Abstract
The regulation of murein hydrolases is a critical aspect of peptidoglycan growth and metabolism. In the present study, we demonstrate that mutations within the Staphylococcus aureus virulence factor regulatory genes, agr and sar, affect autolysis, resulting in decreased and increased autolysis rates, respectively. Zymographic analyses of these mutant strains suggest that agr and sar exert their effects on autolysis, in part, by modulating murein hydrolase expression and/or activity.
Bacterial murein hydrolases have been shown to participate in a number of important biological processes occurring during cell growth and division, including cell wall synthesis, daughter cell separation, and peptidoglycan turnover and recycling (1, 6, 12, 17, 19). Because of their potential to destroy the cell wall, the expression and activity of these enzymes must be tightly controlled. Several studies indicate that murein hydrolase activity and autolysis are controlled at the transcriptional level (2, 3, 5–8, 16). In Bordetella pertussis, autolysis is regulated by the bvg virulence regulatory locus, resulting in the repression of autolysis during the phase shift from avirulence to virulence (6). Thus, in this system only the avirulent population of cells is susceptible to β-lactam antibiotics.
Autolysis assays.
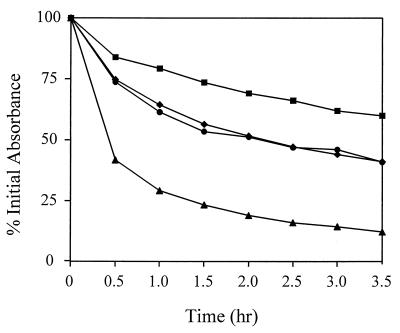
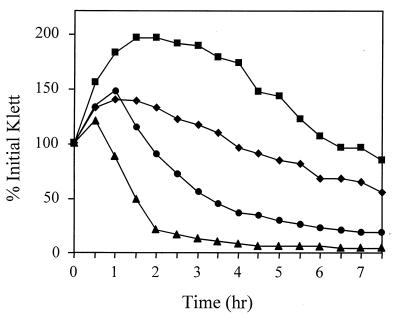
To examine the potential roles of agr and sar in the regulation of autolysis in Staphylococcus aureus, the Triton X-100- and penicillin-induced autolysis of agr, sar, and agr sar mutant strains was examined. To eliminate possible variations in genotype, strains used in this analysis were derived from the same parental strain, RN6390 (agr+ sar+). As shown in Fig. 1, the sar mutant ALC488 had a dramatically increased Triton X-100-induced autolysis rate (50% lysis in about 0.5 h) compared to the parent strain, RN6390 (50% lysis in 3.5 h). This autolysis rate was nearly identical to that exhibited by another sar mutant strain, ALC136 (data not shown). In contrast, the agr mutant, RN6911, exhibited a lower autolysis rate (50% lysis in approximately 5 h). The agr sar mutant, ALC135, exhibited a Triton X-100-induced autolysis rate that was intermediate (50% lysis in about 3.5 h) between those observed for the single-mutant strains. As shown in Fig. 2, the relative penicillin-induced autolysis rates for the respective mutants were similar to those induced by Triton X-100. The sar mutant ALC488 also exhibited a dramatic increase in penicillin-induced autolysis compared to the parental strain, RN6390. As with Triton X-100-induced autolysis, the agr mutant, RN6911, had a lower penicillin-induced autolysis rate than that of the parental strain and the rate for ALC135 was intermediate between those observed for the single-mutant strains.
FIG. 1.
Effect of agr and sar on Triton X-100-induced autolysis. S. aureus RN6390 (wild type) (diamonds), RN6911 (agr mutant) (squares), ALC488 (sar mutant) (triangles), and ALC135 (agr sar mutant) (circles) were grown to an optical density at 580 nm of 0.6 to 0.8. Triton X-100-induced autolysis assays were performed as described by Mani et al. (8) except that the cells were grown in NZY broth (3% NZ amine, 1% yeast extract). Triton X-100-induced autolysis was measured as the decline of optical density versus time and is expressed as the percent of the initial optical density. The data presented are representative of three independent experiments.
FIG. 2.
Effect of agr and sar on penicillin-induced autolysis. Penicillin-induced autolysis of S. aureus RN6390 (wild type) (diamonds), RN6911 (agr mutant) (squares), ALC488 (sar mutant) (triangles), and ALC135 (agr sar mutant) (circles) was measured with a Klett-Summerson colorimeter (filter no. 60; 1 Klett unit = 5 × 106 CFU/ml). Penicillin G (final concentration of 0.4 μg/ml) was added to early-exponential-phase cells (20 Klett units) in tryptic soy broth at 37°C, and the changes in culture turbidity were monitored over an 8-h period. Autolysis was measured as the decline in culture turbidity versus time and is expressed as the percent of the initial Klett reading when penicillin was added. The data presented are representative of three independent experiments.
Penicillin-induced killing.
