Abstract
In plant leaves, starch is composed of glucan polymers that accumulate in chloroplasts as the products of photosynthesis during the day; starch is mobilized at night to continuously provide sugars to sustain plant growth and development. Efficient starch degradation requires the involvement of several enzymes, including β-amylase and glucan phosphatase. However, how these enzymes cooperate remains largely unclear. Here, we show that the glucan phosphatase LIKE SEX FOUR 1 (LSF1) interacts with plastid NAD-dependent malate dehydrogenase (MDH) to recruit β-amylase (BAM1), thus reconstituting the BAM1–LSF1–MDH complex. The starch hydrolysis activity of BAM1 drastically increased in the presence of LSF1–MDH in vitro. We determined the structure of the BAM1–LSF1–MDH complex by a combination of cryo-electron microscopy, crosslinking mass spectrometry, and molecular docking. The starch-binding domain of the dual-specificity phosphatase and carbohydrate-binding module of LSF1 was docked in proximity to BAM1, thus facilitating BAM1 access to and hydrolysis of the polyglucans of starch, thus revealing the molecular mechanism by which the LSF1–MDH complex improves the starch degradation activity of BAM1. Moreover, LSF1 is phosphatase inactive, and the enzymatic activity of MDH was dispensable for starch degradation, suggesting nonenzymatic scaffold functions for LSF1–MDH in starch degradation. These findings provide important insights into the precise regulation of starch degradation.
Phosphatase-inactive LIKE SEX FOUR 1 interacts with moonlighting malate dehydrogenase to form a binary complex acting as a scaffold to recruit β-amylase to promote starch degradation.
IN A NUTSHELL.
Background: Starch is a glucan polymer that accumulates in chloroplasts during the day as a product of photosynthesis and is mobilized at night to continuously provide sugars to sustain plant growth and development. Efficient starch degradation involves multiple enzymes such as glucan phosphatase and β-amylase. LIKE SEX FOUR 1 (LSF1) is a unique multidomain inactive glucan phosphatase, with the lsf1 mutant exhibiting a starch-excess phenotype. LSF1 is thought to interact with other proteins to form a protein complex to regulate starch degradation. However, the composition of this protein complex and how its assembly is regulated remain largely unknown.
Question: How does LSF1 coordinate with other proteins to form a starch degradation complex? What are the molecular functions of LSF1 in starch degradation?
Findings: We discovered that LSF1 interacts with plastid localized NAD-dependent malate dehydrogenase (MDH) to recruit β-amylase (BAM1) and form the BAM1–LSF1–MDH complex. We also demonstrate that the starch hydrolysis activity of BAM1 drastically increases in the presence of the LSF1–MDH complex. A comprehensive analysis of cryo-electron microscopy structure, crosslinking coupled with mass spectrometry, and molecular docking revealed the molecular mechanism by which the LSF1–MDH complex improves the starch degradation activity of BAM1. Specifically, the proximity of the starch-binding domains provided by the dual-specificity phosphatase and the carbohydrate-binding module of LSF1 to BAM1 facilitates the access of BAM1 to the polyglucans of starch, helping in their hydrolysis by BAM1. This study uncovers the scaffold function of the LSF1–MDH complex in starch degradation.
Next steps: Engineering LSF1 to be an enzymatically active glucan phosphatase may allow the BAM1–LSF1–MDH complex to successively catalyze starch dephosphorylation and hydrolysis. Further studies are however needed to explore the effects of active LSF1 on plant growth and development.
Introduction
Starch in plants and glycogen in animals are major storage carbohydrates. In both polymers, glucose residues are linked by α-1,4-glycosidic bonds and branched via α-1,6-glycosidic linkages (Smith et al. 2005). Starch consists of linear (amylose) and highly branched (amylopectin) glucan polymers that are arranged as insoluble semicrystalline starch granules (Smith and Zeeman 2020). In plant leaves, starch is a major product of photosynthetic carbon fixation in chloroplasts, and it is synthesized during the day and hydrolyzed during the following night (Streb and Zeeman 2012). Starch degradation provides the energy required to sustain plant growth and development (Horrer et al. 2016; Flutsch et al. 2020), as well as response to various stresses (Thalmann et al. 2016; Thalmann and Santelia 2017).
Starch degradation requires the synergistic actions of multiple enzymes that are involved in phosphorylation, dephosphorylation, and hydrolysis (Fig. 1A). Phosphorylation disrupts the semicrystalline structure of starch, thus facilitating the hydrolytic enzyme to gain access to the starch granule. A proportion of the glucosyl residues on the granule surface is phosphorylated by the enzymes GLUCAN, WATER DIKINASE (GWD) and PHOSPHOGLUCAN, WATER DIKINASE (PWD; Ritte et al. 2002; Baunsgaard et al. 2005; Kotting et al. 2005). GWD exclusively phosphorylates the C6 positions of glucosyl residues, whereas PWD phosphorylates the C3 positions (Ritte et al. 2006). Mutants lacking either GWD or PWD function exhibit a lower rate of starch degradation, which leads to a starch-excess phenotype (Yu et al. 2001; Baunsgaard et al. 2005).
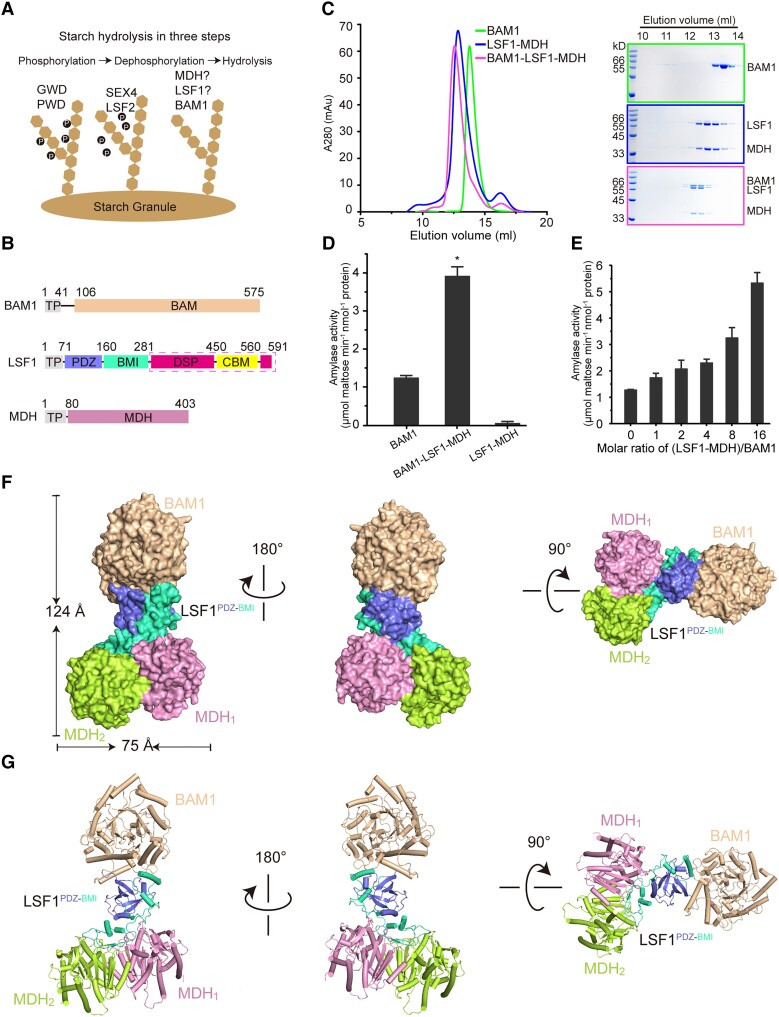
Figure 1.
Structure of the BAM1–LSF1–MDH complex. A) Diagram of starch degradation. Proteins involved in starch phosphorylation, dephosphorylation, and hydrolysis are indicated. B) Diagrams of the domain architecture of BAM1, LSF1, and MDH. TP, chloroplast TP, were omitted for protein production. BAM1 and MDH are colored in beige and pink. For LSF1, the individual domain of PDZ, BMI, DSP, and CBM are colored in slate, green-cyan, hot pink, and yellow, respectively. The domains of DSP-CBM absent from the cryo-EM structure are indicated by the dashed magenta box. C) Reconstitution of the ternary complex by gel filtration. Left, gel filtration chromatography. Right, SDS-PAGE corresponding to the chromatography. The incubation of BAM1 with LSF1–MDH led to the formation and coelution of the ternary BAM1–LSF1–MDH complex. D) Starch hydrolytic activity of the enzymes by detecting the maltose product. n.m., no measured activity. E) Titration assay of the starch-degradation activity of BAM1 upon the addition of the binary LSF1–MDH complex with increased molar ratios. All data reported here were obtained from 3 independent experiments and presented as mean ± Sd of triplicate experiments. Significant differences were determined using 2-tailed Student's t-test (*P < 0.05). F and G) Cryo-EM density of the BAM1–LSF1–MDH complex F) and its cartoon representation G). BAM1 is colored in beige, MDH1 and MDH2 are in pink and lemon, and the PDZ and BMI of LSF1 are in slate and green-cyan, respectively.
Although its phosphorylation renders starch more accessible to hydrolytic enzymes, the presence of phosphates also hinders the movement of the starch degradation enzyme along the glucan chain, thus limiting the release of the maltose products and blocking complete glucan hydrolysis (Silver et al. 2014). Thus, the phosphate groups need to be removed for the successive degradation of linear glucan chains. Starch is dephosphorylated by glucan phosphatase. The mammalian glucan phosphatase laforin regulates the phosphorylation of glycogen (Raththagala et al. 2015), and the dysfunction of laforin causes the accumulation of hyperphosphorylated polyglucosan bodies, thereby leading to the fatal neurodegenerative disorder Lafora disease (Nitschke et al. 2013, 2018). The laforin counterparts in chloroplasts consist of STARCH EXCESS 4 (SEX4), LIKE SEX FOUR 1 (LSF1), and LSF2 in Arabidopsis thaliana (Gentry et al. 2016). These glucan phosphatases exhibit distinct biochemical characteristics. Structurally, they all possess a dual-specificity phosphatase (DSP) domain, and both SEX4 and LSF1 contain a carbohydrate-binding module (CBM). Loss of SEX4 function causes elevated starch levels and accumulation of phosphorylated glucans (Kotting et al. 2009). Although the lsf2 mutant shows normal starch levels, the lsf2 sex4 double mutant exhibits a more severe starch-excess phenotype than the sex4 single mutant (Santelia et al. 2011), suggesting a partial overlapping role for LSF2 in starch degradation. SEX4 removes phosphates from both C6 and C3 positions of polyglucan chains (Hejazi et al. 2010), whereas LSF2 only does so from the C3 position (Santelia et al. 2011). These facts suggest that both SEX4 and LSF2 possess phosphatase activity, and structural analyses of substrate-bound SEX4 and LSF2 have revealed the molecular basis by which these 2 phosphatases dephosphorylate glucans at specific positions (Vander Kooi et al. 2010; Meekins et al. 2013, 2014). Unlike SEX4 and LSF2, LSF1 is an inactive glucan phosphatase (Schreier et al. 2019). Although the lsf1 mutant exhibits a starch-excess phenotype, no decrease in glucan dephosphorylation activity and no accumulation of phospho-oligosaccharides are detected (Comparot-Moss et al. 2010). The lsf1 sex4 double mutant has a more severe starch-excess phenotype than the lsf1 or sex4 single mutant (Comparot-Moss et al. 2010). These findings suggest that LSF1 might play a different function from its counterparts SEX4 and LSF2 in starch degradation. LSF1 additionally contains a unique PDZ domain at its N terminus, and this domain may mediate protein-protein interactions (Silver et al. 2014), suggesting that LSF1 might interact with other enzymes to regulate starch degradation. However, the specific biochemical role of LSF1 in starch degradation awaits further exploration.
