Abstract
We describe the presence of a minisatellite sequence that displays length polymorphisms in the fission yeast Schizosaccharomyces pombe. The minisatellite sequence was found to reside within the propeptide region of the vacuolar carboxypeptidase Y gene. The minisatellite sequence, which was found only at a single locus, was mitotically stable and displayed length polymorphisms between the two varieties of S. pombe (S. pombe var. pombe and S. pombe var. malidevorans). The minisatellite sequence, however, appeared to be species specific and was absent in other members of the Schizosaccharomyces genus. This report constitutes the first experimental demonstration of the presence of such sequences in yeasts.
Minisatellite sequences are found widely dispersed in the genomes of a variety of higher eukaryotic organisms. These minisatellite DNA sequences, also referred to as variable-number tandem repeats (VNTR), are comprised of multiple copies of repeats whose base units are in tens of nucleotides (9 to 60 bp) and are found within the genome in variable numbers. It is this variability which in fact forms the basis of DNA fingerprinting and other genomic analyses (5, 12). In yeasts, however, although length polymorphisms due to microsatellite DNA (repeat units of 1 to 6 bp) have been described (8, 16), length polymorphisms due to naturally occurring nonteleomeric minisatellite DNA have not been observed. In this report, we experimentally demonstrate the existence of such sequences in yeasts.
The vacuolar carboxypeptidase Y (CpY) gene (cpy1+) of Schizosaccharomyces pombe has been cloned independently by several groups. Tabuchi et al. reported a protein of 950 amino acids (accession no. D86560) (19). In contrast to this report however, a protein of 1,002 amino acids was observed from the sequence obtained at the Sanger Centre, Cambridge, United Kingdom, as part of the S. pombe genome sequencing project (accession no. D97209), as well as from our own laboratory, where we have cloned the gene by complementation of a cpy-deficient mutant (1).
The CpY protein of S. pombe is characterized by an unusually long propeptide (554 amino acids in contrast to 91 amino acids for that of Saccharomyces cerevisiae [20] and 106 amino acids for that of Candida albicans [11]) that contains two distinct repeat regions (Fig. 1). The first repeat unit is 13 amino acids long and is repeated 11 times (7 times in the sequence reported by Tabuchi et al. [19]), and the second repeat unit is 9 amino acids long and is repeated 8 times (Tabuchi et al. [19] have described this as seven repeats, opting not to refer to the eighth unit as one of the repeats; there are, however, no discrepancies in the sequences of this region). The repeat regions fail to display significant homology to anything in the GenBank and EMBL databases by comparison using the BLAST program (1a). When we examined the nucleotide sequences of the repeat regions we observed only a few differences, even at the nucleotide level, between the repeat units (Fig. 2). This suggested to us that we might be dealing with a minisatellite sequence. We therefore set about to determine whether these two repeat regions indeed corresponded to minisatellite DNA that could display variable numbers of tandem repeats and to examine whether this region was mitotically stable.
FIG. 1.
Schematic representation of the S. pombe CpY protein indicating the different regions. pCPY-K1 is a KpnI subclone that contains the cpy1+ open reading frame, and pCPY-ID6 is a derivative of pCPY-K1 that contains an in-frame deletion of six repeat units in RS I. The diagram has been drawn to scale (1.0 cm = 195 amino acids). Pre, prepeptide region; Pro, propeptide region.
FIG. 2.
Alignment of nucleotide sequences of the tandem repeat units of RS I and RS II that lie within the CpY propeptide region of S. pombe ABP13 h− ade6-210 ura4-D18 (obtained from Fission Yeast Course, Cold Spring Harbor, N.Y.). Primers P1 (5′-TTGATGACGAGCGTCCCAAGC-3′), P2 (5′-CCTTCATGCTCCTCAAGCTCATGG-3′), P3 (5′-CCATGAGCTTGAGGAGCATGAAGG-3′), and P4 (5′-TTGCATGTTTTGCTCAGGACGTTC-3′) used for PCR analysis of the repeat regions are also indicated schematically. The code represents the code for each of the repeat units and indicates a specific sequence. The BstXI sites at which in the in-frame deletion pCPY-ID6 was constructed are underlined.
