 20 min and centrifuged at 6,000 × g for 5 min. The pellets were dissolved in 300 μL REB buffer. After ultrasonication, the supernatant was labeled with 100 μM BIAM at room temperature for 1 h in the dark and then to remove free BIAM. The BIAM-treated proteins were then dissolved in 500 µL REB buffer. After centrifugation at 6,000 × g for 6 min, the supernatant was added to 20 µL streptavidin beads (Biomag) and incubated at 4°C overnight. Beads were washed 3 times with NEB buffer, and proteins were eluted by 50 µL 2× SDS sample buffer, and the samples were separated on 8% SDS–PAGE gels. An aliquot of proteins before incubation with streptavidin beads was also analyzed as total GmNTL1ΔC protein. The gel blots were probed with anti-His antibody (Proteintech, catalog # 6605-1-1 g, China, 1:5,000 dilution).
In vivo biotin-switch assay
The in vivo biotin-switch assay was performed as previously described (Tian et al. 2018). Briefly, the MYC-GmNTL1, MYC-GmNTL8CS, and MYC-GmNTL1C247S transgenic soybean hairy roots were grown under 16-h light/8-h dark conditions and then treated with or without 150 mm NaCl for 6 h or 1 mm H2O2 for 12 h. Seedlings were harvested and ground to fine powder in liquid nitrogen. Proteins were extracted in EBR buffer (20 mm HEPES, pH 8.0, 40 mm KCl, 5 mm EDTA, 0.5% TritonX-100 [v/v], 1% SDS [v/v], 1 mm PMSF, and 1× protease inhibitor cocktail). The gel blots were probed with mouse monoclonal anti MYC (Proteintech, Wuhan, China; catalog # 60003-2-1 g; 1:7,000 dilution)
RNA-seq assay
For the transcriptome analysis, TRIzol (Thermo Fisher Scientific) was used to extract RNA from the root material of 3 biological replicates of Pro35S:GmNTL1ΔC transgenic plants and wild-type plants grown under normal and salt-stressed conditions. DEGs were selected based on the following criteria: q-value ≤ 0.05. The differentially expressed genes (DEGs) were identified using the NOISeq method (Tarazona et al. 2011). The cDNA libraries were sequenced on an Illumina (San Diego, CA, USA) HiSeq platform by BGI (Wuhan, China). The KEGG pathway of DEG clusters was performed with the program Dr.Tom (https://biosys.bgi.com). The significance of the KEGG was corrected using q-value ≤ 0.05.
ChIP-qPCR assay
The synthetic GmNTL1 peptide (PQDRKYPNGHRLNRAC) was injected into rabbits to generate the corresponding polyclonal antibodies by Mw Biotech (HK) Limited. ChIP was conducted as described previously (Yu et al. 2016). Two-wk-old Ludou 11 seedlings were treated with 150 mm NaCl (or H2O as a mock-treated control) for 12 h. Briefly, 2 g of whole seedlings was fixed by immersion in 1% (w/v) formaldehyde, after which the chromatin was sheared by sonication to a size ranging from 100 to 1,000 bp. After centrifugation at 10,000 × g for 10 min at 4 °C, the complex was immunoprecipitated with the anti-GmNTL1-specific antibody at a dilution of 1:500 (or rabbit serum protein as a mock sample). The primers used to amplify amplicons from the promoters of the candidate GmNTL1 target genes or negative (control) genes were used to detect the corresponding promoters in the ChIP products. The primers used in this study are listed in Supplemental Data Set 3. The enrichment percentages were calculated based on the relative change in anti-GmNTL1 compared to the input samples. The mock samples did not generate enough PCR products for detection. Each immunoprecipitation sample with a pool of several seedlings was considered 2 technical replicate, and 3 biological repeats were used.
Electrophoretic mobility shift assay
The recombinant GmNTL1-ΔC protein fused to MBP-His was produced after cloning the GmNTL1-ΔC coding sequence into the vector pMAL2CGW in E. coli strain Rosetta (WEIDI, China). Protein extraction and electrophoretic mobility shift assay were conducted as described previously (Yu et al. 2016). The oligonucleotide probe sequences are listed in Supplemental Data Set 3.