To find its potential interaction partners, an LSF1 variant was constructed with a C-terminal tandem-affinity purification (TAP) tag and transformed into lsf1 mutant plants. TAP purification followed by mass spectrometry identification revealed that the β-amylases BAM1 and BAM3 copurified with LSF1. Moreover, reciprocal TAP-tagging experiments showed that LSF1 copurifies with either BAM1-TAP or BAM3-TAP (Schreier et al. 2019). The potential interactions between LSF1 and BAM1 were validated by coimmunoprecipitation and biomolecular fluorescence complementation experiments (Schreier et al. 2019). BAM1 and BAM3 are chloroplast-localized β-amylases that facilitate the starch-to-maltose conversion by catalyzing the hydrolysis of 1,4-α-glucosidic linkages. The bam1 mutant retains a normal starch level, whereas the bam3 mutant exhibits an elevated starch level (Fulton et al. 2008). The bam1 bam3 double mutant displays a more severe starch-excess phenotype than either single mutants, suggesting the dominant and functionally overlapping roles of BAM1 and BAM3 in starch degradation (Fulton et al. 2008). In addition to BAM1 and BAM3, there are other 4 β-amylases in Arabidopsis chloroplast, including BAM2, BAM4, BAM6, and BAM9 (Monroe and Storm 2018; Monroe 2020; David et al. 2022). These β-amylases exhibit distinct biochemical characteristics (Sparla et al. 2006; Monroe et al. 2014, 2017). BAM1, BAM2, BAM3, and BAM6 are catalytically active, whereas BAM4 and BAM9 are inactive (Monroe and Storm 2018). The mutants of bam2 and bam6 exhibit normal starch levels (Monroe 2020). Although BAM4 and BAM9 have lost their catalytic activity (Fulton et al. 2008; David et al. 2022), they retain their starch degradation function, since a bam4 single mutant exhibits a starch-excess phenotype (Fulton et al. 2008). The bam3 bam9 double mutant and bam1 bam3 bam9 triple mutant display more severe starch-excess phenotype than the bam3 single mutant and the bam1 bam3 double mutant (David et al. 2022). These findings indicate the diverse biochemical roles of β-amylase members and their complicated regulation on starch degradation.
In multiple TAP experiments, the plastid-localized NAD-dependent malate dehydrogenase (pdNAD-MDH) was consistently identified as an interaction partner of LSF1, BAM1, and BAM3 (Schreier et al. 2019), suggesting the association of MDH with these starch degradation enzymes. MDH is widely distributed in bacteria, plants, and animals, and it can catalyze the reversible interconversion of malate and oxaloacetate. Arabidopsis MDH is present as a small protein family with 9 isoforms distributed in different cellular compartments, including plastids, mitochondria, and peroxisomes (Dao et al. 2022). Among these isomers, the mutation of pdNAD-MDH causes embryo death (Beeler et al. 2014; Selinski et al. 2014; Zhao et al. 2018). A transgenic line expressing an artificial microRNA targeting pdNAD-MDH transcripts in Arabidopsis exhibits pale leaves with a disordered chloroplast ultrastructure (Beeler et al. 2014). Interestingly, a catalytically inactive MDH variant can complement the embryo death phenotype of pdnad-mdh (Schreier et al. 2018). These findings suggest that pdNAD-MDH plays crucial roles in chloroplast biogenesis, but its enzymatic activity may not be essential. An immunoprecipitation experiment coupled with mass spectrometry (CXMS) identified an FtsH family protease as the most abundant interacting partner of pdNAD-MDH, followed by LSF1, BAM1, and BAM3 involved in starch degradation (Schreier et al. 2018). These findings suggest that pdNAD-MDH might exert a moonlighting function by forming protein complexes to facilitate chloroplast development. However, the underlying molecular basis of the nonenzymatic role of pdNAD-MDH beyond its canonical function remains unclear.
In this study, we reconstituted the BAM1–LSF1–MDH complex in vitro and demonstrated its enhanced starch degradation activity. Using an integrated biochemical and structural analytical strategy including enzymatic characterization, cryo-electron microscopy (EM) structure determination, crosslinked mass spectrometry identification, and molecular docking, we deciphered the molecular mechanism by which LSF1–MDH functions as a scaffold to recruit β-amylase to promote starch degradation. This study provides important insights into the regulation of starch degradation.
Results
Assembly of a BAM1–LSF1–MDH ternary complex
Previous studies have revealed that the lsf1 and bam1 bam3 mutants exhibit a starch-excess phenotype, indicating the essential roles of these genes in starch degradation (Fulton et al. 2008; Schreier et al. 2019). TAP and mass spectrometry analysis have identified the possible association of pdNAD-MDH with LSF1 and BAM1 (or BAM3; Schreier et al. 2018, 2019). However, how they coordinate to regulate starch degradation remains unknown. To uncover the underlying molecular mechanism, we explored their interactions here. To this end, we individually produced N-terminally His-tagged BAM1, LSF1, or MDH with their chloroplast transit peptide (TP) removed in Escherichia coli (Fig. 1B). Both BAM1 (Residues 42 to 575) and MDH (Residues 81 to 403) were successfully purified to homogeneity with good solution behavior (Fig. 1C, Supplemental Fig. S1A), whereas the purification of LSF1 (Residues 62 to 591) failed, which might be due to instability (Schreier et al. 2019). To optimize protein production, we tested several LSF1 truncations and investigated the interaction of LSF1 with BAM1 or MDH by coexpression. The LSF1 construct (Residues 71 to 591) was prone to form inclusion bodies (Supplemental Fig. S1B). Upon coexpression, we obtained large quantities of a binary LSF1–MDH complex with good solution behavior, suggesting that the presence of MDH improves the folding and solution stability of LSF1 (Fig. 1C, Supplemental Fig. S1B). To examine whether these proteins can form a higher order complex, we incubated BAM1 with LSF1–MDH and subjected the mixture to gel filtration. Coelution of BAM1 with LSF1–MDH led to the formation of a BAM1–LSF1–MDH ternary complex (Fig. 1C), suggesting their robust interaction.
The individual members in the BAM1–LSF1–MDH complex belong to different enzymatic families, with LSF1 containing a DSP domain. A previous study demonstrated that LSF1 is phosphatase inactive by using a truncation (Residues 281 to 591), containing the C-terminal DSP and CBM (Schreier et al. 2019). Since LSF1 contains conserved catalytic residues, we wondered whether its enzymatic activity might be detected in near-full-length LSF1 (Residues 71 to 591), containing all its functional domains and whether LSF1 might be activated in the presence of MDH and BAM1. We therefore performed a phosphatase assay with the LSF1–MDH and BAM1–LSF1–MDH complexes, using SEX4 and LSF2 (counterparts of LSF1) as positive controls (Supplemental Fig. S1, C and D). In contrast to SEX4 and LSF2 exhibiting clear phosphatase activity, LSF1 displayed no enzymatic activity in the presence of MDH and BAM1 (Supplemental Fig. S2A). These results further demonstrate that LSF1 is an inactive glucan phosphatase, suggesting that LSF1 plays different roles from its counterparts LSF2 and SEX4 in starch degradation.
MDH catalyzes the reversible interconversion of oxaloacetate to malate. We examined MDH enzymatic activity by measuring the rate of NADH consumption, which was manifested by the decrease in absorbance at 340 nm. We confirmed that MDH converts oxaloacetate to malate in our assays. To examine the effect of LSF1 and BAM1 on the catalytic activity of MDH, we performed enzymatic assays on LSF1–MDH and BAM1–LSF1–MDH. Both complexes exhibited enzymatic capacity similar to that of MDH alone, suggesting that the presence of BAM1 and/or LSF1 has no obvious effect on the catalytic activity of MDH (Supplemental Fig. S2B).
BAM1 and BAM3 are 2 of the key β-amylases responsible for starch degradation, promoting the release of maltose from the nonreducing end of starch. Like BAM1, BAM3 was also identified as an interaction partner of LSF1 (Schreier et al. 2019). Moreover, BAM3 exhibits a high sequence similarity to BAM1. Thus, we postulated that BAM3 might also interact with– LSF1–MDH. To test this hypothesis, we incubated BAM3 with LSF1–MDH and subjected the mixture to gel filtration assays. The coelution of BAM3 and LSF1–MDH led to the formation of a BAM3–LSF1–MDH ternary complex (Supplemental Fig. S3A). We detected the starch hydrolysis activity of BAM1 and BAM3 by examining the reducing maltose product (Fig. 1D, Supplemental Fig. S3B). Considering that the lsf1 mutant exhibits an increased starch content (Schreier et al. 2019), we wondered whether LSF1–MDH might regulate the starch degradation efficiency of BAM1 and BAM3. We observed that the BAM1–LSF1–MDH complex exhibits a higher (about 3 times) catalytic activity than BAM1 alone (Fig. 1D). We further performed titration assays by increasing the protein molar ratios of LSF1–MDH to BAM1 or BAM3 and examined their catalytic capacity. The starch hydrolysis activity of BAM1 and BAM3 gradually increased upon the addition of more LSF1–MDH (Fig. 1E, Supplemental Fig. S3B). These results suggest that LSF1–MDH can increase the catalytic efficiency of the β-amylases BAM1 and BAM3.
Structure of the BAM1–LSF1–MDH complex
To uncover the molecular mechanism by which LSF1–MDH improved the starch degradation activity of BAM1 or BAM3, we determined the cryo-EM structure of BAM1–LSF1–MDH at an average resolution of 3.0 Å (Supplemental Fig. S4). Data collection and refinement statistics are given in Supplemental Table S1. The overall structure of the ternary complex exhibited a width of ∼75 Å and a height of ∼124 Å; this structure resembled a dumbbell with LSF1 as the central bar connecting the 2-end bells of BAM1 and MDH (Fig. 1, F and G). In the complex, BAM1, LSF1, and MDH presented a stoichiometry ratio of 1:1:2. In the structure, densities of BAM1 Residues 498 to 509 and 540 to 575, MDH Residues 396 to 403, and LSF1 Residues 226 to 243 and 282 to 591 were missing, suggesting the flexibility of these segments.
BAM1 exhibited a typical (β/a)8 barrel-shaped architecture comprised of 8 β-strands arranged in a central pocket surrounded by 8 α-helices (Supplemental Fig. S5A). Structural superposition of BAM1 with its orthologs from soybean (Glycine max; Kang et al. 2005), barley (Hordeum vulgare; Rejzek et al. 2011), sweet potato (Ipomoea batatas; Vajravijayan et al. 2018), wheat (Triticum aestivum; Hofer et al. 2019), maize (Zea mays; Sun et al. 2022), and the bacterium Bacillus cereus (Mikami et al. 1999) revealed conservation of the (β/a)8-barrel architecture (Supplemental Fig. S5, B and C), whereas bacterial BAM possessed an extra CBM (Supplemental Fig. S5, B and C). The active sites of these β-amylases all contained 2 conserved glutamate residues (Supplemental Fig. S5D). The 2 glutamate residues of BAM1 were E279 and E477, which were located in the deep pocket of the (β/a)8-barrel. Structural alignment of BAM1 with maltotetraose-bound sweet potato β-amylase (PDB: 5wqu) revealed the proximity of the glutamate residues to maltotetraose (Supplemental Fig. S5E). Replacing residues E279 and E477 with alanine (A) abolished catalytic activity (Supplemental Fig. S5F).
In the complex structure, plastid NAD-dependent MDH existed as a dimer (Supplemental Fig. S6A). In chloroplasts, redox-regulated NADP-dependent MDH is also present (Yokochi et al. 2021). Structure superposition of MDH with NADP-dependent MDH (PDB: 1CIV) and NAD-dependent MDH (PDB: 5ZI2) over the Cα atoms resulted in root mean square deviation values of 4.47 and 1.285, respectively (Supplemental Fig. S6, B and C), suggesting structural divergence between NAD- and NADP-dependent MDH. The 2 MDH protomers shared a similar spatial architecture (Supplemental Fig. S6D). The catalytic residues were conserved, and they consisted of the charged residues D115, D231, R162, R168, R234, and H258 coordinating the NAD cofactor and malate substrate (Supplemental Fig. S6E). Replacing Residues R162 and R234 with alanine completely blocked enzymatic activity (Supplemental Fig. S6F).
The LSF1 N terminus mediates ternary complex formation
LSF1 consists of the PDZ, DSP, and CBM. Close inspection of the structure of LSF1 revealed the presence of an N-terminal segment containing the PDZ domain, and the absence of the C-terminal segment containing both DSP and CBM. The LSF1 N terminus contained 8 β-strands and 6 α-helices (Fig. 2, A and B). A compact globular-fold PDZ domain was composed of 5 β-strands (β1β2β3β4β5) and 2 α-helices (α1α2), which was well aligned with the canonical PDZ domain (Fig. 2C). Interestingly, we identified a previously unknown region consisting of 3 β-sheets (β6 to β8), 4 α-helices (α3 to α6), and 5 loops (L1 to L5) that follow and wrap around the globular PDZ domain (Fig. 2, A and B). Since this region played crucial roles in mediating the interaction between LSF1 and BAM1 or MDH, we termed it the BAM and MDH Interaction (BMI) domain. Furthermore, we predicted the structure of LSF1 (Residues 71 to 591) containing all 4 functional domains using AlphaFold2 implemented in the ColabFold notebooks v1.5.2 running on Google Colaboratory (Mirdita et al. 2022). We selected the rank_1 model exhibiting the highest predicted local distance difference test (pLDDT) value for subsequent analyses (Supplemental Table S2). Structure alignment of experimentally determined LSF1PDZ-BMI with the LSF1 structure predicted by AlphaFold2 revealed the identity of the PDZ domain, but with a slight difference in the BMI domain (Supplemental Fig. S7, A and B), indicating the relative flexibility of the BMI domain. Helix α6 was unique, connecting the N-terminal PDZ-BMI and the C-terminal DSP-CBM, and α6 was preceded and followed by long loops, demonstrating the relative flexibility of segments between the N and C terminus of LSF1 (Supplemental Fig. S7, A and B).