Minisatellite analysis was carried out by PCR on different S. pombe strains using primers flanking the repeat regions (Fig. 2). We initially used S. pombe strains obtained from different laboratories, all of which have their ancestral origin in the isolate of S. pombe that was first investigated by Leupold (9).
When PCR was carried out with primers P1 and P2, which flank repeat sequence I (RS I), we were able to detect a band whose size corresponded to the expected 11 repeats. The same size band was observed in all the S. pombe strains that we tested (Fig. 3a, lanes 1 and 4 to 8) and was also identical in size to that seen when the pCpY1 plasmid was used as a template (Fig. 3a, lane 11).
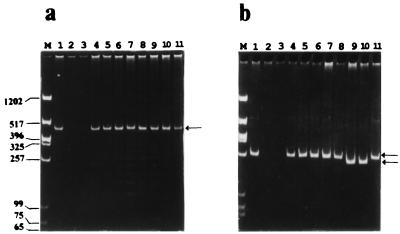
FIG. 3.
PCR analysis of RS I (a) and RS II (b) regions. PCRs were carried out (95°C for 1 min, 60°C for 1 min, and 72°C for 1 min for 30 cycles), and the PCR products were run on 5% acrylamide gels. P1 and P2 were used to amplify RS I; P3 and P4 were used to amplify RS II. Genomic DNA isolated from the different yeast strains by the glass bead method (7) was used as the template for the PCRs. For marker DNA (lanes M) we used a HinfI/XbaI digest of pBSK(−). Lanes: 1, S. pombe ABP13 h− ade6-210 ura4-D18 (obtained from Fission Yeast Course); 2, S. japonicus var. versatilis (obtained from Fission Yeast Course); 3, S. octosporus (obtained from Fission Yeast Course); 4, S. pombe KN1 h− ade6-216 ura4-D18 leu1-32 pmd1::ura4+ (13); 5, S. pombe ABP 320 h+ arg3-D4 ura4-D18 (21); 6, S. pombe GP 25 h− his5-303 lys1-131 (3); 7, S. pombe h90 swi4-1 NCYC 1992; 8, S. pombe NCYC 1990; 9, S. pombe var. malidevorans NCYC 2387; 10, S. malidevorans NCYC 683; and 11, plasmid pCPY1 containing the original cpy1+ clone that was isolated in this laboratory (1). Arrows indicate the repeat regions. Molecular sizes of marker DNA are given (in base pairs) to the left of panel a.
The PCR results with RS II and primers P3 and P4 revealed a band that was detected in all the S. pombe strains that we tested and corresponded in size to the expected eight repeat units (Fig. 3b, lanes 1 and 4 to 8) and was also identical in size to the bands seen with the plasmid control (Fig. 3a, lane 11).
These results indicated that the two repeat regions were mitotically stable and could possibly be used as genetic identification markers. We therefore decided to examine other isolates of S. pombe. The S. pombe species has been subdivided into two varieties: S. pombe var. pombe (which is normally referred to as S. pombe) and S. pombe var. malidevorans (which was previously referred to as S. malidevorans but has subsequently been found to be a variety of S. pombe) (18). We examined two such isolates of S. pombe by the PCR analysis described above. In the case of RS I, an identical band that corresponded to 11 repeats was observed in both strains (Fig. 3a, lanes 9 and 10). However, in the case of RS II, bands migrating faster than would bands of the expected size were observed (Fig. 3b, lanes 9 and 10), indicating a loss of some repeat units. This was also confirmed by Southern blotting (data not shown). To determine exactly how many and which of the repeat units were lacking, we cloned the smaller RS II PCR product from both the above-mentioned S. pombe var. malidevorans isolates and sequenced this region. A loss in one of the internal repeat units, cII (Fig. 2), was observed. Preferential variation in the number of internal repeat units as opposed to variation in the number of the flanking repeats is a feature quite typical of VNTR loci (4).