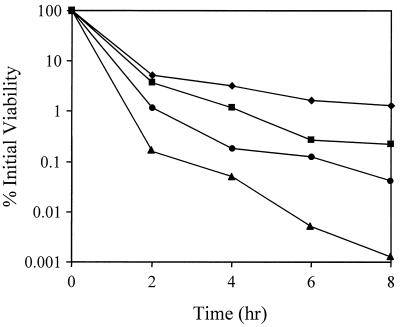
Recently, Piriz-Duran et al. (13) reported that agr and sar mutant strains exhibited reduced resistance to oxacillin, cefoxitin, and imipenem. The demonstration that these mutants also produced altered levels of penicillin-binding proteins led them to speculate that these changes may be responsible for the altered susceptibilities of these strains to β-lactam antibiotics. Initially, the results presented here in Fig. 2, which demonstrate that the agr mutant strain exhibited diminished penicillin-induced lysis compared to the parental strain, appeared to conflict with their data. However, a determination of the number of viable cells present revealed that the measurements of lysis did not accurately reflect cell viability and did not correlate with the observed changes in culture turbidity. In fact, the regulatory mutant strains were all more sensitive than the parental strain to penicillin-induced killing (Fig. 3), in agreement with the findings of Piriz-Duran et al. (13). The RN6911 culture viability 8 h after penicillin treatment was 6.5-fold lower than that of the RN6390 culture (0.2 and 1.3% viability, respectively). The ALC488 (sar mutant) and ALC135 (agr sar mutant) strains both exhibited dramatically increased sensitivity to the killing effects of penicillin (0.001 and 0.04% viability 8 h after penicillin treatment, respectively) compared to RN6390. These data indicate that agr and sar also affect the expression of factors, unrelated to murein hydrolases, that are involved in penicillin-induced killing. These factors could be analogous to the products of the hypothetical cid genes of Streptococcus pneumoniae which are required for the lysis-independent killing effects of penicillin (9).
FIG. 3.
Effect of agr and sar on penicillin-induced killing. Penicillin-induced killing of S. aureus RN6390 (wild type) (diamonds), RN6911 (agr mutant) (squares), ALC488 (sar mutant) (triangles), and ALC135 (agr sar mutant) (circles) was examined by performing viable cell counts every 2 h on the penicillin-treated cultures for which data are presented in Fig. 2. The viability of the cultures was assessed immediately prior to and every 2 h (for 8 h) after the addition of penicillin G by performing serial dilutions and spreading the cells on tryptic soy broth agar medium. The data presented are representative of three independent experiments.
Zymographic analysis.
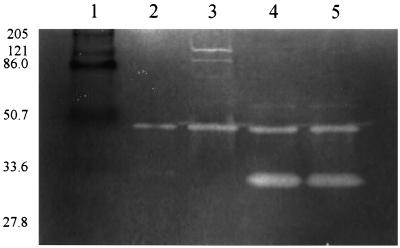
To determine the effects of the different regulatory mutations on the cell wall-associated murein hydrolase activity, the relative amounts and diversity of murein hydrolases produced by the different mutant strains were examined by zymography (Fig. 4). This analysis revealed dramatic differences in the cell wall-associated murein hydrolase profiles produced. Strains ALC488 (sar mutant) and ALC135 (agr sar mutant) produced high levels of a 32-kDa murein hydrolase (Fig. 4, lanes 4 and 5, respectively), unlike the parental strain, which produced barely detectable levels of this murein hydrolase (lane 2). In contrast, strain RN6911 (agr mutant) produced undetectable amounts of the 32-kDa murein hydrolase but produced increased levels of several high-molecular-weight (MW) (>75,000) murein hydrolases. These results suggest that the observed differences in autolysis of these strains might be attributable to changes in the expression of different murein hydrolases.
FIG. 4.
Zymographic analysis of S. aureus murein hydrolases. Cell wall-associated proteins (1.5 μg) were extracted from S. aureus cells grown to early exponential phase (optical density at 580 nm of 0.6 to 0.8) in NZY broth and analyzed as described by Qoronfleh and Wilkinson (14). Murein hydrolase activities were visualized as zones of hydrolysis by staining the gel with methylene blue. Lane 2, RN6390 (wild type); lane 3, RN6911 (agr mutant); lane 4, ALC488 (sar mutant); lane 5, ALC135 (agr sar mutant). The sizes of the prestained broad-range MW standards (Bio-Rad Laboratories, Hercules, Calif.) are given in kilodaltons (lane 1).
A gene that likely encodes the 32-kDa murein hydrolase has recently been identified by Ramadurai and Jayaswal (15). This gene, designated lytM, encodes a 34.4-kDa protein that shares features with a secreted protein. The processed protein was predicted to have a molecular mass of 32 kDa, suggesting that this protein is the same as that which we have observed in our zymographic analysis (Fig. 4). Thus, the observation that the sar mutant exhibited increased levels of the 32-kDa murein hydrolase suggests that the expression of the lytM gene, or a factor that affects the activity of this murein hydrolase, is negatively regulated by Sar. In contrast, the agr mutant strain was observed to have increased levels of high-MW murein hydrolase activities, proteins that have been proposed to be the precursors of the atl-encoded AM and GL murein hydrolases (4, 11). Oshida et al. (11) have demonstrated that the processing of the high-MW murein hydrolases to the mature AM and GL forms can be inhibited with protease inhibitors. Thus, one explanation for the observed increase in the levels of high-MW murein hydrolases in the agr mutant strain is that Agr is required for the expression of a protease(s) involved in the proteolytic processing of these atl-encoded murein hydrolases.