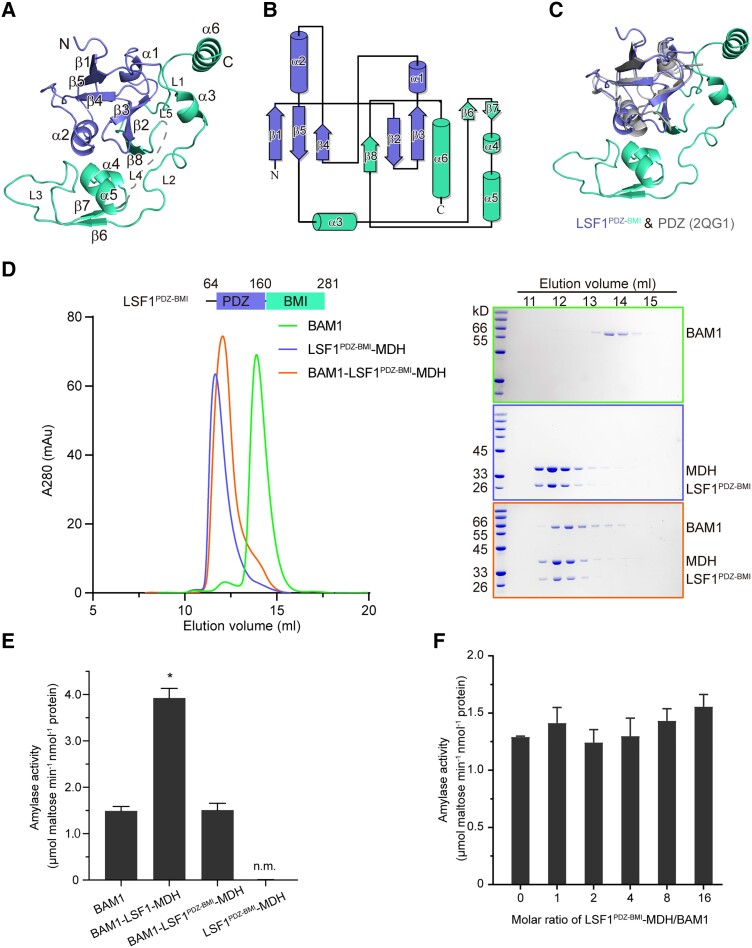
Figure 2.
The LSF1 N terminus is sufficient for ternary complex assembly. A) Structure of the LSF1 N terminus. Secondary structural elements are labeled. The PDZ and BMI domains are colored in slate and green-cyan, respectively. B) Topological model of the LSF1 N terminus. The number of secondary structural elements are labeled. C) Structural superposition of the LSF1 N terminus with the canonical PDZ domain. The structure of 2QG1 is shown in gray. D) The N-terminal PDZ-BMI domain of LSF1 is sufficient to mediate ternary complex formation. Left, a representative gel filtration chromatography; right, SDS-PAGE corresponding to the chromatography. E) Starch hydrolytic activity examined for the BAM1–LSF1PDZ-BMI–MDH complex. n.m., no measured activity. Two-tailed Student's t-tests are used for statistical analysis (*P < 0.05). Error bar, Sd. F) Titration assay of the starch-degradation activity of BAM1 upon the addition of the binary LSF1PDZ-BMI–MDH complex with increased molar ratios. LSF1PDZ-BMI–MDH is unable to promote the starch hydrolytic activity of BAM1. Two-tailed Student's t-tests are used for statistical analysis (*P < 0.05). Error bar, Sd.
To test whether the N-terminal PDZ-BMI segment is sufficient to mediate ternary complex assembly, we generated an LSF1PDZ-BMI truncation containing the N terminus (Residues 64 to 281), and tested its interactions with MDH and BAM1. We observed the formation of a stable binary LSF1PDZ-BMI–MDH complex, and its coincubation with BAM1 led to the formation of a BAM1–LSF1PDZ-BMI–MDH complex (Fig. 2D). These results suggest that the PDZ-BMI of LSF1 is sufficient to mediate formation of the ternary complex.
Further, we asked whether the binary complex LSF1PDZ-BMI–MDH without the DSP-CBM domain was able to promote the starch-hydrolysis activity of BAM1, and examined the starch degradation activity of the BAM1–LSF1PDZ-BMI–MDH complex. We detected no significant improvement in starch degradation activity for BAM1 in the presence of LSF1PDZ-BMI–MDH (Fig. 2E). Further titration experiments revealed that the increase in molar ratio of LSF1PDZ-BMI–MDH to BAM1 or BAM3 has no clear effect on the starch degradation activity of BAM1 and of BAM3 (Fig. 2F, Supplemental Fig. S3B). Thus, the absence of DSP-CBM of LSF1 deprives BAM1 and BAM3 from the abilities to improve their starch hydrolysis activity, suggesting the essential role of DSP-CBM in starch degradation.
A gate-latch-lock recognition mode between LSF1 and MDH
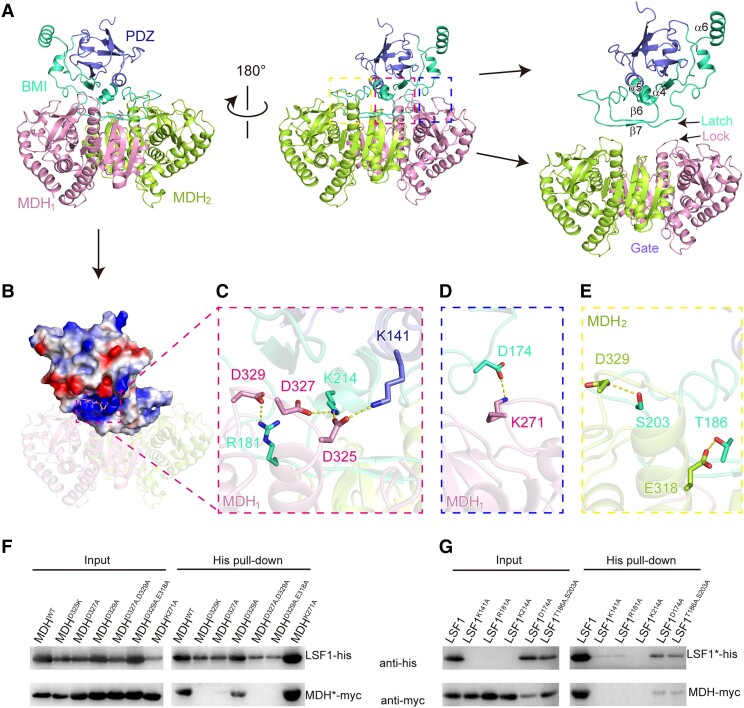
Both protomers of MDH made contact with LSF1, with a buried surface area of 1,247 Å2 for MDH1 and 900 Å2 for MDH2. LSF1 interacted with MDH mainly via the BMI domain (Fig. 3A). The long loops and antiparallel β-sheets β6β7 from the BMI domain of LSF1 extended over the surface of the dimeric MDH-like belts (Fig. 3A). Electrostatic potential analysis revealed that a positively charged pocket is composed of residues mainly from the BMI domain of LSF1 (Fig. 3B). This pocket could accommodate negatively charged residues. Indeed, the 3 acidic aspartates D325, D327, and D329 of MDH were embedded in a positively charged pocket (Fig. 3C, Supplemental Fig. S8A). Salt bridges were formed by the residue pairs D325 (MDH1)–K214 (or K141, LSF1), D327–K214, and D329–R181 (Fig. 3C). In addition, a salt bridge was formed between D174 (LSF1) and K271 (MDH1) (Fig. 3D, Supplemental Fig. S8B), and hydrogen bond networks were formed between residues T186 (LSF1) and E318 (MDH2), between S203 (LSF1) and D329 (MDH2) (Fig. 3E, Supplemental Fig. S8C).
Figure 3.
A gate-latch-lock recognition mode between LSF1 and MDH. A) The interaction surface between LSF1 and dimeric MDH. The PDZ and BMI of LSF1, and the protomers of MDH are shown in slate, green-cyan, pink and lemon, respectively. In the right panel, the location of the gate (dimeric MDH), latch (the sheets β6β7 and its preceding and following loops in LSF1), and lock (a loop) are individually indicated. B) Electrostatic potential analysis of the interaction surface. The positively charged pocket of LSF1 is labeled by a dashed rectangle. The negatively charged residues from MDH are shown in sticks. C) Residues involved in the gate-latch-lock recognition. The interaction residue pairs are shown in sticks and indicated by dashed lines. D) A salt bridge interaction between LSF1 D174 and MDH1 K271. E) Hydrogen-bonded interactions between LSF1 and MDH2. The interaction residue pairs are shown in sticks and indicated by dashed lines. F and G) Validation of critical residues involved in the LSF1–MDH interaction by coexpression and immunoblot analysis. LSF1 and MDH were constructed with His and myc tag, respectively. For each lane, the relative wild-type or mutant constructs are shown above the gel. The star symbol on LSF1 or MDH indicates the relative mutants corresponding to each lane labeled above the gel. Mutations in MDH F) or LSF1 G).
To identify the key residues that are involved in the interaction between LSF1 and MDH, we conducted in vivo coexpression assays using His-tagged LSF1 and myc-tagged MDH, followed by nickel-affinity purification, and immunoblot analysis. Our initial investigation revealed that the introduction of MDH improves the folding and stability of LSF1 by promoting formation of the LSF1–MDH complex (Fig. 1C). Based on this observation, we speculated that a mutation that disrupts the LSF1–MDH interaction might affect the soluble expression and stability of LSF1. We thus performed point mutations of both MDH and LSF1. We validated the thermal stability of all mutant proteins by nano-differential scanning fluorimetry (nanoDSF) assay (Supplemental Fig. S9). We determined that the mutation D325K or D327A of MDH disrupts the LSF1–MDH interaction (Fig. 3F). Although the D329A mutation had little effect, the D329A, E318A double mutation abolished the interaction between LSF1 and MDH (Fig. 3F). K141A, R181A, and K214A mutations affected the soluble production of LSF1 and its ability to interact with MDH (Fig. 3G), suggesting the critical roles of these positively charged residues. However, the LSF1–MDH interaction was not affected by mutations in the residues involved in the salt-bridge interaction D174 (LSF1)–K271 (MDH) and the hydrogen bonding network of T186 (LSF1)–E318 (MDH2), and S203 (LSF1)–D329 (MDH2), suggesting the little contribution of these residues to binary complex formation (Fig. 3, F and G). Based on the above results, we propose a gate-latch-lock recognition model to clarify the interaction between LSF1 and MDH. Specifically, dimeric MDH functions as a gate, and the long belt in the BMI domain of LSF1 resembles a latch. Three adjacent aspartate residues (D325, D327, and D329) in the loop of MDH insert between the gate and the latch to lock LSF1 in a favorable conformation (Fig. 3A), thus improving the solubility and stability of LSF1.
A previous study demonstrated that enzymatically inactive MDH can complement the embryo death phenotype of the pdnad-mdh mutant, suggesting a nonenzymatic function for MDH in chloroplast biogenesis (Schreier et al. 2018). To examine whether enzymatically inactive MDH retained the ability to interact with LSF1 to further promote the starch hydrolysis activity of BAM1, we coexpressed the inactive mutant MDHR162A with LSF1. Like wild-type MDH, mutant MDHR162A also formed a LSF1–MDHR162A binary complex and BAM1–LSF1–MDHR162A ternary complex (Supplemental Fig. S10A). The BAM1–LSF1–MDHR162A ternary complex exhibited a starch degradation capacity comparable with that of the wild-type BAM1–LSF1–MDH complex, but a higher capacity than that of BAM1 alone (Supplemental Fig. S10B). These results provide solid evidence that MDH plays a nonenzymatic role in chloroplast starch degradation.