Transient expression assays
To obtain the Pro35S:GmNTL1-ΔC construct as effector, GmNTL1-ΔC coding sequence was amplified and cloned into the Gateway pDONR221 vector (Thermo Fisher Scientific, Waltham, MA, USA) using a BP reaction. The construct was recombined into the P2FGW7 binary vector (under the control of the 35S promoter) (PSB, Ghent University, Belgium) in an LR reaction. The pGreenII 0800-LUC vector system was used. The 1.5-kb GmRbohB-1, GmRbohB-2, GmNHX1, and GmCHX1 promoter sequences were amplified and cloned into pGreenII 0800-LUC vectors to obtain the Pro GmRbohB-1:LUC (KpnI and SpeI restriction sites), Pro GmRbohB-2:LUC (HindIII and SpeI restriction sites), Pro GmNHX1:LUC (KpnI and SpeI restriction sites), and Pro GmCHX1:LUC (KpnI and SpeI restriction sites) constructs, respectively. The primers used to clone the promoters or to generate the mutation are listed in Supplemental Data Set 3. Each reporter construct, together with 1 of the Pro35S:GmNTL1-ΔC constructs as effector, was individually transformed into Arabidopsis (Col-0) protoplasts. The protoplasts were obtained by enzymatic hydrolysis. The signals of firefly and Renilla LUC were assayed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Physiological measurements
The MDA concentration and POD and SOD activities were measured using a specific detection kit (Beyotime, China), following the manufacturer's instructions.
DAB staining
Whole 5-d-old (for root stain) soybean seedlings were treated with 0 or 150 mm NaCl or 150 mm NaCl coupled with 100 μM DPI in half-strength Hoagland solution for 6 h, 12 h, or 24 h before the roots were detached and stained directly. The samples were submerged in DAB solution (DINGGUO, Beijing, China) overnight and then in 95% (v/v) ethanol overnight to remove chlorophylls. The photographs were taken with a stereomicroscope (SZX16, OLYMPUS, Japan).
Quantification of H2O2 contents
The H2O2 contents were determined using a hydrogen peroxide assay kit (Beyotime, China), following the manufacturer's instructions. Three or more biological replicates were included for each assay. For the roots, 4-cm root tips were collected.
Determination of K+ and Na+ content in roots
Dried roots of plants were ground using a mortar and pestle; 0.2 g of powder was digested with 10 mL of nitric acid and 600 μL 30% (v/v) H2O2 for 1 h, and then, K+ and Na+ concentrations were analyzed using an atomic absorption spectrophotometer (Shimadzu, AA7000, Japan).
Statistical analysis
For comparisons between 2 sample groups, Student's t-test was applied. For multiple comparisons, a significance analysis was performed using a 1-way ANOVA. Two-way ANOVAs were conducted to test the interaction between 2 factors. The statistical analyses were performed using GraphPad Prism software version 8.0. Details of the statistical results are provided in Supplemental Data Set 4.
Accession numbers
Sequence data from this article can be found in the Phytozome (soybean) databases under accession numbers: GmNTL1 (Glyma.02G222300, LOC100778903), GmRbohB-1 (Glyma.10G152200, LOC100799682), GmRbohB-2 (Glyma.20G236200, LOC100787949), GmNHX1 (Glyma.20G229900, LOC732573), GmCHX1 (Glyma.03G171600, LOC10079916), and Gm60S (Glyma.13G318800, LOC100778077). The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Chen et al. 2021) in National Genomics Data Center (CNCB-NGDC Members and Partners 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA012093) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
Supplementary Material
Acknowledgments
The authors thank Haiyan Yu, Xiaomin Zhao, and Sen Wang from SKLMT (State Key Laboratory of Microbial Technology, Shandong University) for the assistance in microimaging of LSCM analysis. We also thank Jiaqi Sun, Shandong University, Shandong, China, for providing the mScarlet-HDEL vector.
Contributor Information
Wenxiao Zhang, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Wenjiao Zhi, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Hong Qiao, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Jingjing Huang, Department of Plant Biotechnology and Bioinformatics, Ghent University, 9052 Ghent, Belgium; Center for Plant Systems Biology, VIB, 9052 Ghent, Belgium.
Shuo Li, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Qing Lu, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Nan Wang, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Qiang Li, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Qian Zhou, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Jiaqi Sun, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Yuting Bai, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Xiaojian Zheng, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Mingyi Bai, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Frank Van Breusegem, Department of Plant Biotechnology and Bioinformatics, Ghent University, 9052 Ghent, Belgium; Center for Plant Systems Biology, VIB, 9052 Ghent, Belgium.
Fengning Xiang, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, People's Republic China.