Although Tabuchi et al. have reported a functional protein that differed in that it contained only seven repeats in RS I (19), we observed no length polymorphism in this region among the strains that we tested. However, despite their mitotic stability VNTR loci do undergo changes in their lengths at low frequencies, and to investigate this point further, we examined whether deletion of a few of the repeat units within this region would still lead to a functional protein. We therefore constructed pCPY-ID6, an in-frame CpY deletion mutant lacking six of the repeat units of RS I (Fig. 1). This was constructed by partial digestion with BstXI followed by religation. The deletion was confirmed by sequencing. This construct was transformed into the cpy1-1 mutant (10), and the transformants were found to exhibit CpY activity similar to the parent plasmid pCpY-K1, as seen by a CpY colorimetric plate assay (6) (data not shown). This indicated that the CpY enzyme can accommodate length polymorphism in RS I. It is possible, therefore, that RS I represents a second minisatellite sequence within this region even though we were not able to observe any polymorphism among the strains that we tested.
We extended the minisatellite analysis to other members of the Schizosaccharomyces genus, namely, S. japonicus and S. octosporus, as we felt it might reflect on the origin of these repeats. However, we failed to detect any bands corresponding to these regions upon PCR (Fig. 3, lanes 2 and 3). This was also confirmed by Southern blotting, by which we failed to observe any bands when RS I and RS II were used as probes (data not shown). This indicated that the repeats were species specific and did not occur in other members of the Schizosaccharomyces genus.
These studies provide a demonstration of the existence of a minisatellite DNA sequence that exhibits the property of possessing a variable number of tandem repeats in yeasts. The limited amount of intergenic and noncoding regions in yeasts may be responsible for the relative lack of these sequences in these unicellular eukaryotes. In retrospect, therefore, it is perhaps not surprising that a minisatellite DNA sequence should be detected in the propeptide region of a precursor protease such as CpY. Although these proregions play an essential role in the maturation of the proteases functioning as intramolecular chaperones, there is a very low level of conservation among the proregions of CpYp. Furthermore, structure-function analysis of the proregion indicates a great deal of structural flexibility (14, 15). It is unclear at this stage whether the minisatellite sequence of S. pombe also has some functional significance, since Shinde and coworkers recently reported that an alteration in the proregion can alter the folding and kinetic properties of the subtilisin enzyme (17).
While the results that we have reported relate to the fission yeast, it is likely that such sequences exist in other yeasts as well. A recent report described the presence of a putative minisatellite sequence in Saccharomyces carlsbergensis based on sequence comparisons with other minisatellite regions (2). Our results, while demonstrating that such sequences do exist in yeasts, also show that they display the property of possessing a variable number of tandem repeats that perhaps are variety specific. These sequences could well provide a useful system for examining how such polymorphisms occur and originate, in addition to possibly adding another dimension to DNA fingerprinting and taxonomic analyses in yeasts and other unicellular eukaryotes.
Acknowledgments
S.S.I. and R.K. contributed equally to this work.
We thank A. Carr for the genomic libraries and M. Yoshida, S. Wadell, and G. Cottarel for the strains. We thank Raj Kumar for technical assistance and K. Ganesan for helpful discussions.
This work was supported in part by a grant-in-aid (project BT/R&D/15/40/93) from the Department of Biotechnology, Government of India. S.S.I., R.K., and P.A. are Senior Research Fellows of the Council of Scientific & Industrial Research, Government of India.
REFERENCES
- 1.Aggarwal, P., and A. K. Bachhawat. Unpublished observations.