The interaction between LSF1 and BAM1
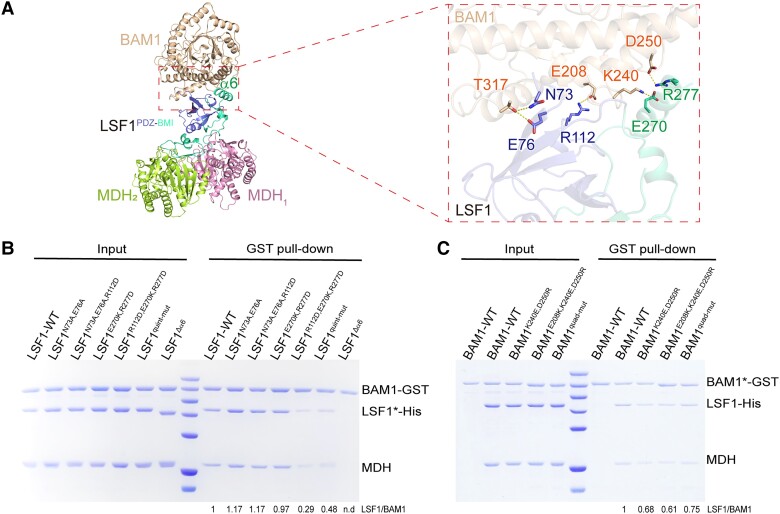
In the BAM1–LSF1–MDH complex structure, both the PDZ and BMI domains of LSF1 were involved in the interaction with BAM1 (Fig. 4A). To test which key residues are involved in this interaction, we performed in vitro glutathione S-transferase (GST) pull-down assays with proteins containing point mutations within BAM1 or LSF1. Specifically, we purified BAM1 with a GST tag, whereas LSF1 was coexpressed with MDH to obtain the binary complex LSF1–MDH. We incubated GST-tagged BAM1 with LSF1–MDH, incubating wild-type BAM1 with LSF1 for use as a positive control.
Figure 4.
Interaction between BAM1 and LSF1. A) The interaction surface between BAM1 and LSF1. BAM1 is shown in beige, PDZ and BMI are colored in slate and green-cyan respectively. The unique α6 is labeled. Key residues involved in the interaction are shown in sticks on the right panel. B and C) Critical residues involved in the BAM1–LSF1 interaction validated by in vitro GST pull-down assay by mutations in either LSF1 B) or BAM1 C). Relative mutants are shown on top of the gel, and the star symbols on LSF* or BAM1* represent relative mutants corresponding to each gel lane. The wild-type binary His-LSF1–MDH or mutant binary His-LSF1*–MDH complexes were obtained by coproduction, and incubated with GST-BAM1. LSF1 quint-mut indicates the quintuple mutant combination N73A, E76A, R112D, E270K, R277D. BAM1 quad-mut indicates the quadruple mutant combination E208K, K240E, D250R, T317A. The band intensities were estimated by ImageJ bundled with 64-bit Java 8. The lane of the wild-type BAM1 and LSF1 was set to 1; the intensities for other lanes of BAM1* and LSF1 were calculated relative to the wild type. The ratio of LSF1/BAM1* for each is calculated and provided below the gel.
In the BMI domain, helix α6 (Residues 268 to 280) of LSF1 was in close proximity to BAM1, and residues E270 and R277 residing in α6 formed salt bridges with residues K240 and D250 of BAM1, respectively (Fig. 4A, Supplemental Fig. S8, D and E). Deletion of α6 had no effect on the formation of the LSF1Δα6–MDH binary complex, but it abolished LSF1Δα6–BAM1 interaction (Fig. 4B), suggesting the essential role of α6 in the LSF1–BAM1 interaction. The double mutation LSF1E270K,R277D had little effect on the binding affinity between BAM1 and LSF1E270K,R277D (Fig. 4B, Supplemental Fig. S8E). In the PDZ domain, residues mediated both salt bridge interaction and hydrogen bond interactions. For example, a salt bridge was formed between LSF1 R112 and BAM1 E208. Residues N73 and E76 of LSF1 formed hydrogen bonds with T317 of BAM1 (Fig. 4A). We performed point mutations in both LSF1 and BAM1. We validated the thermal stability of the LSF1–MDH mutant complex and BAM1 by nanoDSF assay (Supplemental Fig. S9, B and C). The triple mutant variant N73A, E76A, R112D of LSF1 had no effects on the interaction between LSF1 and BAM1. The above results indicate that mutations in either BMI or PDZ do not disrupt the LSF1–BAM1 interaction. We conducted a combined mutation in both PDZ and BMI domains. The triple mutant variant R112D, E270D, R277D, and the quintuple mutant variant N73A, E76A, R112D, E270D, R277D of LSF1 greatly decreased binding affinity between LSF1 and BAM1, which was ∼29% and 48% of the intact proteins, respectively (Fig. 4B), suggesting synergetic action of PDZ and BMI in mediating the interaction between LSF1 and BAM1.
We also generated double, triple, and quadruple mutant variants of BAM1, namely BAM1K240E,D250R, BAM1E208,K240E,D250R, and BAM1E208,K240E,D250R,T317A. GST pull-down results revealed that these mutations do not disrupt the LSF1–BAM1 interactions. These mutants exhibited slightly decreased binding affinity between BAM1 and LSF1, which was ∼68%, 61%, and 75% of intact BAM1 (Fig. 4C). We speculated that there might be a potential interaction interface between BAM1 and LSF1DSP-CBM, in addition to the structurally observed interaction interface between BAM1 and LSF1PDZ-BMI. To test this idea, we performed a pull-down assay using an LSF1 truncation lacking the C-terminal DSP-CBM segment. The above BAM1 mutations disrupted the interactions between BAM1 and LSF1PDZ-BMI–MDH (Supplemental Fig. S11), suggesting the contribution of the DSP-CBM domain to the LSF1–BAM1 interaction. We corroborated this speculation by in vitro crosslinking CXMS assay.
Structural modeling of the BAM1–LSF1–MDH complex
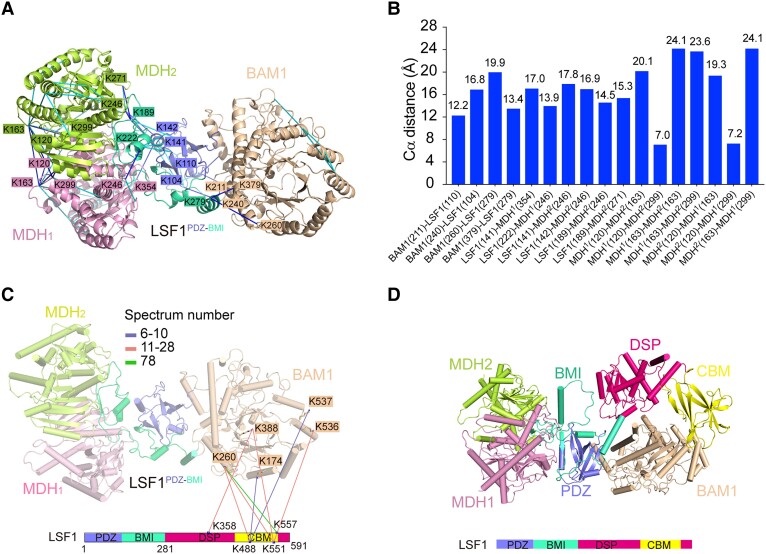
LSF1 contains 4 functional domains, namely the PDZ, BMI, DSP, and CBM domains. Although we used near-full-length LSF171-591 for ternary complex assembly and structural determination, the densities of DSP and CBM were missing. To locate the relative position of DSP-CBM in the ternary complex, we conducted in vitro CXMS characterization and modeling of the complete structure of BAM1–LSF1–MDH on the basis of the solved structure. Through in vitro CXMS, we identified 165 crosslinks (72 unique intersubunit and 93 intrasubunit crosslinks) with a false discovery rate (FDR) <5% and spectrum number ≥6 (Supplemental Fig. S12, Table S3, Supplemental Data Sets S1 and S2). The complete CXMS data were deposited with the ProteomeXchange Consortium (ID: PXD043466). The representative primary data of the MS2 spectra were exemplified by the crosslinked residue pairs (Supplemental Fig. S13). Structure determination and gel filtration results showed that the LSF1 N-terminal PDZ-BMI fragment is sufficient to mediate ternary complex assembly (Figs. 1F, 2D). We identified a total of 10 crosslinked pairs between LSF1PDZ-BMI and BAM1 (or MDH) (Fig. 5, A and B), including 4 pairs between LSF1PDZ-BMI and BAM1, namely, K110 (LSF1)–K211 (BAM1), K104–K240, K279–K260, and K279–K379, and 6 pairs between LSF1PDZ-BMI and MDH, namely, K141–K354 (MDH1), K222–K246 (MDH1), K141–K246 (MDH2), K142–K246 (MDH2), K189–K246 (MDH2), and K189–K271 (MDH2). These crosslinking results are consistent with the complex structure resolution results, confirming the reliability of the CXMS data.
Figure 5.
Structural modeling of the complete BAM1–LSF1–MDH complex on the basis of CXMS data and AlphaFold2 prediction. A) Crosslinks mapped onto the structure of BAM1–LSF1–MDH complex. The blue and cyan lines indicate intersubunit and intrasubunit crosslinked residue pairs, respectively. For clarity, residues involved in the intersubunit interaction are shown, whereas those involved in the intrasubunit interaction are omitted. Residues are colored consistent with the corresponding subunits. B) The distance distribution of all crosslinked residue pairs mapped onto the structure of BAM1–LSF1–MDH complex. The 26 Å cutoff value was used to filter the crosslinks, well below the 35 Å upper limit. C) The intersubunit crosslinks between LSF1DSP-CBM and BAM1. The DSP and CBM domains of LSF1 are shown in hot pink, and yellow, respectively. The identified crosslink pairs are indicated by colored lines based on the spectrum number. For clarity, the LSF1PDZ-BMI-mediated crosslinked residue pairs are omitted. D) Structure model of the complete BAM1–LSF1–MDH complex based on CXMS data and AlphaFold2 predictions. BAM1, MDH1, and MDH2 are colored in beige, pink, and lemon, respectively. The LSF1 domains PDZ, BMI, DSP, and CBM are shown in slate, green-cyan, hot pink, and yellow, respectively. The DSP-CBM of LSF1 was docked in close proximity to BAM1 based on CXMS data.
In addition, we identified 8 crosslinked pairs between LSF1DSP-CBM and BAM1: K358 (LSF1)–K388 (BAM1), K488–K174, K488–K260, K488–537, K551–K260, K551–K388, K551–K536, and K557–K260 (Fig. 5D, Supplemental Table S3). However, we identified no crosslink pairs between LSF1DSP-CBM and MDH. These results suggest that LSF1DSP-CBM is close to BAM1. To localize the DSP-CBM domain of LSF1 relative to BAM1, we performed molecular docking of BAM1 with LSF171-591 by HADDOCK with CXMS data to provide distance restraints. The structure of BAM1 was obtained by the resolution of the ternary complex. The structure of LSF171-591 was predicted using AlphaFold2 (Supplemental Table S2). Of the top 5 clusters from the docking model, we selected Cluster 2 with the highest HADDOCK score and lowest Z-score for subsequent analysis (Supplemental Fig. S14A, Table S4). In addition, we calculated the Cα-Cα distance of the crosslinked residue pairs between BAM1 and LSF1DSP-CBM to evaluate the reliability of the docked complex. The results showed that the distance within each residue pair is below 35 Å (Supplemental Fig. S14B), which is consistent with the CXMS data (Supplemental Table S3), indicating the reliability of the docked complex. In the complex model, the DSP-CBM of LSF1 was in close proximity to BAM1 (Supplemental Fig. S14A). The docked BAM1–LSF171-591 was aligned to the resolved LSF171-280–MDH complex (Supplemental Fig. S14C), to obtain a comprehensive view of the whole BAM1–LSF171-591–MDH complex (Fig. 5D).