Author contributions
F.X. conceived the project. W.Z., F.X., S.L., and Q.L. designed the experiments. W.Z. and F.X. drafted the manuscript. W.Z. performed most of the experiments. W.Z., H.Q., N.W., Q.Z., J.S., Y.B., and X.Z. performed parts of the biochemical, phenotyping, and transformation experiments. F.X., Q.L., F.B., J.H., and S.L. revised the manuscript. F.B. and M.B. provided some suggestions. All authors read and approved of its content.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Subcellular localization of GmNTL1.
Supplemental Figure S2. Mutations of the oxidized cysteines affect the subcellular localization of GmNTL1.
Supplemental Figure S3. Identification of GmNTL1ΔC overexpression and RNAi soybean lines.
Supplemental Figure S4. Physiological parameters of GmNTL1ΔC-overexpressing lines.
Supplemental Figure S5. Analysis of the oxidative modification of GmNTL1ΔC by BIAM-labeling assay.
Supplemental Figure S6. Transcriptome analysis of GmNTL1 OE-1 soybean.
Supplemental Figure S7. Expression patterns of GmRbohB-1, GmRbohB-2, GmNHX1, and GmCHX1 under NaCl stress conditions in WT and GmNTL1-RNAi soybean plants.
Supplemental Figure S8. Expression patterns of GmNHX1 and GmCHX1 under H2O2 conditions in WT, GmNTL1 OE-1, and GmNTL1 RNAi-1 soybean plants.
Supplemental Data Set 1. Differentially transcribed genes derived from the RNA-Seq experiment.
Supplemental Data Set 2. Gene Ontology (GO) analysis of NaCl OE versus NaCl WT cluster.
Supplemental Data Set 3. List of primers used in this article.
Supplemental Data Set 4. Summary of statistical analyses.
Funding
This research was supported by the Joint Funds of the National Natural Science Foundation of China (grant U1906203), the National Key Research and Development Program of China (grant 2021YFF1001201), National Key Research and Development Program of China (grant 2022YFF1001601-4), Key Research and Development Program of Shandong Province (grant 2021LZGC003), Key Research and Development Plan (Agricultural Variety Engineering) (grant 2023LZGC008), the National Transgenic Project of China (grants 2018ZX08009-14B and 2016ZX08010002-009), and the Research Foundation-Flanders senior postdoctoral fellowship (n.1227020N to J.H.). Thanks are due to Plant Editors for language editing.
Data availability
The data underlying this article are available in National Center for Biotechnology Information, at http://dx.doi.org/[koad250].
References
- Baillo EH, Kimotho RN, Zhang Z, Xu P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes (Basel). 2019:10(10):771. 10.3390/genes10100771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G, Hu M, Fu L, Zhang X, Zuo J, Li J, Yang J, Zhou J-M. The cytosolic thiol peroxidase PRXIIB is an intracellular sensor for H2O2 that regulates plant immunity through a redox relay. Nat Plants. 2022:8(10):1160–1175. 10.1038/s41477-022-01252-5 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Hanton S, Dasilva L, Boevink P, Evans D, Oparka K, Denecke J, Hawes C. ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J. 2003:34(3):269–281. 10.1046/j.1365-313X.2003.01728.x [DOI] [PubMed] [Google Scholar]
- Chen T, Chen X, Zhang S, Zhu J, Tang B, Wang A, Dong L, Zhang Z, Yu C, Sun Y, et al. The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics. 2021:19(4):578–583. 10.1016/j.gpb.2021.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K-H, Kim MY, Kwon H, Yang X, Lee S-H. Novel QTL identification and candidate gene analysis for enhancing salt tolerance in soybean (Glycine max (L.) Merr.). Plant Sci. 2021:313:111085. 10.1016/j.plantsci.2021.111085 [DOI] [PubMed] [Google Scholar]