It should be noted that a previous study of factors affecting autolysis and murein hydrolase activity in S. aureus indicated that the agr virulence factor regulatory locus had no effect on these processes (18). Although it is not clear why different results were obtained by our laboratories, it is possible that the wild-type strain that was used in that study had acquired a spontaneous agr mutation, similar to that previously described by Novick et al. (10). The phenotypes of the strains used in our study include the exoprotein and cell wall-associated protein expression profiles that are known to be associated with the agr and sar mutations. The results of our studies, along with those of a study by Piriz-Duran et al. (13), indicate that the agr and sar regulatory loci have a significant effect on the susceptibility of S. aureus to β-lactam antibiotics. Thus, the bacteria may have evolved a regulatory strategy that functions to maximize their ability to evade host immune responses, while at the same time minimizing their intrinsic susceptibility to β-lactam antibiotics. Continued studies of the regulatory mechanisms that are involved in resistance to β-lactam antibiotics could lead to improved methods for the treatment of staphylococcal disease.
Acknowledgments
We thank Ambrose Cheung for providing the sar mutants, Scott Minnick for his careful reading of the manuscript, and Deborah Olson for her technical support.
This work was supported by USDA grant 9304112 and NIH grant R29AI38901.
REFERENCES
- 1.Archibald A R, Hancock I C, Harwood C R. Cell wall structure, synthesis, and turnover. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 381–410. [Google Scholar]
- 2.Brunskill E W, Bayles K W. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunskill E W, Bayles K W. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol. 1996;178:5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster S J. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J Bacteriol. 1995;177:5723–5725. doi: 10.1128/jb.177.19.5723-5725.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster S J. Analysis of the autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1992;174:464–470. doi: 10.1128/jb.174.2.464-470.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holtje J-V, Tuomanen E I. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J Gen Microbiol. 1991;137:441–454. doi: 10.1099/00221287-137-3-441. [DOI] [PubMed] [Google Scholar]
- 7.Kuroda A, Asami Y, Sekiguchi J. Molecular cloning of a sporulation-specific cell wall hydrolase gene of Bacillus subtilis. J Bacteriol. 1993;175:6260–6268. doi: 10.1128/jb.175.19.6260-6268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mani N, Tobin P, Jayaswal R K. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J Bacteriol. 1993;175:1493–1499. doi: 10.1128/jb.175.5.1493-1499.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreillon P, Markiewicz Z, Nachman S, Tomasz A. Two bactericidal targets for penicillin in pneumococci: autolysis-dependent and autolysis-independent killing mechanisms. Antimicrob Agents Chemother. 1990;34:33–39. doi: 10.1128/aac.34.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oshida T, Sugai M, Komatsuzawa H, Hong Y M, Suginaka H, Tomasz A. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc Natl Acad Sci USA. 1995;92:285–289. doi: 10.1073/pnas.92.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perkins H R. The bacterial autolysins. In: Rogers H J, Perkins H R, Ward J B, editors. Microbial cell walls. London, United Kingdom: Chapman and Hall; 1980. pp. 437–456. [Google Scholar]
- 13.Piriz-Duran S, Kayser F H, Berger-Bachi B. Impact of sar and agr on methicillin resistance in Staphylococcus aureus. FEMS Microbiol Lett. 1996;141:255–260. doi: 10.1111/j.1574-6968.1996.tb08394.x. [DOI] [PubMed] [Google Scholar]
- 14.Qoronfleh M W, Wilkinson B J. Effects of growth of methicillin-resistant and -susceptible Staphylococcus aureus in the presence of β-lactams on peptidoglycan structure and susceptibility to lytic enzymes. Antimicrob Agents Chemother. 1986;29:250–257. doi: 10.1128/aac.29.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramadurai L, Jayaswal R K. Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. J Bacteriol. 1997;179:3625–3631. doi: 10.1128/jb.179.11.3625-3631.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekiguchi J, Ezaki B, Kodama K, Akamatsu T. Molecular cloning of a gene affecting the autolysin level and flagellation in Bacillus subtilis. J Gen Microbiol. 1988;134:1611–1621. doi: 10.1099/00221287-134-6-1611. [DOI] [PubMed] [Google Scholar]
- 17.Shockman G D, Holtje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Vol. 27. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 131–166. [Google Scholar]
- 18.Tobin P J, Mani N, Jayaswal R K. Effect of physiological conditions on the autolysis of Staphylococcus aureus strains. Antonie Leeuwenhoek. 1994;65:71–78. doi: 10.1007/BF00878281. [DOI] [PubMed] [Google Scholar]
- 19.Ward J B, Williamson R. Bacterial autolysins: specificity and function. In: Nombela C, editor. Microbial cell wall synthesis and autolysis. Amsterdam, The Netherlands: Elsevier Science Publishers; 1985. pp. 159–166. [Google Scholar]