Molecular insights into the improvement of starch hydrolysis mediated by the BAM1–LSF1–MDH complex
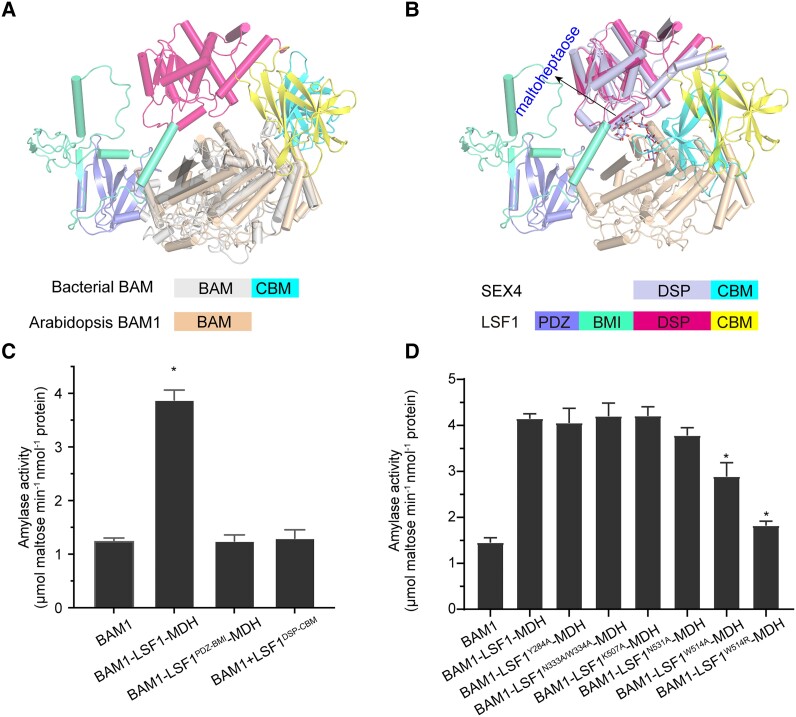
β-Amylases are widely distributed in bacteria and plants. Bacterial β-amylases contain a C-terminal starch-binding domain that was lost in plant BAMs during evolution (Vajravijayan et al. 2018). We performed structural alignment of bacterial BAM with the BAM1–LSF1–MDH complex, revealing a good match between the (β/a)8-barrel of the BAM proteins, whereas the CBM of bacterial BAM was generally aligned to the CBM of LSF1 in a similar position relative to the BAM core barrel (Fig. 6A). Thus, in the BAM1–LSF1–MDH complex, BAM1 employed the CBM from LSF1, resembling bacterial BAM.
Figure 6.
The presence of the C-terminal DSP-CBM of LSF1 is essential for the increased starch degradation of the BAM1–LSF1–MDH complex. A) Structure alignment of bacterial BAM with MDH–LSF1-bound BAM1. For clarity, the structures of MDH are omitted. The bacterial BAM core and CBM are shown in gray and cyan, respectively. Arabidopsis BAM1 is shown in beige. The PDZ, BMI, DSP, and CBM domains of LSF1 are shown in slate, green-cyan, hot pink, and yellow, respectively. B) Structure alignment of maltoheptaose-bound SEX4 with BAM1- and MDH-bound LSF1. For clarity, the structures of MDH are omitted. Maltoheptaose is shown in sticks and indicated. The DSP and CBM segments of SEX4 are shown in light blue and cyan, respectively. The PDZ, BMI, DSP, and CBM segments of LSF1 are shown in slate, green-cyan, hot pink, and yellow, respectively. C) The essential role for both PDZ-BMI and DSP-CBM of LSF1 on the improved starch-hydrolysis activity of BAM1. D) The effect of mutations of the C-terminal DSP and CBM of LSF1 on the starch degradation activity of the BAM1–LSF1–MDH mutant complex. All data reported here were obtained from 3 independent experiments and presented as mean ± Sd of triplicate experiments. Significant differences were determined using 2-tailed Student's t-test (*P < 0.05).
Of the 3 glucan phosphatases in chloroplasts, LSF1 and SEX4 contain DSP-CBM, which exhibit sequence conservation (Supplemental Fig. S15). A previous study reported the crystal structure of maltoheptaose-bound SEX4, revealing a continuous maltoheptaose-binding pocket spanning both DSP and CBM (Meekins et al. 2014). To uncover the potential pathway for maltopolysaccharide binding to the BAM1–LSF1–MDH complex, we performed a structural superposition of maltoheptaose-bound SEX4 with the complete BAM1–LSF1–MDH structure. We observed a good alignment of DSP and CBM, and the close proximity of the maltoheptaose to the gate of BAM1 (β/a)8-barrel, which facilitates BAM1 access to the polyglucan chain (Fig. 6B). This result explains why the presence of DSP-CBM in LSF1 increased the starch degradation efficiency of BAM1 (Fig. 6C). Moreover, the presence of the N-terminal PDZ-BMI of LSF1 was also essential due to its ability to recruit β-amylase (Fig. 6C).
Two tryptophan residues W479 and W514 are located in the CBM of LSF1, and the LSF1W479,W514A double mutant was reported to exhibit a starch-excess phenotype (Schreier et al. 2019), suggesting the critical roles of these residues in starch degradation. To further examine their effect on the starch hydrolysis activity of BAM1, we constructed point mutations at the residues W479 and W514 of LSF1, replacing them with alanine, and attempted to assemble the mutant ternary complex. Through coexpression, we successfully obtained the binary complex LSF1W514A (or LSF1W514R)–MDH, but failed to obtain the binary complex LSF1W479A (or LSF1W479R)–MDH, which might be due to protein instability induced by the W479 mutation (Supplemental Fig. S16A). We then assembled the mutant ternary complex and investigated its starch hydrolysis activity (Supplemental Fig. S16B). We established that the mutant complex BAM1–LSF1W514AMDH exhibits significantly lower starch hydrolysis activity than the intact ternary complex, while the BAM1–LSF1W514R–MDH complex showed even lower catalytic activity, which was comparable with that of BAM1 alone (Fig. 6D). In addition, we generated mutants in other residues of the CBM (K507A, N531A) and the DSP (Y284A, N333A/W334A) of LSF1 (Supplemental Fig. S16, A and B), since these residues were relatively conserved, compared with the corresponding residues that coordinated glucans in SEX4 (Supplemental Fig. S15). Compared with the wild type, the BAM1–LSF1N531A–MDH exhibited slightly decreased activity, whereas the remaining mutant complexes retained normal catalytic activity (Fig. 6D). These results further confirm the critical roles of residue W514 in LSF1 CBM in starch degradation.
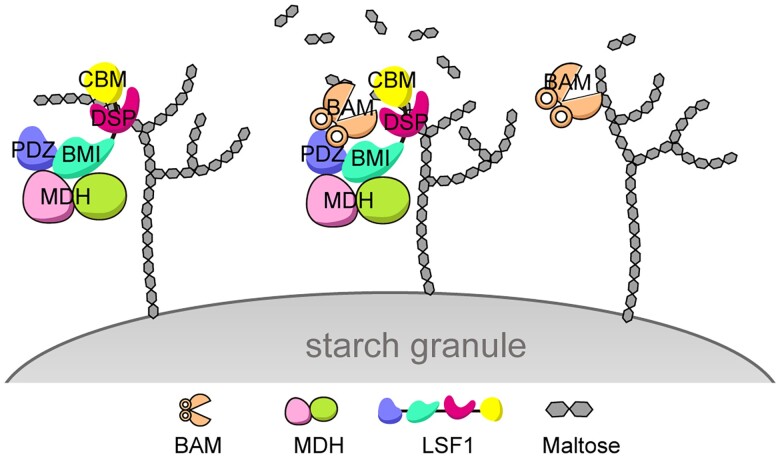
On the basis of these findings, we propose a working model to clarify the role of the BAM–LSF1–MDH complex in starch degradation. MDH plays a nonenzymatic role and acts as a chaperone to interact with and stabilize LSF1 to form a complex, illustrating the moonlighting role of MDH in chloroplast metabolism. The binary complex LSF1–MDH functions as a scaffold to bind polyglucans and anchor to the starch granule via the C-terminal DSP-CBM of LSF1. The N-terminal PDZ-BMI of LSF1 is responsible for the LSF1-MDH interaction to recruit β-amylase in proximity to the polyglucan-binding DSP-CBM, thus facilitating the access of β-amylase to polyglucans, eventually promoting starch degradation (Fig. 7).
Figure 7.
A proposed working model for the ternary BAM–LSF1–MDH complex in starch degradation. Left, LSF1 in complex with MDH functions as a scaffold anchored to the starch granule. Right, BAM1 (or BAM3) alone possesses weak starch hydrolysis activity. Middle panel, BAM1 (or BAM3)–LSF1–MDH exhibits strikingly increased starch degradation activity, generating more maltose products. Specifically, the N-terminal PDZ-BMI domain of LSF1 interacts with MDH and recruits BAM, whereas the C-terminal DSP-CBM segment of LSF1 binds to the polyglucan chain, anchors to the starch granule, and presents the polyglucan to the catalytic pocket of BAM, thus promoting starch degradation.
Overall, this study uncovers the moonlighting role of MDH and the functional divergence of the N- and C-terminal segments of glucan phosphatase-inactive LSF1, providing important insights into the precise regulation of starch degradation orchestrated by the BAM–LSF1–MDH ternary complex on the starch granule.
Discussion
Enzymatic activities of chloroplast β-amylases are under diverse regulations
In this study, we uncovered the molecular mechanism by which LSF1–MDH promotes the starch-degradation activity of BAM1 and BAM3. Among the chloroplast β-amylase members, BAM3 is essential for starch degradation, with BAM1 contributing in the absence of BAM3 or under abiotic stress. The bam3 mutant exhibits a severe starch-excess phenotype, whereas the bam1 bam3 double mutant displays an even more severe starch-excess phenotype (Fulton et al. 2008). Attenuated starch degradation impairs normal plant growth and development. The growth rate of bam1 bam3 plants is severely slower than that of wild type (David et al. 2022). In this study, the structural investigation of the BAM1–LSF1–MDH complex provides solid and direct biochemical evidence related to these phenotypes. Our data showed that both BAM1 and BAM3 possessed the ability to interact with LSF1–MDH to form a ternary complex, hence promoting starch degradation and providing sufficient maltose products to sustain plant normal growth and metabolism. However, in the absence of BAM1 and BAM3, starch breakdown would be severely hindered, thus leading to the accumulation of starch. As a result, plant growth would slow down due to the lack of supply for these metabolic precursors.
The reliance on a CBM from LSF1 by BAM1 or BAM3 is reminiscent of its bacterial homolog, which possesses an extra CBM itself (Vajravijayan et al. 2018). The presence of a CBM provides BAM with access to the polyglucans of the starch granule to promote starch hydrolysis (Fig. 6D). A similar strategy was observed in starch biosynthesis. The catalytically inactive PROTEIN TARGETING TO STARCH (PTST) proteins contain coiled-coil domains and a CBM. PTST1 and PTST2 bind the starch synthases GRANULE-BOUND STARCH SYNTHASE (GBSS) and STARCH SYNTHASE4 (SS4), respectively, via their coiled-coil domains and deliver these enzymes to starch granules through their CBM to facilitate starch granule synthesis (Seung et al. 2015, 2017; Zhang et al. 2022). This finding reveals a general mechanism by which CBM is provided in trans to starch hydrolase or synthase in starch metabolism.
LSF1 functions as a scaffold to recruit β-amylase in starch degradation
In plants, the transient phosphorylation of starch renders the granule surface more accessible to glucan hydrolase, which is required for starch degradation. The reversible removal of phosphate by a glucan phosphatase is also essential for starch breakdown. There are 3 glucan phosphatases in Arabidopsis chloroplasts, namely LSF1, LSF2, and SEX4. The lsf2 mutant displays normal starch levels (Santelia et al. 2011), whereas the mutants lacking either lsf1 or sex4 exhibit a starch-excess phenotype (Kotting et al. 2009; Santelia et al. 2011; Schreier et al. 2019). Both SEX4 and LSF2 are enzymatically active and able to dephosphorylate the glucans to provide access to amylases, whereas LSF1 is a catalytically inactive glucan phosphatase and unable to dephosphorylate the glucans, suggesting a different role for LSF1 from its counterparts LSF2 and SEX4. Unlike LSF2 and SEX4, LSF1 contains a unique PDZ domain followed by an unstructured region prior to the DSP domain, suggesting that LSF1 might participate in protein–protein interactions. Recent studies have indicated that LSF1 might form protein complexes with the major active β-amylases, either BAM1 or BAM3 (Schreier et al. 2019; Feike et al. 2022). Thus, delineation of the composition of the super-complex and deciphering the molecular basis involving LSF1 and BAM are an open question in starch degradation. In this study, we reconstituted the BAM1 (or BAM3)–LSF1–MDH starch degradation complex in vitro, and showed that the ternary complex exhibited increased starch degradation activity compared with BAM1 or BAM3 alone (Fig. 1, Supplemental Fig. S3). Furthermore, our determination of the cryo-EM structure of BAM1–LSF1–MDH (Fig. 1) provided direct evidence for the long-sought issue on the LSF1-mediated starch degradation complex. In the absence of LSF1, BAM proteins exhibit decreased starch degradation activity, providing direct evidence for the starch-excess phenotype of the lsf1 single mutant (Comparot-Moss et al. 2010; Schreier et al. 2019). The bam1 lsf1 and bam3 lsf1 double mutants exhibit comparable or only slightly elevated starch levels relative to the lsf1 single mutant (Schreier et al. 2019), since BAM1 and BAM3 individually retained starch degradation activity even in the absence of LSF1 (Fig. 1, Supplemental Fig. S3). Moreover, this study uncovers the scaffold function of LSF1, which relies on its association with β-amylases BAM1 or BAM3 in starch degradation, explaining why the bam1 bam3 lsf1 triple mutant exhibits comparable starch levels with the bam1 bam3 double mutant, and the bam1 bam3 mutant accumulates more starch than the lsf1 mutant (Schreier et al. 2019).