- CNCB-NGDC Members and Partners . Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022:50(D1):D27–D38. 10.1093/nar/gkab951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Barampuram S, Stacey MG, Hancock CN, Findley S, Mathieu M, Zhang Z, Parrott WA, Stacey G. Tnt1 retrotransposon mutagenesis: a tool for soybean functional genomics. Plant Physiol. 2013:161(1):36–47. 10.1104/pp.112.205369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Backer J, Van Breusegem F, De Clercq I. Proteolytic activation of plant membrane-bound transcription factors. Front Plant Sci. 2022:13:927746. 10.3389/fpls.2022.927746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, Inzé A, Ng S, Ivanova A, Rombaut D, et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell. 2013:25(9):3472–3490. 10.1105/tpc.113.117168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet B, Willems P, Fernandez-Fernandez AD, Alseekh S, Fernie AR, Messens J, Van Breusegem F. In vivo detection of protein cysteine sulfenylation in plastids. Plant J. 2019:97(4):765–778. 10.1111/tpj.14146 [DOI] [PubMed] [Google Scholar]
- Debbarma J, Sarki Y, Saikia B, Boruah H, Singha D, Chikkaputtaiah C. Ethylene response factor (ERF) family proteins in abiotic stresses and CRISPR–Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: a review. Mol Biotechnol. 2019:61(2):153–172. 10.1007/s12033-018-0144-x [DOI] [PubMed] [Google Scholar]
- Djemal R, Khoudi H. The barley SHN1-type transcription factor HvSHN1 imparts heat, drought and salt tolerances in transgenic tobacco. Plant Physiol Biochem. 2021:164:44–53. 10.1016/j.plaphy.2021.04.018 [DOI] [PubMed] [Google Scholar]
- Dong W, Song Y, Zhao Z, Qiu NW, Liu X, Guo W. The Medicago truncatula R2R3-MYB transcription factor gene MtMYBS1 enhances salinity tolerance when constitutively expressed in Arabidopsis thaliana. Biochem Biophys Res Commun. 2017:490(2):225–230. 10.1016/j.bbrc.2017.06.025 [DOI] [PubMed] [Google Scholar]
- Duan M, Zhang R, Zhu F, Zhang Z, Gou L, Wen J, Dong J, Wang T. A lipid-anchored NAC transcription factor is translocated into the nucleus and activates Glyoxalase I expression during drought stress. Plant Cell. 2017:29(7):1748–1772. 10.1105/tpc.17.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fal S, Aasfar A, Rabie R, Smouni A, Arroussi HEL. Salt induced oxidative stress alters physiological, biochemical and metabolomic responses of green microalga Chlamydomonas reinhardtii. Heliyon. 2022:8(1):e08811. 10.1016/j.heliyon.2022.e08811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z-W, Feng Y-R, Gao X, Ding F, Li J-H, Yuan T-T, Lu Y-T. Salt stress-induced chloroplastic hydrogen peroxide stimulates pdTPI sulfenylation and methylglyoxal accumulation. Plant Cell. 2023:35(5):1593–1616. 10.1093/plcell/koad019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesguth M, Sahm A, Simon S, Dietz K-J. Redox-dependent translocation of the heat shock transcription factor AtHSFA8 from the cytosol to the nucleus in Arabidopsis thaliana. FEBS Lett. 2015:589(6):718–725. 10.1016/j.febslet.2015.01.039 [DOI] [PubMed] [Google Scholar]
- Golldack D, Li C, Mohan H, Probst N. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci. 2014:5:151. 10.3389/fpls.2014.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Qu Y, Guo Y, Yu L, Liu Y, Jiang J, Chen J, Ren Y, Liu G, Tian L, et al. Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 2014:80(6):937–950. 10.1111/tpj.12695 [DOI] [PubMed] [Google Scholar]
- Hao Y-J, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011:68(2):302–313. 10.1111/j.1365-313X.2011.04687.x [DOI] [PubMed] [Google Scholar]
- Hoang XLT, Chuong NN, Hoa TTK, Doan H, Van PHP, Trang LDM, Huyen PNT, Le DT, Tran L-SP, Thao NP. The drought-mediated soybean GmNAC085 functions as a positive regulator of plant response to salinity. Int J Mol Sci. 2021:22(16):8986. 10.3390/ijms22168986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T, Rape M, Jentsch S. Membrane-bound transcription factors: regulated release by RIP or RUP. Curr Opin Cell Biol. 2001:13(3):344–348. 10.1016/S0955-0674(00)00218-0 [DOI] [PubMed] [Google Scholar]