- 1a.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andersen T H, Nilsson-Tillgren T A. A fungal minisatellite. Nature. 1997;386:771. doi: 10.1038/386771a0. [DOI] [PubMed] [Google Scholar]
- 3.Cottarel G. The Saccharomyces cerevisiae HIS3 and LYS2 genes complement the Schizosaccharomyces pombe his5-303 and lys1-131 mutations respectively: new selectable markers and new multipurpose, multicopy shuttle vectors pSP3 and pSP4. Curr Genet. 1995;28:380–383. doi: 10.1007/BF00326437. [DOI] [PubMed] [Google Scholar]
- 4.Gray I C, Jeffreys A J. Evolutionary transience of hypervariable minisatellite in man and the primates. Proc R Soc Lond Ser B. 1991;243:241–253. doi: 10.1098/rspb.1991.0038. [DOI] [PubMed] [Google Scholar]
- 5.Jeffreys A J, Wilson V, Thein S L. Hypervariable ‘minisatellite’ regions in human DNA. Nature. 1985;314:67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- 6.Jones E W. Tackling the protease problem in yeast. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics: a laboratory course manual, 1994 edition. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1994. [Google Scholar]
- 8.Kashi Y, King D, Seller M. Simple sequence repeats as a source of genetic variation. Trends Genet. 1997;13:74–78. doi: 10.1016/s0168-9525(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 9.Leupold U. Die Vererbung von Homothallie und Heterothallie bei Schizosaccharomyces pombe. C R Trav Lab Carlsberg Ser Physiol. 1950;24:381–480. [Google Scholar]
- 10.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 11.Mukhtar M, Logan D A, Kaufer N F. The carboxypeptidase Y-encoding gene from Candida albicans and its transcription during yeast-to-hyphae conversion. Gene. 1992;121:173–177. doi: 10.1016/0378-1119(92)90178-r. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura Y, et al. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987;235:1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- 13.Nishi K, Yoshida M, Nishimura M, Nishikawa M, Nishiyama M, Horinouchi S, Beppu T. Leptomycin B resistance gene of Schizosaccharomyces pombe encodes a protein similar to the mammalian P-glycoprotein. Mol Microbiol. 1992;6:761–769. doi: 10.1111/j.1365-2958.1992.tb01526.x. [DOI] [PubMed] [Google Scholar]
- 14.Ramos C, Winther J R, Kielland-Brandt M C. Requirement of the pro-peptide for in vivo formation of active carboxypeptidase Y. J Biol Chem. 1994;269:7006–7012. [PubMed] [Google Scholar]
- 15.Ramos C, Winther J R. Exchange of regions of the carboxypeptidase Y propeptide. Eur J Biochem. 1996;242:29–35. doi: 10.1111/j.1432-1033.1996.0029r.x. [DOI] [PubMed] [Google Scholar]
- 16.Richard G-F, Dujon B. Distribution and variability of trinucleotide repeats in the genome of the yeast Saccharomyces cerevisiae. Gene. 1996;174:165–174. doi: 10.1016/0378-1119(96)00514-8. [DOI] [PubMed] [Google Scholar]
- 17.Shinde U, Liu J J, Inouye M. Protein memory through altered folding mediated by intramolecular chaperones. Nature. 1997;386:771–773. doi: 10.1038/39097. [DOI] [PubMed] [Google Scholar]
- 18.Sipiczki M, Kucsera J, Ulaszewski S, Zsolt J. Hybridization studies by crossing and protoplast fusion within the genus Schizosaccharomyces Lindner. J Gen Microbiol. 1982;128:1989–2000. [Google Scholar]
- 19.Tabuchi M, Iwaihara D, Ohtani Y, Ohuchi N, Sakurai J-I, Morita T, Iwahara S, Takegawa K. Vacuolar protein sorting in fission yeast: cloning, biosynthesis, transport, and processing of carboxypeptidase Y from Schizosaccharomyces pombe. J Bacteriol. 1997;179:4179–4189. doi: 10.1128/jb.179.13.4179-4189.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valls L A, Hunter C P, Rothman J H, Stevens T H. Protein sorting in yeast: the localization of the yeast vacuolar carboxypeptidase Y resides in the pro-peptide. Cell. 1987;48:887–897. doi: 10.1016/0092-8674(87)90085-7. [DOI] [PubMed] [Google Scholar]
- 21.Wadell S, Jenkins J R. arg3+, a new selectable marker system for use in Schizosaccharomyces pombe: application of ura4+ as a removable selectable marker. Nucleic Acids Res. 1995;23:1836–1837. doi: 10.1093/nar/23.10.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]