LSF1 contains 4 functional domains: the N-terminal PDZ-BMI and C-terminal DSP-CBM. Among these domains, the N-terminal PDZ-BMI is unique in LSF1. We identified the N-terminal PDZ-BMI of LSF1 from the structure of the BAM1–LSF1–MDH complex, and found that it mediated ternary complex assembly, revealing the molecular function for this unique segment (Fig. 2). The BMI domain of LSF1 was thought to be an unstructured region with unknown function (Silver et al. 2014). In this study, we discovered that the BMI played an important role in mediating the interaction between LSF1 and BAM1 (or MDH). The PDZ domain of LSF1 is essential for starch breakdown. The starch-excess phenotype of the lsf1 mutant was rescued by the expression of the wild-type LSF1, but the construct LSF1ΔPDZ encoding a variant of LSF1 without the PDZ domain was unable to rescue the starch-excess phenotype (Schreier et al. 2019), suggesting the role of the PDZ domain in starch degradation. Our in vitro biochemical characterization validated this in vivo finding on the function of the PDZ domain.
The C-terminal DSP-CBM of LSF1 is structurally unresolved. Our CXMS assay revealed that the DSP-CBM segment was in close proximity to the catalytic pocket of BAM1. A similar DSP-CBM segment is present in the paralogous SEX4 (Supplemental Fig. S14), and both DSP and CBM domains coordinate glucans (Meekins et al. 2014). The CBM is known to bind carbohydrates. The 2 conserved tryptophan W479 and W514 in the CBM of LSF1 are involved in starch binding, and plants expressing the LSF1W479,W514A mutant exhibit a starch-excess phenotype (Schreier et al. 2019). Here, we reconstituted the BAM1–LSF1W514A (or W514R)–MDH complex, and these 2 complexes exhibited a strikingly lower starch degradation activity than the wild-type complex, validating the critical role of the CBM and residue W514 in starch hydrolysis. All 3 glucan phosphatases members SEX4, LSF1, and LSF2 contain the DSP domain with the catalytic signature of the HCX5R motif. Both SEX4 and LSF2 possess phosphatase activity, whereas LSF1 is enzymatically inactive. Crystal structures of glucan-bound LSF2 and SEX4 revealed the molecular mechanism of how they dephosphorylated glucans at specific positions (Meekins et al. 2013, 2014). In the cryo-EM structure, the densities of DSP-CBM are missing, suggesting the flexibility of these segments relative to the N-terminal PDZ-BMI. Further structural determination of the glucan-bound DSP-CBM of LSF1 or BAM–LSF1–MDH will provide the explanation as to why LSF1 is an inactive glucan phosphatase, which will benefit its potential bioengineering into an active phosphatase.
Starch degradation involves phosphorylation, dephosphorylation, and hydrolysis. The glucan phosphatases LSF2 and SEX4 function in dephosphorylation, whereas LSF1 acts as a scaffold to recruit β-amylase, thus promoting starch degradation in hydrolysis. This study provides new insights into their different roles in starch degradation. It will be of great significance to bioengineer them into a glucan phosphatase-active LSF1* or a chimeric SEX4* containing the PDZ-BMI domain, and to reconstitute an LSF1*(or SEX4*)–BAM–MDH complex possessing both glucan phosphatase activity and starch-hydrolysis activity. It will be of interest to explore the phenotypes of the associated bioengineered plants.
MDH plays a moonlighting role in chloroplast biogenesis
MDH reversibly converts malate to oxaloacetate with NAD(H) or NADP(H) as a cofactor. The mutants lacking pdNAD-MDH are embryo-lethal. Constitutive silencing of pdNAD-MDH (miR-MDH-1) causes a pale and dwarfed phenotype, whereas the inducible silencing of pdNAD-MDH at the rosette stage results in white emerging leaves (Beeler et al. 2014). Interestingly, the expression of enzymatically inactive variants encoding pdNAD-MDH R162Q and R234Q fully complements the pdnad-mdh mutant (Schreier et al. 2018). These findings suggest that pdNAD-MDH might play an essential moonlighting role in plastid development and plant growth. A previous immunoprecipitation experiment of the pdNAD-MDH had identified the FtsH protease as the interaction partner (Schreier et al. 2018). FtsH proteases are transmembrane AAA-type proteases that are associated with the chloroplast protein import machinery in the inner envelope. The biochemical role of pdNAD-MDH in FtsH complex assembly and chloroplast import awaits further investigation.
The pdNAD-MDH coimmunoprecipitation assay indicated that in addition to the FtsH protease members, the proteins involved in the starch degradation including LSF1, BAM1, and BAM3 are also identified as interacting partners. In this study, we found that pdNAD-MDH interacted with LSF1 to improve its solubility and stability in vitro by forming the LSF1–MDH complex (Fig. 1, Supplemental Fig. S1). In pdNAD-MDH-silenced plants, the accumulation of the FtsH12 protein is greatly decreased (Schreier et al. 2018). These findings jointly suggest that the pdNAD-MDH may stabilize the interaction partners. In this study, we showed that the LSF1–MDH binary complex recruited the β-amylase to form a super-complex, thus promoting starch degradation. The enzymatically inactive variant MDHR162A retained the ability to interact with LSF1 to form the BAM1–LSF1–MDHR162A complex, and this mutant complex exhibited comparable starch degradation activity with that of the wild-type complex. Overall, this study uncovers one of the nonenzymatic roles of MDH in starch degradation, and it is perhaps just a beginning to reveal the moonlighting role of plastid NAD-dependent MDH in chloroplast biogenesis.
Materials and methods
Molecular cloning, protein expression, and purification
The coding sequences of BAM1 (At3g23920), BAM3 (At4g17090), LSF1 (At3g01510), and MDH (At3g47520) from Arabidopsis thaliana (Col-0) were amplified and cloned into a modified pET15D vector that adds a 6× His tag at the N terminus of the encoded protein. The point mutants were constructed by overlapping PCR. All constructs were verified by Sanger sequencing. All plasmids were transformed into E. coli strain BL21 (DE3). One liter of LB medium containing 100 mg mL−1 ampicillin was inoculated with a transformed bacterial pre-culture and shaken at 37 °C until the cell density reached an OD600 of ∼1.0 to 1.2. Protein production was induced with 0.2 mm isopropyl-β-d-thiogalactopyranoside at 16 °C for 14 to 16 h. The cells were collected by centrifugation, homogenized in buffer A (25 mm Tris-HCl, pH 8.0, 150 mm NaCl), and lysed using a high-pressure cell disrupter (JNBIO, China). Cell debris was removed by centrifugation at 20,000 × g for 1 h at 4 °C, and the supernatant was loaded onto a column equipped with Ni2+ affinity resin (Ni-NTA, Qiagen), washed with buffer B (25 mm Tris-HCl, pH 8.0, 150 mm NaCl, 15 mm imidazole), and eluted with buffer C (25 mm Tris-HCl, pH 8.0, 250 mm imidazole). Each protein was then separated by cation exchange chromatography (Source 15Q, GE Healthcare) using a linear NaCl gradient in buffer A. The purified protein was concentrated and subjected to gel filtration chromatography (Superdex-200 Increase 10/300, GE Healthcare) in a buffer containing 25 mm Tris-HCl, pH 8.0 and 150 mm NaCl. Purity of the proteins was examined using SDS-PAGE and visualized by Coomassie brilliant blue staining through all purification steps. The peak fractions were collected and stored at −80 °C. The mutant proteins were purified similarly as the wild-type proteins. For protein coexpression, LSF1 was cloned into the pET21 vector adding an 8× His tag at the C terminus of the encoded protein, whereas MDH was cloned into the pBB75 vector with no tag. The 2 plasmids were cotransformed into E. coli BL21 (DE3). Protein production and purification of the LSF1–MDH complex were performed similarly as described above. Primers used for cloning are provided in Supplemental Data Set S3.
Size exclusion chromatograph assays
To examine the interactions between BAM and LSF1–MDH, the purified recombinant LSF1–MDH complex was mixed with BAM at a molar ratio of ∼1:1.2, incubated on ice for 30 min, and further purified by gel filtration chromatography (Superdex 200 increase, GE Healthcare) in a buffer containing 25 mm Tris-HCl, pH 8.0, and 150 mm NaCl. The relevant fractions of each independent injection were collected for SDS-PAGE analysis. The peak fractions were subjected for subsequent biochemical assays and cryo-EM grid preparation.
In vitro pull-down assays
BAM1 interacts with LSF1–MDH to form BAM1–LSF–MDH complex. To validate the interaction interface between BAM1 and LSF1, GST-tagged BAM1 was used to pull down His-tagged LSF1–MDH. For the binary LSF1–MDH complex, His-tagged LSF1 and untagged MDH were coexpressed- in E. coli BL21 (DE3). The mutants of LSF1 were all obtained by coexpression with wild-type MDH. GST-BAM1 and its mutants were produced in E. coli BL21 (DE3). The proteins of GST-BAM1 and His-LSF1–MDH were mixed in lysis buffer (25 mm Tris-HCl, pH 8.0, 150 mm NaCl) and incubated with glutathione agarose beads at 4 °C for 30 min. The GST-resins were washed with lysis buffer and eluted by elution buffer containing 25 mm Tris-HCl, pH 9.0, 150 mm NaCl and 10 mm GSSH. The input and eluent were subjected to SDS-PAGE followed by Coomassie brilliant blue staining.
Enzymatic assays
Activities of β-amylase was examined by the detection of the reducing sugar products using a dinitrosalicylic acid (DNS) assay. Briefly, the reaction was conducted in a mixture (100 µL) containing 50 mm MES, pH 6.5, 0.01 µm BAMs and 5 mg mL−1 soluble starch. After incubation at 30 °C for 15 min, reactions were stopped by adding 100 µL DNS buffer (182 mg mL−1 potassium sodium tartrate, 6.3 mg mL−1 DNS, 0.5 m NaOH, 5 mg mL−1 phenol) and boiling for 5 min at 98 °C. The mixture was then cooled to 25 °C and centrifuged at 14,000 × g for 2 min at room temperature. Absorbance of the supernatant was recorded at 540 nm. Maltose was used to draw a standard curve.
The phosphatase activity of LSF1 was monitored by detecting the hydrolysis of p-nitrophenyl phosphate (pNPP). Various amounts of purified protein were incubated with pNPP buffer (100 mm HEPES-NaOH, pH 7.0; 150 mm NaCl, 1 mm EDTA) and 10 mm DTT at room temperature for 20 min before the reaction. The mixture was incubated with 4 mm pNPP for 30 min at 30 °C and neutralized with an equal volume of 2 m NaOH. Absorbance at 405 nm was recorded.
MDH activity was measured by following NADH oxidation to NAD+ at 340 nm. The assay was carried out in a reaction mixture (250 µL) containing 25 mm Tris-HCl, pH 8.0, 150 mm NaCl, 10 mm MgCl2, 0.4 mm NADH and 0.02 μm MDH. The reaction was initiated by the addition of 5 mm oxaloacetic acid (OAA). OAA is reduced to malic acid and was spectrophotometrically assayed in a Spark multimode microplate reader (Tecan). The baseline rate at 340 nm was acquired for 5 min at 20 °C, measuring absorbance every 30 s. The decline in absorbance at 340 nm was recorded. Activity was determined with 3 replicates for the same protein sample to calculate the mean value.
Grid preparation and data acquisition
Aliquots (3.5 μL) of the BAM1–LSF1–MDH (1.2 mg mL−1) complex were dropped onto glow-discharged Quantifoil R1.2/1.3 300 mesh Cu grids (Quantifoil, MicroTools GmbH, Germany), blotted with a Vitrobot Mark IV (Themo Fisher Scientific) using 3 s blotting time with 100% humidity at 8 °C, and plunged into liquid ethane cooled by liquid nitrogen. The sample was imaged on an FEI Titan Krios transmission electron microscope at 300 kV with a magnification of 130,000×. Images were recorded by a Gatan K3 Summit direct electron detector using the counting mode. Defocus values varied from 1.2 to 2.2 μm. Each image was dose-fractionated to 32 to 42 frames with a total electron dose of 50 to 60 e− Å−2 and a total exposure time of 8.0 s. EPU was used for fully automated data collection (Mastronarde 2005). All stacks were motion corrected using MotionCor2 with a binning factor of 1, resulting in a pixel size of 1.07 Å and dose weighting was performed concurrently (Zheng et al. 2017). The defocus values were estimated using Gctf (Zhang 2016).