- Huang J, Willems P, Wei B, Tian C, Ferreira RB, Bodra N, Martínez Gache SA, Wahni K, Liu K, Vertommen D, et al. Mining for protein S-sulfenylation in Arabidopsis uncovers redox-sensitive sites. Proc Natl Acad Sci U S A. 2019:116(42):21256–21261. 10.1073/pnas.1906768116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji T, Zheng L, Wu J, Duan M, Liu Q, Liu P, Shen C, Liu J, Ye Q, Wen J, et al. The thioesterase APT1 is a bidirectional-adjustment redox sensor. Nat Commun. 2023:14(1):2807. 10.1038/s41467-023-38464-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszt A, Li D, Indrasumunar A, Nguyen CDT, Nontachaiyapoom S, Kinkema M, Gresshoff PM. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat Protoc. 2007:2(4):948–952. 10.1038/nprot.2007.141 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Park M-J, Seo PJ, Song J-S, Kim H-J, Park C-M. Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem J. 2012:448(3):353–363. 10.1042/BJ20120244 [DOI] [PubMed] [Google Scholar]
- Kim S-G, Kim S-Y, Park C-M. A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta. 2007:226(3):647–654. 10.1007/s00425-007-0513-3 [DOI] [PubMed] [Google Scholar]
- Le DT, Aldrich DL, Valliyodan B, Watanabe Y, Ha CV, Nishiyama R, Guttikonda SK, Quach TN, Gutierrez-Gonzalez JJ, Tran L-SP, et al. Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS One. 2012:7(9):e46487. 10.1371/journal.pone.0046487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, Park JH, Wi SD, Kang CH, Chi YH, Chae HB, Paeng SK, Ji MG, Kim WY, Kim MG, et al. Redox-dependent structural switch and CBF activation confer freezing tolerance in plants. Nat Plants. 2021:7(7):914–922. 10.1038/s41477-021-00944-8 [DOI] [PubMed] [Google Scholar]
- Lee S, Seo PJ, Lee H-J, Park C-M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012:70(5):831–844. 10.1111/j.1365-313X.2012.04932.x [DOI] [PubMed] [Google Scholar]
- Leferink NGH, van Duijn E, Barendregt A, Heck AJR, van Berkel WJH. Galactonolactone dehydrogenase requires a redox-sensitive thiol for optimal production of vitamin C. Plant Physiol. 2009:150(2):596–605. 10.1104/pp.109.136929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zheng J-C, Wang T-T, Min D-H, Wei W-L, Chen J, Zhou Y-B, Chen M, Xu Z-S, Ma Y-Z. Expression analyses of soybean VOZ transcription factors and the role of GmVOZ1G in drought and salt stress tolerance. Int J Mol Sci. 2020:21(6):2177. 10.3390/ijms21062177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chen R, Jiang Q, Sun X, Zhang H, Hu Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol Biol. 2021:105(3):333–345. 10.1007/s11103-020-01091-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang N, Ji D, Xue Z, Yu Y, Jiang Y, Liu J, Liu Z, Xiang F. Evolutionary and functional analysis of membrane-bound NAC transcription factor genes in soybean. Plant Physiol. 2016:172(3):1804–1820. 10.1104/pp.16.01132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang N, Ji D, Zhang W, Wang Y, Yu Y, Zhao S, Lyu M, You J, Zhang Y, et al. A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell. 2019a:31(9):2107–2130. 10.1105/tpc.18.00662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-YF, Wong F-L, Tsai S-N, Phang T-H, Shao G, Lam H-M. Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant Cell Environ. 2006:29(6):1122–1137. 10.1111/j.1365-3040.2005.01487.x [DOI] [PubMed] [Google Scholar]
- Li Y, Humbert S, Howell SH. ZmbZIP60 mRNA is spliced in maize in response to ER stress. BMC Res Notes. 2012:5:144. 10.1186/1756-0500-5-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu W, Zhong H, Zhang H-L, Xia Y. Redox-sensitive bZIP68 plays a role in balancing stress tolerance with growth in Arabidopsis. Plant J. 2019b:100(4):768–783. 10.1111/tpj.14476 [DOI] [PubMed] [Google Scholar]
- Liang M, Li H, Zhou F, Li H, Liu J, Hao Y, Wang Y, Zhao H, Han S. Subcellular distribution of NTL transcription factors in Arabidopsis thaliana. Traffic. 2015:16(10):1062–1074. 10.1111/tra.12311 [DOI] [PubMed] [Google Scholar]
- Lin Y, Liu G, Xue Y, Guo X, Luo J, Pan Y, Chen K, Tian J, Liang C. Functional characterization of aluminum (Al)-responsive membrane-bound NAC transcription factors in soybean roots. Int J Mol Sci. 2021:22(23):12854. 10.3390/ijms222312854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Sell S, Müller B, Leister D, Durner J. Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell. 2010:22(8):2894–2907. 10.1105/tpc.109.066464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W-C, Song R-F, Qiu Y-M, Zheng S-Q, Li T-T, Wu Y, Song C-P, Lu Y-T, Yuan H-M. Sulfenylation of ENOLASE2 facilitates H2O2-conferred freezing tolerance in Arabidopsis. Dev Cell. 2022:57(15):1883–1898.e5. 10.1016/j.devcel.2022.06.012 [DOI] [PubMed] [Google Scholar]