Data processing, model building, and refinement
A total of 1,845 good micrographs were selected, from which 2,684,442 particles were auto-picked using RELION (Kimanius et al. 2016). After reference-free 2D classification, 2,634,327 good particles were selected for 3D classification. Multireference 3D classification was performed in RELION. Then, a total of 305,907 particles were selected from good classes and transferred to the cryoSPARC software package for further processing (Punjani et al. 2017), followed by several rounds of ab-initio reconstruction, heterogeneous refinement. Particles belonging to the best class were selected followed by nonuniform refinement and local refinement, applying C2 symmetry, yielding a particle density with an estimated resolution of 3.0 Å based on FSC (Chen et al. 2013). The atomic model was built in COOT (Emsley and Cowtan 2004) and refined with PHENIX (Adams et al. 2010). The structure was validated through the examination of Molprobity scores (Davis et al. 2007). Resolution was estimated with the gold standard Fourier shell correlation 0.143 criterion. High-resolution images were prepared using PyMOL. Data collection and refinement statistics are given in Supplemental Table S1. A diagram of the procedures for data processing is presented in Supplemental Fig. S3.
In vitro crosslinking
For crosslinking experiments, the BAM1–LSF1–MDH complex was purified on a Superdex-200 Increase 10/300 GL column equilibrated with the buffer containing 25 mm HEPES, pH 7.5 and 300 mm NaCl. The crosslinking agent bis-(sulfosuccinimidyl) suberate (BS3, from Thermo Fisher Scientific) was prepared at a concentration of 100 mm in DMSO. BS3 was added to the BAM1–LSF1–MDH complex (∼4 µm) at a molar ratio of 100:1, 200:1, and 400:1, respectively. The reaction was performed at 37 °C for 30 min and then quenched by the addition of 20 mm Tris-HCl (pH 7.5). The experiments were replicated with 3 parallel samples and each sample with 2 technical repeats.
Prediction of LSF1 structure by AlphaFold2
The structure model of LSF171-591 was predicted using the AlphaFold2 implementation in the ColabFold notebooks v1.5.2 running on Google Colaboratory (Jumper et al. 2021; Mirdita et al. 2022), using default settings with Amber relaxation (num_relax = 5, template_mode = pdb100, msa_mode = mmseqs2_uniref_env, num_recycles = 3, pair_mode = unpaired_paired, model_type = alphafold2_ptm, max_msa = auto, recycle_early_stop_tolerance = 0, pairing_strategy = greedy, mnum_seeds = 1). AlphaFold computes the pLDDT score and the predicted template modeling (pTM) score to indicate the accuracy of a prediction. A predicted alignment error (PAE) map between pairs of residues was also calculated for each prediction, which represents confidence in domain positioning. Confidence metrics (pLDDT score, pTM score, PAE maps, per-residue pLDDT, and pLDDT per position) of predictions made in this work are provided in Supplemental Table S2.
Mass spectrometry and data analysis
The crosslinked protein sample was freeze-dried to remove the supernatant. The sample was dissolved in 8 m urea, reduced with 2.5 mm DTT at 37 °C for 30 min, and alkylated with 5 mm iodoacetamide in dark for 30 min. Subsequently, 3 volumes of Tris-HCl (pH 8.5) were added to dilute the sample. Trypsin digestion was carried out at 37 °C overnight with sequencing grade modified trypsin (Promega, mass ratio = 1:50). The reaction was quenched with trifluoroacetic acid to a final concentration of 1.2% (w/v).
Trypsin-digested peptides were purified with C18 Spin Tips (Thermo Fisher Scientific) and were analyzed in a Q Exactive HF Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled to an EASY-nLC 1200 liquid chromatography system, with a 75 μm, 15-cm Acclaim PepMap RSLC column. The peptides were eluted over a 75 min linear gradient from 95% buffer A (water with 0.1% [v/v] formic acid) to 35% buffer B (acetonitrile with 0.1% [v/v] formic acid) with a flow rate of 300 nL/min. Each full MS scan (resolution = 120,000) was followed by 15 data-dependent MS2 (resolution = 15,000), with a stepped normalized collision energy of 10, 25, and 30. The isolation window was set to 1.6 m/z. Precursors of charge states 3 to 6 were collected for MS2 scans. Dynamic exclusion window was set to 40 s.
The crosslinking data were analyzed by pLink2 (Yang et al. 2012; Chen et al. 2019).The following search parameters were used: MS1 accuracy = ±20 ppm; MS2 accuracy = ±20 ppm; enzyme = trypsin (with full tryptic specificity but allowing up to 3 missed cleavages); crosslinker = BS3 (with an assumed reaction specificity for lysine and protein N-termini); fixed modifications = carbamidomethylation on cysteine; variable modifications = oxidation on methionine, hydrolyzed/aminolyzed BS3 from reaction with ammonia or water on a free crosslinker end. The identified candidates were filtered based on the following parameters: FDR <5%, support vector machine score <10−2, identified in at least 2 biological repeat experiments and with total peptide-spectrum matches (PSMs) ≥ 6. The experimental crosslinks were visualized with Crosslink-viewer (Graham et al. 2019). Visualization of the crosslinks on the structures used PyMOL with the PyXlinkViewer plug-in (Schiffrin et al. 2020). The CXMS data are provided in Supplemental Data Sets S1 and S2 and Supplemental Table S3.
CXMS-guided protein–protein docking and complex modeling
CXMS-guided protein–protein docking was carried out using the Expert interface of the HADDOCK server (version 2.4; de Vries et al. 2010). The obtained structure of BAM1 from the BAM1–LSF171-281–MDH ternary complex and the structure of LSF171-591 predicted by AlphaFold were set as input models for molecular docking. Residues M78, E270, R277, and R112 of LSF1, and residues S309, K240, D250, and E208 of BAM1 were set as active residues directly involved in the interaction between the 2 models according to the structure of the BAM1–LSF1–MDH ternary complex. Distance restraints (Orban-Nemeth et al. 2018) with ranges of 0 to 35 Å (Cα–Cα) were applied to the BAM1–LSF1 crosslinked pairs K260–K557, K260–K488, K305–K110, K240–K104, K388–K358, K388–K551, K260–K279, K260–K551, K3536–K551, K379–K279, K174–K488, K211–K110 and K537–K488. Center-of-mass restraints were enabled and other parameters were set as default in HADDOCK. The molecular docking results are provided in Supplemental Table S4.
Statistical analysis
All data were analyzed using a 2-tailed Student's t-test with software Prism9.0. The values are represented as mean ± Sd. Statistical data are provided as Supplemental Data Set S4.
Accession numbers
Sequence data in this article can be found in the Arabidopsis Information Resource or GenBank/EMBL databases under the following accession numbers: BAM1 (At3g23920), BAM3 (At4g17090), LSF1 (At3g01510), LSF2 (At3g10940), SEX4 (At3g52180), MDH (At3g47520), and FtsH12 (At1g79560).
Supplementary Material
Acknowledgments
The authors thank the cryo-EM Facility at the University of Science and Technology of China, Public Laboratory of Electron Microscopy and Center for Protein Research (CPR) of Huazhong Agricultural University, for providing technical support during sample preparation and EM image acquisition. This work was supported by funds from the National Natural Science Foundation of China (32270255 and 32130011), the Foundation of Hubei Hongshan Laboratory (2021hskf003 and 2021hszd011), Fundamental Research Funds for the Central Universities (2662023PY001), and the Open Research Fund of State Key Laboratory of Hybrid Rice (KF202103, Wuhan University).
Contributor Information
Jian Liu, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Xuecui Wang, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Zeyuan Guan, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Menglong Wu, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Xinyue Wang, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Rong Fan, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Fei Zhang, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Junjun Yan, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Yanjun Liu, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Delin Zhang, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Ping Yin, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Junjie Yan, National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.
Author contributions
Junjie Y.: designed and supervised the project. J.L. and X.W.: assembled the complex and performed the biochemical assays. M.W., X.W., R.F., and F.Z.: purified the proteins. Z.G.: acquired EM data, built and refined the atomic model. J.L. and Junjun Y.: performed CXMS. Junjie Y.: wrote the manuscript with all authors contributing to the data analysis.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Fig. S1. Protein purification.
Supplemental Fig. S2. Enzymatic activity of LSF1 and MDH.
Supplemental Fig. S3. LSF1–MDH interacts with and promotes the starch degradation activity of BAM3.
Supplemental Fig. S4. Cryo-EM workflow for structural determination of the ternary BAM–LSF1–MDH complex.
Supplemental Fig. S5. Structure analysis of BAM1.
Supplemental Fig. S6. Structure analysis of MDH.
Supplemental Fig. S7. Structure of LSF1.
Supplemental Fig. S8. EM density map for residues involved in the interaction.
Supplemental Fig. S9. Thermal shift assay performed by nanoDSF.
Supplemental Fig. S10. The enzymatic activity of MDH is dispensable for the complex assembly and the enhanced starch degradation activity of BAM1–LSF1–MDH.
Supplemental Fig. S11. The C-terminal DSP-CBM of LSF1 contributes to the interaction with BAM1.
Supplemental Fig. S12. The in vitro CXMS assay.
Supplemental Fig. S13. Examples of MS2 spectra of the crosslinked residue pairs.
Supplemental Fig. S14. Structural modeling of the BAM1–LSF1–MDH complex.
Supplemental Fig. S15. Sequence alignment of LSF1.
Supplemental Fig. S16. The mutant complex assembly.
Supplemental Table S1. Cryo-EM data collection and refinement statistics.
Supplemental Table S2. Statistics of AlphaFold-predicted LSF171–591.
Supplemental Table S3. Inter-molecular crosslinked residue pairs.
Supplemental Table S4. The top 5 clusters of docked BAM1–LSF1 complex.
Supplemental Data Set S1. The crosslinked residue pairs identified by CXMS.
Supplemental Data Set S2. The intra-molecular crosslinked residue pairs.
Supplemental Data Set S3. Primers used for cloning.