- Liu Y, Yu L, Qu Y, Chen J, Liu X, Hong H, Liu Z, Chang R, Gilliham M, Qiu L, et al. GmSALT3, which confers improved soybean salt tolerance in the field, increases leaf Cl- exclusion prior to Na+ exclusion but does not improve early vigor under salinity. Front Plant Sci. 2016:7:1485. 10.3389/fpls.2016.01485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yu Y, Sun J, Cao Q, Tang Z, Liu M, Xu T, Ma D, Li Z, Sun J. Root-zone-specific sensitivity of K+-and Ca2+-permeable channels to H2O2 determines ion homeostasis in salinized diploid and hexaploid Ipomoea trifida. J Exp Bot. 2019:70(4):1389–1405. 10.1093/jxb/ery461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR. Methods. 2001:25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu S-J, Yang Z-T, Sun L, Sun L, Song Z-T, Liu J-X. Conservation of IRE1-regulated bZIP74 mRNA unconventional splicing in rice (Oryza sativa L.) involved in ER stress responses. Mol Plant. 2012:5(2):504–514. 10.1093/mp/ssr115 [DOI] [PubMed] [Google Scholar]
- Lu Q, Houbaert A, Ma Q, Huang J, Sterck L, Zhang C, Benjamins R, Coppens F, Van Breusegem F, Russinova E. Adenosine monophosphate deaminase modulates BIN2 activity through hydrogen peroxide-induced oligomerization. Plant Cell. 2022:34(10):3844–3859. 10.1093/plcell/koac203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G-Z, Wang H-W, Huang J, Tian A-G, Wang Y-J, Zhang J-S, Chen S-Y. A putative plasma membrane cation/proton antiporter from soybean confers salt tolerance in Arabidopsis. Plant Mol Biol. 2005:59(5):809–820. 10.1007/s11103-005-1386-0 [DOI] [PubMed] [Google Scholar]
- Meng X, Li L, De Clercq I, Narsai R, Xu Y, Hartmann A, Claros DL, Custovic E, Lewsey MG, Whelan J, et al. ANAC017 Coordinates organellar functions and stress responses by reprogramming retrograde signaling. Plant Physiol. 2019:180(1):634–653. 10.1104/pp.18.01603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell. 2006:18(10):2749–2766. 10.1105/tpc.106.044230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. ROS Are good. Trends Plant Sci. 2017:22(1):11–19. 10.1016/j.tplants.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Mittler R, Zandalinas SI, Fichman Y, Van Breusegem F. Reactive oxygen species signalling in plant stress responses. Nat Rev Mol Cell Biol. 2022:23(10):663–679. 10.1038/s41580-022-00499-2 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. NAC Transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012:1819(2):97–103. 10.1016/j.bbagrm.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Nazir F, Fariduddin Q, Khan TA. Hydrogen peroxide as a signalling molecule in plants and its crosstalk with other plant growth regulators under heavy metal stress. Chemosphere. 2020:252:126486. 10.1016/j.chemosphere.2020.126486 [DOI] [PubMed] [Google Scholar]
- Nietzel T, Mostertz J, Ruberti C, Née G, Fuchs P, Wagner S, Moseler A, Müller-Schüssele SJ, Benamar A, Poschet G, et al. Redox-mediated kick-start of mitochondrial energy metabolism drives resource-efficient seed germination. Proc Natl Acad Sci U S A. 2020:117(1):741–751. 10.1073/pnas.1910501117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L, Liao W. Hydrogen peroxide signaling in plant development and abiotic responses: crosstalk with nitric oxide and calcium. Front Plant Sci. 2016:7:230. 10.3389/fpls.2016.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu M, Huang Y, Sun S, Sun J, Cao H, Shabala S, Bie Z. Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure. J Exp Bot. 2018:69(14):3465–3476. 10.1093/jxb/erx386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim Y-S, Kim S-G, Jung J-H, Woo J-C, Park C-M. Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 2011:156(2):537–549. 10.1104/pp.111.177071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Li MW, Xie M, Liu X, Ni M, Shao G, Song C, Kay-Yuen Yim A, Tao Y, Wong FL, et al. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat Commun. 2014:5:4340. 10.1038/ncomms5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Guan R, Yu L, Berkowitz O, David R, Whelan J, Ford M, Wege S, Qiu L, Gilliham M. Enhanced reactive oxygen detoxification occurs in salt-stressed soybean roots expressing GmSALT3. Physiol Plant. 2022:174(3):e13709. 10.1111/ppl.13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed A, Raza A, Jie H, Mahmood A, Ma Y, Zhao L, Xing H, Li L, Hassan MU, Qari SH, et al. Molecular tools and their applications in developing salt-tolerant soybean (Glycine max L.) cultivars. Bioengineering (Basel). 2022:9(10):495. 10.3390/bioengineering9100495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev S, Ansari SA, Ansari MI, Fujita M, Hasanuzzaman M. Abiotic stress and reactive oxygen species: generation. Signaling, and defense mechanisms. Antioxidants (Basel). 2021:10(2):277. 10.3390/antiox10020277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Song J-S, Kim Y-S, Kim H-J, Park C-M. Proteolytic processing of an Arabidopsis membrane-bound NAC transcription factor is triggered by cold-induced changes in membrane fluidity. Biochem J. 2010:427(3):359–367. 10.1042/BJ20091762 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Kim S-G, Park C-M. Membrane-bound transcription factors in plants. Trends Plant Sci. 2008:13(10):550–556. 10.1016/j.tplants.2008.06.008 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park C-M. A membrane-bound NAC transcription factor as an integrator of biotic and abiotic stress signals. Plant Signal Behav. 2010:5(5):481–483. 10.4161/psb.11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setia R, Gottschalk P, Smith P, Marschner P, Baldock J, Setia D, Smith J. Soil salinity decreases global soil organic carbon stocks. Sci.Total Env. 2013:465:267–272. 10.1016/j.scitotenv.2012.08.028 [DOI] [PubMed] [Google Scholar]
- Shao H, Wang H, Tang X. NAC Transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci. 2015:6:902. 10.3389/fpls.2015.00902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H, Jones DP. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat Rev Mol Cell Biol. 2020:21(7):363–383. 10.1038/s41580-020-0230-3 [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Arnaud D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019:221(3):1197–1214. 10.1111/nph.15488 [DOI] [PubMed] [Google Scholar]
- Sun H, Xie Y, Yang W, Lv Q, Chen L, Li J, Meng Y, Li L, Li X. Membrane-bound transcription factor TaNTL1 positively regulates drought stress tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2022:182:182–193. 10.1016/j.plaphy.2022.04.023 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Salazar C, Mondal HA, Shulaev E, Cortes DF, Shuman JL, Luo X, Shah J, Schlauch K, et al. Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell. 2013:25(9):3553–3569. 10.1105/tpc.113.114595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kawakatsu T, Wakasa Y, Hayashi S, Takaiwa F. A rice transmembrane bZIP transcription factor, OsbZIP39, regulates the endoplasmic reticulum stress response. Plant Cell Physiol. 2012:53(1):144–153. 10.1093/pcp/pcr157 [DOI] [PubMed] [Google Scholar]
- Tang Y, Liu M, Gao S, Zhang Z, Zhao X, Zhao C, Zhang F, Chen X. Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol Plant. 2012:144(3):210–224. 10.1111/j.1399-3054.2011.01539.x [DOI] [PubMed] [Google Scholar]
- Tang Y, Zhao C-Y, Tan S-T, Xue H-W. Arabidopsis type II phosphatidylinositol 4-kinase PI4Kγ5 regulates auxin biosynthesis and leaf margin development through interacting with membrane-bound transcription factor ANAC078. PLoS Genet. 2016:12(8):e1006252. 10.1371/journal.pgen.1006252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011:21(12):2213–2223. 10.1101/gr.124321.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Fan M, Qin Z, Lv H, Wang M, Zhang Z, Zhou W, Zhao N, Li X, Han C, et al. Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat Commun. 2018:9(1):1063. 10.1038/s41467-018-03463-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zelm E, Zhang Y, Testerink C. Salt tolerance mechanisms of plants. Annu Rev Plant Biol. 2020:71:403–433. 10.1146/annurev-arplant-050718-100005 [DOI] [PubMed] [Google Scholar]