Supplemental Data Set S4. Results of statistical analysis.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (Chen et al. 2022) with the dataset identifier PXD043466. The EM density map has been deposited in the EM Data Bank (EMDB: 35985) and the Protein Data Bank (PDB: 8J5D). The material and data that support the findings of this study are available from the corresponding authors upon request.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010:66(2):213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baunsgaard L, Lutken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J. 2005:41(4):595–605. 10.1111/j.1365-313X.2004.02322.x [DOI] [PubMed] [Google Scholar]
- Beeler S, Liu HC, Stadler M, Schreier T, Eicke S, Lue WL, Truernit E, Zeeman SC, Chen J, Kotting O. Plastidial NAD-dependent malate dehydrogenase is critical for embryo development and heterotrophic metabolism in Arabidopsis. Plant Physiol. 2014:164(3):1175–1190. 10.1104/pp.113.233866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SH, Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013:135:24–35. 10.1016/j.ultramic.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ma J, Liu Y, Chen Z, Xiao N, Lu Y, Fu Y, Yang C, Li M, Wu S, et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res. 2022:50(D1):D1522–D1527. 10.1093/nar/gkab1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZL, Meng JM, Cao Y, Yin JL, Fang RQ, Fan SB, Liu C, Zeng WF, Ding YH, Tan D, et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat Commun. 2019:10(1):3404. 10.1038/s41467-019-11337-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comparot-Moss S, Kotting O, Stettler M, Edner C, Graf A, Weise SE, Streb S, Lue WL, MacLean D, Mahlow S, et al. A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol. 2010:152(2):685–697. 10.1104/pp.109.148981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao O, Kuhnert F, Weber APM, Peltier G, Li-Beisson Y. Physiological functions of malate shuttles in plants and algae. Trends Plant Sci. 2022:27(5):488–501. 10.1016/j.tplants.2021.11.007 [DOI] [PubMed] [Google Scholar]
- David LC, Lee SK, Bruderer E, Abt MR, Fischer-Stettler M, Tschopp MA, Solhaug EM, Sanchez K, Zeeman SC. BETA-AMYLASE9 is a plastidial nonenzymatic regulator of leaf starch degradation. Plant Physiol. 2022:188(1):191–207. 10.1093/plphys/kiab468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007:35(Web Server):W375–W383. 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010:5(5):883–897. 10.1038/nprot.2010.32 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004:60(12):2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Feike D, Pike M, Gurrieri L, Graf A, Smith AM. A dominant mutation in beta-AMYLASE1 disrupts nighttime control of starch degradation in Arabidopsis leaves. Plant Physiol. 2022:188(4):1979–1992. 10.1093/plphys/kiab603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flutsch S, Wang Y, Takemiya A, Vialet-Chabrand SRM, Klejchova M, Nigro A, Hills A, Lawson T, Blatt MR, Santelia D. Guard cell starch degradation yields glucose for rapid stomatal opening in Arabidopsis. Plant Cell. 2020:32(7):2325–2344. 10.1105/tpc.18.00802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco P, Gil M, Reinhold H, Eicke S, Messerli G, et al. Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell. 2008:20(4):1040–1058. 10.1105/tpc.107.056507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry MS, Brewer MK, Vander Kooi CW. Structural biology of glucan phosphatases from humans to plants. Curr Opin Struct Biol. 2016:40:62–69. 10.1016/j.sbi.2016.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M, Combe CW, Kolbowski L, Rappsilber J. xiView: a common platform for the downstream analysis of Crosslinking Mass Spectrometry data. bioRxiv 561829. 10.1101/561829, 26 February 2019, preprint: not peer reviewed. [DOI]
- Hejazi M, Fettke J, Kotting O, Zeeman SC, Steup M. The Laforin-like dual-specificity phosphatase SEX4 from Arabidopsis hydrolyzes both C6- and C3-phosphate esters introduced by starch-related dikinases and thereby affects phase transition of alpha-glucans. Plant Physiol. 2010:152(2):711–722. 10.1104/pp.109.149914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer G, Wieser S, Bogdos MK, Gattinger P, Nakamura R, Ebisawa M, Makela M, Papadopoulos N, Valenta R, Keller W. Three-dimensional structure of the wheat beta-amylase Tri a 17, a clinically relevant food allergen. Allergy. 2019:74(5):1009–1013. 10.1111/all.13696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrer D, Flutsch S, Pazmino D, Matthews JS, Thalmann M, Nigro A, Leonhardt N, Lawson T, Santelia D. Blue light induces a distinct starch degradation pathway in guard cells for stomatal opening. Curr Biol. 2016:26(3):362–370. 10.1016/j.cub.2015.12.036 [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021:596(7873):583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YN, Tanabe A, Adachi M, Utsumi S, Mikami B. Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism. Biochemistry. 2005:44(13):5106–5116. 10.1021/bi0476580 [DOI] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016:5:e18722. 10.7554/eLife.18722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G. Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 2005:137(1):242–252. 10.1104/pp.104.055954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M, et al. STARCH-EXCESS4 is a laforin-like Phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell. 2009:21(1):334–346. 10.1105/tpc.108.064360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005:152(1):36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Meekins DA, Guo HF, Husodo S, Paasch BC, Bridges TM, Santelia D, Kotting O, Vander Kooi CW, Gentry MS. Structure of the Arabidopsis glucan phosphatase like sex four2 reveals a unique mechanism for starch dephosphorylation. Plant Cell. 2013:25(6):2302–2314. 10.1105/tpc.113.112706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekins DA, Raththagala M, Husodo S, White CJ, Guo HF, Kotting O, Vander Kooi CW, Gentry MS. Phosphoglucan-bound structure of starch phosphatase Starch Excess4 reveals the mechanism for C6 specificity. Proc Natl Acad Sci U S A. 2014:111(20):7272–7277. 10.1073/pnas.1400757111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami B, Adachi M, Kage T, Sarikaya E, Nanmori T, Shinke R, Utsumi S. Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose. Biochemistry. 1999:38(22):7050–7061. 10.1021/bi9829377 [DOI] [PubMed] [Google Scholar]
- Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022:19(6):679–682. 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JD. Involvement of five catalytically active Arabidopsis beta-amylases in leaf starch metabolism and plant growth. Plant Direct. 2020:4(2):e00199. 10.1002/pld3.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JD, Breault JS, Pope LE, Torres CE, Gebrejesus TB, Berndsen CE, Storm AR. Arabidopsis beta-amylase2 is a K(+)-requiring, catalytic tetramer with sigmoidal kinetics. Plant Physiol. 2017:175(4):1525–1535. 10.1104/pp.17.01506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JD, Storm AR. Review: the Arabidopsis beta-amylase (BAM) gene family: diversity of form and function. Plant Sci. 2018:276:163–170. 10.1016/j.plantsci.2018.08.016 [DOI] [PubMed] [Google Scholar]
- Monroe JD, Storm AR, Badley EM, Lehman MD, Platt SM, Saunders LK, Schmitz JM, Torres CE. beta-Amylase1 and beta-amylase3 are plastidic starch hydrolases in Arabidopsis That Seem to Be Adapted for Different Thermal, pH, and Stress Conditions. Plant Physiol. 2014:166(4):1748–1763. 10.1104/pp.114.246421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke F, Ahonen SJ, Nitschke S, Mitra S, Minassian BA. Lafora disease—from pathogenesis to treatment strategies. Nat Rev Neurol. 2018:14(10):606–617. 10.1038/s41582-018-0057-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke F, Wang P, Schmieder P, Girard JM, Awrey DE, Wang T, Israelian J, Zhao X, Turnbull J, Heydenreich M, et al. Hyperphosphorylation of glucosyl C6 carbons and altered structure of glycogen in the neurodegenerative epilepsy Lafora disease. Cell Metab. 2013:17(5):756–767. 10.1016/j.cmet.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Orban-Nemeth Z, Beveridge R, Hollenstein DM, Rampler E, Stranzl T, Hudecz O, Doblmann J, Schlogelhofer P, Mechtler K. Structural prediction of protein models using distance restraints derived from cross-linking mass spectrometry data. Nat Protoc. 2018:13(3):478–494. 10.1038/nprot.2017.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017:14(3):290–296. 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- Raththagala M, Brewer MK, Parker MW, Sherwood AR, Wong BK, Hsu S, Bridges TM, Paasch BC, Hellman LM, Husodo S, et al. Structural mechanism of laforin function in glycogen dephosphorylation and lafora disease. Mol Cell. 2015:57(2):261–272. 10.1016/j.molcel.2014.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejzek M, Stevenson CE, Southard AM, Stanley D, Denyer K, Smith AM, Naldrett MJ, Lawson DM, Field RA. Chemical genetics and cereal starch metabolism: structural basis of the non-covalent and covalent inhibition of barley beta-amylase. Mol Biosyst. 2011:7(3):718–730. 10.1039/C0MB00204F [DOI] [PubMed] [Google Scholar]
- Ritte G, Heydenreich M, Mahlow S, Haebel S, Kotting O, Steup M. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 2006:580(20):4872–4876. 10.1016/j.febslet.2006.07.085 [DOI] [PubMed] [Google Scholar]
- Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M. The starch-related R1 protein is an alpha-glucan, water dikinase. Proc Natl Acad Sci U S A. 2002:99(10):7166–7171. 10.1073/pnas.062053099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelia D, Kotting O, Seung D, Schubert M, Thalmann M, Bischof S, Meekins DA, Lutz A, Patron N, Gentry MS, et al. The phosphoglucan phosphatase like sex Four2 dephosphorylates starch at the C3-position in Arabidopsis. Plant Cell. 2011:23(11):4096–4111. 10.1105/tpc.111.092155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffrin B, Radford SE, Brockwell DJ, Calabrese AN. PyXlinkViewer: a flexible tool for visualization of protein chemical crosslinking data within the PyMOL molecular graphics system. Protein Sci. 2020:29(8):1851–1857. 10.1002/pro.3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier TB, Clery A, Schlafli M, Galbier F, Stadler M, Demarsy E, Albertini D, Maier BA, Kessler F, Hortensteiner S, et al. Plastidial NAD-dependent malate dehydrogenase: a moonlighting protein involved in early chloroplast development through its interaction with an FtsH12-FtsHi protease complex. Plant Cell. 2018:30(8):1745–1769. 10.1105/tpc.18.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier TB, Umhang M, Lee SK, Lue WL, Shen Z, Silver D, Graf A, Muller A, Eicke S, Stadler-Waibel M, et al. LIKE SEX4 1 acts as a beta-amylase-binding scaffold on starch granules during starch degradation. Plant Cell. 2019:31(9):2169–2186. 10.1105/tpc.19.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinski J, Konig N, Wellmeyer B, Hanke GT, Linke V, Neuhaus HE, Scheibe R. The plastid-localized NAD-dependent malate dehydrogenase is crucial for energy homeostasis in developing Arabidopsis thaliana seeds. Mol Plant. 2014:7(1):170–186. 10.1093/mp/sst151 [DOI] [PubMed] [Google Scholar]
- Seung D, Boudet J, Monroe J, Schreier TB, David LC, Abt M, Lu KJ, Zanella M, Zeeman SC. Homologs of PROTEIN TARGETING TO STARCH control starch granule initiation in Arabidopsis leaves. Plant Cell. 2017:29(7):1657–1677. 10.1105/tpc.17.00222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D, Soyk S, Coiro M, Maier BA, Eicke S, Zeeman SC. PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol. 2015:13(2):e1002080. 10.1371/journal.pbio.1002080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DM, Kotting O, Moorhead GB. Phosphoglucan phosphatase function sheds light on starch degradation. Trends Plant Sci. 2014:19(7):471–478. 10.1016/j.tplants.2014.01.008 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC. Starch: a flexible, adaptable carbon store coupled to plant growth. Annu Rev Plant Biol. 2020:71(1):217–245. 10.1146/annurev-arplant-050718-100241 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM. Starch degradation. Annu Rev Plant Biol. 2005:56(1):73–98. 10.1146/annurev.arplant.56.032604.144257 [DOI] [PubMed] [Google Scholar]
- Sparla F, Costa A, Lo Schiavo F, Pupillo P, Trost P. Redox regulation of a novel plastid-targeted beta-amylase of Arabidopsis. Plant Physiol. 2006:141(3):840–850. 10.1104/pp.106.079186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Zeeman SC. Starch metabolism in Arabidopsis. Arabidopsis Book. 2012:10:e0160. 10.1199/tab.0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Palayam M, Shabek N. Structure of maize BZR1-type beta-amylase BAM8 provides new insights into its noncatalytic adaptation. J Struct Biol. 2022:214(3):107885. 10.1016/j.jsb.2022.107885 [DOI] [PubMed] [Google Scholar]
- Thalmann M, Pazmino D, Seung D, Horrer D, Nigro A, Meier T, Kolling K, Pfeifhofer HW, Zeeman SC, Santelia D. Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell. 2016:28(8):1860–1878. 10.1105/tpc.16.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann M, Santelia D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017:214(3):943–951. 10.1111/nph.14491 [DOI] [PubMed] [Google Scholar]
- Vajravijayan S, Pletnev S, Mani N, Pletneva N, Nandhagopal N, Gunasekaran K. Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes. Int J Biol Macromol. 2018:113:329–337. 10.1016/j.ijbiomac.2018.02.138 [DOI] [PubMed] [Google Scholar]
- Vander Kooi CW, Taylor AO, Pace RM, Meekins DA, Guo HF, Kim Y, Gentry MS. Structural basis for the glucan phosphatase activity of Starch Excess4. Proc Natl Acad Sci U S A. 2010:107(35):15379–15384. 10.1073/pnas.1009386107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, et al. Identification of cross-linked peptides from complex samples. Nat Methods. 2012:9(9):904–906. 10.1038/nmeth.2099 [DOI] [PubMed] [Google Scholar]
- Yokochi Y, Yoshida K, Hahn F, Miyagi A, Wakabayashi KI, Kawai-Yamada M, Weber APM, Hisabori T. Redox regulation of NADP-malate dehydrogenase is vital for land plants under fluctuating light environment. Proc Natl Acad Sci U S A. 2021:118(6):6. 10.1073/pnas.2016903118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, Kofler H, Hausler RE, Hille D, Flugge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, et al. The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell. 2001:13(8):1907–1918. 10.1105/TPC.010091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: real-time CTF determination and correction. J Struct Biol. 2016:193(1):1–12. 10.1016/j.jsb.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li N, Zhang J, Zhao L, Qiu J, Wei C. The CBM48 domain-containing protein FLO6 regulates starch synthesis by interacting with SSIVb and GBSS in rice. Plant Mol Biol. 2022:108(4–5):343–361. 10.1007/s11103-021-01178-0 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Luo L, Xu J, Xin P, Guo H, Wu J, Bai L, Wang G, Chu J, Zuo J, et al. Malate transported from chloroplast to mitochondrion triggers production of ROS and PCD in Arabidopsis thaliana. Cell Res. 2018:28(4):448–461. 10.1038/s41422-018-0024-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017:14(4):331–332. 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (Chen et al. 2022) with the dataset identifier PXD043466. The EM density map has been deposited in the EM Data Bank (EMDB: 35985) and the Protein Data Bank (PDB: 8J5D). The material and data that support the findings of this study are available from the corresponding authors upon request.