- Wang N, Zhang W, Qin M, Li S, Qiao M, Liu Z, Xiang F. Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol. 2017:58(10):1764–1776. 10.1093/pcp/pcx111 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhao J, Fang Q, Chang X, Sun M, Li W, Li Y. GmAKT1 is involved in K+ uptake and Na+/K+ homeostasis in Arabidopsis and soybean plants. Plant Sci. 2021:304:110736. 10.1016/j.plantsci.2020.110736 [DOI] [PubMed] [Google Scholar]
- Waszczak C, Akter S, Jacques S, Huang J, Messens J, Van Breusegem F. Oxidative post-translational modifications of cysteine residues in plant signal transduction. J Exp Bot. 2015:66(10):2923–2934. 10.1093/jxb/erv084 [DOI] [PubMed] [Google Scholar]
- Waszczak C, Carmody M, Kangasjärvi J. Reactive oxygen species in plant signaling. Annu Rev Plant Biol. 2018:69:209–236. 10.1146/annurev-arplant-042817-040322 [DOI] [PubMed] [Google Scholar]
- Wu F, Chi Y, Jiang Z, Xu Y, Xie L, Huang F, Wan D, Ni J, Yuan F, Wu X, et al. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature. 2020:578(7796):577–581. 10.1038/s41586-020-2032-3 [DOI] [PubMed] [Google Scholar]
- Xing L, Zhu M, Luan M, Zhang M, Jin L, Liu Y, Zou J, Wang L, Xu M. Mir169q and NUCLEAR FACTOR YA8 enhance salt tolerance by activating PEROXIDASE1 expression in response to ROS. Plant Physiol. 2022:188(1):608–623. 10.1093/plphys/kiab498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Han Y, Wu D, Yong W, Liu M, Wang S, Liu W, Lu M, Wei Y, Sun J. Salt and cadmium stress tolerance caused by overexpression of the Glycine Max Na+/H+ Antiporter (GmNHX1) gene in duckweed (Lemna turionifera 5511). Aquat Toxicol. 2017:192:127–135. 10.1016/j.aquatox.2017.08.010 [DOI] [PubMed] [Google Scholar]
- Yang X, Kim MY, Ha J, Lee S-H. Overexpression of the Soybean NAC Gene GmNAC109 increases lateral root formation and abiotic stress tolerance in transgenic Arabidopsis plants. Front Plant Sci. 2019:10:1036. 10.3389/fpls.2019.01036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Guo Y. Unraveling salt stress signaling in plants. J Integr Plant Biol. 2018:60(9):796–804. 10.1111/jipb.12689 [DOI] [PubMed] [Google Scholar]
- Yarra R, Wei W. The NAC-type transcription factor GmNAC20 improves cold, salinity tolerance, and lateral root formation in transgenic rice plants. Funct Integr Genomics. 2021:21(3–4):473–487. 10.1007/s10142-021-00790-z [DOI] [PubMed] [Google Scholar]
- Yin Y, Qin K, Song X, Zhang Q, Zhou Y, Xia X, Yu J. BZR1 Transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signaling in tomato. Plant Cell Physiol. 2018:59(11):2239–2254. 10.1093/pcp/pcy146 [DOI] [PubMed] [Google Scholar]
- Yu Y, Liu Z, Wang L, Kim S-G, Seo PJ, Qiao M, Wang N, Li S, Cao X, Park C-M, et al. WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 2016:85(1):96–106. 10.1111/tpj.13092 [DOI] [PubMed] [Google Scholar]
- Zhang X, Long Y, Huang J, Xia J. OsNAC45 is involved in ABA response and salt tolerance in rice. Rice (N Y). 2020:13(1):79. 10.1186/s12284-020-00440-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Wen W, Shi Z, Gu Q, Ahammed GJ, Cao K, Shah Jahan M, Shu S, Wang J, et al. Hydrogen peroxide mediates spermidine-induced autophagy to alleviate salt stress in cucumber. Autophagy. 2021:17(10):2876–2890. 10.1080/15548627.2020.1847797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Zhang Q, Liu M, Zhou H, Ma C, Wang P. Regulation of plant responses to salt stress. Int J Mol Sci. 2021:22(9):4609. 10.3390/ijms22094609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Huang J, Willems P, Van Breusegem F, Xie Y. Cysteine thiol-based post-translational modification: what do we know about transcription factors? Trends Plant Sci. 2023:28(4):415–428. 10.1016/j.tplants.2022.11.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in National Center for Biotechnology Information, at http://dx.doi.org/[koad250].